Abstract
Expression of genes encoding starch-degrading enzymes is regulated by glucose repression in the yeast Saccharomyces cerevisiae. We have identified a transcriptional repressor, Nrg1, in a genetic screen designed to reveal negative factors involved in the expression of STA1, which encodes a glucoamylase. The NRG1 gene encodes a 25-kDa C2H2 zinc finger protein which specifically binds to two regions in the upstream activation sequence of the STA1 gene, as judged by gel retardation and DNase I footprinting analyses. Disruption of the NRG1 gene causes a fivefold increase in the level of the STA1 transcript in the presence of glucose. The expression of NRG1 itself is inhibited in the absence of glucose. DNA-bound LexA-Nrg1 represses transcription of a target gene 10.7-fold in a glucose-dependent manner, and this repression is abolished in both ssn6 and tup1 mutants. Two-hybrid and glutathione S-transferase pull-down experiments show an interaction of Nrg1 with Ssn6 both in vivo and in vitro. These findings indicate that Nrg1 acts as a DNA-binding repressor and mediates glucose repression of the STA1 gene expression by recruiting the Ssn6-Tup1 complex.
In yeast, a large number of genes are turned off during growth on glucose (9, 37, 49). These glucose-repressible genes can be divided into three groups: (i) genes for metabolizing other carbon sources; (ii) genes encoding enzymes unique to gluconeogenesis; and (iii) genes involved in the Krebs cycle and in respiration. The Mig1 glucose repressor is a zinc finger protein and binds to the GC-rich motif identified in the promoters of several glucose-repressed genes, including the GAL1, GAL4, SUC2, and MAL genes (10, 13, 28, 29). In the absence of glucose, the Snf1 kinase inhibits the function of Mig1 protein directly or indirectly, leading to derepression of glucose-repressed genes (3, 4). Nuclear translocation of Mig1 is regulated by differential phosphorylation of the protein in response to glucose availability, and recruitment of the general repression complex Ssn6-Tup1 to the DNA-bound Mig1 is required for the repression (5, 17, 48). Disruption of the MIG1 gene, however, only partially relieves glucose repression of SUC2 and has little or no effect on glucose repression of other genes whose promoters contain the Mig1-binding sites (27, 31, 37, 50), indicating the involvement of other repressors in glucose repression. For instance, Mig2 was recently identified as a second repressor responsible for the remaining glucose repression of SUC2 and contains zinc fingers very similar to those of Mig1 (24).
In Saccharomyces cerevisiae var. diastaticus, three unlinked homologous STA genes (STA1, STA2, and STA3) encode glucoamylase isozymes (GAI, GAII, and GAIII), which are responsible for enzymatic degradation of starch to glucose (16, 22, 25, 32, 35, 47, 52). Expression of the STA genes is regulated by complex interactions between positive and negative factors and their cognate elements (1, 19, 21, 33, 41). The negative regulation occurs at three different levels: (i) carbon catabolite repression by glucose (6, 34); (ii) repression by STA10, which is known as a repressor gene in S. cerevisiae (33); and (iii) diploid cell-specific repression (6, 34). The mechanisms underlying repression of the STA genes, however, are not yet understood.
In this study, to identify transcriptional regulators for glucose repression of the STA1 gene, a plasmid was constructed by modifying the strategy used for cloning of the MIG1 gene (29). The plasmid bears the TPK2 gene, encoding a yeast cyclic AMP-dependent protein kinase, whose level of transcription is modulated by upstream regulatory elements (1, 41) of the STA1 promoter. In derepressed conditions, cells with the STA1p-TPK2 construct exhibit a slow-growth phenotype because of the toxic effects of the high level of Tpk2. Taking advantage of this phenotype, we isolated a gene, NRG1, whose presence in a multicopy vector confers normal growth to the cells in derepressed conditions. Here we report that the NRG1 gene encodes a DNA-binding repressor required for the glucose repression of STA1 gene expression.
MATERIALS AND METHODS
Strains and media.
The yeast strains used in this study are listed in Table 1. Yeast transformations were done by a lithium acetate method (14). Genetic methods were performed as described elsewhere (40). Escherichia coli DH5α was used for propagation and selection of recombinant plasmids. To isolate multicopy suppressors of the STA1 promoter, yeast cells were grown in solid synthetic medium containing 0.67% yeast nitrogen base without amino acids, 0.6% Casamino Acids, appropriate amino acids, and 2% Bacto Agar, supplemented with 3% soluble starch, 2% glycerol plus 2% ethanol, and 1% potassium acetate for induction (SC-SGE medium). For enzyme assay, yeast cells were grown at 30°C in synthetic minimal medium. Minimal medium containing Casamino Acids was composed of 0.67% yeast nitrogen base without amino acids plus appropriate amino acids. Carbon sources (2% glucose or 2% glycerol plus 2% ethanol) were added to minimal medium.
TABLE 1.
Yeast strains used
| Strains | Genotype | Reference |
|---|---|---|
| AN20-1a | MATα ura3 ade2 leu1 STA1 sta10 | 1 |
| AN20-5b | MATα ura3 leu1 STA1 sta10 | 1 |
| PHM20-5b | MATα ura3 leu1 nrg1Δ1::URA3 STA1 sta10 | This study |
| ST20-1a | MATα ura3 leu1 UASSTA-TATASTA-TPK2::ADE2 STA1 STA10 | This study |
| MCY829 | MATα hisΔ200 lys2-801 ura3-52 | 48 |
| MCY1974 | MATα hisΔ200 lys2-801 ura3-52 ade2-101 trp1Δ ssn6Δ9 | 48 |
| MCY2437 | MATα hisΔ200 lys2-801 ura3-52 trp1Δ tup1Δ::TRP1 | 48 |
| PJ69-4A | MATa trp1 leu2 ura3 his3 gal4Δ gal80Δ GAL2-ADE2 lys::GAL1-HIS3 met::GAL7-lacZ | 15 |
Isolation and analysis of the NRG1 gene.
The 2.46-kb NaeI-HindIII fragment containing the TPK2 gene from plasmid pJN1 (a gift from H. Ronne) was subcloned into the StuI-HindIII site of pUCSTA-G to construct plasmid pSTATPK2. pUCSTA-G contains upstream regulatory elements, TATA box, and portion of the open reading frame (ORF) of the STA1 gene; the StuI site is located between the TATA box and ATG codon. To construct a yeast integrative plasmid, the 3.2-kb KpnI-StuI fragment containing the STA1 upstream activation sequence (UASSTA)-TATASTA-TPK2 fusion from pSTATPK2 was inserted into the KpnI-BamHI (blunt) of pASZ10 (45). The resulting plasmid (pAST2) was linearized by StuI and used to transform strain AN20-1a. Stable transformants were selected in minimal medium without adenine and confirmed by genomic Southern analysis. The resulting strain (ST20-1a) was transformed with a yeast genomic library made in the multicopy vector pHR81 (a gift from H. Ronne). Transformants were plated on SC-SGE medium for induction of the STA1 promoter and incubated at 30°C for 7 days. Transformants growing faster than the host strain were selected, and plasmids were recovered from these transformants. As a second screening, strain AN20-5b was transformed with the recovered plasmids, and transformants with decreased glucoamylase production were isolated on synthetic medium containing 2% glycerol plus 2% ethanol. Plasmid pRPS50 was isolated as the multicopy suppressor in this genetic screen.
The 4.9-kb ApaI-BamHI fragment of pRPS50 was subcloned into the ApaI-BamHI site of pBlueScript-KS(+), generating pBSM1. The ApaI and BamHI sites of pRPS50 originate from vector pHR81. The 3-kb SpeI (blunt)-HindIII fragment of pBSM1 was subcloned into XbaI (blunt)-HindIII site of pGEM7-Z, resulting in pGM1. The SmaI-EcoRI fragment containing the URA3 gene from YEp24 was inserted into the SmaI-EcoRI site of pBlueScript-KS(+), generating pBSURA. The 1.1-kb XbaI-SalI fragment containing the NRG1 ORF of pGM1 was replaced by the 1.2-kb XbaI-SalI fragment containing the URA3 gene from pBSURA. The resulting plasmid was linearized by SphI-BstXI and transformed into strain AN20-5b. Transformants with the nrg1Δ1::URA3 allele were isolated, and the disruption was confirmed by genomic Southern analysis.
LexA fusions.
pLexA-Nrg1 was constructed by replacement of the MIG1 gene with NRG1 of plasmid pLexA-Mig1 (a gift from M. Carlson) (48). The NRG1 ORF was amplified by using two oligonucleotides (5′-CGGGATCCCCATGTTTTACCCATATAAC-3′ and 5′-AGCTCGAGGATACCGTCAATTATTGTC-3′) and cloned into the BamHI-SalI site of pLexA-Mig1. The expression plasmid encoding only the LexA-binding domain (amino acids 1 to 87) was made by deleting the MIG1 sequence from plasmid pLexA-Mig1. For transcriptional repression by LexA-Nrg1 fusion protein, plasmid JK1621 with four lexA operators (provided by M. Carlson) (48) and pLG669Z without lexA operators were used as reporter plasmids. β-Galactosidase activity was assayed in yeast cells permeabilized with chloroform and sodium dodecyl sulfate as described elsewhere (11).
Northern blot analysis.
Cells were grown in synthetic medium containing 2% glucose or 2% glycerol plus 2% ethanol. Total RNA was isolated as described elsewhere (30), subjected to electrophoresis in a 1.2% agarose–formaldehyde gel, and then transferred onto a nitrocellulose membrane. To detect the STA1 transcript, a 600-bp EcoRI-PvuII internal fragment of the STA1 gene was used as a probe (1). The NRG1 transcript was detected by probing with a 700-bp BamHI-XhoI fragment of plasmid pGEXNrg, constructed for expression of the glutathione S-transferase (GST)–Nrg1 fusion protein. A fragment of the ACT1 gene was used as a control (8).
Gel retardation assay.
The ORF of the NRG1 gene was PCR amplified by using two oligonucleotides (5′-CGGGATCCATGTTTTACCCATATAAC-3′ and 5′-AGCTCGAGGATACCGTCAATTATTGTC-3′) and cloned into the BamHI-XhoI site of pGEX4T-1, generating pGEXNrs. GST-Nrg1 was expressed in E. coli DH5α and purified essentially as described (43). Various amounts of GST-Nrg1 were incubated with a 5 ng of α-32P-labeled probe (approximately 20,000 cpm) and 0.5 to 4 μg of poly(dI-dC) for 15 min at room temperature in a reaction volume of 20 μl containing 20 mM HEPES (pH 7.6), 1 mM MgCl2, 60 mM KCl, 12% glycerol, 6 μg of bovine serum albumin, 10 μM ZnCl2, and 1 mM dithiothreitol. The reaction samples were loaded onto 5% nondenaturing polyacrylamide gel and electrophoresed at 150 V for 2.5 h in a buffer of 22.5 mM Tris base, 22.5 mM boric acid, and 0.63 mM disodium EDTA, adjusted to pH 8.0. The gel was dried and autoradiographed. To confirm that Nrg1 protein is responsible for formation of the binding complex, GST-Nrg1 fusion protein was treated with thrombin, which cleaves specifically at the junction between GST and Nrg1. The amount of thrombin was determined empirically.
DNase I footprinting analysis.
The coding strand of the 112-bp UAS1-1 was end labeled with [α-32P]dATP by using Klenow enzyme. DNase I footprinting was performed as described elsewhere (42). For footprinting, a standard binding reaction mixture containing 0.5 μg of poly(dI-dC) was adjusted to 5 mM CaCl2 and digested with empirically determined amounts of DNase I for 5 min. The reaction was stopped by adding EDTA and sodium dodecyl sulfate to final concentrations of 25 mM and 0.1%, respectively, extracted once with phenol, precipitated with ethanol, and then analyzed on an 8% polyacrylamide gel containing 7 M urea. The gel was dried and exposed to X-ray film at −70°C. The size marker of the footprinting was prepared by G+A sequencing of UAS1-1 by the Maxam-Gilbert method.
Two-hybrid analysis.
To identify Nrg1-interacting proteins, the Nrg1 fusion with the DNA-binding domain (DBD) of Gal4 (DBD-Nrg1) was made by ligating the BamHI-XhoI fragment of the NRG1 ORF from the GST-Nrg1 construct into BamHI-SalI of plasmid pGBDU (a gift from P. James) (15). A library of fusions between the activation domain (AD) of Gal4 and yeast genomic DNA (a gift from P. James) was transformed into strain PJ69-4A containing the DBD-Nrg1-expressing plasmid and plated on minimal medium lacking adenine and histidine, supplemented with 7 mM 3-aminotriazole. From 820,000 transformants, three positive clones were isolated. Plasmids were recovered from the three clones, and the genomic DNA of the plasmids was sequenced. All three plasmids contained regions of the N-terminal tetratricopeptide repeat domain of the SSN6 gene.
In vivo interaction of Nrg1 with Ssn6 was tested with the plasmids expressing DBD-Nrg1 and AD-Ssn6 (Ssn6 residues 1 to 403 fused with the Gal4 AD) (48) and strain Y190 (12). As a positive control, plasmids expressing DBD-Snf1 and AD-Snf4 were used (7). β-Galactosidase activity was assayed as described elsewhere (38).
GST pull-down analysis.
The SSN6 ORF was cloned into the baculovirus transfer vector pBacPAK9 (Clontech) by PCR amplification of the gene with the oligonucleotides 5′-CGCGGATCCATGAATCCGGGCGGTGAACAAACA-3′ and 5′-TGCTCTAGATTAGTCGTCGTAGTTTTCATCTTC-3′. Recombinant baculoviruses were generated and used to infect Spodoptera frugiperda Sf21 insect cells. Insect cell extract was prepared as described previously (18). The Ssn6-containing extract (300 μg) was incubated with 10 μg of GST-Nrg1 for 3 h on ice. Ovalbumin was used as a control for nonspecific aggregation. Gluthione-agarose beads (30 μl) were added and incubated for 1 h at 4°C with constant agitation. Beads were precipitated and washed. Proteins in the pellet were eluted by being boiled in sample buffer and analyzed by Western blotting. Polyclonal rabbit anti-Ssn6 (36), polyclonal rabbit antiovalbumin (Sigma), and monoclonal mouse anti-GST (Santa Cruz) antibodies were used at 1:1,000 dilution. Horseradish peroxidase-conjugated anti-mouse (Pierce) and anti-rabbit (Amersham) secondary antibodies were used at 1:2,000 dilution. Detection was performed by enhanced chemiluminescence as instructed by the manufacturer (Pierce).
Glucoamylase assay.
To measure glucoamylase activities, cells were grown at 30°C for 2.5 days and then pelleted by centrifugation. The culture supernatant was incubated in a total volume of 1 ml containing 100 mM sodium acetate (pH 5.2) and 1.62% soluble starch at 55°C for 30 min. Glucose produced by the action of glucoamylase on soluble starch was assayed by using a coupled glucose oxidase-peroxidase assay kit (Sigma). Amylase activity was determined as micrograms of glucose released per 100 μl of supernatant per A600 cells.
Nucleotide sequence accession number.
The GenBank accession number of the NRG1 gene is Z49812.
RESULTS
Isolation of a multicopy inhibitor of the STA1 promoter.
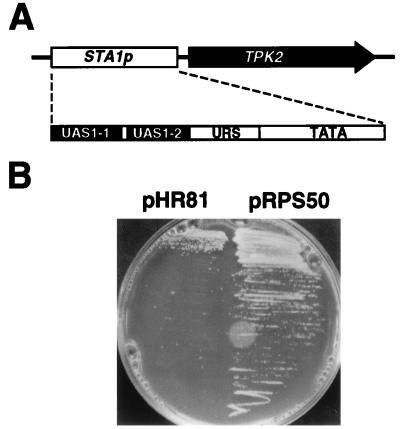
To investigate the negative regulation of STA1 gene expression, we constructed a plasmid in which the TPK2 gene, encoding a yeast cyclic AMP-dependent protein kinase, is transcribed from the STA1 promoter containing upstream regulatory elements and TATA box of the STA1 gene (1, 41) (Fig. 1A). Upon induction of the promoter by glycerol plus ethanol, starch, and 1% potassium acetate, cells harboring the plasmid exhibit a slow-growth phenotype (Fig. 1B), since overexpression of the kinase has toxic effects on cell growth (29). A yeast strain with a chromosomally integrated UASSTA-TATASTA-TPK2 fusion was transformed with a yeast genomic library made in a multicopy vector. To find plasmids that inhibit the STA1 promoter function, we screened cells for the ability to grow normally on SC-SGE medium. Among 25,000 transformants, 57 colonies resumed normal growth. Plasmids were recovered from the 57 colonies and transformed into a Sta+ strain to measure glucoamylase activities. One plasmid, pRPS50, both conferred normal growth to the cells with integrated UASSTA-TATASTA-TPK2 (Fig. 1B) and significantly decreased glucoamylase production to the Sta+ strain (Table 2) under STA1 promoter function-inducing conditions and was considered a candidate for the multicopy inhibitor of STA1 promoter function.
FIG. 1.
Isolation of a multicopy inhibitor of the STA1 promoter. (A) Schematic presentation of UASSTA-TATASTA-TPK2 fusion. Subregions of UASSTA (UAS1-1, UAS1-2, and URS) are described by Ahn et al. (1). (B) ST20-1a cells were transformed with either pHR81 (vector control) or pRPS50, and transformants were grown at 30°C on SC-SGE medium.
TABLE 2.
Suppression of glucoamylase production by plasmid pRPS50a
| Plasmid | Glucoamylase activity (U)b | Sta+ phenotype |
|---|---|---|
| pHR81 | 14.5 | ++++ |
| pRPS50 | 7.2 | ++ |
Strain AN20-5b was transformed with either control plasmid pHR81 or pRPS50. Transformants were grown in synthetic medium lacking uracil and containing 2% glycerol plus 2% ethanol at 30°C.
Average of at least duplicated experiments using three independent transformants with 10% deviation.
The genomic DNA of pRPS50 was sequenced and found to contain the ORF YDR043c, which has the capacity to encode a 231-amino-acid protein with a molecular weight of 25,000 (Fig. 2A). The gene was named NRG1. The predicted Nrg1 protein contains two tandem zinc finger motifs near the C terminus (Fig. 2A). The zinc finger regions are very similar to those of yeast Mig1 and Msn2 and are also closely related to those of mammalian early growth response proteins Egr-1/Zif268/Krox-24 and the Wilms’ tumor locus protein (Fig. 2B).
FIG. 2.
The amino acid sequence of Nrg1. (A) Predicted amino acid sequence of the NRG1 coding region. The two zinc finger motifs are underlined. (B) Alignment of the Nrg1 zinc finger motifs to those of other zinc finger proteins. Numbers in parentheses refer to the number of the motif counted from the amino terminus of the protein. The two zinc fingers of yeast MIG1 and MSN2 genes are aligned. Also shown are the two fingers of Zif268 (Krox-24/Egr-1), Wilms’ tumor protein (WT), Sp1, and the first two fingers of Xenopus transcription factor TFIIIA.
Interestingly, the NRG1 gene has previously been identified as MSS1 in a genetic screen for a multicopy suppressor of elevated glucoamylase production caused by sns1 mutation (1). The fact that the same gene was isolated by two different approaches to identify negative factors for STA1 expression implies that the NRG1 gene may play an important role in the negative regulation process.
Nrg1 is involved in glucose repression of STA1 gene expression.
Since overexpression of Nrg1 does not change the growth rate of the UASSTA-TATASTA-TPK2-integrated cells grown in glucose (data not shown), it is possible that Nrg1 mediates glucose repression of STA1 gene expression. To test this idea, a strain with deletion of the entire NRG1 coding region was generated. Deletion analysis revealed that NRG1 is not essential for cell viability (data not shown). Glucoamylase production in wild-type and nrg1Δ cells was monitored during growth in minimal medium containing glucose as the sole carbon source. Under this repressed condition, wild-type cells produce negligible levels of glucoamylase activity. In contrast, nrg1Δ cells exhibit under the repressed condition dramatically increased glucoamylase activity comparable to the activity in a derepressed condition (Table 3), indicating that deletion of Nrg1 relieves glucose repression of STA1 gene expression.
TABLE 3.
Effects of nrg1 disruption and carbon sources on STA1 gene expression
| Strains | Glucoamylase activity (U)a
|
|
|---|---|---|
| Repressed | Derepressed | |
| AN20-5b (wild type) | 2.2 | 13.5 |
| PHM20-5b (nrg1Δ1)b | 10.8 | 12.6 |
For the glucoamylase assay, cells were grown in synthetic medium containing 2% glucose (repressed condition) or 2% glycerol plus 2% ethanol (derepressed condition) at 30°C. Results are averages of at least duplicate experiments of three independent colonies with 10% deviation.
Derivative of AN20-5b.
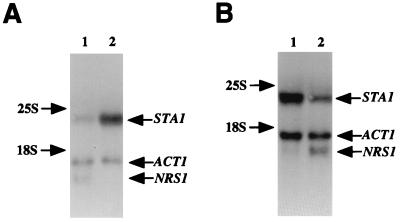
Northern blot analysis was performed to determine whether the increased glucoamylase activity in nrg1Δ cells correlates with the transcription level of the STA1 gene. The results show that the level of the STA1 transcript under the repressed condition was five times higher in nrg1Δ cells than in wild-type cells, compared with band intensity of yeast actin transcript as an internal control (Fig. 3A). Interestingly, the level of the NRG1 transcript was reduced about sixfold under the derepressed condition (Fig. 3B), indicating that the transcription of NRG1 itself is regulated by different carbon sources. Taken together, the results of the glucoamylase assay and Northern analysis indicate that Nrg1 is required for glucose repression of STA1 gene expression.
FIG. 3.
nrg1Δ alleviates glucose repression of STA1 transcription. (A) Effects of nrg1Δ on the STA1 transcription under the repressed condition. Cells were cultured in synthetic medium containing 2% glucose as a carbon source, and then total RNA was prepared for Northern analysis. Lane 1, strain AN20-5b (NRG1 [here designated NRS1]); lane 2, strain PHM20-5b (nrg1Δ). (B) Effects of carbon sources on the NRG1 transcription. Strain AN20-5b (NRG1) was grown in synthetic medium containing either 2% glycerol plus 2% ethanol (derepressed condition; lane 1) or 2% glucose (repressed condition; lane 2), and then total RNA was extracted for Northern analysis. The yeast actin gene (ACT1) was used as an internal control.
Nrg1 binds to an upstream element of the STA1 promoter.
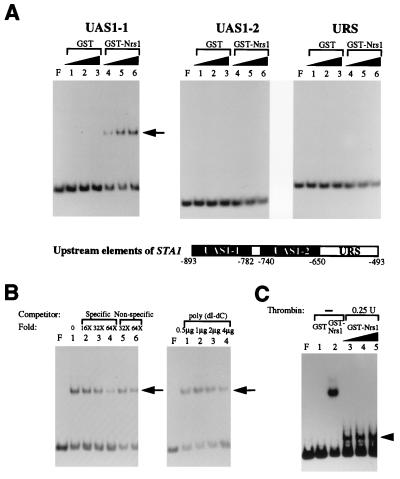
The presence of zinc finger motifs in Nrg1 suggests that this protein may exert its role during glucose repression by binding to a specific DNA sequence of the STA1 promoter. To examine this possibility, we purified recombinant Nrg1 as a fusion with GST and tested its binding to the upstream sequence elements (UAS1-1, UAS1-2, and an unidentified upstream repression sequence [URS]) located between nucleotides −493 and −893 of the STA1 promoter. Gel retardation analysis revealed that GST-Nrg1 binds to UAS1-1 but not to UAS1-2 and URS (Fig. 4A). No binding activity was detected in reactions containing the GST control (Fig. 4A). The specificity of GST-Nrg1 binding to UAS1-1 was tested by competitive gel retardation experiments. Addition of a specific competitor, unlabeled UAS1-1, to the binding reactions reduced the intensity of the shifted band, while the addition of a nonspecific competitor, a DNA fragment of unrelated sequence at up to 64-fold molar excess or poly(dI-dC) at up to 12,000-fold molar excess, had little effect on the binding of GST-Nrg1 to UAS1-1 (Fig. 4B). To exclude the possibility that the DNA-protein complex was formed as a fortuitous consequence of the GST-Nrg1 recombinant protein, the junction between Nrg1 and GST of the purified GST-Nrg1 fusion was cleaved by using thrombin in solution. Gel retardation experiments with the thrombin-treated GST-Nrg1 and UAS1-1 resulted in a reduction in the size of the DNA-protein complex due to the smaller size of Nrg1 than of the GST-Nrg1 fusion protein (Fig. 4C), indicating that Nrg1 itself binds to UAS1-1. These results show that Nrg1 is a protein binding specifically to the UAS1-1 region of the STA1 promoter.
FIG. 4.
Nrg1 binds to the UAS1-1 region of the STA1 promoter. (A) Gel retardation assays were performed with GST-Nrg1 (here designated Nrs1) and labeled upstream elements of the STA1 promoter (UAS1-1, UAS1-2, and unidentified URS). Lanes: F, no added protein; 1 to 3, reactions with increasing amounts of GST (0.05, 0.1, and 0.2 μg); 4 to 6, reactions with increasing amounts of GST-Nrg1 (0.05, 0.1, and 0.2 μg). In all the binding reactions, 0.5 μg of poly(dI-dC) was added. (B) Competitive gel retardation assays were performed with GST-Nrg1 and labeled UAS1-1. Lanes in left panel: F, no added protein; 1, no addition of competitor; 2 to 4, addition of increasing amounts of specific competitor (16-, 32-, and 64-fold molar excess of unlabeled UAS1-1); 5 and 6, addition of increasing amounts of nonspecific competitor (32- and 64-fold molar excess of pBluescript polylinker region). Lanes in right panel: F, no added protein; 1 to 4, addition of increasing amounts poly(dI-dC) (0.5, 1, 2, and 4 μg). (C) Gel retardation assays with thrombin-treated GST-Nrg1 and labeled UAS1-1. GST-Nrg1 was treated with 0.25 U of thrombin. Lanes: F, no added protein; 1, 0.2 μg of GST; 2, 0.2 μg of GST-Nrg1; 3 to 5, 0.1, 0.2, and 0.4 μg of thrombin-treated GST-Nrg1.
Two regions in UAS1-1 are bound to Nrg1.
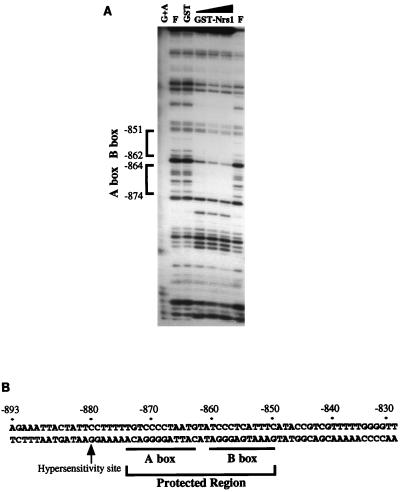
DNase I footprinting analysis was performed with GST-Nrg1 to further characterize the Nrg1-binding regions in UAS1-1. The results show that two regions are protected by Nrg1 binding: one between nucleotides −864 and −874 (A box) and the other between nucleotides −851 and −862 (B box) (Fig. 5). Additionally, a DNase I-hypersensitive site appears concomitantly, as shown in Fig. 5. The same results were obtained from the experiments with thrombin-treated GST-Nrg1 (data not shown). The region from nucleotides −839 to −849 was protected with intact GST-Nrg1 but not with thrombin-treated GST-Nrg1, indicating that the protection in this region is not specific to Nrg1 binding. Although both Nrg1 and Mig1 function in glucose repression, analysis of the two regions protected by Nrg1 reveals no strong homology with the consensus Mig1-binding sequence, (G/C)(C/T)GG(G/A)G (23). Comparison of the Nrg1-binding sites with the consensus Mig1-binding sequence suggests that the nucleotide sequences CCCCT in A box and/or CCCTC in B box may be important for binding of Nrg1 to DNA (Fig. 5B).
FIG. 5.
Two regions in UAS1-1 are protected by Nrg1 binding. (A) DNase I footprinting assay of GST-Nrg1 (here designated Nrs1) on UAS1-1. The amount of DNase I was empirically determined. The assay was performed with either 0.2 μg of GST or increasing amounts of GST-Nrg1 (0.1, 0.2, and 0.4 μg). Lane F, no added protein. The size marker of the footprinting was prepared by G+A sequencing of UAS1-1 by the Maxam-Gilbert method. (B) Sequences of regions of UAS1-1 protected by Nrg1. Protected regions are underlined and named A box and B box. Also indicated is the DNase I hypersensitive site of UAS1-1.
LexA-Nrg1 represses transcription of a target gene.
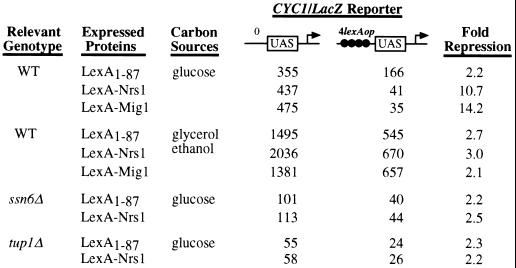
We assayed LexA-Nrg1, a Nrg1 fusion with the LexA DNA-binding domain, for the ability to repress transcription of a target gene with lexA operators. Yeast cells with a reporter gene (CYC1-lacZ) containing either no or four lexA operators was transformed with a LexA-Nrg1 expression plasmid, and transformants were assayed for β-galactosidase activities under both the repressed (glucose) and derepressed (glycerol plus ethanol) conditions. Under the repressed condition, expression of the reporter gene containing LexA-binding sites was 10.7-fold lower than that of the reporter gene with no binding sites (Fig. 6), indicating that DNA-bound LexA-Nrg1 represses transcription. Consistent with previous results (48), LexA-Mig1 represses expression of the reporter gene 14-fold under the repressed condition (Fig. 6). Under the derepressed condition, neither LexA-Nrg1 nor LexA-Mig1 shows significant repression (Fig. 6).
FIG. 6.
Transcriptional repression by LexA-Nrg1. The CYC1-lacZ reporters in this experiment were pLG669Z (no lexA operator) and JK1621 (four lexA operators) (48). LexA1-87 DNA-binding domain, LexA-Nrg1 (here designated Nrs1), or LexA-Mig1 was expressed in either wild-type (wt) or mutant strains. Cells were grown in synthetic media lacking uracil and histidine and containing either 2% glucose (repressed condition) or 2% glycerol plus 2% ethanol (derepressed condition). β-Galactosidase activity values are averages of duplicated experiments using three independent transformants.
Repression by LexA-Nrg1 requires Ssn6 and Tup1.
Both LexA-Mig1 and LexA-Mig2 repress transcription in a Ssn6-Tup1-dependent manner (24, 48). To examine whether the repression by LexA-Nrg1 depends on Ssn6 and/or Tup1, repression in ssn6Δ and tup1Δ mutants was assayed under the repressed condition. LexA-Nrg1 exhibited no significant repression of the reporter gene in both ssn6Δ and tup1Δ mutants (Fig. 6), indicating that repression by LexA-Nrg1 requires Ssn6 and Tup1.
Nrg1 interacts with Ssn6 both in vivo and in vitro.
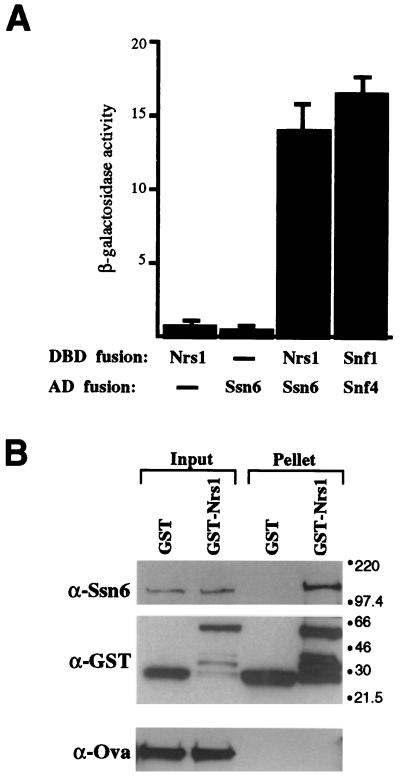
In a two-hybrid screening for detection of Nrg1-interacting proteins, we have identified regions of the N-terminal tetratricopeptide repeat domain of Ssn6 (data not shown). The interaction of Nrg1 with Ssn6 was confirmed by another two-hybrid assay (Fig. 7A). Cells with both DBD-Nrg1 and AD-Ssn6 (48) stimulated reporter gene expression more than 10-fold relative to control cells containing either DBD-Nrs1 or AD-Ssn6 alone. The level of the stimulation was comparable to that for a positive control for Snf1-Snf4 interaction (Fig. 7A).
FIG. 7.
Nrg1 interacts with Ssn6. (A) In vivo interaction. Plasmids expressing DBD-Nrg1 (here designated Nrs1) and AD-Ssn6 fusions were transformed into strain Y190 and assayed for β-galactosidase activity. As a positive control, plasmids expressing DBD-Snf1 and AD-Snf4 were transformed. Negative controls included cells with Gal4 DBD and AD-Ssn6 and cells with DBD-Nrg1 and Gal4 AD. β-Galactosidase activity values are averages of duplicated experiments using three independent transformants. (B) In vitro interaction. Ssn6-containing insect cell extract was incubated with GST or GST-Nrg1, and the GST proteins and their interacting proteins were precipitated by using glutathione-agarose beads. Ovalbumin (Ova) was added to each reaction mixture to serve as a control for specific precipitation. Fractions of input (1/30) and pellet (1/3) were analyzed by Western blotting with specific antibodies. Sizes are indicated in kilodaltons.
These two-hybrid results, together with the genetic interaction between Nrg1 and Ssn6-Tup1, suggested the possibility of direct interaction between Nrg1 and Ssn6. GST pull-down experiments were performed to test the interaction. Insect cell extract containing overexpressed Ssn6 was incubated with purified GST-Nrg1. Proteins interacting with GST-Nrg1 were precipitated, and the pellet was analyzed. GST was used in a parallel reaction to control for specific interaction, and ovalbumin was added to each reaction mixture to control for nonspecific aggregation. The results revealed that Nrg1 interacts with Ssn6 in vitro (Fig. 7B). There was no detectable interaction between Nrg1 and Tup1 (data not shown). The results from the two-hybrid and GST pull-down analyses, therefore, indicate the physical interaction of Nrg1 with Ssn6 both in vivo and in vitro.
DISCUSSION
We provide here several lines of evidence for the identification of Nrg1 as a transcriptional repressor responsible for glucose repression of the STA1 gene. First, nrg1Δ cells, when grown under the repressed conditions, exhibit dramatically increased glucoamylase activity which is comparable to that of cells grown under the derepressed conditions. Second, Northern analyses show that the increased glucoamylase level is correlated with the increased level of STA1 transcript in nrg1Δ cells. Third, gel retardation and DNase I footprinting experiments demonstrate that Nrg1 binds specifically to UAS1-1 element of the STA1 promoter. Fourth, tethering of Nrg1 to DNA via LexA-Nrg1 represses transcription of a target gene in glucose-grown cells, and the repression requires the Ssn6-Tup1 complex, which is needed for repression of diverse genes involved in many different cellular processes. And finally, two-hybrid and GST pull-down experiments demonstrate the physical interaction between Nrg1 and Ssn6 both in vivo and in vitro.
Nrg1 joins Mig1 and Mig2 as a DNA-binding repressor for glucose repression. The putative Nrg1-binding sequence, CCCCT and/or CCCTC, is, however, quite different from the consensus Mig1-binding sequence, (G/C)(C/T)GG(G/A)G, and other known consensus sequences for zinc finger proteins. Mig1 and Mig2 function as repressors of various genes such as GAL1, GAL4, SUC2, and MAL and bind to the same DNA sequence with different affinities (10, 13, 24, 28, 29). Analysis of the promoter regions of the Mig1/2-dependent glucose-repressible genes reveals no Nrg1-binding sequence motif, implying that the main targets of the Nrg1 function are STA genes. Although we cannot exclude the possibility of the involvement of Mig1, Mig2, and/or some unknown repressors, we suggest that Nrg1 is the major repressor responsible for the glucose repression of the STA genes since nrg1Δ cells almost completely alleviate the glucose repression. Consistent with this notion are our observations that the STA promoters do not contain a consensus Mig1-binding sequence and mig1Δ cells do not exhibit increased glucoamylase activity under repressed conditions (our unpublished data). It was also reported that Mig1 is dispensable for glucose repression of STA2 (16). Establishment of a consensus Nrg1-binding sequence, however, awaits more exhaustive investigation; more importantly, consideration should also be given to effects of promoter context since the total output of transcription of a given gene is a consequence of a complicated interaction of both positive and negative regulators. For example, not all genes containing the Mig1-binding site are affected by mig1 deletion (26, 37), and in some promoters more than one repressor is required for complete glucose repression (24, 31).
The binding of Nrg1 to UAS1-1 is consistent with the previous finding that the UAS1 region confers glucose repression when fused to a reporter gene (1). The affinity of Nrg1 for UAS1-1, however, seems to be weak compared with that of Mig1 for SUC2, GAL1, and GAL4 (10, 28, 29), since the addition of a specific competitor in 64-fold molar excess to the binding reactions does not completely abolish the Nrg1-DNA complex (Fig. 5B). This weak binding of Nrs1 may explain why glucose repression of the STA1 genes is not as tight as that of GAL1 or SUC2; Sta+ strains synthesize significant amounts of glucoamylase in rich medium containing glucose, albeit at levels lower than those of the same strains grown in rich medium containing glycerol plus ethanol (34).
Our genetic and biochemical evidence indicates that glucose repression by Nrg1 involves recruitment of the Ssn6-Tup1 complex. The general corepressor Ssn6-Tup1 is required for regulation of a subset of genes that is involved in many cellular processes, including cell type specificity, meiosis, oxygen utilization, and sugar utilization (2, 20, 46, 51), and most DNA-binding repressors involved in these processes, e.g., Mig1, Rox1, and α2, interact with the tetratricopeptide domain of Ssn6 (2, 44, 48). The Nrg1-Ssn6 interaction as well is mediated through the tetretricopeptide domain. Though comparison of the primary amino acid sequence of Nrg1 with those of Mig1, Mig2, Rox1, and α2 reveals no significant homology, it would be interesting to determine the structural requirement for binding of the proteins to the tetratricopeptide domain of Ssn6.
How does the glucose repression of STA1 occur? Based on our observation that the transcription of NRG1 is induced by glucose, we suggest a model in which Nrg1, in the presence of glucose, is highly produced and binds to upstream sequence of the STA1 gene to recruit Ssn6-Tup1 for repression of STA1 expression. In the absence of glucose, the expression level of NRG1 is decreased and hence STA1 expression is derepressed. This repression mechanism of Nrg1 is different from that of Mig1, the function of which is regulated by nuclear translocation through changes in its phosphorylation status (5). How is the expression of NRG1 regulated? One possibility is that NRG1, like HXT genes, is induced in the presence of glucose (31). A possible candidate responsible for the induction of NRG1 is the SNS1 gene product. Previously, NRG1 has been identified as a multicopy suppressor of sns1 mutation which elevates glucoamylase production (1), and our recent data show that the sns1 mutation decreases the expression of the NRG1 gene (unpublished data).
In this report, we suggest that Nrg1 is a key element for glucose repression of the STA1 gene as a DNA-binding repressor. Further genetic and biochemical analysis of Nrg1 together with other regulatory factors should reveal the molecular mechanism for the negative regulations of the STA genes.
ACKNOWLEDGMENTS
We thank H. Ronne for gift of plasmid pHR81 and yeast genomic library, M. Carlson for gift of LexA fusion systems and strains, and P. James for yeast two-hybrid system, respectively.
This work was partially supported by Genetic Engineering Foundation grants from the Ministry of Education (1996 to 1998) and by the Korea Science and Engineering Foundation through the Research Center for Molecular Microbiology at Seoul National University.
REFERENCES
- 1.Ahn J H, Park S H, Kang H S. Inactivation of the UAS1 of STA1 by glucose and STA10 and identification of two loci, SNS1 and MSS1, involved in STA10-dependent repression in Saccharomyces cerevisiae. Mol Gen Genet. 1995;246:529–537. doi: 10.1007/BF00298959. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian B, Lowry C V, Zimoter R S. The Rox1 repressor of the Saccharomyces cerevisiae hypoxic genes is a specific DNA-binding protein with a high-mobility-group motif. Mol Cell Biol. 1993;13:6071–6078. doi: 10.1128/mcb.13.10.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celenza J L, Carlson M. Cloning and genetic mapping of SNF1, a gene required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:49–53. doi: 10.1128/mcb.4.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Celenza J L, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- 5.DeVit M J, Waddle J A, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dranginis M. Regulation of STA1 gene expression by MAT during the life cycle of Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3992–3998. doi: 10.1128/mcb.9.9.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fields, S., and O. Song. A novel genetic system to detect protein-protein interactions. Nature 340:245–246. [DOI] [PubMed]
- 8.Gallwitz D, Seidel R. Molecular cloning of the actin gene from yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1980;8:1043–1059. doi: 10.1093/nar/8.5.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gancedo J M. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griggs D W, Johnston M. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci USA. 1991;88:8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guarente L, Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harper J W, Adam G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 13.Hu Z, Nehlin J O, Ronne H, Michels C A. MIG1-dependent and MIG1-independent glucose regulation of MAL gene expression in Saccharomyces cerevisiae. Curr Genet. 1995;28:258–266. doi: 10.1007/BF00309785. [DOI] [PubMed] [Google Scholar]
- 14.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kartacheva N N, Kuchin S V, Benevolensky S V. Genetic aspects of carbon catabolite repression of the STA2 glucoamylase gene in Saccharomyces cerevisiae. Yeast. 1996;12:1297–1300. doi: 10.1002/(SICI)1097-0061(199610)12:13%3C1297::AID-YEA13%3E3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 17.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 18.Koh, S. S., C. J. Hengartner, and R. A. Young. Baculoviral transfer vectors for expression of Flag fusion proteins in insect cells. BioTechniques 23:622–627. [DOI] [PubMed]
- 19.Kuchin S V, Kartasheva N N, Benevolensky S V. Genes required for derepression of an extracellular glucoamylase gene, STA2, in the yeast Saccharomyces. Yeast. 1993;9:533–541. doi: 10.1002/yea.320090510. [DOI] [PubMed] [Google Scholar]
- 20.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contributes to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambrechts M G, Pretorius I S, D’Aguanno V S, Solliti P, Marmur J. Multiple positive and negative cis-elements of the STA2 gene regulate glucoamylase synthesis in Saccharomyces cerevisiae. Gene. 1994;146:137–144. doi: 10.1016/0378-1119(94)90286-0. [DOI] [PubMed] [Google Scholar]
- 22.Lambrechts M G, Pretorius I S, Solliti P, Marmur J. Primary structure and regulation of a glucoamylase-encoding gene (STA2) in Saccharomyces diastaticus. Gene. 1991;100:95–103. doi: 10.1016/0378-1119(91)90354-e. [DOI] [PubMed] [Google Scholar]
- 23.Lundin M, Nehlin J O, Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol Cell Biol. 1994;14:1979–1985. doi: 10.1128/mcb.14.3.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lutfiyya L L, Johnston M. Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol Cell Biol. 1996;16:4790–4797. doi: 10.1128/mcb.16.9.4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meaden P, Ogden K, Bussey H, Tubb R. A DEX gene conferring production of extracellular amyloglucosidase on yeast. Gene. 1985;34:325–334. doi: 10.1016/0378-1119(85)90141-6. [DOI] [PubMed] [Google Scholar]
- 26.Mercado J J, Gancedo J M. Regulatory regions in the yeast FBP1 and PCK1 genes. FEBS Lett. 1992;311:11–114. doi: 10.1016/0014-5793(92)81379-z. [DOI] [PubMed] [Google Scholar]
- 27.Mercado J J, Vincent O, Gancedo J M. Regions in the promoter of the yeast FBP1 gene implicated in catabolite repression may bind the product of the regulatory gene MIG1. FEBS Lett. 1991;291:97–100. doi: 10.1016/0014-5793(91)81112-l. [DOI] [PubMed] [Google Scholar]
- 28.Nehlin J O, Carlberg M, Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991;10:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nehlin J O, Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms’ tumor finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Connor J P, Peebles C L. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:425–439. doi: 10.1128/mcb.11.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Özcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pardo J, Polaina J, Jimenez A. Cloning of the STA2 and SGA genes encoding glucoamylases in yeasts and regulation of their expression by the STA10 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1986;14:4701–4718. doi: 10.1093/nar/14.12.4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polaina J, Wiggs M. STA10: a gene involved in the control of starch utilization by Saccharomyces. Curr Genet. 1983;7:109–112. doi: 10.1007/BF00365634. [DOI] [PubMed] [Google Scholar]
- 34.Pretorius I, Modena D, Vanoni M, Englard S, Marmur J. Transcriptional control of glucoamylase synthesis in vegetatively growing and sporulating Saccharomyces cerevisiae species. Mol Cell Biol. 1986;6:3034–3041. doi: 10.1128/mcb.6.9.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pretorius I, Chow T, Modena D, Marmur J. Molecular cloning and characterization of the STA2 glucoamylase gene of Saccharomyces diastaticus. Mol Gen Genet. 1986;203:29–35. doi: 10.1007/BF00330380. [DOI] [PubMed] [Google Scholar]
- 36.Redd M J, Arnaud M B, Johnson A D. A complex composed of Tup1 and Ssn6 represses transcription in vitro. J Biol Chem. 1997;272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- 37.Ronne H. Glucose repression in fungi. Trends Genet. 1995;11:12–17. doi: 10.1016/s0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- 38.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 41.Shima H, Inui M, Akada R, Yamashita I. Upstream regions of the yeast glucoamylase gene which are required for efficient transcription. Agric Biol Chem. 1989;53:749–755. [Google Scholar]
- 42.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that bind to both silencer and activator elements. Cell. 1987;51:721–730. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 43.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1989;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 44.Smith R L, Redd M J, Johnson A D. The tetratricopeptide repeats of Ssn6 interacts with the homeo domain of α2. Genes Dev. 1995;9:2903–2910. doi: 10.1101/gad.9.23.2903. [DOI] [PubMed] [Google Scholar]
- 45.Stotz A, Linder P. The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- 46.Surosky R T, Strich R, Esposito R E. The yeast UME5 gene regulates the stability of meiotic mRNAs in response to glucose. Mol Cell Biol. 1994;14:3446–3458. doi: 10.1128/mcb.14.5.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamaki H. Genetic studies of ability to ferment starch in Saccharomyces: gene polymorphism. Mol Gen Genet. 1978;164:205–209. [Google Scholar]
- 48.Treitel M A, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trumbly R J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992;6:15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- 50.Vallier L G, Carlson M. Synergistic release from glucose repression by mig1 and ssn mutation in Saccharomyces cerevisiae. Genetics. 1994;137:49–54. doi: 10.1093/genetics/137.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wahi M, Johnson A D. Identification of genes required for alpha 2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamashita I, Maemura T, Hatano Y, Fukui S. Polymorphic extracellular glucoamylase genes and their evolutionary origin in the yeast Saccharomyces diastaticus. J Bacteriol. 1985;161:574–58. doi: 10.1128/jb.161.2.574-582.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]