Abstract
Recently, we reported the identification of a 55-kDa polypeptide (p55) from Tetrahymena macronuclei as a catalytic subunit of a transcription-associated histone acetyltransferase (HAT A). Extensive homology between p55 and Gcn5p, a component of the SAGA and ADA transcriptional coactivator complexes in budding yeast, suggests an immediate link between the regulation of chromatin structure and transcriptional output. Here we report the characterization of a second transcription-associated HAT activity from Tetrahymena macronuclei. This novel activity is distinct from complexes containing p55 and putative ciliate SAGA and ADA components and shares several characteristics with NuA4 (for nucleosomal H2A/H4), a 1.8-MDa, Gcn5p-independent HAT complex recently described in yeast. A key feature of both the NuA4 and Tetrahymena activities is their acetylation site specificity for lysines 5, 8, 12, and 16 of H4 and lysines 5 and 9 of H2A in nucleosomal substrates, patterns that are distinct from those of known Gcn5p family members. Moreover, like NuA4, the Tetrahymena activity is capable of activating transcription from nucleosomal templates in vitro in an acetyl coenzyme A-dependent fashion. Unlike NuA4, however, sucrose gradient analyses of the ciliate enzyme, following sequential denaturation and renaturation, estimate the molecular size of the catalytically active subunit to be ∼80 kDa, consistent with the notion that a single polypeptide or a stable subcomplex is sufficient for this H2A/H4 nucleosomal HAT activity. Together, these data document the importance of this novel HAT activity for transcriptional activation from chromatin templates and suggest that a second catalytic HAT subunit, in addition to p55/Gcn5p, is conserved between yeast and Tetrahymena.
Multiple lines of evidence demonstrate that chromatin structure plays a pivotal role in the regulation of eukaryotic gene transcription (reviewed in references 9 and 47). For appropriate gene transcription to occur in a chromatin environment, nucleosomal structures are often modified, remodeled, or disrupted to facilitate access of the transcriptional machinery to DNA. Currently, two mechanisms are known to help relieve nucleosomal repression of transcription: chromatin remodeling by ATP-dependent remodeling complexes (reviewed in references 21 and 48) and histone modification, particularly acetylation as regulated by histone acetyltransferases (HATs) and deacetylases (HDACs) (reviewed in references 5, 14, 32, and 43).
Posttranslational acetylation of specific lysine residues in the amino-terminal tails of core histones is closely linked to transcriptional activation in numerous biological settings (reviewed in references 14 and 47). Recently, a surprisingly large number of polypeptides, including p55/Gcn5p (6), PCAF (50), p300/CBP (1, 31), TAFII250 (28), and SRC/ACTR (7, 41), have been shown to possess intrinsic HAT activity (reviewed in reference 26). In addition, several proteins, including SAS2 and SAS3 (35), MOZ (3), Tip60 (20), MOF (16), and Esa1p (38), possess a conserved putative acetyl coenzyme A (acetyl-CoA) binding motif common to GCN5 homologs and other acetyltransferases (29), and Tip60 (49) and Esa1p (38) have recently been shown to acetylate histones in vitro. However, whether histones are physiological substrates for all of these activities and whether the HAT activity of each protein is required for transcriptional activation of target genes in vivo remain unclear. Functional requirements for the HAT activities of yGcn5p, PCAF, and p300 for transcriptional activation in vivo have recently been demonstrated (22, 24, 34). However, with the possible exception of yGcn5p (see references 24 and 46), it remains a formal possibility that nonhistone proteins are the bona fide substrates for these activities in vivo, and evidence supporting this possibility has been described previously (15, 19, 33).
A related issue is to what extent HAT activities acetylate histones within a nucleosome or a polynucleosome array, in comparison to nonnucleosomal (free) histones. For example, yeast Gcn5p alone readily acetylates certain “free” (nonnucleosomal) histones in vitro (23) but nucleosomal histones are not acetylated unless Gcn5p is present in one of several large multisubunit complexes. By using yeast whole-cell extracts as a starting source, several HAT complexes that acetylate nucleosomal histones have been identified (12, 13, 33; reviewed in references 11 and 44). For example, a 1.8-MDa nucleosomal acetylation complex referred to as SAGA (for Spt-Ada-Gcn5 acetyltransferase) contains Gcn5p, Ada3, Spt20, Spt3, Spt7, and a subset of TATA binding protein-associated factors (TAFs) (12, 13). However, not all of these HAT complexes contain Gcn5p as the catalytic subunit. These same yeast extracts contain two nucleosome acetylation complexes, NuA4 (formerly complex II) and NuA3 (formerly complex III), which do not contain Gcn5p (12). These complexes are characterized by distinct and nonoverlapping specificities: NuA3 preferentially acetylates nucleosomal H3, while NuA4 preferentially acetylates nucleosomal H2A and H4 in vitro (12, 45). The catalytic subunit responsible for NuA3 activity has not been identified. Recently, by use of a temperature-sensitive (ts) allele of Esa1p, it has been shown that Esa1p is the catalytic subunit responsible for NuA4 activity (8).
Like yeast Gcn5p, p55 from the ciliated protozoan Tetrahymena thermophila was initially identified as a HAT by virtue of its ability to acetylate free histones in the context of the HAT activity gel assay (5, 6). The nature of this sodium dodecyl sulfate (SDS)-gel-based assay precludes the use of nucleosomal substrates. Thus, we were curious whether, as in yeast, HAT activities with strict nucleosomal substrate specificity that had been missed by the activity gel assay exist in Tetrahymena. Here we report the characterization of second Tetrahymena type A HAT activity that acetylates nucleosomal H2A and H4 but is inactive with free-histone substrates. This activity is not dependent upon p55, suggesting that the catalytic subunit is a novel protein that was not detected in our previous studies. We show here that the substrate specificity and acetylation site usage profiles of this ciliate activity are indistinguishable from those of the yeast NuA4 complex. Moreover, we show that this ciliate activity, like that of the yeast NuA4 complex, activates transcription from a chromatin template in vitro in an acetyl-CoA-dependent manner. These data suggest that this ciliate activity plays an important and conserved role in facilitating transcriptional activation through nucleosomal acetylation.
MATERIALS AND METHODS
Tetrahymena macronuclei preparation and DNase I extraction.
T. thermophila CU 427 or CU 428 (provided by P. Bruns, Cornell University, Ithaca, N.Y.) was grown to log phase (cell density, 2.5 × 105 cells/ml) under standard conditions as previously described (10). Macronuclei were prepared as described previously (4) except for the elimination of iodoacetamide and butyric acid from nucleus isolation buffer A. HAT activity was quantitatively extracted from macronuclei by extensive digestion with DNase I. For DNase I extraction, freshly prepared macronuclei (108 macronuclei/ml) were resuspended in DNase I extraction buffer, which consisted of 25 mM Tris-HCl (pH 8.0), 15 mM NaCl, 10 mM MgCl2, 0.1 mM CaCl2, 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.05 mM dithiothreitol (DTT), and 500 U of DNase I (Gibco BRL) per ml, and incubated on ice for 90 min. After extraction, macronuclear remnants and insoluble debris were removed by centrifugation at 50,500 × g for 30 min at 2°C. The resulting supernatant was used as crude extract for all experiments. Crude DNase I extract, containing 10% glycerol, was stable for several months at −70°C.
Sucrose density gradient and size estimation.
To separate nucleosomal histone acetylation activity from free (nonnucleosomal)-histone acetylation activity, crude DNase I extracts were subjected to centrifugation in sucrose gradients containing 25 mM NaCl, 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 5 to 30% sucrose. After sedimentation at 155,000 × g for 42 h at 2°C and fractionation, gradient fractions were analyzed for HAT activity by using either acid-extracted chicken histones or nucleosomes. Size calibration of parallel gradients under the same conditions was as follows: mononucleosome (220 kDa), fractions 2 to 4; catalase (232 kDa), fractions 2 to 4; phosphorylase b (97.4 kDa), fractions 20 to 22; chicken egg albumin (45 kDa), fractions 32 to 34; carbonic anhydrase (29 kDa), fractions 34 to 36; trypsin inhibitor (20 kDa), fractions 38 to 40; cytochrome c (12.4 kDa), fractions 40 to 42. Typically, Tetrahymena peak I HAT activity was found in fraction 18; peak II was found in fraction 24.
Preparation of substrates.
Chicken core histones and oligonucleosomes were prepared from chicken erythrocytes as described previously (27). Tetrahymena core histones and mononucleosomes were isolated from macronuclei as described before (4).
In vitro acetylation.
A typical 50-μl in vitro acetylation reaction mixture consisted of the following: 50 mM Tris-HCl (pH 8.0), 10% glycerol, 10 mM n-butyrate, 1 mM PMSF, 1 mM DTT, 0.1 μCi of [3H]acetyl-CoA (4 to 5 Ci mmol−1), 1 μg of chicken nucleosome or acid-extracted chicken core histones, and 20 μl of crude DNase I extract or corresponding sucrose gradient fractions. Reaction mixtures were typically incubated at 30°C for 10 min, after which 10 μl of each mixture was absorbed onto a P81 filter (2, 18) for scintillation counting. Where appropriate, the remaining 40 μl of each mixture was precipitated with 20% trichloroacitic acid and resolved on an SDS–12.5% polyacrylamide gel. Histones and acetylated histone products were visualized by Coomassie brilliant blue staining and fluorography, respectively.
Antisera and Western blotting.
For Western blotting, fractionated HAT samples were electrophoresed on SDS–8% polyacrylamide gels, transferred to polyvinylidene difluoride membranes, and processed for immunoblotting. Western blots were probed with rabbit polyclonal antibodies generated against an internal peptide sequence of Tetrahymena p55 (K36) (see reference 6), yAda1p (generously provided by J. Horiuchi and L. Guarente; Massachusetts Institute of Technology, Cambridge), and yAda5p/ySpt20p (generously provided by J. Horiuchi and L. Guarente as well as S. Roberts and F. Winston, Harvard Medical School, Boston, Mass.). With each antiserum, only a single strongly reactive band with the expected molecular mass was observed, as follows: Tetrahymena p55, 55 kDa; yAda1p, 55 kDa (17); yAda5p/ySpt20p, 68 kDa (25, 36). For blots probed with antibodies against the yeast Adaps, we assume that the cross-reacting bands represent homologous proteins from Tetrahymena.
For Esa1p antibodies, we used rabbit polyclonal antibodies generated against an N-terminal peptide sequence (residues 4 to 18) of yeast Esa1p (AN191) (see reference 8) or an internal peptide sequence (residues 172 to 186 of yeast ESA1p) corresponding to a region conserved among MYST family members, including yeast Esa1p, human Tip60, Drosophila MOF, Drosophila MOZ, and yeast SAS2 and SAS3 (16).
Reverse-phase high-performance liquid chromatography (RP-HPLC) and reconstitution of HAT activity.
To better separate the closely sedimenting p55 from the H2A/H4 nucleosomal HAT activity, active fractions from sucrose gradients were chromatographed on a 2-mm by 25-cm Brownlee RP-300 column (Applied Biosystems) eluted at 0.25 ml/min with a linear ascending gradient of acetonitrile (36.5 to 63.5% in 30 min) on a SMART system (Pharmacia). p55 and the H2A/H4 nucleosomal HAT activity were eluted by 45 and 51% acetonitrile, respectively. For reconstitution of HAT activity, appropriate fractions were dried under vacuum, resolubilized in 8 M urea–10 mM Tris HCl (pH 8.0)–1 mM EDTA, and reconstituted by slow sequential dialysis against the same buffer containing 4, 2, 1, and 0 M concentrations of urea (12 h per step). Reconstituted fractions were then analyzed for HAT activity against free-histone and nucleosomal substrates. Assay conditions were the same as those of the standard assay (see above) except that the incubation time was extended to 30 min.
Determination of acetylation sites.
Microsequence analysis of the amino-terminal tails of acetylated histone products was essentially as described earlier (40). For microsequencing of acetylated H2A and H4, chicken nucleosomes were acetylated by using the Tetrahymena peak II fraction recovered from sucrose gradients or yeast NuA4 (partially purified by sequential chromatography on nickel-agarose, Mono Q, Mono S, histone-agarose, and DNA-cellulose) (see reference 8). A typical 50-μl reaction mixture consisted of the following: 50 mM Tris-HCl (pH 8.0), 10% glycerol. 10 mM n-butyrate, 1 mM PMSF, 1 mM DTT, 5 μCi of [3H]acetyl-CoA (4 to 5 Ci mmol−1), 5 μg of chicken nucleosomes, and 20 μl of peak II fraction or 7.5 μl of partially purified yeast NuA4. Reaction mixtures were incubated at 30°C for 120 min, after which labeled histones were extracted by 0.4 N H2SO4 and recovered by RP-HPLC. Histones were chromatographed on a 2-mm by 25-cm Vydac 218TP52 column (Separations Group, Hesperia, Calif.) eluted at 0.20 ml/min with a linear ascending gradient of acetonitrile (29 to 52% in 90 min) on a SMART system (Pharmacia). Chicken core histones were completely resolved under these conditions. Fractions corresponding to H2A and H4 were pooled, dried under vacuum, and subjected to microsequencing following a deblocking step (39). In all cases, the expected amino-terminal sequence of each histone was obtained.
In vitro transcription.
Analysis of transcription in vitro was performed essentially as described by Steger et al. (42). In short, the human immunodeficiency virus (HIV) dinucleosome-5S array template was incubated with enriched HAT fractions for 30 min at 30°C in the absence or presence of 1 μM acetyl-CoA. The amounts of Tetrahymena peak II and yeast NuA4 fractions added to the reaction mixtures were normalized for nucleosomal HAT activity such that each complex transferred an equal amount of [3H]acetate onto HeLa nucleosomes in a standard liquid HAT assay. However, the amounts of yEsa1p and Tetrahymena peak I employed were determined by normalization to the activity of NuA4 with free-histone substrate and protein concentration, respectively, since these activities do not modify nucleosomal histones. Upon completion of histone acetylation, preinitiation complexes were formed by adding HeLa nuclear extract and incubating at room temperature for at least 20 min. G5-E4 DNA, which contains five GAL4 sites upstream of a minimal adenovirus E4 promoter, and GAL4-AH were included to serve as a control for RNA recovery through subsequent steps. Transcription was initiated upon the addition of ribonucleoside triphosphates and terminated after a 30-min incubation at 30°C. RNA transcripts were detected by primer extension.
RESULTS
Identification of a novel nucleosome-specific HAT activity in Tetrahymena.
Recently, an in-gel HAT assay employing free (nonnucleosomal) histones as a substrate was used to identify and purify a 55-kDa polypeptide (p55) as a catalytic subunit of a transcription-associated, nuclear HAT (4). Like p55, yGcn5p displays a strong preference for certain histones in a free form (23), unless it exists as part of native HAT complexes, in which case nucleosomes also serve as a substrate (12). We sought to determine whether extracts prepared from Tetrahymena macronuclei might also contain additional HAT activities which were not revealed in our original in-gel assays with free-histone substrates.
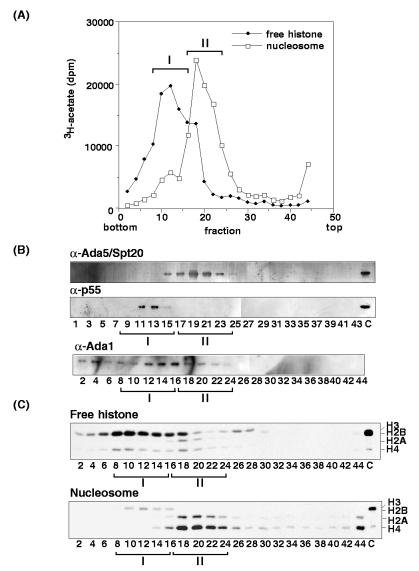
As a starting point for this work, we selected a DNase I extract of macronuclei since preliminary experiments indicated that it contained a significant amount of both a free-histone and a nucleosome acetylation activity (see below). To investigate whether multiple HAT activities might exist in Tetrahymena, DNase I extract was resolved on a 5 to 30% sucrose gradient and assayed with both free and nucleosomal histones (Fig. 1A). Two closely sedimenting activities were evident: (i) a free-histone acetylation activity (hereafter called peak I) that was largely H3 specific and that failed to acetylate nucleosomal histones, and (ii) a nucleosomal activity (peak II) that was largely H2A/H4 specific (Fig. 1C). To examine the potential relationship of p55 and other known interacting proteins (for example, Adaps or Sptps) to these activities, gradient fractions were probed with antibodies against p55 peptide (K36), Ada1p, and Ada5p/Spt20p (Fig. 1B) (see references 6 and 17 for details). Ada1p and Ada5p/Spt20p antibodies are the only available antibodies to known yeast HAT complex subunits that react strongly with Tetrahymena proteins of the appropriate molecular weights (34a). As expected from the distribution of H3-specific free-histone HAT activity across the gradient, immunoblotting analyses show that peak I, but not peak II, contained the majority of the p55. Thus, these data suggested that p55 was not the catalytic subunit responsible for the nucleosomal H2A/H4 acetylation exhibited by peak II. On sucrose gradients, Ada1p and Ada5p/Spt20p sedimented closely with the peak I and peak II activities, respectively.
FIG. 1.
Two distinct type A HAT activities exist in Tetrahymena macronuclei. (A) Sucrose-gradient sedimentation of HAT activities characterized with either a mixture of free (nonnucleosomal) histones (peak I) or nucleosomal histones (peak II); histone substrates were from chickens. (B) Western blot analysis using yAda1p and yAda5p/ySpt20p antisera as well as Tetrahymena p55 (K36) peptide antibodies. (C) Acetylated histone products were examined following gel electrophoresis and fluorography. Lanes C contain DNase I extract as input.
Resolution of distinct HAT activities by RP-HPLC.
Given the large native molecular weights reported for the Gcn5p-dependent HAT complexes in yeast (12, 13, 33), it was surprising that the sizes of the peak I and II HAT activities estimated by sedimentation analyses were ∼110 and ∼80 kDa, respectively (relative to protein standards sedimented in parallel gradients; see Materials and Methods). These molecular sizes suggested that the peak I and II activities observed in Fig. 1, unlike what has been described in yeast, were due to small subcomplexes or even single polypeptides.
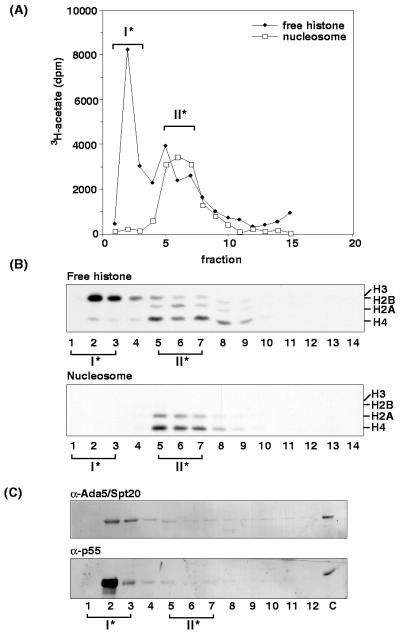
To investigate this possibility further, peak II (prepared by sucrose gradient sedimentation) was subjected to RP-HPLC, under conditions (e.g., acidic buffers and nonpolar solvents) expected to promote dissociation of protein complexes, in an attempt to determine if the nucleosomal HAT activity of peak II required the assembly of subunits for function. After reconstitution by stepwise dialysis, RP-HPLC gradient fractions were assayed for free-histone and nucleosomal HAT activities (Fig. 2A and B) and by Western blot analysis using p55 antibodies and Ada5p/Spt20p antibodies (Fig. 2C). A prominent peak of nucleosomal HAT activity was detected in fractions 5 to 9, with the peak of this activity in fraction 6. Like the ∼80-kDa peak II nucleosomal activity shown in Fig. 1, this activity (hereafter referred to as peak II*) was also specific for H2A/H4 (Fig. 2B). As well, two free-histone HAT activities were detected, and, for the most part, these were well separated from the major H2A/H4 nucleosomal activity. One free-histone HAT activity was H3 specific, with the peak of this activity in fraction 2. As expected, this fraction contained the majority of p55 and was most likely to be the same or closely related to the peak I activity of Fig. 1 (hereafter referred to as peak I*). A second free-histone HAT activity was H2A/H4 specific, and the peak overlapped considerably with nucleosome acetylation activity in fractions 5 to 9. These data suggest that a specific conformation of the catalytic protein and/or correct subcomplex in peak II may be necessary to restrict natural substrate specificity. Consistent with sucrose gradient analyses (Fig. 1), the complete absence of p55 in the peak II* activity (Fig. 2C) suggested that p55 is not the catalytic subunit responsible for the H2A/H4 nucleosomal HAT activity. Ada5p/ Spt20p was detected in fractions 2 and 3 also (Fig. 2C). These data show clearly that these proteins were well separated from peak II* activity (fractions 5 to 9) after RP-HPLC.
FIG. 2.
The H2A/H4 nucleosomal HAT activity in Tetrahymena is not p55 dependent. (A) HAT activity of RP-HPLC fractions following reconstitution by stepwise dialysis. A peak of free, nonnucleosomal HAT activity (I*) was contained in fraction 2. In contrast, a well-separated peak of nucleosomal HAT activity was contained in fractions 5 to 7 (II*). (B) Substrate specificity of these fractions. Note that nucleosomal HAT activity (II*) was H2A/H4 specific; in contrast, free-histone HAT activity was H3 specific. (C) Western blot analysis of the fractions. As expected, free-histone HAT activity (I*) (fractions 2 and 3) contained essentially all of the p55 signal. In contrast, the peak of nucleosomal H2A/H4 HAT activity (fractions 5 to 7) was devoid of p55, suggesting that this polypeptide cannot be responsible for the nucleosomal H2A/H4 HAT activity (see text). Note that fractions 2 and 3 also contained the majority of Ada5/Spt20 signal. Lane C contains DNase I extract as input.
Size and nature of the reconstituted H2A/H4 nucleosomal HAT activity.
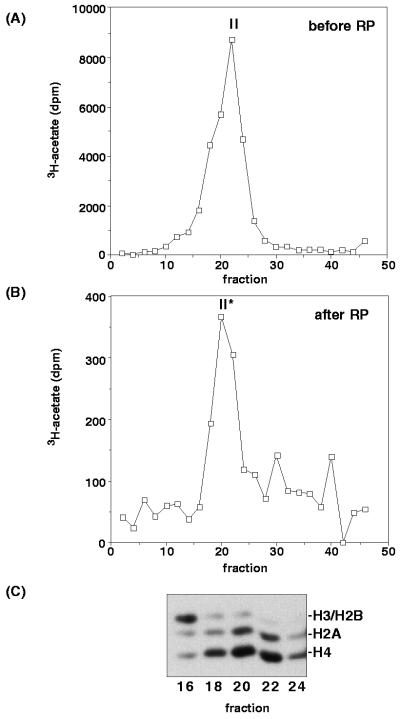
To determine the size of the H2A/H4 HAT activity reconstituted following RP-HPLC (peak II*), fractions displaying activity after reconstitution (fractions 5 to 7) were pooled and sedimented on a 5 to 30% sucrose gradient. Gradient fractions were again assayed for HAT activity by using nucleosomes as substrates (Fig. 3). These data clearly show that the size of the nucleosomal HAT activity was the same, about 80 kDa, before (Fig. 3A) or after (Fig. 3B) RP-HPLC and subsequent reconstitution. Moreover, the H2A/H4 specificity of the reconstituted activity appeared to be identical to that of the native activity (compare Fig. 1C and 3C). When similar analyses were performed with peak I* fractions, free-histone acetylation activity was detected in fractions 30 to 34, corresponding to a size of about 55 to 45 kDa (data not shown). These data suggest that the macronuclear H2A/H4 nucleosome-specific HAT activity results from either a single subunit or a small multisubunit complex whose activity can be reconstituted after RP-HPLC. Our data also indicate that p55 is not likely to be the catalytic subunit responsible for this activity.
FIG. 3.
Following RP-HPLC and renaturation, the size of the H2A/H4 nucleosomal HAT activity and its substrate specificity are unchanged. (A) Sedimentation profile of HAT activity of native DNase I macronuclear extract on a sucrose-density gradient, shown for reference. (B) Sedimentation profile of HAT activity of the nucleosomal H2A/H4 HAT activity (peak II) following RP-HPLC and renaturation. In both cases, the size of the active protein/complex was estimated to be about ∼80 kDa relative to marker proteins resolved in parallel gradients (data not shown). (C) Substrate specificity of nucleosome acetylation by fractions from panel B as indicated. A fluorogram of a SDS–12.5% polyacrylamide gel is shown. Note that nucleosomal HAT activity (II*) remained H2A/H4 specific (compare to Fig. 2B).
Biochemical fractionation of HAT complexes from yeast and human extracts indicate that Gcn5p (12, 13, 33) and a related GCN5 family member, PCAF (30), function in the context of large multisubunit complexes. Interestingly, both the yeast SAGA and human PCAF HAT complexes contain a subset of histone-like TAFs, some of which are also associated with TFIID, a key basal component of the RNA polymerase II transcription machinery (13, 30; reviewed in reference 44). However, the above data suggest that the H2A/H4 nucleosomal HAT from Tetrahymena may be able to function alone as an isolated ∼80-kDa subunit.
To determine whether any of the other known interacting proteins (Adaps or Sptps) cofractionate with this nucleosomal HAT activity, the sucrose and RP-HPLC gradients shown in Fig. 1 and 2 were assayed by using Ada1p and Ada5p/Spt20p antibodies (see Fig. 1B and Fig. 2C). These analyses show clearly that while Ada5p/Spt20p sediments closely to the peak II activity on sucrose gradients, this polypeptide is well separated from peak II* after RP-HPLC. In addition, Ada1p is well separated from the peak II activity on the initial sucrose gradients. Although our analyses may be restricted by limited cross-reactivity of antibodies raised against the yeast proteins, these results indicate that Ada1p and Ada5p/Spt20p are not required for this ciliate activity. Taken together, these data suggest that the polypeptide or subcomplex responsible for this activity in Tetrahymena is distinct from fully assembled ADA and SAGA complexes in yeast.
The p55-independent H2A/H4 nucleosomal HAT from Tetrahymena macronuclei is reminiscent of the Gcn5p-independent H2A/H4 nucleosomal (NuA4) HAT activity recently described in yeast (12). However, the large (1.3-MDa) molecular size of yeast NuA4 suggests the possibility that the Tetrahymena H2A/H4 activity represents a metastable subspecies formed upon disassembly of a larger native complex. Several attempts were made to determine if a H2A/H4 nucleosomal HAT activity with a similar size (∼80 kDa) could be renatured following dissociation and resolution of yNuA4 components by RP-HPLC. Unfortunately, H2A/H4-specific HAT activity was not reconstituted from denatured yeast NuA4 despite numerous attempts.
Identical site usage patterns are catalyzed by the H2A/H4 nucleosomal HAT activities in Tetrahymena and yeast.
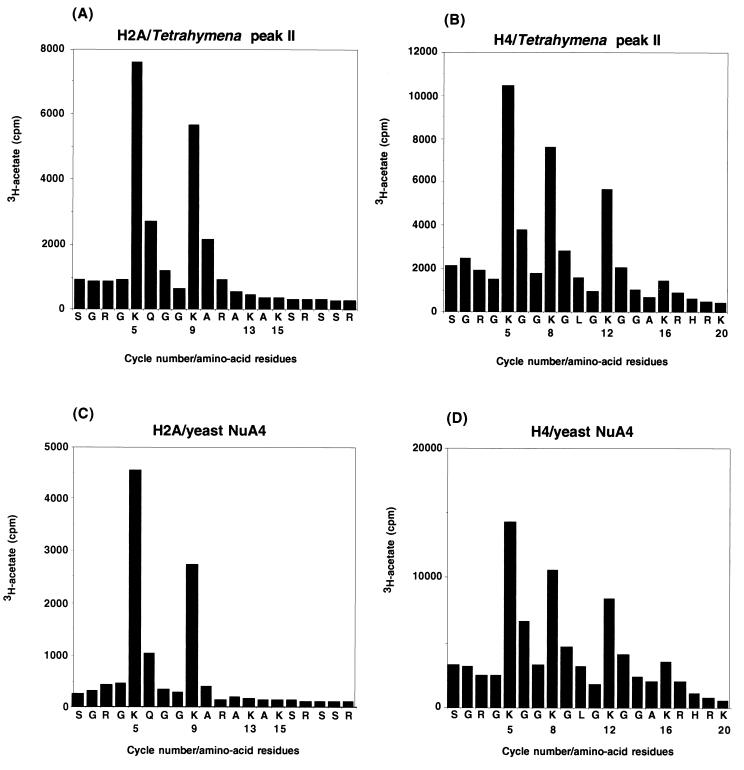
Given the strong similarities in histone preference between the Tetrahymena and yeast H2A/H4 nucleosomal HAT activities, we sought to identify and compare the acetylation site profiles of both activities. To that end, we recovered H2A and H4 following in vitro nucleosome acetylation reactions with these activities and subjected each histone to microsequence analyses with direct determination of [3H]acetate incorporation at each position by liquid scintillation counting. The results show clearly that the acetylation patterns were essentially identical when either the Tetrahymena peak II or yeast NuA4 activities were used to acetylate chicken mononucleosomes: lysines 5 and 9 in H2A (compare Fig. 4A and C) and, considering repetitive yield, lysines 5, 8, 12, and 16 in H4 (compare Fig. 4B and D) were strongly preferred acetylation sites for both of these activities.
FIG. 4.
Identical acetylation site usage by the Tetrahymena and yeast H2A/H4 nucleosomal HAT activities. Chicken nucleosomes were acetylated by partially purified yeast NuA4 or Tetrahymena peak II. RP-HPLC-purified acetylated H2A and H4 reaction products were then deblocked and analyzed by microsequencing. The graphs represent [3H]acetate incorporation determined by liquid scintillation counting plotted as a function of the residue identified in each sequencing cycle. Data for Tetrahymena peak II of H2A (A), Tetrahymena peak II of H4 (B), yeast NuA4 of H2A (C), and yeast NuA4 of H4 (D) are shown.
It is noteworthy that the above pattern of H4 and H2A acetylation usage is distinct from that reported for yeast Gcn5p (23) or for human PCAF and p300 (37), and potentially, other GCN5 family members. The site usage profile shown with H2A and H4 in Fig. 4 is similar to what has recently been reported for a novel H4/H2A/H3-specific HAT, Esa1p (for essential Sas2-related acetyltransferase), in yeast (38). Recent studies taking advantage of a ts mutant of ESA1 are consistent with the possibility that Esa1p is the catalytic subunit responsible for HAT activity in the NuA4 complex (8). Based upon the acetylation site usage profiles, it seems likely that the H2A/H4 nucleosomal activity observed in peaks II and II* from Tetrahymena macronuclear extracts is related to the yeast NuA4 HAT complex. While the predicted molecular mass of Esa1p (∼53 kDa) is reasonably close to the size of the Tetrahymena activity under investigation here, recombinant Esa1p does not acetylate nucleosomal substrates (38). The exact relationship between yEsa1p and the catalytically active protein in Tetrahymena remains unclear (see Discussion).
The H2A/H4 nucleosome acetylation activity in Tetrahymena stimulates transcription from a chromatin template.
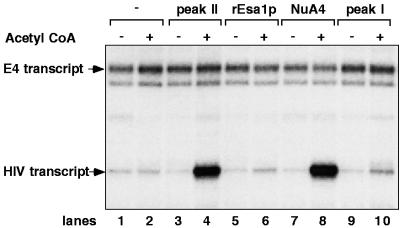
Recently, purified yeast HAT complexes were shown to facilitate transcription in vitro from nucleosomal DNA templates, but not naked DNA templates, containing either the HIV-1 enhancer/promoter region (42) or a minimal adenovirus E4 promoter placed downstream of five GAL4-binding sites (45). In each case, the HAT complexes stimulated transcription only in the presence of acetyl-CoA, suggesting a critical role for acetylation in the effect. Given these observations, we investigated whether the Tetrahymena H2A/H4 nucleosomal activity could also enhance transcription from nucleosomal DNA. Therefore, the HIV type 1 (HIV-1) chromatin template discussed above was incubated with the 80S peak II activity in the absence and presence of acetyl-CoA, and HIV-1 transcription levels were determined by primer extension analysis (Fig. 5). The Tetrahymena peak II fraction stimulated HIV-1 transcription 13-fold in the presence of acetyl-CoA (Fig. 5, compare lanes 2 and 4) but exhibited no ability to induce transcription in the absence of acetyl-CoA (lane 3). Yeast NuA4, which was included in the experiment as a control, increased HIV-1 transcription 18-fold (Fig. 5, compare lanes 2 and 8). Thus, although the subunit composition of the Tetrahymena peak II activity appears to be less complex than that of yeast NuA4, the levels of transcriptional enhancement mediated by the two complexes were quite similar under these experimental conditions. As expected from their inability to catalyze nucleosomal acetylation, recombinant yeast Esa1p (lane 6) and Tetrahymena peak I (lane 10) did not significantly increase transcription in this assay.
FIG. 5.
Tetrahymena H2A/H4 nucleosomal HAT activity facilitates HIV-1 transcription from a chromatin template. An HIV dinucleosome-5S array fragment (10 ng), assembled into an array of 12 nucleosomes, was incubated with acetyl-CoA (1 mM) and peak II, yEsa1p, NuA4, and peak I HAT activities as indicated, and the reaction mixtures were assayed for transcription. GAL4-AH stimulated transcription of a nonnucleosomal G5-E4 DNA was included to serve as a control for RNA recovery. The positions in the gel of the HIV-1 and E4 transcripts are indicated.
DISCUSSION
A novel H2A/H4 nucleosomal HAT in Tetrahymena is active as a ∼80-kDa activity.
We report the existence of a novel nucleosomal H2A/H4-specific HAT from Tetrahymena macronuclei that is clearly distinct from p55, the founding GCN5 family member (6). The estimated size of this novel activity is ∼80 kDa. Although estimation of size by sedimentation analyses is approximate, this result, combined with the fact that this activity resediments at 80 kDa following relatively harsh dissociating treatments (RP-HPLC), indicates that the ciliate activity does not depend upon the assembly of a large multisubunit complex like the activities of the yeast SAGA, NuA4, and ADA HAT complexes do (12, 13, 33). Our data are most consistent with a single protein or small subcomplex possessing the nucleosomal HAT activity detected in our experiments.
By using yeast whole-cell extracts as starting material, NuA4 was reported to acetylate H2A/H4 with nucleosomes as a substrate (12, 45). The substrate specificity, acetylation site profile, and functional data of yeast NuA4 are remarkably similar to those of the Tetrahymena nucleosomal HAT activity. However, the estimated size of yeast NuA4 (∼1.3 MDa [8, 12]) is much larger than the 80 kDa estimated here for the Tetrahymena activity. It seems likely that the yeast activity, as isolated from whole-cell extracts, is present as a multisubunit complex while the ciliate activity described here represents a subset of these polypeptides or even a single catalytically active protein. Whether these apparent size differences reflect fundamental differences between the Tetrahymena and yeast H2A/H4 enzyme and its subunit composition or reflect trivial differences in preparation and extraction, etc., remains to be determined. Despite this uncertainty, our data, together with previous findings (8), underscore the importance of determining the appropriate form of histone substrate (free versus nucleosomal) to use when exploring and characterizing HATs in vitro.
The H2A/H4 nucleosomal HAT activity is distinct from p55/Gcn5p.
With both the yeast and Tetrahymena H2A/H4-specific nucleosomal activities, several lines of evidence suggest that p55/Gcn5p is not the responsible catalytic subunit. Immunoblotting analyses indicate that yeast NuA4 activity does not contain Gcn5p, Ada, Spt, or Swi/Snf-related proteins, and studies with gcn5 disruption strains demonstrate that Gcn5p is not required for NuA4 HAT activity (12). Similarly, in Tetrahymena, what appears to be a comparable activity does not overlap to any large extent with p55, Ada1p, or Ada5p/Spt20p, the only polypeptides to which we have cross-reacting antisera (Fig. 1 and 2).
Substrate specificities are also considerably different between p55/Gcn5p and the H2A/H4 activities reported here. Like Gcn5p, p55 displays a strong preference for H3 with free-histone substrates (23). In agreement, the vast majority of p55 detected in peaks I and I* acetylates H3 and is unable to acetylate nucleosomal substrates. Crude DNase I extracts of Tetrahymena macronuclei also contain H3/H2B-specific nucleosomal HAT activity (Fig. 1C). These data suggest that there may be a SAGA-like HAT complex in Tetrahymena which acetylates nucleosome H3/H2B and whose catalytic enzyme is p55. During the preparation of our extract and/or sucrose gradient sedimentation, this SAGA-like complex may dissociate into several subcomplexes, one of which may correspond to peak I. Taken together, these data suggest that both a SAGA-like activity and a NuA4-like activity exist in macronuclear extracts following extensive DNase I digestion. Our data strongly suggest that the catalytic subunit of the NuA4-like activity is different from p55.
Conservation between yeast and ciliate NuA4-like activities.
The substrate and site specificities of yeast NuA4 are remarkably similar to that described here for the Tetrahymena nucleosome acetylation activity. Both are H2A/H4 specific with essentially identical site specificity. The finding that NuA4 HAT activity is severely reduced in yeast strains bearing ts alleles of ESA1 suggests that Esa1p is the catalytic component in the yeast NuA4 complex (8) and that a similar protein may be responsible for the activity of Tetrahymena peak II. As shown in Fig. 4, lysines 5, 8, 12, and 16 in H4 and lysines 5 and 9 in H2A are strongly preferred sites of nucleosomal acetylation for both the ciliate and yeast activities. Recombinant yESA1p displays a remarkably similar site preference profile with free-histone substrates, acetylating lysines 5, 8, 12, and 16 in H4 (with some differences in preference among H4 sites) and lysines 5, 9, 13, and 15 in H2A (38). The inability of recombinant yESA1p to acetylate nucleosomal substrates is likely to stem from the absence of other components of the NuA4 complex (8, 38). Even though the similarities in acetylation site profiles suggest that an ESA1-like protein is responsible for the Tetrahymena peak II activity, the markedly smaller size of the ciliate activity (80 kDa) compared to that of NuA4 (1.3 MDa) suggests that the ciliate catalytic protein, in contrast to yESA1p, requires few, if any, interacting subunits for nucleosomal acetylation and chromatin transcription activation activity. This notion is supported by the difference in our ability to reconstitute the activities of Tetrahymena peak II and NuA4 following denaturation. Although we do not have any data addressing the minimal composition of the NuA4 complex required for activity with chromatin substrates, it appears that the ciliate catalytic polypeptide may differ from ESA1p in that it possesses features attributable to other proteins that endow the NuA4 complex with nucleosomal acetylation and chromatin transcription activation activity. Similar differences in the activity of related HAT catalytic proteins with free-histone and nucleosomal substrates has been described previously for hPCAF in comparison to yGcn5p and a short form of hGcn5p (12, 50).
The predicted sizes of the ciliate activity and recombinant yEsa1p were ∼80 and ∼53 kDa, respectively. Given the uncertainties in the accuracy of these molecular sizes, potential differences arising through evolution, and the striking similarities in substrate and site specificities and functional properties between these two activities, it seems most likely that an Esa1p-like protein is also the catalytic subunit of the ciliate enzyme. However, when peptide antibodies directed against the N terminus of yeast Esa1p or an internal fragment conserved in MYST family members were used, all attempts to detect a cross-reacting polypeptide in appropriate gradient fractions from Tetrahymena were unsuccessful. Recent studies dramatically underscore a high degree of conservation between yeast and human complexes containing histone-like TAFs, Sptps, Adaps, and p55/Gcn5p family members (13, 30). As suggested by Struhl and Moqtaderi (44), primitive eukaryotes, like ciliates, may have evolved a basal machinery containing TATA binding protein, histones with amino-terminal tails, and relatively simple chromatin-modifying activities that facilitate transcription from chromatin templates. Our data demonstrate that yeast and ciliates have evolved a robust nucleosomal H2A/H4 HAT activity whose essential nature in yeast (i.e., yEsa1p) suggests a conserved and important function. Whether the size and functional differences we observed between the ciliate activity and yeast NuA4 result from distinct activities or represent differences in subunit composition, etc., is not clear. Despite these uncertainties, our finding that what is presumably a single polypeptide or small complex of simple composition is sufficient for this H2A/H4 nucleosomal HAT activity suggests that ciliates have evolved an activity that does not require an extensive collection of additional subunits to perform this function. As far as we are aware, the ciliate activity described here is the smallest nucleosomal HAT known. Understanding the differences between this activity in yeast and in Tetrahymena may yield insights into the minimal requirements for carrying out this acetylation reaction and also its regulation.
ACKNOWLEDGMENTS
We are grateful to J. Horiuchi and L. Guarente as well as S. Roberts and F. Winston for providing the Ada1p and Ada5p/Spt20p antisera used in this study, respectively, and to Tamara Ranalli for her help in characterizing these antibodies by using Tetrahymena samples. We are also grateful to Stéphane Allard for his help in purification of NuA4 samples.
This work was supported by grants from the NIH (GM53512 to C.D.A.), from HHMI (to J.L.W.), and from the Medical Research Council (MRC) of Canada (to J.C.) J.C. is a Canadian MRC Scholar.
REFERENCES
- 1.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 2.Belikoff E, Wong L J, Alberts B M. Extensive purification of histone acetylase A, the major histone N-acetyl transferase activity detected in mammalian cell nuclei. J Biol Chem. 1980;255:11448–11453. [PubMed] [Google Scholar]
- 3.Borrow J, Stanton V P, Jr, Andresen J M, Becher R, Behm F G, Chaganti R S, Civin C I, Disteche C, Dube I, Frischauf A M, Horsman D, Mitelman F, Volinia S, Watmore A E, Housman D E. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 4.Brownell J E, Allis C D. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brownell J E, Allis C D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 6.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 8.Côté, J., R. T. Utley, S. Allard, A. Clarke, P. Grant, J. Savard, L. Pillus, and J. L. Workman. Essential Esa1p functions as the catalytic subunit of NuA4, a multisubunit complex that acetylates nucleosomal histone H4 and enhances transcription. Genes Dev., in press.
- 9.Felsenfeld G. Chromatin unfolds. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 10.Gorovsky M A, Yao M C, Keevert J B, Pleger G L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9:311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- 11.Grant P A, Sterner D E, Duggan L J, Workman J L, Berger S L. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 1998;8:193–197. doi: 10.1016/s0962-8924(98)01263-x. [DOI] [PubMed] [Google Scholar]
- 12.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 13.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates J R, Workman J L. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 14.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 15.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 16.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi J C. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horiuchi J, Silverman N, Pina B, Marcus G A, Guarente L. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horiuchi K, Fujimoto D. Use of phosphocellulose paper disks for the assay of histone acetyltransferase. Anal Biochem. 1975;69:491–496. doi: 10.1016/0003-2697(75)90151-7. [DOI] [PubMed] [Google Scholar]
- 19.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 20.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 21.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 22.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 23.Kuo M H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 24.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcus G A, Horiuchi J, Silverman N, Guarente L. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol. 1996;16:3197–3205. doi: 10.1128/mcb.16.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizzen C A, Allis C D. Linking histone acetylation to transcriptional regulation. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizzen, C. A., J. E. Brownell, and C. D. Allis. Histone acetyltransferases: preparation of substrates and assay procedures. Methods Enzymol., in press. [DOI] [PubMed]
- 28.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 29.Neuwald A F, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 30.Ogryzko V V, Kotani T, Zhang X, Schlitz R L, Howard T, Yang X J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 31.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 32.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 33.Pollard K J, Peterson C L. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puri P L, Sartorelli V, Yang X-J, Hamamori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y J, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1998;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 34a.Ranalli, T., and C. D. Allis. Unpublished data.
- 35.Reifsnyder C, Lowell J, Clarke A, Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- 36.Roberts S M, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiltz R L, Mizzen C A, Vassilev A, Cook R G, Allis C D, Nakatani Y. Overlapping, but distinct, patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 38.Smith E R, Eisen A, Gu W, Sattah M, Pannuti A, Zhou J, Cook R G, Lucchesi J C, Allis C D. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc Natl Acad Sci USA. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobel R E, Cook R G, Allis C D. Non-random acetylation of histone H4 by a cytoplasmic histone acetyltransferase as determined by novel methodology. J Biol Chem. 1994;269:18576–18582. [PubMed] [Google Scholar]
- 40.Sobel R E, Cook R G, Perry C A, Annunziato A T, Allis C D. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 42.Steger D J, Eberharter A, John S, Grant P A, Workman J L. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc Natl Acad Sci USA. 1998;95:12924–12929. doi: 10.1073/pnas.95.22.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 44.Struhl K, Moqtaderi Z. The TAFs in the HAT. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- 45.Utley R T, Ikeda K, Grant P A, Côté J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 46.Wang L, Liu L, Berger S L. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 48.Wu C. Chromatin remodeling and the control of gene expression. J Biol Chem. 1997;272:28171–28174. doi: 10.1074/jbc.272.45.28171. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 50.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]