Abstract
Background: Artemisinin-based combination therapy (ACT) was recommended to treat uncomplicated falciparum malaria. Unlike the situation in Asia where resistance to ACT has been reported, artemisinin resistance has not yet emerged in Africa. However, some rare failures with ACT or patients continuing to be parasitaemic on day 3 after ACT treatment have been reported in Africa or in travellers returning from Africa. Three mutations (G50E, R100K, and E107V) in the pfcoronin gene could be responsible for artemisinin resistance in Africa. Methods: The aims of this study were first to determine the prevalence of mutations in the pfcoronin gene in African P. falciparum isolates by Sanger sequencing, by targeting the 874 samples collected from patients hospitalised in France after returning from endemic areas in Africa between 2018 and 2019, and secondly to evaluate their association with in vitro reduced susceptibility to standard quinoline antimalarial drugs, including chloroquine, quinine, mefloquine, desethylamodiaquine, lumefantrine, piperaquine, and pyronaridine. Results: The three mutations in the pfcoronin gene (50E, 100K, and 107V) were not detected in the 874 P. falciparum isolates. Current data show that another polymorphism (P76S) is present in many countries of West Africa (mean prevalence of 20.7%) and Central Africa (11.9%) and, rarely, in East Africa (4.2%). This mutation does not appear to be predictive of in vitro reduced susceptibility to quinolines, including artemisinin derivative partners in ACT such as amodiaquine, lumefantrine, piperaquine, pyronaridine, and mefloquine. Another mutation (V62M) was identified at low prevalence (overall prevalence of 1%). Conclusions: The 76S mutation is present in many African countries with a prevalence above 10%. It is reassuring that this mutation does not confer in vitro resistance to ACT partners.
Keywords: malaria, Plasmodium falciparum, antimalarial drug, pfcoronin, resistance, in vitro, molecular marker
1. Introduction
Since the introduction of artemisinin-based combination therapy (ACT) in 2005 [1] global malaria cases have dropped. According to the 2020 World Health Organization (WHO) report, there were 229 million malaria cases and 409,000 deaths in 2019 [2] compared to 228 million malaria cases and 405,000 deaths in 2018 [3]. However, the number of cases has been declining since 2000, from 238 million to 229 million [2]. There have been 215 million malaria cases in Africa, representing 94% of the total number of cases [2]. The use of prophylaxis for travellers in endemic areas is strongly recommended. However, and despite these recommendations, France continues to have a high rate of imported malaria, with 82,000 imported malaria cases reported between 2000 and 2015, including 2430 cases in the French Army [4,5]. During this period, most malaria cases (95%) were contracted in sub-Saharan Africa (particularly Côte d’Ivoire, Cameroon, Mali, and Senegal) [5,6]. The number of reported cases (around 50% of estimated cases) to the National Reference Centre for Malaria Surveillance peaked at 3400 cases in 2000, decreased to 1824 cases in 2005 and was around 2895 cases in 2019 [7].
Artemisinin-based combination therapy (ACT) was introduced as a result of reported resistance to other antimalarial drugs in Africa. In most African countries, artemether-lumefantrine or artesunate-amodiaquine are recommended as first-line malaria treatment, and dihydroartemisinin-piperaquine is recommended as second-line treatment in some countries [8]. Unlike the situation in Asia, and more particularly in western Cambodia, Vietnam, Myanmar, Thailand, and Laos, where resistance to ACT has been reported [9,10,11,12], artemisinin resistance has not yet emerged in Africa.
However, some rare failures with ACT or patients who remain parasitaemic on day 3 after ACT treatment have been reported in Africa or in travellers returning from Africa [13,14,15,16,17,18,19,20,21,22]. Moreover, some rare therapeutic efficacy studies (TES) reported a proportion of clinical failures on day 28 above 10% after artemether-lumefantrine treatment in Angola, the Gambia, Malawi, and Uganda [23].
In 2014, the gene coding for the kelch 13 helix (pfk13) was identified as a molecular marker with in vivo and in vitro association for artemisinin resistance in South-East Asia [13,24]. However, until 2020, ACT clinical failures in Africa were not associated with polymorphisms in this gene and more particularly with the mutations described in Asia [13,14,17,19,20,22,25]. Recently, a new mutation on pfk13, R561H, has emerged in P. falciparum parasites collected in Masaka (Rwanda) and has spread rapidly in Rwanda with a high prevalence of up to 22% [26,27,28]. This mutation was found to be associated with delayed parasite clearance after treatment with ACT [28,29].
In 2018, a study on Senegalese P. falciparum strains described three polymorphisms (G50E, R100K, and E107V) in the pfcoronin gene which are thought to be responsible for dihydroartemisinin resistance in Africa [30]. These mutations were identified in two culture-adapted Senegalese field isolates from Pikine and Thies (Senegal) which became resistant in vitro to artemisinin after 13 cycles of discontinued selection under dihydroartemisinin pressure for four years [30]. The role of the mutations in pfcoronin was confirmed by CRISPR/Cas9 editing [31]. These three mutations, initially identified in P. falciparum parasites after drug pressure, were not reported in isolates from Senegal, Gabon, Ghana, Congo, Kenya, or Pakistan [16,17,32,33]. A new mutation, P76S, was detected in parasites from Senegal (prevalence 16.2%), Gabon (11%), Ghana (16%), Kenya (4%), and Congo (17%) [17,32]. However, none of these mutations appears to be associated with artemisinin reduced susceptibility in field isolates [16,17]. The pfcoronin gene is conserved in the different species infecting humans and shows 57% identity with Plasmodium berghei, a rodent-infecting species [34]. This gene codes for an actin-filament binding protein involved in the motility of sporozoites of P. falciparum [34]. It is expressed during schizogony and is located on the periphery of merozoites before and during invasion [35].
The aims of this study were first to determine the prevalence of the mutations in the pfcoronin gene in African P. falciparum isolates, by targeting the 874 samples collected from patients hospitalised in France after returning from endemic areas in Africa between 2018 and 2019, and secondly, to evaluate their association with in vitro reduced susceptibility to standard quinoline antimalarial drugs, including chloroquine, quinine, mefloquine, desethylamodiaquine, lumefantrine, piperaquine, and pyronaridine.
2. Materials and Methods
2.1. Sample Collection
From 2018 to 2019, 874 blood samples from patients hospitalised in France after returning from endemic countries in Africa, were sent within 72 h after collection to the French National Reference Centre for Malaria Surveillance (IRBA, IHU Méditerranée Infection Marseille) by hospitals in the French National Reference Centre for Imported Malaria network. Clinical and epidemiological data were collected for each sample. The diagnosis of P. falciparum was determined on the basis of a stained thin blood smear and confirmed by real-time PCR [36].
2.2. Gene Sequencing
Analysis of pfcoronin (codon positions 43 to 177) was performed using the same venous blood samples used for diagnostic analysis. DNA from each sample was extracted using a QIAamp DNA Blood Kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations.
Amplification of the pfcoronin gene was performed in the same way as described previously [17]. The pfcoronin gene (PF3D7_1251200) was amplified by PCR using the forward primer 5′-TATATCGTTTTATATGATTTGTTC-3′ and reverse primer 5′CCGATATCCCATTGTAAAGA-3′. Sanger sequencing was carried out using Biofidal (Biofidal, Vaulx en Velin, France) and the reverse primer. Sequence analysis of the samples was performed using the CodonCode Aligner software (CodonCode Corporation, Centerville, MA, USA).
2.3. Ex Vivo Assay
Chloroquine, quinine, mefloquine, desethylamodiaquine, lumefantrine, piperaquine, and pyronaridine were obtained from Sigma-Aldrich (St Quentin Fallavier, France). For the ex vivo chemosusceptibility assay, each isolate was aliquoted in 96-well plates previously treated with a gradient of antimalarial drug concentrations without culture adaptation, as previously described [37]. The plates were then incubated for 72 h at 37 °C in a controlled atmosphere with 10% O2, 5% CO2, and 85% N2. An ELISA assay was then performed by targeting the HRP2 protein using the Malaria Ag Celisa kit (ref KM2159, Cellabs PTY LDT, Brookvale, Australia) to estimate parasite growth, as previously described [38].
Each batch of plates was controlled by assessing the chloroquine-resistant P. falciparum strain W2 (MR4, Charlottesville, VA, USA) in between three and six independent experiments.
2.4. Data Management and Statistical Analysis
The IC50 were estimated through nonlinear regression by using ICEstimator version 1.2 [39]. Geometric means of IC50 values were calculated for each antimalarial drug. T-test and Chi-squared test were used to compare groups.
3. Results
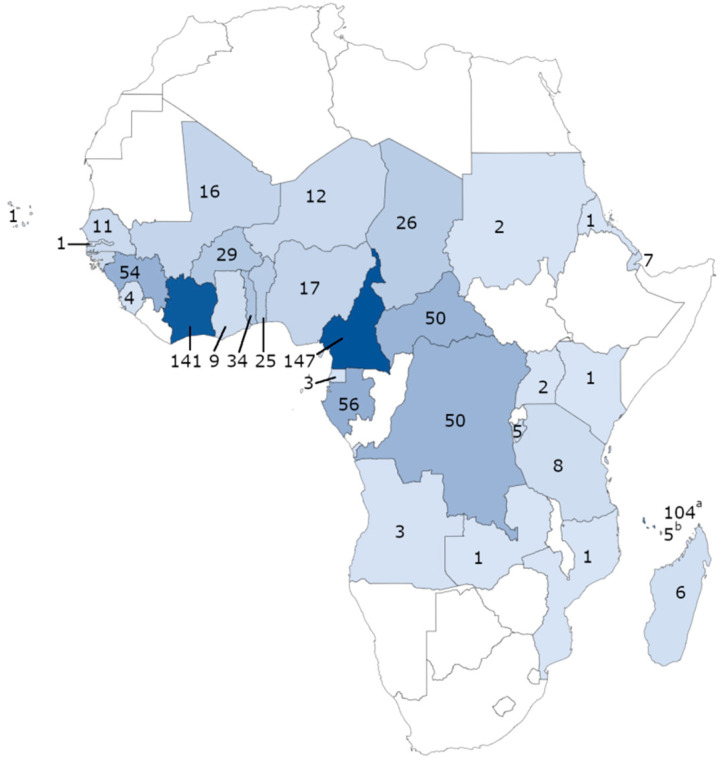
A total of 874 samples of P. falciparum collected from patients hospitalised in France after returning from malaria-endemic areas in Africa between 2018 and 2019, were analysed. For this study, 481 samples were analysed for 2018 and 403 samples for 2019. The samples were sent to the French National Reference Centre for Malaria Surveillance (IRBA, IHU Méditerranée Infection, Marseille) from various civilian or military hospitals in the French National Reference Centre for Imported Malaria network (Aix en Provence, Bordeaux, Lyon, Marseille, Mayotte, Montpellier, Nice, Toulon, Toulouse, and Valence). All the patients had returned from endemic areas in Africa, particularly from Cameroon (147), Côte d’Ivoire (141), Comoros (104), Gabon (56), Guinea (54), Central African Republic (50), Congo (50), Togo (34), Burkina Faso (29), Chad (26), and Benin (25) (Figure 1).
Figure 1.
Geographical distribution and numbers of Plasmodium falciparum isolates collected in 2018 (397 samples) and 2019 (477 samples) from patients hospitalised in France after returning from malaria endemic areas in Africa (39 from an unspecified country in Africa) (a from Comoros and b from Mayotte).
3.1. Analysis of pfcoronin Gene
The results of the pfcoronin gene sequencing on the P. falciparum isolates revealed the absence of the three mutations (G50E, R100K, and E107V) described previously as being associated with artemisinin resistance. From 2018 to 2019, 40.8% of the isolates studied came from West Africa, 38.3% from Central Africa, 16.1% from East Africa, and 0.2% from North Africa. Among the 874 P. falciparum isolates, 125 carried the P76S mutation (76S) (14.3% mean prevalence) and nine carried the V62M mutation (62M) (1% mean prevalence) (Table 1). No other mutation was identified.
Table 1.
Proportion of mutations in the pfcoronin gene according to the geographical origin of the Plasmodium falciparum isolates.
| Areas | Isolates No and (%) | P76S Mutation No and % a | V62M Mutation No and % a |
|---|---|---|---|
| West Africa | 357 (40.8%) | 74 (8.5%) | 3 (0.3%) |
| Central Africa | 335 (38.3%) | 40 (4.5%) | 6 (0.7%) |
| East Africa | 143 (16.3%) | 6 (0.7%) | 0 |
| Unspecified country in Africa | 39 (4.6%) | 5 (0.6%) | 0 |
| Total | 874 | 125 (14.3%) | 9 (1.0%) |
a Proportion of 76S and 62M mutations compared to the total number of isolates from Africa.
The proportion of the 76S mutation was higher for patients who came from West Africa (8.5%) and Central Africa (4.5%) compared to patients from East Africa (0.7%). The 76S mutation was present in 20.7% of the isolates from West Africa, 11.9% of the isolates from Central Africa, and 4.2% of the isolates from East Africa (Table 2). The difference in the proportion of 76S mutation between the three areas was significant (p = 3.1 × 10−6; Pearson’s Chi-squared test). Concerning countries included where at least 50 P. falciparum isolates came from, the proportion of 76S mutation ranged from 6.0% in Congo to 24.1% in Ivory Coast (Table 2). The proportion of 76S mutation was low in East Africa (4.2%). This mutation was not detected in the 104 isolates from Comoros.
Table 2.
Proportion of pfcoronin 76S and 62M mutations in Plasmodium falciparum isolates per country and per year.
| Country | 2018 No and % |
2019 No and % |
2018–2019 No and % |
|||
|---|---|---|---|---|---|---|
| 76S Mutation | 62M Mutation | 76S Mutation | 62M Mutation | 76S Mutation | 62M Mutation | |
| West Africa | 38/179 (21.2) | 2/179 (1.1) | 36/178 (20.2) | 1/178 (0.6) | 74/357 (20.7) | 3/357 (0.8) |
| Côte d’Ivoire | 20/72 (27.8) | 0/20 | 14/69 (20.3) | 0/69 | 34/141 (24.1) | 0/141 |
| Guinea | 4/25 (16.0) | 0/25 | 6/29 (20.7) | 0/29 | 10/54 (18.5) | 0/54 |
| Togo | 1/14 (7.1) | 0/14 | 2/20 (10.0) | 0/20 | 3/34 (8.8) | 0/34 |
| Burkina Faso | 4/19 (21.1) | 2/19 (10.5) | 2/10 (20.0) | 0/10 | 6/29 (20.7) | 2/29 (6.9) |
| Benin | 2/13 (15.4) | 0/13 | 4/12 (33.3) | 0/12 | 6/25 (24.0) | 0/25 |
| Mali | 2/8 (25.0) | 0/8 | 2/8 (25.0) | 0/8 | 4/16 (25.0) | 0/16 |
| Nigeria | 2/5 (40.0) | 0/5 | 2/12 (16.7) | 1/12 (8.3) | 4/17 (23.5) | 1/17 (5.9) |
| Senegal | 1/8 (12.5) | 0/8 | 0/3 | 0/3 | 1/11 (9.1) | 0/11 |
| Niger | 0/3 | 0/3 | 3/9 (33.3) | 0/9 | 3/12 (25.0) | 0/12 |
| Ghana | 1/4 (25.0) | 0/4 | 1/5 (20.0) | 0/5 | 2/9 (22.2) | 0/9 |
| Sierra Leone | 1/4 (25.0) | 0/4 | 0 | 0 | 1/4 (25.0) | 0/4 |
| Guinea Conakry | 0/3 | 0/3 | 0/1 | 0/1 | 0/4 | 0/4 |
| Cape Verde | 0/1 | 0/1 | 0 | 0 | 0/1 | 0/1 |
| Central Africa | 20/154 (13.0) | 1/154 (0.6) | 20/181 (11.0) | 5/181 (2.8) | 40/335 (11.9) | 6/335 (1.8) |
| Cameroon | 8/68 (11.8) | 0/68 | 11/79 (13.9) | 2/79 (2.5) | 19/147 (12.9) | 2/147 (1.4) |
| Gabon | 3/28 (10.7) | 0/28 | 2/28 (7.1) | 1/28 (3.6) | 5/56 (8.9) | 1/56 (1.8) |
| Democratic Republic of Congo | 2/22 (9.1) | 0/22 | 1/28 (3.6) | 0/28 | 3/50 (6.0) | 0/50 |
| Central African Republic | 6/19 (3.2) | 0/19 | 4/31 (12.9) | 2/31 (6.5) | 10/50 (20.0) | 2/50 (4.0) |
| Chad | 1/13 (7.7) | 1/13 (7.7) | 2/13 (15.4) | 0/13 | 3/26 (11.5) | 2/26 (3.8) |
| Angola | 0/2 | 0/2 | 0/1 | 0/1 | 0/3 | 0/3 |
| Equatorial Guinea | 0/2 | 0/2 | 0/1 | 0/1 | 0/3 | 0/3 |
| East Africa | 1/56 (1.8) | 0/56 | 5/87 (5.7) | 0/87 (0.0) | 6/143 (4.2) | 0/143 (0.0) |
| Mayotte | 1/3 (33.3) | 0/3 | 1/2 (50.0) | 0/2 | 2/5 (40.0) | 0/5 |
| Eritrea | 0 | 0 | 1/1 (100.0) | 0/1 | 1/1 (100.0) | 0/1 |
| Tanzania | 0/1 | 0/1 | 2/7 (28.6) | 0/7 | 2/8 (25.0) | 0/8 |
| Sudan | 0 | 0 | 1/2 (50.0) | 0/2 | 1/2 (50.0) | 0/2 |
| Kenya | 0 | 0 | 0/1 | 0/1 | 0/1 | 0/1 |
| Zambia | 0 | 0 | 0/1 | 0/1 | 0/1 | 0/1 |
| Uganda | 0 | 0 | 0/2 | 0/2 | 0/2 | 0/2 |
| Mozambique | 0/1 | 0/1 | 0 | 0 | 0/1 | 0/1 |
| Djibouti | 0/6 | 0/6 | 0/6 | 0/6 | 0/12 | 0/12 |
| Comoros | 0/40 | 0/40 | 0/64 | 0/64 | 0/104 | 0/104 |
| Madagascar | 0/5 | 0/5 | 0/1 | 0/1 | 0/6 | 0/6 |
| Unspecified African country | 2/8 (25.0) | 0/8 | 3/31 (9.7) | 0/31 (0.0) | 5/39 (12.8) | 0/39 (0.0) |
| Total | 31/397 (15.4) | 3/397 (0.8) | 64/477 (13.4) | 6/477 (1.3) | 128/874 (14.3) | 9/874 (1.0) |
Bold represents the African area to which the countries listed below belong.
Only six patients from Central Africa (0.7%) and three patients from West Africa carried the 62M mutation (0.3%) (Table 1). This mutation was reported in Burkina Faso (6.9% of the parasites from Burkina Faso), Nigeria (5.9%), Cameroon (1.4%), Gabon (1.8), Central African Republic (4.0), and Chad (3.8%) (Table 2).
3.2. Drug Susceptibility
Of the 874 samples analysed by genotyping, 330 isolates were successfully assessed in vitro for at least one antimalarial drug: 328 isolates for chloroquine (CQ), 325 isolates for quinine (QN), 329 isolates for lumefantrine (LMF), 306 isolates for desethylamodiaquine (the active metabolite of amodiaquine) (DQ), 321 isolates for mefloquine (MQ), 319 isolates for pyronaridine (PND), and 321 isolates for piperaquine (PPQ). The other isolates failed in culture during the ex vivo assay or were not tested due to low parasitaemia under 0.05% or because samples were only collected as dried blood spots, such as the 104 isolates from Comoros.
The values of 50% inhibitory concentration (IC50) estimates ranged from 2.8 nM to 764.7 nM for CQ, 6.2 nM to 757.2 nM for QN, 0.4 nM to 68.8 nM for LMF, 0.6 nM to 775.9 nM for DQ, 0.7 nM to 98.0 nM for MQ, 0.2 nM to 119.3 nM for PND, and 0.7 nM to 379.3 nM PPQ as shown in Table 3.
Table 3.
Ex vivo susceptibility geometric mean to chloroquine, quinine, lumefantrine, desethylamodiaquine, pyronaridine, and piperaquine according to the pfcoronin genotype (P76 or 76S).
| Drug (Isolates No) | Isolates Wild-Type (P76) | Isolates with 76S Mutation |
p-Value (t-Test) |
||
|---|---|---|---|---|---|
| IC50 Geometric Mean (nM) | Min-Max IC50 | IC50 Geometric Mean (nM) | Min-Max IC50 | ||
| Chloroquine (328) | 84.0 | 2.8–764.7 | 95.2 | 8.0–350.9 | 0.463 |
| Quinine (325) | 281.0 | 6.2–757.2 | 268.7 | 23.3–664.7 | 0.617 |
| Lumefantrine (329) | 6.7 | 0.4–68.8 | 6.5 | 0.5–46.0 | 0.878 |
| Desethylamodiaquine (306) | 40.7 | 0.6–775.9 | 34.3 | 2.0–152.5 | 0.343 |
| Mefloquine (321) | 22.9 | 0.7–98.0 | 26.1 | 3.2–87.8 | 0.295 |
| Pyronaridine (319) | 14.0 | 0.2–119.3 | 12.6 | 1.0–56.9 | 0.410 |
| Piperaquine (321) | 25.1 | 5.5–67.1 | 25.2 | 0.7–379.3 | 0.983 |
3.3. Association between In Vitro Responses (IC50) and pfcoronin Mutations
The values of drug ex vivo susceptibility (IC50) of the wild-type samples (P76) (no = 281 isolates) were compared to those of the mutated parasites (76S) (no = 49 isolates) for each antimalarial drug. There was no significant difference between the two groups (Table 3) (p-values ranged from 0.295 to 0.983). The IC50 geometric means for chloroquine and mefloquine were close to their respective threshold of reduced susceptibility (100 nM and 30 nM, respectively).
Moreover, based on the cut-off values for reduced ex vivo susceptibility to CQ (100 nM), QN (800 nM), LMF (150 nM), DQ (80 nM), MQ (30 nM), DOX (35 nM), PND (60 nM), or PPQ (135 nM) [40,41], we compared the proportion of isolates with reduced susceptibility to each drug according to the genotype of pfcoronin (P76 or 76S). There was no significant difference between the two groups (Table 4).
Table 4.
Proportions of isolates with in vitro reduced susceptibility to chloroquine, quinine, lumefantrine, desethylamodiaquine, pyronaridine, and piperaquine according to the pfcoronin genotype (P76 or 76S).
| Drug (No Isolates) | % of Isolates with Reduced Susceptibility (No of Isolates with Reduced Susceptibility/Total of Isolates) |
p-Value (Chi-Squared Test) |
|
|---|---|---|---|
| Wild-Type Isolates (P76) | Isolates with 76S Mutation | ||
| Chloroquine (328) | 25.1 (70/279) | 32.7 (16/49) | 0.506 |
| Quinine (325) | 0 (0/276) | 0 (0/49) | 0 |
| Lumefantrine (329) | 0 (0/280) | 0 (0/49) | 0 |
| Desethylamodiaquine (306) | 8.0 (21/262) | 9.1 (4/44) | 1 |
| Mefloquine (321) | 23.4 (64/273) | 29.2 (14/48) | 0.631 |
| Pyronaridine (319) | 1.1 (3/273) | 0 (0/46) | 1 |
| Piperaquine (321) | 0.7 (2/273) | 0 (0/48) | 0.999 |
4. Discussion
The aim of this study was to assess the prevalence of the mutations in the pfcoronin gene and their impact on the susceptibility to common antimalarial drugs in vitro. We did not find the three polymorphisms previously described by Demas et al. (G50E, R100K, and E107V) [30] but we identified two polymorphisms, P76S and V62M. The overall prevalence of the 76S mutation for Africa was 14.3%. We found a prevalence of 9.1% for this mutation in imported parasites from Senegal collected between 2018 and 2019. The 76S mutation had already been reported in Dakar, Senegal between 2015 and 2019 in 15.9% of the P. falciparum isolates [17]. More particularly, this mutation was identified in 12.5% of isolates from Dakar in 2016, 12.9% in 2017, 17.0% in 2018, and 25.9% in 2019 [17]. This mutation was not reported in parasites from the centre and the south of Senegal (Diourbel and Kedougou regions, respectively) [16].
The P76S mutation was also reported in Gabon (11%), Ghana (16%), Kenya (4%), and Congo (17%) [32]. Our data were in line with these previous observations. We found a prevalence of 8.9% in isolates from Gabon and 22.2% in isolates from Ghana.
Isolates from East Africa showed a prevalence of the 76S mutation (4.2%) which was lower than that observed in West Africa (20.7%) or Central Africa (11.9%). This mutation was not detected in the 104 isolates from Comoros.
Additionally, the 76S mutation was not reported in a study from Pakistan which analysed 179 Plasmodium falciparum isolates collected between 2018 and 2019 [33].
The prevalence of the 62M mutation (0.8%) was very low in Africa: 1.8% in Central Africa, 0.8% in West Africa and 0% in East Africa. This mutation was only reported in Burkina Faso (6.9%), Nigeria (5.9%), Cameroon (1.4%), Gabon (1.8), Central African Republic (4.0), and Chad (3.8%). These data were consistent with a previous study that showed an overall prevalence of 0.7% in Africa, and more particularly a prevalence of 1% in Gabon, 2% in Ghana, and 0% in Kenya and Congo [32].
It appears that the 76S mutation is not involved in ex vivo quinoline reduced susceptibility, and particularly in reduced susceptibility to artemisinin derivative partners in artemisinin-based combination therapy (ACT), including amodiaquine, lumefantrine, piperaquine, pyronaridine, or mefloquine. A previous study showed that the 76S mutation was observed in 31.3% of parasites collected from patients who continued to be parasitaemic on day 3 after ACT treatment and in 15.3% of isolates from patients who were successfully cured in Senegal [17]. However, the difference was not significant (p = 0.151).
5. Conclusions
The three mutations in the pfcoronin gene which are responsible for artemisinin resistance were not reported in this study. Current data showed that the 76S mutation is present in many countries of West Africa (mean prevalence of 20.7%) and Central Africa (mean prevalence of 11.9%). This mutation does not seem to be predictive of in vitro reduced susceptibility to quinolines, including artemisinin derivative partners in ACT such as amodiaquine, lumefantrine, piperaquine, pyronaridine, or mefloquine. It is reassuring that this mutation does not confer in vitro resistance to ACT partners. One limitation of this study was that it did not assess in vitro susceptibility to artemisinin using the ring survival assay (RSA) to determine the association between in vitro reduced susceptibility to artemisinin and the 76S mutation. These data do not allow the potential involvement of mutations in the pfcoronin gene in artemisinin resistance to be discounted.
Acknowledgments
The authors would like to thank the patients and staff of the hospitals of the French National Reference Centre for Imported Malaria network. The authors would to thank the following members of the French National Reference Centre for Imported Malaria Study Group: V Augis (Groupe Hospitalier Pellegrin, Bordeaux), P Bastien (Centre Hospitalier Universitaire de Montpellier, Montpellier), A Berry (Centre Hospitalier Universitaire de Rangueil, Toulouse), P Brouqui (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), B Cattin (Centre Hospitalier de Mayotte, Mayotte), P Chauvin (Centre Hospitalier Universitaire de Rangueil, Toulouse), M Cividin (Centre Hospitalier du Pays d’Aix, Aix en Provence), L Collet (Centre Hospitalier de Mayotte, Mayotte), P Combe (Centre Hospitalier de Mayotte, Mayotte), F Courtier (Centre Hospitalier de Valence, Valence), P Delaunay (Centre Hospitalier Universitaire de l’Archet, Nice), L Delhaes (Groupe Hospitalier Pellegrin, Bordeaux), M Drancourt (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), V Dumas (Groupe Hospitalier Pellegrin, Bordeaux), B Foucher (Hôpital d’Instruction des Armées Saint-Anne, Toulon), V Fuster (Groupe Hospitalier Pellegrin, Bordeaux), T Gaillard (Hôpital d’Instruction des Armées Desgenettes, Lyon), A Genin (Centre Hospitalier du Pays d’Aix, Aix en Provence), E Garnotel (Hôpital d’Instruction des Armées Laveran, Marseille), F Janvier (Hôpital d’Instruction des Armées Saint-Anne, Toulon), E Javelle (Hôpital d’Instruction des Armées Laveran, Marseille), C L’Ollivier (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), JC Lagier (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), E Ledault (Hôpital d’Instruction des Armées Laveran, Marseille), JF Lepere (Centre Hospitalier de Mayotte. Mayotte), M Leveque (Centre Hospitalier Universitaire de Montpellier, Montpellier), D Malvy (Groupe Hospitalier Pellegrin, Bordeaux), P Marty (Centre Hospitalier Universitaire de l’Archet, Nice), G Ménard (Hôpital d’Instruction des Armées Saint-Anne, Toulon), E Menu (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), P Millet (Groupe Hospitalier Pellegrin, Bordeaux), M Million (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), P Minodier (Hôpital Nord, Marseille), P Munier (Centre Hospitalier de Valence), P Parola (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), G Pasquier (Centre Hospitalier Universitaire de Montpellier, Montpellier), L Peyclit (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), S Picot (Hôpital de la Croix Rousse, Lyon), C Pomares-Estran (Centre Hospitalier Universitaire de l’Archet, Nice), S Ranque (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), M-C Receveur (Groupe Hospitalier Pellegrin, Bordeaux), A Robin (Centre Hospitalier du Pays d’Aix, Aix en Provence), E Sappa (Centre Hospitalier du Pays d’Aix, Aix en Provence), H Savini (Hôpital d’Instruction des Armées Laveran, Marseille), J Sevestre (Institut Hospitalo-Universitaire Méditerranée Infection, Marseille), F Simon (Hôpital d’Instruction des Armées Laveran, Marseille), Y Sterkers (Centre Hospitalier Universitaire de Montpellier, Montpellier), C Surcouf (Hôpital d’Instruction des Armées Laveran, Marseille), N Tayeb (Centre Hospitalier de Mayotte, Mayotte), V Thomas (Centre Hospitalier de Valence, Valence), F Thiebaut (Centre Hospitalier de Valence, Valence), E Varlet (Centre Hospitalier Universitaire de Montpellier, Montpellier), A Wolff (Hôpital d’Instruction des Armées Laveran, Marseille).
Author Contributions
Conceptualisation, O.D., M.M. and B.P.; validation, O.D., M.G. and B.P.; formal analysis, O.D. and B.P.; investigation, O.D., M.G., I.F., J.M., N.B., R.A. and N.G.; writing—original draft preparation, O.D. and B.P.; writing—review and editing, B.P.; supervision, M.M. and B.P.; project administration, M.M. and B.P.; funding acquisition, B.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the French Institute for Public Health Surveillance (Santé Publique France), grant CNR Paludisme and the Délégation Générale pour l’Armement, grant no PDH-2-NRBC-4-B-4104.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Informed consent was not required for this study because this work was performed under the statutory auspices of the French national reference centre for imported malaria, and isolates were anonymised by re-coding. Additionally, bio-banking of human clinical samples used for malaria diagnostics and secondary uses for scientific purposes is possible as long as the corresponding patients are informed and have not indicated any objections. This requirement was fulfilled here by giving verbal information to the patients, and no immediate or later patient opposition was reported to the hospital clinicians.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors have no conflict of interest to declare. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Guidelines for the Treatment of Malaria. [(accessed on 13 July 2021)]; Available online: https://apps.who.int/iris/handle/10665/162441.
- 2.World Malaria Report 2020. [(accessed on 13 July 2021)]; Available online: https://www.who.int/publications-detail-redirect/9789240015791.
- 3.World Malaria Report 2019. [(accessed on 13 July 2021)]; Available online: http://www.who.int/malaria/publications/world-malaria-report-2019/en/
- 4.Kendjo E., Houzé S., Mouri O., Taieb A., Gay F., Jauréguiberry S., Tantaoui I., Ndour P.A., Buffet P., Piarroux M., et al. Epidemiologic trends in malaria incidence among travelers returning to metropolitan France, 1996-2016. JAMA Netw. Open. 2019;2:e191691. doi: 10.1001/jamanetworkopen.2019.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thellier M., Simard F., Musset L., Cot M., Velut G., Kendjo E., Pradines B. Changes in malaria epidemiology in France and worldwide, 2000–2015. Med. Mal. Infect. 2020;50:99–112. doi: 10.1016/j.medmal.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Pradines B., Thellier M., Kendjo E., Houzé S. Epidémiologie du paludisme d’importatation en France. Rev. Prat. 2019;69:150–152. [PubMed] [Google Scholar]
- 7.Rapport d’activité 2020 du Centre National de Référence du Paludisme. [(accessed on 13 July 2021)]; Available online: https://cnr-paludisme.fr/activites-dexpertise/rapports-dactivites/
- 8.Mbacham F.W., Ayong L., Guewo-Fokeng M., Makoge V. Current situation of malaria in Africa. Methods Mol. Biol. 2019;2013:29–44. doi: 10.1007/978-1-4939-9550-9_2. [DOI] [PubMed] [Google Scholar]
- 9.Dondorp A.M., Nosten F., Yi P., Das D., Phyo A.P., Tarning J., Lwin K.M., Ariey F., Hanpithakpong W., Lee S.J., et al. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phyo A.P., Nkhoma S., Stepniewska K., Ashley E.A., Nair S., McRready R., Moo C.L., Al-Saai S., Dondorp A.M., Lwin K.M., et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: A longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imwong M., Suwannasin K., Kunasol C., Sutawong K., Mayxay M., Rekol H., Smithuis F.M., Hlaing T.M., Tun K.M., van der Pluijm R.W., et al. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: A molecular epidemiology observational study. Lancet Infect. Dis. 2017;17:491–497. doi: 10.1016/S1473-3099(17)30048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashley E.A., Dhorda M., Fairhurst R.M., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J.M., Mao S., Sam B., et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ménard D., Khim N., Beghain J., Adegnika A.A., Alam M.S., Amodu O., Rahim-Awab G., Barnadas C., Berry A., Boum Y., et al. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N. Engl. J. Med. 2016;374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madamet M., Kouta M.B., Wade K.A., Lo G., Diawara S., Fall M., Bercion R., Nakoulima A., Fall K.B., Benoit N., et al. Absence of association between polymorphisms in the K13 gene and the presence of Plasmodium falciparum parasites at day 3 after treatment with artemisinin derivatives in Senegal. Int. J. Antimicrob. Agents. 2017;49:754–756. doi: 10.1016/j.ijantimicag.2017.01.032. [DOI] [PubMed] [Google Scholar]
- 15.Dieye B., Affara M., Sangare L., Joof F., Ndiaye Y.D., Gomis J.F., Ndiaye M., Mbaye A., Diakite M., Sy N., et al. West Africa international centers of excellence for malaria research: Drug resistance patterns to artemether-lumefantrine in Senegal, Mali, and The Gambia. Am. J. Trop. Med. Hyg. 2016;95:1054–1060. doi: 10.4269/ajtmh.16-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diallo M.A., Yade M.S., Ndiaye Y.D., Diallo I., Diongue K., Sy S.A., Sy M., Seck M.C., Ndiaye M., Dieye B., et al. Efficacy and safety of artemisinin-based combination therapy and the implications of Pfkelch13 and Pfcoronin molecular markers in treatment failure in Senegal. Sci. Rep. 2020;10:8907. doi: 10.1038/s41598-020-65553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delandre O., Daffe S.M., Gendrot M., Diallo M.N., Madamet M., Kounta M.B., Diop M.N., Bercion R., Sow A., Ngom P.M., et al. Absence of association between polymorphisms in the pfcoronin and pfk13 genes and the presence of Plasmodium falciparum parasites after treatment with artemisinin derivatives in Senegal. Int. J. Antimicrob. Agents. 2020;56:106190. doi: 10.1016/j.ijantimicag.2020.106190. [DOI] [PubMed] [Google Scholar]
- 18.Gobbi F., Buonfrate D., Menegon M., Lunardi G., Angheben A., Severini C., Gori S., Bisoffi Z. Failure of dihydroartemisinin-piperaquine treatment of uncomplicated Plasmodium falciparum malaria in a traveller coming from Ethiopia. Malar. J. 2016;15:525. doi: 10.1186/s12936-016-1572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russo G., L’Episcopia M., Menegon M., Souza S.S., Dongho B.G.D., Vullo V., Lucchi N.W., Severini C. Dihydroartemisinin-piperaquine treatment failure in uncomplicated Plasmodium falciparum malaria case imported from Ethiopia. Infection. 2018;46:867–870. doi: 10.1007/s15010-018-1174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutherland C.J., Lansdell P., Sanders M., Muwanguzi J., van Schalkwyk D.A., Kaur H., Nolder D., Tucker J., Bennett H.M., Otto T.D., et al. pfk13-independent treatment failure in four imported cases of Plasmodium falciparum malaria treated with artemether-lumefantrine in the United Kingdom. Antimicrob. Agents Chemother. 2017;61:e02382-16. doi: 10.1128/AAC.02382-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plucinski M.M., Dimbu P.R., Macaia A.P., Ferreira C.M., Samutondo C., Quivinja J., Afonso M., Kiniffo R., Mbounga E., Kelley J.S., et al. Efficacy of artemether-lumefantrine, artesunate-amodiaquine, and dihydroartemisinin-piperaquine for treatment of uncomplicated Plasmodium falciparum malaria in Angola, 2015. Malar. J. 2017;16:62. doi: 10.1186/s12936-017-1712-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malvy D., Torrentino-Madamet M., L’Ollivier C., Receveur M.C., Jeddi F., Delhaes L., Piarroux R., Millet P., Pradines B. Plasmodium falciparum recrudescence two years after treatment of an uncomplicated infection without return to an area where malaria is endemic. Antimicrob. Agents Chemother. 2018;62:e01892-17. doi: 10.1128/AAC.01892-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Report on Antimalarial Drug Efficacy, Resistance and Responses (2010–2019) [(accessed on 9 July 2021)]; Available online: https://www.who.int/publications/i/item/9789240012813.
- 24.Ariey F., Witkoxski B., Amaratunga C., Beghain J., Langlois A.C., Khim N., Kim S., Duru V., Bouchier C., Ma L., et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foguim Tsombeng F., Gendrot M., Robert M.G., Madamet M., Pradines B. Are k13 and plasmepsin II genes, involved in Plasmodium falciparum resistance to artemisinin derivatives and piperaquine in Southeast Asia, reliable to monitor resistance surveillance in Africa? Malar. J. 2019;18:285. doi: 10.1186/s12936-019-2916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uwimana A., Legrand E., Stokes B.H., Mangala Ndikumana J.L., Warsame M., Umulisa N., Ngamije D., Munyaneza T., Mazarati J.B., Munguti K., et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 2020;26:1602–1608. doi: 10.1038/s41591-020-1005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergmann C., van Loon W., Habarugira F., Tacoli C., Jäger J.C., Savelsberg D., Nshimiyimana F., Rwamugema E., Mbarushimana D., Ndoli J., et al. Increase in Kelch 13 polymorphisms in Plasmodium falciparum, Southern Rwanda. Emerg. Infect. Dis. 2021;27:294–296. doi: 10.3201/eid2701.203527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straimer J., Gandhi P., Czermak Renner K., Schmitt E.K. High prevalence of P. falciparum K13 mutations in Rwanda is associated with slow clearance after treatment with artemether-lumefantrine. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uwimana A., Umulisa N., Venkatesan M., Svigel S.S., Zhou Z., Munyanesa T., Habimana R.M., Rucogoza A., Moriarty L.F., Sandford R., et al. Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: An open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect. Dis. 2021;21:1120–1128. doi: 10.1016/S1473-3099(21)00142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demas A.R., Sharma A.I., Wong W., Early A.M., Redmond S., Bopp S., Neafsey D.E., Volkman S.K., Hartl D.L., Wirth D.F. Mutations in Plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc. Natl. Acad. Sci. USA. 2018;115:12799–12804. doi: 10.1073/pnas.1812317115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A.I., Shin S.H., Bopp S., Volkman S.K., Hartl D.L., Wirth D.F. Genetic background and PfKelch13 affect artemisinin susceptibility of PfCoronin mutants in Plasmodium falciparum. PLoS Genet. 2020;16:e1009266. doi: 10.1371/journal.pgen.1009266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Velavan T.P., Nderu D., Agbenyega T., Ntoumi F., Kremsner P.G. An alternative dogma on reduced artemisinin susceptibility: A new shadow from east to west. Proc. Natl. Acad. Sci. USA. 2019;116:12611–12612. doi: 10.1073/pnas.1907142116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan A.Q., Pernaute-Lau L., Khattak A.A., Luijcx S., Aydin-Schmidt B., Hussain M., Khan T.A., Mufti F.U., Morris U. Surveillance of genetic markers associated with Plasmodium falciparum resistance to artemisinin-based combination therapy in Pakistan, 2018–2019. Malar. J. 2020;19:206. doi: 10.1186/s12936-020-03276-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bane K.S., Lepper S., Kehrer J., Sattler J.M., Singer M., Reining M., Klug D., Heiss K., Baum J., Mueller A.K., et al. The actin filament-binding protein coronin regulates motility in Plasmodium sporozoites. PLoS Pathog. 2016;12:e1005710. doi: 10.1371/journal.ppat.1005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olshina M.A., Angrisano F., Marapana D.S., Riglar D.T., Bane K., Wong W., Catimel B., Yin M.X., Holmes A.B., Frischknecht F., et al. Plasmodium falciparum coronin organizes arrays of parallel actin filaments potentially guiding directional motility in invasive malaria parasites. Malar. J. 2015;14:280. doi: 10.1186/s12936-015-0801-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wurtz N., Fall B., Bui K., Pascual A., Fall M., Camara C., Diatta B., Fall K.B., Mbaye P.S., Diémé Y., et al. Pfhrp2 and Pfhrp3 polymorphisms in Plasmodium falciparum isolates from Dakar, Senegal: Impact on rapid malaria diagnosis. Malar. J. 2013;12:34. doi: 10.1186/1475-2875-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gendrot M., Delandre O., Robert M.G., Foguim F.T., Benoit N., Amalvict R., Fonta I., Mosnier J., Madamet M., Pradines B. Absence of association between methylene blue reduced susceptibility and polymorphisms in 12 genes involved in antimalarial drug resistance in African Plasmodium falciparum. Pharmaceuticals. 2021;14:351. doi: 10.3390/ph14040351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gendrot M., Foguim F.T., Robert M.G., Amalvict R., Mosnier J., Benoit N., Madamet M., Pradines B. The D113N mutation in the RING E3 ubiquitin protein ligase gene is not associated with ex vivo susceptibility to common anti-malarial drugs in African Plasmodium falciparum isolates. Malar. J. 2018;17:108. doi: 10.1186/s12936-018-2252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ICEstimator Version 1.2. [(accessed on 13 July 2021)]; Available online: https://www.antimalarial-icestimator.net.
- 40.Pascual A., Madamet M., Briolant S., Gaillard T., Amalvict R., Benoit N., Travers D., Pradines B. Multinormal in vitro distribution of Plasmodium falciparum susceptibility to piperaquine and pyronaridine. Malar. J. 2015;14:49. doi: 10.1186/s12936-015-0586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pradines B., Bertaux L., Pomares C., Delaunay P., Marty P. Reduced in vitro susceptibility to artemisinin derivatives associated with multi-resistance in a traveller returning from South-East Asia. Malar. J. 2011;10:268. doi: 10.1186/1475-2875-10-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.