Abstract
Kinome from apicomplexan parasites is composed of eukaryotic protein kinases and Apicomplexa specific kinases, such as rhoptry kinases (ROPK). Ropk is a gene family that is known to play important roles in host–pathogen interaction in Toxoplasma gondii but is still poorly described in Eimeria tenella, the parasite responsible for avian coccidiosis worldwide. In the E. tenella genome, 28 ropk genes are predicted and could be classified as active (n = 7), inactive (incomplete catalytic triad, n = 12), and non-canonical kinases (active kinase with a modified catalytic triad, n = 9). We characterized the ropk gene expression patterns by real-time quantitative RT-PCR, normalized by parasite housekeeping genes, during the E. tenella life-cycle. Analyzed stages were: non-sporulated oocysts, sporulated oocysts, extracellular and intracellular sporozoites, immature and mature schizonts I, first- and second-generation merozoites, and gametes. Transcription of all those predicted ropk was confirmed. The mean intensity of transcription was higher in extracellular stages and 7–9 ropk were specifically transcribed in merozoites in comparison with sporozoites. Transcriptional profiles of intracellular stages were closely related to each other, suggesting a probable common role of ROPKs in hijacking signaling pathways and immune responses in infected cells. These results provide a solid basis for future functional analysis of ROPK from E. tenella.
Keywords: rhoptry kinases, gene expression, Eimeria tenella, life-cycle
1. Introduction
The Apicomplexa phylum consists of a large group of parasitic protists of medical and veterinary importance (Plasmodium, Toxoplasma, Cryptosporidium, Eimeria, etc). The genus Eimeria contains more than 1000 parasitic species, which invade the epithelial cell lining in the intestinal tract of their host, causing coccidiosis. All Eimeria spp. are considered to be highly host-specific. Avian coccidiosis is caused by seven Eimeria species, among which Eimeria tenella, E. maxima, and E. acervulina are the main species responsible for coccidiosis in poultry, worldwide. Clinical coccidiosis is associated with a reduction in body-weight gain, a decrease in the feed conversion index, and may result in a high mortality rate. Clinical and sub-clinical infections have a huge economic impact on poultry production. To control coccidiosis, coccidiostats are used as a feed additive in broilers production [1]. Attenuated live vaccines, which are very efficient, but expensive to produce, are mostly used in layers and labels with high economic importance in their production [2]. Natural alternative strategies, such as probiotics and phytochemicals, are becoming more explored [3].
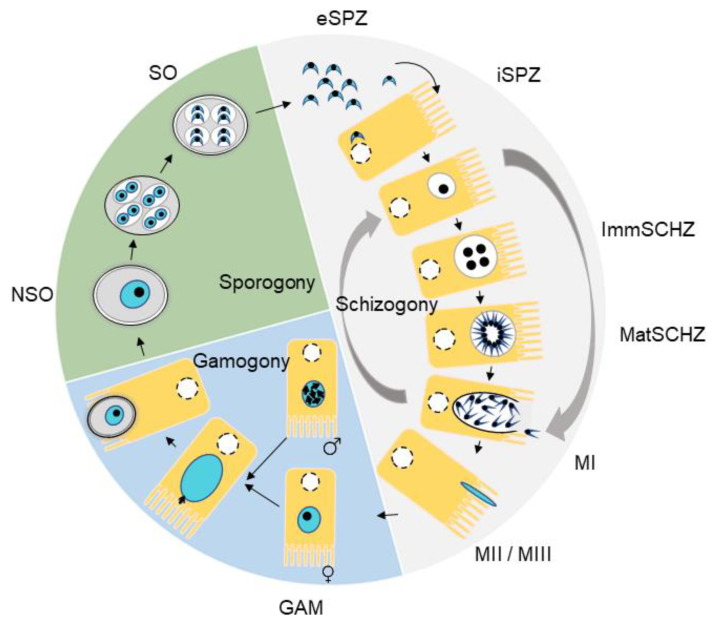
The life-cycle of Eimeria is monoxenous with one phase in the environment and another in the host (asexual multiplication and sexual reproduction, respectively), with several developmental stages, either intracellular or extracellular (Figure 1), characterized by asynchronicity. Free-living stages consist of oocysts, which are very resistant to environmental conditions. After oral contamination of chickens with sporulated oocysts, released sporozoites invade intestinal epithelial cells and undergo three rounds of asexual multiplications (first-, second-, and third-schizogony), each generating a large number of merozoites. Third-generation merozoites differentiate into micro- and macro-gametes. Gamete fecundation results in the production of zygotes (gamogony). After maturation, zygotes evolve into non-sporulated oocysts that are released in the environment. In the environment, the sporogony will result in the production of sporulated oocysts under appropriate hygrometric and temperature conditions. The most pathogenic phase for the host is the merogony, during which successive intracellular multiplications induce digestive mucosal lesions. Live attenuated vaccines correspond to Eimeria strains that have only one merogony, leading to fewer mucosal lesions than wild-type (WT) strains.
Figure 1.
Eimeria tenella life-cycle. Sporogony occurs in the environment, whereas schizogony and gamogony occur in the chicken digestive tract. For the wild-type strain, three rounds of schizogony produce first-, second-, and third-generation merozoites. NSO, non-sporulated oocysts, SO sporulated oocysts, eSPZ extracellular sporozoites, iSPZ intracellular sporozoites, ImmSCHZ immature schizonts, MatSCHZ mature schizonts, MI first-generation merozoites, MII second-generation merozoites, and MIII third-generation merozoites.
The kinome of coccidia (Toxoplasma, Neospora, and Eimeria) is composed of eukaryotic protein kinases (ePKs) [4,5] and a coccidia-specific kinase family (rhoptry kinases, ROPK) [6]. ROPK sequences are highly divergent from ePK sequences [7] and include active kinases (i.e., a kinase domain organized in 12 subdomains that retain all the key residues needed for catalysis, as the catalytic triad KDD), pseudo-kinases (inactive enzymes, characterized by the absence of a complete catalytic domain and/or a mutated catalytic triad), and non-canonical kinases (i.e., active kinases with most of the residues necessary for catalysis, but with some differences in the conserved residues).
Ropks were amplified from an ancestral gene. T. gondii type I, II, and III contain 37, 55, and 38 ropk, respectively [6]. Individual ropk deletion in a type II T. gondii resulted in less virulent strains for 16 ropk genes [8]. Contrary to Eimeria sp., which develops in one host, T. gondii infects intermediate warm-blooded hosts for asexual multiplication (tachyzoite and bradyzoite stages) and reproduces sexually in cats as a definitive host (sporozoites and gametes). In T. gondii, ROPK is associated with host cell manipulation during asexual multiplication (i.e., tachyzoite stage): invasion process, virulence, and host cell modulation have been well documented ([9] for review). Although ROPK expression was also detected to a lesser extent, in merozoites stages [10,11], no clear function was identified due to the difficulty to purify these enteric stages ([12] for review).
This gene family remains mostly unknown in E. tenella [7,13]. Among 90 PKs predicted in the E. tenella genome, 28 are likely to belong to the ROPK family [7,14], among which, 18 ropk are organized in 6 tandem repeat loci. Within E. tenella ROPKs, 12 are predicted to be pseudo-kinases (i.e., inactive), 7 to be active kinases, and 9 to be non-canonical kinases (Table 1). Ropk genes from a tandem repeat locus belong to the same enzymatic class (i.e., active, inactive, or non-canonical kinases), except for the Etrop5170/Etrop5190 locus for which the genes belong to inactive and active classes of kinase, respectively, and for the Etrop28835/Etrop28855 locus which belongs to non-canonical and inactive kinases, respectively (see Table 1). Phylogenetic analysis showed that 19 ROPKs (i.e., 68%) from E. tenella were clustered into one specific clade [6]. This clade contains active (Eten3 sub-clade), inactive (Eten4, 5 and 6 sub-clades), and non-canonical kinases (Eten2a, 2b, and 3 sub-clades) (Figure S1). Several other E. tenella ROPKs were found outside this clade: either in one clade of unique ROPKs (6 ROPKs = 21%) or more closely related to T. gondii ROPK (TgROP21 and TgROP35) (3 ROPKs = 11%). Ropk genes from a tandem repeat locus are phylogenetically closer than other ropk genes (Figure S1). Only two ROPKs have been readily identified so far at the proteomic level in E. tenella sporozoites and four ROPKs at the merozoite stage [15]. In a previous study, the authors characterized the first active ROPK (EtROP1) found in E. tenella sporozoites [16]: EtROP1 interacts and phosphorylates the cellular p53, resulting in the inhibition of host cell apoptosis and in the arrest of the host cell cycle in the G0/G1 phase. Here, we characterized ropk gene expression patterns during the E. tenella life-cycle, generating 9 developmental stages. Two WT strains, EtINRAE and EtWIS, from different geographical origins, were analyzed to describe the biological diversity of ropk transcription between strains. We confirmed the gene expression of those predicted ropk from the E. tenella genome in the various life stages. Their transcription in both extracellular and intracellular stages of E. tenella is consistent with their known functions in host-cell invasion and hijacking of host cell signaling pathways by T. gondii.
Table 1.
Rhoptry kinase genes from the Eimeria tenella genome.
| Activity | Gene ID | Gene Symbol | Phylogeny 1 |
|---|---|---|---|
| active | ETH_00005190 | Etrop5190 | ROPK unique |
| active | ETH_00005905 | Etrop5905 | TgROP35 |
| active | ETH_00014495 | Etrop14495 | TgROP21 |
| active | ETH_00026495 | Etrop26495 | TgROP35 |
| active | ETH_00027695 | Etrop27695 | Eten1 |
| active | ETH_00027700 | Etrop27700 | Eten1 |
| active | ETH_00027705 | Etrop27705 | Eten1 |
| inactive | ETH_00000075 | Etrop75 | Eten4 |
| inactive | ETH_00000080 | Etrop80 | Eten4 |
| inactive | ETH_00002510 | Etrop2510 | Eten6 |
| inactive | ETH_00005170 | Etrop5170 | ROPK unique |
| inactive | ETH_00005335 | Etrop5335 | ROPK unique |
| inactive | ETH_00005400 | Etrop5400 | Eten5 |
| inactive | ETH_00005405 | Etrop5405 | Eten5 |
| inactive | ETH_00005410 | Etrop5410 | Eten5 |
| inactive | ETH_00005415 | Etrop5415 | Eten5 |
| inactive | ETH_00016910 | Etrop16910 | ROPK unique |
| inactive | ETH_00028835 | Etrop28835 | ROPK unique |
| non-canonical | ETH_00005840 | Etrop5840 | Eten3 |
| non-canonical | ETH_00020585 | Etrop20585 | Eten3 |
| non-canonical | ETH_00020590 | Etrop20590 | Eten3 |
| non-canonical | ETH_00020610 | Etrop20610 | Eten3 |
| non-canonical | ETH_00020615 | Etrop20615 | Eten3 |
| non-canonical | ETH_00020620 | Etrop20620 | Eten3 |
| non-canonical | ETH_00021185 | Etrop21185 | Eten3 |
| non-canonical | ETH_00028765 | Etrop28765 | Eten2a |
| non-canonical | ETH_00028855 | Etrop28855 | Eten2b |
1 Adapted from [6]. See text for details about the phylogeny and supplementary data Figure S1.
2. Materials and Methods
2.1. Ethics Statements
All animal experimentations have been performed in the Infectiology of Farm, Model, and Wildlife Animals Facility (PFIE, Centre INRAE Val De Loire: https://doi.org/10.15454/1.5572352821559333E12, accessed on 25 July 2021); member of the National Infrastructure EMERG’IN). Experimental protocols were designed in compliance with French law (2010/63/EU, 2010; Rural Code, 2018; Decree No. 2013-118, 2013) concerning the use of laboratory animals. Care and euthanasia of animals were practiced according to the national ethical guidelines and approved by the local ethics committee for animal experimentation (Comité d’Ethique en Expérimentation Animale Val de Loire, CEA VdL N°19): APAFIS#25884. The authors are committed to the principles of the 3Rs: reduction, refinement, and replacement of experimental animals.
2.2. Parasite Strains and Propagation
The Wisconsin (EtWIS) and INRAE (EtINRAE) strains of E. tenella were used throughout this study. EtWIS was first described in 1974 [17] and is a laboratory strain used in most studies. EtINRAE, initially described as a PAPt38 strain from INRA, was isolated from a poultry farm in France in 1974. The parasites were propagated by oral infection (104 sporulated oocysts) of 3–5-week-old outbred PA12 chickens, reared in a coccidia-free environment with supply ad libitum of filtered water and anticoccidial- and antibiotic-free feed, following the standard protocol for oocyst purification [18].
2.3. Purification of Developmental Stages
We investigated nine distinct developmental stages of the parasites: non-sporulated oocysts (NSO), sporulated oocysts (SO), extracellular sporozoites (eSPZ), intracellular sporozoites (iSPZ), immature schizont (ImmSCHZ), mature schizont (MatSCHZ), first-generation merozoites (MI), second-generation merozoites (MII), and gametes (GAM). NSO were collected directly from the ceca at 7 days post-inoculation (p.i.) and purified as described previously [18]. SO were obtained after incubation in a solution of potassium dichromate 2.5% (w/v) for 48–72 h at 27 °C. Sporozoites were obtained from fresh fully sporulated oocysts. Briefly, after breaking the oocyst walls with 0.5 mm glass beads (Thermo Fisher Scientific), sporocysts were incubated in the excystation medium (0.25% (w/v) trypsin and 0.5% (w/v) biliary salts in 10 mM phosphate-buffered saline (PBS), pH 7.4) at 41 °C for 1 h. eSPZ were purified by a two-step filtration protocol, first on cotton and then on polycarbonate filters (pore size, 5 µm; Whatman). Second-generation merozoites and gametes were recovered from chicken caecal mucosa 112 h (5 d.p.i.) and 136 h (6 d.p.i.), after inoculation with 2.5 × 105 or 1 × 104 sporulated oocysts, respectively, as previously described [18,19]. Briefly, second-generation merozoites and gametes were purified from caecal content and scrapped infected mucosa at necropsies.
To generate the intracellular stages (i.e., iSPZ, ImmSCHZ, and MatSCHZ) and to cope with the asynchronicity of E. tenella development, we purified early stages from in vitro propagation. Freshly excysted, purified sporozoites were incubated with MDBK cells, at an MOI = 3 (5 × 105 cells, 3 × 105 cells and 2 × 105 cells, respectively) in 6-well plates at 41 °C in 5% CO2. Infected cells were washed at 24 h p.i., for iSPZ stage, 48 h p.i., for ImmSCHZ stage, or at 62 h p.i., for MatSCHZ stage. For first-generation merozoites (MI) production, 2 × 106 MDBK were seeded in a Petri dish and infected with 0.5–1 × 107 freshly purified sporozoites. MI were recovered in the supernatant at 72 h p.i. and filtered on polycarbonate filters (pore size, 5 µm, Whatman).
2.4. RNA Extraction
Extraction was performed on 0.5–2 × 108 parasites, whatever the stage. NSO and SO were resuspended in 50 µL of TRIzol and were fully disrupted by vortexing with 0.5 mm glass beads (Thermo Fisher Scientific, Waltham, MA, USA) for 1 min followed by cooling for 1 min in ice; this procedure was repeated 5 times. The lysate was then added to 950 µL of TRIzol provided in the kit (Direct-zol RNA Microprep, Zymo Research Corporation, Irvine, CA, USA), and the RNA extraction was performed according to the manufacturer’s protocol. For other developmental stages, parasites were directly lysed with TRIzol and RNA extraction was performed following the standard protocol of the kit. RNA concentration was measured using a NanoDrop One spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). RNA integrity was checked according to an RNA Integrity Number (RIN > 8) using RNA 6000 Nano chips run on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The samples were stored at −80 °C until further use. All samples were tested for genomic contamination using a negative control, which consisted of RNA extraction and PCR amplification without adding the RT enzyme.
cDNA was synthesized using the M-MLV Reverse Transcriptase (Promega Corporation, Madison, WI, USA), with random hexamer primers and oligo (dT)15 primer (Promega). cDNAs were then amplified by qPCR using the SYBR Green master mix (Bio-Rad Corporation, Hercules, CA, USA). Housekeeping genes used to normalize ropk gene amplification were: Et18s (EF210326) and Etactin (ETH_00009555). Stage-specific gene expression was determined using Etama1 (ETH_00007745) for the sporozoite stage, Etsag3 (ETH_00010755) for the schizont, and merozoite stages, and Etgam56 (ETH_00007320) for macrogametes [20] (Figure S2). All ROPK and host-cell-specific primers are listed in (Table S1).
The protocol used for qPCR was: 95 °C for 5 min and 40 cycles at 95 °C for 10 s and 60 °C (or 62 °C) for 15 s. The absence of primer dimers in the reaction was verified with melting curves, performed at 60 °C (or 62 °C) for 5 s followed by gradual heating (0.5 °C/s) to 95 °C. The absence of cross-reactivity of parasite primers with host cell nucleic acid (and host cell primers with parasite nucleic acid) was verified (data not shown). qPCRs were performed in triplicate for each experiment. ROPK and stage-specific gene expression were normalized to Ct values obtained for Et18s and Etactin using the formula: 2−(Ct Et specific stage gene or ropk–Ct mean of Et housekeeping genes). Gametes, being the stage with the lowest ropk transcription level (present study and [18,19]), gene expression values were standardized to gamete stage expression: genes with a fold change (FC) in expression > 2 were considered as expressed genes at that particular stage. Gene expression values are expressed as the mean ± SD of two independent biological replicates.
2.5. Gene Expression Profiling and Hierarchical Clustering
To examine the global expression profiles of ROP kinase genes among different developmental stages, a principal component analysis (PCA) and a hierarchical clustering analysis (HCA) with average linkage (Pearson distance) were carried out using the R software [21]. The variables located near the origin of the axis were of little value in interpreting the factorial plan of projection, whereas variables found on the circle were highly significant in describing the data. The proximity of variables (and/or data) on a plan showed that they were positively correlated. The interest of each plan of projection was assessed by its inertia (percentage of variance). The regressions were established by the least-squares method. The heatmap was illustrated with Heatmapper software [22].
3. Results
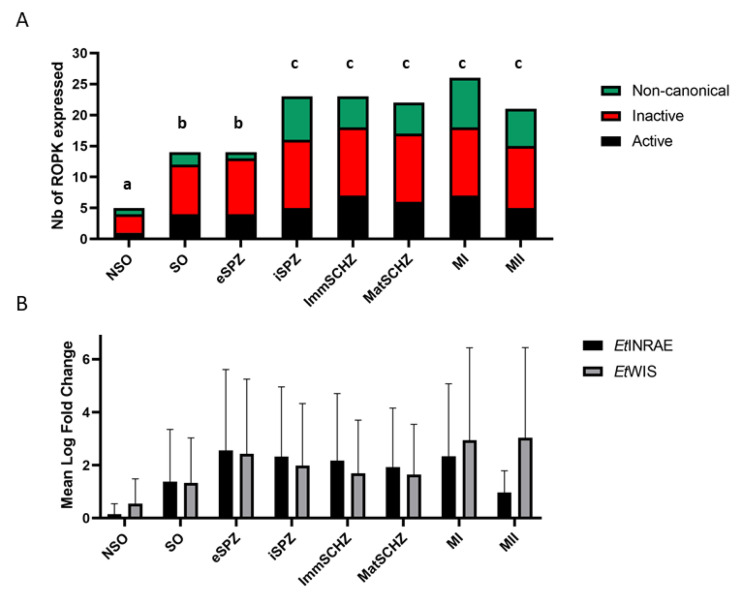
Ropk transcription is regulated across the E. tenella life-cycle in the EtINRAE strain (Figure 2A and Table S2). A significant and gradual increase in the number of transcribed ropk is observed during the parasite cycle. Ropk gene expression is the lowest in free-living stages and gradually increases with sporulation, and even more in the intracellular stages (schizonts and merozoites). The transcription of all three classes of ROPK is similar, independently of the enzymatic status of the kinase (active, inactive, and non-canonical). The level of expression, expressed as log of fold change compared to the gamete stage expression, is the highest in extracellular sporozoites and MI stages (Figure 2B). Although the number of transcribed ropk is not significantly different between SO and eSPZ, the intensity of transcription is 10-fold higher in eSPZ. Those results were confirmed with the EtWIS strain (Figure 2B and Supplementary data, Figure S3) except for the MII stage in which ropk transcription levels tended to be lower in the EtINRAE strain in comparison with EtWIS. As the quality of mRNA samples was checked, the lower transcription of ropk in the EtINRAE MII stage may be explained by biological diversity.
Figure 2.
(A) The number of rhoptry kinases ROPK expressed in each developmental stage for EtINRAE, according to the putative classes of kinase activity (non-canonical in green, inactive in red, and active in black). Gene expression was standardized with the expression level in gametes: genes with a fold change > 2 were considered as transcribed. Two-way ANOVA, different letters (a, b, c) refer to statistical differences between the groups (p < 0.05). “a” means that the number of ROPK expressed is statistically different between NSO and all other stages, “b” means that the number of ROPK expressed is not statistically different between SO and eSPZ stages, but it is statistically different in comparison with all other stages, “c” means that the number of ROPK expressed is not statistically different between iSPZ, ImmSCHZ, MatSCHZ, MI, and MII stages, but it is statistically different in comparison with NSO, SO, and eSPZ. (B) The expression level of ropk was quantified in each developmental stage for EtINRAE (black) and EtWIS (grey) strains, expressed as log fold change (FC) in comparison with the gamete stages. Data are expressed as the mean ± SD of two independent experiments. Two-way ANOVA showed no significant difference between strains and between developmental stages.
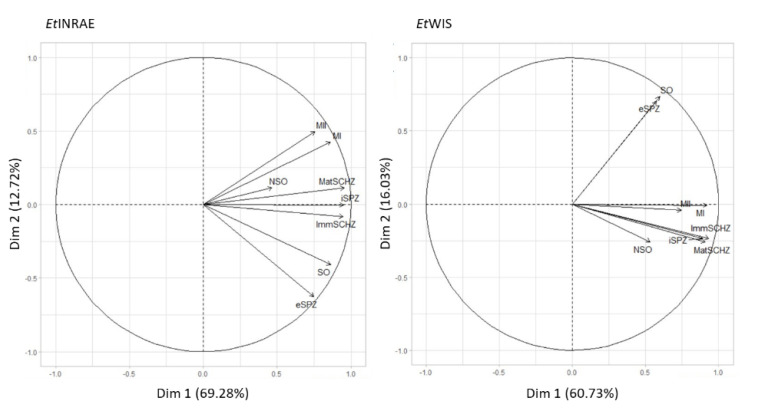
A strong and positive correlation was observed between the transcriptional profile of three groups of developmental stages in EtINRAE (Figure 3): (i) the three intracellular stages (iSPZ, ImmSCHZ, and MatSCHZ) were closely related to each other; whereas, (ii) SO showed an expression pattern similar to eSPZ, and (iii) MI and MII profiles were highly correlated. These correlations are consistent with the nature and the similarity of the developmental stages. In the EtWIS strain, similar correlations were confirmed, except for the merozoite profiles that were more closely related to the intracellular stage profiles than the ones observed in the EtINRAE strain (Figure 3).
Figure 3.
Principal component analysis (PCA) of rhoptry kinase transcription at different stages of development of the Eimeria tenella life-cycle. NSO non-sporulated oocysts, SO sporulated oocysts, eSPZ extracellular sporozoites, iSPZ intracellular sporozoites, ImmSCHZ immature schizont, MatSCHZ mature schizont, MI first-generation merozoites, and MII second-generation merozoites.
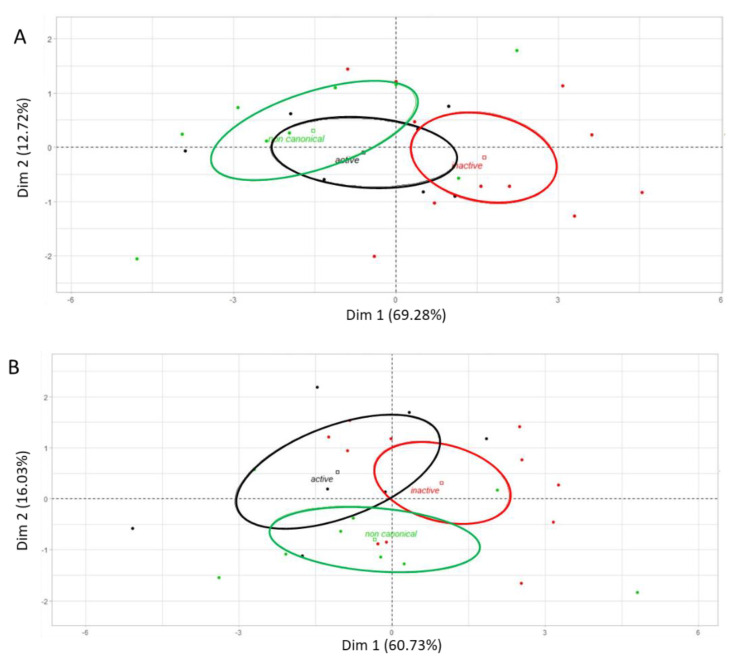
The PCA supports a wide distribution of ROPKs, with a partial overlap of the active kinases with either non-canonical or inactive kinases (Figure 4A). Although active and non-canonical kinase groups were partly differentially represented in the EtWIS strain (Figure 4B), no significant differences were observed between global kinase transcription for both strains (Figure S4).
Figure 4.
Principal component analysis of rhoptry kinase transcription according to their activity status in EtINRAE (A) and EtWIS (B) strains. Active kinases are represented in black, inactive in red, and non-canonical in green.
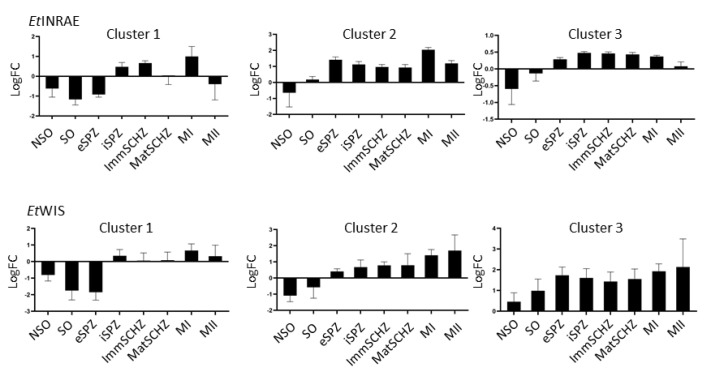
The hierarchical clustering analysis, based on the PCA, showed three clusters according to the stages in which they are transcribed across the life-cycle of E. tenella but no sub-clustering based on ROPKs activity (Figure 5 and Table S2). This is illustrated with the paragon of each cluster (i.e., the nearest ROPK of each cluster barycentre). In the EtINRAE strain, (i) cluster 1 contained ropks that were transcribed neither in oocysts (non-sporulated and sporulated) nor in eSPZ; (ii) cluster 2 presented ropks that were not transcribed in non-sporulated oocysts only, which included the large majority of ROPKs; and finally, (iii) cluster 3 was composed of ropks that were not transcribed in oocysts only (non-sporulated and sporulated). In the EtWIS strain, 3 clusters were also found according to the stages in which ropks were transcribed. Cluster 1 was similarly defined between strains, but clusters 2 and 3 slightly differed between the strains.
Figure 5.
Expression profile clustering of the rhoptry kinases in different stages of development of Eimeria tenella. Histograms show log fold change (Log FC) in the stage of development vs. gamete stage; Etactin and Et18s served as housekeeping genes.
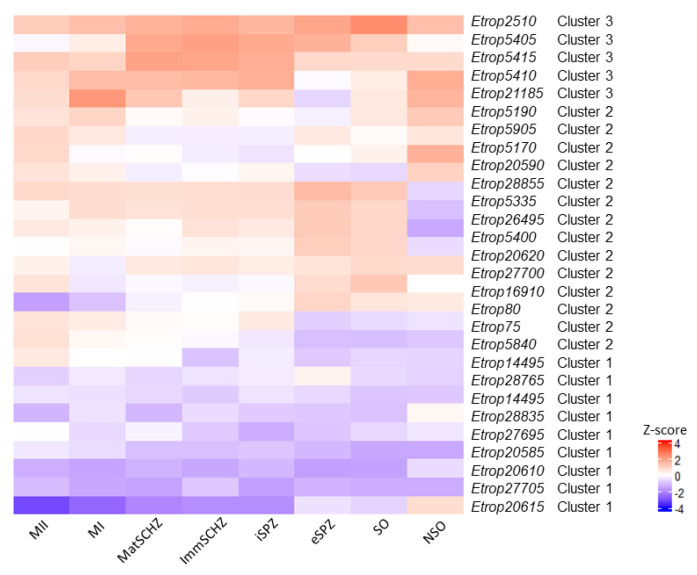
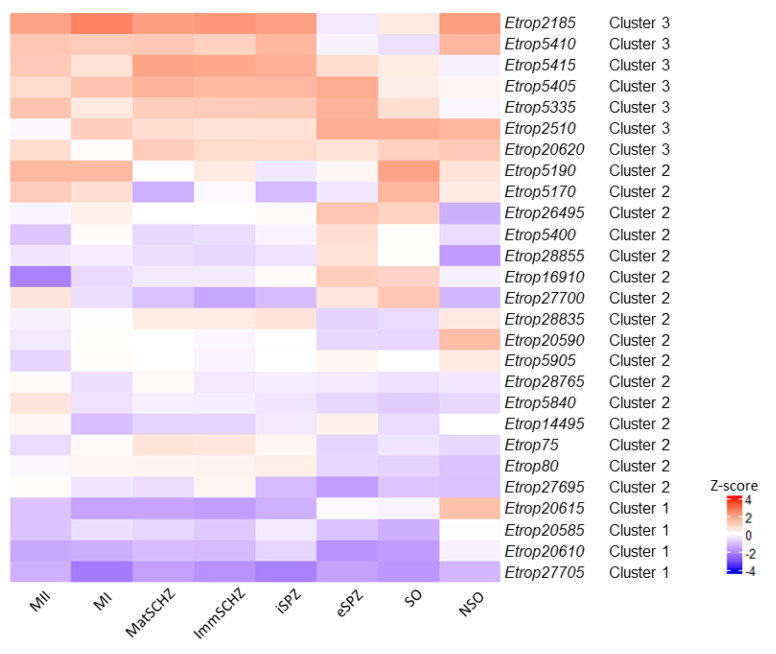
A heatmap representation of the transcription profiles for ropk genes for the EtINRAE and EtWIS strains are shown in Figure 6 and Figure 7, respectively. The clustering of ropks is consistent with the profiles of expression, with a lower transcription of cluster 1 ropks in early stages, a higher transcription of ropks from cluster 3 in intracellular stages, and cluster 2 appears less homogeneous. The heatmap also illustrates the diversity of ropks transcription inside each cluster: a slight difference may be observed between ropks from a cluster and the ropk paragon. The z-score for a given ropk corresponds to this ropk transcription in comparison to all other ropk transcription in this particular stage. So, a given ropk can be described as “overexpressed” (in comparison to the GAM stage, in Figure 5) but it may be “less overexpressed” than the mean of all ropk, and consequently, be illustrated in blue color in the heatmap (Figure 6 and Figure 7). The increase in ropk transcription during sporulation is clearly shown, and a higher transcription of ropk in intracellular stages is obvious.
Figure 6.
Heatmap of differentially transcribed rhoptry kinase genes in the EtINRAE strain. Red color indicates a rhoptry kinase transcription level higher than the mean, and blue color represents a transcription level lower than the mean. NSO non-sporulated oocysts, SO sporulated oocysts, eSPZ extracellular sporozoites, iSPZ intracellular sporozoites, ImmSCHZ immature schizonts, MatSCHZ mature schizonts, MI first-generation merozoites, and MII second-generation merozoites. The cluster resulting from the hierarchical clustering analysis are indicated.
Figure 7.
Heatmap of differentially transcribed rhoptry kinase genes in the EtWIS strain. Red color indicates a rhoptry kinase transcription level higher than the mean and blue colour represents a transcription level lower than the mean. NSO non-sporulated oocysts, SO sporulated oocysts, eSPZ extracellular sporozoites, iSPZ intracellular sporozoites, ImmSCHZ immature schizonts, MatSCHZ mature schizonts, MI first-generation merozoites, and MII second-generation merozoites. The cluster resulting from the hierarchical clustering analysis are indicated.
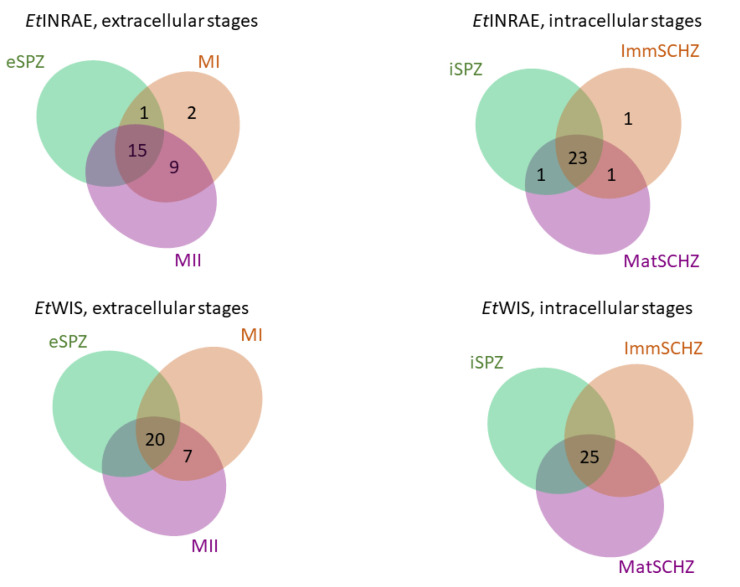
Ropks that were differentially transcribed between parasitic stages (sporozoites, schizonts, and merozoites) were combined in a Venn diagram to identify ropks related either to invasive stages or to intracellular stages (Figure 8 and Table S2). Ropks profiles of intracellular stages were highly similar between strains: the same 23 for EtINRAE and 25 out of 26 ropks for EtWIS (Etrop20615 being not transcribed in those stages) were expressed in iSPZ, ImmSCHZ, and MatSCHZ. More differences were observed between extracellular stages: eSPZ, MI, and MII share the transcription of 15 and 20 out of 27 ropks, in EtINRAE and EtWIS, respectively. The 9 ropk transcribed in both MI and MII, but not in eSPZ, in the EtINRAE strain are partly the same as the 7 ropk in the EtWIS strain: Etrop75, Etrop80, Etrop20585, Etrop20590, and Etrop28835 are common to both strains; Etrop5840, Etrop21185, Etrop27695, and Etrop28765 are shared by EtINRAE MI and MII stages, and Etrop20610 and Etrop27705 are shared by EtWIS MI and MII stages. No ropks were found specific to the MII stage: ropks expressed in MII are either also expressed in eSPZ or in MI. The future characterization of those ROPK would be very interesting to decipher, including how different developmental extracellular stages invade and hijack caecal epithelial cells.
Figure 8.
Venn diagram of differentially transcribed rhoptry kinases in three extracellular stages and three intracellular stages of the EtINRAE and EtWIS strains. eSPZ extracellular sporozoites, iSPZ intracellular sporozoites, ImmSCHZ immature schizonts, MatSCHZ mature schizonts, MI first-ge-neration merozoites, and MII second-generation merozoites. In extracellular stages, 27 ropks were transcribed, in intracellular stages, 26 and 25 ropks were transcribed in EtINRAE and EtWIS, respectively.
4. Discussion
Protein kinases are key regulators of host–pathogen interactions. In particular, the ROP kinase family, mostly described in T. gondii, is required for survival and development throughout the parasite life-cycle. We carried out a comprehensive transcriptional analysis of the ropk gene family, in E. tenella (free-living stages, extracellular and intracellular stages). As observed in T. gondii, where the ropk genes share a similar expression profile [23,24], our data reveal a dynamic and regulated transcription of ropk genes, along the E. tenella life-cycle. We confirmed a global increase in ropk transcription during the sporulation, as described for Plasmodium berghei [25]. We also noticed a stronger ropk gene expression in the intracellular stages of the parasite development (iSPZ, ImmSCHZ, and MatSCHZ), which might be associated with known functions of ROPK on cell invasion and intracellular development of T. gondii [26,27]. The co-transcription of active ROPK with either inactive or non-canonical kinases is consistent with the complex co-expression of ROPK observed in T. gondii. The inactive kinase TgROP5 is a co-factor of active ROPK, regulating TgROP17 and TgROP18 activity [28,29,30,31]. In E. tenella, the inhibition of host cell apoptosis by EtROP1 is independent of EtROP1 kinase activity [16], suggesting the involvement of at least another active kinase.
A very strong correlation was observed between ropk gene transcription from iSPZ, ImmSCHZ, and MatSCHZ. As sporozoites do not engage their development in a synchronous manner, we cannot rule out the possible presence of few intracellular sporozoites in immature schizonts samples. A similar situation may also occur for immature schizonts in mature schizonts samples. To the best of our knowledge, no specific gene has been identified to confirm the absence of developmental stages contamination for these three successive stages. However, Etrop20610, which is transcribed in intracellular sporozoites of the EtINRAE strain, was not detected in immature schizonts. Moreover, the similarity of transcriptional profiles of successive developmental stages was also observed between sporulated oocysts and extracellular sporozoites, through genome-wide transcriptomic analysis (not only the ropk gene family) [32]. For MI and MII, the similarity of ropk expression profiles (also confirmed by [32]) cannot be explained by any contamination, as MI were recovered from the supernatant of cultured cells (in vitro) whereas MII were purified from infected caecal cells (in vivo). Taken together, these results support the fact that even if minor contamination exists between intracellular stages, the ropk gene transcription patterns observed across the developmental stages of E. tenella are correct and reliable.
TgROPK are broadly known to be involved in host cell invasion and hijacking of host signaling pathways and immune responses in T. gondii. Venn diagram allows identification of candidate genes involved into extracellular and intracellular stages in E. tenella. The 9 ropk transcribed in MI and MII but not in eSPZ in the EtINRAE strain are partly the same as the 7 ropk in the EtWIS strain. These data are consistent with the proteomic data identifying merozoites specific ROPK expression [15]. The future functional characterization of those ROPK would be very useful to decipher how different developmental extracellular stages invade and hijack caecal epithelial cells. Concerning intracellular stages, ropk gene transcription was highly similar and no transcriptional signature could be associated with the extracellular or intracellular stages of E. tenella in both strains. To date, only EtROP5190 [16] has been functionally characterized and associated with cell apoptosis inhibition, which is consistent with Etrop5190 transcription in iSPZ (sporozoites early invading epithelial cell) and ImmSCHZ, and MatSCHZ.
Although the main part of the ropk family is transcriptionally regulated, some differences may be observed at the protein level. Indeed, post-transcriptional modifications and translational control are frequent in Apicomplexa (for a recent review [33]). Proteomic data on E. tenella are scarce. To date, no exhaustive proteomic analysis of the ROPK library has been published on the developmental stages analyzed in the present study to support our analysis. However, some proteomic data are available on sporozoites [15,34,35,36,37] and merozoites [36,38,39]. Previous publications demonstrated that the higher transcription of Etrop75, Etrop5400, Etrop5840, Etrop5905, Etrop26495, Etrop27695, and Etrop28765 observed in merozoites in the present study is confirmed at the protein level [36,38], and the higher transcription of Etrop27700 in sporozoites is consistent with proteomic data [15].
Studying two different wild-type E. tenella strains, the results showed a remarkable congruence. The main difference between strains was the greater similarity of MI and MII transcriptional patterns with intracellular stages patterns in the EtWIS strain compared to the EtINRAE strain. Although non-significant, the mean transcription level of ropk tends to be higher in EtWIS MII than in EtINRAE MII. Although both strains have an equivalent virulence status (data not shown), EtINRAE was propagated on PA12 White Leghorn chickens since its isolation from the field, whereas EtWIS was recently obtained from the Royal Veterinary College (London, UK), where it was propagated on Light Sussex, White Leghorn or Cobb500 chickens. This difference may account for a local adaptation of the EtINRAE strain. It is noteworthy that the difference in ropks expression between EtINRAE and EtWIS resides mainly in ropks expression in the extracellular stages. We may speculate that co-evolution acted more on the host-cell invasion step than on intracellular parasite development. To determine whether this strain specificity of ropk transcription may impact the virulence or fitness of E. tenella, supplementary experiments would be necessary. Some loci mapped to chromosome 2 have been associated with precociousness in E. tenella [40], and transcriptomic differences were found between virulent and precocious strains [41]. The comparison of ropk transcription between WT and attenuated (or precocious) strains may be very useful to identify new stage-specific ROPKs, possibly involved in virulence.
5. Conclusions
A comprehensive expression analysis of the ropk gene family in Eimeria tenella was conducted for the first time, all along the life-cycle (free-living, extracellular, and intracellular stages). If ROPK protein expression confirms the transcriptional data, a large number of ROPK could be co-expressed, suggesting some complex interactions between active and inactive, or non-canonical ROPK, as observed in T. gondii. Our results suggest that ropks are a large family of multi-functional genes which are expected to play essential roles in parasite development and hijacking of host cell signaling pathways. This exhaustive inventory of poorly characterized and developmentally regulated ropk genes provides a solid basis for future functional studies of the ROPKs in E. tenella, (as published for Etrop5190 [16]) and paves the way for the development of new strategies/vaccines to control avian coccidiosis.
Acknowledgments
We are grateful to Virginia Marugan-Hernandez, Damer Blake, Fiona Tomley (Royal Veterinary College, London, United Kingdom), for kindly providing us with the Wisconsin E. tenella strain (EtWIS). We thank the Infectiology of Farm, Model and Wildlife Animals Facility (PFIE), for the animal care and help with the animal experiments (Thierry Chaumeil, Fanny Faurie-Sarcé, Olivier Dubes and Mylène Girault). We thank Estelle Godet, within the BOA UMR (INRAE), for technical advice and support with RIN measures, and Tiffany Pezier, for assistance in bioinformatics analysis with R. We are grateful to Sebastien Santini (CNRS/AMU IGS UMR7256) and the PACA Bioinfo platform (supported by IBISA) for the availability and management of the phylo-geny.fr website.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9081621/s1. Table S1: PCR primers ROPK and stage-specific primers. Table S2: Mean ropks transcription in developmental stages. Fold change ropks transcription levels in EtINRAE and EtWIS strains, in comparison to gamete stage. Differential ropks expressed in extracellular and intracellular stages, in EtINRAE and EtWIS strains. Figure S1: Phylogeny of rhoptry kinases (ROPK) from Eimeria tenella. ROPK amino acid sequences from E. tenella (Houghton strain) were recovered from VEuPathDB (release 52, 20 May 2021). Phylogeny software (Muscle program for alignment and maximum-likelihood method) was used [42]. The number at each node represents the percentage bootstrap support, calculated from 1,000 replicates. Figure S2: Stage-specific genes transcription during E. tenella development (EtINRAE (A) and EtWIS (B) strains). Et18s (EF210326) and Etactin (ETH_00009555) were used as housekeeping genes. Stage-specific genes were Etama1 (ETH_00007745) for sporozoites, Etsag3 (ETH_00010755) for schizonts and merozoites and Etgam56 (ETH_00007320) for macrogametes [20]. Data are expressed as the mean ± SD of two independent experiments. Figure S3: Number of rhoptry kinases expressed in each stage of E. tenella development (EtWIS), according to the kinase activity (non-canonical, inactive, and active). Gene expression was standardized with the expression level in gametes: genes with a fold change > 2 were considered as transcribed. Two-way ANOVA, different letters refer to statistical differences (p < 0.05). Figure S4: Principal component analysis of rhoptry kinases expression according to the Eimeria tenella strain. ropk in EtINRAE are represented in black and in EtWIS in red.
Author Contributions
Conceptualization, A.S. (Anne Silvestre) F.I.B., F.L. and S.L.-L.; methodology, A.S. (Anne Silvestre) A.R.E.S. and A.S. (Alix Sausset); formal analysis, A.S. (Anne Silvestre), A.R.E.S. and A.S. (Alix Sausset); investigation, A.R.E.S. and A.S. (Alix Sausset); writing, A.R.E.S. and A.S. (Alix Sausset), F.I.B., F.L., S.L.-L. and A.S. (Anne Silvestre); funding acquisition, A.S. (Anne Silvestre). All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the INRAE, Animal Health division (project 00002230). A.R.E.S. is the grateful recipient of a Ph.D. grant from Région Centre-Val de Loire (32000951).
Institutional Review Board Statement
All animal experimentations have been performed in the Infectiology of Farm, Model, and Wildlife Animals Facility (PFIE, Centre INRAE Val De Loire: https://doi.org/10.15454/1.5572352821559333E12, accessed on 25 July 2021); member of the National Infrastructure EMERG’IN). Experimental protocols were designed in compliance with French law (2010/63/EU, 2010; Rural Code, 2018; Decree No. 2013-118, 2013) concerning the use of laboratory animals. Care and euthanasia of animals were practiced according to the national ethical guidelines and approved by the local ethics committee for animal experimentation (Comité d’Ethique en Expérimentation Animale Val de Loire, CEA VdL N°19): APAFIS#25884.
Informed Consent Statement
Not applicable.
Data Availability Statement
All raw data are available as Supplementary Materials Table S2.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Noack S., Chapman H.D., Selzer P.M. Anticoccidial drugs of the livestock industry. Parasitol. Res. 2019;118:2009–2026. doi: 10.1007/s00436-019-06343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soutter F., Werling D., Tomley F.M., Blake D.P. Poultry Coccidiosis: Design and Interpretation of Vaccine Studies. Front. Vet. Sci. 2020;7:7. doi: 10.3389/fvets.2020.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madlala T., Okpeku M., Adeleke M.A. Understanding the interactions between Eimeria infection and gut microbiota, towards the control of chicken coccidiosis: A review. Parasite. 2021;28:48. doi: 10.1051/parasite/2021047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doerig C., Billker O., Haystead T., Sharma P., Tobin A.B., Waters N.C. Protein kinases of malaria parasites: An update. Trends Parasitol. 2008;24:570–577. doi: 10.1016/j.pt.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Hanks S.K. Genomic analysis of the eukaryotic protein kinase superfamily: A perspective. Genome Biol. 2003;4:111. doi: 10.1186/gb-2003-4-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talevich E., Kannan N. Structural and evolutionary adaptation of rhoptry kinases and pseudokinases, a family of coccidian virulence factors. BMC Evol. Biol. 2013;13:117. doi: 10.1186/1471-2148-13-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peixoto L., Chen F., Harb O., Davis P.H., Beiting D.P., Brownback C.S., Ouloguem D., Roos D.S. Integrative Genomic Approaches Highlight a Family of Parasite-Specific Kinases that Regulate Host Responses. Cell Host Microbe. 2010;8:208–218. doi: 10.1016/j.chom.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox B.A., Rommereim L.M., Guevara R.B., Falla A., Triana M.A.H., Sun Y., Bzik D.J. The Toxoplasma gondii Rhoptry Kinome Is Essential for Chronic Infection. mBio. 2016;7:e00193-16. doi: 10.1128/mBio.00193-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ihara F., Nishikawa Y. Toxoplasma gondii manipulates host cell signaling pathways via its secreted effector molecules. Parasitol. Int. 2021;83:102368. doi: 10.1016/j.parint.2021.102368. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz J.A., Fouts A.E., Cummings C.A., Ferguson D., Boothroyd J.C. A novel rhoptry protein in Toxoplasma gondii bradyzoites and merozoites. Mol. Biochem. Parasitol. 2005;144:159–166. doi: 10.1016/j.molbiopara.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Behnke M.S., Zhang T.P., Dubey J.P., Sibley L.D. Toxoplasma gondii merozoite gene expression analysis with comparison to the life cycle discloses a unique expression state during enteric development. BMC Genom. 2014;15:350. doi: 10.1186/1471-2164-15-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramakrishnan C., Smith N.C. Recent achievements and doors opened for coccidian parasite research and development through transcriptomics of enteric sexual stages. Mol. Biochem. Parasitol. 2021;243:111373. doi: 10.1016/j.molbiopara.2021.111373. [DOI] [PubMed] [Google Scholar]
- 13.Talevich E., Mirza A., Kannan N. Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa. BMC Evol. Biol. 2011;11:321. doi: 10.1186/1471-2148-11-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid A., Blake D., Ansari H.R., Billington K., Browne H., Bryant J., Dunn M., Hung S.S., Kawahara F., Miranda-Saavedra D., et al. Genomic analysis of the causative agents of coccidiosis in domestic chickens. Genome Res. 2014;24:1676–1685. doi: 10.1101/gr.168955.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oakes R.D., Kurian D., Bromley E., Ward C., Lal K., Blake D., Reid A., Pain A., Sinden R.E., Wastling J.M., et al. The rhoptry proteome of Eimeria tenella sporozoites. Int. J. Parasitol. 2013;43:181–188. doi: 10.1016/j.ijpara.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 16.Diallo M.A., Sausset A., Gnahoui-David A., Silva A.R.E., Brionne A., Le Vern Y., Bussière F.I., Tottey J., Lacroix-Lamandé S., Laurent F., et al. Eimeria tenella ROP kinase EtROP1 induces G0/G1 cell cycle arrest and inhibits host cell apoptosis. Cell. Microbiol. 2019;21:e13027. doi: 10.1111/cmi.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doran D.J., Vetterling J.M., Augustine P.C. Eimeria-tenella—in-vivo and in-vitro comparison of wisconsin, weybridge, and beltsville strains. Proc. Helminthol. Soc. Wash. 1974;41:77–80. [Google Scholar]
- 18.Shirley M.W. Eimeria species and strains of chicken. Biotechnology-Guidelines Techniques Coccidiosis Research. Eur. Commission DGXII; Luxembourg: 1995. pp. 1–24. [Google Scholar]
- 19.Walker R.A., Sharman P.A., Miller C.M., Lippuner C., Okoniewski M., Eichenberger R.M., Ramakrishnan C., Brossier F., Deplazes P., Hehl A.B., et al. RNA Seq analysis of the Eimeria tenella gametocyte transcriptome reveals clues about the molecular basis for sexual reproduction and oocyst biogenesis. BMC Genom. 2015;16:94. doi: 10.1186/s12864-015-1298-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker R., Slapetova I., Slapeta J., Miller C.M., Smith N.C. The Glycosylation Pathway of Eimeria tenella Is Upregulated during Gametocyte Development and May Play a Role in Oocyst Wall Formation. Eukaryot. Cell. 2010;9:127–135. doi: 10.1128/EC.00255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R Core Team . R: A Language and Environment for Statistical Computing. R foundation for Statistical Computing; Vienna, Austria: 2020. [(accessed on 31 March 2021)]. Available online: https://www.R-project.org/ [Google Scholar]
- 22.Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., Wishart D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camejo A., Gold D.A., Lu D., McFetridge K., Julien L., Yang N., Jensen K.D.C., Saeij J.P. Identification of three novel Toxoplasma gondii rhoptry proteins. Int. J. Parasitol. 2014;44:147–160. doi: 10.1016/j.ijpara.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim E.W., Nadipuram S.M., Tetlow A.L., Barshop W.D., Liu P.T., Wohlschlegel J.A., Bradley P.J. The Rhoptry Pseudokinase ROP54 Modulates Toxoplasma gondii Virulence and Host GBP2 Loading. mSphere. 2016;1:e00045-16. doi: 10.1128/mSphere.00045-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tokunaga N., Nozaki M., Tachibana M., Baba M., Matsuoka K., Tsuboi T., Torii M., Ishino T. Expression and Localization Profiles of Rhoptry Proteins in Plasmodium berghei Sporozoites. Front. Cell. Infect. Microbiol. 2019;9:316. doi: 10.3389/fcimb.2019.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben Chaabene R., Lentini G., Soldati-Favre D. Biogenesis and discharge of the rhoptries: Key organelles for entry and hijack of host cells by the Apicomplexa. Mol. Microbiol. 2021;115:453–465. doi: 10.1111/mmi.14674. [DOI] [PubMed] [Google Scholar]
- 27.Hakimi M.-A., Olias P., Sibley L.D. Toxoplasma Effectors Targeting Host Signaling and Transcription. Clin. Microbiol. Rev. 2017;30:615–645. doi: 10.1128/CMR.00005-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behnke M., Fentress S.J., Mashayekhi M., Li L.X., Taylor G.A., Sibley L.D. The Polymorphic Pseudokinase ROP5 Controls Virulence in Toxoplasma gondii by Regulating the Active Kinase ROP. PLoS Pathog. 2012;8:e1002992. doi: 10.1371/journal.ppat.1002992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El Hajj H., Lebrun M., Fourmaux M.N., Vial H., Dubremetz J.F. Inverted topology of the Toxoplasma gondii ROP5 rhoptry protein provides new insights into the association of the ROP2 protein family with the parasitophorous vacuole membrane. Cell. Microbiol. 2007;9:54–64. doi: 10.1111/j.1462-5822.2006.00767.x. [DOI] [PubMed] [Google Scholar]
- 30.Etheridge R.D., Alaganan A., Tang K., Lou H.J., Turk B.E., Sibley L.D. The Toxoplasma Pseudokinase ROP5 Forms Complexes with ROP18 and ROP17 Kinases that Synergize to Control Acute Virulence in Mice. Cell Host Microbe. 2014;15:537–550. doi: 10.1016/j.chom.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niedelman W., Gold D.A., Rosowski E., Sprokholt J.K., Lim D., Arenas A., Melo M., Spooner E., Yaffe M.B., Saeij J.P.J. The Rhoptry Proteins ROP18 and ROP5 Mediate Toxoplasma gondii Evasion of the Murine, But Not the Human, Interferon-Gamma Response. PLoS Pathog. 2012;8:e1002784. doi: 10.1371/journal.ppat.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novaes J., Rangel L.T.L.D., Ferro M., Abe R.Y., Manha A.P.D.S., De Mello J.C.M., Varuzza L., Durham A.M., Madeira A.M.B.N., Gruber A. A comparative transcriptome analysis reveals expression profiles conserved across three Eimeria spp. of domestic fowl and associated with multiple developmental stages. Int. J. Parasitol. 2012;42:39–48. doi: 10.1016/j.ijpara.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 33.Sharma J., Rodriguez P., Roy P., Guiton P.S. Transcriptional ups and downs: Patterns of gene expression in the life cycle of Toxoplasma gondii. Microbes Infect. 2020;22:525–533. doi: 10.1016/j.micinf.2020.09.001. [DOI] [PubMed] [Google Scholar]
- 34.de Venevelles P., Chich J.F., Faigle W., Loew D., Labbé M., Girard-Misguich F., Péry P. Towards a reference map of Eimeria tenella sporozoite proteins by two-dimensional electrophoresis and mass spectrometry. Int. J. Parasitol. 2004;34:1321–1331. doi: 10.1016/j.ijpara.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 35.de Venevelles P., Chich J.F., Faigle W., Lombard B., Loew D., Péry P., Labbé M. Study of proteins associated with the Eimeria tenella refractile body by a proteomic approach. Int. J. Parasitol. 2006;36:1399–1407. doi: 10.1016/j.ijpara.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 36.Lal K., Bromley E., Oakes R., Prieto J.H., Sanderson S.J., Kurian D., Hunt L., Yates J.R., Wastling J.M., Sinden R.E., et al. Proteomic comparison of four Eimeria tenella life-cycle stages: Unsporulated oocyst, sporulated oocyst, sporozoite and second-generation merozoite. Proteomics. 2009;9:4566–4576. doi: 10.1002/pmic.200900305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thabet A., Honscha W., Daugschies A., Bangoura B. Quantitative proteomic studies in resistance mechanisms of Eimeria tenella against polyether ionophores. Parasitol. Res. 2017;116:1553–1559. doi: 10.1007/s00436-017-5432-z. [DOI] [PubMed] [Google Scholar]
- 38.Li X.-Y., Liu L.-L., Zhang M., Zhang L.-F., Wang X.-Y., Wang M., Zhang K.-Y., Liu Y.-C., Wang C.-M., Xue F.-Q., et al. Proteomic analysis of the second-generation merozoites of Eimeria tenella under nitromezuril and ethanamizuril stress. Parasites Vectors. 2019;12:592. doi: 10.1186/s13071-019-3841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen X.-J., Li T., Fu J.-J., Zhang K.-Y., Wang X.-Y., Liu Y.-C., Zhang H.-J., Fan C., Fei C.-Z., Xue F.-Q. Proteomic analysis of the effect of diclazuril on second-generation merozoites of Eimeria tenella. Parasitol. Res. 2013;113:903–909. doi: 10.1007/s00436-013-3721-8. [DOI] [PubMed] [Google Scholar]
- 40.Shirley M.W., Harvey D.A. A Genetic Linkage Map of the Apicomplexan Protozoan Parasite Eimeria tenella. Genome Res. 2000;10:1587–1593. doi: 10.1101/gr.149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsubayashi M., Kawahara F., Hatta T., Yamagishi J., Miyoshi T., Anisuzzaman, Sasai K., Isobe T., Kita K., Tsuji N. Transcriptional profiles of virulent and precocious strains of Eimeria tenella at sporozoite stage; novel biological insight into attenuated asexual development. Infect. Genet. Evol. 2016;40:54–62. doi: 10.1016/j.meegid.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 42.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data are available as Supplementary Materials Table S2.