Abstract
Meiosis in Saccharomyces cerevisiae is followed by encapsulation of haploid nuclei within multilayered spore walls. Formation of this spore-specific wall requires the coordinated activity of enzymes involved in the biosynthesis of its components. Completion of late events in the sporulation program, leading to spore wall formation, requires the SWM1 gene. SWM1 is expressed at low levels during vegetative growth but its transcription is strongly induced under sporulating conditions, with kinetics similar to those of middle sporulation-specific genes. Homozygous swm1Δ diploids proceed normally through both meiotic divisions but fail to produce mature asci. Consistent with this finding, swm1Δ mutant asci display enhanced sensitivity to enzymatic digestion and heat shock. Deletion of SWM1 specifically affects the expression of mid-late and late sporulation-specific genes. All of the phenotypes observed are similar to those found for the deletion of SPS1 or SMK1, two putative components of a sporulation-specific MAP kinase cascade. However, epistasis analyses indicate that Swm1p does not form part of the Sps1p-Smk1p-MAP kinase pathway. We propose that Swm1p, a nuclear protein, would participate in a different signal transduction pathway that is also required for the coordination of the biochemical and morphological events occurring during the last phase of the sporulation program.
Sporulation in the yeast Saccharomyces cerevisiae involves a regulated program of cell development that includes premeiotic DNA replication, the two meiotic divisions, and the encapsulation of haploid nuclei within spore walls (15). Sporulation is initiated when MATa/MATα cells are starved of nitrogen in the presence of a nonfermentable carbon source, such as acetate, and is under the control of master regulatory genes that include IME1, IME2, and RME1 (31, 36). In yeast cells, the nuclear envelope remains intact throughout meiosis and the intranuclear segregation of chromosomes generates four bulges in the nucleus, each next to the respective spindle pole body (for a review, see reference 8).
Progression through the sporulation program depends on the sequential expression of at least four temporally distinct groups of sporulation-specific genes, classified as early, middle, mid-late, and late (31, 36). Sporulation in S. cerevisiae thus provides a model system to study the temporal control of gene expression during development. The mid-late and late genes are activated at the time of the meiotic divisions and spore formation, and this activation is accompanied and followed by morphological changes that result in spore formation. Such changes include (i) the formation of a flattened membrane sac (the “prospore wall”) closely apposed to the cytoplasmic faces of the spindle pole bodies, (ii) the extension of the prospore walls along the outer surface of the nuclear envelope, (iii) the separation of the prospore walls from the spindle pole bodies and the nuclear envelope and the movement of cytoplasm and organelles into the intervening space, (iv) the final engulfment of the nuclear lobes (containing the haploid chromosome sets) and associated cytoplasm, and (v) the deposition of spore wall components between the two membranes of the prospore wall (2, 8, 20, 37, 38, 40).
The final differentiated spore wall offers increased protection to stress conditions, compared to the wall of the vegetative cell, and consists of four layers (35). The two inner layers are formed by closely juxtaposed glucans and mannans and often appear as a single layer strongly resembling the vegetative cell wall when examined by electron microscopy (29). The third and fourth layers are spore-specific structures and are composed primarily of chitosan and dityrosine, respectively. They are thought to confer upon the spore wall its protective nature (4, 6, 7). Chitosan, a β-(1,4)-d-glucosamine homopolymer, is produced by deacetylation of nascent chains of chitin, a β-(1,4)-N-acetyl-d-glucosamine homopolymer produced by the action of chitin synthases (14). The deacetylation reaction is catalyzed by the enzyme chitin deacetylase (28), which is encoded by two recently identified sporulation-specific genes (10). The outermost layer of the spore wall is the dityrosine coat, which is composed of an insoluble macromolecule containing a high number of cross-linked tyrosine residues that is synthesized in a two-step reaction catalyzed by the products of the sporulation-specific genes DIT1 and DIT2 (5).
In this report we describe the characterization of a novel yeast gene named SWM1 (spore wall maturation) that is transcribed during vegetative growth but whose expression is strongly induced during the sporulation process with kinetics similar to those of middle sporulation-specific genes. SWM1 is required for normal activation of mid-late and late genes during sporulation and, in consequence, swm1Δ mutants fail to assemble the spore wall, a phenotype similar to that described for sps1 and smk1 mutants (18, 30).
MATERIALS AND METHODS
Plasmids used.
Plasmid pSU4, used to generate the swm1::hisG deletion allele, was constructed by PCR amplification of the 5′-end (from −291 to −1) and the 3′-end (from +524 to +876) flanking regions with specific oligonucleotide primers and cloning the PCR fragments in Bluescript SK(+) vector (Stratagene), yielding plasmid pSU3. The two fragments were separated by a synthetic BamHI site that was used to clone a 3.8-kb BamHI-BglII fragment carrying the URA3::hisG cassette (1) to generate plasmid pSU4.
Plasmids pSU16 and pSU17, used to construct strains carrying the smk1::kanMX4 and sps1::kanMX4 alleles, respectively, were constructed by using the technique described by Wach (47). For pSU16, specific oligonucleotide primers were used to amplify two fragments, one containing the 5′-flanking region (−393 to −1) and the other the 3′-flanking region (309 nucleotides [nt] downstream from the stop codon). These fragments were fused by recombinant PCR to the kanMX4 cassette (48), and the amplified fragment was cloned in Bluescript SK(+) vector digested with EcoRV, resulting in plasmid pSU16. A similar approach was used to construct plasmid pSU17. In this case, the SPS1 flanking regions amplified with specific oligonucleotide primers were −416 to −1 (5′ end) and from the stop codon to 415 nt downstream from it (3′ end). Plasmids pSU35 and pSU37 contain, respectively, the sps1::HIS3 and sps1::URA3 alleles and were constructed by replacing the kanMX4 cassette with a 1.7-kb BamHI fragment containing the HIS3 gene or a 1.2-kb HindIII fragment carrying the URA3 gene.
Plasmids pJC5, pRN30, and pPS47 contain different DNA fragments of the EXG2-YDR260c (SWM1) chromosomal region (Fig. 1). pJC5 contains a 3.5-kb PstI-SacI fragment that carries the whole EXG2 gene and flanking regions cloned in the YEp352 vector (23). Plasmid pRN30 was constructed by cloning a 3.0-kb BamHI-Sau3A fragment that includes the last 364 amino acids of EXG2 and the whole coding sequence of SWM1 in the high-copy-number vector pCGS44. Plasmid pPS47 was created by PCR amplifying a DNA fragment containing SWM1 and flanking regions (from −291 to +876) with specific oligonucleotide primers that generated ClaI and BamHI sites at the ends of the amplified region and by cloning the resulting fragment in the corresponding sites of vector pRS426 (9).
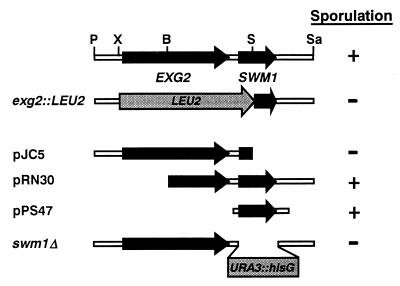
FIG. 1.
Physical and functional map of EXG2 and SWM1 (YDR260c). At the top is the map of the EXG2 and SWM1 chromosomal region, with arrows indicating the coding sequences. Immediately below is a representation of the region replaced in the construction of the exg2::LEU2 allele (YPA69 strain). Constructs used to test for complementation of the sporulation defect of YPA69 are indicated (pJC5, pRN30, and pPS47), with arrows designating the regions of each ORF present in the plasmids. The ability of the strains to sporulate is indicated at the right. The lower line represents the extent of the deletion in the swm1Δ allele, in which the complete coding region was replaced with the URA3 gene flanked by hisG repeats. Restriction sites: B, BamHI; P, PstI; S, SmaI; Sa, Sau3A; X, XhoI.
Plasmid pSU25 contains a GFP-SWM1 fusion in which the green fluorescent protein (GFP) sequence was fused in frame after the fifth codon of SWM1. To construct this plasmid, a NotI site was introduced in frame by recombinant PCR between codons 5 and 6 of the SWM1 open reading frame (ORF) cloned in the centromeric vector Ycplac33 (19). The NotI site was subsequently used to insert a PCR fragment containing the GFP coding sequences (carrying the mutations S65T and V163A) flanked by synthetic NotI sites to generate plasmid pSU25.
Yeast strains and growth conditions.
Table 1 lists the yeast strains used in this study. The wild-type diploid strain YPA24 was constructed by mating strains W303-1A and α131-20 (45). The diploid strain YPA207, homozygous for the swm1Δ::hisG allele, was constructed by transforming strains W303-1A and α131-20 with plasmid pSU4 digested with XhoI and NotI to release a linear fragment containing the deletion cassette. Several independent transformants from each haploid strain were grown on plates containing 5-FOA (3) to select for colonies that had lost the URA3 marker (which is flanked by direct repeats of hisG). Replacement of the SWM1 coding sequence by the hisG fragment was confirmed by Southern blot analysis of genomic DNA obtained from Ura− colonies. Finally, a MATa swm1::hisG strain (YPA203) and a MATα swm1::hisG strain (YPA202) were mated to construct diploid strain YPA207, which is isogenic to the wild-type strain YPA24. Heterozygous diploids YPA205 and YPA206 were constructed by crossing, respectively, YPA202 and YPA203 with the wild-type strains from the opposite mating type.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1A and isogenic derivatives | ||
| W303-1A | MATa ura3 leu2 his3 trp1 ade2 can1 | S. Lindquist |
| YPA203 | MATa ura3 leu2 his3 trp1 ade2 can1 swm1::hisG | This study |
| LS21 | MATa ura3 leu2 his3 trp1 ade2 can1 smk1::kanMX4 | This study |
| LS22 | MATa ura3 leu2 his3 trp1 ade2 can1 sps1::kanMX4 | This study |
| LS27 | MATa ura3 leu2 his3 trp1 ade2 can1 swm1::hisG smk1::kanMX4 | This study |
| LS44 | MATa ura3 leu2 his3 trp1 ade2 can1 sps1::HIS3 smk1::kanMX4 | This study |
| α131-20 and isogenic derivatives | ||
| α131-20 | MATα ade2 ura3 leu1 can1 cyh2 | J. E. Haber |
| YPA202 | MATα ade2 ura3 leu1 can1 cyh2 swm1::hisG | This study |
| LS24 | MATα ade2 ura3 leu1 can1 cyh2 smk1::kanMX4 | This study |
| LS25 | MATα ade2 ura3 leu1 can1 cyh2 sps1::kanMX4 | This study |
| LS30 | MATα ade2 ura3 leu1 can1 cyh2 swm1::hisG smk1::kanMX4 | This study |
| LS42 | MATα ade2 ura3 leu1 can1 cyh2 sps1::URA3 smk1::kanMX4 | This study |
| YPA24 and isogenic derivatives | ||
| YPA24 | MATa/MATα ura3/ura3 leu2/+ his3/+ trp1/+ +/ade2 can1/can1 cyh2/+ | 45 |
| YPA205 | YPA24 but swm1Δ::hisG/SWM1 | This study |
| YPA206 | YPA24 but SWM1/swm1Δ::hisG | This study |
| YPA207 | YPA24 but swm1Δ::hisG/swm1Δ::hisG | This study |
| LS34 | YPA24 but swm1Δ::hisG/swm1Δ::hisG smk1::kanMX4/smk1::kanMX4 | This study |
| LS35 | YPA24 but smk1::kanMX4/smk1::kanMX4 | This study |
| LS36 | YPA24 but sps1::kanMX4/sps1::kanMX4 | This study |
| LS37 | YPA24 but swm1::kanMX4/swm1::kanMX4 sps1::kanMX4/sps1::kanMX4 | This study |
| LS45 | YPA24 but sps1::HIS3/sps1::URA3 smk1::kanMX4/smk1::kanMX4 | This study |
| Other strains | ||
| AP1a/α | MATa/MATα ade1/+ ade2/ade2 ura1/+ his7/+ lys2/+ tyr1/+ +/ura3 +/leu1 +/cyh2 +can1 gal1/+ | J. E. Haber |
| AP1α/α | MATα/MATα ade1/+ ade2/ade2 ura1/+ his7/+ lys2/+ tyr1/+ +/ura3 +/leu1 +/cyh2 +/can1 gal1/+ | J. E. Haber |
| NKY278 | MATa/MATα lys2/lys2 ura3/ura3 ho::LYS2/ho::LYS2 | 30 |
| LAKY30 | NKY278, but smk1-Δ1::URA3/smk1-Δ1::URA3 | 30 |
| YPA69 | MATa/MATα ura3/ura3 leu2/leu2 his3/+ trp1/+ ade2/+ +/ino1 exg2::LEU2/exg2::LEU2 | This study |
To construct isogenic homozygous diploid strains with a deletion of either SMK1 or SPS1, strains W303-1A or α131-20 were transformed with plasmids pSU16 or pSU17 (digested with XhoI and SpeI). After the deletions were verified by Southern blot analysis, the haploid strains were mated to generate the homozygous diploid strains LS35 (smk1::kanMX4/smk1::kanMX4) and LS36 (sps1::kanMX4/sps1::kanMX4). A similar approach was used to construct the double-mutant diploid strains LS34 (swm1::hisG/swm1::hisG smk1::kanMX4/ smk1::kanMX4), LS37 (swm1::hisG/swm1::hisG sps1::kanMX4/sps1::kanMX4), and LS45 (sps1::URA3/sps1::HIS3 smk1::kanMX4/smk1::kanMX4). For the construction of the double mutants, strains YPA202 and YPA203 were transformed with the deletion cassette from pSU16 or pSU17, and strains LS21 and LS24 were transformed with plasmids pSU35 or pSU37, respectively.
Yeast cells were grown vegetatively in YEPD (1% yeast extract, 2% peptone, 2% glucose) or YEPA (0.5% yeast extract, 0.6% yeast nitrogen base, 0.5% peptone, 1% potassium acetate, 1.02% potassium biphtalate [pH 5.5]). Transformants carrying the kanMX4 gene were selected on YEPD plates containing geneticin (200 mg/liter). To induce sporulation, cells were grown in YEPA for at least three generations and harvested at 1 × 107 to 2 × 107 cells/ml, washed twice with sporulation medium (1% potassium acetate), and resuspended at 1.5 × 107 cells/ml in the same sporulation medium supplemented with the appropriate auxotrophic requirements. Asci formation was determined by light microscopy by using phase-contrast optics.
RNA isolation and Northern analysis.
Cells (1.3 × 109) were collected at different time intervals after transfer to sporulation medium, and total RNA was prepared by the method described by Percival-Smith and Segall (44). For Northern blot analysis, 5 μg of RNA was denatured and transferred to Hybond membranes (Amersham) according to the manufacturer’s instructions. The DNA probes used to detect the different transcripts were as follows: SWM1, a 360-bp internal fragment (from +38 to +398) obtained by PCR; SPS1, the 1.02-kb EcoRV-BglII fragment of plasmid pSU9; SMK1, the 1.8-kb SpeI-XhoI fragment of plasmid pSU10 (which includes the coding sequence as well as 5′- and 3′-flanking regions); HOP1, the 1.3-kb BamHI-HindIII fragment of plasmid pNH33-2 (24); SPO12, the 0.45-kb BamHI-EcoRI fragment of plasmid pRE129 (34); SSG1, the 1.2-kb SacI-SalI fragment of plasmid pPS23 (45); DIT1, a 0.7-kb fragment obtained by PCR amplification of the region from −41 to +657; SPS100, a 0.75-kb fragment obtained by PCR amplification of the region from −14 to +745; and ACT1, the 1.7-kb BamHI-HindIII fragment. The probes were radioactively labeled by the random priming method (16). The specificity of the mRNA detected by the SWM1 probe used was confirmed by using a swm1Δ mutant.
Resumption of mitotic growth and resistance to stress.
Resumption of growth was assessed by using cells grown in YEPA to a concentration of 1 × 107 to 2 × 107 cells/ml and then transferred to sporulation medium. At various times during sporulation, samples of wild-type and mutant cells were withdrawn, diluted, plated on YEPD, and then incubated 2 days at 30°C before the numbers of viable colonies were counted. To determine the efficiency of haploidization, we used the appearance of the recessive cycloheximide resistance allele cyh2 by replica plating the germination plates on YEPD plates containing cycloheximide (10 μg/ml). The sensitivity of sporulating cells to heat shock treatment (55°C) or to enzymatic digestion (glusulase) was determined as described by Briza and coworkers (4).
Fluorescence-activated cell sorter (FACS) analysis.
Cells grown on YEPD to the early log phase were fixed and stained with propidium iodide according to the protocol previously described by Hutter and Eipel (27). Flow cytometry analysis was performed on a Becton Dickinson FACSort analyzer.
Microscopy.
For light microscopy, cells were fixed in ethanol and stained with DAPI (4′,6-diamidino-2-phenylindole) as previously described (46). Samples were viewed and photographed as a wet mount with a Zeiss Axiophot microscope equipped for Nomarski optics and epifluorescence. For visualization of the GFP-Swm1 fusion protein, cells that had been in sporulation medium for 10 h were fixed with 3.7% formaldehyde for 1 h, washed briefly with phosphate-buffered saline (PBS), resuspended in PBS buffer containing DAPI, and observed with a Nikon E800 microscope with an FITC-HYQ (Chroma) filter set. Pictures were taken with a Photometrics Sensys CCD camera.
Samples were prepared for electron microscopy according to the protocol described by Wright and Rine (49). In brief, cells from strains YPA24 and YPA207, which had been in sporulation medium for 24 h, were directly fixed by the addition of one-tenth of the volume of 10× fixation solution (10% glutaraldehyde, 2% formaldehyde, 0.4 M potassium phosphate [pH 7]) to the culture medium, pelleted, and then incubated in fixation solution on ice for 30 min. Samples were washed and treated with 1% sodium metaperiodate for 15 min, washed again, and resuspended in 50 mM ammonium phosphate for another 15 min. Fixed cells were embedded in agar, dehydrated through a graded series of ethanol, and then embedded in LR White Resin (London Resin Company Ltd.). Thin sections were stained with uranyl acetate and Reynold’s lead citrate and examined on a Zeiss EM900 electron microscope.
RESULTS
Identification of the SWM1 gene.
In our studies with EXG2, a gene encoding a 1,3-β-glucanase expressed during the S. cerevisiae vegetative cycle, we observed that a diploid strain carrying a homozygous deletion that replaces the whole EXG2 coding region, as well as 466 nt of the 3′ region by the LEU2 gene (exg2::LEU2 in Fig. 1) renders diploid strains unable to sporulate (12). However, this deletion also eliminates the first 58 amino acids of a small ORF located downstream from EXG2, named YDR260c in the yeast genome sequencing project. In order to test whether the sporulation defect was due to the absence of the Exg2 protein or the YDR260c gene product, strain YPA69 (exg2::LEU2) was transformed with plasmids carrying different DNA fragments containing the EXG2-YDR260c region (Fig. 1), and the ability of the cells to sporulate was tested. Our results indicated that plasmid pJC5, carrying the full-length EXG2 gene, was unable to complement the sporulation defect, while plasmids pRN30 (carrying YDR260c and part of the 3′ end of the EXG2 coding sequence) and pPS47 (carrying only YDR260c) restored the ability to form spores in the homozygous diploid mutant strain (Fig. 1). These results thus suggested that the lack of the protein encoded by YDR260c was responsible for the sporulation defect initially attributed by us to the exg2 mutation.
YDR260c, named SWM1 (for spore wall maturation; see below), encodes a small, slightly acidic 170-amino-acid protein with a predicted molecular size of 19,356 Da. Homology searches of databases revealed that the encoded protein is unique in the S. cerevisiae genome and has no significant similarity to any other protein.
SWM1 is required for ascospore development.
To confirm that SWM1 is required for sporulation, a gene replacement that deleted the SWM1 coding region (swm1::URA3::hisG) was performed (Fig. 1). Sporulation and dissection of spores from an swm1Δ/SWM1 heterozygous diploid revealed no differences in growth between swm1Δ and wild-type haploid segregants. SWM1 is therefore not required for vegetative growth or germination.
A swm1Δ homozygous diploid (YPA207) and an otherwise isogenic wild-type strain (YPA24) were transferred to sporulation medium, and their ability to form ascospores was monitored by phase-contrast microscopy. In the wild type, 70% of the cells formed normal ascospores after a 48-h incubation in sporulation medium. By contrast, no examples of mature asci were observed in the swm1Δ mutant, as evidenced by an organized tetrahedral array of birefringent spores. This sporulation defect was rescued by a plasmid carrying the SWM1 gene (pPS47) but not by vector alone (pRS426 [data not shown]). These results thus indicate that SWM1 is required for spore development and that the swm1Δ allele is recessive.
Expression of SWM1 is induced during the sporulation process.
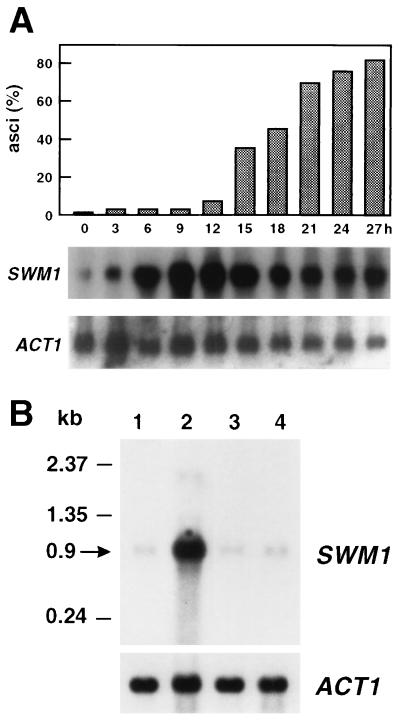
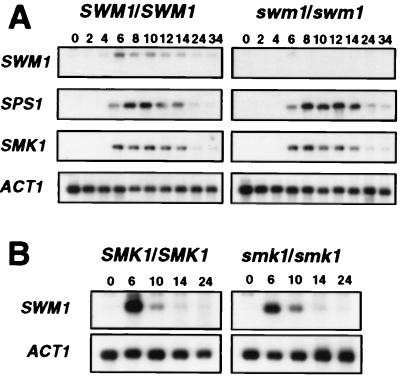
To pinpoint the exact time at which SWM1 serves its role in sporulation, the meiotic time course of SWM1 expression was examined by Northern analysis of wild-type cells at various stages in the differentiation process (Fig. 2A). An RNA molecule of 0.9 kb was detected by using a radiolabeled SWM1 probe, a finding consistent with the predicted size of the gene. SWM1 transcripts were present at the time when the cells were shifted to the nitrogen-deficient medium (0 h), but a sharp rise in the amount of SWM1 RNAs occurred in the middle period of the sporulation process (6 h), with maximal accumulation being observed between 9 and 12 h. SWM1 RNA levels remained high during the time the first mature asci were observed (12 to 15 h) but thereafter slowly declined. This observation indicates that the SWM1 induction profile is similar to that of genes expressed midway through the sporulation process (36), although SWM1 expression is not only restricted to the meiotic process.
FIG. 2.
Expression of the SWM1 gene during the sporulation process. (A) Meiotic time course of SWM1 expression. RNA purified from wild-type cells (strain AP1a/α) at the indicated times after transfer to sporulation medium was hybridized with a radioactively labeled SWM1 probe. The top panel represents the percentage of mature asci at each time point. (B) Expression of SWM1 in MATα/MATα cells. RNA purified from strain AP1a/α at 0 or 10 h (lanes 1 and 2, respectively) after transfer to sporulation medium or from strain AP1α/α at 0 or 10 h (lanes 3 and 4, respectively) was hybridized with the SWM1 probe. In both experiments, the ACT1 probe was used to test for equal loading of RNA in all lanes.
To verify that expression is indeed triggered as a result of the sporulation program and is not a result of nutrient deprivation, RNA samples from strain AP1 a/α and the isogenic nonsporulating derivative AP1 α/α incubated for 10 h in sporulation medium were hybridized with the same SWM1 probe. As shown in Fig. 2B, no transcript accumulation was observed in RNA from strain AP1 α/α, indicating that SWM1 expression is developmentally regulated; it is specifically induced by the sporulation process and not by starvation.
SWM1 is not required for meiotic nuclear divisions.
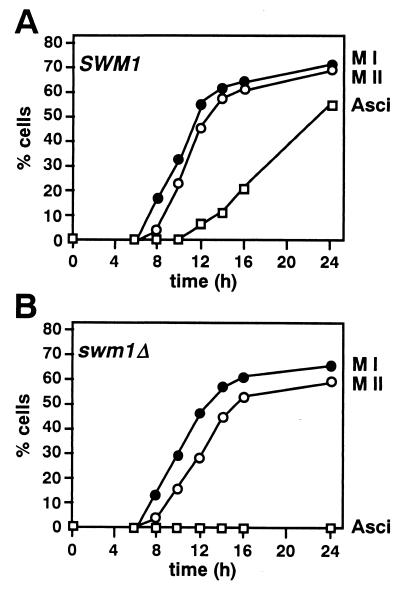
To study the sporulation defect in more detail, the homozygous swm1 diploid YPA207 and the isogenic parental strain YPA24 were transferred to liquid sporulation medium, and the cells were examined by phase-contrast microscopy to monitor asci formation and by fluorescence microscopy with the DNA-specific fluorophore DAPI to follow the meiotic divisions. DAPI staining revealed that both meiosis I and meiosis II occurred at approximately the same time in mutant and wild-type cells, the majority of bi- and tetranucleate cells appearing between 8 and 16 h (Fig. 3). However, we also observed that mutant cells consistently displayed a slight reduction in their ability to complete the second meiotic division (69% in the wild-type cells versus 59% in swm1Δ cells). Despite this, most of the cells that had completed meiosis I went on to complete meiosis II but failed to form spores, as assessed by phase-contrast microscopy.
FIG. 3.
Time course of meiosis in swm1 cells. Samples of cells from sporulating cultures of the wild-type strain YPA24 (A) or the swm1Δ mutant YPA207 (B) were fixed, stained with DAPI, and examined by fluorescence microscopy to determine the percentage of cells that had completed meiosis I (●) or meiosis II (○). Cells that appeared to be binucleate, trinucleate, or tetranucleate by DAPI staining were considered to have completed meiosis I. Cells that appeared to be trinucleate or tetranucleate were considered to have completed meiosis II. Samples were also examined by phase-contrast microscopy to monitor ascus formation (□).
In wild-type cells, birefringent spore walls were first observed after 12 to 14 h in sporulation medium. By contrast, the swm1Δ mutant strain failed to assemble spore walls, even after prolonged incubation in sporulation medium, and some of the mutant cells were unusually large. The DAPI staining profiles in wild-type and mutant cells were very similar after 24 h in the sporulation medium (Fig. 4), but as incubation progressed the DAPI staining in the swm1Δ mutant became more irregular and the number of asci containing diffuse and extranumerary DAPI-staining foci increased (data not shown), perhaps due to the failure to assemble properly the cell wall (see below). Taken together, these results indicate that swm1Δ mutants initiate sporulation and complete meiosis I and meiosis II normally and that the main consequence of the absence of the SWM1 gene product is a defect in proper spore packaging, findings that are in consonance with the expression pattern of the SWM1 gene.
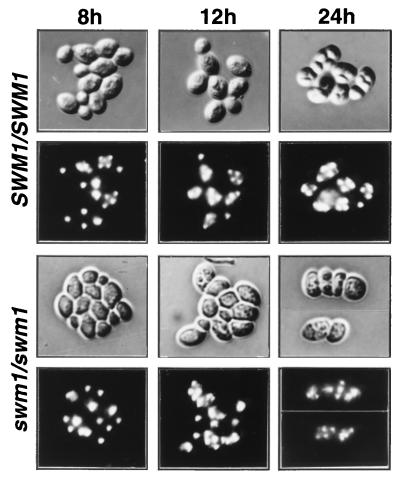
FIG. 4.
Microscopic appearance of wild-type and swm1 mutant asci during the sporulation process. Rows 1 and 3, differential interference contrast microscopy (DIC) photomicrographs; rows 2 and 4, corresponding photomicrographs of DAPI-stained cells. Wild-type cells (strain YPA24) at 8, 12, and 24 h after transfer to sporulation medium present a characteristic tetranuclear organization and birefringent spore walls. swm1Δ cells (strain YPA207) at 8, 12, and 24 h after transfer to sporulation medium do not display discernible spore walls by DIC microscopy. These mutant cells are clearly tetranucleate after 12 h in sporulation medium.
SWM1 is required for assembly of functional spore walls.
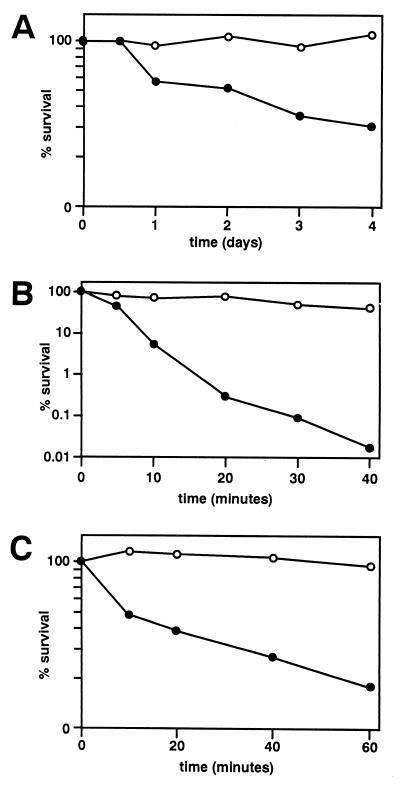
In wild-type cells, birefringent spore walls were first observed after 12 to 14 h in sporulation medium. By contrast, the swm1Δ mutant strain failed to assemble spore walls, even after prolonged incubation in sporulation medium. Microscopic examinations suggested that swm1Δ mutants are blocked in development before the assembly of the spore wall, the structure that confers a high degree of resistance to stress conditions. The viability of the swm1Δ mutant incubated in sporulation medium for 24 h was 56% that of wild-type sporulating cells and 30% that of wild-type sporulating cells when plated 4 days after transfer to the sporulation medium (Fig. 5A). This indicates that swm1 mutant cells are able to resume mitotic growth after incubation in sporulation medium, although their viability decreases after a prolonged incubation in it.
FIG. 5.
Resistance of wild-type and swm1Δ asci to prolonged incubations in sporulation medium, exposure to heat shock, or enzymatic (glusulase) digestion. (A) Wild-type cells from the strain YPA24 (○) or the isogenic swm1Δ strain YPA207 (●) were incubated in sporulation medium for the indicated times (at 30°C) before the plating efficiencies were assayed. (B and C) Wild-type (strain YPA24 [○]) or swm1Δ cells (strain YPA207 [●]) were incubated in sporulation medium for 24 h and then assayed for plating efficiency after exposure to 55°C (B) or to glusulase (C) for the indicated times (see Materials and Methods).
Normal asci, with mature functional spore walls, are resistant to heat shock and enzymatic (glusulase) digestion. To determine whether swm1Δ mutants assemble functional spore walls, wild-type and mutant cells incubated in sporulation medium for 24 h were analyzed for their resistance to heat shock and glusulase treatment. The plating efficiency of swm1 mutants was reduced by at least 4 orders of magnitude after 40 min of incubation at 55°C (41% in wild-type cells versus 0.018% in the mutant) and by at least fivefold after glusulase digestion in comparison with wild-type asci (96 versus 18%) (Fig. 5B and C, respectively). Thus, according to functional criteria, swm1Δ asci are blocked in development before they are able to assemble functional spore walls.
Haploid progeny can be recovered from swm1-arrested cells.
Compartmentalization of the haploid genome is thought to be dependent on the formation of prospore walls around the four nuclear lobes that arise after meiosis. The prospore envelope traps some cytoplasmic material and then serves as the scaffold for the deposition of the spore-wall-specific material (37, 38, 40). Upon germination, the four spores present within the ascal sac are released, and growth of the individual spores gives rise to haploid progeny.
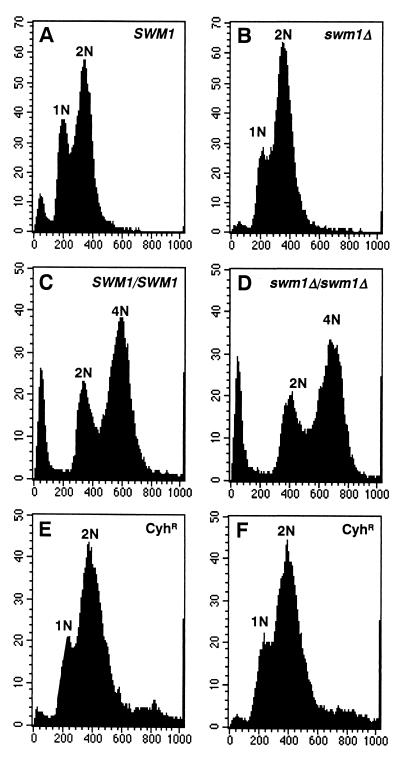
To test for the compartmentalization of meiotic products in swm1 mutant cells, we assessed the ploidy of progeny derived from these cells after incubation in sporulation medium. Our rationale was that if the meiotic nuclei generated in an swm1/swm1 cell did undergo compartmentalization, it would then be possible to recover haploid progeny. First, we tested for the appearance of the recessive drug-resistance marker cyh2 in the progeny derived from the swm1Δ/swm1Δ cyh2/CYH2 diploid strain YPA207. In this experiment, the mutant strain and its isogenic wild type, both of which are sensitive to the drug cycloheximide, were incubated for 24 h in sporulation medium, plated onto rich medium, and then tested for resistance to the drug in the resulting progeny. We found that spores resistant to cycloheximide were present in the progeny derived from both the wild type and from the swm1Δ mutant, although the number of resistant clones was reduced in the progeny derived from the mutants (71% Cyhr colonies in wild type versus 37% Cyhr colonies in swm1Δ).
Cycloheximide-resistant colonies can arise from either haploid segregants obtained by haploidization during the meiosis process or from mitotic gene conversion in diploid strains. To confirm that the cycloheximide-resistant colonies obtained were in fact haploid, the DNA content of the progeny derived from sporulating cells was analyzed by flow cytometry. As controls, FACS analyses were performed on exponentially growing cultures of haploid SWM1 cells (Fig. 6A) or the isogenic haploid swm1Δ strain (Fig. 6B) and on the isogenic pair of diploid strains (wild type shown in Fig. 6C; mutant in Fig. 6D). These control scans revealed a typical distribution of cells. Similar scans were then performed on two of the cycloheximide-resistant clones obtained after sporulation and germination of swm1Δ/swm1Δ cells (Fig. 6E and F). Both scans were similar to the one obtained for the haploid control strains. In sum, both the genetic and DNA content analyses showed that swm1 mutants are able to generate haploid progeny on resumption of growth, an indication that the meiotic nuclei generated in a swm1/swm1 cell do undergo compartmentalization, which is consistent with the normal course of meiotic events in swm1 mutants.
FIG. 6.
FACS analysis shows that swm1Δ/swm1Δ cells are able to generate haploid progeny upon germination. Cells were grown in YEPD, fixed, and stained with propidium iodide prior to analysis in a Becton Dickinson FACSort. (A) Scan of haploid SWM1 cells (W303-1A). The peaks of cells marked 1N and 2N represent cells in the G1 and the G2 phases of the cell cycle, respectively. (B) Scan of haploid swm1Δ cells (YPA203). (C) Scan of diploid wild-type strain YPA24. The peaks of cells marked 2N and 4N represent cells in the G1 and the G2 phases of the cell cycle, respectively. (D) Scan of diploid mutant strain YPA207. (E and F) Scan of two of the cycloheximide-resistant clones obtained after sporulation and germination of the swm1Δ/swm1Δ diploid strain YPA207. The graphs depict relative DNA content (x axis) versus cell number (y axis).
Spore walls are aberrant in swm1 cells.
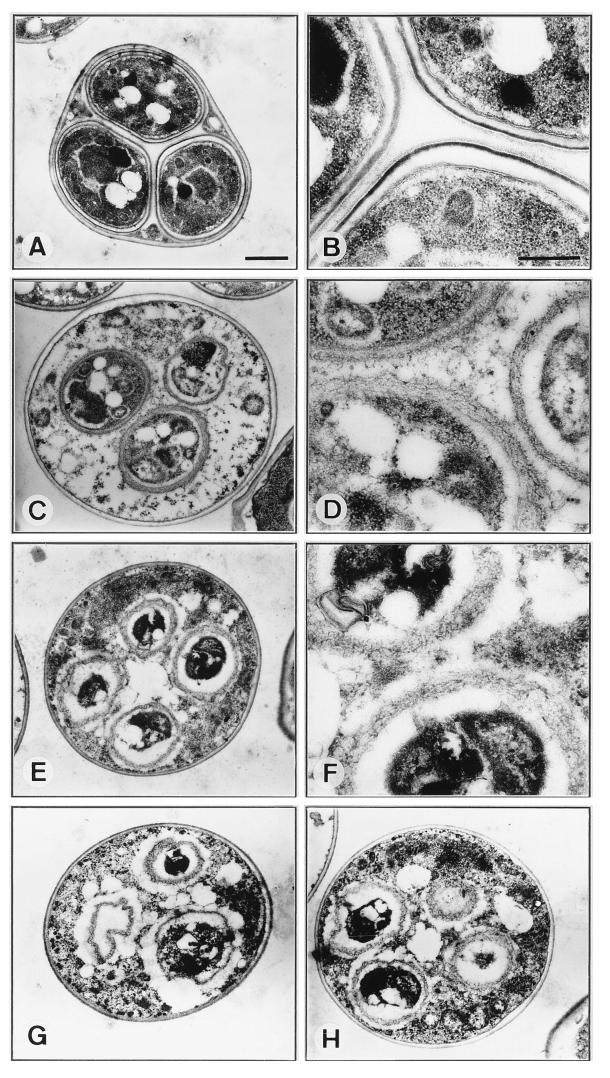
To assess the nature of the sporulation defect in swm1Δ cells at high resolution, we used electron microscopy to examine cells that had been under sporulation conditions for 24 h. The ultrastructure of a typical wild-type ascus is shown in Fig. 7A and B. The innermost layers of the spore wall, which often appear as a single electron-transparent layer, are similar in composition to the vegetative cell wall, which contains mainly glucans and mannans (7, 29). The outermost surface of the spore wall consists of a cross-linked insoluble macromolecule containing a large amount of dityrosine (4, 6) and often appears as a very thin, osmiophilic layer. This layer is responsible for the resistance of spores to degradative enzymes and organic solvents (4) and is closely linked, perhaps by covalent linkages, to an underlying chitin and chitosan layer (7), which appears as a more diffuse osmiophilic layer.
FIG. 7.
Electron microscopy of wild-type and swm1Δ mutant cells incubated for 24 h in sporulation medium. (A and B) wild-type strain YPA24. (C to H) swm1Δ mutant strain YPA207. Single asci at low magnification are depicted in panels A, C, E, G, and H, while higher magnifications of spore walls are shown in panels B, D, and F. In panel A, a typical ascus of the wild-type strain YPA24 showing three of the four spores clearly presents a well-defined spore wall surrounding all of them. In panel B, one portion of the wall of the three spores present in the ascus at higher magnification clearly shows the inner, electron-transparent components, which appear as a single layer followed by the chitin-chitosan layer closely juxtaposed next to the dityrosine coat. In panels C to H, a variety of spore wall defects are apparent in swm1Δ mutant asci. Mutant spores are surrounded by an electron-dense region with a heterogeneous structure (see the text for a detailed description of the aberrant morphology in the mutant). Scale bars, 1 μm (A, C, E, G, and H) and 0.4 μm (B, D, and F).
Electron microscopic examination revealed that the normal spore morphology was completely absent in swm1Δ cells. Instead, we observed multiple patterns of abnormal asci. It was clear that many mutant cells were able to develop prospore-like compartments, in which a single nucleus was apparently enveloped by a single spore wall, but these were generally smaller than wild-type spores. It was also clear that in swm1Δ mutants a substantial amount of cytoplasmic components was present in the ascoplasm compared with wild-type asci. Electron micrographs representative of swm1 mutant asci are shown in Fig. 7C to H. The pictures shown in panels C and D represent an example of the most mature prospore-like compartments observed in mutant cells. Cell wall material had been deposited around the compartment (Fig. 7D), but its appearance and structure were completely different from those of the wild-type cells (Fig. 7B). The image reveals an electron-dense region with a heterogeneous and amorphous structure in which the inner and outer layers cannot be distinguished. A similar pattern in the structure of the components of the spore wall can be observed in the cells shown in micrographs 7E and F. In this case, however, the prospore-like compartments seemed to be even less mature, the nucleus being surrounded by the cell wall but relatively little cytoplasmic material. In the examples shown in Fig. 7G and H, the appearance of the compartments within a single cell differed, and in some cases it was not clear whether the compartment actually contained a nucleus. In sum, swm1 mutant cells were able to enclose meiotic nuclei within distinct compartments surrounded by a heterogeneous and amorphous spore wall that failed to mature, and no distinction between the inner and outer layers could be observed.
SWM1 is required for normal expression of late sporulation-specific genes.
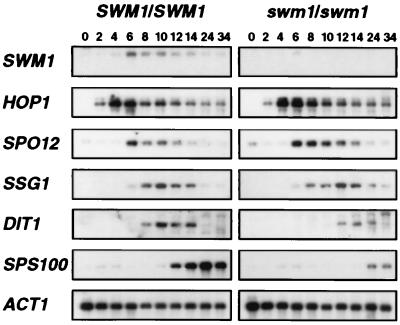
Several classes of temporally distinct sporulation-specific genes, referred to as early, middle, mid-late, and late, are sequentially expressed as the sporulation program proceeds (for a review, see references 31 and 36). To determine whether deletion of the SWM1 gene affects the pattern of sporulation-specific gene expression, we used Northern blot analysis to measure the transcript levels of various genes in isogenic wild-type and swm1 diploid cells incubated in sporulation medium for different periods of time. The timing and relative expression levels of the early sporulation-specific gene HOP1 (24) and the middle sporulation-specific gene SPO12 (34) were indistinguishable between mutant and wild-type cells (Fig. 8). These results are consistent with the microscopic observations indicating that sporulation initiation and the execution of meiotic landmark events are unaffected in swm1Δ cells. However, the timing of induction of several of the mid-late and late genes tested was different between wild-type and mutant cells. Thus, the expression of the mid-late SSG1 gene (also known as SPR1) (39, 45) was slightly delayed in swm1Δ cells, with maximal expression being detected between 12 and 14 h after transfer to nitrogen-deficient medium (in contrast to 10 h in wild-type cells). More-dramatic effects were found in another two genes that are required for spore wall maturation: the expression of DIT1 (5) and SPS100 (32) was delayed and the level of transcript accumulation was greatly reduced in the swm1Δ mutant (Fig. 8). These results therefore suggest that SWM1 is required for the normal expression of mid-late and late sporulation-specific genes, which are involved in spore cell wall formation and maturation, and are consistent with the morphological defects observed by electron microscopy.
FIG. 8.
Expression of sporulation-specific genes in wild-type and swm1Δ mutant cells. RNA was purified from wild-type (strain YPA24) and swm1Δ cells (strain YPA207) at the indicated times after transfer to sporulation medium. RNA blots were sequentially hybridized with the following radioactively labeled gene-specific probes: SWM1, HOP1, SPO12, SSG1, DIT1, and SPS100. The ACT1 gene was used to test for equal loading of RNA in all lanes.
SPS1 and SMK1 expression is not regulated by Swm1p.
Because deletion of the SPS1 or SMK1 genes, two components of a sporulation-specific mitogen-activated protein (MAP) kinase cascade, has similar effects on the expression of SPS100 to those described here (18, 30), it seemed possible that SWM1 might be required to induce the expression of these two genes during the sporulation process. To test this possibility, we used the same set of RNA blots to compare the pattern of expression of SPS1 and SMK1 between isogenic wild-type and swm1Δ diploid strains. We found that the timing and relative expression levels of both protein kinase-encoding genes were indistinguishable between mutant and wild-type cells (Fig. 9A), indicating that the SWM1 is not required for the correct expression of SPS1 and SMK1. Interestingly, the induction of SWM1 expression during the sporulation process coincided in time with the induction of SPS1 and SMK1 transcripts.
FIG. 9.
(A) Expression of SMK1 and SPS1 in wild-type and swm1Δ mutant cells. The same RNA blots used in Fig. 8 were stripped and sequentially hybridized with radioactively labeled probes for SPS1 and SMK1. For a better comparison, the panels corresponding to SWM1 and ACT1 expression are also shown. (B) Expression of SWM1 in wild-type and mutant smk1Δ cells. RNA purified from wild-type (strain NKY278) and smk1Δ (strain LAKY30) at the indicated times after transfer to sporulation medium was hybridized with an SWM1 radioactively labeled probe, and the ACT1 gene was used to test for equal loading of RNA in all lanes.
We also used Northern blotting to analyze the pattern of temporal expression of SWM1 in smk1Δ mutant cells that had been incubated in sporulation medium for different periods of time to test the possibility that SWM1 expression might be regulated by the Sps1p-Smk1p MAP kinase cascade. We found that the timing and relative expression level of SWM1 (normalized against the constitutively expressed ACT1 gene) was indistinguishable between mutant and wild-type cells (Fig. 9B), indicating that SWM1 induction does not require an intact MAP kinase cascade.
Genetic analysis of the relationship between Swm1p and Sps1p-Smk1p.
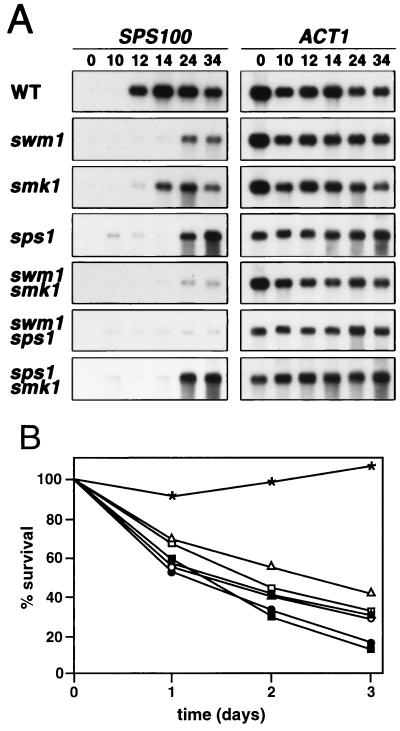
The similarity of the phenotypes observed in swm1Δ mutant cells to those reported for the deletion of SMK1 and SPS1 (18, 30) suggested that the corresponding products might operate in the same pathway. To test this hypothesis, we constructed a set of isogenic strains carrying all possible combinations of deletions in the SWM1, SPS1, and SMK1 genes. If Swm1p is a downstream component of the Sps1p-Smk1p signaling pathway and is involved in functions controlled by this pathway, then deletion of SWM1 would be expected to have a similar effect to the deletion of SMK1 or SPS1; however, if Swm1p operates in a pathway parallel to the sporulation-specific MAP kinase pathway, then smk1Δ or sps1Δ would be expected to enhance the swm1Δ mutant defects. These possibilities were tested by examining the expression of the late gene SPS100 by Northern blot analysis in wild-type and single- and double-mutant homozygous diploid strains (Fig. 10A). Expression of SPS100 in wild-type and swm1Δ mutant cells was similar to the pattern described in Fig. 8. Accumulation of SPS100 transcripts in the smk1Δ or sps1Δ strains was both delayed and reduced, as described previously (18, 30) (Fig. 10A). This analysis clearly shows that SPS100 expression is more severely affected in swm1Δ cells than in isogenic smk1Δ or sps1Δ mutant cells, suggesting that both proteins do not act in the same linear pathway. The timing of induction of SPS100 transcription in the swm1Δ smk1Δ double mutant was similar to the pattern observed in the swm1Δ mutant, although the level of induction was reduced. Similarly, expression in the double sps1Δ swm1Δ mutant was also reduced compared with any of the single mutants. Interestingly, no additive effect was observed when the sps1Δ and smk1Δ mutations were combined. These results indicate that Swm1p does not lie in the same linear pathway as Sps1p-Smk1p. Additional support for this idea was obtained when the resistance of the different diploid strains to incubation in sporulation medium was analyzed (Fig. 10B). The viability of the double mutants swm1Δ smk1Δ and swm1Δ sps1Δ (20 and 18%, respectively) after 72 h of incubation in sporulation medium was more reduced than that of any of the single mutant strains swm1Δ, smk1Δ, or sps1Δ (30, 29, and 43%, respectively). By contrast, the double-mutant smk1Δ sps1Δ showed a viability similar to that of the single smk1Δ mutant (30 versus 29%) after 3 days of incubation in sporulation medium. Together, these results indicate that although Swm1p is required for normal expression of the late sporulation-specific genes, it is not a component of the Smk1p signaling pathway necessary for the late events of the sporulation program.
FIG. 10.
(A) Expression of SPS100 in wild type (WT) and in single- and double-deletion mutants. RNA from strains YPA24 (WT), YPA207 (swm1Δ), LS35 (smk1Δ), LS36 (sps1Δ), LS34 (smk1Δ swm1Δ), LS37 (swm1Δ sps1Δ), and LS45 (smk1Δ sps1Δ) that had been incubated in sporulation medium for the indicated time periods was hybridized with a radioactively labeled probe specific for the SPS100 gene. After the probe stripping, the ACT1 gene was applied to same filters to test for equal loading of RNA in all lanes. (B) Resistance of wild-type and single- and double-mutant cells to prolonged incubations in sporulation medium. Wild-type cells from the strain YPA24 (∗) or the isogenic strains YPA207 (swm1Δ [□]), LS35 (smk1Δ [○]), LS36 (sps1Δ [▵]), LS34 (swm1Δ smk1Δ [■]), LS37 (sps1Δ swm1Δ [●]) and LS45 (sps1Δ smk1Δ [▴]) were incubated in sporulation medium for the indicated times before direct assay of the plating efficiencies.
Localization of Swm1p in sporulating cells.
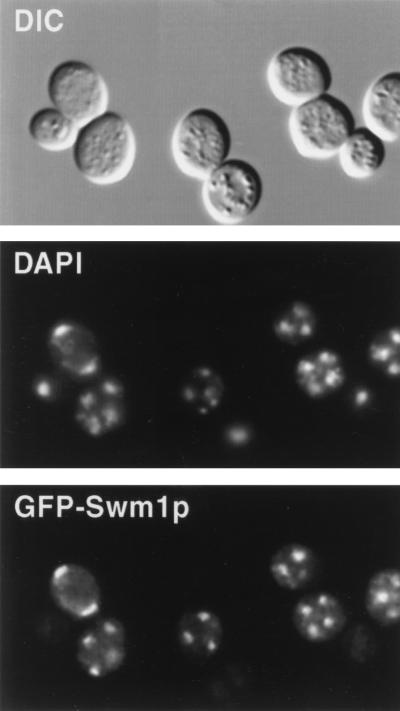
To determine the intracellular localization of Swm1p, we constructed a GFP-SWM1 fusion expressed under the control of the SWM1 promoter and present in a centromeric plasmid. The GFP-Swm1 fusion protein is fully functional and it completely restores sporulation ability when introduced into an swm1Δ mutant (not shown). The localization of Swm1p was examined in diploid cells containing the plasmid that had been in sporulation medium for 10 h. At this time, a fraction of the cells had already undergone meiosis I and II. As shown in Fig. 11, GFP-Swm1p appears to be a nuclear protein, whose GFP fluorescence coincides with the DAPI staining in bi- and tetranucleate cells. The signal was completely absent from cells carrying the untagged wild-type SWM1 gene (not shown). This result indicates that a large fraction of Swm1p appears to be concentrated in the nucleus.
FIG. 11.
Localization of GFP-Swm1p in sporulating cells. Cells from strain YPA207 carrying a GFP-SWM1 fusion in the centromeric vector Ycplac33 were incubated for 10 h in sporulation medium, fixed, stained with DAPI, and photographed.
DISCUSSION
We have characterized a novel gene, SWM1, that is required for the final steps of the sporulation process to be completed. When diploid yeast cells are deprived of nitrogen in the presence of a nonfermentable carbon source, such as acetate, they enter the meiotic program, in which a reductional division (meiosis I) is rapidly followed by a mitosis-like division (meiosis II) to generate four haploid nuclei. The final steps involve encapsulation of the newly formed meiotic products in an extremely resistant spore wall, which consists of four layers: the two inner ones are closely juxtaposed and are similar in composition to the vegetative cell wall, while the two outer ones are spore-specific structures, composed of chitin and chitosan (the third) and dityrosine (the fourth). The genes required to execute these landmark meiotic events exhibit a tight temporal expression pattern and have been classified as early, middle, mid-late, and late (36). Our results indicate that Swm1p functions as a positive effector of the machinery responsible for spore wall component assembly.
SWM1 (YDR260c) codes for a 170-amino-acid protein with a predicted molecular size of 19,356 Da. Similarity searches against sequences present in the databases revealed no homology with any known protein, either from S. cerevisiae or from any other organism. Also, after several databases were searched, no relevant motifs or patterns hinting to the function of the protein were found in the sequence. Analysis of the expression pattern of SWM1 revealed that it is expressed at a low basal level during the vegetative cycle but strongly activated during the sporulation process, the expression peak coinciding with other middle sporulation-specific genes, such as SPO12 or SMK1. Similar expression kinetics have been described for the SPR3 gene, which encodes a sporulation-specific homolog of the yeast Cdc3/10/11/12 family of septins (42). Detailed examination of the promoter region of the SPR3 gene showed that its pattern of expression is regulated by at least two essential elements: a palindromic sequence containing a sporulation-specific element, termed MSE (midsporulation element) and an ABFI element (42, 43). The MSE element alone has been shown to confer sporulation-specific regulation, suggesting that it is directly involved in regulating the timing of expression of the SPR3 gene, and it is also present in other middle sporulation-specific genes, such as SPS4, SPR2, or SMK1 (43). Recently, it has been shown that direct binding of Ndt80p to the MSE is necessary for activating transcription of the middle sporulation genes (11, 21). The ABFI element [consensus sequence CGTNNNN(G/A)(T/C) GA(TC)] is normally found upstream from the MSE element and is also essential for normal regulation of SPR3 expression (42). The promoter region of SWM1 also contains a perfect match to the MSE consensus sequence [gNCRCAAA(A/T)] in an inverted orientation, between positions −77 and −85, which is preceded by a sequence that shows an almost perfect match (7 of 8 nt) with the ABFI consensus sequence (−147 to −135). These two elements, as described for SPR3, may be responsible of the induction of expression of SWM1 that we observed during the sporulation process.
Several lines of evidence indicate that Swm1p is required for completion of the final steps of spore wall maturation. First, although no mature spores are formed after incubation in sporulation medium, swm1Δ cells were able to complete meiosis I and meiosis II with efficiencies similar to that of wild-type cells, as assayed by DAPI staining and microscopic observation. Second, after incubation in sporulation medium, the mutant cells were able to resume mitotic growth upon transfer to rich medium, generating viable haploid progeny, as has been shown for other mutants arrested at different stages of the sporulation process, such as spo14 (25, 26) or sps1 (18). The recovery of haploid progeny from swm1Δ mutants was an indication that postmeiotic compartmentalization had occurred, at least to some degree, in some of the mutant cells. Third, cells lacking the SWM1 gene are more sensitive to heat shock or enzymatic digestion than wild-type cells, indicating that they are unable to assemble a functional spore cell wall, the cellular structure responsible for the extreme resistance to stress conditions characteristic of mature spores. Finally, examination of the morphology of swm1 defective asci by electron microscopy revealed the formation of four distinct compartments inside each cell that were generally smaller than wild-type spores. The appearance of the mutant spores was variable, and they were surrounded by a rudimentary cell wall with a fibrillar aspect. Taken together, all of these results indicate that Swm1p functions late in sporulation, at the time when maturation and assembly of the spore cell wall occurs.
Relation of Swm1p and the Sps1p-Smk1p MAP kinase pathway.
Two yeast genes, SPS1 and SMK1, encode protein kinases that play a regulatory role in spore packaging: Sps1p is a homolog of the protein kinase STE20 (44% identity in the catalytic domain) (18), while Smk1p is a homolog of MAP kinases, sharing 40% identity with FUS3 (30). Both sps1 and smk1 null mutations cause defects in spore wall formation. On the basis of their common functions and structural features, it has been proposed that the two proteins may act in a sporulation-specific MAP kinase pathway. Indeed, by analogy with the pheromone response pathway, Sps1p might govern the activity of a MEKK-MEK-MAP kinase module containing Smk1p as its MAP kinase, although no epistasis analysis has been done to confirm this proposal (18, 22, 30, 31). From these analyses, it was suggested that although Sps1p and Smk1p are required for activation of late sporulation-specific genes, they have opposite roles in the expression of the mid-late gene DIT1 because, whereas a sps1 mutant overexpresses DIT1, an smk1 mutant underexpresses it. This difference was interpreted assuming that Sps1p may have a second function in addition to the presumed stimulation of Smk1p activity (31).
Using a set of isogenic strains, we have found that diploid strains lacking SPS1 present a more severe phenotype as regards the expression of the late sporulation-specific gene SPS100 than smk1-null strains and that no additive effect can be observed in the double-mutant sps1 smk1. These results are in good agreement with published observations (18, 30) and point out the possibility that Sps1p regulates a branched pathway in which Smk1p functions in one branch to activate the expression of mid-late and late sporulation-specific genes and a second branch, which is SMK1 independent, is necessary for maximal expression of late sporulation-specific genes. In this model, loss of any of the branches of the pathway, either the SMK1-dependent branch or the SMK1-independent branch, is not as severe as a loss of the entire pathway (by deletion of SPS1). Alternatively, it is possible that the less-severe phenotype observed in smk1-null mutants could be due to activation of another MAP kinase that partially replaces Smk1p function during the sporulation program.
The fact that swm1Δ mutants also show phenotypes similar to those described previously for sps1Δ or smk1Δ mutants could be taken as an indication of a functional relationship between SWM1 and the components of the Sps1p-Smk1p MAP kinase cascade. Similar to null mutations in SPS1 or SMK1, mutations in SWM1 result in cells that proceed normally through the second meiotic division but produce defective spores with altered walls. As seen in sps1Δ and smk1Δ mutants, cells lacking SWM1 that have been incubated in sporulation medium are more sensitive to stress conditions than are wild-type strains. Finally, as judged by electron microscopy, the morphology of the spore wall is aberrant in mutant cells in which any of the three genes has been deleted. Although the similar phenotypes suggest a functional relationship between Swm1p and the components of the Sps1p-Smk1p MAP kinase cascade, detailed analysis of single and double mutants provided evidence that they do not act in the same linear pathway (Fig. 10). If Swm1p were a downstream component of any of the branches of the Sps1p signaling pathway and involved in functions controlled by this pathway, the deletion of SWM1 would be expected to have an effect similar to that of the deletion of SPS1. However, Northern analysis indicated that the expression of late sporulation-specific genes is more impaired in swm1Δ strains than in smk1Δ or sps1Δ mutant cells, transcription being considerably delayed and reduced in the former. Furthermore, when sps1Δ or smk1Δ null mutations are combined with swm1 deletion, an additive effect can be observed in the expression of the sporulation-specific gene analyzed. Similar results were obtained when survival of the different strains in sporulation medium was analyzed; a decrease in viability was observed when swm1Δ was combined with either sps1Δ or smk1Δ but not in the sps1Δ smk1Δ strain. The synergistic effect of two mutations suggests that the two gene products or the pathways in which they act perform related functions. For example, mutations in BRO1 (41), SPA2 (13), or in the redundant type 1-related protein phosphatases PPZ1 and PPZ2 (33) exacerbate the phenotypes of mutations in the components of the Pkc1p-MAP kinase cascade. A possible explanation for the additive effect of the mutations is that Swm1p and the Sps1p-Smk1p MAP kinase pathway would produce signals that converge to activate one or more targets. A similar convergent model relating the activity of Bro1p and the protein phosphatases Ppz1p and Ppz2p to the Pkc1p-MAP kinase pathway has been proposed for these genes (33, 41). Although our results do not completely rule out the possibility of Swm1p being a member of the Sps1p-Smk1p MAP kinase pathway, we suggest that Swm1p is able to perform a function independently of the sporulation-specific MAP kinase cascade to activate the expression of the sporulation genes required for the late steps of this process, although this does not exclude the possibility that direct signaling between Swm1p and the kinase cascade may also occur. For example, the activity of Swm1p may regulate or be regulated by one of the kinase components.
The pattern of sporulation-specific gene expression during the late portion of the sporulation program requires both an intact Smk1p-MAP kinase signaling pathway and Swm1p, a protein that has been found in the nucleus during the sporulation process. However, the present study does not demonstrate that SWM1 directly regulates the transcription of sporulation-specific genes. A recent study has shown that repression of DIT1 during vegetative growth is controlled by the Ssn6p-Tup1p protein complex through a negative regulatory element (NRE) (17). Derepression of DIT1 during sporulation requires the participation of the NRE and at least two cis-acting elements which bind to several different factors. The model proposed in that study postulates that midway through sporulation, a sporulation-specific event occurs that displaces Ssn6-Tup1 from the NRE, thus allowing some other regulatory factors to interact with the general transcription machinery (17). Based on the nuclear localization of Swm1p and the severe defects observed in the expression of mid-late and late sporulation-specific genes, an intriguing possibility is that Swm1p might act as one of those regulatory factors, either directly or as a part of a protein complex. Alternatively, it is also possible that the severe reduction in the expression of the late genes in the swm1Δ mutant may represent a secondary phenotype resulting from the inability of the cell to assemble the spore wall correctly. Spore maturation is a complex process in which multiple events must be synchronized and in which multiple critical assembly reactions must be coordinated with the transcriptional program. It is therefore possible that in swm1 mutants the failure to perform properly one of the earlier events of the maturation process properly might indirectly affect the transcription of the late sporulation-specific genes. A more precise understanding of the role of Swm1p in the sporulation process should emerge from future studies on the interaction with other proteins.
ACKNOWLEDGMENTS
We are grateful to Edward Winter for the strains provided. We thank Angel Durán and María Molina for helpful comments and discussions on the manuscript, Hortensia Rico and Carlos Belinchón for advice and technical assistance with the electron microscopy, and Nick Skinner for revision of the manuscript.
This research was supported by grants from the Comisión Interministerial de Ciencia y Tecnología (BIO96-1413-C02-02) and from the European Community (CIPA-CT93-0117). S. Ufano is a recipient of a fellowship from Ministerio de Educación y Ciencia (Spain).
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiple disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckett A, Illingworth R F, Rose A H. Ascospore wall development in Saccharomyces cerevisiae. J Bacteriol. 1973;113:1054–1057. doi: 10.1128/jb.113.2.1054-1057.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 4.Briza P, Breitenbach M, Ellinger A, Segall J. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 1990;4:1775–1789. doi: 10.1101/gad.4.10.1775. [DOI] [PubMed] [Google Scholar]
- 5.Briza P, Eckerstorfer M, Breitenbach M. The sporulation-specific enzymes encoded by the DIT1 and DIT2 genes catalyze a two-step reaction leading to a soluble ll-dityrosine-containing precursor of the yeast spore wall. Proc Natl Acad Sci USA. 1994;91:4524–4528. doi: 10.1073/pnas.91.10.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briza P, Ellinger A, Winkler G, Breitenbach M. Characterization of a dl-dityrosine-containing macromolecule from yeast ascospore walls. J Biol Chem. 1990;265:15118–15123. [PubMed] [Google Scholar]
- 7.Briza P, Ellinger A, Winkler G, Breitenbach M. Chemical composition of the yeast ascospore wall: the second outer layer consists of chitosan. J Biol Chem. 1988;263:11569–11574. [PubMed] [Google Scholar]
- 8.Byers B. Cytology of the yeast life cycle. In: Strathern J N, Jones E W, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 59–96. [Google Scholar]
- 9.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 10.Christodoulidou A, Bouriotis V, Thireos G. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J Biol Chem. 1996;271:31420–31425. doi: 10.1074/jbc.271.49.31420. [DOI] [PubMed] [Google Scholar]
- 11.Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- 12.Correa J, Vazquez de Aldana C R, San Segundo P, del Rey F. Genetic mapping of 1,3-β-glucanase-encoding genes in Saccharomyces cerevisiae. Curr Genet. 1992;22:283–288. doi: 10.1007/BF00317922. [DOI] [PubMed] [Google Scholar]
- 13.Costigan C, Gehrung S, Snyder M. A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol Cell Biol. 1992;12:1162–1178. doi: 10.1128/mcb.12.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis L L, Bartnicki-García S. Chitosan synthesis by the tandem action of chitin synthetase and chitin deacetylase from Mucor rouxii. Biochemistry. 1984;23:1065–1073. [Google Scholar]
- 15.Esposito R E, Klapholtz S. Meiosis and ascospore development. In: Strathern J N, Jones E W, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 211–287. [Google Scholar]
- 16.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 17.Friesen H, Hepworth S R, Segall J. An Ssn6-Tup1-dependent negative regulatory element controls sporulation-specific expression of DIT1 and DIT2 in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:123–134. doi: 10.1128/mcb.17.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friesen H, Lunz R, Doyle S, Segall J. Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 1994;8:2162–2175. doi: 10.1101/gad.8.18.2162. [DOI] [PubMed] [Google Scholar]
- 19.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 20.Guth E, Hashimoto T, Conti S F. Morphogenesis of ascospores in Saccharomyces cerevisiae. J Bacteriol. 1972;109:869–880. doi: 10.1128/jb.109.2.869-880.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hepworth S R, Friesen H, Segall J. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5750–5761. doi: 10.1128/mcb.18.10.5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 23.Hill J, Myers A, Koerner T, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 24.Hollingsworth N M, Goetsch L, Byers B. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990;61:73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- 25.Honigberg S M, Conicella C, Esposito R E. Commitment to meiosis in Saccharomyces cerevisiae: involvement of the SPO14 gene. Genetics. 1992;130:703–716. doi: 10.1093/genetics/130.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honigberg S M, Esposito R E. Reversal cell determination in yeast meiosis: postcommitment arrest allows return to mitotic growth. Proc Natl Acad Sci USA. 1994;91:6559–6563. doi: 10.1073/pnas.91.14.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutter K J, Eipel H E. DNA determination of yeast by flow cytometry. J Gen Microbiol. 1979;113:369–375. doi: 10.1099/00221287-113-2-369. [DOI] [PubMed] [Google Scholar]
- 28.Kafetzopoulos D, Martinou A, Bouriotis V. Bioconversion of chitin to chitosan: purification and characterization of chitin deacetylase from Mucor rouxii. Proc Natl Acad Sci USA. 1993;90:2564–2568. doi: 10.1073/pnas.90.7.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katohda S, Konno K, Sasaki Y, Suzuki K, Sakamoto S. Isolation and composition of the spore wall of Saccharomyces cerevisiae. Agric Biol Chem. 1984;48:895–901. [Google Scholar]
- 30.Krisak L, Strich R, Winters R S, Hall J P, Mallory M J, Kreitzer D, Tuan R S, Winter E. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 1994;8:2151–2161. doi: 10.1101/gad.8.18.2151. [DOI] [PubMed] [Google Scholar]
- 31.Kupiec M, Byers B, Esposito R E, Mitchell A P. Meiosis and sporulation in Saccharomyces cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Cell cycle and cell biology. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 889–1036. [Google Scholar]
- 32.Law D T S, Segall J. The SPS100 gene of Saccharomyces cerevisiae is activated late in the sporulation process and contributes to spore wall maturation. Mol Cell Biol. 1988;8:912–922. doi: 10.1128/mcb.8.2.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee K S, Hines L K, Levin D E. A pair of functionally redundant yeast genes (PPZ1 and PPZ2) encoding type 1-related protein phosphatases function within the PKC1-mediated pathway. Mol Cell Biol. 1993;13:5843–5853. doi: 10.1128/mcb.13.9.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malavasic M J, Elder R T. Complementary transcripts from two genes necessary for normal meiosis in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2809–2819. doi: 10.1128/mcb.10.6.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller J J. Sporulation in Saccharomyces cerevisiae. In: Rose A H, Harrison J S, editors. The yeasts. Vol. 3. New York, N.Y: Academic Press, Inc.; 1989. pp. 489–550. [Google Scholar]
- 36.Mitchell A P. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moens P B. Fine structure of ascospore development in the yeast Saccharomyces cerevisiae. Can J Microbiol. 1971;17:507–510. doi: 10.1139/m71-084. [DOI] [PubMed] [Google Scholar]
- 38.Moens P B, Rapport E. Spindles, spindle plaques and meiosis in the yeast Saccharomyces cerevisiae. J Cell Biol. 1971;50:344–361. doi: 10.1083/jcb.50.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muthukumar G, Suhng S H, Magee P T, Jewell R D, Primerano D A. The Saccharomyces cerevisiae SPR1 gene encodes a sporulation-specific exo-1,3-β-glucanase which contributes to ascospore thermoresistance. J Bacteriol. 1993;175:386–394. doi: 10.1128/jb.175.2.386-394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neiman A N. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J Cell Biol. 1998;140:29–37. doi: 10.1083/jcb.140.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nickas M E, Yaffe M P. BRO1, a novel gene that interacts with components of the Pkc1p-mitogen-activated protein kinase pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2585–2593. doi: 10.1128/mcb.16.6.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozsarac N, Bhattacharyya M, Dawes I W, Clancy M J. The SPR3 gene encodes a sporulation-specific homologue of the yeast CDC3/10/11/12 family of bud neck microfilaments and is regulated by ABFI. Gene. 1995;164:157–162. doi: 10.1016/0378-1119(95)00438-c. [DOI] [PubMed] [Google Scholar]
- 43.Ozsarac N, Straffon M J, Dalton H E, Dawes I A. Regulation of gene expression during meiosis in Saccharomyces cerevisiae: SPR3 is controlled by both ABFI and a new sporulation control element. Mol Cell Biol. 1997;17:1152–1159. doi: 10.1128/mcb.17.3.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Percival-Smith A, Segall J. Isolation of DNA sequences preferentially expressed during sporulation in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:142–150. doi: 10.1128/mcb.4.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.San Segundo P, Correa J, Vázquez de Aldana C R, del Rey F. SSG1, a gene encoding a sporulation-specific 1,3-β-glucanase in Saccharomyces cerevisiae. J Bacteriol. 1993;175:3823–3837. doi: 10.1128/jb.175.12.3823-3837.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sherman F, Fink G, Hicks J B. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 47.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in Saccharomyces cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 48.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 49.Wright R, Rine J. Transmission electron microscopy and immunochemical studies of yeast: analysis of HMG-CoA reductase overproduction by electron microscopy. Methods Cell Biol. 1989;31:473–512. doi: 10.1016/s0091-679x(08)61624-6. [DOI] [PubMed] [Google Scholar]