Abstract
Approximately 70% of mRNAs in Caenorhabditis elegans are trans spliced to conserved 21- to 23-nucleotide leader RNAs. While the function of SL1, the major C. elegans trans-spliced leader, is unknown, SL1 RNA, which contains this leader, is essential for embryogenesis. Efforts to characterize in vivo requirements of the SL1 leader sequence have been severely constrained by the essential role of the corresponding DNA sequences in SL1 RNA transcription. We devised a heterologous expression system that circumvents this problem, making it possible to probe the length and sequence requirements of the SL1 leader without interfering with its transcription. We report that expression of SL1 from a U2 snRNA promoter rescues mutants lacking the SL1-encoding genes and that the essential embryonic function of SL1 is retained when approximately one-third of the leader sequence and/or the length of the leader is significantly altered. In contrast, although all mutant SL1 RNAs were well expressed, more severe alterations eliminate this essential embryonic function. The one non-rescuing mutant leader tested was never detected on messages, demonstrating that part of the leader sequence is essential for trans splicing in vivo. Thus, in spite of the high degree of SL1 sequence conservation, its length, primary sequence, and composition are not critical parameters of its essential embryonic function. However, particular nucleotides in the leader are essential for the in vivo function of the SL1 RNA, perhaps for its assembly into a functional snRNP or for the trans-splicing reaction.
In a number of eukaryotes, including trypanosomes and nematodes, an RNA-processing reaction called trans splicing results in the addition of a small (22- to 41-nucleotide [nt]) leader exon-like sequence (referred to here as a leader exon or spliced leader [SL]) onto the 5′ ends of some or all mRNAs (reviewed in references 1, 3, 4, 29, and 30). The leaders are derived from larger (∼100-nt) RNAs (referred to here as SL RNAs), that contain a leader exon at their 5′ ends and a 3′ intron-like domain. These RNAs appear to function in trans splicing of their leader exon following their assembly into an SL ribonucleoprotein (RNP) complex, similar to the small nuclear RNPs (snRNPs) that function in cis splicing (6, 21, 24, 26, 27, 32, 39, 40). In the nematode Caenorhabditis elegans, ∼60% of all messages are trans spliced to the major 22-nt SL1 leader, which is identical in sequence to the leaders found in most other nematodes (18, 29, 43). A minor leader, SL2, appears to be appended specifically to the ∼10% of messages that are downstream in operons (16, 36, 43). A family of additional SL2-like leaders, whose functions are unknown, has also been identified in this organism (10, 34).
Although the mechanism of trans splicing and the sequence requirements for the trans-splicing reaction have been well characterized in vitro and in vivo (reference 19; reviewed in references 1, 3, 4, 29, and 30), the biological functions of spliced leaders in vivo, and the sequences required for these functions, are not as well understood. trans-splicing functions at least in part to process polycistronic messages into individual coding units in trypanosomes and C. elegans (1, 36, 43). However, as only a fraction (∼25%) of messages appear to be organized into operons in C. elegans (43), it is likely that trans-spliced leaders perform additional functions in mRNA metabolism. For example, once a leader is trans spliced onto an mRNA, it may play an active role in controlling the stability, transport, or translation of messages. Indeed, in vitro-translation experiments have shown that the SL leader sequence of the nematode Ascaris lumbricoides (which is identical in sequence to C. elegans SL1), in conjunction with the specialized trimethylguanosine cap structure found on all nematode SL RNAs, results in maximal mRNA translation in vitro (23). However, rather than serving an active role, trans splicing of the leader onto a message may instead function solely to remove inhibitory sequences in the 5′ untranslated region that might otherwise prohibit efficient translation, consistent with the observation that leader sequences are often spliced close to the initiating AUG codon (2).
There appears to be some flexibility in the primary sequence of the leader relative to its potential function in mRNA metabolism in C. elegans, since SL2, which is only ∼45% identical to SL1 (16), has been shown to substitute functionally for SL1 in the embryo, and it can be trans spliced onto SL1 acceptor sites (12). In addition, although the function of the other minor leaders in C. elegans is not known, they are also quite divergent from SL1 in sequence (10, 34). However, some features of SL leaders in nematodes are well conserved: all are 21 to 23 nt in length, and all those examined exhibit a predicted secondary-structure element, a stem-loop involving the leader and a portion of the SL RNA intron-like sequences (6, 10, 14, 29, 34, 42). Although the conservation of at least some of these features may reflect a requirement for the corresponding DNA sequences in transcription of the SL RNAs, as has been shown for the Ascaris SL gene in vitro (15), these evolutionarily conserved features may also reflect structural requirements of the leader exon.
Our previous identification of mutants that lack zygotic SL1 RNA (12) provided the opportunity to address the in vivo requirement for the SL1 leader sequence. These mutants carry deletions of the rrs-1 cluster, which contains ∼110 tandem copies of a 1-kb sequence encoding both 5S rRNA and the 105-nt SL1 RNA (12, 18, 28). An SL1 RNA encoding gene is necessary and sufficient to rescue the embryonic lethality associated with the rrs-1 deletions (12).
In this study, we evaluate the in vivo requirements for the SL1 leader RNA sequence by using rescue of the embryonic lethality of the rrs-1 mutants as an assay. Our approach makes it possible to examine sequences required both for trans splicing and for the function of the trans-spliced leader in vivo. A recent study also took advantage of the rrs-1 mutants as a system to analyze mutant SL1 RNAs in vivo (41). However, since this study relied on the wild-type SL1 promoter to drive expression of various mutant constructs, all major changes in SL1 eliminated or dramatically decreased detectable expression of the mutant SL RNA. While this finding confirmed results in other systems demonstrating that the leader DNA sequence contains elements essential for SL RNA transcription (15), it prevented functional analysis of all changes in the SL1 leader sequence other than single-nucleotide changes at three positions. We developed a heterologous expression system that uncouples the SL1 RNA sequence per se from sequences required for its transcription, allowing extensive manipulation of the leader by using the rrs-1 mutants. We find that SL1 leader variants containing substantial deletions, insertions, or substitutions involving conserved regions are able to rescue embryonic lethality of rrs-1 deletions, suggesting that in spite of the conserved features of the SL1 leader sequence, the precise primary sequence of the leader does not appear to be essential for its embryonic function. The rigid conservation in the length of trans-spliced leaders in nematodes also does not appear to relate to their essential role in embryogenesis, as substantially shorter and longer variants of SL1 support embryonic development. In contrast, several sequence alterations abolish the embryonic function of SL1 and appear to identify limited portions of the leader responsible for this function.
MATERIALS AND METHODS
Plasmid constructions.
A 341-bp fragment containing the promoter region from the C. elegans U2-3 gene was amplified from a plasmid containing the U2-3 snRNA gene (a kind gift from Tom Blumenthal) by PCR with Taq polymerase (Perkin-Elmer) and the buffer provided by the manufacturer; this fragment corresponds to nt 47 to 388 of the U2-3 snRNA GenBank sequence (39). The upstream primer used for PCR contains an XbaI site followed by nt 47 to 67 of the U2-3 sequence (KF 101), and the downstream primer contains a BamHI site followed by nt 388 to 371 of the U2-3 sequence (KF 99). PCR was performed for 25 cycles under the following conditions: 94°C, 30 s; 50°C, 1 min; 72°C, 1 min. The PCR product was digested with XbaI and BamHI and subcloned into the Stratagene pBluescript SKII(−) vector. The wild-type SL1 RNA gene was amplified from a plasmid containing the SL1 gene by PCR with Pfu polymerase (Stratagene) and the buffer provided by the manufacturer. The upstream primer (KF 65) used for PCR corresponds to the first 22 nt of the SL1 RNA (nt 231 to 210 of the GenBank rrs-1 repeat sequence, the 1-kb sequence that encodes SL1 and 5S rRNA [18, 28]), and the downstream primer (KF 102) contains an EcoRI site, followed by nt 27 to 7 of the GenBank sequence of the rrs-1 repeat. These primers amplify a 224-bp fragment that contains the SL1 RNA gene followed by 120 nt of 3′ sequence. This 3′ sequence was included to ensure proper 3′ end formation of the SL1 RNA (15). PCR was performed for 25 cycles under the following conditions: 94°C, 30 s; 40°C, 1 min; 72°C, 1 min. To construct the U2 promoter-SL1 chimera, the U2 promoter plasmid described above was digested with ScaI (which cuts after the last nucleotide of the U2 promoter) and EcoRI. The EcoRI-digested SL1 PCR product was then subcloned into this vector; this resulted in a construct in which the last nucleotide of the U2 promoter was fused to the first nucleotide of the SL1 RNA gene. To construct the SL1 RNA genes with the mutant leaders, upstream primers were used that contained the leader deletions, substitutions, or additions described in the text, followed by 9 to 21 nt of downstream sequence corresponding to the wild-type SL1 gene (Δ3–12, KF 103; Δ11–21, KF 104; 11–20 shuffle, KF 106; 5′ (5) extra As, KF 107; SL2-SL1 chimera, KF 108; G20 to A20, KF 110; ΔGU loop, KF 116; loop sub, KF 123). PCRs were performed with a plasmid containing the SL1 RNA gene, each of the upstream mutant primers, and the downstream SL1 3′ end primer (KF 102) under the conditions described above for the wild-type SL1 PCR. These fragments were then individually subcloned into the U2-3 promoter-containing plasmid as described above for all constructs except the 7U loop insert. For the 7U loop insert construct, the mutant insert was PCR amplified from an SL1 RNA-encoding plasmid under the conditions described above, except an upstream primer containing a 5′ BbsI site was used to facilitate cloning, followed by the mutant leader sequence (KF 144) and the downstream SL1 3′ end primer described above (KF 102). In order to create a vector containing complementary BbsI and EcoRI sites at the 5′ and 3′ ends, respectively, the U2-3 vector sequence described above was PCR amplified with a primer complementary to nt 388 to 369 of the U2-3 snRNA GenBank sequence preceded by a 5′ BbsI site (KF 139) and with a primer specific for nt 711 to 686 of the pSKII(−) vector GenBank sequence (Stratagene) preceded by a 5′ EcoRI site (KF 140). PCR was performed for 20 cycles under the following conditions: cycle 1, 94°C for 3 min, 50°C for 1 min, 72°C for 7 min; cycles 2 to 20, 94°C for 35 s, 50°C for 1 min, 72°C for 7 min. BbsI and EcoRI digestion and subsequent ligation yielded a construct with the last nucleotide of the U2-3 promoter fused to the first nucleotide of the SL1 RNA gene as described above. For each construct generated, appropriate sequences and junctions were confirmed by sequence analysis.
Worm culture, strains, microinjection, and analysis of rescue.
Nematodes were cultured as described previously (12). Microinjection of DNAs was performed as described previously (25). Heterozygous animals of the genotype unc-76(e911) wDf1 / unc-61(e228) dpy-21(e428) were used in the transformation experiments. wDf1, formerly called e2482 (12), is one of two rrs-1 deletion alleles. For scoring of rescue and for determining levels of SL RNAs, hermaphrodites of the genotype pha-1(e2123) III; unc-76(e911) wDf1 / unc-61(e228) dpy-21(e428) V were used. The pha-1(e2123) mutation allows the pha-1(+) gene to be used as a selectable marker for transformation (13). pha-1(e2123) results in 100% embryonic lethality at the nonpermissive temperature of 25°C (35). Introduction of a wild-type pha-1(+) transgene rescues this phenotype and allows for growth of the animals at 25°C. Therefore, only transformed animals are propagated at this temperature. This procedure allows for enrichment of transformed animals in the population, which was useful for subsequent RNA analysis (described below). Mutant leader RNA constructs were injected at a concentration of ∼40 to 100 μg/ml, along with an 8-kb plasmid containing the wild-type pha-1 gene (pBX1 [13]; kindly provided by Peter Barrett) at a concentration of ∼35 μg/ml and the pRF4 plasmid containing the rol-6(su1006dm) gene (a second selectable marker used to confirm that the surviving animals contained the extrachromosomal array) at a concentration of ∼40 μg/ml. Following transformation, F1 Rol animals were shifted from 15°C to 25°C, and stably transformed lines were identified as those F1s which gave rise to surviving F2 Rol progeny. Rescue of the embryonic lethality of wDf1 was scored as described previously after shifting the animals back to 15°C (12). In the case of the 7U loop insertion construct, the heterozygous wDf1 strain without the pha-1(e2123) mutation was used for transformation (since this construct was not analyzed for SL RNA expression). In this experiment, the lines were maintained at 20°C and transformed animals were identified by using the Rol marker only. For each experiment, total progeny were generally counted from several worms obtained from each line. The percent embryonic lethality from each line was calculated; the average percent arrested embryos was determined for all lines from a given experiment and was used in the calculation of percent rescue reported (see Tables 1 to 4). Percent rescue was calculated by the formula 100[1 − (average percent arrested embryos observed/average percent arrested embryos from the parental strain)]. The average percent arrested embryos for the wDf1 parental line was 25.3%, as expected for heterozygotes carrying a recessive lethal mutation. For the constructs that rescue embryonic lethality, the percentage of arrested embryos was found to be significantly different from that of the parental strain, with P values of less than 0.01.
TABLE 1.
SL1 driven by the U2-3 promoter can rescue lethalitya
| Construct | No. of rescued lines/total no. of lines | % Rescue of wDf1 embryonic lethality (n)b | Embryonic rescuec |
|---|---|---|---|
| None | NA | 0 (2,039)e | NA |
| SL1-SL1 promoter | 4/5d | 49 (1,394)e | + |
| SL1-U2 promoter | 6/7d | 54 (2,525) | + |
NA, not applicable. Heterozygous animals of the genotype unc-76 (e911) wDf1 / unc-61 (e228) dpy-21 (e428) were used in transformation experiments (see Materials and Methods).
n, total number of progeny counted, including arrested embryos and larvae. Percent rescue was calculated by the formula 100[1 − (average percent arrested embryos observed/average percent arrested embryos from the parental strain)]. The average percent arrested embryos for the wDf1 parental line was 25.3%, as expected for heterozygotes carrying a recessive lethal mutation (see Materials and Methods).
Embryonic rescue was scored as “+” if the average percentage of arrested embryos was below 20% and arrested elongated embryos and/or extra Unc-76 larvae (defined as significantly greater than the ∼2% of Unc-76 recombinants usually observed in the parental strain) were observed. (Since the unc-76 mutation is linked in cis to the lethal wDf1 mutation, Unc-76 animals are infrequently observed in the parental strain [12].)
Lines that showed no apparent rescue were not used in the calculation of percent rescue.
As reported in reference 12.
TABLE 4.
Analysis of leader insertion and addition constructsa
| Sequence of mutant leaderb | SL RNA expressionc | No. of rescued lines/total no. of lines | % Rescue of wDf1 embryonic lethality (n) | Embryonic rescue |
|---|---|---|---|---|
| No construct injected | NA | NA | 0 (2,039) | NA |
| GGUUUAAUUACCCAAGUUUGAG (wild type) | NDd | 6/7e | 54 (2,525) | + |
| AAAAAGGUUUAAUUACCCAAGUUUGAG [5′ (5) extra As] | 68 | 10/10 | 56 (4,113) | + |
| GGUUUAAUUACCCAAUUUUUUUGUUUGAG (7U loop insert) | ND | 9/20e | 29 (3,948) | + |
NA, not applicable. Percent rescue was calculated and embryonic rescue was scored as described in notes b and c to Table 1 and Materials and Methods.
Nucleotides added to the SL1 leader are underlined. SL RNA expression and secondary structures were determined as described in Table 2 and Materials and Methods.
Expression level is shown as a percentage of the level seen for the highest-expressing mutant (ΔGU loop) (Fig. 4A) after normalizing for loading.
ND, not determined.
Lines that showed no apparent rescue were not used in the calculation of percent rescue.
RNA isolation and primer extension analysis.
Stably transformed wDf1/+ lines were grown at 25°C to enrich for transformants (as described above) on agarose nematode growth medium (NGM) plates. One line for each construct that gave rise to a representative number of arrested embryos or rescued animals was chosen for RNA preparation. The average percentages of arrested embryos from these selected lines were as follows: wild-type SL1, 17.5%; Δ3–12, 26%; Δ11–21, 25%, ΔGU loop, 17.8%; G20 to A20, 13.3%; loop sub, 13.8%; 11–20 shuffle, 33%; SL2-SL1 intron, 32%; 5′ (5) extra As, 11.4%. Mixed-stage worms were harvested, and RNA was prepared with guanidine isothiocyanate, phenol-chloroform, and glass bead disruption as previously described (8), with the following modifications. After disruption by vortexing and centrifugation, the aqueous phase was extracted with 1 volume of acid phenol-chloroform, pH 4.7 (Ambion). After centrifugation, the RNA was precipitated with 0.1 volume of 5 M ammonium acetate and 2.5 volumes of 100% ethanol. After being washed with 70% ethanol, the pellets were resuspended in 0.1 mM EDTA, pH 8.
For primer extension, primers were end labeled with [γ32P]ATP and T4 polynucleotide kinase (Promega) and gel purified on 20% denaturing polyacrylamide gels. Probes were eluted overnight in 300 mM sodium acetate–0.01% sodium dodecyl sulfate (SDS). For analysis of RNAs that differed in length from the endogenous SL1 RNA (see Fig. 4A), ∼10 ng of labeled primers corresponding to nt 190 to 168 (KF 111) of the SL1 RNA gene from the GenBank sequence of the 1-kb rrs-1 repeat (i.e., nt 61 to 39 of the SL1 RNA [18, 28]), and nt 121 to 100 (KF 126) of the U6 snRNA GenBank sequence (nt 41 to 20 of the U6 snRNA [38]) were added to 20 μg of mixed-stage total RNA, heated to 70°C for 10 min, and annealed at 50°C for 1 h. Reverse transcription (RT) was performed at 50°C for 30 min with 400 U of Superscript II reverse transcriptase (Gibco-BRL), with the buffer and conditions specified by the manufacturer. Reactions were run on 12% sequencing gels, and the products were detected by autoradiography. For analysis of RNAs that are the same size as the endogenous SL1 RNA (see Fig. 4B), a dideoxy primer extension experiment was performed. Primers were labeled and purified as described above. The following primers were used for each reaction. For N2 (wild-type), G20 to A20, and loop sub reactions, an SL1 primer (KF 114) corresponding to positions 192 to 210 of the rrs-1 repeat GenBank sequence (nt 40 to 22 of the SL1 RNA [18, 28]) was used; for the SL2-SL1 chimera, a primer (KF 125) corresponding to positions 199 to 211 of the rrs-1 repeat GenBank sequence (nt 33 to 21 of the SL1 RNA) followed by nt 134 to 130 of the SL2α GenBank sequence (nt 20 to 16 of the SL2α RNA) was used; for the 11–20 shuffle, a primer (KF 112) corresponding to positions 200 to 211 of the rrs-1 repeat GenBank sequence (nt 32 to 21 of the SL1 RNA), followed by 6 nt complementary to the shuffled sequence (see Table 3), was used. For each reaction, a U6 control primer (KF 82) corresponding to nucleotide positions 227 to 196 of the U6 snRNA GenBank sequence (nt 70 to 39 of the U6 snRNA, [38]) was used. Total RNA (20 μg) was annealed to ∼10 ng of each labeled primer at 46°C for 1 h. RTs were performed as above, except 10 μM dideoxycytosine was used in place of deoxycytosine. The reactions were run on an 18% sequencing gel and analyzed by autoradiography. Densitometry was performed on an LKB UltroScan XL, using two different exposures to confirm the RNA levels for each experiment. The levels of mutant SL RNAs were normalized to the levels of U6 snRNA to correct for loading differences.
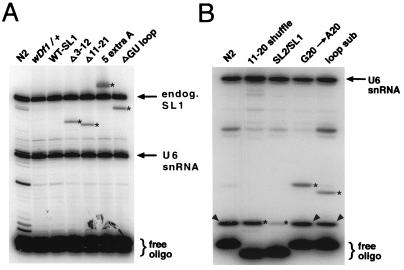
FIG. 4.
Primer extension analysis of mutant SL RNA expression. (A) Analysis of mutant SL RNAs which differ in size from the endogenous (endog.) SL1 RNA. RNA was prepared from mixed-stage animals carrying the indicated mutant SL RNA transgene. A labeled primer that recognizes a portion of the SL1 RNA intron was used (see Materials and Methods). Since the populations contained heterozygous mutant and wild-type animals, primer extension yields both an endogenous SL1 RNA product (upper arrow) and the mutant leader SL RNA product (stars). RNA analysis of wild-type animals (N2) and heterozygous deletion mutant animals (wDf1/+) are shown as controls, demonstrating that the products detected in the mutant strains are specific for those strains. All constructs shown were analyzed in a wDf1/+ genetic background. No additional products were detected in the WT-SL1 sample (SL1 RNA transcribed from the U2-3 promoter), indicating that this RNA is apparently of wild-type length. A labeled primer specific for a portion of the U6 snRNA (lower arrow) was included in the reaction; its extended product serves as a control for loading of comparable amounts of RNA in each lane. (B) Analysis of mutant SL RNAs which are the same size as the endogenous SL1 RNA. RNA was prepared from mixed-stage wild-type or wDf1/+ animals carrying the indicated mutant SL RNA transgene (as in panel A). In order to distinguish mutant from endogenous wild-type SL1 RNA, a dideoxy primer extension reaction was performed. The RNA was extended with a labeled primer specific for a portion of the SL1 RNA intron-like sequence or leader sequence (see Materials and Methods). Dideoxycytosine was included in the reaction mixture to interrupt extension of the SL RNAs; because of the differences in sequence between mutant leaders (stars) and wild-type endogenous SL1 RNA (arrowheads), products are extended to different lengths for each RNA. In the case of the 11–20 shuffle and SL2-SL1 chimeric RNAs, labeled primers were used that recognized these RNAs specifically; thus, the endogenous RNA is not extended and the band migrating at the same position as wild-type SL1 is exclusively the mutant RNA. A U6 RNA primer (arrow) was used to control for loading, as in panel A.
TABLE 3.
Analysis of leader substitution constructsa
| Sequence of mutant leaderb | SL RNA expressionc | No. of rescued lines/ total no. of lines | % Rescue of wDf1 embryonic lethality (n) | Embryonic rescue |
|---|---|---|---|---|
| No construct injected | NA | NA | 0 (2,039) | NA |
| GGUUUAAUUACCCAAGUUUGAG (wild type) | NDd | 6/7e | 54 (2,525) | + |
| GGUUUAAUUACCCAAGUUUAAG (G20 → A20) | 68 | 9/11e | 56 (2,814) | + |
| GGUUUAAUUACCAGGCGAAUAG (loop sub) | 30 | 7/14e | 33 (7,292) | + |
| GGUUUAAUUAAGAUCUCUCGAG (11–20 shuffle) | 100 | 0/12 | 0 (2,606) | − |
| GGUUUUAACCCAGUUACUCAAG (SL2-SL1 intron) | 24 | 0/10f | 0 (1,857) | − |
NA, not applicable. Percent rescue was calculated and embryonic rescue was scored as described in notes b and c to Table 1 and Materials and Methods.
Nucleotides that are different from those in the wild-type SL1 leader are underlined. SL RNA expression and secondary structures were determined as described in Table 2 and Materials and Methods.
Expression level is shown as a percentage of the level seen for the highest-expressing mutant (11–20 shuffle) (Fig. 4B) after normalizing for loading.
ND, not determined.
Lines that showed no apparent rescue were not used in the calculation of percent rescue.
For two lines, the percent embryonic lethality was 19 and 19.4%, respectively. Normally, percentages below 20% are indicative of rescue. However, these lines appear to be atypical of constructs which normally result in rescue. Progeny from only one animal were counted from these lines, and the eight remaining lines exhibited no evidence of rescue, indicating that these percentages are not likely to be significant.
RT-PCR analysis.
Total RNA isolation and RT-PCR from homozygous wDf1 mutant embryos was performed as described previously (12). In these experiments, RNA was isolated from wild-type (N2) embryos, homozygous, arrested wDf1 embryos, embryos derived from stably transformed heterozygous wDf1 animals carrying a wild-type SL1 transgene under the control of the U2-3 snRNA promoter, or an 11–20 shuffle transgene. Single elongated embryos (in the case of the wild-type embryos) or arrested, unelongated embryos (in the case of homozygous wDf1 embryos) were picked ∼10 to 12 h postfertilization. In the case of the embryos rescued with the wild-type transgene, embryos were picked ∼16 to 18 h postfertilization; at this time, the wild-type embryos have hatched, so only elongated embryos which are rescued with the wild-type transgene remain. For the 11–20 shuffle, it was not possible to distinguish homozygous mutant embryos that carry the transgene from those that did not, since the lethal phenotype does not allow scoring of the transgenic marker (extrachromosomal arrays are not transmitted to all progeny [25]). Therefore, ∼20 to 30 unelongated, homozygous mutant embryos were picked, and three independent RNA samples were prepared, to ensure that RNA transcribed from this transgene would be represented in the sample. Annealing of the downstream primer specific to a portion of the myo-3 coding region (200 ng; KF 79, listed below [9]) to the total RNA preparation, followed by RT, was performed as described previously (12), with the following modifications: one-half of each annealing reaction was added to a RT mixture of 50 mM Tris-HCl (pH 8.3), 50 mM KCl, 20 mM MgCl2, 5 mM dithiothreitol, 6.7 mM (each) deoxynucleoside triphosphate, 20 U of RNasin ribonuclease inhibitor (Promega), and 8 U of avian myeloblastosis virus reverse transcriptase (Promega). After RT for 30 min at 42°C, the reaction mixture was diluted to 50 μl with distilled water. Fifty nanograms of each primer (described below) and 5 μl of diluted cDNA were added to a 25-μl PCR mixture, and 35 cycles were performed with the following parameters: 94°C for 30 s, X°C for 1 min, and 72°C for 1 min, where X, the annealing temperature, varied according to the melting temperatures of the primers used in the reactions. In the case of the PCRs with cDNA from wild-type embryos, from embryos carrying wild-type SL1 transgenes, or from homozygous wDf1 arrested embryos, an annealing temperature of 50°C and the primers KF 45 and KF 79 (listed below) were used. In the case of reactions with cDNA from embryos carrying the 11–20 shuffle transgene (primers KF 151 and KF 79), a temperature of 47°C was used. In the case of control reactions performed to amplify an internal portion of the myo-3 message (9) (primers KF 92 and KF 79), 50°C was used. The products were separated on 1.3% agarose gels, the gels were blotted, and the filters were probed with [α-32P]dATP-labeled probes corresponding to an internal portion of the myo-3 message and prepared by PCR (using primers KF 90 and KF 78), as described previously (12).
Oligonucleotide sequences.
The sequences of the oligonucleotides used were as follows: KF 45, GGTTTAATTACCCAAGTTTG; KF 65, GGTTTAATTACCCAAGTTTGAG; KF 78, CTCGAGATTCCCAAGAATGGG; KF 79, CCACGGTCACCTGTTCGCCG; KF 82, CATCCTTGCGCAGGGGCCATGCTAATCTTCTC; KF 90, AGAAATGTCTGGAAATC; KF 92, CCAGACGCATTCGAAA; KF 99, CGGGATCCAGTACTGAATGGAGGAGAGGG; KF 101, GCTCTAGAGGACTCCGGCTTCAGCACGAC; KF 102, CGGAATTCGTTCCCCAATCAATATCATC; KF 103, GGCAAGTTTGAGGTAAACATTGA; KF 104, GGTTTAATTAGGTAAACATTGAAACTG; KF 106, GGTTTAATTAAGATCTCTCGAGGTAAACATTGAAAC; KF 107, AAAAAGGTTTAATTACCCAAGTTTG; KF 108, GGTTTTAACCCAGTTACTCAAGGTAAACATTGAAAC; KF 110, GGTTTAATTACCCAAGTTTAAGGTAAAC; KF 111, AGCTAACGCCAAATTTCTTTGGG; KF 112, CAATGTTTACCTCGAGAG; KF 114, GTCAGTTTCAATGTTTACC; KF 116, GGTTTAATTACCCAAAGGTAAACATTG; KF 123, GGTTTAATTACCAGGCGAATAGGTAAACAT; KF 125, TCAATGTTTACCTTGAGT; KF 126, CTCTGTATTGTTCCAATTTTAG; KF 139, GAAGACTTACTGAATGGAGGAGAGGGTA; KF 140, CGGAATTCGATATCAAGCTTATC; KF 144, GAAGACCTCAGTGGTTTAATTACCCAATTTTTTTGTTTGAGGTAACA; and KF 151, GGTTTAATTAAGATCTCTCG.
RESULTS
Expression of SL1 under the control of the U2 snRNA promoter supports embryonic development.
The 22-nt DNA sequence encoding the SL leader of A. lumbricoides contains an element that is essential for efficient in vitro transcription of the SL RNA (15); moreover, substantial alterations of the SL1 RNA sequence have suggested that it is similarly required for transcription in C. elegans in vivo (41). Because of this requirement for the SL1 leader sequence, it has not been possible to analyze substantial sequence alterations in the leader without eliminating expression of SL1 RNA. In a recent study (41), only very limited information regarding sequences essential for SL1 function, involving changes at three single-nucleotide positions, could be obtained; all deletions of the leader completely blocked its function and generally eliminated detectable levels of SL1 RNA. It could not be determined from this study whether such deletions eliminated expression or destabilized the RNA. To address the sequence requirements for the SL1 leader distinct from the role of the corresponding DNA sequence in its expression, it was necessary to develop a system in which the leader sequence could be altered without affecting its transcription. To accomplish this, we expressed the SL1 RNA gene under the control of the C. elegans U2-3 snRNA gene promoter (38). In other organisms, transcription of U snRNAs does not require downstream transcribed sequences (33), and we found that the U2-3 snRNA promoter similarly does not appear to require downstream sequences in C. elegans (see below). Since the U2 snRNA participates in cis and trans splicing (20, 29), both of which are ubiquitous processes in C. elegans, we reasoned that expression from the U2-3 promoter might be sufficient to drive SL1 expression throughout the animal in a manner similar to that of the SL1 promoter.
We tested a chimeric gene fusion (U2-SL1), in which the SL1 RNA transcription unit is expressed from the U2-3 promoter, for its ability to rescue the pleiotropic embryonic defects of wDf1, a deletion of the rrs-1 cluster (12), by transformation into wDf1 heterozygotes (Fig. 1 and Table 1). Deletions of the rrs-1 cluster result in arrest of embryos as differentiated, unelongated masses of cells and in embryonic lethality; the wild-type SL1 gene rescues this embryonic lethality (12). The U2-SL1 construct was also found to rescue this lethality: approximately the same fraction of mutant embryos were rescued with the U2-SL1 construct as we had found previously for the intact wild-type SL1 gene (Table 1) (12). Rescue was indicated by a significant decrease in the fraction of arrested, unelongated embryos from transgenic wDf1 heterozygotes relative to the ∼25% arrested embryos produced by the same strain lacking the transgene and by the presence of homozygous wDf1 larvae or arrested, but elongated, embryos. (Such embryos and larvae are never observed in progeny of control wDf1 heterozygotes). Rescue of all wDf1 homozygotes is never observed even with the wild-type SL1 RNA gene, since extrachromosomal arrays produced by transformation are inefficiently transmitted to subsequent generations (25) (Table 1). We conclude that SL1 RNA can effectively support embryonic development even when expressed from a heterologous promoter.
FIG. 1.
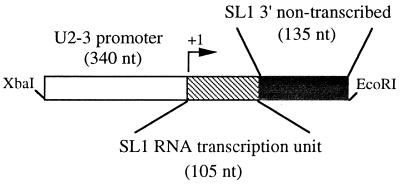
Expression system used to analyze mutant leader RNAs. Shown is a schematic of the U2-SL1 construct. Fragments (∼240 nt) of each mutant SL RNA gene, including 135 nt of 3′ nontranscribed sequence (used to ensure correct 3′ end formation), were cloned directly behind a 340-nt fragment containing the U2-3 snRNA promoter. This latter fragment contains the C. elegans U snRNA consensus proximal sequence element sequence (beginning 65 nt before the start site of transcription [38]), which is likely to be an important transcriptional regulatory element, based on studies in other systems (33).
One observation indicated that expression of SL1 RNA from the U2-SL1 construct was not identical to that of the endogenous SL1 RNA genes. When a 5S ribosomal DNA construct is cotransformed with the wild-type SL1 RNA construct, homozygous viable wDf1 lines can be generated and propagated for many generations (12). In contrast, cotransformation of 5S ribosomal DNA with the U2-SL1 construct failed to produce a viable transgenic line and the majority of rescued animals died during early larval stages (11). Therefore, it is possible that either the levels or spatiotemporal expression of the SL1 RNA produced from the U2-SL1 construct is not sufficient for rescue to adulthood. However, rescue of embryonic lethality was quite efficient. In addition, we were able to detect the SL1 leader on trans-spliced messages (see below).
Deletions identify essential structural elements of SL1.
The U2-SL1 expression system and our rescue assay allowed us to analyze large structural changes in the leader sequence for their effects on the essential in vivo function of the leader. The design of leader sequence alterations was guided by previous studies of the sequence requirements for trans splicing in nematodes (22, 23) and by the proposed secondary structure of the leader (14, 42). All known SL RNAs are predicted to fold into similar secondary structures (6, 8, 10, 16, 29, 31, 37). The first stem-loop of this structure includes the leader sequence, a portion of which can form base pairs with nucleotides surrounding and including the splice donor site; this interaction is predicted to be conserved in all SL RNAs (Fig. 2). The existence of two similar structures in the first stem-loop of the SL1 RNA has been determined by nuclear magnetic resonance (NMR) (14, 42); both structures include the base pairing of the splice donor site nucleotides (Fig. 2). Previous work has suggested that this interaction may be important for trans splicing, perhaps for splice donor site recognition in analogy to the U1 snRNA–pre-mRNA intron interaction required for cis splicing (5, 6). However, experiments with A. lumbricoides extracts suggested that neither the composition nor the length of the leader sequence is a critical parameter for efficient trans splicing in vitro (22). For example, SL leaders carrying deletions of either the 5′ or 3′ half of the leader were trans spliced in vitro with approximately the same efficiency as that of the wild-type leader. This study indicated that neither the primary sequence nor the first stem-loop secondary structure is relevant for efficient trans splicing in vitro (22).
FIG. 2.
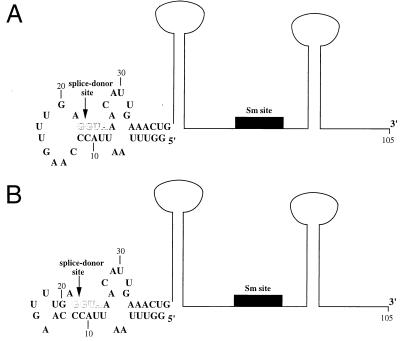
Predicted secondary structures of SL1 RNA. (A) Predicted secondary structure of SL1 RNA (6); the stem loop structure depicted for nucleotides 1 to 38 was determined by NMR (14). An outline of the rest of the structure (67 nt), generated by computer prediction (6), is indicated. The position of the 8-nt Sm binding site is represented by a solid box. The arrow indicates the position of the splice donor site; the sequence 5′ of this site comprises the leader exon. The G/GUA of the splice donor site (outlined letters) is conserved in all known SL RNAs, except in the recently identified minor SL RNAs in C. elegans (SL3, SL4, and SL5), which contain the sequence G/GUU (10, 34). In addition, the predicted base pair interactions that occur between these nucleotides and the leader are also highly conserved in SL RNAs (6). (B) An alternative structure determined by NMR analysis for the first stem-loop of the SL1 RNA (42); an outline of the rest of the structure as predicted (6) is shown and labeled as in panel A. This structure differs from that in panel A in that the nucleotides at positions 13, 14, 19, and 20 are base paired instead of forming part of the single-stranded loop.
We reasoned that if portions of the leader sequence are similarly dispensable for trans splicing in vivo, it might be possible to address whether these sequences performed any essential postsplicing functions on a trans-spliced message. Therefore, we first tested whether SL1 RNAs containing leader sequence deletions identical to those analyzed in in vitro trans-splicing reactions eliminate the essential function of SL1. The Δ3–12 mutation deletes nt 3 to 12 of the leader; in the predicted structure of the SL1 RNA, as well as the structures determined for a portion of the wild-type SL1 RNA, 8 of these 10 nucleotides form base pairs with nucleotides that span the splice donor site (6, 14, 42) (Table 2; and Fig. 2). In addition, 6 of 10 nt are conserved among all known C. elegans leaders (allowing for gaps) (10, 34) (Fig. 3). The Δ11–21 construct deletes nt 11 to 21 of the leader. In the solution structures of a portion of the SL1 RNA, 2 or 4 of 11 deleted nucleotides are base-paired; however, in both cases, one of these nucleotides is predicted to participate in the GU base pair comprising the splice donor site, which is conserved in all SL RNAs (6, 14, 42) (Table 2 and Fig. 2). In addition, three of the deleted nucleotides are conserved among all C. elegans leaders (10, 34) (Fig. 3). We found that neither of these deletion constructs was able to rescue embryonic lethality in several independent transformed lines (Table 2). By analyzing the mutant SL1 RNAs produced by these constructs in transgenic animals, we found that they were present at approximately the same level as other SL RNAs which rescue embryonic lethality (see below) (Tables 2 to 4 and Fig. 4A), demonstrating that the inability of the larger deletion mutants to rescue is not a result of their poor expression or instability per se. Smaller deletions of the leader analyzed by Xie and Hirsh (41), using the SL1 promoter, generally eliminated detectable SL1 RNA levels; our results imply that the deletions in that study probably did not destabilize the mutant RNAs but instead eliminated their transcription, consistent with in vitro transcription studies (15). We conclude that deletion of the 5′ or 3′ half of the leader eliminates the in vivo function of the SL1 RNA or SL1 leader without dramatically altering its expression or stability.
TABLE 2.
Analysis of leader deletion constructsa
| Sequence of mutant leaderb | SL RNA expressionc | No. of rescued lines/total no. of lines | % Rescue of wDf1 embryonic lethality (n) | Embryonic rescue |
|---|---|---|---|---|
| No construct injected | NA | NA | 0 (2,039) | NA |
| GGUUUAAUUACCCAAGUUUGAG (wild type) | NDd | 6/7e | 54 (2,525) | + |
| GG-----------------------CAAGUUUGAG (Δ3–12) | 71 | 0/8 | 0 (3,278) | − |
| GGUUUAAUUA-------------------------G (Δ11–21) | 76 | 0/4 | 0 (1,850) | − |
| GGUUUAAUUACCCAA------------AG (ΔGU loop) | 100 | 3/8e | 26 (2,909) | + |
NA, not applicable. Percent rescue was calculated and embryonic rescue was scored as described in notes b and c to Table 1 and Materials and Methods.
Dashes represent nucleotides deleted from the wild-type sequence.
Expression level is shown as a percentage of the level seen for the highest-expressing mutant (ΔGU loop) (Fig. 4A) after normalizing for loading.
ND, not determined. Presumably, the wild-type RNA under the control of the U2 promoter is efficiently expressed, since this construct rescues lethality. We were not able to determine relative levels of this RNA, since high levels of endogenous SL1 RNA are present in the mixed-stage RNA samples analyzed (Fig. 4) (Materials and Methods).
Lines that showed no apparent rescue were not used in the calculation of percent rescue.
FIG. 3.
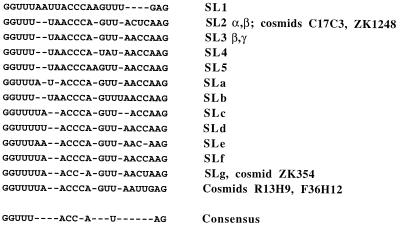
Alignment of C. elegans SLs. The sequences of the C. elegans trans-spliced leaders that have been identified are shown (10, 34). In some cases, SL RNA genes contain identical leader sequences but divergent sequences corresponding to the rest of the RNA (for example, SL genes encoded by the SL2 α and β genes, as well as SL genes encoded within the cosmids C17C3 and ZK1248, have identical leader sequences [10, 16, 34]). Gaps (indicated by dashes) are positioned in the alignment to maximize the degree of identity between the leaders. The consensus sequence for the leaders is shown at the bottom; the AG dinucleotide is also highly conserved in C. elegans cis-splice donor sites (17).
Although removal of half of the SL1 leader sequence abolishes its essential embryonic function, analysis of an additional deletion mutant led to the surprising observation that a substantial portion of the SL1 leader sequence is dispensable for embryogenesis. A deletion mutation (ΔGU loop) in which nt 16 to 20 of the leader were removed and which deletes 5 of the 11 nucleotides that were also deleted in the Δ11–21 construct, did not abolish SL1 function. This ΔGU loop RNA was efficiently expressed and was able to rescue embryonic lethality, albeit at a reduced efficiency compared to that of the wild type (Table 2 and Fig. 4A). This diminished efficiency might reflect a requirement for the precise structure observed by NMR for this region, either for the base pairs of 2 of 5 of these nucleotides, at positions 19 and 20 (42), or for the non-Watson-Crick nucleotide interactions within this loop region (14). The result with this deletion further underscores the importance of uncoupling analysis of RNA requirements from promoter sequence requirements: the same deletion construct expressed from the SL1 promoter was found not to rescue, presumably owing to defects in its transcription, and no definitive conclusion could be made regarding the functional requirements in this region of the leader (41). Our observation that the ΔGU loop mutant SL1 supports embryonic development demonstrates that neither the normal length of the leader nor the sequence of the loop region is essential for trans splicing or for leader function on trans-spliced mRNAs.
Substitutions that identify essential SL1 sequences.
As described above, elimination of either half of the leader sequence abolishes the essential embryonic function of SL1. This defect might result from removal of essential sequence elements per se. Alternatively, as all known trans-spliced leaders are at least 21 nt long, there may be a length requirement for these leaders and this result might indicate that the mutant leaders are simply too short to provide SL1 function. To address possible sequence requirements of the leader without altering its length, we tested the effects of several nucleotide substitutions.
To examine whether any sequence substitutions could be tolerated at all, we first analyzed the effects of a single-nucleotide change, a G-to-A transition at position 20 (Table 3). Position 20 is not predicted to participate in the first stem of SL1 RNA (6, 14) (Fig. 2), although it has been observed to be base paired with the C at position 9 in one of the two solution structures (42) (Fig. 2). However, the nucleotide at this position in the SL2 leader and most of the other minor leaders that have been identified in C. elegans is an A (10, 16, 34) (Table 3 and Fig. 3). We found that this construct rescued embryonic lethality efficiently, suggesting that the identity of the nucleotide at this position is not crucial for SL1 RNA or SL1 leader function (Table 3).
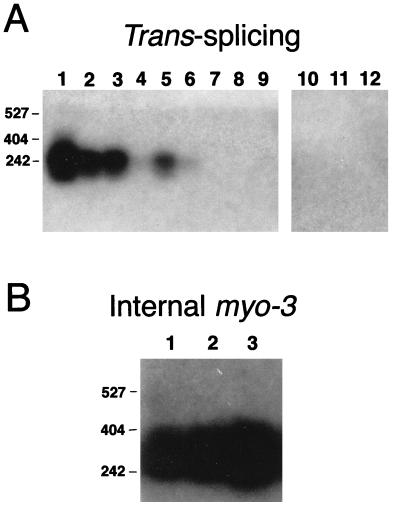
Next, we examined the effects of randomly rearranging the sequence of the second half of the leader (nt 11 to 20) without altering its base composition (11–20 shuffle) (Table 3). One result of this shuffle is that the nucleotides at positions 11 and 12, which are predicted to base pair with the splice donor site (as described above) (6, 14, 42) are rearranged, thereby likely disrupting these base pairs (Table 3 and Fig. 2). Although this mutant SL RNA was expressed at high levels from the U2 promoter, the mutant RNA failed to rescue embryonic lethality (Table 3 and Fig. 4B). We were unable to detect this shuffled leader RNA sequence on trans-spliced mRNAs by RT-PCR, although the normally trans-spliced mRNAs are present in the embryonic extracts (Fig. 5). It appears, therefore, that this leader is not trans spliced onto messages. This result suggests that a portion of the leader sequence or the SL1 RNA secondary structure is essential for efficient trans splicing of the leader in vivo.
FIG. 5.
RT-PCR analysis of trans splicing of mutant leaders. (A) Lanes 1 to 3, trans splicing of SL1 or mutant leaders to the myo-3 message in wild-type (N2) elongated embryos; lanes 4 to 6, homozygous wDf1 embryos rescued with the wild-type SL1 RNA transcribed from the U2-3 promoter (WT-SL1); lanes 7 to 9, homozygous wDf1 embryos (wDf1); lanes 10 to 12, wDf1 embryos carrying the 11–20 shuffle transgene. Three independent RNA samples were prepared and analyzed for each strain. Primers specific for the wild-type SL1 leader or for the 11–20 shuffle leader, together with a downstream primer specific for the myo-3 coding region, were used in the RT-PCRs (see Materials and Methods). The myo-3 message is embryonically transcribed (9), and both the message and protein are expressed robustly in wDf1 mutant embryos (12), although no trans splicing to SL1 is detected (therefore, these reactions serve as negative controls for contamination in the RT-PCR). There are no detectable bands in lanes 7 to 12, as described above, even in overexposed autoradiograms. The bands present in lanes 4 and 6, which appear faint in this exposure, are clearly evident in overexposed autoradiograms (data not shown). (B) RT-PCR analysis of the myo-3 message in wDf1 embryos carrying the 11–20 shuffle (lanes 1 to 3) transgene, using primers specific for an internal portion of the myo-3 message (see Materials and Methods). The positions of the molecular size markers (in base pairs) are shown at the left.
To further investigate primary sequence requirements in this region of the leader, we tested a mutant in which the 8 nucleotides in the loop region were substituted (loop substitution mutant). The nucleotides at each position in this region (positions 13 to 20) were made different from those found in all other spliced leaders in C. elegans; this change also altered the composition of this sequence significantly (e.g., the purine content was increased from 50 to 75%) (Table 3 and Fig. 3). This mutant RNA was expressed, albeit at 30% of the level of the 11–20 shuffle construct, and rescued embryonic lethality at a somewhat reduced efficiency, similar to that of the ΔGU loop mutant (Table 3 and Fig. 4B). This observation demonstrates that the SL1 leader can tolerate certain major changes in primary sequence (i.e., altering more than one-third of the sequence) without abolishing its essential embryonic function.
The SL2 leader cannot substitute for the SL1 leader when conjoined with the SL1 RNA intron.
We showed previously that SL2 RNA, when overexpressed from an extrachromosomal array, can rescue the lethality of embryos lacking zygotic SL1 RNA (12). We also demonstrate that the normally expressed SL2 is trans spliced onto SL1 splice acceptor sites when SL1 RNA is absent. These experiments suggested that, although the leaders are only ∼45% identical (16) (Fig. 3), the 22-nt SL2 leader can perform the essential embryonic function normally carried out by SL1. We further explored whether the SL2 leader can substitute for the SL1 leader by asking whether it could be efficiently trans spliced when coupled to the SL1 RNA intron-like sequence (the SL1 RNA sequence downstream of the leader). A chimeric SL RNA-encoding construct containing the 22-nt SL2 leader fused to the SL1 intron-like sequence was assayed for rescue of embryonic lethality (Table 3). Surprisingly, we found that the SL2-SL1 chimeric construct could not rescue embryonic lethality (Table 3). Since an SL2 leader can apparently functionally substitute for the SL1 leader once it is donated by the SL2 RNA (12), the failure of the SL2-SL1 construct to rescue may result from the inability of the SL2 leader to be trans spliced from the chimeric RNA (see Discussion).
The SL1 leader can tolerate a substantial increase in length.
Though all known nematode SLs are nearly identical in length (21 to 23 nt) (10, 16, 18, 34), our deletion studies demonstrated that SL1 can function when substantially shorter than normal (i.e., as few as 17 nt). To assess whether SL1 can function when substantially longer than normal, we analyzed SL1 RNAs containing additional nucleotides in the leader.
A construct was created in which the SL1 RNA contains five additional adenosine residues upstream of the 22-nt leader (Table 4). This RNA was found to be expressed efficiently in transgenic worms, was larger than the wild type by the expected amount (Fig. 4A), and was able to rescue embryonic lethality effectively (Table 4). This result suggests that the additional adenosine nucleotides do not affect the trans-splicing ability of the SL1 RNA or the function of the leader.
To examine the effects of an insertion of extra nucleotides into the leader, we tested a construct containing seven extra uridine residues within the loop region of the leader. This construct rescued embryonic lethality, although somewhat less efficiently than the wild-type construct (Table 4). This observation demonstrated that there are not stringent requirements for contiguous blocks of sequence within the SL1 leader in this region. In addition, it suggests that a nematode can proceed through embryogenesis normally even when ∼60% of all of its messages, which are normally trans spliced to SL1, contain spliced leaders that are >30% longer than normal.
DISCUSSION
We have exploited mutants lacking zygotic SL1 RNA to analyze the structural requirements of the SL1 leader, allowing the first dissection of major sequences required for its essential in vivo function. We report four major findings: (i) the U2-SL1 expression system is effective for analyzing SL1 leader sequences necessary for trans splicing and leader function, as the embryonic lethality of mutants lacking zygotic SL1 RNA can be rescued by the U2-SL1 chimera; (ii) while all characterized nematode leaders are 21 to 23 nt in length, the length of the leader sequence is apparently not critical for its function in mRNA metabolism; (iii) substantial alterations of the leader sequence do not dramatically affect leader function in vivo, since leaders containing deletions, substitutions, or additions can support embryogenesis; and (iv) certain primary sequence alterations are not tolerated in the SL1 leader, suggesting the importance of the identities of at least some of these nucleotides, or structures involving these nucleotides, for proper SL1 RNA or leader function.
System for analyzing SL1 leader sequence requirements.
The SL1 leader DNA sequence is highly conserved in all nematodes, leading to the suggestion that the precise leader sequence may be important for trans-splicing or leader function. However, the finding that the leader DNA also serves as part of the wild-type promoter in Ascaris and C. elegans (15, 41) suggests an alternative explanation for the conservation of this sequence. The U2-3 promoter used here to express SL1 uncouples the sequence requirements for the leader RNA from sequences required for transcription. This system allowed us to assess the requirement for the SL1 leader for trans-splicing and postsplicing functions without interfering with expression of the RNA. In contrast, in another study it was not possible to examine the effect of large alterations of the SL1 leader sequence when the SL1 promoter was used, since such mutations eliminated detectable SL1 RNA transcription (41). With the U2-SL1 expression system, SL1 RNA was expressed at levels sufficient to rescue embryonic lethality (Table 1) even in mutants with substantial alterations in the SL1 leader sequence. In addition, the SL1 RNAs expressed from this promoter initiate at the correct nucleotide (Fig. 4). The promoters of the U snRNAs may be useful for expression of other transgenes for which downstream sequences promote transcription.
The primary sequence of the leader per se does not appear to be essential for its function.
Although the C. elegans leaders are quite divergent in their primary sequences, there are several blocks of sequences conserved among most of them, suggesting regions that may be required for SL RNA or leader function (10, 34) (Fig. 3). However, mutants with major alterations in the primary sequence, specifically the ΔGU loop deletion, the loop substitution, and the 7U loop insertion, were able to provide the essential embryonic function of SL1 (Tables 2 to 4). These mutant leaders are substantially different in primary sequence from all of the known wild-type C. elegans leaders.
NMR analysis in one study showed that the loop region of the SL1 RNA assumes an ordered conformation, with interactions between the bases in the loop, while another analysis demonstrated that several of the nucleotides in this region appear to be base paired (14, 42). While these bases and this structure may be important for wild-type levels of function, we have found that they are not essential for trans splicing or leader function in vivo. The absence of a stringent requirement for sequence specificity or length in the loop also suggests the intriguing possibility that either splicing factors do not bind to this sequence or they are not required for SL1 RNA function.
The Δ16–20 deletion mutant was also analyzed in another recent study (41). However, it was reported that this mutant SL RNA did not rescue the lethality of the rrs-1 deletion mutants, even at a high concentration of injected DNA (41). Two important differences between these studies may explain these apparently conflicting results. First, in the previous study, this construct was expressed under the control of the wild-type promoter, and therefore this mutation dramatically reduced the amount of this RNA relative to other mutant SL RNAs, presumably due to a debilitated transcriptional regulatory region (41). In our analysis, although the Δ16–20 deletion mutant was also found to be expressed somewhat less efficiently than the endogenous wild-type SL1 RNA (Fig. 4), it was expressed at levels similar to those of other mutant SL RNAs that rescue embryonic lethality (Tables 2 to 4) (Fig. 4). Secondly, the assay used here, unlike that in the previous study, was more sensitive, as it did not require that the mutant constructs rescue rrs-1 mutant embryos through to adulthood (41).
The SL1 leader can tolerate substantial length variation.
We found that a number of additions, insertions, or deletions in the leader sequence do not substantially affect the ability of the SL1 RNA to rescue. The ability of an SL1 RNA to function with a leader as short as 17 nt, i.e., significantly shorter than leaders known in any organism, or containing as many as 7 extra nucleotides, demonstrates that substantial variability in leader length can be tolerated and that alterations in its length do not hinder its interaction with factors that might be required for its function. Thus, there appears not to be an obligatory length for trans-spliced leaders in C. elegans, and the conserved 21- to 23-nt length of the leader may relate to spacing requirements for transcriptional regulatory elements rather than to leader function per se.
Large deletions or rearrangements of the SL1 leader abolish in vivo leader function.
Deletions of the 5′ or 3′ half of the leader and a rearrangement of the 3′ half eliminate rescue of rrs-1 deletion mutants (Tables 2 and 3). Several possibilities could explain the effects of these mutations. For example, these sequences may be required for SL1 RNP assembly or trans splicing. In the case of the 11–20 shuffle construct, we were unable to detect the mutant leader on trans-spliced messages by RT-PCR (Fig. 5), although this construct was expressed at high levels compared to other mutant SL RNAs and the mRNAs were detectable in the extracts by RT-PCR (Table 3 and Fig. 4B and 5). Since 8 of 10 nt were changed in the loop substitution construct, which showed rescue, the additional 2 nt affected in the shuffle construct (positions 11 and 12) may be required for leader function, perhaps because they base pair with splice donor site nucleotides (6, 14, 42). Therefore, the secondary structure of the SL1 RNA may be a critical parameter of its function in vivo; indeed, it has been proposed that these base pair interactions are essential for splice site recognition (5, 6).
In addition, an SL2 leader-SL1 intron chimeric construct does not rescue embryonic lethality (Table 3). This was unexpected in light of our earlier results, which demonstrated that the SL2 leader can supply SL1 leader function when present on a normally SL1 trans-spliced mRNA (12). It is possible that this SL2-SL1 chimeric RNA is not expressed at sufficient levels for rescue (although levels of expression are similar to those observed for the loop substitution mutant, which rescues embryonic lethality). Alternatively, the chimera may assume an inappropriate structure which affects trans splicing.
What is the function of the SL1 leader in mRNA metabolism?
The apparent lack of strict sequence requirement for SL function in C. elegans suggests that the leader sequence might serve a passive role once it is trans spliced to mRNAs. Our results leave open the possibility that C. elegans leaders function in mRNA metabolism or in translational efficiency (as has been demonstrated in vitro in Ascaris [23]), since some altered leader sequences do not rescue as efficiently as the wild-type RNA. As requirements for mRNA metabolism and translation in C. elegans become better defined, a definitive role for trans splicing or SLs in these processes may be revealed.
ACKNOWLEDGMENTS
We thank Tom Blumenthal for the gift of the U2-3 plasmid, Peter Barrett for the gift of the pBX1 (pha-1) plasmid, and Tom Blumenthal, members of the Blumenthal lab, and David Brow for helpful discussions and suggestions.
This work was supported by grants from the National Institutes of Health (AG13736) and the National Science Foundation (IBN-9506089) to J.H.R.
REFERENCES
- 1.Agabian N. Trans-splicing of nuclear pre-mRNAs. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- 2.Bektesh S, Van Doren K, Hirsh D. Presence of the Caenorhabditis elegans spliced leader on different mRNAs and in different genera of nematodes. Genes Dev. 1988;2:1277–1283. doi: 10.1101/gad.2.10.1277. [DOI] [PubMed] [Google Scholar]
- 3.Blumenthal T. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 4.Bonen L. Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J. 1993;7:40–46. doi: 10.1096/fasebj.7.1.8422973. [DOI] [PubMed] [Google Scholar]
- 5.Bruzik J P, Steitz J A. Spliced leader RNA sequences can substitute for the essential 5′ end of U1 RNA during splicing in a mammalian in vitro system. Cell. 1990;62:889–899. doi: 10.1016/0092-8674(90)90264-f. [DOI] [PubMed] [Google Scholar]
- 6.Bruzik J P, Van Doren K, Hirsh D, Steitz J A. Trans-splicing involves a novel form of small nuclear ribonucleoprotein particles. Nature. 1988;335:559–562. doi: 10.1038/335559a0. [DOI] [PubMed] [Google Scholar]
- 7.Conrad R, Thomas J, Spieth J, Blumenthal T. Insertion of part of an intron into the 5′ untranslated region of a Caenorhabditis elegans gene converts it into a trans-spliced gene. Mol Cell Biol. 1991;11:1921–1926. doi: 10.1128/mcb.11.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis R E, Singh H, Botka C, Hardwick C, Ashraf el Meanawy M, Villanueva J. RNA trans-splicing in Fasciola hepatica. Identification of a spliced leader (SL) RNA and SL sequences on mRNAs. J Biol Chem. 1994;269:20026–20030. [PubMed] [Google Scholar]
- 9.Dibb N J, Maruyama I N, Krause M, Karn J. Sequence analysis of the complete Caenorhabditis elegans myosin heavy chain gene family. J Mol Biol. 1989;205:603–613. doi: 10.1016/0022-2836(89)90229-5. [DOI] [PubMed] [Google Scholar]
- 10.Evans D, Zorio D, MacMorris M, Winter C E, Lea K, Blumenthal T. Operons and SL2 trans-splicing exist in nematodes outside the genus Caenorhabditis. Proc Natl Acad Sci USA. 1997;94:9751–9756. doi: 10.1073/pnas.94.18.9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson, K. C., and J. H. Rothman. Unpublished data.
- 12.Ferguson K C, Heid P J, Rothman J H. The SL1 trans-spliced leader RNA performs an essential embryonic function in Caenorhabditis elegans that can also be supplied by SL2 RNA. Genes Dev. 1996;10:1543–1556. doi: 10.1101/gad.10.12.1543. [DOI] [PubMed] [Google Scholar]
- 13.Granato M, Schnabel H, Schnabel R. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 1994;22:1762–1763. doi: 10.1093/nar/22.9.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenbaum N L, Radhakrishnan I, Hirsh D, Patel D. Determination of the folding topology of the SL1 RNA from Caenorhabditis elegans by multidimensional heteronuclear NMR. J Mol Biol. 1995;252:314–327. doi: 10.1006/jmbi.1995.0499. [DOI] [PubMed] [Google Scholar]
- 15.Hannon G J, Maroney P A, Ayers D G, Shambaugh J D, Nilsen T W. Transcription of a nematode trans-spliced leader RNA requires internal elements for both initiation and 3′ end formation. EMBO J. 1990;9:1915–1921. doi: 10.1002/j.1460-2075.1990.tb08318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X Y, Hirsh D. A second trans-spliced RNA leader sequence in the nematode Caenorhabditis elegans. Proc Natl Acad Sci USA. 1989;86:8640–8644. doi: 10.1073/pnas.86.22.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krause M. Transcription and translation. In: Epstein H F, Shakes D, editors. Caenorhabditis elegans: modern biological analysis of an organism. San Diego, Calif: Academic Press, Inc.; 1995. pp. 483–512. [Google Scholar]
- 18.Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lücke S, Xu G-L, Palfi Z, Cross M, Bellofatto V, Bindereif A. Spliced leader RNA of trypanosomes: in vivo mutational analysis reveals extensive and distinct requirements for trans-splicing and cap4 formation. EMBO J. 1996;15:4380–4391. [PMC free article] [PubMed] [Google Scholar]
- 20.Madhani H D, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 21.Maroney P A, Hannon G J, Denker J A, Nilsen T W. The nematode spliced leader RNA participates in trans-splicing as an Sm snRNP. EMBO J. 1990;9:3667–3673. doi: 10.1002/j.1460-2075.1990.tb07578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maroney P A, Hannon G J, Shambaugh J D, Nilsen T W. Intramolecular base pairing between the nematode spliced leader and its 5′ splice site is not essential for trans-splicing in vitro. EMBO J. 1991;10:3869–3875. doi: 10.1002/j.1460-2075.1991.tb04956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maroney P A, Denker J A, Darzynkiewicz E, Laneve R, Nilsen T W. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: a role for spliced leader addition in translational efficiency. RNA. 1995;1:714–723. [PMC free article] [PubMed] [Google Scholar]
- 24.Mattaj I W. U snRNP assembly and transport. In: Birnstiel M L, editor. Structure and function of major and minor small nuclear ribonucleoprotein particles. New York, N.Y: Springer-Verlag; 1988. pp. 100–114. [Google Scholar]
- 25.Mello C C, Kramer J M, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaeli S, Roberts T G, Watkins K P, Agabian N. Isolation of distinct small ribonucleoprotein particles containing the spliced leader and U2 RNAs of Trypanosoma brucei. J Biol Chem. 1990;265:10582–10588. [PubMed] [Google Scholar]
- 27.Miller S I, Wirth D F. trans-splicing in Leishmania enriettii and identification of ribonucleoprotein complexes containing the spliced leader and U2 equivalent RNAs. Mol Cell Biol. 1988;8:2597–2603. doi: 10.1128/mcb.8.6.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson D W, Honda B M. Genes coding for 5S ribosomal RNA of the nematode Caenorhabditis elegans. Gene. 1985;38:245–251. doi: 10.1016/0378-1119(85)90224-0. [DOI] [PubMed] [Google Scholar]
- 29.Nilsen T W. Trans-splicing of nematode premessenger RNA. Annu Rev Microbiol. 1993;47:413–440. doi: 10.1146/annurev.mi.47.100193.002213. [DOI] [PubMed] [Google Scholar]
- 30.Nilsen T W. Trans-splicing: an update. Mol Biochem Parisitol. 1995;73:1–6. doi: 10.1016/0166-6851(94)00107-x. [DOI] [PubMed] [Google Scholar]
- 31.Nilsen T W, Shambaugh J, Denker J A, Chubb G, Faser C, Putnam L, Bennett K. Characterization and expression of a spliced leader RNA in the parasitic nematode Ascaris lumbricoides var. suum. Mol Cell Biol. 1989;9:3543–3547. doi: 10.1128/mcb.9.8.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palfi Z, Günzl A, Cross M, Bindereif A. Affinity purification of Trypanosoma brucei snRNPs reveals common and specific protein components. Proc Natl Acad Sci USA. 1991;88:9097–9101. doi: 10.1073/pnas.88.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parry H D, Scherly D, Mattaj I W. ‘Snurpogenesis’: the transcription and assembly of U snRNP components. Trends Biochem Sci. 1989;14:15–19. [Google Scholar]
- 34.Ross L H, Freedman J H, Rubin C S. Structure and expression of novel spliced leader RNA genes in Caenorhabditis elegans. J Biol Chem. 1995;270:22066–22075. doi: 10.1074/jbc.270.37.22066. [DOI] [PubMed] [Google Scholar]
- 35.Schnabel H, Schnabel R. An organ-specific differentiation gene, pha-1, from Caenorhabditis elegans. Science. 1990;250:686–688. doi: 10.1126/science.250.4981.686. [DOI] [PubMed] [Google Scholar]
- 36.Spieth J, Brooke G, Kuersten S, Lea K, Blumenthal T. Operons in C. elegans: polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell. 1993;73:521–532. doi: 10.1016/0092-8674(93)90139-h. [DOI] [PubMed] [Google Scholar]
- 37.Tessier L-H, Keller M, Chan R L, Fournier R, Weil J-H, Imbault P. Short leader sequences may be transferred from small RNAs to pre-mature mRNAs by trans-splicing in Euglena. EMBO J. 1991;10:2621–2625. doi: 10.1002/j.1460-2075.1991.tb07804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas J, Lea K, Zucker-Aprison E, Blumenthal T. The spliceosomal snRNAs of Caenorhabditis elegans. Nucleic Acids Res. 1990;18:2633–2642. doi: 10.1093/nar/18.9.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas J D, Conrad R C, Blumenthal T. The C. elegans trans-spliced leader RNA is bound to Sm and has a trimethylguanosine cap. Cell. 1988;54:533–539. doi: 10.1016/0092-8674(88)90075-x. [DOI] [PubMed] [Google Scholar]
- 40.Van Doren K, Hirsh D. Trans-spliced leader RNA exists as small nuclear ribonucleoprotein particles in Caenorhabditis elegans. Nature. 1988;335:556–559. doi: 10.1038/335556a0. [DOI] [PubMed] [Google Scholar]
- 41.Xie H, Hirsh D. In vivo function of mutated spliced leader RNAs in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:4235–4240. doi: 10.1073/pnas.95.8.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J, Lapham J, Crothers D M. Determining RNA solution structure by segmental isotopic labeling and NMR: application to Caenorhabditis elegans spliced leader RNA 1. Proc Natl Acad Sci USA. 1996;93:44–48. doi: 10.1073/pnas.93.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zorio D A R, Cheng N N, Blumenthal T, Spieth J. Operons as a common form of chromosomal organization in C. elegans. Nature. 1994;372:270–272. doi: 10.1038/372270a0. [DOI] [PubMed] [Google Scholar]