Abstract
RNA polymerase II transcribes the mRNA-encoding genes and the majority of the small nuclear RNA (snRNA) genes. The formation of a minimal functional transcription initiation complex on a TATA-box-containing mRNA promoter has been well characterized and involves the ordered assembly of a number of general transcription factors (GTFs), all of which have been either cloned or purified to near homogeneity. In the human RNA polymerase II snRNA promoters, a single element, the proximal sequence element (PSE), is sufficient to direct basal levels of transcription in vitro. The PSE is recognized by the basal transcription complex SNAPc. SNAPc, which is not required for transcription from mRNA-type RNA polymerase II promoters such as the adenovirus type 2 major late (Ad2ML) promoter, is thought to recruit TATA binding protein (TBP) and nucleate the assembly of the snRNA transcription initiation complex, but little is known about which GTFs other than TBP are required. Here we show that the GTFs IIA, IIB, IIF, and IIE are required for efficient RNA polymerase II transcription from snRNA promoters. Thus, although the factors that recognize the core elements of RNA polymerase II mRNA and snRNA-type promoters differ, they mediate the recruitment of many common GTFs.
In the past several years, all of the factors required for basal RNA polymerase II transcription from TATA-containing RNA polymerase II mRNA promoters have been identified and purified and most of them have been cloned (61, 70). In vivo, several of these factors may be recruited to promoters as part of large, RNA polymerase II-containing complexes, some of which contain all the factors required for activated transcription in vitro and are referred to as holoenzymes (9, 39, 42, 54). In vitro, however, the assembly of an RNA polymerase II transcription initiation complex on a TATA box can be divided into several steps. TATA binding protein (TBP) or the TBP-containing complex TFIID binds to the TATA box in an association that is greatly stabilized by the subsequent binding of TFIIB, which contacts both TBP and the DNA. The presence of TFIIB allows the recruitment of a TFIIF-RNA polymerase II complex and then of TFIIE and TFIIH. Another general transcription factor (GTF), TFIIA, can join the initiation complex at any stage of assembly. Like TFIIB, TFIIA greatly stabilizes the association of TBP with the TATA box (61, 70).
The role of the various transcription factors in directing transcription initiation is the subject of intense studies. While TBP and TFIIB play central roles in the nucleation of the transcription initiation complex, TFIIF, TFIIE, and TFIIH play roles at later steps. TFIIF interacts directly with RNA polymerase II and TFIIB and is required for stable assembly of RNA polymerase II with the TATA-TBP-TFIIB complex (11, 20). It also inhibits nonspecific binding of RNA polymerase II to nonpromoter sequences (10, 37) and stimulates the rate of transcription elongation (4, 6, 21, 31, 36, 68). TFIIE incorporation into the TATA-TBP-TFIIB-RNA polymerase II-TFIIF complex is required for subsequent assembly of TFIIH (19). TFIIE and TFIIH are involved in promoter melting and promoter clearance (15, 28, 65–67, 82). TFIIE regulates the activities of TFIIH (50), which possesses both ATP-dependent helicase activities and a kinase activity capable of phosphorylating the C-terminal domain of RNA polymerase II (14, 16–18, 50, 71, 74–77). The helicase activity is thought to be involved in promoter melting (27, 29). The C-terminal domain kinase activity may be involved in promoter clearance and transcription elongation (1, 32, 44). In addition, TFIIE plays a direct role in promoter melting, perhaps by binding to the single-stranded region and thereby stabilizing the melted region of the promoter (29), and has been shown to help recruit TBP and TFIIA to the TATA box (93).
TFIIA is required for activation of transcription (see, for example, references 13, 38, 40, 51, 62, 64, 81, and 94). In addition, TFIIA plays a role in basal transcription, although this role varies with the precise in vitro transcription system used. Thus, when transcription reaction mixtures are reconstituted with TBP, addition of TFIIA has no effect (12, 52, 81). However, when transcription reaction mixtures are reconstituted with TFIID, addition of TFIIA is stimulatory (12, 94). This may be attributed in part to the ability of TFIIA to counteract the activities of repressors such as Dr1, Mot1 (also known as TAF-172), and Dr2 (also known as PC3 and topoisomerase 1) (2, 8, 30, 43, 51, 58). However, TFIIA is also capable of stimulating transcription when very pure preparations of TFIID are used (51, 81). This may reflect the ability of TFIIA to counteract the inhibitory effect of TBP-associated factors in TFIID on TFIID binding (41, 63).
Many mRNA promoters lack TATA boxes altogether. In several of these promoters, basal transcription is directed by an initiator (Inr) element (79). The Inr is recognized by some of the TFIID TBP-associated factors (35, 55, 96). In particular, Drosophila TAFII150, or its recently cloned human homolog, CIF150 or hTAFII150, is required for TFIID-dependent, Inr-directed transcription (34, 56, 85, 86). In addition, fractions referred to as TIC-1, TIC-2, and TIC-3, as well as the GTF TFIIA, are required (56). Other GTFs, namely TFIIB, TFIIF, TFIIE, and TFIIH, are assumed to be required, and in the case of the TATA-less DNA polymerase β promoter, which can be transcribed with TBP rather than TFIID, this has been shown directly (90).
The human small nuclear RNA (snRNA) gene family consists of RNA polymerase II and RNA polymerase III genes. Both of the human RNA polymerase II and III snRNA promoters contain a distal sequence element, which serves as a transcriptional enhancer. The basal RNA polymerase II snRNA promoters contain a single essential element, the proximal sequence element (PSE), which is sufficient to nucleate the assembly of an RNA polymerase II transcription initiation complex and to direct basal RNA polymerase II transcription in vitro. The basal RNA polymerase III snRNA promoters contain, in addition to the PSE, a TATA box, which in this context determines the RNA polymerase III specificity of the promoter (25, 47).
The PSE is recognized by a complex named SNAPc (73) or PTF (60), which is composed of five subunits (24). The TATA box in the RNA polymerase III snRNA promoters is recognized by TBP (48, 72, 78, 88). TBP is also required for transcription of the TATA-less RNA polymerase II snRNA genes but not as part of the TBP-containing complexes TFIID or TFIIB (73, 95). Thus, in this case, it is not clear how TBP is recruited to the promoter. The other factors required for assembly of the RNA polymerase II and III snRNA initiation complexes are not well characterized. Most notably, it is not known whether transcription from RNA polymerase II snRNA promoters relies on the same GTFs as those used in transcription from mRNA promoters. Here we compare the requirements of RNA polymerase II snRNA and mRNA promoters for various GTFs. We find that basal RNA polymerase II transcription from snRNA promoters requires TFIIA, TFIIB, TFIIF, and TFIIE. These results suggest that snRNA promoters and TATA-containing mRNA promoters use different pathways to recruit many of the same general transcription factors. They also point to a different function for TFIIA in basal transcription from snRNA promoters and TATA-containing mRNA promoters.
MATERIALS AND METHODS
Sources of proteins.
TBP, TFIIA, TFIIB, TFIIE, and TFIIF were expressed in Escherichia coli and purified as described previously (53). RNA polymerase II and TFIIH were purified from HeLa nuclear extracts also as described previously (53). In some cases, TBP was purchased from Promega and TFIIB was expressed in E. coli BL21 (DE3) cells as a glutathione S-transferase (GST) fusion protein with the T7 expression system (80). The GST-TFIIB fusion protein was purified by chromatography on glutathione-agarose beads (Sigma). TFIIB was released from GST, which remained bound to the beads, by cleavage with thrombin and was dialyzed against buffer D100 (20 mM HEPES [pH 7.9], 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 3 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride, 0.05% Tween 20). Protein concentration was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and staining with Coomassie blue, with bovine serum albumin as a standard.
Generation of anti-peptide antibodies for TFIIB.
Synthetic peptides corresponding to TFIIB amino acids 1 to 18 (peptide CSH505), 50 to 66 (peptide CSH506), and 300 to 316 (peptide CSH508) were coupled to keyhole limpet hemocyanin (Pierce) as described previously (23) and injected into rabbits to generate the polyclonal anti-peptide antibodies α-IIB/1, α-IIB/2, and α-IIB/4, respectively.
Constructs.
The constructs pU1*G−, p119MLP(C2A), pSBM13+VAI, and pU6/Hae/RA.2 have been described previously (46, 49, 73). The construct pU1*G−Oct−, in which the octamer sequence in pU1*G− was mutated into a BamHI site, was constructed by oligonucleotide-directed PCR mutagenesis (Stratagene) with the following oligonucleotides: U1mOctFd, with the sequence 5′-GGACAGGGCGACTTCTGGGATCCAGAGGCAGCGCAGAGG-3′, and U1mOctRv, with the sequence 5′-CCTCTGCGCTGCCTCTGGATCCCAGAAGTCGCCCTGTCC-3′.
Immunodepletions.
Normal rabbit serum, immune sera directed against the various RNA polymerase II GTFs, or, in the experiments whose results are shown in Fig. 2, preimmune sera were incubated with protein A-agarose beads (Boehringer Mannheim) for 1 h at room temperature, washed in phosphate-buffered saline, and cross-linked as described previously (23) except in the experiments whose results are shown in Fig. 2, 3, and 5A and B, where the antibodies were not cross-linked to the beads. The antibody beads were washed in buffer D50 (20% glycerol, 20 mM HEPES [pH 7.9], 50 mM KCl, 0.2 mM EDTA, 3 mM DTT, 0.05% Tween 20, 0.5 mM phenylmethylsulfonyl fluoride), and used for depletions. Whole-cell or nuclear extracts (approximately 30 or 20 μg/μl, respectively) were subjected to two successive incubations with antibody beads, each for 25 min at room temperature with agitation. The beads-to-extract ratios indicated in the figure legends reflect the total amounts of beads used in the two successive incubations. For example, two successive incubations with beads-to-extract ratios of 1:1 are indicated as a beads-to-extract ratio of 2:1. After the second incubation, the supernatants were used for in vitro transcription reactions and depletions were monitored by immunoblotting.
FIG. 2.
TFIIB is required for RNA polymerase II snRNA transcription from the U1 promoter. (A) Parallel in vitro transcription reactions were performed with the Ad2ML, U1 snRNA, U6 snRNA, and VAI promoters with untreated whole-cell extract (WCE; lane 1), WCE treated with increasing amounts of preimmune antibody beads (Pre-I; lanes 2 and 3), or WCE-treated with increasing amounts of anti-TFIIB antibody beads (α-IIB/2; lanes 4 and 5) at beads-to-extract ratios of 2:1 (lanes 2 and 4) and 2.5:1 (lanes 3 and 5). The locations of correctly initiated transcripts are indicated at left, and an asterisk marks readthrough transcripts (73). (B) Immunoblot analysis of the extracts used in the experiments whose results are shown in panel A. The membrane was probed with the α-IIB/4 antibody. Eight microliters of each extract was loaded per lane. (C) In vitro transcription reactions were performed with WCE supplemented with SNAPc (lane 1), WCE supplemented with SNAPc and treated with preimmune antibody beads (lanes 2 and 7), or WCE supplemented with SNAPc and treated with α-IIB/1 (lanes 3 to 6) or α-IIB/2 (lanes 8 to 11) antibody beads at a beads-to-extract ratio of 0.5:1. In lanes 4 to 6 and 9 to 11, 40, 200, and 400 ng, respectively, of E. coli-expressed TFIIB was added back to the TFIIB-depleted reaction mixtures. An asterisk marks readthrough transcripts. (D) Immunoblot analysis of the extracts used in the experiments whose results are shown in panel C. The membrane was probed with the α-IIB/4 antibody. Eight microliters of each extract was loaded per lane.
FIG. 3.
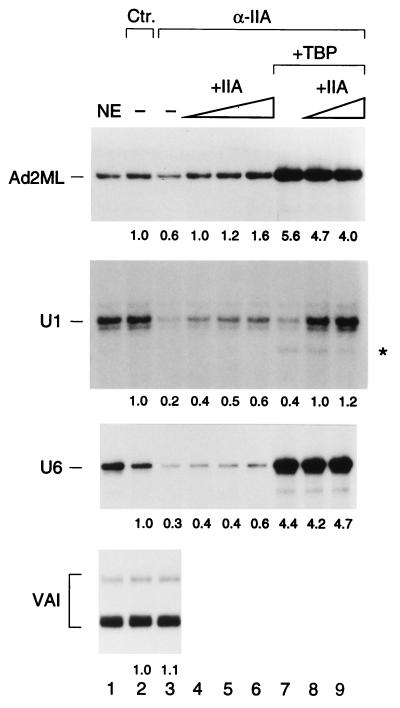
TFIIA is required for efficient RNA polymerase II snRNA transcription from the U1 promoter in vitro. In vitro transcription reactions were performed with untreated nuclear extract (NE; lane 1), NE treated with control (nonimmune) antibody beads (lane 2), or NE treated with antibody beads directed against the α and β subunits of TFIIA (α-IIA; lanes 3 to 9) at a 1:1 beads-to-extract ratio. Two, 4, and 6 μl of E. coli-expressed TFIIA was added to the reaction mixtures in lanes 4 to 6. Recombinant TBP (3 ng) was added to the reaction mixture in lane 7 and a combination of 3 ng of TBP and 2 and 6 μl of TFIIA was added to the reaction mixtures in lanes 8 and 9, respectively. The asterisk marks RNA polymerase III transcripts that are produced in U1 transcription reactions when TBP is added. The signals corresponding to correct transcription initiation in each panel were quantitated with a phosphorimager, the background was subtracted, and the numbers were normalized for the signal obtained in lane 2, which was set at 1. Ctr., control.
FIG. 5.
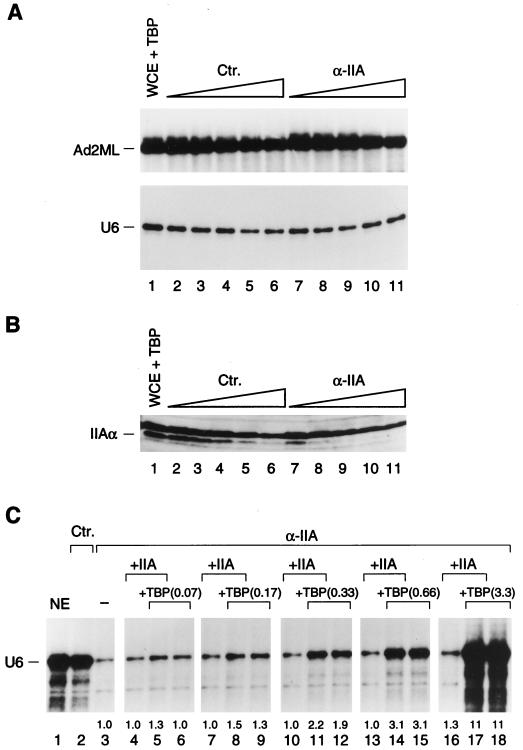
TFIIA is not required for efficient RNA polymerase III snRNA transcription from the U6 promoter in vitro. (A) Parallel in vitro transcription reactions from the Ad2ML and U6 snRNA promoters. Whole-cell extract (WCE) was supplemented with TBP (lane 1) and then treated with increasing amounts of control (nonimmune) antibody beads (lanes 2 to 6) or α-IIA antibody beads directed against the α and β subunits of TFIIA (lanes 7 to 11) at beads-to-extract ratios of 0.5:1, 1:1, 1.5:1, 2:1, and 2.5:1, respectively. (B) Immunoblot analysis of the extracts used in the experiment whose results are shown in panel A with the antibody directed against the α and β subunits of TFIIA. WCE plus TBP (4 μl) was loaded in lane 1, and the depleted extracts (6 μl) were loaded in lanes 2 to 11. The location of the α subunit is indicated at left. (C) Addition of TFIIA to TFIIA-depleted extracts does not stimulate U6 transcription in vitro in the presence of limiting amounts of recombinant TBP. Nuclear extract (NE) was treated with control (nonimmune) (lane 2) or anti-TFIIA (lanes 3 to 18) antibody beads at a 1:1 beads-to-extract ratio. E. coli-expressed TFIIA (2 μl) and TBP (amounts are indicated in nanograms) were added to the reaction mixtures as indicated above the lanes (lanes 4 to 18). The signals corresponding to correct transcription initiation were quantitated with a phosphorimager, the background was subtracted, and the numbers were normalized for the signal obtained in lane 3, which was set at 1. Ctr., control.
In vitro transcriptions.
Transcriptions from the pU1*G− construct, which contains U1 promoter sequences in front of a G-less cassette, were performed in a total volume of 40 μl containing 45 to 50 mM KCl, 12 mM HEPES (pH 7.9), 5 mM MgCl2, 1 mM spermidine trihydrochloride (Sigma), 1 mM DTT, 400 μM ATP, 400 μM UTP, 0.625 μM (1.5 to 2 μl) [α-32P]CTP (20 μCi), 1.2 mM 3′-O-methyl-GTP (Pharmacia), 12% glycerol, 0.01% Tween 20, 0.1 mM EDTA, 2% polyethylene glycol 8000, 4.5 U of RNase T1, and 0.5 to 1.0 μg of pU1*G− template (73) or, in the experiments whose results are shown in Fig. 8 and 9, pU1*G− that had been linearized with HindIII. In addition, reaction mixtures included approximately 360 μg of whole-cell extract or 160 μg of nuclear extract and similar amounts of mock-depleted or depleted extracts. The reaction mixtures were incubated for 90 min at 30°C and stopped by addition of 270 μl of stop buffer containing 0.3 M sodium acetate, 0.5% SDS, 2.5 mM EDTA, 50 μg of tRNA per ml, and 80 μg of proteinase K. After further incubation for 30 min to 1 h at 37°C, the reaction mixtures were extracted with phenol and the nucleic acids were precipitated with ethanol and fractioned on a 4.5% polyacrylamide–urea gel. The dried gels were quantitated with a phosphorimager (Fuji) and exposed to film.
FIG. 8.
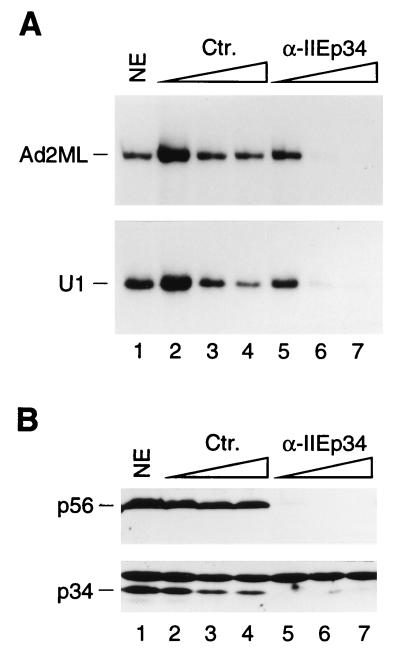
Depletion of TFIIE inhibits RNA polymerase II transcription from the Ad2ML and the U1 snRNA promoters. (A) In vitro transcription reactions from linearized templates were performed with untreated nuclear extract (NE; lane 1) or NE treated with increasing amounts of control (nonimmune) antibody beads (lanes 2 to 4) or with increasing amounts of anti-TFIIE antibody beads directed against the p34 subunit of TFIIE (α-IIEp34; lanes 5 to 7) at beads-to-extract ratios of 0.1:1, 1:1, and 2:1, respectively. (B) Immunoblot analysis of the extracts used to obtain the results shown in panel A with antibodies directed against the p34 and p56 subunits of TFIIE. The locations of the TFIIE subunits are indicated at the left. Ten microliters of extracts was loaded per lane. Ctr., control.
FIG. 9.
TFIIE stimulates transcription from the RNA polymerase II snRNA U1 promoter. (A) Addition of E. coli-expressed TFIIE to TFIIE-depleted extract. Nuclear extract (NE) was treated with control (nonimmune) antibody beads (lane 2) or anti-TFIIE antibody beads directed against the p34 subunit of TFIIE (α-IIEp34; lanes 3 to 6) at a 2:1 beads-to-extract ratio and used in transcription reactions with linearized Ad2ML and U1 templates. In lanes 4 to 6, 0.5, 1.5, and 3 μl of E. coli-expressed TFIIE was added to the reaction mixtures. (B) Immunoblot analysis of the extracts used to obtain the results shown in panel A with antibodies directed against the p34 and p56 subunits of TFIIE. (C) Addition of GTFs to TFIIE-depleted extracts. Nuclear extract was treated with control (nonimmune) antibody beads (lane 2) or α-TFIIEp34 antibody beads (lanes 3 to 10) at a 1:1 beads-to-extract ratio and used in parallel transcription reactions with linearized Ad2ML and U1 templates. In the reaction whose results are shown in lane 4, a combination of recombinant TFIIE (1 μl), purified RNA polymerase II (pol II; 1.5 μl), recombinant TFIIA (1 μl), recombinant TFIIB (2 μg), recombinant TFIIF (1 μl), and purified TFIIH (2 μl) was added. In lanes 5 to 10, the same combination of GTFs was added except for the factor indicated above each lane. (D) Immunoblot analysis of the extracts used to obtain the results shown in panel C with antibodies directed against the p34 and p56 subunits of TFIIE, the largest subunit of RNA polymerase II, and TFIIB. Ten microliters of extract was loaded per lane. Ctr., control.
Transcriptions from the p119MLP(C2A) construct, which contains the adenovirus type 2 major late (Ad2ML) promoter in front of a G-less cassette, were performed under the same conditions as those described above except that the total reaction volume was 25 μl and the reaction mixtures contained 10 mM MgCl2, no Tween 20, 0.5 μg of the p119MLP(C2A) template (49), 90 to 120 μg of whole-cell extract or 50 μg of nuclear extract, and similar amounts of mock-depleted or depleted extracts. In the experiments whose results are shown in Fig. 8 and 9, p119MLP(C2A) was first linearized with Eco0109I.
VAI transcription reactions were performed as described previously (49) in a total volume of 20 μl containing 250 ng of pSBM13+VAI supercoiled template, 20 to 30 μg of whole-cell or nuclear extract, and similar amounts of mock-depleted or depleted extract.
U6 transcription reactions were performed as described previously (49) in a total volume of 20 μl containing 400 ng of the pU6/Hae/RA.2 supercoiled template, approximately 90 to 150 μg of whole-cell extract or 35 μg of nuclear extract, and similar amounts of mock-depleted or depleted extract.
Immunoblots.
Whole-cell or nuclear extract and extracts treated with antibody beads were fractionated by SDS-PAGE on 12.5% polyacrylamide gels. Purified protein preparations and Rainbow markers (Amersham) were used as markers (not shown). The proteins were transferred to nitrocellulose, and the membranes were probed with the antibodies indicated in the figures.
RESULTS
To assess the roles of various RNA polymerase II GTFs in transcription from RNA polymerase II snRNA promoters, we immunodepleted each of these GTFs from extracts and tested the depleted extracts for their ability to direct transcription from four types of promoters, whose structures are illustrated in Fig. 1A; the Ad2ML promoter, a typical mRNA-type RNA polymerase II promoter, the RNA polymerase II U1 and RNA polymerase III U6 snRNA promoters, and the Ad2 VAI promoter, a typical RNA polymerase III promoter with gene-internal A and B boxes. In those cases where transcription was reduced by the immunodepletion, we then tested whether addition of the factor against which the antibody had been raised restored transcription. All of these factors except for RNA polymerase II and TFIIH were recombinant proteins expressed in bacteria, and Fig. 1B shows their polypeptide compositions. Both TBP (lane 1) and TFIIB (lane 3) migrated close to their calculated molecular masses of 38 and 33 kDa, respectively. TFIIA is composed of three subunits, α, β, and γ, two of which (α and β) are derived from a single gene, probably by protein processing (61). The recombinant TFIIA used here contains a 56-kDa polypeptide corresponding to fused α and β subunits as well as the 14-kDa γ subunit (lane 2). Lanes 4 and 5 show the 56- and 34-kDa subunits of TFIIE and the RAP74 and RAP30 subunits of TFIIF, respectively, while lane 6 shows highly purified human RNA polymerase II.
FIG. 1.
(A) Basal promoter elements in the Ad2ML, U1 snRNA, U6 snRNA, and VAI promoters. Pol II and III, RNA polymerases II and III, respectively. (B) Polypeptide compositions of recombinant (TBP, TFIIA, TFIIB, TFIIE and TFIIF) or highly purified (RNA polymerase II) factors. The proteins were fractionated by SDS-PAGE and stained with silver.
TFIIB is required for RNA polymerase II snRNA transcription.
In experiments where anti-TFIIB monoclonal antibodies were added to an extract, transcription from both the U1 and Ad2ML promoters was inhibited and could then be restored by the addition of recombinant TFIIB (5). These results were consistent with TFIIB being required for RNA polymerase II snRNA gene transcription, but because TFIIB was not depleted from the extracts, it remained possible that the anti-TFIIB monoclonal antibodies cross-reacted with another factor required for U1 transcription. Upon addition of recombinant TFIIB, such a factor might have been released from the antibodies and thus become available again to participate in U1 transcription. We therefore tested whether RNA polymerase II transcription from snRNA genes requires TFIIB by depleting a whole-cell extract with anti-TFIIB antibodies (α-IIB/2) bound to protein A-agarose beads. Figure 2A shows the effects of such depletion on transcription from various promoters. Treatment of the extract with anti-TFIIB antibodies inhibited transcription from the Ad2ML promoter much more than treatment with preimmune beads, as expected (compare lanes 4 and 5 with lanes 2 and 3 in the top gel). Significantly, the anti-TFIIB depletion also inhibited transcription from the RNA polymerase II U1 snRNA promoter but not from the RNA polymerase III U6 snRNA and VAI promoters (Fig. 2A, compare lanes 4 and 5 to lanes 2 and 3 in the second, third, and fourth gels). An immunoblot analysis of the extracts, shown in Fig. 2B, indicated that the anti-TFIIB depletion had been efficient.
To determine whether the inhibition of transcription from the U1 snRNA promoter was indeed due to removal of TFIIB rather than, for example, removal of a TFIIB-associated factor, we tested whether addition of recombinant TFIIB to depleted extracts could restore U1 transcription. For this experiment, we first complemented a whole-cell extract with biochemically purified SNAPc to ensure that SNAPc would not be limiting. As shown in Fig. 2C, treatment of the extract with preimmune beads diminished transcription but much less so than treatment with the α-IIB/2 antibody beads (compare lanes 7 and 8) or beads coupled to another anti-TFIIB antibody, α-IIB/1 (compare lanes 2 and 3). Both of these antibodies depleted TFIIB efficiently, as determined by immunoblotting (Fig. 2D). Importantly, in both cases, addition of increasing amounts of recombinant full-length TFIIB restored efficient U1 transcription (lanes 4 to 6 and 9 to 11). Addition of just the C-terminal core domain of TFIIB, which is sufficient for association with a TBP-TATA box complex but cannot sustain basal RNA polymerase II mRNA transcription (see reference 61 for a review), did not restore U1 transcription (data not shown). Together, these results strongly suggest that TFIIB is required for snRNA gene transcription by RNA polymerase II.
TFIIA is required for efficient basal RNA polymerase II transcription from the U1 snRNA promoter.
In in vitro transcription systems that are reconstituted wtih TFIID, but not with TBP, TFIIA is required for basal transcription from RNA polymerase II mRNA-type promoters (12, 52, 81, 94). This finding probably reflects the ability of TFIIA to counteract the effects of factors that interfere with the binding of TBP to the TATA box and that are associated with TFIID or may be present in TFIID fractions (2, 8, 30, 41, 43, 51, 58, 63).
We used polyclonal antibodies directed against the α and β subunits of TFIIA coupled to protein A-agarose beads to deplete extracts and tested the depleted extracts for transcription from the four model promoters. As shown in Fig. 3, treatment with anti-TFIIA antibodies had only a small effect on transcription from the Ad2ML promoter (compare lanes 2 and 3), even though most of the TFIIA present in the extract was removed by such treatment (data not shown, but see Fig. 4B below). Addition of increasing amounts of TFIIA restored transcription from the Ad2ML promoter to levels that were slightly higher than starting levels (compare lanes 4 to 6 with lane 2). Since the role of TFIIA in basal transcription from mRNA-type RNA polymerase II promoters is mainly to counteract inhibitors that disrupt association of TBP with the TATA box, addition of excess recombinant TBP to depleted extracts should overcome the need for TFIIA. Indeed, addition only of recombinant TBP to the TFIIA-depleted extract achieved levels of transcription that were higher than those observed in the mock-depleted extract (lane 7) and that were not increased further by addition of TFIIA (lanes 8 and 9). In contrast, when highly purified TFIID was added, transcription could be further enhanced by addition of TFIIA (data not shown). Together, these results indicate that TFIIA has a small positive effect on basal transcription from the Ad2ML promoter in this system but that this effect is not apparent in the presence of added recombinant TBP. This is consistent with a role for TFIIA in counteracting repressors that inhibit the binding of TFIID to the TATA box.
FIG. 4.
TFIIA is required for basal transcription from the U1 snRNA promoter. (A) In vitro transcription reactions were performed with the pU1*G−Oct− template and untreated whole-cell extract (WCE; lane 1) or WCE treated with either control (nonimmume) antibody beads (lane 2) or beads coupled to antibodies directed against the α and β subunits of TFIIA (α-IIA; lanes 3 to 6) at a 1:1 beads-to-extract ratio. Six microliters of E. coli-expressed TFIIA was added to the reaction mixtures in lanes 4 and 6, and recombinant TBP (3 ng) was added to the reaction mixtures in lanes 5 and 6. The signals corresponding to correct transcription initiation were quantitated with a phosphorimager, the background was subtracted, and the numbers were normalized for the signal obtained in lane 2, which was set at 1. (B) Immunoblot analysis of the extracts used in the experiment whose results are shown in panel A with a polyclonal antibody directed against the α subunit of TFIIA. WCE (5 μl) was loaded in lane 1, and control depleted extract (10 μl) and α-IIA depleted extract (10 μl) were loaded in lanes 2 and 3, respectively. The location of the α-subunit is indicated at left. Ctr., control.
We then tested the same extracts for transcription from the RNA polymerase II U1 snRNA promoter and observed strikingly different results. Depletion with anti-TFIIA antibody beads reduced transcription much more efficiently than depletion with control antibody beads (Fig. 3, compare lanes 2 and 3 in the second panel). Addition of increasing amounts of TFIIA reproducibly restored only a low level of U1 transcription, below that observed in mock-immunodepleted extract (lanes 4 to 6). Unlike with transcription from the Ad2ML promoter, where the requirement for TFIIA was obviated by addition of excess recombinant TBP, addition only of recombinant TBP to the U1 reaction mixtures restored only a low level of transcription (lane 7), which could be increased to levels that were higher than starting levels by further addition of increasing amounts of TFIIA (compare lanes 8 and 9 to lane 2). The combined effect of TBP and TFIIA could not be achieved by simply increasing the amounts of added TBP indeed, this resulted only in stimulation of transcription from an aberrant start site (Fig. 3, asterisk) which is directed by RNA polymerase III (data not shown).
For TATA-containing mRNA promoters, TFIIA is much more important for activated transcription than for basal transcription. To ensure that the signal derived from the U1 promoter reflected basal transcription, we repeated the experiment using a U1 template with a mutated distal sequence element. As shown in Fig. 4A, depletion with anti-TFIIA antibodies again severely and specifically decreased U1 transcription (lanes 2 and 3) and addition of either TFIIA or TBP alone did not reconstitute efficient transcription (lanes 4 and 5). However, as before, addition of both TFIIA and TBP reconstituted high levels of U1 transcription (lane 6). The immunoblot in Fig. 4B shows that TFIIA depletion was efficient. Together, these results indicate that TFIIA plays a role in basal transcription from the U1 snRNA promoter that is different from its role in transcription from the Ad2ML promoter. Indeed, whereas with the Ad2ML promoter addition of recombinant TBP circumvents the need for TFIIA, recombinant TBP cannot be used by the U1 snRNA promoter unless TFIIA is present.
TFIIA is not essential for RNA polymerase III transcription from the U6 snRNA promoter in vitro.
TFIIA has been reported to be required for or to stimulate RNA polymerase III transcription from the U6, VAI, tRNA, and 5S promoters (57, 87). These results contrast with the finding that, unlike RNA polymerase II transcription, RNA polymerase III transcription is unaffected in extracts from Saccharomyces cerevisiae cells carrying temperature-sensitive mutations in the TFIIA subunits (33) and with the recent observation that depletion of TFIIA from a nuclear extract has no effect on VAI transcription (8).
The same depleted extracts tested as described above for RNA polymerase II transcription were also tested for their ability to direct RNA polymerase III transcription. As shown in Fig. 3, treatment with anti-TFIIA antibody beads had no effect on VAI transcription, even though TFIIA had been efficiently depleted as judged from immunoblots (data not shown), suggesting that TFIIA is not required for transcription from RNA polymerase III tRNA-type promoters (compare lanes 2 and 3, bottom panel). In sharp contrast, however, U6 snRNA transcription was significantly reduced in extracts immunodepleted of TFIIA compared to transcription in mock-immunodepleted extracts (lanes 2 and 3, third panel). Addition of recombinant TFIIA had no significant effect (lanes 4 to 6), but U6 transcription could be reconstituted to levels higher than starting levels by addition of recombinant TBP (lane 7). These levels could not be increased by further addition of TFIIA (lanes 8 and 9).
To confirm that TFIIA is dispensable for U6 RNA polymerase III transcription in the presence of exogenous TBP, we supplemented the extracts with recombinant TBP before treatment with the beads. As shown in Fig. 5A and B, treatment with anti-TFIIA antibody beads depleted TFIIA much more efficiently than treatment with control antibody beads (Fig. 5B, compare lanes 7 to 11 with lanes 2 to 6), yet in this case, depletion of TFIIA had no specific deleterious effect on transcription from either the Ad2ML promoter, as expected, or the U6 promoter (Fig. 5A, compare lanes 7 to 11 with lanes 2 to 6). These results suggest that in the presence of excess exogenous TBP, TFIIA is not required for U6 transcription in vitro.
It remained possible, however, that TFIIA helped to recruit limiting amounts of TBP to the U6 promoter. To test this possibility, we supplemented an extract treated with anti-TFIIA antibody beads with decreasing amounts of TBP, with or without TFIIA. As shown in Fig. 5C, addition of TFIIA alone had little effect on U6 transcription (compare lanes 4, 7, 10, 13, and 16 to lane 3). Addition of 3.3 ng of TBP restored transcription to levels higher than those observed in the extract depleted with preimmune antibody beads (lane 18), but this level was not changed by further addition of TFIIA (lane 17). As lower levels of TBP were added, U6 transcription decreased but in no case did addition of TFIIA significantly improve transcription (compare lanes 5 and 6, 8 and 9, 11 and 12, and 14 and 15). Thus, even at levels of TBP too low to restore efficient U6 transcription, addition of TFIIA did not have a stimulatory effect. These results suggest that TFIIA is not required for basal U6 transcription in vitro, even when limiting amounts of TBP are added to the extract. They do not exclude, however, that TFIIA performs a role in U6 transcription in vitro.
TFIIF is required for RNA polymerase II snRNA gene transcription in vitro.
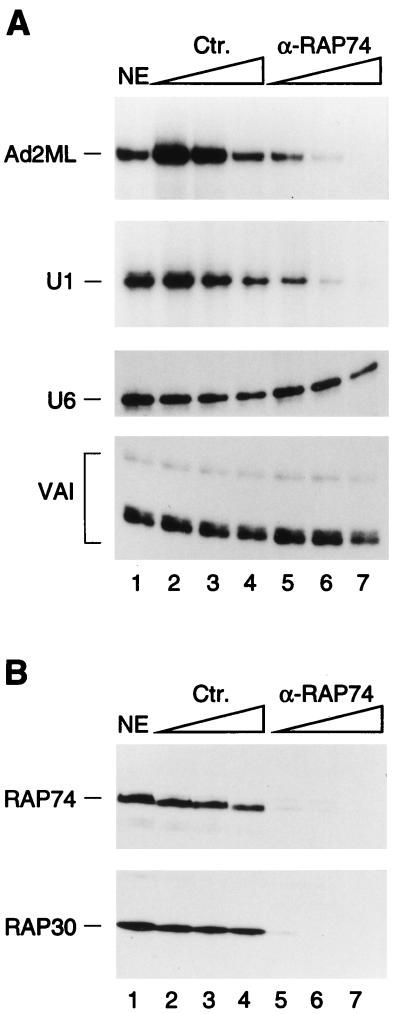
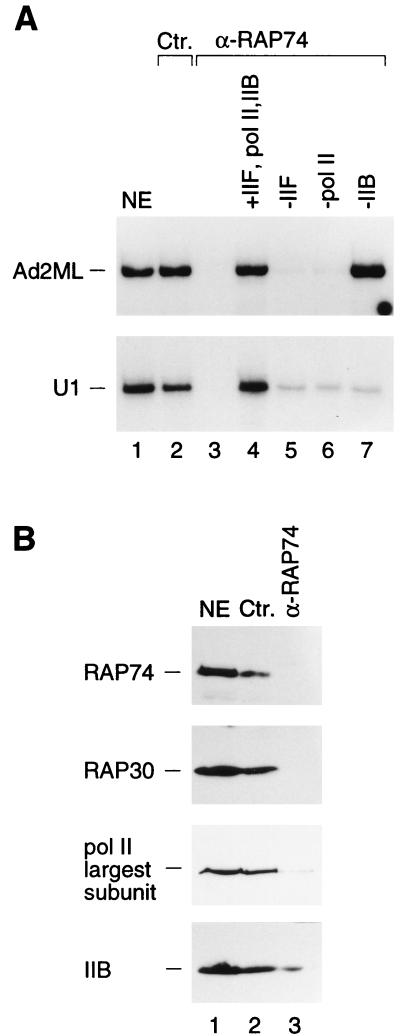
TFIIF complexed with RNA polymerase II interacts directly with TFIIB and is thus essential for the recruitment of RNA polymerase II to mRNA-type RNA polymerase II promoters. To determine if TFIIF participates in RNA polymerase II snRNA gene transcription, we depleted a nuclear extract using polyclonal antibodies directed against the RAP75 subunit of TFIIF. As shown in Fig. 6A, treatment of the extract with the anti-TFIIF antibody beads specifically inhibited transcription from both the Ad2ML and U1 snRNA promoters but had no specific effect on RNA polymerase III transcription from the VAI or U6 snRNA promoters (compare lanes 5 to 7 with lanes 2 to 4). Figure 6B shows an immunoblot of these reactions that confirms that most of the TFIIF RAP74 and RAP30 subunits had been depleted.
FIG. 6.
Depletion of TFIIF inhibits RNA polymerase II snRNA transcription from the U1 promoter. (A) In vitro transcription reactions with the Ad2ML, U1 snRNA, U6 snRNA, and VAI promoters. Transcription reactions were performed with untreated nuclear extract (NE; lane 1), NE treated with increasing amounts of control (nonimmune) antibody beads (lanes 2 to 4), or NE treated with increasing amounts of α-RAP74 antibody beads (lanes 5 to 7) at beads-to-extract ratios of 0.5:1, 1:1, and 2:1, respectively. (B) Immunoblot analysis of the extracts used to obtain the results shown in panel A with α-RAP74 and α-RAP30 antibodies. Ten microliters of every extract was loaded per lane. Ctr., control.
Attempts to reconstitute transcription by addition only of recombinant TFIIF were unsuccessful (data not shown). We reasoned that the anti-TFIIF depletion had probably removed other associated factors such as RNA polymerase II. In preliminary experiments, we therefore supplemented depleted extracts with a mixture of TFIIF and the GTFs TFIIA, TFIIB, TFIIE, TFIIH, and RNA polymerase II or with every possible mixture of all but one of these GTFs (data not shown). Figure 7 shows the results of an experiment in which we supplied only the factors we had identified as necessary for reconstitution of transcription. Addition of a mixture containing TFIIF, RNA polymerase II, and TFIIB reconstituted transcription from both the Ad2ML and U1 snRNA promoters in an extract treated with anti-TFIIF antibody beads (Fig. 7A, lanes 3 and 4). However, a mixture lacking TFIIF did not reconstitute transcription efficiently (lane 5). Thus, TFIIF is not only required for RNA polymerase II transcription from the Ad2ML promoter but also essential for efficient transcription from the U1 snRNA promoter.
FIG. 7.
TFIIF is required for U1 transcription in vitro. (A) Parallel transcription reactions were performed with the Ad2ML and U1 promoters as templates and untreated nuclear extract (NE; lane 1), NE treated with control (nonimmune) antibody beads (lane 2), or NE treated with anti-RAP74 (α-RAP74) antibody beads (lanes 3 to 7) at a 1:1 beads-to-extract ratio. Recombinant TFIIF (1 μl), purified RNA polymerase II (pol II; 1.5 μl), and recombinant TFIIB (2 μg) were added to the reaction mixture in lane 4. In lanes 5 to 7, the same combination of factors was added except for the factor indicated above each lane. (B) Immunoblot analysis of the extracts used in the experiment whose results are shown in panel A with α-RAP74 and α-RAP30 antibodies, anti-C-terminal domain antibodies directed against the largest subunit of RNA polymerase II, and anti-TFIIB antibodies. Ten microliters of extract was loaded per lane. Ctr., control.
When the TFIIF-depleted extract was complemented with a mixture containing TFIIF and TFIIB but lacking RNA polymerase II, we observed little or no transcription from both the Ad2ML and the U1 promoter (Fig. 7A, lane 6). This result suggests that depletion with the anti-TFIIF antibodies also removed RNA polymerase II. Indeed, consistent with the known association of TFIIF with RNA polymerase II, the amounts of both factors were severely reduced in extracts treated with anti-TFIIF antibody beads as determined by immunoblotting (Fig. 7B). More surprisingly, a mixture lacking TFIIB did not restore U1 transcription even though it restored transcription from the Ad2ML promoter efficiently (Fig. 7A, lane 7). As shown in the bottom gel of Fig. 7B, the amounts of TFIIB were reduced in the TFIIF-depleted extract, although much less so than the amounts of either TFIIF or RNA polymerase II. These results indicate that the Ad2ML and U1 promoters differ in their requirements for TFIIB. One possibility is that transcription from the U1 promoter simply requires higher levels of TFIIB. Indeed, even different mRNA promoters require different concentrations of TFIIB for optimal transcription. Thus, at concentrations of TFIIB that still stimulate transcription of the Drosophila Krüppel and Jockey promoters, transcription from the Drosophila alcohol dehydrogenase and Ad2E4 promoters is repressed, probably by a squelching effect (89).
TFIIE stimulates transcription from the RNA polymerase II snRNA U1 promoter.
TFIIE regulates the activity of TFIIH, plays a direct role in promoter melting, and can help recruit TFIIA and TBP to the TATA box of mRNA promoters (61, 93). To determine if TFIIE is involved in RNA polymerase II snRNA gene transcription, we depleted nuclear extracts using polyclonal antibodies directed against the p34 subunit of TFIIE. Because both TFIIE and TFIIH requirements are more apparent with linear templates than with supercoiled templates (66, 83), we linearized the templates by digestion with a restriction enzyme. As shown in Fig. 8, treatment of the extract with the anti-TFIIE antibody beads depleted TFIIE from the extracts (Fig. 8B) and inhibited transcription from linearized templates containing the Ad2ML or the U1 promoter (Fig. 8A, compare lanes 5 to 7 with lanes 2 to 4). We then tested the effect of adding back TFIIE. As shown in Fig. 9A and B, treatment with anti-TFIIE antibody beads inhibited transcription (Fig. 9A, lane 3) and efficiently depleted TFIIE (Fig. 9B, lane 3), as expected, and for both the Ad2ML promoter and the U1 snRNA promoter, addition of recombinant TFIIE resulted in recovery of a significant but low level of transcription (Fig. 9A, lanes 4 to 6). These results suggested that other factors required for U1 transcription had been depleted along with TFIIE. Indeed, as shown in Fig. 9C, addition of RNA polymerase II and all of the GTFs (excluding TBP) to the TFIIE-depleted extract restored full levels of transcription from the Ad2ML and U1 promoters (compare lanes 3 and 4). Importantly, addition of these same GTFs without TFIIE resulted in much lower levels of transcription (lane 5). Together, these results suggest that TFIIE is involved in RNA polymerase II transcription of snRNA genes.
Figure 9D shows the levels of the two TFIIE subunits, the largest RNA polymerase II subunit, and TFIIB, present in the extracts treated with the anti-TFIIE antibodies. As expected, both TFIIE subunits were efficiently depleted (lane 3). In addition, however, there were small decreases in the amounts of the largest RNA polymerase II subunit and TFIIB. Consistent with these small decreases, omission of RNA polymerase II or TFIIB from the combination of GTFs had a deleterious effect on transcription from the Ad2ML and U1 promoters (Fig. 9C, lanes 6 and 8). Notably, however, omission of RNA polymerase II was more detrimental to transcription from the Ad2ML promoter than to transcription from the U1 promoter, whereas the reverse was true for omission of TFIIB. Thus, as observed above for TFIIF-depleted extracts, the Ad2ML and the U1 promoters have different quantitative requirements for various factors, in particular TFIIB.
DISCUSSION
Which of the GTFs are required for RNA polymerase II transcription of snRNA genes is not known. Here we show that efficient transcription from the human U1 snRNA promoter requires TFIIA, TFIIB, TFIIF, and TFIIE.
TFIIB, including the N-terminal domain, is required for U1 transcription.
On mRNA promoters, a platform for recruitment of RNA polymerase II is created by the binding of TBP and TFIIB to the TATA box. TFIIA stabilizes the complex but is not absolutely required. In contrast, TFIIB is essential because it then recruits the RNA polymerase II-TFIIF complex through interactions with both RNA polymerase II and the small subunit of TFIIF (61). TFIIB is composed of two functional domains, a protease-resistant C-terminal domain and a protease-sensitive N-terminal domain. The C-terminal domain is sufficient for association with TBP bound to a TATA box, but it cannot direct basal RNA polymerase II transcription (3, 7, 22, 26, 91) or recruit the RNA polymerase II-TFIIF complex (22). Our data indicate that U1 transcription requires TFIIB, including the N-terminal domain, as well as TFIIF. This suggests that TFIIB performs the same role on both mRNA and RNA polymerase II snRNA promoters, namely, bridging DNA binding factors with the RNA polymerase II-TFIIF complex. It is striking, however, that transcription from the U1 promoter is more sensitive to small decreases in the TFIIB concentration brought about by the anti-TFIIF and anti-TFIIE depletions than transcription from the Ad2ML promoter (Fig. 7B and 9D). This suggests that TFIIB recruitment to the U1 promoter may be less efficient than recruitment to the Ad2ML promoter. The affinity of TFIIB for various mRNA promoters is determined in part by contacts with the DNA sequence upstream of the TATA box (45). The Ad2ML promoter has a good TFIIB binding site, but this may not be the case for the U1 promoter. In addition, the protein-protein contacts that mediate TFIIB recruitment to the Ad2ML and U1 promoters are likely to be different.
TFIIA and TBP are required for U1 transcription.
The RNA polymerase II core snRNA promoters do not contain a TATA box. Instead, they consist of the PSE, which recruits SNAPc and thus presumably nucleates the assembly of the initiation complex. TBP is required for RNA polymerase II snRNA gene transcription, but how it is recruited to the TATA-less RNA polymerase II snRNA promoters is not clear. Neither TFIID nor TFIIIB can complement a TBP-depleted extract (73, 95), and recombinant TBP has some activity but fails to reconstitute efficient transcription (73). In addition, as shown here, recombinant TBP also fails to reconstitute efficient transcription in a TFIIA-depleted extract, even though it does reconstitute efficient transcription from the Ad2ML promoter. Significantly, however, a combination of TBP and TFIIA restores efficient transcription, suggesting that TFIIA somehow allows the utilization of TBP by the U1 promoter. Together, these data are consistent with TFIIA playing a much more crucial role in the recruitment of TBP to basal snRNA promoters than in the recruitment of TBP to basal TATA-containing mRNA promoters. Because TFIIB is also required for U1 transcription, it is likely that SNAPc, TBP, TFIIA, and TFIIB are all involved in the establishment of the platform that can recruit the RNA polymerase II-TFIIF complex.
TFIIE and TFIIH in U1 snRNA gene transcription.
In the stepwise assembly of transcription complexes on mRNA promoters, the recruitment of the RNA polymerase II-TFIIF complex is followed by the recruitment of TFIIE and TFIIH. We have shown that TFIIE is involved in RNA polymerase II transcription of snRNA genes; however, we were unable to show convincingly that TFIIH is required. Indeed, although treatment of extracts with anti-TFIIH antibodies reduced U1 transcription dramatically, we could restore transcription from linear templates nearly as efficiently with a mixture of GTFs missing TFIIH as with a mixture of GTFs containing TFIIH (data not shown). In contrast, addition of TFIIH was needed to reconstitute transcription from the Ad2ML promoter. Because we cannot exclude the presence of trace amounts of TFIIH in the RNA polymerase II preparation, we can only conclude that in our assay, U1 transcription either did not require TFIIH or required much lower levels than transcription from the Ad2ML promoter. If U1 transcription indeed did not require TFIIH, what would then be the role of TFIIE? Two of TFIIE’s functions are to recruit TFIIH and cooperate with TFIIH to bring about promoter melting, late in preinitiation complex assembly. However, TFIIE has also been shown to stimulate basal mRNA transcription from supercoiled templates in the absence of TFIIH (28, 83, 93). Consistent with this observation, TFIIE enhances the binding of TBP to the TATA box as well as the cooperative binding of TFIIA and TBP to the Ad2ML promoter and the small TFIIE subunit interacts directly with TFIIA (93). Thus, TFIIE may also play a role early in preinitiation complex assembly. It is quite possible that for RNA polymerase II transcription of snRNA promoters, TFIIE helps in the recruitment of TFIIA, TBP, and/or SNAPc to the promoter.
TFIIB is not involved in RNA polymerase III transcription from the U6 promoter.
We also tested the roles of many of the GTFs in RNA polymerase III transcription from both the U6 snRNA promoter and the VAI promoter. RNA polymerase III transcription from the VAI promoter and other promoters with gene-internal elements requires the TBP-containing complex TFIIIB, which in mammalian cells consists minimally of TBP and the human TFIIB-related factor (BRF) (59), probably the same protein as another homolog of yeast BRF referred to as human TFIIIB90 (69). However, we found that human BRF is not required for transcription from the human U6 promoter in vitro (59). We therefore wondered whether the U6 promoter recruits instead the related protein TFIIB. Our results clearly indicate that this is not the case, which is consistent with the idea that recruiment of TFIIB to a promoter is the decisive step towards specific recruitment of RNA polymerase II.
A role for TFIIA in U6 transcription?
TFIIA has been reported previously to be required for U6 transcription in vitro, as well as for transcription from several promoters with gene-internal elements, including the VAI promoter (57, 87). We were unable to demonstrate a TFIIA requirement for transcription from the VAI promoter, consistent with other observations from both yeast and mammalian systems (8, 33). With the U6 promoter, however, depletion with anti-TFIIA antibodies severely inhibited transcription but efficient transcription could be reconstituted by addition of recombinant TBP. There are at least two possible interpretations for these results. It is possible that TFIIA plays an antirepressor role for basal U6 transcription in much the same way as it does for basal RNA polymerase II transcription from the Ad2ML promoter, for example, by displacing a repressor such as Mot1. Indeed, Mot1 is capable of repressing transcription from the human U6 promoter in vitro (8). Addition of excess recombinant TBP would then relieve the need for TFIIA. It is also possible that depletion with anti-TFIIA antibodies removes all of the TBP competent for U6 transcription; some TBP associates with TFIIA on the anti-TFIIA beads (data not shown), consistent with the previous observation that endogenous Drosophila IIA associates with TBP in the absence of DNA (12, 84, 92). A removal of the TBP competent for U6 transcription by the anti-TFIIA antibodies would imply that all such TBP is complexed with TFIIA in extracts. This in turn would suggest either that the TFIIA-TBP complex has to be broken apart to liberate TBP for U6 transcription or, more likely, that the complex is recruited as such to the U6 promoter. Thus, although we could not demonstrate a role for TFIIA in U6 transcription in our assay, it remains possible that TFIIA is indeed recruited to the U6 promoter in vivo.
ACKNOWLEDGMENTS
We thank M. Tanaka and W. P. Tansey for GST-TFIIB expression vectors; G. Binns for peptide synthesis; and E. Ford, R. W. Henry, B. Ma, V. Mittal, P. S. Pendergast, and C. L. Sadowski for advice and reagents. We also thank R. Drapkin for providing reagents and advice in the early stages of this work; D. Ma for anti-IIAα and -β antibodies and TFIIA; S. Kim for TFIIB; K. P. Kumar for anti-IIEp34 antibodies; and E. Maldonado for TFIIE, TBP, and anti-RAP74, anti-IIEp56, anti-RAP30, and anti-C-terminal domain antibodies. The monoclonal ERCC3 antibodies were characterized by G. Le Roy. We thank V. Mittal for comments on the manuscript and M. Ockler, J. Duffy, and P. Renna for artwork and photography.
This work was funded in part by NIH grants GM38810 to N.H. and GM37120 to D.R. N.H and D.R. are supported by the Howard Hughes Medical Institute.
REFERENCES
- 1.Akoulitchev S, Makela T P, Weinberg R A, Reinberg D. Requirements for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- 2.Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 3.Barberis A, Müller C W, Harrison S C, Ptashne M. Delineation of two functional regions of transcription factor TFIIB. Proc Natl Acad Sci USA. 1993;90:5628–5632. doi: 10.1073/pnas.90.12.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bengal E, Flores O, Krauskopf A, Reinberg D, Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol. 1991;11:1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernues J, Simmen K A, Lewis J D, Gunderson S I, Polycarpou-Schwarz M, Moncollin V, Egly J-M, Mattaj I W. Common and unique transcription factor requirements of human U1 and U6 snRNA genes. EMBO J. 1993;12:3573–3585. doi: 10.1002/j.1460-2075.1993.tb06031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradsher J N, Tan S, McLaury H-J, Conaway J W, Conaway R C. RNA polymerase II transcription factor SIII. J Biol Chem. 1993;268:25594–25603. [PubMed] [Google Scholar]
- 7.Buratowski S, Zhou H. Functional domains of transcription factor TFIIB. Proc Natl Acad Sci USA. 1993;90:5633–5637. doi: 10.1073/pnas.90.12.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chicca J J, Auble D T, Pugh B F. Cloning and biochemical characterization of TAF-172, a human homolog of yeast Mot1. Mol Cell Biol. 1998;18:1701–1710. doi: 10.1128/mcb.18.3.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho H, Maldonado E, Reinberg D. Affinity purification of human RNA polymerase II complex using monoclonal antibodies against transcription factor IIF. J Biol Chem. 1997;272:11495–11502. doi: 10.1074/jbc.272.17.11495. [DOI] [PubMed] [Google Scholar]
- 10.Conaway J W, Conaway R C. An RNA polymerase II transcription factor shares functional properties with Escherichia coli ς70. Science. 1990;248:1550–1553. doi: 10.1126/science.2193400. [DOI] [PubMed] [Google Scholar]
- 11.Conaway R C, Garrett K P, Hanley J P, Conaway J W. Mechanism of promoter selection by RNA polymerase II: mammalian transcription factors α and β γ promote entry of polymerase into the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:6205–6209. doi: 10.1073/pnas.88.14.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortes P, Flores O, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II: purification and analysis of transcription factor IIA and identification of transcription factor IIJ. Mol Cell Biol. 1992;12:413–421. doi: 10.1128/mcb.12.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeJong J, Bernstein R, Roeder R G. Human general transcription factor TFIIA; characterization of a cDNA encoding the small subunit and requirement for basal and activated transcription. Proc Natl Acad Sci USA. 1995;92:3313–3317. doi: 10.1073/pnas.92.8.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drapkin R, Reardon J T, Ansari A, Huang J-C, Zawel L, Ahn K, Sancar A, Reinberg D. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:169–172. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 15.Dvir A, Garrett K P, Chalut C, Egly J M, Conaway J W, Conaway R C. A role for ATP and TFIIIH in activation of the RNA polymerase II preinitiation complex prior to transcription initiation. J Biol Chem. 1996;271:7245–7258. doi: 10.1074/jbc.271.13.7245. [DOI] [PubMed] [Google Scholar]
- 16.Feaver W J, Gileadi O, Li Y, Kornberg R D. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell. 1991;67:1223–1230. doi: 10.1016/0092-8674(91)90298-d. [DOI] [PubMed] [Google Scholar]
- 17.Feaver W J, Svejstrup J Q, Bardwell L, Bardwell A J, Buratowski S, Gulyas K D, Donahue T F, Friedberg E C, Kornberg R D. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell. 1993;75:1379–1387. doi: 10.1016/0092-8674(93)90624-y. [DOI] [PubMed] [Google Scholar]
- 18.Feaver W J, Svejstrup J Q, Henry N L, Kornberg R D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 19.Flores O, Ha I, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and subunit composition of transcription factor IIF. J Biol Chem. 1990;265:5629–5634. [PubMed] [Google Scholar]
- 20.Flores O, Lu H, Killeen M, Greenblatt J, Burton Z F, Reinberg D. The small subunit of transcription factor IIF recruits RNA polymerase II into the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:9999–10003. doi: 10.1073/pnas.88.22.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flores O, Maldonado E, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Factors IIE and IIF independently interact with RNA polymerase II. J Biol Chem. 1989;264:8913–8921. [PubMed] [Google Scholar]
- 22.Ha I, Roberts S, Maldonado E, Sun X, Green M, Reinberg D. Multiple functional domains of human transcription factor IIB. Distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 1993;7:1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- 23.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 24.Henry R W, Mittal V, Ma B, Kobayashi R, Hernandez N. Assembly of a functional, core promoter complex (SNAPc) shared by RNA polymerase II and III. Genes Dev. 1998;12:2664–2672. doi: 10.1101/gad.12.17.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez N. Transcription of vertebrate snRNA genes and related genes. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 281–313. [Google Scholar]
- 26.Hisatake K, Roeder R G, Horikoshi M. Functional dissection of TFIIB domains required for TFIIB-TFIID-promoter complex formation and basal transcription activity. Nature. 1993;363:744–747. doi: 10.1038/363744a0. [DOI] [PubMed] [Google Scholar]
- 27.Holstege F C, Fiedler U, Timmers H T. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holstege F C P, Tantin D, Carey M, van der Vilet P C, Timmers H T M. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holstege F C P, van der Vliet P C, Timmers H T M. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 30.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 31.Izban M G, Luse D S. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′ to 5′ direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y, Yan M, Gralla J D. A three-step pathway of transcription initiation leading to promoter clearance at an activated RNA polymerase II promoter. Mol Cell Biol. 1996;16:1614–1621. doi: 10.1128/mcb.16.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang J J, Auble D T, Ranish J A, Hahn S. Analysis of the yeast transcription factor TFIIA: distinct functional regions and a polymerase II-specific role in basal and activated transcription. Mol Cell Biol. 1995;15:1234–1243. doi: 10.1128/mcb.15.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaufmann J, Ahrens K, Koop R, Smale S T, Muller R. CIF150, a human cofactor for transcription for IID-dependent initiator function. Mol Cell Biol. 1998;18:233–239. doi: 10.1128/mcb.18.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaufmann J, Smale S T. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 1994;8:821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- 36.Kephart D D, Wang B Q, Burton Z F, Price D H. Functional analysis of Drosophila factor 5 (TFIIF), a general transcription factor. J Biol Chem. 1994;269:13536–13543. [PubMed] [Google Scholar]
- 37.Killeen M, Greenblatt J. The general transcription factor RAP30 binds to RNA polymerase II and prevents it from binding nonspecifically to DNA. Mol Cell Biol. 1992;12:30–37. doi: 10.1128/mcb.12.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 39.Kim Y J, Bjorklund S, Li Y, Sayre H M, Kornberg R d. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi N, Horn P J, Sullivan SM, Triezenberg S J, Boyer T G, Berk A J. DA-complex assembly activity required for VP16C transcriptional activation. Mol Cell Biol. 1998;18:4023–4031. doi: 10.1128/mcb.18.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kokubo T, Swanson M J, Nishikawa J I, Hinnebusch A G, Nakatani Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol Cell Biol. 1998;18:1003–1012. doi: 10.1128/mcb.18.2.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 43.Kretzschmar M, Meisterernst M, Roeder R G. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc Natl Acad Sci USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar K P, Akoulitchev S, Reinberg D. Promoter-proximal stalling results from the inability to recruit transcription factor IIH to the transcription complex and is a regulated event. Proc Natl Acad Sci USA. 1998;95:9767–9772. doi: 10.1073/pnas.95.17.9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lagrange T, Kapanidis A N, Tang H, Reinberg D, Ebright R H. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lobo S, Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- 47.Lobo S M, Hernandez N. Transcription of snRNA gene by RNA polymerases II and III. In: Conaway R C, Conaway J W, editors. Transcription, mechanisms and regulation. New York, N.Y: Raven Press, Ltd.; 1994. pp. 127–159. [Google Scholar]
- 48.Lobo S M, Lister J, Sullivan M L, Hernandez N. The cloned RNA polymerase II transcription factor IID selects RNA polymerase III to transcribe the human U6 gene in vitro. Genes Dev. 1991;5:1477–1489. doi: 10.1101/gad.5.8.1477. [DOI] [PubMed] [Google Scholar]
- 49.Lobo S M, Tanaka M, Sullivan M L, Hernandez N. A TBP complex essential for transcription from TATA-less but not TATA-containing RNA polymerase III promoters is part of the TFIIIB fraction. Cell. 1992;71:1029–1040. doi: 10.1016/0092-8674(92)90397-u. [DOI] [PubMed] [Google Scholar]
- 50.Lu H, Zawel L, Fisher L, Egly J-M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 51.Ma D, Olave I, Merino A, Reinberg D. Separation of the transcriptional coactivator and antirepression functions of transcription factor IIA. Proc Natl Acad Sci USA. 1996;93:6583–6588. doi: 10.1073/pnas.93.13.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma D, Watanabe H, Mermelstein F, Admon A, Oguri K, Sun S, Wada T, Imai T, Shiroya T, Reinberg D, Handa H. Isolation of a cDNA encoding the largest subunit of TFIIA reveals functions important for activated transcription. Genes Dev. 1993;7:2246–2257. doi: 10.1101/gad.7.11.2246. [DOI] [PubMed] [Google Scholar]
- 53.Maldonado E, Drapkin R, Reinberg D. Purification of human RNA polymerase II and general transcription factors. Methods Enzymol. 1996;274:72–100. doi: 10.1016/s0076-6879(96)74009-0. [DOI] [PubMed] [Google Scholar]
- 54.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 55.Martinez E, Chiang C M, Ge H, Roeder R G. TAFs in TFIID function through the initiator to direct basal transcription from a TATA-less class II promoter. EMBO J. 1994;13:3115–3126. doi: 10.1002/j.1460-2075.1994.tb06610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinez E, Ge H, Tao Y, Yuan C X, Palhan V, Roeder R G. Novel cofactors and TFIIA mediate functional core promoter selectivity by the human TAFII150-containing TFIID complex. Mol Cell Biol. 1998;18:6571–6583. doi: 10.1128/mcb.18.11.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meissner W, Holland R, Waldschmidt R, Seifart K H. Transcription factor IIA stimulates the expression of classical pol III-genes. Nucleic Acids Res. 1993;21:1013–1018. doi: 10.1093/nar/21.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merino A, Madden K, Lane W S, Champoux J, Reinberg D. Topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 59.Mital R, Kobayashi R, Hernandez N. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol. 1996;16:7031–7042. doi: 10.1128/mcb.16.12.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murphy S, Yoon J-B, Gerster T, Roeder R G. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol Cell Biol. 1992;12:3247–3261. doi: 10.1128/mcb.12.7.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 62.Ozer J, Lezina L E, Ewing J, Audi S, Lieberman P M. Association of transcription factor IIA with TATA binding protein is required for transcriptional activation of a subset of promoters and cell cycle progression in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:2559–2570. doi: 10.1128/mcb.18.5.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ozer J, Mitsouras K, Zerby D, Carey M, Lieberman P M. Transcription factor IIA derepresses TATA-binding protein (TBP)-associated factor inhibition of TBP-DNA binding. J Biol Chem. 1998;273:14293–14300. doi: 10.1074/jbc.273.23.14293. [DOI] [PubMed] [Google Scholar]
- 64.Ozer J, Moore P A, Bolden A H, Lee A, Rosen C A, Lieberman P M. Molecular cloning of the small (γ) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- 65.Pan G, Greenblatt J. Initiation of transcription by RNA polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J Biol Chem. 1994;269:30101–30104. [PubMed] [Google Scholar]
- 66.Parvin J D, Sharp P A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 67.Parvin J D, Shykind B M, Meyers R E, Kim J, Sharp P A. Multiple sets of basal factors initiate transcription by RNA polymerase II. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 68.Price D H, Sluder A E, Greenleaf A L. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roberts S, Miller S J, Lane W S, Lee S, Hahn S. Cloning and functional characterization of the gene encoding the TFIIIB90 subunit of RNA polymerase III transcription factor TFIIIB. J Biol Chem. 1996;271:14903–14909. doi: 10.1074/jbc.271.25.14903. [DOI] [PubMed] [Google Scholar]
- 70.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 71.Roy R, Adamczewski JP, Seroz T, Vermeulen W, Tassan J P, Schaeffer L, Nigg E A, Hoeijmakers J H, Egly J M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 72.Ruppert S, Wang E H, Tjian R. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature. 1993;362:175–179. doi: 10.1038/362175a0. [DOI] [PubMed] [Google Scholar]
- 73.Sadowski C L, Henry R W, Lobo S M, Hernandez N. Targeting TBP to a non-TATA box cis-regulatory element: a TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes Dev. 1993;7:1535–1548. doi: 10.1101/gad.7.8.1535. [DOI] [PubMed] [Google Scholar]
- 74.Schaeffer L, Moncollin V, Roy R, Staub A, Mezzina M, Sarasin A, Weeda G, Hoeijmakers J H, Egly J-M. The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J. 1994;13:2388–2392. doi: 10.1002/j.1460-2075.1994.tb06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schaeffer L, Roy R, Humbert S, Moncollin V, Vermeulen W, Hoeijmakers J H, Chambon P, Egly J-M. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 1993;260:58–63. doi: 10.1126/science.8465201. [DOI] [PubMed] [Google Scholar]
- 76.Serizawa H, Conaway R C, Conaway J W. A carboxyl-terminal-domain kinase associated with RNA polymerase II transcription factor d from rat liver. Proc Natl Acad Sci USA. 1992;89:7476–7480. doi: 10.1073/pnas.89.16.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shiekhattar R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 78.Simmen K A, Bernues J, Parry H D, Stunnenberg H G, Berkenstam A, Cavallini B, Egly J-M, Mattaj I W. TFIID is required for in vitro transcription of the human U6 gene by RNA polymerase III. EMBO J. 1991;10:1853–1862. doi: 10.1002/j.1460-2075.1991.tb07711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smale S T, Baltimore D. The “Initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 80.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 81.Sun X, Ma D, Sheldon M, Yeung K, Reinberg D. Reconstitution of human TFIIA activity from recombinant polypeptides: a role in TFIID-mediated transcription. Genes Dev. 1994;8:2336–2348. doi: 10.1101/gad.8.19.2336. [DOI] [PubMed] [Google Scholar]
- 82.Tantin D, Carey M. A heteroduplex template circumvents the energetic requirement for ATP during activated transcription by RNA polymerase II. J Biol Chem. 1994;269:17397–17400. [PubMed] [Google Scholar]
- 83.Timmer H T. Transcription initiation by RNA polymerase II does not require hydrolysis of the beta-gamma phosphoanhydride bond of ATP. EMBO J. 1994;13:391–399. doi: 10.1002/j.1460-2075.1994.tb06273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Usuda Y, Kubota A, Berk A J, Handa H. Affinity purification of transcription factor IIA from HeLa cell nuclear extracts. EMBO J. 1991;10:2305–2310. doi: 10.1002/j.1460-2075.1991.tb07767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Verrijzer C P, Chen J L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 86.Verrijzer K, Yokomori K, Chen J L, Tjian R. Drosophila TAFII150: similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science. 1994;264:933–941. doi: 10.1126/science.8178153. [DOI] [PubMed] [Google Scholar]
- 87.Waldschmidt R, Seifart K H. TFIIA is required for in vitro transcription of mammalian U6 genes by RNA polymerase III. J Biol Chem. 1992;267:16359–16364. [PubMed] [Google Scholar]
- 88.Waldschmidt R, Wanadi I, Seifart K H. Identification of transcription factors required for the expression of mammalian U6 genes in vitro. EMBO J. 1991;10:2595–2603. doi: 10.1002/j.1460-2075.1991.tb07801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wampler S L, Kadonaga J T. Functional analysis of Drosophila transcription factor IIB. Genes Dev. 1992;6:1542–1552. doi: 10.1101/gad.6.8.1542. [DOI] [PubMed] [Google Scholar]
- 90.Weis L, Reinberg D. Accurate positioning of RNA polymerase II on a natural TATA-less promoter is independent of TATA-binding-protein-associated factors and initiator-binding proteins. Mol Cell Biol. 1997;17:2973–2984. doi: 10.1128/mcb.17.6.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamashita S, Hisatake K, Kokubo T, Doi K, Roeder R G, Horikoshi M, Nakatani Y. Transcription factor TFIIB sites important for interaction with promoter-bound TFIID. Science. 1993;261:463–466. doi: 10.1126/science.8332911. [DOI] [PubMed] [Google Scholar]
- 92.Yokomori K, Admon A, Goodrich J A, Chen J-L, Tjian R. Drosophila TFIIA-L is processed into two subunits that are associated with the TBP/TAF complex. Genes Dev. 1993;7:2235–2245. doi: 10.1101/gad.7.11.2235. [DOI] [PubMed] [Google Scholar]
- 93.Yokomori K, Verrijzer C P, Tjian R. An interplay between TATA box-binding protein and transcription factors IIE and IIA modulates DNA binding and transcription. Proc Natl Acad Sci USA. 1998;95:6722–6727. doi: 10.1073/pnas.95.12.6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yokomori K, Zeidler M P, Chen J L, Verrijzer C P, Mlodzik M, Tjian R. Drosophila TFIIA directs cooperative DNA binding with TBP and mediates transcriptional activation. Genes Dev. 1994;8:2313–2323. doi: 10.1101/gad.8.19.2313. [DOI] [PubMed] [Google Scholar]
- 95.Yoon J-B, Roeder R G. Cloning of two proximal sequence element-binding transcription factor subunits (γ and δ) that are required for transcription of small nuclear RNA genes by RNA polymerases II and III and interact with the TATA-binding protein. Mol Cell Biol. 1996;16:1–9. doi: 10.1128/mcb.16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou Q, Lieberman P M, Boyer T G, Berk A J. Holo-TFIID supports transcription stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]