Abstract
In this study we further characterized the 3′-5′ exonuclease activity intrinsic to wild-type p53. We showed that this activity, like sequence-specific DNA binding, is mediated by the p53 core domain. Truncation of the C-terminal 30 amino acids of the p53 molecule enhanced the p53 exonuclease activity by at least 10-fold, indicating that this activity, like sequence-specific DNA binding, is negatively regulated by the C-terminal basic regulatory domain of p53. However, treatments which activated sequence-specific DNA binding of p53, like binding of the monoclonal antibody PAb421, which recognizes a C-terminal epitope on p53, or a higher phosphorylation status, strongly inhibited the p53 exonuclease activity. This suggests that at least on full-length p53, sequence-specific DNA binding and exonuclease activities are subject to different and seemingly opposing regulatory mechanisms. Following up the recent discovery in our laboratory that p53 recognizes and binds with high affinity to three-stranded DNA substrates mimicking early recombination intermediates (C. Dudenhoeffer, G. Rohaly, K. Will, W. Deppert, and L. Wiesmueller, Mol. Cell. Biol. 18:5332–5342), we asked whether such substrates might be degraded by the p53 exonuclease. Addition of Mg2+ ions to the binding assay indeed started the p53 exonuclease and promoted rapid degradation of the bound, but not of the unbound, substrate, indicating that specifically recognized targets can be subjected to exonucleolytic degradation by p53 under defined conditions.
The best-studied molecular activity of the tumor suppressor p53 probably is that of a sequence-specific transactivator (3, 9, 19, 28, 44, 69). p53 becomes activated as a transcription factor under various cellular stress situations, including genotoxic stress. This leads to transcriptional upregulation of p53 target genes, which in turn mediate growth arrest or apoptosis (13, 32, 41). In addition to its transactivator function, p53 exerts a variety of other biochemical activities involved in DNA damage recognition and repair, which characterize p53 as a superior control element in maintaining the integrity of the cell genome (2, 26, 37, 49). We recently reported that wild-type (wt) but not mutant p53 exerts a novel intrinsic 3′-5′ exonuclease activity (48). Exonucleases are required in a variety of processes contributing to genomic stability, such as proofreading, mismatch and nucleotide excision repair, and recombination (31, 39). Therefore, we hypothesize that p53, through its exonuclease activity, could be actively involved in such processes, thereby significantly expanding the role of p53 as a “guardian of the genome” (35).
In this study we further characterized the p53 exonuclease activity and specifically addressed the question of how this activity is related to the sequence-specific DNA binding activity of p53. Our previous experiments had suggested that the p53 intrinsic exonuclease activity is exerted by the p53 core domain (48), which also mediates sequence-specific DNA binding by p53 (1, 4, 16, 51, 67). The localization of two such different activities to the same domain of the p53 molecule poses the problem of how these activities are regulated, since one would expect that p53, which regulates transcription in a sequence-specific manner, would not exert an exonuclease activity. The p53 core domain has a complex structure, as it is composed of two alpha-helical loop domains and a beta-sheet domain, compacted via metal (zinc) binding (6). This is an unusual arrangement for a DNA binding domain, which so far has been found only in a few sequence-specific DNA binding proteins, including NFκB (47). Interestingly, a similar structural arrangement has been found for the catalytic domain of Escherichia coli exonuclease III, a multifunctional enzyme exhibiting 3′ phosphatase, endonuclease, and 3′-5′ and 5′-3′ exonuclease activities (45). As a working hypothesis, we therefore assumed that the composite structure of the p53 core domain might allow the execution of different activities through conformational alterations leading to slightly different arrangements of its various structural components.
The data presented in this report confirm that the central domain of wild-type, but not that of mutant, p53 exerts the p53 intrinsic exonuclease activity. Like sequence-specific DNA binding (22, 24), this exonuclease activity is negatively regulated by the C-terminal basic regulatory domain of p53. However, treatments activating sequence-specific DNA binding of full-length p53 strongly inhibited its exonuclease activity, indicating that p53 exonuclease and sequence-specific DNA binding are separate activities of the p53 core domain, regulated in opposing manners. As C-terminally truncated p53 had at least a 10-fold higher specific exonuclease activity than full-length p53, we conclude that under appropriate conditions, wt p53 acts as a bona fide exonuclease. Activation of the p53 exonuclease activity by addition of Mg2+ ions when p53 had bound to a potential in vivo substrate, a three-stranded DNA mimicking a recombination intermediate with a single mismatch (8), resulted in rapid degradation of the bound, but not the unbound, substrate, which is indicative of the activation of the p53 exonuclease within a defined enzyme-substrate complex.
Our data are compatible with a model according to which p53 exerts two complementary functions in maintaining the integrity of the genome. As its basal function in maintaining genetic stability, p53 participates actively in repair processes through activities not related to sequence-specific DNA binding, specifically through its exonuclease activity (48). At another level of control, cellular stress activates the functions of p53 generally associated with its role as a guardian of the genome, namely, growth arrest and apoptosis (34, 35).
(This work was conducted by F. Janus and N. Albrechsten in partial fulfillment of the requirements for a Ph.D. degree at the University of Hamburg, Hamburg, Germany.)
MATERIALS AND METHODS
Bacteria and cells.
Recombinant baculovirus expressing full-length and fragments of murine wt p53 protein with coding information for a His6 tag at the N terminus were kindly provided by P. Tegtmeyer (SUNY, Stony Brook) (67). Murine wt and MethA (7) p53 DNAs coding for amino acids 80 to 280 were inserted into the pH6EX3 vector and expressed in DH5α bacteria. Simian virus 40 (SV40) large-T-antigen (T-Ag) recombinant baculovirus was kindly provided by Ellen Fanning (Department of Molecular Biology, Vanderbilt University, Nashville, Tenn.). Human wt p53 and human oligomerization mutant 1262 recombinant baculoviruses were kindly provided by John Jenkins (Cell Proliferation Laboratory, Marie Curie Research Institute, Oxted, Surrey, United Kingdom).
Protein purification. (i) Immunoaffinity chromatography.
Protein was purified by PAb421 immunoaffinity chromatography as described previously (48).
(ii) Co2+ metal affinity chromatography.
About 109 High Five insect cells were infected with the corresponding recombinant baculovirus and harvested at 44 h postinfection (hpi). Okadaic acid (200 nM) was added as indicated in Results at 41 hpi. To purify histidine-tagged proteins by Co2+ metal affinity column chromatography (TALON; Clontech), cells were lysed by addition of ice-cold lysis buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.5% [wt/vol] Lubrol, 5 mM β-mercaptoethanol, 2 mM phenylmethylsulfonyl fluoride, 2 mM Na2S2O5, 50 μg of leupeptin, per ml, 1% [vol/vol] Trasylol) and rocked for 1 h. Crude lysate was centrifuged at 200,000 × g for 35 min and incubated with column material for 45 min. The column material was washed with buffer A (20 mM Tris-HCl [pH 8.0], 100 mM KCl, 2 mM β-mercaptoethanol) and with 10 mM imidazole in buffer A. Proteins were eluted with 300 mM imidazole in buffer A, and 1-ml fractions were collected. The majority of the bound p53 proteins were recovered in fraction 2 at high purity.
(iii) Ion-exchange and heparin sulfate affinity chromatographies.
After metal affinity chromatography, the p531-320 fragment was further purified by anion-exchange chromatography with UNO Q anion-exchange column or by heparin sulfate-Sepharose affinity chromatography with an ÄKTApurifier fast protein liquid chromatography system (Pharmacia). The p531-320 fragment was purified via Talon metal affinity chromatography as described above, and a >90% pure, exonuclease-active peak fraction was subjected to ion-exchange chromatography. For anion-exchange chromatography, a 1.3-ml UnoQ column (Bio-Rad) was equilibrated with 5 column volumes of buffer A (20 mM Tris, 40 mM KCl, 0.1% mercaptoethanol, pH 8.0) and loaded with 100 μg of protein. For cation-exchange chromatography 140 μg of the TALON-purified p531-320 fragment was loaded onto a 1-ml HiTrap heparin-Sepharose column (Pharmacia) equilibrated with buffer A. The columns were washed with 3 column volumes of buffer A, and the protein was eluted with a linear salt gradient (40 to 800 mM KCl) at a flow rate of 2 ml/min. Fractions of 0.5 ml were collected. All fractions were tested for exonuclease activity, and peak fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining.
SDS-PAGE and Western immunoblotting.
Protein fractions were denatured in SDS sample buffer, separated by SDS-PAGE, and transferred to a polyvinylidine difluoride membrane. Proteins were detected with monoclonal antibodies as indicated in Results.
Exonuclease assays.
The 3′-5′ exonuclease activity was measured by filter binding assays as described previously (48). For inhibition experiments with PAb421 and PAb108, 5 ng to 10 μg of antibody and 150 ng of metal affinity-purified p53 protein were mixed, preincubated for 15 min at 4°C, and tested for exonuclease activity. Proteolytic digestion was done by addition of a dilution series of thermolysin from 4 ng (1:50) to 20 pg (1:10,000) to 150 ng of metal affinity-purified p53 in exonuclease reaction buffer (25 mM Tris [pH 8.5], 10 mM MgCl2, 1 mM dithiothreitol) and incubation for 15 min at 37°C. 3H-labeled DNA was added, and the filter binding assay was performed as described above. Exonuclease III (Boehringer, Mannheim, Germany) with the same exonucleolytic activity as 150 ng of p53 was used as a control.
Gel mobility shift assay.
Synthetic p21 promoter oligonucleotides (10) were end labeled by using T4 polynucleotide kinase and [γ-32P]ATP, gel purified, and used as probes in binding reactions after annealing with unlabeled complementary strand. p53 was purified by metal affinity chromatography or nuclear extraction (29). 32P-labeled three-stranded substrates containing an A-G mismatch were prepared as described previously (8). Binding reactions were carried out as described previously (8, 22).
RESULTS
The p53 exonuclease activity is mediated by the p53 core domain.
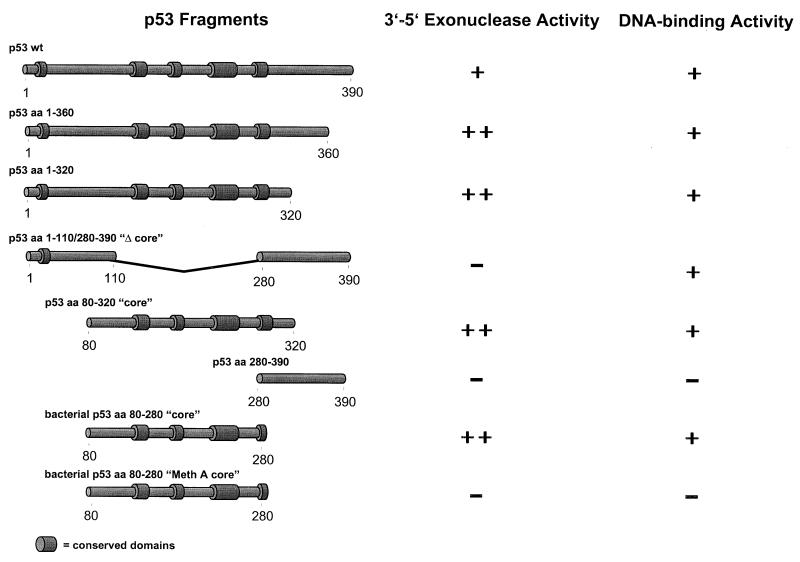
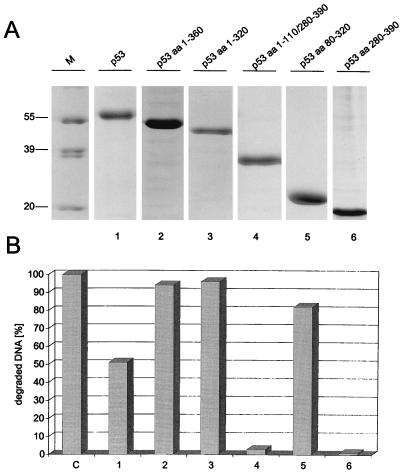
In our previous experiments, a bacterial fragment comprising the p53 core domain (amino acids 80 to 280) was renatured after SDS-PAGE and was shown to exhibit 3′-5′ exonuclease activity (48). To analyze the structural prerequisites of the p53 exonuclease activity on native p53, we analyzed various deletion fragments of mouse p53 (see Fig. 3). With the exception of the bacterial p5380-280 fragments, all of the p53 fragments were expressed in insect cells infected with the respective recombinant baculoviruses. Full-length p53 and all p53 fragments were His tagged at their N termini, which allowed their easy purification via metal affinity chromatography (see Materials and Methods). Figure 1 shows the purified baculovirus-expressed p53 proteins and their corresponding exonuclease profiles. It is evident that all fragments containing the p53 core domain, as well as the isolated p53 core domain itself, exhibited exonuclease activity, whereas those fragments not containing the core domain were exonuclease negative. We conclude that the p53 core domain mediates exonuclease activity. This conclusion was further substantiated by comparing the exonuclease activities of the isolated core domains (amino acids 80 to 280) of wt p53 and MethA mutant p53 (7) (containing the mutations C132F, E168G, and M234I [11]), which were expressed in bacteria and purified by metal affinity chromatography. Figure 2A and B show that these fragments were obtained in high yields and reasonable purity. Whereas the wt p53 core fragment displayed exonuclease activity (Fig. 2C), the MethA mutant p53 core domain was devoid of any exonuclease activity (Fig. 2D). This provides further evidence that structural alterations induced by mutations in the p53 core domain abolish not only sequence-specific DNA binding of p53 but also its exonuclease activity, supporting our previous finding (48) that mutant p53 proteins have lost the p53 exonuclease activity. The results of the exonuclease-mapping experiment are summarized in Fig. 3.
FIG. 3.
Summary of p53 exonuclease activity mapping data. p53 fragments and their corresponding exonuclease activities are shown schematically. aa, amino acids.
FIG. 1.
Exonuclease activities of p53 and p53 deletion mutants. wt p53 and p53 deletion mutants were expressed in High Five insect cells infected with recombinant baculovirus and purified by metal affinity chromatography as described in Materials and Methods. Peak fractions of all proteins were analyzed by SDS-PAGE (A), and 150 ng of each protein was tested for exonuclease activity by the 3H filter retention assay (B). Exonuclease III was used as a control. aa, amino acids. Lane M, markers. Numbers on the left in panel A are molecular masses in kilodaltons.
FIG. 2.
wt but not mutant p53 core domain is necessary and sufficient for p53 exonuclease activity. wt (A and C) and MethA (B and D) p53 core fragments (amino acids 80 to 280) were expressed in bacteria and purified by metal affinity chromatography. Column fractions of both proteins were analyzed by SDS-PAGE (A and B) and tested for exonuclease activity by the 3H filter retention assay (C and D). Exonuclease III (lane C) was used as a control. Numbers on the left in panels A and B are molecular masses in kilodaltons.
Binding of SV40 T-Ag to p53 eliminates its exonuclease activity.
SV40 T-Ag targets p53 in SV40 lytic infection and cellular transformation by binding to the p53 core domain (54). This largely eliminates sequence-specific DNA binding of p53 and its ability to transactivate p53 target genes (5, 27, 42). It was therefore of interest to analyze whether binding of T-Ag to p53 would also affect the p53 exonuclease activity. If so, this would provide further and independent evidence that exonuclease activity and sequence-specific DNA binding of p53 are mediated by the same catalytic domain. Furthermore, targeting of p53 exonuclease activity by SV40 T-Ag might provide hints as to a possible in vivo relevance of this activity (see Discussion).
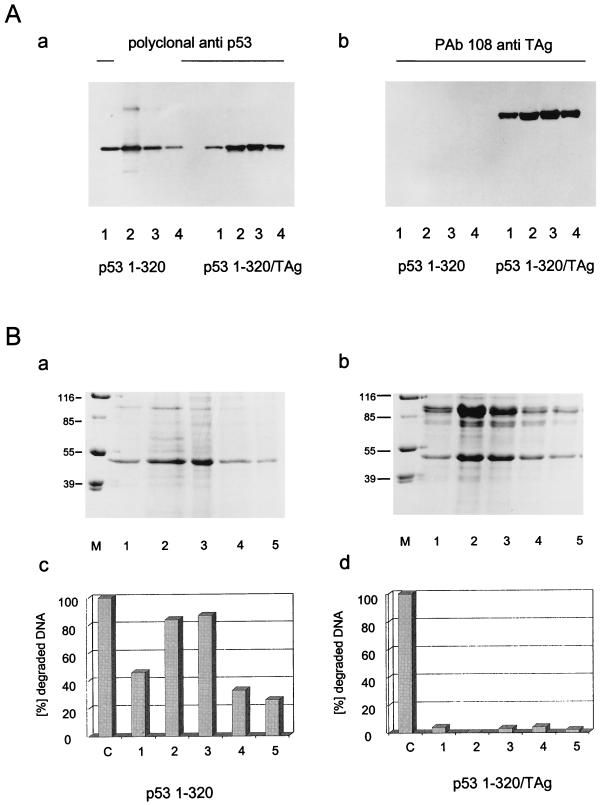
Insect cells were infected in parallel either with a recombinant baculovirus encoding SV40 T-Ag or with a virus encoding the p531-320 fragment. The p531-320 fragment was chosen for complex formation with T-Ag, because this fragment, in contrast to full-length p53, very effectively and quantitatively forms a complex with T-Ag in vitro (unpublished observation). After lysis of the infected cells, the lysate containing the p531-320 fragment was split, and half of it was mixed with the lysate of cells expressing T-Ag to allow formation of the T-Ag/p531-320 complex. The p531-320 fragment and the T-Ag/p531-320 complex were purified from the respective lysates by metal affinity chromatography via the His tag of the p531-320 fragment (for details, see Materials and Methods). Figure 4A shows the analysis of the purified p531-320 fragment and of the T-Ag/p531-320 complex by Western blotting with a polyclonal antiserum for p53 (panel a) or the T-Ag-specific monoclonal antibody PAb108 (panel b). This analysis demonstrates that similar amounts of free p531-320 fragment and of p531-320 fragment complexed to T-Ag were recovered during purification and that the complexed p531-320 fragment had bound a significant amount of T-Ag. Figure 4B shows the Coomassie blue-stained pattern of fractions of the free p531-320 fragment (panel a) and of this fragment in complex with T-Ag after metal affinity chromatography and SDS-PAGE (panel b) and the corresponding exonuclease activities of the p531-320 fragment in the respective fractions (panels c and d). As is evident from Fig. 4B, the free p531-320 fragment had exonuclease activity (panel c), whereas this fragment in complex with T-Ag was devoid of exonuclease activity (panel d).
FIG. 4.
SV40 T-Ag inhibits p53 exonuclease activity. High Five insect cells were infected with recombinant baculovirus coding for the p531-320 fragment or SV40 T-Ag. Cells were lysed at 44 h pi, and the p531-320-containing lysate was split. One half was purified by metal affinity chromatography, and the other half was mixed with SV40 T-Ag-containing lysate and also purified for His-tagged p531-320 by metal affinity chromatography. Column fractions of both preparations were analyzed by SDS-PAGE (B, panels a and b) and Western blotting (A) and tested for exonuclease activity (B, panels c and d). Numbers on the left in panel B (panels a and b) are molecular masses in kilodaltons. Lanes M, markers.
C-terminally truncated p53 exhibits a significantly higher exonuclease activity than full-length p53.
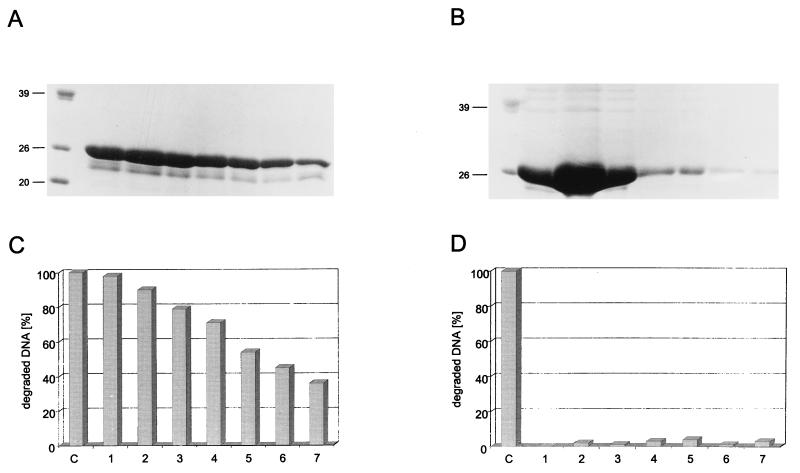
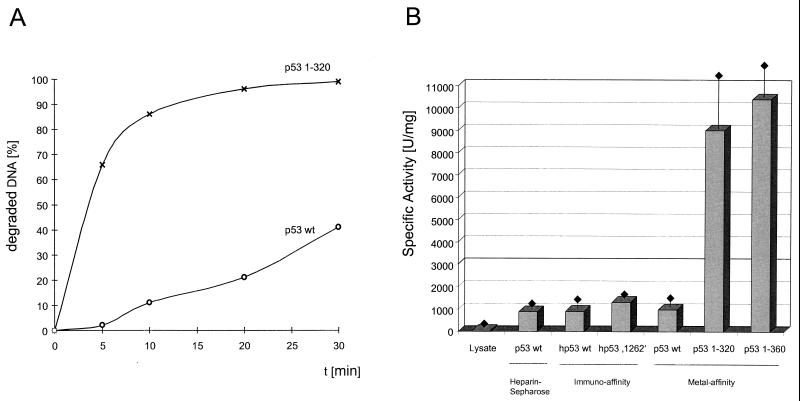
During our analyses of p53 fragments for exonuclease activity, we noticed that the C-terminally truncated p53 fragments seemed to have a higher specific activity than full-length p53. This was verified by comparing the kinetics of substrate degradation for full-length p53 and for the p531-320 fragment. Figure 5A shows that the p531-320 fragment degraded the input substrate DNA at a much higher catalytic rate than did full-length p53. Quantitative evaluation and determination of the specific exonuclease activities of various full-length p53 preparations and of the C-terminally truncated p531-320 and p531-360 fragments (Fig. 5B) revealed that full-length p53, regardless of its mode of purification, had about a 10-times-lower specific exonuclease activity than the C-terminally truncated p531-320 and p531-360 fragments. We conclude that the exonuclease activity of full-length p53 is negatively regulated by the C-terminal basic domain, as the shortest truncation which strongly enhanced the p53 exonuclease was a deletion of the C-terminal 30 amino acids (p531-360 fragment). Interestingly, and in contrast to the case for sequence-specific DNA binding (16, 63), the oligomerization status of p53 did not influence the p53 intrinsic exonuclease activity. This conclusion is based on the findings that the oligomerization-defective p531-320 fragment exhibited the same, activated specific exonuclease activity as the oligomerization-competent p531-360 fragment and, conversely, that the full-length oligomerization-defective p53, 1262 (60, 61), had an exonuclease activity similar to that of oligomerization-competent wt p53.
FIG. 5.
p53 exonuclease activity is negatively regulated by the C-terminal basic regulatory domain. Full-length wt p53 and p531-320 were tested for exonuclease activity, and their relative activities were compared in a time course (A). Specific exonuclease activities were determined for wt p53 purified by heparin-Sepharose, immunoaffinity, or metal affinity chromatography and were compared to those of the oligomerization-defective mutant p53 1262 and deletion mutants p531-320 and p531-360 (B). Standard deviations are indicated by error bars. One unit corresponds to the degradation of 60 pmol of DNA per 10 min at 37°C. hp53, human p53.
Protease treatment of the full-length p53 protein activates exonuclease activity.
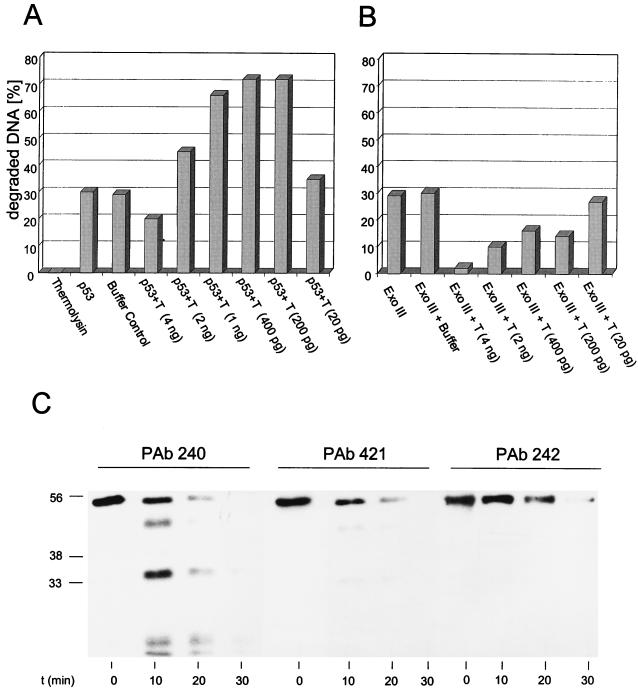
It has been reported that the full-length p53 molecule is rather unstable but that C-terminal truncation enhances the stability of p53 (17). To test the possibility that such an effect might be responsible for the observed higher specific activity of the C-terminally truncated p53 fragments, we designed a control experiment which was based on the previously reported finding that limited proteolytic digestion of p53 with thermolysin preferentially degrades the N- and C-terminal portions of the p53 molecule, whereas the p53 core fragment is more resistant to degradation (4). Constant amounts of purified wt p53 were incubated in parallel with consecutively higher dilutions of thermolysin and subjected to exonuclease assays. If activation of the p53 exonuclease results from C-terminal truncation, then protease treatment at certain thermolysin-to-p53 ratios should lead to higher exonuclease activities. Figure 6A shows that incubation of p53 with thermolysin at certain enzyme dilutions resulted in a significant activation of the p53 exonuclease activity. Analysis of the p53 fragments generated by thermolysin in a time course experiment (Fig. 6C) shows that thermolysin treatment rapidly generated lower-migrating forms of p53; these were detected by PAb240, which reacts with an epitope in the p53 core domain (15, 59, 70), but not by PAb421, which reacts with a C-terminal epitope of p53 (18, 58, 65), or by PAb242, which reacts with an N-terminal epitope of p53 (36, 70). This strongly supports the interpretation that the lower exonuclease activity of full-length p53 compared to C-terminally truncated p53 fragments results from negative regulation by the C-terminal basic regulatory domain of p53. Note that incubation of full-length p53 with reaction buffer alone did not affect the p53 exonuclease activity (Fig. 6A), further demonstrating that this activity was stable under our assay conditions and that its activation by protease treatment resulted from C-terminal truncation. Figure 6B shows a control experiment demonstrating that a comparable protease treatment of the bacterial exonuclease III reduced rather than activated its exonuclease activity.
FIG. 6.
Protease treatment of p53 activates its exonucleolytic activity. (A and B) Wild-type p53 (A) or exonuclease III (Ex. III) (B) (150 ng) was digested with various amounts of thermolysin (40 U/mg), ranging from 4 ng to 20 pg. Exonuclease activity was measured after 15 min of digestion. The amount of exonuclease III was chosen to match the exonuclease activity of 150 ng of wt p53. Incubation of p53 with buffer alone for 15 min (p53 buffer control) did not affect the p53 exonuclease activity. (C) Western blot analysis of p53 fragments resulting from thermolysin digestion after 0, 10, 20, and 30 min of incubation. Digestions were performed with 1 ng of thermolysin, corresponding to bar 6 in panel A. After 10 min of thermolysin digestion, lower-migrating forms of p53 were detected with PAb240, directed against an epitope in the p53 core domain. Those forms could not be detected with monoclonal antibodies directed against the p53 N terminus (PAb242) or the p53 C terminus (PAb 421). Numbers on the left in panel C are molecular masses in kilodaltons.
Copurification of the exonuclease activity with the p531-320 fragment.
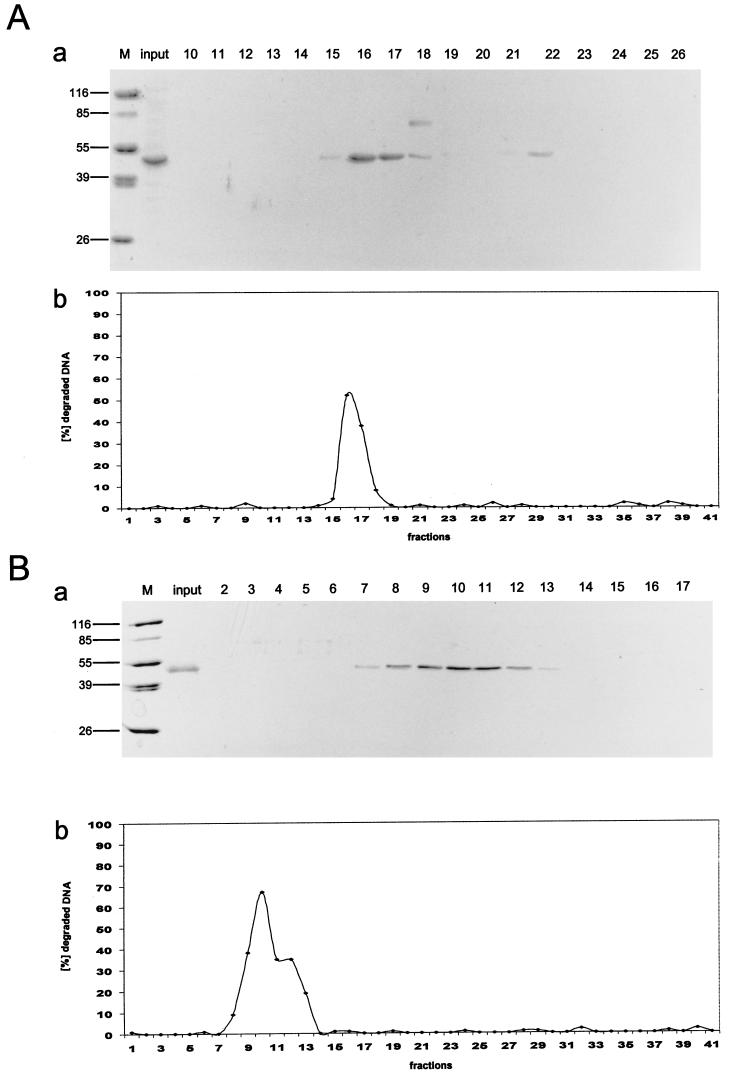
We previously provided strong evidence that the exonuclease activity of p53 is intrinsic to the p53 molecule and is not mediated by an exonuclease associating with p53 (48). The finding that C-terminal truncation of p53 activated the p53 exonuclease by a factor of at least 10 allowed us to address the question of an associated exonuclease by a different approach: in a preparation of highly purified exonuclease-active C-terminally truncated p53, an associated exonuclease should be a major contaminant, which thus should be clearly detectable. Therefore, we monitored the exonuclease activity of already highly pure p531-320 protein, purified by metal chelate chromatography, during subsequent purification steps. The p531-320 fragment was chosen over the p531-360 fragment because first, it lacks the oligomerization domain, thereby greatly facilitating further purification steps due to the absence of p53 protein in different oligomeric forms, and second, its apparent molecular mass (45 kDa) is significantly lower than that of full-length p53, which should allow the easy identification of a copurifying exonuclease. One hundred micrograms of p531-320 fragment, purified to greater than 90% homogeneity by metal affinity chromatography (Fig. 7A, panel a, input) was loaded onto a UNO Q anion-exchange column and eluted by increasing the salt (KCl) concentration (for details, see Materials and Methods). Analysis of the protein eluted from the UNO Q column by SDS-PAGE and Coomassie blue staining (Fig. 7A, panel a) shows that the p531-320 protein was recovered as a single peak at about 250 mM KCl (fractions 14 to 18). Most of the protein ran as a single band corresponding to the p531-320 fragment. However, fraction 18 revealed the presence of another protein with a molecular mass of approximately 60 kDa; a minor portion of the p531-320 fragment eluted at higher salt concentration (300 mM) and was recovered in fraction 22. Fractions 14 to 18 contained about 85% of the input p531-320 protein. These fractions also contained the majority of the input exonuclease activity (Fig. 7A, panel b, fractions 14 to 18). No detectable exonuclease activity was recovered in fractions not containing the p531-320 protein. The exonuclease activity was closely proportional to the amount of p531-320 fragment, as the exonuclease activity of the p531-320 fragment in lane 10, which was contaminated by a protein with a molecular mass of approximately 60 kDa, was proportional to the amount of p531-320, and was independent of the contaminating protein. Alternatively, 140 μg of metal affinity-purified p531-320 (Fig. 7B, panel a, input) was loaded onto a heparin sulfate affinity column. The p531-320 fragment was again eluted by increasing the salt (KCl) concentration and was recovered at 200 to 300 mM KCl. Figure 7B, panel a, shows the analysis of the heparin sulfate-purified p531-320 fragment by SDS-PAGE and Coomassie blue staining. The p531-320 fragment eluted as a broad peak (fractions 7 to 14), with no detectable contaminating proteins and recovering 120 μg of the input protein. Again, the exonuclease activity coeluted with the p531-320 fragment (Fig. 7B, fractions 7 to 14), rendering it extremely unlikely that this exonuclease activity was due to an associated exonuclease. Interestingly, the exonuclease activity profile (Fig. 7B, panel b), in contrast to the protein elution profile (panel a), of the p531-320 protein showed a broad shoulder, suggesting the possibility that the p531-320 protein eluted at a higher salt concentration has a higher specific exonuclease activity than that eluted at lower salt concentrations.
FIG. 7.
Copurification of the p53 exonuclease activity with the p531-320 fragment. The p531-320 fragment was first purified by metal affinity chromatography and then further purified either by anion-exchange chromatography (UNO Q) (A) or by heparin-Sepharose affinity chromatography (B). p531-320 was eluted from the columns with a KCl gradient (see Materials and Methods for details). All fractions were analyzed for exonuclease activity (panels b), and peak fractions were analyzed by SDS-PAGE and Coomassie blue staining (panels a). The purity of the input metal affinity-purified p531-320 protein is shown in panels a, lanes input. Note that the exonuclease activity copurifies with the p531-320 protein in all purification schemes. For quantitative evaluation of these data, see Table 1. Lanes M, markers. Numbers on the left in panels a are molecular masses in kilodaltons.
Table 1 summarizes the purification steps and the specific activities of the recovered p531-320 fragment. As expected, the already highly pure p531-320 protein obtained after metal affinity chromatography had the highest specific exonuclease activity. However, the losses in specific exonuclease activity encountered during further chromatography were in the expected range for the inactivation of such an enzymatic activity.
TABLE 1.
Fast protein liquid chromatography purification of metal affinity-purified p531-320 fragment
| Stage and parameter (unit) | Value after:
|
|
|---|---|---|
| UNO Q AIEX step | Heparin Sulfate step | |
| Inputa | ||
| Total protein (μg) | 100 | 140 |
| Sp act (U/mg) | 8,400 | 9,572 |
| Total activity (U) | 840 | 1,340 |
| Recovery | ||
| Total protein (μg) | 85 (85%) | 120 (85%) |
| Sp act (U/mg) | 4,000 (47%) | 4,700 (49%) |
| Total activity (U) | 340 (40%) | 564 (42%) |
Peak fraction of Co2+ metal affinity-purified murine p531-320.
Opposing regulation of p53 sequence-specific DNA binding and exonuclease activities. (i) Effects of PAb421.
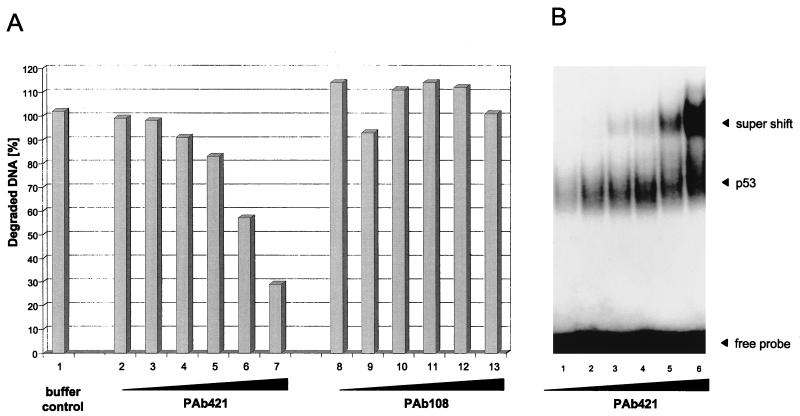
PAb421 binds to an epitope within the basic regulatory domain of p53 and was shown to activate sequence-specific DNA binding of p53 to certain DNA substrates (22, 23, 29). As both sequence-specific DNA binding and exonuclease activities are negatively regulated by the basic regulatory domain of p53, we asked whether and how PAb421 would affect the p53 exonuclease activity compared to sequence-specific DNA binding. Full-length p53 was incubated with PAb421 or p53-unrelated PAb108 and subjected to the exonuclease assay. Figure 8A shows that the exonuclease activity of full-length p53 was strongly inhibited by PAb421 in a concentration-dependent manner. This inhibition was due to binding of PAb421 to the p53 molecule, as even the highest concentration of the unrelated antibody PAb108 had no effect on its exonuclease activity. In contrast, binding of PAb421 to p53 strongly activated DNA binding of p53 to the p21 promoter substrate (Fig. 8B). We conclude that the exonuclease and sequence-specific DNA binding activities of p53 are regulated in opposing manners, although both activities are negatively regulated by the basic regulatory domain of p53.
FIG. 8.
PAb421 influences p53 exonuclease and sequence-specific DNA binding activities in opposite manners. Metal affinity-purified full-length p53 was incubated with increasing amounts of PAb421 and tested for exonuclease activity (A, lanes 2 to 7) or sequence-specific DNA binding (B). p53 was also incubated with a non-p53-specific antibody (PAb108, specific for SV40 T-Ag) and tested for exonuclease activity (A, lanes 8 to 13).
(ii) Influence of phosphorylation status.
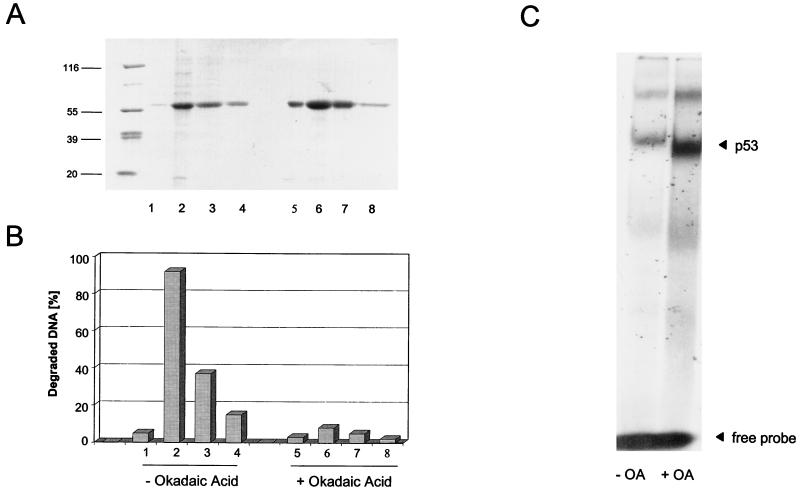
p53 is a phosphoprotein, and there is accumulating evidence that phosphorylation is an important determinant in regulating p53 functions (12, 14, 20, 40, 57). We therefore asked whether and how the phosphorylation status of p53 would influence the p53 exonuclease activity compared to sequence-specific DNA binding. The phosphorylation status of a phosphoprotein is determined by the net result of kinase and phosphatase reactions. Therefore, the phosphorylation status of p53 can be enhanced by inhibiting the activity of phosphatase 2A (PP2A) in infected insect cells with the PP2A inhibitor okadaic acid. A similar approach recently was applied to analyze the influence of an enhanced phosphorylation status of rat p53 on its sequence-specific DNA binding activity (14).
Insect cells were infected with baculovirus encoding full-length p53. Half of the cells were kept in normal growth medium, and the other half were treated with 200 nM okadaic acid at 41 hpi. Cells were harvested at 44 hpi, and p53 from both infections was purified by metal affinity chromatography (for details, see Materials and Methods). Purified p53 proteins obtained from the untreated and the okadaic acid-treated infections (Fig. 9A) were analyzed in parallel for exonuclease activity and for sequence-specific DNA binding in electrophoretic mobility shift assays (EMSA), using the p21 promoter substrate. Figure 9B shows that p53 from the untreated infection displayed exonuclease activity. In contrast, p53 obtained from the okadaic acid-treated infection was exonuclease negative. Conversely, p53 from untreated cells showed only a low affinity for the p21 promoter substrate in EMSA, whereas p53 from okadaic acid-treated cells was highly active in this assay, even in the absence of activating antibody PAb421 (Fig. 9C [compare Fig. 8]). The same results were obtained with a PG oligonucleotide (24) (data not shown). We conclude that phosphorylation can modulate the function of the p53 core domain, endowing it with either an exonuclease or a sequence-specific DNA binding activity.
FIG. 9.
Okadaic acid treatment influences p53 exonuclease and sequence-specific DNA binding activities in opposite manners. High Five insect cells were infected with recombinant baculovirus coding for p53. The cells were split, and one half was treated with okadaic acid (OA). p53 from both samples was purified by metal affinity chromatography. Column fractions were analyzed by SDS-PAGE (A) and tested for exonuclease activity (B) and sequence-specific DNA binding (C). Numbers on the left in panel A are molecular masses in thousands.
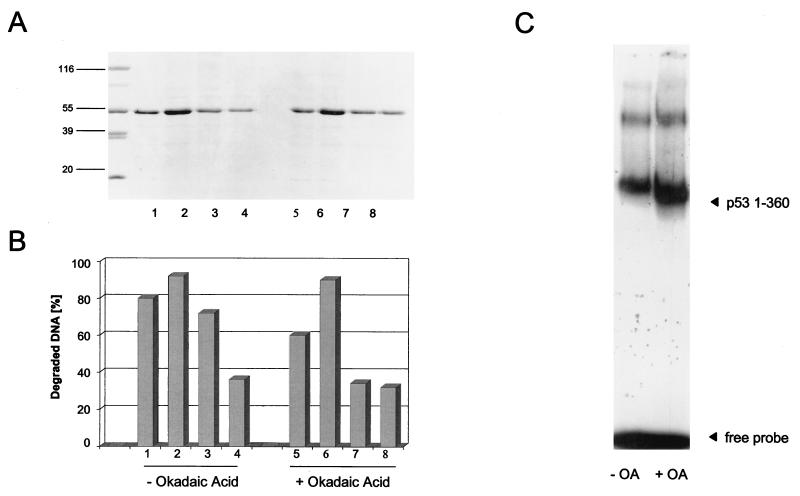
The 30 C-terminal amino acids of p53 are a prime target for regulatory posttranslational events. In mouse p53, this region contains the documented CK II phosphorylation site Ser386, which also is a putative site for the cdk7-cyclin H-p36 complex of TFIIH (30), and two putative protein kinase C type II phosphorylation sites, Ser370 and Ser372 (43), which possibly are also a target for the cdk7-cyclin H-p36 complex. The C-terminal region also is a target for acetylation of p53 by p300 (38, 55) and for glycosylation (56), with both events activating sequence-specific DNA binding of p53. As the C terminus negatively regulates both sequence-specific DNA binding of p53 and its exonuclease activity, we asked how an enhanced phosphorylation status of the C-terminally truncated p531-360 fragment would affect these activities. In an experiment performed in parallel to the one shown in Fig. 9, we compared the 3′-5′ exonuclease and the sequence-specific DNA binding activities of the p531-360 fragment purified from okadaic acid-treated and untreated baculovirus-infected insect cells (Fig. 10). Figure 10A shows that this fragment was obtained in similar yields and similar purities from both preparations. In contrast to the case for full-length p53, okadaic acid treatment did not inhibit the exonuclease activity of the p531-360 fragment (Fig. 10B). We conclude that C-terminal modification negatively regulates the exonuclease activity of p53, either directly by phosphorylation events within the C-terminal region or indirectly by phosphorylation of other sites which affect C-terminal modification. Analysis of the DNA binding activity of the p531-360 fragment (Fig. 10C) showed that truncation as such already significantly enhanced the DNA binding activity compared to that of full-length p53 (compare Fig. 9C and 10C). Enhancing the phosphorylation status of the p531-360 fragment by treating the culture with okadaic acid further enhanced its DNA binding activity towards the waf1/p21 promoter substrate in EMSA, in line with the observation that the phosphorylation status of the cyclin-dependent kinase site at Ser309 modulates sequence-specific DNA binding of p53 (66).
FIG. 10.
The exonuclease activity of C-terminally truncated p53 is not influenced by okadaic acid treatment. High Five insect cells were infected with recombinant baculovirus coding for the C-terminally truncated p531-360 fragment. The cells were split, and one half was treated with okadaic acid (OA). The p531-360 fragment from both samples was purified by metal affinity chromatography, analyzed by SDS-PAGE (A), and tested for exonuclease activity (B) and sequence-specific DNA binding (C). Numbers on the left in panel A are molecular masses in kilodaltons.
Binding of p53 to DNA substrates mimicking recombination intermediates is required for their effective degradation.
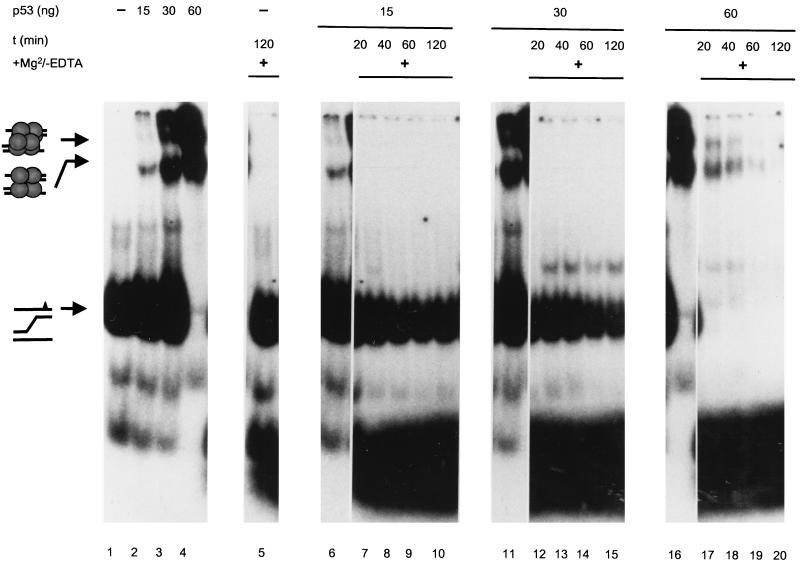
We recently provided evidence that p53 might control fidelity of homologous recombination by specific mismatch recognition in heteroduplex recombination intermediates (8). In this study we showed that wt p53, but not mutant p53, binds specifically and with high affinity to three-stranded DNA substrates mimicking early recombination intermediates. This interaction also requires an intact p53 core domain (8). As such substrates probably are biologically more relevant than the poly(dT) · poly(dA) substrates normally used in our exonuclease assays, we asked whether they might serve as effective substrates for the p53 exonucleolytic activity. Furthermore, as the interaction of p53 with such substrates can be analyzed by EMSA, it seemed possible to analyze the connection between substrate binding and exonucleolytic degradation of the substrate by p53 by switching on the p53 exonuclease after substrate binding.
A three-stranded DNA substrate comprising an A-G mismatch and a radioactively labeled top strand was prepared as described previously (8); its structure is schematically outlined on the left in Fig. 11. The substrate then was incubated with 0, 15, 30, or 60 ng of purified full-length wt p53 in the absence of Mg2+ ions, which are required for activation of the p53 exonuclease activity, and the mixture was subjected to PAGE (Fig. 11, lanes 1 to 4). Two discretely shifted bands were seen in all fractions containing p53 (lanes 2 to 4), corresponding to the interaction of p53 tetramers (lower band) or higher oligomeric forms of p53 (upper band) with the substrate. Both shifted bands increased in intensity with increasing amounts of p53 added, leading to a complete shift of the labeled substrate at the highest p53 concentration (60 ng) (Fig. 11, lane 4). In parallel, 5 mM Mg2+, which is required for activation of the p53 exonuclease (48), was added to the corresponding mixtures and incubated for 20, 40, 60, and 120 min, and p53-substrate complexes were analyzed by EMSA (Fig. 11). It is evident that the Mg2+-induced exonuclease activity of p53 led to a rapid, time- and p53 concentration-dependent degradation of the p53-bound substrate, as indicated by the loss of the shifted bands and the appearance of a radioactive smear at the bottom of the gel in those mixtures containing p53, but not in the one devoid of p53 (lane 5), even after prolonged incubation (120 min). Most importantly, the p53 exonuclease activity paralleled the degree of substrate binding and did not affect the unbound substrate. The slight time-dependent decrease in the amount of free substrate seen in lanes 12 to 15 reflects the recruitment of new substrate by p53 after complete digestion of the bound substrate, consistent with the high on-and-off rates reported by us for this dynamic interaction (8). We conclude that by binding to the three-stranded DNA substrate, p53 forms an enzyme-substrate complex; the Mg2+-induced p53 exonuclease activity then leads to rapid degradation of the p53-bound substrate.
FIG. 11.
DNA recombination intermediates are degraded effectively only in complex with p53. Purified wt p53 (0, 15, 30, and 60 ng) was incubated with a three-stranded DNA substrate mimicking an early recombination intermediate and containing an A-G mismatch (schematically outlined on the left) in the presence (lanes 5, 7 to 10, 12 to 15, and 17 to 20) or absence (lanes 1 to 4, 6, 11, and 16) of 5 mM Mg2+, which is required for switching on the p53 exonuclease (lanes 6, 11, and 16 are identical to lanes 2, 3, and 4, respectively). Incubation reactions were performed for 20, 40, 60, and 120 min at room temperature. Lanes 1 to 4 show a protein concentration-dependent shift of the p53-substrate complexes. Addition of Mg2+ to the binding reaction mixture (lanes 7 to 10, 12 to 15, and 17 to 20) led to complete degradation of the bound (lanes 17 to 20) but not the unbound (lanes 7 to 10 and 12 to 15) substrate in a time- and p53-dependent fashion. A slight decrease in the amount of unbound substrate (lanes 12 to 15) reflects the recruitment of new substrate by p53 after complete digestion of the bound substrate.
DISCUSSION
Our analyses of the exonuclease activities of native p53 fragments, expressed in insect cells infected with the respective recombinant baculoviruses and purified by metal affinity chromatography, demonstrated that the p53 core domain harbors two different activities, i.e., sequence-specific DNA binding activity and exonuclease activity. All p53 fragments containing the core domain were exonuclease positive, whereas all fragments devoid of this domain were exonuclease negative. In support of this, comparison of the exonuclease activities of the bacterially expressed core domains from wt p53 and from MethA mutant p53 showed that only the wt and not the mutant MethA p53 core domain exhibited exonuclease activity. Finally, SV40 T-Ag, bound to the p531-320 fragment via the p53 core domain, completely inhibited its exonuclease activity, which is even more impressive because the p531-320 fragment exhibited about a 10-fold-higher specific exonuclease activity than full-length p53.
The finding that C-terminal truncation of p53 activated the exonuclease activity by at least a factor of 10 suggests that the p53 exonuclease activity is also subject to regulation in vivo. The observed activation was not due to different protein stabilities of the full-length p53 and the C-terminally truncated p53 fragments, as purified full-length p53 could be activated in vitro by specific limited proteolytic digestion with thermolysin, which created p53 fragments lacking the N and C termini of full-length p53. The strongly enhanced exonuclease activity of C-terminally truncated p53 fragments also is not due to a more efficient association of an exogenous exonuclease. When metal affinity-purified p531-320 protein of already high purity (>90%) was further purified by two different chromatography steps, the exonuclease activity always copurified with this p53 fragment. The purity of the p531-320 protein and the fact that the exonuclease activity remained associated with this protein during purification according to quite different separation criteria in all likelihood exclude an associated exonuclease. We therefore conclude that the intrinsic exonuclease activity of full-length p53 is negatively regulated by the basic regulatory domain of p53 comprising the C-terminal 30 amino acids of the p53 molecule.
Despite a negative regulation of both exonuclease and sequence-specific DNA binding activities by the p53 C-terminal regulatory domain, these activities were found to be regulated in different and seemingly opposing manners on full-length p53. Whereas addition of PAb421 activated p53 binding to the waf1/p21 promoter substrate, the same treatment strongly reduced the p53 exonuclease activity. Similarly, hyperphosphorylation of p53 enhanced its sequence-specific DNA binding activity but abolished its exonuclease activity. In this respect, p53 prepared according to protocols optimized for analyzing sequence-specific DNA binding might be negative in exonuclease assays and vice versa. Interestingly, the exonuclease activity of the C-terminally truncated p531-360 fragment was no longer affected by alterations of its phosphorylation status, in contrast to sequence-specific DNA binding of the p531-360 fragment, which could still be further activated by enhancing its phosphorylation status. The most straightforward explanation for the insensitivity of the exonuclease activity of the p531-360 fragment to hyperphosphorylation is that the exonuclease activity of full-length p53 is regulated mainly by the phosphorylation status of the C-terminal phosphorylation site Ser386, Ser370, or Ser372. However, other possibilities cannot be excluded, since phosphorylation sites in other regions of the p53 molecule might control posttranslational modification events within the p53 C terminus and vice versa. Further experiments in our laboratory are aimed at identifying the posttranslational modification(s) inhibiting the p53 exonuclease activity.
So far, an experimental differentiation between mechanisms regulating the exonuclease and the sequence-specific DNA binding activities of full-length p53 could be achieved only by means that activate sequence-specific DNA binding but inactivate the exonuclease. However, it is noteworthy that bacterially expressed, i.e., nonphosphorylated, p53 is virtually devoid of sequence-specific DNA binding activity (22, 24) but exerts exonuclease activity (48), pointing to the possibility that the p53 exonuclease activity might be exerted by hypo- or even nonphosphorylated p53. The specific targeting of distinct phosphorylation sites of p53 by PP2A has been described (53). Furthermore, one could imagine specific dephosphorylation of p53 by other phosphatases. Considering that C-terminal truncation of p53 strongly activated its exonuclease activity, specific in vivo proteolytic cleavage of p53 (46, 50) could also lead to activation. In this regard it is noteworthy that at least in mouse cells p53 exists in two forms, as a regularly spliced full-length protein and as an alternatively spliced p53 missing the C-terminal regulatory domain (33). This alternatively spliced form of p53 is preferentially expressed in the G2 phase of the cell cycle (33), i.e., at a time when the replicated genome is scanned for replication errors prior to mitosis (62). Finally, p53 can interact with a large number of cellular proteins (52), which might induce conformational alterations activating the p53 exonuclease.
We are especially intrigued by the finding that sequence-specific DNA binding and exonuclease activities seem to be mutually exclusive p53 activities. So far, activation of p53 has been considered a prerequisite for p53 function, and there even has been the notion that p53, in the absence of cellular stress, is functionally inactive (64). Stress-mediated activation of p53 function correlates with activation of sequence-specific DNA binding, which in turn correlates with inhibition of the p53 exonuclease activity. Therefore, we hypothesize that p53 in the absence of cellular stress is not an inactive protein but exerts non-stress-induced functions required for maintaining genomic integrity, (e.g., repair of spontaneous DNA damage or the control of homologous recombination) via intrinsic activities not related to sequence-specific DNA binding (e.g., its exonuclease activity) (25). In this regard, it recently has been reported that p53 specifically interacts with DNA polymerase α (32a) and may control fidelity of DNA replication mediated by this enzyme by acting as an external proofreader (21).
To get further clues about a possible in vivo function of the p53 exonuclease, we here followed up our previous observations that p53 negatively regulates homologous recombination, possibly by controlling fidelity of homologous recombination via specific mismatch recognition (8, 68). wt p53 specifically binds to three-stranded DNA substrates mimicking early recombination intermediates and rapidly and efficiently degrades the bound substrate when the exonuclease activity of p53 is switched on by addition of Mg2+ as a cofactor. In addition to suggesting that binding of such a substrate represents the formation of a relevant enzyme-substrate complex, this finding provides further independent evidence for the intrinsic nature of the p53 exonuclease: as the exonuclease activity as well as binding of the three-stranded DNA substrate requires an intact p53 core domain (8), it is extremely unlikely that an exogenous exonuclease will associate with the p53 core domain and still allow binding of the substrate but, on the other hand, will be in a spatial orientation that allows degradation of the substrate attached to the same p53 molecule. This conclusion is drawn from our finding that specifically the bound, and not the unbound, substrate was degraded upon addition of Mg2+ ions as a cofactor, starting the p53 exonuclease activity. Regarding the biological relevance of this interaction, it may be more than a coincidence that binding of SV40 T-Ag to p53 abolished the p53 exonuclease activity in vitro (this study) and enhanced the frequency of recombination in SV40-infected cells by at least 1 order of magnitude (68). However, more direct proof for an in vivo involvement of the p53 exonuclease activity in recombination or other repair events has to be obtained in order to substantiate our model of a dual role for p53 in maintaining genomic integrity (25).
ACKNOWLEDGMENTS
This work was supported by Deutsche Krebshilfe grant 10-0858-De2, German Israeli Foundation (G.I.F.) grant 1044-207.03/96, DFG grant Wi 1376/1-2, Boehringer Mannheim, and the Fonds der chemischen Industrie. F.J. was supported by Boehringer Ingelheim Fonds, Stuttgart, Germany. The Heinrich-Pette-Institut is financially supported by Freie und Hansestadt Hamburg and Bundesministerium für Gesundheit.
F.J. and N.A. contributed equally to this work.
REFERENCES
- 1.Bakalkin G, Selivanova G, Yakovleva T, Kiseleva E, Kashuba E, Magnusson K P, Szekely L, Klein G, Terenius L, Wiman K G. p53 binds single-stranded DNA ends through the C-terminal domain and internal DNA segments via the middle domain. Nucleic Acids Res. 1995;23:362–369. doi: 10.1093/nar/23.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakalkin G, Yakovleva T, Selivanova G, Magnusson K P, Szekely L, Kiseleva E, Klein G, Terenius L, Wiman K G. p53 binds single-stranded DNA ends and catalyzes DNA renaturation and strand transfer. Proc Natl Acad Sci USA. 1994;91:413–417. doi: 10.1073/pnas.91.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild-type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bargonetti J, Manfredi J J, Chen X, Marshak D R, Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev. 1993;7:2565–2574. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- 5.Bargonetti J, Reynisdottir I, Friedman P N, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev. 1992;6:1886–1898. doi: 10.1101/gad.6.10.1886. [DOI] [PubMed] [Google Scholar]
- 6.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 7.DeLeo A B, Shiku H, Takahashi T, John M, Old L J. Cell surface antigens of chemically induced sarcomas of the mouse. I. Murine leukemia virus-related antigens and alloantigens on cultured fibroblasts and sarcoma cells: description of a unique antigen on BALB/c Meth A sarcoma. J Exp Med. 1977;146:720–734. doi: 10.1084/jem.146.3.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudenhoeffer C, Rohaly G, Will K, Deppert W, Wiesmueller L. Specific mismatch recognition in heteroduplex intermediates by p53 suggests a role in fidelity control of homologous recombination. Mol Cell Biol. 1998;18:5332–5342. doi: 10.1128/mcb.18.9.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Deiry W S, Harper J W, O’Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y, Wiman K G, Mercer W E, Kastan M B, Kohn K W, Elledge S J, Kinzler K W, Vogelstein B. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 10.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 11.Eliyahu D, Goldfinger N, Pinhasi-Kimhi O, Shaulsky G, Skurnik Y, Arai N, Rotter V, Oren M. Meth A fibrosarcoma cells express two transforming mutant p53 species. Oncogene. 1988;3:313–321. [PubMed] [Google Scholar]
- 12.Fiscella M, Ullrich S J, Zambrano N, Shields M T, Lin D, Lees-Miller S P, Anderson C W, Mercer W E, Appella E. Mutation of the serine 15 phosphorylation site of human p53 reduces the ability of p53 to inhibit cell cycle progression. Oncogene. 1993;8:1519–1528. [PubMed] [Google Scholar]
- 13.Fritsche M, Haessler C, Brandner G. Induction of nuclear accumulation of the tumor-suppressor protein p53 by DNA-damaging agents. Oncogene. 1993;8:307–318. [PubMed] [Google Scholar]
- 14.Fuchs B, Hecker D, Scheidtmann K H. Phosphorylation studies on rat p53 using the baculovirus expression system—manipulation of the phosphorylation state with okadaic acid and influence on DNA binding. Eur J Biochem. 1995;228:625–639. doi: 10.1111/j.1432-1033.1995.0625m.x. [DOI] [PubMed] [Google Scholar]
- 15.Gannon J V, Greaves R, Iggo R, Lane D P. Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J. 1990;9:1595–1602. doi: 10.1002/j.1460-2075.1990.tb08279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halazonetis T D, Kandil A N. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 1993;12:5057–5064. doi: 10.1002/j.1460-2075.1993.tb06199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen S, Hupp T R, Lane D P. Allosteric regulation of the thermostability and DNA binding activity of human p53 by specific interacting proteins. J Biol Chem. 1996;271:3917–3924. doi: 10.1074/jbc.271.7.3917. [DOI] [PubMed] [Google Scholar]
- 18.Harlow E, Crawford L V, Pim D C, Williamson N M. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harper J W, Adami G R, Wei M, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 20.Hecker D, Page G, Lohrum M, Weiland S, Scheidtmann K H. Complex regulation of the DNA-binding activity of p53 by phosphorylation: differential effects of individual phosphorylation sites on the interaction with different binding motifs. Oncogene. 1996;12:953–961. [PubMed] [Google Scholar]
- 21.Huang P. Excision of mismatched nucleotides from DNA: a potential mechanism for enhancing DNA replication fidelity by the wild-type p53 protein. Oncogene. 1998;17:261–270. doi: 10.1038/sj.onc.1201946. [DOI] [PubMed] [Google Scholar]
- 22.Hupp T R, Lane D P. Allosteric activation of latent p53 tetramers. Curr Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 23.Hupp T R, Meek D W, Midgley C A, Lane D P. Activation of the cryptic DNA binding function of mutant forms of p53. Nucleic Acids Res. 1993;21:3167–3174. doi: 10.1093/nar/21.14.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hupp T R, Meek D W, Midgley C A, Lane D P. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 25.Janus, F., N. Albrechtsen, I. Dornreiter, L. Wiesmüller, F. Grosse, and W. Deppert. The dual role model for p53 in maintaining genomic integrity. Cell. Mol. Life Sci., in press. [DOI] [PMC free article] [PubMed]
- 26.Jayaraman L, Prives C. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 27.Jiang D, Srinivasan A, Lozano G, Robbins P D. SV40 T antigen abrogates p53-mediated transcriptional activity. Oncogene. 1993;8:2805–2812. [PubMed] [Google Scholar]
- 28.Kastan M B, Zhan Q, El-Deiry W D, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 29.Kim E, Albrechtsen N, Deppert W. DNA-conformation is an important determinant of sequence-specific DNA binding by tumor suppressor p53. Oncogene. 1997;15:857–869. doi: 10.1038/sj.onc.1201412. [DOI] [PubMed] [Google Scholar]
- 30.Ko L J, Shieh S Y, Chen X B, Jayaraman L, Tamai K, Taya Y, Prives C, Pan Z Q. p53 is phosphorylated by Cdk7-cyclin H in a p36mat1-dependent manner. Mol Cell Biol. 1997;17:7220–7229. doi: 10.1128/mcb.17.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kornberg A, Baker T. DNA replication. 2nd ed. San Francisco, Calif: W. H. Freeman; 1991. [Google Scholar]
- 32.Kuerbitz S J, Plunkett B S, Walsh W V, Kastan M B. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Kühn, C., F. Müller, C. Meller, H.-P. Nasheue, F. Janus, W. Deppert, and F. Grosse. Surface plasma resonance measurements reveal stable complex formation between p53 and DNA polymerase α. Oncogene, in press. [DOI] [PubMed]
- 33.Kulesz-Martin M F, Lisafeld B, Huang H, Kisiel N D, Lee L. Endogenous p53 protein generated from wild-type alternatively spliced p53 RNA in mouse epidermal cells. Mol Cell Biol. 1994;14:1698–1708. doi: 10.1128/mcb.14.3.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane D P. Cancer: a death in the life of p53. Nature. 1993;362:786–787. doi: 10.1038/362786a0. [DOI] [PubMed] [Google Scholar]
- 35.Lane D P. Cancer: p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 36.Lane D P, Stephen C W, Midgley C A, Sparks A, Hupp T R, Daniels D A, Greaves R, Reid A, Vojtesek B, Picksley S M. Epitope analysis of the murine p53 tumour suppressor protein. Oncogene. 1996;12:2461–2466. [PubMed] [Google Scholar]
- 37.Lee S, Elenbaas B, Levine A, Griffith J. p53 and its 14 kDa C-terminal domain recognize primary DNA damage in the form of insertion/deletion mismatches. Cell. 1995;81:1013–1020. doi: 10.1016/s0092-8674(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 38.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 39.Linn S M, Lloyd R S, Roberts R J. Nucleases. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1993. [Google Scholar]
- 40.Lohrum M, Scheidtmann K H. Differential effects of phosphorylation of rat p53 on transactivation of promoters derived from different p53 responsive genes. Oncogene. 1996;13:2527–2539. [PubMed] [Google Scholar]
- 41.Marx J. How p53 suppresses cell growth. Science. 1993;262:1644–1645. doi: 10.1126/science.8259506. [DOI] [PubMed] [Google Scholar]
- 42.Mietz J A, Unger T, Huibregtse J M, Howley P M. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milne D M, McKendrick L, Jardine L J, Deacon E, Lord J M, Meek D W. Murine p53 is phosphorylated within the PAb421 epitope by protein kinase C in vitro, but not in vivo, even after stimulation with the phorbol ester o-tetradecanoylphorbol 13-acetate. Oncogene. 1996;13:205–211. [PubMed] [Google Scholar]
- 44.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 45.Mol C D, Kuo C F, Thayer M M, Cunningham R P, Tainer J A. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 46.Molinari M, Okorokov A L, Milner J. Interaction with damaged DNA induces selective proteolytic cleavage of p53 to yield 40 kDa and 35 kDa fragments competent for sequence-specific DNA binding. Oncogene. 1996;13:2077–2086. [PubMed] [Google Scholar]
- 47.Muller C W, Harrison S C. The structure of the NF-kappa B p560:DNA-complex: a starting point for analyzing the Rel family. FEBS Lett. 1995;369:113–117. doi: 10.1016/0014-5793(95)00541-g. [DOI] [PubMed] [Google Scholar]
- 48.Mummenbrauer T, Janus F, Mueller B, Wiesmueller L, Deppert W, Grosse F. p53 protein exhibits 3′-to-5′ exonuclease activity. Cell. 1996;85:1089–1099. doi: 10.1016/s0092-8674(00)81309-4. [DOI] [PubMed] [Google Scholar]
- 49.Oberosler P, Hloch P, Ramsperger U, Stahl H. p53-catalyzed annealing of complementary single-stranded nucleic acids. EMBO J. 1993;12:2389–2396. doi: 10.1002/j.1460-2075.1993.tb05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okorokov A L, Ponchel F, Milner J. Induced N- and C-terminal cleavage of p53: a core fragment of p53, generated by interaction with damaged DNA, promotes cleavage of the N-terminus of full-length p53, whereas ssDNA induces C-terminal cleavage of p53. EMBO J. 1997;16:6008–6017. doi: 10.1093/emboj/16.19.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pavletich N P, Chambers K A, Pabo C O. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 52.Pietenpol J A, Vogelstein B. Tumor suppressor genes: no room at the p53 inn. Nature. 1993;365:17–18. doi: 10.1038/365017a0. [DOI] [PubMed] [Google Scholar]
- 53.Scheidtmann K H, Mumby M C, Rundell K, Walter G. Dephosphorylation of simian virus 40 large-T antigen and p53 protein by protein phosphatase 2A: inhibition by small-t antigen. Mol Cell Biol. 1991;11:1996–2003. doi: 10.1128/mcb.11.4.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmieg F I, Simmons D T. Characterization of the in vitro interaction between SV40 T antigen and p53: mapping the p53 binding site. Virology. 1988;164:132–140. doi: 10.1016/0042-6822(88)90628-9. [DOI] [PubMed] [Google Scholar]
- 55.Scolnick D M, Chehab N H, Stavridi E S, Lien M C, Caruso L, Moran E, Berger S L, Halazonetis T D. CREB-binding protein and p300/CBP-associated factor are transcriptional coactivators of the p53 tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- 56.Shaw P, Freeman J, Bovey R, Iggo R. Regulation of specific DNA binding by p53: evidence for a role for O-glycosylation and charged residues at the carboxy-terminus. Oncogene. 1996;12:921–930. [PubMed] [Google Scholar]
- 57.Slingerland J M, Jenkins J R, Benchimol S. The transforming and suppressor functions of p53 alleles: Effects of mutations that disrupt phosphorylation, oligomerization and nuclear translocation. EMBO J. 1993;12:1029–1037. doi: 10.1002/j.1460-2075.1993.tb05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stephen C W, Helminen P, Lane D P. Characterization of epitopes on human p53 using phage-displayed peptide libraries: insights into antibody-peptide interactions. J Mol Biol. 1995;248:58–78. doi: 10.1006/jmbi.1995.0202. [DOI] [PubMed] [Google Scholar]
- 59.Stephen C W, Lane D P. Mutant conformation of p53. Precise epitope mapping using a filamentous phage epitope library. J Mol Biol. 1992;225:577–583. doi: 10.1016/0022-2836(92)90386-x. [DOI] [PubMed] [Google Scholar]
- 60.Stürzbecher H W, Brain R, Addison C, Rudge K, Remm M, Grimaldi M, Keenan E, Jenkins J R. A C-terminal a-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene. 1992;7:1513–1523. [PubMed] [Google Scholar]
- 61.Tarunina M, Jenkins J R. Human p53 binds DNA as a protein homodimer but monomeric variants retain full transcription transactivation activity. Oncogene. 1993;8:3165–3173. [PubMed] [Google Scholar]
- 62.Tavormina P A, Wang Y, Burke D J. Differential requirements for DNA replication in the activation of mitotic check points in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:3315–3322. doi: 10.1128/mcb.17.6.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Unger T, Mietz J A, Scheffner M, Yee C L, Howley P M. Functional domains of wild-type and mutant p53 proteins involved in transcriptional regulation, transdominant inhibition, and transformation suppression. Mol Cell Biol. 1993;13:5186–5194. doi: 10.1128/mcb.13.9.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vogelstein B, Kinzler K W. p53 function and dysfunction. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- 65.Wade-Evans A, Jenkins J R. Precise epitope mapping of the murine transformation-associated protein, p53. EMBO J. 1985;4:699–706. doi: 10.1002/j.1460-2075.1985.tb03686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Reed M, Wang P, Stenger J E, Mayr G, Anderson M E, Schwedes J F, Tegtmeyer P. p53 domain: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- 68.Wiesmüller L, Cammenga J, Deppert W. In vivo assay of p53 function in homologous recombination between simian virus 40 chromosomes. J Virol. 1996;70:737–744. doi: 10.1128/jvi.70.2.737-744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–2232. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 70.Yewdell J W, Gannon J V, Lane D P. Monoclonal antibody analysis of p53 expression in normal and transformed cells. J Virol. 1986;59:444–452. doi: 10.1128/jvi.59.2.444-452.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]