Abstract
POU domain proteins have been implicated as key regulators during development and lymphocyte activation. The POU domain protein T-cell factor β1 (TCFβ1), which binds octamer and octamer-related sequences, is a potent transactivator. In this study, we showed that TCFβ1 is phosphorylated following activation via the T-cell receptor or by stress-induced signals. Phosphorylation of TCFβ1 occurred predominantly at serine and threonine residues. Signals which upregulate Jun kinase (JNK)/stress-activated protein kinase activity also lead to association of JNK with TCFβ1. JNK associates with the activation domain of TCFβ1 and phosphorylates its DNA binding domain. The phosphorylation of recombinant TCFβ1 by recombinant JNK enhances the ability of TCFβ1 to bind to a consensus octamer motif. Consistent with this conclusion, TCFβ1 upregulates reporter gene transcription in an activation- and JNK-dependent manner. In addition, inhibition of JNK activity by catalytically inactive MEKK (in which methionine was substituted for the lysine at position 432) also inhibits the ability of TCFβ1 to drive inducible transcription from the interleukin-2 promoter. These results suggest that stress-induced signals and T-cell activation induce JNK, which then acts on multiple cis sequences by modulating distinct transactivators like c-Jun and TCFβ1. This demonstrates a coupling between the JNK activation pathway and POU domain proteins and implicates TCFβ1 as a physiological target in the JNK signal transduction pathway leading to coordinated biological responses.
The demonstrated importance of octamer and octamer-like motifs in expression of a number of genes suggests that octamer-binding proteins are essential for both constitutive (36, 37) and inducible (3, 18–20, 43) gene expression. In addition, octamer motifs have been shown to regulate both lineage-specific (10, 37, 40) and ubiquitous (35, 38, 40) gene expression. POU proteins are the major transactivators which bind octamer and octamer-related sequences and upregulate transcription in an octamer-dependent manner (for reviews, see references 14 and 36). Spontaneous mutations or targeted disruption of a number of POU proteins has dramatic effects during development (5, 11, 25). We have previously cloned a novel POU domain protein, T-cell factor β1 (TCFβ1), which is the only member of a new class (class VI) of POU domain proteins and whose transcript levels are highest in the thymus and brain (30). Although it is a bonafide POU domain protein, it is distantly related to other known members of the POU family of transactivators. TCFβ1 binds to octamer and octamer-related sequences from a number of genes and is a potent transactivator (30). Until we cloned TCFβ1 (30), only two other POU proteins, Oct1 and Oct2, were known to be expressed in lymphocytes (36). TCFβ1 has also been subsequently cloned by others and variously termed Brn5 (1), pou[c] (16), Emb (33), and mPOU (45). The ability of TCFβ1 to bind an inducible element in the proximal octamer motif in the interleukin-2 (IL-2) promoter (see below) suggests that it might be involved in an activation-dependent pathway. This is consistent with the observation that Oct2-null mice have deficits in lipopolysaccharide-induced secretion of immunoglobulins in the B-cell lineage (5).
Activation of cells by growth factors and other extracellular stimuli is known to result in activation of a set of serine/threonine kinases. These include the extracellular signal-related kinases (ERKs) as well as the stress-activated protein kinases (SAPKs), or Jun kinases (JNKs). A major function of JNK is the phosphorylation of the c-Jun component of the AP-1 transcription factor, thereby regulating its transactivating function in various gene promoters, including that of the IL-2 gene (15, 39). The JNK (SAPK) family members are activated by UV irradiation, tumor necrosis factor alpha, cycloheximide, heat shock, and T-cell activation (6, 8, 15, 17, 22, 31, 39). The JNK family members display a sequence similarity of ∼83% to each other (8, 17, 22) and exhibit ∼40% sequence homology to other mitogen-activated protein kinases, such as ERK2. Three JNK genes have been cloned: JNK1 (46 kDa) and its rat homologue, SAPKγ; JNK2 (55 kDa) and its rat homologue, SAPKαII; and the SAPKβ gene (8, 22). The SAPKαI transcript is an alternatively spliced form of the JNK2 gene (17, 22). These kinases define a subgroup of mitogen-activated protein kinases that share the sequence Thr-Pro-Tyr in their activating phosphorylation sites (8, 17, 22), in contrast to the Thr-Glu-Tyr sequence in the ERK1 and ERK2 genes. The two JNKs are activated identically by a diverse set of stimuli (8, 17, 22). The similarity in regulation of different members of the JNK family suggests that they may have redundant functions. Despite essentially identical regulation, the two JNKs are differ considerably in their ability to bind c-Jun. JNK2 binds c-Jun with a much higher affinity than does JNK1 and thus phosphorylates it more efficiently, and it has been shown to be a better inducer of c-Jun promoter activity (17).
In this study, we investigated the role of JNK in activation-dependent phosphorylation of a POU domain protein, TCFβ1. We showed that TCFβ1 is phosphorylated after activation via the T-cell receptor or by stress-induced signals like UV light. In addition, we demonstrated that JNK binds the activation domain of TCFβ1 and phosphorylates its DNA binding domain. Further, we mapped the phosphorylation site in the DNA binding domain. Phosphorylation of TCFβ1 increased its ability to bind to octamer motifs. We also demonstrated that activation-induced transactivation by TCFβ1 is dependent on JNK activity. This suggests a mechanism by which activation-dependent signals can integrate at octamer motifs implicated in inducible gene expression.
MATERIALS AND METHODS
GST fusion proteins.
The cDNAs encoding TCFβ1 and its fragments were cloned in frame into the pGEX vector. The full-length human TCFβ1 (amino acid residues 1 to 301), the DNA binding domain of TCFβ1 (residues 145 to 301), and the activation domain of TCFβ1 (residues 10 to 145) were expressed as glutathione S-transferase (GST) fusion proteins in Escherichia coli and purified as described previously (12). The GST–c-Jun (1-223) vector was obtained from M. Karin (University of California, San Diego [UCSD]). Site-specific mutagenesis of the TCFβ1 protein was undertaken by PCR, and the target sequence was confirmed at least twice by sequencing. The mutant TCFβ1 GST fusion proteins were made by standard methods and had their predicted molecular weights.
Transient transfections.
Human Jurkat T cells were grown in RPMI 1640 medium (GIBCO) supplemented with 10% fetal bovine serum (GIBCO), 10 mM HEPES (pH 7.3), 2 mM l-glutamine, 1 mM sodium pyruvate, and 100 U of penicillin-streptomycin/ml. The full-length TCFβ1 cDNA (residues 1 to 301) or the activation domain of TCFβ1 (residues 10 to 145) was expressed in Jurkat cells under the control of the SRα promoter. These vectors expressed proteins which were HA epitope (human influenza virus hemagglutinin nonapeptide, Tyr-Pro-Tyr-Asp-Val-Pro-Asp-Tyr-Ala) tagged at their N termini. The expression vectors for HA-JNK1 (8), HA-JNK2 (17), and the dominant-negative mutant of MEKK were obtained from M. Karin (UCSD). Jurkat cells (107) were washed twice with serum-free RPMI 1640 medium, resuspended in 400 μl of serum-free medium, mixed with 15 μg of DNA in a Bio-Rad 0.4-cm-light-path cuvette, and kept on ice for 10 min. The cells were then electroporated at 260 V and 960 μF (Gene-Pulser; Bio-Rad) and kept on ice for an additional 10 min before being resuspended in complete RPMI 1640 medium containing 10% fetal bovine serum. After 48 h, the cells were stimulated for various time periods and then harvested.
Luciferase assays.
Briefly, different IL-2 promoter constructs was cotransfected with various expression vectors. The variations in transfection efficiencies were normalized by using the pCMV β-galactosidase expression vector. Forty hours posttransfection, the cells were activated with phorbol myristate acetate (PMA) plus ionomycin and incubated for another 12 to 18 h. The cells were harvested, washed three times with phosphate-buffered saline (PBS), and lysed in 100 μl of the lysis buffer (see below). Cell debris was removed by centrifugation, and the supernatant was used in the luciferase assay employing a Monolight model 2010 luminometer.
Activation and cell lysis.
Approximately 107 Jurkat cells were washed twice with PBS, resuspended in 100 μl of PBS, and activated at 37°C with PMA (5 ng/ml), phytohemagglutinin (PHA; 5 μg/ml), anti-CD3 (OKT3) antibody (10 μg/ml), anti-CD28 antibody (2 μg/ml), and UV-C (40 J/m2) for various periods of time. Stimulation was terminated by adding 1 ml of cold lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Brij 96, 50 μg of aprotinin/ml, 50 μg of leupeptin/ml, and 1 mM Na3VO4. Nuclei and cell debris were removed by centrifugation at 13,000 × g for 15 min at 4°C. Five-microgram quantities of the control GST protein and the different GST fusion proteins were separately mixed with 1 ml of Jurkat cell lysate and incubated for from 4 h to overnight at 4°C prior to incubation with 30 μl of glutathione-Sepharose beads for 1 h. Immobilized fusion proteins were then washed four times with lysis buffer and once with kinase buffer and then subjected to the kinase assay as described below. Immunoprecipitation of transfected JNK was done as described elsewhere (15). In some experiments, transiently transfected Jurkat cells were resuspended in 0.5 ml of phosphate-free RPMI medium and metabolically labelled for 4 h with 0.5 mCi of 32Pi (9,120 Ci/mmol; DuPont NEN, Boston, Mass.) prior to activation for the last 30 min of labelling.
Immunoprecipitations and kinase assays.
HA epitope-tagged proteins were immunoprecipitated from cell lysates with 5 to 10 μg of anti-HA antibody (12CA5) at 4°C for 4 h and then incubated with 40 μl of protein A-Sepharose for 1 to 2 h. Immune complexes were washed four times with lysis buffer and once in kinase buffer (10 mM HEPES [pH 7.4], 5 mM MnCl2, 5 mM MgCl2, 0.1% Nonidet P-40, 5 mM dithiothreitol) and then resuspended in 30 μl of kinase buffer supplemented with 1 μM ATP and 10 μCi of [γ-32P]ATP (Dupont NEN; 7,000 Ci/mmol). Kinase reactions were carried out at 30°C for 15 min and stopped by washing once with kinase buffer; proteins eluted with 2× sodium dodecyl sulfate (SDS) sample buffer (60 mM Tris [pH 6.8], 2.3% SDS, 10% glycerol, 5% β-mercaptoethanol), resolved by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to Immobilon membranes (Millipore, Bedford, Mass.), and subjected to autoradiography. Recombinant JNK2 was used to phosphorylate TCFβ1 wild-type and mutant peptides. Peptides A, B, and C were synthesized by using the La Jolla Institute for Allergy and Immunology peptide synthesizer and purified to homogeneity by high-performance liquid chromatography. The phosphorylated peptides were then analyzed on 15% Tricine gels.
Tryptic peptide mapping.
Phosphorylated TCFβ1 proteins were resolved by SDS-PAGE, transferred to Immobilon membranes, localized by autoradiography, and excised. The membranes were then treated with polyvinylpyrrolidone 360 and N-tosyl-l-phenylalanine chloromethyl ketone-treated trypsin (27). The resulting phosphopeptides were separated by electrophoresis on cellulose thin-layer chromatography plates at pH 1.9 for 27 min followed by ascending chromatography in n-butanol–pyridine–acetic acid–water (75:50:15:60). Labelled peptides were detected by autoradiography.
Phospho-amino acid (PAA) analysis.
Phosphorylated TCFβ1 proteins were resolved by SDS-PAGE and transferred to Immobilon membranes as described above. Following autoradiography, bands corresponding to labelled proteins were excised, washed extensively with distilled water, and subjected to hydrolysis in 100 μl of 6 N HCl at 110°C for 1 h. The membranes were then discarded, 1 ml of distilled water was added to the tube, and samples were lyophilized and then resuspended in 10 μl of electrophoresis buffer (pH 1.9) containing 1 μg of each amino acid standard (phosphoserine, phosphothreonine, and phosphotyrosine). Samples were spotted on cellulose thin-layer plates and analyzed by two-dimensional thin-layer electrophoresis at pH 1.9 and then at pH 3.5. Nonradioactive standards were detected by staining with 0.25% ninhydrin in acetone, and labelled amino acids were detected by autoradiography.
DNA binding assays.
Purified TCFβ1 fusion proteins were phosphorylated by recombinant JNK2 in the presence or absence of 30 μM exogenous ATP and were then used in a gel shift assay (30). DNA binding reactions were carried out for 20 min at 4°C in a buffer containing 50 mM HEPES (pH 7.8), 20 mM MgCl2, 0.5 mM EDTA, 20 mM spermidine, 500 μg of bovine serum albumin/ml, 10 mM dithiothreitol, 75% glycerol, and 104 cpm of labelled probe. The probe used was a 30-mer double-stranded synthetic oligonucleotide containing the sequence 5′ TTTGAAATATGTGTAATATGTAAAACAT 3′ of the proximal octamer site in the human IL-2 promoter (18). Each strand was labelled separately with T4 polynucleotide kinase (Bethesda Research Laboratories) and [γ-32P]ATP [5,000 Ci/mmol], and the strands were then slowly allowed to reanneal. The samples were analyzed on a 4% nondenaturing acrylamide gel in 0.5× Tris-borate-EDTA. In some experiments, nuclear extracts from transfected Jurkat cells were made as described previously (28). Binding reaction mixtures contained 5 to 10 μg of nuclear extract, 32P-labelled probe (25,000 cpm), and 2 μg of poly(dI-dC) in the binding buffer. For supershift experiments, extracts were preincubated with 1 μl of anti-HA antibody at 4°C for 45 min before addition of the probe.
RESULTS
Activation-dependent phosphorylation of TCFβ1.
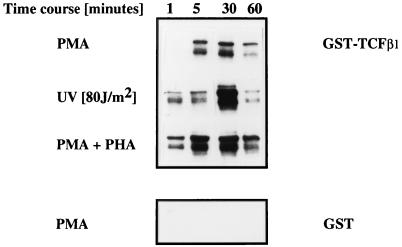
To determine whether TCFβ1 undergoes activation-dependent phosphorylation, we generated and purified a GST fusion protein of TCFβ1 and analyzed the abilities of resting and activated Jurkat cell lysates to phosphorylate GST-TCFβ1. Activation of Jurkat cells with diverse stimuli such as PMA, PMA plus PHA, or stress-inducing signals like UV light (Fig. 1) induced phosphorylation of TCFβ1. In addition, we found that activation by anti-CD3 plus anti-CD28 antibodies also induced phosphorylation of TCFβ1 (data not shown). Incubation of GST-TCFβ1 with cell lysates from Jurkat T cells activated with PMA, PMA plus PHA, or UV light for various lengths of time showed that maximum phosphorylation of TCFβ1 occurred at 30 min after activation, whereas the control GST protein was not phosphorylated (Fig. 1).
FIG. 1.
TCFβ1 is phosphorylated in vitro in an activation-dependent manner. Full-length TCFβ1 (residues 1 to 301) expressed as a GST fusion protein was incubated with cell extracts from Jurkat cells activated with PMA (5 ng/ml), PMA (5 ng/ml) plus PHA (5 μg/ml), or UV for the indicated lengths of time, processed for in vitro kinase assay, analyzed by SDS-PAGE, and visualized by autoradiography. GST alone was used as a negative control.
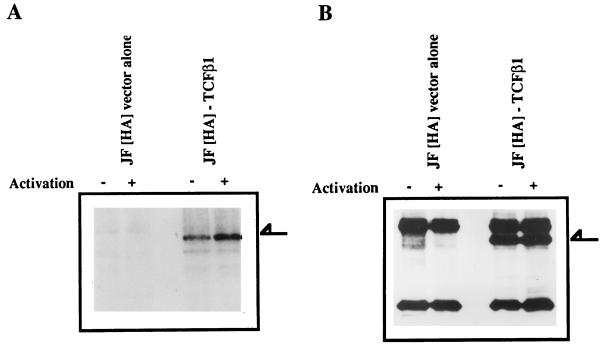
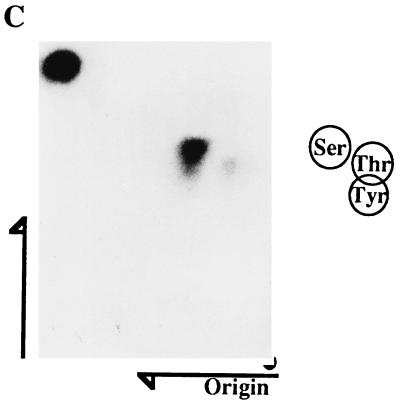
In an attempt to clarify whether TCFβ1 was also phosphorylated in vivo by activation-dependent serine/threonine kinases, we generated TCFβ1 expression vectors. These vectors expressed the full-length TCFβ1 (residues 1 to 301) as HA epitope-tagged proteins. Cells transfected with such vectors expressed the appropriately sized HA epitope-tagged TCFβ1 protein, as determined by Western immunoblotting of lysates from transfected Jurkat cells with 12CA5, an HA-specific monoclonal antibody (Fig. 2B). These experiments were done in Jurkat T cells which were stably transfected with the simian virus 40 large T antigen to ensure a high level of expression by this vector in these cells. To detect phosphorylation of TCFβ1 in vivo, the cells were metabolically labelled with inorganic 32P for the last 4 h of transfection. During the final 30 min of labelling, Jurkat cells were activated with PHA plus PMA. A two- to threefold increase in phosphorylation of full-length TCFβ1 upon activation (Fig. 2A) was reproducibly demonstrated. A Western blot analysis using an antibody generated against the epitope (12CA5) showed that there were equal amounts of proteins in the two lanes (Fig. 2B). A PAA analysis of TCFβ1 phosphorylated in vivo (Fig. 2C) showed predominantly phosphoserine and some phosphothreonine but no phosphotyrosine content. These data suggest that TCFβ1 is phosphorylated in vivo on serine and threonine residues.
FIG. 2.
TCFβ1 is phosphorylated in vivo by serine/threonine kinases. (A) Phosphorylation of epitope-tagged TCFβ1 in Jurkat cells after activation. Jurkat cells that were transfected with pJF-HA vector alone, or the same vector expressing full-length TCFβ1, and then metabolically labelled (32P) were activated with PMA (5 ng/ml) plus PHA (5 μg/ml). TCFβ1 was immunoprecipitated from Jurkat cell lysates with anti-HA antibody, resolved on SDS-PAGE gels, transferred to Immobilon membranes, and visualized by autoradiography. The arrow indicates the TCFβ1 band. (B) Expression of epitope-tagged TCFβ1 in Jurkat cells after transient transfection. Jurkat cells were transfected, and 48 h later, expression of HA-TCFβ1 protein was determined by Western immunoblotting. TCFβ1 was immunoprecipitated from Jurkat cell lysates with anti-HA antibody (12CA5), resolved on SDS-polyacrylamide gels, and Western blot probed with anti-HA antibody. The western immunoblot shows that the levels of expression of HA-TCFβ1 in activated and unactivated Jurkat cells are similar, as evident in panel A. The arrow indicates the TCFβ1 band. (C) PAA analysis of full-length TCFβ1, labelled in vivo, from activated Jurkat cells. Phosphoserine and some phosphothreonine were detectable, but phosphotyrosine was not.
JNK binds the activation domain of TCFβ1.
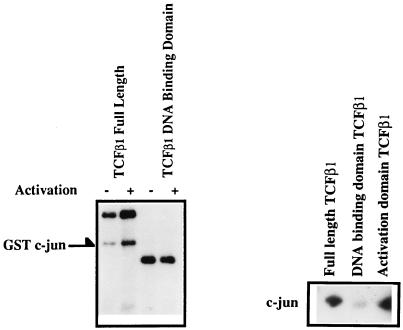
The amino acid sequence of TCFβ1 revealed the presence of potential JNK phosphorylation sites. This suggested the possibility that JNK was a likely candidate for a kinase which could phosphorylate TCFβ1. This was especially relevant since TCFβ1 was phosphorylated by T-cell activation signals and also by stress-inducing signals like UV irradiation (Fig. 1A). Since analysis of JNK activity had been facilitated by its ability to bind the activation domain of c-Jun, we decided to do “fishing” experiments with GST fusion proteins of full-length TCFβ1 or its DNA binding domain and determine whether JNK associates with TCFβ1. This was done by allowing cell lysates from Jurkat cells activated with anti-CD3 and anti-CD28 antibodies to bind GST fusion proteins containing either full-length TCFβ1 or its activation or DNA binding domain. Proteins bound to GST-TCFβ1 were then immobilized on glutathione-agarose beads and washed several times to remove the unbound proteins. Exogenous GST–c-Jun was then added as the substrate in an in vitro kinase assay. Such experiments showed that full-length TCFβ1 and the activation domain of TCFβ1 bound an activation-dependent kinase which phosphorylated c-Jun in vitro (Fig. 3). In contrast, the DNA binding domain of TCFβ1 did not bind this kinase (Fig. 3). Tryptic peptide maps of phosphorylated c-Jun were similar to those previously described after phosphorylation of c-Jun with JNK (data not shown), suggesting association of JNK with the activation domain of TCFβ1.
FIG. 3.
Proteins which associate with TCFβ1 phosphorylate GST–c-Jun in vitro. Bacterially expressed GST fusion proteins of full-length TCFβ1 (residues 1 to 301), the activation domain of TCFβ1 (residues 1 to 144), or the DNA binding domain of TCFβ1 (residues 145 to 301) were incubated with cell lysates from unactivated (−) Jurkat cells (left panel) or Jurkat cells activated (+) with anti-CD3 plus anti-CD28 (left panel) or were incubated with activated recombinant JNK2 (a kind gift from M. Karin) (right panel). The proteins associated with GST-TCFβ1 were immobilized on glutathione-agarose beads. GST–c-Jun was subsequently added to the kinase reaction mixture, and the phosphorylated proteins were resolved on SDS-polyacrylamide gels and analyzed by autoradiography.
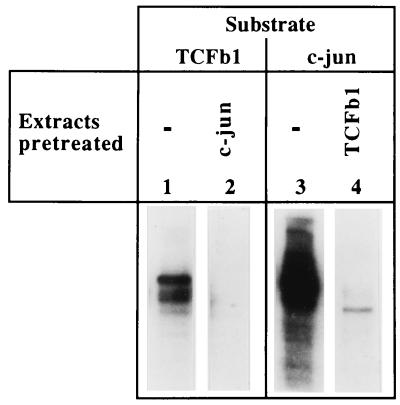
We extended these studies by performing criss-cross depletions with TCFβ1 and c-Jun. Activated extracts phosphorylate both TCFβ1 and c-Jun in an activation-dependent manner. Such extracts were pretreated to remove proteins which bind to the activation domain of c-Jun. These pretreated extracts were no longer able to phosphorylate TCFβ1 (Fig. 4). In the criss-cross experiment, extracts were depleted with immobilized GST-TCFβ1 beads, and such depleted extracts were then unable to phosphorylate c-Jun (Fig. 4). These studies suggested that c-Jun kinases bind and phosphorylate TCFβ1. Consistent with our conclusions that the activation domain of TCFβ1 binds JNK, homologous conserved motifs can be identified in both the δ region (residues 20 to 80) of c-Jun and the activation domain (residues 1 to 145) of TCFβ1 (Table 1).
FIG. 4.
The kinase which phosphorylates TCFβ1 is depleted by using immobilized c-Jun, and the c-Jun kinase activity is depleted by using immobilized GST-TCFβ1 beads. Activated extracts which contained c-Jun kinase activity were first incubated with immobilized GST-TCFβ1, and the ability of the flow-through to phosphorylate c-Jun in vitro was then determined. Alternatively, the ability of extract to phosphorylate TCFβ1 was determined after running it over immobilized GST–c-Jun. These criss-cross absorption studies suggest that the same kinase (JNK) which binds and phosphorylates the activation domain of c-Jun also binds and phosphorylates TCFβ1.
TABLE 1.
Conservation of sequences between JNK binding sites in c-Jun and TCFβ1a
| Motif | Protein | Sequence |
|---|---|---|
| A | TCFβ1 | 130 S.KPHTPSLDED 140 |
| c-Jun | 48 SLKPHLRAKNSD 59 | |
| v-Jun | ........NNAD | |
| Conserved sequence | S-KPH------D | |
| B | TCFβ1 | 67 PVAVRKPSTPE.SP 79 |
| TCFβ1 | 103 PAPAAKPSASAPIP 116 | |
| c-Jun | 51 PHLRAKNSDLLTSP 64 | |
| c-Jun | 31 P.KILKQSMTLNLA 43 | |
| v-Jun | .....NNADILTSP | |
| Conserved sequence | P----K-S----SP | |
| C | TCFβ1 | 128 LVSKPHTPSLDED 140 |
| TCFβ1 | 38 LQVQAVTPQLLLN 50 | |
| c-Jun | 33 ILKQSMTLNLADP 45 | |
| v-Jun | ................ | |
| Conserved sequence | LV----T--L-D- |
Sequence comparisons between the JNK binding sites in c-Jun (amino acids 20 to 80) and the activation domain of TCFβ1 (amino acids 1 to 145) reveal a modest degree of sequence conservation. The residue 20 to 80 region of c-Jun was used as a comparison since c-Jun1–223, c-Jun1–144, and c-Jun1–79 bind JNK activity whereas c-Jun43–223 and v-Jun do not bind JNK activity. The symbol “.” denotes a deletion, whereas the symbol “-” suggests the absence of sequence conservation. The consensus conserved sequence is shown at the bottom of each motif. The numbers denote the amino acid numbers in the native TCFβ1 and c-Jun proteins.
JNK2 phosphorylates TCFβ1.
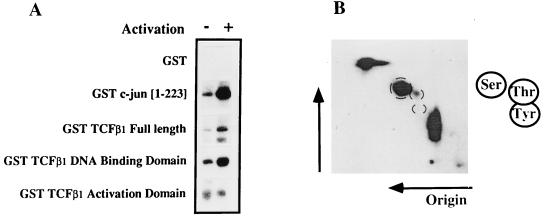
To determine if TCFβ1 is a substrate for JNK2, we transfected Jurkat T cells with JNK2 expression vectors (8, 17). These vectors drive expression of the JNK2 kinase with an N-terminal HA epitope tag. The JNK2 from such cells was activated by UV irradiation and then immunoprecipitated with HA-specific antibodies 48 h after transfection. The ability of immunoprecipitated JNK2 to phosphorylate various substrates was then tested in an in vitro kinase assay. Our results show that JNK2 phosphorylates the TCFβ1 DNA binding domain, but not its activation domain, in an activation-dependent manner (Fig. 5A). As expected, JNK2 phosphorylated GST–c-Jun but not the control GST protein (Fig. 5A). PAA analysis of the DNA binding domain of TCFβ1, phosphorylated in vitro by JNK2, indicated that it contained both phosphoserine and phosphothreonine residues (Fig. 5B).
FIG. 5.
JNK2 phosphorylates the DNA binding domain, on Ser and Thr residues, but not the activation domain of TCFβ1. (A) JNK2 phosphorylates the DNA binding domain of TCFβ1. Jurkat cells were transfected with a vector expressing HA-JNK2, and lysates were prepared from either unactivated cells or cells activated with anti-CD3 and anti-CD28 antibodies. The HA-JNK2 protein was immunoprecipitated with anti-HA antibody immobilized on protein A-Sepharose beads and then used in kinase reactions. Bacterially expressed GST fusion proteins containing full-length TCFβ1 (residues 1 to 301), the DNA binding domain of TCFβ1 (residues 145 to 301), the activation domain of TCFβ1 (residues 10 to 145), and c-Jun (residues 1 to 223) were then added to the HA-JNK2-dependent kinase reactions. GST alone was used as a negative control. The products of the kinase reactions were then analyzed on SDS-polyacrylamide gels and autoradiographed. (B) PAA analysis of the DNA binding domain of TCFβ1, phosphorylated in vitro by activated JNK2. TCFβ1 is primarily phosphorylated at serine and threonine residues. No phosphotyrosine was detected.
Identification of JNK2 phosphorylation sites in TCFβ1.
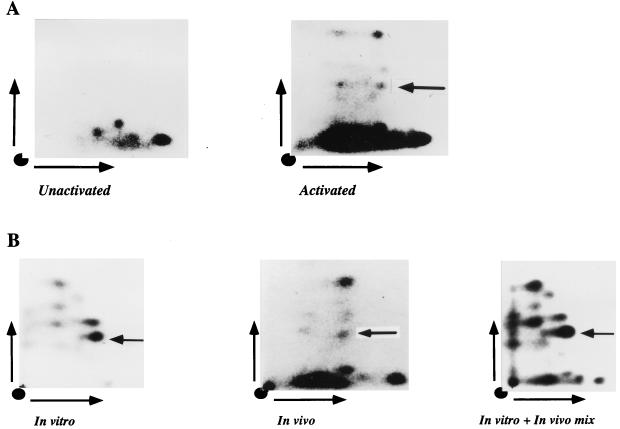
Full-length TCFβ1 was phosphorylated in vivo, as shown in Fig. 2, by transfecting Jurkat cells with vectors driving expression of epitope-tagged TCFβ1. Comparison of tryptic maps generated from unactivated (Fig. 6A, left panel) and activated (Fig. 6A, right panel) cells showed that activated cells contained additional phosphopeptides. If the ability of JNK2 to phosphorylate TCFβ1 in vitro reflects that TCFβ1 is a physiological target of JNK activity, when TCFβ1 is phosphorylated in vivo, it should be possible to identify phosphopeptides identical to those generated by TCFβ1 that has been phosphorylated in vitro. TCFβ1 was phosphorylated in vivo as described above. TCFβ1 was phosphorylated in vitro by JNK2 as described in the legend to Fig. 5. We then compared the two-dimensional chromatographic patterns of tryptic phosphopeptides generated from TCFβ1 protein phosphorylated in vitro with JNK2 (Fig. 6B, left panel) with those generated from TCFβ1 phosphorylated in vivo (Fig. 6B, middle panel). At least one common tryptic phosphopeptide can be identified when the in vitro panel is compared to the in vivo panel. To strengthen this argument, we did experiments in which we mixed tryptic digests of in vitro- and in vivo-labelled TCFβ1. The same peptide was found to be completely overlapping (Fig. 6B, right panel). These data suggest that at least one such site in TCFβ1 is a target of JNK2 in vivo.
FIG. 6.
Identification of phosphopeptides in TCFβ1 phosphorylated in vivo or in vitro with JNK2. (A) Tryptic maps of in vivo-phosphorylated TCFβ1 from unactivated or activated Jurkat cells. Jurkat T cells were transfected with epitope-tagged TCFβ1 expression plasmids. Parallel cultures were either left unactivated or activated and labelled in vivo. The in vivo-phosphorylated TCFβ1 was then analyzed by two-dimensional tryptic mapping. The leftward-pointing arrow identifies a phosphopeptide which is indistinguishable from a phosphopeptide identified when recombinant TCFβ1 is phosphorylated in vitro with JNK2 (see below). (B) Identification of TCFβ1 phosphopeptides phosphorylated in vivo or in vitro by JNK2. The tryptic phosphopeptide maps of TCFβ1 labelled in vitro (left panel) or in vivo (middle panel) or a mix of in vitro- and in vivo-phosphorylated TCFβ1 (right panel) are shown. The three samples were run in parallel. The middle panel is from in vivo-labelled, activated Jurkat cells which had been transiently transfected with epitope-tagged TCFβ1 expression plasmids. Labelled proteins were analyzed as described in Materials and Methods, and the protein of interest was excised from the gels, digested with trypsin, and analyzed by two-dimensional electrophoresis. The leftward-pointing arrows identify the phosphopeptide which is completely superimposable and is present in both in vitro- and in vivo-labelled TCFβ1 maps.
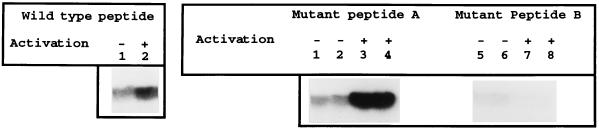
A more careful examination of the primary sequence of TCFβ1 revealed two residues flanking the basic region of the POU homeodomain of TCFβ1 which could be putative targets of JNK2 activity. (It is of interest that the basic region itself is also a target of other protein kinases in two different POU domain proteins [4, 21, 38].) These residues are identified in Table 2. We then determined the ability of JNK2 to phosphorylate in vitro a wild-type peptide from this region of TCFβ1. JNK2 phosphorylated this peptide in an activation-dependent manner (Fig. 7, left panel). We then generated two mutant peptides. In one, termed mutant peptide A [T239S240 to A239 A240], residues phosphorylated by protein kinase A have been mutated to alanine. In mutant peptide B [S232T242 to A232A242], the putative JNK phosphorylation sites in TCFβ1 have been mutated. In vitro phosphorylation experiments with JNK2 revealed that mutant peptide A was still phosphorylated by JNK2 in an activation-dependent manner whereas when the putative JNK sites were mutated (mutant peptide B), JNK2 could not phosphorylate the resultant peptide (Fig. 7, right panel), thus suggesting that we had identified one target of JNK2 activity in the DNA binding domain of TCFβ1.
TABLE 2.
Phosphorylation sites flanking the basic region of the POU homeodomain
| Protein | Region
|
||
|---|---|---|---|
| Linker | Basic | Helix 1 | |
| Pit-1 | QVGALYNEKVGANE | RKRKRRTTI | SIAAKDALERHFG |
| Oct1 | SPSALNSPGIEGLS | RRRKKRTSI | ETNIRVALEKSFL |
| Oct2 | NQLSSPSLGFEPAG | RRRKKRTSI | ETNVRFALEKSFL |
| cfla1 | TGSPTSIDKIAAQG | RKRKKRTSI | EVSVKGALEQHFH |
| Brn-1 | TGSPTSIDKIAAQG | RKRKKRTSI | EVSVKGALESHFL |
| unc-86 | KDTIGDINGILPNT | DKKRKRTSI | AAPEKRELEQFFK |
| Brn-3 | QREKMNKPELFNGG | EKKRKRTSI | AAPEKRSLEAYFA |
| i-pou | KRRDPDAPSVLPAG | EKK--RTSI | AAPEKRSLEAYFA |
| Oct3/4 | NLQEICKSETLVQA | -RKRKRTSI | ENRVRWSLETMFL |
| TCFβ1 | GQQNLMEFVGGEPS | KKRKRRTSF | TPQAIEALNAYFE |
| § | †† | § | |
The POU domain protein sequences in the linker-basic-helix 1 region of the DNA binding homeodomain are shown. The flanking threonine and serine residues in the basic region which have been shown by others (4, 21, 38) to be phosphorylated in Oct1 and Pit1 are identified by daggers. The two sites in TCFβ1, one in the linker region and the other in helix 1, which are phosphorylated by JNK2 are identified by “§”. This was shown by making synthetic peptides corresponding to this region of TCFβ1 and determining whether they were phosphorylated by JNK2 in vitro. We generated three TCFβ1 peptides. The wild-type peptide (VGGEPS KKRKRRTSF TPQAI) was phosphorylated by JNK2. Mutant peptide A (VGGEPS KKRKRRAAF TPQAI), in which residues in the basic region that are phosphorylated in Oct1 and Pit1 are mutated, was found to be still phosphorylated by JNK2 in vitro. Mutant peptide B (VGGEPA KKRKRRTSF APQAI), in which the two potential JNK2 sites are mutated, was not phosphorylated by JNK2 in vitro. The mutated residues (underlined) involve substitution of alanine for serine or threonine.
FIG. 7.
Mapping of a JNK2 phosphorylation site in the DNA binding domain of TCFβ1. The ability of recombinant JNK2 to phosphorylate the wild-type peptide and two mutant peptides from a region spanning the basic region of the POU homeodomain of TCFβ1 was determined (also see Table 1). The wild-type peptide extends from residues 227 to 246 of the TCFβ1 protein (33). In mutant peptide A (T239S240 to A239A240), residues which have been shown to be the target of protein kinase A in other POU domain proteins (21, 38) have been mutated to alanine. In mutant peptide B (S232T242 to A232A242), the putative JNK2 phosphorylation sites in TCFβ1 have been mutated to alanine. JNK2 can phosphorylate the wild-type peptide and mutant peptide A in an activation-dependent manner, whereas mutant peptide B cannot be phosphorylated by JNK2. −, cultures were not activated; +, cultures were activated.
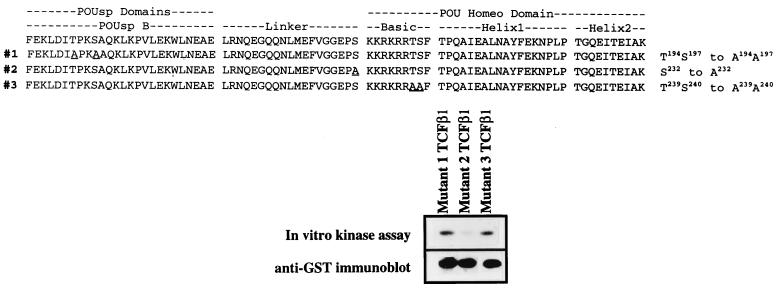
To confirm that these potential sites in TCFβ1 were actually phosphorylated by JNK2, we generated four site-specific mutants of TCFβ1. The abilities of these mutant TCFβ1 proteins to be phosphorylated in vitro by JNK2 were compared to that of wild-type TCFβ1. The T239S240-to-A239A240 TCFβ1 mutant, as expected, was phosphorylated by JNK2 (Fig. 8). In contrast, and as expected from the peptide data, the S232-to-A232 mutant was phosphorylated less efficiently (Fig. 8). These results with site-specific mutant TCFβ1 proteins and the mutant peptide data (Fig. 7 and Table 2) support the conclusion that we had identified one target of JNK2 activity in the DNA binding domain of TCFβ1.
FIG. 8.
Ability of JNK2 to phosphorylate site-specific mutants of TCFβ1. A panel of mutant TCFβ1-GST fusion proteins was tested for the ability to be phosphorylated by activated recombinant JNK2 in vitro. The TCFβ1 mutants 1 (T194S197 to A194A197), 2 (S232 to A232), and 3 (T239S240 to A239A240) were generated, and equal amounts of the mutants were tested as substrates in an in vitro JNK2 kinase assay. The sequences of the different mutants are shown at the top, and the phosphorylation of the mutants described above is shown below. The immunoblot of the same gel, obtained by using anti-GST antibodies, is shown below the in vitro kinase assay results. The underlined amino acids have been mutated to alanine in the mutants. POUsp refers to the POU-specific domain B regions of the POU DNA binding domains.
Phosphorylation of TCFβ1 by JNK2 enhances its ability to bind an octamer motif.
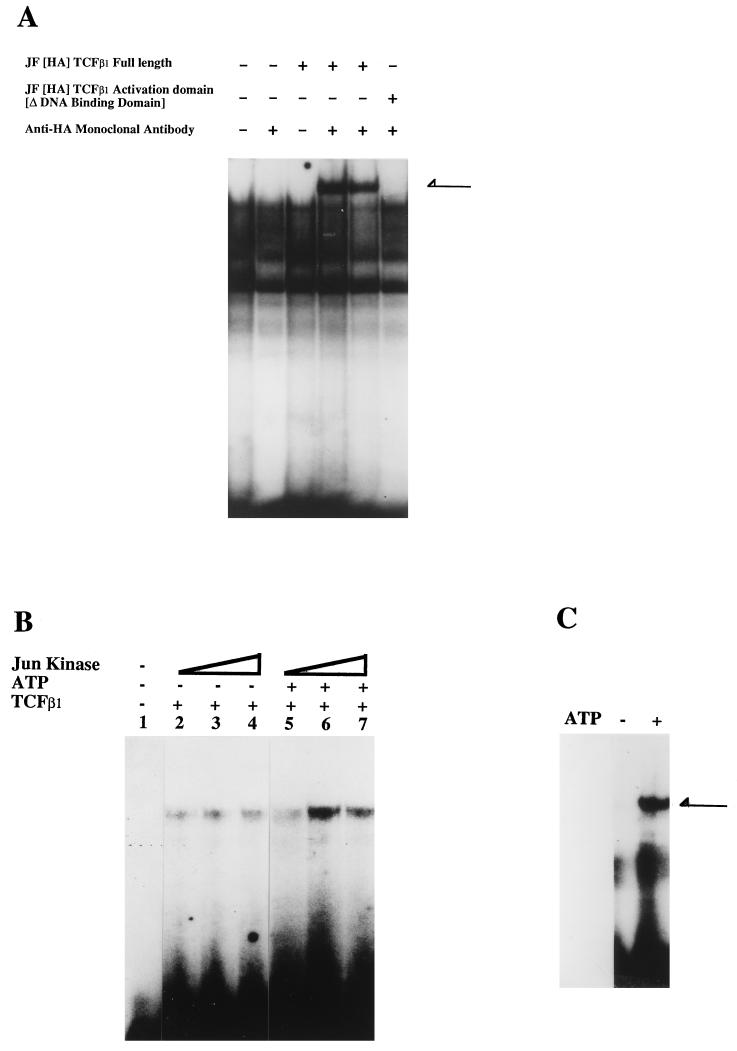
We had shown earlier that recombinant TCFβ1 can bind octamer motifs in a sequence-specific manner (30). We now wanted to determine whether TCFβ1 would bind octamer sites when other, competing POU domain proteins were also present. We addressed this by doing supershifting experiments in Jurkat cells. As shown in Fig. 9A, TCFβ1 can effectively bind an octamer motif in the presence of other POU domain proteins.
FIG. 9.
JNK2 phosphorylation of TCFβ1 enhances its ability to bind octamer motifs from the IL-2 promoter. (A) TCFβ1 binds to the proximal octamer in the IL-2 promoter. Nuclear extracts were prepared, as described previously (29), from Jurkat cells transfected 48 h earlier with a vector expressing HA epitope-tagged full-length TCFβ1. An end-labelled proximal octamer motif from the IL-2 promoter was used as a probe. Binding reactions were done as described previously (30) and analyzed on 4% nondenaturing acrylamide gels. As expected, extracts from cells transfected with vector expressing full-length TCFβ1 bound octamer motifs and were supershifted in the presence of 1 or 2 μl of anti-HA antibody. Extracts containing the epitope-tagged activation domain of TCFβ1 were not supershifted with anti-HA antibody, suggesting that the supershifted band (indicated by the arrow) was indeed due to binding of TCFβ1 to the octamer motif via its DNA binding domain. (B) The ability of the DNA binding domain of TCFβ1 to bind to the octamer motif increases with increasing amounts of JNK2. As described above, the recombinant JNK2 was purified and used in increasing amounts to phosphorylate the DNA binding domain of TCFβ1 in the presence of 30 μM cold ATP. As expected, the ability of the DNA binding domain to bind the octamer motif increased with increasing amounts of the kinase, but only in the presence of exogenous ATP. (C) Phosphorylation of full-length TCFβ1 by JNK2 enhances its ability to bind an octamer motif. COS-1 cells were transfected with an HA-JNK2 expression vector and activated by UV irradiation, and JNK2 was purified from activated Jurkat cell lysates by immunoprecipitation with anti-HA antibodies. The HA-JNK2 was used to phosphorylate TCFβ1. Each preparation of purified HA-JNK2 was first tested for its ability to phosphorylate GST–c-Jun. To ascertain that the DNA binding activity of TCFβ1 was indeed due to JNK phosphorylation, we performed the kinase reaction, using activated kinase in the presence or absence of exogenous ATP. Phosphorylation of TCFβ1 by JNK2 (in the presence of ATP) enhanced the ability of TCFβ1 to bind a consensus octamer motif in a gel shift assay. The arrow indicates the TCFβ1 DNA-protein complex.
Phosphorylation of transcription factors has been found to be an important regulator of transcriptional activity, acting to perturb nuclear translocation, transactivation ability (17), or DNA binding ability (21). To determine whether JNK phosphorylation of TCFβ1 affects its DNA binding ability, we examined the effect of phosphorylation of recombinant TCFβ1 in vitro by JNK2 on its ability to bind an octamer motif (43). COS-1 cells were transfected with a vector expressing HA epitope-tagged JNK2 and, 48 h later, were activated by UV irradiation. The JNK2 was then immunoprecipitated, and the immunoprecipitates were used to phosphorylate recombinant GST-TCFβ1 fusion proteins. To rule out any artifactual effects, we performed our kinase reaction with activated JNK2 in the presence and in the absence of exogenous cold ATP. The phosphorylated TCFβ1 proteins were then used in an electromobility shift assay with an end-labelled octamer motif from the IL-2 promoter (43). Phosphorylation of the TCFβ1 DNA binding domain by JNK2 enhanced its ability to bind to an octamer motif (Fig. 9B) in a JNK2 concentration-dependent manner (Fig. 9C). The ability of full-length TCFβ1 to bind an octamer motif was also dramatically enhanced by JNK2 in the presence of exogenous ATP (Fig. 9C). We therefore conclude that JNK binds to the activation domain of TCFβ1 and phosphorylates its DNA binding domain, which increases the ability of TCFβ1 to bind octamer motifs.
TCFβ1 transactivates the IL-2 promoter in a JNK-dependent manner.
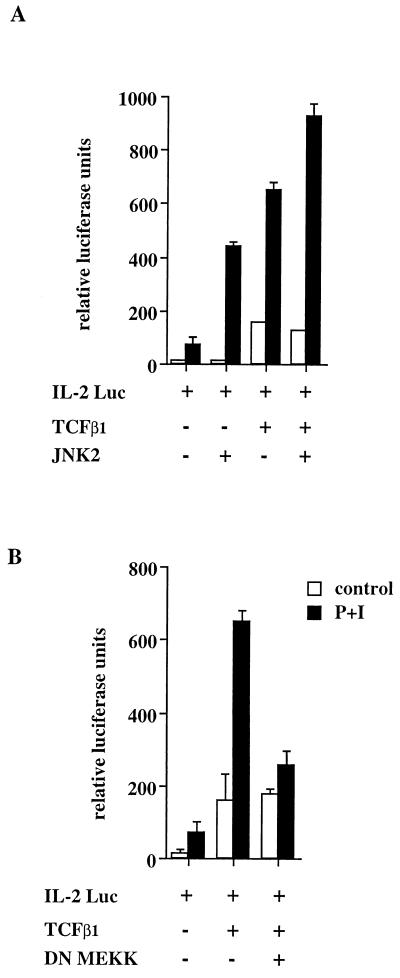
Using reporter genes in which octamer motifs are located upstream of the minimal promoter, we have previously shown that TCFβ1 can transactivate in an octamer-dependent manner (30). The IL-2 promoter contains octamer binding sites (proximal and distal) which are critical for activation-dependent transcription (9). We performed transient-transfection experiments in Jurkat cells to determine whether TCFβ1 could transactivate the IL-2 promoter in an activation-dependent manner. Consistent with our observations that TCFβ1 could bind the IL-2 promoter octamer motifs in supershifting experiments (Fig. 9A), TCFβ1 was found to drive transcription from the IL-2 promoter in an activation-dependent manner (Fig. 10A). Furthermore, consistent with our observation that JNK2 increases the binding of TCFβ1 to octamer motifs, we found that JNK2 enhances the ability of TCFβ1 to transactivate the minimal IL-2 promoter in an activation-dependent manner (Fig. 10A). In previous studies, MEKK has been shown to be the rate-limiting step in the JNK activation pathway. MEKK phosphorylates the JNK kinase, which activates JNK activity. If TCFβ1 is a target of JNK2 activity, it should be possible to demonstrate that a dominant-negative mutant of MEKK (in which methionine has been substituted for the lysine at position 432 [(K432M]) inhibits the activation-dependent induction of the IL-2 promoter by TCFβ1 but has no effect on activation-independent transactivation of the IL-2 promoter by TCFβ1. We found that the catalytically inactive MEKK (K432M), which inhibits the activation-dependent induction of JNK activity, also inhibits the activation-dependent induction of the IL-2 promoter by TCFβ1 (Fig. 10B). The dominant-negative mutant of MEKK had no effect on TCFβ1-dependent basal transcription. The data presented in this paper therefore suggest a mechanism by which the functional activity of TCFβ1 is coupled to the JNK signal transduction pathway.
FIG. 10.
The ability of TCFβ1 to induce activation-dependent transactivation of the IL-2 promoter is JNK dependent. (A) TCFβ1 enhances the activation-inducible transcription of the IL-2 promoter. Jurkat T cells were transfected with a minimal promoter from the IL-2 gene cloned into a luciferase reporter plasmid (9). The activation-dependent transcription from the IL-2 promoter was enhanced by cotransfection with a TCFβ1 expression plasmid or a JNK2 expression plasmid. Cotransfection of both TCFβ1 and JNK2 expression plasmids further enhanced transcription from the cotransfected IL-2 reporter plasmid. Cells were activated with PMA plus ionomycin for 12 to 18 h, harvested, and assayed for luciferase activity as described in Materials and Methods. (B) The dominant-negative mutant of MEKK (K432M) inhibits the ability of TCFβ1 to enhance inducible transcription from the IL-2 promoter. Jurkat T cells were transfected with the IL-2 reporter plasmid and either left unactivated or activated with PMA plus ionomycin (P+I) for 12 to 18 h. Cells were cotransfected with TCFβ1 alone or with the catalytically inactive MEKK (K432M) expression plasmid. The dominant-negative mutant of MEKK inhibits inducible, but not basal, transactivation by TCFβ1.
DISCUSSION
The potent regulatory effect of phosphorylation on the ability of transactivators to bind DNA has been underlined by studies of many families of transactivators. Fibroblast growth factor induces protein kinase C, which phosphorylates the basic region of the DNA binding domain of myogenin and decreases its DNA binding activity (24). Myogenin is a helix-loop-helix protein which is essential for muscle development. Phosphorylation of the PU.1 transactivator (a member of the ETS family of transactivators) by casein kinase II (34) leads to the recruitment of Pip to sites in the immunoglobulin lambda enhancer (10). Pip by itself does not bind to this motif. Pip is probably identical to the previously identified nuclear factor EM5. Pip and PU.1 function as mutually dependent transcriptional activators of the composite element (10).
A number of cases in which the JNK binds and phosphorylates transcriptional activators involved in T-cell costimulation and stress-induced gene expression have been documented. The best-studied example remains the c-Jun transactivator (15, 32). The phosphorylation of c-Jun at Ser-63 and Ser-73 in the activation domain increases c-Jun transactivation. In a similar manner, JNK also increases transactivation by the ATF-2 transactivator (13, 26, 44) and the TCF protein Elk1 transactivator (46). The ATF-2 transcription factor mediates adenovirus E1A-inducible transcriptional activation, and Elk-1 belongs to the subfamily of ETS domain transcription factors and is a key factor in serum response element-dependent gene transcription. We have now extended the model to include POU domain proteins. TCFβ1, a POU protein, is phosphorylated by JNK, and JNK activity determines the ability of TCFβ1 to upregulate inducible transcription from the IL-2 promoter.
The δ domain in c-Jun, which is responsible for binding of c-Jun to JNK1 and JNK2, has been mapped (6, 8, 17). Similarly, a region in ATF-2 flanking the phosphorylated residues Thr69 and Thr71, which bind JNK, has been mapped (13, 26, 44). The region in TCFβ1 that binds JNK2 has also been mapped to the activation domain, and three modestly conserved motifs homologous to the δ region in c-Jun have been identified in the activation domain of TCFβ1 (Table 1). The conservation of such motifs in JNK2 binding sites in c-Jun and TCFβ1 suggests their involvement in JNK binding. These motifs are absent in v-Jun, which does not bind JNK. These studies, while integrating the JNK signaling pathway with multiple transactivators, also suggest that there are two distinct mechanisms by which JNK influences transactivator activity; both involve binding of JNK to the activation domain, but one involves phosphorylation of the activation domain (c-Jun and ATF-2) and enhanced transactivation. The second mechanism involves binding of JNK to the activation domain of TCFβ1 and phosphorylation of the DNA binding domain, which increases the ability of TCFβ1 to bind DNA. These studies also underline the relevance of studying in parallel the multiple transcriptional targets of the JNK signaling pathway.
Phosphorylation of POU transactivators has been shown to be a potent regulatory mechanism which influences DNA binding. Oct1 is phosphorylated at Ser385 during mitosis (38). This residue is conserved in all POU proteins. Pit1 is also phosphorylated at the homologous residue, Thr220, during mitosis (4). This phosphorylation of Pit1 and Oct1 during mitosis inhibits DNA binding (4, 38). These homologous residues in Pit1 (Thr220) and Oct1 (Ser385) are phosphorylated in vitro by protein kinase A (4, 21, 38). Although cyclic AMP agonists induce phosphorylation of Pit1 in vitro (4, 21), there have been conflicting reports as to whether this site is phosphorylated by such agents in vivo (4, 21). The homologous amino acid residue in TCFβ1 is not phosphorylated by JNK2, but a flanking site in TCFβ1 is phosphorylated by that protein (Table 2). This was shown by determining the ability of JNK2 to phosphorylate wild-type and mutated peptides from this region of TCFβ1 (Table 2 and Fig. 7, right panel). This was also supported by the data from studies using TCFβ1 proteins which had been mutagenized in a site-specific manner (Fig. 8). The basic region of the homeodomain of POU proteins is critical for DNA binding, as suggested by the observation that i-pou, which does not bind DNA (41), is generated by alternative splicing of two amino acids in the basic region of the homeodomain of the “twin of pou” gene (42). The contention that this region of the POU homeodomain is critical for binding DNA is further supported by crystal structural analysis of Oct1 and Oct2 (2, 7).
In our studies, we have demonstrated that JNK2 can bind and phosphorylate TCFβ1, thereby increasing its ability to bind an octamer motif. It is possible that after phosphorylating TCFβ1, JNK phosphorylates an adjacent protein even though it does not directly bind to it. It has been reported that E2F associates with p107, cyclin A, and cdk2 (23). This was suggested as a means of recruiting cdk2 to phosphorylate other DNA binding proteins. It has also been demonstrated that the KU antigen associates with DNA-dependent protein kinase (SCID kinase), allowing it to phosphorylate other DNA binding proteins (for a review, see reference 1a). In a similar manner, TCFβ1 may recruit JNK to phosphorylate other components of the transcriptional machinery and thereby influence their activity. One particularly interesting example is the proximal octamer motif in the IL-2 enhancer (43). It is a composite binding site for both Jun-Fos heterodimers and POU proteins (43). The ability of JNK to upregulate c-Jun transactivation (8, 17, 22) and DNA binding of TCFβ1 (this paper) suggests an additional mechanism of synergy between these two transactivators. In this regard, the ability of both proteins to bind JNK and the adjacent location of these sites in the proximal octamer motif suggest that the JNK signaling pathway can act via both AP-1 and POU proteins to ensure IL-2 gene induction after T-cell activation. Our demonstration that TCFβ1 in Jurkat extracts binds the proximal octamer motif from the IL-2 enhancer (Fig. 9), and the expression of TCFβ1 in thymus and T-cell lines (30) as well as the JNK-dependent transactivation of the IL-2 promoter by TCFβ1 (Fig. 10), suggests a role for TCFβ1 in IL-2 gene induction.
In summary, we have shown in this study that the POU domain protein TCFβ1 is phosphorylated after T-cell costimulation or UV treatment. JNK2 binds the activation domain of TCFβ1 and phosphorylates its DNA binding domain. This phosphorylation of TCFβ1 by JNK2 increases its ability to bind octamer motifs. In addition, JNK activity dictates the ability of TCFβ1 to drive inducible transcription from the IL-2 promoter. These results suggest a critical role for JNK in regulating gene transcription via an octamer-binding transcription factor.
ACKNOWLEDGMENTS
We thank Douglas Green (La Jolla Institute of Allergy and Immunology) for invaluable suggestions with regard to the manuscript. We acknowledge the kind gifts of c-Jun, JNK1, JNK2, and DN-MEKK expression plasmids from M. Karin (UCSD). We acknowledge the invaluable help of members of the Fotedar lab, especially Howard Brickner, Vincent Pennaneach, and Patrick Fitzgerald.
This work was supported by grants from the American Cancer Society (DB-107) and the National Institutes of Health (NIH) (AI45301, CA74435] to A.F. and from the NIH (CA35299) to A.A.
Footnotes
Manuscript 138 from the La Jolla Institute for Allergy and Immunology.
REFERENCES
- 1.Andersen B, Schonemann M D, Pearse R V, Jenne K, Sugarman J, Rosenfeld M G. Brn-5 is a divergent POU domain factor highly expressed in layer IV of the neocortex. J Biol Chem. 1993;268:23390–23398. [PubMed] [Google Scholar]
- 1a.Anderson C W, Lees-Miller S. The nuclear serine/threonine protein kinase DNAPK. Crit Rev Eukaryot Gene Expr. 1992;2:282–314. [PubMed] [Google Scholar]
- 2.Assa-Munt N, Mortishire-Smith R J, Aurora R, Herr W, Wright P E. The solution structure of Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage λ repressor DNA-binding domain. Cell. 1993;73:193–205. doi: 10.1016/0092-8674(93)90171-l. [DOI] [PubMed] [Google Scholar]
- 3.Bhargava A K, Li Z, Weissman S M. Differential expression of four members of the POU family of proteins in activated and phorbol 12-myristate 13-acetate treated Jurkat T cells. Proc Natl Acad Sci USA. 1993;90:10260–10264. doi: 10.1073/pnas.90.21.10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caelles C, Hennemann H, Karin M. M-phase-specific phosphorylation of the POU transcription GHF-1 by a cell cycle-regulated protein kinase inhibits DNA binding. Mol Cell Biol. 1995;15:6694–6701. doi: 10.1128/mcb.15.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cochrane L, Karvelas M, Nossal G, Ye Z, Jacks T, Baltimore D. Oct-2, although not required for early B cell development, is critical for later B-cell maturation and for postnatal survival. Genes Dev. 1993;7:570–582. doi: 10.1101/gad.7.4.570. [DOI] [PubMed] [Google Scholar]
- 6.Dai T, Rubie E, Franklin C C, Kraft A, Gillespie D A F, Avruch J, Kyriakis J M, Woodgett J R. Stress-activated protein kinases bind directly to the δ domain of c-jun in resting cells: implications for repression of c-jun function. Oncogene. 1995;10:849–855. [PubMed] [Google Scholar]
- 7.Dekker N, Cox M, Boelens R, Verrijzer C P, vander Vliet P C, Kaptein R. Solution structure of the POU-specific DNA-binding domain of Oct-1. Nature. 1993;362:852–855. doi: 10.1038/362852a0. [DOI] [PubMed] [Google Scholar]
- 8.Derijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 9.Durand D B, Shaw J-P, Bush M R, Replogle R E, Belagaje R, Crabtree G R. Characterization of antigen receptor response elements within the interleukin-2 enhancer. Mol Cell Biol. 1988;8:1715–1724. doi: 10.1128/mcb.8.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenbeis C F, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid specific, PU.1 dependent transactivator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 11.Finney M, Ruvkun G, Horvitz H R. The C. elegans cell lineage and differentiation gene unc-86 encodes a protein containing a homeo domain and extended sequence similarity to mammalian transcription factors. Cell. 1988;55:757–769. doi: 10.1016/0092-8674(88)90132-8. [DOI] [PubMed] [Google Scholar]
- 12.Fotedar R, Fitzgerald P, Rousselle T, Cannella D, Doree M, Messier H, Fotedar A. p21 contains independent binding sites for cyclin and cdk2: both sites are required to inhibit cdk2 kinase activity. Oncogene. 1996;12:2155–2164. [PubMed] [Google Scholar]
- 13.Gupta G, Campbell D, Derijard B, Davis R G. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Nature. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 14.Herr W, Sturm R, Clerc R, Corcoran L, Baltimore D, Sharp P, Ingraham H, Rosenfeld M, Finney M, Ruvkun G, Horvitz H. The POU domain: a large conserved region in the mammalian Pit-1, Oct-1, Oct-2, and C. elegans unc-86 gene products. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 15.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein and UV-responsive protein kinase that binds and phosphorylates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 16.Johansen T, Moens U, Holm T, Fjose A, Krauss S. Zebrafish pou[c], a divergent POU family gene ubiquitously expressed during embryogenesis. Nucleic Acids Res. 1993;21:475–483. doi: 10.1093/nar/21.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kallunki T, Su B, Tsigelny I, Sluss K H, Derijard B, Moore G, Davis R, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 18.Kamps M P, Corcoran L, LeBowitz J H, Baltimore D. The promoter of the human interleukin-2 gene contains two octamer-binding sites and is partially activated by the expression of Oct-2. Mol Cell Biol. 1990;10:5464–5472. doi: 10.1128/mcb.10.10.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang S-M, Beverly B, Tran A-C, Brorson K, Schwartz R H, Lenardo M J. Transactivation by AP-1 is a molecular target of T cell anergy. Science. 1992;257:1134–1138. doi: 10.1126/science.257.5073.1134. [DOI] [PubMed] [Google Scholar]
- 20.Kang S-M, Tsang W, Doll S, Scherle P, Ko H-S, Tran A-C, Lenardo M J, Staudt L M. Induction of the POU domain transcription factor Oct-2 during T-cell activation by cognate antigen. Mol Cell Biol. 1992;12:3149–3154. doi: 10.1128/mcb.12.7.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapiloff M S, Farkash Y, Wegner M, Rosenfeld M G. Variable effects of phosphorylation of Pit-1 dictated by the DNA response elements. Science. 1991;254:786–789. doi: 10.1126/science.1652153. [DOI] [PubMed] [Google Scholar]
- 22.Kyriakis J M, Banerjee P, Nikolakaki E, Dia T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. The stress activated protein kinase subfamily of c-jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 23.Lees E, Faha B F, Dulic V, Reed S I, Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes Dev. 1992;6:1874–1885. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- 24.Li L, Zhou J, James G, Heller-Harrison R, Czech M P, Olson E N. FGF inactivates myogenic helix-loop-helix proteins through phosphorylation of a conserved protein kinase C site in their DNA binding domains. Cell. 1992;71:1181–1194. doi: 10.1016/s0092-8674(05)80066-2. [DOI] [PubMed] [Google Scholar]
- 25.Li S, Crenshaw E B, Rawson E J, Simmons D M, Swanson L W, Rosenfeld M G. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU domain gene Pit-1. Nature. 1990;347:528–533. doi: 10.1038/347528a0. [DOI] [PubMed] [Google Scholar]
- 26.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo K X, Hurley T R, Sefton B M. Transfer of proteins to membranes facilitates both CNBR cleavage and two dimensional proteolytic mapping. Oncogene. 1990;5:921–923. [PubMed] [Google Scholar]
- 28.Messier H, Ratnavongsiri J, Fuller T, Mangal S, Kilgannon P, Fotedar R, Fotedar A. Mapping of an inducible element in the T cell receptor Vβ2 promoter. J Immunol. 1992;149:1980–1986. [PubMed] [Google Scholar]
- 29.Messier H, Fuller T, Mangal S, Brickner H, Igarashi S, Gaikwad J, Fotedar R, Fotedar A. p70 lupus autoantigen binds the TCR β enhancer. Proc Natl Acad Sci USA. 1993;90:2685–2689. doi: 10.1073/pnas.90.7.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messier H, Brickner H, Gaikwad J, Fotedar A. A novel POU domain protein which binds to the T-cell receptor β enhancer. Mol Cell Biol. 1993;13:5450–5460. doi: 10.1128/mcb.13.9.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minden A, Lin A, Claret F X, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 32.Minden A, Lin A, Smeal T, Dérijard B, Cobb M, Davis R, Karin M. c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol Cell Biol. 1994;14:6683–6688. doi: 10.1128/mcb.14.10.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamoto K, Wakamiya M, Noji S, Koyama E, Taniguchi S, Takemura R, Copeland N, Gilbert D, Jenkins N A, Muramatsu M, Hamada H. A novel class of murine POU gene predominantly expressed in central nervous system. J Biol Chem. 1993;268:7449–7457. [PubMed] [Google Scholar]
- 34.Pongubala J M R, van Beveren C, Nagulapalli S, Klemz M J, McKeron S R, Maki R A, Atchison M L. Effect of PU.1 phosphorylation on interaction with NF-EM5 and transcriptional activation. Science. 1993;259:1622–1625. doi: 10.1126/science.8456286. [DOI] [PubMed] [Google Scholar]
- 35.Roberts S B, Segil N, Heintz N. Differential phosphorylation of the transcription factor Oct1 during the cell cycle. Science. 1991;253:1022–1026. doi: 10.1126/science.1887216. [DOI] [PubMed] [Google Scholar]
- 36.Rosenfeld M G. POU-domain transcription factors: pou-er-ful development regulators. Genes Dev. 1991;5:897–907. doi: 10.1101/gad.5.6.897. [DOI] [PubMed] [Google Scholar]
- 37.Scheidereit C, Cromlish J A, Gerster T, Kawakami K, Balmaceda C, Currie R, Roeder R G. A human lymphoid specific transcription factor that activates Ig genes is a homeobox protein. Nature. 1988;336:552–557. doi: 10.1038/336551a0. [DOI] [PubMed] [Google Scholar]
- 38.Segil N, Roberts S B, Heintz N. Mitotic phosphorylation of the Oct-1 homeo domain and regulation of Oct-1 DNA binding activity. Science. 1991;254:1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- 39.Su B, Jacinto E, Hibi M, Kallunki T, Karin M, Ben-Neriah Y. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka M, Lai J-S, Herr W. Promoter selective activation domains in Oct1 and Oct2 direct differential activation of an snRNA and mRNA promoter. Cell. 1992;68:755–767. doi: 10.1016/0092-8674(92)90150-b. [DOI] [PubMed] [Google Scholar]
- 41.Treacy R, He X, Rosenfeld M G. i-pou, a POU domain protein that inhibits neuron specific gene activation. Nature. 1991;350:577–584. doi: 10.1038/350577a0. [DOI] [PubMed] [Google Scholar]
- 42.Treacy R, Neilson L I, Turner E E, He X, Rosenfeld M G. Twin of i-pou: a two amino acid difference in the i-pou homeodomain distinguishes an activator from an inhibitor of transcription. Cell. 1992;68:491–505. doi: 10.1016/0092-8674(92)90186-g. [DOI] [PubMed] [Google Scholar]
- 43.Ullman K, Flanagan W, Edwards C, Crabtree G. Activation of early gene expression in T cells by Oct-1 and an inducible protein, OAP40. Science. 1991;254:558–562. doi: 10.1126/science.1683003. [DOI] [PubMed] [Google Scholar]
- 44.van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wey E, Lyons G E, Schafer B W. A human POU domain gene, mPOU, is expressed in developing brain and specific adult tissues. Eur J Biochem. 1994;220:753–762. doi: 10.1111/j.1432-1033.1994.tb18676.x. [DOI] [PubMed] [Google Scholar]
- 46.Whitmarsh J A, Shore P, Sharrocks D, A, Davis R. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]