Abstract
The gene encoding the Na/I symporter (NIS) is expressed at high levels only in thyroid follicular cells, where its expression is regulated by the thyroid-stimulating hormone via the second messenger, cyclic AMP (cAMP). In this study, we demonstrate the presence of an enhancer that is located between nucleotides −2264 and −2495 in the 5′-flanking region of the NIS gene and that recapitulates the most relevant aspects of NIS regulation. When fused to either its own or a heterologous promoter, the NIS upstream enhancer, which we call NUE, stimulates transcription in a thyroid-specific and cAMP-dependent manner. The activity of NUE depends on the four most relevant sites, identified by mutational analysis. The thyroid-specific transcription factor Pax8 binds at two of these sites. Mutations that interfere with Pax8 binding also decrease transcriptional activity of the NUE. Furthermore, expression of Pax8 in nonthyroid cells results in transcriptional activation of NUE, strongly suggesting that the paired-domain protein Pax8 plays an important role in NUE activity. The NUE responds to cAMP in both protein kinase A-dependent and -independent manners, indicating that this enhancer could represent a novel type of cAMP responsive element. Such a cAMP response requires Pax8 but also depends on the integrity of a cAMP responsive element (CRE)-like sequence, thus suggesting a functional interaction between Pax8 and factors binding at the CRE-like site.
Cell type-specific gene transcription is often dependent on a set of transcription factors whose combination is unique to that cell type. Three transcription factors, TTF-1, TTF-2, and Pax8, have been implicated in such a control in the case of thyroid-specific transcription of the thyroglobulin and thyroperoxidase genes (13). TTF-1 is an homeodomain (HD)-containing protein, present in the developing thyroid, lung, and diencephalon (26). TTF-2, a forkhead protein, has been detected in the endoderm of the developing foregut, including the thyroid anlage, and in the anterior pituitary (41), while the paired-domain (PD) factor Pax8 is present in both the thyroid and kidney (35). The unique combination of these factors in the thyroid follicular cells strongly suggests that their interaction plays an important role in inducing a specific pattern of gene expression in these cells. Cyclic AMP (cAMP), whose intracellular level is elevated by the thyroid-stimulating hormone (TSH), is an important modulator of gene expression in thyroid cells (1, 2, 14, 18, 20, 22, 34, 37). Nevertheless, direct roles for the thyroid-restricted factors TTF-1, TTF-2, and Pax8 in mediating the cAMP effects in thyroid cells have not yet been demonstrated. Interestingly, well-known mediators of transcriptional regulation by cAMP, such as those acting through the cAMP responsive element (CRE) sequence (5) have been proposed to be involved only in the regulation of TSH receptor gene expression (22), but no conclusive evidence on their roles in the control exerted by TSH cAMP or on other thyroid-specific genes has been provided. To gain further insights into the mechanisms responsible for cAMP-dependent transcription in thyroid cells and their relationships to thyroid-specific transcriptional mechanisms, we decided to study the regulation of the gene encoding the Na/I symporter (NIS). Thyroid follicular cells accumulate iodide against a concentration gradient and utilize it for thyroid hormone biosynthesis (40). Iodide transport, which is catalyzed by NIS, is primarily regulated by TSH through cAMP (30, 36, 40) by at least two mechanisms. The first induces the activity of the NIS protein (27, 32), presumably by posttranslational modifications, while the second positively regulates NIS mRNA levels (24) in a cycloheximide-sensitive manner.
In the present study, we isolated the rat NIS (rNIS) gene and searched for a regulatory element(s) capable of thyroid-specific and TSH-regulated transcriptional activation. We identified in the upstream region of rNIS a short enhancer capable of increasing transcription of its own or a heterologous promoter. This regulatory element shows four remarkable features. (i) It responds to cAMP only in differentiated thyroid cells, suggesting that a cell-type-specific mechanism is operating to mediate the transcriptional response of NIS to cAMP. (ii) It can be activated by cAMP even when thyroid cells, as a consequence of chronic stimulation of the cAMP pathway, down-regulate their PKA levels and, hence, are unable to carry out transcriptional activation of CRE-containing promoters (1). (iii) It requires the binding of Pax8, in addition to a degenerate CRE-like sequence, for transcriptional activity. (iv) It can be activated in a cAMP-dependent manner in nonthyroid cells by cotransfection of a Pax8 expression vector. Taken together, these data suggest that the rNIS enhancer mediates thyroid-specific gene expression by the interaction of Pax8 with a novel cAMP-dependent pathway.
MATERIALS AND METHODS
Isolation of the rNIS gene.
A λDASHII rat genomic library (Clontech) was screened with a 32P-labeled 384-bp fragment named P5AP and derived from the NIS cDNA (12) (from +133 to +516 bp, the first nucleotide of ATG being considered +1) by digestion with ApaI and PstI. Among the positive clones, one containing a larger 5′ upstream region was selected by dot blot hybridization with probes P5AP and P3H. P3H contained the DNA sequence extending from +1937 to +2730 bp in the rNIS cDNA in the 3′ untranslated region, and it was obtained by cleaving pNIS/SPORT with HindIII. One genomic clone that hybridized P5AP and not with P3H was selected. DNA was extracted from the clone, and its insert was excised with NotI and cloned into pBluescriptIIKS(−) (Stratagene) to yield plasmid BS39N.
Plasmids. (i) pNISLUC1 to -6.
DNA fragments from the NIS regulatory region were obtained from plasmid BS39N and inserted into pGL3-basic vector (Promega) containing the luciferase (LUC) reporter gene. Fragments were obtained either by cleaving with restriction enzymes (KpnI and NcoI for pNISLUC1; XbaI and NcoI for pNISLUC4) or by PCR with forward primers containing either an NheI or KpnI site and abutting the deletion endpoint and reverse primers downstream of either the XbaI (−2257 for pNISLUC2 and -3) or NcoI (-2 for pNISLUC5 and -6) site.
(ii) pNISLUC7 to -10.
DNA fragments were cleaved from pNISLUC1 (−2946 to −2264 for pNISLUC7) or from pNISLUC3 (−2495 to −2264 for pNISLUC9) by KpnI and XbaI. Alternatively, fragments corresponding to regions −2946 to −2494 (pNISLUC8) and −2386 to −2264 (pNISLUC10) were amplified by PCR with forward primers containing a KpnI site and reverse primers either containing an NheI site or located downstream of the XbaI site at position −2257. All fragments were cleaved by KpnI and either NheI or XbaI and cloned between the KpnI and NheI sites of pNISLUC5.
pNISTKLUC1 to -4.
The thymidine kinase (TK) promoter was amplified from the pBLCAT5 (4) by PCR, using a forward primer with a BglII site (5′-TGTAAAGATCTGGATCCGGCCCCGCCCAGCG) and a reverse primer (5′-ATGCCATTGGGATATATCAA). The PCR product was digested with BglII and cloned into the BglII site of pGL3-basic to obtain pTKLUC. The same promoter fragments described for plasmids pNISLUC7 to -9 were inserted into the KpnI and NheI sites of pTKLUC to construct plasmids pNISTKLUC1 to -3. For pNISTKLUC4, the fragment from −2495 to −2264 was amplified by PCR with primers 5′-GAAGGGTCGACAGATTGCAGCTGGCAAGTGC-3′ and 5′-GTCTCGTCGACTAGAAGAAGGTGTTTGCCTC-3′ which contained a SalI site. This PCR fragment was digested with SalI and cloned into the SalI site downstream of the LUC reporter gene of pTKLUC. All the sequences of the fragments generated by PCR were confirmed by sequencing after cloning.
Scanning mutants of pNISTKLUC3, 2-1 to 16-2.
In order to search the binding sites for transcription factors, every 6 bases in the fragment located from −2483 to −2302 bp, as shown in Fig. 5A, were changed to GATATC, the EcoRV recognition site, by using the QuickChange Site-Directed Mutagenesis Kit (Stratagene) and sequenced. Since this method requires amplifying whole plasmid, to exclude any possibilities of the mutation within other regions of the vector, KpnI-NheI fragments including mutated versions of rNIS enhancer were subcloned into the KpnI and NheI sites in pTKLUC.
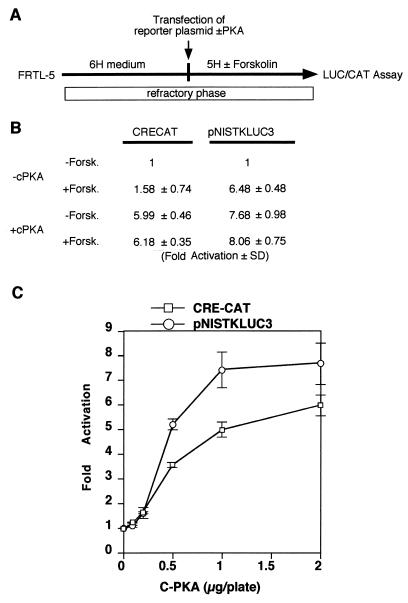
FIG. 5.
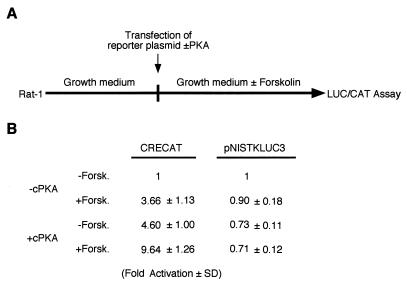
The NUE responds in thyroid cells to PKA-independent and -dependent pathways. (A) Experimental strategy. The entire experiment was performed under conditions that keep FRTL-5 thyroid cells in a state refractory to cAMP induction (6H) of CREB-dependent promoters. (B) Normalized CAT (for CRE-CAT) and LUC (for pNISTKLUC3) activity. Results are expressed as fold activation of the value obtained in the absence (−) of both cPKA and forskolin (Forsk.). The means of three independent experiments in duplicate are shown. (C) Activation of CRE-CAT and pNISTKLUC3 in response to different amounts of cPKA expression vector in the refractory phase of FRTL-5 cells. Means and standard deviations of relative activity of CRE-CAT (squares) and pNISLUC3 (circles) were plotted.
Construction of CMV-TTF1 and CMV-Pax8 was described previously (15, 42).
The expression vector, C-PKA, containing the catalytic subunit of mouse PKA (cPKA), and CRE-CAT containing a fragment from the somatostatin promoter, as well as the TK promoter and the chloramphenicol acetyltransferse (CAT) reporter gene, were all kindly provided by E. V. Avvedimento (6).
Anchored PCR.
Total RNA from FRTL-5 cells was extracted by the acid guanidinium-phenol-chloroform method (7). Anchored PCR (rapid amplification of 5′ cDNA ends [5′-RACE]) was used to amplify the 5′ end of the rNIS cDNA by using the 5′ RACE system (GIBCO BRL), according to the instructions supplied. The amplified products were electrophoresed on 2% agarose gels and subjected to Southern analysis with a 32P-labeled fragment corresponding to −355 to +109 bp of rNIS gene as a probe. The hybridizing fragments were cloned into plasmid pCRII (Invitrogen) and sequenced.
Cells and transient-expression analysis.
The FRTL-5 cell line was grown and transfected by calcium phosphate coprecipitation technique as previously described (19). For promoter studies, 2 pmol of test plasmid and 1 pmol of CMV-CAT, used to monitor for transfection efficiency, were transfected. To study the effects of PKA on CRE-CAT or pNISTKLUC3, 3 μg of either CRE-CAT or pNISTKLUC3, 2 μg of either C-PKA or pBluescriptII (to adjust the quantity of total plasmid), and 1 μg of CMV-LUC or CMV-CAT were used. After 72 h, cells were collected to determine either the levels of CAT protein with a CAT enzyme-linked immunosorbent assay kit (Boehringer Mannheim) or LUC activities as described previously (41). To investigate the effects of TSH and forskolin, transfected cells were cultured in the medium supplemented with 0.2% calf serum for 72 h (starvation medium). After an additional 72 h of incubation in 4H medium (5% calf serum, 3.6 ng of cortisol per ml, 5 μg of transferrin per ml, 10 ng of glycyl-l-histidyl-l-lysine acetate per ml, and 10 ng of somatostatin per ml) with 1 mU of bovine TSH per ml, 10 μg of insulin, or 10 μM forskolin, transfected cells were collected for LUC and CAT assays. Rat-1 and HeLa cells were grown and transfected as previously described (19, 42). For the extracts for band shift assays, 10 μg of CMV-Pax8 was transfected into HeLa cells in 100-mm-diameter dishes and cultured for 48 h to obtain HeLa-Pax8 extracts.
Extract preparation, band shift assays, and DNase I footprinting.
Total cell extracts were prepared from FRTL-5 cells, and HeLa cells were grown to near confluency as previously described (9). Probes and competitors for band shift assays are shown in Fig. 9C. Anti-Pax8 antibody, Pax8 peptide, and anti-TTF1 antibody were previously described (16, 26). Preparation of recombinant TTF-1 HD and Pax-8 PD and band shift and footprinting assays were performed as previously described (42). The footprinting probe from the 5′-flanking fragment of rNIS gene was generated by PCR with a sense primer located from −2634 to −2615 bp, 5′-CCAGAGTGAATCAGGAGGTT, and a 32P-labeled antisense primer, 5′-CAGTGGGTCTCTGTGTCTAG, reverse complementary sequence of −2267 to −2248 bp.
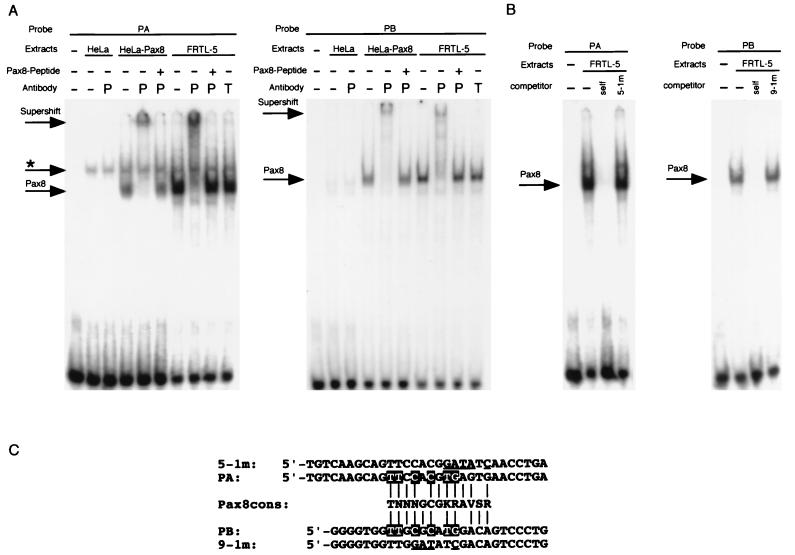
FIG. 9.
Binding of Pax8 protein on rNISA and rNISC. (A) Band shift assays were performed with 32P-labeled oligonucleotides corresponding to the Pax8 binding sites PA and PB. Each probe was incubated without extracts (−) or with extracts from FRTL-5 cells, mock-transfected HeLa cells (HeLa), or HeLa cells transfected with a Pax8 expression vector (HeLa-Pax8) in the presence (+) or absence (−) of either anti-Pax8 (P) or anti-TTF1 (T) antibody. Where indicated, a Pax8 synthetic peptide used to generate the antibody was added. The DNA-Pax8 complexes (Pax8), DNA-ubiquitous factor complex (asterisk), and supershift by anti-Pax8 antibody (Supershift) are indicated by arrows. (B) Competition experiments, using either PA or PB oligonucleotide. The complex formed by FRTL-5 nuclear protein with each oligonucleotide was challenged with 100-fold excess of itself (self) (cold) or cold oligonucleotides carrying either the 5-1m or 9-1m mutation. (C) Alignments between Pax8 consensus binding sequence (Pax8cons) and probes used in this study. Nucleotides in PA and PB matching the consensus sequence are indicated by vertical lines. Identical nucleotides between the PA and PB binding sites are indicated by white letters on a black background, and nucleotides changed by mutations 5-1m and 9-1m are underlined.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases with the accession number AB005653.
RESULTS
Isolation and sequencing of a genomic fragment containing the 5′-flanking region of the rNIS gene.
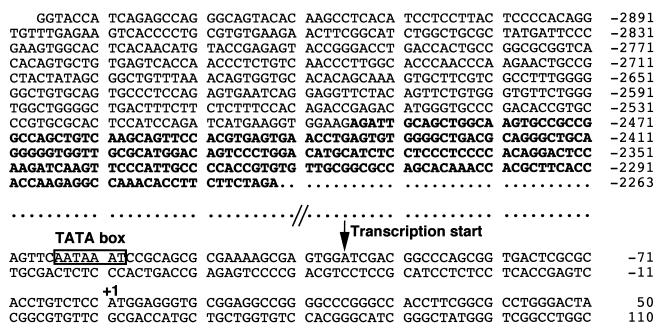
Ten positive clones were isolated by screening a rat genomic library with 32P-labeled P5AP probe, a 384-bp fragment at the 5′ end of rNIS cDNA. One clone (clone 39), extending the most in the 5′-flanking region was selected. DNA was extracted from clone 39 and digested with NotI, yielding three fragments of 8, 5.5, and 3 kb. Sequencing of the entire 5.5-kb fragment demonstrated that it contains the 5′ end of the NIS cDNA and extended for 4 kb into the 5′-flanking region. Our sequence of 2264 bp upstream of the NIS translation initiation site is in agreement with that already reported (39). Figure 1 shows additional sequence of the 5′-flanking region of rNIS, from −2263 to −2947, with the first nucleotide of the NIS translation initiation codon being referred to as +1.
FIG. 1.
DNA sequence of the 5′-flanking region and part of the coding region of the rat NIS gene. The first nucleotide of the translation start codon is designated +1. The enhancer fragment described in detail in this study is shown in bold letters, and the TATA box and major transcription start site are indicated.
To assess the transcriptional initiation site(s) of the rNIS mRNA, we performed 5′-RACE. The results of Southern blot analysis of the amplified cDNA products identified an approximately 0.2-kb fragment that hybridized with a probe containing the NIS gene translation initiation region (corresponding to −355 to +109 bp). After subcloning, 14 positive products were sequenced. The 5′ ends of these products were as follows: −96 (eight clones), −95 (one clone), −94 (two clones), −93 (one clone), −335 (one clone), and −329 (one clone). These data suggest that the major transcription start site of the rNIS gene is located around position −96 and that other, minor, transcriptional start site(s) may be located at −335 and at −329 bp. In keeping with this hypothesis is the presence of a TATA box-like sequence, AATAAAT, at position −124, 23 bp upstream of the major transcriptional start site (Fig. 1).
Identification of a thyroid-specific transcriptional regulatory element in the 5′-flanking region of the rNIS gene.
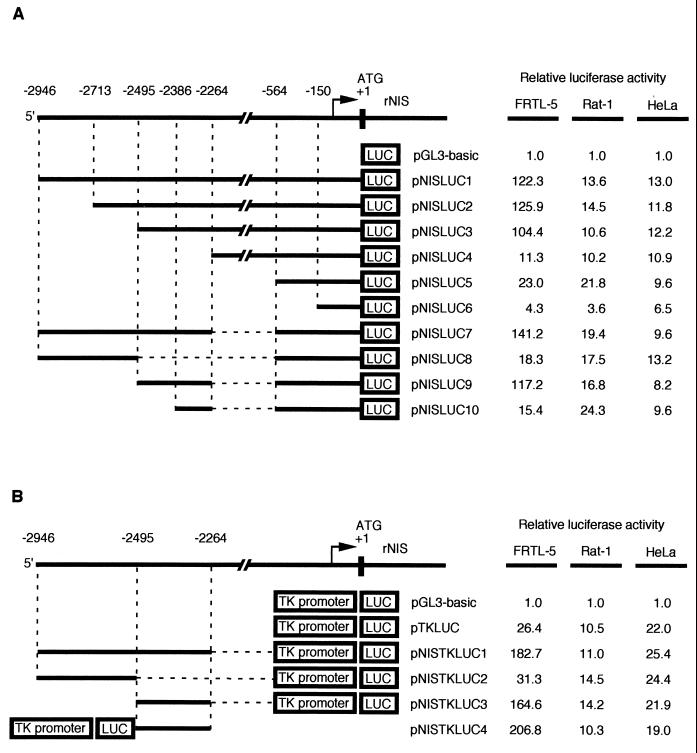
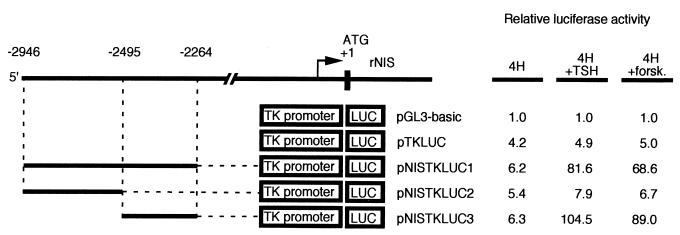
To functionally identify transcriptional regulatory elements in the 5′-flanking region of the rNIS gene, chimeric constructs were made containing either the entire upstream region or deletion derivatives fused to the LUC reporter gene in the vector pGL3-basic (Fig. 2A). Each construct was transiently transfected, together with a CMV-CAT construct used to normalize for transfection efficiency, into either the rat thyroid cell line FRTL-5, the rat fibroblast cell line Rat-1, or human HeLa cells. The transcriptional activity of each construct is reported, in Fig. 2A, as fold induction over the transcription obtained with pGL3-basic, whose value was set at 1.0 in each cell line. A 2.9-kb DNA fragment from the rNIS regulatory region significantly stimulated transcription of the reporter in all three cell lines (pNISLUC1). However, pNISLUC1 was at least 10-fold more active in FRTL-5 cells than in the two nonthyroid cell lines. Deletion of 5′-flanking sequences up to position −2495 (pNISLUC2 and -3) showed no significant decrease in activity, whereas further deletions, such as those in pNISLUC4, -5, and -6 showed drastic reductions in transcription only in FRTL-5 cells, suggesting the presence, immediately downstream of nucleotide −2495, of regulatory information that is relevant exclusively for thyroid-specific transcription. Additional, nonthyroid-specific, regulatory information must be located between positions −564 and −150, as deletion of these sequences causes another dramatic reduction in transcription in all three cell lines used. We conclude that the rNIS regulatory region contains a promoter from −564 to −2 and a tissue-specific regulatory element located downstream of −2494 bp. To further define the tissue-specific rNIS regulatory elements, several fragments from rNIS 5′-flanking sequence were inserted into either pNISLUC5 (pNISLUC7 to -10 [Fig. 2A]) or pTKLUC (pNISTKLUC1 to -4 [Fig. 2B]). The results obtained, mostly with pNISLUC9 and pNISTKLUC3, showed that the fragment from −2262 to −2494 could reproduce most, if not all, of the transcriptional activity of the entire pNISLUC1 construct. Furthermore, the newly identified regulatory element behaved as an enhancer, since it was capable of stimulating transcription of the heterologous TK promoter even if it was placed downstream of the LUC cistron, as in pNISTKLUC4 (Fig. 2B). The sequence from −2495 to −2264 did not cause a significant stimulation of transcription in Rat-1 cells and HeLa cells, strongly indicating that a thyroid specific cis-regulatory element is present in this region of the rNIS gene.
FIG. 2.
Cell-type-specific promoter activity of rNIS-LUC chimeric plasmids. Schematic representations of various chimeric LUC plasmids containing different fragments from rNIS 5′-flanking region in front of its own promoter (A) or of the TK promoter (B). The activity of each construct is expressed relative to that of pGL3-basic. The results presented are the means for at least three separate experiments, with each experiment done in duplicate. In all transfections, a CAT expression vector was introduced to normalize for transfection efficiency.
To verify whether the newly identified thyroid-specific enhancer was TSH and/or cAMP responsive, we transfected the pNISTKLUC constructs and the pTKLUC control in FRTL-5 cells. After transfection, cells were cultured for 72 h in starvation medium (Ham’s F-12 medium containing 0.2% calf serum). Cells were then kept in starvation medium or treated with either TSH or forskolin. After the cells were cultured for an additional 72 h, expression of LUC was measured (Fig. 3). All constructs showed similar transcriptional activity in starvation medium, indicating that under this condition the thyroid-specific enhancer is transcriptionally inactive. Conversely, pNISTKLUC1 and -3 exhibited significant stimulation of transcription by forskolin or TSH, while no effect was observed on the pTKLUC control or on pNISTKLUC2, which lacked the enhancer. No effects were observed when insulin was added (data not shown). Thus, the same region necessary to elicit thyroid-specific transcription is strongly activated by forskolin or TSH, suggesting that thyroid-specific factors could participate in the cAMP response of the rNIS upstream enhancer (NUE).
FIG. 3.
Hormonal regulation of the promoter activity of rNIS-TKLUC chimeric plasmids in FRTL-5 cells. Schematic representations of rNIS-TKLUC chimeric plasmids and their relative LUC activities in FRTL-5 cells cultured in 44 medium containing four hormones with (+) or without TSH or forskolin (forsk.). After transfection, cells were cultured for 72 h in starvation medium containing 0.2% calf serum and then incubated for an additional 72 h in medium containing 5% calf serum and the indicated additives. The activity of each construct is expressed relative to that of pGL3-basic. The results presented are the means for at least three separate experiments, with each experiment done in duplicate. In all transfections, a CAT expression vector was introduced to normalize for transfection efficiency.
To assess whether the cAMP response of the NUE was thyroid specific, we measured the transcriptional activation of pNISTKLUC3 by either forskolin or by the cPKA (21) in Rat-1 cells (Fig. 4). For a control for the treatment, we used a reporter containing the somatostatin CRE, which is known to be activated by cAMP via PKA-mediated phosphorylation of the transcription factor CREB. The data shown in Fig. 4 demonstrate that transcription of pNISTKLUC3 is not activated by either forskolin or cPKA in Rat-1 cells. Conversely, a CRE-containing promoter is activated by either agent. We conclude that cAMP-mediated transcriptional activation of NUE requires thyroid-specific mechanisms.
FIG. 4.
The NUE does not respond to forskolin in nonthyroid cells. (A) CRE-CAT and pNISTKLUC3 were transiently transfected, with or without an expression vector for cPKA in Rat-1 cells. Cells were plated 48 h prior to transfection. After transfection, cells were cultured in Dulbecco modified Eagle medium containing 5% calf serum in the absence or presence of 10 μM forskolin for 48 h. (B) Normalized CAT (for CRE-CAT) and LUC (for pNISTKLUC3) activity. Results are expressed as fold activation of the value obtained in the absence (−) of both cPKA and forskolin (Forsk.). The means of three independent experiments in duplicate are shown.
A novel cAMP-dependent transcription-activating mechanism in thyrocytes.
It has been reported before that FRTL-5 cells exposed chronically to TSH enter a state characterized by lack of response of CRE-containing promoters to further cAMP stimulation. Such a refractory state has been demonstrated to last for several days after removal of TSH from the medium and to depend on down-regulation of the mRNA encoding cPKA (1). We transfected FRTL-5 cells, grown in the presence of TSH and hence in the refractory state, with either CRE-CAT or pNISTKLUC3, in the presence or absence of an expression vector encoding cPKA. After transfection, cells were incubated in the presence or absence of 10 μM forskolin for 72 h (Fig. 5A). In these conditions, both CRE-CAT and pNISTKLUC3 were activated by cPKA expression. However, in the absence of cPKA, pNISTKLUC3 was activated by forskolin, while CRE-CAT was not (Fig. 5B). We suggest that the NUE is either responding to a PKA-independent pathway or is sensitive to the lower concentration of PKA that may be present in thyroid cells induced into the refractory state. To test the latter hypothesis, we transfected either CRE-CAT or pNISTKLUC3 in FRTL-5 thyroid cells in the refractory state, together with increasing amounts of cPKA expression vector (Fig. 5C). Since the results of this experiment do not reveal a preferential activation of NUE at low cPKA concentration, we conclude that the rNIS enhancer is capable of responding to cAMP in both PKA-dependent and -independent manners, while a classical CRE element can only be activated by a PKA-dependent mechanism.
Mutations and localization of binding sites for thyroid-enriched transcription factors within the rNIS enhancer.
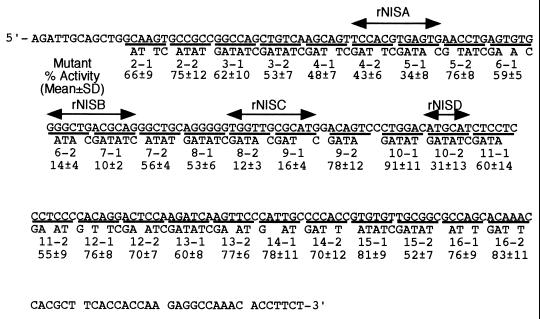
To identify potential protein binding sites responsible for transcriptional regulation, we introduced mutations throughout the NUE in the context of pNISTKLUC3 (Fig. 6). Each mutant was transfected in FRTL-5 cells grown in complete medium, and transcription levels were assessed by the activity of the LUC reporter in cell extracts. Mean values of activities of each mutant from three independent transfections are shown in Fig. 6. Four regions important for NUE activity, corresponding to mutation sites 5-1, 6-2 7-1, 8-2 9-1, and 10-2 were thus identified, and were named rNISA, -B, -C, and -D, respectively.
FIG. 6.
NUE mutants and relative transcriptional activity in FRTL-5 cells. The sequence of wild-type NUE is shown at the top. Mutants are numbered from 2-1 to 16-2, and only the nucleotides that are different from those of the wild-type sequence are indicated for each mutant. Transcriptional activity of each mutant is expressed as a percentage of that of pNISTKLUC3. rNISA, -B, -C, and -D are the regions that showed significantly lower LUC activity upon mutagenesis.
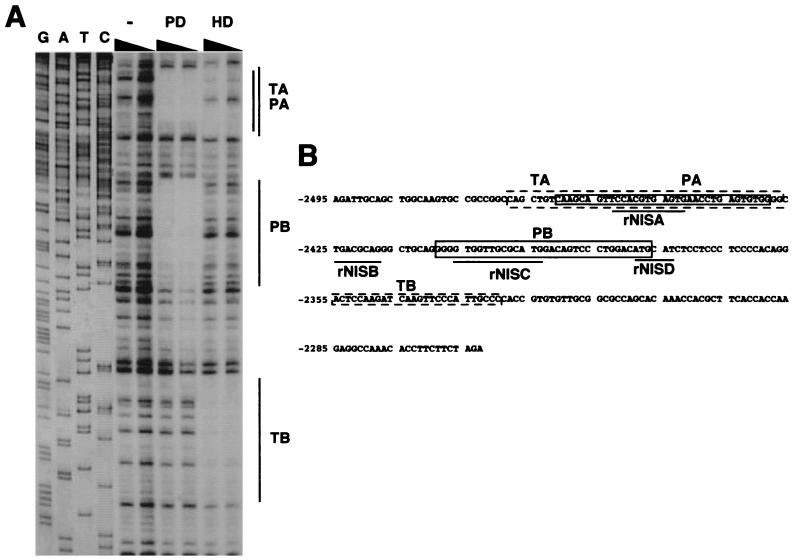
To test whether thyroid-specific transcription factors, such as Pax8, TTF1, and TTF-2, can bind to the important regions of the NUE, DNase I footprinting analysis was performed, using bacterial expressed Pax8 PD, TTF-1 HD, or TTF-2 (Fig. 7A). A sequencing reaction, run along the footprints, was used to precisely map the footprinted regions. Pax8 PD protected two areas in the rNIS enhancer located from −2461 to −2429 bp (PA) and −2409 to −2377 bp (PB), and TTF1 HD protected two areas from −2468 to −2427 bp (TA) and −2355 to −2330 bp (TB). No footprints were detected when TTF-2 was used (data not shown).
FIG. 7.
Binding of known thyroid-specific transcription factors to NUE. (A) DNase I footprinting obtained on the rNIS enhancer fragment in the absence of added proteins (−) or in the presence of either Pax8 PD or TTF1 HD. A sequence ladder of the rNIS fragment (G, A, T, and C) was used as size marker. The regions protected by PD (PA and PB) and HD (TA and TB) are depicted as lines. (B) Summary of the regions on rNIS upstream enhancer protected from DNase I digestion. Protected sequences obtained with PD (PA and PB) or HD (TA and TB) are indicated.
A summary of these data (Fig. 7B) shows that both TTF-1 and Pax8 bind to rNISA, while only Pax8 binds to rNISC. The second binding site of TTF-1 (TB) does not correspond to any relevant region. rNISB does not bind any of the transcription factors tested, but it contains the sequence 5′-TGACGCA-3′, which has been implicated as mediator of cAMP response in other promoters (see Discussion). rNISD was not further investigated in this study.
Pax8, not TTF-1, is required for transcriptional activation and cAMP stimulation of the rNIS enhancer.
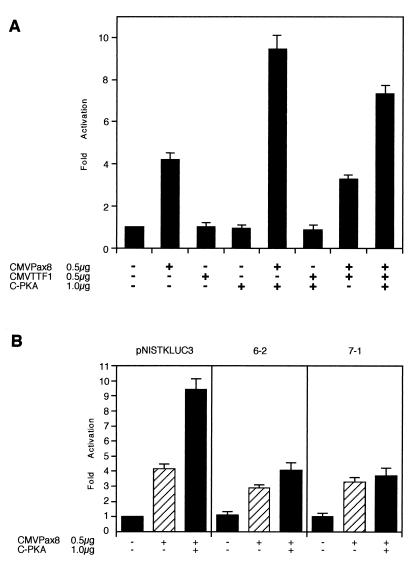
To provide direct evidence on the role of Pax8 and/or TTF-1 in rNIS enhancer activity, we introduced pNISTKLUC3 in HeLa cells together with various combinations of expression vectors encoding Pax8, TTF1, and cPKA and measured LUC activity in each transfection. As shown in Fig. 8A, rNIS enhancer is activated by Pax8. cPKA, which has no effect on its own, further enhances Pax-8-stimulated transcription. TTF-1 has no effect on rNIS transcription, either alone or in combination with Pax8, cPKA, or both. These data suggest that a large part of the rNIS enhancer activity is due to Pax8, which is also necessary to observe a cPKA-dependent increase in transcription. However, a mutation at the CRE-like sequence (7-1 [Fig. 8B]) does not interfere with Pax8 activation but abolishes the further increase in transcription by cPKA. Thus, we conclude that the target of cPKA is not Pax8 itself but an unidentified factor that binds at the CRE-like sequence and that mediates the cAMP or PKA response by cooperating with Pax8.
FIG. 8.
Pax8 stimulates rNIS enhancer activity in HeLa cells. (A) pNISTKLUC3 (3 μg) and CMV-CAT (1 μg) were transfected in HeLa cells with the expression vectors of Pax8 (0.5 μg of CMV-Pax8), cPKA (1 μg of C-PKA), and TTF1 (0.5 μg of CMV-TTF1). Activity of pNISTKLUC3 without Pax8, cPKA, and TTF1 was set at 1, and mean relative activities and standard deviations of cotransfected pNISTKLUC3 with the various expression vectors from three separate experiments are shown. pBluescriptII was used to adjust the total amount of DNA transfected. HeLa cells were incubated 48 h after transfection and harvested, and LUC activity and CAT amount were measured. (B) Comparison of the effects of cPKA and/or Pax8 on pNISTKLUC3 and mutants of the CRE-like sequence in NUE. Three micrograms of mutants 6-2 and 7-1, derived from pNISTKLUC3, and 1 μg of CMV-CAT were transfected in HeLa cells with or without 1 μg of C-PKA and/or 0.5 μg of CMV-Pax8. Activity of pNISTKLUC3 without Pax8 and cPKA was set at 1, and the mean relative activities and standard deviations of cotransfected test plasmids with the various expression vectors from three separate experiments are shown. pBluescriptII was used to adjust the total amount of DNA transfected. HeLa cells were incubated for 48 h after transfection and harvested, and LUC activity and CAT amount were measured.
To gain further support for the role of Pax8 in NUE transcriptional activity, we performed band shift assays. Oligonucleotides derived from both the PA and PB regions of the NUE form the same protein-DNA complex, as judged by the identical electrophoretic mobility, when challenged with nuclear extracts from FRTL-5 cells. A similar protein-DNA complex is absent in mock-transfected HeLa cells but can be easily observed if extracts of HeLa cells transfected with a Pax8 expression vector are used (Fig. 9A). To conclusively demonstrate the presence of Pax8 in the protein-DNA complex specifically formed with both PA and PB oligonucleotides, we performed supershift experiments with an anti-Pax8 specific antibody. These experiments clearly show that only the anti-Pax8 antibody, not an anti-TTF1 antibody, recognizes the main protein-DNA complex present with both oligonucleotides when FRTL-5 and HeLa-Pax8 extracts are used. No other bands were supershifted in the lane. Furthermore, the supershift could be abolished by the addition of the Pax8 synthetic peptide used to generate the antibody (Fig. 9A). Interestingly, mutations 5-1 (within rNISA) and 9-1 (within rNISC), both of which greatly reduce transcriptional activity of the NUE (Fig. 6), strongly interfere with Pax8 binding (Fig. 9B), as indicated by their inability to compete for the formation of the Pax8-DNA complex, thus indicating an important role for this transcription factor in NUE activity. As expected, the binding consensus sequence for Pax8 (11, 25, 33) is present in both PA and PB and is significantly altered by mutations 5-1m and 9-1m.
DISCUSSION
This study reports a structural and functional analysis of the regulatory region of the rNIS gene aimed at identifying regulatory elements responsible for thyroid-specific and TSH- or cAMP-controlled expression of the rNIS gene. We demonstrate the presence of a promoter within 0.56 kb upstream of the translation start site and of an enhancer located between nucleotides −2495 to −2264. In this report, we have focused on the rNIS enhancer, which we have called NUE, for NIS upstream enhancer. The NUE stimulates transcription of both its own and of a heterologous promoter exclusively in thyroid cells. Furthermore, the NUE is strictly controlled in thyroid cells by the TSH via the cAMP pathway. However, the NUE does not respond to forskolin-induced cAMP elevation in nonthyroid cell types, suggesting that the cAMP regulation of this enhancer utilizes thyroid-specific factors. In keeping with such a hypothesis, we show that the thyroid-restricted transcription factor Pax8 binds at two sites, which we call PA and PB, within the NUE. Mutations at these sites result in great reduction of both transcription and Pax8 binding, thus suggesting a functional role for Pax8 in NUE transcriptional activity. TTF-1, another transcription factor previously implicated in thyroid-specific transcription also binds at two sites (TA and TB) within the NUE. Interestingly, TA overlaps with PA. However, while the expression of Pax8 in nonthyroid cells is capable of inducing NUE-dependent transcription that can be further stimulated by cotransfection of a cPKA expression vector, similar experiments with TTF-1 did not induce NUE-dependent transcription. Taken together, these data suggest an important role for Pax8 in controlling NIS expression and add further relevance to the role of this protein in controlling thyroid function, as also demonstrated by the severe hypothyroidism observed in mice homozygous for a null Pax8 allele (29) and by the finding of PAX8 gene mutations in patients with congenital hypothyroidism (28). Another site, containing a degenerate CRE-like sequence (5′-TGACGCA-3′), is of relevance for NUE transcriptional activity. Several lines of evidence suggest that a factor(s) binding at this site plays an important role in the transcriptional activity of NUE. First, mutations at this site almost abolish NUE activity. Second, a NUE mutated at the CRE-like sequence can still be transcriptionally activated by Pax8 but cPKA induction is abolished. Finally, the same CRE-like sequence has been implicated in tissue-specific cAMP response in other promoters. In the case of the dopamine β-hydroxylase gene, a factor(s) binding at the same sequence acts synergistically with the HD protein Arix/Phox2 binding at a nearby site to activate transcription in a cAMP-dependent, cell-type-specific manner (38). This situation is reminiscent of what we describe for the NUE where the CRE-like sequence acts synergistically with Pax8 bound at two flanking sites. The same CRE-like sequence is found in the prohormone convertase 1 promoter, where it is thought to mediate cell-type-specific cAMP response by the binding of a heterodimer containing c-Jun and a novel, as yet unidentified, cell-type-specific protein (23). In the CRE-2 element of the proenkephalin gene promoter, where the sequence TGACGCA was first identified (10), it has been proposed that ATF-3 and the ATF-3-JunD heterodimer bind to stimulate cAMP-induced transcription (8). Thus, it appears that the TGACGCA sequence in many promoters plays an important role in controlling transcriptional regulation by cAMP, even though in most cases the factors binding to it have not been identified. In the case of NUE, we propose that this element acts synergistically with the PD protein Pax8 in order to obtain full, TSH-controlled, transcription.
The response of NUE to cAMP has one intriguing feature, in that it can be observed in thyroid cells cultured under conditions that do not allow activation mediated by a classical CRE octamer. It has been shown that such a condition depends on down-regulation of PKA expression, as a consequence of the chronic stimulation of the cAMP pathway (1). We have confirmed that in these conditions transcription from a CRE-dependent promoter can be restored by cotransfection of a cPKA expression vector. Interestingly, coexpression of cPKA also activates NUE-dependent transcription, suggesting that this enhancer can respond to cAMP both in the presence and absence of PKA. However, the novel feature of the NUE is its ability to respond to agents that elevate the intracellular concentration of cAMP in thyroid cells chronically exposed to TSH where cPKA is absent or greatly reduced. What could be the mechanism responsible for mediating the cAMP response of the NUE under these conditions? One possibility envisages the presence in thyroid cells of a transcription factor that binds to the CRE-like sequence of the NUE and that is sensitive to a low cPKA concentration. However, a titration experiment with a cPKA expression vector did not reveal any differential sensitivity between the NUE and a CRE-dependent promoter. Thus, we propose that the NUE might be sensitive to a factor whose activation by cAMP is PKA independent.
A few reports have recently appeared on the regulatory elements present in the NIS proximal promoter. Regulatory information to confer higher expression in thyroid cells has been reported for both the rat (17) and human (3) proximal promoters. The rat proximal promoter also appears to contain signals for TSH-regulated transcription (31). Surprisingly, however, Tong et al. reported that an 8-kb fragment from the 5′-flanking region of rNIS gene is not sufficient to confer thyroid-specific transcription (39). In this report we have concentrated on an upstream enhancer that we have called NUE. It is likely that the NUE acts synergistically with the elements identified in the proximal promoter to achieve high-level transcription. The NUE is a novel regulatory element that requires the functional interaction between Pax8 and a factor binding at a CRE-like sequence to achieve thyroid-specific transcription in a cAMP-dependent manner. We have also presented evidence suggesting that this enhancer is able to mediate cAMP-dependent transcription with a novel, PKA-independent mechanism. We are actively searching for the mediators of such a regulatory pathway in FRTL-5 cells.
ACKNOWLEDGMENTS
We thank Vittorio Enrico Avvedimento for providing CRE-CAT and C-PKA vectors and Caterina Missero and Maria Ina Arnone for reviewing the manuscript and for helpful discussions.
M.O. was supported by a grant from Japan Society for the Promotion of Science (JSPS), and O.L. was supported by National Institutes of Health Hepatology Research training grant DK-07218. This project was also supported by TELETHON, program no. D67, by Ministero per l’Università e la Ricerca Scientifica, project “Protein-Nucleic Acid Interactions,” by the Associazione Italiana per la Ricerca sul Cancro (A.I.R.C.), and by grants from the National Institutes of Health (DK-41544) and the American Cancer Society (BE-79422) (N.C.).
REFERENCES
- 1.Armstrong R, Wen W, Meinkoth J, Taylor S, Montminy M. A refractory phase in cyclic AMP-responsive transcription requires down regulation of protein kinase A. Mol Cell Biol. 1995;15:1826–1832. doi: 10.1128/mcb.15.3.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aza-Blanc P, Di Lauro R, Santisteban P. Identification of a cis-regulatory element and a thyroid-specific nuclear factor mediating the hormonal regulation of rat thyroid peroxidase promoter activity. Mol Endocrinol. 1993;7:1297–1306. doi: 10.1210/mend.7.10.8264661. [DOI] [PubMed] [Google Scholar]
- 3.Behr M, Schmitt T L, Espinoza C R, Loos U. Cloning of a functional promoter of the human sodium/iodide-symporter gene. Biochem J. 1998;331:359–363. doi: 10.1042/bj3310359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boshart M, Kluppel M, Schmidt A, Schutz G, Luckow B. Reporter constructs with low background activity utilizing the cat gene. Gene. 1992;110:129–130. doi: 10.1016/0378-1119(92)90456-y. [DOI] [PubMed] [Google Scholar]
- 5.Brindle P, Montminy M. The CREB family of transcription activators. Curr Opin Genet Dev. 1992;2:199–204. doi: 10.1016/s0959-437x(05)80274-6. [DOI] [PubMed] [Google Scholar]
- 6.Cassano S, Gallo A, Buccigrossi V, Porcellini A, Cerillo R, Gottesman M E, Avvedimento E V. Membrane localization of cAMP-dependent protein kinase amplifies cAMP signaling to the nucleus in PC12 cells. J Biol Chem. 1996;271:29870–29875. doi: 10.1074/jbc.271.47.29870. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Chu H-M, Tan Y, Kobierski L A, Balsam L B, Comb M J. Activating transcription factor-3 stimulates 3′,5′-cyclic adenosine monophosphate-dependent gene expression. Mol Endocrinol. 1994;8:59–68. doi: 10.1210/mend.8.1.8152431. [DOI] [PubMed] [Google Scholar]
- 9.Civitareale D, Lonigro R, Sinclair A J, Di Lauro R. A thyroid-specific nuclear protein essential for the tissue specific expression of the thyroglobulin promoter. EMBO J. 1989;8:2537–2542. doi: 10.1002/j.1460-2075.1989.tb08391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comb M, Mermod N, Hyman S E, Pearlberg J, Ross M E, Goodman H M. Proteins bound at adjacent DNA elements act synergistically to regulate human proenkephalin cAMP inducible transcription. EMBO J. 1988;7:3793–3805. doi: 10.1002/j.1460-2075.1988.tb03264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- 12.Dai G, Levy O, Carrasco N. Cloning and characterization of the thyroid iodide transporter. Nature. 1996;379:458–460. doi: 10.1038/379458a0. [DOI] [PubMed] [Google Scholar]
- 13.Damante G, Di Lauro R. Thyroid-specific gene expression. Biochim Biophys Acta. 1994;1218:255–266. doi: 10.1016/0167-4781(94)90176-7. [DOI] [PubMed] [Google Scholar]
- 14.Damante G, Rapoport B. TSH stimulates the activity of the c-fos promoter in FRTL5 rat thyroid cells. Mol Cell Endocrinol. 1988;58:279–282. doi: 10.1016/0303-7207(88)90165-7. [DOI] [PubMed] [Google Scholar]
- 15.De Felice M, Damante G, Zannini M, Francis-Lang H, Di Lauro R. Redundant domains contribute to the transcriptional activity of thyroid transcription factor 1 (TTF-1) J Biol Chem. 1995;270:26649–26656. doi: 10.1074/jbc.270.44.26649. [DOI] [PubMed] [Google Scholar]
- 16.De Vita G, Zannini M, Cirafici A M, Melillo R M, Di Lauro R, Fusco A, Santoro M. Expression of the RET/PTC1 oncogene impairs the activity of TTF-1 and Pax-8 thyroid transcription factors. Cell Growth Differ. 1998;9:97–103. [PubMed] [Google Scholar]
- 17.Endo T, Kaneshige M, Nakazato M, Ohmori M, Harii N, Onaya T. Thyroid transcription factor-1 activates the promoter activity of rat thyroid Na+/I− symporter gene. Mol Endocrinol. 1997;11:1747–1755. doi: 10.1210/mend.11.11.0012. [DOI] [PubMed] [Google Scholar]
- 18.Endo T, Shimura H, Saito T, Ikeda M, Ohmori M, Onaya T. Thyrotropin stimulates glucose-regulated protein (GRP78) gene expression in rat functional thyroid epithelial cells, FRTL. Mol Endocrinol. 1991;5:905–910. doi: 10.1210/mend-5-7-905. [DOI] [PubMed] [Google Scholar]
- 19.Francis-Lang H, Zannini M, Felice M D, Berlingieri M, Fusco A, Di Lauro R. Multiple mechanisms of interference between transformation and differentiation in thyroid cells. Mol Cell Biol. 1992;12:5793–5800. doi: 10.1128/mcb.12.12.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen C, Javaux F, Juvenal G, Vassart G, Christophe D. cAMP-dependent binding of a trans-acting factor to the thyroglobulin promoter. Biochem Biophys Res Commun. 1989;160:722–731. doi: 10.1016/0006-291x(89)92493-5. [DOI] [PubMed] [Google Scholar]
- 21.Huggenvick J I, Collard M W, Stofko R E, Seasholtz A F, Uhler M D. Regulation of human enkephalin promoter by two isoforms of the catalytic subunit of cyclic adenosine 3′,5′-monophosphate-dependent protein kinase. Mol Endocrinol. 1991;5:921–930. doi: 10.1210/mend-5-7-921. [DOI] [PubMed] [Google Scholar]
- 22.Ikuyama S, Shimura H, Hoeffler J P, Kohn L D. Role of the cyclic adenosine 3′,5′-monophosphate response element in efficient expression of the rat thyrotropin receptor promoter. Mol Endocrinol. 1992;6:1701–1715. doi: 10.1210/mend.6.10.1333054. [DOI] [PubMed] [Google Scholar]
- 23.Jansen E, Ayoubi T A Y, Meulemans S M P, Van de Ven W J M. Cell type-specific protein-DNA interactions at the cAMP response elements of the prohormone convertase 1 promoter. J Biol Chem. 1997;272:2500–2508. doi: 10.1074/jbc.272.4.2500. [DOI] [PubMed] [Google Scholar]
- 24.Kogai T, Endo T, Saito T, Miyazaki A, Kawaguchi A, Onaya T. Regulation by thyroid-stimulating hormone of sodium/iodide symporter gene expression and protein levels in FRTL-5 cells. Endocrinology. 1997;138:2227–2232. doi: 10.1210/endo.138.6.5189. [DOI] [PubMed] [Google Scholar]
- 25.Kozmik Z, Czerny T, Busslinger M. Alternatively spliced insertions in the paired domain restrict the DNA sequence specificity of Pax6 and Pax8. EMBO J. 1997;16:6793–6803. doi: 10.1093/emboj/16.22.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazzaro D, Price M, Felice M D, Di Lauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113:1093–1104. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- 27.Levy O, Dai G, Riedel C, Ginter C S, Paul E M, Lebowitz A N, Carrasco N. Characterization of the thyroid Na+/I− symporter with an anti-COOH terminus antibody. Proc Natl Acad Sci USA. 1997;94:5568–5573. doi: 10.1073/pnas.94.11.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macchia P E, Lapi P, Krude H, Pirro M T, Missero C, Chiovato L, Souabni A, Baserga M, Tassi V, Pinchera A, Fenzi G, Gruters A, Busslinger M, Di Lauro R. PAX8 mutations associated with congenital hypothyroidism caused by thyroid dysgenesis. Nat Genet. 1998;19:83–86. doi: 10.1038/ng0598-83. [DOI] [PubMed] [Google Scholar]
- 29.Mansouri A, Chowdhury K, Gruss P. Follicular cells of the thyroid gland require Pax8 gene function. Nat Genet. 1998;19:87–90. doi: 10.1038/ng0598-87. [DOI] [PubMed] [Google Scholar]
- 30.Nissim M, Lee K O, Petrick P A, Dahlberg P A, Weintraub B D. A sensitive thyrotropin (TSH) bioassay based on iodide uptake in rat FRTL-5 thyroid cells: comparison with the adenosine 3′,5′-monophosphate response to human serum TSH and enzymatically deglycosylated bovine and human TSH. Endocrinology. 1987;121:1278–1287. doi: 10.1210/endo-121-4-1278. . (Erratum, 124:332, 1989.) [DOI] [PubMed] [Google Scholar]
- 31.Ohmori M, Endo T, Harii N, Onaya T. A novel thyroid transcription factor is essential for thyrotropin-induced up-regulation of Na+/I− symporter gene expression. Mol Endocrinol. 1998;12:727–736. doi: 10.1210/mend.12.5.0101. [DOI] [PubMed] [Google Scholar]
- 32.Paire A, Bernier-Valentin F, Selmi-Ruby S, Rousset B. Characterization of the rat thyroid iodide transporter using anti-peptide antibodies. Relationship between its expression and activity. J Biol Chem. 1997;272:18245–18249. doi: 10.1074/jbc.272.29.18245. [DOI] [PubMed] [Google Scholar]
- 33.Pellizzari L, Fabbro D, Lonigro R, Di Lauro R, Damante G. A network of specific minor-groove contacts is a common characteristic of paired-domain-DNA interactions. Biochem J. 1996;315:363–367. doi: 10.1042/bj3150363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pichon B, Jimenez-Cervantes C, Pirson I, Maenhaut C, Christophe D. Induction of nerve growth factor-induced gene-B (NGFI-B) as an early event in the cyclic adenosine monophosphate response of dog thyrocytes in primary culture. Endocrinology. 1996;137:4691–4698. doi: 10.1210/endo.137.11.8895335. [DOI] [PubMed] [Google Scholar]
- 35.Plachov D, Chowdhury K, Walther C, Simon D, Guenet J-L, Gruss P. Pax8, a murine paired box gene expressed in the developing excretory system and thyroid gland. Development. 1990;110:643–651. doi: 10.1242/dev.110.2.643. [DOI] [PubMed] [Google Scholar]
- 36.Roger P P, Dumont J E. Factors controlling proliferation and differentiation of canine thyroid cells cultured in reduced serum conditions: effects of thyrotropin, cyclic AMP and growth factors. Mol Cell Endocrinol. 1984;36:79–93. doi: 10.1016/0303-7207(84)90087-x. [DOI] [PubMed] [Google Scholar]
- 37.Shimura H, Ikuyama S, Shimura Y, Kohn L D. The cAMP response element in the rat thyrotropin receptor promoter. Regulation by each decanucleotide of a flanking tandem repeat uses different, additive, and novel mechanisms. J Biol Chem. 1993;268:24125–24137. [PubMed] [Google Scholar]
- 38.Swanson D J, Zellmer E, Lewis E J. The homeodomain protein Arix interacts synergistically with cAMP to regulate expression of neurotransmitter biosynthetic genes. J Biol Chem. 1997;272:27382–27392. doi: 10.1074/jbc.272.43.27382. [DOI] [PubMed] [Google Scholar]
- 39.Tong Q, Ryu K Y, Jhiang S M. Promoter characterization of the rat Na+/I− symporter gene. Biochem Biophys Res Commun. 1997;239:34–41. doi: 10.1006/bbrc.1997.7432. [DOI] [PubMed] [Google Scholar]
- 40.Weiss S J, Philp N J, Ambesi-Impiombato F S, Grollman E F. Thyrotropin-stimulated iodide transport mediated by adenosine 3′,5′-monophosphate and dependent on protein synthesis. Endocrinology. 1984;114:1099–1107. doi: 10.1210/endo-114-4-1099. [DOI] [PubMed] [Google Scholar]
- 41.Zannini M, Avantaggiato V, Biffali E, Arnone M I, Sato K, Pischetola M, Taylor B A, Phillips S J, Simeone A, Di Lauro R. TTF-2, a new forkhead protein, shows a temporal expression in the developing thyroid which is consistent with a role in controlling the onset of differentiation. EMBO J. 1997;16:3185–3197. doi: 10.1093/emboj/16.11.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zannini M, Francis-Lang H, Plachov D, Di Lauro R. Pax-8, a paired domain-containing protein, binds to a sequence overlapping the recognition site of a homeodomain and activates transcription from two thyroid-specific promoters. Mol Cell Biol. 1992;12:4230–4241. doi: 10.1128/mcb.12.9.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]