Abstract
Fungi associated with macroalgae are less known when compared with those on wood in the marine environment. In this study, we assessed the diversity of fungi associated with the red alga Pterocladiella capillacea at Chao-Jin Park, Keelung, Taiwan. Algal segments of healthy and dead thalli were washed/sterilized with different solutions (sterile artificial seawater, 70% ethanol, and 4% sodium hypochlorite), plated on three different media (glucose-yeast extract-peptone seawater agar (GYPS), potato dextrose seawater agar (PDAS), and artificial seawater agar (SA)), and isolated as pure cultures. Identification was mainly based on BLAST search analysis of the internal transcribed spacers of rDNA (ITS). The highest isolation frequency (no. of segment with fungi/total no. of segment × 100) was in dead thalli (61.23%), thalli washed with seawater (88.38%), and thalli plated on GYPS (62.10%). A total of 3187 isolates were cultured, representing 129 taxa (in 67 genera); the higher species richness was isolated from healthy thalli (119 species), thalli washed with seawater (111 species), and on GYPS (112 species). Ascomycota (Eurotiales, Hypocreales, Capnodiales, Pleosporales, Xylariales) dominated the fungal community in P. capillacea with many basidiomycetous yeasts and few Mucoromycota. Aspergillus, Cladosporium, Penicillium (Ascomycota), and Rhodosporidium (Basidiomycota) were the dominant genera associated with the alga. The surface washing/sterilization schemes of algal thalli affected fungal diversity, but the isolation media used did not. While these genera are known producers of antimicrobial secondary metabolites, they might form a mutualistic relationship with P. capillacea by exchanging nutrients from photosynthesis for protection from microbial diseases.
Keywords: culture dependent, fungal community, marine fungi, rhodophyta, symbiosis
1. Introduction
Early studies of marine fungi mainly focused on those associated with macroalgae [1,2]. Macroalgae are particularly abundant in coastal environments and a good substrate for growth of microorganisms including marine fungi [3]. Fungi on macroalgae are mainly saprobic, although many species are probably symbiotic, including pathogenic/parasitic associations [4,5]. Saprobic marine fungi of macroalgae form fruiting bodies on the substrate and produce a unique morphology, such as species of the genera Spathulospora, Chadefaudia, Haloguignardia, Retrostium, Histopidicarpomyces, and Pontogenia [4]. Concerning pathogenic species of macroalgae, Mycaureola dilseae is a marine basidiomycete parasitizing the marine alga Dilsea carnosa, forming circular necrotic lesions [6]. Lautitia danica occurs on the red alga Chondrus crispus throughout the whole year, forming ascomata on the fronds [7]. Many filamentous fungi can also asymptomatically colonize the algal inner tissues without causing any apparent damage or disease [8,9]. Protrusions of the cell wall of the ascomycete Verrucaria tavaresiae penetrate walls of the brown alga Petroderma maculiforme, possibly forming a symbiotic relationship [10]. Mycophycias ascophylli and Sigmoidea marina were found to be mutually associated with their algal host and grow within or at the surface of the algal tissue [11,12].
In recent years, fungi have been isolated from surface-sterilized and seawater-washed thalli (pieces) of macroalgae for their production of interesting antimicrobial and/or cytotoxic metabolites [13,14]. Loque et al. [15] and Godinho et al. [16] studied the fungi associated with macroalgae at Antarctic regions, and the common fungi were Geomyces, Penicillium, and Metschnikowia. Meyerozyma guilliermondii was found to be a common yeast [17]. On the red algal genus Kappaphycus (K. alvarezii, K. striatum) in the Philippines, the common fungi were Fusarium sp., Curvularia intermedia, Cladosporium sp., and Phoma sp. [18]. Abdel-Gawad et al. [19] isolated fungi from 20 macroalgae consisting of 8 red algae, 8 brown algae, and 4 green algae collected along the coast of the Red Sea in Egypt, the dominant genera were Penicillium, Alternaria, Cladosporium, Aspergillus, and Acremonium. The above studies used seawater to wash the algae prior to inoculation and resulted numerous typical terrestrial genera, many of which can grow better in the presence of sea salts [18]. However, Zuccaro et al. [20], on the study of fungi associated with Fucus spp., suggested that Emericellopsis spp. and Acremonium spp. on Fucus were distinct from their terrestrial counterparts physiologically and phylogenetically.
Many macroalgae have been examined for their endophytic association with fungi, where the algal surface was sterilized prior to isolation. In the Shetland Islands, UK and Bay of Fundy, Canada, Aspergillus and Penicillium species were common on macroalgae [21,22]. In the same studies, Mycosphaerella ascophylli was not isolated from the brown alga Ascophyllum nodosum, although this is a common marine ascomycete reported to associate with this alga endophytically [7]. Aspergillus spp. were also found to be dominant endophytically in 24 species of red, green, and brown algae collected at Tamilnadu, India, together with an unidentified yeast species [23]. Asexual Ascomycota was abundant on macroalgae, either occurring epiphytically or endophytically [19,23].
Kohlmeyer and Volkmann-Kohlmeyer [24] listed 75 species of marine fungi associated with algae either parasitically or symbiotically. This estimate is restricted to those fungi forming visible fruiting bodies on the algae. Jones et al. [5] provided a more updated figure of 112 taxa, including those parasitic or saprophytic on algae and those forming lichenized relationships with algal hosts. The Ascomycota is the dominant phylum of fungi cultured from microalgae [12,16,21,22,25], such as Acremonium, Alternaria, Arthrinium, Aspergillus, Cladosporium, Emericellopsis, Fusarium, Penicillium, Phoma, Pontogenia, Retrosium, Spathuospora, Sigmoidea, and Trichoderma [7,12,20,24,26,27,28].
Roughly, there are 7000 described species of the Rhodophyta, and only a few have been examined for their associated fungi [29]: Ballia, Ceramium, Chondrus, Dilsea, Gelidiella, Gracilaria, Grateloupia, Halymenia, Hypnea, Porphyra, and Portieria [6,7,23,27]. Pterocladiella capillacea is a red alga growing throughout the year along the rocky shore of northern Taiwan. In this study, we examined the culturable fungal community associated with dead and healthy thalli of P. capillacea washed with sterile seawater, 70% ethanol, or 4% sodium hypochlorite (NaOCl) before inoculation and plated on glucose-yeast extract-peptone seawater agar (GYPS), potato dextrose seawater agar (PDAS), and artificial seawater agar (SA) collected bimonthly over a 1-year period. Sterile seawater, ethanol, and sodium hypochlorite were the most common chemical agents, either individually or in combination, to treat samples of macroalgae before they were inoculated onto isolation media, but concentrations of the chemical agents and duration of treatment differed between studies [15,21,22,23,30]. Sodium hypochlorite was found to be an effective surface sterilizer of samples and has been used in studies targeting endophytic fungal assemblages [31]. However, a combination of 70% ethanol for 5 s and 4% sodium hypochlorite for 60 s and a long surface sterilization time of 60 s caused visible damage of algal tissue [23]. In this study, the use of seawater, 70% ethanol (30 s), and 4% sodium hypochlorite (10 s) to wash/surface sterilize algal samples enabled detection of both epiphytic and endophytic fungi of P. capillacea. Potato dextrose agar, malt extract agar, and glucose-yeast extract-peptone agar have been some of the most commonly used nutritious media for isolation of fungi from macroalgae [15,18,19,21,22,32,33,34,35], although other media have also been used, such as marine agar [16] and cornmeal agar [36]. To broaden the isolation of taxonomically diverse fungi, three media, i.e., potato dextrose agar, glucose-yeast extract-peptone agar, and seawater agar, were used in this study. Fungi were isolated as axenic cultures and identified based on a BLASTn analysis of the internal transcribed spacers of rDNA (ITS), as well as 18S and/or 28S rDNA when the identity provided by ITS alone was ambiguous.
2. Materials and Methods
2.1. Collection of Samples
Algal thalli of Pterocladiella capillacea were collected bimonthly between May 2013 to May 2014 at Chao-Jin Park, located at the east side of the Badouzi Peninsula facing Wanghaixiang Bay (25°08′31.9″ N, 121°48′08.7″ E), Keelung, northern Taiwan (Figure 1). Healthy (red color) and dead or decaying (white color) thalli were collected along with seawater, placed in sterilized polythene bags, and maintained at 4 °C during transportation to the laboratory for immediate isolation.
Figure 1.
Sampling site: (a) Sampling location (star) of Pterocladiella capillacea along Chao-Jin Park, Keelung, northern Taiwan. (b) Photograph of the sampling site.
2.2. Surface Washing/Sterilization of Pterocladiella Capillacea
Fungi isolation was conducted based on the conventional methodology (Figure S1). The collected healthy and dead thalli were cut into segments of approximately 0.5–1.0 cm2. The segments were subjected to three different surface washing/sterilization procedures: (1) washed in sterile artificial seawater for 30 s, (2), immersed in 70% ethanol for 30 s, and (3) immersed in 4% sodium hypochlorite for 10 s. All samples were then further rinsed two times with sterile artificial seawater for 30 s each.
2.3. Fungal Isolation
After washing/sterilization, randomly selected segments were plated onto three different media on Petri dishes using a flame sterilized forceps: seawater agar (SA; 30 g/L artificial sea salt, 15 g/L agar (Bioshop, Burlington, ON, Canada)), glucose-yeast extract-peptone seawater agar (GYPS; 4 g/L glucose (Bioshop, Burlington, ON, Canada), 4 g/L yeast extract (Oxoid, United Kingdom), 4 g/L peptone (Oxoid, United Kingdom), 30 g/L artificial sea salt, 15 g/L agar (Bioshop, Burlington, Ont, Canada)), and potato dextrose seawater agar (PDAS; 39 g/L potato dextrose agar (Becton, Dickinson and Company, Sparks, MD, USA), 30 g/L artificial sea salt) supplemented with 0.5 g/L each of streptomycin sulphate and Penicillin G sodium salt (Bioshop, Burlington, ON, Canada). The inoculated plates were incubated at 25 °C and observed periodically for up to 1 month under a stereomicroscope (Olympus, Tokyo). Fungi with different fungal mycelial morphologies were cut out and subcultured onto cornmeal seawater agar (CMAS; 17 g/L corn meal agar (Himedia, India), 30 g/L artificial sea salt) plates as pure cultures. All cultures are kept at the Institute of Marine Biology, National Taiwan Ocean University (NTOU).
2.4. Fungal Identification
The isolates on CMAS were categorized into morphotypes based on their colony morphology. Total genomic DNA of each morphotype was extracted using a DNeasy® Plant Mini Kit according to the manufacturer’s instructions (Cat. No. 69104, Qiagen, Germany). Using the genomic DNA as the template, the internal transcribed spacer (ITS) regions spanning from the end of the 18S rDNA to the beginning of the 28S rDNA for each morphotype were amplified using the primer pairs ITS5/ITS4 [37] or ITS1-F_KYO1/ITS4_KYO3 [38]. In some cases, the 18S and 28S rDNA were amplified using NS1/NS6 [37] and LR0R/LR6 [39], respectively. Thermal cycling conditions for PCR amplification were set as follows: 95 °C for 2 min, and 35 cycles of 95 °C for 60 s, 54 °C for 60 s, 72 °C for 90 s, and a final extension at 72 °C for 10 min. The presence or absence of a DNA band corresponding to the ITS size (approximately 550 base pairs) was verified through a gel electrophoresis (run time 25 min; 120 V) in an 1% agarose gel (Tris-Acetate EDTA (TAE) buffer, 5 µL SYBR®Safe (Invitrogen, United States)). Positive PCR products were sent to Genomics BioSci and Tech (New Taipei City, Taiwan) for sequencing. The sequences obtained were checked for ambiguity, assembled, and submitted to the National Center for Biotechnology Information (NCBI) for a nucleotide BLAST search.
2.5. Statistics
Isolation frequency (IF) is calculated as the number of algal segments with fungal growth over the total number of algal segments examined and expressed as a percentage [40].
Percentage occurrence (%) is the number of isolates of each fungal taxon divided by the total number of isolates of all taxa at each sampling time.
Diversity of the species cluster was analyzed by the Shannon–Wiener diversity index (H′) [41]; species richness was measured by the Margalef index (d) [42]; Pielou’s evenness index (J′) [43] and Simpson’s dominance index (D) [44] were calculated to determine the heterogeneity of fungal diversity. For comparison of alpha diversity between different isolation conditions, rarefaction analysis was firstly done to check species richness, and the rarefaction and extrapolation curves were constructed based on fungal abundance in the R iNEXT package [45]. Comparisons of the estimated and extrapolated species richness in plots were calculated based on 50 bootstrap replicates at a 95% confidence interval. The significance of the difference between the effects of media, surface washing/sterilization methods, and healthy/dead algal thalli on fungal diversity was tested using the non-parametric Kruskal–Wallis tests (<0.05) and pairwise Wilcoxon test (<0.05). Krona (v.2.6) [46] was adopted to visualize the taxonomic composition of fungi (species richness and percentage occurrence) through the Krona pie chart. The construction of Venn diagrams was performed using a Venn diagram viewer [47].
3. Results
3.1. Culturable Fungal Diversity of Pterocladiella Capillacea
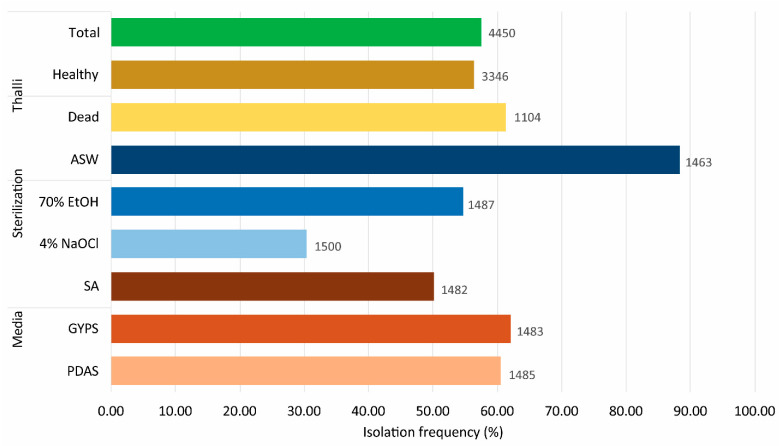
A total of 4450 algal fragments (1104 dead, 3346 healthy) of P. capillacea were inoculated, out of which at least one morphotype grew out from 2562 pieces, giving an isolation frequency of 57.57% (Figure 2, Table S1).
Figure 2.
Isolation frequency (%) of fungi from healthy and dead algal fragments after three surface washing/sterilization methods (ASW—artificial seawater, 70% EtOH—70% ethanol, and 4% NaOCl—4% sodium hypochlorite) on different media (SA—seawater agar, GYPS—glucose-yeast extract-peptone seawater agar, and PDAS—potato dextrose seawater agar). The number shown at the bars represents the total number of algal fragments examined under that treatment (see also Table S1).
Altogether, 3187 fungal isolates (906 from dead, 2281 from healthy) were cultured. These 3187 isolates were categorized into 262 morphotypes based on colony coloration, pigment formation, and mycelial growth patterns on CMAS. ITS sequence analysis referred these 262 fungal morphotypes to a total of 129 species in 67 genera, while 9 morphotypes were not identified to the genus level and referred to a family/order name (Table S1). The % sequence coverage and % sequence similarity of ITS in the BLAST search were above 95%, except for three species. The highest Shannon–Wiener diversity index (H’) was observed from healthy thalli (4.0335), thalli sterilized with 4% sodium hypochlorite (4.0923), and thalli plated on GYPS (4.0107) (Table 1). The Margalef species richness (d) was also the highest in healthy thalli (15.2605) and thalli plated on GYPS (15.6356), followed by thalli sterilized with 70% ethanol (15.0577) (Table 1). However, thalli sterilized with 4% sodium hypochlorite (0.9072) had the highest evenness index (J’) over those sterilized with 70% ethanol (0.8204).
Table 1.
Diversity indices of fungal communities isolated from healthy/dead thalli of Pterocladiella capillacea after surface washing/sterilization by artificial seawater (ASW), 70% ethanol (70% EtOH), and 4% sodium hypochlorite (4% NaOCl) plated on seawater agar (SA), glucose-yeast extract-peptone seawater agar (GYPS), and potato dextrose seawater agar (PDAS).
| . | Thalli | Surface Washing/Sterilization | Medium | Total | |||||
|---|---|---|---|---|---|---|---|---|---|
| Healthy | Dead | ASW | 70% EtOH | 4% NaOCl | SA | GYPS | PDAS | ||
| Total No. of isolates (Total Abundance), N | 2281 | 906 | 1706 | 999 | 482 | 885 | 1211 | 1091 | 3187 |
| Richness (Total number of Taxa in the community), S | 119 | 85 | 111 | 105 | 91 | 86 | 112 | 104 | 129 |
| Species Richness (Margalef): d | 15.2605 | 12.3365 | 14.7812 | 15.0577 | 14.5680 | 12.5265 | 15.6356 | 14.7251 | 15.8674 |
| Shannon-Wiener Diversity Index: H′ | 4.0335 | 3.8510 | 3.9623 | 3.8180 | 4.0923 | 3.7189 | 4.0107 | 3.8901 | 4.0982 |
| Pielou’s Evenness: J′ | 0.8440 | 0.8668 | 0.8413 | 0.8204 | 0.9072 | 0.8349 | 0.8500 | 0.8376 | 0.8433 |
| Simpson’s Dominance Index: D | 0.0349 | 0.0312 | 0.0342 | 0.0489 | 0.0233 | 0.0450 | 0.0379 | 0.0408 | 0.0310 |
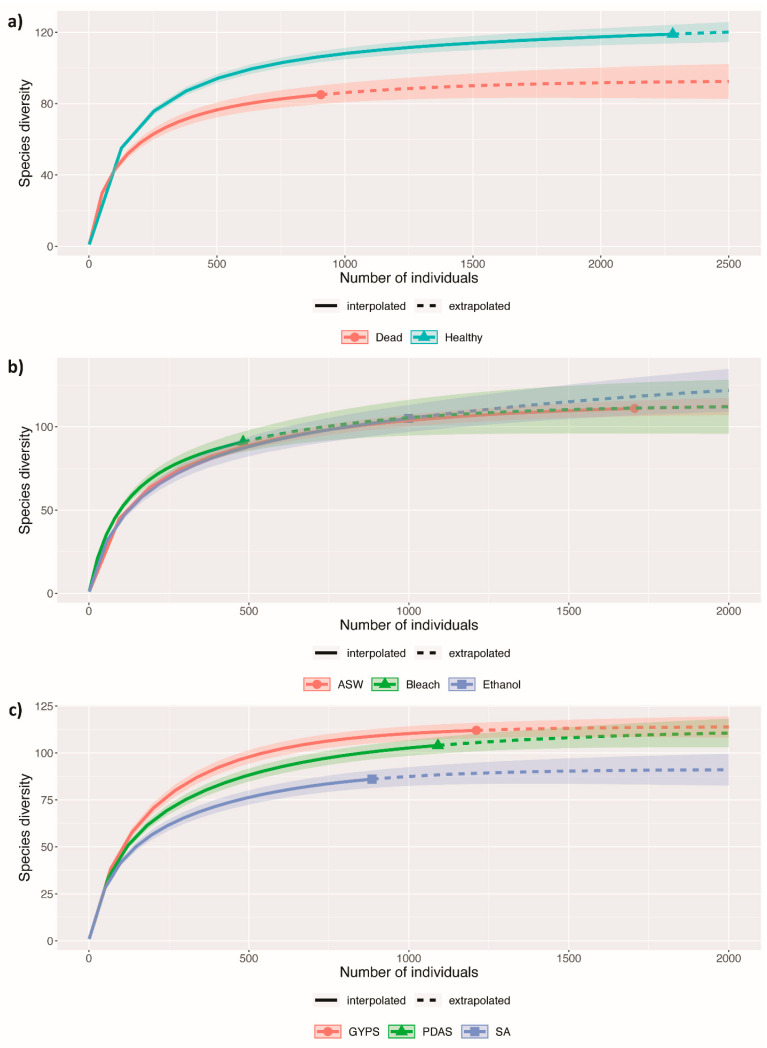
The rarefaction curves calculated from species diversity data shown in Figure 3 revealed species saturation for all treatments (healthy and dead thalli, surface washing/sterilization methods, isolation media)
Figure 3.
Sample-size-based rarefaction and extrapolation curves of fungal diversity from Pterocladiella capillacea for the Hill number (q = 0) for each of the different isolation conditions as following (a) healthy and dead thalli; (b) thalli surface sterilized/washed by artificial seawater (ASW), 70% ethanol (Ethanol), and 4% sodium hypochlorite (Bleach); and (c) thalli plated on seawater agar (SA), glucose-yeast extract-peptone seawater agar (GYPS), and potato dextrose seawater agar (PDAS). The 95% confidence intervals were obtained by a bootstrap method based on 50 replicates.
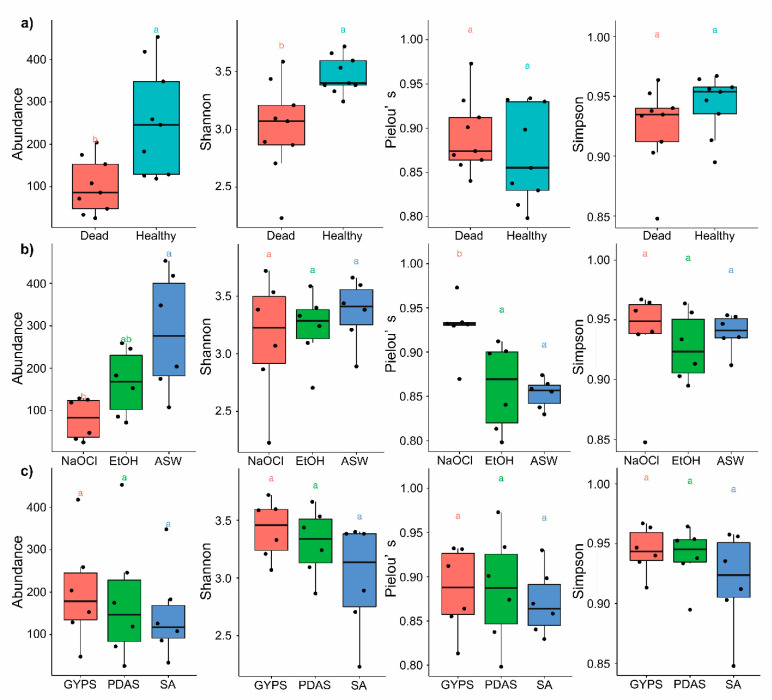
The Kruskal–Wallis analysis showed a statistically significant (p < 0.05, Bonferroni corrected) relationship on fungal diversity between healthy and dead thalli from the abundance (p = 0.0055) and Shannon diversity (p = 0.0056) index, as well as different surface washing/sterilization methods using artificial seawater and 4% sodium hypochlorite from the abundance (p = 0.0066) using 4% sodium hypochlorite and artificial seawater (p = 0.0063), and using 4% sodium hypochlorite and 70% ethanol (p = 0.013) from Pielou’s evenness. However, there was no significant difference on fungal diversity between the different media used (Figure 4).
Figure 4.
Box plot of alpha diversity indices Shannon, Pielou’s, Simpson, and the relative abundance derived from different conditions of Pterocladiella capillacea. The Wilcoxon test was used to determine the significance level between the different groups, while the Kruskal–Wallis test was used to calculate the significance level among the following: (a) healthy and dead thalli; (b) thalli surface sterilized/washed by artificial seawater (ASW), 70% ethanol (EtOH), and 4% sodium hypochlorite (NaOCl); and (c) thalli plated on seawater agar (SA), glucose-yeast extract-peptone seawater agar (GYPS), and potato dextrose seawater agar (PDAS). a, b = same letters indicate no significant differences at p < 0.05.
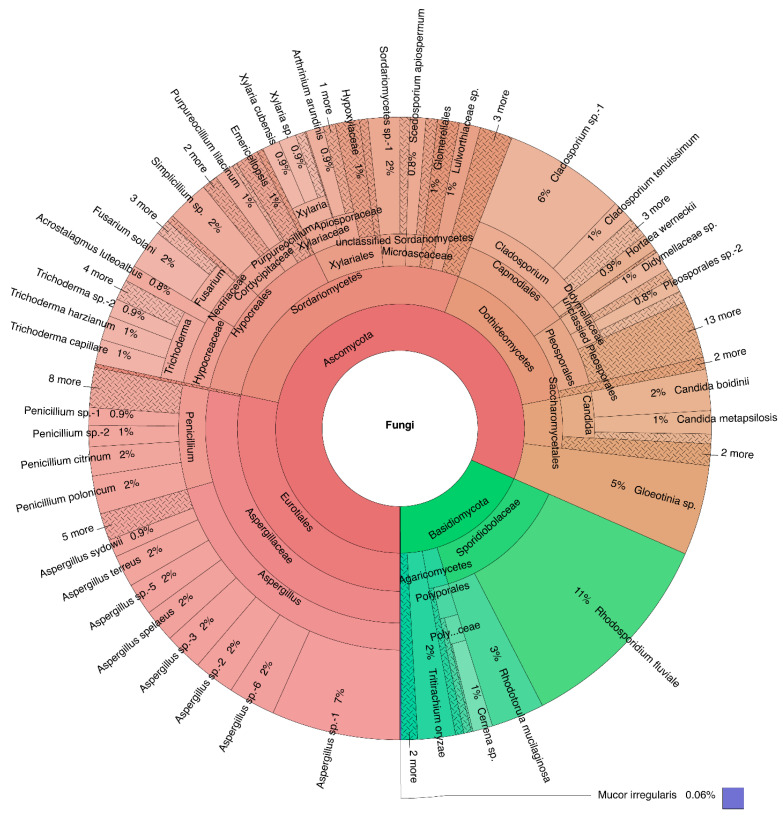
Figure 5 shows the taxonomic classification of fungi cultured from P. capillacea. Ascomycota was represented by 119 species in 57 genera (2600 isolates, 81.58%), Basidiomycota by 9 species in 9 genera (585 isolates, 18.36%), and Mucoromycota by 1 unidentified species (2 isolates, 0.06%). For Ascomycota, the most abundant class was Eurotiomycetes (903 isolates, 28.33%), followed by Sordariomycetes (877 isolates, 27.52%) and Dothideomycetes (513 isolates, 16.10%). The five dominant fungal orders were Eurotiales (903 isolates, 28.33%), Hypocreales (459 isolates, 14.40%), Capnodiales (305 isolates, 9.57%), Pleosporales (188 isolates, 5.90%), and Xylariales (179 isolates, 5.62%). The percentage occurrence of other fungal orders was less than 5%. The most abundant fungal genus was the Aspergillus, with a total 656 isolates that accounted for 20.58% of the overall occurrence, followed by Cladosporium (271 isolates, 8.50%) and Penicillium (241 isolates, 7.56%). Species with the highest overall occurrence were Aspergillus sp. 1 (213 isolates, 6.68%), Cladosporium sp. 1 (201 isolates, 6.31%), and Gloeotinia sp. (149 isolates, 4.68%). In terms of richness, the most speciose genera on P. capillacea were Aspergillus (13 species), Penicillium (12 species), Trichoderma (7 species), and Cladosporium and Xylaria (5 species).
Figure 5.
Krona chart showing the taxonomic classification of fungi isolated from Pterocladiella capillacea based on their relative abundance. The outer circle represents the isolated species and the inner circles represent their higher taxonomic classification. An interactive version of this chart is provided as Figure S2.
For Basidiomycota, Microbotryomycetes and Sporidiobolales (both 13.62%, 434 isolates) were the dominant class and order, respectively. Rhodosporidium (R. fluviale) and Rhodotorula (R. mucilaginosa) accounted for 10.86% (346 isolates) and 2.76% (88 isolates), respectively.
3.2. Fungal Diversity in Healthy and Dead Thalli
The isolation frequencies for healthy and dead thalli were 56.37% and 61.23%, respectively (Figure 2). A significant difference in fungal diversity between the healthy and dead thalli was observed (Figure 4, Table 1).
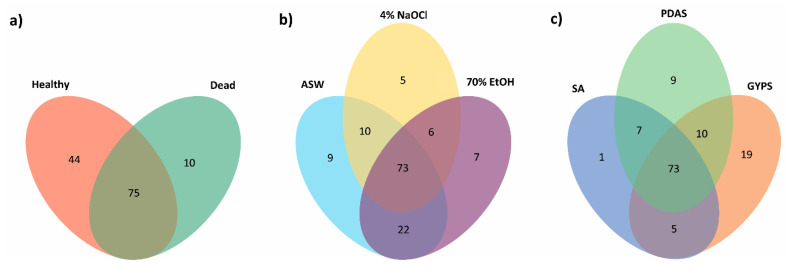
More fungal isolates were cultured from the healthy thalli (2281 isolates, 71.57% occurrence) than the dead thalli (906 isolates, 28.43%) (Table S1). A total of 119 fungal species were obtained from healthy thalli and 85 species from dead thalli (Figure 6a). Additionally, 44 and 10 species were exclusively isolated from healthy and dead thalli, respectively, with 75 common species (Figure 6a).
Figure 6.
Venn diagrams showing fungal species richness (a) in healthy and dead thalli, (b) in thalli under different sterilization regimes (artificial seawater (ASW), 70% ethanol (70% EtOH), and 4% sodium hypochlorite (4% NaOCl)), and (c) in thalli isolated on different media (seawater agar (SA), glucose-yeast extract-peptone seawater agar (GYPS), and potato dextrose seawater agar (PDAS)).
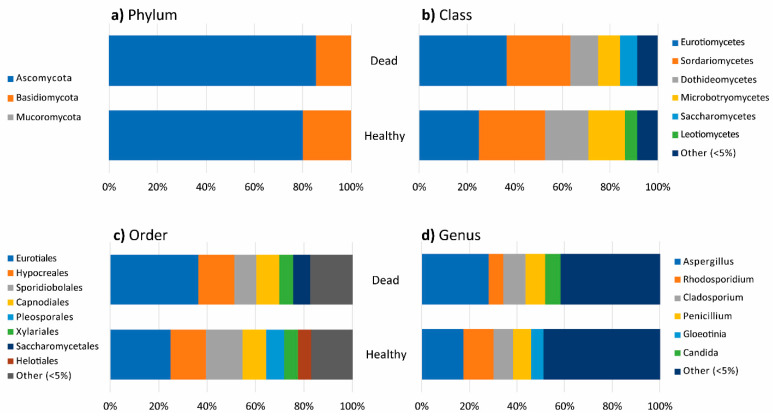
The taxonomic classification of fungi isolated from the healthy and dead thalli of P. capillacea was similar at the phylum, class, and order levels, but with a different percentage occurrence. A slightly higher occurrence of Ascomycota was found in dead than healthy thalli and the reverse was true for Basidiomycota (Figure 7a).
Figure 7.
Bar chart showing diversity of fungi at (a) phylum, (b) class, (c) order, and (d) genus levels in healthy and dead thalli.
Percentage occurrences of Eurotiomycetes and Saccharomycetes were higher in dead thalli, while those of Dothideomycetes and Microbotryomycetes were higher in healthy thalli (Figure 7b). Fungi isolated from the healthy thalli belonged to 23 different fungal orders and the dominant genera were Aspergillus (17.58%), Rhodosporidium (12.71%), and Cladosporium (8.15%). Fungi isolated from the dead thalli belonged to 20 fungal orders and the dominant fungi were Aspergillus spp. (28.15%), Cladosporium spp. (9.38%), and Penicillium spp. (7.95%) (Figure 7c,d).
Lulworthiaceae sp. (1.53%) and Didymellaceae sp. (1.45%) only occurred in the healthy thalli. From the 10 taxa only isolated from the dead thalli, Bisifusarium sp. (0.44%), Gliomastix sp. (0.22%), and Westerdykella sp. (0.11%) had the highest percentage occurrence (Table S1).
3.3. Effect of Algal Surface Washing/Sterilization Methods on Fungal Diversity
The isolation frequencies from the different surface washing/sterilization methods were the highest in seawater (88.38%), followed by 70% ethanol (54.74%) and 4% sodium hypochlorite (30.33%) (Figure 2). Thalli washed with seawater (111 species) had the highest species richness, followed by 70% ethanol (105 species) and 4% sodium hypochlorite (91 species) (Figure 6b).
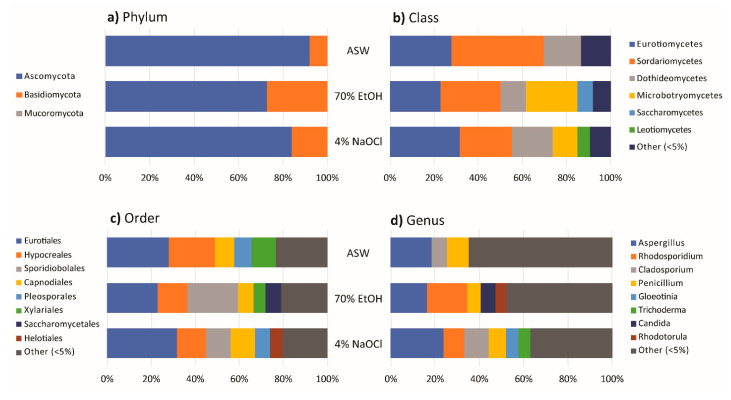
Seventy-three species could be isolated from all sterilization methods. Nine, five, and seven species were only cultured from one washing/sterilization method, being seawater, sodium hypochlorite and ethanol, respectively (Figure 6b). Fungi of the Venturiales were only isolated from the ethanol-sterilized samples; Mucorales were not isolated from the sodium hypochlorite-sterilized thalli (Table S1). At phylum level, a higher occurrence of Basidiomycota was observed on thalli sterilized by 70% ethanol (Figure 8a).
Figure 8.
Bar chart showing diversity of fungi at the (a) phylum, (b) class, (c) order, and (d) genus levels of thalli sterilized with artificial seawater (ASW), 70% ethanol (70% EtOH), and 4% sodium hypochlorite (4% NaOCl).
At the class level, a much higher occurrence of Sordariomycetes (mainly in Hypocreales) was isolated from thalli sterilized with sodium hypochlorite, while Microbotryomycetes’ occurrence (mainly in Sporidiobolales) was the highest in thalli sterilized with ethanol (Figure 8b,c). The dominant genera in seawater-washed thalli were Aspergillus (23.68%), Cladosporium (11.08%), Rhodosporidium (9.38%), and Penicillium (7.74%). Rhodosporidium (17.92%), Aspergillus (16.42%), Candida (6.61%), and Penicillium (6.21%) were the dominant genera in the ethanol-sterilized samples. Aspergillus (18.26%) and Penicillium (9.75%) and Cladosporium were the dominant genera in hypochlorite-sterilized thalli (7.05%) (Figure 8d).
3.4. Effect of Culture Media on Fungal Diversity
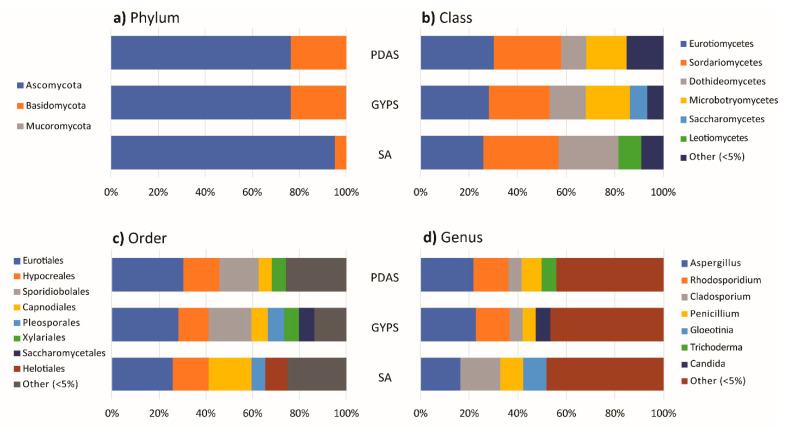
The isolation frequencies on SA, GYPS, and PDAS were 50.13%, 62.10%, and 60.47%, respectively (Figure 2). The highest number of isolates and fungal species were recovered from GYPS with 1211 isolates and 112 species, followed by PDAS (1091 isolates, 104 species) and SA (885 isolates, 86 species) (Figure 6c). Further, 1, 9, and 19 species were only isolated from SA, PDAS, and GYPS, respectively (Figure 6c). On SA, a lower occurrence of Basidiomycota (mainly in Sporidiobolales, Microbotryomycetes) (Figure 9a,b and Table S1), but a higher occurrence of Capnodiales and Helotiales was observed (Figure 9c).
Figure 9.
Bar chart showing diversity of fungi at the (a) phylum, (b) class, (c) order, and (d) genus levels isolated on seawater agar (SA), glucose-yeast extract-peptone seawater agar (GYPS), and potato dextrose seawater agar (PDAS).
The most represented orders were Eurotiales (25.99%), Capnodiales (18.31%), Hypocreales (15.14%), and Helotiales (9.49%), and the dominant fungal genera were Cladosporium (16.61%), Aspergillus (16.38%), and Gloeotinia (9.49%) (Figure 9d). A comparatively high occurrence of Saccharomycetes (i.e., Saccharomycetales) was isolated from GYPS (Figure 9b,c). Additionally, 21 orders were isolated from GYPS with Eurotiales (28.24%), Sporidiales (18.33%), and Hypocreales (12.96%) being the dominant orders (Figure 9c), while the dominant genera were Aspergillus (22.71%), Rhodosporidium (13.71%), Candida (5.78%), Cladosporium, and Penicillium (5.53%) (Figure 9d). Venturiales and Mucorales were only isolated from GYPS. Aspergillus (21.63%), Rhodosporidium (14.67%), and Penicillium (8.34%) were the dominant genera on PDAS with Eurotiales (30.34%), Sporidiales (16.77%), and Hypocreales (15.40%) being the most dominant orders (Figure 9c,d).
Finally, 73 fungal species were common for all media, including Aspergillus spp. (20.58%), Rhodosporidium sp. (10.86%), and Cladosporium (8.50%) with the highest percentage occurrence (Table S1).
4. Discussion
4.1. Fungal Community
This is the first report of culturable fungi associated with Pterocladiella capillacea, and the results revealed a high diversity of culturable fungi with 129 species in 67 genera. The Ascomycota was dominant in P. capillacea, in which Eurotiales, Hypocreales, Pleosporales, and Capnodiales were the most abundant. Fungi of these orders were shown to be halotolerant and halophilic species [48], which may explain their close association with the alga. Xylariales are predominantly terrestrial, while many occur as endophytes of plants, and they produce secondary metabolites against bacterial and fungal pathogens [49]. In the marine environment, the xylarialean fungi also occur as endophytes of seagrasses, mangrove plants, and macroalgae [50]. The xylarialean fungi (as well as other taxa) isolated from P. capillacea in this study might also produce antimicrobial substances to protect the alga from diseases. A Xylaria species isolated from the red alga Bostrychia tenella produced cytochalasin D, an antibiotic compound, which may confirm this conclusion [51]. Nevertheless, Chondrostereum sp. NTOU4196 and Phoma sp. NTOU4195 isolated in this study produced novel chemical structures in spent culture liquid with anti-inflammatory properties, suggesting a prolific source of bioactive compounds in the marine environment [52,53].
Aspergillus, Penicillium (Eurotiales), Cladosporium (Capnodiales), and Trichoderma (Hypocreales) are cosmopolitan in distribution and can be found in terrestrial and aquatic environments. The widespread occurrence of these taxa in the marine environment may suggest their passive migration from terrestrial habitats [54]. Their high occurrence on P. capillacea is not surprising as numerous species of these genera can be found in the marine environment [55]. Some studies suggested that these common genera of fungi were able to tolerate or degrade antifungal compounds in macroalgae, which could explain their wide host colonization [19]. However, it is important to point out that the culture-dependent technique used in this study may bias toward the isolation of these fast-growing taxa [56]. The culture-independent approach revealed a different diversity of fungi associated with the brown alga Fucus serratus from the isolation technique [25].
Species of Aspergillus and Penicillium were among the most common taxa in the marine environment [55,57,58]; they are common endosymbionts of different seaweeds [12,22,59,60] and seagrasses [61]. Aspergillus spp., apart from dominating the endosymbiont assemblage of seaweeds [23], also dominated the fungal assemblages of marine invertebrates [32,62,63]. Aspergillus spp. are known to be halotolerant and have the genetic capacity to adapt to salinity pressure and this plasticity may confer an ecological advantage over others [48,64]. Penicillium is one of the most common genera isolated from macroalgae [13]. Penicillium spp. isolated from red algae in maritime Antarctica displayed carrageenolytic and agarolytic activities, suggesting that they play a role in recycling carbon when the algae die [65]. Both Aspergillus and Penicillium species are known to produce antimicrobial secondary metabolites [32,60] and, thus, may protect macroalgae from microbial infection. In a study of the mycobiota of the red alga Asparagopsis taxiformis, two out of the five species isolated belonged to Cladosporium [66].
Yeast species Rhodosporidium fluviale and Rhodotorula mucilaginosa dominated the occurrence of Basidiomycota in P. capillacea. Basidiomycetous yeasts are common in the marine environment [67]. Few filamentous Basidiomycota can be found in the marine environment, possibly because they are salinity sensitive [68]. The known terrestrial Agaricomycetes isolated from P. capillacea might only represent a superficial association with the alga [69]. Mucoromycota is uncommon in the marine environment [55] and so it seems reasonable that only two isolates were cultured from P. capillacea. Whether isolation of marine Mucoromycota requires special techniques or media needs further study.
The isolation frequencies for healthy and dead thalli were similar but a higher percentage occurrence and a higher species richness were observed in healthy thalli. The fungal community between healthy and dead thalli of P. capillacea was highly similar (75 taxa), while the percentage occurrences of unique species were low (<1%), especially in dead thalli. However, Lulworthiaceae sp. and Didymellaceae sp. (>1%) may play a role in healthy thalli.
Pterocladiella capillacea is a perennial red alga that can be found at the sampling site in Keelung, Taiwan. No disease symptom was observed during sample collection, suggesting the fungi cultured in this study do not cause diseases of the alga. However, Aspergillus ochraceus, A. terreus, and a Phoma species induced bleaching disease symptoms in healthy, non-axenic cultures of Kappaphycus alvarezii [18]. Many cultured taxa in this study, as described above, have been proven to produce antimicrobial compounds. Pterocladiella capillacea might have formed a symbiotic relationship with some of its associated fungi by exchanging nutrients from photosynthesis for protection from microbial diseases.
4.2. Isolation Methods
In this study, sodium hypochlorite (4%) was found to be an effective surface sterilizer based on its low isolation frequency when compared with 70% ethanol. Without surface sterilization, the isolation frequency was three times higher (washed with sterilized seawater). The Kruskal–Wallis test also suggested a significant effect of algal surface washing/sterilization methods on fungal diversity, and consequently, species richness was highest in seawater, followed by ethanol and sodium hypochlorite, but the dominant species were similar for the three methods of surface washing/sterilization. The percentage occurrence of unique species from each of the three methods of surface washing/sterilization was very low. Mucorales might only associate with P. capillacea epiphytically, as this group was not found in sodium hypochlorite-sterilized samples. To study total culturable diversity of fungi associated with P. capillacea, washing of algal thalli with sterile seawater is recommended, otherwise sodium hypochlorite can be used to sterilize the algal surface to study the endophytic assemblage of fungi.
The SA, PDAS, and GYPS used for the isolation in this study are common media to study marine fungi, especially those associated with macroalgae [15,18,19,21,22,32,33,34,35]. For media, high isolation frequencies on PDAS and GYPS were reasonable, as glucose is the major carbon in these media, which favors growth of saprobic species over endophytic species. The yeasts Rhodosporidium fluviale, Rhodotorula mucilaginosa (Microbotryomycetes, Basidiomycota), and Candida spp. (Saccharomycetes, Ascomycota) generally grow well in media rich in glucose [70], and these taxa had high occurrence in PDAS and GYPS. The comparatively lower isolation frequency on SA can be explained by the minimal nutrients it contains [32]. The effect of natural against artificial seawater and ‘aged’ against ‘newly collected’ natural seawater is unknown, but decomposition of organic matter in natural seawater during the ‘aging’ process may enrich the seawater with more readily available nutrients [71]. However, species richness on the three media was similar and only differed in overall occurrence, and the Kruskal–Wallis test also suggested no significant effect of media on fungal diversity. The use of both GYPS and PDAS as the isolation media was sufficient to study the culturable diversity of fungi associated with P. capillacea and possibly other macroalgae.
5. Conclusions
A high diversity of epiphytic and endophytic fungi was associated with the red alga Pterocladiella capillacea, with 129 species in 67 genera. Filamentous Ascomycota (Eurotiales, Capnodiales, Hypocreales, Pleosporales) dominated the community with many basidiomycetous yeasts (Sporidiobolales) and few Mucoromycota. Many cultured fungi (Aspergillus, Penicillium, Xylariales) are known producers of antimicrobial secondary metabolites and may symbiotically associate with the alga. Sodium hypochlorite is an effective surface sterilizer to study the endophytic mycobiota of macroalgae. Media used for fungal isolation did not have a significant effect on species richness, but only an effect on the overall occurrence.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof7080651/s1, Figure S1: Schematic diagram of experimental, Figure S2: An interactive version of Krona chart showing the taxonomic classification of fungi isolated from Pterocladiella capillacea based on their relative abundance, Table S1: Taxonomic information and overall fungal isolates.
Author Contributions
Conceptualization, K.-L.P.; Data curation, H.-J.C., M.W.L.C., S.-Y.G. and K.-L.P.; Formal analysis, M.W.L.C. and S.-Y.G.; Investigation, K.-L.P.; Methodology, H.-J.C. and K.-L.P.; Resources, S.-M.L., K.-L.P.; Visualization, H.-J.C., M.W.L.C. and S.-Y.G.; Writing—original draft, H.-J.C.; Writing—review & editing, H.-J.C., S.-M.L. and K.-L.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequence data associated with this study have been submitted to the NCBI.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sutherland G. New marine fungi on Pelvetia. New Phytol. 1915;14:33–42. doi: 10.1111/j.1469-8137.1915.tb07171.x. [DOI] [Google Scholar]
- 2.Sutherland G. Marine fungi imperfecti. New Phytol. 1916;15:35–48. doi: 10.1111/j.1469-8137.1916.tb07201.x. [DOI] [Google Scholar]
- 3.Vrijmoed L.L.P. Isolation and culture of higher filamentous fungi. In: Hyde K.D., Pointing S.B., editors. Marine Mycology—A Practical Approach. Fungal Diversity Press; Hong Kong, China: 2000. pp. 1–20. [Google Scholar]
- 4.Zuccaro A., Mitchell J.I. Fungal communities of seaweeds. In: Dighton J., Oudemans P., White J., editors. The Fungal Community: Its Organisation and Role in the Ecosystem. Marcel Dekker; New York, NY, USA: 2005. pp. 553–579. [Google Scholar]
- 5.Jones E.B.G., Pang K.L., Stanley S.J. Fungi from marine algae. In: Jones E.B.G., Pang K.L., editors. Marine Fungi and Fungal-Like Organisms. De Gruyter; Berlin, Germany: 2012. pp. 329–344. [Google Scholar]
- 6.Porter D., Farnham W.F. Mycaureola dilseae, a marine basidiomycete parasite of the red alga, Dilsea carnosa. Trans. Br. Mycol. Soc. 1986;87:575–582. doi: 10.1016/S0007-1536(86)80098-5. [DOI] [Google Scholar]
- 7.Stanley S.J. Observations on the seasonal occurrence of marine endophytic and parasitic fungi. Can. J. Bot. 1992;70:2089–2096. doi: 10.1139/b92-259. [DOI] [Google Scholar]
- 8.Porras-Alfaro A., Bayman P. Hidden fungi, emergent properties: Endophytes and microbiomes. Annu. Rev. Phytopathol. 2011;49:291–315. doi: 10.1146/annurev-phyto-080508-081831. [DOI] [PubMed] [Google Scholar]
- 9.Debbab A., Aly A., Proksch P. Endophytes and associated marine derived fungi—ecological and chemical perspectives. Fungal Divers. 2012;57:45–83. doi: 10.1007/s13225-012-0191-8. [DOI] [Google Scholar]
- 10.Sanders W.B., Moe R.L., Ascaso C. The intertidal marine lichen formed by the pyrenomycete fungus Verrucaria tavaresiae (Ascomycotina) and the brown alga Petroderma maculiforme (Phaeophyceae): Thallus organization and symbiont interaction. Am. J. Bot. 2004;91:511–522. doi: 10.3732/ajb.91.4.511. [DOI] [PubMed] [Google Scholar]
- 11.Kohlmeyer J., Volkmann-Kohlmeyer B. Mycophycias, a new genus for the mycobionts of Apophlaea, Ascophyllum and Pelvetia. Syst. Ascomycetum. 1998;16:1–7. [Google Scholar]
- 12.Zuccaro A., Schoch C.L., Spatafora J.W., Kohlmeyer J., Draeger S., Mitchell J.I. Detection and identification of fungi intimately associated with the brown seaweed Fucus serratus. Appl. Environ. Microbiol. 2008;74:931–941. doi: 10.1128/AEM.01158-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bugni T.S., Ireland C.M. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004;21:143–163. doi: 10.1039/b301926h. [DOI] [PubMed] [Google Scholar]
- 14.Kjer J., Debbab A., Aly A.H., Proksch P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc. 2010;5:479–490. doi: 10.1038/nprot.2009.233. [DOI] [PubMed] [Google Scholar]
- 15.Loque C.P., Medeiros A.O., Pellizzari F.M., Oliveira E.C., Rosa C.A., Rosa L.H. Fungal community associated with marine macroalgae from Antarctica. Polar Biol. 2010;33:641–648. doi: 10.1007/s00300-009-0740-0. [DOI] [Google Scholar]
- 16.Godinho V.M., Furbino L.E., Santiago I.F., Pellizzari F.M., Yokoya N.S., Pupo D., Alves T.M., Junior P.A., Romanha A.J., Zani C.L., et al. Diversity and bioprospecting of fungal communities associated with endemic and cold-adapted macroalgae in Antarctica. ISME J. 2013;7:1434–1451. doi: 10.1038/ismej.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duarte A.W.F., Dayo-Owoyemi I., Nobre F.S., Pagnocca F.C., Chaud L.C.S., Pessoa A., Felipe M.G.A., Sette L.D. Taxonomic assessment and enzymes production by yeasts isolated from marine and terrestrial Antarctic samples. Extremophiles. 2013;17:1023–1035. doi: 10.1007/s00792-013-0584-y. [DOI] [PubMed] [Google Scholar]
- 18.Solis M.J.L., Draeger S., dela Cruz T.E.E. Marine-derived fungi from Kappaphycus alvarezii and K. striatum as potential causative agents of ice-ice disease in farmed seaweeds. Bot. Mar. 2010;53:587–594. doi: 10.1515/bot.2010.071. [DOI] [Google Scholar]
- 19.Abdel-Gawad K.M., Hifney A.F., Issa A.A., Gomaa M. Spatio-temporal, environmental factors, and host identity shape culturable-epibiotic fungi of seaweeds in the Red Sea, Egypt. Hydrobiologia. 2014;740:37–49. doi: 10.1007/s10750-014-1935-0. [DOI] [Google Scholar]
- 20.Zuccaro A., Summerbell R.C., Gams W., Schroers H.J., Mitchell J.I. A new Acremonium species associated with Fucus spp., and its affinity with a phylogenetically distinct marine Emericellopsis clade. Stud. Mycol. 2004;50:283–297. [Google Scholar]
- 21.Flewelling A.J., Ellsworth K.T., Sanford J., Forward E., Johnson J.A., Gray C.A. Macroalgal Endophytes from the Atlantic coast of Canada: A potential source of antibiotic natural products? Microorganisms. 2013;1:175–187. doi: 10.3390/microorganisms1010175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flewelling A.J., Johnson J.A., Gray C.A. Isolation and bioassay screening of fungal endophytes from North Atlantic marine macroalgae. Bot. Mar. 2013;56:287–297. doi: 10.1515/bot-2012-0224. [DOI] [Google Scholar]
- 23.Suryanarayanan T.S., Venkatachalam A., Thirunavukkarasu N., Ravishankar J.P., Doble M., Geetha V. Internal muconiota of marine macroalgae from the Tamilnadu coast: Distribution, diversity and biotechnological potential. Bot. Mar. 2010;53:457–468. doi: 10.1515/bot.2010.045. [DOI] [Google Scholar]
- 24.Kohlmeyer J., Volkmann-Kohlmeyer B. Marine ascomycetes from algae and animal hosts. Bot. Mar. 2003;46:285–306. doi: 10.1515/BOT.2003.026. [DOI] [Google Scholar]
- 25.Zuccaro A., Schulz B., Mitchell J.I. Molecular detection of ascomycetes associated with Fucus serratus. Mycol. Res. 2003;107:1451–1466. doi: 10.1017/S0953756203008657. [DOI] [PubMed] [Google Scholar]
- 26.Kohlmeyer J., Kohlmeyer E. Marine Mycology: The Higher Fungi. Academic Press; New York, NY, USA: 1979. [Google Scholar]
- 27.Kohlmeyer J., Volkmann-Kohlmeyer B. Illustrated key to the filamentous marine fungi. Bot. Mar. 1991;34:1–61. doi: 10.1515/botm.1991.34.1.1. [DOI] [Google Scholar]
- 28.Wainwright B.J., Zahn G.L., Zushi J., Lee N.L.Y., Ooi J.L.S., Lee J.N., Huang D. Seagrass-associated fungal communities show distance decay of similarity that has implications for seagrass management and restoration. Ecol. Evol. 2019;9:11288–11297. doi: 10.1002/ece3.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guiry M.D. How many species of Algae are there? J. Phycol. 2012;48:1057–1063. doi: 10.1111/j.1529-8817.2012.01222.x. [DOI] [PubMed] [Google Scholar]
- 30.Teixeira T.R., Santos G.S., Turatti I.C.C., Paziani M.H., von Zeska Kress M.R., Colepicolo P., Debonsi H.M. Characterization of the lipid profile of Antarctic brown seaweeds and their endophytic fungi by gas chromatography–mass spectrometry (GC–MS) Polar Biol. 2019;42:1431–1444. doi: 10.1007/s00300-019-02529-w. [DOI] [Google Scholar]
- 31.Sieber T.N. Fungal root endophytes. In: Waisel Y., Eshel A., Kafkafi U., editors. Plant Roots: The Hidden Half. Marcel Dekker; New York, NY, USA: 2002. pp. 887–917. [Google Scholar]
- 32.Wong Chin J.M., Puchooa D., Bahorun T., Jeewon R. Antimicrobial properties of marine fungi from sponges and brown algae of Mauritius. Mycology. 2021;12:1–14. doi: 10.1080/21501203.2021.1895347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deutsch Y., Gur L., Berman Frank I., Ezra D. Endophytes from Algae, a Potential Source for New Biologically Active Metabolites for Disease Management in Aquaculture. Front. Mar. Sci. 2021;8:333. doi: 10.3389/fmars.2021.636636. [DOI] [Google Scholar]
- 34.Kamat S., Kumari M., Taritla S., Jayabaskaran C. Endophytic fungi of marine alga from Konkan coast, India—A rich source of bioactive material. Front. Mar. Sci. 2020;7:31. doi: 10.3389/fmars.2020.00031. [DOI] [Google Scholar]
- 35.Lee S., Park M.S., Lee H., Kim J.J., Eimes J.A., Lim Y.W. Fungal diversity and enzyme activity associated with the macroalgae, Agarum clathratum. Mycobiology. 2019;47:50–58. doi: 10.1080/12298093.2019.1580464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gnavi G., Garzoli L., Poli A., Prigione V., Burgaud G., Varese G.C. The culturable mycobiota of Flabellia petiolata: First survey of marine fungi associated to a Mediterranean green alga. PLoS ONE. 2017;12:e0175941. doi: 10.1371/journal.pone.0175941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White T.J., Bruns T., Lee S.J.W.T., Taylor J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990;18:315–322. [Google Scholar]
- 38.Toju H., Tanabe A.S., Yamamoto S., Sato H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE. 2012;7:e40863. doi: 10.1371/journal.pone.0040863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beenken L., Lutz M., Scholler M. DNA barcoding and phylogenetic analyses of the genus Coleosporium (Pucciniales) reveal that the North American goldenrod rust C. solidaginis is a neomycete on introduced and native Solidago species in Europe. Mycol. Prog. 2017;16:1073–1085. doi: 10.1007/s11557-017-1357-2. [DOI] [Google Scholar]
- 40.Petrini O., Fisher P.J., Petrini L.E. Fungal endophytes of bracken (Pteridium aquilinum), with some reflections on their use in biological control. Sydowia. 1992;44:282–293. [Google Scholar]
- 41.Shannon C.E., Weaver W. The Mathematical Theory of Communication. The University of Illinois Press; Urbana, IL, USA: 1949. pp. 3–24. [Google Scholar]
- 42.Margalef R. Perspectives in Ecological Theory. University of Chicago Press; Chicago, IL, USA: 1968. [Google Scholar]
- 43.Pielou E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966;13:131–144. doi: 10.1016/0022-5193(66)90013-0. [DOI] [Google Scholar]
- 44.Simpson E.H. Measurement of Diversity. Nature. 1949;163:688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- 45.Hsieh T.C., Ma K.H., Chao A. iNEXT: An R package for interpolation and extrapolation of species diversity (Hill numbers) Methods Ecol. Evol. 2016;7:1451–1456. doi: 10.1111/2041-210X.12613. [DOI] [Google Scholar]
- 46.Ondov B.D., Nicholas H.B., Adam M.P. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bardou P., Mariette J., Escudié F., Djemiel C., Klopp C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014;15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gunde-Cimerman N., Ramos J., Plemenitaš A. Halotolerant and halophilic fungi. Mycol. Res. 2009;113:1231–1241. doi: 10.1016/j.mycres.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Becker K., Stadler M. Recent progress in biodiversity research on the Xylariales and their secondary metabolism. J. Antibiot. 2021;74:1–23. doi: 10.1038/s41429-020-00376-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakayaroj J., Preedanon S., Phongpaichit S., Buatong J., Chaowalit P., Rukachaisirikul V. 16 Diversity of endophytic and marine-derived fungi associated with marine plants and animals. In: Gareth Jones E.B., Pang K.-L., editors. Marine Fungi. De Gruyter; Berlin, Germany: 2012. pp. 291–328. [Google Scholar]
- 51.De Felício R., Pavão G.B., de Oliveira A.L.L., Erbert C., Conti R., Pupo M.T., Furtado N.A., Ferreira E.G., Costa-Lotufo L.V., Young M.C.M., et al. Antibacterial, antifungal and cytotoxic activities exhibited by endophytic fungi from the Brazilian marine red alga Bostrychia tenella (Ceramiales) Rev. Bras. Farmacogn. 2015;25:641–650. doi: 10.1016/j.bjp.2015.08.003. [DOI] [Google Scholar]
- 52.Lee M.S., Wang S.W., Wang G.J., Pang K.L., Lee C.K., Kuo Y.H., Cha H.J., Lin R.K., Lee T.H. Angiogenesis inhibitors and anti-inflammatory agents from Phoma sp. NTOU4195. J. Nat. Prod. 2016;79:2983–2990. doi: 10.1021/acs.jnatprod.6b00407. [DOI] [PubMed] [Google Scholar]
- 53.Hsiao G., Chi W.C., Pang K.L., Chen J.J., Kuo Y.H., Wang Y.K., Cha H.J., Chou S.C., Lee T.H. Hirsutane-type sesquiterpenes with inhibitory activity of microglial nitric oxide production from the red alga derived fungus Chondrostereum sp. NTOU4196. J. Nat. Prod. 2017;80:1615–1622. doi: 10.1021/acs.jnatprod.7b00196. [DOI] [PubMed] [Google Scholar]
- 54.Alva P., McKenzie E.H.C., Pointing S.B., Pena-Muralla R., Hyde K.D. Do sea grasses harbour endophytes? Fungal Divers. Res. Ser. 2002;7:167–178. [Google Scholar]
- 55.Jones E.B.G., Suetrong S., Sakayaroj J., Bahkali A.H., Abdel-Wahab M.A., Boekhout T., Pang K.L. Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Divers. 2015;73:1–72. doi: 10.1007/s13225-015-0339-4. [DOI] [Google Scholar]
- 56.Hyde K.D., Soytong K. Understanding microfungal diversity: A critique. Cyptogamie Mycologie. 2007;28:281–289. [Google Scholar]
- 57.Das S., Lyla P.S., Khan S.A. Filamentous fungal population and species diversity from the continental slope of Bay of Bengal, India. Acta Oecol. 2009;35:269–279. doi: 10.1016/j.actao.2008.11.003. [DOI] [Google Scholar]
- 58.Matallah-Boutiba A., Ruiz N., Sallenave-Namont C., Grovel O., Amiard J.C., Pouchus Y.F., Boutiba Z. Screening for toxigenic marine-derived fungi in Algerian mussels and their immediate environment. Aquaculture. 2012;342:75–79. doi: 10.1016/j.aquaculture.2012.02.016. [DOI] [Google Scholar]
- 59.König G.M., Kehraus S., Seibert S.F., Abdel-Lateff A., Müller D. Natural products from marine organisms and their associated microbes. ChemBioChem. 2006;7:229–238. doi: 10.1002/cbic.200500087. [DOI] [PubMed] [Google Scholar]
- 60.Sarasan M., Job N., Puthumana J., Ravinesh R., Prabhakaran M.P., Thomas L.C., Philip R. Exploration and profiling of hidden endophytic mycota of marine macroalgae with potential drug leads. FEMS Microbiol. Lett. 2020;367:fnaa078. doi: 10.1093/femsle/fnaa078. [DOI] [PubMed] [Google Scholar]
- 61.Venkatachalam A., Thirunavukkarasu N., Suryanarayanan T.S. Distribution and diversity of endophytes in seagrasses. Fungal Ecol. 2015;13:60–65. doi: 10.1016/j.funeco.2014.07.003. [DOI] [Google Scholar]
- 62.Höller U., Wright A.D., Matthee G.F., Konig G.M., Draeger S., Hans-Jürgen A.U.S.T., Schulz B. Fungi from marine sponges: Diversity, biological activity and secondary metabolites. Mycol. Res. 2000;104:1354–1365. doi: 10.1017/S0953756200003117. [DOI] [Google Scholar]
- 63.Baker P.W., Kennedy J., Dobson A.D., Marchesi J.R. Phylogenetic diversity and antimicrobial activities of fungi associated with Haliclona simulans isolated from Irish coastal waters. Mar. Biotechnol. 2009;11:540–547. doi: 10.1007/s10126-008-9169-7. [DOI] [PubMed] [Google Scholar]
- 64.Pang K.L., Chiang M.W.-L., Guo S.-Y., Shih C.-Y., Dahms H.U., Hwang J.-S., Cha H.-J. Growth study under combined effects of temperature, pH and salinity and transcriptome analysis revealed adaptations of Aspergillus terreus NTOU4989 to the extreme conditions at Kueishan Island Hydrothermal Vent Field, Taiwan. PLoS ONE. 2020;15:e0233621. doi: 10.1371/journal.pone.0233621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Furbino L., Pellizzari F.M., Neto P.C., Rosa C.A., Rosa L.H. Isolation of fungi associated with macroalgae from maritime Antarctica and their production of agarolytic and carrageenolytic activities. Polar Biol. 2017;41:527–535. doi: 10.1007/s00300-017-2213-1. [DOI] [Google Scholar]
- 66.Garzoli L., Gnavi G., Varese G.C., Picco A.M. Mycobiota associated with the rhodophyte alien species Asparagopsis taxiformis (Delile) Trevisan de Saint-Léon in the Mediterranean Sea. Mar. Ecol. 2015;36:959–968. doi: 10.1111/maec.12189. [DOI] [Google Scholar]
- 67.Fell J.W., Boekhout T., Fonseca A., Scorzetti G., Statzell-Tallman A. Biodiversity and systematics of basidiomycetous yeasts as determined by large subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- 68.Jones E.B.G., Choeyklin R. Ecology of marine and freshwater basidiomycetes. In: Boddy L., Frankland J.C., van West P., editors. Ecology of Saprotrophic Basidiomycetes. Elsevier; London, UK: 2008. pp. 301–324. [Google Scholar]
- 69.Pang K.L., Guo S.Y., Chen I.A., Burgaud G., Luo Z.H., Dahms H.U., Hwang J.S., Lin Y.L., Huang J.S., Ho T.W., et al. Insights into fungal diversity of a shallow-water hydrothermal vent field at Kueishan Island, Taiwan by culture-based and metabarcoding analyses. PLoS ONE. 2019;14:e0226616. doi: 10.1371/journal.pone.0226616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zaky A.S., Tucker G.A., Daw Z.Y., Du C. Marine yeast isolation and industrial application. FEMS Yeast Res. 2014;14:813–825. doi: 10.1111/1567-1364.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson T.W., Sparrow F.K. Fungi in Oceans and Estuaries. Cramer; Weinheim, Germany: 1961. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data associated with this study have been submitted to the NCBI.