Abstract
Replication fidelity is controlled by DNA polymerase proofreading and postreplication mismatch repair. We have genetically characterized the roles of the 5′→3′ Exo1 and the 3′→5′ DNA polymerase exonucleases in mismatch repair in the yeast Saccharomyces cerevisiae by using various genetic backgrounds and highly sensitive mutation detection systems that are based on long and short homonucleotide runs. Genetic interactions were examined among DNA polymerase ɛ (pol2-4) and δ (pol3-01) mutants defective in 3′→5′ proofreading exonuclease, mutants defective in the 5′→3′ exonuclease Exo1, and mismatch repair mutants (msh2, msh3, or msh6). These three exonucleases play an important role in mutation avoidance. Surprisingly, the mutation rate in an exo1 pol3-01 mutant was comparable to that in an msh2 pol3-01 mutant, suggesting that they participate directly in postreplication mismatch repair as well as in other DNA metabolic processes.
Chromosome replication fidelity is generally considered to be determined by a combination of base selection and error correction activities of DNA polymerases along with postreplication mismatch repair (MMR). The combined effect in Escherichia coli results in an overall error rate that is as low as 10−10 errors per replicated nucleotide (27). In E. coli, the α polymerase subunit of DNA polymerase III holoenzyme, encoded by the dnaE gene, determines base selection and the ɛ subunit (dnaQ) provides proofreading. These subunits are tightly bound together with the θ subunit to form the polymerase III core (16). The errors left by the polymerase III holoenzyme are corrected by the MutHLS MMR system (28).
In eukaryotes the three polymerases required for chromosome replication are polymerase α, δ, and ɛ, encoded by the POL1, POL3, and POL2 genes, respectively. Polymerase α (Polα) is responsible for the synthesis of primers for Okazaki fragments in the lagging strand, and the δ and ɛ polymerases, it has been proposed, are responsible for lagging and leading DNA strand replication (35), although their specific roles have not been established. Unlike Polα, the δ and ɛ polymerases also have a 3′→5′ proofreading exonuclease activity in their N-terminal regions (14, 22, 32). In the yeast Saccharomyces cerevisiae, the point mutations pol3-01 and pol2-4, which eliminate the proofreading activities of the δ and ɛ polymerases, respectively, result in a frameshift and base substitution mutator phenotype (22, 23).
The postreplication MMR system is responsible for the correction of errors generated during replication. Based on in vitro analysis with purified proteins, the steps in E. coli MMR include mismatch recognition by MutS and MutL proteins, methyl-directed strand discrimination, incision by the MutH endonuclease in the unmethylated nascent strand at sites opposite methylated GATC, degradation from a nick towards the mismatched site by exonucleases (ExoI, ExoVII, or RecJ), and gap filling by polymerase III holoenzyme followed by DNA ligation (21).
Except for mismatch recognition, relatively little is known about the MMR mechanism in eukaryotes. Many genes homologous to E. coli mutS and mutL have been cloned and studied (21). Strand-specific MMR was demonstrated in human cell extracts when a single-strand nick was introduced on either side of a mismatch (7, 37), suggesting that both 5′→3′ and 3′→5′ exonucleases may be involved. The 5′→3′ exonuclease Exo1 of the yeast S. cerevisiae has been implicated in MMR. The EXO1 gene, which is homologous to the Schizosaccharomyces pombe EXO1 gene (36), was isolated in a two-hybrid interaction screen with yeast MSH2 (38). Tishkoff et al. (38) concluded that EXO1 and MSH2 are in one epistasis group; however, other interpretations are possible. In an exo1-deficient S. cerevisiae strain, the canavanine resistance (Canr) forward and hom3-10 reverse mutation (−1 frameshifts) rates are increased only eight- and sixfold, respectively, in comparison with a wild-type strain. This is much lower than the corresponding 25- and 850-fold increases observed in msh2 mutants (38), and the msh2 exo1 double mutant exhibits rates comparable to that of the single msh2 mutant. However, it cannot be ascertained from these data whether MSH2 and EXO1 have an epistatic or additive interaction (i.e., single versus separate pathways), since the rate in the msh2 mutant differs little from the sum of the rates in the msh2 and exo1 mutants. Since both Exo1 and Msh2 are involved in homologous recombination (9, 26, 34), their physical interaction could be related to this process rather than MMR.
We have examined the role of various exonucleases in MMR as part of our efforts to screen and characterize MMR genes in the yeast S. cerevisiae. For example, the small mutator effect of an exo1 mutation as compared to that of an msh2 mutation suggests that there are additional exonucleases involved in MMR. A synergistic mutator effect was found for defects in the Exo1 nuclease and the Polɛ 3′→5′ proofreading exonuclease, suggesting that these activities participate in postreplication steps. Cells deficient in both DNA Polδ 3′→5′ proofreading exonuclease and either Exo1, Msh2, or DNA Polɛ 3′→5′ proofreading exonuclease (24) were inviable, implying a strong synergistic interaction. On the other hand, diploid strains defective in both DNA Polδ 3′→5′ proofreading exonuclease and Exo1 or Msh2 were viable. Surprisingly they exhibit comparable levels of hypermutability. We propose that the Polδ and Polɛ proofreading exonucleases as well as Exo1 may play a major role in mutation prevention.
MATERIALS AND METHODS
General genetic and molecular methods.
Yeast standard media (30) and yeast-extract-peptone-dextrose (YPD) medium with G418 (45) have been described previously. Yeast cells were grown at 30°C. Yeast transformations were performed according to the methods of Gietz and Schiestl (10). The preparation of bacterial media and general molecular methods have been described previously (25).
Strains and plasmids.
A series of isogenic strains were constructed from the original CG379 (MATα ade5-1 his7-2 leu2-3,112 trp1-289 ura3-52) (22) and pol2-4 and pol3-01 derivatives (40). The pol2-4 and pol3-01 mutations are point mutations in the exonuclease domain of the POL2 and POL3 genes, respectively, resulting in the loss of 3′→5′ proofreading exonuclease activities in the corresponding polymerases (22–24). These strains contain modified insE inserts in the chromosomal LYS2 gene, where the A4 run was changed to A5, A12, or A14 (40). Mutations of the following DNA metabolism genes were introduced into these strains: MMR genes MSH2, MSH3, and MSH6 and the 5′→3′ exonuclease gene EXO1.
Strains containing combinations of pol2-4 lys2::insE-A12, pol2-4 lys2::insE-A14, pol3-01 lys2::insE-A12, and pol3-01 lys2::insE-A14 were constructed by transferring lys2::insE-A12 and lys2::insE-A14 inserts from the Pol+ strains (40) to strains with pol2-4 or pol3-01 mutations by plasmid gap-filling and allele replacement techniques as described previously (40, 41).
A mismatch repair gene tester strain (MATa pms1::LEU2 msh2::ADE2 mlh1::URA3) was derived from our strain 1036 (lys2-BX MATa ade2-1 arg4-8 leu2-3,112 lys2-BX thr1-4 trp1-1 ura3-52 cup1-1) lys2-BX is a deletion of the BamHI-XhoI region covering the insE inserts in the LYS2 gene). The 1036 lys2-BX exo1::kanMX strain was obtained from 1036 (lys2-BX) by deleting the EXO1 gene by PCR disruption with a kanMX cassette (see below).
The replicative pBL304 plasmid containing the POL3 and URA3 markers was constructed by P. Burgers and has been described previously (24).
Mutator screening with a disruption library.
Plasmid DNA of a gene disruption library, kindly provided by M. Snyder (4), was digested with NotI to release yeast genomic DNA fragments containing Tn3::lacZ-LEU2 inserts and transformed into Pol+ lys2::insE-A14 or pol2-4 lys2::insE-A14 strains, and Leu+ transformants were selected. The transforming DNA fragments randomly knock out different nonessential genes through homologous recombination. The Leu+ transformants were replicated to complete medium without lysine in order to identify clones that could yield Lys+ papillae. These potential mutators were verified and crossed with tester strain 1036 (lys2-BX pms1 msh2 mlh1) to identify possible MMR mutators. The nature of the disrupted genes in the remaining clones was determined by using an inverse PCR technique instead of the rescue plasmid technique described by Burns et al. (4). First, genomic DNA from the clones was isolated and digested with the frequently cutting HpaII or TaqI endonucleases. These enzymes will cut inside the Tn3-lacZ-LEU2 insert as well as nearby regions. Digested DNA fragments were circularized by DNA ligation. After circularization, the junction region between the yeast genomic DNA and Tn3-lacZ-LEU2 insert was amplified with the following primers: lacZ, 5′-GCGGGCCTCTTCGCTATTACG-3′, and lacZ-2, 5′-TGAATGGCGAATGGCGCTTTG-3′. The PCR products were sequenced with the lacZ-1 primer, 5′-GTCACGACGTTGTAAAACGACG-3′.
DNA and mutation analysis.
An ABI sequencer was used for DNA sequencing. Mutation rates were determined by a fluctuation test by the method of the median (19) with at least 12 independent cultures. The nature of the Lys+ revertants was identified by sequencing the reversion window of the lys2::insE insert as described previously (41). The DNA regions sequenced between the Tn3-lacZ-LEU2 inserts and the yeast genome were identified by using the S. cerevisiae DNA database (26a), and Swissprot was used to search the protein database (24a).
Gene replacement and disruption.
The following genes were disrupted: MSH2, MSH3, MSH6, and EXO1. For MSH2 disruption we used a SacI-PstI msh2::LEU2 fragment from p203 (39). The BamHI-AatII fragment from pmsh3::LEU2 (29) was used to disrupt the MSH3 gene.
The entire open reading frames of the EXO1 and MSH6 genes were deleted by the PCR disruption technique with the kanMX module (45) and primers described below. Lowercase letters indicate nucleotide sequences that belong to the kanMX cassette; DNA sequences belonging to the genes are written in uppercase. For the EXO1 gene we amplified the kanMX cassette with EXO1-kanMX-3′ (5′-TTGGCTTGACTTAGTAGTTTCGATGTCCCTTTTCTTACTTatcgatgaattcgagctcg-3′) and EXO1-kanMX-5′ (5′-AGGTATGAAGGAGAAGTGTTAGCCATTGATGGCTATGCATcgtacgctgcaggtcgac-3′). For the MSH6 gene the following primers were used: MSH6-kanMX-5′ (5′-CTACCCCTAA AACTTCTAAGACTGCACACTTCGAAAATGGatcgatgaattcgagctcg-3′) and MSH6-kanMX-3′ (5′-GTCCATCTCCGTACGCAATTCGAACGAAATCACTTTGTAAcgtacgctgcaggtcgac-3′). YPD medium with G418 was used for the selection of transformants (45). To verify the disruption of the EXO1 gene we used the following pair of primers for the EXO1 gene: EXO1-test-3 (5′-ATTGGGAAAGCAAGGAGATAG-3′) and EXO1-test-5 (5′-TCTTCTTCCTCAGTTAAAGC-3′). The disruption of the MSH6 gene was verified by PCR with primers MSH6-test-5 (5′-CAGCTACCCCTAAAACTTC-3′) and MSH6-test-3 (5′-TTCCAATCATAGTTCAAGACCCC-3′). The EXO1 gene was also disrupted with the HindIII-KpnI fragment (exo1::URA3) from plasmid p244. The p244 plasmid was constructed by first generating a PCR product of the chromosome EXO1 gene with primers EXO1-test-3 and EXO1-test-5. A BglII-NsiI fragment from the PCR product was cloned into the BamHI-PstI sites of the pUC19 plasmid, resulting in p243. The URA3 gene BglII-BglII fragment from pFL34 (3) was cloned into the BamHI site inside the EXO1 gene of p243, resulting in plasmid p244. To verify the disruption of MSH2 and MSH3 genes, PCR primers and conditions were used as described previously (42).
Tetrad analysis.
The MATα pol3-01 lys2::insE-A4 and MATα pol3-01 lys2::insE-A5 strains were mated with strain 1036 (lys2-BX exo1::kanMX). In the diploid strains the second copy of the EXO1 gene was disrupted with the HindIII-KpnI (exo1::URA3) fragment from p244. The diploid strains were sporulated, and tetrad analysis was performed. The spore colonies with the wild-type POL3 gene were distinguished from pol3-01 mutants by using PCR. Genomic DNA from viable spores was isolated and analyzed by PCR with two primers, p3 (5′-GGAGATACCAAATTACCA-3′) (785-802) and d8 (5′-CTTGTACCATAAGCCTTC-3′) (1512-1495). The PCR product covers the pol3-01 mutation. The PCR products were digested with EcoRV; the pol3-01 mutation lacks an EcoRV site (23).
Construction of homozygous diploid strains.
Haploid leu2 strains were transformed with plasmid YEpHO (a gift from Y. Chernoff) carrying the LEU2 marker and the HO endonuclease gene. The HO endonuclease will induce mating type switching in haploid strains from MATa to MATα or vice versa. Haploid strains with the opposite mating types could form MATa/MATα homozygous diploid strains. Transformants with YEpHO were grown on YPD media to allow loss of the plasmid. Single Leu− clones were isolated. Diploid clones were identified as nonmating with both MATa his3 and MATα his3 testers as well as giving a low forward mutation rate to Canr due to the presence of two CAN1 gene copies. For strains in which the LEU2 marker could not be used we utilized plasmid pGHO-TRP1 (1) containing the TRP1 marker and the HO gene under the GAL1-10 promoter. Mating type switching was induced in galactose media for 8 h, and diploid clones were identified as described above.
Plasmid loss rate measurement.
To establish the requirement for the POL3 gene in strains with pol3-01 msh2 or pol3-01 exo1 combinations, the rate of loss of the plasmid-borne POL3 gene on pBL304 was determined. By using the URA3 marker on pBL304, the plasmid loss rate was determined from the median in a fluctuation test of 10 independent clones on 5-FOA (5-fluoro-orotic acid) media (2). For each fluctuation test, 10 5-FOA-resistant clones were analyzed by PCR to determine the loss of the POL3 gene as described above (see the paragraph on tetrad analysis).
RESULTS
Isolation of mutators for a long A14 homonucleotide run.
The MMR process is conserved in E. coli and eukaryotes. There are several E. coli MutS and MutL homologs that have been identified in yeast (21). While genes involved in mismatch recognition are well characterized, genes involved in later MMR steps such as strand discrimination, mismatch excision, and DNA resynthesis remain to be identified in S. cerevisiae. Previously a lys2::insE-A14 mutation system based on a homonucleotide run of 14 A that is hypersensitive to defects in MMR has been described (40). For example, an msh2 mutant exhibits a 10,000-fold increase in the Lys+ reversion rate over the wild type. Thus, even a relatively small impact of mutators can be detected. Using this sensitive system we tried to identify additional mutators affecting the instability of a long homonucleotide run through a saturation inactivation screen of nonessential genes.
The Pol+ lys2::insE-A14 (S1-A14) or pol2-4 lys2::insE-A14 (S3-A14) mutator detection strain containing the homonucleotide run A14 in the LYS2 gene was transformed by a gene disruption library (4), and colonies that exhibited higher Lys+ reversion rates were identified. As shown below, the pol2-4 background increases the magnitude of a mutator effect. Among more than 100,000 transformants of the Pol+ strain and 50,000 transformants of the pol2-4 mutant, 44 and 25, respectively, exhibited markedly increased Lys+ reversion frequencies. Of these hypermutable transformants, 34 and 19, respectively, carried mutations in the MMR gene PMS1, MSH2, or MLH1. The frequency of repeat mutants indicates that we have efficiently inactivated most nonessential genes that were likely to lead to an increase of the mutation rate in a sensitive system. Included in the remaining 10 hypermutable transformants of the Pol+ strain were 5 exo1-dhs1 mutants (ORF-YOR033C; the DHS1 gene also has been referred to as EXO1 [38]). The Tn3::lacZ-LEU2 inserts were at nucleotide positions 13, 104, 131, 150, and 163 from the EXO1 start codon. Among six hypermutable transformants of pol2-4, two were exo1 mutants and three were msh6 mutants.
To establish that the increased reversion rate of the lys2::insE-A14 allele in the exo1 mutant strains was due to the disruption of the EXO1 gene and not to additional mutations elsewhere in the genome, the EXO1 gene was deleted from the S1-14A strain by using a kanMX cassette in combination with PCR (reference 45; see Materials and Methods). The disruption of EXO1 led to a 100-fold increase in the Lys+ reversion rate that was due to mutations in the A14 run (Table 1). In the six mutator clones lacking alterations in EXO1 or MMR genes (five from the Pol+ strain and one from the pol2-4 strain), disruptions were identified in the OXA1, CSD3, PMR2, ENA2, and 25S rRNA genes. Since the direct disruption of these genes (except 25S rRNA) in the S1-14A strain did not lead to a mutator phenotype, the mutator phenotype of these transformant clones is likely to be due to secondary mutator mutations elsewhere in the genome.
TABLE 1.
Relative increases in mutation rates in homonucleotide runs (A4, A5, A12, and A14 of lys2::insE alleles) and CAN1
| Strain genotype | Relative rate of mutationa (mutants vs wild type)b
|
||||
|---|---|---|---|---|---|
| A4 (−1) | A14 (−1) | A5 (+1) | A12 (+1) | Canr | |
| Wild type | |||||
| Absolute (109) | 0.4 | 186 | 1.1 | 140 | 240 |
| Relative | 1 (4/55)c | 1 (10/10)c | 1 (7/21)c | 1 (8/8)c | 1 |
| exo1 | 2.5 (4/34) | 97 (8/8) | 2 (19/29) | 2 (5/5) | 6.5 |
| msh2 | 77.5 (20/30)c | 9,460 (8/8) | 35 (16/30)c | 1,240 (8/8) | 27.4 |
| exo1 msh2 | 90 (14/22) | 8,980 (6/6) | 47 (16/21) | 544 (5/5) | 32.2 |
| pol2-4 | 6 (7/20)c | 2 (10/10) | 7 (13/27)c | 1 (10/10) | 2.3 |
| pol2-4 exo1 | 278 (29/38) | 925 (6/6) | 769 (33/35) | 116 (6/6) | 37 |
| pol2-4 msh2 | 2,870 (26/29)c | 23,390 (10/10) | 10,820 (30/30) | 2,040 (9/9) | 475 |
| pol2-4 exo1 msh2 | NDd | 16,666 | ND | 3,150 | 729 |
| msh3 | ND | 6 (6/6) | ND | 2 (6/6) | 2.0 |
| pol2-4 msh3 | ND | 15 (6/6) | ND | 2 (6/6) | 4.3 |
| msh6 | ND | 189 (7/7) | ND | 32 (7/7) | 9.2 |
| pol2-4 msh6 | ND | 317 (7/7) | ND | 60 (7/7) | 480 |
| msh3 msh6 | ND | 18,820 (6/6) | ND | 1,670 (6/6) | 29.2 |
| pol2-4 msh3 msh6 | ND | 46,450 (6/6) | ND | 3,170 (6/6) | 1,440 |
| pol3-01 | 37.5 (4/37)c | 54 (10/10) | 93 (19/22)c | 20 (10/10) | 44.2 |
The mutation rate in the run (Rr) was calculated as follows: Rr = Rt (Nr/Nt) where Rt is the total rate of reversion, which was determined from the median in a fluctuation test of more than 12 independent cultures (19). The fold increase in the mutation rate was determined as the ratio of the mutation rate of the mutant strain to the mutation rate of the wild-type strain. The nature of the Canr mutants was not determined. The Canr mutation rate was determined from the median in a fluctuation test of more than 12 independent cultures.
The ratio of the number of revertants with a frameshift mutation in the run (Nr) to the total number of revertants (Nt) analyzed by sequencing is given in parentheses. As observed previously (40) all the frameshift mutations in the A4 and A14 runs that were analyzed were due to −1 nucleotide deletions; all frameshift mutations in the A5 and A12 runs were due to +1 nucleotide insertions.
Data from the work of Tran et al. (40), with permission.
ND, not determined.
Based on the number of repeated isolations of different mutants we propose that we have nearly exhausted the number of single mutants that can lead to small (see below) as well as large increases in the mutation rate in a long homonucleotide run. It is possible that additional mutants might be lost in this screen if growth defects reduce the likelihood of detection and/or if the genes have only small open reading frames so that there is less likelihood of disruption. (These reasons could account for our lack of detection of a rad27 mutant in this screen although it is known to be a mutator [13]. The role of RAD27 in MMR and interactions between rad27, MMR, and polymerase mutants will be presented elsewhere.)
Mutator activity of an exo1 mutant.
Based on genetic and in vitro observations, exonucleases play an important role in mutation avoidance in E. coli (5, 18, 44). We, therefore, examined the role of EXO1 by using several assays that reveal −1 and +1 frameshift mutations in or near short (A4 and A5) and long (A14 and A12) homonucleotide runs within the lys2::insE sequence (40). Forward mutation to canavanine resistance was also examined in these strains (see reference 30). Many of the lys2::insE-A4 and lys2::insE-A5 revertants were not due to frameshift mutations in homonucleotide A4 and A5 runs, respectively (Table 1). Instead, most mutations were frameshifts located elsewhere in the reversion window (i.e., the small region where mutations occur; see references 40 and 42). As expected, all lys2::insE-A14 and lys2::insE-A12 revertants were due to −1 and +1 nucleotide frameshift mutations in the A14 and A12 runs, respectively (Table 1). Forward mutation rates in the CAN1 gene were determined, but Canr mutations were not sequenced.
Inactivation of the EXO1 gene led to only a weak mutator phenotype for the A4 and A5 homonucleotide runs and a sevenfold increase in the appearance of Canr forward mutations. However, we observed a strong effect (a 100-fold increase) on a long homonucleotide A14 run that was specific for −1 frameshift mutations; there was only a twofold increase in +1 insertions in the A12 run (Table 1). The preference of the mutator effect for deletions over insertions is characteristic of defects in MMR genes such as MSH2, MSH3, and MSH6 (references 31 and 33; also see Table 1). The pattern of a relatively weak effect on short runs and a strong effect on long runs is typical for an msh2 defect (reference 40 and Table 1). These observations led us to determine the consequences of combining these two mutations (i.e., exo1 and msh2). Since there was no synergy for the combined exo1 msh2 mutants as compared to msh2, it appears either that msh2 is epistatic to exo1 or that their effects are additive (Table 1). These modes of interaction cannot be distinguished because of the relatively small effect of the exo1 mutant on mutation rates.
Mutator phenotype resulting from a lack of DNA 3′→5′ Polɛ proofreading and 5′→3′ Exo1 exonucleases.
The mutator phenotype of the Exo1 defects suggests that this protein might be involved in MMR. Since DNA polymerase proofreading and postreplication MMR act in series, we examined the mutator phenotype of double mutants defective in DNA Polɛ proofreading (pol2-4) and the 5′→3′ exonuclease Exo1. The double mutant lacking proofreading and Exo1 activities exhibited a synergistic mutator effect for all mutation rates examined (Tables 1 and 2). The pol2-4 mutant is a weak mutator (less than a sevenfold increase over the wild type; Table 1). In the pol2-4 exo1 double mutant, mutation rates were increased by as much as 10- to 100-fold over those for either single mutant (Tables 1 and 2).
TABLE 2.
Relative mutation rates in short and long homonucleotide runs and the CAN1 gene in various pol2-4 mutants
| Strain genotype comparison | Relative mutation rate
|
||||
|---|---|---|---|---|---|
| A4 (−1) | A14 (−1) | A5 (+1) | A12 (+1) | Canr | |
| pol2-4 vs wild type | 6 | 2.2 | 7 | 1 | 2.3 |
| exo1 pol2-4 vs exo1 | 111 | 10 | 385 | 55 | 5.7 |
| msh2 pol2-4 vs msh2 | 37 | 2.5 | 309 | 1.6a | 17 |
| msh3 pol2-4 vs msh3 | NDb | 2.6 | ND | 1.1a | 2.2 |
| msh6 pol2-4 vs msh6 | ND | 1.7 | ND | 1.9 | 52 |
| msh3 msh6 pol2-4 vs msh3 msh6 | ND | 1 | ND | 1 | 49 |
No significant difference; the 95% confidence intervals for the mutation rate in these two strains overlap.
ND, not determined.
Previously, the 3′→5′ proofreading exonuclease activity of DNA Polɛ had been shown to have almost no effect on frameshift mutations in long homonucleotide runs (≥8 nucleotides) during replication (40). The lack of proofreading in long homonucleotide runs accounts for the absence of a multiplicative mutator effect when the proofreading exonuclease mutant is combined with a complete (msh2) or partial (msh3 and msh6) defect in the MMR system (Tables 1 and 2). The synergistic effect of exo1 with the pol2-4 defect for mutations in long homonucleotide runs suggests that the Polɛ-exonuclease can prevent errors by a process other than replication proofreading (see Discussion).
Mutation in DNA Polδ proofreading and Exo1 or Msh2 is lethal.
The synergism of mutations in exo1 with pol2-4 led us to examine the double mutant phenotype for exo1 with pol3-01, a 3′→5′ exonuclease deficient mutation of DNA Polδ. The pol3-01 mutant exhibited a much stronger mutator effect than the pol2-4 strain in all assays examined (Table 1). We failed to obtain an exo1 pol3-01 double mutant by the deletion of the EXO1 gene in a pol3-01 strain. To analyze the apparent synthetic lethality of the double mutant, we crossed a pol3-01 and an exo1 mutant (see Materials and Methods) and subsequently inactivated the second copy of the EXO1 gene with an exo1::URA3 DNA fragment. Among 14 tetrads analyzed, all segregated as 2 viable:2 nonviable. Microscopic analysis after 4 days revealed that the two spores that did not yield colonies underwent several divisions, resulting in microcolonies containing approximately 100 cells. Using PCR and restriction analysis (see Materials and Methods), we established that the viable spores contained the wild-type copy of the POL3 gene. Thus, exo1 pol3-01 mutants are inviable.
We transformed the pol3-01 lys2::insE-A14 strain with plasmid pBL304 (24) carrying the POL3 and the URA3 genes. The POL3 gene on the plasmid suppresses the mutator phenotype of the chromosomal pol3-01 mutation (data not shown). Subsequently the EXO1 gene was deleted from the pol3-01 strain carrying the pBL304 plasmid to assess the compatibility of pol3-01 and exo1. Since the pol3-01 mutation was previously shown to be lethal in combination with the MMR pms1 mutant (23), we also deleted MSH2 from the pol3-01 pBL304 background. While the pol3-01 pBL304 strain lost the plasmid at a high rate when measured on 5-FOA medium, which selects against the Ura+ phenotype (∼10−2 loss events per cell/generation), the pol3-01 msh2 pBL304 or pol3-01 exo1 pBL304 strain demonstrated an undetectable level of plasmid loss (5-FOAr colonies formed at a rate lower than 10−5 loss events per cell/generation). Based on PCR analysis, the 5-FOAr clones derived from the pol3-01 msh2 pBL304 or pol3-01 exo1 pBL304 strain still retain the wild-type copy of the POL3 gene. Thus, the pol3-01 msh2 and pol3-01 exo1 haploid double mutants are inviable.
Viability of msh2 pol3-01 and exo1 pol3-01 diploids.
A likely explanation for the inviability of the pol3-01 msh2 and pol3-01 exo1 haploid strains is that unedited replication errors generated by the proofreading-deficient DNA Polδ are lethal to a haploid cell. If this were the case, then diploid strains might be viable due to the fact that most mutations are recessive. We constructed diploid strains that contained the pBL304 plasmid and had the following homozygous mutations: pol3-01/pol3-01 msh2/msh2 and pol3-01/pol3-01 exo1/exo1 (see Materials and Methods). The plasmid loss rates in the diploid strains were measured on 5-FOA media. Unlike the haploid strain, all diploid strains were able to lose the plasmid rapidly (3 × 10−3 loss events/cell/generation).
Synergistic interaction between a DNA Polδ 3′→5′ proofreading defect and exo1 or msh2 mutations in diploid strains.
The genetic interaction between pol3-01 and msh2 or exo1 in diploid strains was studied by examining the reversion of homozygous lys2::insE-A14 and his7-2 mutations. (While the his7-2 molecular defect is not known, it has been used in several studies investigating proofreading and MMR defects [22, 23, 24].) As shown in Table 3, the pol3-01/pol3-01 msh2/msh2 double mutant exhibits a His+ reversion rate 47-fold higher than that of the pol3-01/pol3-01 strain and 250-fold higher than that of the msh2/msh2 diploid. The rate of Lys+ reversion in the A14 homonucleotide run in the pol3-01/pol3-01 msh2/msh2 strain is similar to that in an msh2/msh2 strain (Table 3). The data are consistent with previous suggestions based on in vitro and in vivo data (17, 40) that DNA polymerase exonucleolytic proofreading becomes ineffective during replication of long repetitive sequences (e.g., ≥8 nucleotides in a homonucleotide run). The strong synergistic interaction between exo1 and pol3-01 was observed for Lys+ as well as His+ reversion (Table 3); the reversion rates were multiplicative for the double versus single mutants. The rate of Lys+ reversion for the A14 homonucleotide run in the pol3-01/pol3-01 exo1/exo1 strain was 23-fold higher than the rate in the pol3-01/pol3-01 strain and 120-fold higher than the rate in an exo1/exo1 strain. The reversion rates to His+ and Lys+ in the pol3-01/pol3-01 exo1/exo1 strain are approximately the same as those found in the pol3-01/pol3-01 msh2/msh2 double mutant (Table 3). In contrast, in a Pol+ strain the exo1 deficiency increases the mutation rate in the A14 run 100-fold less than an msh2 deficiency (Table 1). Since DNA Polδ exonucleolytic proofreading is inefficient during replication of a long homonucleotide run (A14), the synergistic effect between exo1 and pol3-01 for mutations in long homonucleotide runs suggests that the Polδ-exonuclease can prevent errors by processes other than replication proofreading.
TABLE 3.
Spontaneous mutation rates in homozygous diploid strains measured as the rate of reversion of his7-2 or lys2::insE-A14 to His+ or Lys+, respectively
| Strain genotype | Relative reversion ratea
|
|
|---|---|---|
| his7-2 | lys2::insE-A14 | |
| POL+/POL+ | ||
| Absolute (109) | 13 | 600 |
| Relative | 1 | 1 |
| pol3-01/pol3-01 | 454 | 200 |
| exo1/exo1 | 10 | 38 |
| msh2/msh2 | 85 | 6,000 |
| pol3-01/pol3-01 exo1/exo1 | 8,460 | 4,500 |
| pol3-01/pol3-01 msh2/msh2 | 21,310 | 5,670 |
Fold increase over the wild type.
DISCUSSION
We have investigated mutator effects due to the inactivation of the 5′→3′ Exo1 and 3′→5′ DNA polymerase exonuclease activities in yeast. To investigate possible genetic networks of mutation avoidance, we combined exo1 with mutations in MMR genes and/or deficiencies in the 3′→5′ proofreading activities of the DNA polymerase genes. Based on the phenotypes of the mutants and double or triple mutants, we propose that the exonuclease activities of Exo1, Polδ, and Polɛ participate directly in MMR.
Participation of the Exo1 5′→3′ exonuclease in MMR.
It has been proposed that Exo1 may function in MMR (38). The exo1 defect caused a nearly 100-fold increase in −1 frameshift mutations within the A14 homonucleotide run but had a smaller effect in other assays (two- to threefold increases for A4, A5, and A12 runs and a 6.5-fold increase for the Canr forward mutation; see Table 1). In all cases, the mutation rate in the exo1 msh2 mutant was comparable to the rate in the msh2 mutant (Table 1 and reference 38). This suggests that msh2 is epistatic to exo1 and that Exo1 is involved in the MSH2-dependent MMR pathway. (However, separate pathways resulting in the additivity of mutation rates cannot be excluded because the single exo1 mutant exhibits a low mutation rate.)
The conclusion that Exo1 and Msh2 are epistatic is consistent with the following properties of the exo1 mutant: (i) Msh2 interacts physically with the C-terminal region of Exo1 (38); (ii) defects in MMR or in Exo1 lead to a stronger mutator effect in long versus short homonucleotide runs (Table 1; references 11 and 40); and (iii) proofreading interacts with the MMR system based on the synergy between the double mutant pol2-4 pms1 (24) and pol2-4 msh2 (40) strains. The observed synergy between exo1 and the proofreading defect (pol2-4) for all mutation detection systems examined (Table 1) is also consistent with a role for Exo1 in MMR. In addition, as reported previously for pms1 (23) and for msh2 (this work), the combination of exo1 with the DNA Polδ 3′→5′ proofreading exonuclease defect (pol3-01) results in lethality for haploid strains, which is likely due to excessive errors. Diploid pol3-01 msh2 and pol3-01 exo1 strains are viable and exhibit synergistic mutator effects (Table 3).
Participation of 3′→5′ exonucleases of Polɛ and Polδ as well as the 5′→3′ exonuclease Exo1 in error avoidance.
Proofreading decreases with the increased size of the homonucleotide run that is replicated (17, 40). Previously it has been shown that both MMR and DNA Polɛ proofreading are efficient at preventing mutations in short runs (synergy had been shown for pol2-4 and pms1 or msh2 double mutants [24, 40], while only MMR prevents frameshift mutations in homonucleotide runs larger than 7 nucleotides [40]). It has been proposed that frameshift intermediates generated in long runs escape Polɛ proofreading but are correctable by MMR. This would explain the lack of synergy for mutations in long homonucleotide runs when proofreading (pol2-4) and msh2, msh3, or msh6 mutants are combined (Table 2 and reference 40).
Unlike the situation with the double mutant pol2-4 msh2, pol2-4 msh3, and pol2-4 msh6 strains, synergy was clearly observed for mutations in long homonucleotide runs when pol2-4 was combined with an exo1 mutation (Table 2). The combination of pol2-4 and exo1 led to an increase in the mutation rate of up to 55-fold over that found for either single mutant, consistent with the finding that MMR in eukaryotes may be bidirectional (7, 37). Since Polɛ-exonuclease proofreading is absent in long homonucleotide runs, we propose that this synergy is a manifestation of both gene products (Exo1 and Polɛ) participating in error avoidance. While other mechanisms might be involved we suggest that the Exo1 and Polɛ exonucleases function in the MMR of long homonucleotide runs and compete for substrates when mismatch recognition components are present (i.e., Msh2, Msh3, and Msh6). Another possibility is that the MMR system is saturated in a pol2-4 mutant due to the accumulation of mismatches. Thus, a partial defect in MMR due to loss of Exo1 might lead to a synergistic increase in the instability of long homonucleotide runs. However, this contradicts the lack of synergy for mutation of long homonucleotide runs in double mutants between pol2-4 and either msh3 or msh6, which are partially defective in MMR (Table 2).
The proposed role for the Exo1 and Polɛ exonucleases could occur if the two types of exonucleases participate in the excision of mismatches in a manner similar to that found for the RecJ and ExoVII 5′→3′ exonucleases and the Exo1 3′→5′ exonuclease in E. coli MMR (21). However, in E. coli the recJ xseA double mutant defective in RecJ and ExoVII is not hypermutable, implying that the system functions efficiently even when restricted to a unidirectional mode of action (5). Recently, Viswanathan and Lovett (44) reported that frameshift mutations were stimulated in a RecJ− Exo1− ExoVII− triple mutant, although base substitution mutations were not increased in several assays. This mutator effect was primarily due to a synergistic interaction between the Exo1− and ExoVII− mutations.
Based on in vitro data, Longley et al. (20) have suggested that DNA Polδ participates in MMR at the resynthesis step. Our genetic data suggest that DNA Polδ proofreading exonuclease may also be directly involved in the excision step of MMR. Similar to the DNA Polɛ proofreading exonuclease, DNA Polδ-exonuclease also appears inefficient during replication of the homonucleotide run A14, so there is no synergistic interaction between proofreading defects and msh2 (Tables 2 and 3). While the haploid pol3-01 exo1 strain is inviable, the homozygous diploid strain is viable and exhibits a strong synergistic mutator interaction both for the instability of the A14 run and the reversion of his7-2 (Table 3). The synergism between exo1 and pol3-01 for mutations in the A14 run suggests that the Polδ-exonuclease prevents errors by processes other than proofreading. Since Exo1 is likely to be involved in MMR (reference 38 and this work) we propose that the mutator synergy between exo1 and pol3-01 is due to these exonucleases being able to act on the same substrate. Therefore, an essential step in MMR (such as removal of the mismatch) could not occur if both exonuclease functions were inactivated. This view is supported by the observation that the rate of reversion to Lys+ and His+ in the pol3-01/pol3-01 exo1/exo1 double mutant is comparable to the rate in the pol3-01/pol3-01 msh2/msh2 double mutant (Table 3).
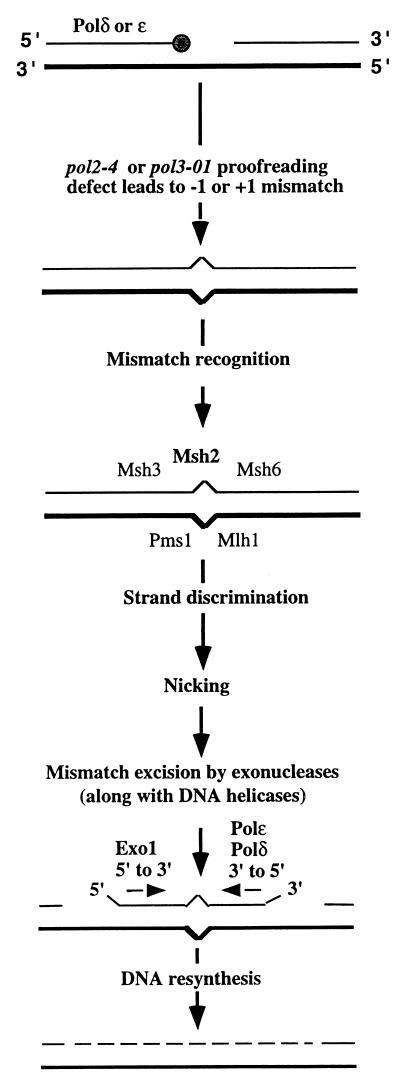
Presented in Fig. 1 is a model describing how Exo1 along with DNA Polδ and/or DNA Polɛ could act to reduce frameshift mutations in homonucleotide runs. The DNA Polδ and Polɛ 3′→5′ proofreading exonuclease reduces the incidence of frameshift intermediates at the time of replication. Frameshift intermediates that escape proofreading are subsequently corrected by MMR. The synergy between the pol2-4 (or pol3-01) and the exo1 mutation in short homonucleotide runs (or in the his7-2 reversion assay) can be explained by the participation of Exo1 in the removal of mismatches generated during replication by the proofreading-deficient Polɛ- or Polδ-exonuclease (i.e., acting in series with proofreading). Although the proofreading exonucleases of Polδ and Polɛ have no function in long homonucleotide runs, we propose that they function along with the 5′→3′ exonuclease of Exo1 to excise mismatches in the MMR pathway. The degradation process would be facilitated by DNA helicases, since Exo1, Polɛ, and Polδ nucleases degrade single-stranded DNA efficiently (9, 22, 32). (In an E. coli reconstituted MMR system, both a DNA helicase and exonucleases are required for mismatch removal [5, 18].) In the framework of this model, the stronger synergy of the pol2-4 or the pol3-01 mutation with the msh2 defect in MMR for mutations in short as compared to long homonucleotide runs can be due to pol2-4 or pol3-01 affecting both proofreading and MMR in short homonucleotide runs, but only MMR in the longer runs.
FIG. 1.
Model of interaction between 3′→5′ and 5′→3′ nucleases in the appearance and prevention of frameshift mutations. The small shaded circle represents DNA polymerase. As discussed in the text, frameshift intermediates escape proofreading in long homonucleotide runs. The DNA Polɛ and DNA Polδ 3′→5′ exonucleases as well as the Exo1 5′→3′ exonuclease, in conjunction with a DNA helicase similar to the helicase involved in E. coli MMR (5, 18), could participate in mismatch removal in opposite directions, so that multiple enzyme deficiencies cause synergistic mutator effects for frameshift mutations.
The importance of the combination of Polδ 3′→5′ exonuclease with Exo1 in error prevention is apparent for the long homonucleotide run and provides strong support for our model, in which these nucleases are involved in a late step of strand-specific mismatch removal. For the long homonucleotide runs A12 and A14, in which Polɛ-exonuclease proofreading is inefficient, a defect in Exo1, Polɛ-exonuclease, or Polδ-exonuclease alone results in a mutation rate that is <1% of that for an msh2 strain. The pol2-4 exo1 double mutant exhibits mutation rates that are 10% of those seen with the pol2-4 msh2 strains (Table 1). However the combination of pol3-01 with exo1 in a diploid strain (the haploid strain is inviable) has an even more profound effect, resulting in mutation rates that are comparable to the most mutation-prone genetic combination, pol3-01 msh2 (mutation rates of the pol3-01 exo1 mutant are from 40 to 80% of those seen in the pol3-01 msh2 mutant; Table 3). Assuming that their impact on the mutation rate is directly due to a role in MMR, our genetic results suggest that Polδ-exonuclease and Exo1 may be responsible for much of the mismatch excision during MMR. The synergistic interaction between Polδ-exonuclease and Exo1 in MMR is similar to that found for Msh3 and Msh6. While the single mutant msh3 and msh6 strains have a small effect on the mutation rate, the msh3 msh6 double mutant exhibits a mutator phenotype comparable to that of the msh2 mutant (Table 1). We therefore suggest that Exo1 and Polδ-exonucleases are major exonucleases involved in the mismatch excision step.
Formally, it is possible that there are two separate pathways (polymerase exonucleases plus Exo1 versus Msh2) of error avoidance, so that defects in both pathways might lead to additive increases in mutation rates. Because of the observed high error rates, it would be difficult to distinguish epistasis (same pathway) from additivity (separate pathways). Consistent with a single pathway is the fact that the Lys+ frameshift reversion spectra in the msh2, msh2 pol2-4, and exo1 pol2-4 strains are similar in that they exhibit hotspots in the A4 and A5 homonucleotide runs of the lys2::insE-A4 and lys2::insE-A5 alleles, respectively (Table 1).
Roles of 5′→3′ and 3′→5′ nucleases in DNA metabolism.
We and others have found that several combinations of nuclease mutations can lead to lethality for haploid yeast cells (Table 4). The inviability of a double mutant has been attributed to a high mutation rate resulting in error catastrophe (8, 23, 24). In E. coli the dnaQ926 mutation appears to cause lethality as a result of the loss of proofreading and the subsequent saturation of DNA MMR. Consistent with this, dnaQ926 strains are viable if they carry a dnaE antimutator allele or a multicopy plasmid with the E. coli mutL gene (8).
TABLE 4.
Effects on viability and mutation rates in haploid strains that are double mutants for deficiency in nuclease and/or MMR activities
| Mutation | Effect on viability and mutation rates when combined with:
|
||
|---|---|---|---|
| msh2 | pol2-4 | pol3-01 | |
| exo1 | Epistasis | Synergy | Inviablea |
| pol3-01 | Inviablebc | Inviabled | |
| pol2-4 | Synergyc | ||
The pol3-01 exo1 haploid double mutant is not viable. The diploid strain exhibits a synergistic hypermutable phenotype (Table 3).
The pol3-01 mutant is also inviable in combination with a strain with another MMR defect, pms1; the pol3-01 pms1 (23) and pol3-01 msh2 diploid strains are viable and exhibit a synergistic hypermutable phenotype (Table 3).
The synergistic interactions between msh2 and proofreading defects (pol2-4 and pol3-01) are observed only for short runs and base substitutions but not in the long homonucleotide runs A12 and A14 for pol2-4 (Table 2) or in the A14 run for pol3-01 (Table 3).
Morrison and Sugino (24). The pol2-4 pol3-01 haploid double mutant is not viable. The diploid strain exhibits a synergistic hypermutable phenotype.
The pol2-4 exo1 double mutant is viable and exhibits a synergistic increase in mutation rate relative to the single mutants. The pol3-01 msh2 and pol3-01 exo1 haploid strains are inviable. However, pol3-01 msh2 and pol3-01 exo1 double mutants are viable as diploid strains. The inviability of the haploid double mutant pol3-01 msh2 and pol3-01 exo1 strains is most likely due to an accumulation of excessive replication errors based on the hypermutability of the homozygous diploid strains (Table 3).
Our results suggest that both DNA Polɛ-exonuclease and DNA Polδ-exonuclease participate in the removal of mismatches during MMR and that DNA Polδ is associated with MMR (20). It is interesting that both DNA Polδ and DNA Polɛ interact with PCNA (6), which is also involved in MMR (12, 43). DNA Polδ and DNA Polɛ might be recruited to the MMR complex by an interaction with PCNA or with other MMR proteins. Thus, mutations in the putative MMR protein interaction domains of DNA Polɛ and DNA Polδ could lead to a defect in the excision step of MMR. Such mutations might exhibit phenotypes similar to pol2-4 and pol3-01 in combination with exo1 (i.e., instability of long homonucleotide runs but normal proofreading). Recently we identified such a mutation in the POL2 gene located between the polymerase and checkpoint domains (14a). A search for similar mutations in the POL3 gene is in progress.
Previously, it was reported that the pol2-4 pol3-01 haploid double mutant was also inviable, but diploid double mutants are viable and exhibit a mutator synergy (24). It was proposed that even if MMR is functional, a complete deficiency in proofreading of both polymerases can lead to high mutation rates, resulting in error catastrophe. As an alternative, we propose that the double defect in pol2-4 pol3-01 leads not only to a loss of proofreading but also to a defect in MMR; this reflects the participation of polymerase-associated exonucleases in MMR as described above (Fig. 1).
As summarized in Table 4, this and other studies demonstrate that while defects in the individual Exo1, DNA Polδ, and DNA Polɛ nucleases may have a small effect, the combination of mutations can have a profound impact on genome stability. Single mutations in these exonucleases have only a moderate mutator phenotype in comparison with a mutation in MSH2. We have shown that double mutations in each pair of these three nuclease genes can lead to strong, synergistic mutator effects or lethality for haploid yeast. The deletion of another 5′→3′ exonuclease gene, RAD27, in combination with pol3-01 or exo1 mutations results in lethality (15, 38). In recent studies with this flap endonuclease, we have shown that it also exhibits synergy with either pol2-4 or msh2, suggesting a possible role in mismatch excision (unpublished data).
Synergistic interactions affecting genome instability, such as those described in this work, have important implications for the human genome, in which genetic polymorphisms are common. Given the close relationship between yeast and human genes involved in replication and repair, particularly MMR, similar relationships may apply to the equivalent pathways in human cells. For example, synergistic effects might be expected for partially defective gene products that alone cause a weak phenotype. These alleles would then cause a much more serious deficiency and phenotype when combined with other weakly defective proteins that act in overlapping genetic pathways.
ACKNOWLEDGMENTS
We thank M. Snyder for providing the disruption library, Y. Chernoff for the YEpHO plasmid, and J. Westmoreland and O. Kozyreva for help with experiments. We thank S. Bennett, J. Drake, R. Schaaper, and M. Sanders for helpful comments on the manuscript.
REFERENCES
- 1.Bennett C, Lewis A, Baldwin K, Resnick M. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc Natl Acad Sci USA. 1993;90:5613–5617. doi: 10.1073/pnas.90.12.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 3.Bonneaud N, Ozier K O, Li G Y, Labouesse M, Minvielle S L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 4.Burns N, Grimwade B, Ross M P, Choi E Y, Finberg K, Roeder G S, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 5.Cooper D L, Lahue R S, Modrich P. Methyl-directed mismatch repair is bidirectional. J Biol Chem. 1993;268:11823–11829. [PubMed] [Google Scholar]
- 6.Eissenberg J C, Ayyagari R, Gomes X V, Burgers P M. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase δ and DNA polymerase ɛ. Mol Cell Biol. 1997;17:6367–6378. doi: 10.1128/mcb.17.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang W H, Modrich P. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J Biol Chem. 1993;268:11838–11844. [PubMed] [Google Scholar]
- 8.Fijalkowska I J, Schaaper R M. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiorentini P, Huang K N, Tishkoff D X, Kolodner R D, Symington L S. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz R D, Schiestl R H. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 11.Greene C N, Jinks R S. Frameshift intermediates in homopolymer runs are removed efficiently by yeast mismatch repair proteins. Mol Cell Biol. 1997;17:2844–2850. doi: 10.1128/mcb.17.5.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson R E, Kovvali G K, Guzder S N, Amin N S, Holm C, Habraken Y, Sung P, Prakash L, Prakash S. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J Biol Chem. 1996;271:27987–27990. doi: 10.1074/jbc.271.45.27987. [DOI] [PubMed] [Google Scholar]
- 13.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 14.Kesti T, Syvaoja J E. Identification and tryptic cleavage of the catalytic core of HeLa and calf thymus DNA polymerase ɛ. J Biol Chem. 1991;266:6336–6341. [PubMed] [Google Scholar]
- 14a.Kirchner, J., H. T. Tran, and M. A. Resnick. Unpublished data.
- 15.Kokoska R J, Stefanovic L, Tran H T, Resnick M A, Gordenin D A, Petes T D. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase δ (pol3-t) Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kornberg A, Baker T. DNA replication. 2nd ed. New York, N.Y: Freeman; 1992. [Google Scholar]
- 17.Kroutil L C, Register K, Bebenek K, Kunkel T A. Exonucleolytic proofreading during replication of repetitive DNA. Biochemistry. 1996;35:1046–1053. doi: 10.1021/bi952178h. [DOI] [PubMed] [Google Scholar]
- 18.Lahue R S, Au K G, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 19.Lea D E, Coulson C A. The distribution of the number of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 20.Longley M J, Pierce A J, Modrich P. DNA polymerase δ is required for human mismatch repair in vitro. J Biol Chem. 1997;272:10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 21.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 22.Morrison A, Bell J B, Kunkel T A, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′ to 5′ exonuclease activity. Proc Natl Acad Sci USA. 1991;88:9473–9477. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison A, Johnston A L, Johnston L H, Sugino A. Pathway correcting DNA replication errors in S. cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison A, Sugino A. The 3′ to 5′ exonucleases of both DNA polymerase δ and ɛ participate in correcting errors of DNA replication in S. cerevisiae. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 24a.Protein Data Base. 1998, copyright date. [Online.] http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-blast?Jform=1. [13 January 1999, last date accessed.]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Saparbaev M, Prakash L, Prakash S. Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics. 1996;142:727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.S. cerevisiae DNA Data Base. 1998, copyright date. [Online.] http://genome-www2.stanford.edu/cgi-bin/SGD/nph-blast2sgd. [13 January 1999, last date accessed.]
- 27.Schaaper R M. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J Biol Chem. 1993;268:23762–23765. [PubMed] [Google Scholar]
- 28.Schaaper R M. Mechanisms of mutagenesis in the Escherichia coli mutator mutD5: role of DNA mismatch repair. Proc Natl Acad Sci USA. 1988;85:8126–8130. doi: 10.1073/pnas.85.21.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selva E M, New L, Crouse G F, Lahue R S. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherman F, Fink G F, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 31.Sia E A, Kokoska R J, Dominska M, Greenwell P, Petes T D. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon M, Giot L, Faye G. The 3′ to 5′ exonuclease activity located in the DNA polymerase δ subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991;10:2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strand M, Earley M C, Crouse G F, Petes T D. Mutations in the MSH3 gene preferentially lead to deletions within tracts of simple repetitive DNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:10418–10421. doi: 10.1073/pnas.92.22.10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugawara N, Paques F, Colaiacovo M, Haber J E. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugino A. Yeast DNA polymerases and their role at the replication fork. Trends Biochem Sci. 1995;20:319–323. doi: 10.1016/s0968-0004(00)89059-3. [DOI] [PubMed] [Google Scholar]
- 36.Szankasi P, Smith G R. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 37.Thomas D C, Roberts J D, Kunkel T A. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- 38.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, Kane M F, Kolodner R D. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran H, Degtyareva N, Gordenin D, Resnick M A. Altered replication and inverted repeats induce mismatch repair-independent recombination between highly diverged DNAs in yeast. Mol Cell Biol. 1997;17:1027–1036. doi: 10.1128/mcb.17.2.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran H T, Keen J D, Kricker M, Resnick M A, Gordenin D A. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran T H, Degtyareva N P, Koloteva N N, Sugino A, Masumoto H, Gordenin D A, Resnick M A. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol Cell Biol. 1995;15:5607–5617. doi: 10.1128/mcb.15.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran T H, Gordenin D, Resnick M. The prevention of repeat-associated deletions in Saccharomyces cerevisiae by mismatch repair depends on size and origin of deletions. Genetics. 1996;143:1579–1587. doi: 10.1093/genetics/143.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 44.Viswanathan M, Lovett S. Single-strand DNA-specific exonucleases in Escherichia coli: roles in repair and mutation avoidance. Genetics. 1998;149:7–16. doi: 10.1093/genetics/149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]