Abstract
Mammalian Ras GTPase-activating protein (GAP), p120 Ras-GAP, has been implicated as both a downregulator and effector of Ras proteins, but its precise role in Ras-mediated signal transduction pathways is unclear. To begin a genetic analysis of the role of p120 Ras-GAP we identified a homolog from the fruit fly Drosophila melanogaster through its ability to complement the sterility of a Schizosaccharomyces pombe (fission yeast) gap1 mutant strain. Like its mammalian homolog, Drosophila RasGAP stimulated the intrinsic GTPase activity of normal mammalian H-Ras but not that of the oncogenic Val12 mutant. RasGAP was tyrosine phosphorylated in embryos and its Src homology 2 (SH2) domains could bind in vitro to a small number of tyrosine-phosphorylated proteins expressed at various developmental stages. Ectopic expression of RasGAP in the wing imaginal disc reduced the size of the adult wing by up to 45% and suppressed ectopic wing vein formation caused by expression of activated forms of Breathless and Heartless, two Drosophila receptor tyrosine kinases of the fibroblast growth factor receptor family. The in vivo effects of RasGAP overexpression required intact SH2 domains, indicating that intracellular localization of RasGAP through SH2-phosphotyrosine interactions is important for its activity. These results show that RasGAP can function as an inhibitor of signaling pathways mediated by Ras and receptor tyrosine kinases in vivo. Genetic interactions, however, suggested a Ras-independent role for RasGAP in the regulation of growth. The system described here should enable genetic screens to be performed to identify regulators and effectors of p120 Ras-GAP.
The Ras family of small GTPases play pivotal roles in the regulation of signal transduction pathways downstream of receptor tyrosine kinases (RTKs), G protein-coupled receptors, and cytokine receptors. Ras proteins cycle between the inactive GDP-bound state and the active GTP-bound conformation that interacts with downstream effector proteins such as the Raf family of protein kinases and phosphoinositide 3′-kinase (42). This GTPase cycle is regulated by guanine nucleotide exchange factors (GEFs) that activate Ras by stimulating release of GDP and binding to GTP and by GTPase-activating proteins (GAPs) that inhibit Ras by increasing the intrinsic rate of GTP hydrolysis (5).
Three types of GAP protein for Ras have been identified in mammalian cells: p120 Ras-GAP (64, 68), neurofibromin (the product of the human tumor suppressor gene NF1) (46, 71), and two closely related proteins, Gap1m (45) and Gap1IP4B (16). Although it is clear that all three types can act as GAPs and thus inhibit the activity of Ras proteins, p120 Ras-GAP has been also implicated as an effector of Ras. Notably, p120 Ras-GAP has an amino-terminal region similar in structural organization to “adapter” proteins. This region contains Src homology 2 and 3 (SH2 and SH3) domains involved in interactions with other proteins and pleckstrin homology (PH) and C2 (or CalB) domains that may promote membrane association by binding to phospholipids (31, 56). Studies using mammalian tissue culture cells suggest a role for the SH2 and SH3 domains of p120 Ras-GAP in cell transformation, changes in gene expression, K+ channel opening, and cytoskeletal rearrangements, and in Xenopus oocytes there is evidence that the SH3 domain of p120 Ras-GAP is required for meiotic maturation in response to Ras or insulin (63).
The identification of proteins that bind to the amino-terminal adapter-like region of p120 Ras-GAP has provided further evidence that it has roles independent of its GAP activity. The SH2 domains bind to certain plasma membrane-associated RTKs such as the platelet-derived growth factor β receptor (2) and to the cytosolic proteins p190 Rho-GAP (59) and p62dok, a “docking” protein of unknown function (10, 72). Proteins that bind to the SH3 domain of p120 Ras-GAP have proved difficult to identify but G3BP, a cytoplasmic protein that contains RNA binding motifs, may be such an SH3 ligand (55). The biochemical and physiological consequences of the interaction of p120 Ras-GAP with these proteins are poorly understood.
An important advance in understanding the physiological roles of p120 Ras-GAP has come from the analysis of mice carrying a null mutation in the gene for p120 Ras-GAP (Gap) (36). Embryos homozygous for this mutation die in utero with abnormal vasculature and increased apoptosis in the nervous system. Fibroblasts from Gap−/− embryos can proliferate in vitro and show enhanced accumulation of Ras-GTP upon growth factor stimulation (67). This genetic analysis shows that p120 Ras-GAP is not necessary for mitogenesis, at least for most cell types, but is required to downregulate Ras following exposure to growth factors and to protect neuronal cells from apoptosis. Whether p120 Ras-GAP is involved in other Ras-regulated processes such as cell migration or differentiation is unresolved.
To study the role of p120 Ras-GAP in a genetically tractable organism we set out to identify a homolog from the fruit fly Drosophila melanogaster, which has proved to be a powerful model system for the genetic analysis of Ras signaling pathways (19, 41). Using an expression cloning strategy in the fission yeast Schizosaccharomyces pombe we cloned a cDNA encoding Drosophila RasGAP, which is similar in both sequence and biochemical properties to mammalian p120 Ras-GAP. Although loss-of-function mutations in the Drosophila RasGAP gene are not available we have begun to analyze its function in vivo by ectopic expression. We show that overexpression of RasGAP in the wing imaginal disc downregulates signaling through RTKs and inhibits wing growth. The system we have developed will prove useful in understanding the physiological role of RasGAP and the function of each of its modular domains.
MATERIALS AND METHODS
Methods for S. pombe.
Yeast extract supplements (YES) and Edinburgh minimal medium (EMM) were used for growth of S. pombe cultures (49), and synthetic sporulation agar (SSA) was used to promote mating and sporulation (22). The gap1 mutant strain JZ446 (h90 gap1::ura4+ ade6-M216 leu1 ura4-D18) (40) was used in the screen for the identification of novel GTPase-activating proteins and was transformed by the lithium acetate method (53). Plasmids were rescued from yeast into Escherichia coli as described previously (49).
Isolation of a full-length RasGAP cDNA and plasmid constructions.
The RasGAP cDNA isolated in the yeast screen (RasGAP78) was truncated at the 5′ end and predicted to initiate translation at Met266. Through screening a λgt10 embryonic cDNA library (Stratagene) and by rapid amplification of cDNA ends (RACE) PCR with Drosophila polyA+ RNA (Clontech) as template for the Marathon cDNA Amplification Kit (Clontech), a cDNA including 291 bp of 5′ untranslated sequence was assembled. A myc tag epitope recognized by the 9E10 monoclonal antibody, followed by two TAA termination codons, was engineered at the C terminus of the open reading frame by using the Exsite Mutagenesis Kit (Stratagene). To generate the GAP catalytic domain construct, RasGAPCat, the C-terminal 443 codons were amplified with a sense primer incorporating an NcoI site which was used to fuse the resulting PCR product in-frame to the first 85 codons of the RasGAP cDNA, just N terminal to the first SH2 domain. The full-length cDNA and the C terminus myc-tagged constructs were subcloned as NotI fragments into the Drosophila expression vector pUAST (6) and the S. pombe expression vector pREPCD1 (24), a derivative of the pREP1 plasmid with a NotI site in the polylinker (47). The C-terminal part of RasGAP (amino acids 429 to 954) was cloned as a ClaI/BamHI fragment into the E. coli expression vector pGEX-KG (33). The SH2-SH3-SH2 region (amino acids 83 to 343) and the N-terminal SH2 domain (amino acids 83 to 182) were amplified by PCR and cloned as XbaI/XhoI fragments into pGEX-KG. Missense mutations were introduced by PCR-mediated mutagenesis (37) into each SH2 domain, changing a highly conserved Arg residue to Leu (amino acid 110 in the N-terminal domain and amino acid 278 in the C-terminal domain) to give the GST-SH2*32* mutant and the RasGAPSH2* mutant. Sequences of the oligonucleotides used for PCR and site-directed mutagenesis are available upon request from the corresponding author. All PCR-derived fragments were sequenced to ensure that no mutations were introduced. Sequences were determined with oligonucleotide primers with the TaqFS Terminator Cycle Sequencing Kit on an ABI 377 DNA sequencer (Applied Biosystems) and were analyzed using the Genetics Computer Group package (32). The UAS-tor4021-btl and UAS-tor4021-htl constructs were generated by using KpnI and XbaI to remove the fusion genes from P[sE-tor4021-FGFR1] and P[sE-tor4021-FGFR2], respectively (57), and the inserts were subcloned into pUAST.
General biochemical and immunological techniques.
Protein samples were separated by sodium dodecyl sulfate (SDS)-polyacrylamide electrophoresis (PAGE) and electroblotted to Immobilon membranes (NEN) as previously described (35). Anti-RasGAP antibody incubations were for 1 to 2 h in 5% skimmed milk–TBST (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Tween-20) with washes in TBST. Antiphosphotyrosine blots were blocked in 3% bovine serum albumin (BSA)–TBST at room temperature for 1 to 2 h, incubated overnight at 4°C with the primary antibody 4G10 (Upstate Biotechnology) at a concentration of 0.3 μg/ml in 3% BSA–TBST, and washed in TBST. Horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence substrates were used as recommended by the manufacturer (Pierce). Protease inhibitors were the Complete mix (Boehringer Mannheim) together with 1 mM phenylmethylsulfonyl fluoride.
Purification of GST fusion proteins and preparation of antibodies.
Glutathione S-transferase (GST) fusion proteins were expressed in E. coli by induction with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside for 2 to 4 h at 37°C. To purify wild-type and Val12 H-Ras GST fusion proteins (69), cells were resuspended in TMD buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 1 mM dithiothreitol) plus protease inhibitors and lysozyme added to a concentration of 1 mg/ml, and the cell suspension was incubated for 20 min at 4°C. Cells were lysed by adding Triton X-100 and DNase I to concentrations of 0.5% and 20 μg/ml, respectively, and the lysate was incubated for 20 min at 4°C. Following centrifugation at 12,000 × g for 15 min, the supernatants were mixed with glutathione-Sepharose beads and washed extensively with TMD, and the fusion proteins were eluted with 50 mM Tris-HCl (pH 8.0)–5 mM MgCl2–10 mM reduced glutathione. The purified proteins were dialyzed against TMD and stored in aliquots at −70°C. The GST-SH232 and GST-NSH2 fusion proteins were purified by glutathione-Sepharose affinity chromatography following solubilization with N-lauroylsarcosine (26). The purified GST-NSH2 fusion protein was dialyzed against phosphate-buffered saline and used to raise rat monoclonal antibodies by using standard protocols (35). The GST-RasGAP C-terminal region was purified with glutathione-Sepharose beads following solubilization with N-lauroylsarcosine, and the insoluble RasGAP polypeptide was released from the fusion protein by cleavage with thrombin. Following SDS-PAGE purification the RasGAP polypeptide was used to immunize a rabbit by using standard protocols (35). The rabbit polyclonal antibody was purified by applying the crude antiserum to the GST-fusion protein cross-linked to glutathione-Sepharose resin (3) and eluting the bound immunoglobulin with 100 mM glycine, pH 2.5 (35). The purified antibody was titered to determine its optimum concentration for Western blotting and immunoprecipitation.
Immunoprecipitations and GST pull-downs from Drosophila cell extracts.
Soluble extracts from Drosophila embryos, larvae, pupae, and adults in Nonidet P-40 (NP-40) lysis buffer plus protease inhibitors were prepared as previously described (15). For immunoprecipitations, 0.1% SDS–0.5% deoxycholate were added to the extracts and 1 mg of protein was incubated with the purified anti-RasGAP antibody for 2 h followed by incubation with protein G-Sepharose for 1 h. The beads were washed three times in the same buffer before analysis by SDS-PAGE and Western blotting. For pull-downs with GST-SH232 fusion proteins, cell extracts (0.5 mg of protein) were incubated with the fusion proteins (10 μg) bound to glutathione-Sepharose for 1 h at 4°C, washed three times in lysis buffer, and analyzed by SDS-PAGE and Western blotting.
Immunostaining of embryos.
Embryos were dechorionated, fixed with 4% paraformaldehyde, devitellinized in methanol, and incubated with antibodies as previously described (73). Incubation was overnight with the anti-RasGAP monoclonal antibody (1/2 diluted tissue culture supernatant) and for 2 h with Cy3-conjugated anti-rat immunoglobulin G (IgG) (Jackson ImmunoResearch). Images were collected with a BioRad MRC-1024 confocal laser scanning microscope and assembled in Adobe Photoshop 4.0.
GAP assay.
S. pombe cell extracts were prepared from the gap1 mutant strain JZ446 transformed with a vector plasmid (pREPCD1) or the full-length RasGAP cDNA in pREPCD1 by vortexing with glass beads in 50 mM HEPES [pH 7.4]–100 mM NaCl–5 mM MgCl2–1 mM sodium phosphate (pH 7.4)–1-mg/ml BSA plus protease inhibitors. Following centrifugation at 15,000 × g for 15 min, the supernatant was concentrated to ∼20-mg/ml protein by ultrafiltration (Centricon 30; Amicon). GAP assays were performed by a method described previously (20) with [α-32P]GTP-loaded GST-Ras or GST-RasV12 fusion proteins and separation of GTP and GDP bound to Ras by thin-layer chromatography, except that following the incubation with cell extract the GST-Ras fusion proteins were purified with glutathione-Sepharose. A PhosphorImager (Molecular Dynamics) was used to image and quantify the [α-32P]-labeled guanine nucleotides, and the percent GTP remaining was calculated as 100(GTP/GDP + GTP).
Drosophila stocks and phenotypic analysis.
Flies were raised and crossed at 25°C according to standard procedures. Transgenic lines were made by P element-mediated transformation into a y w stock. The GAL4 lines used were MS-1096 (X chromosome) for expression in the dorsal pouch of the wing disc (9) and hsp70-GAL4 (3rd chromosome) for expression upon heat shocking (51). The Ras1e2F mutant allele was used (60). Wings were dissected, dehydrated in ethanol, and mounted in Euparal (Agar Scientific). Photographs of wings were digitized with a flat-bed scanner, processed in Adobe Photoshop 4.0, and analyzed using NIH Image 1.60 software.
Nucleotide sequence accession number.
The RasGAP cDNA nucleotide sequence has been submitted to the EMBL database under accession no. AJ012609.
RESULTS
Expression cloning in fission yeast of a Drosophila p120 Ras-GAP homolog.
An expression cloning screen with S. pombe was devised to identify inhibitors of Ras function from D. melanogaster. In S. pombe, mutation of the gap1 gene (also known as sar1), which encodes a member of the RasGAP family of proteins, results in hypersensitivity to mating pheromones, formation of extended mating projections, and inefficient mating (Fig. 1A, i, and 1B, i) (40, 70). These phenotypes result from hyperactivation of Ras1, the S. pombe homolog of mammalian Ras oncoproteins, because mutations in Ras1 that inhibit its GTPase activity cause identical effects (28, 50). We reasoned that expression of inhibitors of Ras function from higher organisms in a gap1 mutant strain would restore efficient mating by downregulating the activities of Ras1-regulated signaling pathways. Such a screen might identify both proteins that bind directly to Ras1, such as GAPs and effectors, and proteins that interfered with the function of downstream components of the Ras1 signaling pathways.
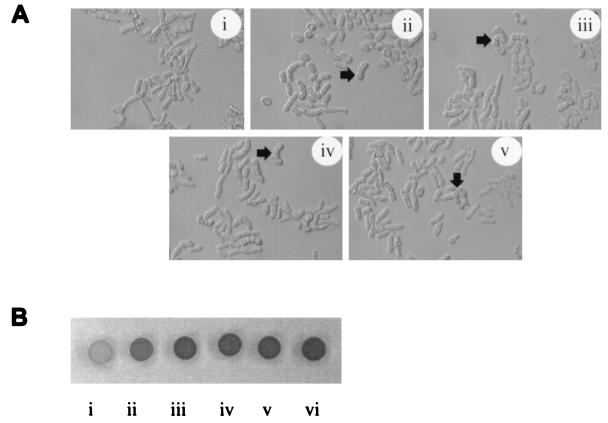
FIG. 1.
Complementation of the S. pombe gap1 mutant phenotype by the Drosophila RasGAP cDNA. The homothallic h90 gap1 mutant strain (JZ446) was transformed with the vector pREPCD1 (i), pST200-1(gap1+) carrying the S. pombe gap1+ gene (ii), pREPRasGAP78 containing the original cDNA isolated from the library screen (iii), pREPRasGAP containing the full-length cDNA (iv), and pREPRasGAPCat containing the GAP catalytic domain (v). A wild-type homothallic h90 strain was used as control (vi). Transformants were grown on sporulation agar (SSA) at 30°C for 3 days. Differential interference contrast microscopy (A) and iodine staining (B), which stains spores, revealed that the three RasGAP cDNA constructs allowed the gap1 mutant strain to undergo mating and sporulation. The arrows in panel A indicate typical zygotic asci.
To identify inhibitors of Ras from D. melanogaster, a gap1 mutant strain (JZ446) was transformed with a Drosophila embryonic cDNA library constructed in the pREP expression vector (21). Sixty-thousand transformant colonies were screened by iodine vapor staining, a qualitative indicator of mating efficiency. Dark-staining colonies were then checked microscopically for the presence of zygotic asci. Two transformant colonies showing efficient mating were identified, and plasmids were rescued from them into E. coli. We confirmed that both plasmids could suppress the gap1 mutant phenotype (Fig. 1A, iii and 1B, iii; and data not shown). One of the plasmids carried a cDNA insert encoding the carboxy-terminal 263 amino acids of the Drosophila moesin homolog, a member of the ERM (ezrin, radixin, and moesin) family (21, 66). This cDNA could rescue the sterility of an S. pombe strain carrying an activated ras1 allele (ras1-Val17), suggesting that it inhibits signaling downstream of Ras1; the properties of this cDNA will be described elsewhere (24). The second plasmid could not suppress the sterility of the activated ras1-Val17 mutant strain (24), suggesting that it was acting either upstream of or directly upon Ras1. Sequence analysis of the cDNA insert revealed that it encoded a putative protein with significant similarity to members of the RasGAP family, particularly to human and bovine p120 Ras-GAPs (65, 68). Sequence alignment of the Drosophila cDNA with mammalian p120 Ras-GAP indicated that the cDNA was truncated at its 5′ end, and a full-length cDNA was subsequently identified (see Materials and Methods). Analysis of this Drosophila cDNA confirmed that it encoded a homolog of mammalian p120 Ras-GAP, which we have named RasGAP. The predicted RasGAP protein consists of 954 amino acids with a predicted Mr of ∼108,000.
Structure of Drosophila RasGAP.
Drosophila RasGAP and human p120 Ras-GAP share 47% amino acid identity (68% similarity) overall (Fig. 2A). Like its mammalian homologs, the Drosophila protein can be divided into two regions. The amino-terminal part consists of protein-protein (SH2 and SH3) and protein-lipid (PH and C2/CalB) interaction domains. The carboxy-terminal part contains the GAP catalytic domain which is conserved in RasGAPs from yeast to mammals (5).
FIG. 2.
RasGAP amino acid sequence and genomic organization of the RasGAP gene. (A) Alignment of the predicted amino acid sequence of RasGAP (top) with human p120 Ras-GAP (bottom). The comparison was generated with the GAP program of the Genetics Computer Group package (32). |, identical amino acids; :, amino acid pairs with Dayhoff matrix scores greater than 0.5; ., nonidentical amino acids having a positive Dayhoff matrix score of <0.5. The SH2 domains are shown by shading and the SH3 domain is boxed. The PH domain is underlined by a solid line and the C2/CalB domain is underlined by a dotted line. The asterisk indicates the conserved tyrosine that is phosphorylated in human p120 Ras-GAP. (B) The exon-intron structure of the RasGAP transcription unit. The white boxes represent noncoding regions, and the black boxes represent coding regions.
Mammalian p120 Ras-GAP is tyrosine phosphorylated upon activation of certain RTKs and in cells expressing non-RTKs (23, 48). The major site of tyrosine phosphorylation in vitro by the epidermal growth factor (EGF) receptor and the non-RTKs Src and Lck is tyrosine 460 in human p120 Ras-GAP, lying between the most carboxy-terminal SH2 domain and the PH domain (1, 44, 54). A tyrosine residue in a similar position with similar flanking sequences (IYATLR in Drosophila RasGAP and IYNTIR in human p120 Ras-GAP) is found in the Drosophila protein, suggesting that this is a conserved phosphorylation site (Fig. 2A).
The structure of the RasGAP transcription unit was determined by a comparison of cDNA and genomic sequence (Fig. 2B). It consists of eight exons and seven introns; six of the introns are small (<200 nucleotides [nt]) but one that splits the 5′ untranslated region is much larger than the others (1,535 nt). The RasGAP gene was mapped to cytological position 14A-B1 on the first (X) chromosome by in situ hybridization to polytene chromosomes (24).
GAP activity of RasGAP.
The original RasGAP cDNA (RasGAP78) identified in the expression cloning screen was amino-terminally truncated and predicted to initiate at Met266. We examined whether constructs carrying the full-length cDNA or just the GAP catalytic domain (amino acids 553 to 954, designated RasGAPCat) could also rescue the gap1 mutant strain (JZ446). Both constructs could restore efficient mating to the gap1 mutant strain (Fig. 1A, iv and v and B, iv and v) and in fact both rescued better than overexpression of the S. pombe gap1+ gene (Fig. 1A, ii and B, ii), restoring mating to nearly wild-type levels (Fig. 1B, vi). These results show that the RasGAP catalytic domain is responsible for downregulation of Ras1 activity in S. pombe. The ability of RasGAP to rescue the Ras “hyperactivation” phenotype caused by loss of gap1 function, but not that caused by a GAP-insensitive ras1 mutant (ras1-Val17), suggested that RasGAP was acting as a GAP on the S. pombe Ras1 protein in vivo, rather than as a “dominant negative” by binding to Ras1-GTP and preventing interaction with Ras1 effector proteins.
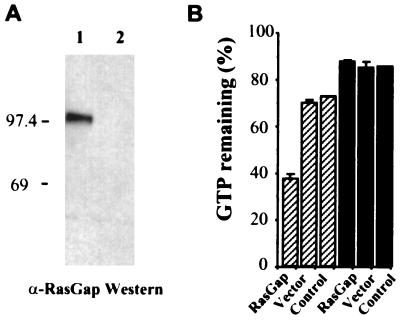
To confirm that RasGAP has GAP activity we tested the ability of the full-length protein to stimulate the GTPase activity of mammalian H-Ras in vitro. Cell extracts prepared from the S. pombe gap1 mutant strain expressing full-length RasGAP or a vector control were assayed for GAP activity on [α-32P]GTP-loaded wild-type or Val12 (GTPase-deficient) H-Ras. The results (Fig. 3) show that the extract expressing RasGAP stimulated GTP hydrolysis on wild-type H-Ras but not on the Val12 mutant protein, consistent with the properties of the mammalian GAPs (5).
FIG. 3.
GAP activity of RasGAP. (A) Expression of full-length RasGAP in S. pombe. Cell extracts prepared from a gap1 mutant strain transformed with pREPRasGAP (lane 1) or a vector control (lane 2) were subjected to SDS-PAGE and Western blotted with an anti-RasGAP polyclonal antibody (α-RasGAP). Expression of the full-length RasGAP protein migrating close to the predicted molecular weight (108 kDa) can be detected in lane 1. Migration of molecular weight markers (in kilodaltons) is shown. (B) The RasGAP and vector cell extracts were assayed for GAP activity on wild-type H-Ras (hatched bars) or Val12 H-Ras (filled bars); a control assay without extract was done to determine the intrinsic GTPase activity of the Ras proteins. The percent GTP remaining after a 15-min incubation was measured and plotted in a bar graph. Error bars show standard deviations of assays done in triplicate. Results are representative of three independent experiments.
Characterization of the RasGAP protein.
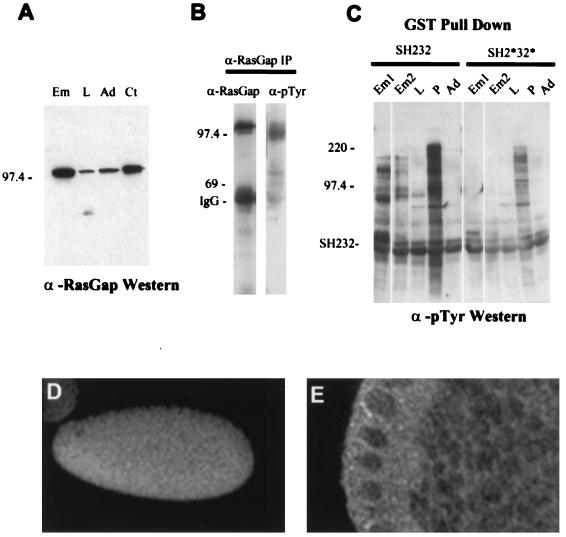
A monoclonal antibody raised against the N-terminal SH2 domain of RasGAP detected the 108-kDa RasGAP protein by immunoblotting. The levels of expression of RasGAP in embryos, larvae, and adults were assessed by immunoblotting equal amounts of total protein from each developmental stage. This showed that the protein is expressed throughout development but with significantly higher levels of embryos (Fig. 4A). Immunostaining of embryos with the anti-RasGAP monoclonal antibody detected staining in precellularized embryos (0 to 2 h), before zygotic transcription begins, indicating that the RasGAP protein is supplied maternally (Fig. 4D). The RasGAP protein was cytoplasmic and appeared to be present in all cells at the cellular blastoderm stage (Fig. 4E).
FIG. 4.
Characterization of RasGAP and interacting proteins. (A) Developmental expression of RasGAP protein. Equal amounts of total cell lysates from embryos (0 to 18 h) (Em), mixed-stage larvae (L), and adults (Ad) were Western blotted with anti-RasGAP monoclonal antibody. A control lysate (Ct) was made from adult flies expressing a heat-shock-inducible RasGAP transgene after heat shock for 1 h at 37°C. (B) Tyrosine phosphorylation of RasGAP in vivo. RasGAP was immunoprecipitated from embryonic extracts and Western blotted with either an anti-RasGAP polyclonal antibody (α-RasGAP) or a monoclonal antiphosphotyrosine antibody (α-pTyr). Migration of molecular weight markers (in kilodaltons) is shown. (C) In vitro association of the SH2 domains of RasGAP with tyrosine-phosphorylated proteins from different developmental stages. Extracts were prepared from 0 to 4-h embryos (Em1), 12- to 24-h embryos (Em2), mixed-stage larvae (L), pupae (P), and adult females (Ad). The wild-type GST-SH232 or mutant GST-SH2*32* fusion proteins bound to glutathione-Sepharose were used to precipitate proteins which were subsequently Western blotted with the antiphosphotyrosine antibody. (D) Immunostaining of precellularized embryo with anti-RasGAP monoclonal antibody showing early expression of RasGAP protein during embryonic development. (E) Immunostaining of cellularized blastoderm embryo showing cytoplasmic localization of RasGAP protein.
Mammalian p120 Ras-GAP is tyrosine phosphorylated upon activation of several RTKs and in cells expressing oncogenic non-RTKs. To determine whether Drosophila RasGAP was tyrosine phosphorylated in vivo, the protein was immunoprecipitated from embryonic extracts with a polyclonal antibody raised against the GAP domain and Western blotted with an antiphosphotyrosine monoclonal antibody. The result (Fig. 4B) showed that RasGAP is tyrosine phosphorylated at a low level in vivo. However, not all the bands detected by the antiphosphotyrosine antibody were detected with the anti- RasGAP polyclonal antibody so that it is possible that some of these bands are from proteins that coimmunoprecipitate with RasGAP.
The SH2 domains of mammalian p120 Ras-GAP bind to phosphotyrosine residues on RTKs, p190 Rho-GAP and the docking protein p62dok (2, 10, 39, 72). To investigate whether the SH2 domains of Drosophila RasGAP could bind to tyrosine-phosphorylated proteins, the region encompassing the two SH2 domains and the intervening SH3 domain (designated SH232) was expressed as a fusion protein in bacteria and purified to give the GST-SH232 fusion protein. As a control a mutant version of this protein, designated GST-SH2*32*, with an inactivating single amino acid substitution in each SH2 domain was also expressed in bacteria and purified. The wild-type GST-SH232 and mutant GST-SH2*32* fusion proteins were then used to pull down proteins from cell extracts of different developmental stages. The precipitated proteins were separated by SDS-PAGE, and phosphotyrosine-containing proteins were detected by immunoblotting with an antiphosphotyrosine antibody. The results of this experiment (Fig. 4C) show that the wild-type GST-SH232 but not the mutant GST-SH2*32* pulls down a small number of tyrosine-phosphorylated proteins in embryos and larvae which appear to be absent or much less abundant in adult flies. Some of the associating proteins clearly change in abundance or phosphorylation state during development. The band at ∼75 kDa present in larvae and pupae in both the wild-type and mutant SH232 pull-downs was visible on Coomassie blue staining of the blot; peptide microsequencing showed that this protein is the γ-subunit of larval serum protein 1 (Lsp1γ) (24), a very abundant protein in third instar larvae and pupae (8).
Inhibition of wing growth by ectopic RasGAP expression.
To gain insight into the in vivo role of RasGAP, we generated transgenic Drosophila expressing the full-length RasGAP cDNA, tagged with the myc epitope, under the control of the yeast GAL4 upstream activating sequence (UAS) (6). Several transgenic lines were obtained by P element germ-line transformation. To determine which lines were able to express RasGAP in a GAL4-dependent manner, we crossed the UAS-RasGAP lines with a line expressing GAL4 under the control of a heat-shock-regulated promoter from the Hsp70 gene. Progeny carrying both UAS-RasGAP and hsp70-GAL4 were heat shocked, and the expression levels of the ectopically expressed RasGAP were analyzed by Western blotting of whole-fly lysates with both the anti-RasGAP polyclonal antibody and the anti-myc antibody (24). On the basis of this analysis, we selected three independent UAS-RasGAP lines expressing the transgene at different levels for further experiments. The UAS-RasGAP lines were crossed to a number of stocks expressing GAL4 in various spatial and temporal patterns throughout development, and the progeny from these crosses were analyzed for phenotypes dependent on RasGAP expression.
Signaling through Ras1 and RTKs has been particularly extensively studied in the Drosophila eye, but we found that overexpression of RasGAP in the eye-antennal imaginal disc with eyeless (ey)-GAL4 or GMR-GAL4 lines did not cause any detectable phenotype (24). Furthermore, expression of RasGAP with the sevenless (sev) enhancer did not affect photoreceptor differentiation in otherwise wild-type flies nor did it suppress the sevenless gain-of-function allele, sevS11 (4, 24). It is known from genetic studies that Gap1, a related but distinct putative GAP, inhibits Ras1 signaling in the eye-antennal disc (30). Already high-level expression of Gap1 might explain the insensitivity of this tissue to overexpression of RasGAP.
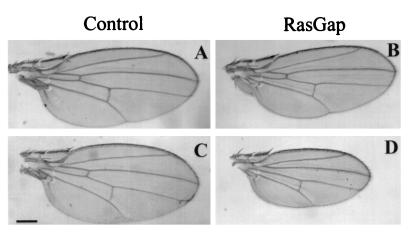
Another adult tissue known to require Ras1 function during its development is the wing. Therefore we expressed high levels of RasGAP in the wing imaginal disc with the MS-1096 GAL4 line, which drives expression throughout the disc with highest levels in the dorsal pouch (9). This resulted in a decrease in the size of the adult wing and an upward curvature of the wing blade, presumably because the dorsal surface of the wing was more strongly affected than the ventral surface (Fig. 5 and Table 1). With the strongest-expressing UAS-RasGAP line (line 1), wings from males were about 45% smaller than the wings from control flies not carrying the transgene, whereas the wings of females were reduced in size by 10 to 20%. The difference between males and females can be explained by the fact that GAL4 is expressed from the X chromosome in the MS-1096 line and will be subject to dosage compensation in males. This suggests that the reduction in wing size caused by ectopic RasGAP expression is dosage sensitive, and we observed that the two lines expressing RasGAP at a lower level (lines 2 and 3) had a correspondingly lesser effect on wing size (Table 1).
FIG. 5.
Ectopic expression of RasGAP in the wing imaginal disc leads to a decrease in size of the adult wing. GAL4 and RasGAP were expressed in the dorsal pouch of the wing with the MS-1096 GAL4 line. The genotypes are GAL4/+ (A), GAL4/+ UAS-RasGAP/+ (B), GAL4/Y (C), and GAL4/Y UAS-RasGAP/+ (D). (A and B) Wings from females; (C and D) wings from males. Bar, 250 μm.
TABLE 1.
Effect of RasGAP expression on wing size
| Transgenic linea | Wing area (mm2)b
|
Wing area (% of control)
|
||
|---|---|---|---|---|
| Male | Female | Male | Female | |
| Control | 1.48 ± 0.02 | 1.49 ± 0.05 | 100 | 100 |
| Line 1 | 0.80 ± 0.04 | 1.09 ± 0.04 | 54.7 | 77.9 |
| Line 2 | 1.03 ± 0.02 | 1.30 ± 0.04 | 69.9 | 87.2 |
| Line 3 | NDc | 1.29 ± 0.05 | ND | 86.7 |
The transgenic lines were hemizygous (males) or heterozygous (females) for the MS-1096 GAL4 insertion on the X chromosome. Lines 1 and 2 carry independent insertions of UAS-RasGAP on the second chromosome; line 3 was heterozygous for UAS-RasGAP insertions on the second and third chromosomes.
Values are means ± SD (n = 4).
ND, not determined.
A decrease in wing size could be the result of a decrease in cell number or a decrease in cell size or a combination of both effects. Each cell on the wing blade produces a single hair so that measuring the density of hairs gives a measure of cell size. However, it was difficult to measure surface areas accurately because the curvature of the wing blade caused folding of the wing surface when mounted on slides for microscopy. This effect was less pronounced at the wing margins so the number of hairs (and therefore cells) on the dorsal margin of the wing between longitudinal veins LIII and LIV was counted and the length of the wing margin measured. Ectopic expression of RasGAP led to a reduction in the number of hairs from 19.50 ± 0.95 to 16.33 ± 0.47 in females of line 1 (mean ± standard deviation [SD]), without significantly affecting cell diameter along the wing margin (14.2 μm in controls and 14.8 μm in RasGAP-expressing females). From these measurements, we attribute the small wing phenotype primarily to a decrease in cell number rather than cell size, although we cannot rule out the possibility that cell size is affected in other parts of the wing.
From the in vitro biochemical properties and effect of its expression in S. pombe it seemed likely that the effect of ectopic RasGAP expression on growth of the wing imaginal disc was the result of downregulating the activity of Ras1, the Drosophila homolog of mammalian Ras proteins. It is known that decreasing the strength of signaling through the Ras1–mitogen-activated protein kinase (MAPK) pathway can inhibit proliferation of wing disc cells causing a reduction in adult wing size (18). If inhibition of Ras1 function was responsible for the small wing phenotype caused by RasGAP overexpression then lowering the Ras1 gene dosage would be expected to enhance the small wing phenotype. However, we found that heterozygosity for Ras1 did not enhance the RasGAP-induced small wing phenotype: in this experiment the wings from GAL4(MS-1096) UAS-RasGAP females were 80.6 ± 2.3% and wings from GAL4(MS-1094) UAS-RasGAP Ras1/+ females were 81.7 ± 3.1% the size of GAL4(MS-1096) UAS-lacZ controls.
Downregulation of receptor tyrosine kinase signaling by ectopic RasGAP expression.
Inhibition of the Ras1-MAPK pathway also disrupts the formation of wing veins, a process that requires the Drosophila EGF receptor homolog (DER) (18). Ectopic expression of RasGAP, however, did not disrupt the longitudinal wing veins (Fig. 5B and D). (Loss of the anterior cross vein as seen in Fig. 5D is also seen at a variable frequency in males hemizygous for the MS-1096 GAL4 insertion but not carrying UAS-RasGAP.) However, differentiation of wing veins occurs during the pupal stage when the level of expression from the MS-1096 GAL4 line might be lower than during the larval stages (11).
To investigate the effect of RasGAP overexpression on wing vein differentiation in a genetically sensitized system we examined whether RasGAP overexpression could suppress the ectopic veins caused by gain-of-function mutations in RTKs or by overexpression of Ras1. Activated mutants of the EGF receptor homolog DER and the fibroblast growth factor (FGF) receptor homologs encoded by the breathless (btl) and heartless (htl) genes have been made by fusing their intracellular part carrying the tyrosine-kinase catalytic domain to the extracellular and membrane-spanning domains of a gain-of-function torso mutant, tor4021 (57). At 25°C no adult flies eclosed when UAS constructs of Tor4021-DER, Tor4021 itself, or Ras1 were expressed under control of the MS-1096 GAL4 line; at 18°C a few adult flies expressing these UAS constructs eclosed but had wings that were too abnormal to analyze further (24). Expression of activated Tor4021-btl or Tor4021-htl, however, gave a phenotype characteristic of activation of the DER-Ras1-MAPK pathway: extra wing vein material and “deltas” at the wing margins were present and the wings were larger (Fig. 6A and C). Coexpression of RasGAP suppressed the formation of extra wing vein material and reduced the size of the wings (Fig. 6B and D). These effects required the SH2 domains of RasGAP because we found that expression of the RasGAPSH2* mutant, with inactivating substitutions in both SH2 domains, was unable to suppress the activated FGF receptor phenotypes (Fig. 6E). The RasGAPSH2* mutant also failed to inhibit wing growth when expressed alone, showing that this effect also requires intact SH2 domains (24). Consistent with signaling from the FGF receptors leading to Ras1 activation, heterozygosity for a Ras1 loss-of-function mutation suppressed the extra wing vein material caused by ectopic RTK expression (Fig. 6F), though not as efficiently as RasGAP overexpression. These results show that RasGAP can attenuate activation of the Ras1-MAPK pathway by the Breathless and Heartless RTKs, presumably by inhibiting the accumulation of Ras1-GTP, and that the SH2 domains of RasGAP are essential for its ability to inhibit both RTK signaling and wing growth.
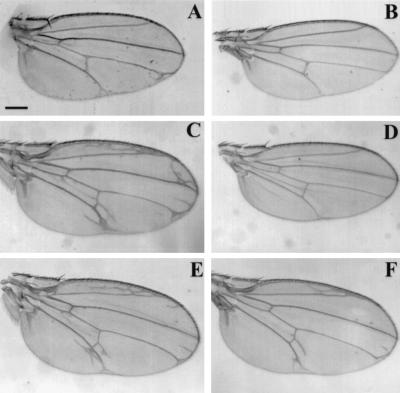
FIG. 6.
Ectopic expression of RasGAP inhibits signaling by activated FGF receptor homologs (Breathless and Heartless) in the wing. The genotypes are GAL4/+ UAS-tor4021-btl/+ (A), GAL4/+ UAS-tor4021-btl/+ UAS-RasGAP/+ (B), GAL4/+ UAS-tor4021-htl/+ (C), GAL4/+ UAS-tor4021-htl/+ UAS-RasGAP/+ (D), GAL4/+ UAS-tor4021-htl/+ UAS-RasGAPSH2*/+ (E), and GAL4/+ UAS-tor4021-htl/+ Ras1e2F/+ (F). The GAL4 line was MS-1096. Bar, 250 μm.
DISCUSSION
The p120 Ras-GAP homolog we have identified, RasGAP, is the third GAP for Ras proteins to be identified in Drosophila and shows that this invertebrate has one of each of the types of RasGAPs found in mammals (30, 62). The most conserved region of the protein encompasses the SH2 and SH3 protein-protein interaction domains in the amino-terminal adapter-like region, which is 57% identical in amino acid sequence between the Drosophila and human proteins. The SH2 domains of mammalian p120 Ras-GAP mediate binding to certain RTKs, p190 Rho-GAP, and the docking protein p62dok (see the introduction). In work described elsewhere we show that the SH2 domains of RasGAP bind in vitro to an autophosphorylation site on Torso (14), a RTK required during early embryonic development (61). The sequences surrounding the phosphotyrosine residue (phosphoYLEP) on Torso match the optimal binding site for the SH2 domains of mammalian p120 Ras-GAP (38, 39). The RasGAP SH2 domains bind in vitro to a small number of tyrosine-phosphorylated proteins from different developmental stages. Whether these are RTKs and the Drosophila homologs of p190 Rho-GAP and p62dok remains to be established.
One notable difference between RasGAP and the mammalian p120 Ras-GAP isoform is that the latter has a proline-rich sequence near the amino terminus which is absent in the Drosophila protein. This region appears to be required for in vitro association of p120 Ras-GAP with Src family tyrosine kinases (Src, Hck, Lck, and Fyn) via their SH3 domain (7) and could be involved in the in vivo association of p120 Ras-GAP with Src family tyrosine kinases (13). Drosophila RasGAP also lacks an ∼70-amino-acid hydrophobic region found at the amino terminus of p120 Ras-GAP. The p100 Ras-GAP isoform expressed in placenta does not have the polyproline region or a hydrophobic amino terminus (65).
We have begun to study the function of RasGAP in vivo by ectopic expression of the full-length protein in the wing imaginal disc with the GAL4-UAS system. Overexpression of RasGAP inhibits growth of the wing, resulting in a decrease in size of the adult wing of up to 45% as a consequence of having fewer rather than smaller cells. This suggests that RasGAP overexpression reduces the rate of proliferation or increases the rate of apoptosis in the wing imaginal disc. Is this effect due to inhibition of Ras1 function? This could occur either by reducing the amount of Ras1-GTP, which would be consistent with the biochemical properties of RasGAP in vitro, or by competition between RasGAP and Ras1 effectors for binding to Ras1-GTP.
The ability of RasGAP to suppress ectopic wing vein formation caused by hyperactivation of the Breathless and Heartless FGF receptor homologs strongly suggests that RasGAP can indeed downregulate Ras1 activation by these RTKs. Both Drosophila FGF receptors have exact matches to the optimal binding site (phospho YxxPxD, where x is any amino acid) for the SH2 domains of mammalian p120 Ras-GAP (38, 39). Given our demonstration in this study that the SH2 domains of RasGAP are essential for its activity in vivo it is likely that the activated FGF receptors directly recruit RasGAP to the plasma-membrane where it is well placed to interact with its substrate, Ras1-GTP.
The DER-Ras1-MAPK pathway regulates both cell proliferation and wing vein differentiation in the wing disc (18). It was puzzling, therefore, why RasGAP overexpression did not affect wing vein formation in otherwise wild-type flies. One possible explanation is that the MS-1096 GAL4 line does not drive sufficient expression of RasGAP in the pupal disc when vein differentiation occurs. However, the ectopic veins caused by the activated FGF receptors were efficiently suppressed by coexpression of RasGAP. We favor the explanation that RasGAP is not efficiently recruited to the plasma membrane by the DER RTK, and this is consistent with our failure to detect an effect of RasGAP overexpression on photoreceptor development in the eye, a process controlled by the DER and Sev RTKs, neither of which contains the RasGAP SH2 binding consensus. The insulin receptor (Inr), another RTK, is also implicated in the control of cell proliferation in the wing disc (12, 25), but we did not see any enhancement of the RasGAP overexpression phenotype in flies heterozygous for an inr mutation (24). The insensitivity of the RasGAP-induced small wing phenotype to the gene dosage of Ras1 suggests that RasGAP could be inhibiting proliferative signals that are not dependent on Ras1.
One important and unresolved issue is the specificity of RasGAPs in vivo. Mammalian p120 Ras-GAP can act as a GAP in vitro for the Ras-related protein R-Ras (29), and also, more surprisingly, for the much more distantly related small GTPase Rab5 (43), which is involved in the regulation of endocytosis. A further twist is that the Ras-related protein Rap1/Krev-1 binds to p120 Ras-GAP with high affinity but is not a substrate (27). Drosophila homologs of R-Ras (Ras2), Rap1 (Roughened), and Rab5 (Drab5) have been identified (34, 52, 58). Their roles in imaginal disc growth and differentiation are not clear, although it has been observed that expression of a constitutively active Ras2 mutant in the wing disc gives rise to ectopic veins (6, 17). Therefore, it is possible that some of the effects of RasGAP overexpression are the consequence of interactions with Ras2, Roughened, or Drab5.
Alternatively, the functions of proteins that interact with the amino-terminal adapter region of RasGAP may be affected by RasGAP overexpression and cause growth inhibition. However, overexpressing just the SH2-SH3-SH2 region of RasGAP by using the same UAS-GAL4 system does not affect wing growth (24). Further studies of the effects of expressing RasGAP transgenes with mutations in the modular domains and the isolation of RasGAP mutants should help to clarify the physiological roles of this important signaling protein. The extensive structural and functional similarities between the Drosophila and mammalian RasGAPs indicates that insights gained from genetic approaches in flies will also be relevant to the role of p120 Ras-GAP in mammals.
ACKNOWLEDGMENTS
We thank members of the Hughes and Hafen labs, Chris Marshall, Vaughn Cleghon, and Kevin O’Hare for advice and encouragement; Jacky Cordell for generating the monoclonal antibody; Fiona Mitchell for careful reading of the manuscript; and Jonathan Cooper and Chris Norbury for materials.
This work was supported by The Wellcome Trust, the Cancer Research Campaign, the Swiss National Science Foundation, and the Ludwig Institute. P.F. was supported by a short-term EMBO fellowship while visiting E.H.’s lab. S.J.L. is a Royal Society University Research Fellow.
REFERENCES
- 1.Amrein K E, Panholzer B, Molnos J, Flint N A, Scheffler J, Lahm H W, Bannwarth W, Burn P. Mapping of the p56lck-mediated phosphorylation of GAP and analysis of its influence on p21ras-GTPase activity in-vitro. Biochim Biophys Acta. 1994;1222:441–446. doi: 10.1016/0167-4889(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 2.Anderson D, Koch C A, Grey L, Ellis C, Moran M F, Pawson T. Binding of SH2 domains of PLC-γ1, GAP and src to activated growth factor receptors. Science. 1990;250:979–982. doi: 10.1126/science.2173144. [DOI] [PubMed] [Google Scholar]
- 3.Barpeled M, Raikhel N V. A method for isolation and purification of specific antibodies to a protein fused to the GST. Anal Biochem. 1996;241:140–142. doi: 10.1006/abio.1996.0390. [DOI] [PubMed] [Google Scholar]
- 4.Basler K, Christen B, Hafen E. Ligand-independent activation of the sevenless receptor tyrosine kinase changes the fate of cells in the developing Drosophila eye. Cell. 1991;64:1069–1081. doi: 10.1016/0092-8674(91)90262-w. [DOI] [PubMed] [Google Scholar]
- 5.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 6.Brand A H, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 7.Briggs S D, Bryant S S, Jove R, Sanderson S D, Smithgall T E. The Ras GTPase-activating protein (GAP) is an SH3 domain-binding protein and substrate for the Src-related tyrosine kinase, Hck. J Biol Chem. 1995;270:14718–14724. doi: 10.1074/jbc.270.24.14718. [DOI] [PubMed] [Google Scholar]
- 8.Brock H W, Roberts D B. Comparison of the larval serum proteins of Drosophila melanogaster using one and two-dimensional peptide mapping. Eur J Biochem. 1980;106:129–135. doi: 10.1111/j.1432-1033.1980.tb06003.x. [DOI] [PubMed] [Google Scholar]
- 9.Capdevila J, Guerrero I. Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J. 1994;13:4459–4468. doi: 10.1002/j.1460-2075.1994.tb06768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpino N, Wisniewski D, Strife A, Marshak D, Kobayashi R, Stillman B, Clarkson B. p62dok: a constitutively tyrosine-phosphorylated, GAP-associated protein in chronic myelogenous leukemia progenitor cells. Cell. 1997;88:197–204. doi: 10.1016/s0092-8674(00)81840-1. [DOI] [PubMed] [Google Scholar]
- 11.Casares F, Calleja M, Sanchez-Herrero E. Functional similarity in appendage specification by the Ultrabithorax and abdominal-A Drosophila HOX genes. EMBO J. 1996;15:3934–3942. [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C, Jack J, Garofalo R S. The Drosophila insulin-receptor is required for normal growth. Endocrinology. 1996;137:846–856. doi: 10.1210/endo.137.3.8603594. [DOI] [PubMed] [Google Scholar]
- 13.Cichowski K, McCormick F, Brugge J S. p21ras GAP association with Fyn, Lyn, and Yes in thrombin-activated platelets. J Biol Chem. 1992;267:5025–5028. [PubMed] [Google Scholar]
- 14.Cleghon V, Feldmann P, Ghiglione C, Copeland T D, Perrimon N, Hughes D A, Morrison D K. Opposing actions of CSW and RasGAP modulate the strength of Torso RTK signaling in the Drosophila terminal pathway. Mol Cell. 1998;2:719–727. doi: 10.1016/s1097-2765(00)80287-7. [DOI] [PubMed] [Google Scholar]
- 15.Cleghon V, Gayko U, Copeland T D, Perkins L A, Perrimon N, Morrison D K. Drosophila terminal structure development is regulated by the compensatory activities of positive and negative phosphotyrosine signaling sites on the Torso RTK. Genes Dev. 1996;10:566–577. doi: 10.1101/gad.10.5.566. [DOI] [PubMed] [Google Scholar]
- 16.Cullen P J, Hsuan J J, Truong O, Letcher A J, Jackson T R, Dawson A P, Irvine R F. Identification of a specific Ins(1,3,4,5)P-4-binding protein as a member of the Gap1 family. Nature. 1995;376:527–530. doi: 10.1038/376527a0. [DOI] [PubMed] [Google Scholar]
- 17.de Celis J F. Expression and function of decapentaplegic and thick veins during the differentiation of the veins in the Drosophila wing. Development. 1997;124:1007–1018. doi: 10.1242/dev.124.5.1007. [DOI] [PubMed] [Google Scholar]
- 18.Diaz-Benjumea F J, Hafen E. The Sevenless signaling cassette mediates Drosophila EGF receptor function during epidermal development. Development. 1994;120:569–578. doi: 10.1242/dev.120.3.569. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez M, Hafen E. Genetic dissection of cell fate specification in the developing eye of Drosophila. Semin Cell Dev Biol. 1996;7:219–226. [Google Scholar]
- 20.Downward J. Measurement of nucleotide exchange and hydrolysis activities in immunoprecipitates. Methods Enzymol. 1995;255:110–117. doi: 10.1016/s0076-6879(95)55013-5. [DOI] [PubMed] [Google Scholar]
- 21.Edwards K A, Montague R A, Shepard S, Edgar B A, Erikson R L, Kiehart D P. Identification of Drosophila cytoskeletal proteins by induction of abnormal cell shape in fission yeast. Proc Natl Acad Sci USA. 1994;91:4589–4593. doi: 10.1073/pnas.91.10.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egel R. Physiological aspects of conjugation in fission yeast. Planta. 1971;98:89–96. doi: 10.1007/BF00387025. [DOI] [PubMed] [Google Scholar]
- 23.Ellis C, Moran M, McCormick F, Pawson T. Phosphorylation of GAP and GAP-associated proteins by transforming and mitogenic tyrosine kinases. Nature. 1990;343:377–381. doi: 10.1038/343377a0. [DOI] [PubMed] [Google Scholar]
- 24.Feldmann, P., and D. A. Hughes. Unpublished data.
- 25.Fernandez R, Tabarini D, Azpiazu N, Frasch M, Schlessinger J. The Drosophila insulin-receptor homolog—a gene essential for embryonic development encodes 2 receptor isoforms with different signaling potential. EMBO J. 1995;14:3373–3384. doi: 10.1002/j.1460-2075.1995.tb07343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frangioni J V, Neel B G. Solubilization and purification of enzymatically active glutathione-S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 27.Frech M, John J, Pizon V, Chardin P, Tavitian A, Clark R, McCormick F, Wittinghofer A. Inhibition of GTPase activating protein stimulation of Ras-p21 GTPase by the Krev-1 gene product. Science. 1990;249:169–171. doi: 10.1126/science.2164710. [DOI] [PubMed] [Google Scholar]
- 28.Fukui Y, Kozasa T, Kaziro Y, Takeda T, Yamamoto M. Role of a ras homolog in the life-cycle of Schizosaccharomyces pombe. Cell. 1986;44:329–336. doi: 10.1016/0092-8674(86)90767-1. [DOI] [PubMed] [Google Scholar]
- 29.Garrett M D, Self A J, van Oers C, Hall A. Identification of distinct cytoplasmic targets for ras/R-ras and rho regulatory proteins. J Biol Chem. 1989;264:10–13. [PubMed] [Google Scholar]
- 30.Gaul U, Mardon G, Rubin G M. A putative Ras GTPase activating protein acts as a negative regulator of signaling by the Sevenless receptor tyrosine kinase. Cell. 1992;68:1007–1019. doi: 10.1016/0092-8674(92)90073-l. [DOI] [PubMed] [Google Scholar]
- 31.Gawler D J, Zhang L J, Reedijk M, Tung P S, Moran M F. CaLB: a 43 amino acid calcium-dependent membrane/phospholipid binding domain in p120 Ras GTPase-activating protein. Oncogene. 1995;10:817–825. [PubMed] [Google Scholar]
- 32.Genetics Computer Group. Program manual for the Wisconsin package, version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 33.Guan K L, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 34.Hariharan I K, Carthew R W, Rubin G M. The Drosophila Roughened mutation—activation of a Rap homolog disrupts eye development and interferes with cell determination. Cell. 1991;67:717–722. doi: 10.1016/0092-8674(91)90066-8. [DOI] [PubMed] [Google Scholar]
- 35.Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 36.Henkemeyer M, Rossi D J, Holmyard D P, Puri M C, Mbamalu G, Harpal K, Shih T S, Jacks T, Pawson T. Vascular system defects and neuronal apoptosis in mice lacking Ras GTPase-activating protein. Nature. 1995;377:695–701. doi: 10.1038/377695a0. [DOI] [PubMed] [Google Scholar]
- 37.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 38.Holland S J, Gale N W, Gish G D, Roth R A, Zhou S Y, Cantley L C, Henkemeyer M, Yancopoulos G D, Pawson T. Juxtamembrane tyrosine residues couple the Eph family receptor EphB2/Nuk to specific SH2 domain proteins in neuronal cells. EMBO J. 1997;16:3877–3888. doi: 10.1093/emboj/16.13.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu K Q, Settleman J. Tandem SH2 binding sites mediate the RasGAP-RhoGAP interaction: a conformational mechanism for SH3 domain regulation. EMBO J. 1997;16:473–483. doi: 10.1093/emboj/16.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imai Y, Miyake S, Hughes D A, Yamamoto M. Identification of a GTPase-activating protein homolog in Schizosaccharomyces pombe. Mol Cell Biol. 1991;11:3088–3094. doi: 10.1128/mcb.11.6.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karim F D, Rubin G M. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development. 1998;125:1–9. doi: 10.1242/dev.125.1.1. [DOI] [PubMed] [Google Scholar]
- 42.Katz M E, McCormick F. Signal transduction from multiple Ras effectors. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 43.Liu K, Li G. Catalytic domain of the p120 Ras GAP binds to Rab5 and stimulates its GTPase activity. J Biol Chem. 1998;273:10087–10090. doi: 10.1074/jbc.273.17.10087. [DOI] [PubMed] [Google Scholar]
- 44.Liu X Q, Pawson T. The epidermal growth factor receptor phosphorylates GTPase-activating protein (GAP) at Tyr-460, adjacent to the GAP SH2 domains. Mol Cell Biol. 1991;11:2511–2516. doi: 10.1128/mcb.11.5.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maekawa M, Li S W, Iwamatsu A, Morishita T, Yokota K, Imai Y, Kohsaka S, Nakamura S, Hattori S. A novel mammalian Ras GTPase-activating protein which has phospholipid-binding and Btk homology regions. Mol Cell Biol. 1994;14:6879–6885. doi: 10.1128/mcb.14.10.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin G A, Viskochil D, Bollag G, McCabe P C, Crosier W J, Haubruck H, Conroy L, Clark R, Oconnell P, Cawthon R M, Innis M A, McCormick F. The GAP-related domain of the neurofibromatosis type-1 gene product interacts with ras p21. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- 47.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 48.Molloy C J, Bottaro D P, Fleming T P, Marshall M S, Gibbs J B, Aaronson S A. PDGF induction of tyrosine phosphorylation of GTPase activating protein. Nature. 1989;342:711–714. doi: 10.1038/342711a0. [DOI] [PubMed] [Google Scholar]
- 49.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 50.Nadin-Davis S A, Nasim A, Beach D. Involvement of ras in sexual differentiation but not in growth control in fission yeast. EMBO J. 1986;5:2963–2971. doi: 10.1002/j.1460-2075.1986.tb04593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 52.Neuman-Silberberg F S, Schejter E, Hoffmann F M, Shilo B Z. The Drosophila ras oncogenes: structure and nucleotide sequence. Cell. 1994;37:1027–1033. doi: 10.1016/0092-8674(84)90437-9. [DOI] [PubMed] [Google Scholar]
- 53.Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by transcomplementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park S, Liu X Q, Pawson T, Jove R. Activated Src tyrosine kinase phosphorylates Tyr-457 of bovine GTPase-activating protein (GAP) in vitro and the corresponding residue of rat GAP in vivo. J Biol Chem. 1992;267:17194–17200. [PubMed] [Google Scholar]
- 55.Parker F, Maurier F, Delumeau I, Duchesne M, Faucher D, Debussche L, Dugue A, Schweighoffer F, Tocque B. A Ras-GTPase-activating protein SH3-domain-binding protein. Mol Cell Biol. 1996;16:2561–2569. doi: 10.1128/mcb.16.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 57.Reichman-Fried M, Dickson B, Hafen E, Shilo B Z. Elucidation of the role of Breathless, a Drosophila FGF receptor homolog, in tracheal cell migration. Genes Dev. 1994;8:428–439. doi: 10.1101/gad.8.4.428. [DOI] [PubMed] [Google Scholar]
- 58.Sasamura T, Kobayashi T, Kojima S, Qadota H, Ohya Y, Masai I, Hotta Y. Molecular cloning and characterization of Drosophila genes encoding small GTPases of the rab and rho families. Mol Gen Genet. 1997;254:486–494. doi: 10.1007/s004380050443. [DOI] [PubMed] [Google Scholar]
- 59.Settleman J, Narasimhan V, Foster L C, Weinberg R A. Molecular cloning of cDNAs encoding the GAP-associated protein p190—implications for a signaling pathway from Ras to the nucleus. Cell. 1992;69:539–549. doi: 10.1016/0092-8674(92)90454-k. [DOI] [PubMed] [Google Scholar]
- 60.Simon M A, Bowtell D D L, Dodson G S, Laverty T R, Rubin G M. Ras1 and a putative guanine-nucleotide exchange factor perform crucial steps in signaling by the Sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 61.Sprenger F, Stevens L M, Nusslein-Volhard C. The Drosophila gene torso encodes a putative receptor tyrosine kinase. Nature. 1989;338:478–483. doi: 10.1038/338478a0. [DOI] [PubMed] [Google Scholar]
- 62.The I, Hannigan G E, Cowley G S, Reginald S, Zhong Y, Gusella J F, Hariharan I K, Bernards A. Rescue of a Drosophila NF1 mutant phenotype by protein kinase A. Science. 1997;276:791–794. doi: 10.1126/science.276.5313.791. [DOI] [PubMed] [Google Scholar]
- 63.Tocque B, Delumeau I, Parker F, Maurier F, Multon M C, Schweighoffer F. Ras-GTPase activating protein (GAP): a putative effector for Ras. Cell Signalling. 1997;9:153–158. doi: 10.1016/s0898-6568(96)00135-0. [DOI] [PubMed] [Google Scholar]
- 64.Trahey M, McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987;238:542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- 65.Trahey M, Wong G, Halenbeck R, Rubinfeld B, Martin G A, Ladner M, Long C M, Crosier W J, Watt K, Koths K, McCormick F. Molecular cloning of two types of GAP complementary DNA from human placenta. Science. 1988;242:1697–1700. doi: 10.1126/science.3201259. [DOI] [PubMed] [Google Scholar]
- 66.Tsukita S, Yonemura S, Tsukita S. ERM (ezrin/radixin/moesin) family: from cytoskeleton to signal transduction. Curr Opin Cell Biol. 1997;9:70–75. doi: 10.1016/s0955-0674(97)80154-8. [DOI] [PubMed] [Google Scholar]
- 67.van der Geer P, Henkemeyer M, Jacks T, Pawson T. Aberrant Ras regulation and reduced p190 tyrosine phosphorylation in cells lacking p120-Gap. Mol Cell Biol. 1997;17:1840–1847. doi: 10.1128/mcb.17.4.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vogel U S, Dixon R A F, Schaber M D, Diehl R E, Marshall M S, Scolnick E M, Sigal I S, Gibbs J B. Cloning of bovine GAP and its interaction with oncogenic Ras-p21. Nature. 1988;335:90–93. doi: 10.1038/335090a0. [DOI] [PubMed] [Google Scholar]
- 69.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, Boguski M, Riggs M, Rodgers L, Wigler M. Sar1, a gene from Schizosaccharomyces pombe encoding a protein that regulates Ras1. Cell Regul. 1991;2:453–465. doi: 10.1091/mbc.2.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu G F, O’Connell P, Viskochil D, Cawthon R, Robertson M, Culver M, Dunn D, Stevens J, Gesteland R, White R, Weiss R. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- 72.Yamanashi Y, Baltimore D. Identification of the abl- and rasGAP-associated 62 kDa protein as a docking protein, dok. Cell. 1997;88:205–211. doi: 10.1016/s0092-8674(00)81841-3. [DOI] [PubMed] [Google Scholar]
- 73.Young P E, Pesacreta T C, Kiehart D P. Dynamic changes in the distribution of cytoplasmic myosin during Drosophila embryogenesis. Development. 1991;111:1–14. doi: 10.1242/dev.111.1.1. [DOI] [PubMed] [Google Scholar]