Abstract
Ras plays a key role in regulating cellular proliferation, differentiation, and transformation. Raf is the major effector of Ras in the Ras > Raf > Mek > extracellular signal-activated kinase (ERK) cascade. A second effector is phosphoinositide 3-OH kinase (PI 3-kinase), which, in turn, activates the small G protein Rac. Rac also has multiple effectors, one of which is the serine threonine kinase Pak (p65Pak). Here we show that Ras, but not Raf, activates Pak1 in cotransfection assays of Rat-1 cells but not NIH 3T3 cells. We tested agents that activate or block specific components downstream of Ras and demonstrate a Ras > PI 3-kinase > Rac/Cdc42 > Pak signal. Although these studies suggest that the signal from Ras through PI 3-kinase is sufficient to activate Pak, additional studies suggested that other effectors contribute to Pak activation. RasV12S35 and RasV12G37, two effector mutant proteins which fail to activate PI 3-kinase, did not activate Pak when tested alone but activated Pak when they were cotransfected. Similarly, RacV12H40, an effector mutant that does not bind Pak, and Rho both cooperated with Raf to activate Pak. A dominant negative Rho mutant also inhibited Ras activation of Pak. All combinations of Rac/Raf and Ras/Raf and Rho/Raf effector mutants that transform cells cooperatively stimulated ERK. Cooperation was Pak dependent, since all combinations were inhibited by kinase-deficient Pak mutants in both transformation assays and ERK activation assays. These data suggest that other Ras effectors can collaborate with PI 3-kinase and with each other to activate Pak. Furthermore, the strong correlation between Pak activation and cooperative transformation suggests that Pak activation is necessary, although not sufficient, for cooperative transformation of Rat-1 fibroblasts by Ras, Rac, and Rho.
Ras is one of the most commonly mutated oncogenes and is found activated in 20 to 30% of tumors (29). Ras encodes a small G protein that binds GTP and GDP and possesses intrinsic GTPase activity. In its oncogenic form, Ras acquires a point mutation that inactivates the GTPase activity and causes it to be locked into its activated GTP-bound state. Normally, Ras is activated by growth factor receptors through its guanine nucleotide exchange factors (GEF) (12).
The major oncogenic signal from Ras utilizes the serine threonine kinase Raf as the effector (52, 54). GTP-bound Ras binds and activates Raf and simultaneously recruits it to the membrane. Upon activation, Raf phosphorylates and activates another kinase, Mek, which, in turn, activates extracellular signal-activated kinase (ERK) (mitogen-activated protein kinase [MAPK]). This Ras > Raf > Mek > ERK signal is usually referred to as the MAPK cascade (34). In recent years, Ras has also been shown to bind other effectors besides Raf and activate other signaling pathways that cooperate with the Raf > ERK signal (56). The other pathways are not as well defined as the Raf cascade. The three effectors that have been most widely studied are Rin, Ral GDS, and phosphoinositide 3-OH kinase (PI 3-kinase) (1, 39, 46). Each binds Ras-GTP and can, in some experimental systems, cooperate with partially activated Raf mutants to transform cells (Rin cooperates with Abl). Further evidence of the importance of these effectors in Ras signaling comes from new Ras point mutants, known as effector mutants, that bind and activate only subsets of Ras effectors (56). RasV12S35 binds and activates Raf, RasV12G37 binds and activates Ral GDS and Rin 1, and RasV12C40 binds and activates PI 3-kinase (21, 46, 56, 57). These mutants are deficient in signaling when tested alone but cooperate to transform cells when introduced together. Signals from Ras through the alternate effectors utilize other small G proteins. Ral GDS uses Ral, and PI 3-kinase uses the small G protein Rac (46, 57).
Rho and two related proteins, Rac and Cdc42, are members of the Rho family of small G proteins. These proteins are about 50% identical to Ras and regulate the actin cytoskeleton. Rho induces stress fibers and focal adhesions (44), Rac induces accumulation of actin-rich ruffles or lamellipodia at the periphery of cells (45), and Cdc42 induces microspikes or filopodia (37). Each Rho family member also activates a kinase cascade that leads to transcriptional activation similar to the MAPK cascade but not as well defined. Rho activates the ternary complex factors, and Rac and Cdc42 activate the Jun N-terminal kinase cascade JNK(SAPK) (8, 15, 19, 35). Dominant negative mutants of Rac, Rho, and Cdc42 each inhibit Ras transformation, and activated mutants cooperate with Raf to transform cells (23, 40–42). These observations suggest that the signals through the Rho family of small G proteins play essential roles in Ras transformation.
The signals through Rac are directly connected to Ras (4, 45). This is because Ras and Rac both cause membrane ruffling when microinjected into cells and a dominant negative Rac mutant inhibits Ras-induced ruffling. The signal from Ras to Rac is likely to be mediated by PI 3-kinase, since RasV12C40, which activates PI 3-kinase, and activated mutants of PI 3-kinase both induce ruffles (21, 46). The mechanism that PI 3-kinase uses to activate Rac probably involves stimulation of Rac GEF by PI 3-kinase products such as phosphatidylinositol-3,4,5-triphosphate (17, 36). The immediate effector downstream of Rac in Ras signal transduction to both the JNK and actin pathways has remained elusive. One candidate has been the serine threonine kinase p65Pak (32). Pak was first isolated as a protein that binds both Rac and Cdc42 in their GTP-bound forms. Pak is homologous to Ste20, a protein kinase in the yeast Saccharomyces cerevisiae regulated by Cdc42 (28, 47). Some of the activities of Pak resemble those of Ras and Rac. For example, microinjection of Pak into some cells causes ruffling and breaks up stress fibers and membrane targeting of Pak in PC12 cells induces extension of neurites (10, 31, 48). Although microinjection of Pak can cause membrane ruffling, ruffling does not require kinase activity (48). Moreover, direct signals from Ras to Pak have not been reported. Finally, RacV12H40, an effector mutant that does not bind to Pak, still cooperates with Raf to transform cells and causes membrane ruffling when it is microinjected (20, 26, 55). These studies suggest that there may be no role for Pak in Ras transformation or signaling and even question the existence of any direct signals from Ras to Pak.
We recently reported that Pak mutants that lack kinase activity behave as dominant negative mutants and inhibit Ras transformation of Rat-1 cells and Schwann cells but not of NIH 3T3 cells (50, 51). Inhibition was not the result of Rac/Cdc42 sequestering, since kinase-deficient mutants that fail to bind Rac and Cdc42 also inhibited transformation. Studies of downstream effectors suggested that Pak mutants blocked the signal from Ras to ERK. More recently, Pak was shown to facilitate ERK kinase activation by phosphorylating Mek (14, 30). Together, these data suggest that there is a Ras > Rac > Pak > ERK signal that is essential to sustain transformation in some cells.
We demonstrate here that Ras activates Pak in transfection assays of Rat-1 cells but not NIH 3T3 cells. The primary signal from Ras to Pak was mediated by PI 3-kinase to Rac and Cdc42. However, effector mutants of both Ras and Rac which could not activate Pak by themselves cooperated with each other and with Raf to activate Pak. All combinations of Ras, Rac, and Rho mutants that transformed cells were capable of activating Pak and were also inhibited by Pak dominant negative mutants. These studies developed an assay system for studying the signals from Ras to Pak and suggest that activation of Pak may be essential for transformation by Ras, Rac, and Rho.
MATERIALS AND METHODS
Plasmids.
cDNA expression plasmids utilizing the cytomegalovirus promoter to express myc-tagged Pak1, Pak1R299, Pak1L83,L86, and Pak1L83,L86,R299 based on the plasmid pCMV6M (a modified version of pCMV5) have been described elsewhere (48). RacV12, RacV12L37, and RacV12H40 that utilized the PCGT vector were gifts from Dafna Bar-Sagi and Linda Van Aelst (20). RacL61, RafD340, and human K-ras4B were gifts from Channing Der. RhoAV14 and RhoAN19 were gifts from Marc Symons. p110-CAAX, which consists of the p110 catalytic subunit of PI 3-kinase targeted to the membrane through fusion with the CAAX sequence of H-ras was as previously described (25). v-Raf, JNK, and ERK plasmids have been described elsewhere (50).
Cell culture and transformation assays.
Rat-1 cells were grown at 37°C in 5% CO2–95% air in high-glucose (4.5 g/liter) Mediatech Dulbecco’s modified Eagle medium purchased from Fisher Scientific (Pittsburgh, Pa.) supplemented with 10% fetal bovine serum (Fisher), penicillin (100 U/ml), and streptomycin (100 mg/ml). rv68BUR cells were a gift from Jim Stone (49). DNA transfections were performed by the calcium phosphate precipitation technique as described previously (50). Twenty micrograms of total DNA (7 μg of each test DNA and 6 μg of Pak test DNA; the total DNA content in all transfections was brought to 20 μg, if necessary, with plasmid pUC19 was transfected for each dish. Soft agar assays were performed as previously described (9). We plated 103 posttransfection cells on 60-mm-diameter dishes. After 18 to 21 days, colonies were examined under a Nikon DIAPhot microscope using phase-contrast optics and the dishes were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide overnight at 37°C.
Pak and ERK/MAP kinase assays.
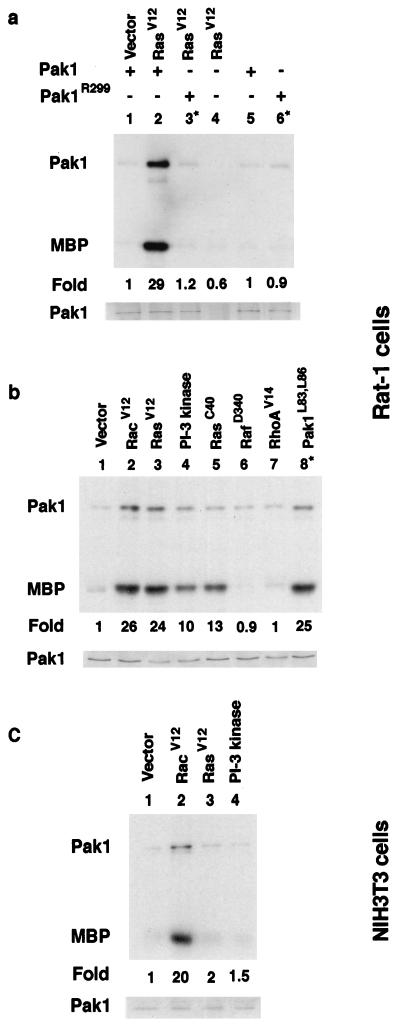
Transfections of Rat-1 cells were performed as described above for the transformation assays. For ERK kinase assays, 5 μg of total DNA (1 μg of hemagglutinin-ERK1, 1.5 μg of each test DNA, and 1 μg of Pak DNA; the total DNA content was brought to 5 μg with plasmid pUC19) was transfected for each dish. The procedure for the ERK kinase assay has been described elsewhere (50). For the Pak kinase assays, a total of 5 μg of DNA was transfected into cells (1 μg of Pak DNA and 1.3 μg of each test DNA; the total DNA content was brought to 5 μg with pUC19 plasmid DNA, if necessary). Cells were lysed 24 to 48 h after transfection. Transfected cells were washed with cold phosphate-buffered saline and lysed in 40 mM HEPES (pH 7.4)–1% Nonidet P-40–100 mM NaCl–1 mM EDTA–25 mM NaF–1 mM sodium orthovanadate–10-mg/ml leupeptin–10-mg/ml aprotinin and centrifuged at 12,000 × g for 25 min at 4°C. Protein concentrations ranged from 2.9 to 6.6 mg/ml. The extracts were routinely tested for Pak expression on Western blots probed with antibody 9E10. An example of a Western blot is shown in Fig. 1.
FIG. 1.
Ras and PI 3-kinase activation of Pak in Rat-1 cells. Rat-1 cells were transfected with the indicated plasmids along with a myc-tagged Pak1 construct; extracts were then prepared and used in immune kinase assays. Fold indicates fold activation compared to a vector control (lane 1) as determined by PhosphorImager analysis. MBP, myelin basic protein.
Pak kinase assays were performed on anti-Myc immunoprecipitates from cell extracts as follows. Extracts were incubated with antibody 9E10 and protein A beads for 2 h at 4°C. Precipitates were washed three times with lysis buffer. Immunoprecipitates were washed twice in 2× phosphorylation buffer (10 mM MgCl2, 40 mM HEPES, pH 7.4) and then incubated with 5 μg of myelin basic protein (Sigma) for 5 min on ice. Kinase assays were initiated by the addition of 10 μCi of [γ-32P]ATP (3,000 Ci/mmol) and 20 μM (final concentration) ATP, followed by incubation for 10 min at 22°C (3). Reactions were stopped by the addition of 2× sodium dodecyl sulfate sample buffer and heated to 95°C, and the products were resolved by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis and visualized by autoradiography. All experiments were performed two or three times with similar results.
RESULTS
Ras activates Pak primarily through PI 3-kinase.
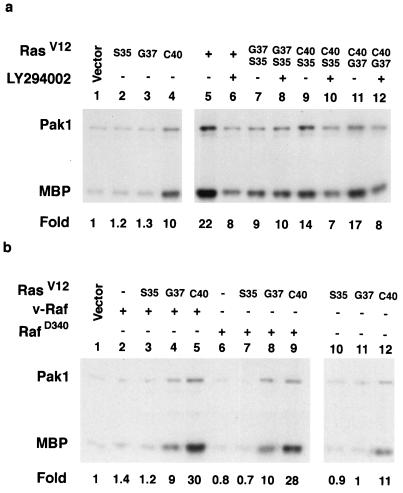
Since previous data showed that Pak dominant negative mutants inhibit Ras but not Raf transformation in Rat-1 fibroblast cells, we designed experiments to determine if Ras activates Pak (50). We cotransfected Pak into cells with Ras test plasmids, prepared extracts, and then performed immune complex kinase assays taking advantage of the myc tag fused to the N terminus of Pak to precipitate Pak from cell lysates. Kinase assays were carried out by using [γ-32P]ATP to label myelin basic protein as a substrate. Pak also autophosphorylates and is seen as an ∼65-kDa band. We found that activated Ras stimulated Pak activity more than 20-fold over basal levels (Fig. 1a). As controls for the assay, we tested the kinase-deficient mutant PakR299 or omitted Pak altogether from the transfections. In each case, no activity was observed over basal levels. Thus, we concluded that Ras activates Pak. In direct comparisons, we found that Ras activation was equivalent to both Rac activation and Pak1L83,L86, an activated mutant (Fig. 1b, lanes 1 to 3 and 8). Rho failed to activate Pak (lane 7). In contrast, when we performed similar experiments with NIH 3T3 cells, we failed to detect any activation by Ras or PI 3-kinase under conditions in which activated Rac stimulated Pak (Fig. 1c).
To determine if Ras activation of Pak might be mediated by Raf, we tested RafD340, a partially activated Raf, and found that it failed to activate Pak (Fig. 1b, lane 6). We also tested RasV12S35, an effector mutant that activates only Raf and RasV12G37, an effector mutant which binds and activates Ral GDS, and found that both failed to activate Pak in the transfection assays (not shown in Fig. 1). Thus, Ras activates Pak through an effector distinct from Raf and Ral GDS. As PI 3-kinase has been shown to mediate Ras activation of Rac (46), we tested if an activated PI 3-kinase mutant stimulated Pak (Fig. 1b, lane 4). An activated PI 3-kinase, p110-CAAX, stimulated Pak to approximately half of the level achieved by Ras. We also observed a similar level of partial activation by RasV12C40, a Ras effector mutant that activates PI 3-kinase but not Raf (Fig. 1b, lane 5). These data suggest that RasV12 activation of PI 3-kinase is sufficient to activate Pak. The partial activation may result because RasV12C40 and p110-CAAX are not fully activated. However, evidence will be presented later suggesting that other signals from Ras participate in Pak activation.
To trace the signals from Ras and pI 3-kinase to Pak, we tested the effects of various inhibitors on Pak activation by Ras, Rac, and PI 3-kinase (Fig. 2 and 3). Substitution of Asn for the amino acid at position 17 of small G proteins (position 19 for Rho) reduces their affinity for GTP, creating dominant negative mutants (13). When we tested dominant negative mutants, we found that Ras activation of Pak was almost completely inhibited by RasN17, RacN17, Cdc42N17, and RhoN19. RasN17 inhibited RasV12 activation of Pak but not activation by PI 3-kinase or RacV12 (data not shown for Ras). While this may suggest that RasV12 uses endogenous Ras to activate Pak, the Ras dominant negative data must be interpreted cautiously since in some experimental systems, RasV12 shows partial dependence on GEF activity, while in other systems there is very little effect of RasN17 on RasV12 (7, 13). RhoN19 also appeared to act near the Ras step, since it inhibited wild-type RasV12 activation of Pak but failed to block either PI 3-kinase or RasV12C40 activation of Pak (compare Fig. 2a, lane 5, with lane 9 and Fig. 2b, lane 6). Rac and Cdc42 were farthest downstream, since the corresponding dominant negative mutants inhibited activation by RasV12, RasV12C40, and PI 3-kinase. As many GEF activate both Rac and Cdc42, our experiments with the dominant negative mutants did not allow us to distinguish between them. To further dissect the signal from Ras to Pak, we tested the effect of LY294002, a PI 3-kinase inhibitor, on Pak activation (53). We found that LY294002 inhibited Pak activation by RasV12, RasV12C40, and PI 3-kinase (Fig. 3a). LY294002 did not inhibit Rac activation of Pak, nor did it inhibit Pak activation by RacV12L37, a Rac effector mutant that activates only Pak. RacV12H40, which fails to bind Pak, did not activate Pak in this assay (Fig. 3b). Dose-response curves for LY294002 showed similar inhibition profiles for both Ras and PI 3-kinase. The 50% inhibitory concentrations were 5 to 10 μM for both genes, which is comparable to the reported in vitro 50% inhibitory concentration of 1.4 μM; antiproliferative effects are observed at ∼10 μM (53) (Fig. 3c). Thus, we placed PI 3-kinase between Ras and Rac/Cdc42. Together, these experiments allowed us to trace a Ras > PI 3-kinase > Rac/Cdc42 > Pak signal. Ras, but not PI 3-kinase, appears to require Rho as well to activate Pak; although Rho could not activate Pak when tested alone (see above), the dominant negative Rho mutant prevented Ras activation of Pak.
FIG. 2.
Effect of dominant negative mutants on Pak activation. (a) Ras activation of Pak is inhibited by dominant negative Rac, Cdc42, and Rho. (b) PI 3-kinase activation of Pak is sensitive to dominant negative Rac and Cdc42 but not dominant negative Rho. MBP, myelin basic protein
FIG. 3.
(a) Ras and PI 3-kinase activation of Pak is sensitive to LY294002. Cells were incubated for 90 min with 20 μM LY294002 prior to lysis. (b) Rac activation of Pak is not sensitive to LY294002 (20 μM). (c) Ras and PI 3-kinase dose-response curves for LY294002. MBP, myelin basic protein.
Dominant negative Pak mutants inhibit transformation by Ras, Rac, and Rho.
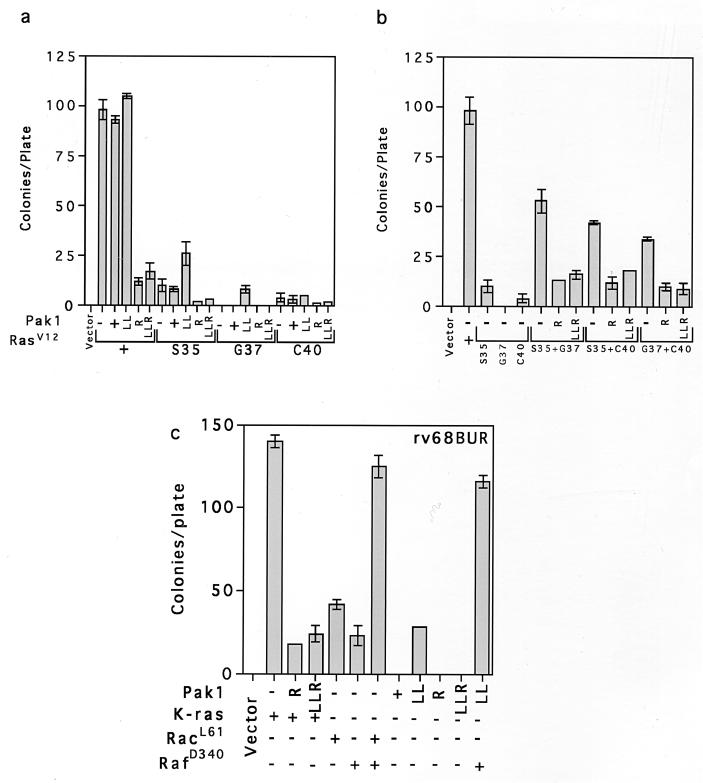
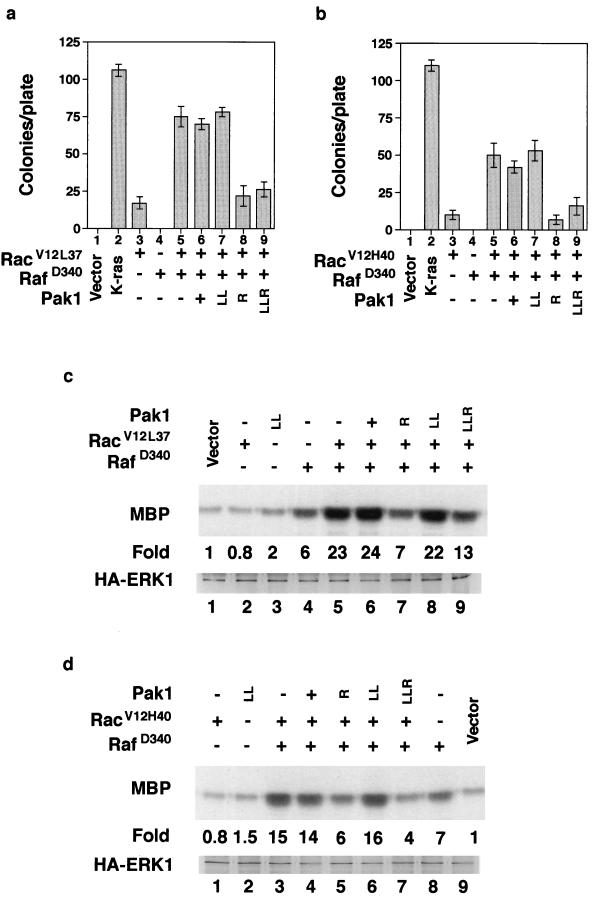
The Ras effector mutants described above transform poorly when transfected into cells individually. However, when they are introduced together, they transform at much higher frequencies. We tested the effect of activated Pak on transformation by transfecting cells with plasmids and counting the number of colonies that grew on soft agar or staining cells grown in tissue culture dishes with crystal violet to visualize transformed foci (data not shown for focus assays). We found that a small (about twofold) but reproducible stimulation of RasV12S35 and RasV12G37 transformation by Pak1L83,L86 activated Pak (Fig. 4a). No stimulation of RasV12C40 was observed, which is consistent with the observation that RasV12C40 activates Pak by itself. As expected from earlier studies, all combinations of the Ras effector mutants cooperatively transformed cells. That is, the combination of two mutants yielded more colonies than the sum of the two effector mutants. As previously reported, Ras transformation was strongly inhibited by the Pak dominant negative mutants. Interestingly, all combinations of effector mutants were also inhibited by Pak dominant negative mutants, including the combination of RasV12S35 and RasV12G37, although neither mutant activated Pak when tested alone (Fig. 4b). The small stimulation of RasV12S35 transformation by Pak1L83,L86 suggested that Pak might transform cells in cooperation with Raf. We observed no reliable stimulation of transformation with wild-type Pak in Rat-1 cells. However, Pak1L83,L86 (abbreviated as LL) transformed strain rv68BUR. This cell line is hypersensitive to transformation because it expresses an activated mutant form of Mek1. RafD340, which normally does not transform Rat-1 cells, also transformed rv68BUR (Fig. 4c). The transformation of rv68BUR was markedly increased when Pak1L83,L86 and RafD340 were tested together. In this strain, Pak1L83,L86 was about as effective as Rac in transforming and cooperating with RafD340. Furthermore, transformation of both Ras and Pak/RafD340 was inhibited by the kinase-deficient Pak mutants (data not shown for Pak inhibition). These experiments demonstrate that Pak can recapitulate most of Rac’s effects; the reason why the activated Pak mutant usually transforms poorly may be that it is not fully activated (5).
FIG. 4.
Pak dominant negative mutants inhibit transformation by Ras effector mutants. Cell transformation was measured by determining growth on soft agar as described in Materials and Methods. (a) Effects of Pak mutants on cell transformation by Ras effector mutants. (b) Effects of Pak mutants on cooperative transformation by Ras effector mutants. (c) Activated Pak cooperates with Raf to transform rv68BUR, a hypersensitive Rat fibroblast cell line. Abbreviations for the Pak mutants: LL, Pak1L83,L86 (hyperactive Pak1); R, Pak1R299 (kinase-deficient Pak1); LLR, Pak1L83,L86,R299 (both kinase-deficient and Rac/Cdc42 binding-deficient Pak1). Ras mutants are abbreviated as S35, G37, and C40, which denote mutations in the effector binding loop of RasV12.
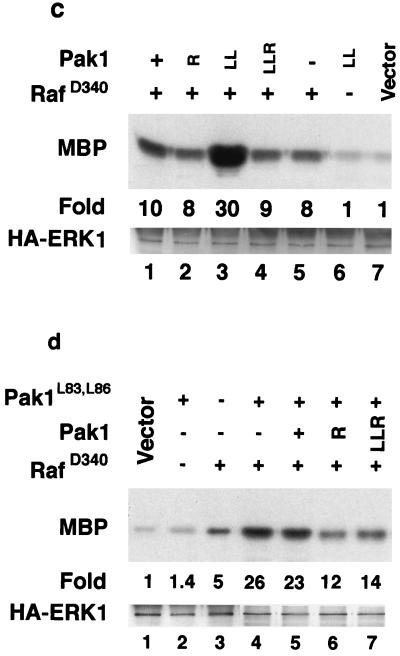
We also tested the effects of Pak mutants on RacV12 and two Rac effector mutants in cell transformation and ERK kinase assays. Rac transforms poorly by itself but cooperates with Raf to transform cells with much higher efficiency. We found that Pak dominant negative mutants inhibited Rac/Raf transformation about as effectively as they inhibited Ras transformation (Fig. 5a). Although Rac and Pak are not usually associated with ERK activation, they have recently been shown to cooperate with Raf to activate ERK in a cross-cascade activation (14). We also found that Rac did not activate ERK when tested by itself but cooperated with RafD340 to activate ERK (Fig. 5b, lanes 6 to 9). Furthermore, cooperative activation was inhibited by Pak dominant negative mutants, including Pak1L83,L86,R299, a mutant that fails to bind Rac or Cdc42 (Fig. 5b, lanes 2 to 6). The dominant negative mutants did not inhibit the partial activation of ERK by RafD340, while Pak1L83,L86 cooperated with RafD340 to activate ERK (Fig. 5c). In addition, Pak/RafD340 cooperative activation of ERK was inhibited by both Pak dominant negative mutants (Fig. 5d). Together, these data support a role for Pak in the cooperative activation of ERK by Rac.
FIG. 5.
Effects of Pak mutants on Rac/Raf cooperative. (a) Rac/RafD340 cooperative transformation is inhibited by Pak dominant negative mutants. (b) Rac/RafD340 cooperative activation of ERK is inhibited by Pak dominant negative mutants. (c) Activated Pak cooperates with Raf to stimulate ERK. (d) Pak dominant negative mutants inhibit Pak/RafD340 cooperative activation of ERK. MBP, myelin basic protein. Other abbreviations are as in Fig. 4.
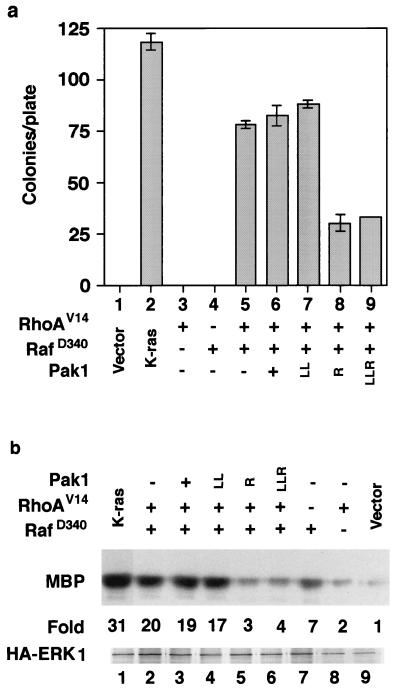
Because we had found that dominant negative Rho inhibited Ras activation of Pak (Fig. 2), we also tested the effects of Pak mutants on Rho transformation and Rho activation of ERK. Rho does not transform cells and does not activate ERK by itself but cooperates with Raf to both transform cells and activate ERK. As seen with other cells, we found that RhoV14 (activated Rho) did not transform our Rat-1 cells but cooperated with RafD340 to transform cells. Transformation was inhibited by the two dominant negative Pak mutants (Fig. 6a). We also observed cooperation between Rho and RafD340 in ERK kinase assays, and again the cooperative activation was inhibited by dominant negative Pak (Fig. 6b). These experiments suggest that Rho requires Pak for cell transformation and ERK activation.
FIG. 6.
Pak dominant negative mutants inhibit Rho/RafD340 cooperation. (a) Transformation assays. (b) ERK kinase assays. MBP, myelin basic protein; HA, hemagglutinin. Other abbreviations are as in Fig. 4.
We next tested the effects of two Rac effector mutants, RacV12L37 (Fig. 7a) and RacV12H40 (Fig. 7b), in transformation assays and ERK kinase assays (Fig. 7c and d). As discussed above, RacL12L37 binds and activates Pak, while RacV12H40 does not bind Pak. Both mutants cooperated with Raf to transform cells, although transformation by RacV12H40 was about 25% reduced. As was observed with Ras and Rho, Pak dominant negative mutants inhibited transformation by both combinations of Rac effector mutants. Surprisingly, both Rac effector mutants were equally effective in activating ERK in cooperation with RafD340 (Fig. 7c and d). Cooperative activation of ERK by both mutants was also inhibited by the dominant negative Paks. These observations suggest that transformation and ERK activation by Ras, Rho, and Rac require a common signal through Pak.
FIG. 7.
Effects of Pak mutants on Rac effector mutants. (a) Pak dominant negative mutants inhibit RacV12L37/RafD340 cooperative transformation. (b) Pak dominant negative mutants inhibit RacV12H40/RafD340 cooperative transformation. (c) RacV12L37 and RafD340 cooperate to activate ERK and are inhibited by Pak dominant negative mutants. (d) RacH40 and RafD340 cooperate to activate ERK and are inhibited by Pak dominant negative mutants. MBP, myelin basic protein; HA, hemagglutinin. Other abbreviations are as in Fig. 4.
Rho, Rac, and Ras effector mutants can cooperate to activate Pak.
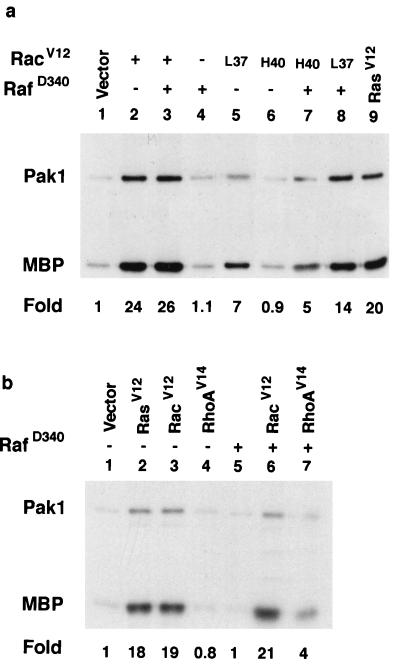
The above results indicate that while PI 3-kinase and Cdc42/Rac play a dominant role in Ras-mediated activation of Pak, other Ras- and Rho-dependent signals may also be required. This is because we observed that many Rho, Ras, and Rac effector mutants were inhibited by Pak dominant negative mutants in transformation assays and in ERK activation assays. Therefore, we tested if these proteins could activate Pak in transfection assays under the same conditions we used to perform the transformation and ERK assays. We tested combinations of the Ras effector mutants and, as expected, found that Pak was activated by RasV12C40 alone (Fig. 8a, lane 4) and in combination with all of the other effector mutants (Fig. 8a, lanes 9 to 12). Interestingly, RasV12G37 and RasV12S35, neither of which activated Pak alone, stimulated Pak when they were tested together (Fig. 8a, lane 7). Two lines of evidence suggested that RasV12G37 and Ras12S35 were activating Pak independently of PI 3-kinase. First, RasV12G37 and RasV12S35 cooperative activation was not inhibited by LY294002 (Fig. 8a, lane 8). Second, we found that two Raf mutants, v-Raf and RafD340, could substitute for RasV12S35 to activate Pak in cooperation with RasV12G37 (Fig. 8b, lanes 4 and 8). RasV12S35, v-Raf, and RafD340 cooperation with RasV12G37 consistently activated Pak to levels about half of those achieved with RasV12 (Fig. 8a, lanes 5 and 7, and b, lanes 4 and 8). Similarly, RasV12C40, the mutant capable of activating Pak by itself, only activated Pak about half as well as did Ras. However, when we cotransfected RafD340 or v-Raf with RasV12C40, we observed maximum levels of Pak activation (Fig. 8b, lanes 5 and 9). Together, these data suggest that multiple Ras effectors cooperate to activate Pak.
FIG. 8.
Cooperative activation of Pak by Ras effector mutants through a PI 3-kinase-independent mechanism. (a) Ras effector mutants cooperate with each other to activate Pak. (b) Ras effector mutants cooperate with Raf to activate Pak. MBP, myelin basic protein. Other abbreviations are as in Fig. 4.
When we cotransfected Pak and the Rac effector mutants, we found that as expected, RacV12L37, but not RacV12H40, activated Pak (Fig. 9a, lanes 5 and 6). However, we found that addition of RafD340 to transfections with RacV12H40 led to significant activation of Pak (Fig. 9a, lane 7). We failed to detect cooperative Pak activation by RacV12H40 and RafD340 in NIH 3T3 cells (data not shown). Thus, a mutant Rac which failed to bind and activate Pak could cooperate with Raf to activate Pak, activate ERK, and transform cells (Fig. 7b and d).
FIG. 9.
Activation of Pak by Rac and Rho effector mutants. (a) RacV12H40 cooperates with Raf to activate Pak. (b) RhoV14 cooperates with Raf to activate Pak. L37 is RacV12L37, and H40 is RacV12H40. MBP, myelin basic protein.
Since we found that Rho cooperation with RafD340 was inhibited by Pak dominant negative mutants (Fig. 6) and dominant negative Rho inhibited Ras activation of Pak (Fig. 2a), we tested if RhoV14 could activate Pak. As seen in Fig. 1, both RhoV14 and RafD340 failed to activate Pak when tested alone (Fig. 9b, lanes 4 and 5). However, cotransfection of RhoV14 and RafD340 together activated Pak to approximately the same levels attained by RacV12H40 and RafD340. To determine if the signals to Pak were mediated by an autocrine loop, we prepared medium from cells transfected with Ras or Raf and added it to cells expressing wild-type Pak. We failed to detect any stimulation of Pak, even when cells were transfected with RacV12H40 (data not shown). Thus, we concluded that our cells did not secrete any growth factors that can stimulate Pak through an autocrine loop. Together, these studies suggest that multiple signals from Ras, Rac, and Rho facilitate Pak activation, which can then lead to ERK activation and high-efficiency transformation in Rat-1 fibroblasts.
DISCUSSION
Pak protein kinases have been candidates for signaling proteins downstream of Ras because they are activated by Rac and Cdc42, but direct signals from Ras to Pak have not been reported. We describe here an assay to measure Ras activation of Pak through immune complex kinase assays. Four lines of evidence suggest that Ras activates Pak through a Raf-independent signal. First, RasV12 activated Pak in our assay system. Second, a PI 3-kinase-specific effector mutant, RasV12C40, also activated Pak, while RasV12S35, which only activates Raf, failed to activate Pak. Third, activated PI 3-kinase stimulated Pak while activated Raf did not. Fourth, the PI 3-kinase inhibitor LY294002 blocked Pak activation by PI 3-kinase and Ras but not that by Rac. Furthermore, activation of Pak by both Ras and PI 3-kinase was inhibited by dominant negative Rac and Cdc42. These data allowed us to delineate a Ras > PI 3-kinase > Rac/Cdc42 > Pak signal. Ras can activate PI 3-kinase directly, but PI 3-kinase probably activates Rac and Cdc42 through activation of specific GEF by second messengers synthesized by PI 3-kinase (17, 36). Products of PI 3-kinase can bind a modular protein domain, the PH domain, found on almost all Rac/Cdc42 exchange factors (43). However, a recent study comparing exchange factors showed that they differentially transduce signals to Pak and JNK and concluded that each exchange factor confers specificity on downstream signals and does not behave as a universal small G protein activator (58). Since we found that Ras and PI 3-kinase activate Pak in Rat-1 but not NIH 3T3 cells, our data suggest that a level of signal specificity exists from upstream inputs.
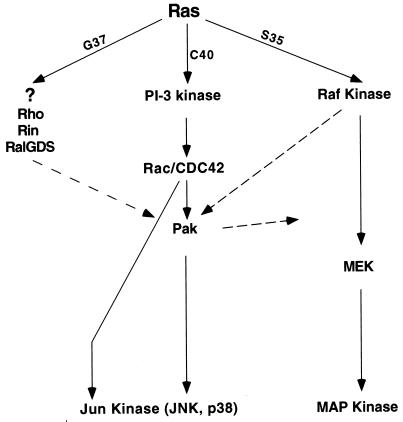
While the PI 3-kinase signal is sufficient to activate Pak, we also provide evidence suggesting that cross talk from other Ras effectors is necessary to maintain Pak activation. Figure 10 shows a model summarizing the proposed signals from Ras through Pak to ERK and JNK. Our evidence for cross talk, or indirect signals, is supported by the observation that only partial activation was observed when RasV12C40 or PI 3-kinase was used by itself; maximal activation was observed when Raf was transfected along with either RasV12C40 or PI 3-kinase. Furthermore, dominant negative Rho also inhibits Ras activation of Pak. Rho is not likely to act through PI 3-kinase because the dominant negative Rho mutant does not inhibit PI 3-kinase or RasV12C40 activation of Pak. Multiple mechanisms of Pak activation are supported by the observation that three small G proteins that fail to activate Pak when tested alone, RacV12H40, RasV12G37, and RhoV14, activate Pak when cotransfected with either activated Raf or RasV12S35, a mutant that activates Raf. At least one of these alternate inputs to Pak, RasV12G37/Raf, is likely to be PI 3-kinase independent since it is resistant to LY294002.
FIG. 10.
Model of Ras signaling to Pak and ERK. Dashed lines show possible indirect signals.
Candidate transducers of indirect signals to Pak include Raf (because it cooperates with RacV12H40, RasV12G37, and RhoV14), Ral GDS, Rin (both of which bind RasV12G37), and Ras-GAP (which can regulate the cytoskeleton through Rho) (18, 22, 24, 27). Rho kinase (p160Rock) is another candidate because it binds Rho, Rac, and RacV12C40 (another effector mutant that fails to bind Pak) (26). Indirect activation of Pak need not occur through protein kinases, since Pak can be activated by two types of translocation events mediated by SH3 proteins that bind proline-rich regions in the amino terminus. The first is mediated by the SH3 adapter protein Nck (6, 16, 30), and the second is mediated by a newly discovered Cdc42/Ras GEF, PIX. Manser et al. found that coexpression of PIX with Cdc42H40 allowed this effector mutant to activate Pak, presumably by translocating Pak to the cell surface, where it could be activated by endogenous GTPases or membrane lipids (5, 11, 33). These two mechanisms may be relevant to our studies, since membrane targeting of Pak is sufficient to bring about biological responses similar to those caused by Ras, including ERK activation and, in the PC12 cell line, extension of Neurites (10, 30). Finally, although we have not found evidence of secreted autocrine factors, Pak is activated by many growth factors which may contribute to the indirect signals we have observed. Thus, there are multiple ways to activate Pak without direct stimulation of Rac or Cdc42.
Rac is downstream of Ras in the membrane ruffling signal (45). Furthermore, RasV12C40 is the only effector mutant that can stimulate ruffling, and PI 3-kinase is the only Ras effector that can induce ruffling (21, 46). Pak also stimulates reorganization of the actin cytoskeleton (31, 48). This suggested that the major route to Pak is through PI 3-kinase and Rac. Our studies demonstrated the presence of a ‘Ras > Rac > Pak signal in at least one cell. However, as discussed above, we also found that several combinations of Ras and Rac effector mutants activated Pak. These include RasV12S35 plus RasV1G37, two mutants that fail to activate Pak on their own. It was recently reported that most combinations of Ras effector mutants that fail to activate Raf produce colonies with Rho-type morphology and are sensitive to dominant negative Rho mutants (24), This suggested that Rho family members were mediating the major signals in the Raf-independent pathways. Our observation that dominant negative Pak inhibited Rho transformation suggests a role for Pak in this signal.
While many cells require signals from Raf and are sensitive to agents that inactivate the Raf pathway, Ras does not activate ERK in several cell lines, such as Wistar rat thyroid cells (2). Other cells, such as rat intestinal epithelial cells, can be transformed by Ras but not Raf (38). This has led to efforts to identify the Raf-independent signals for transformation. The commonly used NIH 3T3 cell is likely to mask some Raf-independent signals, since it is readily transformed by Raf. Our data suggest that Pak protein kinases mediate key signals through the Rho GTPase family independently of Raf. This contrasts with several studies with Rac effector mutants that rule out Pak as a mediator of transformation by Rac. Specifically, the other studies found that RacV12L37, which binds Pak but not other effectors, transforms cells poorly. Our Rat-1 cells transformed almost as well with RacV12L37 as with RacV12. The simplest explanation for the differences lies in the cells used in each study. The other studies were carried out with NIH 3T3 cells (20, 26, 55), which do not respond to Pak dominant negative mutants in transformation assays (50) and, as we have shown here (Fig. 1), do not express the necessary intermediates to permit Ras to activate Pak, not even through cross talk. Thus, Rat-1 cells are likely to rely more heavily on the Ras-to-Pak pathway than on the Ras-to-Raf pathway. The Ras-to-Pak signals are likely to be found in many other cells, since Pak dominant negative mutants inhibit Ras transformation in rat Schwann cells and a Ras-sensitive neurofibrosarcoma.
Rac, Rho, and Cdc42 all transform cells, especially in the presence of weakly activated Raf, and dominant negative mutants of each of these small GTPases inhibit Ras transformation suggesting that each plays an essential role in cell transformation (23, 40–42). As all have profound effects on the actin cytoskeleton, it is possible that they contribute to Ras transformation by maintaining the transformed cell morphology. However, a recent study of stable cell lines reverted with dominant negative mutants of all three GTPases found no reliable coorelation between morphology and transformation (41). Perhaps most relevant to our study is the finding that cells expressing dominant negative Rac maintained their transformed morphology yet failed to grow on soft agar. This suggests that the actin cytoskeleton plays a role secondary to other activities, such as regulation of kinase cascades. Another group found that dominant negative Rho, Rac, and Cdc42 attenuate ERK activation and that activated mutants all cooperate with Raf to activate ERK (14, 15). Pak was proposed as a downstream gene for all three GTPases, since Pak dominant negative mutants inhibited cooperative ERK activation. We have observed that activated Pak stimulates both Raf and Mek about two- to fourfold (unbublished observations), suggesting that either one may be the direct target. Because activated Pak phosphorylates Mek, this kinase is more likely the direct site of the cross talk to ERK; the effects of Raf may be indirect. Our data support the proposed cross talk signal from Rho and Rac through Pak to ERK, since we found that Rho and Rac can cooperate to activate ERK and that activation by each is sensitive to Pak dominant negative mutants. The observation that Rho family members and Pak are all required to sustain ERK activation suggests that this common property is the one most critical for maintenance of cell transformation.
The studies described here often used kinase-deficient Pak mutants to inhibit Pak, so we have performed experiments to test their specificity. We have demonstrated that the Pak mutants do not inhibit Raf (both v-Raf and RafD340) or Mek (unpublished observations), two other Ras-regulated kinases, when tested for effects on ERK activation and cell transformation. However, the two kinase-deficient mutants both inhibit Pak activation of ERK (Fig. 5d). Furthermore, a functional Rac/Cdc42 binding domain is not required for inhibition, which rules out the possibility that the mutants merely sequester Rac and Cdc42. Finally, we have now documented the biological effects of Pak on cell transformation and ERK activation in both directions; we have conditions under which activated Pak stimulates ERK and promotes cell transformation and can reverse both with both kinase-deficient Pak mutants. Together, these data suggest that the two kinase-deficient Paks are specific inhibitors of Pak–hence, dominant negative mutants.
The observation that Rho, Rac, and Cdc42 all play central roles in Ras transformation has prompted several searches for key downstream effectors. Pak emerged as one candidate because it was the first protein kinase found to bind Rac/Cdc42 and it was homologous to Ste20, a Cdc42 effector in yeast (32). Subsequent studies with effector mutants suggested that Pak does not play a role in transformation. However, we now present three types of experiments suggesting that Pak is an essential downstream gene for Rho, Rac, and Ras. First, transformation by Rac, Rho, Ras, and several Rac and Ras effector mutants was inhibited by dominant negative Pak mutants. Second, cooperative ERK activation by all three GTPases was inhibited by the Pak dominant negative mutants. Third, all combinations of Ras, Rho, and Rac mutants that yielded high-efficiency transformations also activated Pak. It should be noted that these correlations suggest that Pak activation is necessary for high-efficiency transformation, but Pak activation is clearly not sufficient for transformation, since RasV12C40, RacV12, RacV12L37, and Pak1L83,L86, all of which activate Pak, transform poorly when tested individually. In conclusion, Pak dominant negative mutants inhibit many forms of Ras, Rac, and Rho both in transformation assays and in ERK activation assays, suggesting that signals from all three GTPases converge on Pak. Hence, Pak becomes part of a growing list of proteins, such as Ras and Raf, that may be targets for novel antineoplastic drugs.
ACKNOWLEDGMENTS
We thank Jim Stone for rv68BUR, Linda Van Aelst for Rac effector mutants, Mike White for Ras effector mutants, and all of the above for helpful comments on the manuscript. We thank Jonathan Chernoff for Pak mutants and Channing Der for Ras and Raf plasmids. We also thank Amita Sehgal, Margaret Chou, and members of the Field lab for helpful discussions and for comments on the manuscript.
This work is supported by grants to J.F. from the NIH (GM48241), the Lucille P. Markey Charitable Trust, and the Neurofibromatosis Foundation.
ADDENDUM IN PROOF
While this paper was under review, Pak3 was shown to phosphorylate and positively regulate Raf-1 (A. J. King, H. Sun, B. Diaz, D. Barnard, W. Miao, S. Bagrodia, and M. S. Marshall, Nature 396:180–183, 1998). This study supports our conclusion that Pak kinases are key regulators of the ERK cascade.
REFERENCES
- 1.Afar D E H, Han L, McLaughlin J, Wong S, Dhaka A, Parmar K, Rosenberg N, Witte O N, Colicelli J. Regulation of the oncogenic activity of BCR-ABL by a tightly bound substrate protein RIN1. Immunity. 1997;6:773–782. doi: 10.1016/s1074-7613(00)80452-5. [DOI] [PubMed] [Google Scholar]
- 2.Al-Alawi N, Rose D W, Buckmaster C, Ahn N, Rapp U, Meinkoth J, Feramisco J R. Thyrotropin-induced mitogensis is Ras dependent but appears to bypass the Raf-dependent cytoplasmic kinase cascade. Mol Cell Biol. 1995;15:1162–1168. doi: 10.1128/mcb.15.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagrodia S, Taylor S J, Creasy C L, Chernoff J, Cerione R A. Identification of a mouse p21Cdc42/Rac activated kinase. J Biol Chem. 1995;270:22731–22737. doi: 10.1074/jbc.270.39.22731. [DOI] [PubMed] [Google Scholar]
- 4.Bar-Sagi D, Feramisco J R. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science. 1986;233:1061–1068. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- 5.Bokoch G M, Reilly A M, Daniels R H, King C C, Olivera A, Spiegel S, Knaus U G. A GTPase-independent mechanism of p21-activated kinase activation. J Biol Chem. 1998;273:8137–8144. doi: 10.1074/jbc.273.14.8137. [DOI] [PubMed] [Google Scholar]
- 6.Bokoch G M, Wang Y, Bohl B P, Sells M A, Quilliam L A, Knaus U G. Interaction of the Nck adapter protein with p21-activated kinase (PAK1) J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- 7.Broek D, Toda T, Michaeli T, Levin L, Birchmeier C, Zoller M, Powers S, Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987;48:789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- 8.Coso O A, Chiariello M, Yu J-C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 9.Cox A D, Der C J. Biological assays for cellular transformation. Methods Enzymol. 1994;238:277–294. doi: 10.1016/0076-6879(94)38026-0. [DOI] [PubMed] [Google Scholar]
- 10.Daniels R H, Hall P S, Bokoch G M. Membrane targeting of p21-activated kinase 1 (PAK1) induces neurite outgrowth from PC12 cells. EMBO J. 1998;17:754–764. doi: 10.1093/emboj/17.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dharmawardhane S, Sanders L, Martin S, Daniels R, Bokoch G. Localization of p21-activated kinase 1 (PAK1) to pinocytic vesicles and cortical actin structures in stimulated cells. J Cell Biol. 1997;138:1265–1278. doi: 10.1083/jcb.138.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egan S E, Weinberg R A. The pathway to signal achievement. Nature. 1993;365:781–783. doi: 10.1038/365781a0. [DOI] [PubMed] [Google Scholar]
- 13.Feig L A, Cooper G M. Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost J A, Steen H, Shapiro P, Lewis T, Ahn N, Shaw P E, Cobb M H. Cross-cascade activation of ERKs and ternary complex factors by Rho family proteins. EMBO J. 1997;16:6426–6438. doi: 10.1093/emboj/16.21.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frost J A, Xu S, Hutchison M R, Marcus S, Cobb M H. Actions of Rho family small G proteins and p21-activated protein kinases on mitogen-activated protein kinase family members. Mol Cell Biol. 1996;16:3707–3713. doi: 10.1128/mcb.16.7.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galisteo M L, Chernoff J, Su Y-C, Skolnik E Y, Schlessinger J. The adaptor protein Nck links receptor tyrosine kinases with the serine-threonine kinase Pak1. J Biol Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- 17.Han J, Luby-Pelps K, Das B, Shu X, Xia Y, Mosteller R D, Krishna U M, Falk J R, White M A, Broek D. Role for substrates and products of PI3-kinase in regulating activation of Rac-related guanosine triphopsphatases by Vav. Science. 1998;279:558–560. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]
- 18.Han L, Wong D, Dhaka A, Afar D, White M, Xie W, Herschman H, Witte O, Colicelli J. Protein binding and signaling properties of RIN1 suggest a unique effector function. Proc Natl Acad Sci USA. 1997;94:4954–4959. doi: 10.1073/pnas.94.10.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 20.Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. RAC regulation of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science. 1996;274:1374–1376. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- 21.Joneson T, White M A, Wigler M H, Bar-Sagi D. Stimulation of membrane ruffling and MAP kinase activation by distinct effectors of RAS. Science. 1996;271:810–812. doi: 10.1126/science.271.5250.810. [DOI] [PubMed] [Google Scholar]
- 22.Jullien-Flores V, Dorseuil O, Romero F, Letourneur F, Saragosti S, Berger R, Tavitian A G, Gacon G, Camonis J H. Bridging Ral GTPase to Rho pathways. J Biol Chem. 1995;38:22473–22477. doi: 10.1074/jbc.270.38.22473. [DOI] [PubMed] [Google Scholar]
- 23.Khosravi-Far R, Solski P A, Clark G J, Kinch M S, Der C J. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol Cell Biol. 1995;15:6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khosravi-Far R, White M A, Westwick J K, Solski P A, Chrzanowska-Wodnicka M, Van Aelst L, Wigler M H, Der C J. Oncogenic Ras activation of Raf/mitogen-activated protein kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol Cell Biol. 1996;16:3923–3933. doi: 10.1128/mcb.16.7.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khwaja A, Rodriguez-Viciana P, Wennstrîm S, Warne P H, Downward J. Matrix adhesion and Ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/Akt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamarche N, Tapon N, Stowers L, Burbelo P D, Aspenstrom P, Bridges T, Chant J, Hall A. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell. 1996;67:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 27.Leblanc V, Tocque B, Delumeaum I. Ras-GAP controls Rho-mediated cytoskeletal reorganization through its SH3 domain. Mol Cell Biol. 1998;18:5567–5578. doi: 10.1128/mcb.18.9.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim L, Manser E, Leung T, Hall C. Regulation of phosphorylation by p21 GTPases. The p21 Ras-related Rho subfamily and its role in phosphorylation signalling pathways. Eur J Biochem. 1996;242:171–185. doi: 10.1111/j.1432-1033.1996.0171r.x. [DOI] [PubMed] [Google Scholar]
- 29.Lowry D R, Willumsen B R. Function and regulation of ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 30.Lu W, Katz S, Gupta R, Mayer B J. Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr Biol. 1997;7:85–94. doi: 10.1016/s0960-9822(06)00052-2. [DOI] [PubMed] [Google Scholar]
- 31.Manser E, Huang H-Y, Loo T-H, Chen X-Q, Dong J-M, Leung T, Lim L. Expression of constitutively active α-Pak reveals effects of the kinase on actin and focal complexes. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manser R, Leung T, Salihuddin H, Zhao Z-S, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Naure. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 33.Manser E, Loo T-S, Koh C-G, Zhao Z-S, Chen X-Q, Tan L, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 34.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extacellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 35.Minden A, Lin A, Claret F-X, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 36.Nimnual A S, Yatsula B A, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 37.Nobes C D, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 38.Oldham S M, Clark G J, Gangarosa L M, Coffey R J, Der C J. Activation of the Raf-1/Map kinase cascade is not sufficient for Ras transformation of RIE-1 epithelial cells. Poc Natl Acad Sci USA. 1996;93:6924–6928. doi: 10.1073/pnas.93.14.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson S N, Trabalzini L, Brtva T, Fischer T, Altschuler D L, Martelli P, Lapetina E G, Der C J, White G C. Identification of a novel RalGDS-related protein as a candidate effector for Ras and Rap1. J Biol Chem. 1996;271:29903–29908. doi: 10.1074/jbc.271.47.29903. [DOI] [PubMed] [Google Scholar]
- 40.Qiu R, Chen J, McCormick F, Symons M. A role for Rho in Ras transformation. Proc Natl Acad Sci USA. 1995;92:11781–11785. doi: 10.1073/pnas.92.25.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qiu R-G, Abo A, McCormick F, Symons M. Cdc42 regulates anchorage-independent growth and is necessary for Ras transformation. Mol Cell Biol. 1997;17:3449–3458. doi: 10.1128/mcb.17.6.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu R-G, Chen J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature. 1995;374:457–459. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- 43.Quilliam L A, Khosravi-Far R, Huff S Y, Der C J. Guanine nucleotide exchange factors: activators of the Ras superfamily of proteins. Bioessays. 1995;17:395–404. doi: 10.1002/bies.950170507. [DOI] [PubMed] [Google Scholar]
- 44.Ridley A J, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 45.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Viciana P, Warne P H, Khwaja A, Marte B M, Pappin D, Das P, Waterfield M D, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 47.Sells M A, Chernoff J. Emerging from the Pak: p21-activated protein kinase family. Trends Cell Biol. 1997;7:162–167. doi: 10.1016/S0962-8924(97)01003-9. [DOI] [PubMed] [Google Scholar]
- 48.Sells M A, Knaus U G, Bagrodia S, Ambrose D M, Bokoch G M, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7:202–210. doi: 10.1016/s0960-9822(97)70091-5. [DOI] [PubMed] [Google Scholar]
- 49.Stang S, Bottorff D, Stone J C. Interaction of activated Ras with Raf-1 alone may be sufficient for transformation of rat2 cells. Mol Cell Biol. 1997;17:3047–3055. doi: 10.1128/mcb.17.6.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang Y, Chen Z, Ambrose D, Liu J, Gibbs J B, Chernoff J, Field J. Kinase-deficient Pak1 mutants inhibit Ras transformation of Rat-1 fibroblasts. Mol Cell Biol. 1997;17:4454–4464. doi: 10.1128/mcb.17.8.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang Y, Marwaha S, Rutkowski J L, Tennekoon G I, Phillips P C, Field J. A role for Pak protein kinases in Schwann cell transformation. Proc Natl Acad Sci USA. 1998;95:5139–5144. doi: 10.1073/pnas.95.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M H. Complex formation between RAS and RAF and other protein kinases. Proc Natl Acad Sci USA. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vlahos C J, Matter W F, Hui K Y, Brown R F. A Specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-enzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 54.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 55.Westwick J K, Lambert Q T, Clark G J, Symons M, Van Aelst L, Pestell R G, Der C J. Rac regulation of transformation, gene expression, and actin organization by multiple, PAK-independent pathways. Mol Cell Biol. 1997;17:1324–1335. doi: 10.1128/mcb.17.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White M A, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler M H. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 57.White M A, Vale T, Camonis J H, Schaefer E, Wigler M H. A role for the Ral guanine nucleotide dissociation stimulator in mediating Ras-induced transformation. J Biol Chem. 1996;271:16439–16442. doi: 10.1074/jbc.271.28.16439. [DOI] [PubMed] [Google Scholar]
- 58.Zhou K, W Y, Gorski J L, Nomura N, Collard J, Bokoch G. Guanine nucleotide exchange factors regulate specificity of downstream signaling from Rac and Cdc42. J Biol Chem. 1998;273:16782–16786. doi: 10.1074/jbc.273.27.16782. [DOI] [PubMed] [Google Scholar]