Abstract
Background: The mTOR signaling pathway is inactivated by AMPK’s tumor-suppressing function. It is recognized that ubiquitin conjugating enzyme 2O (UBE2O), which directly targets AMPK for ubiquitination and degradation, is intensified in human cancers. Methods: This study investigated the clinical data about prostate cancer. Examination was also carried out into tissue microarrays (TMA) of human prostate cancer (n = 382) and adjacent non-neoplastic tissues around prostate cancer (n = 61). The TMA slides were incubated with antibodies against UBE2O, and the cores were scored by the pathologist blind to cancer results. Results: Very strong positive correlations were identified between the expression of UBE2O staining and high PSA and pathological stage of prostate cancer. Cox’s proportional hazard analysis established correlations between the following: (1) positive surgical margin and biochemical recurrence free survival, (2) PSA grade and clinical recurrence free survival, (3) regional lymph node positive and clinical recurrence free survival, (4) adjuvant treatment and overall survival, and (5) pathological T stage and overall survival. Conclusion: There is a positive correlation between the expression of UBE2O staining and prognosis for prostate cancer. Thus, a prostate cancer prognosis can be assessed with the expression of UBE2O staining.
Keywords: UBE2O, ubiquitin, prostate cancer
1. Introduction
In eukaryotes, the ubiquitin proteasome system (UPS) targets cell cycle regulators for proteasome-mediated degradation, thereby strictly controlling the cell cycle at major checkpoints. In order to enable ubiquitination of target proteins, the UPS necessitates the ubiquitin-activating enzyme (E1), the ubiquitin-conjugating enzyme (E2), and the ubiquitin ligases (E3) to work harmoniously. Mono-ubiquitination controls the ubiquitin-reliant endocytosis, rearrangement of protein complexes, repair of DNA, and transcriptional regulation. Poly-ubiquitination, which is a chain of a minimum of four ubiquitin’s added to an individual lysine (Lys) residue, is necessary for the labelling of target proteins for degradation [1].
In terms of the ubiquitin enzymes, the E1 activates ubiquitin by attaching the molecule to an active site cystine (Cys) and then using a thioester linkage, moves the ubiquitin to the E2 active site Cys. Subsequently, via E3-mediated specificity, the E2 gives the ubiquitin from its Cys to Lys of the target protein. The E3 enzyme binds to the target protein that will be degraded [1]. E3 enzymes are part of the domain group that includes the anaphase-promoting complex (also referred to as cyclosome, APC/C) [2]. As their deregulation is linked with cancer, the majority of studies have primarily addressed E3s. It is only of late that the critical part E2s play in the regulation of cell cycle progression and in specific cancer development and progression has begun to receive more attention.
AMP-activated protein kinase (AMPK) is a key monitor of cellular energy and nutrient levels. Associations have been identified between cancer and AMPK loss or activity deregulation. More specifically, it has been established that decreased AMPK activity is found in human breast and kidney cancers [3,4]. It is recognised that UBE2O, which directly targets AMPK for ubiquitination and degradation, is intensified in human cancers. UBE20 is a comparatively large E2 ubiquitin-conjugation enzyme. It is magnified in a subset of human cancers [5,6,7,8,9,10]; however, its contribution in tumorigenesis remains partially undefined. Overexpressed in numerous human cancers, UBE2O targets AMPK for ubiquitination and degradation, which subsequently fosters the activation of the mTOR-HIF1a pathway. Tumorigenesis is impeded by the genetic deletion of UBE2O via the regeneration of AMPKa2 [11].
In UBE2O-deficient TRAMP mice, it was found that there is reduced formations of invasive prostate carcinoma and metastasis [11]. Obstructing UBE2O treatment resulted in decreased prostate lobe enlargement and high G-PIN prostate cancer development in TRAMP in UBE2O positive mice. Additionally, the hindering of UBE2O decreases tumorigenesis to degrees in line with UBE2O deficiency cases [11]. UBE2O plays a key role in the initiation, progression, invasion, and metastasis of prostate cancer. In mouse models of breast and prostate cancers, decreased tumor growth and metastasis rates were observed when one or both UBE2O alleles were lost [11]. The purpose of this study is to examine the relationships between the expression of immunohistochemical UBE2O staining and the progression factors in prostate cancer patients.
2. Results
2.1. Clinicopathological Characteristics of Prostatic Cancer Patients
Total TMA (n = 382) was comprised of 202 prostate cancer patients from Soonchunhyang University Hospital and 180 purchased externally; the surgical margin could be assessed in only the 200 prostate cancer patients from Soonchunhyang University Hospital out of the total 382 patients (Table 1).
Table 1.
Clinicopathological properties of patient and UBE2O expression in prostate cancer.
| Prognosis Factors |
n (%) (n = 382) |
|---|---|
| Age (years) | 67.0 ± 0.4 |
| PSA (ng/mL) | |
| <10 | 220 (57.6%) |
| 10–20 | 97 (25.4%) |
| >20 | 65 (17.0%) |
| Gleason | |
| ≤6 | 52 (13.6%) |
| 7 | 155 (40.6%) |
| 8–10 | 175 (45.8%) |
| Pathological stage | |
| ≤T2 | 162 (42.4%) |
| ≥T3 | 220 (57.6%) |
| Seminal vesicle invasion | |
| Negative | 319 (83.5%) |
| Positive | 63 (16.5%) |
| Lymph node involvement | |
| Negative | 370 (96.9%) |
| Positive | 12 (3.1%) |
| Surgical margin * | |
| Negative | 115 (57.5%) |
| Positive | 85 (42.5%) |
| Expression of UBE2O # | |
| 0 | 0 |
| 1 | 120 (33.8%) |
| 2 | 154 (43.4%) |
| 3 | 81 (22.8%) |
UBE2O grade—0: negative 1: weak 2: moderate 3: strong. Age was expressed as Mean ± standard error. * The number of surgical margin could be evaluated in only 200 with prostate cancer of our hospital from 382 patients. # The number of UBE2O immunohistochemical expression could be evaluated in only 355 from 382 patients with prostate cancer of our hospital and purchased TMA.
2.2. UBE2O Expression
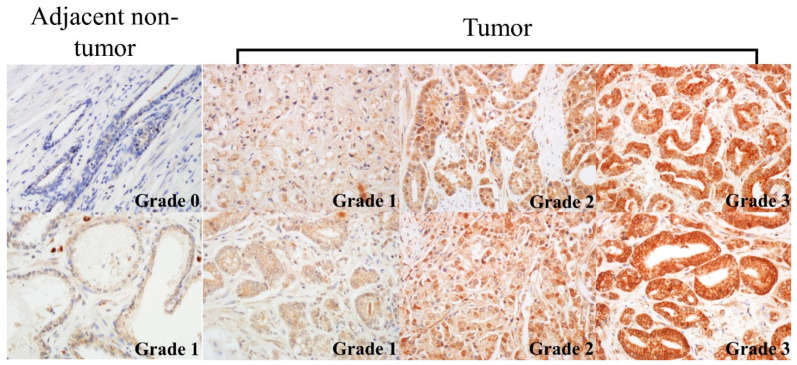
UBE2O was expressed in cytoplasm of the luminal epithelial cell in the non-neoplastic prostatic tissue. UBE2O in prostatic cancer primarily materialized in the cytoplasm of tumor cells (Figure 1). The classifications for the 355 prostate cancer patients’ expression of UBE2O were as follows: grade 0 (n = 0), grade 1 (n = 120), grade 2 (n = 154), and grade 3 (n = 81). Additionally, for the 61 non-neoplastic prostate tissue, expressed UBE2O were as follows: grade 0 (n = 22), grade 1 (n = 15), grade 2 (n = 23), and grade 3 (n = 1). Furthermore, prostate cancer tissues and non-neoplastic prostate tissues showed higher and lower grades of UBE2O, respectively (Table 2, Figure 1, Figure 2, Suppl. Figure S1) (p < 0.001).
Figure 1.
Different expression of UBE20 in non-neoplastic and neoplastic tissues. UBE2O grade—0: negative, 1: weak 2: moderate, 3: strong. (×400).
Table 2.
UBE2O Immunohistochemical staining of prostate cancer and adjacent non-neoplastic tissues for radical prostatectomy due to prostate cancer.
| UBE2O | Adjacent Non-Neoplastic Tissues | Prostate Cancer | Total | p Value |
|---|---|---|---|---|
| 0 | 22 | 0 | 22 | <0.001 |
| 1 | 15 | 120 | 135 | |
| 2 | 23 | 154 | 177 | |
| 3 | 1 | 81 | 82 | |
| Total | 61 | 355 | 416 |
UBE2O grade—0: negative 1: weak 2: moderate 3: strong. The number of UBE2O immunohistochemical staining of prostate cancer could be evaluated in 416 from patients including 355 prostate cancer tissues and 61 adjacent non-neoplastic tissues around prostate cancer in 382 patients with prostate cancer. 382 patients included 202 patients with prostate cancer of our hospital and 180 from purchased TMA. Adjacent non-neoplastic tissues were 39 from TMA of our hospital, 22 from purchased TMA.
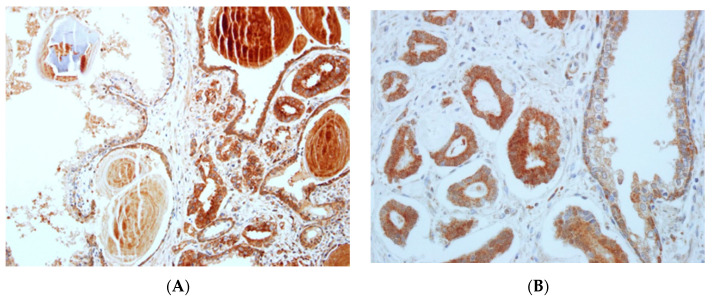
Figure 2.
Different expression of UBE20 in neoplastic tissue and non-neoplastic tissue. (A) Expression score of UBE20 in right side (neoplastic tissue) was 3 and was 1 in left side (non-neoplastic tissue) with preservation of basal cell. (×200) (B) Expression score of UBE20 in left side (non-neoplastic tissue) was 3 and was 1 in right side (neoplastic tissue) with preservation of basal cell (×400).
2.3. Association of UBE2O Expression with Clinicopathologic Parameters
As mentioned, of the total 382 study population, the UBE2O immunohistochemical staining of prostate cancer could be assessed in 200 patients. It was only possible to assess the surgical margin in 200 patients from Soonchunhyang University Hospital. Of those 200, 173 could be assessed with UBE2O immunohistochemical staining.
Cox’s proportional hazard analysis was performed on the 200 prostate cancer patients from Soonchunhyang University Hospital, and it was found that patients with a higher grade of UBE2O demonstrated higher PSA than those of lower grade UBE2O (p < 0.001). Additionally, patients with higher UBE2O grades demonstrated greater pathologic stage than those with lower UBE2O grades (p < 0.001) (Table 3).
Table 3.
UBE2O immunohistochemical staining of prostate cancer for radical prostatectomy due to prostate cancer.
| UBE20 | Total (n = 355) |
p Value | |||
|---|---|---|---|---|---|
| 1 (n = 120) | 2 (n = 154) | 3 (n = 81) | |||
| Age | 65 (60, 68) | 65 (60, 69) | 66 (62, 72) | 65 (61, 69) | 0.029 |
| PSA | <0.001 | ||||
| <10 | 28 | 45 | 39 | 112 | |
| 10–20 | 92 | 109 | 42 | 243 | |
| >20 | 0 | 0 | 0 | 0 | |
| Gleason sum | 0.168 | ||||
| ≤6 | 17 | 18 | 8 | 43 | |
| 7 | 52 | 67 | 25 | 144 | |
| 8–10 | 51 | 69 | 48 | 168 | |
| Pathological stage | <0.001 | ||||
| ≤T2 | 25 | 42 | 39 | 106 | |
| ≥T3 | 95 | 112 | 42 | 249 | |
| Seminal vesicle invasion | 0.772 | ||||
| Negative | 101 | 126 | 65 | 292 | |
| Positive | 19 | 27 | 16 | 62 | |
| Lymph node involvement | 0.015 | ||||
| Negative | 120 | 149 | 75 | 344 | |
| Positive | 0 | 5 | 5 | 10 | |
| Surgical margin # | 0.555 | ||||
| Negative | 19 | 40 | 35 | 94 | |
| Positive | 12 | 32 | 35 | 79 | |
UBE2O grade—0: negative, 1: weak, 2: moderate, 3: strong. Age was expressed as Median (lower quadrant, upper quadrant). The number of UBE2O immunohistochemical staining of prostate cancer could be evaluated in 355 from our hospital from 200 patients with prostate cancer of our hospital and 180 from purchased TMA. # The number of surgical margin could be evaluated in only 200 of our hospital from 382 patients with prostate cancer excluding purchased TMA. Out of 202 patients with prostate cancer of our hospital, the 173 patients could be evaluated with UBE2O immunohistochemical staining.
However, patients with higher UBE2O grades did not present with greater involvement of seminal vesicle than those with lower UBE2O grades. Patients with higher UBE2O grades demonstrated greater involvement of lymph nodes than those with lower UBE2O grades (p < 0.05). Finally, patients with higher UBE2O grades did not demonstrate higher positive surgical margins than those with lower UBE2O grades (Table 3).
2.4. Survival Analysis
Of the 335 prostate cancer patients, it was possible to evaluate 173 for survival. Biochemical recurrence, clinical recurrence, and overall survival were described in Table 4. Biochemical recurrence (mean ± standard error (SE)) of UBE2O grade 1, UBE2O grade 2, and UBE2O grade 3 were 7.77 ± 0.66 year, 11.39 ± 0.68 year, and 9.43 ± 0.57 year, respectively. Clinical recurrence (mean ± SE) of UBE2O grade 1, UBE2O grade 2, and UBE2O grade 3 were 10.23 ± 0.51 year, 11.82 ± 0.45 year, and 11.31 ± 0.44 year, respectively. Overall survival (mean ± SE) UBE2O grade 1, UBE2O grade 2, and UBE2O grade 3 were 8.47 ± 0.70 year, 9.65 ± 0.60 year, and 10.63 ± 0.46 year, respectively.
Table 4.
Biochemical recurrence free, clinical recurrence free and overall survival time for UBE2O expression—173 from 335 patients with prostate cancer could be evaluated for survival.
| UBE2O Grade | Total | Event | Censored | Survival Time | Survival Time | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean ± SE | 95% CI | Median ± SE | 95% CI | ||||||
| Lower | Upper | Lower | Upper | ||||||
| Biochemical recurrence free survival | |||||||||
| 1 | 31 | 5 | 26 | 7.77 ± 0.66 | 6.47 | 9.06 | |||
| 2 | 72 | 10 | 62 | 11.39 ± 0.68 | 10.07 | 12.72 | |||
| 3 | 70 | 14 | 56 | 9.43 ± 0.57 | 8.32 | 10.55 | |||
| Total | 173 | 29 | 144 | 10.77 ± 0.54 | 9.71 | 11.83 | |||
| Clinical recurrence free survival | |||||||||
| 1 | 31 | 1 | 30 | 10.23 ± 0.51 | 9.23 | 11.23 | |||
| 2 | 72 | 5 | 67 | 11.82 ± 0.45 | 10.94 | 12.70 | 12.42 ± 3.87 | 4.84 | 20.00 |
| 3 | 70 | 5 | 65 | 11.31 ± 0.44 | 10.45 | 12.18 | |||
| Total | 173 | 11 | 162 | 11.74 ± 0.34 | 11.08 | 12.40 | 12.42 ± 2.40 | 7.72 | 17.11 |
| Overall survival | |||||||||
| 1 | 31 | 7 | 24 | 8.47 ± 0.70 | 7.10 | 9.84 | |||
| 2 | 72 | 17 | 55 | 9.65 ± 0.60 | 8.47 | 10.84 | 10.97 ± 1.72 | 7.61 | 14.33 |
| 3 | 70 | 11 | 59 | 10.63 ± 0.46 | 9.74 | 11.53 | 12.11 ± 3.07 | 6.09 | 18.13 |
| Total | 173 | 35 | 138 | 10.17 ± 0.39 | 9.41 | 10.94 | 12.11 ± 1.11 | 9.93 | 14.29 |
UBE2O grade—0: negative, 1: weak, 2: moderate, 3: strong.
Cox’s proportional hazard analysis in patients with prostate cancer found the following (Table 5 and Table 6): positive surgical margin correlated with biochemical recurrence free survival (p = 0.003); clinical recurrence free survival of prostate cancer correlated with PSA grade (p = 0.004); clinical recurrence free survival of prostate cancer correlated with regional lymph node positive (p < 0.001). Cox’s proportional hazard modelling in patients with prostate cancer found a correlation between adjuvant treatment (p = 0.030) and pathological T stage (p = 0.020), and overall survival (OS) of prostate cancer (Table 7).
Table 5.
Cox proportional hazard modelling of UBE2O after accounting for biochemical recurrence free survival—173 from 335 patients with prostate cancer could be evaluated for survival.
| Univariate | Multivariate | |||||||
|---|---|---|---|---|---|---|---|---|
| HR | Lower | Upper | p-Value | HR | Lower | Upper | p-Value | |
| Age | 0.998 | 0.946 | 1.054 | 0.950 | ||||
| Height | 0.947 | 0.896 | 1.001 | 0.053 | ||||
| Weight | 0.985 | 0.951 | 1.020 | 0.394 | ||||
| Adjuvant treatment: no | Reference | Reference | ||||||
| Adjuvant treatment: yes | 20.171 | 7.139 | 56.993 | <0.001 | 8.765 | 2.829 | 27.153 | <0.001 |
| UBE2O grade 1 | Reference | Reference | ||||||
| UBE2O grade 2 | 0.619 | 0.207 | 1.854 | 0.392 | 0.649 | 0.203 | 2.074 | 0.466 |
| UBE2O grade 3 | 1.035 | 0.372 | 2.883 | 0.947 | 0.629 | 0.216 | 1.832 | 0.395 |
| PSA grade 0 | Reference | |||||||
| PSA grade 1 | 2.170 | 0.983 | 4.789 | 0.055 | ||||
| PSA grade 2 | 2.769 | 1.273 | 6.022 | 0.010 | ||||
| Gleason score grade 0 | Reference | |||||||
| Gleason score grade 1 | 2.037 | 0.432 | 9.617 | 0.369 | ||||
| Gleason score grade 2 | 4.923 | 1.162 | 20.86 | 0.031 | ||||
| Pathologic T stage grade 0 | Reference | Reference | ||||||
| Pathologic T stage grade 1 | 3.880 | 1.974 | 7.627 | <0.001 | 2.165 | 0.859 | 5.459 | 0.102 |
| Seminal vesicle invasion: negative | Reference | |||||||
| Seminal vesicle invasion: positive | 3.757 | 1.919 | 7.357 | <0.001 | ||||
| Regional lymph node: negative | Reference | |||||||
| Regional lymph node: positive | 5.712 | 2.483 | 13.142 | <0.001 | ||||
| Surgical margin: negative | Reference | Reference | ||||||
| Surgical margin: positive | 5.987 | 2.732 | 13.122 | <0.001 | 5.261 | 1.737 | 15.931 | 0.003 |
UBE2O grade 0: negative, 1: weak, 2: moderate, 3: strong. PSA grade 0: PSA < 10ng/mL, 1: PSA 10–20 ng/mL, 2: PSA > 20 ng/mL. Gleason score grade—0: Gleason score ≤ 6, 1: Gleason score 7, 2: Gleason score ≥ 8. Pathologic T stage grade—0: ≤T2, 1: ≥T3.
Table 6.
Cox proportional hazard modelling of UBE2O after accounting for clinical recurrence free survival—173 from 335 patients with prostate cancer could be evaluated for survival.
| Univariate | Multivariate | |||||||
|---|---|---|---|---|---|---|---|---|
| HR | Lower | Upper | p-Value | HR | Lower | Upper | p-Value | |
| Age | 1.074 | 0.976 | 1.182 | 0.142 | ||||
| Height | 0.936 | 0.854 | 1.026 | 0.155 | 1.276 | 1.134 | 1.435 | <0.001 |
| Weight | 0.933 | 0.875 | 0.995 | 0.036 | 0.807 | 0.710 | 0.918 | <0.001 |
| Adjuvant treatment: no | Reference | |||||||
| Adjuvant treatment: yes | 19.624 | 2.544 | 151.376 | 0.004 | ||||
| UBE2O grade 1 | Reference | |||||||
| UBE2O grade 2 | 1.356 | 0.151 | 12.169 | 0.786 | 0.028 | 0.008 | 0.106 | <0.001 |
| UBE2O grade 3 | 1.644 | 0.191 | 14.150 | 0.651 | 0.023 | 0.006 | 0.084 | <0.001 |
| PSA grade 0 | Reference | |||||||
| PSA grade 1 | 2.473 | 0.715 | 8.546 | 0.153 | 6.938 | 1.847 | 26.064 | 0.004 |
| PSA grade 2 | 1.798 | 0.429 | 7.537 | 0.422 | 0.213 | 0.027 | 1.708 | 0.145 |
| Gleason score grade 0 | ||||||||
| Gleason score grade 1 | ||||||||
| Gleason score grade 2 | ||||||||
| Pathologic T stage grade 0 | Reference | |||||||
| Pathologic T stage grade 1 | 4.154 | 1.272 | 13.562 | 0.018 | ||||
| Seminal vesicle invasion: negative | Reference | |||||||
| Seminal vesicle invasion: positive | 5.392 | 1.735 | 16.756 | 0.004 | ||||
| Regional lymph node: negative | Reference | |||||||
| Regional lymph node: positive | 12.474 | 3.933 | 39.567 | <0.001 | 90.544 | 22.747 | 360.410 | <0.001 |
| Surgical margin: negative | Reference | |||||||
| Surgical margin: positive | 3.169 | 0.967 | 10.381 | 0.057 | ||||
UBE2O grade—0: negative, 1: weak, 2: moderate, 3: strong. PSA grade 0: PSA < 10 ng/mL, 1: PSA 10–20 ng/mL, 2: PSA > 20 ng/mL. Gleason score grade—0: Gleason score ≤ 6, 1: Gleason score 7, 2: Gleason score ≥ 8. Pathologic T stage grade—0: ≤T2, 1: ≥T3.
Table 7.
Cox proportional hazard modelling of UBE2O after accounting for overall survival—173 from 335 patients with prostate cancer could be evaluated for survival.
| Univariate | Multivariate | |||||||
|---|---|---|---|---|---|---|---|---|
| HR | Lower | Upper | p-Value | HR | Lower | Upper | p-Value | |
| Height | 0.980 | 0.931 | 1.031 | 0.434 | ||||
| Weight | 0.973 | 0.939 | 1.009 | 0.139 | ||||
| Age | 1.032 | 0.981 | 1.087 | 0.225 | 1.052 | 0.99 | 1.117 | 0.102 |
| Adjuvant treatment: no | Reference | Reference | ||||||
| Adjuvant treatment: yes | 0.733 | 0.379 | 1.417 | 0.356 | 0.399 | 0.174 | 0.915 | 0.030 |
| UBE2O grade 1 | Reference | Reference | ||||||
| UBE2O grade 2 | 0.776 | 0.320 | 1.883 | 0.575 | 0.503 | 0.197 | 1.289 | 0.152 |
| UBE2O grade 3 | 0.473 | 0.182 | 1.230 | 0.125 | 0.310 | 0.112 | 0.856 | 0.024 |
| PSA grade 0 | Reference | |||||||
| PSA grade 1 | 1.332 | 0.633 | 2.805 | 0.451 | ||||
| PSA grade 2 | 1.573 | 0.748 | 3.309 | 0.233 | ||||
| Gleason score grade 0 | Reference | |||||||
| Gleason score grade 1 | 0.869 | 0.349 | 2.159 | 0.762 | ||||
| Gleason score grade 2 | 0.707 | 0.298 | 1.678 | 0.431 | ||||
| Pathologic T stage grade 0 | Reference | Reference | ||||||
| Pathologic T stage grade 1 | 1.818 | 0.981 | 3.370 | 0.058 | 2.458 | 1.155 | 5.232 | 0.020 |
| Seminal vesicle invasion: negative | Reference | |||||||
| Seminal vesicle invasion: positive | 1.915 | 0.954 | 3.843 | 0.067 | ||||
| Regional lymph node: negative | Reference | |||||||
| Regional lymph node: positive | 2.098 | 0.817 | 5.386 | 0.123 | ||||
| Surgical margin: negative | Reference | |||||||
| Surgical margin: positive | 1.183 | 0.638 | 2.191 | 0.594 | ||||
UBE2O grade—0: negative, 1: weak, 2: moderate, 3: strong. PSA grade 0: PSA < 10 ng/mL, 1: PSA 10–20 ng/mL, 2: PSA > 20 ng/mL. Gleason score grade—0: Gleason score ≤ 6, 1: Gleason score 7, 2: Gleason score ≥ 8. Pathologic T stage grade—0: ≤T2, 1: ≥T3.
In addition, Cox’s proportional hazard analysis of UBE2O expression in patients with prostate cancer from Soonchunhyang University Hospital identified a correlation with between UBE2O immunohistochemical staining and the following (see Table 5, Table 6 and Table 7): clinical recurrence free (HR = 0.028[95% CI: 0.008–0.106], HR= 0.023[95% CI: 0.006–0.841], p < 0.001); Overall survival (HR = 0.503[95%CI: 0.197–1.289], HR = 0.310[95% CI: 0.112–0.856], p < 0.05).
Kaplan–Meier survival analysis was performed and subsequently the log-rank test for UBE2O expression in patients with prostate cancer was run for the purposes of comparison. Results showed that there was no correlation between the grade of UBE2O immunohistochemical staining and prostate cancers’ biochemical recurrence (p = 0.45), clinical recurrence (p = 0.89), or OS (p = 0.24).
3. Discussion
In this study, it was found that adjacent non-neoplastic tissues around prostate cancer and prostate cancer itself showed lower and higher grades of UBE2O, respectively (p < 0.05). This result signified that UBE2O expression enables distinction between benign and malignant prostate tumors, as immunohistochemical studies in prostate cancer display a substantial disparity of UBE2O staining cells between benign and malignant lesions. The results of this study underscore the importance of the ubiquitination process in the prostate carcinogenesis and in the propagation of prostate cancer cells.
One AMPK tumor-suppressive functions is hindering the synthesis of most cellular macromolecules by deactivating the mTOR signalling pathway [12]. Other functions include the downregulation of the glycolytic pathway to impose an anti-Warburg impact [13,14], halt the cell cycle in conjunction with the stabilisation of p53 and p27Kip1 [15], and combat the epithelial–mesenchymal transition related to tumor invasion and metastasis [16].
Per cBioPortal’s (www.cbioportal.org (accessed on 4 April 2021)) TCGA datasets, UBE2O is upregulated in human breast, bladder, liver, and lung carcinomas. UBE2O expression is amplified and relates to AMPKa2/mTOR/HIF1a in human cancers, using the existing microarray database: Liu’s BCa, breast carcinoma (GEO: GSE22820, n = 176) [17]; Stephenson’s CaP, prostate carcinoma (n = 97) [8]; Taylor’s CaP (GEO: GSE21032, n = 179) [18].
Past studies of microarray-based gene expression have identified a high UBE2O expression rate in a variety of human cancer subsets [11]. Furthermore, immunohistochemical analysis of breast cancer samples determined high UBE20 expression [11]. Another database [19] analyzing cancer patient survival rates shows the clinical significance of UBE2O overexpression. In a substantial proportion of human cancers, UBE2O expression could impact both neoplastic malignancies and clinical outcomes. As UBE2O is upregulated, it may play a role in the regulation of the AMPKa2-mTOR-HIF1a pathway.
In mouse models of breast and prostate cancers, deferred tumor initiation and reduced tumor growth and metastasis rates were found to be attributable to the loss of one or both UBE2O alleles [11]. In cancer cells, UBE2O positively regulates aerobic glycolysis and cellular biosynthesis; the deactivation of UBE2O can switch off the tumor cells’ glycolytic and biosynthetic programs. Therefore, UBE2O can function as an oncogene that initiates cancer progression and removes important metabolic checkpoints that provoke pro-growth cellular metabolism [11]. In cancer, the presence of UBE2O causes upregulation in the mTOR-HIF1a pathway, which is strongly correlated with pro-growth, glycolytic, and biosynthetic programs [11]. In this study, a higher PSA was demonstrated in patients with a higher UBE2O grade (p < 0.001). A higher pathologic stage was identified in patients with a higher UBE2O grade (p < 0.001). The expression of UBE2O immunohistochemical staining facilitates prostate cancer prognosis.
Furthermore, in mouse cancer models, the removal of UBE2O weakened the intra-tumoral vascularization and expression of neovascularization genes, including HIF1a targets. This underscores the importance of UBE2O’s regulation of the mTOR-HIF1a pathway for both the angiogenic signaling pathway, and the neovascularization necessary for cancer growth and metastasis [11]. It has been found that drugs regulating UBE2O activity function as a form of cancer therapy, as the obstructing of UBE2O with ATO has an impact on tumor biology comparable to that of UBE2O deficiency Human Tumor TMA Analysis [11].
In this study, it was found that patients with higher UBE2O grades did not demonstrate greater seminal vesicle involvement than the lower UBE2O grade patients. Additionally, patients with higher UBE2O grades presented greater lymph node involvement than the UBE2O lower grade (p < 0.05). With prostate cancer, lymphatic metastasis can be anticipated with the expression of UBE2O immunohistochemical staining.
In human breast cancer, tumor tissue microarrays (TMAs) presented a high expression of UBE2O staining [11]. UBE2O is often intensified or mutated in many cancers, and its high expression is correlated with low survival rates of breast, gastric, lung, and prostate cancers. In this study, Cox’s proportional hazard analysis ascertained the following correlations: between positive surgical margin and biochemical recurrence free survival; between PSA grade and clinical recurrence free survival between regional lymph node positive and clinical recurrence free survival; between adjuvant treatment and overall survival; between pathological T stage and overall survival. Cox’s proportional hazard modelling for UBE2O expression also identified that UBE2O immunohistochemical staining correlated with clinical recurrence free survival (p < 0.001) and overall survival (p < 0.05) in prostate cancers (Table 6 and Table 7).
However, the Kaplan–Meier test and the subsequent log-rank comparison test establish that there was no correlation between the grade of UBE2O immunohistochemical staining and prostate cancers’ biochemical recurrence, clinical recurrence, or OS. An assumption is made that in this study, the number of prostate cancer patients was low, as was the prostate cancer mortality rate; thus, there were minimal cancer deaths within the study’s duration. Hence, the expression of UBE2O immunohistochemical staining, PSA, Gleason score, and pathological stage can all predict prostate cancer prognosis.
Although we have explored pilot UBE20 expression using human TMA prostate cancer tissue, UBE2O/antibody used validated antibody from Sigma-Aldrich but could not detect UBE2O expression in any of the 494 prostate cancer tissues. This could be due to the difference of antibody characteristics, however, needs validation study using large human tissue. Recently, a study using the same UBE2O antibody (GTX108039) as we used in mice model was conducted. Vila IK, et al. generated a UBE2O knockout mouse line that has been cross-bred in two transgenic mouse models of spontaneous cancer (transgenic adenocarcinoma mouse prostate (TRAMP) for prostate cancer) [11]. In their result, it has been shown that high expression of UBE2O promotes tumor initiation in mouse models of prostate cancers. The rates of distant metastasis of UBE2O-deficient mice were much lower than those of UBE2O-proficient mice of prostate cancers [11].
4. Materials and Methods
4.1. Patients and Specimen
A group of 202 patients with prostatic adenocarcinoma diagnoses were recruited. All diagnoses were confirmed both histologically and immunohistochemically, and surgical resection had been performed on each of the 202 patients between January 2002 and December 2012 in the Soonchunhyang University Hospital. Preparation of tumor tissues was conducted via formalin-fixed and paraffin-embedded processing. In order to determine the representative area, hematoxylin-eosin (H&E) slides were retrospectively reassessed by two skilled pathologists. Pathological reports and other medical records were also reviewed to gather clinicopathological information. The criteria from the International Union Against Cancer and World Health Organizations/International Society of Urological Pathology was employed to establish the tumor stage and Gleason score. Following the radical prostatectomy (RP), patient follow-ups were carried out via regular measurement of the serum PSA. Average follow-up time was 132 months (range 1–252). Adjuvant treatment was defined as a case of hormonal or radiation therapy without an increase in PSA within 6 months after surgery.
We graded PSA and Gleason scores according to the American Joint Committee on Cancer Eighth Edition Cancer Staging system. PSA is classified as low risk when <10 ng/mL and high risk when >20 ng/mL [20]. The Gleason score was divided into three categories: low, intermediate, and high grade. Prostate cancers with a Gleason score of 6 or less may be called well-differentiated or low-grade and Gleason score of 7 may be called moderately-differentiated or intermediate-grade. Prostate cancers with Gleason scores of 8 to 10 may be called poorly-differentiated or high-grade [20]. Clinically, stage T2 or less is an organ confined disease and above T3 is classified as locally advanced disease in prostatic cancer. Therefore, we classified the T stage into two grades.
Local scientific ethics committees approved this study (Seoul hospital: 2017-02-002, Bucheon hospital: 2017-03-004, Cheonan hospital: 2017-03-031-024, Gumi hospital: 2017-03-031-002). Other factors evaluated were age, Gleason score, seminal vesicle invasion, lymph node invasion, plasm PSA level, and stage.
4.2. Construction of Tissue Microarray (TMA)
As mentioned above, tissue microarrays were built from formalin-fixed paraffin-embedded blocks. The H&E slides were meticulously examined under light microscopy in order to obtain the most representative viable tumor portions (n = 202). A 3-mm-diameter cylinder was used to core the corresponding areas of each paraffin block twice, and they were subsequently transferred to a recipient paraffin block via a trephine apparatus (Superbiochips Laboratories, Seoul, Korea). Also included were control or adjacent non-neoplastic tissues (n = 39). For tissue validation purposes, one section of the block was stained with H&E.
4.3. Purchased Additional Tissue Microarray
There was a shortage of patients with prostate cancer with more than T3 pathological stage in the hospital, therefore TMA containing human prostate cancer (n = 180) and adjacent non-neoplastic tissues (n = 22) were purchased from AccuMax (ISU ABXIS Co., LTD, Seongnam, Korea). All samples were anonymous, and the pathologist reconfirmed the diagnoses of normal or tumor tissue. The TMA encompassed a range of 180 malignant RP specimens. The two previously mentioned pathologists reassessed the hematoxylin and eosin-stained slides to pinpoint the representative areas. Again, the criteria of the International Union Against Cancer and World Health Organization/International Society of Urological Pathology was applied to establish the tumor stage and Gleason score. As previously, local scientific ethics committees approved the study, and the factors investigated were age, Gleason score, seminal vesicle invasion, lymph node invasion, plasm PSA level, and stage.
4.4. Immunohistochemistry and Interpretation
Immunohistochemical analysis of the expression of UBE2O was carried out using anti-UBE2O primary antibodies. Successive 4-μM sections were cut from the TMA tissue blocks. Normal prostatic tissue was selected as positive control. A summary of the process is as follows: the TMA sections were moved to adhesive-coated slides, which were subsequently heated at 60 °C for 60 min to deparaffinize them; the slides were washed in xylene three times; the slides were treated in 5% hydrogen peroxide in methanol at 37 °C for 15 min to prevent endogenous peroxidase activity; the antigens were retrieved through microwave treatment in a pH 6.0 epitope retrieval solution for 20 min; the anti-UBE2O antibody (GTX108039, Genetex, Irvine, CA,, USA) was diluted 1:100 and the sections were incubated overnight with the primary antibody in a humidified chamber at 4 °C; a bond polymer refine detection kit (Leica Biosystem, Wetzlar, Germany) and diaminobenzidine as a chromogen were applied to treat the secondary antibody. Incubations were employed as negative controls (both excluding and preabsorbing the specific antibody), and UBE2O expression was only evaluated on tissue cores that had been well-preserved.
The pathologist scored the TMA cores without any knowledge of the patients’ clinicopathological information. Based on histological scoring, the expression of staining was classified as follows: grade 3—strong (+++); grade 2—moderate (++); grade 1—weak (+); and grade 0—negative. It was determined that 355 of the 382 prostate cancer patients could be successfully stained with UBE2O. Additionally, 61 adjacent non-neoplastic tissues from the 355 patients could be successfully stained with UBE2O.
4.5. Statistical Analyses
Initially, the patients’ baseline variables were evaluated. The correlation between the UBE2O group and the established prognostic factors was examined. Chi-square tests or Fisher’s exact test were employed to analyze the classified data, and results were expressed as n (%) in descriptive statistics. Analysis of variance analysis (ANOVA) or the Kruskal–Wallis test were carried out on the continuous data and expressed as mean ± standard error (SE).
It was possible to assess 173 of the 382 prostate cancer patients for survival. Biochemical recurrence was established if the following three criteria were met: (a) a PSA increase of a minimum of 0.2 ng/mL, (b) a minimum of two distinct consecutive measurements, (c) that are a minimum of three months apart. Clinical recurrence was clarified as lesions within the bone observable on a radionuclide bone scan and lymphadenopathy or visceral lesions observable via computed tomography imaging of the abdomen, pelvis, and chest. Patients were deemed at risk from the surgery date until recurrence or until the date of the final PSA test. Patients that were not available for follow-up were cut from the date of their last follow-up or PSA test.
In order to assess the prevalence of each outcome stratified by the UBE2O group, Kaplan–Meier survival analysis was conducted. OS, which is defined as the time from surgery until death (from any cause), was applied. OS curves were plotted based on the Kaplan–Meier approach, and the log-rank test was employed to facilitate comparison. Additionally, in order to evaluate their independent link with OS, Cox’s proportional hazard regression analyses were conducted on UBE2O expression.
Following consideration of patient age at diagnosis, histological grade, pathological stage, PSA category, margin status, and lymph node involvement, Cox’s proportional hazard modelling was carried out in order to evaluate the independent prognostic impact of the UBE2O group. For multivariate analysis, the fixed variable was UBE2O, and other aspects were analyzed via the variable selection method so as to assess the link between the UBE2O variable and the results via Cox’s proportional risk regression analysis. In addition, statistical analysis was performed utilizing SPSS Software (version 26) and Rex (Version 3.5.0, RexSoft Inc., Seoul, Korea), and the statistical significance level was set at p < 0.05.
5. Conclusions
For prostate cancer, there is a positive correlation between the expression of UBE2O staining and high PSA, pathological stage, and lymph node involvement. Hence, it can be concluded that the expression of UBE2O staining can facilitate the assessment of a prediction for prostate cancer prognosis.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ph14080778/s1, Figure S1: Representative photographs of high UBE2O expression in prostate cancer samples and low UBE2O expression in non-cancerous prostate.
Author Contributions
Conceptualization: J.-H.K. (Jae-Heon Kim), H.-J.Y., K.-W.L., J.-J.P., J.-H.K. (Jae-Ho Kim), and A.-R.M.; Methodology: J.-H.K. (Jae-Heon Kim), K.-W.L., S.-J.S., and Y.-S.S.; Software: J.-H.K. (Jae-Heon Kim) and S.-Y.P.; Validation: C.-H.L., Y.-S.J., S.-Y.P., S.-J.S., and Y.-S.S.; Formal analysis: J.-H.K. (Jae-Heon Kim), K.-W.L., Y.-H.K., J.-H.L., and Y.-S.S.; Investigation: H.-J.Y., K.-W.L., J.-J.P., C.-H.L., Y.-S.J., and J.-H.K. (Jae Ho Kim); Resources: K.-W.L., Y.-H.K., and J.-H.L.; Writing—original draft preparation: J.-H.K. (Jae-Heon Kim) and H.-J.Y.; Writing—review and editing: J.-H.L. and Y.-S.S.; Visualization: S.-Y.P. and A.-R.M.; Supervision: Y.-S.J., J.-H.L., and Y.-S.S.; Project administration: J.-H.K. (Jae-Heon Kim), H.-J.Y., J.-H.L., and Y.-S.S.; Funding acquisition: Y.-S.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grant from Soonchunhyang University Research Fund and the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2018R1A2B6001693).
Institutional Review Board Statement
This study was performed in line with the principles of the Declaration of Helsinki. Approval was obtained from local scientific ethics committees (Soonchunhyang University Seoul hospital: 2017-02-002, Bucheon Hospital: 2017-03-004, Cheonan Hospital: 2017-03-031-024, Gumi Hospital: 2017-03-031-002, Approval date: 27 April 2017). Consent to participate: We have been granted a waiver of informed consent from local scientific ethics committees for the following reasons; (1) The subject of the study does not conduct genetic testing as a material of human origin. (2) The subject of the study is human-derived, and the identification of the sample is not possible because the information of the subject is not provided or cannot be identified to all research-related person. (3) It is coded and provided to the researcher without the subject identification record, or the personal information is processed and secured so that it is impossible to identify it by an impartial third party. (4) We have appropriate management guidelines and procedures for samples to prevent exposure of personal information.
Informed Consent Statement
The authors affirm that human research participants provided informed consent for publication of the images in Figure 1, Figure 2A,B, Figure S1A–D.
Data Availability Statement
Data is contained within the article and supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ye Y., Rape M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer H.-J., Rape M. Seminars in Cell & Developmental Biology. Elsevier; Amsterdam, The Netherlands: 2011. Processive ubiquitin chain formation by the anaphase-promoting complex; pp. 544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadad S.M., Baker L., Quinlan P.R., Robertson K.E., Bray S.E., Thomson G., Kellock D., Jordan L.B., Purdie C.A., Hardie D.G., et al. Histological evaluation of ampk signalling in primary breast cancer. BMC Cancer. 2009;9:1–9. doi: 10.1186/1471-2407-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briffa R., Um I., Faratian D., Zhou Y., Turnbull A.K., Langdon S.P., Harrison D.J. Multi-scale genomic, transcriptomic and proteomic analysis of colorectal cancer cell lines to identify novel biomarkers. PLoS ONE. 2015;10:e0144708. doi: 10.1371/journal.pone.0144708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin M., Smith L., Smiraglia D., Kazhiyur-Mannar R., Lang J., Schuller D., Kornacker K., Wenger R., Plass C.J.O. DNA copy number gains in head and neck squamous cell carcinoma. Oncogene. 2006;25:1424–1433. doi: 10.1038/sj.onc.1209166. [DOI] [PubMed] [Google Scholar]
- 7.Rice K.L., Lin X., Wolniak K., Ebert B.L., Berkofsky-Fessler W., Buzzai M., Sun Y., Xi C., Elkin P., Levine R., et al. Analysis of genomic aberrations and gene expression profiling identifies novel lesions and pathways in myeloproliferative neoplasms. Blood Cancer J. 2011;1:e40. doi: 10.1038/bcj.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephenson A.J., Smith A., Kattan M.W., Satagopan J., Reuter V.E., Scardino P.T., Gerald W.L. Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy. Cancer. 2005;104:290–298. doi: 10.1002/cncr.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toffoli S., Bar I., Abdel-Sater F., Delrée P., Hilbert P., Cavallin F., Moreau F., Van Criekinge W., Lacroix-Triki M., Campone M., et al. Identification by array comparative genomic hybridization of a new amplicon on chromosome 17q highly recurrent in brca1 mutated triple negative breast cancer. Breast Cancer Res. 2014;16:1–15. doi: 10.1186/s13058-014-0466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., Li X., Cheng Y., Sun X., Sun X., Self S., Kooperberg C., Dai J.Y. Copy number alterations detected by whole-exome and whole-genome sequencing of esophageal adenocarcinoma. Hum Genom. 2015;9:1–15. doi: 10.1186/s40246-015-0044-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vila I.K., Yao Y., Kim G., Xia W., Kim H., Kim S.J., Park M.K., Hwang J.P., González-Billalabeitia E., Hung M.C., et al. A ube2o-ampkα2 axis that promotes tumor initiation and progression offers opportunities for therapy. Cancer Cell. 2017;31:208–224. doi: 10.1016/j.ccell.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gwinn D.M., Shackelford D.B., Egan D.F., Mihaylova M.M., Mery A., Vasquez D.S., Turk B.E., Shaw R.J. Ampk phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faubert B., Boily G., Izreig S., Griss T., Samborska B., Dong Z., Dupuy F., Chambers C., Fuerth B.J., Viollet B., et al. Ampk is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faubert B., Vincent E.E., Griss T., Samborska B., Izreig S., Svensson R.U., Mamer O.A., Avizonis D., Shackelford D.B., Shaw R.J., et al. Loss of the tumor suppressor lkb1 promotes metabolic reprogramming of cancer cells via hif-1α. Proc. Natl. Acad. Sci. USA. 2014;111:2554–2559. doi: 10.1073/pnas.1312570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardie D.G., Alessi D.R. Lkb1 and ampk and the cancer-metabolism link-ten years after. BMC Biol. 2013;11:1–11. doi: 10.1186/1741-7007-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu C., Zhang W., Zheng G., Zhang Z., Yin J., He Z. Metformin reverses multidrug resistance and epithelial–mesenchymal transition (emt) via activating amp-activated protein kinase (ampk) in human breast cancer cells. Mol. Cell Biochem. 2014;386:63–71. doi: 10.1007/s11010-013-1845-x. [DOI] [PubMed] [Google Scholar]
- 17.Liu R.Z., Graham K., Glubrecht D.D., Germain D.R., Mackey J.R., Godbout R. Association of fabp5 expression with poor survival in triple-negative breast cancer: Implication for retinoic acid therapy. Am. J. Pathol. 2011;178:997–1008. doi: 10.1016/j.ajpath.2010.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor B.S., Schultz N., Hieronymus H., Gopalan A., Xiao Y., Carver B.S., Arora V.K., Kaushik P., Cerami E., Reva B., et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ullah K., Zubia E., Narayan M., Yang J., Xu G. Diverse roles of the e2/e3 hybrid enzyme ube 2o in the regulation of protein ubiquitination, cellular functions, and disease onset. FEBS J. 2019;286:2018–2034. doi: 10.1111/febs.14708. [DOI] [PubMed] [Google Scholar]
- 20.Buyyounouski M.K., Choyke P.L., McKenney J.K., Sartor O., Sandler H.M., Amin M.B., Kattan M.W., Lin D.W. Prostate cancer-major changes in the american joint committee on cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017;67:245–253. doi: 10.3322/caac.21391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article and supplementary material.