Abstract
Pseudouridine (Ψ) residues were localized in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (UsnRNAs) by using the chemical mapping method. In contrast to vertebrate UsnRNAs, S. cerevisiae UsnRNAs contain only a few Ψ residues, which are located in segments involved in intermolecular RNA-RNA or RNA-protein interactions. At these positions, UsnRNAs are universally modified. When yeast mutants disrupted for one of the several pseudouridine synthase genes (PUS1, PUS2, PUS3, and PUS4) or depleted in rRNA-pseudouridine synthase Cbf5p were tested for UsnRNA Ψ content, only the loss of the Pus1p activity was found to affect Ψ formation in spliceosomal UsnRNAs. Indeed, Ψ44 formation in U2 snRNA was abolished. By using purified Pus1p enzyme and in vitro-produced U2 snRNA, Pus1p is shown here to catalyze Ψ44 formation in the S. cerevisiae U2 snRNA. Thus, Pus1p is the first UsnRNA pseudouridine synthase characterized so far which exhibits a dual substrate specificity, acting on both tRNAs and U2 snRNA. As depletion of rRNA-pseudouridine synthase Cbf5p had no effect on UsnRNA Ψ content, formation of Ψ residues in S. cerevisiae UsnRNAs is not dependent on the Cbf5p-snoRNA guided mechanism.
Introns are universally present in the nuclear genes transcribed by RNA polymerase II. Introns with GU and AG terminal dinucleotides and some introns with AU and AC terminal dinucleotides are removed by spliceosomal complexes containing the U1, U2, U4, U5, and U6 small nuclear RNAs (UsnRNAs) (for reviews, see references 54 and 59), the remaining part of introns with AU and AC terminal dinucleotides being excised by complexes containing the U11, U12, U4atac, U5, and U6atac UsnRNAs (32, 79, 106, 105). In yeast cells, only introns with GU and AG borders have been detected, and their excision is catalyzed by ribonucleoprotein complexes containing UsnRNAs homologous to the vertebrate U1, U2, U4, U5, and U6 snRNAs (for a review, see reference 31). However, compared to their counterparts in other eukaryotes, the Saccharomyces cerevisiae spliceosomal UsnRNAs differ by their larger size. For example, U2 snRNA is 1,175 nucleotides (nt) long in S. cerevisiae versus 187 nt in humans, and U1 snRNA is 568 nt long in S. cerevisiae versus 164 nt in humans (for a review, see reference 31).
In spite of this difference, the splicing machineries for the elimination of the GU-AG type of introns, in both vertebrates and S. cerevisiae, share several common properties. In particular, UsnRNPs are assembled in the same sequential order (17, 27; for a review, see reference 59), and the same kinds of bi- and multimolecular RNA-RNA interactions are implicated. The picture that now emerges from a large body of experiments in several laboratories is rather complex. First, upon U1 snRNP association, the 5′ extremity of U1 forms a base-pair interaction with the intron 5′ extremity (62, 94, 95, 96, 123). Then, the U2 snRNP is associated and a base-pair interaction is formed between U2 and the intron branch-point sequence (71, 72, 116, 124). A U4/U6 RNA duplex is present in the U4/U6 snRNP (34, 86) and a tri-snRNP is generated by association of the U4/U6 snRNP with the U5 snRNP (11). When this tri-snRNP particle joins the prespliceosomal complex, several conformational changes take place and the U1 snRNA interaction at the 5′ end of the intron is replaced by a U6 snRNA interaction (41, 52, 91). The U4/U6 RNA duplex is disrupted and replaced by a U2/U6 RNA duplex (20, 35, 53, 102, 117), and the terminal loop I of U5 contacts the 3′ extremity of the upstream exon (19, 65, 66, 67, 99, 118). These structural rearrangements are required for the first trans-esterification step to occur. Following this first step, other structural rearrangements take place that reveal the catalytic activity for the second step of the reaction. In particular, the highly conserved terminal loop I of U5 then interacts with the two exon extremities for proper alignment and ligation (65–67, 99). Although several proteins play an essential role in spliceosome assembly and function (for reviews, see references 45, 109, and 114), the general idea is that some of the UsnRNAs, in particular U2 and U6, may be directly involved in catalysis (for reviews, see references 18, 30, 40, 101).
The first determinations of UsnRNA sequences were made at the RNA level, and several posttranscriptional modifications were identified. This analysis was done on U1, U2, U4, U5, and U6 snRNAs from HeLa, chicken, mouse (13, 14, 33, 42, 47, 48), rat hepatoma (for a review, see reference 81), Drosophila melanogaster (63), and plant (for a review, reference 98) cells. Then, for a long period of time, UsnRNA sequences were deduced essentially from the corresponding gene sequences so that posttranscriptional modifications were only investigated for a limited number of UsnRNAs: the Physarum polycephalum U1 and U5 snRNAs (63, 103) and, more recently, the Schizosaccharomyces pombe U1, U2, U4, U5, and U6 snRNAs (28).
Except for the cap and cap-related modifications, the two most frequently found posttranscriptional modifications at internal positions of UsnRNAs are methylation at the 2′-O position of ribose and isomerization of uridine to pseudouridine (Ψ). Only a few base methylations (m5C, m6A, and m2G) were detected (for a review, see reference 55). Since Ψ residues and 2′-O-methylated residues stabilize RNA double helices (for reviews, see references 2, 6, and 21), the functional importance of these modified nucleotides in UsnRNAs may be, at least in part, linked to the necessity to form base-pair interactions between RNA molecules at one or the other steps of the splicing process. Furthermore, in the case of Ψ residues, the presence of an additional free NH group at position 3 of the ring, as compared to uridine, generates the possibility to form an additional hydrogen bond with RNA or proteins. It is noteworthy that the modified residues of spliceosomal UsnRNAs are clustered in the segments involved in intermolecular interactions, and several of these modifications are conserved in all of the species that were studied so far (103; for a review, see reference 55). This is the case for one of the two Ψ residues located in the U1 region that base pairs with the intron, for the two Ψ residues present in the U2 region, which interacts with the branch site, and for the numerous posttranscriptional modifications of the U5 snRNA terminal loop I (28, 103).
The number of posttranscriptionally modified nucleotides present in the interacting regions of U6 and U2 snRNAs, supposed to form at least a part of the spliceosome active site, is rather impressive in vertebrate species. All of these observations strongly suggest an important role of UsnRNA posttranscriptional modifications in spliceosome assembly and function. An experimental evidence for a role of posttranscriptional modifications in splicing was obtained for the human U2 snRNA (93, 120). Indeed, whereas fully active U2 snRNPs were obtained upon in vitro reconstitution with HeLa cell U2 snRNP proteins and the authentic human U2 snRNA, the in vitro-produced human U2 snRNA lacking all of the posttranscriptional modifications failed to form functional splicing complexes (93). Replacement of the authentic Xenopus laevis U2 snRNA by chimeric U2 snRNAs, in which some sequences are from cellular-derived U2 and others are from in vitro-transcribed U2, demonstrated that the essential posttranscriptional modifications of the vertebrate U2 snRNA are restricted to the 27-nt 5′ terminus (120).
The biogenesis of Ψ in U1, U2, U4, U5, and U6 snRNAs from HeLa cells have been the subject of several reports (121; reference 122 and references therein; for a review, see reference 76). Using in vitro-transcribed snRNAs and nuclear or S100 extracts from HeLa cells, the existence of multiple RNA-pseudouridine synthase activities that specifically recognize U1, U2, and U5 snRNAs was demonstrated (73–75, 77). However, to date, none of the implicated RNA-pseudouridine synthases has been identified, nor was their precise specificity elucidated.
The splicing machineries of HeLa cells and S. cerevisiae are the two most extensively studied, with respect to understanding how the spliceosome is assembled and how it functions. However, in the case of S. cerevisiae snRNAs, due to the low amounts of spliceosomal UsnRNAs and their unusual lengths, no internal nucleotide modifications have been identified so far. We started such a study by investigating the presence of Ψ residues in the S. cerevisiae full-length U4, U5, and U6 snRNAs and in the regions of the S. cerevisiae U1 and U2 snRNAs that have a counterpart with modified residues in the vertebrate U1 and U2 snRNAs. We also looked for the pseudouridine synthases that may catalyze the formation of the identified residues. Based on in vitro experiments, a case of dual-specificity was already described for an E. coli RNA-pseudouridine synthase (the RluAp enzyme that catalyzes the site-specific formation of Ψ at position 32 of tRNA anticodon and at position 746 of 23 S rRNA) (115). We therefore asked whether some of the already characterized yeast pseudouridine synthases, which act on tRNAs or rRNAs, can also modify UsnRNAs. To this end, we performed the mapping of the Ψ residues in UsnRNAs extracted from yeast strains carrying separate disruptions of the genes coding for the already-characterized RNA-pseudouridine synthases Pus1p, Pus3p, and Pus4p acting on tRNAs (10, 51, 60, 97); the putative Pus2p enzyme, whose substrate has not been identified so far (97); and Cbf5p acting on rRNA complexed with snoRNA guides (50).
In this study, we report the mapping of Ψ residues in the S. cerevisiae spliceosomal UsnRNAs and demonstrate that one of the previously identified tRNA-pseudouridine synthases (Pus1p) is directly implicated in the pseudouridylation of U2 snRNA in vivo.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The following S. cerevisiae strains were used in this study: S. cerevisiae FL100 (ATCC 28383) (49); S. cerevisiae strains carrying disruptions in the PUS1, PUS2, and PUS3 genes that were described previously (51, 97); an S. cerevisiae strain with a disrupted PUS4 gene that was kindly provided by R. Planta (University of Amsterdam) (10); and an S. cerevisiae strain carrying a deletion in the PUS1 gene transformed with a plasmid expressing an active recombinant protein ProtA-Pus1p (pUN100-PUS1) (97). All of these S. cerevisiae strains were grown at 30°C on YPD liquid medium (1% [wt/vol] yeast extract, 1% [wt/vol] Bacto Peptone, and 2% [wt/vol] glucose). The essential gene CBF5 was fused on the chromosome to a GAL-repressible promoter (50). Transcription driven from GAL-regulated promoters is strongly repressed when strains are grown on glucose medium, allowing the effects of depletion of essential proteins to be monitored. For depletion of Cbf5p, cells growing exponentially in permissive conditions (2% galactose, 2% sucrose, and 2% raffinose minimal medium) at 30°C were harvested by centrifugation, washed, and resuspended in 2% glucose minimal medium. During growth, cells were diluted with prewarmed medium and constantly maintained in exponential phase. For the experiment described in Fig. 4, two independently isolated GAL::cbf5 strains (YDL521-1 and YDL521-3) (50) were used. RNA was extracted from these strains after transfer to nonpermissive conditions for up to 70 h. At this time point of transfer, Ψ formation in rRNA is strongly inhibited and no H+ACA snoRNA were detected (50).
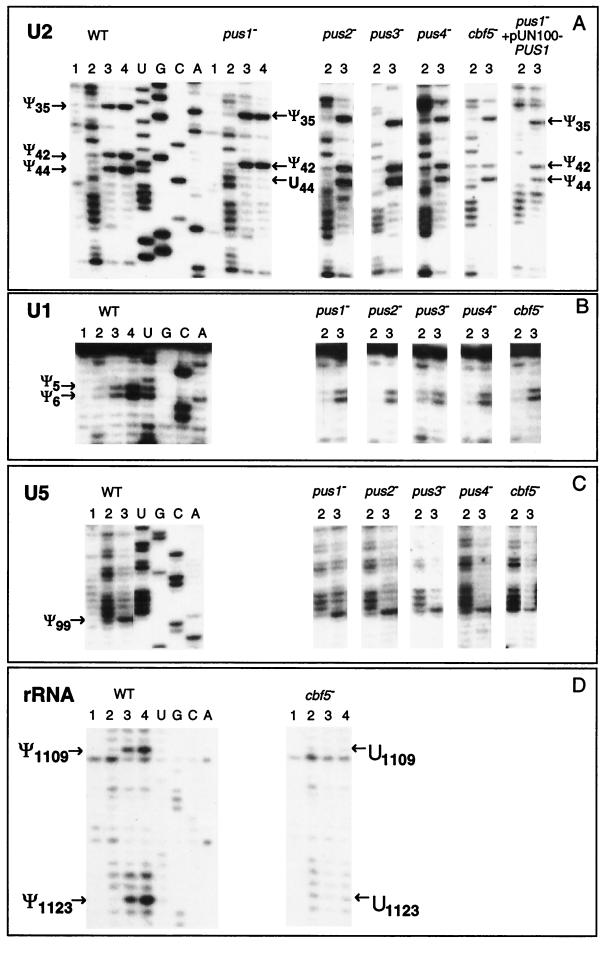
FIG. 4.
UsnRNA and rRNA pseudouridylation pattern in yeast mutant strains. Mapping of Ψ residues in U2 (A), U1 (B), and U5 (C) snRNAs by the CMCT-RT method, was performed as indicated in Fig. 1. The total RNAs used were extracted from the wild-type S. cerevisiae (WT) and from strains carrying a disruption in one of the PUS1, PUS2, PUS3, or PUS4 genes or from GAL::cbf5 strain (cbf5−) grown as described in Materials and Methods. In the case of U2 snRNA (last row of panel A), an additional analysis was performed with total RNA extracted from the pus1− strain transformed with plasmid pUN100-PUS1 containing a wild-type PUS1 gene. Lanes 1, 2, 3, and 4 correspond to the CMCT analysis according to the legend of Fig. 1. U, G, C, and A is the sequencing ladder. To minimize the figure size, only lanes 2 and 3 are shown for strains that show no variation as compared to wild type. The detected Ψ residues are indicated by arrows. The control experiment showing the absence of Ψ residues in the 26S rRNA domain II (positions 1010 to 1140) from the GAL::cbf5 strain is shown in panel D. For the wild-type and the GAL::cbf5 (cbf5−) strains, Ψ residues in this 26S region were mapped by CMCT as described in Materials and Methods. To minimize the figure size, only the portion of 26S rRNA containing the two Ψ residues 1109 and 1123 in the wild-type strain are shown.
Preparation of RNA from S. cerevisiae.
The soluble RNA fraction (containing mainly tRNAs and snRNAs) from wild-type, disrupted yeast strains and the GAL::cbf5 strain was prepared as follow. Cells grown on YPD liquid medium until the late stationary phase were harvested by centrifugation and resuspended in twice their volume of lysis buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 10% glycerol, and 10 mM β-mercaptoethanol. The suspension was frozen in dry ice and passed through a French Press at about 3,000 lb/in2. The homogenate obtained was centrifuged for 10 min at 10,000 × g at 4°C. The supernatant was further centrifuged in a high-speed centrifuge TL100 (Beckman) for 2 h at 80,000 rpm and 4°C. The resulting S100 extract was successively treated by equal volumes of phenol, phenol-chloroform (1:1), and chloroform-isoamyl alcohol (24:1). The extracted RNA was ethanol precipitated and used for further analysis by reverse transcription or Northern hybridization.
In vitro transcription of snRNA genes.
Plasmids containing the S. cerevisiae U1, U2, U4, U5, and U6 snRNA coding sequences under the control of a T7 promoter were kindly provided by P. Fabrizio. Synthetic U1, U2, U4, U5, and U6 snRNAs were prepared by in vitro transcription of PvuII-linearized pT7U1 plasmid, XhoI-linearized pT7U2 plasmid, StyI-linearized pT7U4 plasmid, DraI-linearized pT7U5 plasmid, and DraI-linearized pT7U6 plasmid, respectively (23, 57).
Synthesis of transcripts was carried out in 30 μl of buffer containing 40 mM Tris-HCl (pH 8.1), 20 mM MgCl2, 5 mM dithiothreitol (DTT), 1 mM spermidine, 0.01% Triton X-100, 80 mg of polyethylene glycol 8000 per ml, 2 μg of the linearized plasmid, 4 mM concentrations of each ribonucleoside triphosphate, 19 U of RNase Guard (Pharmacia), and 138 U of T7 RNA polymerase (Pharmacia). After 1 h of incubation at 37°C, the template DNA was digested by using 7.5 U of RNase-free DNase I (Pharmacia) for 30 min at 37°C. After phenol extraction and ethanol precipitation, the RNA was dissolved in 100 μl of sterile water.
Localization of Ψ residues in S. cerevisiae UsnRNAs and 26S rRNA by primer extension analysis.
Total RNA (10 μg) from the wild-type or mutant S. cerevisiae strains was used for reverse transcription. The CMCT [N-cyclohexyl-N′-(2-morpholinoethyl)-carbodiimid metho-p-toluolsulfonate] modification protocol was adapted from Bakin and Ofengand (8) with the following modifications: the CMCT treatment was performed for 2, 10, or 20 min; the treatment in bicarbonate buffer at pH 10.4 was done for 3 h; and all precipitations were done by using 0.3 M sodium acetate buffer (pH 5.3). The hydrazine reaction was performed essentially as described by Peattie (78).
Positions of CMCT and hydrazine modifications were identified by primer extension analysis with AMV reverse transcriptase (RT; Life Sciences) as described by Mougin et al. (61). The oligonucleotides complementary to the following regions of UsnRNAs and 26S rRNA were used as primers for reverse transcription: U1 (nt 57 to 72), U2 (nt 104 to 126), U4 (nt 68 to 90 and nt 134 to 160), U5 (nt 159 to 182), U6 (nt 93 to 112), and 26S rRNA (nt 1144 to 1163). Oligonucleotides were 5′ end labeled with [γ-32P]ATP (3,000 Ci/mmol) and T4 polynucleotide kinase (90).
In vivo analysis of rp51A pre-mRNA and pre-U3 snoRNA splicing.
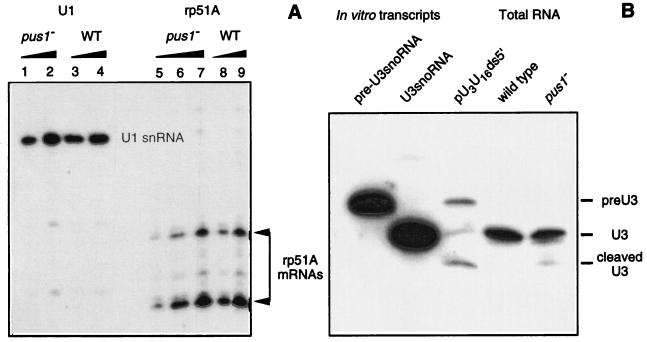
The yield of rp51A pre-mRNA in the absence of an active PUS1 gene was evaluated by primer extension analysis. Primer extension analysis on the U1 snRNA was used as a control. The oligonucleotide primer complementary to the rp51A pre-mRNA and its two splicing products were described by Teem and Rosbash (107). The oligonucleotide primer used for U1 snRNA was that described above for Ψ residue identification. Primer extension analysis was done in the conditions described by Mougin et al. (61). For quantitative cDNA synthesis, the labeled primers were always added in excess, 8 ng per assay. At the end of the incubation period, the elongation mixtures were treated with 20 μg of RNase A per ml for 30 min at 37°C and analyzed by electrophoresis on a 5% sequencing gel. To verify that the amount of synthesized cDNAs was proportional to the amount of rp51A mRNAs in the total RNA extract, the experiments on the pus1− strain were made in triplicate by using 10, 20, and 50 μg of total RNA. The linearity of the 32P amount in cDNA bands versus the total RNA amount used as the template was verified by PhosphorImager measurement. For the wild-type strain, 10 and 50 μg of total RNA were used for rp51A mRNA quantification, and for the U1 snRNA control assays, 5 and 15 μg of total RNA were used. The amounts of synthesized U1 and rp51A cDNAs, in each assay, were estimated by PhosphorImager measurement. Based on the values obtained, the relative yields of rp51A mRNAs versus U1 snRNA were established for the wild-type and pus1− strains.
The efficiency of pre-U3 snoRNA splicing in the absence or the presence of an active PUS1 gene was analyzed by Northern blot analysis. A 5′-end 32P-labeled oligonucleotide complementary to the S. cerevisiae U3 snoRNA (nt 1 to 16) was used as the probe. Total RNA (10 μg) from the wild-type and the pus1− disrupted strains (prepared as described above) was fractionated on a 5% polyacrylamide gel. In vitro transcripts (10 ng) corresponding to the spliced and unspliced pre-U3 snoRNAs were used as controls. The plasmid pVs51:snR17A (92) was used to produce the U3A snoRNA transcript, while the pre-U3 snoRNA transcript was obtained by using the construct described by Mougin et al. (61). Total RNA (10 μg) from the JH84 S. cerevisiae strain (37) transformed by the pU3U14ds5′ plasmid (61) was also applied on the gel. The total RNA fractions from the JH84 strain were prepared as described by Méreau et al. (58). Plasmid pU3U14ds5′ contains a U3 snoRNA gene carrying mutations that lead to an accumulation of unspliced pre-U3 snoRNA and of a degraded form of the pre-U3 snoRNA in vivo (24).
In vitro pseudouridine formation in yeast U2 snRNA transcripts.
Two micrograms of S. cerevisiae U2 transcript dissolved in 9 μl of buffer (100 mM Tris-HCl buffer [pH 8.0] containing 100 mM ammonium acetate, 5 mM MgCl2, 2 mM DTT, and 0.1 mM EDTA) was heated for 3 min at 80°C and cooled down to 37°C. The purified Pus1p enzyme (2.5 μg), prepared from the recombinant E. coli strain (97) as described by Motorin et al. (60), was added, and the reaction was performed for 30 min at 37°C. After incubation, the modified transcript was phenol extracted and ethanol precipitated. The presence of pseudouridine residues was analyzed by the CMCT-RT technique as described above.
RESULTS
Identification of Ψ residues in S. cerevisiae UsnRNAs.
Two independent approaches were used for mapping Ψ residues in the S. cerevisiae spliceosomal UsnRNAs. The first approach was based on the CMCT-RT technique developed by Bakin and Ofengand (8). This method depends on the efficient chemical reaction of U and Ψ residues in RNA with CMCT (very strong for U residues, less strong for Ψ residues). Upon alkaline treatment with bicarbonate buffer at pH 10.4, the bulky CMC group linked to the U residues can be selectively hydrolyzed, while it remains bound to the Ψ residues. Stops of reverse transcription at CMCT-modified Ψ allow localization of Ψ residues by primer extension analysis. Guanosine residues also react with CMCT but less efficiently than do U and Ψ. Moreover, the CMC groups bound to G residues are easily removed by the alkaline treatment.
The second complementary approach was based on the observation that Ψ (and also m5U) residues are resistant to hydrazine treatment under conditions developed for chemical sequencing of RNA (15, 78). Thus, with this analysis, the absence of reactivity indicates the presence of one of these two posttranscriptional modifications. The absence of hydrazine reactivity can be detected directly by observing the cleavage pattern of purified 3′-end-labeled RNA after aniline treatment or indirectly, as described above, by primer extension analysis with RT.
In both methods, primer extension was performed on total, unfractionated yeast RNA prepared as described in Materials and Methods. Oligonucleotide primers allowed to explore different regions of the yeast UsnRNA molecules. For the long U1 (568 nt in length) and U2 snRNAs (1,175 nt), only the 5′-terminal regions (positions 1 to 50 of U1 and positions 1 to 100 of U2) were analyzed. These regions were chosen taking into account the already localized modified nucleotides in the U1 and U2 snRNAs of vertebrates, plants (reviewed in references 81 and 98), and S. pombe (28) (Fig. 1A and 2A). Indeed, vertebrate U1 snRNA contains five modified nucleotides, four of which are found, respectively, at positions 1 (Am), 2 (Um), 5 (Ψ), and 6 (Ψ), and the fifth one, located at position 70, is a 2′-O-methylated adenosine residue (for a review, see reference 81). Likewise, the 5′-terminal 100-nt region of rat hepatoma U2 snRNA contains all of the posttranscriptional modifications that were found in this snRNA (12 Ψ, 9 2′-O-methylated, and 1 m6Am residues) (84). For the shorter U4 (160 nt), U5S (179 nt), U5L (214 nt), and U6 (112 nt) snRNAs, only a short 3′-terminal region was not explored due to the constraint of primer extension analysis: the 30 nt at the 3′ end of the U4 snRNA, the 61 and 26 nt at the 3′ extremity of the U5L and U5S, respectively, and the 23 nt at the 3′ end of the U6 snRNA.
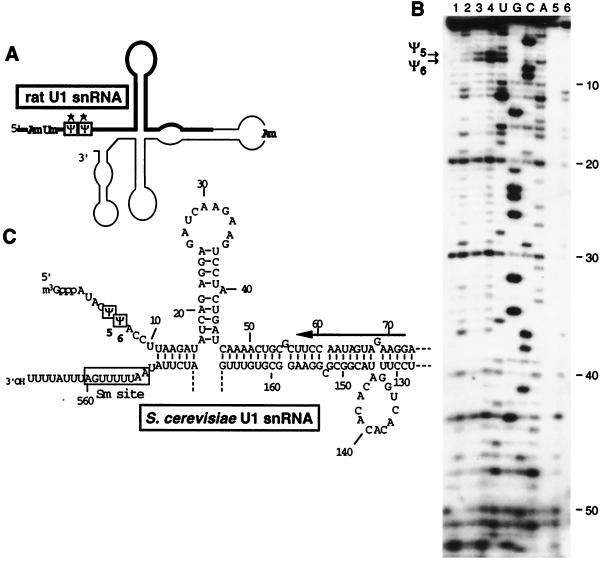
FIG. 1.
Localization of Ψ residues in S. cerevisiae U1 snRNA. (A) Schematic representation of the secondary structure of vertebrate U1 snRNA (12, 62). The Ψ residues are boxed, and the 2′-O-methylated residues are also indicated (for a review, see reference 55). The thick line shows the region of rat hepatoma U1 snRNA that corresponds to the analyzed region of S. cerevisiae U1 snRNA. The Ψ residues that were conserved in S. cerevisiae U1 snRNA are indicated by stars. (B) Primer extension analysis of the S. cerevisiae U1 snRNA modified by CMCT in a total RNA fraction for 2, 10, and 20 min (lanes 2, 3, and 4, respectively). Experimental conditions were as described in Materials and Methods. In lanes 3 and 4, the CMCT-modified RNA was subjected to an alkaline treatment at pH 10.4 as described in Materials and Methods. A control extension experiment was made without CMCT treatment (lane 1). The two reverse-transcription stops, corresponding to residues Ψ5 and Ψ6, are indicated by arrows on the left of panel B. U1 snRNA in a total RNA mixture was also treated by hydrazine under the conditions described in Materials and Methods (lane 6). A control extension experiment was made in absence of hydrazine (lane 5). The absence of hydrazine reactivity at positions 5 and 6 indicates the presence of a Ψ residue at these positions. Lanes U, G, C, and A correspond to the RNA sequencing ladder. Nucleotide positions, starting from the 5′-terminal nucleotide bound to the cap structure, are indicated on the right. (C) Nucleotide sequence of the analyzed region of the S. cerevisiae U1 snRNA. The oligonucleotide used as the primer for reverse transcription is indicated by a horizontal arrow. The two detected Ψ residues are boxed. The secondary structure was taken from Kretzner et al. (46).
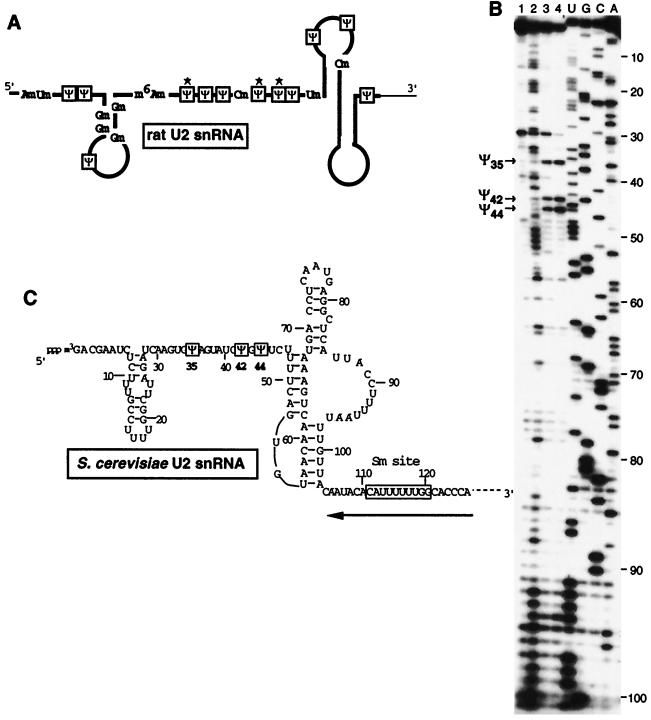
FIG. 2.
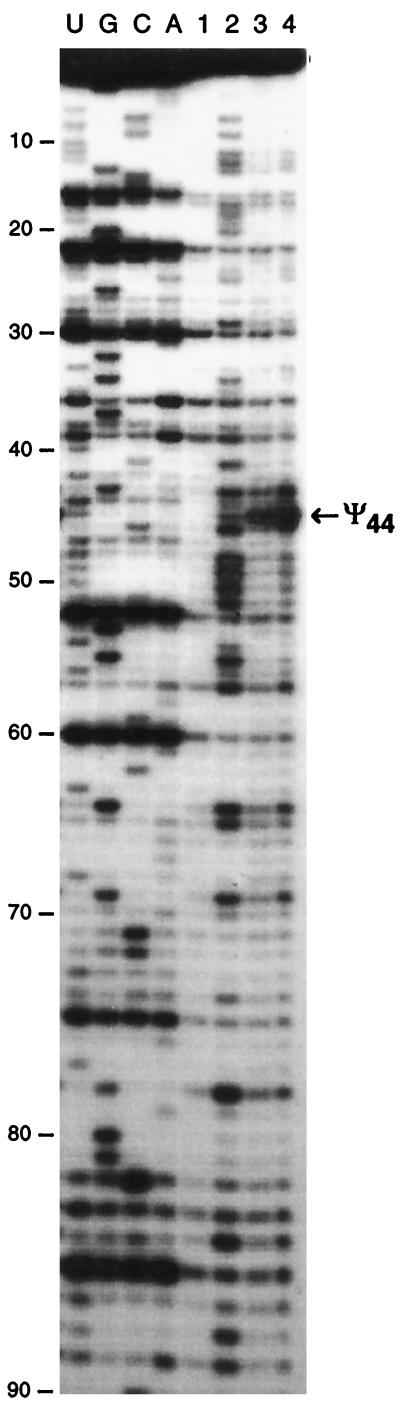
Localization of Ψ residues in S. cerevisiae U2 snRNA. (A) Rat hepatoma U2 snRNA, folded according to the U2 snRNA secondary structure model of Ares and Igel (4). The Ψ residues are boxed, the 2′-O-methylated residues and base-methylated residues are indicated (for a review, see reference 81). The thick line shows the region of rat hepatoma U2 snRNA that corresponds to the analyzed region of S. cerevisiae U2 snRNA. The Ψ residues that are conserved in the S. cerevisiae U2 snRNA are marked by stars. (B) Primer extension analysis of the S. cerevisiae U2 snRNA modified by CMCT (lanes 2, 3, 4, and 1 [for the control]). The conditions are the same as those for Fig. 1. The alkaline-resistant RT stops in lanes 3 and 4 revealed the presence of Ψ residues at positions 35, 42, and 44. The nucleotide sequence (3) and the secondary structure (4, 38) of the analyzed region of S. cerevisiae U2 snRNA are shown in panel C. The position of the primer oligonucleotide is shown by the horizontal arrow. The identified Ψ residues are boxed.
Only a few Ψ residues are found in the S. cerevisiae UsnRNAs.
The primer extension analysis of U1, U2, U4, U5, and U6 snRNAs modified by CMCT was used to map the pseudouridine residues. Incubation of the RNAs with CMCT was performed in each case for 2, 10, or 20 min, followed or not (control experiment) by alkaline treatment at pH 10.4. The cleavage patterns upon hydrazine treatment were also analyzed in each case. Both approaches gave essentially the same conclusions. Representative examples of primer extension patterns are shown.
Figure 1B illustrates the analysis of the S. cerevisiae U1 snRNA modified by hydrazine or CMCT. As described for vertebrate UsnRNAs (Fig. 1A and 3B) (12, 83), two Ψ residues are found at positions 5 and 6 (Fig. 1B and C and 3A). Only one of these two Ψ residues was detected in S. pombe (28). Analysis of the 5′-terminal region of S. cerevisiae U2 snRNA by CMCT (Fig. 2B) and hydrazine modification (data not shown) revealed the presence of three Ψ residues, respectively, at positions 35, 42, and 44 (Fig. 2C and 3A). Three Ψ residues were found at the same positions in the rat hepatoma (Fig. 2A and 3B) and the S. pombe U2 snRNAs, but additional Ψ residues were also detected in these two RNAs (Fig. 3B) (28, 84). Only one Ψ residue was detected in the S. cerevisiae U5 snRNAs (Fig. 4C). It corresponds to one of the two phylogenetically highly conserved Ψ residue found in the U5 snRNA terminal loop I (Fig. 3B) (103). At the other conserved pseudouridylation site of this terminal loop, the uridine residue is replaced by a cytidine residue in S. cerevisiae (Fig. 3A). The same situation was found for S. pombe (28). As found for U4 and U6 snRNAs from S. pombe (28), no Ψ residues were detected in the examined regions of U4 and U6 snRNAs (data not shown), while in the vertebrates, three Ψ residues were detected in both U4 (47, 82) and U6 snRNAs (Fig. 3B) (33). Hence, altogether only 6 Ψ residues were detected in the S. cerevisiae spliceosomal UsnRNAs (Fig. 3A), compared to 9 in the UsnRNAs from S. pombe (28) and 23 in the UsnRNAs from rat hepatoma (for a review, see reference 81).
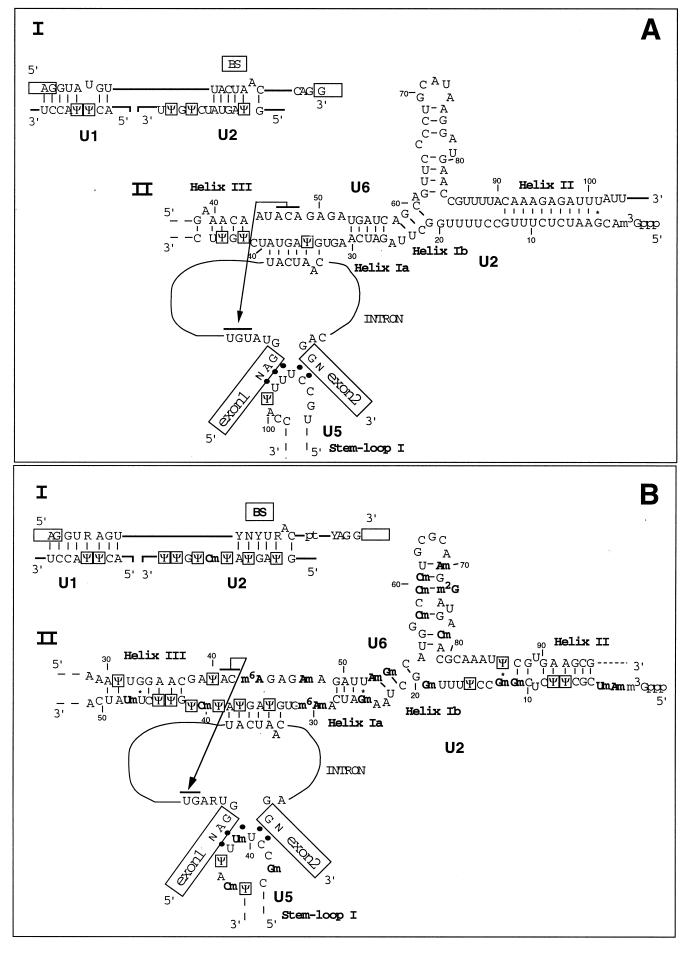
FIG. 3.
RNA-RNA interactions in the S. cerevisiae spliceosomes (A) and in the GU-AG spliceosomes of higher eukaryotes (B). The interaction between the 5′ and 3′ splice sites and branch-site (BS) consensus sequences with U1 and U2 snRNAs are shown in scheme I of panels A and B. UsnRNA-UsnRNA and UsnRNA-pre-mRNA interactions at the catalytic center of the spliceosome are shown in scheme II of panels A and B. Helices Ia, Ib, II, and III between U2 and U6 snRNAs are represented, as well as the base-pairing interaction between U2 snRNA and the branch-site sequence. The interaction between U6 snRNA and the UGU trinucleotide, close to the 5′ splice site, is indicated by overlined residues joined by an arrow; the interaction between the terminal loop I of U5 snRNA and the exon extremities is also shown (for a review of all of these interactions, see reference 54). The Ψ residues are boxed, and the base and ribose methylations are indicated in boldface (this study; for a review, see reference 55). Nucleotide positions, starting from the 5′ extremity of each UsnRNA, are indicated.
Disruption of the PUS1 gene results in the absence of Ψ44 in U2 snRNA.
Based on sequence homology with known E. coli tRNA-pseudouridine synthases, several potential RNA-pseudouridine synthases were identified in the S. cerevisiae genome (44). Three genes (PUS1, PUS3, and PUS4) were shown to code for tRNA-pseudouridine synthases (respectively, Pus1p, Pus3p, and Pus4p [10, 51, 60, 97]), and a fourth one (CBF5) was shown to be involved in pseudouridine formation in rRNA (protein Cbf5p [50]). The target RNA for a putative pseudouridine synthase Pus2p has not yet been determined (97).
Total RNA was extracted from S. cerevisiae strains disrupted for one of the nonessential PUS1, PUS2, PUS3, or PUS4 genes. Depletion of the Cbf5p enzyme was achieved in a GAL::cbf5 conditional strain after transfer to nonpermissive conditions for 70 h (50; see Materials and Methods). The pseudouridine residues in UsnRNAs were mapped by the CMCT-RT method. Since the previous results showed that only U1, U2, and U5 snRNAs contain Ψ residues in S. cerevisiae, the analysis was limited to these three snRNAs.
The results of the mapping analysis indicate that only the disruption of the PUS1 gene affects Ψ formation in the spliceosomal UsnRNAs (Fig. 4). Conversion of U into Ψ residues in U1, U2, and U5 UsnRNAs was unaffected by Cbf5p depletion under conditions where Ψ formation in pre-rRNA was strongly inhibited. This is illustrated by the analysis of the 26S rRNA segment (positions 1010 to 1140) that we performed together with the UsnRNA analysis (Fig. 4D) (50). Compared to a wild-type strain (WT), the pus1− strain differed by the disappearance of Ψ44 in U2 snRNA (Fig. 4A, compare lanes 3 and 4 [WT] to lanes 3 and 4 [pus1−]). No apparent change at the other positions (35 and 42) in U2 snRNA (Fig. 4A), as well as at positions 5 and 6 of U1 snRNA (Fig. 4B) and position 99 in U5 snRNA (Fig. 4C), was detected.
Pus1p catalyzes the pseudouridylation of U44 in U2 snRNA in vivo and in vitro.
To verify that Pus1p directly catalyzes Ψ formation at position 44 of U2 snRNA, the pus1− disrupted strain was transformed with plasmid pUN100-PUS1 (97) containing a wild-type PUS1 gene. As shown in Fig. 4A (lanes 2 and 3 pus1− + pUN100-PUS1), Ψ formation at position 44 of U2 snRNA was completely restored in the transformed yeast cells.
The capacity of the yeast Pus1p enzyme to catalyze U to Ψ conversion at position 44 in U2 snRNA was also tested in vitro. To this end, we used a recombinant Pus1p enzyme purified from E. coli (60) and an in vitro-produced T7 transcript of the S. cerevisiae U2 snRNA. The results of CMCT mapping demonstrate the efficient formation of Ψ44 in the U2 snRNA transcript and show that only this residue is modified in vitro (Fig. 5).
FIG. 5.
In vitro pseudouridylation of U2 snRNA transcript by using recombinant Pus1p pseudouridine synthase. After incubation of the transcript with the purified Pus1p enzyme (as described in Materials and Methods), the phenol-extracted RNA was analyzed by the CMCT-RT method (lanes 1, 2, 3, and 4) as described in the legend to Fig. 1. The strong alkaline-resistant RT stop at position 44 is indicated by an arrow. Lanes U, G, C, and A correspond to the sequencing ladder.
The absence of Ψ44 in U2 snRNA does not influence pre-mRNA and pre-U3 snoRNA splicing.
Although no apparent growth defect was found for the PUS1 gene deletion (97), it was interesting to test whether the absence of a Ψ at position 44 in U2 snRNA could affect the efficiency of pre-mRNA splicing. We tested this possibility on the rp51A pre-mRNA, the precursor for the mRNA of the S. cerevisiae rp51A ribosomal protein (87). To this end, using primer-extension analysis in conditions allowing quantification of the RNA template, we compared the relative yields of rp51A mRNAs and pre-mRNA in total RNA extracted from the wild-type and the pus1− strains grown until mid-exponential phase. U1 snRNA, which was not spliced in S. cerevisiae, was used as an internal control. In both strains, no rp51A pre-mRNA was accumulated (not shown). Only the two cDNA products corresponding to the two rp51A mRNAs previously described (1, 107) were detected (Fig. 6A). As shown in Fig. 6, only a very slight decrease of the rp51A mRNA concentrations in total RNA was observed in the absence of residue Ψ44 in U2 snRNA. Based on the rp51A mRNA/U1 snRNA ratio, estimated by PhosphorImager measurement, the rp51A mRNA concentration in total RNA was only reduced by 10% in the absence of residue Ψ44 in U2 snRNA. The observed difference is at the limit of the accuracy of mRNA quantification by primer extension analysis.
FIG. 6.
No marked effect of the PUS1 gene deletion on rp51A pre-mRNA and pre-U3 snoRNA splicing. In panel A, total RNA extracted from wild-type (WT) and the pus1− strains were analyzed by primer extension analysis under the conditions described in Materials and Methods that allow rp51A pre-mRNA and mRNA quantification. The pus1− strain primer extension analyses with the rp51A primer were performed on 10 μg (lane 5), 20 μg (lane 6), or 50 μg (lane 7) of total RNA, and for the wild-type strain the experiments were performed with 10 (lane 8) or 50 μg (lane 9) of total RNA, respectively. Control extension experiments with the U1 primer were performed with 5 μg (lanes 1 and 3) or 15 μg (lanes 2 and 4) of total RNA. The cDNA fractionation was performed on a 5% sequencing gel; only the portions of the gel corresponding to the two expected rp51A mRNA extension products are shown. The cDNA amounts in the U1 and rp51A bands of gel were estimated by PhosphorImager measurement. In panel B, total RNA extracted from the wild-type and the disrupted pus1− strains was analyzed by Southern blot hybridization by using a labeled oligonucleotide complementary to the yeast U3 snoRNA from position 1 to 16 (92). In vitro transcripts corresponding to the pre-U3 snoRNA and the U3 snoRNA, respectively, were used as controls (the two lanes on the left), as well as the total RNA extracted from a pU3U16ds5′ strain that shows the accumulation of unspliced and degraded pre-U3 snoRNA (24). The 32P amounts in the U3 signals obtained for the wild-type and pus1− strains were compared by PhosphorImager analysis.
Since the S. cerevisiae U3 snoRNA is produced from a precursor RNA that is spliced in a spliceosome and since the branch-point sequence of the U3 snRNA intron shows a substitution at the 5′ extremity compared to the consensus branch-site sequence (64), we tested whether splicing of this substrate may be more dramatically alterated by the absence of a Ψ residue at position 44 of U2 snRNA. For this purpose, Northern blot analysis was performed with total RNA extracted from the wild-type and the pus1− strains (Fig. 6B). An S. cerevisiae strain carrying a mutant U3 snoRNA gene that produces a pre-U3 snoRNA with a splicing defect (plasmid pU3U14ds5′) (61) was used as a control. In this strain, the pre-U3 snoRNA and its main degradation product accumulate (24). As shown in Fig. 6B, the level of mature U3 snoRNA was very slightly decreased in the absence of Ψ44 in U2 snRNA. However, here again it was at the limit of the accuracy of this kind of experiment. Furthermore, no accumulation of the U3 snoRNA precursor was detected. Hence, at least for the two pre-RNAs tested, the presence of a Ψ residue at position 44 of U2 snRNA did not show a strong effect on splicing efficiency. As no precursor accumulation was detected, the slight differences observed were perhaps not due to a splicing defect but to differences in RNA stability in the pus1− strain.
DISCUSSION
S. cerevisiae spliceosomal UsnRNAs contain a few Ψ residues located at sites of RNA-RNA or RNA-protein interactions.
We detected only six Ψ residues in the studied regions of S. cerevisiae UsnRNAs. This is significantly lower than the 23 and 20 residues found in the corresponding parts of rat hepatoma and plant UsnRNAs, respectively (for a review, see reference 55). A low number of Ψ residues in RNAs seems to be a general feature of yeasts, as only nine Ψ residues were detected in the S. pombe spliceosomal UsnRNAs (28), and a low level of Ψ residues was also observed for rRNAs and tRNAs in yeasts (7, 9, 16, 100).
Despite the absence of phenotype when there was a lack of Ψ formation at position 44 of the S. cerevisiae U2 snRNA and the very low effect that we detected on the level of rp51A mRNAs and U3 snoRNA, which are among the very few S. cerevisiae RNAs processed in a spliceosome, it is noteworthy that the six Ψ residues that we detected in S. cerevisiae UsnRNAs are evolutionarilly conserved. Furthermore, they are all located in or very close to the UsnRNA segments involved in intermolecular RNA-RNA interactions within the spliceosomes (Fig. 3A). Altogether, this suggests that at least some of them may have a functional importance. In U1 snRNA, the two Ψ residues detected are located in the segment that base pairs with the 5′ extremity of introns. Posttranscriptional modifications at position 5 and 6 of the U1 snRNA are nearly universal. Two Ψ residues are found at these positions in vertebrates and insects (Fig. 3B), in plants a Ψm residue is present at position 5 and a Um residue is found at position 6. Surprisingly, only position 5 shows a posttranscriptional modification in S. pombe (28). In the S. cerevisiae U1 snRNA-intron interaction, residue Ψ5 of U1 snRNA faces residue U4 of the intron (Fig. 3A). The presence of a U residue at position 4 of yeast introns is required for the interaction with U6 snRNA in the active conformation of the spliceosome (Fig. 3A) (41, 52, 91), and the conversion of the Ψ5-U4 pair into a canonical Watson-Crick pair in the U1 snRNA-intron interaction results in a 50% increase of the cell doubling time (96). In this context, Ψ formation at position 5 of U1 snRNA may be of functional importance.
Among the 12 Ψ residues detected in the rat hepatoma U2 snRNA (Fig. 2A and 3B), only 3 are conserved in the S. cerevisiae U2 snRNA (Fig. 2C and 3A). None are found in the 27-nt 5′-terminal segment, whose modification at the posttranscriptional level was found to be essential for vertebrate U2 snRNA function (120). However, residue Ψ35 may be of high functional importance, since it is involved in one of the two canonical base pairs that bulge out the A residue responsible for the nucleophilic attack in the first step of the splicing reaction. Helix stabilization by the presence of a Ψ-A pair (reviewed in references 2, 6, and 21) may be the reason for the universal conservation of a Ψ residue at this position. In addition, since it was shown for vertebrates that the branch-point bulged structure is recognized by proteins (26, 80; for a review, see reference 85), residue Ψ35 may also be involved in this recognition. The presence of a second Ψ-A pair in the vertebrate U2 snRNA-branch site interaction (Fig. 3B) may be needed to compensate for the high degree of degeneracy of the vertebrate branch-site sequence (for reviews, see references 39 and 89).
Using in vitro splicing assays, the effect of base substitutions on splicing efficiency was tested for almost all residues of the 5′-terminal region of the S. cerevisiae U2 snRNA (56). Mutations at position 35 resulted in a strong decrease in the splicing efficiency. Pascolo and Séraphin (72) tested the effect of compensatory mutations in the branch-site sequence and its U2 snRNA recognition element in vivo. However, the data obtained for the U2 Ψ35-branch site A5 pair are not sufficient to determine whether the presence of a Ψ residue at position 35 is required for high splicing efficiency.
The S. cerevisiae Prp5, Prp9, Prp11, Prp21, and Cus1 proteins, which are homologous to proteins from the human splicing factors SF3a and SF3b, associate with the S. cerevisiae U2 snRNA region from position 40 to 87 (36, 88, 110, 111, 119). This region contains the Ψ residues 42 and 44. In addition, the segments from positions 42 to 46 in the S. cerevisiae U2 snRNA and from positions 42 to 49 in the vertebrate U2 snRNA were found to form a base-pair interaction with U6 snRNA (Fig. 3). Such interaction delineates helix III, which is needed for active spliceosome formation in HeLa cells (102). In S. cerevisiae, no growth defect was detected for base substitution at positions 42 of U2 snRNA (119). This argues against an essential role of the U-to-Ψ conversion at position 42. Mutations at position 44 strongly affected growth (119). However, based on the absence of phenotype after deletion of the PUS1 gene (reference 97 and this study), the defect was not related to the absence of a U-to-Ψ conversion at this position. Indeed, a proposed explanation of the defect was that the replacement of U44 (in fact, Ψ44) by an A or a G residue favors an alternative conformation of U2 snRNA which impairs active spliceosome formation (119).
The Ψ residue at position 99 in the 5′-terminal loop of U5 snRNA is at the border of the segment that interacts with the exon extremities. Crystallographic and nuclear magnetic resonance studies of the yeast tRNAAsp and tRNAGln showed an essential role for Ψ32 and Ψ38 residues in the anticodon loop for maintaining the conformation required for proper anticodon recognition (5, 21, 22, 112, 113). As referred to this model, it may be that the U5 snRNA Ψ99 favors a conformation of the U5 snRNA 5′-terminal loop, which facilitates the correct alignment of exons. In connection with this hypothesis, it should be noticed that mutations affecting the 5′-terminal loop of S. cerevisiae U5 snRNA block splicing after the first step of the reaction (68).
In conclusion, based upon available genetic data and the present work, four of the six Ψ residues detected in the S. cerevisiae spliceosomal UsnRNAs belong to RNA segments involved in intermolecular interactions and may be of high functional importance. The other two are not essential taken individually. However, their conservation from yeasts to humans suggests that they may provide some selective advantage, which is difficult to test in laboratory conditions.
Among several characterized yeast pseudouridine synthases, only the Pus1p tRNA-pseudouridine synthase acts on spliceosomal UsnRNAs.
In yeast tRNAs, Ψ residues are found at 15 different locations, and at least five or six distinct enzymes are required for complete pseudouridylation of tRNAs. Only three of them, Pus1p (60), Pus3p (51), and Pus4p (10) have been characterized. The S. cerevisiae 17S and 26S rRNAs contain, respectively, 13 and 30 Ψ residues (16, 69, 70). The recent discovery of the snoRNA-guided mechanism of eukaryotic rRNA pseudouridylation has allowed to assign the Cbf5p (in yeast) and NAP57 (in higher eukaryotic) enzymes as the ones which act on the rRNA-snoRNP complex (25, 50). The S. cerevisiae pseudouridine synthases responsible for U isomerization in UsnRNAs have not been investigated so far. Studies of UsnRNA-specific pseudouridine synthases have been restricted to human cells. The results obtained revealed the presence of several distinct pseudouridine synthases; however, none of them was identified or cloned (73–75, 77, 121, 122).
Since no decrease of the level of Ψ residues was observed in UsnRNAs upon Cbf5p protein depletion, whereas Ψ synthesis in rRNA was strongly affected, our results strongly suggest that the Cbf5p enzyme is not involved in the pseudouridylation of UsnRNAs in S. cerevisiae. This implies that the U-to-Ψ conversion in the S. cerevisiae spliceosomal UsnRNAs is not based on the snoRNA guide mechanism involving the Cbf5p enzyme (50). This is in contrast to the recently reported mechanism of 2′-O-methylation of the vertebrate U6 snRNA (43, 108). Our results, obtained for yeasts, do not rule out the possibility that some pseudouridylation in the vertebrate U6 snRNA is generated by the snoRNA-guided mechanism. The nonimplication of Cbf5p in S. cerevisiae spliceosomal UsnRNA pseudouridylation may simply be due to the absence in S. cerevisiae U6 snRNA of an appropriate U residue that can be modified by the Cbf5p-snoRNA system. A larger number of Ψ residues are present in the vertebrate U6 snRNA (33), and some of them may be generated by a snoRNA-guided mechanism.
Two of the characterized S. cerevisiae pseudouridine synthases were found to act on tRNAs: Pus3p catalyzes the U isomerization at positions 38 and 39 in tRNAs (51), and Pus4p catalyzes the isomerization at position 55 (10). Our data show that these enzymes are not involved in the modification of UsnRNAs. Both of them are highly dependent on the global tRNA three-dimensional structure (10, 51), which probably explains their absence of reactivity on UsnRNAs. In contrast, Pus1p catalyzes Ψ formation at eight different sites in various tRNAs (positions 26 to 28, 34 to 36, 65, and 67) (60). Pseudouridine formation at the anticodon positions 34, 35, and 36 depends on the presence of an intron in tRNA, but it does not require the intact three-dimensional tRNA architecture (97, 104). These results obtained with tRNA substrates allowed the authors of these studies to conclude that Pus1p recognizes only a limited structural domain in RNA. The results presented here demonstrate that Pus1p is also capable of modifying, both in vivo and in vitro, one of the uridine residues (U44) in U2 snRNA. Evidently, this enzyme shows a dual specificity for both tRNA and snRNA.
The only well-documented case of a pseudouridine synthase with substrate dual specificity concerns the E. coli pseudouridine synthase RluAp. This enzyme catalyzes U-to-Ψ conversion at position 32 in several tRNAs and at position 746 in 23S rRNA (115). This dual specificity towards tRNA and rRNA was observed in vitro with synthetic RNA transcripts. In this case, the dual specificity was attributed to the recognition of a consensus sequence (UUNAAAA, where N is any of the four nucleotides) that is present in both the tRNA anticodon loop and in the loop of 23S rRNA bearing residue U746. Similarly, the E. coli tRNA–U54-methyltransferase catalyzes the in vitro formation of m5U54 in tRNA and m5U788 in a 16S rRNA fragment produced by in vitro transcription (29). However, no m5U residue has been found in naturally occurring E. coli rRNA, attesting that in vitro assays do not necessarily reflect the in vivo situation.
How does Pus1p act on substrates with different architectures? From the present results, we can conclude that Pus1p does not require an external guide RNA for the formation of Ψ44 in U2 snRNA, since we obtained Ψ44 formation in vitro by using an RNA transcript and the purified enzyme. Previous results on tRNA modification suggest that Pus1p requires a double-stranded RNA portion for binding, while the target uridine should be present in a rather flexible RNA structure (internal loop or even single-strand) (60). The target nucleotide in U2 snRNA is in a single-stranded region, which was confirmed by secondary-structure probing on the U2 snRNA transcript (data not shown), and no common sequence element was found for Pus1p target sites in tRNAs and in U2 snRNA. To identify the determinants required for U-to-Ψ conversion at position 44 by the Pus1p enzyme, a mutational analysis of the U2 snRNA target site is underway.
The presence of numerous Ψ and 2′-O-methylated residues in eukaryotic tRNAs, rRNAs, and UsnRNAs raises the important question of how many enzymes are required to account for all of these posttranscriptional modifications. To minimize the number of required enzymes, the cell seems to use two main strategies: (i) the involvement of guide RNAs conferring different specificities to a unique enzymatic machinery and (ii) the utilization of a unique multisite-specific enzyme that allows the formation of posttranscriptional modifications at different locations in a given type of RNA, as well as in different RNA substrates, and this is exemplified by our observation with the Pus1p enzyme.
However, the observation that Pus1p catalyzes only one of the Ψ residues detected in the S. cerevisiae UsnRNAs is in agreement with the previous observation for vertebrate UsnRNAs showing the involvement of several pseudouridine synthases (73–75). Other putative pseudouridine synthase genes are present in the S. cerevisiae genome, and we have initiated systematic gene disruption experiments in order to identify the enzymes responsible for the formation of the five other Ψ residues that were detected in this study.
ACKNOWLEDGMENTS
This work was supported by laboratory funds from the Ministère de la Recherche et de l’Enseignement Supérieur, the CNRS, specific research grants from the CNRS (Programme Physique et Chimie du Vivant 1997) and the Action de Recherche sur le Cancer. S. Massenet is a predoctoral fellow supported by a fellowship from the Ministère de la Recherche et de l’Enseignement Supérieur. Y. Motorin is the recipient of an Associated Researcher position at the CNRS (Poste Rouge). D. L. J. Lafontaine was the recipient of a fellowship from the European Commission.
The plasmids containing yeast UsnRNA genes were kindly provided by P. Fabrizio (Institut für Molekularbiologie and Tumorforschung, Marburg, Germany). We also thank R. Planta (University of Amsterdam, Amsterdam, The Netherlands) for providing the yeast strain with a disrupted PUS4 gene and G. Simos for providing the PUS1, PUS2, and PUS3-disrupted strains. D. Tollervey (University of Edinburgh, Edinburgh, United Kingdom) is thanked for the GAL::cbf5 RNA preparation, prepared in his laboratory. J. Ugolini is thanked for technical assistance. A. Mougin is acknowledged for useful advice at the beginning of this work. We also thank J.-P. Waller (CNRS, Gif-sur-Yvette, France) for critical reading of the manuscript and for useful comments.
REFERENCES
- 1.Abovich N, Rosbash M. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosomes. Mol Cell Biol. 1984;4:1871–1879. doi: 10.1128/mcb.4.9.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agris P F. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol. 1996;53:79–129. doi: 10.1016/s0079-6603(08)60143-9. [DOI] [PubMed] [Google Scholar]
- 3.Ares M. U2 RNA from yeast is unexpectedly large and contains homology to vertebrate U4, U5, and U6 small nuclear RNAs. Cell. 1986;47:49–59. doi: 10.1016/0092-8674(86)90365-x. [DOI] [PubMed] [Google Scholar]
- 4.Ares M, Igel A H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990;4:2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- 5.Arnez J G, Steitz T A. Crystal structure of unmodified tRNAGln complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudouridines in stabilization of RNA structure. Biochemistry. 1994;33:7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 6.Auffinger P, Westhof E. Effects of pseudouridylation on tRNA hydratation and dynamics: a theoretical approach. In: Grosjean H, Benne R, editors. The modification and editing of RNA. Washington, D.C: ASM Press; 1998. pp. 103–111. [Google Scholar]
- 7.Bakin A, Lane B G, Ofengand J. Clustering of pseudouridine residues around the peptidyltransferase center of yeast cytoplasmic and mitochondrial ribosomes. Biochemistry. 1994;33:13475–13483. doi: 10.1021/bi00249a036. [DOI] [PubMed] [Google Scholar]
- 8.Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 1993;32:9754–9762. doi: 10.1021/bi00088a030. [DOI] [PubMed] [Google Scholar]
- 9.Bakin A, Ofengand J. Mapping of the 13 pseudouridine residues in Saccharomyces cerevisiae small subunit ribosomal RNA to nucleotide resolution. Nucleic Acids Res. 1995;23:3290–3294. doi: 10.1093/nar/23.16.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker H F, Motorin Y, Planta R J, Grosjean H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of Ψ55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 1997;25:4493–4499. doi: 10.1093/nar/25.22.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behrens S E, Lührmann R. Immunoaffinity purification of a [U4/U6.U5] tri-snRNP from human cells. Genes Dev. 1991;5:1439–1452. doi: 10.1101/gad.5.8.1439. [DOI] [PubMed] [Google Scholar]
- 12.Branlant C, Krol A, Ebel J P, Gallinaro H, Lazar E, Jacob M. The conformation of chicken, rat and human U1A RNAs in solution. Nucleic Acids Res. 1981;9:841–58. doi: 10.1093/nar/9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branlant C, Krol A, Ebel J P, Lazar E, Gallinaro H, Jacob M, Sri-Widada J, Jeanteur P. Nucleotide sequences of nuclear U1A RNAs from chicken, rat, and man. Nucleic Acids Res. 1980;8:4143–4154. doi: 10.1093/nar/8.18.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branlant C, Krol A, Ebel J P, Lazar E, Haendler B, Jacob M. U2 RNA shares a structural domain with U1, U4, and U5 RNAs. EMBO J. 1982;1:1259–1265. doi: 10.1002/j.1460-2075.1982.tb00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branlant C, Krol A, Machatt M A, Ebel J P. Structural study of ribosomal 23S RNA from Escherichia coli. FEBS Lett. 1979;107:177–181. doi: 10.1016/0014-5793(79)80490-1. [DOI] [PubMed] [Google Scholar]
- 16.Branlant C, Krol A, Veldman G M, Klootwijk J, Regt V, Planta R J, Ebel J. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981;9:6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brody E, Abelson J. The “spliceosome”: yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985;228:963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- 18.Cech T R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986;44:207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- 19.Cortes J J, Sontheimer E J, Seiwert S D, Steitz J A. Mutations in the conserved loop of human U5 snRNA generate use of novel cryptic 5′ splice sites in vivo. EMBO J. 1993;12:5181–5189. doi: 10.1002/j.1460-2075.1993.tb06213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datta B, Weiner A M. Genetic evidence for base-pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature. 1991;352:821–824. doi: 10.1038/352821a0. [DOI] [PubMed] [Google Scholar]
- 21.Davis D R. Biophysical and conformational properties of modified nucleosides in RNA (nuclear magnetic resonance studies) In: Grosjean H, Benne R, editors. The modification and editing of RNA. Washington, D.C: ASM Press; 1998. pp. 85–102. [Google Scholar]
- 22.Davis D R, Poulter C D. 1H-15N NMR studies of Escherichia coli tRNA (Phe) from HisT mutants: a structural role for pseudouridine. Biochemistry. 1991;17:4223–4231. doi: 10.1021/bi00231a017. [DOI] [PubMed] [Google Scholar]
- 23.Fabrizio P, McPheeters D S, Abelson J. In vitro assembly of yeast U6 snRNP: a functional assay. Genes Dev. 1989;3:2137–2150. doi: 10.1101/gad.3.12b.2137. [DOI] [PubMed] [Google Scholar]
- 24.Fournier, R., et al. Unpublished data.
- 25.Ganot P, Bortolin M L, Kiss T. Site-specific pseudouridine formation in pre-ribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 26.Gaur R K, Valcàrcel J, Green M R. Sequential recognition of the pre-mRNA branch point by U2AF65 and a novel spliceosome-associated 28-kDa protein. RNA. 1995;1:407–417. [PMC free article] [PubMed] [Google Scholar]
- 27.Grabowski P J, Seiler S R, Sharp P A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985;42:345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- 28.Gu J, Patton J R, Shimba S, Reddy R. Localisation of modified nucleotides in Saccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions. RNA. 1996;2:909–918. [PMC free article] [PubMed] [Google Scholar]
- 29.Gu X G, Ofengand J, Santi D V. In vitro methylation of Escherichia coli 16S rRNA by tRNA (m5U54)-methyltransferase. Biochemistry. 1994;33:2255–2261. doi: 10.1021/bi00174a036. [DOI] [PubMed] [Google Scholar]
- 30.Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991;253:157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- 31.Guthrie C, Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- 32.Hall S L, Padgett R A. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J Mol Biol. 1994;239:357–365. doi: 10.1006/jmbi.1994.1377. [DOI] [PubMed] [Google Scholar]
- 33.Harada F, Kato N, Nishimura S. The nucleotide sequence of nuclear 4.8S RNA of mouse cells. Biochem Biophys Res Commun. 1980;95:1332–1340. doi: 10.1016/0006-291x(80)91620-4. [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto C, Steitz J A. U4 and U6 RNAs coexist in a single small nuclear ribonucleoprotein particle. Nucleic Acids Res. 1984;12:3283–3293. doi: 10.1093/nar/12.7.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hausner T P, Giglio L M, Weiner A M. Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes Dev. 1990;4:2146–2156. doi: 10.1101/gad.4.12a.2146. [DOI] [PubMed] [Google Scholar]
- 36.Hodges P E, Beggs J D. RNA splicing. U2 fulfills a commitment. Curr Biol. 1994;4:264–267. doi: 10.1016/s0960-9822(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 37.Hughes J M X, Ares M. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Igel A H, Ares M. Internal sequences that distinguish yeast from metazoan U2 snRNA are unnecessary for pre-mRNA splicing. Nature. 1988;334:450–453. doi: 10.1038/334450a0. [DOI] [PubMed] [Google Scholar]
- 39.Jackson I J. A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 1991;19:3794–3798. doi: 10.1093/nar/19.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacquier A. Self-splicing group II and nuclear pre-mRNA introns: how similar are they? Trends Biochem Sci. 1990;15:351–354. doi: 10.1016/0968-0004(90)90075-m. [DOI] [PubMed] [Google Scholar]
- 41.Kandels-Lewis S, Séraphin B. Role of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 42.Kato N, Harada F. Nucleotide sequence of nuclear 5S of mouse cells. Biochem Biophys Res Commun. 1981;99:1468–1476. doi: 10.1016/0006-291x(81)90784-1. [DOI] [PubMed] [Google Scholar]
- 43.Kiss-Làszlò Z, Henry Y, Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koonin E V. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 1996;24:2411–2415. doi: 10.1093/nar/24.12.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 46.Kretzner L, Krol A, Rosbash M. Saccharomyces cerevisiae U1 small nuclear RNA secondary structure contains both universal and yeast-specific domains. Proc Natl Acad Sci USA. 1990;87:851–855. doi: 10.1073/pnas.87.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krol A, Branlant C, Lazar E, Gallinaro H, Jacob M. Primary and secondary structures of chicken, rat and man nuclear U4 RNAs. Homologies with U1 and U5 RNAs. Nucleic Acids Res. 1981;9:2699–2716. doi: 10.1093/nar/9.12.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krol A, Gallinaro H, Lazar E, Jacob M, Branlant C. The nuclear 5S RNAs from chicken, rat and man. U5 RNAs are encoded by multiple genes. Nucleic Acids Res. 1981;9:769–787. doi: 10.1093/nar/9.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lacroute F. Regulation of pyrimidine biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1968;95:824–832. doi: 10.1128/jb.95.3.824-832.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lafontaine D L J, Bousquet-Antonelli C, Henry Y, Caisergues-Ferrer M, Tollervey D. The box H+ACA snoRNAs carry the rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lecointe F, Simos G, Sauer A, Hurt E C, Motorin Y, Grosjean H. Characterization of yeast protein Deg 1 as pseudouridine synthase (Pus 3) catalysing the formation of Ψ38 and Ψ39 in tRNA anticodon loop. J Biol Chem. 1998;273:1316–1323. doi: 10.1074/jbc.273.3.1316. [DOI] [PubMed] [Google Scholar]
- 52.Lesser C F, Guthrie C. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 53.Madhani H D, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 54.Madhani H D, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 55.Massenet S, Mougin A, Branlant C. Posttranscriptional modifications in the U small nuclear RNAs. In: Grosjean H, Benne R, editors. The modification and editing of RNA. Washington, D.C: ASM Press; 1998. pp. 201–227. [Google Scholar]
- 56.McPheeters D S, Abelson J. Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell. 1992;71:819–831. doi: 10.1016/0092-8674(92)90557-s. [DOI] [PubMed] [Google Scholar]
- 57.McPheeters D S, Fabrizio P, Abelson J. In vitro reconstitution of functional yeast U2 snRNPs. Genes Dev. 1989;3:2124–2136. doi: 10.1101/gad.3.12b.2124. [DOI] [PubMed] [Google Scholar]
- 58.Méreau A, Fournier R, Grégoire A, Mougin A, Fabrizio P, Lührmann R, Branlant C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interactions with the pre-ribosomal RNA. J Mol Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- 59.Moore M J, Query C C, Sharp P A. Splicing of precursors to messenger RNAs by the spliceosome. In: Gesteland R, Athuis J, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–358. [Google Scholar]
- 60.Motorin Y, Keith G, Simon C, Foiret D, Simos G, Hurt E, Grosjean H. The yeast tRNA:pseudouridine synthase Pus1p displays a multisite substrate specificity. RNA. 1998;4:856–869. doi: 10.1017/s1355838298980396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mougin A, Grégoire A, Banroques J, Ségault V, Fournier R, Brulé F, Chevrier-Miller M, Branlant C. Secondary structure of the yeast Saccharomyces cerevisiae pre-U3A snoRNA and its implication for splicing efficiency. RNA. 1996;2:1079–1093. [PMC free article] [PubMed] [Google Scholar]
- 62.Mount S M, Steitz J A. Sequence of U1 RNA from Drosophila melanogaster: implications for U1 secondary structure and possible involvement in splicing. Nucleic Acids Res. 1981;9:6351–6368. doi: 10.1093/nar/9.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Myslinski E, Branlant C, Wieben E D, Pederson T. The small nuclear RNAs of Drosophila. J Mol Biol. 1984;180:927–945. doi: 10.1016/0022-2836(84)90264-x. [DOI] [PubMed] [Google Scholar]
- 64.Myslinski E, Ségault V, Branlant C. An intron in the genes for U3 small nucleolar RNAs of the yeast Saccharomyces cerevisiae. Science. 1990;247:1213–1216. doi: 10.1126/science.1690452. [DOI] [PubMed] [Google Scholar]
- 65.Newman A J, Norman C. Mutations in yeast U5 snRNA alter the specificity of 5′ splice site cleavage. Cell. 1991;65:115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- 66.Newman A J, Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 67.Newman A J, Teigelkamp S, Beggs J D. SnRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- 68.O’Keefe R T, Newmann A J. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J. 1998;17:565–574. doi: 10.1093/emboj/17.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ofengand J, Bakin A. Mapping to nucleotide resolution of pseudouridine residues in a large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J Mol Biol. 1997;17:1993–2005. doi: 10.1006/jmbi.1996.0737. [DOI] [PubMed] [Google Scholar]
- 70.Ofengand J, Bakin A, Wrzesinski J, Nurse K, Lane B G. The pseudouridine residues of ribosomal RNA. Biochem Cell Biol. 1995;73:915–924. doi: 10.1139/o95-099. [DOI] [PubMed] [Google Scholar]
- 71.Parker R, Siliciano P G, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49:229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 72.Pascolo E, Séraphin B. The branchpoint residue is recognized during commitment complex formation before being bulged out of the U2 snRNA-pre-mRNA duplex. Mol Cell Biol. 1997;17:3469–3476. doi: 10.1128/mcb.17.7.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patton J R. Pseudouridine modification of U5 RNA in ribonucleoprotein particles assembled in vitro. Mol Cell Biol. 1991;11:5998–6006. doi: 10.1128/mcb.11.12.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patton J R. Multiple pseudouridine synthase activities for small nuclear RNAs. Biochem J. 1993;290:595–600. doi: 10.1042/bj2900595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Patton J R. Formation of pseudouridine in U5 small nuclear RNA. Biochemistry. 1994;33:10423–10427. doi: 10.1021/bi00200a025. [DOI] [PubMed] [Google Scholar]
- 76.Patton J R. Pseudouridine formation in small nuclear RNAs. Biochimie. 1994;76:1129–1132. doi: 10.1016/0300-9084(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 77.Patton J R, Jacobson M R, Pederson T. Pseudouridine formation in U2 small nuclear RNA. Proc Natl Acad Sci USA. 1994;91:3324–3328. doi: 10.1073/pnas.91.8.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peattie D A. Direct chemical method for sequencing RNA. Proc Natl Acad Sci USA. 1979;76:1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qiang W, Krainer A R. Splicing of a divergent subclass of AT-AC introns requires the major spliceosomal snRNAs. RNA. 1997;3:586–601. [PMC free article] [PubMed] [Google Scholar]
- 80.Query C C, Strobel S A, Sharp P A. Three recognition events at the branch-site adenine. EMBO J. 1996;15:1392–1402. [PMC free article] [PubMed] [Google Scholar]
- 81.Reddy R. Compilation of small RNA sequences. Nucleic Acids Res. 1988;16(Suppl.):71–85. doi: 10.1093/nar/16.suppl.r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reddy R, Henning D, Busch H. The primary nucleotide sequence of U4 RNA. J Biol Chem. 1981;256:3532–3538. [PubMed] [Google Scholar]
- 83.Reddy R, Henning D, Busch H. Pseudouridine residues in the 5′-terminus of uridine-rich nuclear RNA I (U1 RNA) Biochem Biophys Res Commun. 1981;98:1076–1083. doi: 10.1016/0006-291x(81)91221-3. [DOI] [PubMed] [Google Scholar]
- 84.Reddy R, Henning D, Epstein P, Busch H. Primary and secondary structure of U2 snRNA. Nucleic Acids Res. 1981;9:5645–5658. doi: 10.1093/nar/9.21.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reed R. Initial splice-site recognition and pairing during pre-mRNA splicing. Curr Opin Gen Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 86.Rinke J, Appel B, Digweed M, Lührmann R. Localization of a base-paired interaction between small nuclear RNAs U4 and U6 in intact U4/U6 ribonucleoprotein particles by psoralen cross-linking. J Mol Biol. 1985;185:721–731. doi: 10.1016/0022-2836(85)90057-9. [DOI] [PubMed] [Google Scholar]
- 87.Rosbash M, Harris P K W, Woolford J L, Teem J L. The effect of temperature-sensitive mutants on the transcription products from cloned ribosomal protein genes of yeast. Cell. 1981;24:679–686. doi: 10.1016/0092-8674(81)90094-5. [DOI] [PubMed] [Google Scholar]
- 88.Ruby S W, Chang T H, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- 89.Rymond B C, Rosbash M. Yeast pre-mRNA splicing. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 143–192. [Google Scholar]
- 90.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 91.Sawa H, Shimura Y. Association of U6 snRNA with the 5′ splice site region of pre-mRNA in the spliceosome. Genes Dev. 1992;6:244–254. doi: 10.1101/gad.6.2.244. [DOI] [PubMed] [Google Scholar]
- 92.Ségault V, Mougin A, Grégoire A, Banroques J, Branlant C. An experimental study of Saccharomyces cerevisiae U3 snRNA conformation in solution. Nucleic Acids Res. 1992;20:3443–3451. doi: 10.1093/nar/20.13.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ségault V, Will C L, Sproat B S, Lührmann R. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 1995;14:4010–4021. doi: 10.1002/j.1460-2075.1995.tb00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Séraphin B, Kretzner L, Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Séraphin B, Rosbash M. Exon mutations uncouple 5′ splice site selection from U1 snRNA pairing. Cell. 1990;63:619–629. doi: 10.1016/0092-8674(90)90457-p. [DOI] [PubMed] [Google Scholar]
- 96.Siliciano P G, Guthrie C. 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988;2:1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- 97.Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt E C. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- 98.Solymosy F, Pollàk T. Uridylate-rich small nuclear RNAs (UsnRNAs), their genes and pseudogenes, and UsnRNPs in plants: structure and function. A comparative approach. Crit Rev Plant Sci. 1993;12:275–369. [Google Scholar]
- 99.Sontheimer E J, Steitz J A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 100.Sprinzl S, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Steitz T A, Steitz J A. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun J S, Manley J L. A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev. 1995;9:843–854. doi: 10.1101/gad.9.7.843. [DOI] [PubMed] [Google Scholar]
- 103.Szkukàlek A, Myslinski E, Mougin A, Lührmann R, Branlant C. Phylogenetic conservation of modified nucleotides in the terminal loop 1 of the spliceosomal U5 snRNA. Biochimie. 1995;77:16–21. doi: 10.1016/0300-9084(96)88099-0. [DOI] [PubMed] [Google Scholar]
- 104.Szweykowska-Kulinska Z, Senger B, Keith G, Fasiolo F, Grosjean H. Intron-dependent formation of pseudouridines in the anticodon of Saccharomyces cerevisiae minor tRNAIle. EMBO J. 1994;13:4636–4644. doi: 10.1002/j.1460-2075.1994.tb06786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tarn W Y, Steitz J A. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science. 1996;273:1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- 106.Tarn W Y, Steitz J A. A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84:1–20. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 107.Teem J L, Rosbash M. Expression of a β-galactosidase gene containing the ribosomal protein 51 intron is sensitive to the rna2 mutation in yeast. Proc Natl Acad Sci USA. 1983;80:4403–4407. doi: 10.1073/pnas.80.14.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tycowski, K. T., Z. H. You, P. J. Graham, and J. A. Steitz. Personal communication.
- 109.Valcàrcel J, Green M R. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- 110.Wells S E, Ares M. Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6337–6349. doi: 10.1128/mcb.14.9.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wells S E, Neville M, Haynes M, Wang J, Igel H, Ares M. CUS1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP 145. Genes Dev. 1996;10:220–232. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- 112.Westhof E, Dumas P, Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985;184:119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- 113.Westhof E, Dumas P, Moras D. Hydration of transfer RNA molecules: a crystallographic study. Biochimie. 1988;70:145–165. doi: 10.1016/0300-9084(88)90056-9. [DOI] [PubMed] [Google Scholar]
- 114.Will C L, Lührmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 115.Wrzesinski J, Nurse K, Bakin A, Lane B G, Ofengand J. A dual-specificity pseudouridine synthase: an Escherichia coli synthase purified and cloned on the basis of its specificity for Psi 746 in 23S RNA is also specific for Psi 32 in tRNAphe. RNA. 1995;1:437–448. [PMC free article] [PubMed] [Google Scholar]
- 116.Wu J, Manley J L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base-pairing. Genes Dev. 1989;3:1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- 117.Wu J, Manley J L. Base-pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature. 1991;352:818–821. doi: 10.1038/352818a0. [DOI] [PubMed] [Google Scholar]
- 118.Wyatt J R, Sontheimer E J, Steitz J A. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- 119.Yan D, Ares M J. Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3A and SF3B subunits. Mol Cell Biol. 1996;16:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu Y, Shu M, Steitz J A. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zerby D B, Patton J R. Metabolism of pre-messenger RNA splicing cofactors: modification of U6 RNA is dependent on its interaction with U4 RNA. Nucleic Acids Res. 1996;24:3583–3589. doi: 10.1093/nar/24.18.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zerby D B, Patton J R. The modification of U4 RNA requires U6 RNA and multiple pseudouridine synthases. RNA. 1997;25:4808–4815. doi: 10.1093/nar/25.23.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhuang Y, Weiner A M. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 124.Zhuang Y, Weiner A M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989;3:1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]