Abstract
To investigate the mode of action of the p16INK4a tumor suppressor protein, we have established U2-OS cells in which the expression of p16INK4a can be regulated by addition or removal of isopropyl-β-d-thiogalactopyranoside. As expected, induction of p16INK4a results in a G1 cell cycle arrest by inhibiting phosphorylation of the retinoblastoma protein (pRb) by the cyclin-dependent kinases CDK4 and CDK6. However, induction of p16INK4a also causes marked inhibition of CDK2 activity. In the case of cyclin E-CDK2, this is brought about by reassortment of cyclin, CDK, and CDK-inhibitor complexes, particularly those involving p27KIP1. Size fractionation of the cellular lysates reveals that a substantial proportion of CDK4 participates in active kinase complexes of around 200 kDa. Upon induction of p16INK4a, this complex is partly dissociated, and the majority of CDK4 is found in lower-molecular-weight fractions consistent with the formation of a binary complex with p16INK4a. Sequestration of CDK4 by p16INK4a allows cyclin D1 to associate increasingly with CDK2, without affecting its interactions with the CIP/KIP inhibitors. Thus, upon the induction of p16INK4a, p27KIP1 appears to switch its allegiance from CDK4 to CDK2, and the accompanying reassortment of components leads to the inhibition of cyclin E-CDK2 by p27KIP1 and p21CIP1. Significantly, p16INK4a itself does not appear to form higher-order complexes, and the overwhelming majority remains either free or forms binary associations with CDK4 and CDK6.
In mammalian cells, the activity of cyclin-dependent kinases (CDKs) is regulated in part by the expression of specific inhibitory proteins, termed CDK inhibitors (CKIs) (65). Although their precise roles have yet to be established, the prevailing view is that specific CKIs may be responsible for diverting cells out of the proliferative cycle to facilitate terminal differentiation or to maintain quiescence (14), and there is considerable interest in their potential to act in tumor suppression (64). Based on primary sequence comparisons, two families of CKIs, commonly referred to as the CIP/KIP and the INK4 proteins, respectively, have been identified in mammalian cells, (14, 65); as well as having distinct structural characteristics (46, 59), it is clear that these families have distinct modes of action (19, 20).
The CIP/KIP proteins, p21CIP1/WAF1/sdi1 p27KIP1, and p57KIP2 are grouped together largely because they share common sequence motifs that mediate interaction between the CKI and cyclin-CDK complexes (8, 35, 36, 43, 50, 53, 71). It is in fact possible to demonstrate direct interactions between p21CIP1 and p27KIP1 with cyclins in the absence of a kinase subunit (8, 16, 17, 20, 36, 71), although this may be an intermediate step in the formation of a ternary cyclin-CDK-CKI complex (25, 43). The CIP/KIP proteins can bind to a variety of cyclins and CDKs and in assays based on expressing active complexes in insect cells, using baculovirus vectors, it is clear that all three are capable of inhibiting the kinase activity of CDK4 and CDK6 complexed to D cyclins and of CDK2 complexed to either cyclin E or cyclin A (24, 25, 35, 43, 53, 71, 76). They therefore have the potential to block cell cycle progression at multiple points, but the situation is complicated by the fact that CIP/KIP proteins can also participate in active kinase complexes, both in vivo and in vitro (5, 8, 16, 25, 33, 66, 78). This may reflect their ability to serve as assembly factors in some circumstances, so that at low concentrations they may facilitate the association of cyclins and CDKs whereas at higher concentrations they function as inhibitors (33, 78). There are also indications that p27KIP1, for example, is a more potent inhibitor of CDK2 than CDK4 (5), suggesting that there are important subtleties to these interactions that have yet to be explored.
In contrast to the CIP/KIP family, the INK4 proteins, p16INK4a, p15INK4b, p18INK4c, and p19INK4d, are highly related, each comprising between three and five ankyrin-type repeats with minimal amino- and carboxy-terminal extensions (reviewed in reference 58). All four INK4 proteins bind directly to CDK4 and CDK6, and there is no evidence that they associate with other CDKs (7, 18, 19, 21, 28, 29, 52, 62). They therefore serve as specific inhibitors of cyclin D-dependent kinases and have the capacity to induce a G1 cell cycle arrest by preventing the phosphorylation of the retinoblastoma gene product, pRb (18, 28, 32, 40, 47, 51, 61, 63, 69). The prototypic and most illustrious member of the family, p16INK4a, is a bona fide tumor suppressor that is incapacitated by homozygous deletion, mutation, or promoter methylation in a wide variety of sporadic human tumors (58). Germ line mutations in the p16INK4a gene are also associated with familial melanoma (58).
In trying to determine why it is p16INK4a, rather than other members of the INK4 family, that acts as a tumor suppressor, we and others have noted that p16INK4a levels increase significantly as primary cells reach the end of their finite life span in culture (2, 22, 37, 57, 75). This would be consistent with a role for p16INK4a in establishing the G1 arrest associated with replicative senescence. Indeed, ectopic expression of p16INK4a in primary fibroblasts and some established cell lines elicits many of the phenotypic characteristics associated with senescence (45, 72, 74). A need to escape senescence would therefore provide a strong selection against p16INK4a function during the establishment of an immortal cell clone.
However, the mechanisms through which the INK4 family induce a cell cycle arrest are still a matter of debate. Most of the evidence favors the competitive binding model in which the direct association of INK4 proteins with CDK4 or CDK6 prevents them from interacting with their regulatory D cyclins. As well as supporting in vitro data (19, 52), this mode of action has the logical appeal that in vivo, the relatively unstable D-type cyclins bound to CDK4/CDK6 would be progressively replaced by the more stable INK4 proteins (52). This would explain why INK4 proteins are rarely found in immune complexes with the D cyclins (19, 20, 28, 52, 60). However, the opposing model holds that in some circumstances it is indeed possible to find ternary complexes of INK4 proteins, D cyclins, and CDKs. The strongest evidence is that recombinant INK4 proteins are capable of inhibiting the kinase activity of cyclin D-CDK complexes assembled by using baculovirus systems without quantitatively disrupting these complexes (28, 55). Similarly, in mammalian cells arrested by induced expression of p15INK4b or p19INK4d, the INK4 proteins have been reported to coprecipitate with D cyclins and to invoke a cell cycle arrest without displacing CDK4/CDK6 from the cyclin D complexes (1, 55).
Because of these conflicting views, and the importance of p16INK4a as a senescence regulator and tumor suppressor, we have investigated what happens to cyclin-CDK-CKI complexes in cells engineered for inducible expression of p16INK4a. Most of the previous studies have described the results simply in terms of binary and ternary associations, with little focus on the stoichiometry of the complexes or the proportions of the individual components involved in each interaction. By using gel filtration chromatography to distinguish complexes of different size, we show that the G1 arrest imposed by p16INK4a is accompanied by a reassortment of components. By sequestering CDK4 and CDK6 in inactive binary complexes, p16INK4a allows CDK2 to associate with cyclin D1 and releases enough p27KIP1 to inhibit cyclin E-CDK2. The data effectively eliminate the possibility that p16INK4a forms stable ternary or higher-order complexes with D-type cyclins.
MATERIALS AND METHODS
Inducible cell lines.
The human osteosarcoma cell line U2-OS was maintained at 37°C in Dulbecco modified Eagle’s medium supplemented with 10% fetal calf serum. Isopropyl-β-d-thiogalactopyranoside (IPTG)-regulatable p16INK4a was obtained by using the Stratagene LAC-SWITCH system. The DNA fragment encoding p16INK4a was excised from a pcDNA3-based plasmid by using XbaI and BamHI and subcloned into pBluescript to exploit the NotI sites in the polylinker. The insert was then recovered by digestion with NotI and subcloned into the NotI site in pOPRSVI. The orientation of the insert was determined by DNA sequencing. U2-OS cells were cotransfected with a 1:1 mixture of p16INK4a in pOPRSVI and the regulatory plasmid p3′SS. Stable transfectants were selected in hygromycin (150 μg/ml; Sigma) and geneticin (300 μg/ml; Gibco) followed by the isolation of single colonies. Of 30 clones tested, two (designated EH1 and EH2) were found to express p16INK4a under the control of the lac promoter. EH1 cells were maintained under selective conditions, but to ensure that the cells were not metabolically challenged during various assays, the antibiotics were removed from the medium 24 h prior to initiation of experiments. To induce expression, EH1 cells were split at a ratio of 1:2 and 1 mM IPTG was added to the fresh medium.
Antibodies.
Immunoblotting of the D cyclins was done with monoclonal antibodies DCS6 for cyclin D1, DCS3.1 for cyclin D2, and DCS22 for cyclin D3 (3, 39). Corresponding immunoprecipitations used the polyclonal antibody 287.3 for cyclin D1 (4) and the monoclonal antibodies DCS5.2 and DCS28, respectively, for cyclins D2 and D3. Monoclonal antibodies HE12 and HE172, used for Western blot detection and immunoprecipitation of cyclin E, were provided by E. Lees. p16INK4a was detected with monoclonal antibody JC8 supplied by J. Koh, and pRb was detected with the Pharmingen monoclonal antibody 14001A. pRb phosphorylated at S780 was detected with a polyclonal antibody provided by Y. Taya (31). The following polyclonal antibodies from Santa Cruz were used for immunoprecipitation and/or Western blotting: CDK2 (sc-163), CDK4 (sc-601), p21CIP1 (sc-397), p27KIP1 (sc-528), and cyclin A (sc-751).
Immunoprecipitation and immunoblotting.
Cells were recovered from the culture flasks by treatment with trypsin and washed once with phosphate-buffered saline by centrifugation and resuspension. Cells were then lysed by resuspension in Tween lysis buffer (50 mM HEPES [pH 8.0], 10 mM MgCl2, 0.1% Tween 20, 1 mM dithiothreitol, 1 mM sodium fluoride, 0.1 mM sodium orthovanadate, 5 μg of aprotinin per ml, 100 μg of phenylmethylsulfonyl fluoride per ml) followed by sonication for two 5-s pulses. Lysates were clarified by centrifugation at 10,000 × g for 10 min, and the protein concentrations were determined by using the Pierce bicinchoninic acid protein assay reagents according to the manufacturer’s instructions.
Immunoprecipitations were performed with 800 μg of lysate plus 0.5 μg of antibody and 25 μl of a 50% slurry of protein A- or protein A/G-Sepharose beads (Pierce) by rotating overnight at 4°C. For Western blotting, samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto Immobilon-P membrane (Millipore). Immunodetection was carried out by enhanced chemiluminescence (Amersham).
Analysis of 35S-labeled proteins.
To metabolically label the cellular proteins, EH1 cells (approximately 50% confluent) were incubated for 30 min in Dulbecco modified Eagle’s medium lacking methionine and cysteine and supplemented with 10% dialyzed fetal calf serum. The cells were then labeled for 2 h in the same medium containing 50 μCi of [35S]methionine-cysteine (Promix; Amersham plc) per ml. Cell lysates were prepared by using Nonidet P-40 lysis buffer and processed as described previously (4). Labeled proteins were detected by autoradiography.
Sequential immunoprecipitations.
To determine the proportion of p27KIP1 associated with different cyclins and CDKs, sequential immunoprecipitations were performed with antibodies covalently coupled to agarose beads. Typically, 450 μg of lysate was subjected to two rounds of precipitation with 30 μl of beads (50% slurry) at 4°C for 2 h. After each incubation, the beads were pelleted and the supernatants were transferred to the next set of beads. Each precipitate was then subjected to SDS-PAGE and immunoblotted for p27KIP1. Mock immunoprecipitations were carried out in parallel with anti-rabbit immunoglobulin G bound to beads (Sigma), and the supernatant from this control set of precipitations served as the control for the total amount of p27KIP1 present in the lysate.
Gel filtration chromatography.
Gel filtration chromatography was carried out by using a Superdex 200 HR 10/30 column (Pharmacia) with a fast protein liquid chromatography system (BioLogic system; Bio-Rad). Samples of 4 mg of cell lysate in 250 μl of Tween lysis buffer were loaded onto the column and separated at a flow rate of 0.4 ml per min. The molecular mass standards (Sigma) used to calibrate the column were thyroglobulin (669 kDa), apoferritin (443 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), myoglobin (17.6 kDa), and cytochrome c (12.4 kDa). Lysates from untreated and IPTG-treated cells were analyzed sequentially on the same column followed by a subset of molecular mass standards to ensure that the efficiency of separation and calibration of the column was being maintained.
CDK assays.
Kinase assays were carried out essentially as described by Matsushime et al. (44) except that the substrate was a glutathione S-transferase (GST) fusion protein containing only the carboxy-terminal region of pRb (48). Lysates from untreated and IPTG-treated EH1 cells were immunoprecipitated with antisera to CDK4, CDK6, or CDK2 as described above except that the incubation time was reduced to 5 h. Immune complexes were washed three times with Tween lysis buffer and twice with kinase reaction buffer (50 mM HEPES, pH 8.0, 10 mM MgCl2, 2.5 mM EGTA, 1 mM dithiothreitol, 25 mM ATP); 25 μl of kinase assay buffer containing 2 μg of GST-Rb and 10 μCi of [γ-32P]ATP was added to each sample, and after incubation at 30°C for 20 min, reactions were stopped by adding 25 μl of 2× dissociation buffer (23). Samples were boiled for 2 min and resolved by SDS-PAGE in a 10% gel. The phosphorylated substrate was detected initially by autoradiography and quantitated by PhosphorImager (Molecular Dynamics) analysis using Imagequant software.
RESULTS
Cell cycle arrest by inducible p16INK4a.
To examine the effect of p16INK4a on G1 CDK activity and investigate its mode of action, stable cell clones that express p16INK4a under the control of an IPTG-inducible promoter were derived. The osteosarcoma cell line U2-OS was chosen as the recipient because it retains functional p53 and pRb (11) and does not express endogenous p16INK4a due to methylation of the exon 1α promoter (70). The clone selected for these studies, EH1, showed a low basal level of p16INK4a expression in the absence of IPTG, reflecting leakiness in the regulatable system (Fig. 1A). However, the levels were comparable to the low amounts of p16INK4a expressed in early-passage human diploid fibroblasts and had no discernible effect on the growth of the cells (data not shown). Upon addition of IPTG, the expression of p16INK4a increased in a dose-dependent manner, reaching a maximum level at around 1 mM IPTG (70). Under these conditions, the cells expressed p16INK4a at levels equivalent to those in senescent human fibroblasts (Fig. 1A and data not shown).
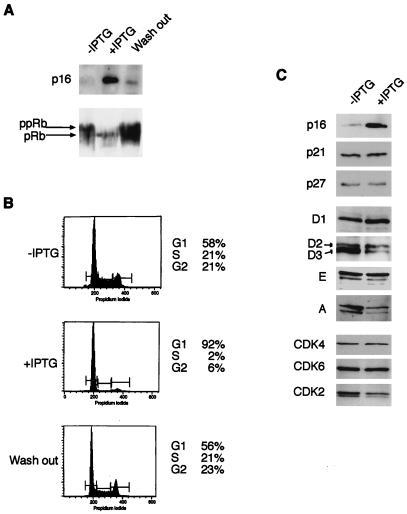
FIG. 1.
Reversible growth arrest and effects on cell cycle regulators after induction of p16INK4a. EH1 cells were grown from 48 h in the absence or presence of 1 mM IPTG, as indicated, or for an additional 48 h following removal of IPTG (Wash out). (A) The levels of p16INK4a and the phosphorylation status of pRb were evaluated by direct immunoblotting of cell lysates. (B) Cell cycle profiles were determined by staining the cells with propidium iodide followed by fluorescence-activated cell sorting. (C) Equal amounts of protein from EH1 cells grown in the absence or presence of IPTG were fractionated by SDS-PAGE and immunoblotted for the indicated cell cycle regulatory proteins. D1, D2, etc., refer to the different cyclins analyzed.
As expected, EH1 cells treated with 1 mM IPTG arrested in the G1 phase of the cell cycle (Fig. 1B). The effects were apparent within 24 h and maximal by 48 h, at which time over 90% of the cells had accumulated with a G0/G1 DNA content. This arrest was reversible; upon removal of IPTG, an asynchronous cell cycle profile was restored within 48 h (Fig. 1B), correlating with reduced expression of p16INK4a (Fig. 1A). The p16INK4a-mediated arrest also correlated directly with the phosphorylation status of pRb. Induction of p16INK4a caused pRb to accumulate in the faster-migrating, hypophosphorylated state, whereas removal of IPTG restored pRb to its hyperphosphorylated state (Fig. 1A). However, despite the analogies to senescent fibroblasts, EH1 cells maintained in IPTG did not express other phenotypic markers of senescent cells, such as increased size and granularity or expression of senescence associated β-galactosidase activity (12, 45).
We next asked whether the induction of p16INK4a had altered the steady-state levels of other cell cycle components that might have contributed to the G1 arrest. Compared to the marked change in p16INK4a expression, no significant changes were observed in the levels of the CKIs p21CIP1 and p27KIP1 (Fig. 1C). Likewise, the steady-state levels of cyclins D1, D2, and E were unaffected, but substantially lower amounts of cyclins D3 and A were detected in the IPTG-treated cells (Fig. 1C). No significant changes were observed in CDK4 and CDK6 levels, but there was a modest down regulation of CDK2 (Fig. 1C). The reduction was more striking for the active, phosphorylated form of CDK2 (faster-migrating band) so that in the IPTG-treated cells, CDK2 appeared to be primarily in its inactive, unphosphorylated state (slower-migrating band).
Inhibition of CDK-associated kinase activity by inducible p16INK4a.
Since p16INK4a binds directly to CDK4 and CDK6, and not to CDK2 (21, 52, 62), induction of p16INK4a in EH1 cells should principally affect the kinase activity associated with cyclin D-CDK complexes. CDK4 immunoprecipitates from EH1 cells, before and after IPTG treatment, were therefore tested for the ability to phosphorylate the carboxy-terminal domain of pRb in vitro. The specificity of the signal was confirmed by blocking the precipitation of CDK4 with an excess of the cognate peptide antigen. Upon induction of p16INK4a with IPTG, there was a reduction of the CDK4 activity to almost background levels (Fig. 2A).
FIG. 2.
Inhibition of CDK4 and CDK2 by induction of p16INK4a. (A) EH1 cells were grown for 48 h with or without IPTG and CDK4 immunoprecipitates, prepared with or without competing peptide antigen, were tested for the ability to phosphorylate the carboxy-terminal domain of pRb in vitro. (B) Thirty-microgram aliquots of lysates from EH1 cells grown in the presence or absence of IPTG were immunoblotted with an antibody specific for pRb phosphorylated at serine 780 (31). (C) Lysates from IPTG-treated and untreated cells were immunoprecipitated with a control nonimmune serum (immunoglobulin G [IgG]) or with antisera against CDK2, cyclin E, or cyclin A as indicated. The immune complexes were then tested for kinase activity using the carboxy-terminal domain of pRb as the substrate. The results were quantitated by phosphorimaging and expressed as the percentage reduction in kinase activity after induction of p16INK4a. The values presented are from two independent experiments.
To substantiate this finding, we exploited an antibody that specifically recognizes pRb phosphorylated on serine 780, a specific target for cyclin D-dependent kinases (31). Whereas the S780 phosphorylation was clearly detected in asynchronously growing EH1 cells, induction of p16INK4a resulted in a complete loss of signal, consistent with a direct effect on cyclin D-associated kinase activity (Fig. 2B).
In view of the altered pattern of CDK2 isoforms noted in IPTG-treated EH1 cells, we also measured the kinase activity associated with CDK2 immunoprecipitates. Total CDK2 activity, measured by using the carboxy-terminal region of pRb as the substrate, was drastically reduced by the induction of p16INK4a (Fig. 2C). Moreover, the effects were observable with both cyclin E- and cyclin A-associated CDK2.
Rearrangement of cyclin-CDK complexes upon expression of p16INK4a.
Since p16INK4a does not bind directly to CDK2, we were interested in exploring the mechanisms behind its impact on CDK2 activity. In the case of cyclin A, the most likely explanation would be the reduced expression of the cyclin (Fig. 1C), but since cyclin E levels remained constant in IPTG-treated cells, the reduced kinase activity presumably occurred through a change in its association with other proteins, such as CIP/KIP type CKIs (55, 56). To address this possibility, selected cyclins and CKIs were immunoprecipitated from EH1 cells before and after p16INK4a induction, and the associated CDKs were detected by immunoblotting (Fig. 3A). As expected, although the total levels of CDK4 remained the same (lanes 1 and 2), there was significantly less CDK4 associated with cyclin D1 and significantly more CDK4 associated with p16INK4a in cells treated with IPTG. This would be consistent with the down regulation of CDK4-associated kinase activity. The redistribution of CDK4 was also apparent in the p27KIP1 immunoprecipitates. Thus, in untreated EH1 cells, CDK4 and p27 participated in a complex, presumably including D cyclins, that appeared to be substantially disrupted upon the induction of p16INK4a. Although a similar trend was apparent for complexes containing p21CIP1, the effects were less pronounced (Fig. 3A).
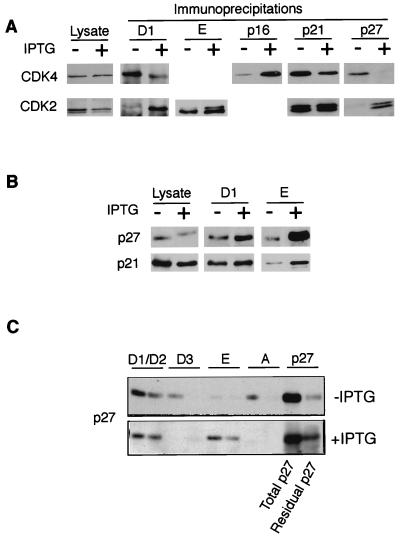
FIG. 3.
Altered cyclin-CDK-CKI associations upon induction of p16INK4a. (A) Equal amounts of cell lysates (∼1 mg) prepared from EH1 cells grown with (+) or without (−) IPTG were immunoprecipitated with antisera against cyclin D1, cyclin E, p16INK4a, p21CIP1, and p27KIP1 as indicated. After fractionation by SDS-PAGE, the precipitated proteins were immunoblotted for CDK4 (upper panels) and CDK2 (lower panels). The total amounts of CDK4 and CDK2 were monitored by direct immunoblotting of a portion of the cell lysates (Lysate). (B) Levels of p27KIP1 or p21CIP1 associated with cyclin D1 or cyclin E were determined as for panel A. (C) Lysates from untreated or IPTG-treated cells were subjected to sequential immunoprecipitation with antibodies against cyclins D1 plus D2, cyclin D3, cyclin E, and cyclin A as indicated. Each antibody was covalently coupled to agarose beads so that the supernatant from each immunoprecipitation could be quantitatively recovered and used in the subsequent precipitation. Two rounds of precipitation were conducted with each antibody. The final supernatant (Residual p27) was then precipitated with antiserum against p27KIP1. As a control, a sample of the starting lysate was subjected to repeated cycles of precipitation with nonimmune serum (Total p27). The amounts of p27KIP1 in the various precipitates were then compared by immunoblotting.
Interestingly, these changes in CDK4-containing complexes were largely mirrored by changes in CDK2-containing complexes. For example, the reduced association of CDK4 with cyclin D1 was accompanied by an increased interaction between CDK2 and cyclin D1. Likewise, the p27KIP1 displaced from CDK4 complexes appeared to move into CDK2 complexes. Although the amounts of CDK2 bound to cyclin E were unaltered, the activation state of CDK2 associated with cyclin E changed, with a higher proportion of the unphosphorylated, inactive form in IPTG-treated cells.
This change in the activation state of CDK2 was accompanied by a substantial increase in the amount of p27KIP1 associated with cyclin E, consistent with the formation of inactive cyclin E-CDK2-p27KIP1 complexes (Fig. 3B). However, there was no concomitant loss of p27KIP1 from cyclin D1-containing complexes as might have been expected from the release of p27KIP1 from CDK4. A possible explanation would be that the disruption of cyclin D1-CDK4-p27KIP1 complexes by p16INK4a is balanced by the formation of cyclin D1-CDK2-p27KIP1 complexes. There was also an increased association of p21CIP1 with cyclin E, but again the effects with p21CIP1 were less striking than those with p27KIP1.
An obvious question posed by these findings was the source of the p27KIP1 that had entered the cyclin E-CDK2 complex, given that there was no apparent diminution in the amount bound to cyclin D1. To address this issue, lysates from induced and uninduced cells were subjected to sequential immunoprecipitations with antisera to cyclins D1 plus D2, cyclin D3, cyclin E, and cyclin A. Two rounds of immunoprecipitation were performed with each antibody, and the final supernatant was then precipitated with an antibody against p27KIP1 to determine the residual fraction of the protein not bound to these cyclins. The amount of p27KIP1 in each precipitate was directly compared by immunoblotting. In asynchronously growing EH1 cells, p27KIP1 was found associated with all of the cyclins, although the major proportions were bound to cyclins D1 plus D2 and cyclin A. Less than 10% seemed to be associated with cyclin E complexes. In the presence of IPTG, the pattern changed dramatically with little if any p27KIP1 associated with either cyclin D3 or cyclin A, and a substantial fraction now precipitating with cyclin E. Again, there was no appreciable decrease in the amount associated with cyclins D1 and D2. Although there was a considerable amount of p27KIP1 in the supernatant that was not precipitated with the cyclin antisera, it is not clear whether this residual fraction reflects binding to other, as yet unidentified proteins or the fact that the immunodepletions were not exhaustive.
Size fractionation of cyclin-CDK-CKI complexes.
One of the limitations of this simple coprecipitation approach is that it does not provide information about the stoichiometry of the various multicomponent complexes. To try to address this question, protein lysates from IPTG-treated or untreated EH1 cells were subjected to gel filtration to separate complexes of different sizes. The fractionated lysates were first analyzed directly by immunoblotting to determine the proportions of each protein in different size fractions (Fig. 4). In asynchronously proliferating EH1 cells, p21CIP1 and p27KIP1 were found exclusively in high-molecular-mass complexes of around 200 kDa, with the p21CIP1 complex consistently appearing marginally larger than the p27KIP1 complex. These complexes appeared unchanged when p16INK4a expression was induced with IPTG. Interestingly, the induced p16INK4a was distributed between two major peaks, the smaller of which would be consistent with free p16INK4a. The larger peak, running at approximately 50 kDa, would be compatible with binary complexes between p16INK4a and either CDK4 or CDK6, but not with the formation of ternary complexes incorporating a D cyclin. However, since this main peak tailed into higher-molecular-mass fractions, it remains possible that a small percentage of the induced p16INK4a entered complexes containing both cyclins and CDKs (see below).
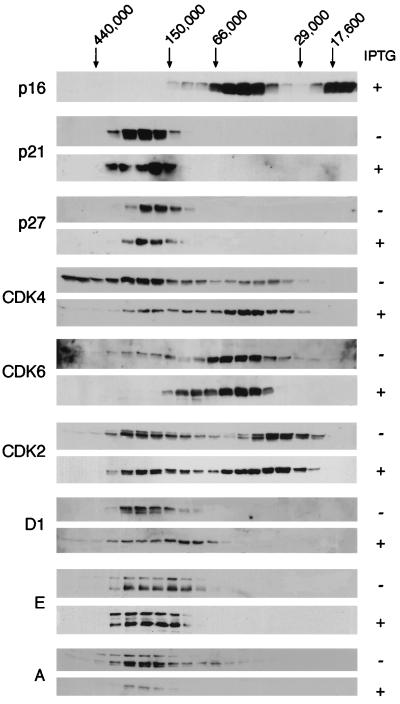
FIG. 4.
Altered size distribution of cyclins, CDKs, and CKI after induction of p16INK4a. Equal amounts (4 mg) of lysate from EH1 cells before (−) and after (+) treatment with IPTG were subjected to gel filtration chromatography. Individual fractions were separated by SDS-PAGE and immunoblotted directly with antibodies against various cell cycle components, as indicated on the left. The elution positions of molecular weight standards are indicated at the top.
As expected, the most striking differences were in the distribution of CDK4 and CDK6. In untreated EH1 cells, CDK4 was found in three distinct size ranges. A significant proportion eluted close to the void volume of the column with an estimated size of over 450 kDa, but the major peak was around 200 kDa, the same size range as the peaks of p21CIP1, p27KIP1, and cyclins. There was also a considerable amount of CDK4 in 50-kDa complexes that would be consistent with binary associations with INK4 family members. In the case of CDK6, this peak was much more prominent than the 200-kDa fraction, as previously shown in a mouse T-cell line (42). Upon induction of p16INK4a, there was a noticeable shift in the CDK4 distribution into the 50-kDa complex, presumably reflecting its increased association with p16INK4a. A similar effect was observed with CDK6. With CDK2, on the other hand, there was little change in the size distribution, and the most obvious difference was the increased ratio of inactive to active configurations. In contrast to CDK4 and CDK6, there appeared to be a considerable amount of free CDK2 in the cells, irrespective of the induction of p16INK4a.
Cyclins D1, E, and A were also predominantly in ∼200-kDa complexes, but induction of p16INK4a resulted in a broadening of the cyclin D1 peak. The change in molecular mass would be compatible with removal of CDK4 from a proportion of the complexes, suggesting that the replacement of CDK4 by CDK2 may be incomplete. Cyclin E did not shift in its distribution, continuing to remain in a broad-molecular-mass peak, ranging from 150 to 250 kDa. However, the ratio of the different forms of cyclin E recognized by the antibody did change, with a higher proportion of the more slowly migrating band being present when cells were arrested with p16INK4a.
Changes in composition of CDK complexes upon induction of p16INK4a.
Direct immunoblotting provided information about the distribution of cell cycle components in complexes of different sizes but not about the composition or kinase activity of the various complexes. Individual fractions were therefore immunoprecipitated with antibodies against either CDK4 or CDK2, and the associated cyclins and CDK inhibitors were visualized by immunoblotting. As well as separating equal amounts of lysates by gel filtration, care was taken to analyze the immunoblots from IPTG-treated and untreated cells under identical conditions (simultaneously) in order to assess quantitative differences in the proteins bound to the CDKs. The CDK4 and CDK2 immunoprecipitates from untreated cells were also assayed for kinase activity on pRb as the substrate. The immunoprecipitated CDK4 recapitulated the distributions observed in Fig. 4, with three major peaks at ≥450, 200, and 50 kDa (Fig. 5A). Kinase activity was predominantly associated with the 200-kDa peak, corresponding with peaks in the distribution of D cyclins, p21CIP1, and p27KIP1 that coprecipitated with CDK4.
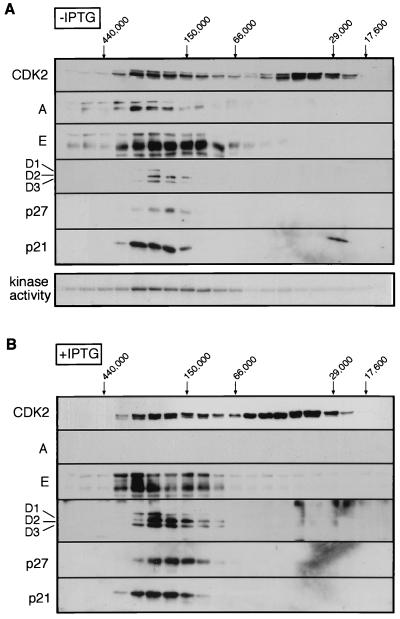
FIG. 5.
Reassortment of CDK4 complexes upon induction of p16INK4a. Following gel filtration as for Fig. 4, individual fractions were immunoprecipitated with antibodies against CDK4. The precipitates were separated by SDS-PAGE and immunoblotted with antisera against different components, as indicated on the left. Fractions from untreated (A) and IPTG-treated (B) cells were immunoblotted under identical conditions to allow quantitative comparisons. CDK4-associated kinase activity directed against the carboxy-terminal domain of pRb was determined for fractions from untreated cells. No kinase activity was detected in the IPTG-treated cells (Fig. 2).
Upon treatment of the cells with IPTG, the majority of the CDK4 shifted to a 50-kDa complex which coprecipitation confirmed as a binary complex with p16INK4a (Fig. 5B). As a consequence, the amounts of D cyclins associated with CDK4 were significantly reduced (Fig. 5B). No p27KIP1 was detected in CDK4 complexes after induction of p16INK4a (as in Fig. 3A), but there was little change in either the amount or the size distribution of p21CIP1 associated with CDK4 (Fig. 5B). Note that although the D cyclins and CDK4 can be found in complexes of 70 to 100 kDa, these do not appear to include p16INK4a, ruling out the existence of stable ternary complexes.
In the parallel analysis of CDK2 complexes, kinase activity was again predominantly in the 200- to 250-kDa size range, but the peak of activity was noticeably broad (Fig. 6A). This peak colocalized with the cyclin E, cyclin A, and p21CIP1 that coimmunoprecipitated with CDK2. As expected, no kinase activity was associated with the free pool of CDK2, which represented at least 50% of the total CDK2 in the lysates and contained both the phosphorylated and unphosphorylated isoforms. When p16INK4a was expressed, cyclin A was no longer detectable in the immunoprecipitates (as previously shown in Fig. 1 and 4), whereas the amounts of cyclin E associated with CDK2 remained unchanged (Fig. 6B). As in Fig. 3A, the levels of p27KIP1 coprecipitated with CDK2 were significantly increased in the peak fractions. In addition, much higher amounts of the D cyclins were associated with CDK2 in cells treated with IPTG.
FIG. 6.
Reassortment of CDK2 complexes upon induction of p16INK4a. Following gel filtration as for Fig. 4, individual fractions were immunoprecipitated with antibodies against CDK2. The precipitates were separated by SDS-PAGE and immunoblotted with antisera against different components, as indicated on the left. Fractions from untreated (A) and IPTG-treated (B) cells were immunoblotted under identical conditions to allow quantitative comparisons. CDK2-associated kinase activity directed against the carboxy-terminal domain of pRb was determined for fractions from untreated cells. No kinase activity was detected in the IPTG-treated cells (Fig. 2).
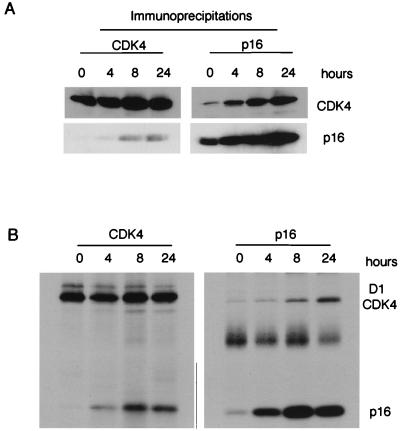
Time course of p16INK4a-induced changes.
Although we found no evidence for ternary complexes at 48 h postinduction, it remained formally possible that a transient association of p16INK4a with cyclin D1 and CDK4 was responsible for the inhibition of CDK4-associated kinase activity and G1 arrest. To explore this possibility, we prepared lysates from EH1 cells at various times after addition of IPTG and examined the association of CDK4 with either cyclin D1 or p16INK4a by immunoprecipitation and Western blotting. Upon addition of IPTG, the steady-state levels of p16INK4a increased over the first 6 to 8 h and then remained relatively constant (Fig. 7A and additional data not shown). This was paralleled by an increase in the association of CDK4 with p16INK4a as assayed by immunoprecipitation of either CDK4 or p16INK4a (Fig. 7A). However, no cyclin D1 was detected in the p16INK4a immunoprecipitates at any time point, and vice versa (negative data not shown).
FIG. 7.
Association of CDK4 and p16INK4a at different times after induction. (A) EH1 cells were harvested at 0, 4, 8, and 24 h after addition of IPTG, and the lysates were immunoprecipitated with antisera to CDK4 (left) and p16INK4a (right). After fractionation by SDS-PAGE, coprecipitating proteins were detected by immunoblotting with antisera against CDK4 and p16INK4a as indicated on the right. (B) EH1 cells were induced with IPTG for 0, 4, 8, and 24 h and metabolically labeled with [35S]methionine-cysteine for 2 h prior to harvesting. The cell lysates were immunoprecipitated with antisera against CDK4 (left) or p16INK4a (right), and the labeled proteins were detected by autoradiography. The positions of cyclin D1, CDK4, and p16INK4a are as shown on the right.
A similar situation prevailed when metabolic labeling was used to examine the fate of newly synthesized components. In this experiment, cells were treated with IPTG for different times and pulse-labeled with [35S]methionine-cysteine for the 2 h prior to harvesting. The synthesis of p16INK4a increased to maximum levels by about 8 h, whereas the synthesis of CDK4 remained constant throughout the time course. Immunoprecipitation of p16INK4a revealed an increased association with newly synthesized CDK4; conversely, there was an increase in the amount of labeled p16INK4a entering complexes with CDK4. Since a relatively small proportion of the total CDK4 in the cell is likely to be in a complex with cyclin D1 (Fig. 4), the signal for coprecipitating cyclin D1 was relatively faint in this experiment. Nevertheless, there was a significant decrease in the amount of cyclin D1 coprecipitating with CDK4 at 24 h. No labeled cyclin D1 was detected in the p16INK4a immunoprecipitates.
DISCUSSION
The induction of p16INK4a in EH1 cells had a number of striking effects on the composition of cyclin-CDK complexes, the combined result of which was a G1 arrest. A major aim of this work was to determine whether the primary effect of p16INK4a was to divert CDK4 from active complexes, as predicted by the competitive binding model, or to inhibit preexisting cyclin D-CDK4 by joining ternary or higher-order complexes. In this respect, the gel filtration results effectively preclude the latter model since the overwhelming majority of p16INK4a in IPTG-treated EH1 cells was found in fractions in the 50- and 15- to 20-kDa size ranges. Selective immunoprecipitation and Western blotting confirmed that these peaks correspond to CDK-bound and free p16INK4a, respectively, and attempts to find cyclin D1 and p16INK4a in the same fractions were consistently negative. Although we cannot formally exclude the transient formation of higher-order complexes, we estimate that in a steady-state situation, 48 h postinduction, less that 1% of the total p16INK4a in the cells eluted in complexes larger than 50 kDa.
Previous studies using similar inducible expression systems demonstrated coprecipitation of D cyclins and either p15INK4b or p19INK4d but did not address the proportions of the INK4 proteins in such complexes (1, 55). While it is possible that different INK4 proteins have different properties in this regard, the structural data suggest that they adopt similar three-dimensional conformations (6, 38, 46, 73). Another possibility is that the rapid induction of artificially high levels of INK4 protein leads to a transient association with cyclin-CDK complexes that is rarely seen under physiological conditions. In the previous studies, the cells were harvested relatively soon after induction (1, 55). However, we found no evidence for coprecipitation of cyclin D1 and p16INK4a during a time course of p16INK4a induction (Fig. 7 and data not shown). On the contrary, the interactions of newly synthesized components observed at various time points would be entirely consistent with a competitive binding model in which the unstable cyclin is gradually replaced by the more stable inhibitor. The rate at which the different complexes appear to change would of course depend on the stoichiometry of the different components.
Another feature of our data that contrasts with conclusions drawn elsewhere (55) is that the induction of p16INK4a demonstrably affected the composition of cyclin D-CDK complexes. In this respect, our data are more in line with those reported by Adachi et al. and Sandhu et al., both of whom showed a reduction in cyclin D-CDK4 interactions when INK4 levels were elevated (1, 60). We are also in complete agreement with a recent report by Jiang et al. based on inducible expression of p16INK4a (30). Thus, in IPTG-treated EH1 cells a substantial proportion of the CDK4 relinquished its association with cyclin D1 to form complexes with p16INK4a. One consequence was the broadening of the cyclin D1 peak on the gel filtration column, and it will be interesting to determine whether the smaller cyclin D1 complexes, at around 120 to 150 kDa, contain any kinase subunits. However, much of the cyclin D1 remains in the 200-kDa peak and the total amounts in the cell are unchanged, ruling out the increased turnover of cyclin D1 when it is dissociated from CDK4. A likely reason is that the CDK4 is replaced in the complex by CDK2.
Cyclin D1 has been previously shown to interact with CDK2 in some cell types (4, 77), and this association is increased in senescent fibroblasts (13). However, it is predominantly the unphosphorylated, inactive form of CDK2 that coprecipitates with cyclin D1, consistent with the change in the ratio of the two forms when p16INK4a is induced in EH1 cells. It has been formally demonstrated that cyclin D1-CDK2 complexes are inactive as kinases and insensitive to activation by cyclin H-CDK7 (27), but the situation is less clear for cyclins D2 and D3; coexpression with CDK2 in baculovirus-based systems has been shown to produce active kinases (15).
Replacement of CDK4 by CDK2 puts a different complexion on the reassortment of the various components in IPTG-treated EH1 cells. It has been previously surmised that induction of INK4 proteins causes p27KIP1 to move off cyclin D1-CDK4 complexes and onto cyclin E-CDK2 (30, 55, 56, 60), but our data show little if any displacement of p27KIP1 from cyclin D1. A more likely interpretation, therefore, is that p27KIP1 remains associated with cyclin D1, to which it can bind directly in the absence of a CDK (8, 16, 17, 20, 36, 71). The substitution of CDK2 for CDK4 in this complex would then explain the dramatic shift in allegiance of p27KIP1 from CDK4 to CDK2 shown in Fig. 3 and elsewhere (30, 55, 56, 60). The one facet of the data not explained by this model is the increase in the amount of p27KIP1 and p21CIP1 bound to cyclin E after p16INK4a induction (Fig. 3 and reference 30). Several possibilities can be considered. The first is that the levels of cyclin E are generally much lower than those of cyclin D1 so that a severalfold increase in the amount of p27KIP1 that coprecipitates with cyclin E may represent a very small proportion of the amount normally bound to cyclin D1. Another is that there is an appreciable reduction in the expression of cyclin D3 in IPTG-treated EH1 cells which, together with the inability to activate cyclin A expression, leads to a surplus of p27KIP1 that accumulates on cyclin E-CDK2 complexes. The immunodepletion experiments shown in Fig. 3C favor the latter interpretation since there is a substantial reduction in cyclin D3-p27KIP1 complexes in the arrested cells. What causes this reduction remains a matter of conjecture, but one possibility would be that, at least in some cell systems, cyclin D3 expression is delayed relative to that of cyclins D1 and D2. Cyclin D3 expression may therefore be partially dependent on the activation of CDK4 or CDK6.
The net effect of these changes is a marked reduction in the kinase activity associated with CDK2, whether it is bound to cyclin E or to cyclin A. However, it remains unclear whether this inhibition of CDK2 activity is an essential factor in the cell cycle arrest imposed by p16INK4a or is simply an adjunct to the direct inhibition of cyclin D-associated CDK4 and CDK6. Our data are clearly at odds with a previous report that cotransfection of p16INK4a can prevent the phosphorylation of pRb without inhibiting the activity of cyclin E-CDK2 (41) but agree with those recorded for inducible or ectopic expression of p15INK4b (55, 56) and p16INK4a (30). The IPTG-treated EH1 cells arrest with pRb in its hypophosphorylated state, consistent with the observed inhibition of CDK2, -4, and -6, but this would also be consistent with models in which the hyperphosphorylation of pRb by cyclin E-CDK2 is dependent on prior phosphorylation by cyclin D-dependent kinases (26, 41). Nevertheless, it is clear from Fig. 2 that upon p16INK4a-mediated arrest, pRb is not phosphorylated on Ser780, a site specifically targeted by cyclin D-dependent kinases (31).
It is now well established that cyclins and CDKs are found in multicomponent complexes within mammalian cells, although the exact composition of these complexes is not fully understood (10, 42, 49, 54, 67). Most cyclins show a major peak of around 150 to 220 kDa, although size estimates vary in different studies, and our data are in broad agreement. CDK4 and CDK6 are found primarily in three distinct complexes of very different sizes. As recently reported for CDK6 (42), a significant proportion of the CDK4 eluted with an apparent molecular mass in excess of 400 kDa. The other components of this complex have not been identified but may include the molecular chaperones HSP90 and CDC37 (9, 34, 42, 68). In the case of CDK6, Mahony et al. noted that the minor peak at 170 to 200 kDa corresponded to active kinase (42), and our data for EH1 cells concur (not shown). However, a much higher proportion of the CDK4 is present in the active kinase complex at the 170- to 200-kDa peak, and in contrast to CDK6, only a minor fraction of the CDK4 is found in binary complexes with INK4 proteins at ∼50 kDa. Since current methods do not allow quantitative comparisons of the CDK4- and CDK6-associated kinase activity, it is not possible to assess the relative contributions of the two kinases to G1 progression. Nevertheless, our data indicate that both active complexes are affected by the induction of p16INK4a.
It is clear that the interactions between cell cycle regulatory components are integrally connected. By making one alteration, such as increasing the levels of p16INK4a, we find that several multicomponent complexes undergo reassortment and that the impact of the inhibitor is more pleiotropic than simply altering the activity of the proteins to which it directly binds. Moreover, the debate over the ability of INK4 proteins to associate with or disrupt binary cyclin D-CDK4 complexes in vitro may be largely irrelevant since there is no evidence that such binary complexes ever exist in vivo.
ACKNOWLEDGMENTS
We are indebted to Jim Koh, Emma Lees, and Yoichi Taya for providing monoclonal antibodies and to David Parry and Emma Lees for communicating results prior to publication. We also thank Harmut Land and Nic Jones for helpful discussions and comments on the manuscript.
REFERENCES
- 1.Adachi M, Roussel M F, Havenith K, Sherr C J. Features of macrophage differentiation induced by p19INK4d, a specific inhibitor of cyclin D-dependent kinases. Blood. 1997;90:126–137. [PubMed] [Google Scholar]
- 2.Alcorta D A, Xiong Y, Phelps D, Hannon G, Beach D, Barrett J C. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence. Proc Natl Acad Sci USA. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartkova J, Zemanova M, Bartek J. Abundance and subcellular localisation of cyclin D3 in human tumours. Int J Cancer. 1996;65:323–327. doi: 10.1002/(SICI)1097-0215(19960126)65:3<323::AID-IJC8>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 4.Bates S, Bonetta L, MacAllan D, Parry D, Holder A, Dickson C, Peters G. CDK6 (PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin D1. Oncogene. 1994;9:71–79. [PubMed] [Google Scholar]
- 5.Blain S W, Montalvo E, Massagué J. Differential interaction of the cyclin-dependent kinase (Cdk) inhibitor p27KIP1 with cyclin A-Cdk2 and cyclin D2-Cdk4. J Biol Chem. 1997;272:25863–25872. doi: 10.1074/jbc.272.41.25863. [DOI] [PubMed] [Google Scholar]
- 6.Byeon I-J L, Li J, Ericson K, Selby T L, Tevelev A, Kim H-J, O’Maille P, Tsai M-D. Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4. Mol Cell. 1998;1:421–431. doi: 10.1016/s1097-2765(00)80042-8. [DOI] [PubMed] [Google Scholar]
- 7.Chan F K M, Zhang J, Cheng L, Shapiro D N, Winoto A. Identification of human and mouse p19, a novel CDK4 and CDK6 inhibitor with homology to p16ink4. Mol Cell Biol. 1995;15:2682–2688. doi: 10.1128/mcb.15.5.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Saha P, Kornbluth S, Dynlacht B D, Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai K, Kobayashi R, Beach D. Physical interaction of mammalian CDC37 with CDK4. J Biol Chem. 1996;271:22030–22034. doi: 10.1074/jbc.271.36.22030. [DOI] [PubMed] [Google Scholar]
- 10.Della Ragione F, Russo G L, Oliva A, Mercurio C, Mastropietro S, Della Pietra V, Zappia V. Biochemical characterization of p16INK4- and p18-containing complexes in human cell lines. J Biol Chem. 1996;271:15942–15949. doi: 10.1074/jbc.271.27.15942. [DOI] [PubMed] [Google Scholar]
- 11.Diller L, Kassel J, Nelson C E, Gryka M A, Litwak G, Gebhardt M, Bressac B, Ozturk M, Baker S J, Vogelstein B, Friend S H. p53 functions as a cell cycle control point in osteosarcoma. Mol Cell Biol. 1990;10:5772–5781. doi: 10.1128/mcb.10.11.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimri G P, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano E E, Linskens M, Rubelj I, Pereira-Smith O, Peacocke M, Campisi J. A novel biomarker identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dulic V, Drullinger L F, Lees E, Reed S I, Stein G H. Altered regulation of G1 cyclins in senescent human diploid fibroblasts: accumulation of inactive cyclin E-Cdk2 and cyclin D1-Cdk2 complexes. Proc Natl Acad Sci USA. 1993;90:11034–11038. doi: 10.1073/pnas.90.23.11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elledge S J, Winston J, Harper J W. A question of balance: the role of cyclin-kinase inhibitors in development and tumorigenesis. Trends Cell Biol. 1996;6:388–392. doi: 10.1016/0962-8924(96)10030-1. [DOI] [PubMed] [Google Scholar]
- 15.Ewen M E, Sluss H K, Sherr C J, Matsushime H, Kato J, Livingston D M. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73:487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 16.Fotedar R, Fitzgerald P, Rousselle T, Cannella D, Dorée M, Messier H, Fotedar A. p21 contains independent binding sites for cyclin and cdk2: both sites are required to inhibit cdk2 kinase activity. Oncogene. 1996;12:2155–2164. [PubMed] [Google Scholar]
- 17.Goubin F, Ducommun B. Identification of binding domains on the p21Cip1 cyclin-dependent kinase inhibitor. Oncogene. 1995;10:2281–2287. [PubMed] [Google Scholar]
- 18.Guan K-L, Jenkins C W, Li Y, Nichols M A, Wu X, O’Keefe C L, Matera A G, Xiong Y. Growth suppression by p18, a p16INK4/MTS1 and p14INK4/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 1994;8:2939–2952. doi: 10.1101/gad.8.24.2939. [DOI] [PubMed] [Google Scholar]
- 19.Guan K-L, Jenkins C W, Li Y, O’Keefe C L, Noh S, Wu X, Zariwala M, Matera A G, Xiong Y. Isolation and characterization of p19INK4d, a p16-related inhibitor specific to CDK4 and CDK6. Mol Biol Cell. 1996;7:57–70. doi: 10.1091/mbc.7.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall M, Bates S, Peters G. Evidence for different modes of action of cyclin-dependent kinase inhibitors: p15 and p16 bind to kinases, p21 and p27 bind to cyclins. Oncogene. 1995;11:1581–1588. [PubMed] [Google Scholar]
- 21.Hannon G J, Beach D. p15INK4B is a potential effector of TGF-β-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 22.Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harlow E, Lane D. Antibodies. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 24.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 25.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L-H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, Fox M P, Wei N. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatakeyama M, Brill J A, Fink G R, Weinberg R A. Collaboration of G1 cyclins in the functional inactivation of the retinoblastoma protein. Genes Dev. 1994;8:1759–1771. doi: 10.1101/gad.8.15.1759. [DOI] [PubMed] [Google Scholar]
- 27.Higashi H, Suzuki-Takahashi I, Saitoh S, Segawa K, Taya Y, Okuyama A, Nishimura S, Kitagawa M. Cyclin-dependent kinase-2 (Cdk2) forms an inactive complex with cyclin D1 since Cdk2 associated with cyclin D1 is not phosphorylated by Cdk7-cyclin H. Eur J Biochem. 1996;237:460–467. doi: 10.1111/j.1432-1033.1996.0460k.x. [DOI] [PubMed] [Google Scholar]
- 28.Hirai H, Roussel M F, Kato J-Y, Ashmun R A, Sherr C J. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol Cell Biol. 1995;15:2672–2681. doi: 10.1128/mcb.15.5.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jen J, Harper J W, Bigner S H, Bigner D D, Papadopoulos N, Markowitz S, Willson J K V, Kinzler K W, Vogelstein B. Deletion of p16 and p15 genes in brain tumors. Cancer Res. 1994;54:6353–6358. [PubMed] [Google Scholar]
- 30.Jiang H, Chou H S, Zhu L. Requirement of cyclin E-Cdk2 inhibition in p16INK4a-mediated growth suppression. Mol Cell Biol. 1998;18:5284–5290. doi: 10.1128/mcb.18.9.5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitagawa M, Higashi H, Jung H-K, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J, Segawa K, Yoshida E, Nishimura S, Taya Y. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- 32.Koh J, Enders G H, Dynlacht B D, Harlow E. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature. 1995;375:506–510. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 33.LaBaer J, Garrett M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 34.Lamphere L, Fiore F, Xu X, Brizuela L, Keezer S, Sardet C, Draetta G F, Gyuris J. Interaction between Cdc37 and Cdk4 in human cells. Oncogene. 1997;14:1999–2004. doi: 10.1038/sj.onc.1201036. [DOI] [PubMed] [Google Scholar]
- 35.Lee M-H, Reynisdóttir I, Massagué J. Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 1995;9:639–649. doi: 10.1101/gad.9.6.639. [DOI] [PubMed] [Google Scholar]
- 36.Lin J, Reichner C, Wu X, Levine A J. Analysis of wild-type and mutant p21WAF-1 gene activities. Mol Cell Biol. 1996;16:1786–1793. doi: 10.1128/mcb.16.4.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loughran O, Malliri A, Owens D, Gallimore P H, Stanley M A, Ozanne B, Frame M C, Parkinson E K. Association of CDKN2A/p16INK4A with human head and neck keratinocyte replicative senescence: relationship of dysfunction to immortality and neoplasia. Oncogene. 1996;13:561–568. [PubMed] [Google Scholar]
- 38.Luh F Y, Archer S J, Domaille P J, Smith B O, Owen D, Brotherton D H, Raine A R C, Xu X, Brizuela L, Brenner S L, Laue E D. Structure of the cyclin-dependent kinase inhibitor p19Ink4d. Nature. 1997;389:999–1003. doi: 10.1038/40202. [DOI] [PubMed] [Google Scholar]
- 39.Lukas J, Bartkova J, Welcker M, Petersen O W, Peters G, Strauss M, Bartek J. Cyclin D2 is a moderately oscillating nucleoprotein required for G1 phase progression in specific cell types. Oncogene. 1995;10:2125–2134. [PubMed] [Google Scholar]
- 40.Lukas J, Parry D, Aagaard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma-protein-dependent cell cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 41.Lundberg A S, Weinberg R A. Functional inactivation of the retinobalstoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahony D, Parry D A, Lees E. Active cdk6 complexes are predominantly nuclear and represent only a minority of the cdk6 in T cells. Oncogene. 1998;16:603–611. doi: 10.1038/sj.onc.1201570. [DOI] [PubMed] [Google Scholar]
- 43.Matsuoka S, Edwards M C, Bai C, Parker S, Zhang P, Baldini A, Harper J W, Elledge S J. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995;9:650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 44.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J-Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McConnell B B, Starborg M, Brookes S, Peters G. Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr Biol. 1998;8:351–354. doi: 10.1016/s0960-9822(98)70137-x. [DOI] [PubMed] [Google Scholar]
- 46.McDonald N Q, Peters G. Ankyrin for clues about the function of p16INK4a. Nat Struct Biol. 1998;5:85–88. doi: 10.1038/nsb0298-85. [DOI] [PubMed] [Google Scholar]
- 47.Medema R, Herrera R E, Lam F, Weinberg R A. Growth suppression by p16INK4 requires functional retinoblastoma protein. Proc Natl Acad Sci USA. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musgrove E A, Swarbrick A, Lee C S L, Cornish A L, Sutherland R L. Mechanism of cyclin-dependent kinase inactivation by progestins. Mol Cell Biol. 1998;18:1812–1825. doi: 10.1128/mcb.18.4.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakanishi M, Robetorye R S, Adami G R, Pereira-Smith O M, Smith J R. Identification of the active region of the DNA synthesis inhibitory gene p21SdiI/CIP1/WAF1. EMBO J. 1995;14:555–563. doi: 10.1002/j.1460-2075.1995.tb07031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okamoto A, Demetrick D J, Spillare E A, Hagiwara K, Hussain S P, Bennett W P, Forrester K, Gerwin B, Serrano M, Beach D H, Harris C C. Mutations and altered expression of p16INK4 in human cancer. Proc Natl Acad Sci USA. 1994;91:11045–11049. doi: 10.1073/pnas.91.23.11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parry D, Bates S, Mann D J, Peters G. Lack of cyclin D-Cdk complexes in Rb-negative cells correlates with high levels of p16INK4/MTS1 tumour suppressor gene product. EMBO J. 1995;14:503–511. doi: 10.1002/j.1460-2075.1995.tb07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polyak K, Lee M-H, Erdjument-Bromage H, Koff A, Roberts J M, Tempst P, Massagué J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 54.Poon R Y C, Toyoshima H, Hunter T. Redistribution of the CDK inhibitor p27 between different cyclin-CDK complexes in the mouse fibroblast cell cycle and in cells arrested with lovastatin or ultraviolet irradiation. Mol Biol Cell. 1995;6:1197–1213. doi: 10.1091/mbc.6.9.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reynisdóttir I, Massagué J. The subcellular locations of p15Ink4b and p27Kip1 coordinate their inhibitory interactions with cdk4 and cdk2. Genes Dev. 1997;11:492–503. doi: 10.1101/gad.11.4.492. [DOI] [PubMed] [Google Scholar]
- 56.Reynisdóttir I, Polyak K, Iavarone A, Massagué J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-β. Genes Dev. 1995;9:1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 57.Reznikoff C A, Yeager T R, Belair C A, Savelieva E, Puthenveettil J A, Stadler W M. Elevated p16 at senescence and loss of p16 at immortalization in human papillomavirus 16 E6, but not E7, transformed human uroepithelial cells. Cancer Res. 1996;56:2886–2890. [PubMed] [Google Scholar]
- 58.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta Rev Cancer. 1998;1378:115–177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 59.Russo A A, Jeffrey P D, Patten A K, Massagué J, Pavletich N P. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 60.Sandhu C, Garbe J, Bhattacharya N, Daksis J, Pan C-H, Yaswen P, Koh J, Slingerland J M, Stampfer M R. Transforming growth factor β stabilizes p15INK4B protein, increases p15INK4B-cdk4 complexes, and inhibits cyclin D1-cdk4 association in human mammary epithelial cells. Mol Cell Biol. 1997;17:2458–2467. doi: 10.1128/mcb.17.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serrano M, Gómez-Lahoz E, DePinho R A, Beach D, Bar-Sagi D. Inhibition of Ras-induced proliferation and cellular transformation by p16INK4. Science. 1995;267:249–252. doi: 10.1126/science.7809631. [DOI] [PubMed] [Google Scholar]
- 62.Serrano M, Hannon G J, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 63.Shapiro G I, Park J E, Edwards C D, Mao L, Merlo A, Sidransky D, Ewen M E, Rollins B J. Multiple mechanisms of p16INK4A inactivation in non-small cell lung cancer cell lines. Cancer Res. 1995;55:6200–6209. [PubMed] [Google Scholar]
- 64.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 65.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 66.Soos T J, Kiyokawa H, Yan J S, Rubin M S, Giordano A, DeBlasio A, Bottega S, Wong B, Mendelsohn J, Koff A. Formation of p27-CDK complexes during the human mitotic cell cycle. Cell Growth Differ. 1996;7:135–146. [PubMed] [Google Scholar]
- 67.Steiner P, Philipp A, Lukas J, Godden-Kent D, Pagano M, Mittnacht S, Bartek J, Eilers M. Identification of a Myc-dependent step during the formation of active G1 cyclin-cdk complexes. EMBO J. 1995;14:4814–4826. doi: 10.1002/j.1460-2075.1995.tb00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stepanova L, Leng X, Parker S B, Harper J W. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 1996;10:1491–1502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- 69.Stone S, Dayanath P, Jiang P, Weaver-Feldhaus J M, Tavtigian S V, Cannon-Albright L, Kamb A. Genomic structure, expression and mutational analysis of the P15 (MTS2) gene. Oncogene. 1995;11:987–991. [PubMed] [Google Scholar]
- 70.Stott F J, Bates S, James M C, McConnell B B, Starborg M, Brookes S, Palmero I, Hara E, Vousden K H, Peters G. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 72.Uhrbom L, Nistér M, Westermark B. Induction of senescence in human malignant glioma cells by p16INK4A. Oncogene. 1997;15:505–514. doi: 10.1038/sj.onc.1201227. [DOI] [PubMed] [Google Scholar]
- 73.Venkataramani R, Swaminathan K, Marmorstein R. Crystal structure of the CDK4/6 inhibitory protein p18INK4c provides insights into ankyrin-like repeat structure/function and tumor-derived p16INK4 mutations. Nat Struct Biol. 1998;5:74–81. doi: 10.1038/nsb0198-74. [DOI] [PubMed] [Google Scholar]
- 74.Vogt M, Haggblom C, Yeargin J, Christiansen-Weber T, Haas M. Independent induction of senescence by p16INK4a and p21CIP1 in spontaneously immortalized human fibroblasts. Cell Growth Differ. 1998;9:139–146. [PubMed] [Google Scholar]
- 75.Wong H, Riabowol K. Differential CDK-inhibitor gene expression in aging human diploid fibroblasts. Exp Gerentol. 1996;31:311–325. doi: 10.1016/0531-5565(95)00025-9. [DOI] [PubMed] [Google Scholar]
- 76.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 77.Xiong Y, Zhang H, Beach D. D type cyclins associate with multiple protein kinases and the DNA replication and repair factor PCNA. Cell. 1992;71:505–514. doi: 10.1016/0092-8674(92)90518-h. [DOI] [PubMed] [Google Scholar]
- 78.Zhang H, Xiong Y, Beach D. Proliferating cell nuclear antigen and p21 are components of multiple cell cycle kinase complexes. Mol Biol Cell. 1993;4:897–906. doi: 10.1091/mbc.4.9.897. [DOI] [PMC free article] [PubMed] [Google Scholar]