Abstract
The COVID-19 pandemic has generated an overuse of antimicrobials in critically ill patients. Acinetobacter baumannii frequently causes nosocomial infections, particularly in intensive care units (ICUs), where the incidence has increased over time. Since the WHO declared the COVID-19 pandemic on 12 March 2020, the disease has spread rapidly, and many of the patients infected with SARS-CoV-2 needed to be admitted to the ICU. Bacterial co-pathogens are commonly identified in viral respiratory infections and are important causes of morbidity and mortality. However, we cannot neglect the increased incidence of antimicrobial resistance, which may be attributed to the excess use of antimicrobial agents during the COVID-19 pandemic. Patients with COVID-19 could be vulnerable to other infections owing to multiple comorbidities with severe COVID-19, prolonged hospitalization, and SARS-CoV-2-associated immune dysfunction. These patients have acquired secondary bacterial infections or superinfections, mainly bacteremia and urinary tract infections. This review will summarize the prevalence of A. baumannii coinfection and secondary infection in patients with COVID-19.
Keywords: Acinetobacter baumannii, infections, antimicrobial resistance, COVID-19
1. Introduction
After its recognition in Wuhan, China, in December 2019, coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), quickly spread across the globe. It became a global health threat [1,2,3]. As of 2 July 2021, SARS-CoV-2 had infected exceeding 180 million people and caused almost 4 million deaths around the world, according to the COVID-19 Weekly Epidemiological Update for that date [4]. Commonly, the clinical manifestations of COVID-19 are less severe, such as a mild upper respiratory tract disease or even asymptomatic infection [5,6]. However, SARS-CoV-2 infection can lead to severe and critical respiratory failure requiring mechanical ventilation, septic shock, or other organ dysfunction or loss that requires intensive care treatment [7]. Moreover, evidence suggests that coinfections and secondary infections can play an essential role in higher mortality risk during COVID-19 among a significant number of hospitalized patients [3,8,9].
Previous experiences with other coronaviruses and other respiratory pathogens have indicated this possibility of coinfection and secondary infections, especially by bacteria [10,11,12,13,14]. For example, Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus are the most commonly reported bacteria associated with co/secondary influenza infections [15,16]. In COVID-19 cases, while the prevalence of coinfections varied, the prevalence of secondary infections could be as high as 50% among non-surviving patients [2].
Hospitalization of COVID-19 patients, especially in intensive care units (ICUs), predisposes them to severe consequences such as healthcare-associated infections (HAIs) and/or secondary infections [17]. For example, COVID-19 patients admitted to ICUs with severe pulmonary symptoms can require the use of mechanical ventilation as part of supportive care. This use of mechanical ventilation can lead to ventilator-associated pneumonia (VAP), especially with multidrug-resistant bacteria such as A. baumannii [18]. A. baumannii infection cases have been reported in COVID-19 patients [19].
A. baumannii is an opportunistic pathogen mainly associated with HAI. Opportunistic pathogens can also cause superinfections, especially in combination with viral respiratory tract infections in hospitalized patients [19]. Belonging to the ESKAPE group (which includes Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter spp.), A. baumannii stands out with its ability to effectively escape antibiotic treatments, affecting mainly immunocompromised and critically ill patients in ICUs [20]. Most commonly, A. baumannii infections manifest as VAP and bloodstream infections [21]. Given the clinical relevance and the antimicrobial resistance, the World Health Organization (WHO) appointed A. baumannii as a critical-priority pathogen that poses a significant threat to human health in 2017 [4]. Thus, we reviewed the medical literature to summarize and discuss the role played by A. baumannii coinfection and secondary infection in patients with COVID-19.
2. Acinetobacter baumannii Presentation
2.1. Characteristics of the Genus Acinetobacter
The genus Acinetobacter is a large and diverse group of biochemically, physiologically, and naturally multi-skilled bacteria. Acinetobacter spp., a ubiquitous coccobacillus genus, is characterized as glucose non-fermentative, non-motile, catalase-positive, oxidase-negative, and non-fastidious Gram-negative bacteria [22,23,24]. The taxonomy of this genus is complicated due to the numerous and closely related species, which are often impossible to distinguish from each other by phenotypic and chemotaxonomic methods [22,25]. Since the first description by Beijerinck in 1911 [25], this bacterial taxonomy has been reclassified under various names [21,26]. The Acinetobacter genus today contains 65 species with validly published names [27]. Currently, six species, namely, A. calcoaceticus, A. baumannii, A. pittii, A. nosocomialis, A. seifertii, and A. lactucae (a later heterotypic synonym of A. dijkshoorniae) [28,29] belonging to the Acb complex (Acinetobacter calcoaceticus–Acinetobacter baumannii complex) have been associated with human diseases [30]. Even though these species differ in their pathogenicity, antimicrobial resistance, and epidemiology [31], the Acb complex is physiologically and genetically highly related, making it difficult to distinguish them phenotypically with standard laboratory methods [32].
Highly found in the environment, bacteria of this genus can be recovered from different habitats, such as soil, surface water, foods, vegetables, and arthropods [33,34]. Acinetobacter can be retrieved as commensals of skin, wounds, and respiratory and gastrointestinal tracts [24,33,34]. In the hospital environment, Acinetobacter is also easily isolated, especially the A. baumannii species. When recovered from clinical samples, most species may have some significance as human pathogens [34].
Of all of the Acinetobacter species, A. baumannii is the most critical pathogenic member. A. baumannii has become a successful opportunistic pathogen and has emerged as a major cause of healthcare-associated infections (HAIs), mainly in critical and immunocompromised patients [23,35]. Furthermore, A. pittii, A. nosocomialis, A. seifertii, and A. lactucae are widespread and are occasionally associated with emerging important nosocomial pathogens involved in hospital-acquired infections [28,29,36].
2.2. Clinical Importance of A. baumannii
A. baumannii is mainly recognized as causing an extended range of HAIs, including pneumonia, bacteremia, urinary tract infections, wound infections, and meningitis [37]. A previous study estimated a global incidence of more than 1,000,000 cases of A. baumannii infections annually, of which 50% are carbapenem-resistant cases [38]. The National Healthcare Safety Network (NHSN) data for 2009–2010 indicate that A. baumannii was found in catheter-associated urinary tract infection, central line-associated bloodstream infection, surgical site infection, and ventilator-associated pneumonia [39]. These infections due to A baumannii may be associated with considerable mortality, varying from 8% to 35% [40]. Among these nosocomial infections, ventilator-associated pneumonia and bloodstream infections are the most important, with the highest mortality rates [41]. Risk factors associated with A. baumannii infections include prolonged hospitalization, intensive care unit admission, presence of devices or previous invasive procedures, prior use of antimicrobial agents, prior hospitalization, nursing home residence, older age, and prior colonization with A. baumannii [42,43,44,45,46,47].
According to previous studies, infected and colonized patients represent major reservoirs for the horizontal transmission and spread of A. baumannii in a hospital environment [48,49]. Furthermore, contaminated healthcare worker hands may also play a decisive role in this transmission. Thom et al. (2018) found that healthcare workers who provide care for patients known to be infected or colonized with A. baumannii exit the room with A. baumannii on their hands or gloves 30% of the time [50]. While infected and colonized patients are major reservoirs, A baumannii can be isolated from other hospital environmental sources. In the literature, different studies have been reported recovering A. baumannii isolates from various sources: sink and water taps [51], healthcare worker hand samples [52], hospital furniture, medical devices, and gloves [53].
According to previous studies, the ability of A. baumannii to persist and survive for long periods on surfaces and under dry conditions has made it a critical frequent cause of HAIs worldwide [54,55,56,57,58]. Moreover, outbreaks caused by A. baumannii isolates that displayed resistance to the vast majority of available antimicrobial agents have been increasingly reported during the last decades [26,59]. This ability to acquire antibiotic resistance determinants has propelled its clinical relevance [60].
While A. baumannii is an important nosocomial pathogen that can cause various diseases, community-acquired infections by this microorganism (including pneumonia and bacteremia) are less common, but it is associated with relatively high mortality [61]. The majority of community-acquired infection cases were reported in Australia and Asia: Australia [62,63,64], Hong Kong [65], India [66], Singapore [67], Korea [68], and Taiwan [69,70,71]. Risk factors for community-acquired infections caused by A. baumannii include chronic obstructive pulmonary disease, renal disease, diabetes mellitus, excessive alcohol consumption, and smoking [72,73]. Previous reports have also described the association of A. baumannii with infections consequent to war injuries and natural disaster victims [74,75,76].
2.3. Antimicrobial Resistance in A. baumannii
The drug-resistant A. baumannii and its susceptibility patterns have made empirical and therapeutic decisions even more complex [44]. High antimicrobial resistance rates are observed in Eastern and Southern Europe, Latin America, and many Asian countries [77]. This pathogen exhibits intrinsic resistance to different antibiotic classes, such as penicillins, macrolides, trimethoprim, and fosfomycin [78,79,80]. Moreover, A. baumannii displays a successful ability to acquire antimicrobial resistance [22]. Several studies have reported cephalosporin, carbapenem, aminoglycoside, and fluoroquinolone resistance in A. baumannii strains [78,81,82,83,84,85,86]. Mainly, carbapenem resistance in A. baumannii is an essential concern because this type of antimicrobial is the last line of defense used to treat infections caused by multidrug-resistant Gram-negative bacteria. Infections caused by carbapenem-resistant Acinetobacter baumannii (CRAb) cause more extended hospitalization, adverse outcomes, and increased costs than infections caused by carbapenem-susceptible strains [87,88,89].
Carbapenem resistance in Acinetobacter spp. is often associated with acquired carbapenemase production [81]. Among carbapenemases, Class D beta-lactamases—also called oxacillinases (OXAs)—are more frequent. Currently, the main groups of OXA-type carbapenemases identified in A. baumannii are OXA-23-like, OXA-24/40-like, OXA-58-like, OXA-143-like, and OXA-235-like groups, and the intrinsic chromosomal OXA-51-like group [90]. Acquired resistance to aminoglycosides (plasmid-borne aminoglycosides-modifying enzymes and 16S rRNA methylases) and fluoroquinolones (mutations in gyrA and/or parC) have been described in carbapenemase-producing A. baumannii strains [91]. Because of the increasing carbapenem resistance, second-line agents such as polymyxins and tigecycline have been considered for treating carbapenem-resistant A. baumannii infections [77]. However, resistance to polymyxins emerged and has been reported [92,93].
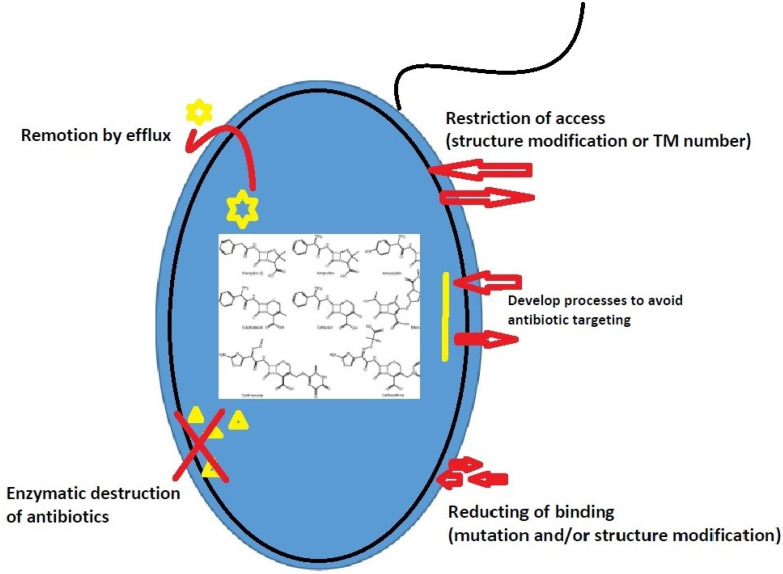
Antibiotic resistance mechanisms can be classified into three groups [94]: (1) Restriction or impediment of target access by reducing membrane permeability or increasing antibiotic efflux. (2) Direct antibiotic inactivation by a genetic mutation, post-translational modification, or enzymatic hydrolysis. (3) Access restriction through a structural change of membrane components or number of transmembrane segments.
The antibiotic resistance mechanisms of A. baumannii based on this classification are summarized in Figure 1.
Figure 1.
Schematic representation of the different mechanisms of resistance developed by A. baumannii. In the middle of the cell are the antibiotics.
3. SARS-CoV-2 and A. baumannii
3.1. Carbapenem-Resistant A. baumannii in Hospitals
Carbapenem-resistant Acinetobacter baumannii (CRAb), an opportunistic pathogen primarily associated with hospital-acquired infections, is an urgent public health threat [95]. CRAb readily contaminates the hospital environment and health care providers’ hands, can survive for prolonged periods on dry surfaces, and can be spread by asymptomatic colonization; these factors make CRAb outbreaks in acute care hospitals challenging to control [34,96,97,98]. It is resistant to common disinfectants, leading to outbreaks that are hard to contain and affect the most vulnerable and critically ill patients [34].
CRAb is the leading cause of morbi-mortality from infection in several European countries, with Italy being one of the top affected countries [99]. Compared to the pre-COVID-19 period, some authors observed a decreased antibiotic susceptibility in local pathogens during the first wave [100]. Others reported an increased risk of carbapenem-resistant infections in patients hospitalized with COVID-19 [18,99]. In both ICU and non-ICU settings, rigorous adherence to infection control practices is vital to discontinue the transmission of CRAb in hospitals [99,101,102,103]. Zhang et al. found that 55.6% of COVID-19 patients were coinfected with carbapenem-resistant A. baumannii (CRAb) in the ICU [104].
A recent study from a Mexican ICU with COVID-19 patients identified and characterized ESKAPE bacterias, detecting their possible clonal spread on medical devices, inert surfaces, medical personnel, and patients. A. baumannii was the most predominant ESKAPE member (52%) with all strains showing multidrug resistance (MDR). Moreover, the analysis of intergenic regions revealed a critical clonal distribution of A. baumannii (AdeABCRS+) in the ICU [105].
The outbreak of A. baumannii was described as one of the main determinants of severity and mortality of ICU patients in [106]. A New Jersey hospital experienced a large multidrug-resistant OXA-23 CRAb outbreak, primarily involving ICU patients, which extended across multiple units during a surge in COVID-19 cases [98]. A Japanese tertiary hospital reported an OXA-23-producing A. baumannii outbreak in 5 of 10 ICU beds, and all isolates had similar antibiotic susceptibility patterns with resistance to all β-lactams, including imipenem [107]. Of interest, an outbreak of A. baumannii was a determining factor in the increases of the incidence of infection and the morbi-mortality of ICU patients, with all strains multidrug resistant and only sensitive to colistin [108]. Gottesman et al. described a monoclonal outbreak of CRAb in two wards of a dedicated hospital to treat COVID-19 patients. All clinical (five cases) and environmental specimens (n = 24) belonged to international clonal lineage 2 (blaOXA-66 allele) and harbored the blaOXA-24-like carbapenemase [109]. Shinohara et al. also report a monoclonal outbreak of an endemic strain of CRAb (14 cases) in a new COVID-19 ICU of a tertiary teaching hospital in southern Brazil [18]. Interestingly, CRAb outbreaks in COVID-19 patients seem to be effectively treated with a pre-optimized two-phage cocktail [110].
3.2. COVID-19 and A. baumannii Coinfections
The COVID-19 pandemic caused many immunocompromised individuals to be hospitalized, and some reports indicated that some COVID-19 patients were diagnosed with coinfections and secondary infections [93,94,111,112]. The incidence, prevalence, and characteristics of bacterial infection in these patients were not well understood and were raised as an important knowledge gap [113,114]. The specific nature and source of these infections have not yet been fully investigated; however, evidence indicates that multidrug-resistant bacteria are among the microbes responsible for developing these infections. The prevalence of coinfection was variable among COVID-19 patients in different studies. However, it could be up to 50% among non-survivors [2]. A systematic review and meta-analysis of bacterial coinfection and secondary infection in patients with COVID-19 reported 3.5% and 14.3% for coinfection and secondary infection, respectively. However, in general, bacterial infection was 6.9%, varying slightly in the patient population, ranging from 5.9% in hospitalized patients to 8.1% in critically ill patients [115]. Similarly, another study concluded that only 7% of hospitalized patients presented a bacterial coinfection with a high degree of heterogeneity, although this increased to 14% in studies that only included ICU patients [116,117].
Coinfection with A. baumannii secondary to SARS-CoV-2 infections has been reported multiple times in literature during the COVID-19 pandemic including Wuhan (China), France, Spain, Iran, Egypt, New York (USA), Italy, and Brazil (Table 1). The incidence of secondary infections (mostly lower respiratory tract infections) due to A. baumannii was said to be as high as 1% of hospitalized COVID-19 patients in an Italian hospital [118]. A descriptive study reported the exact incidence (1%) among hospitalized patients from Wuhan, China [119]. A simultaneous survey from Wuhan reported coinfection with A. baumannii in 1 out of 69 hospitalized patients (1.4%) with COVID-19 [120]. In addition, a recent study from a French ICU reported a 28% rate of bacterial coinfection with A. baumannii at 1 out of 92 (1.1%) in severe SARS-CoV-2 pneumonia patients [121]. A study conducted by Siyuan et al. (2021) investigated the frequency and characteristics of respiratory coinfections in COVID-19 patients in the ICU; they detected that A. baumannii and S. aureus were more frequently identified during late ICU admission [122].
Table 1.
Incidence of co- and secondary infection of A. baumannii during COVID-19 pandemic reported in various countries.
| Country/City | COVID-19 Patients |
A. baumannii Coinfection n (%) |
Other Pathogenic Organisms Found | Reference |
|---|---|---|---|---|
| China/Wuhan | 102 | 57 (35.8%) | K. pneumoniae (30.8%), Stenotrophomonas maltophilia (6.3%) and others | [8] |
| Iran/Qom | 90 | 17 (90%) | S. aureus (10%) | [19] |
| China/Wuhan | 221 | 5 (55.6%) | Escherichia coli, P. aeruginosa, and Enterococcus (data not shown) | [103] |
| Spain/Valladolid | 712 | 25 (18.7%) | E. faecium (17.2%) and others | [106] |
| Brazil/Minas Gerais | 212 | 21 (32.8%) | Staphylococcus spp. (45.3%), Pseudomonas spp. (32.8%), Stenotrophomonas spp. (14.06%), Klebsiella spp. (12.5%), Enterobacter spp. (9.4%), Enterococcus spp. (9.4%), E. coli (6%). | [108] |
| France/Argenteuil | 92 | 1 (3%) | S. aureus (31%), Haemophilus influenzae (22%), Streptococcus pneumoniae (19%), Enterobacteriaceae (16%), P. aeruginosa (6%), Moraxella catarrhalis (3%) | [121] |
| Egypt/Alrajhrt | 260 | 28 (16.6%) | S. aureus (11.9%), S. pneumoniae (4.7%), E. faecalis (2.3%), K. pneumoniae (28.5%), E. coli (9.5%), P. aeruginosa (9.5%), Enterobacter cloacae (4.7%) | [129] |
| Italy/Milan | 731 | 7 (30.4%) | S. aureus (69.7%), E. coli (21.7%) | [118] |
| China/Wuhan | 99 | 1 (1%) | K. pneumoniae (1%), Aspergillus flavus (1%) | [119] |
| China/Wuhan | 69 | 1 (1.4%) | Candida albicans (2.8%), E. cloacae (2.8%) | [120] |
| China/Beijing | 20 | 10 (20%) | Stenotrophomonas maltophilia (28%), P. aeruginosa (28%) | [122] |
| France/Paris | 5 | 1 (20%) | A. flavus (20%) | [127] |
| Italy/Naples | 32 | 4 (19%) | K. pneumoniae (32%), P. aeruginosa (14%), E. cloacae (9%), S. aureus (4%), E. faecium (9%), S. maltophilia (9%), E. faecalis (4%) | [128] |
| Italy/Ferrara | 28 | 17 (13.6%) | E. faecalis (14.2%), E. faecium (8%), S. epidermidis (33.6%), S. maltophilia (10.4%), C. albicans (23.2%) | [130] |
| Taiwan/Tainan | 18 | 2 (11.1%) | Streptococcus dysgalactiae (11.1%), Influenza virus B (5.55%) | [131] |
Within the first few days of SARS-CoV-2 infection, critically ill patients with COVID-19 often develop pulmonary dysbiosis or respiratory tract distortion, which can progress further to a secondary bacterial or fungal infection a few weeks later [123,124,125]. A retrospective cohort study in a secondary care setting in Cambridge, UK, reported that a high percentage of patients with COVID-19 (9 out of 14) in the ICUs had confirmed secondary ventilator-associated pneumonia (VAP) [126]. Lescure et al. (2020) identified A. baumannii as the responsible agent in a COVID-19 patient of VAP [127]. A retrospective observational study of all patients admitted for COVID-19 in a University Hospital in Spain showed that 16% presented with fungal or bacterial coinfection/superinfection, and multidrug-resistant A. baumannii was the leading agent in respiratory infections and bacteremia with an outbreak contributing to this result [106]. Chen et al. (2020) also reported that bacterial and fungal coinfections in COVID-19 patients, including one patient that presented with an A. baumannii infection that was highly resistant to antibiotics, cause difficulties with anti-infective treatment, leading to the higher possibility of developing septic shock [119]. In a cohort study, the authors evaluated data from 212 severely ill COVID-19 patients admitted in a public tertiary hospital exclusively dedicated to attending COVID-19 patients during the pandemic and analyzed the association between fungal/bacterial coinfections and the mortality of these patients. Acinetobacter spp. was the second most isolated of the patients with positive bacterial cultures and was responsible for the third-highest mortality rate of COVID-19 patients suffering from these coinfections [108].
A study in an Iranian ICU reported coinfection with MDR A. baumannii in 17 out of 19 COVID-19 patients with high-level resistance to all antimicrobials tested, except colistin, which showed a resistance rate of 52%, although none of the patients survived [19]. Among 1495 COVID-19 patients hospitalized in Wuhan, 102 (6.8%) patients had acquired secondary bacterial infections, mainly due to A. baumannii (35.8%) with high resistance rates (91.2%), and almost half of them (49.0%, 50/102) died during hospitalization [8]. An Italian retrospective analysis of 32 COVID-19 ICU patients showed that 50% of patients developed an MDR infection during ICU stay. Overall, >80% of MDR bacteria isolated were Gram-negative bacilli, and the second most common pathogen isolated was carbapenem-resistant A. baumannii (CRAb). Four patients were coinfected with an extensively drug-resistant A. baumannii phenotype (12.5%) [128]. A. baumannii was detected in 20% of samples acquired from COVID-19 patients in an ICU in Beijing, China, during late ICU admission [122]. The highest incidence of MDR A. baumannii coinfection was documented in an Egyptian study (2.7%; 7 out of 260 patients hospitalized with COVID-19, susceptible only to tigecycline and fluoroquinolones, having resistance genes NDM-1, TEM, and CTX-M) [129]. Another study in hospitalized COVID-19 patients from Spain showed that coinfection with A. baumannii was apparent in 2.4% of hospitalized patients (17 out of 712; 16 of these 17 patients in ICU); it was the strongest determinant of mortality [106]. Cultrera et al. (2021) compared coinfections in critically ill patients with or without COVID-19, and A. baumannii was the most frequently isolated bacterium found in the three ICUs. In addition, CRAb infections in COVID-19-positive patients admitted to the ICU were more frequent compared to those that were COVID-19 negative [130]. The rapid expansion of the ICU to manage SARS-CoV-2 can potentially increase nosocomial infection rates in the hospital environment. As a result, there is great concern with bacterial coinfections in patients with COVID-19 as they significantly increase the morbidity and mortality of these patients. Thus, the early identification of bacterial infections will help to identify high-risk patients and determine the correct interventions to reduce mortality.
4. Conclusions
The worldwide outbreak of COVID-19 emerged in late 2019, and caused severe public health crises with considerably more significant morbidities and mortality rates, particularly in individuals with severe medical comorbidities and elderly populations. COVID-19 disease symptoms are diverse and may have varied manifestations among patients. In extreme cases, patients may develop pneumonia, acute respiratory distress syndrome, and multi-organ dysfunction. The screening and understanding of the proportion of COVID-19 patients with acute respiratory bacterial coinfection, especially VAP, is crucial for treating patients with COVID-19 and assisting in the responsible use of antibiotics, minimizing damaging consequences of overuse, and ensuring a better clinical outcome. In addition, this knowledge could have a significant impact on refining empirical antibiotic management guidelines for patients with COVID-19. The coinfection of SARS-CoV-2 and secondary infections with other microorganisms, such as A. baumannii, is an essential factor in COVID-19, and it can aggravate the progression and prognosis, especially in hospitalized patients. However, there are few reports about secondary infections and SARS-CoV-2 coinfections with bacteria; it is necessary to understand and strengthen the investigation of them. While there are many guidelines for diagnosing COVID-19 patients, bacterial coinfections and secondary infections have gaps and require further study.
This summarized review was designed to offer insight into A. baumannii coinfections and secondary infection in patients with COVID-19 using the most recent information available. In hospital settings, COVID-19 patients may face another health threat: antimicrobial-resistant bacteria such as A. baumannii and its carbapenem resistance pattern, which poses a considerable challenge to the health system. The isolation and the recognition of A. baumannii from COVID-19 patients highlights the importance of appropriate prevention and control practices. This microorganism potentially has important and severe implications for the clinical outcomes of these COVID-19 patients. Further studies are necessary to investigate the incidence, prevalence, and characteristics of COVID-19 coinfection and microbiological (especially A. baumannii) distribution.
Acknowledgments
K.R. was a postdoctoral fellow from FIOCRUZ/CDTS-CAPES.
Author Contributions
Conceived and designed the experiments: K.R., T.P.G.C.; writing—original draft: K.R.; review and editing: K.R., S.G.D.-S.; funding: S.G.D.-S. All authors have read and agreed to the published version of the manuscript.
Funding
S.G.S. receives support from the Brazilian Council for Scientific Research (CNPq # 467.488.2014-2; #3075732011; #3013322015-0), Carlos Chagas Filho Foundation for Research Support of the State of Rio de Janeiro (FAPERJ # 110.198-13; # 202.841-2018).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chung J.Y., Thone M.N., Kwon Y.J. COVID-19 vaccines: The status and perspectives in delivery points of view. Adv. Drug Deliv. Rev. 2021;170:1–25. doi: 10.1016/j.addr.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai C.C., Wang C.Y., Hsueh P.R. Co-infections among patients with COVID-19: The need for combination therapy with non-anti-SARS-Cov-2 agents? J. Microbiol. Immunol. Infect. 2020;53:505–512. doi: 10.1016/j.jmii.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirzaei R., Goodarzi P., Asadi M., Soltani A., Aljanabi H.A.A., Jeda A.S., Dashtbin S., Jalalifar S., Mohammadzadeh R., Teimoori A., et al. Bacterial co-infections with SARS-CoV-2. UBMB Life. 2020;72:2097–2111. doi: 10.1002/iub.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO 2021 Word and Health Organization. [(accessed on 2 July 2021)]; Available online: https://www.Who.Int/emergencies/diseases/novel-coronavirus-2019/situation-reports.
- 5.Bengoechea J.A., Bamford C.G.G. SARS-CoV-2, bacterial coinfections, and AMR: The deadly trio in COVID-19? EMBO Mol. Med. 2020;12:e12560. doi: 10.15252/emmm.202012560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng M.P., Papenburg J., Desjardins M., Kanjilal S., Quach C., Libman M., Dittrich S., Yansouni C.P. Diagnostic test for Severe Acute Respiratory Syndrome-Related Coronavirus 2: A narrative review. Ann. Intern. Med. 2020;172:726–734. doi: 10.7326/M20-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., Cuomo-Dannenburg G., Thompson H., Walker P.G., Fu H., et al. Estimates of the severity of coronavirus disease 2019: A model-based analysis. Lancet Infect. Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J., Wang J., Yang Y., Cai P., Cao J., Cai X., Zhang Y. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: A retrospective analysis. Antimicrob. Resist. Infect. Control. 2020;9:153. doi: 10.1186/s13756-020-00819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zahariadis G., Gooley T.A., Ryall P., Hutchinson C., Latchford M.I., Fearon M.A. Risk of ruling out severe acute respiratory syndrome by ruling in another diagnosis: Variable incidence of atypical bacteria coinfection based on diagnostic assays. Cancer Res. J. 2006;13:17–22. doi: 10.1155/2006/862797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee N., Chan P.K., Yu I.T., Tsoi K.K., Lui G., Sung J.J. Co-circulation of human metapneumovirus and SARS-associated coronavirus during a major nosocomial SARS outbreak in Hong Kong. J. Clin. Virol. 2007;40:333–337. doi: 10.1016/j.jcv.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alfaraj S.H., Al-Tawfiq J.A., Altuwaijri T.A., Memish Z.A. Middle East respiratory syndrome coronavirus and pulmonary tuberculosis coinfection: Implications for infection control. Intervirology. 2017;60:53–55. doi: 10.1159/000477908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alfaraj S.H., Al-Tawfiq J.A., Altuwaijri T.A., Memish Z.A. The impact of coinfection of influenza A virus on the severity of middle east respiratory syndrome coronavirus. J. Infect. 2017;74:521–523. doi: 10.1016/j.jinf.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arabi Y.M., Al-Omari A., Mandourah Y., Al-Hameed F., Sindi A.A., Alraddadi B. Critically ill patients with the middle east respiratory syndrome: A multicenter retrospective cohort study. Crit. Care Med. 2017;45:1683–1695. doi: 10.1097/CCM.0000000000002621. [DOI] [PubMed] [Google Scholar]
- 15.Morris D.E., Cleary D.W., Clarke S.C. Secondary bacterial infections associated with influenza pandemics. Front. Microbiol. 2017;8:1041. doi: 10.3389/fmicb.2017.01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta R.K., George R., Nguyen-Van-Tam J.S. Bacterial pneumonia and pandemic influenza planning. Emerg. Infect. Dis. 2008;14:1187–1192. doi: 10.3201/eid1408.070751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khurana S., Singh P., Sharad N., Kiro V.V., Rastogi N., Lathwal A., Malhotra R., Trikha A., Mathur P. Profile of coinfections & secondary infections in COVID-19 patients at a dedicated COVID-19 facility of a tertiary care Indian hospital: Implication on antimicrobial resistance. Ind. J. Med. Microbiol. 2021;39:147–153. doi: 10.1016/j.ijmmb.2020.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinohara D.R., Saalfeld S.M.S., Martinez H.V., Altafini D.D., Costa B.B., Fedrigo N.H., Tognim M.C.B. Outbreak of endemic carbapenem-resistant Acinetobacter baumannii in a coronavirus disease 2019 (|COVID-19)-specific intensive care unit. Infect. Cont. Hosp. Epidemiol. 2021;9:1–3. doi: 10.1017/ice.2021.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharifipour E., Shams S., Esmkhani M., Khodadadi J., Fotouhi-Ardakani R., Koohpaei A., Doosti Z., Golzari S.E. Evaluation of bacterial coinfections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect. Dis. 2020;20:646. doi: 10.1186/s12879-020-05374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monem S., Furmanek-Blaszk B., Łupkowska A., Kuczyńska-Wiśnik D., Stojowska-Swędrzyńska K., Laskowska E. Mechanisms protecting Acinetobacter baumannii against multiple stress triggered by the host immune response, antibiotics and outside-host environment. Int. J. Mol. Sci. 2020;21:5498. doi: 10.3390/ijms21155498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harding C.M., Hennon S.W., Feldman M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018;16:91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C.R., Lee J.H., Park M., Park K.S., Bae I.K., Kim Y.B., Cha C.J., Jeong B.C., Lee S.H. Biology of Acinetobacter baumannii: Pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 2017;7:55. doi: 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin M.F., Lan C.Y. Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside. World J. Clin. Cases. 2014;2:787–814. doi: 10.12998/wjcc.v2.i12.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Looveren M., Goossens H. Antimicrobial resistance of Acinetobacter spp. in Europe. Clin. Microbiol. Infect. 2004;10:684–704. doi: 10.1111/j.1469-0691.2004.00942.x. [DOI] [PubMed] [Google Scholar]
- 25.Dijkshoorn L., Nemec A. The diversity of the genus Acinetobacter. In: Gerischer U., editor. Acinetobacter Molecular Biology. 1st ed. Caister Academic Press; Norwich, UK: 2008. pp. 1–34. [Google Scholar]
- 26.Towner K.J. Acinetobacter: An old friend, but a new enemy. J. Hosp. Infect. 2009;73:355e63. doi: 10.1016/j.jhin.2009.03.032. [DOI] [PubMed] [Google Scholar]
- 27.Alexandr Nemec. [(accessed on 22 May 2021)];2021 Available online: https://apps.szu.cz/anemec/anemec.htm.
- 28.Cosgaya C., Mari-Almirall M., van Assche A., Fernandez-Orth D., Mosqueda N., Telli M., Huys G., Higgins P.G., Seifert H., Lievens B., et al. Acinetobacter dijkshoorniae sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex mainly recovered from clinical samples in different countries. Int. J. Syst. Evol. Microbiol. 2016;66:4105–4111. doi: 10.1099/ijsem.0.001318. [DOI] [PubMed] [Google Scholar]
- 29.Nemec A., Krizova L., Maixnerova M., Sedo O., Brisse S., Higgins P.G. Acinetobacter seifertii sp. nov., a member of the Acinetobacter calcoaceticus–Acinetobacter baumannii complex isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 2015;63:934–942. doi: 10.1099/ijs.0.000043. [DOI] [PubMed] [Google Scholar]
- 30.Vijayakumar S., Biswas I., Veeraraghavan B. Accurate identification of clinically important Acinetobacter spp.: An update. Future Sci. OA. 2019;5:FSO395. doi: 10.2144/fsoa-2018-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen T.L., Lee Y.T., Kuo S.C., Yang S.P., Fung C.P., Lee S.D. Rapid identification of Acinetobacter baumannii, Acinetobacter nosocomialis, and Acinetobacter pittii with a multiplex PCR assay. J. Med. Microbiol. 2014;63:1154–1159. doi: 10.1099/jmm.0.071712-0. [DOI] [PubMed] [Google Scholar]
- 32.Marí-Almirall M., Cosgaya C., Higgins P.G., Van Assche A., Telli M., Huys G., Lievens B., Seifert H., Dijkshoorn L., Roca I., et al. MALDI-TOF/MS identification of species from the Acinetobacter baumannii (Ab) group revisited: Inclusion of the novel A. seifertii and A. dijkshoorniae species. Clin. Microbiol. Infect. 2017;23:210.e1–210.e9. doi: 10.1016/j.cmi.2016.11.020. [DOI] [PubMed] [Google Scholar]
- 33.Munoz–Price L.S., Robert A., Weinstein M.D. Acinetobacter infection. N. Engl. J. Med. 2008;358:1271–1281. doi: 10.1056/NEJMra070741. [DOI] [PubMed] [Google Scholar]
- 34.Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergogne-Bérézin E., Towner K.J. Acinetobacter spp. as nosocomial pathogens: Microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 1996;9:148–165. doi: 10.1128/CMR.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X., Li J., Cao X., Wang W., Luo Y. Isolation, identification, and characterization of an emerging fish pathogen, Acinetobacter pittii, from diseased loach (Misgurnus anguillicaudatus) in China. Antonie Van Leeuwenhoek. 2019;113:21–32. doi: 10.1007/s10482-019-01312-5. [DOI] [PubMed] [Google Scholar]
- 37.Maragakis L.L., Perl T.M. Acinetobacter baumannii: Epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 2008;46:1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 38.Spellberg B., Rex J.H. The value of single-pathogen antibacterial agents. Nat. Rev. Drug Discov. 2013;12:963. doi: 10.1038/nrd3957-c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sievert D.M., Ricks P., Edwards J.R., Schneider M., Patel J., Srinivasan A., Kallen A., Limbago B., Fridkin S. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Cont. Hosp. Epidemiol. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 40.Falagas M.E., Rafailidis P.I. Attributable mortality of Acinetobacter baumannii: No longer a controversial issue. Crit. Care. 2007;11:134. doi: 10.1186/cc5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dijkshoorn L., Nemec A., Seifert H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007;5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 42.Zhou H., Yao Y., Zhu B., Ren D., Yang Q., Fu Y., Yu Y., Zhou J. Risk factors for acquisition and mortality of multidrug-resistant Acinetobacter baumannii bacteremia: A retrospective study from a Chinese hospital. Medicine. 2019;98:e14937. doi: 10.1097/MD.0000000000014937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaye K.S., Pogue J.M. Infections caused by resistant Gram-Negative bacteria: Epidemiology and management. Pharmacotherapy. 2015;5:949–962. doi: 10.1002/phar.1636. [DOI] [PubMed] [Google Scholar]
- 44.Fishbain J., Peleg A.Y. Treatment of Acinetobacter infections. Clin. Infect. Dis. 2010;51:79–84. doi: 10.1086/653120. [DOI] [PubMed] [Google Scholar]
- 45.Jang T., Lee S., Huang C., Lee C., Chen W. Risk factors and impact of nosocomial Acinetobacter baumannii bloodstream infections in the adult intensive care unit: A case-control study. J. Hosp. Infect. 2009;73:143–150. doi: 10.1016/j.jhin.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Mahgoub S., Ahmed J., Glatt A.E. Underlying characteristics of patients harboring highly resistant Acinetobacter baumannii. Am. J. Infect. Control. 2002;30:386–390. doi: 10.1067/mic.2002.122648. [DOI] [PubMed] [Google Scholar]
- 47.Wisplinghoff H., Perbix W., Seifert H. Risk factors for nosocomial bloodstream infections due to Acinetobacter baumannii: A case-control study of adult burn patients. Clin. Infect. Dis. 1999;28:59–66. doi: 10.1086/515067. [DOI] [PubMed] [Google Scholar]
- 48.Da Silva K.E., Maciel W.G., Croda J., Cayô R., Ramos A.C., de Sales R.O., Kurihara M.N.L., Vasconcelos N.G., Gales A.C., Simionatto S. A high mortality rate associated with multidrug-resistant Acinetobacter baumannii ST79 and ST25 carrying OXA-23 in a Brazilian intensive care unit. PLoS ONE. 2018;13:e0209367. doi: 10.1371/journal.pone.0209367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moghnieh R., Siblani L., Ghadban D., El Mchad H., Zeineddine R., Abdallah D., Ziade F., Sinno L., Kiwan O., Kerbaj F., et al. Extensively drug-resistant Acinetobacter baumannii in a Lebanese intensive care unit: Risk factors for acquisition and determination of a colonization score. J. Hosp. Infect. 2016;92:47–53. doi: 10.1016/j.jhin.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 50.Thom K.A., Rock C., Jackson S.S., Johnson J.K., Srinivasan A., Magder L.S., Roghmann M.C., Bonomo R.A., Harris A.D. Factors leading to transmission risk of Acinetobacter baumannii. Crit. Care Med. 2017;45:e633–e639. doi: 10.1097/CCM.0000000000002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong K.B., Oh H.S., Song J.S., Lim J.H., Kang D.K., Son I.S., Park J.D., Kim E.C., Lee H.J., Choi E.H. Investigation and control of an outbreak of imipenem-resistant Acinetobacter baumannii infection in a pediatric intensive care unit. Pediatr. Infect. Dis. J. 2012;31:685–690. doi: 10.1097/INF.0b013e318256f3e6. [DOI] [PubMed] [Google Scholar]
- 52.Enoch D.A., Summers C., Brown N.M., More L., Gillham M.I., Burnstein R.M., Thaxter R., Enoch L.M., Matta B., Sule O. Investigation and management of an outbreak of multidrug-carbapenem-resistant Acinetobacter baumannii in Cambridge, UK. J. Hosp. Infect. 2008;70:109–118. doi: 10.1016/j.jhin.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 53.Raro O.H.F., Gallo S.W., Ferreira C.A.S., Oliveira S.D. Carbapenem-resistant Acinetobacter baumannii contamination in an intensive care unit. Rev. Soc. Bras. Med. Trop. 2017;50:167–172. doi: 10.1590/0037-8682-0329-2016. [DOI] [PubMed] [Google Scholar]
- 54.Zeighami H., Valadkhani F., Shapouri R., Haghi F. Virulence characteristics of multidrug-resistant biofilm-forming Acinetobacter baumannii isolated from intensive care unit patients. BMC Infect. Dis. 2019;19:629. doi: 10.1186/s12879-019-4272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghasemi E., Ghalavand Z., Goudarzi H., Yeganeh F., Hashemi A., Hossein Dabiri H., Mirsamadi E.S., Foroumand M. Phenotypic and genotypic investigation of biofilm formation in clinical and environmental isolates of Acinetobacter baumannii. Arch. Clin. Infect. Dis. 2018;13:e12914. doi: 10.5812/archcid.12914. [DOI] [Google Scholar]
- 56.Vijayakumar S., Rajenderan S., Laishram S., Anandan S., Balaji V., Biswas I. Biofilm formation and motility depend on the nature of the Acinetobacter baumannii clinical isolates. Front. Public Health. 2016;4:105. doi: 10.3389/fpubh.2016.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernards A.T., Frénay H.M., Lim B.T., Hendriks W.D., Dijkshoorn L., Van Boven C.P. Methicillin-resistant Staphylococcus aureus and Acinetobacter baumannii: An unexpected difference in epidemiologic behavior. Am. J. Infect. Control. 1998;26:544–551. doi: 10.1053/ic.1998.v26.a84555. [DOI] [PubMed] [Google Scholar]
- 58.Allen K.D., Green H.T. Hospital outbreak of multi-resistant Acinetobacter anitratus: An airborne mode of spread? J. Hosp. Infect. 1987;9:110–119. doi: 10.1016/0195-6701(87)90048-X. [DOI] [PubMed] [Google Scholar]
- 59.Bianco A., Quirino A., Giordano M.V., Marano V., Rizzo C., Liberto M.C., Focà A., Pavia M. Control of carbapenem-resistant Acinetobacter baumannii outbreak in an intensive care unit of a teaching hospital in Southern Italy. BMC Infect. Dis. 2016;16:747. doi: 10.1186/s12879-016-2036-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anane Y.A., Aptala T., Vasaikar S., Okuthe G.E., Songca S. Prevalence and molecular analysis of multidrug-resistant Acinetobacter baumannii in the extra-hospital environment in Mthatha, South Africa. Braz. J. Infect. Dis. 2019;23:371–380. doi: 10.1016/j.bjid.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chusri S., Chongsuvivatwong V., Silpapojakul K., Singkhamanan K., Hortiwakul T., Charernmak B., Doi Y. Clinical characteristics and outcomes of community and hospital-acquired Acinetobacter baumannii bacteremia. J. Microbiol. Immunol. Infect. 2019;52:796–806. doi: 10.1016/j.jmii.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 62.Anstey N.M., Currie B.J., Withnall K.M. Community-acquired Acinetobacter pneumonia in the Northern Territory of Australia. Clin. Infect. Dis. 1992;14:83–91. doi: 10.1093/clinids/14.1.83. [DOI] [PubMed] [Google Scholar]
- 63.Anstey N.M., Currie B.J., Hassell M., Palmer D., Dwyer B., Seifert H. Community-acquired bacteremic Acinetobacter pneumonia in tropical Australia is caused by diverse strains of Acinetobacter baumannii, with carriage in the throat in at-risk groups. J. Clin. Microbiol. 2002;40:685–686. doi: 10.1128/JCM.40.2.685-686.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davis J.S., McMillan M., Swaminathan A., Kelly J.A., Piera K.E., Baird R.W., Currie B.J., Anstey N.M. A 16-year prospective study of community-onset bacteremic Acinetobacter pneumonia: Low mortality with appropriate initial empirical antibiotic protocols. Chest. 2014;146:1038–1045. doi: 10.1378/chest.13-3065. [DOI] [PubMed] [Google Scholar]
- 65.Leung W.S., Chu C.M., Tsang K.Y., Lo F.H., Lo K.F., Ho P.L. Fulminant community-acquired Acinetobacter baumannii pneumonia as a distinct clinical syndrome. Chest J. 2006;129:102–109. doi: 10.1378/chest.129.1.102. [DOI] [PubMed] [Google Scholar]
- 66.Sharma A., Shariff M., Thukral S.S., Shah A. Chronic community-acquired Acinetobacter pneumonia that responded slowly to rifampicin in the anti-tuberculous regime. J. Infect. 2005;51:149–152. doi: 10.1016/j.jinf.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 67.Ong C.W.M., Lye D.C.B., Khoo K.L., Chua G.S.W., Yeoh S.F., Leo Y.S., Tambyah P.A., Chua A.C. Severe community-acquired Acinetobacter baumannii pneumonia: An emerging highly lethal infectious disease in the Asia–Pacific. Respirology. 2009;14:1200–1205. doi: 10.1111/j.1440-1843.2009.01630.x. [DOI] [PubMed] [Google Scholar]
- 68.Oh Y.J., Song S.H., Baik S.H., Lee H.H., Han I.M., Oh D.H. A case of fulminant community-acquired Acinetobacter baumannii pneumonia in Korea. Korean J. Intern. Med. 2013;28:486–490. doi: 10.3904/kjim.2013.28.4.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen M.Z., Hsueh P.R., Lee L.N., Chen M.Z., Hsueh P.R., Lee L.N., Yu C.J., Yang P.C. Severe community-acquired pneumonia due to Acinetobacter baumannii. Chest J. 2001;120:1072–1077. doi: 10.1378/chest.120.4.1072. [DOI] [PubMed] [Google Scholar]
- 70.Wang J.T., McDonald L.C., Chang S.C., Ho M. Community-acquired Acinetobacter baumannii bacteremia in adult patients in Taiwan. J. Clin. Microbiol. 2002;40:1526–1529. doi: 10.1128/JCM.40.4.1526-1529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang W.N., Lu C.H., Huang C.R., Chuang Y.C. Community-acquired Acinetobacter meningitis in adults. Infection. 2000;28:395–397. doi: 10.1007/s150100070013. [DOI] [PubMed] [Google Scholar]
- 72.Dexter C., Murray G.L., Paulsen I.T., Peleg A.Y. Community-acquired Acinetobacter baumannii: Clinical characteristics, epidemiology, and pathogenesis. Exp. Rev. Anti Infect. Ther. 2015;13:567–573. doi: 10.1586/14787210.2015.1025055. [DOI] [PubMed] [Google Scholar]
- 73.Falagas M.E., Karveli E.A., Kelesidis I., Kelesidis T. Community-acquired Acinetobacter infections. Eur. J. Clin. Microb. Infect. Dis. 2007;26:857–868. doi: 10.1007/s10096-007-0365-6. [DOI] [PubMed] [Google Scholar]
- 74.Falagas M.E., Karveli E.A. The changing global epidemiology of Acinetobacter baumannii infections: A development with major public health implications. Clin. Microb. Infect. 2007;13:117–119. doi: 10.1111/j.1469-0691.2006.01596.x. [DOI] [PubMed] [Google Scholar]
- 75.Davis K.A., Moran K.A., McAllister C.K., Gray P.J. Multidrug-resistant Acinetobacter extremity infections in soldiers. Emerg. Infect. Dis. 2005;11:1218–1224. doi: 10.3201/1108.050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Öncül O., Keskin Ö., Acar H.V., Küçükardalı Y., Evrenkaya R., Atasoyu E.M., Top C., Nalbant S., Özkan S., Emekdaş G., et al. Hospital-acquired infections following the 1999 Marmara earthquake. J. Hosp. Infect. 2002;51:47–51. doi: 10.1053/jhin.2002.1205. [DOI] [PubMed] [Google Scholar]
- 77.Wong D., Nielsen T.B., Bonomo R.A., Pantapalangkoor P., Luna B., Spellberg S. Clinical and pathophysiological overview of Acinetobacter infections: A century of challenges. Clin. Microbiol. Rev. 2017;30:409–447. doi: 10.1128/CMR.00058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gales A.C., Seifert H., Gur D., Castanheira M., Jones R.N., Sader H.S. Antimicrobial Susceptibility of Acinetobacter calcoaceticus-Acinetobacter baumannii Complex and Stenotrophomonas maltophilia Clinical Isolates: Results from the SENTRY Antimicrobial Surveillance Program (1997–2016) Open Forum Infect. Dis. 2019;15:S34–S46. doi: 10.1093/ofid/ofy293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lupo A., Haenni M., Madec J.Y. Antimicrobial resistance in Acinetobacter spp. and Pseudomonas spp. Microbiol. Spectr. 2018;6 doi: 10.1128/microbiolspec.ARBA-0007-2017. [DOI] [PubMed] [Google Scholar]
- 80.Gil-Marqués M.L., Moreno-Martínez P., Costas C., Pachón J., Blázquez J., McConnell M.J. Peptidoglycan recycling contributes to intrinsic resistance to fosfomycin in Acinetobacter baumannii. J. Antimicrob. Chemother. 2018;73:2960–2968. doi: 10.1093/jac/dky289. [DOI] [PubMed] [Google Scholar]
- 81.Rodríguez C.H., Nastro M., Famiglietti A. Carbapenemases in Acinetobacter baumannii. Review of Their Dissemination in Latin America. Rev. Argent. Microbiol. 2018;50:327–333. doi: 10.1016/j.ram.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 82.Ayobami O., Willrich N., Harder T., Okeke I.N., Eckmanns T., Markwart R. The incidence and prevalence of hospital-acquired (carbapenem-resistant) Acinetobacter baumannii in Europe, Eastern Mediterranean and Africa: A systematic review and meta-analysis. Emerg. Microbes Infect. 2019;8:1747–1759. doi: 10.1080/22221751.2019.1698273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nowak P., Paluchowska P.M., Budak A. Co-occurrence of carbapenem and aminoglycoside resistance genes among multidrug-resistant clinical isolates of Acinetobacter baumannii from Cracow, Poland. Med. Sci. Monit. Basic Res. 2014;27:9–14. doi: 10.12659/MSMBR.889811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bakour S., Touati A., Sahli F., Ameur A.A., Haouchine D., Rolain J.M. Antibiotic resistance determinants of multidrug-resistant Acinetobacter baumannii clinical isolates in Algeria. Diagn. Microbiol. Infect. Dis. 2013;76:529–531. doi: 10.1016/j.diagmicrobio.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 85.Moniri R., Farahani R.K., Shajari G., Shirazi M.H.N., Ghasemi A. Molecular epidemiology of aminoglycosides resistance in Acinetobacter spp. with emergence of multidrug-resistant strains. Iran. J. Public Health. 2010;39:63–68. [PMC free article] [PubMed] [Google Scholar]
- 86.Nemec A., Dolzani L., Brisse S., Van den Broek P., Dijkshoorn L. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J. Med. Microbiol. 2004;53:1233–1240. doi: 10.1099/jmm.0.45716-0. [DOI] [PubMed] [Google Scholar]
- 87.Boinett C.J., Cain A.K., Hawkey J., Do Hoang N.T., Khanh N.N.T., Thanh D.P., Dordel J., Campbell J.I., Lan N.P.H., Mayho M., et al. Clinical and laboratory-induced colistin-resistance mechanisms in Acinetobacter baumannii. Microb. Genom. 2019;5:e000246. doi: 10.1099/mgen.0.000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Friedman N.D., Temkin E., Carmeli Y. The negative impact of antibiotic resistance. Clin. Microbiol. Infect. 2016;22:416–422. doi: 10.1016/j.cmi.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 89.Lautenbach E., Synnestvedt M., Weiner M.G., Bilker W.B., Vo L., Schein J., Kim M. Epidemiology and impact of imipenem resistance in Acinetobacter baumannii. Infect. Control. Hosp. Epidemiol. 2009;30:1186–1192. doi: 10.1086/648450. [DOI] [PubMed] [Google Scholar]
- 90.Oliveira E.A., Paula G.R., Mondino P.J.J., Chagas T.P.G., Mondino S.S.B., Mendonça-Souza C.R.V. High rate of detection of OXA-23-producing Acinetobacter from two general hospitals in Brazil. Rev. Soc. Bras. Med. Trop. 2019;52:e20190243. doi: 10.1590/0037-8682-0243-2019. [DOI] [PubMed] [Google Scholar]
- 91.Ruppé É., Woerther P.L., Barbier F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann. Intensive Care. 2015;5:61. doi: 10.1186/s13613-015-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zilberberg M.D., Kollef M.H., Shorr A.F. Secular trends in Acinetobacter baumannii resistance in respiratory and bloodstream specimens in the United States, 2003 to 2012: A survey study. J. Hosp. Med. 2016;11:21–26. doi: 10.1002/jhm.2477. [DOI] [PubMed] [Google Scholar]
- 93.Qureshi Z.A., Hittle L.E., O’Hara J.A., Rivera J.I., Syed A., Shields R.K., Pasculle A.W., Ernst R.K., Doi Y. Colistin-resistant Acinetobacter baumannii: Beyond carbapenem resistance. Clin. Infect. Dis. 2015;60:1295–1303. doi: 10.1093/cid/civ048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kyriakidis I., Vasileiou E., Pana Z.D., Tragiannidis A. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens. 2021;10:373. doi: 10.3390/pathogens10030373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.CDC . Antibiotic Resistance Threats in the United States 2019. US Department of Health and Human Services; Atlanta, GA, USA: 2019. [Google Scholar]
- 96.Nutman A., Lerner A., Schwartz D., Carmeli Y. Evaluation of carriage and environmental contamination by carbapenem-resistant Acinetobacter baumannii. Clin. Microbiol. Infect. 2016;22:949.e5–949.e7. doi: 10.1016/j.cmi.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 97.Chen C.H., Lin L.C., Chang Y.J., Chen Y.M., Chang C.Y., Huang C.C. Infection control programs and antibiotic control programs to limit transmission of multidrug-resistant Acinetobacter baumannii infections: Evolution of old problems and new challenges for institutes. Int. J. Environ. Res. Public Health. 2015;12:8871–8882. doi: 10.3390/ijerph120808871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gootz T.D., Marra A. Acinetobacter baumannii: An emerging multidrug-resistant threat. Exp. Ver. Anti. Infect. Ther. 2008;6:309–325. doi: 10.1586/14787210.6.3.309. [DOI] [PubMed] [Google Scholar]
- 99.Perez S., Innes G.K., Walters M.S., Mehr J., Arias J., Greeley R., Chew D. Increase in hospital-acquired carbapenem-resistant Acinetobacter baumannii infection and colonization in an acute care hospital during a surge in COVID-19 admissions—New Jersey, February–July 2020. Morb. Mort. Weekly Rep. 2020;69:1827–1831. doi: 10.15585/mmwr.mm6948e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nori P., Cowman K., Chen V., Bartash R., Szymczak W., Madaline T., Katiyar C.P., Jain R., Aldrich M., Weston G., et al. Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York City pandemic surge. Infect. Cont. Hosp. Epidemiol. 2020;42:84–88. doi: 10.1017/ice.2020.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Porretta A.D., Baggiani A., Arzilli G., Casigliani V., Mariotti T., Mariottini F., Scardina G., Sironi D., Totaro M., Barnini S., et al. Increased risk of acquisition of new Delhi metallo-beta-lactamase-producing carbapenem-resistant enterobacterales (NDM-CRE) among a Cohort of COVID-19 Patients in a teaching hospital in Tuscany, Italy. Pathogens. 2020;9:635. doi: 10.3390/pathogens9080635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tiri B., Sensi E., Marsiliani V., Cantarini M., Priante G., Vernelli C., Martella L.A., Costantini M., Mariottini A., Andreani P., et al. Antimicrobial stewardship program, COVID-19, and infection control: Spread of carbapenem-resistant Klebsiella Pneumoniae colonization in ICU COVID-19 patients. What did not work? J. Clin. Med. 2020;9:2744. doi: 10.3390/jcm9092744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tacconelli E., Cataldo M.A., Dancer S.J., De Angelis G., Falcone M., Frank U., Kahlmeter G., Pan A., Petrosillo N., Rodríguez-Baño J., et al. ESCMID guidelines for the management of the infection control measures to reduce transmission of multidrug-resistant Gram-negative bacteria in hospitalized patients. Clin. Microbiol. Infect. 2014;20:1–55. doi: 10.1111/1469-0691.12427. [DOI] [PubMed] [Google Scholar]
- 104.Zhang G., Hu C., Luo L., Fang F., Chen Y., Li J., Peng Z., Pan H. Clinical features and outcomes of 221 patients with COVID-19 in Wuhan, China. J. Clin. Virol. 2020;127:104364. doi: 10.1016/j.jcv.2020.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Durán-Manuel E.M., Cruz-Cruz C., Ibáñez-Cervantes G., Bravata-Alcantará J.C., Sosa-Hernández O., Delgado-Balbuena L., León-García G., Cortés-Ortíz I.A., Cureño-Díaz M.A., Castro-Escarpulli G., et al. Clonal dispersion of Acinetobacter baumannii in an intensive care unit designed to patients COVID-19. J. Infect. Dev. Ctries. 2021;15:58–68. doi: 10.3855/jidc.13545. [DOI] [PubMed] [Google Scholar]
- 106.Nebreda-Mayoral T., Miguel-Gómez M.A., March-Rossellóa G.A., Puente-Fuertes L., Cantón-Benito E., Martínez-García A.M., Muñoz-Martín A.B., Orduña-Domingo A. Infección bacteriana/fúngica en pacientes con COVID-19 ingresados en un hospital de tercer nivel de Castilla y León, Spain. Enferm. Infecc. Microbiol. Clin. 2020 doi: 10.7883/yoken.JJID.2020.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Duployez C., Guern R.L., Milliere L., Caplan M., Loïez C., Ledoux G., Jaillette E., Favory R., Mathieu D., Wallet F. An outbreak can hide another. Jpn. J. Infect. Dis. 2020 doi: 10.7883/yoken.JJID.2020.705. [DOI] [PubMed] [Google Scholar]
- 108.Silva D.L., Lima C.M., Magalhães V.C.R., Baltazar L.M., Peres N.T.A., Caligiorne R.B., Moura A.S., Fereguetti T., Martins J.C., Rabelo L.F., et al. Fungal and bacterial coinfections increase mortality of severely ill COVID-19 patients. J. Hosp. Infect. 2021;113:145–154. doi: 10.1016/j.jhin.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gottesman T., Fedorowsky R., Yerushalmi R., Lellouche J., Nutman A. An outbreak of carbapenem-resistant Acinetobacter baumannii in COVID-19 dedicated hospital. Infect. Prev. Pract. 2021;3:100113. doi: 10.1016/j.infpip.2021.100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu N., Dai J., Guo M., Li J., Zhou X., Li F., Gao Y., Qu H., Lu H., Jin J., et al. Pre-optimized phage therapy on secondary Acinetobacter baumannii infection in four critical COVID-19 patients. Emerg. Microbes Infect. 2021;10:612–618. doi: 10.1080/22221751.2021.1902754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rasmussen S.A., Smulian J.C., Lednicky J.A., Wen T.S., Jamieson D.J. Coronavirus disease 2019 (COVID-19) and pregnancy: What obstetricians need to know. Am. J. Obstet. Gynecol. 2020;222:415–426. doi: 10.1016/j.ajog.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ritchie A.I., Singanayagam A. Immunosuppression for hyper inflammation in COVID-19: A double-edged sword? Lancet. 2020;395:1111. doi: 10.1016/S0140-6736(20)30691-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huttner B., Catho G., Pano-Pardo J.R., Pulcini C., Schouten J. COVID-19: Don’t neglect antimicrobial stewardship principles! Clin. Microbiol. Infect. 2020;26:808–810. doi: 10.1016/j.cmi.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cox M.J., Loman N., Bogaert D., O’Grady J. Co-infections: Potentially lethal and unexplored in COVID-19. Lancet Microbe. 2020;1:e11. doi: 10.1016/S2666-5247(20)30009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Langford B.J., So M., Raybardhan S., Leung V., Westwood D., MacFadden D.R., Soucy J.P.R., Daneman N. Bacterial coinfection and secondary infection in patients with COVID19: A living rapid review and meta-analysis. Clin. Microb. Infect. 2020;26:1622–1629. doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fu Y., Yang Q.X.M., Kong H., Chen H., Fu Y., Yao Y., Zhou H., Zhou J. Secondary bacterial infections in critically ill patients with coronavirus disease 2019. Open Forum Infect. Dis. 2020;7:ofaa220. doi: 10.1093/ofid/ofaa220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lansbury L., Lim B., Baskaran V., Lim W.S. Coinfections in people with COVID-19: A systematic review and meta-analysis. J. Infect. 2020;81:266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ripa M., Galli L., Poli A., Oltolini C., Spagnuolo V., Mastrangelo A., Muccini C., Monti G., De Luca G., Landoni G., et al. Secondary infections in patients hospitalized with COVID-19: Incidence and predictive factors. Clin. Microbiol. Infect. 2021;27:451–457. doi: 10.1016/j.cmi.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang Z., Yang B., Li Q., Wen L., Zhang R. Clinical Features of 69 Cases with Coronavirus Disease 2019 in Wuhan, China. Clin. Infect. Dis. 2020;71:769–777. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Contou D., Claudinon A., Pajot O., Micaëlo M., Longuet Flandre P., Dubert M., Cally R., Logre E., Fraissé M., Mentec H., et al. Bacterial and viral coinfections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann. Int. Care. 2020;10:119. doi: 10.1186/s13613-020-00736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang S., Hua M., Liu X., Du C., Pu L., Xiang P., Wang L., Liu J. Bacterial and fungal coinfections among COVID-19 patients in intensive care unit. Microbes Infect. 2021;23:104806. doi: 10.1016/j.micinf.2021.104806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: A descriptive study. Lancet Infect. Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rawson T.M., Moore L., Zhu N., Ranganathan N., Skolimowska K., Gilchrist M., Satta G., Cooke G., Holmes A. Bacterial and fungal coinfection in individuals with coronavirus: A rapid review to support COVID-19 antimicrobial prescribing. Clin. Infect. Dis. 2020;71:2459–2468. doi: 10.1093/cid/ciaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu C.P., Adhi F., Highland K. Recognition and management of respiratory coinfection and secondary bacterial pneumonia in patients with COVID-19. Clin. J. Med. 2020;87:658–663. doi: 10.3949/ccjm.87a.ccc015. [DOI] [PubMed] [Google Scholar]
- 126.Hughes S., Troise O., Donaldson H., Mughal N., Moore L. Bacterial and fungal coinfection among hospitalized patients with COVID-19: A retrospective cohort study in a UK secondary-care setting. Clin. Microbiol. Infect. 2020;26:1395–1399. doi: 10.1016/j.cmi.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Hingrat Q.L., et al. Clinical and virological data of the first cases of COVID-19 in Europe: A case series. Lancet Infect. Dis. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Karruli A., Boccia F., Gagliardi M., Patauner F., Ursi M.P., Sommese P., De Rosa R., Murino P., Ruocco G., Corcione A., et al. Multidrug-resistant infections and outcome of critically Ill patients with coronavirus disease 2019: A single-center experience. Microb. Drug Resist. 2021 doi: 10.1089/mdr.2020.0489. [DOI] [PubMed] [Google Scholar]
- 129.Ramadan H.K.-A., Mahmoud M.A., Aburahma M.Z., Elkhawaga A.A., El-Mokhtar M.A., Sayed I.M., Hosni A., Hassany S.M., Medhat M.A. Predictors of severity and coinfection resistance profile in COVID-19 patients: First report from upper Egypt. Infect. Drug Resist. 2020;13:3409–3422. doi: 10.2147/IDR.S272605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cultrera R., Barozzi A., Libanore M., Marangoni E., Pora R., Quarta B., Spadaro S., Ragazzi R., Marra A., Segala D., et al. Coinfections in critically Ill patients with or without COVID-19: A Comparison of clinical microbial culture findings. Int. J. Environ. Res. Public Health. 2021;20:4358. doi: 10.3390/ijerph18084358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lai C.C., Shey-Ying Chen S.Y., Wen-Chien Ko W.C., Hsueh P.R. Increased antimicrobial resistance during the COVID-19 pandemic. Int. J. Antim. Agents. 2021;57:106324. doi: 10.1016/j.ijantimicag.2021.106324. [DOI] [PMC free article] [PubMed] [Google Scholar]