Abstract
Metabolic diseases caused by disorders in amino acids, glucose, lipid metabolism, and other metabolic risk factors show high incidences in young people, and current treatments are ineffective. N6-methyladenosine (m6A) RNA modification is a post-transcriptional regulation of gene expression with several effects on physiological processes and biological functions. Recent studies report that m6A RNA modification is involved in various metabolic pathways and development of common metabolic diseases, making it a potential disease-specific therapeutic target. This review explores components, mechanisms, and research methods of m6A RNA modification. In addition, we summarize the progress of research on m6A RNA modification in metabolism-related human diseases, including diabetes, obesity, non-alcoholic fatty liver disease, osteoporosis, and cancer. Furthermore, opportunities and the challenges facing basic research and clinical application of m6A RNA modification in metabolism-related human diseases are discussed. This review is meant to enhance our understanding of the molecular mechanisms, research methods, and clinical significance of m6A RNA modification in metabolism-related human diseases.
Keywords: m6A RNA modification, metabolic disease, glucose metabolism, lipid metabolism, amino acid metabolism
Graphical abstract
N6-methyladenosine (m6A) RNA modification plays critical roles in various metabolic pathways and development of common metabolic diseases. Zhang et al. summarize components, mechanisms, and research methods of m6A RNA modification, and they also review research progress and clinical significance and discuss future directions of m6A RNA modification in human metabolic diseases.
Introduction
More than 100 types of chemical modifications in cellular RNAs have been reported during the past 6 decades.1 Several methylation modifications occur in eukaryotic messenger RNA (mRNA), including N7-methylguanosine, N6-methyl-2′-O-methyladenosine, 2′-O-methylation, N6-methyladenosine (m6A), and 5-methylcytosine.2 Notably, m6A is the most common RNA modification type. It was first reported in mRNAs from eukaryotes in the early 1970s.3 m6A RNA modification mainly occurs in the RRACH sequence,4,5 which is mainly found near stop codons, and in 3′ untranslated regions (UTRs) and long internal exons in mRNAs.6,7 Advances in high-throughput sequencing technology and gradual progress of epigenetic research have enabled the study of m6A RNA modifications in a variety of non-coding RNAs (ncRNAs), including ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), microRNA (miRNA), and long ncRNA (lncRNA).8,9 Notably, m6A RNA modification is a dynamic and reversible event that is catalyzed by a collection of enzymes, including methyltransferase “writers,” demethylase “erasers,” and “readers” that recognize such modifications.10,11 m6A modifications are implicated in most steps of target RNA metabolism, including RNA maturation, splicing, export and folding, translation, and stability of RNA, thus modulating the downstream signaling pathway and physiological function.12,13
Metabolic diseases caused by disorders in amino acids, glucose, lipid metabolism, and other metabolic disorders, including obesity, type 2 diabetes (T2D), non-alcoholic fatty liver disease (NAFLD), hypertension, atherosclerosis, chronic kidney disease, cardiovascular disease, are a global health burden.14 Several studies report that cancer is a type of metabolic disease, which may shift metabolic pathways to facilitate uptake and incorporation of nutrients into cell building blocks, such as nucleotides, amino acids, and lipids required by highly proliferating cells.15,16 Although various approaches have been developed to prevent and treat these metabolic diseases, they have limited efficacy. The potentially important role of the m6A RNA modification in the development and progression of human metabolic disease is an emerging field of study.17
In this review, we summarize the recent progress in the study on the role and molecular mechanisms of m6A RNA modification in diseases associated with metabolism.
Mechanisms of m6A RNA modification
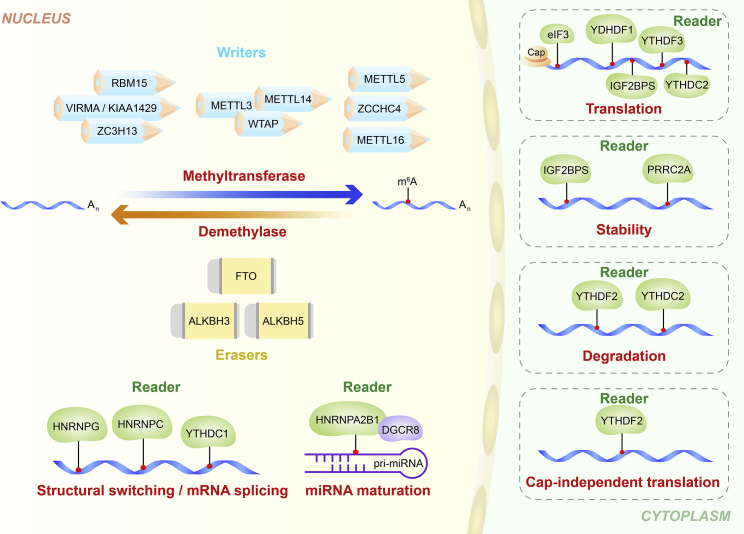
Regulators of m6A RNA modification can be classified into three types: writers, erasers, and readers. These enzymatic proteins are implicated in installing, removing, or recognizing m6A on mRNAs or ncRNAs, respectively (Figure 1).
Figure 1.
Mechanisms of m6A RNA modification
m6A methylation is catalyzed by the writer enzyme complex, which includes METTL3, METTL5, METTL14, METTL16, WTAP, RBM15, VIRMA/KIAA1429, ZC3H13, and ZCCHC4. The m6A modification is removed by the demethylase action of FTO, ALKBH3, and ALKBH5. Reader proteins recognize m6A and affect multiple downstream reactions, and they mainly include members of the YTH domain family (YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2), the HNRNP family (HNRNPA2B1 and HNRNPC), the IGF2BP family (IGF2BP1, IGF2BP2, and IGF2BP3), the eukaryotic initiation factor eIF3, and the proline-rich coiled-coil protein PRRC2A.
Writers comprise m6A methyltransferases, which catalyze m6A modification through a multicomponent methyltransferase complex that co-regulates transfer of methyl groups from S-adenosylmethionine to adenine bases in RNA.18 The main components of this complex include methyltransferase-like 3 (METTL3), methyltransferase-like 5 (METTL5), methyltransferase-like 14 (METTL14), methyltransferase-like 16 (METTL16), WT1-associated protein (WTAP), RNA-binding motif protein 15 (RBM15), Vir-like m6A methyltransferase associated (VIRMA, also known as KIAA1429), zinc finger CCCH-type containing 13 (ZC3H13), and zinc finger CCHC-type containing 4 (ZCCHC4). METTL3 was the first protein to be identified as an “m6A writer.”19,20 METTL14 structurally supports METTL3, and they form the core methyltransferase complex for m6A modification.21 WTAP stabilizes the core complex and facilitates m6A by recruiting the complex to nuclear speckles.22 RBM15 promotes binding of METTL3 and WTAP, thus guiding the two proteins to their target sites.20,23 VIRMA mainly promotes mRNA methylation modifications near the 3′ UTR and stop codon regions.24 Alternatively, ZC3H13 and CBLL1 control nuclear m6A methylation by combining with other cofactors such as WTAP.25 Recent studies report that ZCCHC4 is a methyltransferase involved in modification of the 28S rRNA.26,27 In addition, METTL16 is an independent mRNA methyltransferase, implicated in maintaining mRNA stability and regulation of splicing, and its binding sites do not overlap with those of METTL3/METTL14 methylation complexes.28 In addition, METTL16 can function alone and catalyze m6A on U6 snRNA, precursor (pre-)mRNAs, and ncRNAs.29,30 METTL5 is implicated in 18S rRNA m6A modification (Table 1).31
Table 1.
Role of regulators in m6A RNA modification
| Regulator | Function | Reference |
|---|---|---|
| m6A writers | ||
| METTL3 | catalyzes m6A modification | 19 |
| METTL14 | helps METTL3 to recognize the substrate | 21 |
| WTAP | contributes to localization of the METTL3-METTL14 heterodimer to the nuclear speckle | 22 |
| RBM15 | binds the m6A-methylation complex and recruits it to specific sites in RNA | 20,23 |
| VIRMA/KIAA1429 | VIRMA recruits the catalytic core components METTL3/METTL14/WTAP to guide methylation of a particular region | 24 |
| ZC3H13 | anchors WTAP to the mRNA-binding factor in the nucleus | 25 |
| ZCCHC4 | methylates human 28S rRNA and also interacts with a subset of mRNAs | 26,27 |
| METTL16 | catalyzes m6A modification U6 snRNA and pre-mRNAs and ncRNAs independently | 28,29 |
| METTL5 | promotes 18S rRNA m6A modification with TRMT112 | 31 |
| m6A erasers | ||
| FTO | catalyzes m6A demethylation | 32 |
| ALKBH5 | reverses m6A modifications oxidatively | 33 |
| ALKBH3 | removes m6A modification on tRNA | 34 |
| m6A readers | ||
| YTHDF1 | increases mRNA translation efficiency | 35 |
| YTHDF2 | promotes mRNA degradation | 36 |
| YTHDF3 | enhances translation and degradation by interacting with YTHDF1 and YTHDF2 | 35 |
| YTHDC1 | contributes to RNA splicing and export | 37 |
| YTHDC2 | enhances translation of target RNA and decreases abundance of target RNA | 38 |
| HNRNPA2B1 | mediates mRNA splicing and primary microRNA processing | 39 |
| HNRNPC | affects abundance and alternative splicing of target mRNAs | 40 |
| HNRNPG | regulates transcriptome-wide alternative splicing | 41 |
| IGF2BPs | enhances mRNA stability and translation | 42 |
| eIF3 | enhances mRNA translation | 43 |
| PRRC2A | stabilizes Olig2 mRNA | 44 |
Erasers comprise m6A demethylases, which remove m6A methyl groups from RNA. Three m6A demethylases have been reported, including fat mass and obesity-associated protein (FTO) and ALKB homologs ALKBH5 and ALKBH3.32,34,45 The demethylation process involves oxidation of m6A to form N6-hydroxymethyladenosine (hm6A), conversion of the hm6A to N6-formyladenosine (f6A), and finally conversion of f6A to adenosine. FTO was the first protein to be identified as an “m6A eraser,” and it catalyzes m6A demethylation.32 ALKBH5 was the second RNA demethylase to be reported, and it reverses m6A modifications.33 In addition, FTO can mediate m6Am (N6,2′-O-dimethyladenosine) demethylation. However, ALKBH5 is an m6A-specific demethylase in mRNA.46 Recently, Chen et al.47 reported that ALKBH3 plays a role as a demethylase of m6A modifications, and that ALKBH3 preferentially modifies tRNA over mRNA or rRNA (Table 1).34
m6A readers comprise the YTH domain-containing family (YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2),37,38,48,49 the heterogeneous nuclear ribonucleoprotein (HNRNP) family (HNRNPA2B1, HNRNPC, and HNRNPG),39, 40, 41,50 insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1, IGF2BP2, and IGF2BP3),42 eukaryotic initiation factor eIF3,43,51 and proline-rich coiled-coil protein PRRC2A.44 YTHDF1 interacts with initiation factors, thus promoting RNA translation initiation in cytosol.52 YTHDF2 regulates RNA degradation by binding to m6A-modified mRNA.36 YTHDF3 promotes translation by enhancing protein synthesis together with YTHDF1 and regulates mRNA decay mediated by YTHDF2.35,53 Furthermore, YTHDF1 and YTHDF2 recognize m6A-modified circRNAs and modulate circRNA expression.54,55 YTHDC1 can affect subcellular localization of circRNA in an m6A-dependent manner.56 Furthermore, IGF2BP1–IGF2BP3 enhance stability and translation of the target mRNAs by recognizing m6A modifications.42 HNRNPA2B1 recognizes primary (pri-)miRNA m6A marks and interacts with DiGeorge syndrome critical region 8 (DGCR8), a critical component of the canonical microprocessor complex, to stimulate miRNA processing, whereas HNRNPC recognizes m6A-dependent splicing in mRNA secondary structures.39 Wu et al.44 reported a novel m6A reader, PRRC2A, which stabilizes Olig2 mRNA by binding to a consensus GGACU motif in the Olig2 coding sequence in an m6A-dependent manner (Table 1).
Methods for m6A RNA modification research
Levels of m6A in RNA are determined by two-dimensional thin layer chromatography, m6A dot blots, and high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS).57,58 Transcriptome-wide distribution of m6A is profiled by methylated RNA immunoprecipitation followed by high-throughput sequencing (MeRIP-seq or m6A-seq).6 In this method, mRNA or ncRNA is fragmented into 100-nt-long oligonucleotides and immunoprecipitated with a specific antibody against m6A. The precipitated RNAs are then subjected to high-throughput sequencing. In addition, methods with higher resolution, such as photo-crosslinking-assisted m6A sequencing (PA-m6A-seq) and site-specific cleavage and radioactive labeling followed by ligation-assisted extraction and thin-layer chromatography (SCARLET), have been developed.59 The m6A individual nucleotide resolution crosslinking immunoprecipitation (miCLIP) method can detect m6A at a precise position, and it is a major advance in m6A research.60 Recent studies developed antibody-free methods for global m6A detection, including MAZTER-seq,61 DART-seq (deamination adjacent to RNA modification targets),62 and m6A-SEAL (an m6A selective chemical labeling method).63
Advances in CRISPR-based genome engineering allow determination of the functional role of changing the m6A modification site in many organisms.64 Furthermore, Zhou et al.65 developed an online tool, named the sequence-based m6A modification site predictor, for prediction of m6A modification sites on RNA sequences of interests. These methods allow exploration of the function and mechanisms of m6A modification.
Role of m6A modification in glucose metabolism-related diseases
Glucose metabolism is a complex and important source of energy in organisms through anaerobic fermentation, aerobic oxidation, and the pentose phosphate pathway. T2D is a complex metabolic disease characterized by hyperglycemia and dyslipidemia.66 Recent studies report that m6A modification plays a critical role in the pathogenesis of T2D. In patients with T2D, glucose levels affect dynamic regulation of m6A. High glucose levels can simultaneously decrease FTO mRNA expression and increase expression of METTL3, METTL14, and WTAP methyltransferases. Forkhead box O1 (FOXO1) and glucose-6-phosphatase catalytic subunit 1 (G6PC) are key regulators in glucose homeostasis.67,68 Diacylglycerol O-acyltransferase 2 (DGAT2) is required for synthesis and storage of intracellular triglycerides, which play a central role in lipid accumulation.69 Notably, a high expression level of FTO induces mRNA expression of FOXO1, G6PC, and DGAT2, resulting in abnormal glucose and lipid metabolism.70,71 Recent studies report that m6A modification is associated with β cell survival and insulin secretion, which are important for regulation of glucose levels in T2D patients. METTL3 suppresses hepatic insulin sensitivity through modification of fatty acid synthase (FASN) mRNA by m6A and promotion of fatty acid metabolism.72 METTL3 is downregulated under inflammatory and oxidative stress conditions. Deletion of METTL3 induces islet β cell failure and hyperglycemia.73 METTL14 is implicated in β cell survival, differentiation, and insulin secretion. Knockdown of METTL14 in mice increases β cell death, alters β cell differentiation, and decreases β cell mass and insulin secretion, resulting in glucose intolerance.74 Furthermore, depletion of m6A in-EndoC-βH1 decreases insulin secretion by decreasing AKT phosphorylation and pancreatic and duodenal homeobox 1 (PDX1) protein levels, which have been explored using β cell-specific METTL14 knockout mice.75 These studies provide a theoretical basis for development of m6A-based molecular therapies to promote β cell survival and function in patients with diabetes. In summary, m6A modification plays an important role in glucose metabolism, which is associated with several human diseases, and is a potential therapeutic target (Table 2).
Table 2.
Functions of m6A RNA modification in glucose metabolism
| m6A regulators | Function | Disease | Reference |
|---|---|---|---|
| FTO | high expression of FTO induced mRNA expression of FOXO1, G6PC, and DGAT2, which are involved in abnormal glucose | type 2 diabetes | 70 |
| METTL3 | suppresses hepatic insulin sensitivity through m6A modification of FASN (fatty acid synthetase) mRNA and promoting fatty acid metabolism | type 2 diabetes | 72 |
| METTL14 | knockdown of METTL14 decreases β cell mass and insulin secretion, eventually resulting in glucose intolerance | type 2 diabetes | 74 |
| METTL14 | knockdown of METTL14 in β cells decreases AKT phosphorylation and PDX1 protein levels, resulting in a decrease in insulin secretion | type 2 diabetes | 75 |
Role of m6A modification in lipid metabolism-related diseases
Lipid metabolism is a complex physiological process that is associated with nutrient adjustment, hormone regulation, and homeostasis, and it involves multiple molecular factors and signaling pathways.76 Abnormal lipid metabolism is implicated in several diseases,71 with recent studies reporting that m6A modification plays a role in this relationship.
Low METTL3 activity in cell cultures decreases m6A modification of the peroxisome proliferator-activated receptor (PPAR)α gene, thus increasing its mRNA expression and extending the life of transcripts, which ultimately reduces lipid accumulation. Analysis shows that YTHDF2 binds to PPARα mRNA, thus mediating its stability and regulates lipid metabolism.77 The zinc finger protein 217 (Zfp217) binds to YTHDF2 to activate transcription of the m6A demethylase FTO, thus promoting its interaction with m6A sites on various mRNAs, and it ultimately promotes adipose differentiation. These activities were confirmed through decreased levels of oil red O staining and lower mRNA expression of key adipogenic genes encoding PPARγ, lipoprotein lipase, and adiponectin in mouse embryonic fibroblasts.78 Translation of mitochondrial carrier homology 2 (MTCH2) is mediated by m6A modification through an YTHDF1-dependent pathway, and it plays a role in regulating adipogenesis in intramuscular preadipocytes.79 For m6A erasers, FTO enhances expression of JAK2 and promotes phosphorylation of STAT3, thus enhancing transcription of C/EBPβ implicated in early stages of adipocyte differentiation. Alternatively, YTHDF2 accelerates decay of JAK2 mRNA and attenuates JAK2-STAT3-C/EBPβ signaling.80 RUNX1 partner transcriptional co-repressor 1 (RUNX1T1) is a regulator of adipogenesis.81 FTO controls splicing of RUNX1T1 exons by regulating m6A levels of its transcripts, thus regulating adipogenesis.82 Angiopoietin-like 4 (ANGPTL4) plays a role in the regulation of triglyceride clearance from the blood serum and in lipid metabolism.83 FTO binds to ANGPTL4 mRNA and promotes its translation, thus enhancing intracellular lipolysis in mouse adipocytes.84 The roles of m6A modification in lipid metabolism are summarized in Table 3.
Table 3.
Functions of m6A RNA modification in lipid metabolism
| Regulators | Function | Mechanism | Reference |
|---|---|---|---|
| METTL3 | METTL3 decreases PPARα m6A abundance and increases PPARα mRNA half-life and expression, reducing lipid accumulation | mRNA stability | 77 |
| YTHDF2/FTO | promotes adipose differentiation | gene expression | 78 |
| YTHDF1 | YTHDF1 promotes mitochondrial carrier homology 2 (MTCH2) translation to regulate adipogenesis | translation | 79 |
| FTO/YTHDF2 | FTO enhances expression of JAK2 and YTHDF2 directly targets JAK2 and accelerates mRNA decay | gene expression, mRNA decay | 80 |
| FTO | FTO controls splicing of RUNX1T1 mRNA by regulating m6A levels, regulating adipogenesis | mRNA splicing | 82 |
| FTO | FTO binds to Angptl4 to encode an adipokine that stimulates intracellular lipolysis in adipocytes | translation | 84 |
| ime4Δ | ime4Δ (yeast m6A methyltransferase gene deletion) cells showed a significant decrease in expression of the key genes involved in peroxisomal β-oxidation in yeast | gene expression | 85 |
| YTHDF1 | facilitates translation of Wnt signaling effectors TCF7L2 and TCF4, which are required for maintenance of intestinal stem cells (ISCs) during regeneration and tumorigenesis | translation | 86 |
Obesity is a common contributor to metabolic syndrome. At the cellular level, obesity is characterized by an increase in both cell size (hypertrophy) and the number of fat cells (hyperplasia).87,88 Several studies report that m6A modification is involved in the development of obesity. The m6A writers WTAP, METTL3, and METTL14 promote cell cycle transition in mitotic clonal expansion; however, these methyltransferases are negatively correlated with adipogenesis.89,90 Risk alleles in the m6A eraser FTO are common among people with high body mass index, and some of these single nucleotide polymorphisms are positively correlated with obesity.91, 92, 93, 94, 95 In addition, FTO promotes adipogenesis by repressing the Wnt/β-catenin signaling pathway in porcine intramuscular preadipocytes.96
However, several studies report that m6A writers, erasers, and readers do not work alone. For example, FTO increases expression of autophagy-related 5 (ATG5) and autophagy-related 7 (ATG7) expression to repress formation of autophagosomes, thus inhibiting autophagy and adipogenesis. Alternatively, YTHDF2 decreases expression of ATG5 and ATG7 by shortening the half-life of their m6A-modified mRNAs.97 Moreover, FTO regulates adipogenesis by controlling cell cycle progression in an m6A-YTHDF2-dependent manner. FTO knockdown significantly decreases expression of cell cycle regulators such as cyclin A2 (CCNA2) and cyclin-dependent kinase 2 (CDK2). YTHDF2 recognizes and destabilizes these mRNAs, leading to reduced protein expression, prolonged cell cycle progression, and suppressed adipogenesis.98 Induced expression of YTHDF2 reverses demethylation of CCNA2 and CDK2 mRNAs induced by FTO.99 Low levels of ZFP217, which is implicated in adipogenesis, increase expression of METTL3. Furthermore, knockdown of METTL3 rescues mitotic clonal expansion inhibited by ZFP217 small interfering RNA and promotes cyclin D1 (CCND1) expression. Moreover, YTHDF2 recognizes and degrades m6A-methylated CCND1 mRNA, resulting in downregulation of CCND1 and inhibition of adipogenesis.100
Currently, prevalence of NAFLD is at epidemic proportions and is a common cause of chronic liver disease worldwide.101 NAFLD is characterized by hepatic steatosis, ballooning degeneration, and fatty retention of liver parenchyma cells, with no history of excessive alcohol intake.102 Hepatic steatosis, the unique pathological feature of NAFLD, is caused by metabolic dysregulation of de novo lipogenesis, fatty acid uptake and oxidation, and triglyceride transport.103,104 Previous studies report that development of NAFLD is highly correlated with m6A alteration.105 m6A modification is associated with fat accumulation both in vivo and in vitro. The m6A eraser FTO decreases mitochondrial content and increases triglyceride deposition by downregulating overall m6A levels. FTO activity is essential for fat metabolism in hepatocytes, indicating the importance of RNA modification in fat deposition.106 FTO is implicated in hepatic oxidative stress and lipid deposition, which participate in the development of NAFLD.107 Curcumin, the active ingredient in dietary turmeric, affects mRNA expression of the m6A writers METTL3 and METTL14 in weaned piglets. Analysis showed that increased m6A methylation levels alleviate lipopolysaccharide-induced liver injury and reverses disruption of lipid metabolism in the liver.108 In another study, knockdown of METTL3 or YTHDF2 increases expression of PPARα mRNA and reduces lipid accumulation.77 In summary, m6A modulators are potential therapeutic targets for NAFLD.
Osteoporosis is a common bone metabolic disease in older populations.109 Recent studies report that m6A modification is involved in development of osteoporosis.110,111 Conditional knockout of the m6A writer METTL3 in bone marrow mesenchymal stem cells in mice decreases bone mass with low osteogenic potential, and it increases marrow adiposity associated with enhanced adipogenic potential by reducing m6A methylation levels.111 METTL3 knockdown in porcine bone marrow stem cells promotes adipogenesis and adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway in an m6A-YTHDF2-dependent manner.112 Upregulation of the m6A eraser FTO by growth differentiation factor 11 promotes differentiation of bone marrow mesenchymal stem cells into adipocytes and osteoblasts through demethylation of PPARγ (Table 4).110
Table 4.
Functions of m6A RNA modification in lipid metabolism-related human diseases
| m6A regulators | Function | Disease | Reference |
|---|---|---|---|
| WTAP | WTAP, METTL3, and METTL14 are negatively related to adipogenesis | obesity | 89 |
| METTL3 | |||
| METTL14 | |||
| FTO | FTO alleles and some SNPs are positively associated with obesity | obesity | 91,93, 94, 95 |
| FTO | promotes adipogenesis by repressing the Wnt/β-catenin signaling pathway in porcine intramuscular preadipocytes | obesity | 96 |
| FTO | increases the autophagy-related protein expression of ATG5/ATG7 and weaken the formation of autophagosomes, thereby inhibiting autophagy and adipogenesis | obesity | 97 |
| FTO/YTHDF2 | FTO knockdown markedly decreases the expression of the cell cycle regulators CCNA2 and CDK2; YTHDF2 recognizes and degrades these mRNAs and reduces their protein expression to suppress adipogenesis | obesity | 98 |
| METTL3 | downregulates CCND1 and inhibits adipogenesis | obesity | 100 |
| FTO | downregulates overall m6A levels and decreases mitochondrial content and triglyceride deposition | NAFLD | 106 |
| FTO | associates with hepatic oxidative stress and lipid deposition | NAFLD | 107 |
| METTL3/METTL14 | Increases m6A methylation level, thus improving lipopolysaccharide-induced liver injury and hepatic lipid metabolism disruption in the liver of piglets | NAFLD | 108 |
| METTL3/YTHDF2 | knockdown of METTL3 or YTHDF2 increases the expression of PPARα mRNA, thereby decreasing lipid accumulation | NAFLD | 77 |
| METTL3/YTHDF2 | deletion of METTL3 promotes adipogenesis and adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway via an m6A-YTHDF2-dependent manner | osteoporosis | 112 |
| FTO | promotes the differentiation of adipocyte and osteoblasts from bone marrow mesenchymal stem cells via GDF11 | osteoporosis | 110 |
| METTL3 | increases the m6A methylated level of fatty acid synthase (Fasn), thereby promoting fatty acid metabolism and enhancing hepatic insulin sensitivity | type 2 diabetes | 72 |
Dysregulated fatty acid metabolism is associated with insulin resistance in diabetes.113 Recent studies report that m6A modification is involved in fatty acid metabolism. For instance, Xie et al.72 reported that the m6A writer METTL3 increases m6A methylation level of FASN, which promotes fatty acid metabolism and enhances hepatic insulin sensitivity. Their study provides key information on blood glucose homeostasis and provides information on potential therapeutic targets for T2D patients. m6A modification of mRNA is well characterized in yeast cells, where peroxisomes are the only sites for fatty acid β-oxidation. A deletion strain of the yeast m6A methyltransferase ime4 showed significant decrease in expression of key genes involved in peroxisomal β-oxidation compared with wild-type yeast. This study provides a basis for exploring the role of m6A methylation in peroxisomal biology (Table 3).85
Role of m6A modification in cancer metabolic reprograming
Reprogramming metabolism is an important feature of cancer pathogenesis.114 Cancer cells activate or inhibit metabolic pathways such as aerobic glycolysis (also known as the Warburg effect),115 disordered lipid metabolism,116 and glutamine-dependent anaplerosis to accelerate cell proliferation.117 Recent studies report that m6A modification plays an important role in regulation of metabolic reprogramming of cancer.
m6A modification modulates glycolysis in cancer cells
The Warburg effect is a key metabolic hallmark of cancer cells that is characterized by elevated activation of glycolysis followed by increased lactate fermentation.118 In colorectal cancer (CRC), the m6A methyltransferase METTL3 interacts with the 3′ UTR regions of hexokinase 2 (HK2) and glucose transporter GLUT1 mRNA and enhances their stability, thus activating the glycolysis pathway.119 Moreover, WTAP enhances stability of HK2 mRNA by binding with the 3′ UTR m6A site in gastric cancer.120 Casein kinase 2 (CK2) is associated with glycolysis, and CK2α is an essential catalytic subunit of the holoenzyme. In bladder cancer, ALKBH5 specifically recognizes the m6A sites of the 3′ UTR in CK2α mRNA and reduces its stability, thus inhibiting cell glycolysis and proliferation.121 Furthermore, FTO triggers m6A demethylation of PKM2 mRNA and accelerates its translation, thus promoting hepatocellular carcinoma tumorigenesis.122 In addition, YTHDF1 facilitates translation of the Wnt signaling effectors TCF7L2 and TCF4, which are required for maintenance of intestinal stem cells during regeneration and tumorigenesis.86 YTHDF2 weakens EGFR mRNA stability in an m6A-dependent manner and inhibits the MEK/ERK pathway, consequently impeding cell proliferation.123 METTL3 enhances translation of IKBKB and RELA and activates the nuclear factor κB (NF-κB) pathway, thus promoting bladder cancer progression.124 METTL14 overexpression inactivates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway in high glucose-treated HK2 cells through PTEN-affected, HDAC5-mediated epithelial-to-mesenchymal transition (EMT) of renal tubular cells in diabetic kidney disease.125 YTHDF2 binds directly to the m6A modification site in the 3′ UTR of 6-phosphogluconate dehydrogenase (6PGD) gene, thus promoting 6PGD mRNA translation in lung cancer cells.126 Moreover, Li et al.127 reported that m6A modification of the 5′ UTR region of PDK4 upregulates its translation, elongation, and mRNA stability by binding with the YTHDF1/eEF-2 complex and IGF2BP3, thus enhancing tumor growth and progression of cervical and liver cancers (Table 5). These studies indicate that m6A modification plays an important role in glucose metabolism by modulating glycolytic enzymes or associated signaling pathways.
Table 5.
Functions of m6A RNA modification in metabolic reprogramming of cancer
| m6A regulators | Molecules | Function | Reference |
|---|---|---|---|
| Glucose metabolism | |||
| METTL3 | HK2, GLUT1 | METTL3 interacts with the 3′ UTR regions of HK2 and GLUT1 mRNA and enhances its stability, thereby activating the glycolysis pathway | 119 |
| ALKBH5 | CK2α | ALKBH5 specifically recognizes the m6A sites of the 3′ UTR in CK2α mRNA and reduces its stability to inhibit cell glycolysis and proliferation | 121 |
| FTO | PKM2 | FTO triggers the m6A demethylation of PKM2 mRNA and accelerates its translation, thus promoting hepatocellular carcinoma tumorigenesis | 122 |
| YTHDF1 | Wnt signaling | YTHDF1 facilitates the translation of the Wnt signaling effectors TCF7L2 and TCF4, which are required for the maintenance of intestinal stem cells during regeneration and tumorigenesis | 86 |
| YTHDF2 | MEK/ERK | YTHDF2 weakens EGFR mRNA stability in an m6A-dependent manner and thus impairs the MEK/ERK pathway and consequently impedes cell proliferation and growth | 123 |
| METTL3 | NF-κB pathway | METTL3 enhances the translation of IKBKB and RELA and activates NF-κB pathway to promote bladder cancer progression | 124 |
| METTL14 | PI3K/Akt | METTL14 overexpression inactivates the PI3K/Akt pathway in high glucose-treated HK2 cells via PTEN-regulated HDAC5-mediated EMT of renal tubular cells in diabetic kidney disease | 125 |
| YTHDF2 | 6PGD | YTHDF2 binds directly to the m6A modification site in the 3′ UTR of the 6-phosphogluconate dehydrogenase (6PGD) gene to promote 6PGD mRNA translation in lung cancer cells | 126 |
| Lipid metabolism | |||
| METTL3 | miR-3619-5p/HDGF | METTL3 promotes the stability of lncRNA LINC00958 and activates the miR-3619-5p/HDGF axis to facilitate lipogenesis in HCC | 128 |
| FTO | SREBP1c/CIDEC | FTO increases lipid accumulation by activating the SREBP1c/CIDEC signaling pathway in an m6A-dependent manner in HepG2 cells | 129 |
| Glutamine metabolism | |||
| FTO, ALKBH5 | D2-HG | FTO and ALKBH5 are α-KG-dependent dioxygenases that are competitively inhibited by structurally related metabolite d-2-hydorxyglutarate (D2-HG), leading to abnormal expression of isocitrate dehydrogenase 1 or 2 (IDH1/2)-mutant tumors | 130,131 |
| YTHDF1 | GLS1 | YTHDF1 accelerates GLS1 translation, a key enzyme of glutamine metabolism, and promotes colon cancer development | 132 |
m6A modification affects lipid metabolism in cancer cells
Cancer cells change lipid metabolism to meet the malignant development demands, including synthesis of macromolecules, the main lipids for biogenesis of membranes and various signaling factors.133 METTL3 promotes stability of lncRNA LINC00958 and activates the miR-3619-5p/HDGF axis, thus inducing lipogenesis in HCC. In addition, LINC00958 affects the expression of sterol regulatory element binding transcription factor 1 (SREBP1), FASN, stearoyl-coenzyme A (CoA) desaturase (SCD1), and acetyl-CoA carboxylase 1 (ACC1), which are implicated in lipogenesis.128 FTO increases lipid accumulation by activating the SREBP1c/CIDEC signaling pathway in an m6A-dependent manner in liver hepatocellular carcinoma HepG2 cells (Table 5).129 These findings show that m6A RNA modification is involved in the regulation of lipogenesis in cancer cells.
m6A modification affects amino acid metabolism in cancer cells
Dysregulation of metabolism of amino acids, including glutamine, serine, and glycine, which play a role as metabolic regulators, is implicated in cancer cell growth.134,135 Glutamine, the most abundant free amino acid, participates in several of pathways in energy generation, macromolecular synthesis, and signal transmission in cancer cells by donating its nitrogen and carbon atoms.136 Glutamine can be converted into α-KG to replenish the tricarboxylic acid (TCA) cycle through glutamate dehydrogenase (GLUD1) or transaminases.137 FTO and ALKBH5 are α-KG-dependent dioxygenases and are competitively inhibited by the structurally related metabolite, d-2-hydorxyglutarate (D2-HG), leading to accumulation in isocitrate dehydrogenase 1 or 2 (IDH1/2) mutant tumors.130,131 In addition, YTHDF1 accelerates glutaminase GLS1 translation, a key enzyme in glutamine metabolism, therefore promoting development of colon cancers (Table 5).132 These findings imply that m6A modification affects glutamine metabolism, resulting in cancer pathogenesis; however, further studies should explore the role of glutamine metabolism of development of different cancers. Moreover, mores studies should explore the role of m6A on other amino acid metabolism.
Conclusions and perspectives
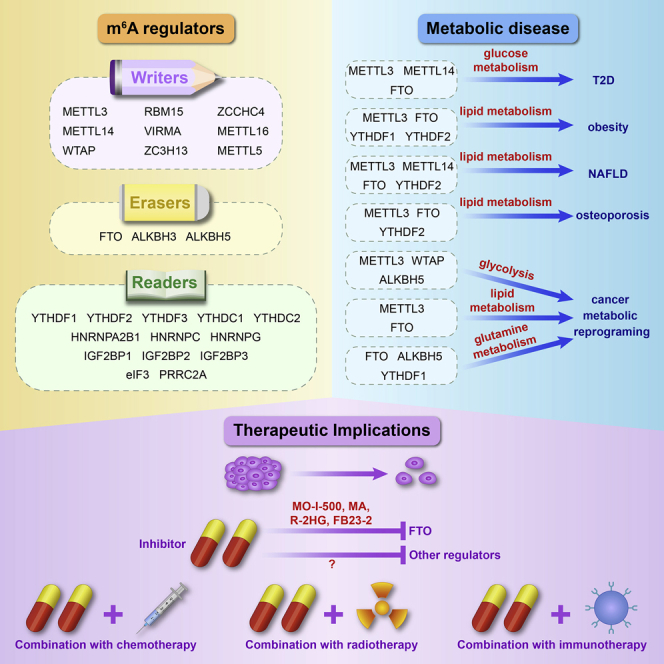
Advances in RNA immunoprecipitation sequencing, high-throughput sequencing, and liquid chromatography have led to the identification of several novel RNA modifications, implicated in metabolic diseases and tumors. In this review, we explored the roles and mechanisms of m6A RNA modifications in the occurrence and development of diseases by regulating glucose, lipid, and amino acid metabolism. The m6A-mediated regulation of glucose, lipid, and amino acid metabolism is associated with metabolic diseases, including T2D, NAFLD, obesity, and cancer (Figure 2). The stability and translation of mRNA of key regulators involved in these metabolic pathways are regulated by m6A modification and various m6A readers, thus promoting metabolic disease progression. In addition, m6A modification of lncRNA participates in progression of metabolic diseases.138 In turn, metabolites and metabolic pathways are involved in regulation of m6A RNA modification; for example, TCA cycle metabolites affect FTO activity, whereas iron and NADPH (the reduced form of nicotinamide adenine dinucleotide phosphate) affect ALKBH5 activity.139
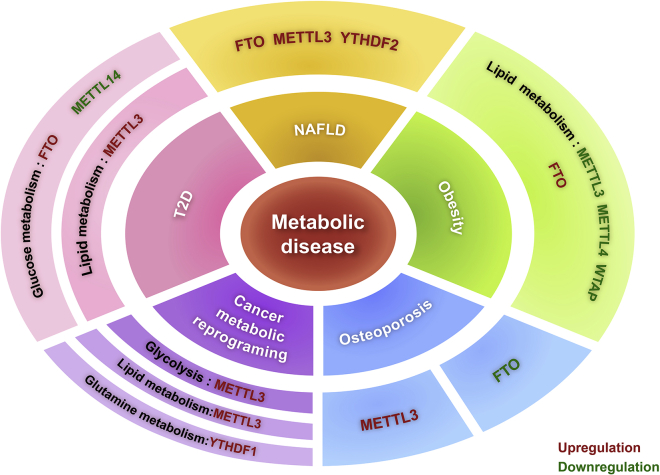
Figure 2.
Role of m6A regulators in human metabolic diseases
Obesity mainly involves dysregulation of lipid metabolism, in which the expression of METTL3, METTL14, and WTAP are downregulated, while FTO is upregulated. In T2D, the expression of METTL14 is downregulated and FTO is upregulated in glucose metabolism, with increased expression of METTL3 in fatty acid metabolism. Metabolic disruption in human cancers mainly involves glycolysis and RNA metabolism. High METTL3 activates glycolysis to promote cancer development. The expression of FTO, METTL3, and YTHDF2 are high in the process of NAFLD. In osteoporosis, METTL3 expression is high, while FTO is low.
Incidence of metabolic diseases in young people are increasing exponentially. Current therapies for these diseases are ineffective, and therefore there is need to urgently explore and develop disease-specific therapeutic targets.17,140 FTO is an attractive target for cancer treatment.141 Rhein, radicicol, epigallocatechin gallate, entacapone, and meclofenamic acid (MA) are a group of compounds that inhibit FTO regulation of m6A levels and affect fat formation.99,142, 143, 144 MO-I-500 inhibits m6A demethylase activity of FTO and inhibits colony formation of a triple-negative inflammatory breast cancer cell line.145 Meclofenamic acid, a nonsteroidal anti-inflammatory drug, selectively inhibits FTO and inhibits GBM cell growth and survival.146 R-2HG, a major metabolic product of mutant IDH1/2, inhibits FTO activity, thus inhibiting leukemic cell growth/survival and leukemia progression.147 A more potent FTO inhibitor, FB23-2, inhibits acute myeloid leukemia (AML) progression in xenotransplanted mice.148 In addition, IOX3, an inhibitor of hypoxia-inducible factor prolyl hydroxylases, can be used to treat immune-related diseases caused by the m6A eraser ALKBH5.45,149 The methylation inhibitors cycloleucine and S-adenosylhomocysteine can be used to reduce m6A levels,106,150 whereas a methyl donor, betaine, increases m6A levels.151
Resistance to chemoradiotherapy is a major challenge in tumor therapy. m6A RNA modification regulators can be used as prediction markers for individualized cancer therapy and for providing clues for overcoming therapeutic resistance in cancer. In addition, silencing of METTL3 leads to an increase in the sensitivity of glioblastoma stem cells (GSCs) to gamma irradiation and pancreatic cancer cells to anticancer reagents.152 Recent studies report that m6A modification is related to development of the immune system. Silencing of METTL3 inhibits interleukin (IL)-7 signaling in CD4+ T cells. METTL3-mediated m6A modification enhances TLR4/NF-κB signaling-induced cytokine production and stimulates T cell activation.153 YTHDF1 is associated with cross-presentation of tumor antigens, cross-priming of CD8+ T cells, and PD-L1 checkpoint inhibition in dendritic cells.154 Knockdown of FTO in melanoma cells sensitizes tumor cells to interferon gamma (IFNγ) in vitro and promotes melanoma response to anti-PD-1 antibody in mice.155 These findings imply that m6A regulators can be combined with anti-PD-1/PD-L1 inhibitors to improve anticancer immunotherapy. The relationship between m6A RNA modification and metabolism should be further explored. Therefore, use of m6A methylation-related inhibitors is a potential strategy for treatment of obesity and other complex metabolic diseases.
In addition, variations in gut microbiota are correlated with m6A modifications in the cecum and the liver, which affect metabolism, inflammation, and antimicrobial responses in mice.156 Molecular interactions among human gut microbiota, m6A methylation, and metabolic diseases should be explored in the future.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (nos. 81872210, 81802948, and 82073101), the Shanxi Province Scientific and Technological Achievements Transformation Guidance Foundation (no. 201804D131043), the Applied Basic Research Project of Shanxi Province (nos. 201801D221421 and 201901D211490), the Youth Foundation of The First Hospital Affiliated with Shanxi Medical University (no. YQ1503), the Fund of Shanxi “1331” Project, Research Project Supported by Shanxi Scholarship Council of China (no. 2020165), the Fund Program for the Scientific Activities of Selected Returned Overseas Professionals in Shanxi Province (no. 20200034), the Open Fund from Key Laboratory of Cellular Physiology (Shanxi Medical University), Ministry of Education, China (nos. KLMEC/SXMU-202008 and KLMEC/SXMU-202009).
Author contributions
Y.W. and W.G. conceived the review. Y.Z. wrote the first version of the manuscript. Y.Z., W.C., X.Z., and Y.G. organized the figures. W.C., Y.Z., J.C., S.W., Y.W., and W.G. revised the manuscript. All of the authors approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Wei Gao, Email: gaoweisxent@sxent.org.
Yongyan Wu, Email: wuyongyan@sxent.org.
References
- 1.Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J., Jia G. Methylation modifications in eukaryotic messenger RNA. J. Genet. Genomics. 2014;41:21–33. doi: 10.1016/j.jgg.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Dubin D.T., Taylor R.H. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975;2:1653–1668. doi: 10.1093/nar/2.10.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 5.Wei C.M., Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977;16:1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- 6.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 7.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem. Sci. 2013;38:204–209. doi: 10.1016/j.tibs.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berulava T., Rahmann S., Rademacher K., Klein-Hitpass L., Horsthemke B. N6-adenosine methylation in miRNAs. PLoS ONE. 2015;10:e0118438. doi: 10.1371/journal.pone.0118438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore L.D., Le T., Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X.Y., Zhang J., Zhu J.S. The role of m6A RNA methylation in human cancer. Mol. Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He L., Li H., Wu A., Peng Y., Shu G., Yin G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer. 2019;18:176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H., Weng H., Chen J. m6A modification in coding and non-coding RNAs: Roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–288. doi: 10.1016/j.ccell.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hotamisligil G.S., Erbay E. Nutrient sensing and inflammation in metabolic diseases. Nat. Rev. Immunol. 2008;8:923–934. doi: 10.1038/nri2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 16.Soga T. Cancer metabolism: Key players in metabolic reprogramming. Cancer Sci. 2013;104:275–281. doi: 10.1111/cas.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Wang J., Huang C., Shen M., Zhan H., Xu K. RNA N6-methyladenosine: A promising molecular target in metabolic diseases. Cell Biosci. 2020;10:19. doi: 10.1186/s13578-020-00385-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bokar J.A., Rath-Shambaugh M.E., Ludwiczak R., Narayan P., Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994;269:17697–17704. [PubMed] [Google Scholar]
- 19.Wang P., Doxtader K.A., Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knuckles P., Lence T., Haussmann I.U., Jacob D., Kreim N., Carl S.H., Masiello I., Hares T., Villaseñor R., Hess D. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415–429. doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Śledź P., Jinek M. Structural insights into the molecular mechanism of the m6A writer complex. eLife. 2016;5:e18434. doi: 10.7554/eLife.18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., Adhikari S., Shi Y., Lv Y., Chen Y.S. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patil D.P., Chen C.K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., Cheng T., Gao M., Shu X., Ma H. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen J., Lv R., Ma H., Shen H., He C., Wang J., Jiao F., Liu H., Yang P., Tan L. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 2018;69:1028–1038.e6. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma H., Wang X., Cai J., Dai Q., Natchiar S.K., Lv R., Chen K., Lu Z., Chen H., Shi Y.G. N6-methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 2019;15:88–94. doi: 10.1038/s41589-018-0184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinto R., Vågbø C.B., Jakobsson M.E., Kim Y., Baltissen M.P., O’Donohue M.F., Guzmán U.H., Małecki J.M., Wu J., Kirpekar F. The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 2020;48:830–846. doi: 10.1093/nar/gkz1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doxtader K.A., Wang P., Scarborough A.M., Seo D., Conrad N.K., Nam Y. Structural basis for regulation of METTL16, an S-adenosylmethionine homeostasis factor. Mol. Cell. 2018;71:1001–1011.e4. doi: 10.1016/j.molcel.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pendleton K.E., Chen B., Liu K., Hunter O.V., Xie Y., Tu B.P., Conrad N.K. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–835.e14. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warda A.S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Höbartner C., Sloan K.E., Bohnsack M.T. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–2014. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Tran N., Ernst F.G.M., Hawley B.R., Zorbas C., Ulryck N., Hackert P., Bohnsack K.E., Bohnsack M.T., Jaffrey S.R., Graille M., Lafontaine D.L.J. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719–7733. doi: 10.1093/nar/gkz619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vågbø C.B., Shi Y., Wang W.L., Song S.H. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueda Y., Ooshio I., Fusamae Y., Kitae K., Kawaguchi M., Jingushi K., Hase H., Harada K., Hirata K., Tsujikawa K. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci. Rep. 2017;7:42271. doi: 10.1038/srep42271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., Ma J., Wu L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roundtree I.A., Luo G.Z., Zhang Z., Wang X., Zhou T., Cui Y., Sha J., Huang X., Guerrero L., Xie P. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife. 2017;6:e31311. doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu P.J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., Qi M., Lu Z., Shi H., Wang J. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alarcón C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou K.I., Shi H., Lyu R., Wylder A.C., Matuszek Ż., Pan J.N., He C., Parisien M., Pan T. Regulation of co-transcriptional pre-mRNA splicing by m6A through the low-complexity protein hnRNPG. Mol. Cell. 2019;76:70–81.e9. doi: 10.1016/j.molcel.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B.S., Mesquita A., Liu C., Yuan C.L. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.B., Jaffrey S.R. 5′ UTR m6A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu R., Li A., Sun B., Sun J.G., Zhang J., Zhang T., Chen Y., Xiao Y., Gao Y., Zhang Q. A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29:23–41. doi: 10.1038/s41422-018-0113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y., Yan J., Li Q., Li J., Gong S., Zhou H., Gan J., Jiang H., Jia G.F., Luo C., Yang C.G. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373–384. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei J., Liu F., Lu Z., Fei Q., Ai Y., He P.C., Shi H., Cui X., Su R., Klungland A. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell. 2018;71:973–985.e5. doi: 10.1016/j.molcel.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z., Qi M., Shen B., Luo G., Wu Y., Li J., Lu Z., Zheng Z., Dai Q., Wang H. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533–2545. doi: 10.1093/nar/gky1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer K.D., Jaffrey S.R. Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017;33:319–342. doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu N., Zhou K.I., Parisien M., Dai Q., Diatchenko L., Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45:6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choe J., Lin S., Zhang W., Liu Q., Wang L., Ramirez-Moya J., Du P., Kim W., Tang S., Sliz P. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 53.Li A., Chen Y.S., Ping X.L., Yang X., Xiao W., Yang Y., Sun H.Y., Zhu Q., Baidya P., Wang X. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27:444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee Y., Choe J., Park O.H., Kim Y.K. Molecular mechanisms driving mRNA degradation by m6A modification. Trends Genet. 2020;36:177–188. doi: 10.1016/j.tig.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y.G., Chen R., Ahmad S., Verma R., Kasturi S.P., Amaya L., Broughton J.P., Kim J., Cadena C., Pulendran B. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell. 2019;76:96–109.e9. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen R.X., Chen X., Xia L.P., Zhang J.X., Pan Z.Z., Ma X.D., Han K., Chen J.W., Judde J.G., Deas O. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bochner B.R., Ames B.N. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- 58.Nagarajan A., Janostiak R., Wajapeyee N. Dot blot analysis for measuring global N6-methyladenosine modification of RNA. Methods Mol. Biol. 2019;1870:263–271. doi: 10.1007/978-1-4939-8808-2_20. [DOI] [PubMed] [Google Scholar]
- 59.Shu X., Cao J., Cheng M., Xiang S., Gao M., Li T., Ying X., Wang F., Yue Y., Lu Z. A metabolic labeling method detects m6A transcriptome-wide at single base resolution. Nat. Chem. Biol. 2020;16:887–895. doi: 10.1038/s41589-020-0526-9. [DOI] [PubMed] [Google Scholar]
- 60.Sugimoto Y., König J., Hussain S., Zupan B., Curk T., Frye M., Ule J. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 2012;13:R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia-Campos M.A., Edelheit S., Toth U., Safra M., Shachar R., Viukov S., Winkler R., Nir R., Lasman L., Brandis A. Deciphering the “m6A code” via antibody-independent quantitative profiling. Cell. 2019;178:731–747.e16. doi: 10.1016/j.cell.2019.06.013. [DOI] [PubMed] [Google Scholar]
- 62.Meyer K.D. DART-seq: An antibody-free method for global m6A detection. Nat. Methods. 2019;16:1275–1280. doi: 10.1038/s41592-019-0570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y., Xiao Y., Dong S., Yu Q., Jia G. Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine. Nat. Chem. Biol. 2020;16:896–903. doi: 10.1038/s41589-020-0525-x. [DOI] [PubMed] [Google Scholar]
- 64.Zhao J., Li B., Ma J., Jin W., Ma X. Photoactivatable RNA N6-methyladenosine editing with CRISPR-Cas13. Small. 2020;16:e1907301. doi: 10.1002/smll.201907301. [DOI] [PubMed] [Google Scholar]
- 65.Zhou Y., Zeng P., Li Y.H., Zhang Z., Cui Q. SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferrannini E., DeFronzo R.A. Impact of glucose-lowering drugs on cardiovascular disease in type 2 diabetes. Eur. Heart J. 2015;36:2288–2296. doi: 10.1093/eurheartj/ehv239. [DOI] [PubMed] [Google Scholar]
- 67.Matsumoto M., Pocai A., Rossetti L., Depinho R.A., Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 68.Roseman D.S., Khan T., Rajas F., Jun L.S., Asrani K.H., Isaacs C., Farelli J.D., Subramanian R.R. G6PC mRNA therapy positively regulates fasting blood glucose and decreases liver abnormalities in a mouse model of glycogen storage disease 1a. Mol. Ther. 2018;26:814–821. doi: 10.1016/j.ymthe.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chitraju C., Walther T.C., Farese R.V., Jr. The triglyceride synthesis enzymes DGAT1 and DGAT2 have distinct and overlapping functions in adipocytes. J. Lipid Res. 2019;60:1112–1120. doi: 10.1194/jlr.M093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Y., Shen F., Huang W., Qin S., Huang J.T., Sergi C., Yuan B.F., Liu S.M. Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. J. Clin. Endocrinol. Metab. 2019;104:665–673. doi: 10.1210/jc.2018-00619. [DOI] [PubMed] [Google Scholar]
- 71.Li Y., Ma Z., Jiang S., Hu W., Li T., Di S., Wang D., Yang Y. A global perspective on FOXO1 in lipid metabolism and lipid-related diseases. Prog. Lipid Res. 2017;66:42–49. doi: 10.1016/j.plipres.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 72.Xie W., Ma L.L., Xu Y.Q., Wang B.H., Li S.M. METTL3 inhibits hepatic insulin sensitivity via N6-methyladenosine modification of Fasn mRNA and promoting fatty acid metabolism. Biochem. Biophys. Res. Commun. 2019;518:120–126. doi: 10.1016/j.bbrc.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 73.Li X., Jiang Y., Sun X., Wu Y., Chen Z. METTL3 is required for maintaining β-cell function. Metabolism. 2021;116:154702. doi: 10.1016/j.metabol.2021.154702. [DOI] [PubMed] [Google Scholar]
- 74.Liu J., Luo G., Sun J., Men L., Ye H., He C., Ren D. METTL14 is essential for β-cell survival and insulin secretion. Biochim. Biophys. Acta Mol. Basis Dis. 2019;1865:2138–2148. doi: 10.1016/j.bbadis.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 75.De Jesus D.F., Zhang Z., Kahraman S., Brown N.K., Chen M., Hu J., Gupta M.K., He C., Kulkarni R.N. m6A mRNA methylation regulates human β-cell biology in physiological states and in type 2 diabetes. Nat. Metab. 2019;1:765–774. doi: 10.1038/s42255-019-0089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosen E.D., Spiegelman B.M. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhong X., Yu J., Frazier K., Weng X., Li Y., Cham C.M., Dolan K., Zhu X., Hubert N., Tao Y. Circadian clock regulation of hepatic lipid metabolism by modulation of m6A mRNA methylation. Cell Rep. 2018;25:1816–1828.e4. doi: 10.1016/j.celrep.2018.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song T., Yang Y., Wei H., Xie X., Lu J., Zeng Q., Peng J., Zhou Y., Jiang S., Peng J. Zfp217 mediates m6A mRNA methylation to orchestrate transcriptional and post-transcriptional regulation to promote adipogenic differentiation. Nucleic Acids Res. 2019;47:6130–6144. doi: 10.1093/nar/gkz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang Q., Sun B., Liu Q., Cai M., Wu R., Wang F., Yao Y., Wang Y., Wang X. MTCH2 promotes adipogenesis in intramuscular preadipocytes via an m6A-YTHDF1-dependent mechanism. FASEB J. 2019;33:2971–2981. doi: 10.1096/fj.201801393RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu R., Guo G., Bi Z., Liu Y., Zhao Y., Chen N., Wang F., Wang Y., Wang X. m6A methylation modulates adipogenesis through JAK2-STAT3-C/EBPβ signaling. Biochim. Biophys. Acta. Gene Regul. Mech. 2019;1862:796–806. doi: 10.1016/j.bbagrm.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 81.Deng K., Ren C., Liu Z., Gao X., Fan Y., Zhang G., Zhang Y., Ma E.S., Wang F., You P. Characterization of RUNX1T1, an adipogenesis regulator in ovine preadipocyte differentiation. Int. J. Mol. Sci. 2018;19:1300. doi: 10.3390/ijms19051300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao X., Yang Y., Sun B.F., Shi Y., Yang X., Xiao W., Hao Y.J., Ping X.L., Chen Y.S., Wang W.J. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mysling S., Kristensen K.K., Larsson M., Kovrov O., Bensadouen A., Jørgensen T.J., Olivecrona G., Young S.G., Ploug M. The angiopoietin-like protein ANGPTL4 catalyzes unfolding of the hydrolase domain in lipoprotein lipase and the endothelial membrane protein GPIHBP1 counteracts this unfolding. eLife. 2016;5:e20958. doi: 10.7554/eLife.20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang C.Y., Shie S.S., Wen M.S., Hung K.C., Hsieh I.C., Yeh T.S., Wu D. Loss of FTO in adipose tissue decreases Angptl4 translation and alters triglyceride metabolism. Sci. Signal. 2015;8:ra127. doi: 10.1126/scisignal.aab3357. [DOI] [PubMed] [Google Scholar]
- 85.Yadav P.K., Rajvanshi P.K., Rajasekharan R. The role of yeast m6A methyltransferase in peroxisomal fatty acid oxidation. Curr. Genet. 2018;64:417–422. doi: 10.1007/s00294-017-0769-5. [DOI] [PubMed] [Google Scholar]
- 86.Han B., Yan S., Wei S., Xiang J., Liu K., Chen Z., Bai R., Sheng J., Xu Z., Gao X. YTHDF1-mediated translation amplifies Wnt-driven intestinal stemness. EMBO Rep. 2020;21:e49229. doi: 10.15252/embr.201949229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Y.C., McPherson K., Marsh T., Gortmaker S.L., Brown M. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet. 2011;378:815–825. doi: 10.1016/S0140-6736(11)60814-3. [DOI] [PubMed] [Google Scholar]
- 88.Hinnouho G.M., Czernichow S., Dugravot A., Nabi H., Brunner E.J., Kivimaki M., Singh-Manoux A. Metabolically healthy obesity and the risk of cardiovascular disease and type 2 diabetes: the Whitehall II cohort study. Eur. Heart J. 2015;36:551–559. doi: 10.1093/eurheartj/ehu123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang X., Zhu L., Chen J., Wang Y. mRNA m6A methylation downregulates adipogenesis in porcine adipocytes. Biochem. Biophys. Res. Commun. 2015;459:201–207. doi: 10.1016/j.bbrc.2015.02.048. [DOI] [PubMed] [Google Scholar]
- 90.Kobayashi M., Ohsugi M., Sasako T., Awazawa M., Umehara T., Iwane A., Kobayashi N., Okazaki Y., Kubota N., Suzuki R. The RNA methyltransferase complex of WTAP, METTL3, and METTL14 regulates mitotic clonal expansion in adipogenesis. Mol. Cell. Biol. 2018;38:e00116-18. doi: 10.1128/MCB.00116-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cecil J.E., Tavendale R., Watt P., Hetherington M.M., Palmer C.N. An obesity-associated FTO gene variant and increased energy intake in children. N. Engl. J. Med. 2008;359:2558–2566. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- 92.Haupt A., Thamer C., Staiger H., Tschritter O., Kirchhoff K., Machicao F., Häring H.U., Stefan N., Fritsche A. Variation in the FTO gene influences food intake but not energy expenditure. Exp. Clin. Endocrinol. Diabetes. 2009;117:194–197. doi: 10.1055/s-0028-1087176. [DOI] [PubMed] [Google Scholar]
- 93.Prakash J., Srivastava N., Awasthi S., Agarwal C.G., Natu S.M., Rajpal N., Mittal B. Association of FTO rs17817449 SNP with obesity and associated physiological parameters in a north Indian population. Ann. Hum. Biol. 2011;38:760–763. doi: 10.3109/03014460.2011.614278. [DOI] [PubMed] [Google Scholar]
- 94.Qureshi S.A., Mumtaz A., Shahid S.U., Shabana N.A. rs3751812, a common variant in fat mass and obesity-associated (FTO) gene, is associated with serum high- and low-density lipoprotein cholesterol in Pakistani individuals. Nutrition. 2017;39-40:92–95. doi: 10.1016/j.nut.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 95.Karra E., O’Daly O.G., Choudhury A.I., Yousseif A., Millership S., Neary M.T., Scott W.R., Chandarana K., Manning S., Hess M.E. A link between FTO, ghrelin, and impaired brain food-cue responsivity. J. Clin. Invest. 2013;123:3539–3551. doi: 10.1172/JCI44403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen X., Luo Y., Jia G., Liu G., Zhao H., Huang Z. FTO promotes adipogenesis through inhibition of the Wnt/β-catenin signaling pathway in porcine intramuscular preadipocytes. Anim. Biotechnol. 2017;28:268–274. doi: 10.1080/10495398.2016.1273835. [DOI] [PubMed] [Google Scholar]
- 97.Wang X., Wu R., Liu Y., Zhao Y., Bi Z., Yao Y., Liu Q., Shi H., Wang F., Wang Y. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2020;16:1221–1235. doi: 10.1080/15548627.2019.1659617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu R., Liu Y., Yao Y., Zhao Y., Bi Z., Jiang Q., Liu Q., Cai M., Wang F., Wang Y., Wang X. FTO regulates adipogenesis by controlling cell cycle progression via m6A-YTHDF2 dependent mechanism. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018;1863:1323–1330. doi: 10.1016/j.bbalip.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 99.Wu R., Yao Y., Jiang Q., Cai M., Liu Q., Wang Y., Wang X. Epigallocatechin gallate targets FTO and inhibits adipogenesis in an mRNA m6A-YTHDF2-dependent manner. Int. J. Obes. 2018;42:1378–1388. doi: 10.1038/s41366-018-0082-5. [DOI] [PubMed] [Google Scholar]
- 100.Liu Q., Zhao Y., Wu R., Jiang Q., Cai M., Bi Z., Liu Y., Yao Y., Feng J., Wang Y., Wang X. ZFP217 regulates adipogenesis by controlling mitotic clonal expansion in a METTL3-m6A dependent manner. RNA Biol. 2019;16:1785–1793. doi: 10.1080/15476286.2019.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alisi A., Feldstein A.E., Villani A., Raponi M., Nobili V. Pediatric nonalcoholic fatty liver disease: A multidisciplinary approach. Nat. Rev. Gastroenterol. Hepatol. 2012;9:152–161. doi: 10.1038/nrgastro.2011.273. [DOI] [PubMed] [Google Scholar]
- 102.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.C., Torbenson M.S., Unalp-Arida A., Nonalcoholic Steatohepatitis Clinical Research Network Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 103.Liu W., Cao H., Yan J., Huang R., Ying H. “Micro-managers” of hepatic lipid metabolism and NAFLD. Wiley Interdiscip. Rev. RNA. 2015;6:581–593. doi: 10.1002/wrna.1295. [DOI] [PubMed] [Google Scholar]
- 104.Lawlor D.A., Callaway M., Macdonald-Wallis C., Anderson E., Fraser A., Howe L.D., Day C., Sattar N. Nonalcoholic fatty liver disease, liver fibrosis, and cardiometabolic risk factors in adolescence: A cross-sectional study of 1874 general population adolescents. J. Clin. Endocrinol. Metab. 2014;99:E410–E417. doi: 10.1210/jc.2013-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Luo Z., Zhang Z., Tai L., Zhang L., Sun Z., Zhou L. Comprehensive analysis of differences of N6-methyladenosine RNA methylomes between high-fat-fed and normal mouse livers. Epigenomics. 2019;11:1267–1282. doi: 10.2217/epi-2019-0009. [DOI] [PubMed] [Google Scholar]
- 106.Kang H., Zhang Z., Yu L., Li Y., Liang M., Zhou L. FTO reduces mitochondria and promotes hepatic fat accumulation through RNA demethylation. J. Cell. Biochem. 2018;119:5676–5685. doi: 10.1002/jcb.26746. [DOI] [PubMed] [Google Scholar]
- 107.Guo J., Ren W., Li A., Ding Y., Guo W., Su D., Hu C., Xu K., Chen H., Xu X. Fat mass and obesity-associated gene enhances oxidative stress and lipogenesis in nonalcoholic fatty liver disease. Dig. Dis. Sci. 2013;58:1004–1009. doi: 10.1007/s10620-012-2516-6. [DOI] [PubMed] [Google Scholar]
- 108.Lu N., Li X., Yu J., Li Y., Wang C., Zhang L., Wang T., Zhong X. Curcumin attenuates lipopolysaccharide-induced hepatic lipid metabolism disorder by modification of m6 A RNA methylation in piglets. Lipids. 2018;53:53–63. doi: 10.1002/lipd.12023. [DOI] [PubMed] [Google Scholar]
- 109.Canalis E., Giustina A., Bilezikian J.P. Mechanisms of anabolic therapies for osteoporosis. N. Engl. J. Med. 2007;357:905–916. doi: 10.1056/NEJMra067395. [DOI] [PubMed] [Google Scholar]
- 110.Shen G.S., Zhou H.B., Zhang H., Chen B., Liu Z.P., Yuan Y., Zhou X.Z., Xu Y.J. The GDF11-FTO-PPARγ axis controls the shift of osteoporotic MSC fate to adipocyte and inhibits bone formation during osteoporosis. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864:3644–3654. doi: 10.1016/j.bbadis.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 111.Wu Y., Xie L., Wang M., Xiong Q., Guo Y., Liang Y., Li J., Sheng R., Deng P., Wang Y. Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 2018;9:4772. doi: 10.1038/s41467-018-06898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yao Y., Bi Z., Wu R., Zhao Y., Liu Y., Liu Q., Wang Y., Wang X. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway via an m6A-YTHDF2-dependent manner. FASEB J. 2019;33:7529–7544. doi: 10.1096/fj.201802644R. [DOI] [PubMed] [Google Scholar]
- 113.Hodson L., Gunn P.J. The regulation of hepatic fatty acid synthesis and partitioning: The effect of nutritional state. Nat. Rev. Endocrinol. 2019;15:689–700. doi: 10.1038/s41574-019-0256-9. [DOI] [PubMed] [Google Scholar]
- 114.Madhavan S., Nagarajan S. GRP78 and next generation cancer hallmarks: An underexplored molecular target in cancer chemoprevention research. Biochimie. 2020;175:69–76. doi: 10.1016/j.biochi.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 115.Gupta A., Ajith A., Singh S., Panday R.K., Samaiya A., Shukla S. PAK2-c-Myc-PKM2 axis plays an essential role in head and neck oncogenesis via regulating Warburg effect. Cell Death Dis. 2018;9:825. doi: 10.1038/s41419-018-0887-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu Q., Luo Q., Halim A., Song G. Targeting lipid metabolism of cancer cells: A promising therapeutic strategy for cancer. Cancer Lett. 2017;401:39–45. doi: 10.1016/j.canlet.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 117.Meijer T.W.H., Peeters W.J.M., Dubois L.J., van Gisbergen M.W., Biemans R., Venhuizen J.H., Span P.N., Bussink J. Targeting glucose and glutamine metabolism combined with radiation therapy in non-small cell lung cancer. Lung Cancer. 2018;126:32–40. doi: 10.1016/j.lungcan.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 118.Hsu P.P., Sabatini D.M. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 119.Shen C., Xuan B., Yan T., Ma Y., Xu P., Tian X., Zhang X., Cao Y., Ma D., Zhu X. m6A-dependent glycolysis enhances colorectal cancer progression. Mol. Cancer. 2020;19:72. doi: 10.1186/s12943-020-01190-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu H., Zhao K., Zeng H., Li Z., Chen K., Zhang Z., Li E., Wu Z. N6-methyladenosine (m6A) methyltransferase WTAP accelerates the Warburg effect of gastric cancer through regulating HK2 stability. Biomed. Pharmacother. 2021;133:111075. doi: 10.1016/j.biopha.2020.111075. [DOI] [PubMed] [Google Scholar]
- 121.Yu H., Yang X., Tang J., Si S., Zhou Z., Lu J., Han J., Yuan B., Wu Q., Lu Q., Yang H. ALKBH5 inhibited cell proliferation and sensitized bladder cancer cells to cisplatin by m6A-CK2α-mediated glycolysis. Mol. Ther. Nucleic Acids. 2020;23:27–41. doi: 10.1016/j.omtn.2020.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li J., Zhu L., Shi Y., Liu J., Lin L., Chen X. m6A demethylase FTO promotes hepatocellular carcinoma tumorigenesis via mediating PKM2 demethylation. Am. J. Transl. Res. 2019;11:6084–6092. [PMC free article] [PubMed] [Google Scholar]
- 123.Zhong L., Liao D., Zhang M., Zeng C., Li X., Zhang R., Ma H., Kang T. YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett. 2019;442:252–261. doi: 10.1016/j.canlet.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 124.Cheng M., Sheng L., Gao Q., Xiong Q., Zhang H., Wu M., Liang Y., Zhu F., Zhang Y., Zhang X. The m6A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene. 2019;38:3667–3680. doi: 10.1038/s41388-019-0683-z. [DOI] [PubMed] [Google Scholar]
- 125.Xu Z., Jia K., Wang H., Gao F., Zhao S., Li F., Hao J. METTL14-regulated PI3K/Akt signaling pathway via PTEN affects HDAC5-mediated epithelial-mesenchymal transition of renal tubular cells in diabetic kidney disease. Cell Death Dis. 2021;12:32. doi: 10.1038/s41419-020-03312-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sheng H., Li Z., Su S., Sun W., Zhang X., Li L., Li J., Liu S., Lu B., Zhang S., Shan C. YTH domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mRNA translation. Carcinogenesis. 2020;41:541–550. doi: 10.1093/carcin/bgz152. [DOI] [PubMed] [Google Scholar]
- 127.Li Z., Peng Y., Li J., Chen Z., Chen F., Tu J., Lin S., Wang H. N6-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat. Commun. 2020;11:2578. doi: 10.1038/s41467-020-16306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zuo X., Chen Z., Gao W., Zhang Y., Wang J., Wang J., Cao M., Cai J., Wu J., Wang X. m6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020;13:5. doi: 10.1186/s13045-019-0839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen A., Chen X., Cheng S., Shu L., Yan M., Yao L., Wang B., Huang S., Zhou L., Yang Z., Liu G. FTO promotes SREBP1c maturation and enhances CIDEC transcription during lipid accumulation in HepG2 cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018;1863:538–548. doi: 10.1016/j.bbalip.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 130.Elkashef S.M., Lin A.P., Myers J., Sill H., Jiang D., Dahia P.L.M., Aguiar R.C.T. IDH mutation, competitive inhibition of FTO, and RNA methylation. Cancer Cell. 2017;31:619–620. doi: 10.1016/j.ccell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Su R., Dong L., Li C., Nachtergaele S., Wunderlich M., Qing Y., Deng X., Wang Y., Weng X., Hu C. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 2018;172:90–105.e23. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen P., Liu X.Q., Lin X., Gao L.Y., Zhang S., Huang X. Targeting YTHDF1 effectively re-sensitizes cisplatin-resistant colon cancer cells by modulating GLS-mediated glutamine metabolism. Mol. Ther. Oncolytics. 2021;20:228–239. doi: 10.1016/j.omto.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ray U., Roy S.S. Aberrant lipid metabolism in cancer cells—The role of oncolipid-activated signaling. FEBS J. 2018;285:432–443. doi: 10.1111/febs.14281. [DOI] [PubMed] [Google Scholar]
- 134.Li Z., Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell. Mol. Life Sci. 2016;73:377–392. doi: 10.1007/s00018-015-2070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tabe Y., Lorenzi P.L., Konopleva M. Amino acid metabolism in hematologic malignancies and the era of targeted therapy. Blood. 2019;134:1014–1023. doi: 10.1182/blood.2019001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Matés J.M., Segura J.A., Martín-Rufián M., Campos-Sandoval J.A., Alonso F.J., Márquez J. Glutaminase isoenzymes as key regulators in metabolic and oxidative stress against cancer. Curr. Mol. Med. 2013;13:514–534. doi: 10.2174/1566524011313040005. [DOI] [PubMed] [Google Scholar]
- 137.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yin L., Zhu X., Novák P., Zhou L., Gao L., Yang M., Zhao G., Yin K. The epitranscriptome of long noncoding RNAs in metabolic diseases. Clin. Chim. Acta. 2021;515:80–89. doi: 10.1016/j.cca.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 139.Kim J., Lee G. Metabolic control of m6A RNA modification. Metabolites. 2021;11:80. doi: 10.3390/metabo11020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shi S., Kong N., Feng C., Shajii A., Bejgrowicz C., Tao W., Farokhzad O.C. Drug delivery strategies for the treatment of metabolic diseases. Adv. Healthc. Mater. 2019;8:e1801655. doi: 10.1002/adhm.201801655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Deng X., Su R., Weng H., Huang H., Li Z., Chen J. RNA N6-methyladenosine modification in cancers: Current status and perspectives. Cell Res. 2018;28:507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen B., Ye F., Yu L., Jia G., Huang X., Zhang X., Peng S., Chen K., Wang M., Gong S. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J. Am. Chem. Soc. 2012;134:17963–17971. doi: 10.1021/ja3064149. [DOI] [PubMed] [Google Scholar]
- 143.Wang R., Han Z., Liu B., Zhou B., Wang N., Jiang Q., Qiao Y., Song C., Chai J., Chang J. Identification of natural compound radicicol as a potent FTO inhibitor. Mol. Pharm. 2018;15:4092–4098. doi: 10.1021/acs.molpharmaceut.8b00522. [DOI] [PubMed] [Google Scholar]
- 144.Peng S., Xiao W., Ju D., Sun B., Hou N., Liu Q., Wang Y., Zhao H., Gao C., Zhang S. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 2019;11:eaau7116. doi: 10.1126/scitranslmed.aau7116. [DOI] [PubMed] [Google Scholar]
- 145.Singh B., Kinne H.E., Milligan R.D., Washburn L.J., Olsen M., Lucci A. Important role of FTO in the survival of rare panresistant triple-negative inflammatory breast cancer cells facing a severe metabolic challenge. PLoS ONE. 2016;11:e0159072. doi: 10.1371/journal.pone.0159072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., Sun G., Lu Z., Huang Y., Yang C.G. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Panneerdoss S., Eedunuri V.K., Yadav P., Timilsina S., Rajamanickam S., Viswanadhapalli S., Abdelfattah N., Onyeagucha B.C., Cui X., Lai Z. Cross-talk among writers, readers, and erasers of m6A regulates cancer growth and progression. Sci. Adv. 2018;4:eaar8263. doi: 10.1126/sciadv.aar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang Y., Su R., Sheng Y., Dong L., Dong Z., Xu H. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019;35:677–691. doi: 10.1016/j.ccell.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Aik W., Scotti J.S., Choi H., Gong L., Demetriades M., Schofield C.J., McDonough M.A. Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014;42:4741–4754. doi: 10.1093/nar/gku085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kloor D., Osswald H. S-Adenosylhomocysteine hydrolase as a target for intracellular adenosine action. Trends Pharmacol. Sci. 2004;25:294–297. doi: 10.1016/j.tips.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 151.Zhang L., Qi Y., ALuo Z., Liu S., Zhang Z., Zhou L. Betaine increases mitochondrial content and improves hepatic lipid metabolism. Food Funct. 2019;10:216–223. doi: 10.1039/c8fo02004c. [DOI] [PubMed] [Google Scholar]
- 152.Zhou S., Bai Z.L., Xia D., Zhao Z.J., Zhao R., Wang Y.Y., Zhe H. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol. Carcinog. 2018;57:590–597. doi: 10.1002/mc.22782. [DOI] [PubMed] [Google Scholar]
- 153.Li H.B., Tong J., Zhu S., Batista P.J., Duffy E.E., Zhao J., Bailis W., Cao G., Kroehling L., Chen Y. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Han D., Liu J., Chen C., Dong L., Liu Y., Chang R., Huang X., Liu Y., Wang J., Dougherty U. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang S., Wei J., Cui Y.H., Park G., Shah P., Deng Y., Aplin A.E., Lu Z., Hwang S., He C., He Y.Y. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019;10:2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jabs S., Biton A., Bécavin C., Nahori M.A., Ghozlane A., Pagliuso A., Spanò G., Guérineau V., Touboul D., Giai Gianetto Q. Impact of the gut microbiota on the m6A epitranscriptome of mouse cecum and liver. Nat. Commun. 2020;11:1344. doi: 10.1038/s41467-020-15126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]