Abstract
We have previously found that epidermal growth factor (EGF) mediates growth through the Jun N-terminal kinase/stress-activated kinase (JNK/SAPK) pathway in A549 human lung carcinoma cells. As observed here, EGF treatment also greatly enhances the tumorigenicity of A549 cells, suggesting an important role for JNK in cancer cell growth (F. Bost, R. McKay, N. Dean, and D. Mercola, J. Biol. Chem. 272:33422–33429, 1997). Several isoforms families of JNK, JNK1, JNK2, and JNK3, have been isolated; they arise from alternative splicing of three different genes and have distinct substrate binding properties. Here we have used specific phosphorothioate oligonucleotides targeted against the two major isoforms, JNK1 and JNK2, to discriminate their roles in EGF-induced transformation. Multiple antisense sequences have been screened, and two high-affinity and specific candidates have been identified. Antisense JNK1 eliminated steady-state mRNA and JNK1 protein expression with a 50% effective concentration (EC50) of <0.1 μM but did not alter JNK2 mRNA or protein levels. Conversely, antisense JNK2 specifically eliminated JNK2 steady-state mRNA and protein expression with an EC50 of 0.1 μM. Antisense JNK1 and antisense JNK2 inhibited by 40 and 70%, respectively, EGF-induced total JNK activity, whereas sense and scrambled-sequence control oligonucleotides had no effect. The elimination of mRNA, protein, and JNK activities lasted 48 and 72 h following a single Lipofectin treatment with antisense JNK1 and JNK2, respectively, indicating sufficient duration for examining the impact of specific elimination on the phenotype. Direct proliferation assays demonstrated that antisense JNK2 inhibited EGF-induced doubling of growth as well as the combination of active antisense oligonucleotides did. EGF treatment also induced colony formation in soft agar. This effect was completely inhibited by antisense JNK2 and combined-antisense treatment but not altered by antisense JNK1 alone. These results show that EGF doubles the proliferation (growth in soft agar as well as tumorigenicity in athymic mice) of A549 lung carcinoma cells and that the JNK2 isoform but not JNK1 is utilized for mediating the effects of EGF. This study represents the first demonstration of a cellular phenotype regulated by a JNK isoform family, JNK2.
The Jun kinase/stress-activated protein kinase (JNK/SAPK) pathway has been implicated in major cellular functions, such as cell proliferation and transformation (4, 7, 13, 57, 78, 79). It is also implicated in DNA repair and cellular stress response (2, 37, 44, 66, 77), including apoptosis (42). The c-Jun–NH2 terminal kinase enzyme (JNK) is responsible for the phosphorylation of c-Jun on serine residues 63 and 73 (23, 37). C-Jun, ATF-2, and Elk-1 are the three major substrates of JNK (9, 23, 30, 37, 48, 87). The JNK pathway consists of several kinases, including mitogen-activated/extracellular response kinase kinase (MEK) kinase 1 (MEKK1) and mitogen-activated protein kinase (MAPK) kinases 7 and 4 (MKK7 and MKK4), which form a kinase cascade (22, 28, 29, 55, 61, 95, 96). When these kinases are activated, they lead to the activation of JNK. So far, 10 different isoforms of human JNK have been characterized (31). These forms result from alternative splicing of three major genes encoding Jun kinase 1 (JNK1), Jun kinase 2 (JNK2), and Jun kinase 3 (JNK3). The apparent molecular masses of these isoforms are classically 46, 55, and 57 kDa, respectively (23, 37, 41, 77). However, some JNK1 isoforms have molecular masses of 55 kDa and some JNK2 isoforms migrate at 46 kDa (31). The three major substrates of JNK, c-Jun, ATF-2, and Elk-1, are phosphorylated to a similar extent by JNK isoforms in vitro (31) even though there are differences in the affinity of JNK for these substrates (31). For example, JNK2 isoforms have a much higher affinity for ATF-2 and c-Jun in vitro (31). Kallunki et al. (41) had similar results showing that JNK2 binds c-Jun 25 times more efficiently than JNK1.
No physiological role has been associated with the difference in substrate affinity of the JNKs. However, several recent studies suggested that JNK1 and JNK2 have distinct functions and are not redundant (8, 10, 91, 98). JNK1 has been more specifically involved in the regulation of the apoptotic response of small-cell lung cancer cells following UV radiation (8). JNK1 was also shown to be activated preferentially by tumor necrosis factor α in mouse macrophages (10). Furthermore, Sluss et al. (77) were able to complement a defect in the expression of HOG-1 (a MAPK analog in yeast) by expressing human JNK1 but not JNK2. Recently, Zhang et al. (99) reported a predominant activation of JNK1 over JNK2 in type 2 astrocytes and oligodendrocytes due to a differential expression of these proteins. Conversely, JNK2 has a crucial role in the survival of inner-medullary collecting duct cells in a hypertonic environment (91). These observations raise the question of a differential role for JNK1 and JNK2 in one or more of the major physiological functions associated with JNKs: cell growth, transformation, DNA repair, and stress response. However, it is not known if one of the JNK isoforms has a predominant role in mediating any of these effects.
Little is known about the role of JNK in human cancer. Most of the studies were performed in animal model cell lines, where it was shown that activated JNK promotes a transformed phenotype (4, 39, 68, 70, 83). Nevertheless, recent studies in HeLa cells, derived from human cervical epidermoid carcinoma, have shown that epidermal growth factor (EGF) activates the JNK pathway (14, 49, 55, 56). Moreover, it was shown that Jun kinase kinase may be mutated in certain tumors, suggesting a suppressor function (86). However, these studies do not show any phenotypic consequences of JNK activation on human cancer cells.
We have shown in a recent study that JNK is constitutively active in a variety of human tumor lines (7, 66, 66a). Furthermore, EGF-induced doubling of proliferation of A549 human lung carcinoma cells requires the activation of JNK (7), suggesting an essential role for this signal transduction pathway in mediating the growth of human cancer cells.
EGF is a very potent autocrine growth factor in human lung cancer (32, 53, 67, 69, 73, 75, 85, 88) and is involved in cancer cell proliferation and tumorigenicity of human glioblastomas (26, 46, 62, 92), breast cancer (1, 25, 27, 58), and keratinocytes (33, 69). Taken together, these data show that EGF and its receptor play significant roles in the genesis and/or progression of many human cancers (84).
In order to test the roles of the two major isoforms, JNK1 and JNK2, we specifically inhibited the expression of JNK1 or JNK2 protein in A549 by using highly specific phosphorothioate antisense oligonucleotides directed against JNK1 isoforms or JNK2 isoforms. Antisense oligonucleotides have the potential advantage of complexing with specific mRNAs, leading to destruction via RNase H action, thereby inhibiting the expression of a single protein (15, 21, 24, 34, 52). Recent reports showed that phosphorothioate antisense oligonucleotides directed against PKCα (17–19, 57), c-Raf (60), c-Fos (20), and ErB2 (47) specifically eliminate the expression of the target protein in cells at low concentrations. Here we showed by Northern analysis and Western analysis that the oligonucleotides complementary to each JNK isozyme were able to inhibit the expression of their respective target isoform without altering the other. We found that EGF treatment caused a doubling of colony formation as well as tumorigenicity in athymic mice. Oligonucleotide-specific depletion of JNK2 in A549 cells greatly reduced total JNK activity induced by EGF and completely blocked both EGF-induced cell proliferation and colony growth in soft agar. Conversely, JNK1 elimination had only a moderate effect on JNK activity and had almost no effect on A549 EGF-induced proliferation or colony formation. Therefore, we conclude that JNK2 and not JNK1 is required in A549 human lung carcinoma cells to mediate EGF induction of growth and transformation.
MATERIALS AND METHODS
Cell culture and treatment.
Non-small-cell lung cancer A549 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco BRL) supplemented with 5% (vol/vol) fetal bovine serum (FBS). For treatment with recombinant human EGF (rhEGF) (lot CE118020; R&D Systems Inc.) and treatment with UV-C, the cells were seeded 2 days before treatment and were maintained in 0.5% FBS for 17 h prior to treatment.
Inhibition of JNK mRNA and protein expression by JNKAS oligonucleotides.
All oligonucleotides used in this study were phosphorothioate oligonucleotides prepared following the same procedure described by Dean et al. and Hélène and Toulme (17, 34) and Monia et al. (60) and were gifts of ISIS Pharmaceuticals, Inc. In this study we used two kinds of phosphorothioate oligonucleotides: unaltered 20-nucleotide phosphorothioate oligonucleotides (see Fig. 1 to 3, 4A, 7, and 8) and phosphorothioate 2′-O-methoxyethyl-modified oligonucleotides (and their respective controls; see Fig. 4B and 6). These latter oligonucleotides have 2′-O-methoxyethyl modifications on nucleotides 1 to 5 and 15 to 20 at their respective 5′ and 3′ ends. Control oligonucleotides were prepared and consisted of the sense phosphorothioate oligonucleotides (JNK1SeISIS14320 and JNK2SeISIS14318) or were of the same base composition as the antisense (JNKAS) oligonucleotides but in arbitrary order (scrambled oligonucleotides JNK1ScrISIS14321 and JNK2ScrISIS14319 or JNK1ScrISIS18076 and JNK2ScrISIS18078).
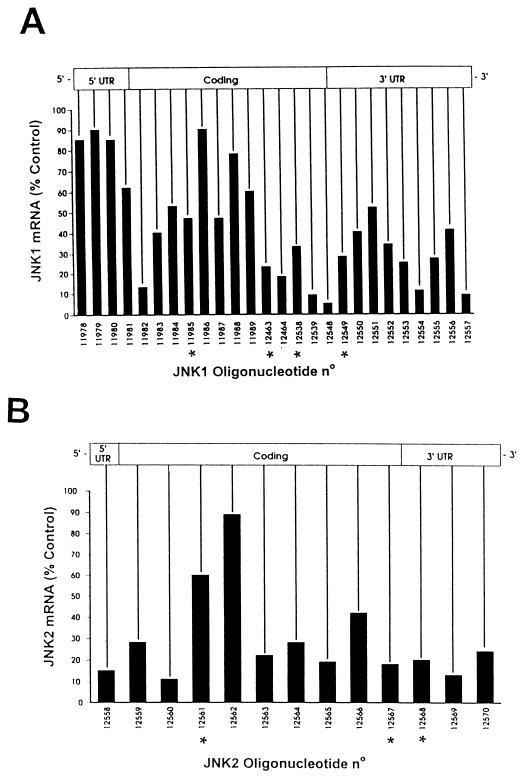
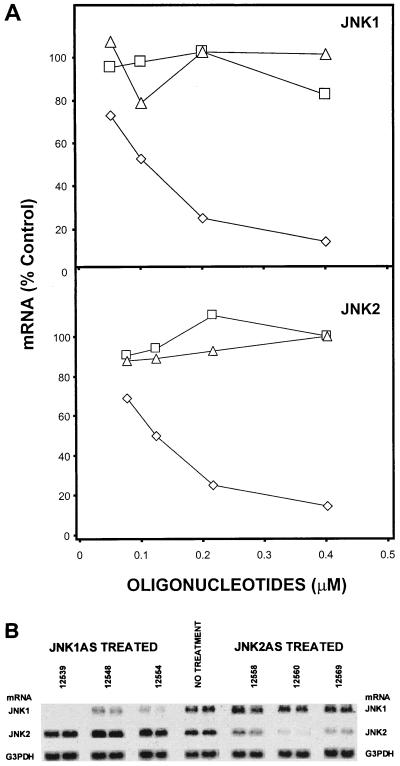
FIG. 1.
Messenger walk survey for the elimination of JNK1 and JNK2 mRNA levels following treatment with phosphorothioate antisense oligonucleotides targeted to JNK1 or JNK2 mRNA. (A) JNK1 mRNA steady-state levels in A549 cells following treatment with 26 different phosphorothioate antisense oligonucleotides targeted to JNK1 mRNA. JNK1ASISIS12539 gave the most consistent results for the elimination of JNK1 mRNA steady-state levels. (B) A similar experiment was performed with 13 phosphorothioate antisense oligonucleotides complementary to the indicated regions of the JNK2 mRNA. JNK2ASISIS12560 yielded the most consistent results for the elimination of JNK2 mRNA steady-state levels. All oligonucleotides are arrayed relative to their complementary sequence along the JNK transcript. The asterisks indicate the oligonucleotides having a nucleotide composition (2 to 4 bases) very similar to that of the leading compound. The screenings were repeated two times with similar results.
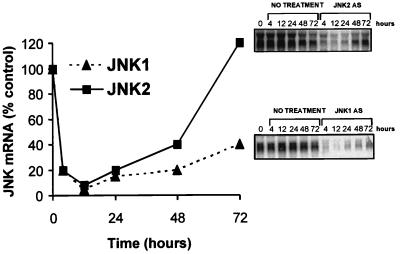
FIG. 3.
Duration of elimination of JNK1 and JNK2 mRNA in A549 cells. A549 cells were transfected with 0.4 μM JNK1ASISIS12539 or JNK2ASISIS12560. The mRNAs were prepared at the indicated times after 4, 12, 24, 48, and 72 h and examined by Northern analysis. Quantification was performed as described in Material and Methods.
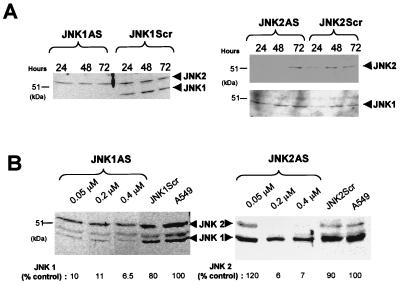
FIG. 4.
Determination of the duration of antisense effect and dose-dependent inhibition of expression of JNK1 and JNK2 protein after treatment with antisense oligonucleotides. (A) Duration of antisense effect. A549 cells were transfected with 0.4 μM JNK1ASISIS12539 or JNK1ScrISIS14321 for 4.5 h, and cell extracts were prepared at the indicated times. Western analysis was performed with anti-JNK1 antibodies (SC-571). A similar experiment was carried out with JNK2ASISIS12560 and JNK2ScrISIS14319 as a control. Western analysis was performed with anti-JNK2 antibodies (SC-827), and the same membrane was reprobed with JNK1 antibodies (SC-571) (bottom). (B) A549 cells were treated with the indicated concentrations of phosphorothioate 2′-O-methoxyethyl-modified antisense oligonucleotides JNK1ASISIS15346 or JNK2ASISIS15353 and 0.4 μM their respective control oligonucleotides, JNK1ScrISIS18076 and JNK2ScrISIS18078. Cell extracts were prepared 36 h after the transfection and examined by Western analysis with anti-JNK1 antibodies (SC-571). The protein levels (shown below the gels) were determined by comparison to the respective protein levels in untreated cells by using the Electrophoresis Documentation and analysis System 120 (Kodak Digital Science).
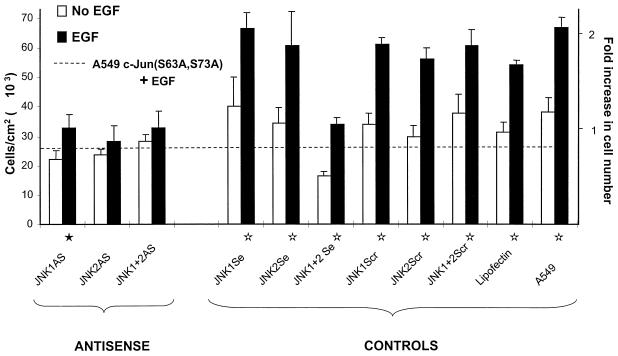
FIG. 7.
JNK2AS preferentially inhibits EGF-induced proliferation in A549. Proliferation assays were performed as described in Materials and Methods with the unmodified oligonucleotides. The cells were maintained in 0.5% FBS during the experiment. Five days after the treatment with 0.1 μM rhEGF, the cells were counted with a Coulter counter. The dashed line indicates the growth attained by A549 cells that stably express the dominant-negative inhibitor c-Jun(S63A,S73A) which is known to be inhibited from responding to rhEGF (7). The proliferation data shown here are the average of two identical and independent experiments, each carried out in triplicate. Fold increase in cell number is given considering 1 as the average number of cells grown in the absence of rhEGF. The standard errors (error bars) are given as √(ς12 + ς22), where ς1 and ς2 are the standard errors of the replicate experiments. Statistical analyses were carried out with the combined data of both replicates by analysis of variance implemented with Systat software. The statistical significance for EGF-induced growth was P < 0.002 (⋆) or P < 0.05 (★).
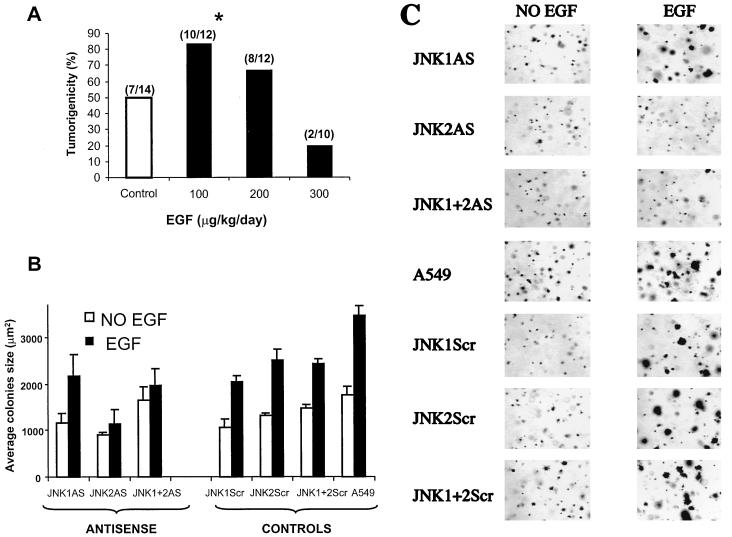
FIG. 8.
EGF-induced tumorigenicity in nude mice and inhibition of growth on soft agar by antisense oligonucleotides. (A) A549 cells (5 × 106) were injected into nude mice and analyzed for their ability to develop tumors in animals treated daily with saline solution (control) (open bar) or 100, 200, or 300 μg of rhEGF/kg (solid bars). After 8 days, visible tumors were scored as the ratio of the number of tumors over the number of sites injected (indicated above the bars). The asterisk indicates that the tumorigenicity increase at the 100-μg/kg/day dose is significant, with a P value of <0.021 (χ2 test). (B) A549 cells were transfected with the indicated unmodified oligonucleotides (0.4 μM or 0.2 plus 0.2 μM [for the combination]). Eighteen hours after transfection, the cells were transferred to 0.3% agarose containing 0.8 or 0.4 μM (for the combination) the indicated oligonucleotides. The cells were stained with p-iodotetrazolium violet and analyzed with IPLabSpectrum software. The error bars indicate the standard deviation. (C) Morphologies of A549 colonies treated with 0.1 μM rhEGF (right) or untreated (left) and transfected with the indicated oligonucleotides.
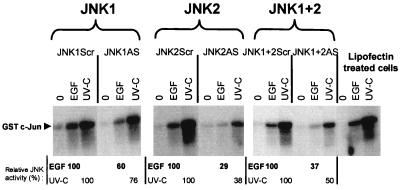
FIG. 6.
Differential inhibition of JNK activity by JNK1AS and JNK2AS. A549 cells were transfected for 4.5 h with the same active antisense or control phosphorothioate 2′-O-methoxyethyl-modified oligonucleotides (0.2 μM) used in the experiments shown in Fig. 4B. Thirty-six hours after transfection, the cells were stimulated with 0.1 μM rhEGF or UV-C at 100 J/m2 or were not stimulated. Twenty minutes after stimulation, cell extracts were prepared and used for a Jun kinase assay, as described in Materials and Methods. The activation is given relative to the activation obtained with rhEGF or UV-C in the scrambled-oligonucleotide-treated cells.
The transfection of cells was performed as described previously (7, 17). Briefly, oligonucleotide at the concentration indicated in the text was added to minimum essential medium containing 10 μl of Lipofectin (Gibco BRL)/ml at 1 mg/ml original concentration. This preparation was added to 50 to 80% confluent A549 cells. After 4.5 h, the transfection medium was generally replaced with DMEM with 5% FBS, except for the JNK kinase assay and the proliferation assay, where we used 0.5% FBS.
For the determination of mRNA levels by Northern analysis, total RNA was prepared from cells by the guanidium isothiocyanate procedure followed by application to a cesium chloride gradient. Total RNA was then examined by Northern analysis, as described by Dean et al. (17–19). RNA was quantified and normalized to glucose-3-phosphate dehydrogenase (G3PDH) mRNA levels with a Molecular Dynamics PhosphorImager.
For determination of protein levels, cell extracts were prepared with cell lysis buffer (25 mM HEPES [pH 7.7], 0.3 M NaCl, 1.5 mM MgCl2, 0.1% Triton X-100, phenylmethylsulfonyl fluoride [100 μg/ml], tolylsulfonyl phenylalanyl chloromethyl ketone [100 μg/ml], EDTA [1 μM], leupeptin [2 μg/ml], aprotinin [2 μg/ml], 20 mM β-glycerophosphate, 0.1 mM Na3VO4). The protein concentration of the cell extracts was determined by the Bradford dye method (Bio-Rad Laboratories Inc.). An equal amount of protein (50 μg) was resolved on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels and visualized by Western analysis with anti-JNK1 antibodies (SC-571) or anti-JNK2 antibodies (SC-827) (Santa Cruz Biotechnology Inc.).
Jun kinase assays.
Cells were transfected with the oligonucleotides as described above. Treatment with 0.1 μM rhEGF or UV-C (100 J/m2) was performed 36 h after transfection and at the indicated times the cells were washed twice with ice-cold phosphate-buffered saline (PBS) and suspended in the same lysis buffer used for the Western analysis described above. The protein concentrations of the cell extracts were determined by the Bradford dye method. Typically all preparations yielded very similar protein concentrations and were adjusted to provide equal amounts of cellular protein in all samples prior to analysis. The kinase assay was performed as described by Hibi et al. (37). Briefly, 50 μg of whole-cell extract was mixed with 10 μg of glutathione S-transferase (GST)–c-Jun(1–223) for 3 h at 4°C. GST–c-Jun fusion proteins were previously expressed and purified from Escherichia coli and bound to glutathione Sepharose 4B beads (Pharmacia Biotech Inc.). After four washes, the beads were incubated with 30 μl of kinase reaction buffer (20 mM HEPES [pH 7.7], 20 mM MgCl2, 20 mM β-glycerophosphate, 20 mM p-nitrophenyl phosphate, 0.1 mM Na3VO4, 2 mM dithiothreitol, 20 μM ATP, and 5 μCi of [γ32P]ATP) for 20 min at 30°C. The reaction was stopped by addition of 20 μl of Laemmli sample buffer as described previously (93). The phosphorylated GST–c-Jun protein was eluted by boiling the sample for 5 min and resolved by a SDS–10% polyacrylamide gel electrophoresis.
For the in-gel Jun kinase assay, 100 μg of cell extracts was resolved on a SDS–12% polyacrylamide gel containing 200 μg of purified GST–c-Jun(1–79) protein. The gel was then treated according to the procedure described by Hibi et al. (37). The quantification of 32P-phosphorylated GST–c-Jun was carried out by digitalization of the autoradiograph of the dried gel. Background values were subtracted in all cases.
Proliferation assay.
Cells were seeded in 24-well tissue culture plates in the presence of DMEM supplemented with 5% FBS. One day later, they were transfected with 0.4 μM JNKAS, each JNK scrambled oligonucleotide, and each JNK sense oligonucleotide. As described above, after the 4.5 h of lipofection, the cells were transferred to DMEM supplemented with 0.5% FBS. The next day, the desired volume of rhEGF was added to the cells to a final concentration of 0.1 μM, and the medium was not changed during the experiment. Five days later, the cells were counted with a Coulter counter. For each experiment and all conditions, triplicate wells were counted.
Tumorigenicity in mice.
A549 cells (5 × 106) were injected subcutaneously into groups of five to six female nude mice (nu/nu; Harlan Sprague Dawley) on two sites on the backs of the animals. rhEGF (lot 913377; Collaborative Biomedical Products) was injected intraperitoneally daily at 100 to 300 μg of rhEGF/kg of body weight/day dissolved in PBS containing 0.1% bovine serum albumin (PBS-BSA). Control animals were injected with an equal volume of the PBS-BSA vehicle solution. Eight days after the injection, the presence of palpable tumors was scored.
Growth on soft agar.
Soft-agar assays were carried out in 12-well plates previously lined with DMEM (0.6% agar) (38). Twenty-four hours after transfection with oligonucleotides, 2 × 103 A549 cells were seeded in DMEM containing 0.3% agarose and 0.8 μM JNKAS or JNKScr oligonucleotides or 0.4 μM (both) oligonucleotides. Next, 0.5 ml of DMEM, with or without 0.1 μM rhEGF, was layered over the cell-containing agar layer. Every 5 days 0.1 μM fresh rhEGF was added to the medium. After 21 days of incubation in 5% CO2, the colonies were stained overnight at 37°C with 0.5 mg of p-iodotetrazolium violet/ml and measured by determination of their area with the IPLabSpectrum software (Scanalytics Inc.).
RESULTS
Antisense inhibition of JNK1 and JNK2 mRNA expression.
To identify effective antisense oligonucleotides capable of inhibiting specifically JNK1 or JNK2 mRNA expression in A549 cells, 26 phosphorothioate oligonucleotides complementary to JNK1 and 13 phosphorothioate oligonucleotides complementary to JNK2 were prepared (Fig. 1). All oligonucleotides were used at 0.2 μM average concentration to transfect A549 cells, as described in Materials and Methods. Steady-state mRNA levels were determined by Northern analysis, as previously described (17, 60). The most potent oligonucleotide of the array complementary to JNK1 is ISIS 12539 (JNK1ASISIS12539) (5′-CTCTCTGTAGGCCCGCTTGG-3′), located in the 3′ end of the coding region. This oligonucleotide decreased the steady-state mRNA level by 95% of the steady-state level of JNK1 mRNA of untreated control cells (Fig. 1A).
ISIS 12560 (JNK2ASISIS12560) (5′-GTCCGGGCCAGGCCAAAGTC-3′), located in the 5′ end of the coding region of JNK2 genes, was the most efficient oligonucleotide in reducing JNK2 steady-state mRNA levels. Lipofection with this oligonucleotide led to a decrease in the steady-state JNK2 message level by 92% compared to the control JNK2 steady-state mRNA level.
Figure 1 suggests that inhibition of steady-state mRNA requires specific features of both the optimal oligonucleotide sequence and the structure of the target mRNA. First, the oligonucleotides targeted to the sequences flanking either side of the sequences for the optimal oligonucleotides, JNK1ASISIS12539 and JNK2ASISIS12560, inhibited JNK1 and JNK2 steady-state mRNA, but in a manner that decreased with increasing distance from the optimum sequence (Fig. 1). This suggested that discrete regions of the target mRNA are accessible and sensitive to the antisense-mediated elimination of steady-state mRNA, consistent with previous observations (15, 16). Second, five oligonucleotides have very similar nucleotide compositions, within 2 to 4 of 20 bases, to that of JNK1ASISIS12539. However, the average expression of mRNA by cells treated identically with these oligonucleotides (Fig. 1A) is 31%, compared to 9% with JNK1ASISIS12539. In the case of JNK2, the optimal oligonucleotide exhibits mRNA suppression to 11% whereas the one with the closest composition, differing by 2 nucleotides of 20 (Fig. 1B), is one of the least efficient, with posttreatment mRNA levels remaining at 60% of the basal levels. These observations suggest that optimal suppression of steady-state mRNA depends upon strict sequence-specific requirements as well as structure requirements. In order to confirm that the optimal oligonucleotides work in a strictly sequence-specific way, dose-response and cross-inhibition tests were carried out.
Specificity of JNK1 and JNK2 antisense inhibition.
A549 cells were transfected with JNK1ASISIS12539 or JNK2ASISIS12560 oligonucleotides at various concentrations from 0.05 to 0.4 μM. Control oligonucleotides consisting of the scrambled-sequence version of each antisense oligonucleotide were prepared: JNK1ScrISIS14321 (5′-CTTTCCGTTGGACCCCTGGG-3′) and JNK2ScrISIS14319 (5′-GTGCGCGCGAGCCCGAAATC-3′). Additional controls consisting of oligonucleotides with the target or sense sequence were synthesized as JNK1SeISIS14320 (5′-CCAAGCGGGCCTACAGAGAG-3′) and JNK2SeISIS14318 (5′-GACTTTGGCCTGGCCCGGAC-3′). Treatment of A549 cells with JNK1ASISIS12539 or JNK2ASISIS12560 caused a dose-dependent reduction of JNK1 and JNK2 steady-state mRNA levels, respectively, with a 50% effective concentration (EC50) of 0.1 μM in both cases (Fig. 2A). Greater inhibition of about 85% of the control steady-state level was observed at 0.2 μM, with a maximum of near-complete elimination of steady-state target sequences at 0.4 μM (Fig. 2A). In contrast, treatment with four different control oligonucleotides (scrambled and sense) had no systematic effect on the steady-state mRNA levels of JNK even when applied at the highest concentrations for the antisense oligonucleotides used here (Fig. 2A).
FIG. 2.
Dose-dependent reduction of JNK1 and JNK2 mRNAs and specific elimination of JNK1AS and JNK2AS steady-state mRNA levels. (A) Dose-response curve for JNK1 mRNA level after treatment of A549 cells with different concentrations of JNK1ASISIS12539 (◊), JNK1ScrISIS14321 (□), and JNK1SeISIS14320 (▵). The mRNAs were prepared 24 h after a 4-h transfection, examined by Northern analysis, quantified, and normalized to JNK1 mRNA levels in untreated A549 cells. A similar experiment was carried out for JNK2 mRNA with JNK2ASISIS12560 (◊), JNK2ScrISIS14319 (□) and JNK2SeISIS14318 (▵). (B) A549 cells were treated with three different antisense oligonucleotides complementary to JNK1 mRNA (including the active antisense oligonucleotide JNK1ASISIS12539) and three different antisense oligonucleotides complementary to JNK2 mRNA (including the active antisense oligonucleotide JNK2ASISIS12560). Twenty-four hours after a 4-h transfection with 0.4 μM antisense oligonucleotide, mRNA was prepared and examined by Northern analysis; the same membrane was hybridized successively with JNK1 probe, JNK2 probe, and the G3PDH probe.
These results indicate that JNK1ASISIS12539 and JNK2ASISIS12560 effectively promote elimination of their respective target mRNAs at low concentration. Furthermore, the results showing that the sense and scrambled control sequence have no effect on JNK mRNA levels indicate that the effects of the antisense oligonucleotides are specific.
JNK1ASISIS12539 and JNK2ASISIS12560 are not complementary to any portion of the JNK2 and JNK1 mRNA sequences, respectively, and therefore, neither is expected to alter the mRNA levels of the other isoform. Indeed, the optimum JNK1AS oligonucleotide and two other randomly chosen antisense oligonucleotides complementary to different regions of JNK1 mRNA did not alter JNK2 mRNA levels (Fig. 2B). Similarly, JNK2ASISIS12560 and two other antisense oligonucleotides complementary to JNK2 mRNA and identified in the initial screening did not alter JNK1 mRNA levels (Fig. 2B). Thus, these cross-inhibition studies confirmed that the antisense oligonucleotides specifically induced the elimination of their respective mRNAs with high affinity (an EC50 of <0.1 μM).
Long-term elimination of JNK1 and JNK2 steady-state mRNA levels by JNKAS oligonucleotides.
Since JNK1ASISIS12539 and JNK2ASISIS12560 appear to specifically eliminate steady-state mRNA, we asked how long this inhibition lasted following a single treatment. We performed a time course experiment after a 4.5-h lipofection of A549 cells with 0.4 μM JNK1ASISIS12539 or JNK2ASISIS12560. mRNA levels were determined by Northern analysis at 4, 12, 24, 48, and 72 h after lipofection. Application of JNK1ASISIS12539 and JNK2ASISIS12560 rapidly decreased JNK1 and JNK2 mRNAs, respectively, with a maximum inhibition of 95% of the control level at 12 h (Fig. 3).
JNK1 mRNA remained under 25% of the control level for up to 48 h and stayed below 50% for 72 h. JNK2 messenger remained below 25 and 50% at 24 and 48 h, respectively, before it returned to normal steady-state levels at 72 h (Fig. 3). These results show that the duration of suppression of steady-state mRNA following a single treatment with antisense oligonucleotides, up to 3 days, is sufficient for carrying out a variety of studies in order to determine if elimination of either mRNA is correlated with protein suppression and phenotypic changes.
Long-term and specific inhibition of JNK1 and JNK2 protein expression by antisense JNK oligonucleotides.
Since JNK1ASISIS12539 and JNK2ASISIS12560 appear to specifically eliminate steady-state mRNA for prolonged periods, we asked (i) whether these compounds specifically eliminated their respective proteins and (ii) how long this inhibition lasted following a single treatment.
Several previous studies of A549 cells showed that a 4-h transfection is sufficient to totally inhibit protein expression for at least 48 h (17, 60). Therefore, we performed a time course experiment to determine the duration of inhibition of JNK1 and JNK2 in A549 cells. The cells were transfected for 4.5 h and then harvested at 24, 48, and 72 h. Western analysis was performed as describe in Materials and Methods.
JNK1ASISIS12539 completely inhibited JNK1 (46 kDa) for up to 72 h, whereas the JNK2 level was not affected (Fig. 4A). At the same time, JNK1 scrambled oligonucleotides had no effect on JNK1 and JNK2. Conversely, JNK2ASISIS12560 specifically inhibited JNK2 protein expression for up to 48 h. However, the JNK2 protein returned to normal levels by 72 h (Fig. 4A). Neither JNK2ASISIS12560 nor JNK2ScrISIS14319 had any effect on the expression of JNK1.
Taken together, these results showed that it is possible to eliminate JNK1 and JNK2 protein in A549 cells for up to 48 h, with a prolonged effect for JNK1 up to 72 h. In addition, the results of the mRNA study (Fig. 2B) were confirmed at the protein level, showing that each antisense oligonucleotide specifically eliminated the corresponding enzyme target (Fig. 4).
In order to determine the relative affinity and optimal concentration for the antisense oligonucleotides, we transfected A549 cells with 0.05, 0.2, and 0.4 μM JNK1ASISIS15346 or JNK2ASISIS15353 following the same procedure used for the studies described above. Thirty-six hours after transfection, cell extracts were examined by Western analysis (Fig. 4B). We observed a 90% inhibition of JNK1 protein with 0.05 μM JNK1ASISIS15346 in A549 cells (Fig. 4B) and obtained a slightly better inhibition of 93% at 0.4 μM. Treatment with 0.4 μM JNK1 scrambled oligonucleotide had no effect on JNK1 and JNK2 protein levels (Fig. 4B), similar to results for JNK1 sense oligonucleotides (data not shown). These results correlate with the mRNA and time course experiments showing a strong inhibition of JNK1 protein at 0.4 μM.
Western analysis revealed a slight decrease of the 55-kDa band with JNK1ASISIS15346, corresponding to the major form of JNK2 (Fig. 4B). It has been shown by numerous studies that the two major forms of JNK, JNK1 and JNK2, have apparent molecular masses of 46 and 55 kDa, respectively (23, 31, 37, 77). However, Gupta et al. (31) described a total of 10 isoforms for JNK and found that some isoforms of JNK1 have a molecular mass of 55 kDa and some isoforms of JNK2 migrate at 46 kDa. In this study we used commercial antibodies raised against JNK1 (SC-571) and recognizing JNK2. Since no change in the JNK2 mRNA level was observed when we used JNK1ASISIS15346 oligonucleotides, we attributed the decrease of the 55-kDa band intensity to the inhibition of an isoform of JNK1 migrating at 55 kDa.
A similar experiment was carried out with JNK2ASISIS15353 oligonucleotides to block JNK2 protein expression. JNK2ASISIS15353 had almost no effect on the JNK1 protein level, whereas it greatly reduced the expression of JNK2 protein, by up to 94% compared to the control level (Fig. 4B). JNK2 scrambled oligonucleotides had no effect on any of the isoforms (Fig. 4B). Thus, the sum of results shows that JNK1 and JNK2 maximum protein inhibition is already achieved with 0.2 μM JNK1ASISIS15346 or JNK2ASISIS15353 in A549 cells. Similar results of complete elimination of JNK1 or JNK2 were obtained for human glioblastoma T98G cells (65a) and human prostate carcinoma PC3 cells (97). These results support the observations based on Northern analysis that suppression occurs with an EC50 of ≤0.1 μM, indicating that the antisense oligonucleotides selected following the “messenger walk” sampling approach (Materials and Methods) have high affinity for their respective target mRNAs. Thus, these oligonucleotides effectively and specifically eliminate their target mRNAs and proteins.
JNK2 has a predominant role in EGF- and UV-C-induced Jun kinase activity in A549 cells.
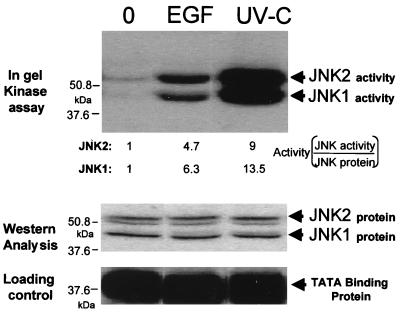
We have previously shown that rhEGF stimulates total JNK activity in A549 cells incubated with 0.1 μM rhEGF (7). In order to determine whether JNK1 and JNK2 are both activated upon addition of rhEGF, we performed an in-gel JNK assay with cell extracts from A549 cells treated with 0.1 μM rhEGF. Figure 5 shows that JNK1 and JNK2 are activated by treatment with rhEGF and UV-C. Treatment with rhEGF led to large and approximately equal activations of JNK1 and JNK2 of 6.3 and 4.7-fold induction, respectively (Fig. 5). The difference in favor of JNK1 may be due to a slightly higher basal level of activity exhibited by JNK2 compared to JNK1 (Fig. 5). Treatment with UV-C led to even larger activations, approximately two times greater than that for the treatment with rhEGF, with 13.5 and 9-fold induction, respectively. These results clearly showed that both JNK1 and JNK2 are activated to similar degrees by rhEGF and UV-C irradiation in A549 cells. We asked, therefore, whether the specific antisense oligonucleotides characterized here could be used to dissect the contributions of the JNK isoforms to total JNK activity and proliferation.
FIG. 5.
Activation of JNK1 and JNK2 by rhEGF and UV-C. A549 cells were stimulated with 0.1 μM rhEGF or UV-C at 100 J/m2. Twenty minutes after stimulation, cell extracts were prepared and used for in-gel kinase assay with GST–c-Jun as a substrate, as described in Materials and Methods. The activation of each isoform (Activity) was calculated by normalization to the corresponding amount of JNK protein shown in the lower panel (Western Analysis).
A549 cells were transfected with 0.2 μM JNK1ASISIS15346 or JNK2ASISIS15353. As a control, JNK1ScrISIS18076 and JNK2ScrISIS18078 oligonucleotides were transfected at the same concentration. For the combination of the two oligonucleotides we used 0.1 μM each to have the same final total concentration of 0.2 μM. Thirty-six hours after the lipofection, a JNK kinase assay was performed on cells treated or not treated with 0.1 μM rhEGF or UV-C (100 J/m2).
Figure 6 shows that in A549 cells JNK kinase is activated with the same intensity whether the cells were treated with Lipofectin, JNK1ScrISIS18076, JNK2ScrISIS18078, or the combination of the two scrambled control oligonucleotides. The difference in JNK activation between rhEGF and UV-C and the level of activation we observed was similar to previous results (7). Cells transfected with JNK1ASISIS15346 showed a 60 and 76% activation by rhEGF and UV-C, respectively, compared to the activation obtained with the control oligonucleotide, reflecting an inhibition of 40 and 34% of total JNK activity. JNK2ASISIS15353 had a more dramatic effect, inhibiting 71 and 62% of the rhEGF and UV-C-induced activity, respectively, and showing about twice the inhibition observed with JNK1ASISIS15346. Combined antisense treatment led to 63 and 50% inhibition of JNK activity induced by rhEGF and UV-C, respectively. Thus, combined antisense treatment yielded a smaller inhibition than JNK2ASISIS15353 alone. This may be expected, since in the combined-treatment case, each oligonucleotide is used at half of the usual concentration (0.1 μM) and only one oligonucleotide, JNK2ASISIS15353, has a major effect. Complete inhibition can be obtained upon use of 0.2 μM (each) oligonucleotide (7). In addition, we noticed that the treatment with either of the JNK scrambled control oligonucleotides did not affect rhEGF and UV-C JNK activation compared to A549 cells treated with Lipofectin (Fig. 6) or untreated cells (data not shown).
These results suggest a predominant role of JNK2 in A549 cells and further suggest that JNK2 may be the major JNK isoform mediating EGF-stimulated growth.
JNK2 inhibition and not JNK1 inhibition blocks EGF-induced proliferation in A549 cells.
To determine the effects of JNK1ASISIS12539 and JNK2ASISIS12560 on cell growth, we treated A549 cells with either JNK1ASISIS12539 or JNK2ASISIS12560 or their respective scrambled control oligonucleotides. Since at 72 after the transfection JNK2 mRNA and protein levels are back to normal, we have a limited window of 3 days (Fig. 3 and 4A) for observing the effect of the suppression of JNK on phenotype. Methods of assessing growth such as tritium uptake and DNA synthesis may well exhibit recovery after 72 h, whereas direct proliferation measurement by cell counting retains the consequences of any previously limited rounds of cell doubling. Moreover, proliferation is a direct expression of the phenotype we want to assess. Therefore, all cells were counted 5 days after transfection with antisense oligonucleotides or Lipofectin. In the absence of rhEGF, i.e., basal proliferation, no difference in proliferation was observed for cells transfected with either antisense oligonucleotides or scrambled and sense sequence controls or for cells treated with Lipofectin (Fig. 7). Moreover, these values are similar to the basal proliferation of A549 cells that stably express a dominant-negative inhibitor, c-Jun(S63A,S73A), of the JNK pathway (Fig. 7) (7). This confirms the previous observation that JNK1 and JNK2 are not required by A549 cells in the absence of EGF. Furthermore, transfection with the oligonucleotides as well as mock transfection or exposure to phosphorothioate had very little effect on basal cell growth, showing that the antisense oligonucleotides and other conditions have no toxic effects on A549 cell growth.
Addition of 0.1 μM rhEGF stimulated proliferation by 164 to 200% in parental cells, cells treated with Lipofectin alone, and cells treated with JNK1Scr, JNK2Scr, JNK1Se, JNK2Se, and combined scrambled or sense oligonucleotides. In all cases, the increase in growth is significant (P < 0.002). As previously shown (7), cells transfected with JNK1AS and JNK2AS (0.2 plus 0.2 μM) failed to respond to the presence of rhEGF (Fig. 7). More interestingly, for cells treated with 0.4 μM JNK2ASISIS12560 alone, the effect of rhEGF on growth is also completely inhibited (Fig. 7). In contrast, for cells treated with an equal amount of JNK1ASISIS12539, the effect of rhEGF led to a 145% increase in proliferation. The growth increase upon addition of rhEGF in JNK1ASISIS12539-treated cells is significant (P < 0.05) (Fig. 7).
This result shows that specifically blocking JNK2 alone caused a readily detectable decrease in proliferation, suggesting that JNK2 mediates signal transduction in EGF-stimulated growth. Moreover when JNK2 protein and activity are specifically eliminated, EGF-induced proliferation is completely inhibited, indicating that JNK2 is the predominant mediator of EGF-induced growth in A549 cells.
JNK2 and not JNK1 is required for EGF-stimulated growth of colonies in soft agar.
Numerous studies have shown that growth in soft agar is a very good model to study tumorigenicity and is closely associated with the transformed property of cells (6, 36, 72). In order to determine whether EGF-dependent colony formation likely reflects transformation of A549 cells, the effect of rhEGF on tumor growth in vivo was examined. We injected A549 cells into nude mice and treated the animals daily with or without different concentration rhEGF. Animals treated with 100 μg of rhEGF/kg/day developed tumors earlier than control animals injected with saline solution (PBS containing 0.1% BSA). As shown in Fig. 8A, 83% of the 12 injection sites exhibited visible tumors only 8 days after the injection whereas only 50% of the 14 sites in control animals developed tumors (P < 0.021). Moreover, the tumors grew approximately twice as fast in animals treated with 100 μg of rhEGF/kg/day (data not shown). It is well known that high doses of EGF can down regulate the expression of its receptor or lead to its internalization in vitro and in vivo (35, 43, 63, 64, 82). Indeed, at higher doses of rhEGF tumorigenicity declined (Fig. 8A).
Taken together, these results show that the optimal dose of 100 μg of rhEGF/kg/day injected into animals promotes A549 tumorigenicity in nude mice, showing that EGF promotes the transformed phenotype of these cells.
To determine the effect of JNK isoforms on anchorage-independent growth, we performed a soft-agar colony formation assay in the absence or presence of rhEGF with cells treated with oligonucleotides. In previous studies (5, 12, 65), it has been shown that the direct application of oligonucleotides to cells grown in soft agar promotes efficient and specific regulation of growth. We combined this method (see Materials and Methods) with our usual lipofection procedure (Fig. 8B and C). Parental A549 cells do not make many colonies in soft agar, and those produced are very small and a little ragged (Fig. 8C). The addition of rhEGF stimulated increased colony size (Fig. 8B and C) but did not greatly affect the number of colonies. We next tested the effects of the unmodified oligonucleotides JNK1ASISIS12539 and JNK2ASISIS12560 on EGF-induced colony growth. Twenty-four hours before the cells were seeded in agarose they were transfected with 0.4 μM oligonucleotides and then transferred to 0.3% agarose–DMEM containing 0.8 μM oligonucleotide. Then colonies were grown in the presence or absence of 0.1 μM rhEGF, which was renewed every 5 days. After 3 weeks, the cells were stained by addition of a tetrazolium dye and incubation at 37°C. The vast majority of all cells were stained positively with the tetrazolium dye, indicating that all cells remained viable over the course of the experiment independent of the differences in the treatment regimens. The colonies were analyzed by computer with IPLab3 Spectrum software.
We noticed fewer colonies in wells treated with either scrambled or antisense oligonucleotides or Lipofectin alone (data not shown) and also that the colonies were smaller (Fig. 8B), suggesting a small toxic effect of all such additions in these prolonged 3-week assays. No significant difference was noticed in colony size in the antisense-oligonucleotide-treated wells of the no-EGF group and the EGF-treated wells. Despite this effect, cells transfected with JNK1ASISIS12539, JNK1ScrISIS14321, JNK2ScrISIS14319, or the combination of scrambled oligonucleotides and rhEGF made significantly larger colonies, up to 195% (P < 0.006) (Fig. 8B). A similar increase of 197% was obtained with A549 parental cells following treatment with rhEGF (P < 0.001) (Fig. 8B and C). Conversely, cells transfected with JNK2ASISIS12560 or the combination of antisense oligonucleotides did not exhibit any significant effect of rhEGF on colony size (P < 0.25 for JNK2ASISIS12560 and P < 0.3 for combined treatment) (Fig. 8B and C).
The sum of results shows a parallel approximately twofold increase in tumorigenicity and colony formation in soft agar upon treatment of A549 cells with rhEGF. Moreover, inhibition of the expression of JNK2 and not JNK1 largely eliminated the ability of EGF to promote growth and transformed phenotype in A549 cells. These results strongly indicate that JNK2 plays a critical role in EGF-induced proliferation, including the growth of colonies in soft agar, suggesting an essential role in mediating the transformed phenotype of these cells.
DISCUSSION
We have shown in the present study that selected phosphorothioate antisense oligonucleotides specifically inhibited the expression of JNK1 or JNK2 mRNA and proteins. The use of antisense oligonucleotides is attractive because they provide a way of inhibiting the expression of any protein of choice through their potential for specific hybridization (97). However, controversies have developed regarding the specificity of oligonucleotides and their interaction with cellular proteins (71, 80, 81).
We selected antisense oligonucleotides following procedures successfully used in previous studies (17–19, 57, 60). The 2 most potent antisense inhibitors against JNK1 and JNK2 were identified from a total of 39 oligonucleotides screened by Northern analysis for their ability to inhibit JNK1 or JNK2 mRNA. Secondly, once selected, the two antisense oligonucleotides were used in parallel with two different kinds of control oligonucleotides, scrambled oligonucleotides that have the same nucleotide composition but in a different order and sense oligonucleotides. We showed that JNK1ASISIS12539 and JNK2ASISIS12560 decreased steady-state mRNA levels in a dose-dependent manner whereas the scrambled and sense control oligonucleotides had no effect. Thirdly, JNKAS oligonucleotides were shown to eliminate specifically their respective target mRNAs and proteins but not closely related isoforms. Thus, JNK1ASISIS12539 greatly reduced the level of JNK1 but not JNK2 and JNK2ASISIS12560 did not interfere with JNK1 but dramatically affected JNK2 levels. Finally, it was confirmed that the JNKAS oligonucleotides are highly effective in reducing JNK activity whereas scrambled oligonucleotides did not affect EGF and UV-C induction of total JNK activity in A549 cells.
Recently, classical phosphorothioate oligonucleotides have been optimized by the introduction of 2′-O-methoxyethyl groups in the first and last 5 nucleotides. These second-generation phosphorothioate oligonucleotides have improved biophysical characteristics that result in enhanced affinity for the target mRNA and, therefore, can be used at a lower concentration (45, 59). This effect is illustrated in Fig. 4B for JNK1AS, where we obtained elimination of JNK1 protein with 0.05 μM modified phosphorothioate oligonucleotides whereas the EC50 is 0.1 μM with classical phosphorothioate oligonucleotides. Taken together, these characteristics demonstrate that the antisense oligonucleotides described here represent a very specific tool for discrimination between the roles of JNK1 and JNK2 in EGF-induced proliferation.
We have shown in a previous study with independent inhibitors that EGF mediates growth through the JNK pathway in A549 cells (7). In this paper we demonstrate that JNK2 plays a preferential role in A549 cells to mediate EGF effects. First, the use of specific antisense oligonucleotides effectively eliminated approximately equal contributions to total JNK activity (Fig. 6), and this is strongly supported by direct observation of the activation of JNK1 and JNK2 by in-gel kinase assays (Fig. 5), indicating that these isoforms are approximately equally activated by EGF. Second, we confirmed that the combined use of antisense JNK1 and JNK2 oligonucleotides blocked the effect of EGF on A549 cell growth. Moreover, we found that JNK2 elimination is sufficient to block EGF-induced proliferation whereas inhibition of expression of JNK1 did not significantly alter EGF-stimulated growth. Third, this conclusion was validated with soft-agar growth assays. A549 cells do not make large colonies in soft agar; however, the addition of EGF leads to colonies that are ∼200% larger. We showed that this effect could be blocked by the inhibition of JNK2 but not JNK1. Similarly, combined-antisense treatment alone appeared to be less inhibitory in one case (Fig. 8B). This effect is likely related to the use of half the usual concentration of each oligonucleotide, thereby diluting the major active factor, JNK2AS. Thus, we demonstrated in a second growth model that JNK2 preferentially mediates the effect of EGF on cancer cell growth. Indeed, this result showed that JNK2 is more important than JNK1 in mediating EGF stimulation of growth and EGF-induced transformation in A549 cells.
Autocrine and paracrine growth stimulation by EGF and related molecules has been implicated in the transformation of a variety of human tumors. EGF stimulates the proliferation of several kinds of human tumor cells, such as gliomas (50), mammary tumor cells (1), and keratinocytes (33). Interestingly, EGF plays a major role in autocrine growth of human non-small-cell lung cancer cells (73, 85) and is effective in inducing cell proliferation in lung primary tumor cells (76). The ability of many cell lines to form colonies when suspended in semisolid medium (anchorage independence) is strongly correlated with their ability to produce tumors when inoculated into animals (36, 72). In this study we have investigated A549 tumorigenicity in nude mice treated daily with or without EGF. EGF-treated animals developed earlier and bigger tumors, showing that EGF is a tumorigenic factor for A549 cells in vivo. Therefore, we have confirmed that colony growth in soft agar is a good in vitro model to mimic cancer cell proliferation in a tumor, as previously described (36, 97).
A recent report showed that EGF induction of the c-Jun promoter is mediated by MEKK, which in turn activates JNK (14). c-Jun is a transcription factor implicated in various cellular events ranging from cell proliferation and differentiation to transformation (3, 7, 78, 79). Phosphorylation of c-Jun on its N-terminal site by JNK is directly implicated in mediating EGF growth effects on A549 lung carcinoma cells. In this study we demonstrated a correlation between JNK2 preferential phosphorylation of c-Jun and EGF-induced proliferation.
It is likely that divergent roles for highly homologous members of the JNK proteins are dictated by a difference in binding affinity or phosphorylation of the cellular target. In fact, it was shown that JNK2 has higher affinity than JNK1 for c-Jun and ATF-2 (31, 41), but no difference in the phosphorylation of these substrates was observed in vitro. However, these experiments were done with CHO cell extracts from cells transfected with expression vectors bearing various JNK isoforms and therefore represent an artificial way to check endogenous protein activity. In our model we measured endogenous JNK activity, showing that JNK2 inhibition reduces c-Jun phosphorylation much more than JNK1 inhibition (Fig. 7). This suggests that c-Jun is one preferential target for JNK2 in A549 cells. Nevertheless, we cannot exclude the possibility that JNK has other crucial targets in A549 cells. Other potential candidates are ATF-2 and Elk-1, and these transcription factors are also activated by growth factors (9, 31, 48, 87). Some others targets were also reported, such as BRCA2, which has an N-terminal region highly homologous with that of c-Jun (74), and p53, found to be activated by JNK after UV irradiation of C57MG cells (54).
To our knowledge, this study demonstrates for the first time differential roles for JNK1 and JNK2 in human cancer cell growth. Based on the elimination of JNK2 in the cell, we suggest that this isozyme plays a dominant role in EGFinduced growth and activation of the JNK pathway. Using the same oligonucleotides, we found that JNK2 is critical for T98G glioblastoma cell basal growth (65a). In the present case, JNKAS had almost no effect on A549 basal cell growth, strongly indicating that these compounds, including JNK2AS, have little toxic effect on cells. It is likely that this result is due in part to the high apparent affinity and specificity of compounds characterized here, leading to their effective use at 0.1 μM. Furthermore, using nine different human prostate carcinoma cells, we have shown that JNK2 plays a preferential role over JNK1 in prostate cancer cell growth in the presence of 10% FBS (97).
Many studies have postulated a role for JNK in apoptosis (11, 40, 89, 90, 94), whereas others have shown that JNK is involved in DNA repair (2, 66) and proliferation, as shown here. In this study, using highly specific antisense oligonucleotides, we have demonstrated that JNK2 has a predominant role in mediating EGF effects on growth and transformation. Therefore, we suggest that JNK1 is under a separate regulation that may involve the control of apoptosis. This prediction and other roles of the JNKs may be testable by using methods outlined here.
ACKNOWLEDGMENTS
We thank Mary Andahazy and Wilfried Charbono for their help and useful advice. We are grateful to Eileen Adamson for critically reading the manuscript. We thank Michael Karin and Javier Piedrafita for providing GST–c-Jun and M. Weiner and D. Carlson for gifts of rhEGF. We thank Pamala Nero for technical help and Leanne Shawler for excellent editorial assistance.
This work was supported by grants CA56834 and CA76173 from the National Institutes of Health (to D.M.), la Ligue Nationale Contre le Cancer (to D.M. and F.B.), and le Conseil Régional de Haute Normandie (to F.B.). This research was also supported in part by funds from the California Breast Cancer Research Program of the University of California (3CB-0246) to D.M., the American Cancer Society, Ray and Estelle Spehar Fellowship, to F.B., and the fellowship program of the Sidney Kimmel Cancer Center.
REFERENCES
- 1.Adamson E D, Wiley L M. The EGFR gene family in embryonic cell activities. Curr Top Dev Biol. 1997;35:71–120. doi: 10.1016/s0070-2153(08)60257-4. [DOI] [PubMed] [Google Scholar]
- 2.Adler V, Polotskaya A, Kim J, Dolan L, Davis R, Pincus M, Ronai Z. Dose rate and mode of exposure are key factors in JNK activation by UV irradiation. Carcinogenesis. 1996;9:2073–2076. doi: 10.1093/carcin/17.9.2073. [DOI] [PubMed] [Google Scholar]
- 3.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochem Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 4.Antonyak M A, Moscatello D K, Wong A J. Constitutive activation of c-Jun N-terminal kinase by a mutant epidermal growth factor receptor. J Biol Chem. 1998;273:2817–2822. doi: 10.1074/jbc.273.5.2817. [DOI] [PubMed] [Google Scholar]
- 5.Baldassarre G, Bianco C, Tortora G, Ruggiero A, Moasser M, Dmitrovsky E, Bianco A R, Ciardiello F. Transfection with a cripto antisense plasmid suppresses endogenous cripto expression and inhibits transformation in a human embryonal carcinoma cell line. Int J Cancer. 1996;66:538–543. doi: 10.1002/(SICI)1097-0215(19960516)66:4<538::AID-IJC19>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Barrett J C, Crawford B D, Mixter L O, Schechtman L M, Ts’o P O, Pollack R. Correlation of in vitro growth properties and tumorigenicity of Syrian hamster cell lines. Cancer Res. 1979;39:1504–1510. [PubMed] [Google Scholar]
- 7.Bost F, McKay R, Dean N, Mercola D. The Jun Kinase/stress activated protein kinase pathway is required for epidermal growth factor stimulation of growth of human A549 lung carcinoma cells. J Biol Chem. 1997;272:33422–33429. doi: 10.1074/jbc.272.52.33422. [DOI] [PubMed] [Google Scholar]
- 8.Butterfield L, Storey B, Maas L, Heasley L E. C-Jun NH2-terminal kinase regulation of the apoptotic response of small cell lung cancer cells to ultraviolet radiation. J Biol Chem. 1997;272:10110–10116. doi: 10.1074/jbc.272.15.10110. [DOI] [PubMed] [Google Scholar]
- 9.Cavigelli M, Dolfi F, Claret F X, Karin M. Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J. 1995;14:5957–5964. doi: 10.1002/j.1460-2075.1995.tb00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan E D, Winston B W, Jarpe M B, Wynes M W, Riches D W H. Preferential activation of the p46 isoform of JNK/SAPK in mouse macrophages by TNF-α. Proc Natl Acad Sci USA. 1997;94:13169–13174. doi: 10.1073/pnas.94.24.13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y R, Meyer C F, Tan T H. Persistent activation of c-Jun N-terminal kinase 1 (JNK1) in gamma radiation-induced apoptosis. J Biol Chem. 1996;271:631–634. doi: 10.1074/jbc.271.2.631. [DOI] [PubMed] [Google Scholar]
- 12.Ciardiello F, Caputo R, Bianco R, Damiano V, Pomatico G, Pepe S, Bianco A R, Agrawal S, Mendelsohn J, Tortora G. Cooperative inhibition of renal cancer growth by anti-epidermal growth factor receptor antibody and protein kinase A antisense oligonucleotide. J Natl Cancer Inst. 1998;90:1087–94. doi: 10.1093/jnci/90.14.1087. [DOI] [PubMed] [Google Scholar]
- 13.Clark G J, Westwick J K, Der C J. p120 GAP modulates Ras activation of Jun kinases and transformation. J Biol Chem. 1997;272:1677–1681. doi: 10.1074/jbc.272.3.1677. [DOI] [PubMed] [Google Scholar]
- 14.Clarke N, Arenzana N, Hai T, Minden A, Prywes R. Epidermal growth factor induction of the c-Jun promoter by a Rac pathway. Mol Cell Biol. 1998;18:1065–1073. doi: 10.1128/mcb.18.2.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crooke S T. Molecular mechanism of antisense drugs: RNase H. Antisense Nucleic Acid Drug Dev. 1998;8:133–134. doi: 10.1089/oli.1.1998.8.133. [DOI] [PubMed] [Google Scholar]
- 16.Crooke S T. Therapeutic applications of oligonucleotides. Annu Rev Pharmacol Toxicol. 1993;32:329–376. doi: 10.1146/annurev.pa.32.040192.001553. [DOI] [PubMed] [Google Scholar]
- 17.Dean N M, McKay R, Condon T P, Bennett C F. Inhibition of protein kinase C-a expression in human A549 cells by antisense oligonucleotides inhibits induction of intracellular adhesion molecule 1 (ICAM-1) mRNA by phorbol esters. J Biol Chem. 1994;269:16146–16424. [PubMed] [Google Scholar]
- 18.Dean N M, McKay R. Inhibition of protein kinase C-a expression in mice after systemic administration of phosphorothioate antisense oligonucleotides. Proc Natl Acad Sci USA. 1994;91:11762–11766. doi: 10.1073/pnas.91.24.11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dean, N. M., R. McKay, L. Miraglia, R. Howard, S. Cooper, J. Giddings, P. Nicklin, L. Meister, R. Zeil, T. Geiger, M. Muller, and D. Fabbro. Inhibition of human tumor cell lines in nude mice by an antisense oligonucleotide inhibitor of protein kinase C-α expression. 1996. Cancer Res. 56:3499–3507. [PubMed]
- 20.Dean N M, McKay R, Holmlund J. Antisense oligonucleotides as inhibitors of genes that regulate AP-1: pharmacology and clinical development. Antisense Nucleic Acid Drug Dev. 1998;8:147–151. doi: 10.1089/oli.1.1998.8.147. [DOI] [PubMed] [Google Scholar]
- 21.Dean N M, McKay R, Miraglia L, Geiger T, Muller M, Fabbro D, Bennett C F. Antisense oligonucleotides as inhibitors of signal transduction: development from research tools to therapeutic agents. RNA interactions: ribozymes and antisense. Biochem Soc Trans. 1996;24:623–629. doi: 10.1042/bst0240623. [DOI] [PubMed] [Google Scholar]
- 22.Dérijard B, Raingeaud J, Barrett T, Wu I H, Han J, Ulevitch R J, Davis R J. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 23.Dérijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 24.Dolnick B J. Antisense agents in cancer research and therapeutics. Cancer Investig. 1991;9:185–194. doi: 10.3109/07357909109044229. [DOI] [PubMed] [Google Scholar]
- 25.Dotzlaw H, Miller T, Karvelas J, Murphy L C. Epidermal growth factor gene expression in human breast cancer biopsy samples: relationship to estrogen and progesterone receptor gene expression. Cancer Res. 1990;50:4204–4208. [PubMed] [Google Scholar]
- 26.Ekstrand A J, Sugawa N, James C D, Collins V P. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci USA. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falette N S, Artagaveytia N, Rostan M C, Garin E, Bobin J Y, Saez N. Analysis of epidermal growth factor receptor mRNA expression by polymerase chain reaction assay in 94 human breast adenocarcinoma tumors. Breast Cancer Res Treat. 1994;30:275–282. doi: 10.1007/BF00665968. [DOI] [PubMed] [Google Scholar]
- 28.Foltz I N, Gerl R E, Wieler J S, Luckach M, Salmon R A, Schrader J W. Human mitogen-activated protein kinase kinase 7 (MKK7) is highly conserved c-Jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) activated by environmental stresses and physiological stimuli. J Biol Chem. 1998;273:9344–9351. doi: 10.1074/jbc.273.15.9344. [DOI] [PubMed] [Google Scholar]
- 29.Gerwins P, Blank J L, Johnson G L. Cloning of a novel mitogen-activated protein kinase kinase kinase, MEKK4, that selectively regulates the c-Jun amino terminal kinase pathway. 1997. J Biol Chem. 1997;272:8288–8295. doi: 10.1074/jbc.272.13.8288. [DOI] [PubMed] [Google Scholar]
- 30.Gupta S, Campbel D, Dérijard B, Davis R J. Transcription factor ATF-2 by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 31.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H K, Dérijard B, Davis R J. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 32.Haeder M, Rotsch M, Bepler G, Hennnig C, Havemann K, Heimann B, Moelling K. Epidermal growth factor receptor expression in human lung cancer cell lines. Cancer Res. 1988;48:1132–1136. [PubMed] [Google Scholar]
- 33.Hashimoto K, Higashiyama S, Asada H, Hashimura E, Kobayashi Y, Sudo K, Nakagawa T, Damm D, Yoshikawa K, Taniguchi N. Heparin-binding epidermal growth factor-like growth factor is an autocrine growth factor for human keratinocytes. J Biol Chem. 1994;269:20060–20066. [PubMed] [Google Scholar]
- 34.Hélène C, Toulme J J. Specific regulation of gene expression by antisense, sense and antigene nucleic acids. Biochim Biophys Acta. 1990;1049:99–125. doi: 10.1016/0167-4781(90)90031-v. [DOI] [PubMed] [Google Scholar]
- 35.Herbst J J, Opresko L K, Walsh B J, Lauffenburger D A, Wiley H S. Regulation of postendocytic trafficking of the epidermal growth factor receptor through endosomal retention. J Biol Chem. 1994;269:12865–12873. [PubMed] [Google Scholar]
- 36.Herschman H R, Brankow D W. Colony size, cell density and nature of the tumor promoter are critical variables in expression of a transformed phenotype (focus formation) in co-culture of UV-TDTX and C3H10T1/2 cells. Carcinogenesis. 1987;8:993–998. doi: 10.1093/carcin/8.7.993. [DOI] [PubMed] [Google Scholar]
- 37.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein and UV-responsive protein kinase that bind and potentiate the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 38.Huang R P, Liu C, Fan Y, Mercola D, Adamson E D. Egr-1 negatively regulates human tumor cell growth via the DNA-binding domain. Cancer Res. 1995;55:5054–5062. [PubMed] [Google Scholar]
- 39.Jarvis W D, Auer K L, Spector M, Kunos G, Grant S, Hylemon P, Mikkelsen R, Dent P. Positive and negative regulation of JNK1 by protein kinase C and p42(MAP kinase) in adult rat hepatocytes. FEBS Lett. 1997;412:9–14. doi: 10.1016/s0014-5793(97)00705-9. [DOI] [PubMed] [Google Scholar]
- 40.Johnson N L, Gardner A M, Diener K M, Lange-Carter C A, Gleavy J, Jarpe M B, Minden A, Karin M, Zon L I, Johnson G L. Signal transduction pathways regulated by mitogen-activated/extracellular response kinase kinase kinase induce cell death. J Biol Chem. 1996;271:3229–3237. doi: 10.1074/jbc.271.6.3229. [DOI] [PubMed] [Google Scholar]
- 41.Kallunki T, Su B, Tsigelny I, Sluss H K, Dérijard B, Moore G, Davis R, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 42.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 43.Kubota T, Josui K, Fukutomi T, Kitajima M. Growth regulation by estradiol, progesterone and recombinant human epidermal growth factor of human breast carcinoma xenografts grown serially in nude mice. Anticancer Res. 1995;15:1275–1278. [PubMed] [Google Scholar]
- 44.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 45.Levesque L, Dean N M, Sasmor H, Crooke S T. Antisense oligonucleotides targeting human protein kinase C-alpha inhibit phorbol ester-induced reduction of bradykinin-evoked calcium mobilization in A549 cells. Mol Pharmacol. 1997;51:209–216. doi: 10.1124/mol.51.2.209. [DOI] [PubMed] [Google Scholar]
- 46.Libermann T A, Nusbaum H R, Razon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield M D, Ulrich A, Schlessinger J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature. 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 47.Liu X, Pogo B G T. Inhibition of erb-2 positive breast cancer cell growth by erb-2 antisense oligonucleotides. Antisense Nucleic Acid Drug Dev. 1996;6:9–16. doi: 10.1089/oli.1.1996.6.9. [DOI] [PubMed] [Google Scholar]
- 48.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Logan S K, Falasca M, Hu P, Schlessinger J. Phosphatidylinositol 3-kinase mediates epidermal growth factor-induced activation of the c-Jun N-terminal kinase signaling pathway. Mol Cell Biol. 1997;10:5784–5790. doi: 10.1128/mcb.17.10.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lund-Johansen M, Bjerkvig R, Humphrey P A, Bigner S H, Bigner D D, Laerum O D. Effect of epidermal growth factor on glioma cell growth, migration, and invasion in vitro. Cancer Res. 1990;50:6039–6044. [PubMed] [Google Scholar]
- 51.McKay R, Cummins L L, Graham M J, Lesnik E A, Owens S R, Winniman M, Dean N M. Enhanced activity of an antisense oligonucleotide targeting murine protein kinase C-α by the incorporation of 2′-O-propyl modifications. Nucleic Acids Res. 1996;24:411–417. doi: 10.1093/nar/24.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mercola D, Cohen J S. Antisense approaches to cancer gene therapy. Cancer Gene Ther. 1995;2:47–59. [PubMed] [Google Scholar]
- 53.Merlino G T, Xu Y, Richert N, Clarck A J, Ishii S, Banks-Schlegel S, Pastan I. Elevated epidermal growth factor receptor gene copy number and expression in a squamous carcinoma cell line. J Clin Investig. 1985;75:1077–1079. doi: 10.1172/JCI111770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milne D M, Campbell L E, Campbell D G, Meeks D W. P53 is phosphorylated in vitro and in vivo by an ultraviolet radiation-induced protein kinase characteristic of the c-Jun kinase, JNK1. J Biol Chem. 1995;270:5511–5518. doi: 10.1074/jbc.270.10.5511. [DOI] [PubMed] [Google Scholar]
- 55.Minden A, Lin A, Claret F X, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 56.Minden A, Lin A, Smeal T, Derijard B, Cobb M, Davis R, Karin M. c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol Cell Biol. 1994;10:6683–6688. doi: 10.1128/mcb.14.10.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 58.Monaghan P, Ormerod M G, O’Hare M J. Epidermal growth factor receptors and EGF-responsiveness of the human breast-carcinoma cell line PMC42. Int J Cancer. 1990;46:935–943. doi: 10.1002/ijc.2910460531. [DOI] [PubMed] [Google Scholar]
- 59.Monia B P. First- and second-generation antisense oligonucleotide inhibitors targeted against human c-raf kinase. Ciba Found Symp. 1997;209:107–119. doi: 10.1002/9780470515396.ch9. [DOI] [PubMed] [Google Scholar]
- 60.Monia B P, Johnston J F, Geiger T, Muller M, Fabbro D. Antitumor activity of a phosphorothioate antisense oligonucleotide targeted against C-raf kinase. Nat Med. 1996;2:668–674. doi: 10.1038/nm0696-668. [DOI] [PubMed] [Google Scholar]
- 61.Moriguchi T, Toyoshima F, Masuyama N, Hanafusa H, Gotoh Y, Nishida E. A novel SAPK/JNK kinase, MKK7, stimulated by TNFalpha and cellular stresses. EMBO J. 1997;16:7045–7053. doi: 10.1093/emboj/16.23.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagane M, Coufal F, Lin H, Bogler O, Cavenee W K, Su Huang H J. A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Cancer Res. 1996;56:5079–5086. [PubMed] [Google Scholar]
- 63.Okuda T, Onda M, Tokunaga A, Sugisaki Y. Stimulatory effect of EGF and inhibitory effect of sialoadenectomy on growth of an EGF receptor-hyperproducing human gastric cancer xenograft in nude mice. Surg Today. 1994;24:725–733. doi: 10.1007/BF01636779. [DOI] [PubMed] [Google Scholar]
- 64.Opresko L K, Chang C P, Will B H, Burke P M, Gill G N, Wiley H S. Endocytosis and lysosomal targeting of epidermal growth factor receptors are mediated by distinct sequences independent of the tyrosine kinase domain. J Biol Chem. 1995;270:4325–4333. doi: 10.1074/jbc.270.9.4325. [DOI] [PubMed] [Google Scholar]
- 65.Perez J R, Higgins-Sochaski K A, Maltese J Y, Narayanan R. Regulation of adhesion and growth of fibrosarcoma cells by NF-kappa B RelA involves transforming growth factor beta. Mol Cell Biol. 1994;8:5326–5332. doi: 10.1128/mcb.14.8.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65a.Potapova, O. Unpublished data.
- 66.Potapova O, Haghighi A, Bost F, Liu C T, Birrer M J, Gjerset R, Mercola D. The Jun kinase/stress-activated protein kinase pathway functions to regulate DNA repair and inhibition of the pathway sensitizes tumor cells to cisplatin. J Biol Chem. 1997;271:14041–14044. doi: 10.1074/jbc.272.22.14041. [DOI] [PubMed] [Google Scholar]
- 66a.Potapova, O., et al. Unpublished data.
- 67.Rabiasz G J, Langdon S P, Bartlett J M, Crew A J, Miller E P, Scott W N, Smyth J F, Miller W R. Growth control by epidermal growth factor and transforming growth factor-alpha in human lung squamous carcinoma cells. Br J Cancer. 1992;66:254–259. doi: 10.1038/bjc.1992.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raitano A B, Halpern J R, Hambuch T M, Sawyers C L. The Bcr-Abl leukemia oncogene activates Jun kinase and requires Jun for transformation. Proc Natl Acad Sci USA. 1995;92:11746–11750. doi: 10.1073/pnas.92.25.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reiss M, Stash E B, Vellucci V F, Zhou Z L. Activation of the autocrine transforming growth factor alpha pathway in human squamous carcinoma cells. Cancer Res. 1991;51:6254–6262. [PubMed] [Google Scholar]
- 70.Rodrigues G A, Park M, Schlessinger J. Activation of the JNK pathway is essential for transformation by the Met oncogene. EMBO J. 1997;16:2634–2645. doi: 10.1093/emboj/16.10.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roush W. Antisense aims for a renaissance. Science. 1997;276:1192–1193. doi: 10.1126/science.276.5316.1192. [DOI] [PubMed] [Google Scholar]
- 72.Rubin H, Arnstein P, Chu B M. High-frequency variation and population drift in a newly transformed clone of BALB/3T3 cells. Cancer Res. 1984;44:5242–5248. [PubMed] [Google Scholar]
- 73.Rusch V, Klimstra D, Venkatraman E, Pisters P W T, Langenfeld J, Dmitrovski E. Overexpression of the epidermal growth factor receptor and its ligand transforming growth factor alpha is frequent in resectable non-small cell lung cancer but does not predict tumor progression. Clin Cancer Res. 1997;3:515–522. [PubMed] [Google Scholar]
- 74.Sharan S K, Morimatsu M, Albretch U, Lim D S, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 75.Sherwin S A, Minna J D, Gazdar A F, Todaro G J. Expression of epidermal and nerve growth factor receptors and soft agar growth factor production by human lung cancer cells. Cancer Res. 1981;41:3538–3542. [PubMed] [Google Scholar]
- 76.Siegfried J M. Culture of primary lung tumors using medium conditioned by lung carcinoma cell line. J Cell Biochem. 1989;41:91–95. doi: 10.1002/jcb.240410205. [DOI] [PubMed] [Google Scholar]
- 77.Sluss H K, Barrett T, Dérijard B, Davis R J. Signal transduction by tumor necrosis factor mediated by JNK protein kinases. Mol Cell Biol. 1994;14:8376–8384. doi: 10.1128/mcb.14.12.8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smeal T, Binetruy B, Mercola D, Birrer M, Karin M. Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature. 1991;354:494–496. doi: 10.1038/354494a0. [DOI] [PubMed] [Google Scholar]
- 79.Smeal T, Binetruy B, Mercola D, Grover-Bardwick A, Heidecker G, Rapp U R, Karin M. Oncogenic-mediated signalling cascade stimulates c-Jun activity by phosphorylation of serines 63 and 73. Mol Cell Biol. 1992;12:3507–3513. doi: 10.1128/mcb.12.8.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stein C A, Cheng Y C. Antisense oligonucleotides as therapeutic agents—is the bullet really magical? Science. 1993;261:1004–1012. doi: 10.1126/science.8351515. [DOI] [PubMed] [Google Scholar]
- 81.Stein C A, Krieg A M. Problems in interpretation of data derived from in vitro and in vivo use of antisense oligodeoxynucleotides. Antisense Res Dev. 1994;4:67–69. doi: 10.1089/ard.1994.4.67. [DOI] [PubMed] [Google Scholar]
- 82.Sturtevant M A, O’Neill J W, Bier E. Down-regulation of Drosophila Egf-r mRNA levels following hyperactivated receptor signaling. Development. 1994;120:2593–2600. doi: 10.1242/dev.120.9.2593. [DOI] [PubMed] [Google Scholar]
- 83.Tanaka S, Ouchi T, Hanafusa H. Downstream of Crk adaptor signaling pathway: activation of Jun kinase by v-Crk through the guanine nucleotide exchange protein C3G. Proc Natl Acad Sci USA. 1997;94:2356–2361. doi: 10.1073/pnas.94.6.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang C K, Lippman M E. EGF family receptors and their ligands in human cancer. In: O’Malley B W, editor. Hormones and signaling. Vol. 1. San Diego, Calif: Academic Press; 1998. pp. 113–165. [Google Scholar]
- 85.Tateishi M, Ishida T, Mitsudomi T, Kaneko S, Sugimachi K. Immunohistochemical evidence of autocrine growth factors in adenocarcinoma of the human lung. Cancer Res. 1990;50:7077–7080. [PubMed] [Google Scholar]
- 86.Teng D H, Perry W L, Hogan J K, Baumgard M, Bell R, Berry S, Davis T, Frank D, Frye C, Hattier T, Hu R, Jammulapati S, Janecki T, Leavitt A, Mitchell J T, Pero R, Sexton D, Schroeder M, Su P H, Swedlund B, Kyriakis J M, Avruch J, Bartel P, Wong A K, Tavtigian S V. Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor. Cancer Res. 1997;57:4177–4182. [PubMed] [Google Scholar]
- 87.Van Dam H, Wilhemm D, Rher I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-Jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Veale D, Kerr N, Gibson G J, Harris A L. Characterization of epidermal growth factor receptor in primary human non-small cell lung cancer. Cancer Res. 1989;49:1313–1317. [PubMed] [Google Scholar]
- 89.Verheij M, Bose R, Lin X H, Yao B, Jarvis W D, Grant S, Birrer M J, Szabo E, Zon L I, Kyriakis J M, Haimovitz-Friedman A, Fuks Z, Kolesnick R N. Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature. 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 90.Wilson D J, Fortner K A, Lynch D H, Mattingly R R, Macara I G, Posada J A, Budd R C. JNK, but not MAPK, activation is associated with Fas-mediated apoptosis in human T cells. Eur J Immunol. 1996;26:989–994. doi: 10.1002/eji.1830260505. [DOI] [PubMed] [Google Scholar]
- 91.Wojtaszek P A, Heasley L E, Siriwardana G, Berl T. Dominant-negative c-Jun NH2-terminal kinase 2 sensitizes renal inner medullary collecting duct cells to hypertonicity-induced lethality independent of organic osmolyte transport. J Biol Chem. 1998;273:800–804. doi: 10.1074/jbc.273.2.800. [DOI] [PubMed] [Google Scholar]
- 92.Wong A J, Ruppert J M, Bigner S H, Grzeschik C H, Humphrey P A, Bigner D S, Vogelstein B. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci USA. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu J X, Carpenter P M, Gresens C, Keh R, Niman H, Morris J W S, Mercola D. The proto-oncogene c-fos is over-expressed in the majority of human osteosarcoma. Oncogene. 1990;5:989–1000. [PubMed] [Google Scholar]
- 94.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 95.Xu S, Robbins D, Frost J, Dang A, Lange-Carter C, Cobb M H. MEKK1 phosphorylates MEK1 and MEK2 but does not cause activation of mitogen-activated protein kinase. Proc Natl Acad Sci USA. 1995;92:6808–6812. doi: 10.1073/pnas.92.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu S, Cobb M H. MEKK1 binds directly to the c-Jun N-terminal kinases/stress-activated protein kinases. J Biol Chem. 1997;272:32056–32060. doi: 10.1074/jbc.272.51.32056. [DOI] [PubMed] [Google Scholar]
- 97.Yang Y-M, Bost F, Liu C T, Mercola D. The Jun kinase pathway promotes growth of prostate carcinoma cell lines, abstr. PD-96. Cancer Gene Ther. 1998;5:S31. [Google Scholar]
- 98.Yao R, Yoshihara M, Osada H. Specific activation of a c-Jun NH2-terminal kinase isoform and induction of neurite outgrowth in PC-12 cells by staurosporine. J Biol Chem. 1997;272:18261–18266. doi: 10.1074/jbc.272.29.18261. [DOI] [PubMed] [Google Scholar]
- 99.Zhang P, Hogan E L, Bhat N R. Activation of JNK/SAPK in primary glial cultures. II. Differential activation of kinase isoforms corresponds to their differential expression. Neurochem Res. 1998;23:219–225. doi: 10.1023/a:1022489127107. [DOI] [PubMed] [Google Scholar]