Abstract
Fas (CD95) and Fas ligand (CD95L) are an interacting receptor-ligand pair required for immune homeostasis. Lymphocyte activation results in the upregulation of Fas expression and the acquisition of sensitivity to FasL-mediated apoptosis. Although Fas upregulation is central to the preservation of immunologic tolerance, little is known about the molecular machinery underlying this process. To investigate the events involved in activation-induced Fas upregulation, we have examined mRNA accumulation, fas promoter activity, and protein expression in the Jurkat T-cell line treated with phorbol myristate acetate and ionomycin (P/I), pharmacological mimics of T-cell receptor activation. Although resting Jurkat cells express Fas, Fas mRNA was induced approximately 10-fold in 2 h upon P/I stimulation. Using sequential deletion mutants of the human fas promoter in transient transfection assays, we identified a 47-bp sequence (positions −306 to −260 relative to the ATG) required for activation-driven fas upregulation. Sequence analysis revealed the presence of a previously unrecognized composite binding site for both the Sp1 and NF-κB transcription factors at positions −295 to −286. Electrophoretic mobility shift assay (EMSA) and supershift analyses of this region documented constitutive binding of Sp1 in unactivated nuclear extracts and inducible binding of p50-p65 NF-κB heterodimers after P/I activation. Sp1 and NF-κB transcription factor binding was shown to be mutually exclusive by EMSA displacement studies with purified recombinant Sp1 and recombinant p50. The functional contribution of the κB-Sp1 composite site in P/I-inducible fas promoter activation was verified by using κB-Sp1 concatamers (−295 to −286) in a thymidine kinase promoter-driven reporter construct and native promoter constructs in Jurkat cells overexpressing IκB-α. Site-directed mutagenesis of the critical guanine nucleotides in the κB-Sp1 element documented the essential role of this site in activation-dependent fas promoter induction.
Lymphocyte activation results in the transcription of a number of gene products that mobilize and maintain a functional immune response. Such gene products regulate cellular differentiation, effector function, and clonal expansion of the responding lymphocyte population. One such gene product is Fas (CD95), a cell surface protein known to induce apoptosis in activated lymphocytes upon binding to its cognate ligand (FasL). The essential roles of Fas and FasL in the maintenance of peripheral self-tolerance underscore their prominence as homeostatic regulators of the immune system (reviewed in reference 41). Within the T-cell compartment, Fas is expressed on immature thymocytes, but does not appear to be involved in thymic deletion (13, 17). Fas expression on resting peripheral blood lymphocytes is low to absent (26, 37, 41, 44). Primary T-cell activation results in upregulation of fas mRNA and protein followed by the gradual acquisition of sensitivity to Fas-mediated apoptosis (37, 44). Although the regulation of Fas-mediated apoptosis is complex, involving assembly of the intracellular proteins necessary to initiate the apoptotic cascade (7, 40) and an absence or low abundance of inhibitory proteins, such as FAP-1 (53), FLIP (22), bcl-2, and bcl-xL (6, 23, 25), Fas receptor cross-linking is fundamentally required (24). Fas expression is maintained on activated lymphocytes for several weeks (44), and upon secondary exposure to antigen, FasL is upregulated (1, 14, 26). Autocrine or paracrine interactions of Fas and FasL result in the apoptotic elimination of responding CD4+ lymphocytes, a process termed “activation-induced cell death” (AICD) (1, 14, 26). AICD can also be induced in CD4+ T-cell lines by T-cell receptor (TCR) engagement or pharmacological mimics of TCR signaling, such as anti-CD3 antibodies or phorbol myristate acetate (PMA) plus ionomycin (P/I), respectively. In direct contrast, activated CD8+ T cells do not appear to undergo apoptosis as a result of Fas ligation, but rather are stimulated to undergo clonal expansion by reverse signaling through FasL (59). Thus, while Fas appears to be equivalently upregulated in both CD4+ and CD8+ T cells upon activation (37), the signals transduced by both Fas and FasL have dissimilar biological effects on the two lymphocyte subsets.
The transcriptional machinery controlling Fas expression is largely unknown. Genomic organization studies showed that human fas is a single-copy gene containing nine exons and eight introns, spanning approximately 25 kb (5). Sequences proximal to the Fas translational start site have a relatively high GC content (61%) between −590 and −1, a number of CpG dinucleotides (28 CpGs between −590 and −1), and an absence of conventional TATA and CAAT boxes (5). These are properties characteristic of a class of polymerase II-dependent promoters that include housekeeping genes (reviewed in reference 3) and other members of the tumor necrosis factor (TNF)-nerve growth factor receptor superfamily (28, 51, 52, 54). Although Fas is constitutively expressed on a variety of nonhematopoietic cells and hematopoietic cell lines (33, 43), multiple reports have documented Fas inducibility in response to such stimuli as UV irradiation, viral infection, wild-type p53, hypoxia, and chemotherapeutic agents (2, 34, 39, 45, 47, 61). To date, however, the molecular events regulating constitutive and inducible fas expression have not been elucidated.
In the present study, we have investigated the transcriptional machinery required for activation-dependent induction of Fas expression in the CD4+ Jurkat T-cell line. Although constitutively expressed on Jurkat cells, Fas can be markedly upregulated by P/I activation. Using deletion constructs of the fas promoter in luciferase reporter assays, we demonstrate the presence of a previously unrecognized, activation-responsive 47-bp sequence containing a composite binding site for both Sp1 and NF-κB transcription factors. Characterization of this noncanonical site (located at positions −295 to −286) by electrophoretic mobility shift assay (EMSA) and supershift analyses revealed that Sp1 was constitutively bound to this site in untreated Jurkat cells, while p50-p65 NF-κB heterodimers were bound after P/I activation. Site-directed mutagenesis of the critical guanine nucleotides in the κB-Sp1 site, as well as inhibition of IκB degradation or NF-κB translocation, inhibited inducible Fas transcription. Taken together, our results indicate a critical requirement for NF-κB translocation to the κB-Sp1 site at positions −295 to −286 in activation-driven fas promoter induction.
MATERIALS AND METHODS
Cell culture and cellular activation.
The T-lymphoma cell line Jurkat-E6 was obtained from the American Type Culture Collection (Rockville, Md.) and cultured in complete medium (RPMI 1640 with 10% fetal bovine serum [FBS] and 2 mM l-glutamine) at 0.4 × 106 to 0.7 × 106 cells/ml. For activation studies, Jurkat cells were removed from culture and washed once with RPMI 1640, plated in 10 ml of complete medium, and rested overnight. The following day, cells were activated with PMA (50 ng/ml) and ionomycin (3 μg/ml) for the times indicated (Sigma, St. Louis, Mo.).
Flow cytometric analysis.
Indirect antibody staining and flow cytometric analyses were carried out as described previously (43). Briefly, 106 cells in 100 μl of Dulbecco’s phosphate-buffered saline (D-PBS) containing 2% FBS and 0.1% sodium azide (fluorescence-activated cell sorter [FACS] buffer) were incubated with either 150 ng of UB2 (Kaimaya Biomedical Co., Tukwila, Wash.) or isotype-matched, control mouse anti-human immunoglobulin G1 (IgG1) antibodies (Sigma) at 4°C for 30 min. Samples were washed twice in FACS buffer, and phycoerythrin-conjugated goat anti-mouse IgG secondary antibodies were added in 100 μl of FACS buffer for 30 min at 4°C. Samples were washed twice with FACS buffer, fixed overnight in PBS containing 1% paraformaldehyde, and analyzed with a FACScan (Becton Dickinson and Co., Mountain View, Calif.). Ten thousand cells were analyzed for each sample, with the gate set to exclude dead cells and debris.
RNA isolation and RPAs.
At the indicated time points, P/I-treated Jurkat cells were pelleted and washed once in D-PBS. Control Jurkat cells were similarly cultured but were not treated with P/I. Total RNA from 107 cells per sample was extracted with the RNeasy kit (Qiagen, Chatsworth, Calif.) according to the manufacturer’s instructions, and RNase protection assays (RPA) were performed with the HybSpeed RPA kit (Ambion, Austin, Tex.). Samples (25 μg of total RNA) were hybridized to a riboprobe spanning Fas exons 3 to 6 (cDNA sequence 510 to 750) and a control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) riboprobe. Following RNAse digestion, protected products were boiled, separated by electrophoresis on a 5% urea–polyacrylamide gel electrophoresis (PAGE) gel, and analyzed by autoradiography. Scanning densitometry (Molecular Dynamics, Sunnyvale, Calif.) was used to quantitate the fas and GAPDH band intensities of each sample. Relative fold induction was calculated by dividing each normalized fas/GAPDH ratio against the zero time point.
Generation of fas promoter constructs and site-directed mutagenesis.
A human placental genomic phage library (Clonetech, Palo Alto, Calif.) was screened with a PCR-generated, [α-32P]dCTP-labeled Fas cDNA probe containing 5′ untranslated exon 1 and 2 sequences: forward, 5′ GGACCCGCTCAGTACGGAGTT; reverse, 5′ TTCACCTGGAGGACAGGGCTTATG (Life Technologies, Grand Island, N.Y.). Plaque hybridizations were carried out as previously described (52), and filters were washed sequentially in 6× SSC (0.9 M NaCl, 0.09 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at 42°C for 20 min, 2× SSC–0.1% SDS at 60°C for 20 min, and 1× SSC–0.1% SDS for 20 min before autoradiography. Approximately 1.35 × 106 plaques were screened, followed by sequential hybridizations of the positive clones. Subsequent subcloning of SacI or HindIII inserts into pGEM11Zf(+) (Promega, Madison, Wis.) identified a 2.16-kb HindIII fragment containing 1.74 kb proximal to the Fas translational start site (FasH3). Sanger dideoxy sequencing (U.S. Biochemical Corp., Cleveland, Ohio) confirmed that FasH3 contained 5′ flanking sequences identical to those previously reported for human fas (5).
Reporter constructs were generated with the 2.16-kb FasH3 fragment and primers corresponding to sequences −1739 to −1717 (5′ AATAATCACTCATCTCACTGGGC) and −43 to −19 (5′ CGAAGTGAAAGAGCTTCCCCAACTC) by PCR to generate a 1.72-kb product. This PCR product was then subcloned into the pcR2 expression vector (Invitrogen, San Diego, Calif.) and digested with EcoRI, and the insert was shuttled into pGEM7+ (Promega) (pFPR7+). A partial XhoI digest and complete HindIII digest of pFPR7+ were made to insert the 1.7-kb fragment into the pGL2Basic luciferase reporter construct (Promega) in either orientation. The final constructs, FPR1-Luc(+) and FPR1-Luc(−), spanning the region (1739 to −19) upstream of the fas ATG were sequenced to verify orientation and identity.
The thymidine kinase promoter reporter construct tk-Luc was generated by subcloning the BglII-HindIII fragment of the thymidine kinase promoter from pRL-tk (Promega) into pGEM-11, followed by insertion of the SacI-HindIII fragment into pGL2B. The Δ5/Δ6-tk-Luc reporter was constructed by subcloning the SacI fragment from FPR-Luc(+) into tk-Luc, digesting it with XhoI-PstI, treating it with a Klenow fragment to fill recessed ends, and performing blunt end ligation.
Deletion constructs were generated as follows. Δ5-Luc (−460 to −19) is the product of a religated XhoI digest of FPR-Luc(+); Δ6-Luc (−236 to −19) is the product of a religated complete SacI digest of FPR-Luc(+). Constructs within the Δ5-Luc region were generated by PCR with the following oligonucleotides, digested with KpnI (underlined)-HindIII (double underlined), and subcloned directly into pGL2B: Δ5.4+ (5′ GGGTACC−429GCCACTGCAGGAACGCCCCGGGACAG); Δ5.5+ (5′GGGTACC−366CACCCTGACTTCTCCCCCTCCCTACC); Δ5.7+ (5′GGGTACC−305TCCCCAACCCGGGCGTTCCCCAGCG); Δ5.8+ (5′GGGTACC−259GACCACCGGGGCTTTTCGTGAGCTCGTCT); FasR1H− (5′GGGGGAAGCTT−19CGAAGTGAAAGAGCTTCCCCAAC). The mutagenized reporter constructs Δ5M5.7-Luc and Δ5M5.8-Luc were generated by PCR with the appropriate primers (Δ5.7M, 5′CTCCCCAACCCtttCGTTCCCCAGCG [forward] and 5′CCTCGCTGGGGAACGaaaGGGTTGG [reverse]; Δ5.8M, 5′CCGccyCTTTTCGTGAGCTCGTCT [forward] and 5′AGACGAGCTCACGAAAAGrggCGG [reverse]; mutated nucleotides are in lowercase) by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer’s recommendations. All constructs were confirmed by dideoxy sequencing with vector and internal primers.
Luciferase reporters containing κB-Sp1 concatemers were constructed with oligonucleotides having two copies of the element at position −295 to position −286. These oligonucleotides were synthesized with either KpnI (underlined) or SacI (double underlined) 3′ overhangs (forward, 5′CGGGCGTTCCCAAAAAGGGCGTTCCCGAGCT and reverse, 5′CGGGAACGCCCTTTTTGGGAACGCCCGGTAC; forward, 5′CGGGCGTTCCCAAAAAGGGCGTTCCCGGTAC and reverse, 5′CGGGAACGCCCTTTTTGGGAACGCCCGAGCT). The corresponding 5′ KpnI-3′ SacI or 5′ SacI-3′ KpnI oligonucleotides were annealed (10 min at 85°C and then cooled to room temperature), and timed ligations were prepared with T4 ligase with various ratios of the double-stranded primers. Aliquots of the reaction mixtures were heated to 75°C for 10 min to inactivate the ligase before subcloning into the KpnI-SacI-digested tk-Luc vector (see above). Various clones were screened by sequence analysis to determine the number of κB-Sp1 elements present. The resultant constructs containing two, four, and six copies of the κB-Sp1 site were used for transient transfection and reporter assays.
Transient transfections and reporter assays.
A DEAE-dextran-chloroquine mixture was used for transient transfection of Jurkat cells. Briefly, 5 × 106 to 7.5 × 106 exponentially growing cells were washed and resuspended in 1 ml of transfection solution (RPMI plus 250 μg of DEAE-dextran per ml, 0.1 M Tris [pH 7.2], and 0.1 mM chloroquine) together with 10 μg of reporter construct and 10 ng of the pRL-tk Renilla normalizing luciferase vector (Promega). The cells were incubated for 1.5 h at 37°C, washed, and plated in 10 ml of complete media. After a 20-h recovery, transfected cells were activated with P/I for 8 h. Cell extracts and luciferase reagents were prepared with the Promega Dual-Luciferase Reporter Assay System. Both firefly and Renilla luciferase activities were monitored with a luminometer (Turner Industries, Sunnyvale, Calif.). Normalized reporter activity is expressed as the firefly luciferase value divided by the Renilla luciferase value. Relative fold induction is calculated as the normalized reporter activity of the test sample divided by either the untreated FPR-Luc or Δ5-Luc reporter construct, as indicated below. The Rous sarcoma virus promoter-driven IκBα construct (RSV-IκBα) and vector control were obtained from Tse-Wa Tan (Baylor College of Medicine, Houston, Tex.).
Nuclear extract preparation, EMSAs, supershift analyses, and displacement studies.
Nuclear extracts were prepared by a modified method of Dignam et al. (15). In brief, 7.5 × 106 Jurkat cells were washed in PBS and pelleted before resuspension in 200 μl of cold buffer A (10 mM HEPES (pH 7.9), 50 mM NaCl, 1 mM dithiothreitol, 0.1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride [PMSF]) and incubated for 20 min on ice prior to the addition of 200 μl of cold buffer B (buffer A with 0.1% Nonidet P-40). Cells were gently pipetted and returned on ice for another 20 min. Nuclei were pelleted (5,000 × g, 2 min), washed in buffer A, and pelleted again, and nuclear proteins were extracted in 25 μl of buffer D (400 mM NaCl, 20 mM HEPES (pH 7.9), 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol 1 mM PMSF). The tubes were iced for at least 30 min, followed by centrifugation at 4°C for 10 min. The supernatant was recovered, snap-frozen in liquid nitrogen, and stored at −80°C prior to use.
Probes used for EMSAs were radiolabeled by [γ-32P]ATP end labeling with T4 polynucleotide kinase. Briefly, 30 μg of the Δ5-Luc plasmid was digested with XhoI, dephosphorylated with calf intestine phosphatase, and both enzymes were inactivated at 85°C for 10 min. An aliquot of the digest (7.5 μg) was end labeled with [γ-32P]ATP for 60 min before the kinase was inactivated at 75°C for 10 min. The end-labeled plasmid was then digested overnight with SacI, and the released probe A (−460 to −236) was gel purified as described previously (11). The oligonucleotide probe B (−306 to −278; 5′−306CTCCCCAACCCGGGCGTTCCCCAGCGAGG) or its mutated counterpart was annealed to the corresponding complementary sequence before 1 pmol was end labeled with [γ-32P]ATP for 30 min. The labeled probe was purified from unincorporated nucleotides by using a Chroma Spin 10 column (Clonetech) following the manufacturer’s instructions. The specific activities of all 32P-labeled oligoprobes were routinely 5 × 106 to 6 × 106 cpm per pmol.
Protein concentration was determined with the Bio-Rad protein assay reagent (Bio-Rad, Richmond, Calif.). For EMSAs, 6 μg of nuclear extract was incubated in a total volume of 19 μl of binding buffer [50 mM NaCl, 10 mM Tris (pH 7.9), 0.5 mM EDTA, 1 μg of poly(dI-dC), and 5% glycerol] for 15 min at room temperature before 50,000 cpm of the indicated γ-32P-end-labeled probe was added for another 15 min. For competition assays, excess unlabeled oligonucleotides were preincubated for 15 min prior to the addition of the radiolabeled probe (50,000 cpm). For antibody-mediated supershift assays, extracts were preincubated with 1 μl of either anti-p50, -p65, -c-Rel (Oncogene Research, Cambridge, Mass.) -relB, -p52, or -Sp1 (Santa Cruz Biotechnology, Santa Cruz, Calif.) antibodies at 4°C for 45 min before the addition of the radiolabeled probe. The reactions were loaded on a 4.5% polyacrylamide nondenaturing PAGE gel in 0.5× Tris-borate-EDTA electrophoresed for 2.5 h at 100 V before drying and exposed to autoradiographic film.
For displacement studies, 4 footprinting units (fpu) of Sp1 (∼30 ng; Promega) was preincubated in a total volume of 19 μl of binding buffer for 5 min prior to addition of 12,500 cpm of labeled probe B. After 15 min at room temperature, increasing amounts of recombinant p50 NF-κB (Promega) subunits were added for an additional 15 min before the mixture was loaded on a 4.5% polyacrylamide PAGE gel as described above.
RESULTS
Expression of Fas mRNA and protein is upregulated in Jurkat cells by pharmacological agents that mimic T-cell activation.
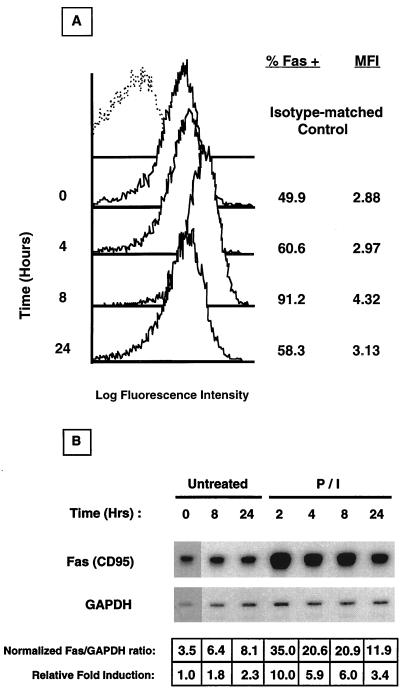
While resting T cells express low to undetectable levels of the Fas protein (26, 37, 41, 44), it is rapidly upregulated upon exposure to specific antigens, mitogens, cytokines, TCR antibodies, and various pharmacological agents such as P/I (6, 37, 44). The Jurkat T-cell line has an activated T-cell phenotype, in that it constitutively expresses cell surface Fas (Fig. 1A). Because we were interested in the transcriptional mechanisms controlling Fas expression, we asked whether Fas could be upregulated in Jurkat cells upon activation. To this end, Jurkat cells were stimulated with P/I for various times, and mRNA and protein expression was determined with RPAs and FACS analyses, respectively. As shown in Fig. 1A, Jurkat cells transiently upregulated surface Fas after P/I stimulation. Increased Fas was detectable as early as 4 h poststimulation, reached maximal levels by 8 h, and declined to near-baseline levels by 24 h. A 1.5- to 2.0-fold increase in both the percentage of cells expressing Fas as well as the mean fluorescence intensity (MFI), representative of antigenic density on a per-cell basis, was observed. Correspondingly, as shown in Fig. 1B, Fas mRNA levels were upregulated by 10-fold in Jurkat cells activated for 2 h; mRNA levels declined subsequently over the time period examined (24 h). These results demonstrate an immediate-early, activation-dependent upregulation of Fas in P/I-stimulated Jurkat cells, suggesting a likely transcriptional control.
FIG. 1.
Time course induction of Fas upregulation in P/I-treated Jurkat cells. (A) Cells were harvested at the indicated time, and Fas expression was analyzed by flow cytometry. Isotype-matched control antibody staining is represented as a dashed line, and Fas staining is represented as solid lines. The percentage of specific Fas staining and the mean fluorescence intensity (MFI) of Jurkat cells at each time point are shown to the right. (B) Cells were harvested at the indicated time, total RNA was isolated, and Fas mRNA accumulation was determined by RNAse protection analysis. Scanning densitometry was used to normalize Fas mRNA levels and to calculate the relative fold induction compared to that of the untreated, time zero sample.
A 220-bp region in the Fas promoter is required for activation-dependent upregulation.
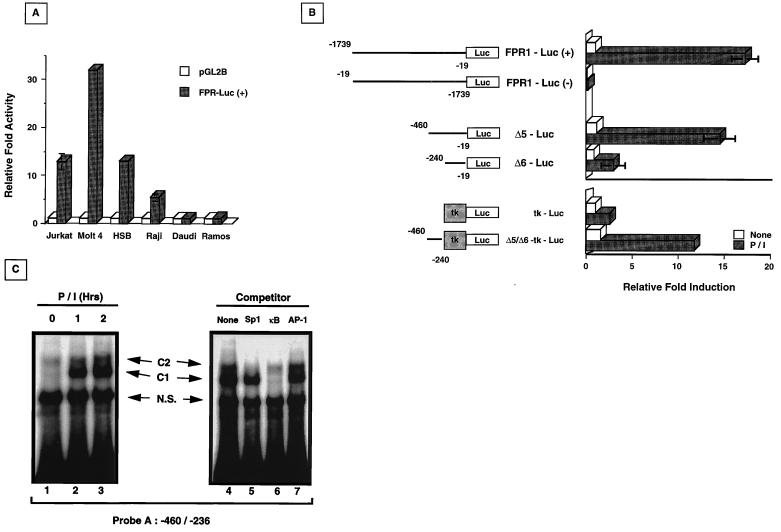
To test the premise that activation-dependent Fas upregulation was modulated at the level of transcription, approximately 1.7 kb of the 5′ region flanking the fas translational start site (nucleotides −1739 to −19) were isolated from a human placental genomic library and subcloned into the pGL2-Basic luciferase reporter vector in both orientations relative to the luciferase gene. The basal activity of the FPR1-Luc(+) construct was first determined by transient transfections into various T- and B-cell lines, followed by extract preparations and luciferase activity measurements. As shown in Fig. 2A, the FPR1-Luc(+) construct demonstrated basal activity in the Jurkat (13-fold over empty vector), Molt-4 (32-fold), and HSB (14-fold) T-cell lines. In B-cell lines, modest basal activity was observed only in Raji cells (fivefold over empty vector alone), but not in Daudi or Ramos cells. These results document that the 1.7-kb region upstream of the fas ATG contains a functional promoter operative in T-cell lines and certain B-cell lines.
FIG. 2.
Basal and inducible fas promoter activity and in vitro EMSA analysis. (A) FPR1-Luc(+) containing the sequence from −1739 to −19 is active in T-cell lines and the B-lymphocyte line Raji, but inactive in Daudi or Ramos cells. Cells were transfected with 10 μg of either the empty reporter (pGL2B [□]) or the fas promoter reporter vector (FPR1-Luc [■]); 10 ng of pRL-tk vector was cotransfected for normalization. Extracts were taken after 40 h, and luciferase activity was determined. Relative fold activity was calculated as the normalized FPR1-Luc(+) activity divided by the normalized pGL2B value. (B) fas promoter sequences −460 to −240 are required for inducible fas reporter activity. FPR1-Luc in both orientations (forward and reverse), Δ5-Luc, Δ6-Luc, tk-Luc, and Δ5/Δ6-tk-Luc constructs were transiently transfected into Jurkat cells with pRL-tk for normalization, and the cells were then left untreated (□) or were activated with P/I (■) for 8 h before harvesting of the extracts for measurement of luciferase activity. The values shown are averages of three independent experiments (± standard deviation) and are expressed as relative fold induction over normalized luciferase values from untreated Jurkat cells containing FPR1-Luc(+). For tk-Luc and Δ5/Δ6-tk-Luc, the relative fold induction was calculated separately as the normalized luciferase value divided by the untreated, normalized tk-Luc luciferase value. (C) Specific protein complexes bind the enhancer region between −460 and −236. 32P-labeled probe A (50,000 cpm) was mixed with nuclear extracts from untreated or P/I-activated Jurkat cells (left panel, lanes 1 to 3). Specific complexes (C1 and C2) are indicated. N.S., nonspecific complexes. Cold competition mixtures with a 25-fold excess of the indicated consensus elements (right panel, lanes 4 to 7) were mixed with extracts from Jurkat cells activated with P/I for 1 h prior to addition of labeled probe A.
To determine whether FPR1-Luc(+) was responsive to activation, transiently transfected Jurkat cells were treated with P/I as described in Materials and Methods, and the relative luciferase activities of untreated and P/I-stimulated cells were compared. As shown in Fig. 2B, FPR1-Luc(+) activity increased approximately 17-fold in response to P/I treatment compared to the basal activity of unstimulated Jurkat cells. The fas promoter is orientation dependent, since FPR1-Luc(−) was inactive in both resting and stimulated cells. To more precisely map the fas promoter region(s) responsive to P/I activation, two deletion constructs containing the sequences from −460 to −19 (Δ5-Luc) and −240 to −19 (Δ6-Luc) were prepared as described in Materials and Methods. As shown in Fig. 2B, P/I inducibility of the fas promoter was maintained when the sequences between −1739 and −460 were deleted. However, when sequences between −460 and −240 were deleted, P/I-induced upregulation was virtually absent. As shown, FPR1-Luc(+) and Δ5-Luc were upregulated 17- and 15-fold after P/I treatment, respectively, while the Δ6-Luc construct was upregulated by less than 4-fold. Interestingly, no differences were noted in the activity of these constructs in unstimulated Jurkat cells, suggesting that the fas promoter sequences between −460 and −240 are critically important for activation-dependent, but not basal, transcription. Sequence analysis within this region demonstrated the presence of consensus elements for several transcription factors, including AP-2, GAS, NF-κB, and NF-AT.
To investigate whether the 220-bp region between −460 and −240 contained enhancer elements responsive to P/I activation, this region was subcloned proximal to a thymidine kinase promoter in a luciferase reporter construct (Δ5/Δ6-tk-Luc), and transient transfection reporter assays were carried out. As shown in Fig. 2B, the Δ5/Δ6-tk-Luc construct was highly inducible by P/I (12-fold relative to tk-Luc), at levels comparable to that of the native Fas promoter. These results suggest that the fas promoter region between −460 and −240 contains enhancer element(s) necessary for activation responsiveness. To analyze whether transcription factors may be involved in the activation-dependent upregulation of the fas promoter within this region, EMSA analyses were carried out. Nuclear extracts, prepared from unactivated and P/I-stimulated Jurkat cells, were incubated with a 32P-labeled probe spanning the fas promoter region between −460 and −236 (probe A). A time course analysis revealed the presence of two complexes binding to this region. An upper complex, designated C2, was constitutively bound in unactivated Jurkat extracts throughout the course of activation (Fig. 2C, lanes 1 to 3). A second lower complex, designated C1, bound only upon P/I activation (Fig. 2C, Lane 2). To determine the nature of the C1 and C2 complexes, cold-target competition was carried out with 25-fold excesses of the Sp1, NF-κB, and AP-1 consensus elements. As shown (Fig. 2C, lane 5), C2 binding was inhibited by Sp1-specific oligonucleotides, indicating the presence of an Sp1-like transcription factor at this site. C1 binding was specifically competed by using κB consensus oligonucleotides derived from the human immunodeficiency virus long terminal repeat (HIV LTR) (Fig. 2C, lane 6). Other cold-target consensus elements tested, including AP-1, AP-2, NF-ATp, and GAS, were incapable of inhibiting either C1 or C2 binding (Fig. 2C, lane 7, and data not shown). Thus, in the promoter region required for activation-dependent Fas upregulation, Sp1 bound constitutively, while NF-κB complex formation occurred only in response to stimulation.
Activation-dependent Fas promoter induction requires nucleotides −306 to −260.
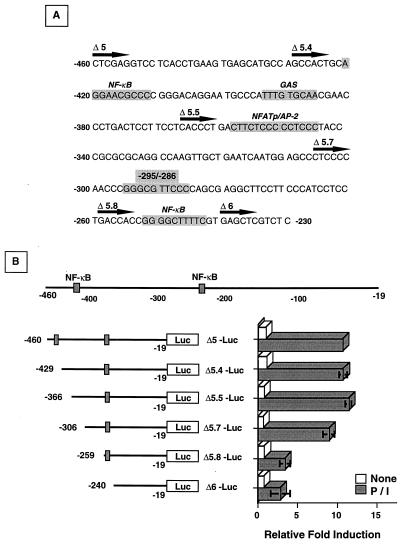
To analyze the region between −460 and −240 in further detail, sequential deletions of Δ5-Luc spanning 40- to 60-bp intervals were generated by PCR. The precise sequences contained in the various constructs are shown in Fig. 3A. Basal and inducible promoter activities in transiently transfected, unactivated, and P/I-stimulated Jurkat cells, respectively, are shown in Fig. 3B. Deletion of nucleotides −460 to −306 and −259 to −240 had little effect on the activation-dependent induction of the fas promoter (Δ5 to Δ5.7, Δ5.8 to Δ6), ruling out the contribution of the consensus NF-κB sites located at −421 to −412 and −252 to −243, the GAS element at −394 to −386, and the NF-ATp–AP-2 site at positions −358 to −345. When nucleotides −306 to −260 (Δ5.7 to Δ5.8) were deleted, however, inducible reporter activity was reduced to that observed with the Δ6 construct. The sequences between −306 and −260 were devoid of any mapped transcription factor consensus sites, except for a κB-like motif present at positions −295 to −286. This motif contains the sequence GGGCGTTCCC and differs from the derived consensus κB p50-p65 heterodimer motif GGGRNNYYCC (20, 30, 48) at position 4 (change of R to C). Notably, however, the critical G1–3 nucleotides necessary for p50 binding are maintained. Comparison of this motif to a consensus Sp1 site, KRGGCGKRRY (35), also revealed homology at 8 of 10 residues, indicating a possible interaction with this transcription factor. Taken together with results from our previous studies showing that complexes that could be competed with Sp1 and NF-κB oligonucleotides bound to the fas promoter region at −406 to −236, it seemed likely that these transcription factors, acting alone or in combination, might be required for activation-dependent fas promoter induction.
FIG. 3.
Localization of activation-dependent fas promoter activity. (A) The fas nucleotide sequence between −460 and −230 is shown with known consensus transcription factor elements indicated (shaded areas). The location and designation of the various deletion constructs are indicated by the solid arrow. (B) P/I inducibility of the fas promoter localizes between nucleotides −306 and −260. Jurkat cells transiently transfected with 10 μg of the various deletion constructs and 10 ng of pRL-tk as the normalizing vector were left untreated (□) or activated with P/I (■) for 8 h before extracts were prepared for luciferase activity measurements. The values shown are averages of three independent experiments ± standard deviation and are expressed as the relative fold induction of the various deletion constructs over the untreated, normalized Δ5-Luc luciferase value.
Sp1 and NF-κB conjointly bind at the κB-Sp1-like motif (−295 to −286) during activation-driven fas promoter induction.
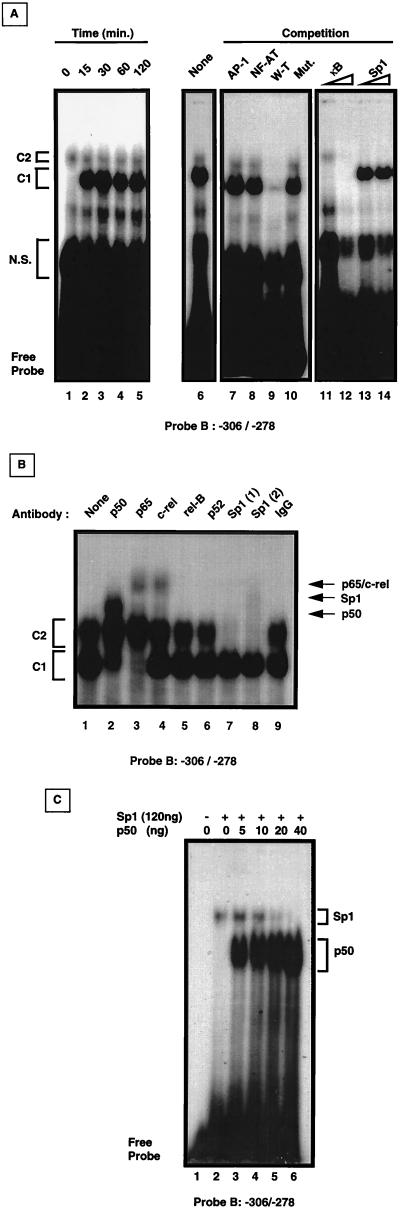
To further investigate the transcription factors involved in fas promoter upregulation in P/I-activated Jurkat cells, EMSA analyses of nuclear extracts were performed with a 29-bp probe spanning the noncanonical κB-Sp1-like motif (−306 to −278; probe B). In concert with our previous observations with probe A, spanning nucleotides −460 to −236 (Fig. 2C), we observed constitutive binding of the C2 complex in both unactivated and P/I-stimulated extracts and inducible binding of the C1 complex upon activation (Fig. 4A). C1 was induced de novo within 15 min of P/I addition (Fig. 4A, lane 2) and persisted for up to 2 h postactivation (Fig. 4A, lane 5). The specificities of C1 and C2 binding to probe B were first demonstrated by cold-competition assays using excess unlabeled probe B (Fig. 4A, lane 9). Jurkat extracts were coincubated with radiolabeled probe B and 25-fold excesses of either unlabeled AP-1- or NF-AT-specific oligonucleotides or a mutated probe B (GGG to TTT [see below]). Under these conditions, no displacement was observed in either C1 or C2 binding complexes (Fig. 4A, lanes 7, 8, and 10). To investigate Sp1–NF-κB binding to probe B, Jurkat extracts were incubated with labeled probe B and increasing concentrations of unlabeled NF-κB consensus oligonucleotides. With a 25-fold excess, NF-κB oligonucleotides effectively competed the inducible C1 complex, but not the C2 complex. However, at 100-fold excess, both C2 and nonspecific binding were inhibited (Fig. 4A, lanes 11 to 12). Increasing concentrations of Sp1 oligonucleotides, on the other hand, specifically competed the constitutive C2 complex, but not the inducible C1 binding (Fig. 4A, lanes 13 and 14).
FIG. 4.
In vitro analysis of a potential κB-Sp1 site at −295 to −286. (A) Time course analysis of complex binding to probe B (−306 to −278). Jurkat cells were activated with P/I for the indicated time, nuclear extracts were harvested, and EMSA analysis was performed with 32P-labeled probe B (left panel, lanes 1 to 5). Cold competition was performed with 1-h-activated Jurkat cell nuclear extracts with a 25-fold excess of consensus oligonucleotides for the transcription factors AP-1, NF-AT, wild-type probe B (W-T), or mutated probe B (Mut.) added prior to incubation with 32P-labeled probe B (lanes 7 to 10). For NF-κB and Sp1 competition, a 25-fold excess (lanes 11 and 13) and 100-fold excess (lanes 12 and 14) of consensus oligonucleotides were added to equivalent amounts of P/I-activated nuclear extracts prior to addition of 32P-labeled probe B. N.S., nonspecific binding. (B) Supershift analyses of P/I-activated Jurkat cell nuclear extracts. Nuclear extracts were subjected to EMSA analysis in the absence (lane 1) or presence of the indicated specific antisera (lanes 2 to 8), or preimmune IgG (lane 9). With two different anti-Sp1 antisera, probe B binding to the upper C2 complex was shown to be either shifted or inhibited (lanes 7 and 8). Arrows indicate the identity of the shifted complexes. (C) Mutual exclusive binding of Sp1 and NF-κB p50 to probe B. Recombinant Sp1 (4 fpu [120 ng]) was preincubated with 12,500 cpm of radiolabeled probe B in the absence (lane 2) or presence of increasing amounts (0 to 40 ng) of recombinant NF-κB p50 (lanes 3 to 6) prior to EMSA analysis. Brackets indicate the Sp1-probe B and p50-probe B complexes.
We next addressed whether extracts from P/I-activated Jurkat cells bound to probe B could be supershifted by using antibodies against various NF-κB–Rel family members, including p50, p52, p65, c-Rel, and RelB. As shown in Fig. 4B, the inducible C1 complex was supershifted with antibodies against p50, p65, and to a lesser extent, c-Rel, indicating the presence of these heterodimers in the shifted complex (Fig. 4B, lanes 2 to 4). No such shifted complexes were observed in extracts from unactivated Jurkat cells lacking the inducible C1 complex (data not shown). Antibodies against Sp1, on the other hand, shifted the constitutive C2 complex in both activated (Fig. 4B, lanes 7 and 8) and unactivated Jurkat extracts (data not shown). These results imply that Sp1 is bound to the κB-Sp1 site at −295 to −286 during normal basal transcription, whereas NF-κB p50-p65 heterodimers occupy the κB-Sp1 site during activation-driven fas promoter induction.
Mutually exclusive binding of Sp1 versus NF-κB at nucleotides −295 to −286.
To assess whether binding of Sp1 and NF-κB to the −295 to −286 site in the fas promoter occurs concurrently or as a mutually exclusive event, EMSA analyses were performed with probe B with recombinant Sp1 and increasing amounts of recombinant p50. Consistent with our observations using extracts from unactivated Jurkat cells, a single complex is observed when probe B is mixed with Sp1 alone (Fig. 4C, lane 2). As increasing amounts of recombinant p50 are added, however, the intensity of the Sp1 complex is decreased, with a concomitant increase in the lower p50 binding complex (Fig. 4C, lanes 3 to 6). These results indicate that the fas promoter region at −295 to −286 contains a single κB-Sp1 binding motif to which either Sp1 or NF-κB is exclusively bound. Upon cellular activation, constitutively bound Sp1 is likely displaced by NF-κB to drive fas promoter upregulation.
NF-κB recruitment regulates activation-dependent Fas promoter induction.
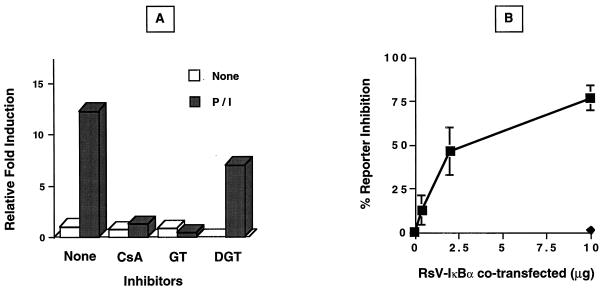
Although the above in vitro analyses document that NF-κB heterodimers are rapidly recruited to the fas promoter region controlling activation-dependent induction, these studies do not address the functional consequences of the effects of NF-κB recruitment on fas promoter activity. To investigate the role of NF-κB recruitment in activation-driven fas upregulation, the effects of various inhibitors of NF-κB function were examined. Jurkat cells transiently transfected with the Δ5-Luc reporter were pretreated with two pharmacological inhibitors of NF-κB and then activated with P/I, and luciferase activity was determined. As shown in Fig. 5A, cyclosporin A pretreatment of Jurkat cells completely prevented P/I-dependent upregulation of the fas promoter. Because this calcineurin inhibitor can inhibit NF-AT translocation and disrupt Ca2+ signals leading to NF-κB induction (36), the fungal metabolite gliotoxin, a specific inhibitor of IκB-α degradation and NF-κB translocation, was also used (46). Gliotoxin, but not the inactive gliotoxin derivative bis-dethio-bis(methylthio)gliotoxin, completely abolished P/I inducibility of the Δ5-Luc reporter, consistent with the notion that NF-κB recruitment was functionally required for activation-dependent fas promoter induction. The critical requirement for NF-κB recruitment was additionally verified in Jurkat cells overexpressing wild-type IκB-α. The overexpression of IκB-α, a natural inhibitor of NF-κB translocation, has been shown to effectively block κB-dependent transactivation (31, 32). As shown in Fig. 5B, P/I-activated Jurkat cells transiently transfected with the Δ5-Luc reporter and RsV-IκB-α expression plasmid showed a dose-dependent decrease in fas promoter activity with increasing IκB-α concentration. Taken together, these results strongly support the premise that NF-κB translocation is essential for activation-dependent fas promoter induction.
FIG. 5.
Inhibition of activation-dependent fas promoter upregulation. (A) Pharmacological inhibition of Δ5-Luc induction by P/I. Jurkat cells transiently transfected with 10 μg of Δ5-Luc and 10 ng of pRL-tk were preincubated in the absence (None) or presence of cyclosporin A (CsA [1 μg/ml]), gliotoxin (GT [1 μg/ml]), or bis-dethio-bis(methylthio)gliotoxin (DGT [3 μg/ml]) for 30 min prior to P/I stimulation for 8 h. Following incubation, cells were harvested for extract preparation, and luciferase activity was determined. The values shown are representative of two independent experiments ± standard deviation and are expressed as the relative fold induction over normalized untreated Δ5-Luc activity. (B) Dose-dependent inhibition of P/I-inducible Δ5-Luc reporter activity by IκBα. Increasing amounts of wild-type RsV-IκBα (■) or vector control (⧫) expression plasmid were cotransfected into Jurkat cells with 10 μg of Δ5-Luc and 10 ng of pRL-tk 40 h prior to P/I activation. Cells were harvested for extract preparation after 8 h of P/I exposure, and luciferase activity was measured. The values shown are averages from two independent experiments ± standard deviation and are expressed as percent inhibition of P/I-induced, normalized Δ5-Luc reporter activity.
The κB-like motif at −295 to −286 controls activation-dependent fas promoter responses.
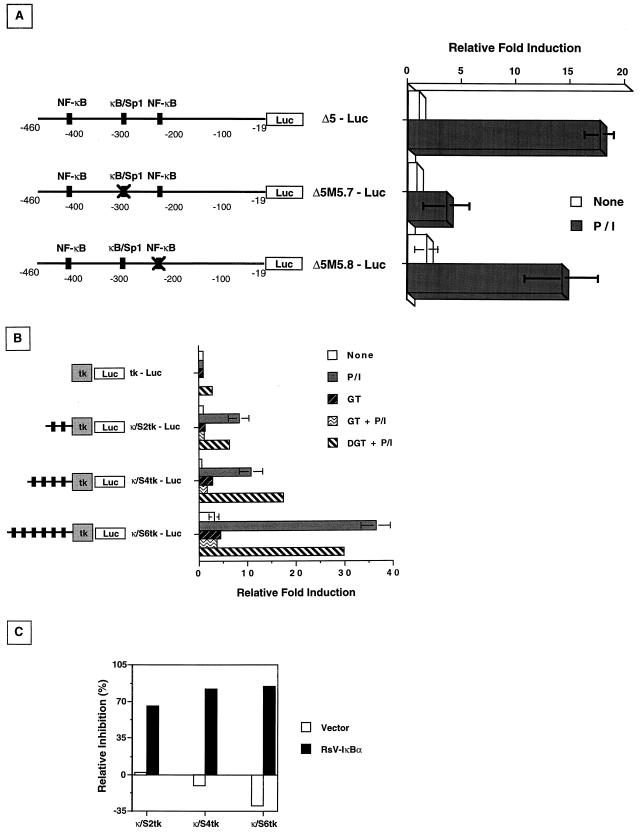
While previous experiments indicated that NF-κB translocation was essential for fas promoter induction, the functional significance of the κB element at positions −295 to −286 remained to be tested. To determine the specific contribution of this κB-Sp1 element, oligonucleotides were synthesized with the composite site altered (as underlined) (GGGCGTTCCC to TTTCGTTCCC) and with a mutagenized Δ5 fas reporter construct generated by PCR. This construct was designated Δ5M5.7-Luc. As a control, a second Δ5 reporter construct containing the κB site at −262 to −253 was also mutagenized (Δ5M5.8; GGGGCTTTTC to GCCYCTTTTC) and used in reporter assays. Jurkat cells transiently transfected with the wild-type Δ5-Luc construct, Δ5M5.7-Luc, or the Δ5M5.8-Luc control were activated with P/I, and reporter activity was measured. Similar to previous experiments with the Δ5 deletion constructs (Fig. 3B), both wild-type Δ5-Luc and Δ5M5.8-Luc showed an approximate 15-fold induction following activation (Fig. 6). In contrast, Δ5M5.7-Luc reporter activity increased only threefold after P/I treatment. These results document the functional contribution of the κB-Sp1 site at positions −295 to −286, but not the consensus NF-κB site at −262 to −243, in activation-dependent fas promoter induction. Interestingly, while Δ5M5.7-Luc induction was significantly suppressed compared to that of the wild-type Δ5-Luc construct (79.6% ± 10.61%, respectively) P/I inducibility was not entirely abolished. Such findings suggest that an additional site or sites, perhaps in the region from −19 to −240 (compare Fig. 3B to Fig. 6), may be required for maximal fas promoter induction during T-cell activation.
FIG. 6.
The κB-Sp1 enhancer element is required for P/I-inducible Δ5-Luc reporter activity. (A) Nucleotide mutagenesis of Δ5-Luc at either the κB-Sp1 element (GGG→CCC; −295 to −293 [Δ5M5.7-Luc]) or the downstream NF-κB element (GGG→CCY; −252 to −249 [Δ5M5.8-Luc]) was introduced by PCR. Wild-type or mutagenized constructs were cotransfected into Jurkat cells with 10 ng of pRL-tk, and the cells were activated with P/I for 8 h prior to being harvested for extract preparation and luciferase activity measurements. The values shown are averages of three independent experiments ± standard deviations and are expressed as the relative fold increase over untreated, normalized Δ5-Luc luciferase activity. (B) Concatemer constructs containing either none (tk-Luc), two (κ/S2tk-Luc), four (κ/S4tk-Luc), or six (κ/S6tk-Luc) copies of the κB-Sp1 element (−295 to −286) were juxtaposed upstream of a thymidine kinase-driven luciferase reporter and cotransfected into Jurkat cells as mentioned above before activation with P/I and luciferase measurements. Inhibition of NF-κB activity were analyzed by preincubation of transfected cells with either gliotoxin (GT [1 μg/ml]) or bis-dethio-bis(methylthio)gliotoxin (DGT [1 μg/ml]) as described in the legend to Fig. 5 prior to activation with P/I. Relative fold induction is calculated against the untreated, normalized tk-Luc reporter luciferase activity. (C) Specific inhibition of NF-κB-dependent upregulation of the κB-Sp1 motif concatemers by coexpression of RsV-IκBα. Multimerized constructs were cotransfected with 5 μg of either RsV-IκBα or vector-only expression plasmids before activation with P/I as described above. Extracts were taken for luciferase measurements, and relative inhibition is calculated as the percentage of P/I-induced luciferase activity of the corresponding reporter constructs.
To further verify the transcriptional contribution of the −295 to −286 κB-Sp1 composite site, concatemer constructs containing multiple copies of this element upstream of a thymidine kinase promoter-driven luciferase reporter were generated (Fig. 2B). Constructs containing none, two, four, or six copies of the κB-Sp1 element were transiently transfected into Jurkat cells, and the reporter activity was measured after P/I activation (Fig. 6B). As expected, the inducibility of the concatemer-reporter constructs increased with the number of the κB-Sp1 elements multimerized, with six copies showing a >35-fold specific induction over that of the thymidine kinase promoter alone. As observed previously with the Δ5-Luc construct (Fig. 5), P/I inducibility of the κB-Sp1 concatemers was specifically eliminated by inhibitors of NF-κB translocation. Figures 6B and C show the effects of gliotoxin and IκB-α overexpression on P/I inducibility of the κB-Sp1-containing constructs. Furthermore, in cells transfected with six copies of the κB-Sp1 element (κ/S6tk-Luc), basal reporter activity was fourfold higher in unstimulated Jurkat cells than that of the tk-Luc reporter alone, suggesting a possible enhancement of constitutive fas promoter regulation by multiple Sp1 sites. Taken together, these experiments confirm the functional significance of the κB-Sp1 site at −295 to −286 in the control of activation-driven fas promoter induction.
DISCUSSION
In recent years, much attention has been focused on how the immune response is downregulated. Because self-reactive T cells are ineffectively deleted by negative selection in the thymus, the preservation of immune homeostasis requires self-regulatory mechanisms for the maintenance of tolerance and the control of excessive T-cell proliferation to foreign antigens in the periphery. Apoptosis is responsible for maintaining T-cell homeostasis by at least two mechanisms-AICD that occurs after TCR ligation by antigen and passive cell death that occurs in the absence of antigen or cytokine stimulation (1, 14, 26, 63). The Fas-FasL-mediated pathway of apoptosis appears to be central in the regulation of peripheral tolerance, because mutations in either the receptor or the ligand can induce lymphadenopathy and autoimmune disease in mice and humans caused by defects in AICD and peripheral T- and B-cell deletion (18, 60, 65). Fas is constitutively expressed on most cultured T cells, whereas freshly isolated, naïve (CD45RO+) peripheral blood T cells have little to no surface Fas. Early after T-cell activation, Fas is rapidly upregulated, but these Fas-positive cells remain resistant to Fas-induced apoptosis (26, 37, 41, 44). With the progression of an immune response, Fas-positive T cells acquire sensitivity to Fas-induced killing (37, 44). Although Fas upregulation is known to be a prerequisite for the elicitation of Fas-FasL-mediated AICD (1, 14, 26), the molecular mechanisms underlying this process have not been well investigated. In this report, we have identified the immediate transcriptional events and cis-acting elements required for activation-induced fas expression in the human Jurkat cell line. Using transient transfection reporter assays, we have localized a 47-bp sequence (−306 to −260) upstream of the translational start site that confers P/I inducibility of the fas promoter (Fig. 3B). Within this region, a critical 10-bp enhancer (−295 to −286) was shown to bind Sp1 during basal transcription expression and NF-κB p50-p65 heterodimers after P/I activation (Fig. 4). When binding of NF-κB was inhibited by the blockade of κB translocation or mutagenesis of the κB-Sp1 site, activation-dependent Fas upregulation was lost. Our findings provide the first molecular evidence for differential control of constitutive and inducible Fas expression and suggest an intervention strategy for inducible Fas upregulation.
Sp1 is a zinc-finger transcription factor constitutively expressed in a variety of cell types that binds the GC-rich consensus sequences (KGGGCGGRRY or KRGGCGKRRY) present in many cellular and viral promoters. The N terminus of Sp1 contains glutamine- and serine/threonine-rich domains required for transactivation, while the C terminus is involved in synergistic activation and interaction with other transcription factors (12). Although Sp1 has been shown to independently initiate transcription in TATA-less promoters (reviewed in reference 3), it can also form homo- or heteromultimeric complexes to transactivate a number of disparate promoters, including interleukin 2, VCAM-1, and the HIV LTR (42, 50, 56). Several lines of circumstantial evidence suggest that Sp1 may be involved in basal fas transcription. First, sequences proximal to the fas translational start site have a relatively high GC content (61% between positions −590 and −1) and an absence of conventional TATA and CAAT boxes (5), typical of a subclass of polymerase II-dependent promoters controlled by the interactions of the glutamine-rich activation domains of Sp1 with TFIID components (9). Second, the TATA-less, GC-rich area of the fas promoter (−590 to −1) has been shown to be functional in analyses of transcriptional start sites (5). Third, the TATA-less Δ5 fas promoter construct was sufficient in driving basal transcription. Fourth, Sp1 bound to the κB-Sp1 site at positions −295 to −286 in resting, but not activated, Jurkat cell extracts (data not shown). Finally, multimers of the κB-Sp1 element at positions −295 to −286 showed increase basal transcription in Jurkat cells (Fig. 6B). Although Sp1 may be involved in the control of fas transcription in resting Jurkat cells, it is likely that multiple Sp1 binding elements are required for optimal basal levels of Fas expression. In this regard, site-directed mutagenesis of the κB-Sp1 site at −295 to −286 alone inhibited basal fas transcription by ∼20%, while disruption of a second consensus Sp1 site at −140 to −131 repressed basal fas transcription by greater than 50% (data not shown).
Nuclear translocation of the NF-κB–Rel transcription factor family occurs in T lymphocytes in response to TCR engagement (reviewed in reference 20). The prototypic form of NF-κB is a heterodimer complex containing NF-κB1–p50 or NF-κB2–p52 in combination with a transactivating subunit such as c-Rel or RelA (p65) (reviewed in reference 64). Each NF-κB–Rel family member contains a conserved N-terminal region responsible for decameric DNA binding, dimerization, and IκB interaction; NF-κB-responsive genes are transactivated by the C-terminal domain of dimeric NF-κB–Rel family members. In resting T lymphocytes, various NF-κB subunits are sequestered in the cytoplasm by virtue of their association with by the IκB family of inhibitors (IκBα, IκBβ, IκBɛ, IκBγ, and bcl-3), which masks their nuclear localization signal (reviewed in reference 64). During T-cell activation, intracellular signaling triggers multiple kinase pathways that converge in the activation of the IκB kinase complex (IKK); IKK then phosphorylates IκBα or IκBβ at conserved serines, resulting in the targeting of the κB inhibitor for ubiquitin-proteasome-dependent destruction and permitting the nuclear translocation of NF-κB (10). Binding sites for NF-κB have been found in numerous genes involved in effector T-cell function, including those regulating cytokine production, cell adhesion molecules, and apoptosis. In this report, we have demonstrated that NF-κB p50-p65 mediates inducible fas promoter activation in T lymphocytes at −460 to −240 and confers the majority of P/I responsiveness at a single, noncanonical κB-Sp1-like motif (GGGCGTTCCC) located at −295 to −286. Interestingly, although two consensus NF-κB sites as well as GAS, NF-ATp, AP-2, and Egr-1 sites are also present within this region, the contribution of these elements to either basal or inducible fas promoter induction in Jurkat cells was insignificant (Fig. 3). Notably, however, such motifs may be functional in other cell types, because Egr-1 binding at −371 to −338 has been reported to repress fas transcription during B-cell activation (16).
The mutually exclusive binding of either Sp1 or NF-κB to the novel composite κB-Sp1 site at −295 to −286 in the fas promoter differs from the previously described cross-coupling of NF-κB with the transcription factor AP-1 or HMG I(Y) (58, 62). Our observations are not unexpected, however, in light of reports documenting the functional interference of Sp1 and NF-κB at selected NF-κB binding sites (21) and crystallographic data showing that Sp1 and p50 homodimers both interact with DNA at the major groove (19, 38, 49). In this report, we provide multiple lines of evidence that NF-κB p50-p65 heterodimers displace Sp1 at the composite κB-Sp1 site to drive activation-dependent interaction of the fas promoter. First, P/I-dependent promoter induction requires the sequences between −306 and −260 containing the κB-Sp1 site (Fig. 3B). Second, NF-κB p50-65 heterodimers bound to this region of the fas promoter only during activation (Fig. 4A and B). Third, recombinant NF-κB p50 displaced Sp1 binding to the κB-Sp1 site in a dose-dependent manner (Fig. 4C). Fourth, recruitment of NF-κB to the fas promoter region at −295 to −286 was required for activation-dependent fas promoter induction, but not for basal activity (Fig. 5). Fifth, specific mutagenesis of the NF-κB site at −295 to −286 significantly diminished P/I-dependent fas promoter induction (Fig. 6A). Finally, multimers of the κB-Sp1 site juxtaposed to a minimal thymidine kinase promoter were functionally responsive to P/I stimulation in a dose-dependent manner (Fig. 6B). In further support of our observations that NF-κB is critically required for activation-induced fas upregulation, several inducers of NF-κB, including UV irradiation, hypoxia, cytokines, and TNF, have been reported to rapidly upregulate Fas (24, 44, 61). It is interesting to speculate that Sp1, bound to the κB-Sp1 site during basal transcription, may facilitate rapid activation-induced fas promoter responsiveness mediated by NF-κB translocation as a consequence of its role in DNA bending (57).
If Fas is similarly regulated in Jurkat and fresh T cells, our findings may be relevant to HIV pathogenesis, in which increased Fas expression and sensitivity to FasL-induced apoptosis have been reported during infection and disease progression (2, 4, 27). Interestingly, both HIV-1 binding and gp120-mediated CD4 cross-linking can induce NF-κB (8). Because NF-κB can drive both HIV-1 expression (50) and fas upregulation, inhibition of NF-κB may be one major strategy to suppress both viral replication (31) and premature execution of the AICD pathway resulting in T-cell depletion. Indeed, inhibitors of NF-κB may also effectively repress Fas-driven apoptosis in response to metabolic stress, genotoxic insults, and viral infections.
Although our studies clearly demonstrate a critical role for NF-κB in fas upregulation, other transcriptional or posttranscriptional processes may also be operative, depending upon the cell type and cellular microenvironment. For example, the proline-, glutamine-, and histidine-rich protein TDAG51 has been reported to be critical for Fas mRNA induction after TCR engagement (47). Our laboratory has demonstrated that wild-type p53 can upregulate fas expression in both hematopoietic and nonhematopoietic cells (45). Taken with our current observations, it is plausible that p53 may act with NF-κB to increase fas transcription, particularly under conditions of metabolic and genotoxic stress. mRNA stability may also regulate fas expression. In this regard, the 3′ untranslated region of fas contains several AU-rich elements reported to induce selective mRNA degradation (29, 55). Given the potent biological function of the Fas protein, multiple levels of gene regulation involving both cis-acting transcription and mRNA stability would not be unexpected.
In summary, our studies have identified a novel κB-Sp1 enhancer motif in the fas promoter in which NF-κB p50-p65 dimers bind to drive activation-dependent Fas upregulation in T cells. The critical involvement of NF-κB further underscores the importance of this transcription factor in immunohomeostasis and prompts consideration for strategies aimed toward its manipulation in various pathophysiological states involving inappropriate Fas and FasL interactions, including malignant disease, autoimmunity, and AIDS.
ACKNOWLEDGMENTS
This work was supported in part by NIH Cancer Center Support Core grant CA16672, NIH predoctoral fellowship (H.C.), and American Cancer Society grant CIM 88929 (L.B.O.-S.).
We thank Karen Ramirez for providing technical expertise with flow cytometric analyses and Kathleen McAveney and David McConkey for critical reading of the manuscript.
REFERENCES
- 1.Alderson M R, Tough T W, Davis-Smith T, Braddy S, Falk B, Schooley K A, Goodwin R G, Smith C A, Ramsdell F, Lynch D H. Fas ligand mediates activation-induced cell death in human T lymphocytes. J Exp Med. 1995;181:71–77. doi: 10.1084/jem.181.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aries S, Scgaaf B, Muller C, Dennin R, Dalhoff K. Fas (CD95) expression of CD4+ T cells from HIV-infected patients increases with disease progression. J Mol Med. 1995;73:591–593. doi: 10.1007/BF00196352. [DOI] [PubMed] [Google Scholar]
- 3.Azizkhan J C, Jensen D E, Pierce A J, Wade M. Transcription from TATA-less promoters: dihydrofolate reductase as a model. Crit Rev Eukaryot Gene Expr. 1993;3:229–254. [PubMed] [Google Scholar]
- 4.Banda N K, Bernier J, Kurahara D K, Kurrle R, Haligwood N, Sekaly R P, Finkel T H. Crosslinking CD4 by human immunodeficiency virus gp120 primes T cells for activation-induced apoptosis. J Exp Med. 1992;176:1099–1106. doi: 10.1084/jem.176.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrmann I, Walczak H, Krammer P H. Structure of the human APO-1 gene. Eur J Immunol. 1994;24:3057–3062. doi: 10.1002/eji.1830241221. [DOI] [PubMed] [Google Scholar]
- 6.Boise L H, Minn A J, Noel P J, June C H, Accavitti M A, Lindsten T, Thompson C B. CD28 costimulation can promote T cell survival by enhancing the expression of Bcl-xL. Immunity. 1995;3:87–98. doi: 10.1016/1074-7613(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 7.Boldin M P, Goncharou T M G, Goltsev Y V, Wallach D. Involvement of MACH1, a novel MORT1/FADD interacting protease, in Fas/APO-1 and TNF receptor-induced cell death. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 8.Briant L, Coudronniere N, Hebmann-Robert V, Benkirane M, Devaux C. Binding of HIV-1 virions or gp120-anti-gp120 immune complexes to HIV-1-infected quiescent peripheral blood mononuclear cells reveals latent infection. J Immunol. 1996;156:3994–4004. [PubMed] [Google Scholar]
- 9.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Hagler J P, Melandri V F, Scherer B D, Maniatis T. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 11.Chodish L A. DNA-protein interactions. In: Ausubel F M, et al., editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1989. pp. 12.2.1–12.2.10. [Google Scholar]
- 12.Courey A J, Holtzman D A, Jackson S P, Tjian R. Synergistic activation by the glutamine-rich domains of human transcription factor Sp1. Cell. 1989;59:827–836. doi: 10.1016/0092-8674(89)90606-5. [DOI] [PubMed] [Google Scholar]
- 13.Debatin K-M, Süss D, Krammer P H. Differential expression of APO-1 on human thymocytes: implications for negative selection? Eur J Immunol. 1994;24:753–758. doi: 10.1002/eji.1830240339. [DOI] [PubMed] [Google Scholar]
- 14.Dhein J, Walczak H, Bäumler C, Debatin K M, Krammer P H. Autocrine T-cell suicide mediated by APO-1/(Fas/CD95) Nature. 1995;373:438–441. doi: 10.1038/373438a0. [DOI] [PubMed] [Google Scholar]
- 15.Dignam J, Lebovitz R, Roeder R. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinkel A, Aicher W K, Haas C, Zipfel P F, Peter H-H, Eibel H. Transcription factor Egr-1 activity down-regulates Fas and CD23 expression in B cells. J Immunol. 1997;159:2678–2684. [PubMed] [Google Scholar]
- 17.Drappa J, Brot N, Elkon K B. The Fas protein is expressed at high levels on CD4+CD8+ thymocytes and activated mature lymphocytes in normal mice but not in the lupus-prone strain, MRL lpr/lpr. Proc Natl Acad Sci USA. 1993;90:10340–10344. doi: 10.1073/pnas.90.21.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher G H, Rosenberg F J, Straus S E, Dale J K, Middleton L A, Lin A Y, Strober W, Lenardo M J, Puck J M. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell. 1995;81:935–946. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh G, Duyne G V, Ghosh S, Sigler P B. Structure of NF-κB p50 homodimer bound to a κB site. Nature. 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh S, May M J, Kopp E B. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 21.Hirano F, Tanaka H, Hirano Y, Hiramoto M, Handa H, Makino I, Scheidereit C. Functional interference of Sp1 and NF-κB through the same DNA binding site. Mol Cell Biol. 1998;18:1266–1274. doi: 10.1128/mcb.18.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer J-L, Schröter M, Burns K, Mattmann C, Rimoldi D, French L E, Tschopp J. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 23.Itoh N, Tsujimoto Y, Nagata S. Effect of bcl-2 on Fas antigen-mediated cell death. J Immunol. 1993;151:621–627. [PubMed] [Google Scholar]
- 24.Itoh N, Yonehara S, Ishii A, Yonehara M, Mizushima S, Sameshima M, Hase A, Seto Y, Nagata S. The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell. 1991;66:233–243. doi: 10.1016/0092-8674(91)90614-5. [DOI] [PubMed] [Google Scholar]
- 25.Jäättelä M, Benedict M, Tewari M, Shayman J A, Dixit V M. Bcl-x and bcl-2 inhibit TNF and Fas-induced apoptosis and activation of phospholipase A2 in breast carcinoma cells. Oncogene. 1995;10:2297–2305. [PubMed] [Google Scholar]
- 26.Ju S T, Panka D J, Cul H, Ettinger R, El-Khatib M, Sherr D H, Stanger B Z, Marshak-Rothstein A. Fas (CD95)/FasL interactions required for programmed cell death after T-cell activation. Nature. 1995;373:444–448. doi: 10.1038/373444a0. [DOI] [PubMed] [Google Scholar]
- 27.Katsikis P D, Wunderlich E S, Smith C A, Herzenberg L A, Herzenberg L A. Fas antigen stimulation induces marked apoptosis of T lymphocytes in human immunodeficiency virus-infected individuals. J Exp Med. 1995;181:2029–2036. doi: 10.1084/jem.181.6.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemper O, Wallach D. Cloning and partial characterization of the promoter for the human p55 tumor necrosis factor (TNF) receptor. Gene. 1993;134:209–216. doi: 10.1016/0378-1119(93)90095-k. [DOI] [PubMed] [Google Scholar]
- 29.Kruys V, Marinx O, Shaw G, Deschamps J, Huez G. Translational blockade imposed by cytokine-derived UA-rich sequences. Science. 1989;245:852–855. doi: 10.1126/science.2672333. [DOI] [PubMed] [Google Scholar]
- 30.Kunsch C, Ruben S M, Rosen C A. Selection of optimal κB/Rel DNA-binding motifs: interaction of both subunits of NF-κB with DNA is required for transcriptional activation. Mol Cell Biol. 1992;12:4412–4421. doi: 10.1128/mcb.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon H, Pelletier N, DeLuca C, Genin P, Cisternas S, Lin R, Wainberg M A, Hiscott J. Inducible expression of IκBα repressor mutants interferes with NF-κB activity and HIV-1 replication in Jurkat T cells. J Biol Chem. 1998;273:7431–7440. doi: 10.1074/jbc.273.13.7431. [DOI] [PubMed] [Google Scholar]
- 32.Lai J-H, Horvath G, Subleski J, Bruder J, Ghosh P, Tan T-H. RelA is a potent transcriptional activator of the CD28 response element within the interleukin 2 promoter. Mol Cell Biol. 1995;15:4260–4271. doi: 10.1128/mcb.15.8.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leithäuser F, Dhein J, Mechtersheimer G, Koretz K, Brüderlein S, Henne C, Schmidt A, Debatin K-M, Krammer P H, Möller P. Constitutive and induced expression of APO-1, a new member of the nerve growth factor/tumor necrosis factor receptor superfamily, in normal and neoplastic cells. Lab Investig. 1993;69:415–429. [PubMed] [Google Scholar]
- 34.Leverkus M, Yaar M, Gilchrest B A. Fas/Fas ligand interaction contributes to UV-induced apoptosis in human keratinocytes. Exp Cell Res. 1997;232:255–262. doi: 10.1006/excr.1997.3514. [DOI] [PubMed] [Google Scholar]
- 35.Locker J. Transcription factors—essential data. Chichester, West Sussex, United Kingdom: John Wiley & Sons; 1996. [Google Scholar]
- 36.Marienfeld R, Neumann M, Chuvpilo S, Escher C, Kneitz B, Avots A, Schimpl A, Serfling E. Cyclosporin A interferes with the inducible degradation of NF-κB inhibitors, but not with the processing of p105/NFκB1 in T cells. Eur J Immunol. 1997;27:1601–1609. doi: 10.1002/eji.1830270703. [DOI] [PubMed] [Google Scholar]
- 37.Miyawaki T, Uehara T, Nibu R, Tsuji T, Yachie A, Yonehara S, Taniguchi N. Differential expression of apoptosis-related Fas antigen on lymphocyte subpopulations in human peripheral blood. J Immunol. 1992;149:3753–3758. [PubMed] [Google Scholar]
- 38.Müller C W, Rey F A, Sodeoka M, Verdine G L, Harrison S C. Structure of the NF-κB p50 homodimer bound to DNA. Nature. 1995;373:311–317. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- 39.Müller M, Strand S, Hug H, Heinemann E-M, Walczak H, Hoffmann W J, Stremmel W, Krammer P H, Galle P R. Drug-induced apoptosis in hepatoma cells is mediated by the CD95 (APO-1/Fas) receptor/ligand system and involves activation of wild-type p53. J Clin Investig. 1997;99:403–413. doi: 10.1172/JCI119174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. FLICE, a novel FADD-homologous ICE/Ced-3-like protease, is recruited to the CD95 (Fas/APO-1) death inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 41.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 42.Neish A S, Khachigian L M, Park A, Baichwal V R, Collins T. Sp1 is a component of the cytokine-inducible enhancer in the promoter of vascular cell adhesion molecule-1. J Biol Chem. 1995;270:28903–28909. doi: 10.1074/jbc.270.48.28903. [DOI] [PubMed] [Google Scholar]
- 43.Owen-Schaub L B, Radinsky R, Kruzel E, Berry K, Yonehara S. Anti-Fas on nonhematopoietic tumors: levels of Fas/APO-1 and bcl-2 are not predictive of biological responsiveness. Cancer Res. 1994;54:1580–1586. [PubMed] [Google Scholar]
- 44.Owen-Schaub L B, Yonehara S, Crump W L, Grimm E A. DNA fragmentation and cell death is selectively triggered in activated human lymphocytes by Fas antigen engagement. Cell Immunol. 1992;140:197–205. doi: 10.1016/0008-8749(92)90187-t. [DOI] [PubMed] [Google Scholar]
- 45.Owen-Schaub L B, Zhang W, Cusack J C, Angelo L S, Santee S M, Fujiwara T, Roth J A, Deisseroth A B, Zhang W-W, Kruzel E, Radinsky R. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol Cell Biol. 1995;15:3032–3040. doi: 10.1128/mcb.15.6.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pahl H L, Kraub B, Schulze-Osthoff K, Decker T, Traenckner E B-M, Vogt M, Myers C, Parks T, Warring P, Mühlbacher A, Czernilofsky A P, Baeuerle P A. The immunosuppressive fungal metabolite gliotoxin specifically inhibits transcription factor NF-κB. J Exp Med. 1996;183:1829–1840. doi: 10.1084/jem.183.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park C G, Lee S Y, Kandala G, Lee S Y, Choi Y. A novel gene product that couples TCR signaling to Fas(CD95) expression in activation-induced cell death. Immunity. 1996;4:583–591. doi: 10.1016/s1074-7613(00)80484-7. [DOI] [PubMed] [Google Scholar]
- 48.Parry G C, Mackman N. A set of inducible genes expressed by activated human monocytic and endothelial cells contain κB-like sites that specifically bind c-Rel-p65 heterodimers. J Biol Chem. 1994;269:20823–20825. [PubMed] [Google Scholar]
- 49.Pavletich N P, Pabo C O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 50.Perkins N D, Edwards N L, Duckett C S, Agranoff A B, Schmid R M, Nabel G J. A cooperative interaction between NF-κB and Sp1 is required for HIV-1 enhancer activation. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothe J, Bluethmann H, Gentz R, Lesslauer W, Steinmetz M. Genomic organization and promoter function of the murine tumor necrosis factor receptor β gene. Mol Immunol. 1993;30:165–175. doi: 10.1016/0161-5890(93)90088-s. [DOI] [PubMed] [Google Scholar]
- 52.Santee S M, Owen-Schaub L B. Human tumor necrosis factor receptor p75/80 (CD120b) gene structure and promoter characterization. J Biol Chem. 1996;271:21151–21159. doi: 10.1074/jbc.271.35.21151. [DOI] [PubMed] [Google Scholar]
- 53.Sato T, Irie S, Kitada S, Reed J C. FAP-1: A protein tyrosine phosphatase that associates with Fas. Science. 1995;268:411–415. doi: 10.1126/science.7536343. [DOI] [PubMed] [Google Scholar]
- 54.Sehgal A, Patil N, Chao M. A constitutive promoter directs expression of the nerve growth factor receptor gene. Mol Cell Biol. 1988;8:3160–3167. doi: 10.1128/mcb.8.8.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 56.Sherka C, Decker E L, Zipfel P F. A regulatory element in the human interleukin 2 gene promoter is a binding site for the zinc finger proteins Sp1 and EGR-1. J Biol Chem. 1995;270:22500–22506. doi: 10.1074/jbc.270.38.22500. [DOI] [PubMed] [Google Scholar]
- 57.Sjottem E, Andersen C, Johansen T. Structural and functional analyses of DNA bending induced by Sp1 family transcription factors. J Mol Biol. 1997;267:490–504. doi: 10.1006/jmbi.1997.0893. [DOI] [PubMed] [Google Scholar]
- 58.Stein B, Baldwin A S, Jr, Ballard D W, Greene W C, Angel P, Herrlich P. Cross-coupling of the NF-κB p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J. 1993;12:3879–3891. doi: 10.1002/j.1460-2075.1993.tb06066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki I, Fink P J. Maximal proliferation of cytotoxic T lymphocytes requires reverse signaling through Fas ligand. J Exp Med. 1998;187:123–128. doi: 10.1084/jem.187.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takahashi T, Tanaka M, Brannan C I, Jenkins N A, Copeland N G, Suda T, Nagata S. Generalized lymphoproliferative disease in mice, caused by a point mutation in the Fas ligand. Cell. 1994;76:969–976. doi: 10.1016/0092-8674(94)90375-1. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka M, Ito H, Adachi S, Akimoto H, Nishikawa T, Kasajima T, Marumo F, Hiroe M. Hypoxia induces apoptosis with enhanced expression of Fas antigen messenger RNA in cultured neonatal rat cardiomyocytes. Circ Res. 1994;75:426–433. doi: 10.1161/01.res.75.3.426. [DOI] [PubMed] [Google Scholar]
- 62.Thanos D, Maniatis T. The high mobility group protein HMG I(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 63.van Parijs L, Ibraghimov A, Abbas A K. The roles of costimulation and Fas in T-cell apoptosis and peripheral tolerance. Immunity. 1996;4:321–326. doi: 10.1016/s1074-7613(00)80440-9. [DOI] [PubMed] [Google Scholar]
- 64.Verma I M, Stevenson J K, Schwartz E M, Van Antwerp D, Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe-Fukunaga R, Brannan C I, Copeland N G, Jenkins N A, Nagata S. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature. 1992;356:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]