Abstract
Neonatal HIV-1 infection is associated with rapidly progressive and frequently fatal immune deficiency if left untreated. Immediate institution of antiretroviral therapy (ART), ideally within hours after birth, may restrict irreversible damage to the developing neonatal immune system and possibly provide opportunities for facilitating drug-free viral control during subsequent treatment interruptions. However, the virological and immunological effects of ART initiation within hours after delivery have not been systematically investigated. We examined a unique cohort of neonates with HIV-1 infection from Botswana who started ART shortly after birth and were followed longitudinally for about 2 years in comparison to control infants started on treatment during the first year after birth. We demonstrate multiple clear benefits of rapid antiretroviral initiation, including an extremely small reservoir of intact proviral sequences, a reduction in abnormal T cell immune activation, a more polyfunctional HIV-1–specific T cell response, and an innate immune profile that displays distinct features of improved antiviral activity and is associated with intact proviral reservoir size. Together, these data offer rare insight into the evolutionary dynamics of viral reservoir establishment in neonates and provide strong empirical evidence supporting the immediate initiation of ART for neonates with HIV-1 infection.
INTRODUCTION
Despite effective interventions to prevent mother-to-child transmission of HIV-1 (1), neonatal HIV-1 infection continues to represent an enormous global health challenge, with about 300 to 500 infants being infected each day in sub-Saharan Africa (2). Clinical studies suggest that HIV-1 disease in neonates with developing immune systems progresses markedly faster than in adults, with 25 to 50% of infected children dying within the first 2 years of life if combination antiretroviral therapy (ART) is not initiated (3). These observations, along with data from randomized controlled studies (4), have led to guidelines by the World Health Organization (5) advocating for rapid initiation of ART within weeks after birth. Even earlier initiation of ART, preferably within hours after birth, is impeded by numerous diagnostic and infrastructural challenges in resource-limited settings but may translate into marked morbidity and mortality reductions relative to the current standard of care. In addition, provocative observations in individual cases of neonatal and pediatric HIV-1 infection (6–10) have raised the possibility that immediate institution of ART after birth may induce distinct virological and immunological characteristics that support and facilitate a transient or permanent drug-free remission of viral infection when treatment is interrupted after a period of suppressive ART (11, 12). However, a systematic analysis of residual reservoirs of HIV-1–infected cells and antiviral immune responses in infected neonates requires collection of blood samples from dedicated clinical studies and is technically challenging due to the limited amounts of blood that can be collected from newborns.
Although the establishment of viral reservoirs during early stages of HIV-1 infection in adults is well documented (13), very little is known about the type, diversity, and longitudinal evolution of HIV-1–infected CD4+ T cells in neonates who start ART during acute stages of HIV-1 infection. The phenotypes, compositions, and functional profiles of CD4+ T cells in neonates and infants differ profoundly from adults and may alter cellular susceptibility to viral infection, affect the selection of proviral chromosomal integration sites, and modify clonal turnover of infected cells, all of which have been shown to affect viral reservoir dynamics in adult HIV-1 infection (14–17) and in infant simian immunodeficiency virus (SIV)–infected rhesus macaques (18). In addition, the specific influence of antiviral immune responses on viral reservoir size and structure during neonatal HIV-1 infection is uncertain given that HIV-1–specific T cells, frequently considered as the hallmark of antiviral immunity in adults (19), are likely to be underdeveloped in neonates because of the characteristic weaknesses of T helper 1 (TH1)–polarized immunity in developing infants (20). Here, we describe a detailed evaluation of neonates with HIV-1 infection who started ART within the first days after birth as part of the Early Infant Treatment Study (EIT Study, NCT02369406). This prospective clinical trial was conducted at two major maternity hospitals in the Francistown and Gaborone regions of Botswana, a country with the third highest HIV-1 prevalence in the world, in which about 24% of pregnant women are living with HIV-1 (21). Using very small amounts of blood obtained from the rare study patients participating in this clinical trial and samples from control infants who started antiretroviral treatment later, we cross-sectionally and longitudinally determined the diversity and evolution of intact and defective HIV-1 DNA sequences and characterized innate and adaptive immune responses to HIV-1.
RESULTS
Viral reservoirs in early-treated infants with HIV-1 infection
To investigate viral reservoir dynamics in the neonatal immune system, we focused on the first 10 infants with HIV-1 infection identified through a comprehensive HIV-1 screening program of over 10,600 neonates in Botswana (table S1). Nine of these infants exhibited positive HIV-1 DNA polymerase chain reaction (PCR) assays performed within a median of 18.5 hours (range, 6.6 to 40.1 hours) after birth, consistent with intrauterine HIV-1 infection (22); the remaining infant initially had a negative HIV-1 DNA PCR assay at birth but exhibited a positive assay 31 days later, compatible with intrapartum HIV-1 infection (22). HIV prophylaxis consisting of nevirapine and zidovudine (±lamivudine) was started at a median of 7 hours (range, 0.2 to 114.5 hours) after birth and then converted to treatment doses of nevirapine, zidovudine, and lamivudine upon EIT enrollment, followed by a combination of ritonavir-boosted lopinavir, lamivudine, and zidovudine after at least 2 weeks (and 40 weeks’ gestational age equivalence) according to the study protocol; the infant with intrapartum infection was started on the lopinavir-containing regimen 31 days after birth. Preterm neonates (<35 weeks of gestational age) were not eligible to participate in the study because of limited information about the safety of antiretroviral agents in this age group. All study participants continued ART indefinitely and were seen for follow-up visits at weeks 4, 8, 12, 24, 48, 72, and 84/96 (fig. S1A). At baseline and most subsequent study visits, blood was collected from all participants for virological and immunological testing. For comparative purposes, we also analyzed a control cohort consisting of 10 children from Botswana who were diagnosed with HIV-1 infection and started ART at a median of 4 months (range, 2.6 to 11.7 months) after birth and from whom blood samples were collected at a median of 93 weeks (range, 65 to 126 weeks) after ART initiation. Suppression of plasma viremia during ART was achieved in all study participants, although intermittent viral rebound, likely related to medication noncompliance, was observed in several instances (fig. S1B).
To profile proviral HIV-1 DNA in these patients, we conducted droplet digital PCR (ddPCR) assays, which provide an absolute quantification of cell-associated HIV-1 5′ long terminal repeat (5′-LTR)–gag copy numbers in a given number of peripheral blood mononuclear cells (PBMCs). These studies revealed a progressive decline in the number of viral DNA copies during the initial 2 years of therapy, resulting in notably lower HIV-1 DNA copies in early-treated children at week 84/96, as compared with control children treated for a similar duration (5.3 versus 981.4 copies per 106 PBMC, P < 0.0001) (Fig. 1A). Proviral DNA levels in the EIT group were also substantially lower compared with a reference cohort of adults with chronic HIV-1 infection who underwent long-term ART (median, 16 years; range, 6 to 25 years) (5.3 versus 134.5 copies per 106 PBMC, P = 0.003) (Fig. 1A). Because proviral DNA contains a substantial proportion of defective viral species (23, 24), we subsequently performed near–full-length single-template next-generation proviral HIV-1 sequencing assays that allow for selective identification of genome-intact proviral DNA sequences, including those that remain undetectable by viral outgrowth assays but contribute to the replication-competent viral reservoir (24). These studies, involving 125 individual intact and 251 individual defective sequences amplified from longitudinally collected cell samples, demonstrated a continuous decrease in the frequency of genome-intact and defective proviral sequences during ART in the EIT group (Figs. 1B and 2A). At week 84/96, the number of intact proviral sequences per million PBMCs was below the detection threshold in all but one child, whereas significantly higher numbers of genome-intact proviral sequences were observed in control children (0.34 versus 1.57 copies per 106 PBMC, P = 0.0165) and in long-term–treated adults (0.34 versus 2.22 copies per 106 PBMC, P = 0.003). In addition, the frequency of defective proviral species was also reduced in the EIT group compared with controls (0.85 versus 19.07 copies per 106 PBMC, P = 0.0355) and to the cohort of adults undergoing long-term ART (0.85 versus 36.5 copies per 106 PBMC, P < 0.0001) (Figs. 1C and 2, A and B). Decline of intact and defective proviral sequences in EIT infants occurred primarily during the first 24 weeks and subsequently plateaued. Although roughly equal numbers of intact and defective proviral sequences were detected at baseline, the longitudinal decay kinetics of intact proviruses during the first 24 weeks were significantly faster than those of defective proviruses (half-life of 3.1 versus 3.98 weeks, P < 0.0001, Wilcoxon test) (Fig. 1D), which translated into a progressively increasing contribution of defective proviral species to the total number of viral sequences (Fig. 1E). Intact proviral sequences detected more than once at a given time point, possibly as a result of clonal proliferation of infected CD4+ T cells (15, 25, 26), were relatively rare in the analyzed pediatric patients, compared with our cohort of patients with HIV-1 undergoing long-term ART (Fig. 1, F and G) and to other adult patients with HIV-1 described in previous studies (25, 26). Although this finding may indicate a more limited role of clonally expanded HIV-1–infected CD4+ T cells in the neonatal and infantile immune system, we cannot exclude that reduced cell frequencies available for analysis from pediatric patients may have contributed to this result.
Fig. 1. Immediate antiretroviral treatment initiation in neonates with HIV-1 infection leads to markedly reduced HIV-1 reservoirs.

(A and B) Longitudinal analysis of cell-associated HIV-1 DNA (A) (determined by ddPCR) and genome-intact or defective proviral sequences (B) (determined by single-genome, near–full-length next-generation sequencing) in neonates with antepartum (AP; n = 9) and peripartum (PP; n = 1) HIV-1 infection at indicated time points after initiation of ART. Data from children with delayed treatment initiation (Controls; median of 93 weeks after treatment initiation, n = 10) and from long-term ART–treated adults (median of 16 years after treatment initiation, n = 31) are shown for comparison in (A). (C) Cross-sectional comparison of the relative frequency of genome-intact and defective proviral sequences in children with early treatment initiation (EIT; week 84/96 after beginning of ART, n = 9), delayed treatment initiation (Controls; median of week 93 after beginning of ART, n = 10), and ART-treated adults with chronic HIV-1 infection (median of 16 years after beginning of ART, n = 31). Box-and-whisker plots in (A) reflect median, minimum, maximum, and interquartile ranges. Dot plots with mean and SEM are shown in (B) and (C). Significance was tested using a two-sided Kruskal-Wallis with post hoc Dunn’s multiple comparison test between longitudinal time points or groups and a Wilcoxon test between pairs. LOD, limit of detection, calculated as 0.2 (ddPCR) or 0.5 (single-genome near–full-length PCR) copies per maximum number of cells tested without target identification (see Materials and Methods for details). (D) Decay kinetics of intact and defective proviral HIV-1 sequences during the first 24 weeks after treatment initiation. Significance was calculated using a Wilcoxon signed-rank test. (E) Longitudinal analysis of viral reservoir composition in EIT neonates, as determined by single-genome near–full-length, next-generation sequencing in samples from indicated time points after treatment initiation. Data from control children (Controls) and ART-treated adults with chronic HIV-1 infection are shown for comparison. Each pie chart reflects relative contribution of HIV-1 amplification products with defined defects or genome-intact sequences. Total number of individual sequences included in each diagram is listed in the middle of each pie chart. (F and G) Bar diagrams reflecting proportions of individual proviral sequences detected once (nonclonal sequences) or detected more than once (clonal sequences) at a given time point. (F) Data from all (intact and defective) proviral sequences. (G) Data from genome-intact proviral sequences only. The total number of individual proviral sequences analyzed at each time point is listed above each bar.
Fig. 2. Viral sequence diversity and chromosomal integration sites in EIT study participants.

(A and B) Diagrams summarizing viral reservoir sequences aligned to the HXB2 reference from indicated study participants. Color coding reflects presence of intact and defective proviral species. Numbers of individual analyzed sequences are listed for each patient on the vertical axis. (A) All analyzed sequences (from any time point) from the EIT group. (B) All analyzed sequences (collected cross-sectionally at median of week 93) from children with delayed treatment initiation. (C) Circular maximum likelihood phylogenetic tree including all genome-intact sequences (n = 135) obtained from infants with early infant treatment (EIT) (“AP,” n = 102 or “PP,” n = 23; data from all time points shown) and from control children with delayed treatment initiation (“C,” n = 10). Identical proviruses are highlighted by gray boxes. w, week. (D) Circos plot reflecting chromosomal positioning of intact and defective proviral sequences from three EIT infants from whom sufficient PBMCs were available for proviral integration site analysis. All integration sites were determined using cell samples collected at baseline. Number of clonal sequences is highlighted by circular symbols. (E to H) Pie charts summarizing features of chromosomal integration sites for intact and defective proviruses described in (D). Data reflect proportions of intact and defective proviruses located in genic versus nongenic elements (E), in same or opposite orientation to host genes (genic sites only) (F), in introns versus exons (genic sites only) (G), and in indicated repetitive genome components [short interspersed nuclear element (SINE), long interspersed nuclear element (LINE), and DNA transposon (DNA)] (H). (I) Comparison between integration sites of intact proviruses from three EIT study participants and three long-term–treated adults described previously (17). Data reflect positions of intact proviruses in genic versus nongenic regions (top); the bottom panel indicates orientation of integrated proviruses relative to host genes (for proviruses in genic locations). Significance was tested using a two-sided Fisher’s exact test in (E) to (I). In (E) to (I), clonal sequences were only counted once. (J) Venn diagram reflecting the overlap between genes harboring HIV-1 integration sites described in this study and previously described independent integration sites by Wagner et al. (n = 288) (28), Maldarelli et al. (n = 1230) (27), Marini et al. (n = 156 hotspot genes) (29), and Einkauf et al. (n = 131) (17). (K) Sunburst chart representing the functional classification of genes targeted for retroviral integration by type and predicted location of encoded gene products. N/A, not available.
Phylogenetic diversity and chromosomal location of intact HIV-1 proviral sequences
A subsequent phylogenetic analysis of viral sequences demonstrated that individual genome-intact proviral species from our study participants showed extremely limited intraindividual variation during cross-sectional and longitudinal comparisons, consistent with early initiation of treatment that precludes viral sequence diversification through ongoing viral replicative activity (Fig. 2, A to C). In contrast, interindividual phylogenetic differences were clearly detectable between most of the study participants, although infection with phylogenetically related viral species was observed in some patients, possibly reflecting geographically localized viral transmission patterns (fig. S2, A and B). In several instances, we observed completely identical genome-intact proviral sequences at multiple longitudinal follow-up time points within the same study participant, strongly suggesting longitudinal persistence of durable HIV-1–infected cell clones (Fig. 2C). Using an experimental technique that allows for simultaneous analysis of proviral sequences and their corresponding chromosomal integration sites after phi29-catalyzed multiple displacement amplification (17), we were able to map the chromosomal locations of 25 intact and 5 defective proviral sequences isolated from the earliest available analytical time point (Fig. 2D and table S2). This analysis was limited by the relatively small number of integration sites that could be evaluated from the modest amount of blood available from neonates with HIV-1 infection, but critically advances prior studies (17, 27, 28) by showing chromosomal integration site locations of intact HIV-1 proviruses during the earliest stages of viral infection (Fig. 2, E to I and K). In one study subject, these experiments identified two identical intact proviral sequences (retrieved from a sample collected at baseline) that shared the same chromosomal integration site (Fig. 2, C and D and fig. S2C), indicating that clonal expansion of infected CD4+ T cell clones can indeed occur during early stages after infection in neonates. Moreover, we noted that about 70% of genome-intact proviral sequences were located in genes covering a diverse range of functions (Fig. 2, E to H and K), some of which were identified as targets for retroviral integration in adults in prior studies (Fig. 2J) (27–29). In the EIT group, intact proviral sequences in genic regions were preferentially (>60%) integrated in the same direction as their respective host genes (Fig. 2F), which represented a marked contrast to long-term–treated adult patients with HIV-1 in whom the majority of intact proviruses (>70%) displayed an opposite orientation to host genes (Fig. 2I) (17). This observation suggests that intact proviruses with the same directional configuration as host genes may be more likely to be selected against during prolonged ART.
Innate immune responses in early-treated infants with HIV-1 infection
Given that natural killer (NK) cells and other components of the innate immune system may be critical for host immune protection in the postnatal period of infants with HIV-1 infection (30), we subsequently focused on a detailed analysis of innate immune responses that may distinguish infants with HIV-1 infection and influence evolution of the viral reservoir. For these experiments, immunological data from EIT infants were compared to HIV-negative infants from Botswana born to mothers with [HIV-1 exposed–uninfected (HEU)] or without [HIV-1 unexposed–uninfected (HUU)] HIV-1 infection from whom samples were available from week 12 after birth; a group of HIV-negative adults from Botswana was also recruited as a reference cohort. We observed that the proportion of NK cells was reduced in the EIT group at week 12 after birth, relative to HUU infants and healthy adults (Fig. 3A). No difference was noted between the EIT group and controls with a delayed treatment onset (Fig. 3B). Subclassification of NK cells into six subgroups according to previously described patterns (31) revealed that relative proportions of CD56dim CD16dim NK cells, which are functionally characterized by potent cytotoxic activity against virally infected cells, were expanded in the EIT group compared with the HEU and HUU groups and reached frequencies otherwise seen in healthy adults (Fig. 3C). CD56dim CD16− NK cells, primarily recognized as abundant cytokine producers (32), were also significantly increased in the EIT group (P = 0.0007 versus HUU; P = 0.0004 versus HEU; P = 0.0037 versus adults; and P = 0.001 versus controls), representing a distinguishing feature of the NK cell response in EIT children relative to all comparison cohorts, including control children with delayed initiation of ART (Fig. 3, C and D). CD56−CD16dim NK cells are a functionally impaired NK cell subgroup that physiologically account for the majority of NK cells in neonates (33) and are pathologically expanded during chronic viral infections in adults (34, 35); these cells were significantly reduced in EIT infants (23.1%) as compared with HUU (69%) and HEU (72%) (P = 0.0008) infants at week 12, and with control children with HIV-1 infection analyzed after about 2 years of ART (24.2% versus 37.8%, P = 0.0006) (Fig. 3, C and D). The differential dynamics of NK cell subgroups in the EIT group were also observed when NK cell subsets expressing the activating NK cell receptors, NKp30 and NKG2D, were selectively analyzed and when expression of CD161, a marker for NK cells endowed with higher responsiveness to innate cytokines and greater proliferative activity (36), was considered (Fig. 3, E to G, and figs. S3 and S4).
Fig. 3. Association between NK cell responses and intact proviral reservoirs in EIT neonates.
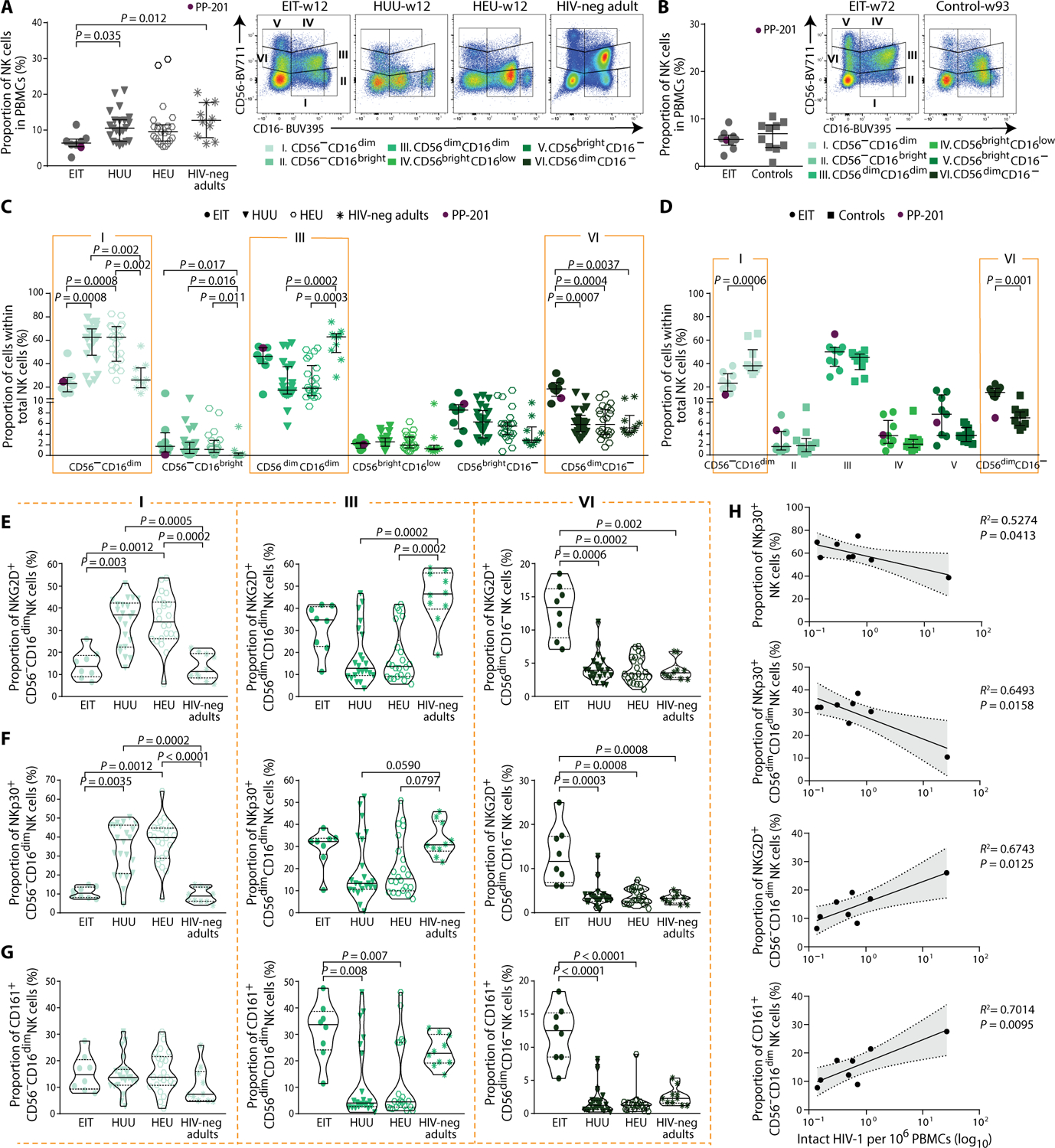
(A and B) Left: Proportion of total NK cells in PBMCs. Right panels: Representative flow cytometry dot plots displaying subclassification of NK cells according to CD56 and CD16 expression in six subsets: I_CD56−CD16dim, II_CD56−CD16bright, III_CD56dimCD16dim, IV_CD56brightCD16low, V_CD56brightCD16−, and VI_CD56dimCD16−. Data from unstimulated PBMCs from EIT study participants (n = 8), HIV-1 unexposed–uninfected infants (HUU; n = 22), HIV-1 exposed–uninfected infants (HEU; n = 22) (all at week 12 after birth), and HIV-1–negative adults from Botswana (HIV-neg adults; n = 10) are shown in (A). Data from the EIT group (week 72, n = 9) and from controls with delayed time of treatment initiation (Controls; week 93, n = 10) are shown in (B). (C and D) Fractional abundance of defined NK cell populations in indicated study groups. Dot plots with median and interquartile ranges are indicated. PP-201, neonate with peripartum HIV-1 infection in the EIT study. (E to G) Violin plots reflecting proportions of NKG2D+ (E), NKp30+ (F), and CD161+ (G) NK cell subpopulations (defined by CD56 and CD16 expression) within the entire NK cell pool. Data from EIT, HEU, and HUU infants (all at week 12) and a reference cohort of adults with HIV-1 infection are shown. Significance was tested using a two-sided Kruskal-Wallis with post hoc Dunn’s multiple comparison test between groups or a two-sided Mann-Whitney U test between two groups. (H) Associations between proportions of indicated NK cell subsets relative to total NK cells and frequency of intact HIV-1 proviral sequences at week 12 after initiation of treatment among EIT infants. Linear regression coefficients are shown. Shaded areas represent 95% confidence intervals.
As our next analysis step, we examined associations between NK cell subsets and the frequency of intact proviral sequences in the EIT study group. NKp30+ CD56dimCD16dim NK cells and total NKp30+ NK cells were both negatively associated with the frequency of intact proviral sequences, whereas CD56−CD16dim NK cell subpopulations expressing NKG2D were reduced in the EIT group and showed a positive correlation with intact proviral species (Fig. 3H). CD161+ CD56−CD16dim NK cells, which displayed a similar fractional abundance in the EIT group as in HUU/HEU infants and adults, were also positively associated with the frequency of intact proviral sequences. Although these findings involve relatively few study subjects and, therefore, require further investigations in larger cohorts, these statistical linkages suggest a role for NK cell responses in shaping and structuring the viral reservoir in the developing neonatal immune system, at a time when adaptive immune responses are immature.
To further explore innate immune activity in our study population, we investigated myeloid immune cell populations that have critical roles in antimicrobial immune defense by acting as professional antigen-presenting cells and by executing antiviral activities through phagocytosis and cytokine secretion. Myeloid and plasma-cytoid dendritic cells demonstrated no detectable numeric or phenotypic differences between study cohorts (fig. S5), and the proportion of total monocytes was significantly lower in the EIT group relative to HIV-negative adults and the control children (Fig. 4, C and E). However, the relative proportion of classical CD14dimCD16− monocytes, characterized by improved phagocytic activity (37), was significantly larger (P = 0.001) in the EIT group compared with the HEU group and reached frequencies that were almost equivalent to HIV-negative adults (Fig. 4D). In contrast, the CD14dimCD16dim intermediate and CD14low/− CD16bright nonclassical subsets of monocytes, typically associated with immune activation and inflammation (38), were reduced in EIT infants relative to HIV-1–negative counterparts (Fig. 4D). No difference in monocyte subset distribution was noted between the EIT group and controls (Fig. 4F). Among EIT study participants at baseline, we noted an inverse statistical association between classical monocytes and the corresponding intact proviral reservoir size, whereas proportions of intermediate and nonclassical monocytes were positively associated with the frequency of intact proviral sequences (Fig. 4G). Together, these data show that rapid initiation of ART in neonates results in a relative expansion of classical monocytes that is associated with decreasing intact viral reservoir size.
Fig. 4. Frequency and phenotype of monocytes in EIT infants.

(A and B) Representative flow cytometry dot plots reflecting classification of monocytes according to CD16 and CD14 expression. (C and D) Proportion of total monocytes (C) and monocyte subsets (D) in EIT (n = 8), HUU (n = 22), and HEU (n = 22) infants (all at week 12) and HIV-1–negative adults (n = 10). (E and F) Proportion of total monocytes (E) and monocyte subsets (F) in EIT children at week 72 (n = 9) and from Controls at median of week 93 (n = 10). PP-201, neonate with peripartum HIV-1 infection in the EIT study. Dot plots with median and interquartile ranges are indicated. Significance was tested using a two-sided Kruskal-Wallis with post hoc Dunn’s multiple comparison test between groups or a two-sided Mann-Whitney U test between two groups. (G) Statistical association between frequency of intact HIV-1 proviruses and proportion of indicated monocyte subsets. Linear regression coefficients are shown. Shaded areas reflect 95% confidence intervals.
Immune activation and antiviral T cell responses
In a subsequent phenotypic and functional characterization of adaptive cellular immune responses, we observed that naïve T cells dominated the CD4+ and CD8+ T cell compartments in neonates and progressively declined longitudinally, as expected based on prior studies involving uninfected infants (39, 40). Proportions of naïve and memory CD4+ and CD8+ T cells in EIT study participants did not significantly differ from control children with delayed antiretroviral initiation or from HEU and HUU infants (Fig. 5A). However, a global analysis of T cell immune activation markers—including human leukocyte antigen–DR (HLA-DR), CD38, programmed cell death protein-1 (PD-1), and CD127—differed profoundly between EIT children and other comparator groups (Fig. 5B and fig. S6, A and B). Most pronounced differences were observed for the CD38+ HLA-DR+ CD4+ and CD38+HLA-DR+ CD8+ T cell subsets, which have been previously identified as independent predictors of HIV-1 disease progression and mortality (41, 42); these cells showed substantial proportional increases in EIT children relative to HUU/HEU infants and HIV-negative adults, but considerable decreases compared with control children with HIV-1 infection (Fig. 5, C and D).
Fig. 5. Early initiation of ART in neonates with HIV-1 infection promotes a more polyfunctional CD8+ T cell response.

(A) Proportion of indicated CD4+ and CD8+ T cell subsets in different patient groups (CM, central memory; EM, effector memory; and TEMRA, effector-memory T cells expressing CD45RA). Numbers after each study cohort reflect time (week) of cell sampling. Data from EIT study participants include n = 9 subjects at week 0, n = 8 subjects at week 12, n = 5 subjects at week 24, and n = 9 subjects at week 72; data from n = 10 HIV-negative adults, n = 10 Controls, n = 25 HUU, and n = 29 HEU were also included. (B) Heat map summarizing the surface expression of indicated phenotypic markers in CD4+ and CD8+ T cells from individual study groups. For each surface expression marker(s), data from the total T cell compartment and from naïve, central memory, effector memory, and effector-memory T cells expressing CD45RA are sequentially shown. (C and D) Box-and-whisker plots indicating the proportion of CD38+HLA-DR+–positive cells within the CD4+ and CD8+ T cell compartment. Left panels show data from EIT, HUU, and HEU infants (all at week 12) and HIV-1–negative adults; right panels indicate data from EIT children and Controls at weeks 72 to 93 after initiation of ART, respectively. Box-and-whisker plots reflect median, minimum, maximum, and interquartile ranges. PP-201, neonate with peripartum HIV-1 infection in the EIT study (E and F) Pie charts generated with SPICE reflecting proportions of CD4+ and CD8+ SEB− (E) or HIV-1 Gag–specific (F) T cell responses with indicated functional profile (determined by Boolean combination gating) in children from the EIT study (n = 10, week 84), Controls (n = 8, week 93), and HIV-1–negative adults (n = 10). Pie chart colors represent number of effector molecules, whereas each effector function is represented by an arc. Significance was tested using a permutation test implemented in SPICE software. (G) Flow cytometry dot plots reflecting the expression of Tbet and Eomes in CD8+ T cells from a representative EIT study participant at indicated time points. (H) Bubble diagrams indicating the longitudinal evolution of Tbet and Eomes expression in CD8+ T cells from indicated time points in EIT study participants, control children, and HIV-1–negative adults. Pooled data from all analyzed samples in each study group are shown; data from n = 10 EIT subjects at week 1 (w1), n = 7 EIT subjects at week 8 (w8), n = 4 EIT subjects at week 48 (w48), n = 10 EIT subjects at week 84 (w84), n = 8 Controls (median of 93 weeks of ART), and n = 10 HIV-negative adults were included. (I) Bar diagram summarizing the expression of perforin and granzyme B (Grz B) in CD8+ T cells classified according to Tbet and Eomes expression in EIT children at week 84 and control children at a median of 93 weeks. Values are expressed as median and interquartile range. P values were calculated using the Mann-Whitney U test.
In this relatively small dataset, there was no statistically significant association between activated CD4+ and CD8+ T cells and the corresponding frequencies of total, intact, or defective proviruses. The functional profile of antigen-specific CD4+ and CD8+ T cell responses to staphylococcus enterotoxin B (SEB) did not differ between EIT and control children (Fig. 5E and fig. S6C), but longitudinally increased in diversity for both groups (fig. S6D). Although the magnitude of HIV-1–specific T cells in both study groups was low, we noted that HIV-1 Gag–specific CD4+ and CD8+ T cells in the EIT group displayed a more polyfunctional profile relative to the control group; this profile was particularly pronounced for interleukin-2 (IL-2)–secreting HIV-1–specific CD8+ T cells, which accounted for a substantial proportion of all HIV-1–specific cytotoxic T cells in early-treated children but were practically undetectable in children with delayed onset of therapy (Fig. 5F and fig. S6E). CD8+ T cells at birth showed little or no expression of Tbet and Eomes, two transcription factors required for effective antiviral immune responses (43). During the ensuing time of follow-up, a distinct population of Tbethi and Eomeshi CD8+ T cells emerged that strongly expressed perforin and granzyme B (Fig. 5, G to I) and eventually represented the dominant T cell population at week 84 of follow-up. In contrast to innate immune parameters, there were no statistical associations between HIV-1–specific T cell responses and the size or structural composition of viral reservoir cells.
DISCUSSION
Neonates and infants are among the individuals who are most vulnerable to infection with HIV-1 and in whom the benefits of modern combination ART are most obvious. Empiric postdelivery ART for neonates at a higher risk for vertical HIV-1 transmission is now recommended in the United States (44). However, the initiation of ART in neonates immediately after birth remains a substantial logistical and infrastructural challenge in areas most affected by the HIV-1 epidemic and is currently not routinely performed in most sub-Saharan countries (45). A better understanding of the clinical, immunological, and virological effects induced by early initiation of ART will likely be instrumental for developing tailored and optimized treatment interventions for neonates with HIV-1 infection and may translate into improved treatment guidelines and management practices. The present study demonstrates a multitude of advantages of immediate ART after birth, including a substantially reduced viral reservoir size, an innate immune profile with multiple features suggestive of improved antiviral activity, and a decrease in abnormal T cell activation that represents an independent predictor of HIV-1–associated morbidity (46).
In addition to highlighting distinct immunological advantages of early treatment initiation, our longitudinal evaluation of intact and defective viral sequences also allows for rare insight into the dynamics of viral reservoir establishment immediately after birth. We here used a near–full-length HIV-1 sequencing approach, which captures all intact proviral sequences, including the considerable number of sequences that do not respond to reactivation stimuli in single-round viral outgrowth assays but nevertheless represent an important component of the functional viral reservoir (24). Our data demonstrate a markedly faster decline of intact HIV-1 DNA sequences relative to their defective counterparts, which may, at least, partially reflect immune-mediated selection mechanisms that favor long-term persistence of cells harboring genetically impaired proviruses. CD4+ T cells encoding intact, replication-competent HIV-1 may be more susceptible to host immune activity compared with cells harboring defective viral sequences that typically display more limited viral transcriptional activity and may be less exposed to immune recognition (47). Together, these results suggest that virally infected cells are subject to strong selection mechanisms from the early onset of ART and that only a small subset of infected cells have the ability to develop into a stable, long-lasting reservoir. This point is further supported by our analysis of chromosomal integration sites of intact proviral sequences in neonates, which showed that intact proviruses detected immediately after birth frequently shared the same directional configuration as host genes, as opposed to intact proviral sequences from long-term–treated patients with HIV-1 in whom the majority of intact proviruses were integrated in the opposite direction to host genes (17). This observation is most compatible with selection mechanisms that favor long-term persistence of intact proviruses in opposite orientation to host genes during prolonged ART.
Our study showed that the decay of intact and defective proviral sequences mostly occurred within the first 24 weeks after treatment initiation and subsequently plateaued. These dynamics of viral reservoir decline may reflect preferential elimination of short-lived viral reservoir cells during early stages of ART, whereas long-lasting reservoir cells persist. The definition of individual CD4+ T cell subsets that serve as viral reservoirs will therefore be a critical next step for exploring evolutionary patterns of viral reservoirs in neonates. In contrast to adults, neonatal CD4+ T cells consist, to a large extent, of naïve cells; naïve cells are less susceptible to HIV-1 in adults (48) but were frequently found to harbor replication-competent HIV-1 in ART-treated infant rhesus macaques (18). In addition, neonatal CD4+ T cells typically display a marked deficiency in TH1-polarized cells (49), which represent an important component of viral reservoir cells in adults (25). Moreover, CCR5 expression, denoting a substantial proportion of the adult viral reservoir cell pool (50), is diminished in circulating (51) but not intestinal (52) neonatal CD4+ T cells. Last, regulatory CD4+ T cells, frequently associated with viral reservoir persistence in adult HIV-1 infection (53), can be expanded during fetal and neonatal development (54). A more detailed analysis of individual CD4+ T cell subsets that constitute the viral reservoir in neonates and infants is technically difficult, given the low number of cells available from such research subjects, but represents a high-priority future research topic (55).
An important finding in this manuscript is the distinct innate immune response profile that was selectively observed in EIT study participants. We observed that ART initiation at birth resulted in substantially increased frequencies of NK cell subsets with phenotypic features of higher functionality, whereas NK cell subpopulations associated with impaired functional properties were decreased. This represents an apparent contrast to the neonatal immune system of HIV-1–uninfected newborns, in whom we and others (56) found an enrichment of NK cells with phenotypic properties suggestive of lower cytotoxic potency. Because antiviral neonatal immune defense relies, to a considerable extent, on innate immune mechanisms due to a frequent TH2 polarization bias of adaptive cellular immune responses (57–59), these data suggest that early treatment initiation can make a substantial contribution to stabilizing and enhancing antimicrobial immune protection during the initial weeks after birth, when neonates are highly vulnerable to microbial pathogens. We also observed that NK and monocyte subsets with phenotypic features of higher antiviral activity were inversely related to intact proviral reservoir size, in notable contrast to adults with hyperacute HIV-1 infection in whom viral reservoir size seemed to be primarily associated with HIV-1–specific T cell responses (13). However, in our analysis of neonates with HIV-1 infection, HIV-1–specific T cells were weak and did not correlate with frequencies of either intact, defective, or total proviral sequences. Although antiviral T cell responses can be primed in utero in infected infants (60), it is likely that rapid initiation of ART reduced viral antigen exposure and dampened the evolution of antiviral T cell responses. Together, these findings raise the possibility that innate immune activity in neonates can influence the size and composition of the evolving viral reservoir, possibly resonating with the recent identification of innate NK cell–mediated immune responses as possible drivers of low viral reservoirs in subgroups of adults with HIV-1 infection who develop a sustained drug-free remission after ART initiation in acute HIV-1 infection (61).
Recent observations in animal models of SIV-infected rhesus macaques suggest that extremely early ART initiation may provide a distinct window of opportunity for curative strategies of HIV-1 infection (62). Likewise, in adults with HIV-1 infection, spontaneous posttreatment control is more frequently encountered in individuals with early treatment initiation than in individuals with delayed ART onset (63, 64). The additional follow-up of our study participants, some of whom have elected to participate in an interventional clinical trial with broadly neutralizing antibodies, might reveal additional benefits of rapid ART initiation that are not immediately evident with the relatively short follow-up data that are currently available. Although our study cohort is too small to be considered as a basis for practice-changing implementations of immediate, postdelivery “test and treat” strategies in all infants at risk for vertical transmission in sub-Saharan Africa, it provides strong scientific support for advocating immediate initiation of ART in neonates with HIV-1 infection and may, if confirmed and expanded in other studies, have an important influence on the management of neonatal HIV-1 infection.
MATERIAL AND METHODS
Study design
Samples from 10 children with HIV-1 infection were collected in Botswana as part of the EIT Study (NCT02369406). In addition, PBMC samples were collected in Botswana from 10 control children with HIV-1 infection and ART onset later in the first year of life, from HIV-negative infants (25 HUU and 29 HEU), and from 10 HIV-uninfected adults. Study protocols were approved by the Botswana Ministry of Health, the Harvard School of Public Health, and the Partners Human Research Committee (the local institutional review board for MGH and BWH). PBMCs from an additional cohort of adults with HIV-1 infection (n = 31) were recruited at the Massachusetts General Hospital and the Brigham and Women’s Hospital (both in Boston, MA) according to protocols approved by the Partners Human Research Committee. Clinical and demographical characteristics of study participants are summarized in table S1. Written informed consent was documented from all adult study participants; for underage children, written consent was obtained from their legal caregivers in accordance with the Declaration of Helsinki. Primary data are reported in data file S1.
Sample processing
Blood samples from neonates and infants were collected using heel sticks or venipuncture; samples from adults were obtained by venipuncture. Blood samples were subjected to PBMC isolation using standard Ficoll-Paque density gradient centrifugation. Cryopreserved PBMC samples were sent by air cargo transportation from Botswana to Boston for analysis. Timeline of collected PBMC samples binned into immunology and virology assays is shown in fig. S1.
Droplet digital PCR
PBMCs were subjected to DNA extraction using commercial kits (QIAGEN AllPrep DNA/RNA Mini Kit or QIAGEN DNeasy kit) according to the manufacturer’s instructions. Total HIV-1 DNA was amplified using the QX100 Droplet Digital PCR System (ddPCR; Bio-Rad) using primers and probes described previously (65) [127–base pair (bp) 5′-LTR–gag amplicon; coordinates 684 to 810 in HIV-1 reference strain HXB2) and normalized to the RPP30 gene. PCR was performed using the following program: 95°C for 10 min, 45 cycles of 94°C for 30 s and 60°C for 1 min, 72°C for 1 min. Data were analyzed using the QuantaSoft software (Bio-Rad). When viral copies where undetectable, data were reported as “limit of detection” (LOD), calculated as 0.2 copies per maximum number of cells tested without target identification.
HIV near–full-genome sequencing
Genomic DNA diluted to single HIV-1 genome levels based on Poisson distribution statistics and ddPCR results or whole-genome amplification products was subjected to HIV-1 near–full-genome amplification using a one-amplicon (25) or five-amplicon approach (17) with primer sets adjusted to clade C sequences. Briefly, 1 U per 20-μl reaction of Invitrogen Platinum Taq (#11304-029) was incubated with 1× reaction buffer, 2 mM MgSO4, 0.2 mM dNTP, and 0.4 μM each of forward and reverse primers; PCR programs were run as described previously. PCR products were visualized by agarose gel electrophoresis. Amplification products were subjected to Illumina MiSeq sequencing at the Massachusetts General Hospital (MGH) DNA Core facility. Resulting short reads were de novo assembled using Ultracycler v1.0 and aligned to HXB2 to identify large deleterious deletions (<8000 bp of the amplicon aligned to HXB2), out-of-frame indels, premature/lethal stop codons, internal inversions, or 5′-LTR defect (≥15 bp insertions and/or deletions relative to HXB2), using an automated in-house pipeline written in R scripting language (66, 67). Presence/absence of APOBEC3G/3F–associated hypermutations was determined using the Los Alamos HIV Sequence Database Hypermut 2.0 program (68). Viral sequences that lacked all mutations listed above were classified as “genome-intact.” Multiple sequence alignments were performed using MUSCLE (69). Phylogenetic analyses were conducted using MEGA X, applying maximum likelihood approaches (70). Viral sequences were considered clonal if they had completely identical consensus sequences; single-nucleotide variations in primer binding sites were not considered for clonality analysis. Viral sequences were deposited in GenBank (accession numbers MK457765–MK458272). When viral DNA sequences were undetectable, data were reported as LOD, calculated as 0.5 copies per maximum number of cells tested without target identification.
Whole-genome amplification
For selected participants from whom sufficient cells were available, extracted DNA was diluted to single viral genome levels according to ddPCR results so that one provirus was present in about 20 to 30% of wells. Subsequently, DNA in each well was subjected to multiple displacement amplification (MDA) with phi29 polymerase (QIAGEN REPLI-g Single Cell Kit) per the manufacturer’s protocol. After this unbiased whole-genome amplification, DNA from each well was split and separately subjected to viral sequencing and integration site analysis, as described below. If necessary, a second-round MDA reaction was performed to increase the amount of available DNA.
Integration site analysis
Integration sites associated with each viral sequence were obtained using ligation-mediated PCR [Lenti-X Integration Site Analysis Kit (Clontech #631263)] and/or integration site loop amplification (28). No modifications were made to these protocols except in Lenti-X: In addition to the 5′-LTR–associated integration site amplification, we added a 3′-LTR–associated integration site amplification reaction using nested LTR1 (5′-CTTAAGCCTCAATAAAGCTTGCCTTGAG-3′, HXB2 9602 forward) and LTR2 (5′-AGACCCTTTTAGTCAGTGTGGAAAATC-3′, HXB2 9686 forward) primers. Resulting PCR products were subjected to next-generation sequencing using Illumina MiSeq. Control cells (8E5/LAV cell line; NIH AIDS Reagent Program #95) were included in most experimental conditions and consistently retrieved integration site chr13-67485907 (3′-LTR) and 67485903 (5′-LTR). MiSeq paired-end FASTQ files were demultiplexed and analyzed using the following bioinformatics approach: Small reads (142 bp) were aligned simultaneously to human reference genome GRCh38 patch 12 and HIV-1 reference genome HXB2 using bwa-mem (71). Chimeric reads containing both human and HIV-1 sequences were evaluated for mapping quality based on (i) absolute counts of chimeric reads, (ii) percentages of chimeric reads per total, (iii) depth of sequencing coverage in the host genome adjacent to the viral integration site, and (iv) HIV-1 coordinates mapping to the terminal nucleotides of the viral genome. The final list of integration sites was checked against GENCODE v28 (72). Repetitive genomic sequences harboring HIV-1 integration sites were identified using RepeatMasker (73). A functional categorization of targeted genes was performed using Ingenuity Pathway Analysis (74).
Flow cytometry
PBMCs were thawed, stained with LIVE/DEAD Blue Viability Dye (Invitrogen) for 20 min and subsequently preincubated for 10 min with 2 μl of FcR blocking reagent. Afterward, cells were incubated for 20 min with different combinations of appropriately titrated antibodies directed against CD19 (HIB19; BioLegend), HLA-DR (TU36; BD Bioscience), CCR7 (G043H7; BioLegend), CD45RA (HI100, BioLegend), CD3 (UCHT1, BioLegend), CD38 (HIT2, BioLegend), CD127 (A019D5, BioLegend), CD8a (RPA-T8, BioLegend), PD1 (EH12.2H7, BioLegend), CD4 (RPA-T4, BD Bioscience), CD14 (HCD14, BioLegend), NKG2D (1D11, BioLegend), NKp30 (P30–15, BioLegend), CD56 (HCD56, BioLegend), CD161 (HP-3G10, BioLegend), CD123 (6H6, BioLegend), CD11c (3.9, BioLegend), and CD16 (3G8, BD Bioscience). Subsequently, the cells were fixed in 2% paraformaldehyde in phosphate-buffered saline (PBS) and acquired on a BD 5LSRFortessa cytometer (BD Bioscience) at the Ragon Institute Imaging Core Facility at MGH.
To analyze HIV-1–specific T cell responses, PBMCs were rested in R10 medium for 4 hours at 37°C in 5% CO2 and then incubated with an HIV-1 consensus clade C Gag peptide pool (mix of 121 overlapping 15-mer peptides at 2 μg/ml for each peptide, NIH AIDS Reagent Program #12756) or SEB at 0.4 μg/ml (Sigma-Aldrich) in the presence of anti-CD28 and anti-CD49d at 1 μg/ml (BD Bioscience) and antibodies against CD107a and CD107b (clones H4A3 and H4B4, respectively, BD Bioscience). After 1 hour of incubation, brefeldin A at 5 μg/ml (BioLegend) and monensin at 1 μg/ml (BD Bioscience) were added, and cells were incubated for an additional 12 hours. An unstimulated negative control that lacked antigenic peptides but was otherwise treated identically was included for each patient. After stimulation, cells were washed and stained with LIVE/DEAD Blue Viability Dye (Invitrogen), followed by preincubation with 2 μl of FcR blocking reagent and surface staining with antibodies against CD4 (RPA-T4; BD Bioscience), CD8a (RPA-T8, BioLegend), and CD3 (UCHT1, BioLegend). After a 20 min of incubation, cells were washed, fixed, and permeabilized using the FoxP3 transcription factor buffer kit (eBioscience) for 30 min at 4°C. Intracellular cytokine staining was performed with antibodies against interferon-γ (IFN-γ) (4S.B3, BioLegend), IL-2 (MQ1-17H12, BioLegend), perforin (B-D48, BioLegend), tumor necrosis factor–α (TNF-α; MAb11, BioLegend), granzyme B (GB11, BD Bioscience), and monoclonal antibodies against Tbet (4B10, BioLegend) and Eomes (WD1928, eBioscience) for 30 min at 4°C. Subsequently, the cells were fixed in 2% paraformaldehyde in PBS and acquired on a BD 5LSRFortessa cytometer (BD Bioscience). Unstimulated controls were run for each sample and subtracted as background. Data were analyzed using FlowJo v.10.5.3 software (Tree Star LLC) and using the Simplified Presentation of Incredibly Complex Evaluations (SPICE) software (version 6.0) (75); gating strategies are summarized in figs. S7 and S8.
Statistical analyses
Experimental variables between two groups of participants were analyzed using a two-sided Mann-Whitney U test or a Wilcoxon matched-pair rank test, as appropriate. Differences were tested for statistical significance between three or more groups using the two-sided Kruskal-Wallis nonparametric test with post hoc Dunn’s multiple comparison test. Statistical associations were assessed using linear regression tests. The decrease of intact or defective viral sequences over time was fitted to an exponential decay model, and the rate of the decay was estimated using a nonlinear least-squares method; decay rates were compared using Wilcoxon signed-rank test. Data are presented as circos plots, pie charts, violin plots, bar charts, box-and-whisker plots, and heat maps. All statistical analyses were performed using GraphPad Prism 8.0.1 and SPICE software.
Supplementary Material
Acknowledgments:
We gratefully acknowledge the support of all study individuals and their families. We also acknowledge the Botswana Ministry of Health, the members of the Institutional Review Boards in Botswana and at the Harvard School of Public Health, the members of the NIH Data Monitoring and Safety Board, all staff members of the Botswana Harvard HIV Partnership Program, and the MGH DNA Core facility.
Funding:
This project was supported by NIH grant U01 AI114235 (to R.S., D.R.K., and M.L.) and NIH grant AI120850 (to K.M.P.). M.L. is supported by NIH grants AI117841, AI120008, AI124776, DK120387, AI130005, AI122377, and AI135940. X.G.Y. is supported by NIH grants AI116228, AI078799, HL134539, AI125109, and DA047034. S.M. was partially supported by the Sub-Saharan African Network for TB/HIV Research Excellence/DELTAS DEL-15- 006 and by the New Partnership for Africa’s Development Planning and Coordinating Agency/Wellcome Trust #107752/Z/15/Z.
Footnotes
SUPPLEMENTARY MATERIALS
stm.sciencemag.org/cgi/content/full/11/520/eaax7350/DC1
Fig. S1. Biological specimen sampling time points and HIV-1 plasma RNA in EIT study participants.
Fig. S2. Viral sequence diversity in early-treated neonates with HIV-1 infection.
Fig. S3. NK cell responses in EIT study participants.
Fig. S4. Longitudinal evolution of NK cell responses in EIT study participants.
Fig. S5. Myeloid cell populations in EIT children.
Fig. S6. Longitudinal evolution of T cell populations in EIT study participants.
Fig. S7. Gating strategy for innate immune cell profiling.
Fig. S8. Gating strategy for T cell immune profiling.
Table S1. Clinical and demographical characteristics of the study cohorts.
Table S2. HIV-1 integration sites identified in neonates with HIV-1 infection.
Data file S1. Primary data.
Competing interests: D.R.K. has received consulting honoraria and/or research support from Gilead, Merck, and ViiV. M.L. has received speaking and consulting honoraria from Merck and Gilead. The remaining authors declare that they have no competing interests.
Data and materials availability:
All data associated with this study are present in the paper or the Supplementary Materials. Proviral sequences have been deposited in GenBank accession numbers MK457765 to MK458272.
REFERENCES AND NOTES
- 1.Siegfried N, van der Merwe L, Brocklehurst P, Sint TT, Antiretrovirals for reducing the risk of mother-to-child transmission of HIV infection. Cochrane Database Syst. Rev 2011, CD003510 (2011). [DOI] [PubMed] [Google Scholar]
- 2. http://aidsinfo.unaids.org/.
- 3.Newell ML, Coovadia H, Cortina-Borja M, Rollins N, Gaillard P; Francois Dabis for the Ghent International AIDS Society (IAS) working group on HIV infection in women and children, Mortality of infected and uninfected infants born to HIV-infected mothers in Africa: A pooled analysis. Lancet 364, 1236–1243 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Violari A, Cotton MF, Gibb DM, Babiker AG, Steyn J, Madhi SA, Jean-Philippe P, McIntyre JA; CHER Study Team, Early antiretroviral therapy and mortality among HIV-infected infants. N. Engl. J. Med 359, 2233–2244 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization, Antiretroviral Therapy for HIV Infection in Infants and Children: Towards Universal Access—Recommendations for a Public Health Approach: 2010 Revision (World Health Organization, 2010); www.who.int/hiv/pub/paediatric/infants2010/en/index.html. [PubMed] [Google Scholar]
- 6.Persaud D, Gay H, Ziemniak C, Chen YH, Piatak M Jr., Chun T-W, Strain M, Richman D, Luzuriaga K, Absence of detectable HIV-1 viremia after treatment cessation in an infant. N. Engl. J. Med 369, 1828–1835 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luzuriaga K, Gay H, Ziemniak C, Sanborn KB, Somasundaran M, Rainwater-Lovett K, Mellors JW, Rosenbloom D, Persaud D, Viremic relapse after HIV-1 remission in a perinatally infected child. N. Engl. J. Med 372, 786–788 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frange P, Faye A, Avettand-Fenoël V, Bellaton E, Descamps D, Angin M, David A, Caillat-Zucman S, Peytavin G, Dollfus C, Le Chenadec J, Warszawski J, Rouzioux C, Saez-Cirión A; ANRS EPF-CO10 Pediatric Cohort and the ANRS EP47 VISCONTI study group, HIV-1 virological remission lasting more than 12 years after interruption of early antiretroviral therapy in a perinatally infected teenager enrolled in the French ANRS EPF-CO10 paediatric cohort: A case report. Lancet HIV 3, e49–e54 (2016). [DOI] [PubMed] [Google Scholar]
- 9.McMahon JH, Chang J, Tennakoon S, Dantanarayana A, Solomon A, Cherry C, Doherty R, Cameron P, Lewin SR, Post-treatment control in an adult with perinatally acquired HIV following cessation of antiretroviral therapy. AIDS 31, 1344–1346 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Violari A, Cotton M, Kuhn L, Schramm D, Paximadis M, Loubser S, Shalekoff S, da Costa Dias B, Otwombe K, Liberty A, McIntyre J, Babiker A, Gibb D, Tiemessen C, Viral and host characteristics of a child with perinatal HIV-1 following a prolonged period after ART cessation in the CHER trial, paper presented at the9th International AIDS Society Conference on HIV Science, Paris, France, 23 to 26 July 2017. [Google Scholar]
- 11.Luzuriaga K, Early combination antiretroviral therapy limits HIV-1 persistence in children. Annu. Rev. Med 67, 201–213 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Rainwater-Lovett K, Luzuriaga K, Persaud D, Very early combination antiretroviral therapy in infants: Prospects for cure. Curr. Opin. HIV AIDS 10, 4–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takata H, Buranapraditkun S, Kessing C, Fletcher JLK, Muir R, Tardif V, Cartwright P, Vandergeeten C, Bakeman W, Nichols CN, Pinyakorn S, Hansasuta P, Kroon E, Chalermchai T, O’Connell R, Kim J, Phanuphak N, Robb ML, Michael NL, Chomont N, Haddad EK, Ananworanich J, Trautmann L; RV254/SEARCH010 and the RV304/SEARCH013 Study Groups, Delayed differentiation of potent effector CD8+ T cells reducing viremia and reservoir seeding in acute HIV infection. Sci. Transl. Med 9, eaag1809 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saez-Cirion A, Hamimi C, Bergamaschi A, David A, Versmisse P, Mélard A, Boufassa F, Barré-Sinoussi F, Lambotte O, Rouzioux C, Pancino G; ANRS CO18 Cohort, Restriction of HIV-1 replication in macrophages and CD4+ T cells from HIV controllers. Blood 118, 955–964 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bui JK, Sobolewski MD, Keele BF, Spindler J, Musick A, Wiegand A, Luke BT, Shao W, Hughes SH, Coffin JM, Kearney MF, Mellors JW, Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLOS Pathog. 13, e1006283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy J-P, Haddad EK, Sékaly R-P, HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med 15, 893–900 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Einkauf KB, Lee GQ, Gao C, Sharaf R, Sun X, Hua S, Chen SMY, Jiang C, Lian X, Chowdhury FZ, Rosenberg ES, Chun T-W, Li JZ, Yu XG, Lichterfeld M, Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J. Clin. Invest 129, 988–998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mavigner M, Habib J, Deleage C, Rosen E, Mattingly C, Bricker K, Kashuba A, Amblard F, Schinazi RF, Lawson B, Vanderford TH, Jean S, Cohen J, McGary C, Paiardini M, Wood MP, Sodora DL, Silvestri G, Estes J, Chahroudi A, Simian immunodeficiency virus persistence in cellular and anatomic reservoirs in antiretroviral therapy-suppressed infant rhesus macaques. J. Virol 92, e00562–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veenhuis RT, Blankson JN, The antiviral immune response and its impact on the HIV-1 reservoir. Curr. Top. Microbiol. Immunol 417, 43–67 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Basha S, Surendran N, Pichichero M, Immune responses in neonates. Expert Rev. Clin. Immunol 10, 1171–1184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.UNAIDS, Ending AIDS: Progress towards 90-90-90 targets (2017); www.unaids.org/en/resources/documents/2017/20170720_Global_AIDS_update_2017.
- 22.Bryson YJ, Luzuriaga K, Sullivan JL, Wara DW, Proposed definitions for in utero versus intrapartum transmission of HIV-1. N. Engl. J. Med 327, 1246–1247 (1992). [DOI] [PubMed] [Google Scholar]
- 23.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho Y-C, Richman DD, Deeks SG, Siliciano JD, Siliciano RF, Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med 22, 1043–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho Y-C, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, Siliciano RF, Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, Kuo H-H, Hua S, Chen H-R, Ouyang Z, Reddy K, Dong K, Ndung’u T, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M, Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J. Clin. Invest 127, 2689–2696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosmane NN, Kwon KJ, Bruner KM, Capoferri AA, Beg S, Rosenbloom DIS, Keele BF, Ho Y-C, Siliciano JD, Siliciano RF, Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J. Exp. Med 214, 959–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH, Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner TA, McLaughlin S, Garg K, Cheung CYK, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM, Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marini B, Kertesz-Farkas A, Ali H, Lucic B, Lisek K, Manganaro L, Pongor S, Luzzati R, Recchia A, Mavilio F, Giacca M, Lusic M, Nuclear architecture dictates HIV-1 integration site selection. Nature 521, 227–231 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Bernstein HB, Kinter AL, Jackson R, Fauci AS, Neonatal natural killer cells produce chemokines and suppress HIV replication in vitro. AIDS Res. Hum. Retroviruses 20, 1189–1195 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Poli A, Michel T, Theresine M, Andres E, Hentges F, Zimmer J, CD56bright natural killer (NK) cells: An important NK cell subset. Immunology 126, 458–465 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S, Functions of natural killer cells. Nat. Immunol 9, 503–510 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Jacobson A, Bell F, Lejarcegui N, Mitchell C, Frenkel L, Horton H, Healthy neonates possess a CD56-negative NK cell population with reduced anti-viral activity. PLOS ONE 8, e67700 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mavilio D, Lombardo G, Benjamin J, Kim D, Follman D, Marcenaro E, O’Shea MA, Kinter A, Kovacs C, Moretta A, Fauci AS, Characterization of CD56-/CD16+ natural killer (NK) cells: A highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc. Natl. Acad. Sci. U.S.A 102, 2886–2891 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bjorkstrom NK, Ljunggren HG, Sandberg JK, CD56 negative NK cells: Origin, function, and role in chronic viral disease. Trends Immunol. 31, 401–406 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Kurioka A, Cosgrove C, Simoni Y, van Wilgenburg B, Geremia A, Björkander S, Sverremark-Ekstrom E, Thurnheer C, Günthard HF, Khanna N; Swiss HIV Cohort Study; Oxford IBD Cohort Investigators, Walker LJ, Arancibia-Cárcamo CV, Newell EW, Willberg CB, Klenerman P, CD161 defines a functionally distinct subset of pro-inflammatory natural killer cells. Front. Immunol 9, 486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee R, Kanti Barman P, Kumar Thatoi P, Tripathy R, Kumar Das B, Ravindran B, Non-classical monocytes display inflammatory features: Validation in sepsis and systemic lupus erythematous. Sci. Rep 5, 13886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziegler-Heitbrock L, The CD14+ CD16+ blood monocytes: Their role in infection and inflammation. J. Leukoc. Biol 81, 584–592 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Schelonka RL, Infante AJ, Neonatal immunology. Semin. Perinatol 22, 2–14 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Garcia AM, Fadel SA, Cao S, Sarzotti M, T cell immunity in neonates. Immunol. Res 22, 177–190 (2000). [DOI] [PubMed] [Google Scholar]
- 41.Hunt PW, Cao HL, Muzoora C, Ssewanyana I, Bennett J, Emenyonu N, Kembabazi A, Neilands TB, Bangsberg DR, Deeks SG, Martin JN, Impact of CD8+ T-cell activation on CD4+ T-cell recovery and mortality in HIV-infected Ugandans initiating antiretroviral therapy. AIDS 25, 2123–2131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roider J, Muenchhoff M, Goulder PJR, Immune activation and paediatric HIV-1 disease outcome. Curr. Opin. HIV AIDS 11, 146–155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knox JJ, Cosma GL, Betts MR, McLane LM, Characterization of T-bet and eomes in peripheral human immune cells. Front. Immunol 5, 217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.US Department of Health and Human Services, Recommendations for the Use of Antiretroviral Drugs in Pregnant Women with HIV Infection and Interventions to Reduce Perinatal HIV Transmission in the United States (2018).
- 45.Kuhn L, Shiau S, The pharmacological treatment of acute HIV infections in neonates. Expert Rev. Clin. Pharmacol 10, 1353–1361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Utay NS, Hunt PW, Role of immune activation in progression to AIDS. Curr. Opin. HIV AIDS 11, 131–137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiegand A, Spindler J, Hong FF, Shao W, Cyktor JC, Cillo AR, Halvas EK, Coffin JM, Mellors JW, Kearney MF, Single-cell analysis of HIV-1 transcriptional activity reveals expression of proviruses in expanded clones during ART. Proc. Natl. Acad. Sci. U.S.A 114, E3659–E3668 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shearer WT, Rosenblatt HM, Gelman RS, Oyomopito R, Plaeger S, Stiehm ER, Wara DW, Douglas SD, Luzuriaga K, McFarland EJ, Yogev R, Rathore MH, Levy W, Graham BL, Spector SA; Pediatric AIDS Clinical Trials Group, Lymphocyte subsets in healthy children from birth through 18 years of age: The Pediatric AIDS Clinical Trials Group P1009 study. J. Allergy Clin. Immunol 112, 973–980 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Levy O, Innate immunity of the newborn: Basic mechanisms and clinical correlates. Nat. Rev. Immunol 7, 379–390 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Shan L, Deng K, Gao H, Xing S, Capoferri AA, Durand CM, Rabi SA, Laird GM, Kim M, Hosmane NN, Yang H-C, Zhang H, Margolick JB, Li L, Cai W, Ke R, Flavell RA, Siliciano JD, Siliciano RF, Transcriptional reprogramming during effector-to-memory transition renders CD4+ T cells permissive for latent HIV-1 infection. Immunity 47, 766–775. e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shalekoff S, Gray GE, Tiemessen CT, Age-related changes in expression of CXCR4 and CCR5 on peripheral blood leukocytes from uninfected infants born to human immunodeficiency virus type 1-infected mothers. Clin. Diagn. Lab. Immunol 11, 229–234 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bunders MJ, van der Loos CM, Klarenbeek PL, van Hamme JL, Boer K, Wilde JCH, de Vries N, van Lier RAW, Kootstra N, Pals ST, Kuijpers TW, Memory CD4+CCR5+ T cells are abundantly present in the gut of newborn infants to facilitate mother-to-child transmission of HIV-1. Blood 120, 4383–4390 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Cesana D, Santoni de Sio FR, Rudilosso L, Gallina P, Calabria A, Beretta S, Merelli I, Bruzzesi E, Passerini L, Nozza S, Vicenzi E, Poli G, Gregori S, Tambussi G, Montini E, HIV-1-mediated insertional activation of STAT5B and BACH2 trigger viral reservoir in T regulatory cells. Nat. Commun 8, 498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahata Y, Nomura A, Takada H, Ohga S, Furuno K, Hikino S, Nakayama H, Sakaguchi S, Hara T, CD25+CD4+ T cells in human cord blood: An immunoregulatory subset with naive phenotype and specific expression of forkhead box p3 (Foxp3) gene. Exp. Hematol 32, 622–629 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Goulder PJ, Lewin SR, Leitman EM, Paediatric HIV infection: The potential for cure. Nat. Rev. Immunol 16, 259–271 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dalle J-H, Menezes J, Wagner É, Blagdon M, Champagne J, Champagne MA, Duval M, Characterization of cord blood natural killer cells: Implications for transplantation and neonatal infections. Pediatr. Res 57, 649–655 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Adkins B, Leclerc C, Marshall-Clarke S, Neonatal adaptive immunity comes of age. Nat. Rev. Immunol 4, 553–564 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Zhang X, Zhivaki D, Lo-Man R, Unique aspects of the perinatal immune system. Nat. Rev. Immunol 17, 495–507 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Hebel K, Weinert S, Kuropka B, Knolle J, Kosak B, Jorch G, Arens C, Krause E, Braun-Dullaeus RC, Brunner-Weinzierl MC, CD4+ T cells from human neonates and infants are poised spontaneously to run a nonclassical IL-4 program. J. Immunol 192, 5160–5170 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Ramduth D, Thobakgale CF, Mkhwanazi NP, De Pierres C, Reddy S, van der Stok M, Mncube Z, Mphatswe W, Blanckenberg N, Cengimbo A, Prendergast A, Tudor-Williams G, Dong K, Jeena P, Coovadia HM, Day CL, Kiepiela P, Goulder PJ, Walker BD, Detection of HIV type 1 gag-specific CD4+ T cell responses in acutely infected infants. AIDS Res. Hum. Retroviruses 24, 265–270 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Essat A, Scott-Algara D, Monceaux V, Avettand-Fenoel V, Didier C, Caillat-Zucman S, Orr S, Theodorou I, Goujard C, Boufassa F, Lambotte O, Rouzioux C, Hocqueloux L, Meyer L, Saez-Cirion A, Association between immunogenetic factors and post-treatment control of HIV-1 infection, in Abstracts of the 22nd International AIDS Conference, Amsterdam, Netherlands (abstract number TUPDA0101) (2018). [Google Scholar]
- 62.Okoye AA, Hansen SG, Vaidya M, Fukazawa Y, Park H, Duell DM, Lum R, Hughes CM, Ventura AB, Ainslie E, Ford JC, Morrow D, Gilbride RM, Legasse AW, Hesselgesser J, Geleziunas R, Li Y, Oswald K, Shoemaker R, Fast R, Bosche WJ, Borate BR, Edlefsen PT, Axthelm MK, Picker LJ, Lifson JD, Early antiretroviral therapy limits SIV reservoir establishment to delay or prevent post-treatment viral rebound. Nat. Med 24, 1430–1440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sáez-Cirión A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, Potard V, Versmisse P, Melard A, Prazuck T, Descours B, Guergnon J, Viard J-P, Boufassa F, Lambotte O, Goujard C, Meyer L, Costagliola D, Venet A, Pancino G, Autran B, Rouzioux C; ANRS VISCONTI Study Group, Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLOS Pathog. 9, e1003211 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Namazi G, Fajnzylber JM, Aga E, Bosch RJ, Acosta EP, Sharaf R, Hartogensis W, Jacobson JM, Connick E, Volberding P, Skiest D, Margolis D, Sneller MC, Little SJ, Gianella S, Smith DM, Kuritzkes DR, Gulick RM, Mellors JW, Mehraj V, Gandhi RT, Mitsuyasu R, Schooley RT, Henry K, Tebas P, Deeks SG, Chun T-W, Collier AC, Routy J-P, Hecht FM, Walker BD, Li JZ, The Control of HIV After Antiretroviral Medication Pause (CHAMP) Study: Posttreatment controllers identified from 14 clinical studies. J. Infect. Dis 218, 1954–1963 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, Pereyra F, Zurakowski R, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M, HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat. Med 20, 139–142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.R Core Team, R: A language and environment for statistical computing. R Foundation for Statistical Computing; (2014);www.R-project.org/. [Google Scholar]
- 67.Lee GQ, Reddy K, Einkauf KB, Gounder K, Chevalier JM, Dong KL, Walker BD, Yu XG, Ndung’u T, Lichterfeld M, HIV-1 DNA sequence diversity and evolution during acute subtype C infection. Nat. Commun 10, 2737 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rose PP, Korber BT, Detecting hypermutations in viral sequences with an emphasis on G → A hypermutation. Bioinformatics 16, 400–401 (2000). [DOI] [PubMed] [Google Scholar]
- 69.Edgar RC, MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar S, Stecher G, Li M, Knyaz C, Tamura K, MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. https://genome.ucsc.edu/
- 73. www.repeatmasker.org.
- 74. www.qiagenbioinformatics.com.
- 75. https://niaid.github.io/spice.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the paper or the Supplementary Materials. Proviral sequences have been deposited in GenBank accession numbers MK457765 to MK458272.


