Abstract
Background
The mycobiome is the fungal component of the gut microbiome and is implicated in several autoimmune diseases. However, its role in MS has not been studied.
Methods
In this case-control observational study, we performed ITS sequencing and characterised the gut mycobiome in people with MS (pwMS) and healthy controls at baseline and after six months.
Findings
The mycobiome had significantly higher alpha diversity and inter-subject variation in pwMS than controls. Saccharomyces and Aspergillus were over-represented in pwMS. Saccharomyces was positively correlated with circulating basophils and negatively correlated with regulatory B cells, while Aspergillus was positively correlated with activated CD16+ dendritic cells in pwMS. Different mycobiome profiles, defined as mycotypes, were associated with different bacterial microbiome and immune cell subsets in the blood. Initial treatment with dimethyl fumarate, a common immunomodulatory therapy which also has fungicidal activity, did not cause uniform gut mycobiome changes across all pwMS.
Interpretation
There is an alteration of the gut mycobiome in pwMS, compared to healthy controls. Further study is required to assess any causal association of the mycobiome with MS and its direct or indirect interactions with bacteria and autoimmunity.
Funding
This work was supported by the Washington University in St. Louis Institute of Clinical and Translational Sciences, funded, in part, by Grant Number # UL1 TR000448 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award (Zhou Y, Piccio, L, Lovett-Racke A and Tarr PI); R01 NS102633-04 (Zhou Y, Piccio L); the Leon and Harriet Felman Fund for Human MS Research (Piccio L and Cross AH). Cantoni C. was supported by the National MS Society Career Transition Fellowship (TA-1805-31003) and by donations from Whitelaw Terry, Jr. / Valerie Terry Fund. Ghezzi L. was supported by the Italian Multiple Sclerosis Society research fellowship (FISM 2018/B/1) and the National Multiple Sclerosis Society Post-Doctoral Fellowship (FG- 1907-34474). Anne Cross was supported by The Manny & Rosalyn Rosenthal-Dr. John L. Trotter MS Center Chair in Neuroimmunology of the Barnes-Jewish Hospital Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Keywords: mycobiome, multiple sclerosis, gut microbiome, immune system, fungi
Research in context.
Evidence before this study
Many studies reported that the gut bacterial microbiome plays an important role in MS. The gut mycobiome is the fungal component of the microbiome, which characteristics – such as compositions, abundances, and interaction with the immune system – in pwMS have not been studied yet.
Added value of this study
This is the first study that comprehensively characterised the gut mycobiome in pwMS and healthy controls, at baseline and after six months. We found altered fungal compositions and diversity in pwMS, and we evaluated mycobiome short-term stability. We also found specific fungi-bacteria communities associations, and a number of novel fungi-immune, and fungi-diet correlations.
Implication of all the available evidence
Our findings bring a new perspective in studying the entire microbiome in MS, highlighting a potentially important role of mycobiome in MS and its interactions with host's immune and dietary factors.
Alt-text: Unlabelled box
1. Introduction
Multiple sclerosis (MS) is a chronic autoimmune disease of the central nervous system (CNS) with an elusive aetiology. The gut microbiome has a strong capacity to modulate host's local and systemic immune responses, and has been postulated to be an essential player in MS pathogenesis as well as a novel therapeutic target [1]. Previous work on the role of the microbiome largely focused on the bacterial microbiota, while the characteristics of other important components of the microbiome, such as fungi, remain undefined in MS [2], [3], [4].
The diverse population of fungi residing in the gastrointestinal tract is collectively termed the gut mycobiome. It accounts for ∼0·1% of gut microbiota and is ubiquitous in all human populations [5,6]. Saccharomyces, Malassezia and Candida are the three major fungal genera in the healthy human gut [5]. Although the mycobiome co-colonizes with bacterial microbiome, mycobiome-bacteria and mycobiome- host interactions have not been well studied. Several studies suggest mutual or competitive relationships between gut mycobiome and bacteria [7]. The mycobiome produces alcohol, antimicrobial peptides, and other metabolites that affect bacteria colonization [8,9]. Bacteria, in turn, generate fatty acids that regulate fungal germination and hyphal growth [10,11]. The commensal mycobiome, such as Candida albicans or Saccharomyces cerevisiae, can alleviate gut mucosal injuries from dextran sodium sulphate (DSS)-induced colitis in mice and exhibit immune modulatory properties [12], suggesting the mycobiome ability to maintain gut homeostasis. In contrast, Saccharomyces can exacerbate gut inflammation and increase gut permeability in an animal model of inflammatory bowel disease (IBD) through overproduction of uric acid [13].
The gut mycobiome profile varies, depending on age, gender, diet, medication, and disease status. Dysbiosis of the gut mycobiome has been implicated in various human metabolic or autoimmune conditions including IBD [14], [15], [16], irritable bowel syndrome (IBS) [17,18], cancer [19], [20], [21], hepatitis B and HIV infections [22], [23], [24], [25], and alcoholic liver disease [26]. It is also linked to neurological disorders including Rett syndrome (RTT) [27,28], autism spectrum disorders [29], [30], [31], and schizophrenia [32], [33], [34]. Multiple lines of evidence suggest the possibility that fungi may be involved in the pathogenesis of MS. For example, a conserved epitope within fungi may trigger autoimmune response in the host through molecular mimicry or epitope spreading mechanisms [35]. In addition, HLA-DRB1*15 allele group, the most important genetic risk factor of MS, is associated with increased sensitivity to infection by Aspergillus fumigatus [36], a commensal fungus commonly found in the gut [37]. Furthermore, C. albicans induces encephalitogenic cytokines production in the periphery and CNS, resulting in severe disease course in a murine MS model [38]. Interestingly, dimethyl fumarate, a fungicide repurposed as a disease-modifying therapy (DMT) for MS, reduces the MS relapse rate [39]. These findings support the view of fungi as infectious agents that are potentially involved in the development of MS. However, to date, a comprehensive characterization of gut commensal mycobiome is lacking in people with MS (pwMS). It is also unclear if the gut mycobiome is affected by diet or other clinical characteristics of pwMS, and how they interact with gut bacterial microbiome and host immune response.
We hypothesise that the gut mycobiome is altered in pwMS considering the evidence of mycobiome changes in other autoimmune diseases, immune regulatory properties of the mycobiome, and bacteria-fungi interaction in the gut. We longitudinally characterised the mycobiome in a cohort of pwMS and healthy controls over six months. Herein, we report altered gut mycobiome composition and diversity in pwMS not on DMT compared to healthy controls at baseline, and mycobiome stability over time in pwMS. We identified correlations of the gut mycobiome with bacterial microbiota, peripheral immune responses, and dietary patterns. We demonstrated a disrupted mycobiome-autoimmune pattern in pwMS compared to healthy controls.
2. Methods
2.1. Ethics
The study was approved by the Human Research Protection Office at Washington University in St. Louis School of Medicine (WUSM) (approval number: 201502105). All patients gave informed consent to participation.
Subject enrolment and sample collections
People with MS were recruited at the John L. Trotter MS Center at WUSM from October 2015 to June 2017. Inclusion criteria for participating in the study were:
(1) diagnosis of MS using the 2010 revision of the McDonald criteria; (2) no DMT or steroid treatments in the past 3 months; (3) ages 18 to 50 years; and (4) not in clinical relapse at study enrolment. Exclusion criteria were: (1) coexistence of other chronic inflammatory (e.g. asthma, chronic hepatitis, inflammatory bowel disease, celiac disease) and autoimmune (e.g. rheumatoid arthritis, systemic lupus erythematosus, type I diabetes), or metabolic (e.g. type II diabetes, familial hypercholesterolemia) diseases; (2) antibiotics or steroid therapy in the past 3 months; (3) history of immunosuppressive or chemotherapeutic treatment;
(4) history of chronic infectious disease (e.g. TBC, HIV, HBV, HCV); (5) neoplastic disease not in complete remission, and (6) pregnancy. Age, gender, BMI, and ethnicity matched healthy controls were enrolled using the same exclusion criteria. After six months, pwMS gave another sample (Supplementary Fig. 1). No relapse was reported in all MS patients during the study period. This was a pilot study and since no previous study was done on the mycobiome in MS, no power calculation was performed.
2.2. Stool collection, mycobiome sequencing, and data processing
Stools were self-collected and shipped on frozen gel packs overnight to the research laboratory. The stools were stored in -80°C until DNA extraction. DNA extraction of the stool samples was conducted using the MOBIO PowerSoil DNA Extraction kit. The gut mycobiome was profiled by ITS1 amplicon sequencing using primer sets (ITS: F’ (18S-F) – GTAAAAGTCGTAACAAGGTTTC, R’ (5·8S-1R) – GTTCAAAGAYTCGATGATTCAC) with inbuilt
Illumina adapters and barcodes. Dual indexed paired-end ITS1 libraries were made using AccuPrime HiFi Taq (Cat # 12346094, Thermofisher Scientific). ITS1 libraries were prepared and sequenced on the Illumina MiSeq sequencing platform using v3-600 cycle reagents (cat # MS-102–3003, Illumina Inc.) paired end sequencing protocol with a target read depth of 10,000 reads/sample.
Sequences from each sample were trimmed of primers and low-quality bases using Trimmomatic. Paired end trimmed reads were assembled using FLASH. Host contaminant/Chimera sequences were filtered out using BMTagger and Usearch/Uchime respectively. The processed and cleaned ITS1 sequences were classified from phylum to genus level using UNITE database (https://unite.ut.ee/repository.php).
2.3. Blood collection and immunophenotyping
Blood was collected in heparinized tubes, insulated, and shipped at room temperature overnight. Peripheral blood mononuclear cells (PBMCs) were isolated immediately on arrival for immunophenotyping. We used seven flow cytometry antibody panels to analyse lymphocyte and monocyte populations. Panel A included: V450-CD3 (UCHT1), PECy7-CD19 (SJ25C1), FITC-CD56 (NCAM16·2), and APC-H7-CD27 (M-T271). Panel B included: V450-CD3 (UCHT1), V500- CD4 (L200), PECy7-CD8 (RPA-T8), APC-CD45RA (HI100), and APC-H7-CD27 (M-T271). Panel C included: V450-CD3 (UCHT1), V500-CD4 (L200), FITC-CD25 (M-A251), and PE-FOXP3 (PCH101). Panel D included: V450-CD3 (UCHT1), V500-CD4 (L200), PECy7-CD8 (RPA-T8), APC-CD45RA (HI100), FITC-Tbet (4B10), PE- IFNγ (4S.B3), PerCP-Cy5·5-GMCSF (BVD2-21C11), and APC-Cy7- IL17 (BL168). Panel E included: V450-CD3 (UCHT1), PECy7-CD19 (SJ25C1), PerCP-Cy5·5-CD5 (L17F12), PE-CD1d (42·1), APC-H7-CD27 (M-T271), FITC-IL10 (BT-10), and APC-IL35/27(EB13). Panel F included: V450-CD3 (UCHT1), V450-CD19 (SJ25C1), PE-HLA-DR (G46-6), PE-Cy7-CD11c (B- ly6), PerCP-CD14 (MφP9), APC-CD123 (7G3), and FITC-BDCA2 (201A). Panel G included: V450-CD3 (UCHT1), V450-CD19 (SJ25C1), PE-HLA-DR (G46-6), PE-Cy7-CD11c (B-ly-6), PerCP-CD14 (MφP9), APC-CD16 (B73·1), and APC-H7-CD80 (L307·4). For panel D, the PBMCs were stimulated with 50 ng/ml of PMA (Sigma cat#16561-29-8) and 1 μg/ml of ionomycin (Sigma cat#I0634), and treated with 0·2ul of Golgi plug (BD cat#555029) for four hours before staining. For Panel E, we stimulated PBMCs with 50 ng/ml of PMA (Sigma cat#16561-29-8), 1 μg/ml of ionomycin (Sigma cat#I0634), and CpG for four hours before staining. Gating strategy can be found in (Supplementary Fig. 2).
2.4. Food frequency questionnaire
A food-frequency questionnaire was self-recorded on four consecutive days (two weekend days and two weekdays) before the stool sample collection at baseline and at six months. The food diary was converted to daily food serving based on the Nutrition Coordinating Center (NCC) Food Group Serving Count System.
2.5. Statistics
ITS1 sequences in each sample were rarefied to 10,000 reads to correct for uneven sample depths and then converted to relative abundances. Descriptive and formal statistical tests were performed to understand the mycobiome compositions, their difference between groups, correlations with diet and immune-phenotype.
Descriptive analysis. The mean relative abundances of the mycobiome were used to show the overall compositions of the stool mycobiome at genus level and phylum by barplot. The most abundant 20 genera were shown, including Unclassified fungi genera as one group. The barplots of MS patients from baseline to six months later were also shown to depict which and how the fungi genera changed. The fungi difference in obesity boxplots were determined by classifying patients’ BMI as normal (BMI < 25), overweight (BMI between 25 and 30), and obese (BMI > 30). The top 25 fungi were tested to find a difference between obese (BMI > 30) and non-obese (BMI < 30) samples.
Mycobiome diversity. Alpha diversity was evaluated by richness and Shannon diversity at genus level. To test inter-subject variation between MS and controls, beta diversity was measured using beta-disper function in vegan package in R.
High dimension data analysis. Nonmetric multidimensional scaling (NMDS) was used to view sample cluster patterns (similarity between samples). The metaMDS function from the vegan package and the isoMDS from the MASS package found the nonmetric multidimensional scaling (NMDS) of our samples’ Bray-Curtis dissimilarity matrices. Statistical significance of the overall mycobiome structure between compared groups or variables were conducted by PERMANOVA using Adonis function in Vegan package.
Wilcoxon sum rank test. Wilcoxon sum rank test was conducted to compare alpha diversity of MS and controls, identification of specific mycobiome (at least 1% mean abundance) difference between MS and controls at baseline, and identification of bacterial difference (at least 5% bacterial mean abundance) or immune cells population or cytokine difference between two mycotypes. To compare the mycobiome changes (at least 1% mean abundance) from baseline to six months, a paired sample t-test was conducted.
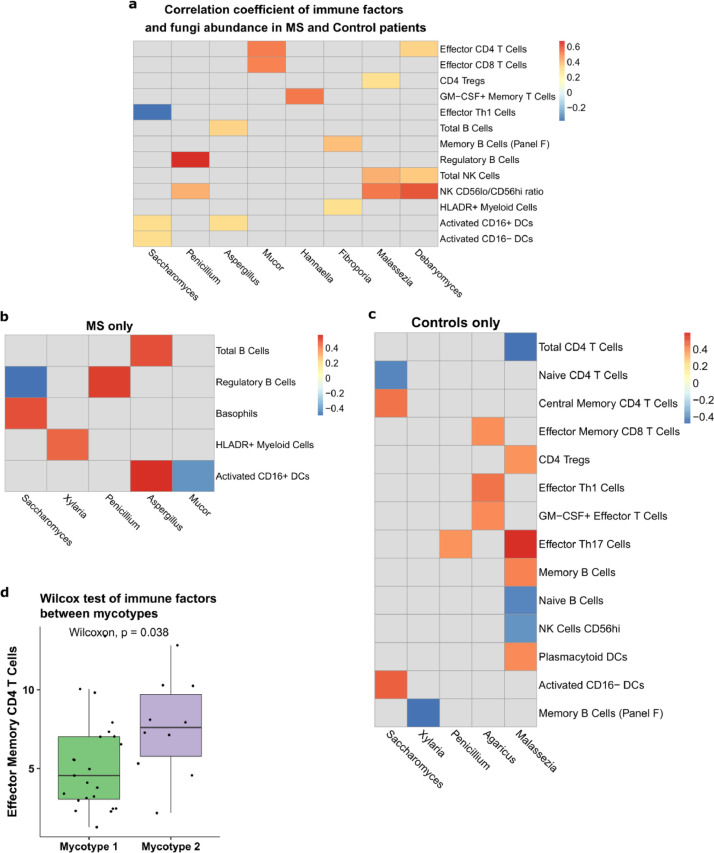
Pearson correlations. Correlation analysis were performed for two continuous variables correlation. Correlations with correlation coefficient r < -0·3 or r > 0·3 were reported. The p-values were less than 0·05 in all reported correlations. All the correlations were manually inspected by plotting raw data and correlations driven by one or two data points were not reported. P-values for correlations were found with the cor.test from the stats package. Relative abundances of fungi genera were correlated with bacteria genera and dietary servings, using both baseline MS and controls samples without discriminating. Immune correlations were done with combined MS and healthy controls as well as separately for MS and controls since MS as an autoimmune disease was likely to have different immune factors from controls (Fig. 6a– c). Heatmap was used to view correlations.
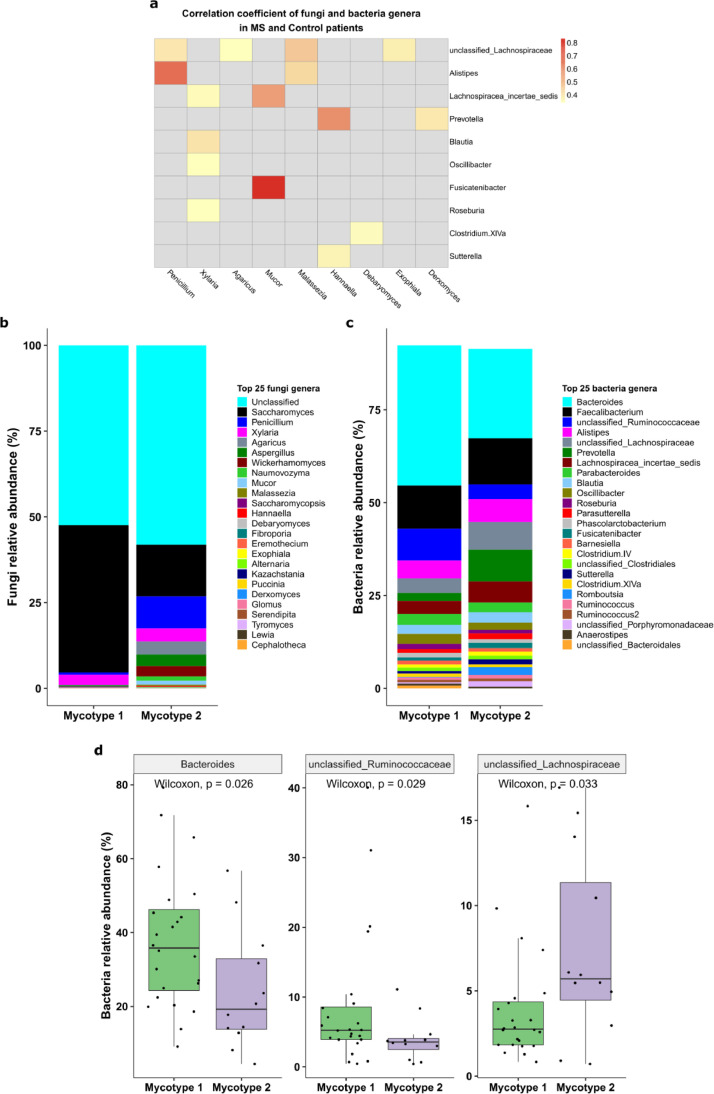
Fig. 6.
Correlation between the mycobiome and blood immune profiles. Pearson correlations between immune cell populations and fungi genera at baseline for (a) pwMS and controls, (b) pwMS only, and (c) controls only. Nonsignificant correlations are omitted. (d) Relative abundance of effector memory CD4 T cells between mycotypes.
Gut mycotype identification. Mycotypes were determined with the “mclust” package based on Gaussian mixture models. The two mycotypes were selected based on Bayesian Information Criteria [40]. Top 20 most abundant mycobiome genera were used to build the model. Barplots were used to show the mean relative abundances of mycobiome in each mycotype.
All plots were made with “ggplot2” and formatted with “ggpubr” except for heatmaps made with “pheatmap”. “Vegan” (v. 2·5–6) was used for count rarefaction, Shannon diversity, specnumber, beta dispersion, Adonis, mantel, and Bray-Curtis dissimilarity. All p-values from multiple comparisons were adjusted with false discovery rate method. All analysis was done in RStudio version 1·2·1.
2.6. Role of funding source
The funders had no role in the conceptualization, study design, data collection, analysis, interpretation of data, in writing the paper, or in the decision to submit the paper for publication.
3. Results
3.1. Study population
Twenty-five pwMS and 22 healthy controls included in this analyses were recruited for the study at Washington University School of Medicine in St. Louis. None of the pwMS was having a relapse at time of enrolment or had received any DMTs or steroid treatment in the previous three months. Of the 25 pwMS, 21 were diagnosed with relapsing-remitting MS (RRMS), 2 with primary progressive MS (PPMS), 1 with secondary progressive MS (SPMS), and 1 with clinical isolated syndrome (CIS). Mean disease duration at study entry was 4·9 (SD 6·6) years. Except for the MS group using more tobacco (P < 0·0005, chi-square test), the characteristics of the control and MS groups were similar (Supplementary Table 1). Stool and blood samples were collected at entry (baseline) and after six months for the gut mycobiome and blood immune cell analyses, respectively. A four-day food-frequency questionnaire was also recorded to provide qualitative dietary information at baseline and at six months for all participants.
3.2. Altered mycobiome compositions and diversity in non-treated MS patients
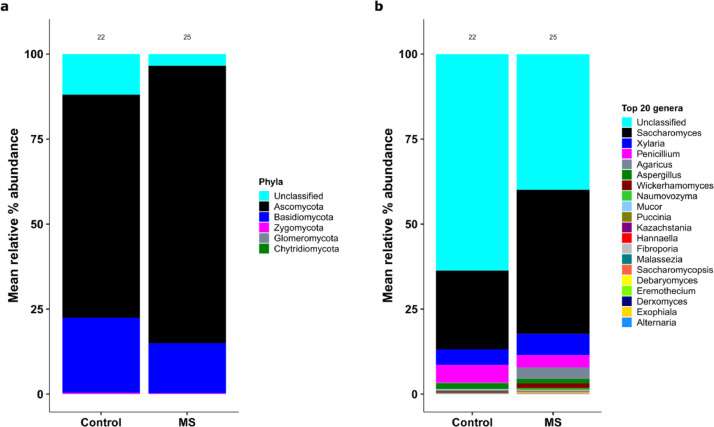
ITS1 sequencing yielded a range of 1146–2,517,870 reads/sample (mean = 61,986 and median = 11,863). We first examined the gut mycobiome composition in pwMS and healthy control individuals at baseline. We considered pwMS in one single group. Separate analysis with exclusion of non-RRMS patients in the study did not change the results. At the phylum level, 92·5% of phyla were identified. Ascomycota and Basidiomycota were the two predominant phyla in both pwMS and controls, together accounting for over 80% of total mycobiome population. At the genus level, we identified 59 genera, of which 25 (excluding the unclassified fungi shared between the two groups) made up 48% of the total relative abundances (Fig. 1). Of the unclassified genera, 61·7% belonged to the phylum Ascomycota and 36·2% belonged to Basidiomycota. Saccharomyces, Xylaria, Pencillium, Agaricus, and Aspergillus were the top five most abundant classified fungi in the gut in both groups. On average, Saccharomyces composed 23% and 42% of the gut mycobiome in control and pwMS, respectively.
Fig. 1.
Mycobiome compositions in pwMS and control groups, based on ITS1 sequencing analysis. (a) Mean relative abundances of fungi phyla. “Unclassified” represent unknown fungal phyla. (b) Mean relative abundances of the 20 most abundant fungi genera. “Unclassified” represents unknown fungal genera.
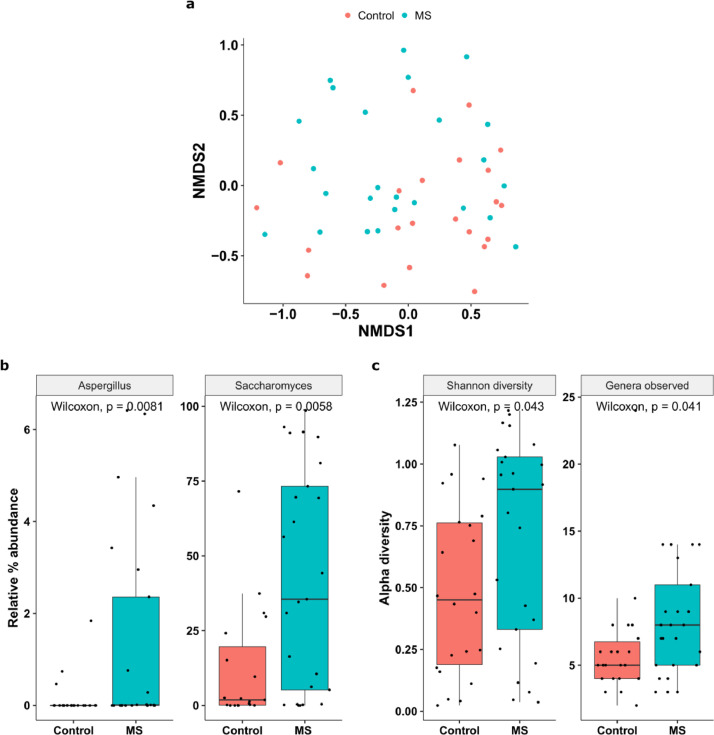
To assess overall mycobiome community differences between pwMS and controls, we performed non- metric multidimensional scaling (NMDS, Fig. 2a) at the genus level. Permutational multivariate analysis of variance (PERMANOVA) analysis showed that the overall mycobiome community differed between pwMS and controls (p=0·04). Further dispersion analysis revealed that the difference in mycobiome between pwMS and the healthy controls is likely due to pwMS having higher inter-subject variation than control participants (beta disper, p < 0·03). Specific analysis indicated that pwMS samples had greater proportions of Aspergillus (p=0·008, padj=0·02, Wilcoxon) and Saccharomyces (p=0·005, padj=0·02, Wilcoxon) (Fig. 2b). The variance of Saccharomyces was significantly higher in pwMS than those in controls (p=0·0004, Levene test). Notably, Unclassified genera were overall more abundant in healthy controls than MS samples (p=0·05, padj=0·088, Wilcoxon, data not shown). No phylum was significantly different between samples from pwMS and controls.
Fig. 2.
Mycobiome diversity in pwMS and control groups. (a) NMDS plot of the Bray-Curtis dissimilarity of control and pwMS based on the mycobiome profile (p = 0•04). (b) Relative abundance of Aspergillus (p = 0•008, padj= 0•02) and Saccharomyces (p = 0•005, padj = 0•02) in the two groups. (c) Alpha diversity variation between the two groups, expressed as Shannon diversity and observed richness.
Stools from pwMS had significantly greater mycobiome richness (p=0·041, Wilcoxon) and Shannon diversity (p=0·043, Wilcoxon) (Fig. 2c) than stools from controls, suggesting overgrowth of different types of fungi in the gut mycobiome in pwMS. There were 18 genera exclusive to pwMS samples. For example, Phlebia and Rhizopus were each detected at very low abundances, but in pwMS samples only. Our findings are consistent with the data in a study of subjects with Crohn's disease, another putative autoimmune disorder, in which was also shown to have increased fungal diversity and richness compared to healthy controls [14].
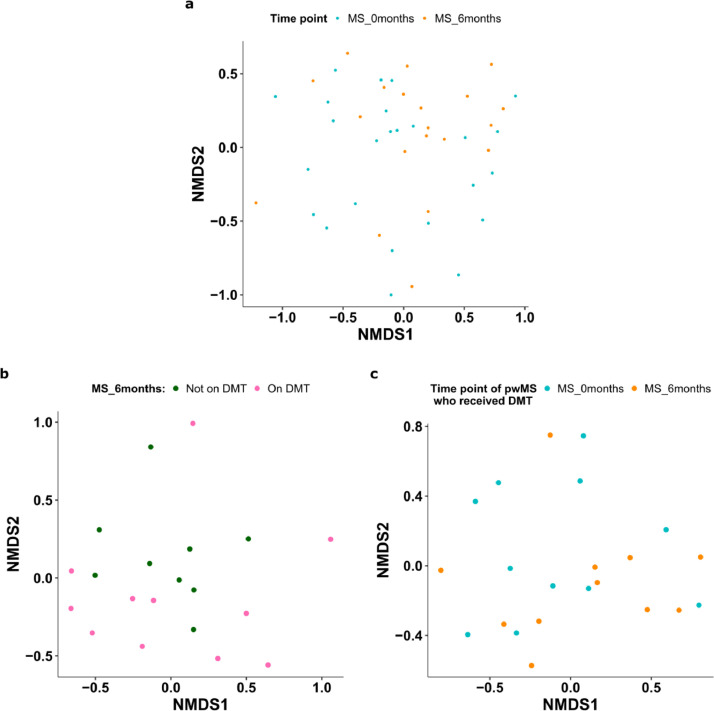
3.3. Mycobiome remains stable over six months
We next investigated the stability of the mycobiome in pwMS. We found that overall mycobiome structure did not change significantly over six months (Fig. 3a, p=0·085, PERMANOVA). The alpha diversity did not significantly change between the two time points (Shannon index p=0·36, richness p=0·25, Wilcoxon). However, there were high inter-individual variations in the dynamics of the mycobiome (Supplementary Fig. 3a). On average, we found that Saccharomyces decreased in six months (p=0·01, padj=0·076, Paired T- test). Our findings are consistent with the notion that the most abundant and common species continue to dominate over time even in samples with high fungal variability [5].
Fig. 3.
Longitudinal stability of the mycobiome. (a) NMDS plot of Bray-Curtis dissimilarity of the mycobiome profile of pwMS samples taken at baseline and at six months (p = 0•085). (b) NMDS of Bray-Curtis dissimilarity of pwMS treated or not treated with DMTs at six months (n = 20, p = 0•79). (c) NMDS of baseline (n = 11) and six- months (n = 11) for pwMS who received DMT treatments within six months (p = 0•22).
Eleven pwMS started treatment with DMTs in the six months after baseline samples were obtained. We did not identify a distinct mycobiome between patients treated with DMTs (n=11) compared to those who were not-treated (n=9) at six months (Fig. 3b, p=0·79, PERMANOVA). We also did not find a difference in the mycobiome between baseline and six months for pwMS who began DMT treatments within six months (p=0·22, PERMANOVA, Fig. 3c). We additionally compared the mycobiome changes in three pwMS who received dimethyl fumarate (DMF), a DMT with fungicidal properties, but found no consistent changes in genus abundances nor alpha diversity after treatment (at the six-month point) (Supplementary Fig. 3b). Further research into DMF's effect on the gut mycobiome with a larger sample size would be warranted.
3.4. Interactions between the gut mycobiome and bacterial microbiome
To investigate relationships between fungi and bacteria in the gut, we performed Pearson correlations with all participants at baseline. We found a high correlation between Mucor and Fusicatenibacter (Fig. 4a, r=0·81, p < 0·001). Fusicatenibacter is a bacterial genus belonging to Clostridium cluster XIV and Lachnospiraceae incertae sedis. The abundance of the bacterial genus Prevotella highly correlated with the fungi Hannaella (r=0·62, p < 0·001) and Derxomyces (r=0·39, p=0·019), both of which are low in abundances. Alistipes, one bacterial genus that is associated with MS [41], was positively correlated with Penicillium (r = 0·68, p < 0·001). When MS and control samples were separated, Saccharomyces which had notably greater abundance in MS samples, was negatively correlated with Lachnospiracea incertae sedis (r=-0·48, p=0·04) in pwMS. In controls alone, Saccharomyces and Oscillibacter were positively correlated (r = 0·51, p=0·029).
Fig. 4.
Association between fungal and bacterial microbiome. (a) Heatmap shows fungi-bacteria Pearson correlations. Nonsignificant correlations are omitted. (b) Mean relative abundances of the major fungi in two mycotypes identified by Mclust Gaussian Mixture analysis (n1 = 24, n2 = 12). (c) Mean relative abundances of the gut bacteria in participants belonging to the two mycotypes. (d) Significantly over-represented gut bacteria between two mycotypes: Bacteroides (p = 0•026, padj = 0•055), unclassified Ruminococcaceae (p = 0•029, padj = 0•055), and Lachnospiraceae (p = 0•033, padj = 0•055).
We next asked if different mycobiome profiles are associated with specific microbiome compositions. The gut microbiome can be classified into three types (termed enterotypes) based on their relative abundances [42,43], which are associated with different health and disease conditions. Using the Mclust method, we classified the mycobiome profile into two fungal clusters (“mycotypes”) which had a similar ratio of MS and controls (Supplementary Fig. 4). Mycotype 1 consists of 11 controls and 13 pwMS, whereas Mycotype 2 has seven controls and five pwMS. Mycotype 1 was dominated by unclassified genera and Saccharomyces, whereas Mycotype 2 shows greater diversity and reduced Saccharomyces, and a higher average proportion of Penicillium, Malassezia, and Mucor (Fig. 4b). Comparison of the relative abundances of the bacteria in participants from the two mycotypes (Fig. 4c) demonstrated that the Bacteroides (p=0·026, padj =0·055, Wilcoxon) and Unclassified Ruminococcaceae (p=0·029, padj =0·055, Wilcoxon) were greater in participants from Mycotype 1 whereas Unclassified Lachnospiraceae was significantly greater in participants from Mycotype 2 (p=0·033, padj =0·055, Wilcoxon, Fig. 4d). The latter finding is consistent with data presented in Fig. 4a that reports a positive correlation between Penicillium and Malassezia and Unclassified Lachnospiraceae family. These results suggest that distinct mycobiome profiles are associated with distinct bacterial compositions.
3.5. Correlations of the diet and body mass index with the gut mycobiome
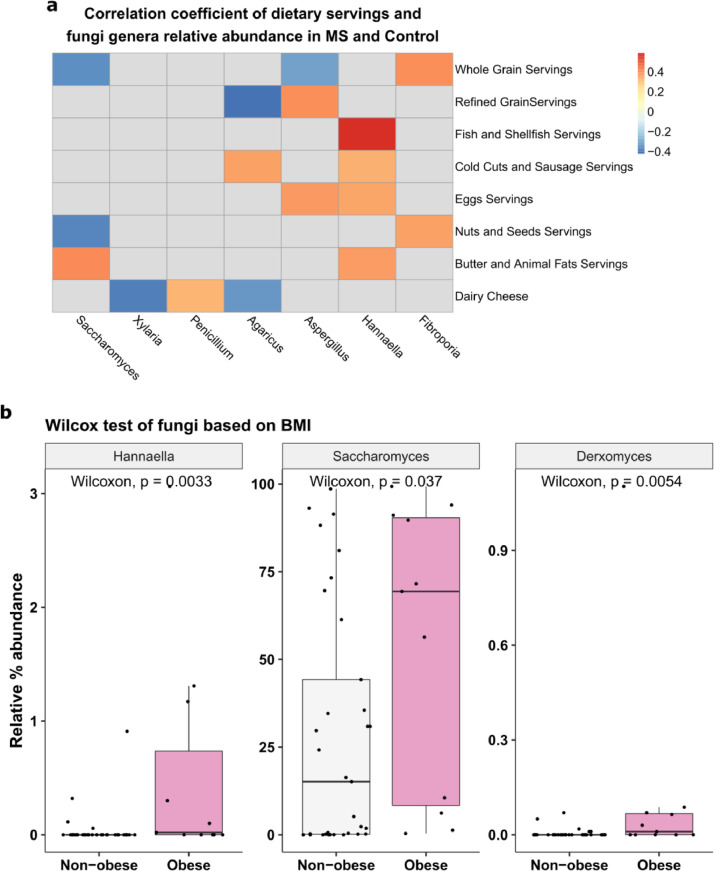
We further determined the effect of diet on the gut mycobiome using baseline data from all participants. All types of food intake were similar in MS and control groups except that the MS group had significantly more meat (p=0·019, Wilcoxon). However, meat had no significant impact on the gut mycobiome variation (p=0·17 padj=0·47, PERMANOVA). We performed PERMANOVA analysis and identified four types of food intakes that are potentially associated with the gut mycobiome community structure (Fig. 5a): ‘butter and animal fats’ (p=0·003, padj =0·069), ‘nuts and seeds’ (p=0·016, padj =0·18), ‘refined grain’ (p=0·04, padj=0·18), and ‘whole grain’ (p=0·042, padj =0·18). Specifically, the Pearson correlations of food servings and fungal genera abundances showed that Saccharomyces (r = 0·42, p=0·008, Pearson) and Hannaella (r = 0·38, p=0·02, Pearson) each had a moderately strong positive correlation with ‘butter and animal fats’, whereas Saccharomyces has a negative correlation with ‘nuts and seeds’ (r = -0·38, p=0·022, Pearson) and ‘whole grain’ servings (r = -0·35, p= 0·037). Hannaella also has a moderately strong correlation with ‘fish and shellfish’ servings (r = 0·59, p= 0·0005, Pearson). Aspergillus had a positive correlation with ‘eggs’ (r= 0·39, p= 0·002) and ‘refined grain’ serving (r = 0·41, p= 0·011) and negative correlation with ‘whole grain’ servings (r= -0·31, p=0·04). In pwMS only, some of the aforementioned correlations became stronger: Saccharomyces and ‘nuts and seeds’ (r = -0·51, p=0·012); Hannaella and ‘butter and animal fats’ (r= 0·59, p=0·003); Aspergillus and ‘eggs’ (r= 0·55, p=0·006) and ‘refined grain’ (r= 0·57, p=0·005). In controls only, Hannaella strengthened its correlation with fish (r= 0·75, p= 0·0007) but lost it with ‘eggs’ servings (r= 0, p=0·93).
Fig. 5.
Diet and the mycobiome. (a) Heatmap shows Pearson correlations between food servings and fungi genera. Nonsignificant correlations are omitted. (b) Relative abundance of Hannaella, Saccharomyces, and Derxomyces in obese (BMI ≥ 30) and non-obese participants (BMI < 30).
Obesity is associated with mycobiome dysbiosis [44]. We found that Hannaella had a moderately strong positive correlation with BMI (r = 0·52, p < 0·001, Pearson). In our study, 21 subjects were classified as normal weight (BMI < 25), 11 were overweight (25 ≤ BMI < 30), and 12 were obese (BMI ≥ 30). The proportions of pwMS and controls who were obese, overweight, and normal weight were similar between the two groups (p=0.803, chi-squared test). The relative abundance of Hannaella (p=0·0033, Wilcoxon), Saccharomyces (p=0·037), and Derxomyces (p=0·005) was significantly higher in obese subjects than non- obese (Fig. 5b). No genus was found to be different between normal and overweight individuals, indicating that mycobiome alteration occurred primarily in the obese vs non-obese level (Supplementary Fig. 5).
3.6. Correlation of the gut mycobiome with peripheral blood immune profiles
We have previously shown in the cohort of subjects studied herein, that the percentages of peripheral blood IL-10+ memory B cells, T-bet+ memory and effector T cells, memory and effector Th17 cells were significantly greater in MS than in control individuals (Supplementary Table 2, and [Cantoni, unpublished]). We next asked if and to what extent the gut mycobiome is associated with peripheral blood immune profile. We determined the correlations between the mycobiome compositions and the peripheral immune cell profiles in the whole cohort of pwMS and controls, or in the pwMS or control groups separately. Pearson correlation of fungi genera and immune cell subsets using all baseline samples (Fig. 6a) showed that Mucor had a positive correlation with effector CD4+ T Cells (r=0·52, p=0·0003, Pearson) and effector CD8+ T Cells (r=0·51, p=0·0004). Hannaella had a positive correlation with memory CD4+ T cells producing GM- CSF (r=0·53, p=0·0003). Additional positive correlations were found between NK cells, CD56low/CD56high NK cells and genera Debaryomyces (r=0·59, p=0·0005), Malassezia (r=0·54, p=0·0001), and between regulatory B cells and Penicillium (r=0·67, p=0·005). Saccharomyces showed a negative correlation with effector Th1 cells (r=-0·37, p=0·02) and a positive correlation with activated CD16+/− DCs (r=0·31, p=0·03). Aspergillus was positively correlated with total B cells (r=0·35, p=0·01) and CD16+ dendritic cells (r=0·32, p=0·03).
Analyses in pwMS and controls separately revealed group specific correlations between the mycobiome compositions and peripheral immune profiles. Interestingly, the number of total correlations (positive or negative) identified in MS were 1·7 fold less, compared with controls, indicating a disrupted immune- mycobiome interaction. We showed that Saccharomyces and Aspergillus were over-represented in pwMS compared to controls (Fig. 2b). In the MS group, Saccharomyces was positively correlated with the frequency of basophils (r=0·50, p=0·015, Pearson) and negatively correlated with regulatory B cells (r=- 0·49, p=0·03). Aspergillus was positively correlated with activated DCs (r=0·57, p=0·004) and total B cells (r=0·50, p=0·014). Penicillium was positively correlated with regulatory B cells (r=0·54, p=0·018, Fig. 6b). In the control group, Saccharomyces was negatively correlated with CD4+ naïve T cells (r=-0·43, p=0·045) and positively correlated with CD16− DCs (r=0·50, p=0·018). Malassezia had the greatest positive correlation with Th17 cells (r=0·59, p=0·004, Pearson) and positively or negatively correlations with six additional immune cell populations (Fig. 6c). Malassezia was the fungal genus with the most frequent interaction with the immune profiles in healthy controls. These connections disappeared completely in pwMS.
Comparison of the immune profile difference between the two mycotypes revealed that effector memory CD4+ T cells were greater in mycotype 2 than in mycotype 1 (p=0·038, Wilcoxon, Fig. 6d). Mycotype 2 was characterised by greater fungi diversity (Fig. 6d).
4. Discussion
This study is the first to define the gut mycobiome of pwMS. Both pwMS and controls were dominated by Saccharomyces, Xylaria, and Penicillium. Nash et al. identified Saccharomyces (∼20% relative abundance), Malassezia (4% relative abundance), and Candida (2% relative abundance) as the dominant genera in healthy samples [5], while Hoffman et al. identified Saccharomyces, Candida, and Cladosporium as the most abundant in healthy control samples [45]. In our study, Saccharomyces was most abundant genera (23% in control and 42% in MS), while Candida and Malassezia only account for 0·26% and 0·86%, respectively, and Cladosporium was not detected at all. Environment, diet, and exposure to different climates could also play a role in the fungi colonization of the gut [46,47]. We recruited participants from St Louis, whereas Nash et al. recruited from Pennsylvania, and Hoffman et al. from Houston. In addition, DNA extraction and sequencing regions may also contribute to mycobiome differences across studies, suggesting the need for a standardized mycobiome characterization.
We compared mycobiome diversity and compositions in pwMS and healthy control samples. Overall, the results indicate that samples from pwMS had higher mycobiome diversity and greater inter-subject variation compared to the mycobiome of healthy controls. These findings are in contrast with most of the studies on the bacterial microbiome in MS patients including our own study from the same study cohort [Cantoni, unpublished], in which bacterial diversity was similar between MS patients and healthy controls. Microbial diversity is an indicator of gut health, and it is generally determined by the bacterial diversity. Higher bacterial diversity is associated with healthier gut ecosystem. Infection or disease conditions, high fat diet or inflammatory immune response in the gut decreases microbial diversity [48]. Our data suggest that the mycobiome diversity is an important indicator of the gut microbial diversity in MS patients, and may be more sensitive than the bacterial diversity in response to MS associated gut microbiome changes.
The gut mycobiome accounts for less than one percent of the entire microbiome, therefore it may be considered rare relative to the bacterial microbiome [49]. However, rare biosphere can have disproportionate effects on health and diseases [50]. We found two commensal gut fungal genera Saccharomyces and Aspergillus that were more abundant in MS. Saccharomyces is called the baker and brewer's yeast for its role in food fermentation, as well as commonly being used as a gut probiotics [51]. However, the biological role of Saccharomyces in health and diseases is inconsistent across studies, with either protective or detrimental effect on the gut inflammation in animal model of IBD [12,13]. MS and IBD share similar epidemiological, immunological, and gut bacterial microbiome characteristics [52]. Our results support a pathogenic correlation of Saccharomyces with MS. In addition, Saccharomyces had positive correlations with ‘butter and milk’ servings and higher relative abundance in obese participants. However, these dietary factors and BMI associated with Saccharomyces were not significantly different between MS and controls, suggesting that observed Saccharomyces difference in MS and controls is not due to differential dietary intake or BMI between the two groups. However, diet compositions in our study were obtained from a food-frequency questionnaire. These self-report food intakes may introduce a certain degree of inaccuracy. Interestingly, Saccharomyces had a positive correlation with basophils and negative correlation with regulatory B cells in pwMS’ blood. Basophils activation by Saccharomyces has only been studied in beer allergy [53]. Whether Saccharomyces affects pathogenesis of MS and its interaction with basophils need to be further investigated. So far, very few studies have investigated the interaction of commensal fungi and peripheral immune response in humans. Our study also opens a new research question in terms of the possible relationship between Saccharomyces and regulatory B cells in MS.
Aspergillus is a genus consisting of several mould species. It is a member of respiratory and gut mycobiome. Aspergillus produce aflatoxins and can cause opportunistic infections in humans [54]. Except direct infection in CNS, it is possible that the gut Aspergillus activates a gut immune response that indirectly affects systemic or CNS inflammation. This gut response was demonstrated by the study that prolonged treatment with anti-fungal drug fluconazole showing decreased level of Candida in the gut, but increased Aspergillus amstelodami, resulting in elevated colitis severity [55]. Aspergillus had a positive correlation with B cells and activated CD16+ DCs in pwMS in our study. While we all acknowledge that causal connection between the microbiome in diseases is important, we first need to establish the association between the mycobiome in MS, and since currently no other study reported the role of gut mycobiome in MS. The correlation analysis in pwMS in our study is crucial to generate future hypotheses to test mycobiome-immune interactions in animal models and MS patients.
Fungi can have direct impact on the immune response or indirectly through their interactions with bacteria. Our study revealed a disrupted correlation pattern between fungi and peripheral immune profile in pwMS. Correlations identified with pwMS have 1·7 fold fewer correlations between mycobiome and immune profiles than controls’. The lack of correlation can be due to higher inter-subject variation of the mycobiome in pwMS or loss of mycobiome-immune homeostasis in disease status. We found Saccharomyces had a negative correlation with Lachnospiraceae incertae sedis in pwMS, raising the possibility that the mycobiome may affect autoimmune through regulation of bacteria. However, future studies are warranted to determine the mechanism governing the relationship among bacteria microbiome, mycobiome, and autoimmunity.
The stability of the fungal microbiome is hard to conclude upon. Some studies have found that the human mycobiome is not very stable, but some species do remain prevalent over different time points. Nash et al. found high inter- and intra-volunteer variability in faecal fungi over one year [5], whereas Cohen et al. did not find changes in the fungal flora in the human small intestine over several months [56]. In our study, samples showed stability over time: 55% of pwMS had the 11 most abundant genera at both time points. For comparison, Hallen-Adams et al. detected common genera 27% of the time over 13–16 weeks [57]. We also found that the most common genus Saccharomyces was detected in all MS samples at both baseline and six-month time points. However, the changes of Saccharomyces are not consistent across all pwMS. Instead, it showed a personalized mycobiome dynamics. We have shown that the gut microbiome over time may exhibit a personalized changing trajectory [58]. In addition, we did not find significant alterations of mycobiome in pwMS before and after DMTs treatment. DMF, which has fungicide property, also showed no consistent effect on the gut mycobiome because of high-intrasubject variation of the mycobiome. Together, these findings suggest a personalized mycobiome dynamics and responsiveness to treatment.
Further studies with larger sample size and more longitudinal time points are needed to discover the precise characteristics of the gut mycobiome dynamics in the short and long-term in pwMS. These will further characterise the underlying mechanisms that govern the pattern of changes associated with DMF treatment, diet, disease course and beyond.
The study has several limitations. It is a single centre study with a small sample size. Future study with larger sample size and a validation cohort are warranted to confirm findings on mycobiome changes and their associations with host and bacterial microbiome. We could not identify uniformed mycobiome changing signal after disease modifying therapy, which can also be due to the limited number of patients who underwent treatment during the study. In addition, RRMS patients were mostly in the remission phase and only a few patients were treated. Thus, we could not determine how the mycobiome could change in the active state of the disease. An additional limitation is that the ITS Sequencing had large unclassified fungi due to poor annotations in the fungal database.
Contributors
YZ, LP, ALR designed the study, obtained funding for the project and supervised the entire study. LP, LG, AHC, EE enrolled patients, reviewed clinical charts, collected samples. SS, AL, YZ, LP, ALR, YD, CC, LG, QL, SB, HP, CS, MG, YL, EE, RM, KO, AS prepared the samples for sequencing, performed data analysis, and contributed to the interpretation of the data. SS, YZ, and QL verified the underlying data. YZ, SS, AL, LP, ALR, PIT, and AHC wrote or reviewed the manuscript. All authors read and approved the final manuscript and have had access to the raw data.
Data sharing statement
Raw sequences were deposited in the NCBI Sequence Read Archive under accession number PRJNA676148.
Declaration of Competing Interest
Dr. Evans has been a paid consultant and/or speaker for the following: Biogen, EMD Serono, National MS Society, Genentech/Roche, Novartis, Sanofi/Genzyme, and Teva.
Dr. Cross has done paid consulting for: Biogen, Celgene, EMD Serono, Genentech/Roche, Greenwich Biosciences, Janssen and Novartis, and has contracted research funded by EMD Serono and Genentech.
Dr. Tarr is a consultant to, a member of the Scientific Advisory Board of, and a holder equity in, MediBeacon Inc., which is developing a method to test intestinal permeability in humans. He might receive royalty payments if the product generates revenues.
All other authors have nothing to declare.
Acknowledgements
This work was supported by the Washington University in St. Louis Institute of Clinical and Translational Sciences, funded, in part, by Grant Number # UL1 TR000448 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award (Zhou Y, Piccio, L, Lovett-Racke A and Tarr PIs); R01 NS102633-04 (Zhou Y, Piccio L); the Leon and Harriet Felman Fund for Human MS Research (Piccio L and Cross AH). Cantoni C. was supported by the National MS Society Career Transition Fellowship (TA-1805-31003) and by donations from Whitelaw Terry, Jr. / Valerie Terry Fund. Ghezzi L. was supported by the Italian Multiple Sclerosis Society research fellowship (FISM 2018/B/1) and the National Multiple Sclerosis Society Post-Doctoral Fellowship (FG-1907-34474). Anne Cross was supported by The Manny & Rosalyn Rosenthal-Dr. John L. Trotter MS Center Chair in Neuroimmunology of the Barnes-Jewish Hospital Foundation.
We thank the Microbial Genomic Services Core at Jackson Laboratory and Metabolome core at University of Massachusetts for data generation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Laura Piccio, Email: picciol@wustl.edu.
Yanjiao Zhou, Email: yazhou@uchc.edu.
References
- 1.Uchiyama K., Naito Y., Takagi T. Intestinal microbiome as a novel therapeutic target for local and systemic inflammation. Pharmacol Ther. 2019;199:164–172. doi: 10.1016/j.pharmthera.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Cantarel B.L., Waubant E., Chehoud C., Kuczynski J., DeSantis T.Z., Warrington J. Gut microbiota in multiple sclerosis: possible influence of immunomodulators. J Investig Med. 2015;63:729–734. doi: 10.1097/JIM.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J., Chia N., Kalari K.R., Yao J.Z., Novotna M., Paz Soldan M.M. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016;6:28484. doi: 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyake S., Kim S., Suda W., Oshima K., Nakamura M., Matsuoka T. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to clostridia XIVa and IV clusters. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nash A.K., Auchtung T.A., Wong M.C., Smith D.P., Gesell J.R., Ross M.C. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. 2017;5:153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin J., Li R., Raes J., Arumugam M., Burgdorf K.S., Manichanh C. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sam Q.H., Chang M.W., Chai L.Y.A. The Fungal Mycobiome and its interaction with gut bacteria in the Host. Int J Mol Sci. 2017;18(2):330–341. doi: 10.3390/ijms18020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kombrink A., Tayyrov A., Essig A., Stöckli M., Micheller S., Hintze J. Induction of antibacterial proteins and peptides in the coprophilous mushroom Coprinopsis cinerea in response to bacteria. ISME J. 2019;13:588–602. doi: 10.1038/s41396-018-0293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernández de Ullivarri M., Arbulu S., Garcia-Gutierrez E., Cotter PD. Antifungal peptides as therapeutic agents. Front Cell Infect Microbiol. 2020;10:105–127. doi: 10.3389/fcimb.2020.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noverr M.C., Huffnagle G.B. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect Immun. 2004;72:6206–6210. doi: 10.1128/IAI.72.11.6206-6210.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García C., Tebbji F., Daigneault M., Liu N.N., Köhler J.R., Allen-Vercoe E. The human gut microbial metabolome modulates fungal growth via the TOR signaling pathway. mSphere. 2017;2 doi: 10.1128/mSphere.00555-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang T.T., Shao T.Y., Ang W.X.G., Kinder J.M., Turner L.H., Pham G. Commensal fungi recapitulate the protective benefits of intestinal bacteria. Cell Host Microbe. 2017;22:809–816. doi: 10.1016/j.chom.2017.10.013. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiaro T.R., Soto R., Zac Stephens W., Kubinak J.L., Petersen C., Gogokhia L. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aaf9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q., Wang C., Tang C., He Q., Li N., Li J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn's disease. J Clin Gastroenterol. 2014;48:513–523. doi: 10.1097/MCG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liguori G., Lamas B., Richard M.L., Brandi G., da Costa G., Hoffmann T.W. Fungal Dysbiosis in mucosa-associated Microbiota of Crohn's disease patients. J Crohns Colitis. 2016;10:296–305. doi: 10.1093/ecco-jcc/jjv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai T., Inoue R., Kawada Y., Morita Y., Inatomi O., Nishida A. Characterization of fungal dysbiosis in Japanese patients with inflammatory bowel disease. J Gastroenterol. 2019;54:149–159. doi: 10.1007/s00535-018-1530-7. [DOI] [PubMed] [Google Scholar]
- 17.Botschuijver S., Roeselers G., Levin E., Jonkers D.M., Welting O., Heinsbroek S.E.M. Intestinal fungal dysbiosis is associated with visceral hypersensitivity in patients with irritable bowel syndrome and rats. Gastroenterology. 2017;153:1026–1039. doi: 10.1053/j.gastro.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Hong G., Li Y., Yang M., Li G., Qian W., Xiong H. Gut fungal dysbiosis and altered bacterial- fungal interaction in patients with diarrhea-predominant irritable bowel syndrome: an explorative study. Neurogastroenterol Motil. 2020;32:e13891. doi: 10.1111/nmo.13891. [DOI] [PubMed] [Google Scholar]
- 19.Zhai B., Ola M., Rolling T., Tosini N.L., Joshowitz S., Littmann E.R. High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat Med. 2020;26:59–64. doi: 10.1038/s41591-019-0709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aykut B., Pushalkar S., Chen R., Li Q., Abengozar R., Kim J.I. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574:264–267. doi: 10.1038/s41586-019-1608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anandakumar A., Pellino G., Tekkis P., Kontovounisios C. Fungal microbiome in colorectal cancer: a systematic review. Updates Surg. 2019;71:625–630. doi: 10.1007/s13304-019-00683-8. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y., Chen Z., Guo R., Chen N., Lu H., Huang S. Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagn Microbiol Infect Dis. 2011;70:492–498. doi: 10.1016/j.diagmicrobio.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Preveden T., Scarpellini E., Milić N., Luzza F., Abenavoli L. Gut microbiota changes and chronic hepatitis C virus infection. Expert Rev Gastroenterol Hepatol. 2017;11:813–819. doi: 10.1080/17474124.2017.1343663. [DOI] [PubMed] [Google Scholar]
- 24.Chauhan A., Kumar R., Sharma S., Mahanta M., Vayuuru S.K., Nayak B. Fecal microbiota transplantation in hepatitis b e antigen-positive chronic hepatitis b patients: a pilot study. Dig Dis Sci. 2020;66(3):873–880. doi: 10.1007/s10620-020-06246-x. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee P.K., Chandra J., Retuerto M., Tatsuoka C., Ghannoum M.A., McComsey GA. Dysbiosis in the oral bacterial and fungal microbiome of HIV-infected subjects is associated with clinical and immunologic variables of HIV infection. PLoS One. 2018;13 doi: 10.1371/journal.pone.0200285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang A.M., Inamine T., Hochrath K., Chen P., Wang L., Llorente C. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest. 2017;127:2829–2841. doi: 10.1172/JCI90562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strati F., Cavalieri D., Albanese D., De Felice C., Donati C., Hayek J. Altered gut microbiota in Rett syndrome. Microbiome. 2016;4:41. doi: 10.1186/s40168-016-0185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strati F., Calabrò A., Donati C., De Felice C., Hayek J., Jousson O. Intestinal Candida parapsilosis isolates from Rett syndrome subjects bear potential virulent traits and capacity to persist within the host. BMC Gastroenterol. 2018;18:57. doi: 10.1186/s12876-018-0785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strati F., Cavalieri D., Albanese D., De Felice C., Donati C., Hayek J. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 2017;5:24. doi: 10.1186/s40168-017-0242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bojović K., Ignjatović Ð.D.I., Soković Bajić S., Vojnović Milutinović D., Tomić M., Golić N. Gut microbiota dysbiosis associated with altered production of short chain fatty acids in children with neurodevelopmental disorders. Front Cell Infect Microbiol. 2020;10:223. doi: 10.3389/fcimb.2020.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou R., Wang Y., Duan M., Guo M., Zhang Q., Zheng H. Dysbiosis of gut fungal microbiota in children with autism spectrum disorders. J Autism Dev Disord. 2020 doi: 10.1007/s10803-020-04543-y. [DOI] [PubMed] [Google Scholar]
- 32.Severance E.G., Gressitt K.L., Stallings C.R., Katsafanas E., Schweinfurth L.A., Savage C.L. Candida albicans exposures, sex specificity and cognitive deficits in schizophrenia and bipolar disorder. NPJ Schizophr. 2016;2:16018. doi: 10.1038/npjschz.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cryan J.F., O'Riordan K.J., Cowan C.S.M., Sandhu K.V., Bastiaanssen T.F.S., Boehme M. The microbiota-gut-brain axis. Physiol Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X., Pan L.Y., Zhang Z., Zhou Y.Y., Jiang H.Y., Ruan B. Analysis of gut mycobiota in first- episode, drug-naïve Chinese patients with schizophrenia: A pilot study. Behav Brain Res. 2020;379 doi: 10.1016/j.bbr.2019.112374. [DOI] [PubMed] [Google Scholar]
- 35.Dendrou C.A., Fugger L., Friese M.A. Immunopathology of multiple sclerosis. Nat Rev Immunol. 2015;15:545–558. doi: 10.1038/nri3871. [DOI] [PubMed] [Google Scholar]
- 36.Chauhan B., Santiago L., Hutcheson P.S., Schwartz H.J., Spitznagel E., Castro M. Evidence for the involvement of two different MHC class II regions in susceptibility or protection in allergic bronchopulmonary aspergillosis. J Allergy Clin Immunol. 2000;106:723–729. doi: 10.1067/mai.2000.109913. [DOI] [PubMed] [Google Scholar]
- 37.Raimondi S., Amaretti A., Gozzoli C., Simone M., Righini L., Candeliere F. Longitudinal survey of fungi in the human gut: its profiling, phenotyping, and colonization. Front Microbiol. 2019;10:1575. doi: 10.3389/fmicb.2019.01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraga-Silva T.F.C., Mimura L.A.N., Marchetti C.M., Chiuso-Minicucci F., França T.G.D., Zorzella- Pezavento S.F.G. Experimental autoimmune encephalomyelitis development is aggravated by Candida albicans infection. J Immunol Res. 2015;2015 doi: 10.1155/2015/635052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benito-León J., Laurence M. The role of fungi in the etiology of multiple sclerosis. Front Neurol. 2017;8:535. doi: 10.3389/fneur.2017.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fraley C., Raftery A.E. Model-based clustering, discriminant analysis, and density estimation. J Am Stat Assoc. 2002;97:611–631. [Google Scholar]
- 41.Takewaki D., Suda W., Sato W., Takayasu L., Kumar N., Kimura K. Alterations of the gut ecological and functional microenvironment in different stages of multiple sclerosis. Proc Natl Acad Sci U S A. 2020;117:22402–22412. doi: 10.1073/pnas.2011703117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y., Mihindukulasuriya K.A., Gao H., La Rosa P.S., Wylie K.M., Martin J.C. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014;15:R66. doi: 10.1186/gb-2014-15-5-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mar Rodríguez M., Pérez D., Javier Chaves F., Esteve E., Marin-Garcia P., Xifra G. Obesity changes the human gut mycobiome. Sci Rep. 2015;5:14600–14615. doi: 10.1038/srep14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffmann C., Dollive S., Grunberg S., Chen J., Li H., Wu G.D. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PloS One. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis J.D., Chen E.Z., Baldassano R.N., Otley A.R., Griffiths A.M., Lee D. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric crohn's disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amato K.R., Jeyakumar T., Poinar H., Gros P. Shifting climates, foods, and diseases: the human microbiome through evolution. Bioessays. 2019;41 doi: 10.1002/bies.201900034. [DOI] [PubMed] [Google Scholar]
- 48.Heisel T., Montassier E., Johnson A., Al-Ghalith G., Lin Y.W., Wei L.N. High-fat diet changes fungal microbiomes and interkingdom relationships in the murine gut. mSphere. 2017;2 doi: 10.1128/mSphere.00351-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huffnagle G.B., Noverr M.C. The emerging world of the fungal microbiome. Trends Microbiol. 2013;21:334–341. doi: 10.1016/j.tim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jousset A., Bienhold C., Chatzinotas A., Gallien L., Gobet A., Kurm V. Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 2017;11:853–862. doi: 10.1038/ismej.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelesidis T., Pothoulakis C. Efficacy and safety of the probiotic Saccharomyces boulardii for the prevention and therapy of gastrointestinal disorders. Ther Adv Gastroenterol. 2012;5:111–125. doi: 10.1177/1756283X11428502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forbes J.D., Bernstein C.N., Tremlett H., Van Domselaar G., Knox N.C. A fungal world: could the gut mycobiome be involved in neurological disease? Front Microbiol. 2018;9:3249. doi: 10.3389/fmicb.2018.03249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Entrala A., Dominguez-Ortega J., Gonzalez-Muñoz M., Fiandor A., Quirce S. Usefulness of the basophil activation test to confirm beer allergy. J Investig Allergol Clin Immunol. 2018;28:279–280. doi: 10.18176/jiaci.0266. [DOI] [PubMed] [Google Scholar]
- 54.Pérez-Torrado R., Querol A. Opportunistic strains of saccharomyces cerevisiae: a potential risk sold in food products. Front Microbiol. 2015;6:1522. doi: 10.3389/fmicb.2015.01522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wheeler M.L., Limon J.J., Bar A.S., Leal C.A., Gargus M., Tang J. Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe. 2016;19:865–873. doi: 10.1016/j.chom.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen R., Roth F.J., Delgado E., Ahearn D.G., Kalser MH. Fungal flora of the normal human small and large intestine. N Engl J Med. 1969;280:638–641. doi: 10.1056/NEJM196903202801204. [DOI] [PubMed] [Google Scholar]
- 57.Hallen-Adams H.E., Suhr M.J. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8:352–358. doi: 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou W., Sailani M.R., Contrepois K., Zhou Y., Ahadi S., Leopold S.R. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature. 2019;569:663–671. doi: 10.1038/s41586-019-1236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]