Abstract
Circular RNAs (circRNAs), an emerging family member of RNAs, have gained importance in research due to their new functional roles in cellular physiology and disease progression. circRNAs are usually available in a wide range of cells and have shown tissue-specific expression as well as developmental specific expression. circRNAs are characterized by structural stability, conservation, and high abundance in the cell. In this review, we discuss the different models of biogenesis. The properties of circRNAs such as localization, structure and conserved pattern, stability, and expression specificity are also been illustrated. Furthermore, we discuss the biological functions of circRNAs such as microRNA (miRNA) sponging, cell cycle regulation, cell-to-cell communication, transcription regulation, translational regulation, disease diagnosis, and therapeutic potential. Finally, we discuss the recent research progress and future perspective of circRNAs. This review provides an understanding of potential diagnostic markers and the therapeutic potential of circRNAs, which are emerging daily.
Keywords: circular RNAs, biogenesis, properties, therapeutic potential
Graphical abstract

Circular RNAs (circRNAs) perform several biological functions and are an emerging family of RNAs. They can be promising biomarkers and have significant roles in therapeutic purposes for human diseases. Sharma et al. review the recent research progress and future perspective of circRNAs that might help explore scientific endeavors in RNA biology.
Introduction
Circular RNAs (circRNAs), an enigmatic and exciting family member of RNAs, have started to gain recognition in recent years.1,2 Progress in research on circRNAs is moving at a rapid pace (Figure 1). These molecules are single-stranded transcripts that are covalently closed and are generated from precursor mRNA (pre-mRNA) through the backsplicing process, also known as alternative splicing.3,4 During this non-canonical process, a splice donor site (downstream 5′ splice site) is covalently linked to an upstream region (upstream 3′ splice site).1 Previously, circRNAs were thought to be a by-product of unusual splicing with no function or minute functional activity, including circRNAs from the Sry (sex-determining region Y) gene in adult mouse testes.4 However, due to rapid progress in the field of high-throughput RNA sequencing (RNA-seq) and circRNA-specific computational biology, a wide array of circRNAs have been reported in eukaryotes, and their functions are being explored.5, 6, 7, 8, 9, 10, 11 The identified circRNAs in eukaryotes have been found in different metazoans, namely, fungi, worms, fish, insects, mammals, and plants.7,8,11, 12, 13, 14, 15 Although circRNA is a single-stranded RNA, it varies from linear RNA for being closed covalently, which imparts circRNA some fascinating properties such as parental gene modulation, protein complex scaffolding, microRNA (miRNA) sponging, and RNA-protein interaction.2,16 Despite not having polyadenylation (poly(A)) and caps, usually, circRNAs can localize to the cytoplasm and thus this might be the reason for their unique functional abilities.17 However, through backsplicing from pre-mRNA exons, it is not easy to locate or annotate through an experiment such as polyadenylated RNA-seq. Due to the lack of a 3′ end in circRNAs, these non-coding RNAs are resistant to digestion by ribonuclease R (RNase R). Thus, in a whole RNA pool, circRNAs remain undigested, while other RNAs are digested through RNase R; hence, it helps to easily access these circRNA molecules for next-generation RNA-seq followed by specific computational studies.18,19
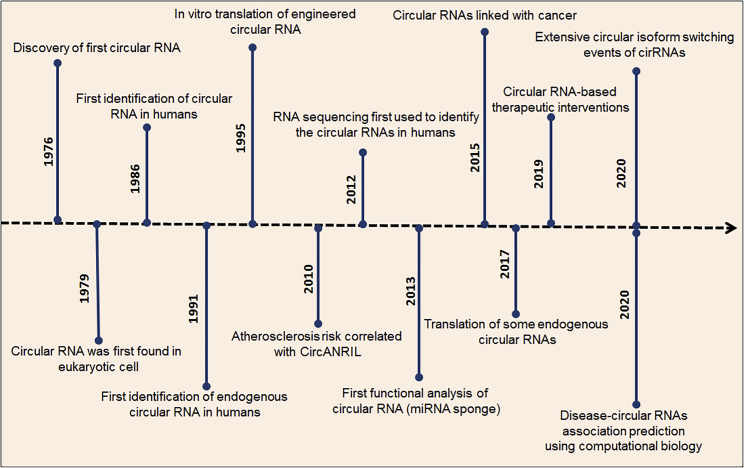
Figure 1.
Recent research progress as shown by a circular RNA PubMed search was performed using the term “circular RNA” (2016–2020)
It is imperative to mention that among several circRNA subtypes, four main subtypes of circRNAs are present. One of them includes exonic circRNAs (ecircRNAs), which are derived from single or several exons, as well as intronic RNAs. Another circRNA is a pre-tRNA intronic circRNA, which is generated by splicing pre-tRNA introns.20 Exon-intron circRNAs are produced for the retention of the internal intron. The production of circular intronic RNAs (ciRNAs) is generated during canonical splicing, generated due to the failure of the debranching of intronic lariats.21,22 By interacting with snRNP (small nuclear ribonucleoprotein), ciRNAs and intron-exon circRNAs can perform the transcription of an ancestral gene in the nucleus.
After the discovery of the first circRNA (plant viroid) through an electron microscope, the aspect of backsplicing model circRNA formation was changed thoroughly with the mounting evidence of thousands of circRNAs in several mice and human cell lines.23,24 After that, several different discoveries of circRNAs created a new line of research (Figure 2). This review highlights a few interesting facts about circRNAs, including their biogenesis and functional characterization. Moreover, the review will help to understand different models of the biogenesis process of circRNAs and types of circRNAs concerning the biogenesis process. The review also discusses different properties of circRNAs such as localization after biogenesis, structure and conserved nature, stability, and expression specificity. Furthermore, a discussion on the biological functions of circRNAs such as miRNA sponging, cell cycle regulation, cell-to-cell communication, transcription regulation, translational regulation, disease diagnosis, and therapeutic potential is also been included. Finally, we discuss the future perspective and conclusions from a future research perspective. This discussion, in turn, will help researchers to develop a new line of scientific endeavor in the field of RNA biology, which is emerging day by day.
Figure 2.
Milestone discoveries in circular RNA research
Biogenesis of circRNAs
circRNAs are generated mainly from pre-mRNA through a phenomenon called backsplicing of exons.25,26 Most circRNAs contain complete exons and originate from protein-coding genes. Nevertheless, circRNAs might also consist of non-coding, antisense, intronic, 3′ untranslated region (UTR), 5′ UTR, or intergenic genomic regions.22,27 The spliceosome mediates backsplicing of pre-mRNA by connecting a downstream splice donor site (5′ splice site) to an upstream acceptor splice site (3′ splice site).28 The splicing mechanism of circRNA is controlled by cis-acting regulatory and trans-acting regulatory factors.29 Canonical splicing sites and regular spliceosomal machinery are essential for the production of circRNAs. Only after transcriptional completion of their host pre-mRNAs has a significant amount of nascent circRNAs been observed.30 Moreover, when the polyadenylation signal is mutated in circRNA-producing linear genes in manipulatable vectors, circRNA production is eliminated, implying that circularization is usually a post-transcriptional event.31 These pieces of evidence suggest that circularization might be a post-transcriptional event, but this is not feasible at all times. Recently, a poly(A) signal has been demonstrated not to be required for circRNA production from minigene vectors, suggesting the possibility of the co-transcription process during circRNA formation.32 Recently, backsplicing of circRNAs from exons has been shown to possibly occur post- or co-transcriptionally.30 Thus, we may assume that in the case of circRNA biogenesis, both post- and co-transcriptional splicing are involved. However, it may not depend on just 3′ end processing; instead, other factors such as transcriptional elongation rate, availability of spliceosome components, and repetitive elements, among others, might also play a critical role.30,32,33 Recent pieces of evidence favor the notion that circRNA processing might largely depend on post-transcription rather than co-transcription.30,31 Metabolic tagging of nascent RNAs with 4-thiouridine (4sU) during circRNA processing revealed that mainly circRNAs are post-transcriptionally processed and are stable.30 For circRNA biogenesis, intronic repeats and exonic sequences must cooperate, and a functional 3′ end processing signal is vital.31 Moreover, their formation might be restricted and controlled by cis-complementary elements.30 Thus, circRNA biogenesis appears to depend mostly on post-transcriptional processes rather than co-transcription, but clear insight into this process is still a prerequisite, and in the near future, researchers might further decipher this dilemma.
Types of circRNAs with respect to the biogenesis process
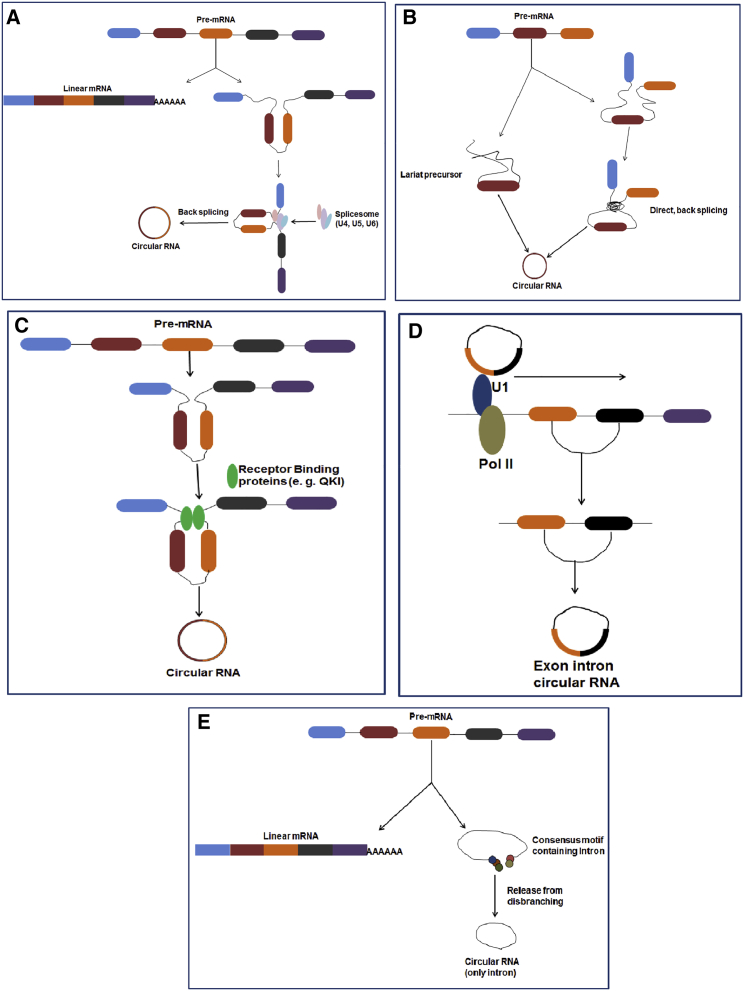
Several types of circRNAs are formed from various biogenesis processes, including exon circRNAs (EcircRNAs), intrinsic circRNAs or ciRNAs, and intron-exon circRNAs (also called exonic-intronic circRNAs (EIciRNAs)) (Figures 3A–3E).34,35
Figure 3.
Different proposed biogenesis mechanisms of circular RNA
(A) Spliceosome-dependent biogenesis of circular RNA. (B) Circular RNA biogenesis occurs through direct backsplicing from pre-mRNA. (C) Circular RNA biogenesis occurs by using RNA-binding proteins (RBPs). (D) Exon-intron circRNA (EIciRNA) biogenesis produced by backsplicing. EIciRNAs contain both exons and introns. (E) Circular intronic RNA (ciRNA) biogenesis process. This ciRNA was derived from lariat introns during canonical splicing.
EcircRNAs are the most common type of circRNA. Most EcircRNAs are formed from the coding gene of pre-mRNA without involving protein-coding genes (Figure 3A).9,12 Certain EcircRNAs have the ability to interact with miRNAs and/or RNA-binding proteins (RBPs), and several EcircRNAs interact with other exons containing canonical translation start codons.2 ciRNA formation depends largely on the conserved sequences at both ends of the introns. ciRNAs contain 2′–5′ phospholipid-linked nucleotides instead of 3′−5′ phospholipid-linked nucleotides (EcircRNAs) and are capable of regulating the expression of genes.36,37 EIciRNAs, however, contain exons along with introns. As a result, EIciRNAs demonstrate the characteristics and functions of both exonic and intronic circRNAs. Similar to ciRNAs, EIciRNA is abundant in the nucleus.29 When EIciRNA is bound to RNA polymerase II (Pol II), EIciRNA usually regulates parental gene transcriptional activity by interacting with snRNP.21
Different models of the circRNA biogenesis process
Spliceosomes or a group of ribozymes I and II mediates the backsplicing of circRNAs from pre-mRNAs (Figure 3D).25,38,39 A reduced level of both circRNAs and linear transcripts after inhibition of canonic spliceosomes further confirms the involvement of spliceosomes in the biogenesis of circRNA.25 Irrespective of orthodox splicing of linear mRNAs, circRNAs can usually arise from a single gene locus. The process usually occurs either through alternative backsplice site selection and/or alternative splice sites. A detail of this process is available in the CIRCpedia database.40 Certain models of the biogenesis process for circRNAs have been proposed. The most common type of model is exon skipping or the lariat-driven circularization model. In this case, a large lariat comprising the exon(s) is synthesized and successively undergoes internal cleavage to excise the intron and produce ecircRNA or EIciRNA (Figure 3B).41 In human umbilical vein endothelial cells stimulated by transforming growth factor (TGF)-β or tumor necrosis factor (TNF)-α, RNA-seq datasets showed that most of the skipped exons could produce ecircRNA.42 The other proposed model is known as the intron pairing-driven circularization or direct backsplicing model. As per this model, exons are split in noncanonical order. Introns containing reciprocal complementary sequences are linked to exons involved in circularization. These introns are spatially close to end-to-end spliced exons that generate circRNAs. However, complementary sequences on either side of an exon do not always produce circRNAs.43 An in vitro minigene constructed model proved that the production of circRNAs does not necessarily require reciprocal complementation between upstream or downstream exon-flanking introns.28 Various types of models of the circRNA biogenesis process are summarized in Figure 4.
Figure 4.
Different types of circular RNA regarding the biogenesis model
Different aspects of circRNA biogenesis
Recent mutational studies with circRNA expression vectors and HeLa cells treated with the splicing inhibitor (blocks spliceosome assembly) isoginkgetin showed that circRNA biogenesis is mainly dependent on canonical splicing.25,26
The efficacy of canonical splicing is much higher than the efficacy of backsplicing. However, the stable mode of circRNA is lower than the stable mode of the mature form of linear RNA. One of the main reasons behind this low efficiency of backsplicing is the unfavorable assembly of spliceosomes, which catalyze the 5′ donor sites of ligation with 3′ acceptor sites of circRNA. However, the exact mechanism involved in backsplicing through the spliceosome is still under investigation. Recently, an analysis of the cryo-electron microscopy structure of the yeast spliceosome E complex revealed that the spliceosome, involving the U1 snRNP and related 3′ splice site (SS) factors (U2AF1, U2AF2, and SF1), can assemble across an exon to define the exon for splicing in a mechanism called exon definition. Splicing through exon definition, rather than the traditional intron definition, leads to backsplicing and circRNA formation.44 Apart from that, the choice of the circRNA expression pattern, as well as the production pattern of linear RNA and circRNA in the cell, remains elusive. These mechanistic insights may reveal more about circRNA biogenesis.
Although the presence of repetitive elements or RBP interactions can facilitate the looping of the flanking introns to enhance backsplicing, these elements are regarded as dispensable. Studies have demonstrated that backsplicing can occur on substrates with minimal introns (∼100 nt) containing only the major splicing signals (5′ SS, 3′ SS, branch site [BS], and polypyrimidine tract [PPT]). Therefore, looping might not be necessary. Nevertheless, the presence of canonical splicing sites has been shown to be indispensable for circRNA formation.26,44 The looping process is regulated by base pairing between the repeat sequence, such as Alu elements, which are present both in the upstream introns and downstream introns.39,45 However, looping can also be facilitated through RBP dimerization. In this case, the RBPs may bind to particular motifs within the flanking introns.
Thus, in addition to cis-elements, RBPs such as FUS (fused in sarcoma) and QKI (quaking; encoded by the QKI gene) can regulate the biogenesis of circRNA (Figure 3C).46,47 FUS has been shown to regulate the production of a considerable number of circRNAs in in vitro-derived mouse motor neurons (MNs). FUS accomplishes this regulation by binding the introns flanking the backsplicing junctions during circRNA biogenesis.46 To enhance the production of numerous circRNAs, QKI binds to recognition elements. During the epithelial-to-mesenchymal transition (EMT), QKI was observed to be involved in the upregulation of circRNAs in humans.47 QKI, which itself is regulated during EMT, is responsible for producing more than one-third of abundant circRNAs. QKI motifs, when added, are enough to induce de novo circRNA synthesis from linearly spliced transcripts. Hence, the insertion process of synthetically QKI-binding sites related to introns may be adequate for the production of circRNAs. The study concluded that cell type-specific mechanisms could purposefully synthesize and regulate circRNAs and play explicit biological functions during EMT.47 The specific adenosine deaminase enzyme related to double-stranded RNA (dsRNA) inhibits base pairing of repetitive elements flanking introns, which in turn inhibits looping and affects the biogenesis of circRNAs and expression. That specific adenosine deaminase enzyme related to dsRNA is coupled with the editing of adenosine to inosine in endogenous dsRNA48 and an enzyme called ATP-dependent RNA helicase A.49 This complex prevents the activation of the innate immune system. Thus, the looping of intron sequences is prevented by this unwinding of dsRNA helical configuration as well as the editing of adenosine to inosine.8,50
Additionally, the biogenesis and expression of circRNAs are regulated by epigenetic regulation of histone proteins and genetic material. Recent studies have shown that silencing the DNMT3B gene, which produces DNA methyltransferase, can affect circRNA expression.51, 52, 53, 54 Thus, the role of gene methylation in the biogenesis and expression of circRNAs indicates circRNA formation. However, the epigenetic regulation status of the promoter region of the host gene can be influenced by circRNAs. The friend leukemia integration 1 transcription factor (FLI1) gene-encoded protein in breast cancer cells is one of the crucial examples of this event.55
Different properties of circRNAs
Localization after biogenesis
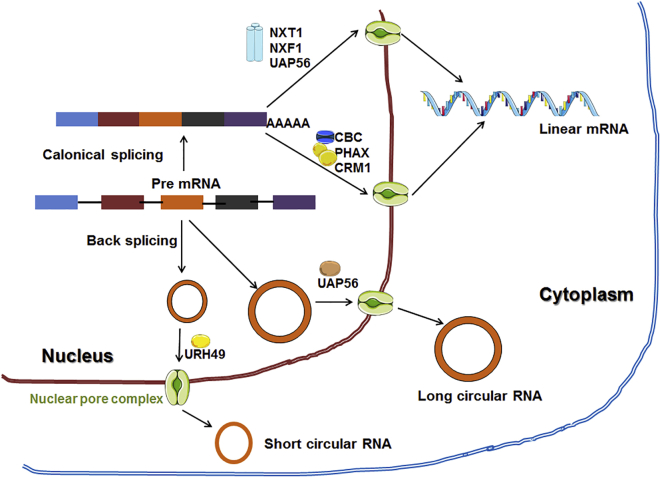
Except for intron-containing circRNAs, most circRNAs are generally exported from the nucleus to the cytoplasm just after their biogenesis.1,9,56 Intronic circRNAs and EIciRNAs have been noted to be extensively found in the nucleus.52,53 This transportation depends on the length of circRNAs and is facilitated by UAP56, a spliceosome RNA helicase, and URH49, an ATP-dependent RNA helicase (Figure 5).54 The export phenomena of circRNAs vary from species to species based on their different length requirements for exporting circRNAs. One example is the export of short circRNAs (<400 nt) related to URH49 in humans. Likewise, the exports of long circRNAs (>1,200 nt) are related to U2AF65-associated protein 56 (UAP56). The conserved K-K/SL-N motif of UAP56 is associated with the export phenomenon of long circRNA through the nuclear pore to the cytoplasm. At the same time, the R-S-F-S motif of URH49 is responsible for the export process of short circRNAs. Interestingly, when URH49 is mutated to contain the Hel25E/UAP56 motif and vice versa, the K-K/SL-N motif for UAP56 and the R-S-F-S motif of URH49 are observed to be sufficient and necessary to dictate circRNA length preferences. This phenomenon subsequently results from the change of behavior of URH49 to Hel25E/UAP56 and vice versa due to the presence of swapped motifs.57 How 7-methylguanosine (m7G)-capped RNAs are exported in cells can explain the role of RNA length in controlling the nuclear export of circRNAs. Capped small nuclear RNAs (snRNAs) (<200 nt) are exported via the PHAX (phosphorylated adaptor RNA export protein)/CRM1 (chromosome region maintenance 1)-mediated pathway but become exportable via NXF1 (nuclear RNA export factor 1)-NXT1 (nuclear transport factor 2-like export factor 1) when their length is increased. Similarly, long capped mRNAs are exported via NXF1-NXT1, but when their length is shortened (intron-less mRNAs), they are exported via the PHAX/CRM1-mediated pathway.58 Localization of circRNAs thus depends on their types (Table 1). Due to the variable lengths, sequences, and structures of circRNAs, they can be fed into specific nuclear export pathways, which further depend on the evolutionarily conserved factors responsible for measuring the length of the mature circRNAs. However, this hypothesis needs further experimental validation.57
Figure 5.
Export mechanism of the circular RNA from the nucleus to the cytoplasm
The export mechanism has occurred through the length-dependent mode.
Table 1.
Different circRNAs: localization, mode of action, and biological functions
| circRNA | Mode of action | Biological functions | References |
|---|---|---|---|
| Cytoplasm | |||
| ciRS-7 (also called CDR1as) | miRNA sponge for miR-7 | positive regulator of insulin secretion, neuronal development, oncogenic functions | 1,2,76,131,170, 171, 172 |
| circHIPK3 | miRNA sponge for multiple miRNAs | tumor suppressor, positive regulator of insulin secretion | 132,172,173 |
| circZNF91 | miRNA sponge or decoy for miR-23b-3p | differentiation of epidermal stem cells | 51 |
| circBIRC6 | miRNA sponge or decoy for miR-34a and miR-145 | pluripotency maintenance | 174 |
| circMbl | protein sponge or decoy for Mbl and template for translation | regulator of neuronal functions | 26,27 |
| circCCDC66 | miRNA sponge or decoy for multiple tumor suppressor miRNAs | oncogenic functions | 175 |
| circPVT1 | miRNA sponge or decoy for miR-497-5p | positive regulator of cell cycle progression | 176 |
| circANRIL | protein sponge or decoy for PES1 | impairs pre-rRNA processing and ribosome biogenesis to induce nucleolar stress and activate p53 | 177 |
| circPABPN1 | protein sponge or decoy for HUR | suppresses PABPN1 translation and decreases cellular proliferation | 178 |
| circ- Amotl1 | protein scaffold (facilitates PDK1-dependent phosphorylation of AKT1) | cardioprotective role in doxorubicin-induced cardiomyopathy | 179 |
| circ-Foxo3 | protein scaffold (facilitates MDM2-dependent ubiquitylation of p53) and sponge for MDM2 (to prevent ubiquitylation of FOXO3) | induces the apoptosis of cancer cells | 103 |
| circ-ZNF609 | template for translation | regulates myoblast proliferation | 114 |
| circPINTexon2 | template for translation | tumor suppressor | 180 |
| circ-FBXW7 | template for translation | tumor suppressor | 181 |
| circ-SHPRH | template for translation | tumor suppressor | 182 |
| Nucleus | |||
| circPAIP2 | enhances protein function (positive regulator of RNA Pol II transcription) | positively regulates the expression of its parental gene | 21 |
| circEIF3J | enhances protein function (positive regulator of RNA Pol II transcription) | positively regulates the expression of its parental gene | 21 |
| ci- ankrd52 | enhances protein function (positive regulator of RNA Pol II transcription) | positively regulates the expression of its parental gene | 22 |
| FECR1 | protein recruitment (recruits TET1 to the promoter region of its own host gene) | oncogenic functions through upregulation of FLI1 | 55 |
| cia-cGAS | protein sponge or decoy for cGAS | protects long-term hematopoietic stem cells from exhaustion | 133 |
FECR1, FLI1 exonic circular RNA; MDM2, mouse double minute 2; PABPN1, poly(A) binding protein 1; PDK1, phosphoinositide-dependent protein kinase 1; PES1, pescadillo homolog 1; TET1, tet methylcytosine dioxygenase 1.
Structure and conserved nature of circRNAs
PSTVd (potato spindle tuber viroid) was the first discovered circRNA and contained rod-shaped, monomeric structures with several distinct loops.59 Liu et al.60 demonstrated 26 circRNA structures (16–26 bp long) from diverse cell lines, except circDHX34 and circPPP1CB. circDHX34 is 29 bp long, and circPPP1CB is 32 bp long. Researchers have shown that endogenous circRNAs can form imperfect RNA duplexes to inhibit dsRNA-activated protein kinase R (PKR). In an autoimmune disease such as systemic lupus erythematosus (SLE) or viral infection, circRNAs undergo massive and rapid degradation by RNase L. This process is essential for PKR activation in the early stage of the innate immune response.
Secondary structures similar to circRNAs may recognize endoribonuclease dicer proteins for the biogenesis of miRNAs. However, the probable molecular mechanism of miRNA production from circRNAs has yet to be discovered. Recent studies have shown that lengthy circRNAs from a plant species (Oryza sativa ssp. indica) include multiple miRNA sequences.61 Apart from that, internal modification may also facilitate mRNA processing. The most abundant internal modification of mRNAs is N6-methyladenosine (m6A). This internal modification has been implicated in all aspects of post-transcriptional RNA metabolism associated with mRNA and long non-coding RNAs (lncRNAs).62 Recently, m6A modifications have also been found in circRNAs. m6A circRNAs mediate mRNA stability via YTHDF2 proteins. Nevertheless, degradation of circRNAs is not promoted by m6A circRNAs as it does for mRNAs.63
Recent studies informed the conservation of circRNA using RNA sequences. The studies include genomic loci and inform the orthologous and paralogous nature of evolution. The conserved nature of circRNAs has been depicted in several species of plants and animals. The conserved nature of circRNAs originates from different genes such as PVT1, HIPK2, IPW, and KIAA0182.9,10 Similarly, the conserved nature of circMBNL1 was expressed in Drosophila heads and humans.24 In addition to animals, approximately 700 conserved ecircRNAs were noted in both plant species (A. thaliana and O. sativa).64 A screening of circRNAs from humans, macaques, and mice also revealed the evolutionarily conserved nature of circRNAs that could be used for prioritization and functional screening. The study tried to utilize the conserved nature and co-expression networks to prioritize circRNAs for the involvement in liver tumerogenisis.65 Another evolutionarily conserved circRNA, GW182, from Drosophila to humans, is shown to have a role in the degradation of other circRNAs.66
Stability and expression specificity
Due to the unique covalent bond between the 5′ and 3′ ends, circRNAs are resistant to RNA exonuclease degradation. Moreover, the absence of a 2′–5′ linkage of an RNA lariat makes the circRNAs resistant to RNA debranching enzymes. circRNAs have long half-lives because of these characteristics. circRNAs have been shown to be stable in cells and exhibit a half-life of more than 48 h compared to mRNAs (10 h).3,9 However, due to the presence of circulating RNA endonucleases in serum, stability for circRNAs is only <15 s.67
The expression profile of circRNAs depends on several factors such as tissue, type of isoforms, and developmental stages, reported in several cases such as WI-38 fibroblast cells, O. sativa, and A. thaliana.1,10,64,68, 69, 70 During drought stress in Triticum aestivum,71 cold tolerance in Vitis vinifera,72 and phosphate imbalance in O. sativa,64 the expression of some stress-specific circRNAs has been reported. In several developmental stages of a plant (Phyllostachys edulis), the differential expression profile of circbHLH93 has been documented.73 One of the most reported tissue/cell-specific circRNAs is circSRY, which shows its expression in adult mouse testes. However, linear Sry mRNA is present only in the tissue of the developing genital ridge.4,74 Another example is the DCC gene, where expression of the circular isoform varies among various human tissues, but its cognate linear mRNA expression is not correlated. In some instances, despite high levels of mRNA expression, no circRNA was detected.23
Meanwhile, it is also true that the expression of some circRNAs is more abundant than the expression of their linear counterparts. In several cell lines, the expression of circCAMSAP1 is several-fold higher than the expression of its linear counterpart. A similar kind of overexpression of circRNA of approximately 50 different genes was observed in different cell lines, i.e., HeLa, AG04450, and A549 cell lines.10 Similarly, in O. sativa ssp. indica, approximately 20% of circRNAs were found to be expressed at higher levels than in their linear counterparts.9
Function
The functional mechanism of circRNAs includes mainly acting as miRNA sponges, regulating gene splicing or transcription, translation, interacting with proteins, and epigenetic regulation. Usually, due to genetic code redundancy, the third position is not highly conserved in a codon. However, in some circRNAs, the third position is more conserved than in exons (not part of circRNAs), suggesting that these highly stable circRNAs have significant non-coding functions. circRNAs can act as post-transcriptional regulators by competing with other RNAs (for binding with miRNAs and RBPs).1 If functional classification is broadly performed according to the target of circRNAs, then they can generally be divided into two categories: regulating the target site and regulating its host gene. However, a recent investigation suggested that circRNAs show different physiological functions and regulate gene expression at various levels.75
miRNA sponges: A novel function of circRNAs
miRNA sponging is one of the well-established functions of circRNAs, where they can inhibit the function of miRNA by binding the target miRNAs directly or indirectly (Table 2). The relationship between the circRNA interaction site and miRNA as well as the mRNA target site is very significant for this functional consideration. circRNAs possess miRNA response elements as linear RNAs, allowing them to compete with miRNA binding to linear RNAs, suggesting a regulatory role in miRNA functioning and gene expression.9,93 Moreover, this study indicated that RNA interference (RNAi) may target circRNAs. Therefore, to support this model, one study was carried out on ciRS-7, which is a special type of circRNA sponge for miR-7. The ciRS-7 was produced from a CDR1 antisense transcript with approximately 74 miR-7 binding sites in humans as well as approximately 63 conserved binding sites from different vertebrates (32 vertebrates). In particular, ciRS-7 has highly stable expression in several tissues, specifically in the brain.1,94,95 The capacity of ciRS-7 to bind almost 20,000 miR-7s in each cell is due to its high expression potential. ciRS-7 can influence the binding capacity of miR-7 with target mRNAs, which are involved in various cancers95,96 and neurodegenerative disorders such as Alzheimer’s disease77 and Parkinson’s disease.97 Hence, if the expression of ciRS-7 can be controlled, then it can regulate mRNA expression along with the binding sites of miR-7. This study further suggests that there is competition between ciRS-7 and mRNAs for miR-7 binding.2 Apart from that, ciRS-7 has a complementary binding site with miR-671, which causes the cleavage of ciRS-7 by Argonaute-2 (AGO-2).98 In addition to ciRS-7, another circRNA (testis-specific circRNA), circSRY, is an example that has several target sites for miR-138 (16 target sites in mice) and might play a crucial role in the progression of several diseases (cancer and Parkinson’s disease).2,99
Table 2.
Diseases regulation of circular RNAs through miRNA sponging
| SI no. | Circular RNAs | Sponged miRNA | Target gene for miRNA | Different diseases regulation | References |
|---|---|---|---|---|---|
| 1 | CDR1as/ciRS-7 | miR-7 | Pax6 and Myrip | diabetes | 76 |
| 2 | CDR1as/ciRS-7 | miR-7 | UBE2A | Alzheimer’s disease | 77 |
| 3 | circRNA-CER | miR-136 | MMP13 | osteoarthritis | 78 |
| 4 | circTCF25 | miR-103a-3p and miR-107 | CDK6 | bladder cancer | 79 |
| 5 | circRNA-MYLK | miR-29a | VEGFA | bladder cancer | 80 |
| 6 | hsa_circ_000984 | miR-106b | CDK6 | colorectal cancer | 81 |
| 7 | hsa_circ_0020397 | miR-138 | TERT and PD-L1 | colorectal cancer | 82 |
| 8 | ciRS-7 | miR-7 | YY1 | colorectal cancer | 22,83 |
| 9 | ciR-SRY | miR-138 | TWIST2 | colorectal cancer | 84 |
| 10 | ciR-SRY | miR-138 | PCNA; Bcl-2 | ovarian cancer | 85,86 |
| 11 | ciRS-7 | miR-7 | IGF1R | gastric cancer | 87 |
| 12 | CDR1as/ciRS-7 | miR-7 | PI3K | gastric cancer | 88 |
| 13 | circRNA_LARP4 | miR-424 | LATS1 | gastric cancer | 89 |
| 14 | ciR-SRY | miR-138 | H2AX | lung cancer | 90,91 |
| 15 | ciR-SRY | miR-138 | APT1/2 | chronic lymphocytic leukemia | 92 |
APT1/2, acyl protein thioesterase 1/2; Bcl-2, B cell lymphoma-2; CDK6, cyclin-dependent kinase 6; H2AX, histone family 2A variant, member X; IGF1R, insulin-like growth factor 1 receptor; LATS1, large tumor suppressor kinase 1; MMP13, matrix metalloproteinase 13; Myrip, myosin VIIA and Rab interacting protein; Pax6, paired box 6; PCNA, proliferating cell nuclear antigen; PD-L1, programmed death-ligand 1; PI3K, phosphoinositide 3-kinase; TERT, telomerase reverse transcriptase; TWIST2, twist basic helix-loop-helix transcription factor 2 gene; UBE2A, ubiquitin protein ligase A; VEGFA, vascular endothelial growth factor A; YY1, Yin Yang 1.
A single circRNA can bind to not only a single miRNA but also many miRNAs at one or more sites. One example is circFOXO3, which can bind with different miRNAs such as miR-138, miR-22, miR-762, miR-136, miR-433, miR-3622b-5p, miR-149, and miR-3614-5p. Another example is circITCH, which can bind to miR-124, miR-17, and miR-138.61,99 However, from 16 different plants, 115,171 circRNAs were observed. This work demonstrated that the circRNA sponging function is not only observed in humans but is also documented in different plants. However, circRNA sponging is observed more often in different diseases.
Cell cycle regulation and cell-to-cell communication
Numerous circRNAs show different mechanisms of action due to various binding sites to miRNAs as well as proteins related to cell cycle regulation. For example, upon binding to miR-762, miR-138, miR-433, miR-22, miR-149, miR-3622b-5p, miR-96, miR-136, and miR-3614-5p, circFOXO3 sponges the binding of these miRNAs to the linear variant of FOXO3. This binding relives its suppression.100,101 Similarly, the binding sites of circFOXO3 control cell cycle regulatory proteins such as p53, p27, p21, mouse double minute 2 (MDM2), and cyclin-dependent kinase (CDK)-2. Using pull-down assays, the role of circRNA-protein interactions was studied for cell cycle regulation.102,103 Studies by Du et al.102 demonstrated that CDK-2 and p21 bind to adjacent sites of circFOXO3 to form a ternary complex. This complex arrests the cell cycle in the G1 phase by inhibiting the activation of the complex (cyclin E/CDK-2), which is required for the G1/S transition.
In recent studies, overexpression of circRASSF2 was identified in circulating exosomes from patients suffering from laryngeal squamous cell carcinoma (LSCC).104 Within the different lengths of circulating exosomal circRNAs, tiny-sized circRNAs, encircled by exosomes circulating in the blood of animals, can act as significant biomarkers as well as cell-signaling molecules. Additionally, changes in the miRNA levels in producer cells may regulate the sorting of associated circRNAs to exosomes. This, in turn, might result in the transmission of the biological activity to recipient cells.105
Transcription regulation
circRNAs are able to regulate transcription and alternate splicing. Recent studies have shown that circRNAs can regulate transcription in both cis and trans manners. Most circRNAs are located in the cytoplasm,6,9 and intron-containing circRNAs are found in the nucleus of cells in humans. Linear RNAs with retained introns are passaged into the cytoplasm, and those with detained introns stay in the nucleus.21,22,106 For example, circPAIP2, circMCM5, circEIF3J, circANKRD52, and circSIRT7 are introns containing circRNAs that may interact with the elongated RNA Pol II complex to regulate the expression of their genes through a positive feedback mechanism.22
Alternative splicing by an intron retention (IR) mechanism can form different isoforms from a single gene. Usually, fully spliced introns in mature mRNA isoforms are passaged out to the nucleus for translation. The presence of premature termination codons (PTCs) in introns causes rapid degradation of intron-retaining isoforms (IRIs) (including circRNAs) mediated by the nonsense-mediated decay (NMD) pathway.107 Sometimes IRIs can avoid the NMD pathway108 and get translated to protein isoforms but are often truncated109 and are toxic to cells. Thus, retained introns after splicing may lead to gene regulation by introducing a stop codon in an ORF and forcing premature termination of translation without affecting the transcriptional activity. At the same time, IRs that are not exported to the cytoplasm but remain in the nucleus are known as detained introns (DIs). A study has shown that CLK1-DIs can serve as a reservoir for the CLK1 mRNA. CLK1 is a dual-specificity kinase responsible for phosphorylating the spliceosomal complex regulating RNA splicing. The study concluded that polyadenylated RNAs having DIs could serve as a reservoir of pre-mRNAs or lead to a dead end by degrading them in the nucleus.110
EiciRNAs can express their genes after binding to the U1 spliceosome complex, along with post-transcriptional regulation.111 However, some circRNAs (e.g., circMBL, circFMN, and circDMD) can suppress expression by binding directly to their cognate mRNAs (mRNA traps).112 Due to incomplete splicing, some EIciRNAs retain introns that allow them to interact with U1 snRNP and promote their host gene transcription.21 In A. thaliana, exon 6 of a key developmental regulatory gene, SEPALLATA3 (SEP3), generates circRNA, of which ∼15% of the circular transcript is retained in the nucleus. This circRNA forms an R-loop (an RNA:DNA hybrid) at endogenous SEP3 genomic loci. The formation of this R-loop results in elevated linear SEP3 splice variant production with exon 6 skipped. R-loop formation has been suggested to result in a pause of SEP3 transcription, allowing recruitment of alternative splicing regulators to promote exon skipping.113 Similar kinds of transcriptional regulation might be expected in mammalian cells, and further research might decipher this process. Thus, circRNAs can play a pivotal role in diverse transcriptional regulations.
Translation of circRNAs
As circRNAs are classified as non-coding RNAs, their role in translation has never caught attention. Several studies have reported the protein-coding function of circRNAs (endogenous) due to polysomal association.27,114,115 Owing to the absence of the 5′ cap and the poly(A) tail, circRNAs are unable to undergo cap-dependent translation. However, due to the presence of internal ribosome entry sites (IRESs) and the ability to bind to open reading frames (ORFs), circRNAs have been shown to code for proteins/polypeptides.116 One of the examples is circZNF609, which is composed of two in-frame start codons that are separated by 150 nt and are found in myoblast cells. This circRNA has a 5′ IRES (conserved IRES). Thus, cap-independent translation can produce two similar intense proteins, which shows the functional ability of circRNAs in translation. However, certain evidence signifies its role in myogenesis, which helps in the propagation of mouse and human myoblast cell lines and can be suppressed through the silencing of this circRNA (circZNF609) using small interfering RNA (siRNA).114 Methylation of the IRES of circZNF609 might be a possible reason for cap-independent translation, which has been investigated through an RNA-wide analysis of the m6A pattern.117 A single m6A site has been observed to be sufficient rather than two to induce circRNA translation with the same efficiency in IRES.115
As listed in circRNADb, protein expression was recorded from 72 circRNAs in humans.118 Apart from this, 250 circRNAs were found to have translatable coding potential, and they can be linked with polysomes.119 circMBL3 encodes a protein (∼37 kDa) that is an endogenous circRNA found in Drosophila. From the functional catalog of circRNAs in protein translation, circRNAs have been reported to have a possible role in cancer progression.120 For example, a product of the circPPP1R12A (hsa_circ_0000423) gene (circPPP1R12A-73aa, having a 216-nt short ORF) is implicated in cancer progression. Protein from circPPP1R12A-73aa has a unique conserved peptide at the C terminus. In nude mice and from 20 different patient samples, circPPP1R12A-73aa protein was shown to be able to regulate colon cancer progression invasion and metastasis. Several colon cancer cell lines (e.g., SW620, HT-116, LoVo, DLD-1, HCT-15, HT-29, SW480, SW48, and Caco-2) also showed expression of circPPP1R12A-73 aa protein compared to the normal cell line (NCM460).119
The functional role of most circRNA-derived peptides has yet to be elucidated. However, peptides from circRNAs have been proposed to often be truncated versions of canonical proteins and lack vital functional sites. Therefore, they might function either as dominant-negative protein variants or decoys or modulators of alternative protein complexes.114 Most circRNAs are synthesized from protein-coding genes. Thus, ORFs in circRNAs often have similarities to the ORFs of the corresponding mRNAs. Hence, the best possible way to detect whether a protein is synthesized from the linear or circular transcript is to characterize the sequence downstream of the back splice junction (BSJ) and before the putative stop codon.32 A detailed methodology for studying the translation of circRNAs has been reviewed by Kristensen et al.32 Authors have suggested using several molecular techniques, such as luciferase reporters or analysis of m6A methylation, to identify the presence of ORF functional IRES-like elements. Mass spectrometry has been applied for detecting circRNA-specific peptides spanning the BSJ and inserting a tag protein in the circRNA ORF (upstream of the putative stop codon) using a minigene setup or CRISPR-Cas9.121 This tag can be detected by western blotting by using an antibody against the circRNA-specific part of the peptide if available. The tag can also help to study localization by using peptide-specific immunofluorescence. Finally, polysome profiling or ribosome footprinting can be used to study the translated products of circRNAs.
circRNAs and disease diagnosis
For disease diagnosis, next-generation biomarker discovery is an urgent need. In recent years, due to the tissue-specific expression, longevity, and high abundance of circRNAs, many studies have characterized circRNAs, which highlights their potential as biomarkers for certain diseases. Previously, miRNAs and several RNA transcript regions, which can act as stable biomarkers, were explored. Recently, after the discovery of circRNAs, scientists have attempted to establish this molecule as a suitable biomarker through RNA profiling and have studied its sensitivity.120 The capability of circRNAs to act at the transcriptional and post-transcriptional levels makes them ideal candidates for biomarkers for various diseases such as cardiovascular diseases, neurological disorders, and cancer. circRNAs tend to be released from cells to the circulatory system, which has the potential to serve as a biomarker. circRNAs found in exosomes can be used as potential biomarkers for cancer diagnosis.105 In colorectal cancer (CRC) patients, exo-circRNAs within serum exosomes constitute a novel and stable class of circRNAs.105 A correlation between the expression of circ_05075 (hsa_circ_0005075) and tumor size in hepatocellular carcinoma (HCC) suggests that it is a potential HCC biomarker. Although a correlation has been identified between HCC and circRNAs, further research is required to understand the role of circRNAs in HCC development.122 Stable small-sized circRNAs are found in the circulatory system. circRNAs have also been found to have the potential to cross the blood-brain barrier (BBB), and the circRNAs can be detected upon entering the cerebrospinal fluid (CSF). Thus, these circRNAs indicate central nervous system (CNS) disorders and can act as potential biomarkers.123
circRNAs and therapeutic potential
circRNAs are being extensively studied in human diseases. Several circRNAs are differentially expressed in different diseases (Figure 6). Among these circRNAs, some were upregulated, and some were downregulated. Due to their ability to regulate gene expression, circRNAs have come into sight as potential therapeutic molecules. Scientists are targeting different circRNAs that have therapeutic potential. Several circRNAs are being used as therapeutic targets in different diseases. Furthermore, tumor-suppressive circRNAs have been observed to have the potential to act as therapeutic molecules in cancer.124 However, to understand the therapeutic potential of circRNAs, it is necessary to perform functional circRNA knockdown experiments and/or circRNA overexpression experiments. Using self-splicing introns125, 126, 127 or splint ligation approaches,39 circRNAs can be generated and added to cells for functional analysis. Overexpression of circRNA can be performed through circRNA overexpression plasmids with the circRNA sequence.74
Figure 6.
Different circular RNAs associated with different diseases
Utilizing endogenous spliceosome machinery, a number of plasmid- and virus-based methods are available for expressing circRNAs of interest. Even in some cases, the production of linear RNAs is minimal.128, 129, 130 Expression of circRNA genes can be achieved using a minigene setup whereby canonical splice sites and inverted complementary sequences flank a circularizing exon26,31 or the entire gene can be expressed.74 Naturally occurring repeat elements such as Alu can act as complementary sequences, or these elements can be produced by inserting a region (>30 nt) of the upstream intron downstream of the circularizing exon in an inverted position. Inserting the binding domains of certain RBPs in the flanking introns (promoting circularization) can further enhance the expression of circRNA transcripts from plasmids.47 Alternatively, the knockdown of specific circRNAs can be attained using short hairpin RNAs (shRNAs)/siRNAs. These molecules can be used to target the backsplicing junction. CRISPR-Cas9 genome editing can be used to remove flanking repeat elements (or the entire locus in the case of CDR1as/ciRS-7)131, 132, 133. Recently, for the first time, circRNA knockout animals were generated. Knockout of CDR1as/ciRS-7 caused impaired sensorimotor gating in mice, a phenotype associated with neuropsychiatric disorders.131 At the same time, cia-cyclic guanosine monophosphate-AMP synthase (cGAS) knockout in mice caused the number of long-term hematopoietic stem cells (associated with severe anemia and death) to be reduced.133 However, the most successful model will be when phenotypes observed after the knockout of a circRNA can be compensated by a plasmid or viral vector reexpressing the circRNA. For instance, overexpression of a mouse Fndc3b gene circRNA via adeno-associated virus (AAV) resulted in better cardiac repair in a myocardial infarction model.130 Due to the presence of its genome, mitochondria play crucial roles in immuno-metabolic syndromes. Steatohepatitis-associated circRNA ATP5B regulator (SCAR), located in mitochondria, was observed to inhibit mitochondrial reactive oxygen species output and activation of fibroblasts from patients with nonalcoholic steatohepatitis (NASH). In vivo study revealed that targeting of circRNA SCAR alleviates insulin resistance and high-fat diet-induced cirrhosis.134
New delivery strategies of therapeutic-based circRNAs may further help to explore the therapeutic potential of circRNAs. The different circRNAs with therapeutic potential for different diseases are listed in Table 3.
Table 3.
Different diseases and related circular RNAs that can be used as their probable therapeutic potential
| Sl no. | Different diseases | Related circular RNAs (probable therapeutic potential) | Remark/inhibition/regulation type | References |
|---|---|---|---|---|
| 1 | lung cancer | cirITCH | downregulation | 135 |
| 2 | gastric cancer | hsa_circ_002059 | downregulation | 136 |
| 3 | hepatocellular carcinoma | circ_0067934 | – | 137 |
| 4 | cervical squamous cell carcinoma | circCDKN2B-AS1 | interacts with IMP3 | 138 |
| 5 | renal cell carcinoma | circRNA_001287 | targets through miR-144-targeted CEP55 | 139 |
| 6 | bladder carcinoma | hsa_circRNA_100876 | interacts with mir-136-5p, and with mRNA-chromobox 4 (CBX4) subsequently | 140 |
| 7 | coronary artery disease | hsa_circ_0124644 | upregulated | 141 |
| 8 | alzheimer’s disease | ciRS-7 | upregulates UBE2A | 77 |
| 9 | diabetes | ciRS-7 | inhibits miR-7 function in islet β cells | 142 |
| 10 | allergic asthma | hsa_circ_0002594 | upregulated in T helper 2-mediated allergic asthma | 143 |
| 11 | schizophrenia | hsa_circRNA_104597 | downregulated | 144 |
| 12 | metabolic dysfunction-associated fatty liver disease (MAFLD) | circScd1 | upregulated and circScd1 influences JAK2/STAT5 signaling | 145 |
| 13 | hepatocellular carcinoma (HCC) | has_circ_0078710 | upregulated and interacts with miR-31/HDAC and CDK2 signaling | 146 |
| 14 | hepatocellular carcinoma (HCC) | circMAT2B | upregulated and interacts with miR-338-3p/PKM2 signaling | 147 |
| 15 | fibrosis | mmu_circ_34116 | downregulation; increases α-SMA expression | 148 |
| 16 | fibrosis | hsa_circ_0070963 | downregulation; regulates miR-223-3p/LEMD3 signaling | 149 |
| 17 | fibrosis | circMTO1 | downregulation; regulates miR-17-5p/Smad7 signaling | 150 |
| 18 | fibrosis | circ-PWWP2A | upregulated; regulates miR-203 and miR-223/TGF-β and LPS signaling | 151 |
| 19 | axial spondyloarthritis | hsa_circ_0079787 | it is positively correlated with the plateletcrit (PCT) and negatively correlated with the mean platelet volume (MPV) | 152 |
| 20 | atherosclerosis | hsa_circ_0003575 | upregulated; regulates oxLDL-induced vascular endothelial cells | 153 |
Recent research progress on circRNAs
Several breakthroughs in research that have been performed on circRNAs are illustrated in the following sections.
Recent basic research progress on circRNAs
Although many eukaryotic genes generate linear mRNAs and circRNAs, the extent to which the ratio of linear RNA to circRNA is generated and how it is modulated remain unknown. In Drosophila cells, utilizing RNAi screening, many transcription termination factors and core spliceosomes that regulate the RNA outputs of the reporter and endogenous genes have been identified. The steady-state output of protein-coding genes was observed to be diverted toward the synthesis of circRNAs when canonical pre-mRNA processing is slowed down or inhibited. circRNA production is increased due to the activation of alternate pathways (backsplicing), leading to the formation of circRNAs from nascent RNAs. This methodology may provide an alternative pathway for circRNA production.154 Wu et al.155 generated a full-length assembly tool, namely, circAST. Depending on the internal structural characteristics of circRNAs, this tool can help analyze the expression of circRNA by using multiple splice graphs and assemble the transcripts of alternatively spliced circRNA. A limitation of the short half-life makes mRNA use in biological systems infeasible. Recently, to extend the duration of protein extension from linear RNA, exogenous circRNA was synthesized. This circRNA was developed by utilizing engineered self-splicing introns. Engineered exogenous circRNAs can produce robust, stable, and potent protein expression in eukaryotic cells and might offer an alternative to linear RNAs.126 Dong et al.156 developed a database entitled CIRCpediav2 for circRNA annotation as well as their expression assessment. For annotation, the database used nearly 180 RNA-seq datasets from six different species. Moreover, this web interface provides computational tools for the comparison of the expression of circRNAs. In the developing human frontal cortex, circSLC45A4 is one of the most highly expressed RNA splice isoforms. Knockdown of circSLC45A4 pushes the human neuroblastoma cell line to undergo spontaneous neuronal differentiation. Thus, to maintain the progenitor state of neural cells in the mammalian group brain, circSLC45A4 is necessary.157 Recent progress in the field of circRNAs would further pave the way for the development of new strategies that could be utilized to address the molecular pathogenesis of several diseases.
For identifying circRNAs, short-read RNA-seq is the most commonly used technique. However, it suffers from the limitation that experimentally it is unable to determine full-length sequences and exact exonic compositions of circRNAs. Due to this, the function of very few circRNAs has been characterized. Knowledge of full-length circRNA sequences could substantially benefit the functional studies of circRNAs.158 Thus, strategies for reading long-read RNA-seq are being developed. IsoCirc is one of those strategies that can sequence full-length circRNA isoforms. The strategy involves rolling circle amplification (RCA) followed by a nanopore long-read sequencing. The strategy successfully prepared a comprehensive catalog of full-length circRNA isoforms from 12 human tissues and one human cell line (HEK293).159 Another study utilized nanopore technology for long-read sequencing of circRNAs from the brain of mice and humans. The study concluded that for mapping the specific exon composition of circRNA, the nanopore sequencing technology is fast and reliable.160 Recently, to assemble and quantify circRNAs, an algorithm called circRNA identifier (CIRI) has been developed. CIRI is a group of software that includes CIRI-AS and CIRI-long. CIRI-AS utilizes a spliced junction signature-based algorithm for high-throughput detection of internal components of circRNAs based on short-read sequencing, whereas CIRI-long uses a high-throughput approach to construct full-length circRNAs and circular isoforms. CIRI-long has the advantage that other than providing complete sequences of circRNAs, it permits the analysis of circRNAs at the isoform level. Hence, by utilizing CIRI-long or CIRI-full, differences among the circRNAs at the isoform level were studied in human liver tumor and normal tissues.161 Another study also utilized CIRI-long for analyzing the data from full-length sequencing of circRNA isoforms using nanopore technology. The strategy was able to identify a new type of circRNA (intronic self-ligated) that exhibited unique splicing and expression patterns.162
Recent research progress on the disease association, progression, and therapeutic potential of circRNAs
Recently, a cancer-specific circRNA database was created that contains several identified circRNAs from normal samples as well as both tumor samples. For each circRNA, researchers have predicted RBP sites. Researchers have also predicted the splicing events in each circRNA to understand the association between backsplicing and linear splicing.163 Another study by Chen et al.164 found that one intronic circRNA developed from the AGO2 gene (circAGO2) regulates cancer progression. At the same time, they found that the interaction between circAGO2 and HuR (human antigen R) may facilitate the activation of circAGO2. Sun et al.165 found a circRNA-based next-generation biomarker of gastric carcinoma, hsa_circ_0000520. Yu et al.166 observed that a circRNA called cSMARCA5 regulates metastasis and progression of HCC, and this circRNA was generated from the SMARCA5 gene (exons 15 and 16). Chen et al.167 discovered a next-generation biomarker for the diagnosis of gastric cancer, i.e., circRNA hsa_circ_0000190. Recently, circRNA circUHRF1 was observed to be associated with oral squamous cell carcinoma. This circRNA is derived from splicing factors and controls tumorigenesis through a feedback loop.168 Similarly, circRNA and circ-CCAC1 were observed to have a role in cholangiocarcinoma progression and to have a role in metastasis and tumorigenesis. This circRNA can be a therapeutic target or next-generation biomarker for this disease.117 Similarly, Xu et al. found another circRNA for this disease that was downregulated. This circRNA controls cell invasion, cell migration, and cell proliferation in cholangiocarcinoma.169
Future perspectives and conclusions
Just some peculiarities or errors of pre-mRNA splicing do not form circRNAs, but instead, they are tightly regulated transcripts. In some instances, circRNA can perform critical biological functions. Many recent studies in the area of circRNA have enlightened RNA biology in a new direction. circRNAs, unlike linear mRNAs, can possess different functionalities due to their circular dimensions. With the discovery of novel circRNAs, many diverse functions have attracted the attention of scientists. circRNAs are critical players in the appearance of different diseases such as cancer, diabetes, neurological diseases, cardiovascular disease, and many other diseases. Mainly, circRNAs can act as a promising biomarker and have a significant role in therapeutic purposes for different diseases. This emerging area could further show the way for functional diagnosis and successful treatment. Based on unique structural features, biological properties, and biological functions, circRNAs have become the modern research hotspot today, especially in the areas of effective diagnostics and therapeutics. However, more research in this direction is needed to establish the therapeutic potential and use of circRNAs.
More functional characterization can be studied by designing exclusive modern tools such as bioinformatics tools for circRNAs, where the possible target identification and interaction of circRNAs with their possible targets can be studied very efficiently. However, many bioinformatics tools with different signature algorithms are needed that can efficiently detect circRNAs, their structures, and their interactions. Moreover, strategies that will allow determining spatial and temporal gene expression patterns in specific cell types, specifically in single cells, can be more realistic for understanding the biological functions of circRNAs. Also, the development of new and rapid genome-wide circRNA sequencing techniques are required in the near future to annotate the roles and biological functions of circRNAs.
Another intriguing fact is that the secondary structure of circRNA may affect miRNA sponging ability. However, more detailed studies are needed on miRNA sponging. circRNAs in developmental biology should be explored as another area of research. Apart from this exploration, many areas need to be investigated to validate the activities and regulation of circRNAs in both plant and animal systems. However, state-of-the-art methodologies such as CRISPR-Cas9 technology will help in more in-depth research and will help to solve the different research questions.
Acknowledgments
This study was supported by the Hallym University Research Fund and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2020R1C1C1008694 and NRF-2020R1I1A3074575).
Author contributions
A.R.S., M.B., and C.C. researched data for the article. A.R.S., M.B., and C.C. wrote the draft manuscript, substantively contributed to the discussion, and edited the article. S.B. and A.S. contributed to generating the tables and editing and reviewing the manuscript. S.-S.L. and C.C. reviewed and edited the manuscript. A.R.S. and S.-S.L. managed funding and provided management.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Sang-Soo Lee, Email: 123sslee@gmail.com.
Chiranjib Chakraborty, Email: drchiranjib@yahoo.com.
References
- 1.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 2.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 3.Cocquerelle C., Mascrez B., Hétuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 4.Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 5.Gaiti F., Calcino A.D., Tanurdžić M., Degnan B.M. Origin and evolution of the metazoan non-coding regulatory genome. Dev. Biol. 2017;427:193–202. doi: 10.1016/j.ydbio.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan X., Zhang X., Wu X., Guo H., Hu Y., Tang F., Huang Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015;16:148. doi: 10.1186/s13059-015-0706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C., Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 9.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westholm J.O., Miura P., Olson S., Shenker S., Joseph B., Sanfilippo P., Celniker S.E., Graveley B.R., Lai E.C. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L., Duff M.O., Graveley B.R., Carmichael G.G., Chen L.-L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu T., Cui L., Zhou Y., Zhu C., Fan D., Gong H., Zhao Q., Zhou C., Zhao Y., Lu D. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–2087. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P.L., Bao Y., Yee M.-C., Barrett S.P., Hogan G.J., Olsen M.N., Dinneny J.R., Brown P.O., Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng L., Chen G., Zhu Z., Shen Z., Du C., Zang R., Su Y., Xie H., Li H., Xu X. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget. 2017;8:808–818. doi: 10.18632/oncotarget.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu M.-T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki H., Zuo Y., Wang J., Zhang M.Q., Malhotra A., Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vincent H.A., Deutscher M.P. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 20.Lu Z., Filonov G.S., Noto J.J., Schmidt C.A., Hatkevich T.L., Wen Y., Jaffrey S.R., Matera A.G. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21:1554–1565. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Zhang X.-O., Chen T., Xiang J.-F., Yin Q.-F., Xing Y.-H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 23.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 24.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L.-H., Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E. Translation of circRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mumtaz P.T., Taban Q., Dar M.A., Mir S., Haq Z.U., Zargar S.M., Shah R.A., Ahmad S.M. Deep insights in circular RNAs: From biogenesis to therapeutics. Biol. Proced. Online. 2020;22:10. doi: 10.1186/s12575-020-00122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.-L., Yang L., Chen L.L. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 31.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 33.Huang C., Shan G. What happens at or after transcription: Insights into circRNA biogenesis and function. Transcription. 2015;6:61–64. doi: 10.1080/21541264.2015.1071301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao X., Cai Y., Xu J. Circular RNAs: Biogenesis, mechanism, and function in human cancers. Int. J. Mol. Sci. 2019;20:3926. doi: 10.3390/ijms20163926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greene J., Baird A.-M., Brady L., Lim M., Gray S.G., McDermott R., Finn S.P. Circular RNAs: Biogenesis, function and role in human diseases. Front. Mol. Biosci. 2017;4:38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holdt L.M., Kohlmaier A., Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell. Mol. Life Sci. 2018;75:1071–1098. doi: 10.1007/s00018-017-2688-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petkovic S., Müller S. Synthesis and engineering of circular RNAs. Methods Mol. Biol. 2018;1724:167–180. doi: 10.1007/978-1-4939-7562-4_14. [DOI] [PubMed] [Google Scholar]
- 38.Vicens Q., Westhof E. Biogenesis of circular RNAs. Cell. 2014;159:13–14. doi: 10.1016/j.cell.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X.-O., Wang H.-B., Zhang Y., Lu X., Chen L.-L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X.-O., Dong R., Zhang Y., Zhang J.-L., Luo Z., Zhang J., Chen L.L., Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–1287. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petkovic S., Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang A., Zheng H., Wu Z., Chen M., Huang Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics. 2020;10:3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Lu T., Wang Q., Liu J., Jiao W. Circular RNAs: Crucial regulators in the human body (Review) Oncol. Rep. 2018;40:3119–3135. doi: 10.3892/or.2018.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X., Liu S., Zhang L., Issaian A., Hill R.C., Espinosa S., Shi S., Cui Y., Kappel K., Das R. A unified mechanism for intron and exon definition and back-splicing. Nature. 2019;573:375–380. doi: 10.1038/s41586-019-1523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelly S., Greenman C., Cook P.R., Papantonis A. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015;427:2414–2417. doi: 10.1016/j.jmb.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 46.Errichelli L., Dini Modigliani S., Laneve P., Colantoni A., Legnini I., Capauto D., Rosa A., De Santis R., Scarfò R., Peruzzi G. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 48.Eisenberg E., Levanon E.Y. A-to-I RNA editing—Immune protector and transcriptome diversifier. Nat. Rev. Genet. 2018;19:473–490. doi: 10.1038/s41576-018-0006-1. [DOI] [PubMed] [Google Scholar]
- 49.Koh H.R., Xing L., Kleiman L., Myong S. Repetitive RNA unwinding by RNA helicase A facilitates RNA annealing. Nucleic Acids Res. 2014;42:8556–8564. doi: 10.1093/nar/gku523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aktaş T., Avşar Ilık İ., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., Manke T., Backofen R., Akhtar A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 51.Kristensen L.S., Okholm T.L.H., Venø M.T., Kjems J. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2018;15:280–291. doi: 10.1080/15476286.2017.1409931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rinaldi L., Datta D., Serrat J., Morey L., Solanas G., Avgustinova A., Blanco E., Pons J.I., Matallanas D., Von Kriegsheim A. Dnmt3a and Dnmt3b associate with enhancers to regulate human epidermal stem cell homeostasis. Cell Stem Cell. 2016;19:491–501. doi: 10.1016/j.stem.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 55.Chen N., Zhao G., Yan X., Lv Z., Yin H., Zhang S., Song W., Li X., Li L., Du Z. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19:218. doi: 10.1186/s13059-018-1594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang C., Liang D., Tatomer D.C., Wilusz J.E. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masuyama K., Taniguchi I., Kataoka N., Ohno M. RNA length defines RNA export pathway. Genes Dev. 2004;18:2074–2085. doi: 10.1101/gad.1216204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.López-Carrasco A., Flores R. Dissecting the secondary structure of the circular RNA of a nuclear viroid in vivo: A “naked” rod-like conformation similar but not identical to that observed in vitro. RNA Biol. 2017;14:1046–1054. doi: 10.1080/15476286.2016.1223005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu C.-X., Li X., Nan F., Jiang S., Gao X., Guo S.-K., Xue W., Cui Y., Dong K., Ding H. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–880.e21. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 61.Guria A., Velayudha Vimala Kumar K., Srikakulam N., Krishnamma A., Chanda S., Sharma S., Fan X., Pandi G. Circular RNA profiling by illumina sequencing via template-dependent multiple displacement amplification. BioMed Res. Int. 2019;2019:2756516. doi: 10.1155/2019/2756516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gilbert W.V., Bell T.A., Schaening C. Messenger RNA modifications: Form, distribution, and function. Science. 2016;352:1408–1412. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou C., Molinie B., Daneshvar K., Pondick J.V., Wang J., Van Wittenberghe N., Xing Y., Giallourakis C.C., Mullen A.C. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20:2262–2276. doi: 10.1016/j.celrep.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye C.Y., Chen L., Liu C., Zhu Q.H., Fan L. Widespread noncoding circular RNAs in plants. New Phytol. 2015;208:88–95. doi: 10.1111/nph.13585. [DOI] [PubMed] [Google Scholar]
- 65.Ji P., Wu W., Chen S., Zheng Y., Zhou L., Zhang J., Cheng H., Yan J., Zhang S., Yang P., Zhao F. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 2019;26:3444–3460.e5. doi: 10.1016/j.celrep.2019.02.078. [DOI] [PubMed] [Google Scholar]
- 66.Jia R., Xiao M.S., Li Z., Shan G., Huang C. Defining an evolutionarily conserved role of GW182 in circular RNA degradation. Cell Discov. 2019;5:45. doi: 10.1038/s41421-019-0113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haupenthal J., Baehr C., Kiermayer S., Zeuzem S., Piiper A. Inhibition of RNAse A family enzymes prevents degradation and loss of silencing activity of siRNAs in serum. Biochem. Pharmacol. 2006;71:702–710. doi: 10.1016/j.bcp.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 68.Zhao W., Cheng Y., Zhang C., You Q., Shen X., Guo W., Jiao Y. Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci. Rep. 2017;7:5636. doi: 10.1038/s41598-017-05922-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao Y., Wang J., Zhao F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Panda A.C., Grammatikakis I., Kim K.M., De S., Martindale J.L., Munk R., Yang X., Abdelmohsen K., Gorospe M. Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor circPVT1. Nucleic Acids Res. 2017;45:4021–4035. doi: 10.1093/nar/gkw1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y., Yang M., Wei S., Qin F., Zhao H., Suo B. Identification of circular RNAs and their targets in leaves of Triticum aestivum L. under dehydration stress. Front. Plant Sci. 2017;7:2024. doi: 10.3389/fpls.2016.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao Z., Li J., Luo M., Li H., Chen Q., Wang L., Song S., Zhao L., Xu W., Zhang C. Characterization and cloning of grape circular RNAs identified the cold resistance-related Vv-circATS1. Plant Physiol. 2019;180:966–985. doi: 10.1104/pp.18.01331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y., Gao Y., Zhang H., Wang H., Liu X., Xu X., Zhang Z., Kohnen M.V., Hu K., Wang H. Genome-wide profiling of circular RNAs in the rapidly growing shoots of Moso bamboo (Phyllostachys edulis) Plant Cell Physiol. 2019;60:1354–1373. doi: 10.1093/pcp/pcz043. [DOI] [PubMed] [Google Scholar]
- 74.Barrett S.P., Salzman J. Circular RNAs: Analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salzman J. Circular RNA expression: Its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu H., Guo S., Li W., Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lukiw W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD) Front. Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Q., Zhang X., Hu X., Dai L., Fu X., Zhang J., Ao Y. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a miR-136 “sponge” in human cartilage degradation. Sci. Rep. 2016;6:22572. doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhong Z., Lv M., Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci. Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhong Z., Huang M., Lv M., He Y., Duan C., Zhang L., Chen J. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi: 10.1016/j.canlet.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 81.Xu X.-W., Zheng B.-A., Hu Z.-M., Qian Z.-Y., Huang C.-J., Liu X.-Q., Wu W.-D. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8:91674–91683. doi: 10.18632/oncotarget.21748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang X.L., Xu L.L., Wang F. hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol. Int. 2017;41:1056–1064. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 83.Zhang N., Li X., Wu C.W., Dong Y., Cai M., Mok M.T., Wang H., Chen J., Ng S.S.M., Chen M. MicroRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene. 2013;32:5078–5088. doi: 10.1038/onc.2012.526. [DOI] [PubMed] [Google Scholar]
- 84.Long L., Huang G., Zhu H., Guo Y., Liu Y., Huo J. Down-regulation of miR-138 promotes colorectal cancer metastasis via directly targeting TWIST2. J. Transl. Med. 2013;11:275. doi: 10.1186/1479-5876-11-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ayaz L., Çayan F., Balci Ş., Görür A., Akbayir S., Yıldırım Yaroğlu H., Doğruer Unal N., Tamer L. Circulating microRNA expression profiles in ovarian cancer. J. Obstet. Gynaecol. 2014;34:620–624. doi: 10.3109/01443615.2014.919998. [DOI] [PubMed] [Google Scholar]
- 86.Yeh Y.M., Chuang C.M., Chao K.C., Wang L.H. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1α. Int. J. Cancer. 2013;133:867–878. doi: 10.1002/ijc.28086. [DOI] [PubMed] [Google Scholar]
- 87.Zhao X., Dou W., He L., Liang S., Tie J., Liu C., Li T., Lu Y., Mo P., Shi Y. MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene. 2013;32:1363–1372. doi: 10.1038/onc.2012.156. [DOI] [PubMed] [Google Scholar]
- 88.Pan H., Li T., Jiang Y., Pan C., Ding Y., Huang Z., Yu H., Kong D. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J. Cell. Biochem. 2018;119:440–446. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 89.Zhang J., Liu H., Hou L., Wang G., Zhang R., Huang Y., Chen X., Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol. Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang H., Tang Y., Guo W., Du Y., Wang Y., Li P., Zang W., Yin X., Wang H., Chu H. Up-regulation of microRNA-138 induce radiosensitization in lung cancer cells. Tumour Biol. 2014;35:6557–6565. doi: 10.1007/s13277-014-1879-z. [DOI] [PubMed] [Google Scholar]
- 91.Fu D., Yu W., Li M., Wang H., Liu D., Song X., Li Z., Tian Z. MicroRNA-138 regulates the balance of Th1/Th2 via targeting RUNX3 in psoriasis. Immunol. Lett. 2015;166:55–62. doi: 10.1016/j.imlet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 92.Berg V., Rusch M., Vartak N., Jüngst C., Schauss A., Waldmann H., Hedberg C., Pallasch C.P., Bastiaens P.I.H., Hallek M. miRs-138 and -424 control palmitoylation-dependent CD95-mediated cell death by targeting acyl protein thioesterases 1 and 2 in CLL. Blood. 2015;125:2948–2957. doi: 10.1182/blood-2014-07-586511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Taulli R., Loretelli C., Pandolfi P.P. From pseudo-ceRNAs to circ-ceRNAs: A tale of cross-talk and competition. Nat. Struct. Mol. Biol. 2013;20:541–543. doi: 10.1038/nsmb.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li R.-C., Ke S., Meng F.-K., Lu J., Zou X.-J., He Z.-G., Wang W.-F., Fang M.H. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9:838. doi: 10.1038/s41419-018-0852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kefas B., Godlewski J., Comeau L., Li Y., Abounader R., Hawkinson M., Lee J., Fine H., Chiocca E.A., Lawler S., Purow B. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 96.Reddy S.D.N., Ohshiro K., Rayala S.K., Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008;68:8195–8200. doi: 10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Junn E., Lee K.-W., Jeong B.S., Chan T.W., Im J.-Y., Mouradian M.M. Repression of α-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. USA. 2009;106:13052–13057. doi: 10.1073/pnas.0906277106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qu S., Zhong Y., Shang R., Zhang X., Song W., Kjems J., Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–999. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Han C., Seebacher N.A., Hornicek F.J., Kan Q., Duan Z. Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget. 2017;8:64622–64637. doi: 10.18632/oncotarget.19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stefanetti R.J., Voisin S., Russell A., Lamon S. Recent advances in understanding the role of FOXO3. F1000Res. 2018;7(F1000 Faculty Rev):1372. doi: 10.12688/f1000research.15258.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tian L., Cao J., Jiao H., Zhang J., Ren X., Liu X., Liu M., Sun Y. circRASSF2 promotes laryngeal squamous cell carcinoma progression by regulating the miR-302b-3p/IGF-1R axis. Clin. Sci. (Lond.) 2019;133:1053–1066. doi: 10.1042/CS20190110. [DOI] [PubMed] [Google Scholar]
- 105.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Braunschweig U., Barbosa-Morais N.L., Pan Q., Nachman E.N., Alipanahi B., Gonatopoulos-Pournatzis T., Frey B., Irimia M., Blencowe B.J. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24:1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ge Y., Porse B.T. The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. BioEssays. 2014;36:236–243. doi: 10.1002/bies.201300156. [DOI] [PubMed] [Google Scholar]
- 108.Lykke-Andersen S., Jensen T.H. Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 109.Lindeboom R.G., Supek F., Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet. 2016;48:1112–1118. doi: 10.1038/ng.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pendleton K.E., Park S.K., Hunter O.V., Bresson S.M., Conrad N.K. Balance between MAT2A intron detention and splicing is determined cotranscriptionally. RNA. 2018;24:778–786. doi: 10.1261/rna.064899.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wilusz J.E. Circular RNAs: Unexpected outputs of many protein-coding genes. RNA Biol. 2017;14:1007–1017. doi: 10.1080/15476286.2016.1227905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li Y., Zheng F., Xiao X., Xie F., Tao D., Huang C., Liu D., Wang M., Wang L., Zeng F., Jiang G. circHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Conn V.M., Hugouvieux V., Nayak A., Conos S.A., Capovilla G., Cildir G., Jourdain A., Tergaonkar V., Schmid M., Zubieta C., Conn S.J. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 114.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e29. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Abe N., Matsumoto K., Nishihara M., Nakano Y., Shibata A., Maruyama H., Shuto S., Matsuda A., Yoshida M., Ito Y., Abe H. Rolling circle translation of circular RNA in living human cells. Sci. Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhao X., Yang Y., Sun B.-F., Shi Y., Yang X., Xiao W., Hao Y.-J., Ping X.-L., Chen Y.-S., Wang W.-J. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen X., Han P., Zhou T., Guo X., Song X., Li Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zheng X., Chen L., Zhou Y., Wang Q., Zheng Z., Xu B., Wu C., Zhou Q., Hu W., Wu C., Jiang J. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer. 2019;18:47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Y., Liu B., Shao C., Xu H., Xue A., Zhao Z., Shen Y., Tang Q., Xie J. Evaluation of the inclusion of circular RNAs in mRNA profiling in forensic body fluid identification. Int. J. Legal Med. 2018;132:43–52. doi: 10.1007/s00414-017-1690-7. [DOI] [PubMed] [Google Scholar]
- 121.Sharma G., Sharma A.R., Bhattacharya M., Lee S.S., Chakraborty C. CRISPR-Cas9: A preclinical and clinical perspective for the treatment of human diseases. Mol. Ther. 2021;29:571–586. doi: 10.1016/j.ymthe.2020.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shang X., Li G., Liu H., Li T., Liu J., Zhao Q., Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore) 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 124.Kristensen L.S., Hansen T.B., Venø M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen Y.G., Kim M.V., Chen X., Batista P.J., Aoyama S., Wilusz J.E., Iwasaki A., Chang H.Y. Sensing self and foreign circular RNAs by intron identity. Mol. Cell. 2017;67:228–238.e5. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wesselhoeft R.A., Kowalski P.S., Anderson D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ford E., Ares M., Jr. Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc. Natl. Acad. Sci. USA. 1994;91:3117–3121. doi: 10.1073/pnas.91.8.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Meganck R.M., Borchardt E.K., Castellanos Rivera R.M., Scalabrino M.L., Wilusz J.E., Marzluff W.F., Asokan A. Tissue-dependent expression and translation of circular RNAs with recombinant AAV vectors in vivo. Mol. Ther. Nucleic Acids. 2018;13:89–98. doi: 10.1016/j.omtn.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Litke J.L., Jaffrey S.R. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol. 2019;37:667–675. doi: 10.1038/s41587-019-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Garikipati V.N.S., Verma S.K., Cheng Z., Liang D., Truongcao M.M., Cimini M., Yue Y., Huang G., Wang C., Benedict C. Circular RNA circFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 2019;10:4317. doi: 10.1038/s41467-019-11777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Piwecka M., Glažar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C.A., Fenske P. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 132.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xia P., Wang S., Ye B., Du Y., Li C., Xiong Z., Qu Y., Fan Z. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity. 2018;48:688–701.e7. doi: 10.1016/j.immuni.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 134.Zhao Q., Liu J., Deng H., Ma R., Liao J.Y., Liang H., Hu J., Li J., Guo Z., Cai J. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell. 2020;183:76–93.e22. doi: 10.1016/j.cell.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 135.Wang F., Nazarali A.J., Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am. J. Cancer Res. 2016;6:1167–1176. [PMC free article] [PubMed] [Google Scholar]
- 136.Li P., Chen S., Chen H., Mo X., Li T., Shao Y., Xiao B., Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin. Chim. Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 137.Zhou C., Li R., Mi W. circ_0067934: A potential biomarker and therapeutic target for hepatocellular carcinoma. Ann. Clin. Lab. Sci. 2020;50:734–738. [PubMed] [Google Scholar]
- 138.Zhang Y., Zhao L., Yang S., Cen Y., Zhu T., Wang L., Xia L., Liu Y., Zou J., Xu J. circCDKN2B-AS1 interacts with IMP3 to stabilize hexokinase 2 mRNA and facilitate cervical squamous cell carcinoma aerobic glycolysis progression. J. Exp. Clin. Cancer Res. 2020;39:281. doi: 10.1186/s13046-020-01793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Feng J., Guo Y., Li Y., Zeng J., Wang Y., Yang Y., Xie G., Feng Q. Tumor promoting effects of circRNA_001287 on renal cell carcinoma through miR-144-targeted CEP55. J. Exp. Clin. Cancer Res. 2020;39:269. doi: 10.1186/s13046-020-01744-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li S., Zhao Y., Chen X. Microarray expression profile analysis of circular RNAs and their potential regulatory role in bladder carcinoma. Oncol. Rep. 2021;45:239–253. doi: 10.3892/or.2020.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhao Z., Li X., Gao C., Jian D., Hao P., Rao L., Li M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 2017;7:39918. doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang J.-R., Sun H.-J. Roles of circular RNAs in diabetic complications: From molecular mechanisms to therapeutic potential. Gene. 2020;763:145066. doi: 10.1016/j.gene.2020.145066. [DOI] [PubMed] [Google Scholar]
- 143.Huang Z., Fu B., Qi X., Xu Y., Mou Y., Zhou M., Cao Y., Wu G., Xie J., Zhao J. Diagnostic and therapeutic value of hsa_circ_0002594 for T helper 2-mediated allergic asthma. Int. Arch. Allergy Immunol. 2021;182:388–398. doi: 10.1159/000511612. [DOI] [PubMed] [Google Scholar]
- 144.Yao G., Niu W., Zhu X., He M., Kong L., Chen S., Zhang L., Cheng Z. hsa_circRNA_104597: A novel potential diagnostic and therapeutic biomarker for schizophrenia. Biomarkers Med. 2019;13:331–340. doi: 10.2217/bmm-2018-0447. [DOI] [PubMed] [Google Scholar]
- 145.Li P., Shan K., Liu Y., Zhang Y., Xu L., Xu L. circScd1 promotes fatty liver disease via the Janus kinase 2/signal transducer and activator of transcription 5 pathway. Dig. Dis. Sci. 2019;64:113–122. doi: 10.1007/s10620-018-5290-2. [DOI] [PubMed] [Google Scholar]
- 146.Xie B., Zhao Z., Liu Q., Wang X., Ma Z., Li H. circRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene. 2019;683:253–261. doi: 10.1016/j.gene.2018.10.043. [DOI] [PubMed] [Google Scholar]
- 147.Li Q., Pan X., Zhu D., Deng Z., Jiang R., Wang X. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70:1298–1316. doi: 10.1002/hep.30671. [DOI] [PubMed] [Google Scholar]
- 148.Zhou Y., Lv X., Qu H., Zhao K., Fu L., Zhu L., Ye G., Guo J. Differential expression of circular RNAs in hepatic tissue in a model of liver fibrosis and functional analysis of their target genes. Hepatol. Res. 2019;49:324–334. doi: 10.1111/hepr.13284. [DOI] [PubMed] [Google Scholar]
- 149.Ji D., Chen G.-F., Wang J.-C., Ji S.-H., Wu X.-W., Lu X.-J., Chen J.-L., Li J.-T. hsa_circ_0070963 inhibits liver fibrosis via regulation of miR-223-3p and LEMD3. Aging (Albany NY) 2020;12:1643–1655. doi: 10.18632/aging.102705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang W., Dong R., Guo Y., He J., Shao C., Yi P., Yu F., Gu D., Zheng J. circMTO1 inhibits liver fibrosis via regulation of miR-17-5p and Smad7. J. Cell. Mol. Med. 2019;23:5486–5496. doi: 10.1111/jcmm.14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu W., Feng R., Li X., Li D., Zhai W. TGF-β- and lipopolysaccharide-induced upregulation of circular RNA PWWP2A promotes hepatic fibrosis via sponging miR-203 and miR-223. Aging (Albany NY) 2019;11:9569–9580. doi: 10.18632/aging.102405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Luo Q., Fu B., Zhang L., Guo Y., Huang Z., Li J. Expression and clinical significance of circular RNA hsa_circ_0079787 in the peripheral blood of patients with axial spondyloarthritis. Mol. Med. Rep. 2020;22:4197–4206. doi: 10.3892/mmr.2020.11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li C.-Y., Ma L., Yu B. Circular RNA hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells proliferation and angiogenesis. Biomed. Pharmacother. 2017;95:1514–1519. doi: 10.1016/j.biopha.2017.09.064. [DOI] [PubMed] [Google Scholar]
- 154.Liang D., Tatomer D.C., Luo Z., Wu H., Yang L., Chen L.-L., Cherry S., Wilusz J.E. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol. Cell. 2017;68:940–954.e3. doi: 10.1016/j.molcel.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wu J., Li Y., Wang C., Cui Y., Xu T., Wang C., Wang X., Sha J., Jiang B., Wang K. CircAST: Full-length assembly and quantification of alternatively spliced isoforms in circular RNAs. Genomics Proteomics Bioinformatics. 2019;17:522–534. doi: 10.1016/j.gpb.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dong R., Ma X.-K., Li G.-W., Yang L. CIRCpedia v2: An updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteomics Bioinformatics. 2018;16:226–233. doi: 10.1016/j.gpb.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Suenkel C., Cavalli D., Massalini S., Calegari F., Rajewsky N. A highly conserved circular RNA is required to keep neural cells in a progenitor state in the mammalian brain. Cell Rep. 2020;30:2170–2179.e5. doi: 10.1016/j.celrep.2020.01.083. [DOI] [PubMed] [Google Scholar]
- 158.Gao Y., Zhao F. Computational strategies for exploring circular RNAs. Trends Genet. 2018;34:389–400. doi: 10.1016/j.tig.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 159.Xin R., Gao Y., Gao Y., Wang R., Kadash-Edmondson K.E., Liu B., Wang Y., Lin L., Xing Y. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat. Commun. 2021;12:266. doi: 10.1038/s41467-020-20459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rahimi K., Venø M.T., Dupont D.M., Kjems J. Nanopore sequencing of full-length circRNAs in human and mouse brains reveals circRNA-specific exon usage and intron retention. bioRxiv. 2019 doi: 10.1101/567164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zheng Y., Ji P., Chen S., Hou L., Zhao F. Reconstruction of full-length circular RNAs enables isoform-level quantification. Genome Med. 2019;11:2. doi: 10.1186/s13073-019-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang J., Hou L., Zuo Z., Ji P., Zhang X., Xue Y., Zhao F. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-00842-6. Published online March 11, 2021. [DOI] [PubMed] [Google Scholar]
- 163.Xia S., Feng J., Chen K., Ma Y., Gong J., Cai F., Jin Y., Gao Y., Xia L., Chang H. CSCD: A database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46(D1):D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen Y., Yang F., Fang E., Xiao W., Mei H., Li H., Li D., Song H., Wang J., Hong M. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sun H., Tang W., Rong D., Jin H., Fu K., Zhang W., Liu Z., Cao H., Cao X. hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21:299–306. doi: 10.3233/CBM-170379. [DOI] [PubMed] [Google Scholar]
- 166.Yu J., Xu Q.G., Wang Z.G., Yang Y., Zhang L., Ma J.Z., Sun S.H., Yang F., Zhou W.-P. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 2018;68:1214–1227. doi: 10.1016/j.jhep.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 167.Chen S., Li T., Zhao Q., Xiao B., Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin. Chim. Acta. 2017;466:167–171. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 168.Zhao W., Cui Y., Liu L., Qi X., Liu J., Ma S., Hu X., Zhang Z., Wang Y., Li H. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 2020;27:919–933. doi: 10.1038/s41418-019-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xu Y., Yao Y., Zhong X., Leng K., Qin W., Qu L., Cui Y., Jiang X. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem. Biophys. Res. Commun. 2018;496:455–461. doi: 10.1016/j.bbrc.2018.01.077. [DOI] [PubMed] [Google Scholar]
- 170.Kleaveland B., Shi C.Y., Stefano J., Bartel D.P. A network of non-coding regulatory RNAs acts in the mammalian brain. Cell. 2018;174:350–362.e17. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., Cai S., Qin H., Ma Y., Goel A. Circular RNA ciRS-7—A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stoll L., Sobel J., Rodriguez-Trejo A., Guay C., Lee K., Venø M.T., Kjems J., Laybutt D.R., Regazzi R. Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol. Metab. 2018;9:69–83. doi: 10.1016/j.molmet.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Okholm T.L.H., Nielsen M.M., Hamilton M.P., Christensen L.-L., Vang S., Hedegaard J., Hansen T.B., Kjems J., Dyrskjøt L., Pedersen J.S. Circular RNA expression is abundant and correlated to aggressiveness in early-stage bladder cancer. NPJ Genom. Med. 2017;2:36. doi: 10.1038/s41525-017-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yu C.-Y., Li T.-C., Wu Y.-Y., Yeh C.-H., Chiang W., Chuang C.-Y., Kuo H.-C. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017;8:1149. doi: 10.1038/s41467-017-01216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hsiao K.-Y., Lin Y.-C., Gupta S.K., Chang N., Yen L., Sun H.S., Tsai S.-J. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Verduci L., Ferraiuolo M., Sacconi A., Ganci F., Vitale J., Colombo T., Paci P., Strano S., Macino G., Rajewsky N., Blandino G. The oncogenic role of circPVT1 in head and neck squamous cell carcinoma is mediated through the mutant p53/YAP/TEAD transcription-competent complex. Genome Biol. 2017;18:237. doi: 10.1186/s13059-017-1368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Abdelmohsen K., Panda A.C., Munk R., Grammatikakis I., Dudekula D.B., De S., Kim J., Noh J.H., Kim K.M., Martindale J.L., Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by circPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zeng Y., Du W.W., Wu Y., Yang Z., Awan F.M., Li X., Yang W., Zhang C., Yang Q., Yee A. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7:3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Huang S. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., Chen W., Gao X., Zhao K., Zhou H. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]