Abstract
In the Caenorhabditis elegans hermaphrodite germ line, the sex-determining gene fem-3 is repressed posttranscriptionally to arrest spermatogenesis and permit oogenesis. This repression requires a cis-acting regulatory element in the fem-3 3′ untranslated region; the FBF protein, which binds to this element; and at least six mog genes. In this paper, we report the molecular characterization of mog-1 as well as additional phenotypic characterization of this gene. The mog-1 gene encodes a member of the DEAH-box family. Three mog-1 alleles possess premature stop codons and are likely to be null alleles, and one is a missense mutation and is likely to retain residual activity. mog-1 mRNA is expressed in both germ line and somatic tissues and appears to be ubiquitous. The MOG-1 DEAH-box protein is most closely related to proteins essential for splicing in the yeast Saccharomyces cerevisiae, but splicing appears to occur normally in a mog-1-null mutant. In addition to its involvement in the sperm-oocyte switch and control of fem-3, zygotic mog-1 is required for robust germ line proliferation and for normal growth during development. We suggest that mog-1 plays a broader role in RNA regulation than previously considered.
Posttranscriptional controls of gene expression are critical for many aspects of animal development but are poorly understood at the molecular level (see, e.g., reference 45). In many cases, posttranscriptional controls are exerted through cis-acting elements in the untranslated regions (UTRs) of regulated mRNAs. Such regulatory cis-acting elements have been identified in a variety of organisms ranging from yeasts to vertebrates. Therefore, the mechanisms by which UTR-mediated controls of gene expression are exerted may be conserved throughout evolution.
Posttranscriptional controls are pivotal for sex determination in the Caenorhabditis elegans hermaphrodite germ line. Hermaphrodites make about 300 sperm before switching to oogenesis. The specification of a germ cell as sperm or oocyte is controlled by virtually the same sex determination pathway that regulates sexual fates throughout the organism (for a review, see reference 40). For the purposes of this report, only two sex-determining genes need to be mentioned. The tra-2 gene promotes female development and is repressed at the translational level to permit spermatogenesis in hermaphrodites (11, 17, 21); the fem-3 gene promotes male development and is repressed posttranscriptionally for the switch from spermatogenesis to oogenesis (1, 4, 20). This minicascade of posttranscriptional controls is essential for the transient generation of sperm.
The posttranscriptional repression of fem-3 relies on a cis-acting regulatory element in its 3′ UTR, dubbed the PME (for point mutation element) (1, 4, 15). In fem-3 gain-of-function (gf) mutants, single nucleotide changes in the PME relieve fem-3 repression and abrogate the normal switch from spermatogenesis to oogenesis. Therefore, in fem-3(gf) mutants, no oocytes are produced; instead, sperm are made continuously and to great excess. To identify trans-acting factors that repress fem-3 via its 3′ UTR, both genetic and molecular approaches have been taken. A genetic screen for recessive mutations that cause failure of the sperm-oocyte switch identified six mog (for masculinization of the germ line) genes (18, 19), while a yeast three-hybrid screen for PME-binding proteins yielded FBF (for fem-3 binding factor), a homolog of Drosophila Pumilio that binds specifically to the fem-3 PME regulatory element (46). FBF is encoded by one of two nearly identical genes, fbf-1 and fbf-2, and therefore is not likely to have been identified in the genetic screens for mog mutants.
Among the various mog genes, mog-1 is best characterized genetically (18). Four mog-1 alleles were isolated, and null mutants were identified by genetic tests. The mog-1 gene is required zygotically for the sperm-oocyte switch, a rather specific developmental function, and maternally for embryogenesis (18). Using double-mutant analysis, mog-1 was placed upstream of fem-3 and, therefore, in a reasonable genetic position to function as a fem-3 repressor. However, the genetic position of mog-1 also lies upstream of fem-1, fem-2, fog-1, and fog-3, four other genes required for the specification of germ cells as sperm (12, 18). Therefore, mog-1 might regulate any of these genes to effect the sperm-oocyte switch. To more carefully examine the relationship between the mog genes and fem-3 repression, a reporter transgene bearing the fem-3 3′ UTR was constructed (15). In wild-type animals, this reporter is poorly expressed, whereas it is derepressed in a mog mutant background. Therefore, the mog genes are required for fem-3 3′ UTR repression, and their further analysis is likely to provide insight into the mechanism by which the fem-3 3′ UTR repression is exerted.
In this study, we determined that the mog-1 gene encodes a protein with a DEAH motif typical of RNA helicases and that this protein is most similar to the yeast splicing factors PRP2, PRP16, PRP22, and PRP43 and the human splicing factors hPRP16 and HRH1 (2, 7, 9, 32, 42, 47). However, we found that mog-1 does not appear to be necessary for general pre-mRNA splicing.
MATERIALS AND METHODS
Transformation rescue of mog-1.
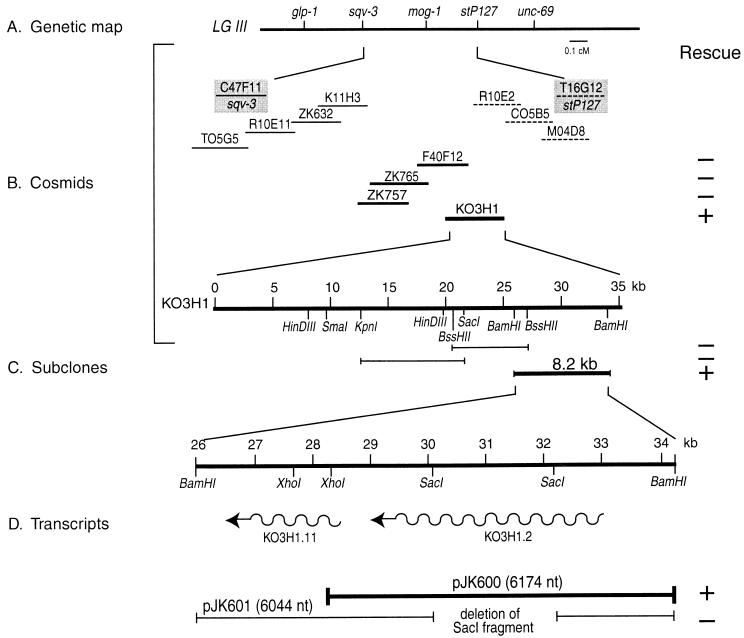
Cosmids within the region spanning sqv-3 and stP127 were microinjected in pools (10 ng of the cosmids to be tested/ml plus 100 ng of pRF4 roller DNA/ml) into young hermaphrodites of genotype mog-1(q223) unc-69(e587)/eT1 (cosmids were kindly provided by A. Coulson). F1 progeny were scored for rollers as well as for fertile Uncs. Since the Unc-69 phenotype is very strong, Unc rollers were difficult to score and had to be backcrossed into wild type (N2) to verify the presence of the roller array. A pool containing cosmids ZK757, ZK765, F40F12, and K03H1 rescued the Mog phenotype, although rescue was poor (approximately 5% of the Unc rollers were fertile). Single-cosmid transformations led to the identification of K03H1 as the rescuing cosmid. Subclones of K03H1 were used to identify an 8.2-kb fragment with two predicted genes; further subcloning and deletion analysis identified pJK600, a 6.17-kb genomic fragment that contains mog-1 rescuing activity. pJK600 is predicted to encode one transcript of 3.6 kb. Rescue by pJK600 was confirmed by coinjection of 70 ng of sonication-fragmented Haemophilus influenzae genomic DNA/ml (13), 30 ng of pRF4/μl, and 8 ng of pJK600/μl. H. influenzae carrier DNA improved mog-1 rescue of fertile Unc-69 rollers from 5 to 80% in F1 and led to the development of stable lines.
Isolation of cDNA clones.
A C. elegans cDNA library (λRB1; provided by R. Barstead) was screened by standard procedures (39) with a 1,014-nucleotide (nt) genomic fragment of mog-1 covering the portion corresponding to amino acids 38 to 341 of the predicted polypeptide (see Fig. 2A). Of 150,000 clones screened, a single positive clone corresponding to a full-length cDNA was isolated. The 3.6-kb cDNA was cloned into the pBluescript KS(+) vector (Stratagene) and sequenced by the chain termination reaction. Not a single difference was found between the sequence of the actual cDNA clone and that predicted by the C. elegans genome project.
FIG. 2.
The MOG-1 protein. (A) Predicted amino acid sequence of MOG-1 and comparison with that of PRP16. Identical amino acids are boxed. We chose to compare MOG-1 to PRP16 and not PRP43 because of their similar sizes (PRP43 is only 767 amino acids long). In the N-terminal part of the protein is the arginine-serine (RS)- and arginine-aspartic acid (RD)-rich region (white letters on black background). The conserved region of each member of the broad family including DEAD- and DEXH-box proteins spans amino acids 474 to 773 and includes five stretches of conserved amino acids (underlined) (33). The first of these stretches, GETGSGKTTQ, contains the A site of the nucleoside triphosphate-binding and ATPase motif; the ATPase B site corresponds to the DEAH motif. In addition to these broadly conserved regions, numerous amino acids (gray shading) are conserved in all 12 DEAH-box proteins examined: human (HRH1 and hPRP16), S. cerevisiae (PRP2, PRP16, PRP22, and PRP43), Saccharomyces pombe (Cdc28), C. elegans (MOG-1, F56D2.6, and EEED8.5), and Arabidopsis thaliana (accession no. X98130 and Z97341). This region of high-level conservation spans amino acids 434 to 1051 in MOG-1. The 12 selected sequences for this comparison correspond to the 12 best scores obtained by the BLASTp program, using MOG-1 as the query. Black dots indicate the positions of mutations in mog-1. The three nonsense mutations, q151, q370, and q223, are at positions 221 (ochre), 330 (opal), and 473 (amber), respectively, and the one missense mutation, q473, is at position 777 (arginine to histidine). Putative nuclear localization signals in MOG-1 are indicated as full lines overlying the sequence. (B) Comparison of MOG-1 to other DEAH-box proteins. The mog-1 amino acid sequence was compared to GenBank-PDB-SwissProt-PIR database sequences by using the BLASTp program. Only DEAH-box proteins had a high level of similarity. We have provided data for the best-understood ones as well as for one DEIH protein, the Drosophila protein Maleless (27). Other proteins with a similar degree of sequence identity but no functional information are not listed; two are from C. elegans (F56D2.6 and EEED8.5) and two are from A. thaliana (accession no. X98130 and Z97341). EEED8.5, F56D2.6 (C. elegans), HRH1 (human), hPRP16 (human), and Z97341 (A. thaliana) each contains an extensive RS domain located in the N terminus (data not shown). Like Maleless (a DEIH-box protein), other DEXX putative helicases are much less similar to MOG-1 than DEAH-box proteins; Xenopus laevis eIF4A (a DEAD-box protein) and Drosophila Homeless (a DEVH-box protein) do not align with MOG-1, indicating a distant relationship. The conserved region consists of amino acids corresponding to residues 434 to 1051 of MOG-1.
RNA extraction, reverse transcription (RT)-PCR, and Northern analysis.
C. elegans was synchronized and grown at 20°C on seeded plates until the desired stage was reached, washed with M9 medium, pelleted, frozen in liquid nitrogen, and stored at −70°C. The different developmental stages were checked by monitoring vulval or germ line development (23). The frozen pellet was quickly homogenized in a solution containing 20 mM Tris-HCl (pH 7.5), 0.5% sodium dodecyl sulfate (SDS), 1 mM EDTA, 100 mM NaCl, and 200 μg/ml of proteinase K (Boehringer Mannheim), using a Polytron homogenizer, and incubated for 1 h at 37°C. The salt concentration was adjusted to 500 mM, and 50 to 100 mg of oligo(dT) cellulose (Pharmacia) was added. After a 2-h incubation at room temperature under conditions of gentle rotation, the resin was washed in a solution consisting of 20 mM Tris-HCl (pH 7.5), 0.5% SDS, 1 mM EDTA, and 500 mM NaCl and the poly(A)+ RNA was eluted in a solution containing 20 mM Tris-HCl (pH 7.5), 0.5% SDS, 1 mM EDTA. Typical yields were 100 μg of poly(A)+-enriched RNA per 1 ml of worm pellet. The RNA was precipitated overnight at −20°C in 0.1 volume of 3 M sodium acetate (pH 5.5) and 2.5 volumes of absolute ethanol.
To extract total RNA, worms were frozen as described above and homogenized, with a Polytron homogenizer, in a mixture containing 4 M guanidine thiocyanate, 25 mM EDTA, 1% sodium N-lauroyl sarcosine, and 50 mM Tris-HCl at pH 7.5. After several extractions with phenol-chloroform, nucleic acids were precipitated as described above. The pellet was resuspended in 10 mM EDTA, and total RNA was precipitated in 2.5 M lithium acetate for 2 h on ice. The RNA pellet was resuspended in 10 mM EDTA and reprecipitated.
Single-stranded cDNA was synthesized by using 2 μg of poly(A)+ or total RNA and 2 μg or 100 ng of oligo(dT) or random primers (Gibco BRL), respectively. The RNA and primers were heated to 70°C and shifted to 37 to 40°C. A mixture containing 50 mM Tris-HCl (pH 7.0), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 2.5 mM deoxynucleoside triphosphates, and Superscript II reverse transcriptase (200 to 400 U; Gibco BRL) was added, and the samples were incubated at 37 to 40°C for 1 h.
PCR was performed on 750 pg of genomic DNA or 2.2 ng of oligo(dT)-primed single-stranded cDNA from wild-type worms for positive controls (see Figure 6B, lanes 2, 3, 6 and 7). One-twentieth of a sample corresponding to, initially, 1.5 μg of total RNA from smg-1; mog-1 adults was used as a template for controls. PCR conditions were 33 cycles of 94°C for 1 min, 57°C for 2 min, 72°C for 2 min. All reaction products were run on a 1.5% agarose gel in Tris-borate-EDTA. The sequences of the primers used are as follows: 5′ ATGGAGGTGGATCCGGGTT 3′ and 5′ TCGTTTCCTGGAGCAATCAG 3′ for fem-3 and 5′ AGGGATTGACAGTTATCCG 3′ and 5′ GCCGTGACTACCGGAAG 3′ for fbf-2.
FIG. 6.
Analysis of splicing in mog-1(0) background. (A) Northern blotting. Ten-microgram quantities of total RNA derived from mog-1 or wild-type worms were run on a denaturing agarose gel. Lanes: 1, smg-1; mog-1(q151) double-mutant adults; 2, dpy-19 mog-1(q223) adults; 3, wild-type adults; 4, wild type at mixed stages (st.). The blot was probed for fbf-1, fbf-2, and act-1. Blots were also overexposed to ensure that no secondary products were present. The expected transcripts for smg-1; mog-1(+) adults could also be seen in a Northern blot (data not shown). (B) RT-PCR. Wild-type (wt) C. elegans genomic DNA (D) (lanes 1 and 5) or cDNA prepared from poly(A)+ RNA (A+) (lanes 2 and 6) were used as positive controls for PCR amplification with oligonucleotides that are specific for either fem-3 (lanes 1 to 4) or fbf-2 (lanes 5 to 8). Total RNA (T) from smg-1; mog-1(q151) hermaphrodites (mog-1) was used as a template for PCR without (−) (lanes 3 and 7) or with (+) (lanes 4 and 8) reverse transcriptase. The 1.17- and 1.83-kb bands correspond to totally spliced fem-3 and fbf-2 mRNAs, respectively.
For Northern analysis, 3 μg of poly(A)+ or 10 μg of total RNA was denatured, run on a 1.2% denaturing agarose-glyoxal gel in 10 mM sodium phosphate (pH 7.0), and transferred to a Hybond-N nylon membrane (Amersham) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Blots were UV cross-linked, baked for 2 h at 80°C, and prehybridized overnight at 42°C in a solution containing 50% formamide, 5× SSC, 0.1% SDS, 5× Denhardt’s solution, and 150 μg of denatured salmon sperm DNA/ml. Hybridization was performed for 16 h at 42°C in a solution consisting of 20 to 35% formamide, 5× SSC, 1% SDS, 2× Denhardt’s solution, and 150 μg of denatured salmon sperm DNA/ml. Blots were washed in 1× to 2× SSC at 42 to 50°C and exposed at −70°C.
GFP reporter experiments.
An EcoRV site was engineered in pJK600 by site-directed mutagenesis of the hexanucleotide GGTCTT located 3 nt upstream of the mog-1 stop codon. A 1,010-nt genomic fragment of green fluorescent protein (GFP) was cloned in frame into the EcoRV site. The actual stop codon of the fusion construct is the GFP stop codon. The 3′ UTR and its flanking sequences were those of mog-1. N2 worms or mog-1 heterozygotes were injected with the gene fusion construct at a concentration of 10 μg/ml along with 30 μg of pRF4/ml and 70 μg of fragmented H. influenzae genomic DNA/ml. Rollers were isolated, and GFP expression was monitored by epifluorescence and confocal microscopy. This MOG-1::GFP fusion protein did not rescue mog-1 (data not shown).
Germ cell counts.
To visualize germ line and somatic nuclei, unmarked adult (24 h after the L4 stage) Mog worms were stained with diamidinophenylindole (DAPI) (0.5 μg/ml in ethanol) for 10 min and washed in M9 buffer. Nuclei from immature germ cells and sperm were counted under a fluorescence microscope. To obtain the total number of germ cells, the numbers of sperm nuclei were divided by four and added to the counts of immature germ cell nuclei. n represents the number of gonadal arms counted.
Growth analyses.
mog-1(x)/balancer hermaphrodites were allowed to lay embryos for 2 h at the appropriate temperature. Embryos were placed individually onto petri dishes and grown at 20 or 25°C for 72 or 48 h, respectively. The developmental stage was determined by monitoring differentiation of the vulva. An animal was scored as an L4 if the vulva was invaginated and as an adult if the vulva had everted. Ten (q370), 24 (q151), 17 (q223), and 11 (q473) mog-1 homozygous animals were grown at 20°C and scored 70 h after laying embryos. For comparison, 110 mog-1/+ (wild-type) heterozygote siblings were analyzed in parallel. The development of 18 fem-3(gf)/+ (q96/+) heterozygotes and 67 wild-type (+/+) siblings was examined 48 h after embryos were laid. Because q96 is a temperature-sensitive allele, q96/+ and +/+ embryos were both grown at 25°C.
Construction and analysis of a smg-1; mog-1 double mutant.
mog-1(q151)/+ males were crossed into smg-1(r861); dpy-19(e1259) unc-69(e587) heterozygotes. Individual cross progeny were placed on petri dishes, and their self-progeny were scored for Smg, Mog, and Dpy Unc phenotypes. Smg non-Mog non-Dpy non-Unc animals were picked to make a stable smg-1(r861); mog-1(q151)/dpy-19(e1259) unc-69(e587) strain. To test whether smg-1 might partially suppress mog-1, we examined smg-1; mog-1 double mutants under Nomarski optics and counted sperm and immature germ cells. No significant difference between the mog-1(q151) and smg-1(r861); mog-1(q151) phenotypes was observed. We also singled out 75 fertile animals and scored their progeny to show that all were mog-1(q151)/dpy-19 unc-69 heterozygotes. Therefore, smg-1 does not appear to either enhance or suppress the Mog-1 phenotype.
Nucleotide sequence accession number.
The mog-1 cDNA sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession no. AF120269.
RESULTS
Cloning the mog-1 gene.
The mog-1 gene maps genetically to linkage group III between sqv-3 and stP127 (Fig. 1A) (18, 44). The physical map of this region has been covered with cosmids by the C. elegans genome consortium (10). To identify mog-1 by mutant rescue, we microinjected pools of cosmids into the germ line of mog-1(q223) unc-69(e587)/eT1 heterozygotes and scored the F1 progeny for rescued fertile mog-1 unc-69 animals. A cosmid pool containing K03H1 rescued mog-1, as did the single K03H1 cosmid (Fig. 1B) (see Materials and Methods). Upon subcloning K03H1, we observed rescue with an 8.2-kb fragment but not with others (Fig. 1C). The rescuing 8.2-kb fragment possesses two predicted open reading frames with the respective transcripts oriented in the same direction from 5′ to 3′ (Fig. 1D). To further narrow the mog-1 region, we tested fragments of the 8.2-kb subclone for mog-1 mutant rescue. pJK600 deleted the entire smaller transcript, whereas pJK601 deleted about 2 kb from the larger transcript (Fig. 1D). pJK600 rescued mog-1, whereas pJK601 failed to rescue mog-1. Therefore, mog-1 is likely to lie within pJK600.
FIG. 1.
Cloning mog-1. (A) Genetic map. mog-1 is located between one cloned gene, sqv-3, and the stP127 Tc1 insertion on chromosome III. (B) Cosmids spanning the sqv-3–stP127 interval were injected in three pools: two pools (single cosmids represented by either dashed or thin lines) failed to rescue mog-1; one pool (single cosmids represented by thick lines) rescued mog-1. Among the single cosmids in the rescuing pool, only K03H1 (thickest line) rescued mog-1 (+); the others failed to rescue mog-1 (−). (C) Subclones of K03H1. Cosmid K03H1 was subcloned, and three fragments were tested for mog-1 rescue. Only the BamHI fragment of 8.2 kb (thick line) supported mog-1 rescue. (D) Transcripts on the rescuing subclone. The 8.2-kb fragment contains two predicted genes (K03H1.2 and K03H1.11 [wavy lines]), which are transcribed in the same direction (arrows) and might be polycistronic. pJK601 deletes an internal 2,110-nt SacI fragment from the K03H1.2 region and fails to rescue mog-1, whereas pJK600 deletes all of K03H1.11 and retains mog-1 rescuing activity.
mog-1 encodes a protein with a DEAH box.
The rescuing genomic fragment, pJK600, bears a single open reading frame (K03H1.2) that is altered in the four existing mog-1 mutant alleles (Fig. 2) (see below). Therefore, this predicted transcript is likely to encode the MOG-1 protein. To examine the actual mog-1 coding region, a 3,599-nt cDNA (pJK602) was isolated from an oligo(dT)-primed cDNA library (a gift from R. Barstead). This mog-1 cDNA confirmed all of the splice junctions predicted by Genefinder and the sequence obtained by the C. elegans sequencing consortium (10). It carries 15 nt of the trans-spliced leader SL1 at its 5′ end, the complete predicted coding region, and a poly(A) tail at the 3′ end. Therefore, pJK602 is likely to represent a nearly full-length transcript. The first AUG lies 3 nt downstream of SL1 and is likely to encode the initiator methionine; it lies in a region with a reasonable translational initiation sequence: in C. elegans that consensus is A/CA/GAAAUG (6), and the mog-1 sequence is GUAAAUG. The stretch of amino acids following this methionine bears similarity to the N-terminal region of another member of the DEAH-box family (Fig. 2A) (see below). The 3′ end of the mog-1 cDNA carries a 130-nt 3′ UTR and an acceptable cleavage and polyadenylation site (CAUAAA [6]), located 13 nt upstream of the poly(A) tail.
The MOG-1 protein is a member of a superfamily of RNA helicases that includes DEAD- and DEAH-box proteins. The defining elements of the DEAH-box protein family are found in a region spanning residues 464 to 770 of MOG-1 (Fig. 2A) (33). Within the DEAH-box family, MOG-1 is most similar to DEAH-box proteins; in the region spanning amino acids 434 to 1051 of MOG-1, many residues are conserved among the 12 DEAH-box family members with the highest BLAST scores to MOG-1 (Fig. 2A). MOG-1 is most similar to four Saccharomyces cerevisiae DEAH proteins, PRP2, PRP16, PRP22, and PRP43, that are required for pre-mRNA splicing (2, 7, 9, 41, 42) and to HRH1 (32) and hPRP16 (31, 47), predicted human homologs of PRP22 and PRP16, respectively (Fig. 2B). The identities among these proteins vary from 30.9 to 53.6% for the whole peptide and from 40 to 74.7% for the conserved region only (amino acids 434 to 1051 in MOG-1), with the strongest homolog being hPRP16. In addition, MOG-1 shares an RS domain with HRH1, hPRP16, and DEAH proteins from other multicellular organisms (Fig. 2A). Such arginine-serine (RS) or arginine-aspartic acid (RD) dipeptides are typical of many pre-mRNA processing factors, including HRH1 and hPRP16 (for a review, see reference 14), but they are not present in yeast PRP2, PRP16, PRP22, and PRP43 proteins. Finally, we note that a search of the virtually complete C. elegans genome does not reveal any other DEAH proteins, suggesting that MOG-1 may be the true PRP16 ortholog.
Molecular basis of mog-1 mutations.
To begin to link the mog-1 molecular characterization with phenotypic effects of various mog-1 mutants, we sequenced the existing four mog-1 alleles. Three of them, q151, q223, and q370, contain nonsense mutations in the first one-third of the predicted cDNA sequence (Fig. 2A). The 5′-most nonsense mutation, mog-1(q151), places a stop codon at residue 221, and all three are predicted to produce truncated fragments of the MOG-1 protein that do not contain the conserved region typical of members of the DEAH family. In C. elegans, mRNAs containing premature stop codons are normally degraded by nonsense-mediated decay (36). Therefore, mog-1 nonsense mutants are predicted not only to lack most of the MOG-1 protein but also to possess only a small fraction of the normal amount of mog-1 mRNA. A truncated nonsense fragment containing partial MOG-1 activity might not be detected phenotypically because of mRNA degradation. To test this possibility, we examined a smg-1; mog-1(q151) double mutant, since in a smg-1 mutant background, mRNAs containing premature stop codons are stabilized (36). We found the mog-1; smg-1 double mutant to be phenotypically indistinguishable from the mog-1 single mutant. Furthermore, since we were able to build a healthy smg-1; mog-1(q151)/dpy-19 unc-69 strain, the truncated fragment does not have a strong dominant-negative effect. We suggest that the three nonsense alleles are molecular nulls. The fourth mog-1 allele (q473) contains a missense mutation predicted to change an arginine to a histidine (amino acid 777) (Fig. 2A). This particular arginine is likely to be of critical importance to MOG-1 function since it is conserved among DEAH-box family members (Fig. 2B).
We next asked whether a phenotypic difference between the null and missense mog-1 mutants could be observed. Previous work focused on the role of mog-1 in the specification of germ line sexual fates (18). Upon further examination of mog-1 germ lines, we found that mog-1(q473) missense mutants have a significantly larger number of mature sperm than do mog-1(q151) null mutants: mog-1(q473) mutants produce 1,073 ± 341 mature sperm per gonadal arm (n = 18), while the mog-1(q151) mutants make only 309 ± 115 mature sperm per gonadal arm (n = 16). The difference between the mog-1 null and missense mutant phenotypes suggests that missense mutants retain partial mog-1 activity. This idea is consistent with the fact that mog-1 null alleles are fully penetrant while mog-1(q473) mutants can occasionally make oocytes (18). During this study, we further noted that mog-1 mutant germ lines are smaller than those of the wild type. Whereas a wild-type or mog-1(q151)/+ adult gonadal arm contains about 1,200 total germ line cells (reference 24 and unpublished data), a mog-1(q151) arm has only about 400 total germ cells (395 ± 179 per gonadal arm; n = 16) and a mog-1(q473) arm contains only about 650 germ cells (638 ± 224 per gonadal arm; n = 6). Therefore, mog mutants produce significantly fewer germ line cells than the wild type. We conclude that the mog-1 gene is required not only for the sperm-oocyte switch but also for robust germ line proliferation.
mog-1 is expressed in germ line and somatic tissues.
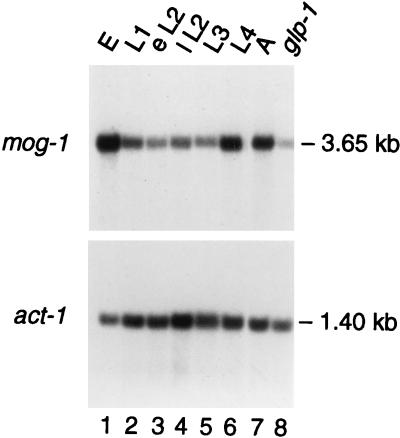
To determine when mog-1 mRNA is expressed during development, we extracted poly(A)+ RNAs from synchronized animals and analyzed them by Northern blotting. C. elegans embryos hatch, develop through four larval stages (L1 to L4), and then molt to adulthood. On a Northern blot probed with the mog-1 cDNA, pJK602, one band of 3.65 kb was observed throughout development (Fig. 3, lanes 1 to 7). The steady-state level of mog-1 mRNA varied during development in a manner predictable by its functions. It peaks in L4 just before the germ line switches from spermatogenesis to oogenesis at the molt from L4 to adult, and it continues in the adult and embryo, as predicted by the maternal-effect lethality of mog-1 homozygotes (18).
FIG. 3.
Northern analysis of mog-1 RNA. Three micrograms of poly(A)+-enriched RNA was loaded per lane. Lanes 1 to 7 contain RNA from staged worms (E, embryos; L1 to L4, four larval stages; A, adults; eL2 and IL2, early and late L2, respectively). Lane 8 was loaded with RNA from adult glp-1(q224) mutants, which contain virtually no germ line. Molecular sizes were determined by comparison to RNA markers (Promega). The same blot was probed with C. elegans act-1 (a gift from M. Krause) as a loading control.
The germ line roles of mog-1 (sperm-oocyte switch, proliferation, and maternal contribution to the embryo) suggest that mog-1 might be expressed specifically in germ line tissue. To investigate this possibility, we compared poly(A)+ RNAs from wild-type adults, which possess over 2,000 germ cells, and glp-1(q224) mutant adults, which contain only 16 to 32 mature sperm (3). Whereas the mog-1 transcript is abundant in wild-type adults (Fig. 3, lane 7), its quantity is dramatically reduced in glp-1 mutants (Fig. 3, lane 8). The ratio of the steady-state level of mog-1 RNA in the wild type to that in glp-1 mutants is 6 to 1 (normalized to actin); the same ratio was found reproducibly in several independent experiments as well as in a comparison of wild-type and glp-4(bn2) RNAs (data not shown). Therefore, most mog-1 RNA appears to be in the germ line, but a fraction of mog-1 RNA is present in somatic tissues.
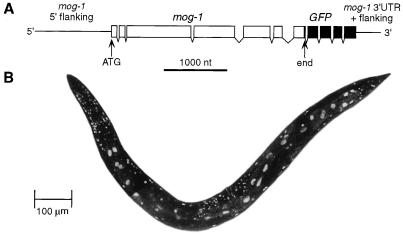
In C. elegans, expression of transgenes is easily obtained in somatic tissues but not in the germ line. The apparent somatic expression of mog-1 prompted the investigation of the expression pattern of a mog-1::GFP fusion construct (Fig. 4A) in transgenic worms. The MOG-1::GFP fusion protein was detected in all somatic tissues, including the intestine, suggesting that the MOG-1 protein is ubiquitous (Fig. 4B). Furthermore, MOG-1::GFP was found to be localized to the nucleus (Fig. 4B), a site within the cell that was predicted by at least three putative nuclear localization signals in the MOG-1 amino acid sequence (Fig. 2A). The lack of MOG-1::GFP fusion protein in germ line tissues was most likely due to its poor expression in the C. elegans germ line.
FIG. 4.
Expression of mog-1::GFP fusion. (A) The MOG-1::GFP fusion construct encodes the GFP protein fused to the C terminus of MOG-1. The 5′ and 3′ flanking regions are those of pJK600, the smallest genomic fragment that was able to rescue mog-1. (B) MOG-1::GFP is found in the nuclei of all somatic tissues. Transgenic worms were examined by epifluorescence and confocal microscopy. Intestinal nuclei can be distinguished by their elongated shape and large size.
mog-1 mutants develop more slowly than the wild type.
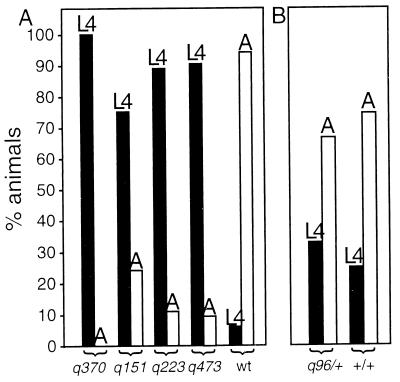
Given the ubiquitous expression of the MOG-1 protein, we wondered about additional phenotypic consequences that might not have been previously detected. Graham and Kimble (18) reported that no sexual transformation occurred in somatic tissues, and they did not find any other specific somatic defect, but they noticed the slower growth of mog-1 unc-69 homozygotes. To verify and extend this initial observation, we compared the growth of unmarked mog-1 mutants to their phenotypically wild-type siblings. To this end, animals were synchronized at hatching and allowed to grow at either 20°C [mog-1(q151, q223, q370, q473) and mog-1(x)/+] or 25°C [fem-3(q96)/+ and wild type]. After 72 h at 20°C or 48 h at 25°C, the developmental stage was assessed by monitoring vulvar differentiation. fem-3(gf) mutants were examined as a control because, like mog-1 mutants, they fail in the sperm-oocyte switch (4). Whereas most mog-1/+ animals had reached adulthood after 72 h at 20°C, most mog-1 homozygotes had only reached the L4 stage after the same period of time (Fig. 5A). By contrast, fem-3(gf)/+ animals were similar to the wild type (Fig. 5B). Since mog-1 mutants develop slowly, we asked whether their life span is different from that of mog-1/+ heterozygotes. At 20°C, heterozygotes survived 13.4 days (standard deviation, 6.7 days; n = 19) and mog-1(0) homozygotes survived 13.2 (standard deviation, 5.7 days; n = 8). Therefore, in spite of their slow growth, mog-1 homozygotes do not live longer than mog-1/+ heterozygotes.
FIG. 5.
mog-1 mutants grow more slowly than the wild type. Black bars represent L4 larvae, and white bars indicate adults. L4 larvae were distinguished from adults (A) by standard features of vulval development (43). The y axis represents the percentage of animals that have reached adulthood or the fourth larval stage. For the numbers of animals tested, refer to Materials and Methods. (A) Four different mog-1 mutants and the wild type (wt) were examined for the developmental stage reached after 72 h at 20°C. Whereas most wild-type animals had reached adulthood, most mog-1 mutants had only reached the fourth larval stage. (B) As a control, fem-3(gf) mutants (which have a Mog phenotype) and fertile wild-type animals were compared after 48 h at 25°C. No significant difference was observed, suggesting that the slow-growth phenotype is associated with mog-1 rather than with the production of sperm instead of oocytes.
General splicing appears to be normal in a mog-1-null mutant.
Since the MOG-1 protein is similar to general splicing factors in S. cerevisiae (PRP2, PRP16, PRP22, and PRP43), we asked whether mog-1 is required for pre-mRNA splicing in C. elegans. To test this hypothesis, total RNA was extracted from 5,000 to 10,000 hand-picked mog-1 homozygotes and analyzed by Northern blotting. To ensure that all RNAs would be represented, we deliberately did not select for poly(A)+ RNA but instead used total RNA. Furthermore, to maximize the chances that incompletely or incorrectly spliced products would be observed, we isolated RNA from a smg-1; mog-1(q151) double mutant, in which mRNAs with premature nonsense codons are stabilized (36).
The RNAs examined included act-1, fem-3, lag-1, and fbf. act-1 encodes body wall actin (26), fem-3 is a sex-determining gene (see the introduction) (38); lag-1 encodes a ubiquitous transcription factor (8, 25), and fbf encodes a trans-acting repressor of fem-3 (46). For actin, lag-1, and fbf, we saw in both wild-type and mog-1 mutants the expected bands corresponding to completely spliced mRNAs, with no additional products in either mog-1 or smg-1; mog-1 mutant extracts (Fig. 6A and data not shown). The light bands above and below the fbf transcript correspond to RNA trapped by 28S and 18S ribosomal RNAs. Although minor differences in transcript size might have escaped Northern analysis, totally unspliced fbf, actin, and lag-1 mRNAs would migrate to positions corresponding to 2.8 kb (with 7 introns), 1.5 kb (with 2 introns), and 15.2 kb (with 11 introns), respectively, and would be distinguishable from the spliced mRNAs in our analysis.
For fem-3, we reproducibly were unable to detect any signal by Northern blot analysis, probably because the fem-3 transcript is relatively rare and is primarily expressed in the germ line, which is small in mog-1 (0) mutants. Therefore, we looked for fem-3 mRNA by RT-PCR. The primers used for PCR span the entire coding region of fem-3 and cover five splice junctions. PCR generated a product of the expected size (1.17 kb) from single-stranded cDNA obtained by using RNA prepared from either wild-type or smg-1; mog-1 worms (Fig. 6B, lanes 2 and 4). The introns removed from this region range from 80 to 380 nt and should have been detected if present. In addition to the 1.17-kb product, a 2.15-kb product was also observed; this larger band corresponded to genomic fem-3 DNA, since it was also obtained without the use of reverse transcriptase or if genomic DNA was used as a template (lanes 1 and 3) but was absent if the cDNA was generated from poly(A)+ RNA (lane 2). The 1.17-kb product was absent if no reverse transcriptase was used, indicating that this band corresponds to reverse-transcribed RNA.
To substantiate the results of the Northern blot analysis (Fig. 6A), RT-PCR was also used for fbf-2 (Fig. 6B, lanes 5 to 8). The expected sizes of the genomic and cDNA products were 2.1 and 1.83 kb, respectively. The 1.83-kb product was detected if RT-PCR was performed on RNA from either wild-type or smg-1; mog-1 worms (lanes 6 and 8). The additional 2.1-kb product corresponded to genomic fbf-2 DNA (lanes 5, 7, and 8) and was not present if the cDNA was made from poly(A)-enriched RNA (lane 6).
Our results suggest that general splicing can occur normally, although we cannot exclude small defects (e.g., skipping of a small exon or retention of a small intron) that occur at low frequencies. Given that the splicing of at least four different mRNAs appears to be normal in a mog-1-null mutant, we suggest that mog-1 is not essential for general splicing. Possible caveats to this conclusion are discussed below.
DISCUSSION
We report the cloning and initial molecular characterization of mog-1, the first of six mog genes involved in the sperm-oocyte switch in the C. elegans hermaphrodite. The mog-1 gene encodes a member of the DEAH-box family of RNA helicases, which includes the yeast splicing factors PRP2, PRP16, PRP22, and PRP43 and their human homologs hPRP16 and HRH1. Three mog-1 alleles bear nonsense codons and are likely to be null mutations; one is a missense mutation and appears to have residual mog-1 activity. The mog-1 gene is expressed not only in the germ line but also in somatic tissues. Consistent with its presence in the soma, we found a slow-development defect in mog-1 mutants; we also report a reduced germ line proliferation in mog-1 mutants. These findings suggest that MOG-1 may be a ubiquitous protein with more general functions than the sperm-oocyte switch. Given its similarity to well-characterized splicing factors, we tested mog-1 for a general role in splicing but were unable to obtain data supporting that idea. We discuss the implications of these results for both mog-1 biology and biochemistry below.
Biological functions of the mog-1 gene.
Our understanding of the biological functions of mog-1 derives from genetic, phenotypic, and molecular analyses (reference 18 and this work). Early studies showed that mog-1 mutants fail in the germ line switch from spermatogenesis to oogenesis and that mog-1; fem-3 mothers, which make oocytes instead of sperm due to the lack of fem-3, produce dead embryos (18). In addition to the phenotypes identified by Graham and Kimble (18), we found two additional defects. First, mog-1 is required for robust germ line proliferation. The adult germ line of mog-1 mutants contains only about half of the number of germ cells present in the wild type; this smaller germ line is organized like the wild type, with mitotic stem cells at one end and differentiating gametes at the other. The germ line defect therefore does not mimic known glp phenotypes: glp-1 mutants lack mitotic stem cells (3), glp-2 mutants have very few germ cells, which fail to differentiate (35), glp-3 mutants fail to progress through meiosis (22), and glp-4 mutants are blocked in mitotic prophase (5). Instead, the mog-1(0) mutation’s effect on germ line proliferation is more similar to that observed after laser ablation of the sheath-spermathecal precursor cells (30). A speculative idea arising from that similarity is that mog-1 may regulate some RNAs in the sheath-spermathecal precursor cell to promote germ line proliferation, in addition to its role in fem-3 regulation. Alternatively, mog-1 may be required for some more general function within the germ line which results in slower cell division. Second, mog-1 is required for growth; mog-1 null mutants grow more slowly than the wild type. However, many mutations retard larval growth, and the significance of this latter phenotype remains unknown. In sum, maternal mog-1 is required for embryogenesis, and zygotic mog-1 is required for the sperm-oocyte switch, normal germ line proliferation, and normal larval growth (reference 18 and this work). This spectrum of phenotypes suggests that mog-1 is broadly required during development.
Possible molecular functions of MOG-1 protein.
What is the molecular function of the MOG-1 protein? Although we cannot yet answer this question in detail, the present work and a complementary study (15) provide important insights. Here we show that MOG-1 is a member of the DEAH family of RNA helicases, that it is expressed in both somatic and germ line tissues, and that it may be nuclear. Gallegos et al. (15) used a reporter transgene that is regulated by the fem-3 3′ UTR to demonstrate that mog-1 activity is required in somatic tissues for the posttranscriptional repression of fem-3. Taking the data together, we can conclude that MOG-1 is involved in RNA regulation and that it is crucial for control mediated via the fem-3 3′ UTR.
The identification of MOG-1 as a DEAH-box protein links it to RNA regulation. Among the yeast DEAH-box proteins, PRP2, PRP16, PRP22, and PRP43 are most similar to MOG-1, with PRP16 showing the highest degree of similarity (Fig. 2). However, none of the yeast proteins contains an RS domain. The presence of an RS domain in MOG-1, HRH1, hPRP16, and at least two other DEAH-box proteins, from C. elegans and Arabidopsis thaliana (see the legend to Fig. 2B), suggests that these proteins might have additional roles in metazoans. DEAD-box and other DEXH-box proteins, such as Drosophila Homeless and Maleless, are only distantly related to MOG-1 (16, 27). PRP2, PRP16, PRP22, and PRP43 have all been implicated in pre-mRNA splicing (2, 7, 9, 41, 42); all four are essential genes in S. cerevisiae, since null mutations in any one are lethal. By contrast with its yeast homologs, mog-1 null mutants are viable and do not appear to be required generally for splicing.
The significance of MOG-1’s similarity to yeast and human splicing factors remains to be determined. Many possibilities exist. If mog-1 is indeed used for general splicing, perhaps some other gene supplies that function or maternal mog-1 is sufficient in mog-1 mutants. Alternatively, the function of mog-1 may have become specialized for splicing selected genes, or this gene may have diverged sufficiently from its closest homologs and not be critical for splicing at all. Finally, the requirement of mog-1 for fem-3 3′ UTR regulation (15) may indicate that the four PRP genes are involved in other aspects of RNA regulation in addition to their roles in splicing.
MOG-1, fem-3 3′ UTR regulation, and the sperm-oocyte switch.
How does mog-1 effect repression by the fem-3 3′ UTR and control the sperm-oocyte switch? Two models are presented in Fig. 7. In both models, fem-3 mRNA bears the cis-acting PME regulatory element which is bound by the FBF repressor. The first model (Fig. 7A) shows an indirect role for MOG-1. MOG-1 may regulate the production of some component of the posttranscriptional regulatory machinery that in turn controls fem-3, here represented by an X (Fig. 7A). A simple example is that MOG-1 might control the splicing of a component needed for fem-3 3′ UTR regulation. With FBF being localized to the cytoplasm, as evidenced by antibody staining (46), a nuclear location for MOG-1 might indicate such an indirect mechanism. However, we have no functional evidence for MOG-1 activity in the nucleus or for FBF activity in the cytoplasm, and therefore this apparent paradox cannot be given too much weight. The second model (Fig. 7B) shows a more direct role for the MOG-1 protein in fem-3 3′ UTR regulation. MOG-1 may either bind FBF directly or be part of a complex that regulates 3′ UTR regulation via the PME. At the present time, we consider both models viable. The identification of MOG-1 in a complex at the fem-3 3′ UTR or the identification of mRNAs that are abnormally spliced in mog-1 mutants could distinguish the two models.
FIG. 7.
Models for the role of mog-1 in fem-3 3′ UTR control. These working models take into account the findings that MOG-1 appears be a nuclear protein and that the splicing of the fem-3 and fbf mRNAs appears to be unaffected by the loss of mog-1. (A) This model suggests that MOG-1 regulates the generation of a specific transcript (mRNA X) that encodes a factor (X) which is required for the fem-3 3′ UTR control. In this diagram, we suggest that MOG-1 may regulate the splicing of a specific pre-mRNA, but it may instead affect translation, stability, or localization of that RNA. (B) This model suggests that MOG-1 may interact directly or indirectly with a PME-binding complex to influence the fem-3 3′ UTR control. Two-headed arrows represent protein-protein or protein-RNA interactions. The PME is shown as a black full circle.
Another possibility is that MOG-1 influences RNA translation, stability, transport, and/or localization. These mechanisms have been implicated for other proteins with DEAD or DEXH motifs. For instance, the Escherichia coli degradosome is a multienzymatic complex containing at least one DEAD-box protein, named RhIB (37). RNA transport from the nucleus to the cytoplasm is dependent on Mtr4p, a nuclear protein with a DEAD-box motif (29). Finally, in D. melanogaster, Vasa, a DEAD-box protein (28), is required for proper localization of nanos mRNA during oogenesis. Similarly, the Drosophila homeless gene is required for transport of specific messages during early development and encodes a protein with a DEVH motif (16). At present we cannot rule out any of these possibilities for mog-1 function.
How does the apparently ubiquitous MOG-1 protein control the sperm/oocyte switch in the C. elegans hermaphrodite germ line? We favor the idea that MOG-1 is part of a regulatory machinery that is present in both somatic and germ line tissues and that influences multiple transcripts, including fem-3. Examples of general regulators that also regulate specific genes are emerging. For example, the SWI-SNF complex appears to be a general transcriptional regulator that plays a role in the induction of specific genes (see, e.g., reference 34). Perhaps MOG-1 and a MOG complex function as a general regulator at the posttranscriptional level and fem-3 is targeted for MOG control by FBF.
ACKNOWLEDGMENTS
We gratefully acknowledge Beth Westlund and Tim Schedl for sharing unpublished mapping data on mog-1. We thank Maria Gallegos for sharing unpublished data and comments on the manuscript. Thanks go to Marv Wickens for fruitful discussions, Niki Gray for helpful comments on the manuscript, and Timothy James for sequencing the mog-1 cDNA. We are grateful to Alan Coulson and his team for providing C. elegans cosmids and for sequencing of the worm genome, as well as to Bob Barstead for cDNA libraries.
A.P. was supported by fellowships from the European Molecular Biology Organization and the Fonds national suisse de la recherche scientifique. J.K. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ahringer J, Kimble J. Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3′ untranslated region. Nature. 1991;349:346–348. doi: 10.1038/349346a0. [DOI] [PubMed] [Google Scholar]
- 2.Arenas J E, Abelson J N. An RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci USA. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- 4.Barton M K, Schedl T B, Kimble J. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics. 1987;115:107–119. doi: 10.1093/genetics/115.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beanan M J, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116:755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- 6.Blumenthal T, Steward K. RNA processing and gene structure. In: Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. C. elegans II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 117–145. [PubMed] [Google Scholar]
- 7.Chen J-H, Lin R-J. The yeast PRP2 protein, a putative RNA-dependent ATPase, shares extensive sequence homology with two other pre-mRNA splicing factors. Nucleic Acids Res. 1990;18:6447. doi: 10.1093/nar/18.21.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen S, Kodoyianni V, Bosenberg M, Friedman L, Kimble J. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H) Development. 1996;122:1373–1383. doi: 10.1242/dev.122.5.1373. [DOI] [PubMed] [Google Scholar]
- 9.Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 10.Coulson A, Huynh C, Kozono Y, Shownkeen R. The physical map of the Caenorhabditis elegans genome. Methods Cell Biol. 1995;48:534–550. doi: 10.1016/s0091-679x(08)61402-8. [DOI] [PubMed] [Google Scholar]
- 11.Doniach T. Activity of the sex-determining gene tra-2 is modulated to allow spermatogenesis in the C. elegans hermaphrodite. Genetics. 1986;114:53–76. doi: 10.1093/genetics/114.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis R E, Kimble J. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics. 1995;139:561–577. doi: 10.1093/genetics/139.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fire, A. Personal communication.
- 14.Fu X-d. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 15.Gallegos M, Ahringer J, Crittenden S, Kimble J. Repression by the 3′ UTR of fem-3, a sex-determining gene, relies on a ubiquitous mog-dependent control in Caenorhabditis elegans. EMBO J. 1998;17:6337–6347. doi: 10.1093/emboj/17.21.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillespie D E, Berg C A. homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev. 1995;9:2495–2508. doi: 10.1101/gad.9.20.2495. [DOI] [PubMed] [Google Scholar]
- 17.Goodwin E B, Okkema P G, Evans T C, Kimble J. Translational regulation of tra-2 by its 3′ untranslated region controls sexual identity in C. elegans. Cell. 1993;75:329–339. doi: 10.1016/0092-8674(93)80074-o. [DOI] [PubMed] [Google Scholar]
- 18.Graham P L, Kimble J. The mog-1 gene is required for the switch from spermatogenesis to oogenesis in Caenorhabditis elegans. Genetics. 1993;133:919–931. doi: 10.1093/genetics/133.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham P L, Schedl T, Kimble J. More mog genes that influence the switch from spermatogenesis to oogenesis in the hermaphrodite germ line of Caenorhabditis elegans. Dev Genet. 1993;14:471–484. doi: 10.1002/dvg.1020140608. [DOI] [PubMed] [Google Scholar]
- 20.Hodgkin J. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics. 1986;114:15–52. doi: 10.1093/genetics/114.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodgkin J, Brenner S. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics. 1977;86:275–287. [PMC free article] [PubMed] [Google Scholar]
- 22.Kadyk L C, Lambie E J, Kimble J. glp-3 is required for mitosis and meiosis in the Caenorhabditis elegans germ line. Genetics. 1997;145:111–121. doi: 10.1093/genetics/145.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- 24.Kimble J E, White J G. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 1981;81:208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- 25.Kodoyianni, V., S. Crittenden, and J. Kimble. Unpublished data.
- 26.Krause M, Wild M, Rosenzweig B, Hirsh D. Wild-type and mutant actin genes in Caenorhabditis elegans. J Mol Biol. 1989;208:381–392. doi: 10.1016/0022-2836(89)90503-2. [DOI] [PubMed] [Google Scholar]
- 27.Kuroda M I, Kernan M J, Kreber R, Ganetzky B, Baker B S. The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell. 1991;66:935–947. doi: 10.1016/0092-8674(91)90439-6. [DOI] [PubMed] [Google Scholar]
- 28.Lasko P F, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- 29.Liang S, Hitomi M, Hu Y-H, Liu Y, Tartakoff A M. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol Cell Biol. 1996;16:5139–5146. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCarter J, Bartlett B, Dang T, Schedl T. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol. 1997;181:121–143. doi: 10.1006/dbio.1996.8429. [DOI] [PubMed] [Google Scholar]
- 31.Nagase T, Seki N, Ishikawa K, Ohira M, Kawarabayasi Y, Ohara O, Tanaka A, Kotani H, Miyajima N, Nomura N. Prediction of the coding sequences of unidentified human genes. VI. The coding sequences of 80 new genes (KIAA0201–KIAA0280) deduced by analysis of cDNA clones from cell line KG-1 and brain. DNA Res. 1996;3:321–329. doi: 10.1093/dnares/3.5.321. [DOI] [PubMed] [Google Scholar]
- 32.Ohno M, Shimura Y. A human RNA helicase-like protein, HRH1, facilitates nuclear export of spliced mRNA by releasing the RNA from the spliceosome. Genes Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- 33.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pazin M J, Kadonaga J T. SWI1/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 35.Petcherski, A., E. Lambie, and J. Kimble. Unpublished data.
- 36.Pulak R, Anderson R P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 37.Py B, Higgins C F, Krisch H M, Carpousis A J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 38.Rosenquist T A, Kimble J. Molecular cloning and transcript analysis of fem-3, a sex-determination gene in Caenorhabditis elegans. Genes Dev. 1988;2:606–616. doi: 10.1101/gad.2.5.606. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Schedl T. Developmental genetics of the germ line. In: Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. C. elegans II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 241–269. [PubMed] [Google Scholar]
- 41.Schwer B, Gross C H. prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwer B, Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991;349:494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- 43.Sulston J E, Horvitz H R. Post-embryonic cell lineages of the nematode Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 44.Westlund, E., and T. Schedl. Personal communication.
- 45.Wickens M, Kimble J, Strickland S. Translational control of developmental decisions. In: Hershey J, Mathews M, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 411–450. [Google Scholar]
- 46.Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens M P. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Z, Reed R. Human homologs of yeast Prp16 and Prp17 reveal conservation of the mechanism for catalytic step II of pre-mRNA splicing. EMBO J. 1998;17:2095–2106. doi: 10.1093/emboj/17.7.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]