Abstract
Breast cancer (BC), a heterogeneous, aggressive illness with high mortality, is essentially a genomic disease. While the high-penetrance genes BRCA1 and BRCA2 play important roles in tumorigenesis, moderate- and low-penetrance genes are also involved. Single-nucleotide polymorphisms (SNPs) in microRNA (miRNA) genes have recently been identified as BC risk factors. miRNA genes are currently classified as low-penetrance. SNPs are the most common variations in the human genome. While the role of miRNA SNPs in BC susceptibility has been studied extensively, results have been inconsistent. This review analyzes the results of association studies between miRNA SNPs and BC risk from countries around the world. We conclude that: (a) By continent, the largest proportion of studies to date were conducted in Asia (65.0 %) and the smallest proportion in Africa (1.8 %); (b) Association studies have been completed for 67 different SNPs; (c) 146a, 196a2, 499, 27a, and 423 are the most-studied miRNAs; (d) The SNPs rs2910164 (miRNA-146a), rs11614913 (miRNA-196a2), rs3746444 (miRNA-499) and rs6505162 (miRNA-423) were the most widely associated with increased BC risk; (e) The majority of studies had small samples, which may affect the precision and power of the results; and (f) The effect of an SNP on BC risk depends on the ethnicity of the population. This review also discusses potential explanations for controversial findings.
Keywords: Polymorphisms, miRNAs, Breast cancer risk, Association studies
Introduction
Breast cancer (BC) has a high mortality rate and is the most common type of cancer among women worldwide. The disease is characterized by expression of aberrant genes that confer tumors with heterogeneous morphology and aggressiveness, producing diverse clinical manifestations [1, 2]. BC susceptibility genes and variants are currently classified into three categories that reflect the probability of developing the disease (high, moderate, or low penetrance) [3]. The most common and well-known high-penetrance susceptibility genes, BRCA1 and BRCA2, account for only about 16 % of cases. There is consensus that moderate- and low-penetrance genes are likely responsible for a significant percentage of familial BC in BRCA1/2-negative families [4]. Recent findings suggest that microRNAs (miRNAs) are low-penetrance genes [5]. miRNAs are small, non-coding, single-stranded RNAs approximately 18–25 nucleotides in length [6]. These molecules have drawn the attention of researchers given their numerous roles in cellular, physiological, and pathological processes. miRNAs regulate gene expression by degrading or blocking translation of targets [7, 8] and are specific to different mRNAs. Approximately 30 % of all human genes are regulated by miRNAs [9, 10]. Current data supports the assertion that these RNAs play important and diverse roles in many molecular pathways and biological processes, including development, apoptosis, differentiation, and cell proliferation [11, 12]. Furthermore, miRNAs have been implicated in various human diseases, including cancer. Genome-wide miRNA expression profiling studies have demonstrated that almost all cancer types show specific profiles of up- and downregulated miRNAs [13, 14]. Growing evidence also indicates that miRNAs can function both as oncogenes and tumor suppressors [15, 16]. In 2005, Iorio et al. [17] described an association between miRNAs and BC for the first time, and evidence of their contribution to disease etiology has mounted in the 16 years since their discovery. Several environmental and genetic elements are involved in the various types of BC, and genetic variations in tumor-suppressor and oncogenes are associated with carcinogenesis [18].
Single-nucleotide polymorphisms (SNPs) are the most common form of variation present in the human genome. SNPs in miRNA regions can alter expression of the gene, provoke aberrant maturation, and alter target-binding affinity and specificity [19]. Many epidemiological studies have examined the association between SNPs in miRNAs and cancer [20], concluding that some of these polymorphisms contribute to BC susceptibility in different populations. Research on miRNA genes is critical for understanding the biology of breast tumors, developing new diagnostic strategies, and identifying more effective therapies [21].
SNPs are ethnicity-specific; as a result, findings for a specific population are not always applicable to other groups. Moreover, many countries have several ancestral lineages. Therefore, we conducted an extensive literature review to clarify the wealth of findings on this important topic in the international context. This review discusses the implications of the many association studies between miRNA genetic variations (SNPs) and BC susceptibility published between 2009 and 2020.
The landscape of breast cancer predisposition: past and present
Risk factors for BC include gender, age, hormonal factors, and, most significantly, genetic predisposition (family history). Characteristics of genetic predisposition include dominant autosomal inheritance, high penetrance (that is, a carrier has a 67 % risk of developing BC by 70 years of age and an 80 % risk by 80 years), a genetic frequency of 0.003, and a carrier frequency of 0.006 [22]. The data suggest that 1 in 20 women with BC, and 1 in 200 women in the general population, carry a genetic predisposition, making BC one of the most widely-distributed heritable pathologies. The existence of a gene or genes responsible for a heritable predisposition to breast and ovarian cancer was suggested more than a century ago [23] and has been supported by a large quantity of epidemiological literature over the past 80 years [22, 24–28]. Segregation studies have indicated the existence of one or more genes that determine predisposition for BC.
The discovery of the tumor-suppressor genes BRCA1 (MIM 113,705) [29] and BRCA2 (MIM 600,185) was a major advance in elucidating the genetic etiology of BC [30, 31]. BRCA1/2 are considered high-penetrance BC susceptibility genes [32, 33]. As noted above, the literature indicates that mutations in BRCA1/2 are responsible for an average of 16–20 % of the risk for hereditary BC [3, 34, 35]. However, genome-wide linkage analyses using large samples of BRCA1/2-negative families have failed to map additional high-penetrance susceptibility loci [36]. It is likely, therefore, that moderate- and low-penetrance genes are responsible for a significant percentage of cases in BRCA1/2-negative families [4]. These low-penetrance genes include miRNAs [5].
General features of miRNAs and their relationship with cancer
As mentioned above, miRNAs are small, non-coding, single-stranded RNAs that have drawn the attention of researchers given their roles in many biological processes [6]. miRNAs regulate gene expression mainly by binding to the 3’-UTR of the target mRNA [7, 8]. However, some studies have reported that miRNAs can also bind to the 5’-UTR [20, 37]. It has been proposed that, depending on the base pairing between the miRNA and target, the negative regulatory effect could vary from weak repression of protein translation to complete cleavage of the mRNA [38]. Since their initial discovery in C. elegans by Lee et al. (1993) [7], more than 1200 miRNAs have been identified in humans, although the specific functions of most remain unknown [39]. A better understanding of how miRNAs regulate their targets would likely yield a great deal of insight into the genetic complexity underlying human health and disease [10]. Many miRNAs have already been implicated in various human diseases such as cardiovascular pathologies, psychiatric disorders, neurodegenerative conditions, and cancers [10]. There is increasing evidence for a vital role of aberrant miRNA expression in the complex and multistep process of carcinogenesis, with miRNA genes acting both as tumor suppressors and oncogenes [40]. As cancer is the second-leading cause of death worldwide [41], understanding its pathogenesis is critical; delineating the role of miRNA in this process would be extremely helpful. One of the first direct links between miRNA and cancer was reported by Callin et al. [42], who found decreased miR-15a and miR-16-1 levels in patients with chronic lymphocytic leukemia. In solid tumors, Michael et al. (2003) [43], identified 28 miRNAs that were differentially expressed in colonic adenocarcinoma vs. normal mucosal tissue, reporting that miR-143 and miR-145 levels were significantly lower in tumors than normal tissues.
As noted, nearly all cancer types have specific profiles of up- and downregulated miRNAs [13, 14]. Several studies have described specific miRNA expression signatures in breast carcinomas [17], primary glioblastomas [44], hepatocellular carcinomas [45], papillary thyroid carcinomas [46], and lung cancer [47]. A large profiling analysis of 540 samples from solid tumors in the lung, breast, stomach, prostate, colon, and pancreas demonstrated that 43 miRNAs were deregulated compared to matched normal tissues [48].
miRNAs can likely function as oncogenes when their targets are onco-suppressor molecules and as tumor-suppressor genes when their targets are oncogenes [15, 16]. Furthermore, a miRNA can function as both a tumor-suppressor and an oncogene depending on the cancer type and cellular context [49]. In fact, a duality of function in different types of cancers has been reported for many miRNAs. One example is miR-125b, which plays opposite roles in different cancer types and cell lines. As a tumor suppressor, miR-125b is downregulated in ovarian, thyroid, breast, and oral squamous-cell carcinomas, promoting cell proliferation and cell-cycle progression [50]. On the other hand, miR-125b is an oncogene in prostate cancer, glioblastomas, and neuroblastomas. miR-125b inhibits apoptosis in a p53-dependent manner in neuroblastoma cells and promotes cell proliferation and invasion in prostate cancer cells [51, 52]. After early studies suggested a role for miRNA genes in the pathogenesis of human cancers, platforms were developed to assess global miRNA expression. The goal of these analyses was to assess the potential of miRNAs in tumor classification and as diagnostic, predictive, or prognostic biomarkers [12].
miRNAs and breast cancer
Microarrays containing all known human miRNAs can be used to identify miRNAs that are differentially expressed in normal and tumor samples, and this approach may be used to determine which miRNA molecules are involved in human cancer. In BC, miRNA microarrays have been used to evaluate miRNA expression profiles in 10 normal and 76 neoplastic breast tissues, identifying 29 miRNAs whose expression was significantly deregulated (p < 0.05) and a smaller set of 15 miRNAs that were able to predict whether a sample was tumor or normal breast tissue with 100 % accuracy [17, 53]. Among the differentially-expressed miRNAs, miR-10b, miR-125b, miR-145, miR-21 and miR-155 were the most consistently deregulated in BC. miR-10b, miR-125b and miR-145 were downregulated, while miR-21 and miR-155 were up-regulated, suggesting that they may act as tumor-suppressor or oncogenes, respectively. In addition, it was possible to identify miRNAs whose expression was correlated with specific BC histopathologic features, such as estrogen and progesterone receptor expression (miR-30), lymph node metastasis (let-7f-1, let-7a-3, let-7a-2) or high proliferative index (let-7c, let-7d) in tumor samples. Therefore, several expression profiling studies have demonstrated that there is a large number of deregulated miRNAs in human BC.
Association studies between miRNA SNPs and breast cancer susceptibility
We conducted a literature review of association studies between miRNA genetic variations (SNPs) and BC susceptibility. PubMed, EBSCO, SciELO, and Google Scholar databases were searched for all studies involving SNPs in miRNAs related to BC risk around the world. The search terms included: “SNPs in miRNA and breast cancer susceptibility;” “association of SNPs in miRNAs with breast cancer risk;” “South America;” “North America;” “Latin America;” “Europe;” “Asia;” “Oceania;” and other terms associated with different countries. Manuscripts published between the years 2009 and 2020 were considered. Only papers published in English were reviewed. Non-human studies, in vitro or in vivo studies, and studies focused on topics other than SNPs in miRNAs and BC susceptibility were excluded. Inclusion criteria were: (a) association studies between SNPs in miRNAs and BC susceptibility; (b) country of origin for BC cases was specified; (c) the miRNAs and SNPs studied were identified. After the search was completed, studies were organized in a Google Sheets spreadsheet. Out of a total of 72 studies, 15 studies were removed due to lack of information regarding the inclusion criteria and 57 were included in this review.
Association studies were found in 17 countries (Australia, Brazil, Chile, China, France, Germany, India, Iran, Ireland, Israel, Italy, Pakistan, Saudi Arabia, Spain, Tunisia, USA, and Vietnam) on 5 continents. Of the 57 association studies included in this review, one was conducted in Africa (1.8 %), 37 in Asia (65.0 %), 9 in Europe (15.8 %), 3 in North America (5.3 %), 3 in Oceania (5.3 %), and 4 in South American countries (7.0 %). In total, 16,906 cases and 19,263 controls were included in the 57 studies. Table 1 shows the studies included, indicating the miRNAs and SNPs studied, the case and control sample sizes, and the continent and country where the study was conducted.
Table 1.
International association studies between miRNA SNPs and breast cancer risk, by continent
| Continent | Country/ countries |
Cases | Controls | miRNA | SNP(s) | References |
|---|---|---|---|---|---|---|
| Africa | Tunisia | 83 | 50 | 146a | rs2910164 | Belaiba et al. 2018 [54] |
| Asia | China | 321 | 290 |
499 27a 196a2 146a |
rs3746444 rs895819 rs11614913 rs2910164 |
Qi et al. 2015 [55] |
| 450 | 450 |
499 149 146a 423 196a2 27a |
rs3746444 rs2292832 rs2910164 rs6505162 rs11614913 rs895819 |
He et al. 2015 [56] | ||
| 560 | 583 |
196a2 499 608 |
rs11614913 rs3746444 rs4919510 |
Dai et al. 2016 [57] | ||
| 264 | 255 | 27a | rs895819 | Zhang et al. 2013 [58] | ||
| 1009 | 1093 |
146a 149 196a2 499 |
rs2910164 rs2292832 rs11614913 rs3746444 |
Hu et al. 2009 [59] | ||
| 252 | 248 |
618 605 149 27a 196a2 |
rs2682818 rs2043556 rs2292832 rs895819 rs11614913 |
Zhang et al. 2012 [60] | ||
| 191 | 192 |
146a 373 373 27a 423 492 124-1 603 604 26a-1 605 608 100 105-1 105-2 1206 1274-a 125b-1 943 196a2 30c-1 Let-7f-2 149 |
rs2910164 rs12983273 rs10425222 rs895819 rs6505162 rs2289030 rs531564 rs11014002 rs2368392 rs7372209 rs2043556 rs4919510 rs1834306 rs5970293 rs5970292 rs2114358 rs318039 rs2081443 rs1077020 rs11614913 rs16827546 rs17276588 rs2292832 |
Ma et al. 2013 [61] | ||
| 114 | 189 | 423 | rs6505162 | Zhao et al. 2015 [62] | ||
| 1138 | 1434 | 608 | rs4919510 | Huang et al. 2012 [63] | ||
| 301 | 310 | Let-7 |
rs10877887 rs13293512 |
Sun et al. 2019 [64] | ||
| 1064 | 1073 | 101-2 |
rs462480 rs1053872 |
Chen et al. 2014 [65] | ||
| 1064 | 1073 |
30a 30a 30a 30c-1 30c-1 30c-1 30c-2 30c-2 30d 30d |
rs763354 rs852963 rs852964 rs928508 rs12743517 rs3767950 rs12208417 rs16881192 rs17709260 rs7846345 |
Zhou et al. 2020 [66] | ||
| India | 121 | 164 |
146a 196a2 499 |
rs2910164 rs11614913 rs3746444 |
Bansal et al. 2014 [67] | |
| 100 | 100 |
146a 196a2 |
rs2910164 rs11614913 |
Bodal et al. 2017 [68] | ||
| Iran | 353 | 353 |
27a 196a2 146a |
rs895819 rs11614913 rs2910164 |
Mashayekhi et al. 2018 [69] | |
| 236 | 203 |
146a 499 196a2 |
rs2910164 rs3746444 rs11614913 rs185070757 |
Omrani et al. 2014 [70] | ||
| 100 | 100 |
499 196a2 |
rs3746444 rs11614913 |
Doulah et al. 2018 [71] | ||
| 200 | 200 |
196a2 146a |
rs11614913 rs2910164 |
Najeti-Azar et al. 2018 [72] | ||
| 161 | 162 | 323b | rs56103835 | Naderi et al. 2018 [73] | ||
| 266 | 288 |
100 124-1 218-2 301b 605 4293 |
rs1834306 rs531564 rs11134527 rs384262 rs2043556 rs12220909 |
Danesh et al. 2018 [74] | ||
| 162 | 180 | 605 | rs2043556 | Kazemi et al. 2020 [75] | ||
| 240 | 231 |
146a 27a |
rs2910164 rs895819 |
Parchami Barjui et al. 2017 [76] | ||
| 100 | 150 |
196a2 499 146a |
rs11614913 rs3746444 rs2910164 |
Afsharzadeh et al. 2017 [77] | ||
| 100 | 100 | 196a2 | rs11614913 | Eslami-S et al. 2018 [78] | ||
| 86 | 96 | 499 | rs3746444 | Kabirizadeh et al. 2016 [79] | ||
| 160 | 192 | 608 | rs4919510 | Hashemi et al. 2016 [80] | ||
| 129 | 153 | 599 | rs58450758 | Baherini et al. 2019 [81] | ||
| 129 | 144 | 520f | rs75598818 | Meshkat et al. 2018 [82] | ||
| 82 | 70 | 146a | rs2910164 | Meshkat et al. 2016 [83] | ||
| 263 | 221 | 34 b/c | rs4938723 | Sanaei et al. 2016 [84] | ||
| 50 | 50 | 146a | rs2910164 | Siasi et al. 2020 [85] | ||
| Israel | 198 | 290 | 27a | rs895819 | Kontorovich 2010 [86] | |
| Saudi Arabia | 100 | 100 |
196a2 146a 499 |
rs11614913 rs2910164 rs3746444 |
Alshatwi et al. 2012 [87] | |
| 100 | 124 | 423 | rs6505162 | Mir et al. 2018 [88] | ||
| Vietnam | 106 | 117 | 27a | rs895819 | Nguyen et al. 2016 [89] | |
| 113 | 127 | 196a2 | rs11614913 | Minh et al. 2018 [90] | ||
| Pakistan | 300 | 230 | 146a | rs2910164 | Ahmad et al. 2019 [91] | |
| Europe | France | 1130 | 596 | 146a | rs2910164 | Garcia et al. 2011 [92] |
| Germany | 1217 | 1422 | 27a | rs895819 | Yang et al. 2010 [93] | |
| 1134 | 1517 |
196a2 499 146a |
rs11614913 rs3746444 rs2910164 |
Catucci et al. 2010 [94] | ||
| 1217 | 1422 |
126 335 |
rs463297 rs41272366 |
Yang et al. 2011 [95] | ||
| Ireland | 523 | 724 | 146a | rs2910164 | McVeigh et al. 2017 [96] | |
| Italy | 760 | 1243 |
196a2 499 146a |
rs11614913 rs3746444 rs2910164 |
Catucci et al. 2010 [94] | |
| 1025 | 1593 | 27a | rs895819 | Catucci et al. 2012 [97] | ||
| 81 | 155 | 146a | rs2910164 | Pastrello et al. 2010 [98] | ||
| Spain | 538 | 189 | 146a | rs2910164 | Cardeñosa 2012 [99] | |
| North America | USA | 441 | 479 | 196a2 | rs11614913 | Hoffman et al. 2009 [100] |
| USA (African-American) | 474 | 412 |
106b 100 331 758 544 487 659 513a-2 |
rs1527423 rs1834306 rs11107973 rs12586258 rs10144193 rs1951032 rs5750504 rs2018562 |
Yao et al. 2013 [101] | |
| USA (European-American) | 329 | 310 |
106b 100 331 758 544 487 659 513a-2 |
rs1527423 rs1834306 rs11107973 rs12586258 rs10144193 rs1951032 rs5750504 rs2018562 |
||
| USA (African-American) | 894 | 788 |
185 9 − 1 9 − 2 16 − 1/15a 34b/c 206 |
rs2008591 rs887205 rs2078749 rs12239077 rs1501672 rs9535416 rs4938723 rs6920648 rs16882131 |
Bensen et al. 2013 [102] | |
| USA (Caucasian) | 1417 | 1234 |
185 9 − 1 9 − 2 16 − 1/15a 34b/c 206 |
rs2008591 rs887205 rs2078749 rs12239077 rs1501672 rs9535416 rs4938723 rs6920648 rs16882131 |
||
| Oceania | Australia | 173 | 187 | 145 | rs353291 | Chacon-Cortes et al. 2015 [103] |
| 193 | 193 | 423 | rs6505162 | Smith et al. 2012 [104] | ||
| 193 | 190 | 196a2 | rs11614913 | Jedlinski et al. 2011 [105] | ||
| South America | Chile | 440 | 807 |
196a2 423 27a 618 608 |
rs11614913 rs6505162 rs895819 rs2682818 rs4919510 |
Morales et al. 2016 [106] |
| 440 | 1048 |
146a 499 125a 605 182 |
rs2910164 rs3746444 rs12975333 rs2043556 rs4541843 |
Morales et al. 2018 [107] | ||
| Brazil | 388 | 388 | 196a2 | rs11614913 | Linhares et al. 2012 [108] | |
| 326 | 411 | 146a | rs2910164 | Brincas et al. 2020 [109] |
When the results were analyzed by continent, we found that Asia had the highest proportion of studies (65.0 %) and Africa the lowest (1.8 %). Within Asia, 45.9 % of the studies were conducted in Iran, 35.1 % in China, 5.4 % in Saudi Arabia, 5.4 % in India, 5.4 % in Vietnam, 2.7 % in Pakistan, and 2.7 % in Israel. The continent with the second-highest number of studies was Europe (15.8 %), where studies were carried out in 5 countries: France (11.1 %), Germany (33.3 %), Ireland (11.1 %), Italy (33.3 %), and Spain (11.1 %). Studies in South America accounted for 7.0 % of studies around the world and were performed only in Chile (50 %) and Brazil (50 %). In Oceania, studies have only been carried out in Australia, corresponding to 5.3 % of the total. In North America, the only country with association studies between miRNA SNPs and BC risk is the USA, representing 5.3 % of total studies. Finally, only one study was available for Africa, conducted in Tunisia, accounting for 1.8 % of studies worldwide. Figure 1 shows the scope of association studies between miRNA SNPs and BC risk in countries around the world.
Fig. 1.
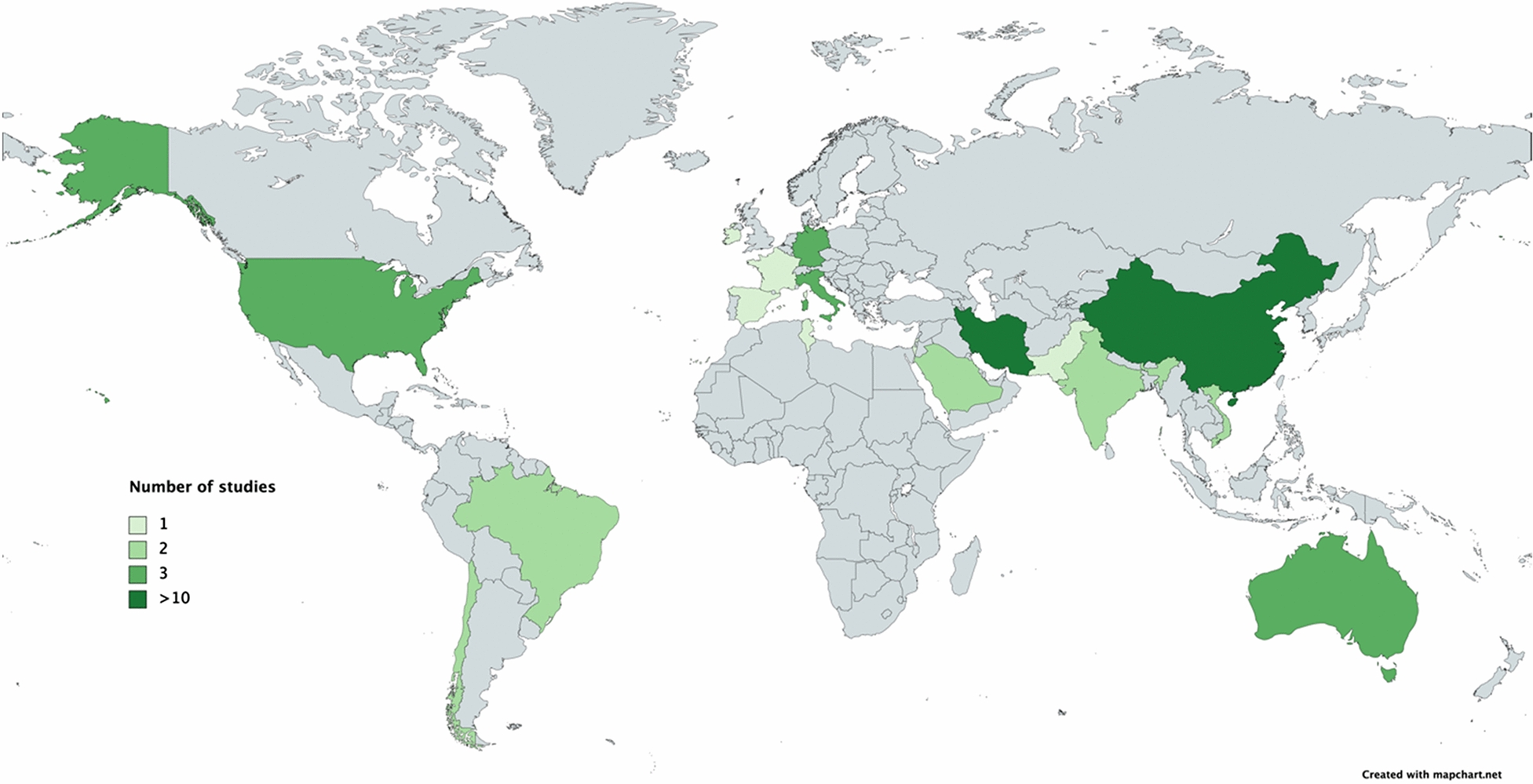
Scope of association studies between miRNA SNPs and breast cancer around the world. Green areas correspond to countries with studies included in this review. The color gradient represents the number of studies in each country
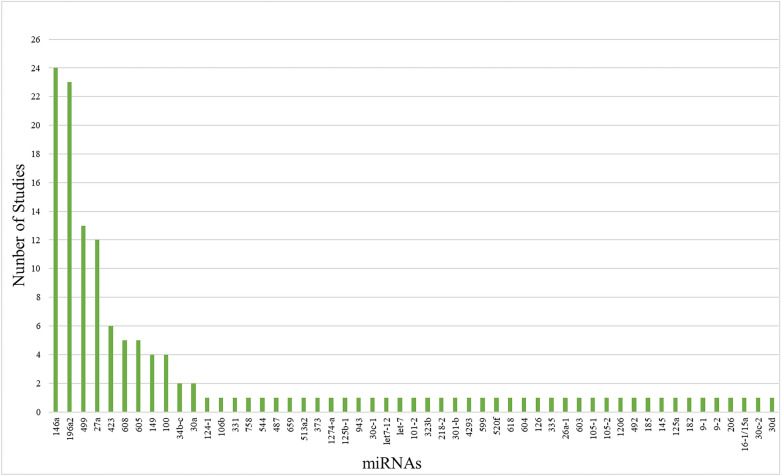
The SNPs studied were located in 53 different miRNAs (Table 1). Figure 2 shows the number of studies for each miRNA evaluated in this review. 146a, 196a2, 499, 27a, and 423 were the most-studied miRNAs, included at least 6 studies (Fig. 2). The most-studied miRNA was miRNA 146a, with reports from 4 to 5 continents (Africa, Asia, Europe, and America). In Africa, this miRNA only has been studied in Tunisia; in Asia, there are studies in China, India, Iran, Saudi Arabia, and Pakistan; in Europe, France, Ireland, Italy, and Spain have studies; and in the Americas, there are only studies from Chile and Brazil. The miRNA-196a2 was studied in 4 of 5 continents, (Asia, Europe, America, and Oceania). In Asia, miRNA-196a2 was studied in China, India, Iran, Saudi Arabia, and Vietnam; in Europe, in Germany and Italy; in North America, only in the USA; in South America, in Chile and Brazil; and in Oceania, only in Australia. The miRNA499 was studied in China, India, Iran, and Saudi Arabia in Asia; in Europe, there are reports from Germany and Italy; and only in Chile in South America. miRNA 27a was studied in 3 of 5 continents (Asia, Europe, and America). In Asia, studies were carried out in China, Iran, and Israel; in Europe, in Germany and Italy; and only in Chile in South America. Finally, miRNA 423 was studied in Asia, Oceania, and America. In Asia, there are reports in China and Saudi Arabia; in Oceania, in Australia; and in South America, only in Chile.
Fig. 2.
Number of studies performed for each miRNA included in this review
Table 2 summarizes the SNPs studied by miRNA, indicating the countries where the studies were conducted.
Table 2.
Summary of SNPs studied by miRNA and country
| miRNA | SNP(s) | Country/countries |
|---|---|---|
| 196a2 | rs11614913 | Australia, Brazil, Chile, China, Germany, India, Iran, Italy, Saudi Arabia, USA, Vietnam |
| rs185070757 | Iran | |
| 146a | rs2910164 | Brazil, Chile, China, France, India, Iran, Ireland, Italy, Pakistan, Saudi Arabia, Spain, Tunisia |
| 499 | rs3746444 | Chile, China, Germany, India, Iran, Italy, Saudi Arabia |
| 27a | rs895819 | Chile, China, Germany, Iran, Israel, Italy, Vietnam |
| 423 | rs6505162 | Australia, Chile, China, Saudi Arabia |
| 608 | rs4919510 | Chile, China, Iran |
| 605 | rs2043556 | Chile, China, Iran |
| 149 | rs2292832 | China |
| 100 | rs1834306 | China, Iran, USA |
| 373 |
rs12983273 rs10425222 |
China |
| 124-1 | rs531564 | China, Iran |
| 618 | rs2682818 | Chile, China |
| 106-b | rs1527423 | USA |
| 331 | rs11107973 | USA |
| 758 | rs12586258 | USA |
| 544 | rs10144193 | USA |
| 487 | rs1951032 | USA |
| 659 | rs5750504 | USA |
| 513a-2 | rs2018562 | USA |
| 1274a | rs318039 | China |
| 125b-1 | rs2081443 | China |
| 943 | rs1077020 | China |
| 30c-1 |
rs16827546 rs928508 rs12743517 rs3767950 |
China |
| Let7-f2 | rs17276588 | China |
| Let7 |
rs10877887 rs13293502 |
China |
| 101-2 |
rs462480 rs105387 |
China |
| 30a |
rs763354 rs852963 rs852964 |
China |
| 30c-2 |
rs12208417 rs16881192 |
China |
| 30d |
rs17709260 rs7846345 |
China |
| 323b | rs56103835 | China |
| 218-2 | rs11134527 | Iran |
| 301-b | rs384262 | Iran |
| 4293 | rs12220909 | Iran |
| 599 | rs58450758 | Iran |
| 520f | rs75598818 | Iran |
| 34b/c | rs4938723 | Iran, USA |
| 604 | rs2368392 | China |
| 126 | rs463297 | Germany |
| 335 | rs41272366 | Germany |
| 26a-1 | rs7372209 | China |
| 603 | rs11014002 | China |
| 105-1 | rs5970293 | China |
| 105-2 | rs5970292 | China |
| 1206 | rs2114358 | China |
| 492 | rs2289030 | China |
| 185 |
rs2008591 rs887205 rs2078749 |
USA |
| 145 | rs353291 | Australia |
| 125a | rs12975333 | Chile |
| 182 | rs4541843 | Chile |
| 9 − 1 | rs12239077 | USA |
| 9 − 2 | rs1501672 | USA |
| 16 − 1/15a | rs9535416 | USA |
| 206 |
rs6920648 rs16882131 |
USA |
Sixty-seven SNPs were studied in the 53 miRNAs. In 85.0 % of the miRNAs, a single SNP was studied, and in 10.4 %, two SNPs were studied. Three different SNPs were studied in the miRNAs 185 and 30a, and 4 in the 30c-1 (Table 2). Forty different SNPs were studied in China, 14 in Iran, 18 in USA, 10 in Chile, 5 in Germany, 4 in Saudi Arabia and Italy, 3 in Australia and India, 2 in Vietnam and Brazil, and only one SNP was studied in France, Ireland, Pakistan, Spain, Tunisia, and Israel.
Table 3 shown the results of the association studies between miRNA SNPs and BC risk according to risk category: increased risk, protective effect, and no association. Of the 53 miRNAs and 67 SNPs included in this review, only 18 miRNAs (33.3 %) and 19 SNPs (28.4 %) (Table 3) were associated with increased risk. The Asian ethnic group had the highest number of SNPs associated with risk (16.4 %). In the USA, 6 SNPs (9 %) were associated with increased BC risk in both African- and European-American women. In South America, 5 different SNPs (7.5 %) in the miRNAs 146a, 196a2, 423, 618, and 182 were associated with increased BC risk, and in Europe (Italy) only one SNP, rs2910164 in miRNA 146a, was associated with risk (1.5 %).
Table 3.
Association categories for miRNA SNPs and breast cancer risk
| BC risk category | miRNA | SNP(s) | Country/countries | Continent(s) |
|---|---|---|---|---|
| Increased risk | 146a | rs2910164:G > C | Brazil, China, Iran, Italy, Pakistan | Asia, Europe, South America |
| 196a2 | rs11614913:C > T | Brazil, China, India, Iran, Saudi Arabia | Asia, South America | |
| 499 | rs3746444:T > C | China, Iran, Saudi Arabia | Asia | |
| 218-2 | rs11134527:A > G | Iran | Asia | |
| 301-b | rs384262G > A | Iran | Asia | |
| 605 | rs2043556T:A > G | Iran | Asia | |
| 599 | rs58450758:C > T | Iran | Asia | |
| 423 | rs6505162:C > A | Chile and Saudi Arabia | Asia, South America | |
| 513a-2 | rs2018562 | USA (African-American) | North America | |
| 106b | rs1527423:A > G | USA (European-American) | North America | |
| 182 | rs4541843:C > T | Chile | South America | |
| 101-2 |
rs462480:A > C rs105387:C > G |
China | Asia | |
| Let-7 | rs13293512:T > C | China | Asia | |
| 331 | rs1110793:A > G | USA (European-American) | North America | |
| 544 | rs10144193:A > T | USA (European-American) | North America | |
| 487 | rs1951032G > A | USA (European-American) | North America | |
| 659 | rs5750504:T > A | USA (European-American) | North America | |
| 618 | rs2682818C > A | Chile (early-onset BC) | South America | |
| Decreased risk | 27a | rs895819:A > G | Chile, China, Germany, Iran, Israel | Asia, Europe, South America |
| 499 | rs3746444:T > C | Iran | Asia | |
| 608 | rs4919510:C > G | Iran | Asia | |
| 520f | rs75598818:G > A | Iran | Asia | |
| 196a2 | rs11614913:C > T | Brazil, China, USA, Vietnam | Asia, North America, South America | |
| 758 | rs12586258:G > A | USA (African-American) | North America | |
| 100 | rs1834306:G > A | USA (European-American) | North America | |
| 185 |
rs2008595:C > T rs887205:A > G |
USA (African-American) | North America | |
| 423 | rs6505162:A > C | Australia | Oceania | |
| 605 | rs2043556:T > C | Chile | South America | |
| 146a | rs2910164:G > C | India | Asia | |
| 149 | rs2292832:T > C | China | Asia | |
| 30a | rs763354:G > A | China | Asia | |
| No association | 196a2 | rs11614913:C > T | Australia, Chile, China, Germany, Iran, India, Italy | Asia, Europe, South America, Oceania |
| 196a2 | rs185070757 | Iran | Asia | |
| 146a | rs2910164:G > C | Chile, China, France, Germany, Iran, Ireland, Italy, Saudi Arabia, Spain, Tunisia | Africa, Asia, Europe, South America | |
| 323b | rs56103835:T > C | Iran | Asia | |
| 100 | rs1834306:T > C | China, Iran, USA (African-American) | Asia, North America | |
| 124-1 | rs531564:G > C | China, Iran | Asia | |
| 605 | rs2043556:T > C | China, Iran | Asia | |
| 4293 | rs12220909:G > C | Iran | Asia | |
| 34b/c | rs4938723: | Iran | Asia | |
| 27a | rs895819:A > C | China, Italy, Vietnam | Asia and Europe | |
| 499 | rs3746444:T > C | Chile, China, Germany, India, Italy | Asia, Europe, South America | |
| 126 | rs463297 | Germany | Europe | |
| 335 | rs41272366 | Germany | Europe | |
| 106b | rs1527423:A > G | USA (African-American) | North America | |
| 331 | rs11107973:A > G | USA (African-American) | North America | |
| 758 | rs12586258:G > A | USA (African-American) | North America | |
| 513a-2 | rs2018563:A > G | USA (African-American) | North America | |
| 185 | rs2078749:A > G | USA (African-American) | North America | |
| 145 | rs353291:T > C | Australia | Oceania | |
| 608 | rs4919510:C > G | Chile, China | Asia, South America | |
| 125a | rs12975333:A > G | Chile | South America | |
| 423 | rs6505162:C > A | China | Asia | |
| 149 | rs2292832:T > C | China | Asia | |
| 9−1 | rs12239077:A > G | USA (African- and European-American) | North America | |
| 9−2 | rs1501672:T > C | USA (African- and European-American) | North America | |
| 16−1/15a | rs9535416:G > A | USA (African- and European-American) | North America | |
| 34b/c | rs4938723:T > C | USA (European-American) | North America | |
| 206 |
rs6920648:A > G rs16882131:C > T |
USA (European-American) | North America | |
| 185 |
rs28591:C > T rs887205:A > G rs2078749:A > G |
USA (European-American) | North America | |
| 618 | rs2682818:C > A | China | Asia | |
| 373 |
rs12983273:C > T rs1042522:C > A |
China | Asia | |
| 492 | rs2289030:C > G | China | Asia | |
| 603 | rs11014002:C > T | China | Asia | |
| 604 | rs2368392:C > T | China | Asia | |
| 26a-1 | rs7372209:C > T | China | Asia | |
| 105-1 | rs5970293:G > C | China | Asia | |
| 105-2 | rs5970292:G > A | China | Asia | |
| 1206 | rs2114358:T > C | China | Asia | |
| 1274a | rs318039:C > T | China | Asia | |
| 125b-1 | rs2081443:T > G | China | Asia | |
| 943 | rs1077020:C > T | China | Asia | |
| 30c-1 |
rs16827546:C > T rs928508:A > G rs12743517:C > A rs3767950:C > A |
China | Asia | |
| Let-7f-2 | rs17276588:G > A | China | Asia | |
| Let-7 | rs10877887:T > C | China | Asia | |
| 30a |
rs852963:G > A rs852964:G > A |
China | Asia | |
| 30c-2 |
rs12208417:C > A rs16881192:A > C |
China | Asia | |
| 30d |
rs17709260:A > G rs7846345:G > C |
China | Asia |
Thirteen miRNAs (24.1 %) and 15 SNPs (22.4 %) were associated with decreased BC risk. In Asia, 8 SNPs (12 %) had a protective effect, and in Europe only one SNP (1.5 %) rs895819:A > G (miRNA-27a) was associated with decreased risk, in a German population. In North America (USA), 4 SNPs (6 %) located in four different miRNAs were associated with decreased BC risk in African- and European-American women. In South America, 3 SNPs (4.5 %) were protective, and in Oceania (Australia), the only SNP associated with decreased BC was rs6505162:A > C in miRNA-423 (1.5 %). Of the total miRNAs included in this review, 28 different miRNAs (52.8 %) and 31 different SNPs (46.3 %) were associated with BC risk.
Genetic variations are ethnicity-specific; therefore, results of association studies between a miRNA SNP and BC risk may diverge depending on ethnicity. The most-studied SNP, rs2910164:G > C in miRNA-146a, was found to increase the risk of developing breast cancer in Brazilian, Chinese, Iranian, Italian, and Pakistani populations (Table 3) but showed no association in Chilean, Chinese, French, German, Iranian, Irish, Italian, Saudi Arabian, Spanish, or Tunisian populations (Table 3). The heterozygous variant showed a protective effect in a North Indian population (Table 3). With respect to the second most-common SNP, rs11614913:C > T in miRNA-196a2 was associated with increased risk in Brazilian, Chinese, Iranian, and Saudi Arabian populations but showed no association in Chilean, Caucasian Australian, Chinese, Iranian, Indian, Italian, or German populations and was protective in Brazil, China, USA, and Vietnam. The studies in China were conducted in different regions of the country, and the Brazilian study by Linhares et al. (2012) [108], showed that for the SNP rs11614913:C > T, the allele T increased risk, while the allele C had a protective effect (Table 4). The SNP rs3746444:A > G in miRNA-499 increased risk in Chinese, Iranian and Saudi Arabian populations, and showed a protective effect in an Iranian population (genotype CC and CT) (Table 4), but showed no association in Chilean, Chinese, German, North Indian, or Italian populations. The rs895819:A > G in miRNA-27a was protective in Chilean, Chinese, German, Iranian, and Israeli populations, but showed no association in Chinese, Italian, or Vietnamese populations. Another relatively common SNP was rs6505162, located in miRNA-423. This SNP showed an association with increased BC risk in Chilean and Saudi Arabian populations but had a protective effect in a Caucasian Australian population.
Table 4.
Allele or genotype associated with breast cancer risk in miRNA SNPs included in this review
| BC risk category | miRNA | SNP(s) | Risk allele or genotype | p-value | Countries | References |
|---|---|---|---|---|---|---|
| Increased risk | 146a | rs2910164:G > C |
C CG and GG C CC C C GC and CC CC |
0.03 < 0.05 0.04 < 0.001 0.03 0.0037 0.033 and 0.028 < 0.0001 |
Brazil China China Iran Iran Iran Italy Pakistan |
Brincas et al. 2020 [109] He et al. 2015 [56] Qi et al. 2015 [55] Mashayekhi et al. 2018 [69] Parchami Barjui et al. 2017 [76] Meshkat et al. 2016 [83] Pastrello et al. 2010 [98] Ahmad et al. 2019 [91] |
| 196a2 | rs11614913:C > T |
T CT C C C CT |
0.024 0.04 0.01 0.011 0.02236 0.01 |
Brazil India China China Iran Saudi Arabia |
Linhares et al. 2012 [108] Bodal et al. 2017 [68] Qi et al. 2015 [55] Hu et al. 2009 [59] Najeti-Azar et al., 2018 [72] Alshtawi et al. 2012 [87] |
|
| 499 | rs3746444:T > C or A > G |
AG and GG G C C G C |
0.008 0.025 0.001 0.034 0.02952 0.001 |
China China Iran Iran Iran Saudi Arabia |
Dai et al. 2016 [57] Hu et al. 2009 [59] Omrani et al. 2014 [70] Afsharzadeh et al., 2017 [77] Kabirizadeh et al., 2016 [79] Alshtawi et al. 2012 [87] |
|
| 218-2 | rs11134527:A > G | G | < 0.0001 | Iran | Danesh et al. 2018 [74] | |
| 301-b | rs384262G > A | A | < 0.0001 | Iran | Danesh et al. 2018 [74] | |
| 605 | rs2043556T:A > G | G | 0.00003 | Iran | Kazemi et al. 2020 [75] | |
| 599 | rs58450758:C > T | CT and TT | < 0.0001 | Iran | Bahreini et al. 2019 [81] | |
| 423 | rs6505162:C > A |
A T |
0.02 0.0001 |
Chile Saudi Arabia |
Morales et al. 2016 [106] MiR et al. 2018 [88] |
|
| 513a-2 | rs2018562:A > G | G | 0.03 | USA | Yao et al. 2013 [101] | |
| 106b | rs1527423:A > G | G | 0.02 | USA | Yao et al. 2013 [101] | |
| 182 | rs4541843:C > T | T | 0.01 | Chile | Morales et al. 2018 [107] | |
| 101-2 |
rs462480:A > C rs105387:C > G |
C G |
0.017 0.010 |
China | Chen et al. 2014 [65] | |
| Let-7 | rs13293512:T > C | C | 0.013 | China | Sun et al., 2019 [64] | |
| 331 | rs1110793:A > G | G | 0.02 | USA | Yao et al. 2013 [101] | |
| 544 | rs10144193:A > T | T | 0.004 | USA | Yao et al. 2013 [101] | |
| 487 | rs1951032G > A | A | 0.001 | USA | Yao et al. 2013 [101] | |
| 659 | rs5750504:T > A | A | 0.03 | USA | Yao et al. 2013 [101] | |
| 618 | rs2682818C > A | CA | 0.03 | Chile | Morales et al. 2016 [106] | |
| Decreased risk | 27a | rs895819:A > G or T > C |
GG G G G G T |
0.01 0.032 0.0287 < 0.001 0.001 0.013 |
Chile China Germany Iran Iran Israel |
Morales et al. 2016 [106] Zhang et al. 2013 [58] Yang et al. 2010 [93] Mashayekhi et al. 2018 [69] Parchami Barjui et al. 2017 [76] Kontorovich et al. 2010 [86] |
| 499 | rs3746444:T > C | C | 0.003 | Iran | Doulah et al. 2018 [71] | |
| 608 | rs4919510:C > G | G | 0.024 | Iran | Hashemi et al. 2016 [80] | |
| 520f | rs75598818:G > A | GA | 0.041 | Iran | Meshkat et al. 2018 [82] | |
| 196a2 | rs11614913:C > T |
CC T T T |
0.009 0.0005 0.002 0.00295 |
Brazil China USA Vietnam |
Linhares et al. 2012 [108] Dai et al. 2016 [57] Hoffman et al. 2009 [100] Mihn et al. 2018 [90] |
|
| 758 | rs12586258:G > A | A | 0.01 | USA | Yao et al. 2013 [101] | |
| 100 | rs1834306:G > A | A | 0.02 | USA | Yao et al. 2013 [101] | |
| 185 |
rs2008595:C > T rs887205:A > G |
TT GG |
0.04 0.03 |
USA | Bensen et al. 2013 [102] | |
| 423 | rs6505162:A > C | CC | 0.035 | Australia | Smith et al. 2012 [104] | |
| 605 | rs2043556:T > C | C | 0.02 | Chile | Morales et al. 2018 [107] | |
| 146a | rs2910164:G > C | C | 0.01 | India | Bansal et al. 2014 [67] | |
| 149 | rs2292832:T > C | CC | 0.053 | China | He et al. 2015 [56] | |
| 30a | rs763354:G > A | A | 0.022 | China | Zhou et al. 2020 [66] |
Table 4 shows the allele or genotype associated with BC risk in the miRNA SNPs included in this review. For the most-studied SNPs, which were analyzed in at least 6 studies, controversial results are observed. For the miRNA-146a rs2910164:G > C, the C allele was the MAF and risk allele in Italy, Pakistan, Iran, and Brazil; in China, however, the risk genotypes were CG and homozygous GG, and allele G was the MAF and risk allele. For the miRNA-196a2 rs11614913:C > T, the risk allele was C in two studies from China as well as studies from Iran and India. Nevertheless, in Brazil and Saudi Arabia, two ethnically-different countries, the risk allele was T. Other discrepancies are shown in Table 4. In sum, association results from a single study should be interpreted and analyzed with caution. Factors to consider include cohort size, ethnicity, and ancestral lineage, especially in countries with more than one lineage.
Discussion
The majority of the association studies between miRNA SNPs and BC risk were carried out in Asia. To paint a more complete view of the influence of miRNA SNPs on BC risk, it will therefore be necessary to perform this type of study in more American, Oceanic, and African countries. Most of the studies had small sample sizes, which, as is well known, may influence the precision of the results and the power of the studies to draw conclusions. Although multiple meta-analyses in recent years have attempted to define the association between certain miRNA polymorphisms and BC risk more precisely, there still seems to be no clear consensus. It has been established that SNPs are the most common source of variability in the human genome and that these variations are ethnicity-specific. Thus, the effect of a specific SNP on BC risk may differ depending of the ethnicity of a specific population. Chen, Q. et al. 2014, observed that miR-196a-2 rs11614913*T, miR-499 rs3746444*T, and miR-605 rs2043556*A alleles predicted a decreased risk of breast cancer among Asians but not Caucasians [19]. Fejerman et al. [110], performed a study comparing genetic variants in Hispanic and non-Hispanic white women based on the fact that Hispanic women in the USA have been shown to have a lower incidence of BC [110]. The authors observed that 3 of 5 variants were associated with BC risk in Hispanic women but not in non-Hispanic women and suggested that the proportion of indigenous American ancestry modified the magnitude and direction of risk associations in 3 of the 10 variants studied. Therefore, the authors concluded that genetic ancestry is a factor to consider when performing association studies in women of mixed descent [110].
Controversial results were observed for the most-studied SNPs, each analyzed in at least 6 studies: miRNA-146a rs2910164:G > C, miRNA-192a2 rs11614913:C > T, miRNA-499 rs3746444:T > C, and miRNA-27a rs895819:A > G.
The miRNA-146a rs2910164 showed controversial results in China, Iran, and Italy. Four studies performed in China included this SNP. The SNP was associated with increased risk in two of these studies [55, 56], but not in the other two [59, 61]. In the articles by He et al. [56], Qi et al. [55], and Ma et al. [61], the case and control sample sizes were small (Table 1). In the Hu et al. [59], study, which included 1009 cases and 1093 controls, rs2910164:G > C was not associated with BC risk. China is a country with many different ethnicities. The study by Qi et al. [55], was conducted in Henan province, which is widely recognized as the place where Chinese civilization originated. In this study, the SNP was associated with increased BC risk. The other 3 studies were conducted in the same region or nearby provinces. However, the discrepancies between these studies may be due to the fact that He et al. identified the risk association in a sample of postmenopausal women with BC, while Qi et al. [55], simply indicated that there was an increased BC risk without further specifications. Regarding the two studies that did not find an association, the Ma et al. study assessed a sample of women with triple-negative BC, while Hu et al. [59], indicated no association with BC risk without further specifications. Therefore, the divergent results for this SNP in the populations studied could be a consequence of the characteristics of the cases.
miRNA-192a2 rs11614913:C > T showed controversial results in Brazil, China, and Iran. In Brazil, Linhares et al. [108], reported an increased risk for the T-allele but a protective effect for the wild-type CC genotype, which is not a discrepancy. In China, 5 authors studied this SNP. In two publications, the SNP was associated with increased BC risk [55, 56]; in one study it was associated with decreased risk [57]; and in two studies there was no association between the SNP and risk [60, 61]. These findings could be the consequence of ethnic differences. The Hu et al. [59], study included a Nanjing population, where the main ethnic group is Han, but 50 other official ethnic groups are also present. In this population, the SNP was associated with increased BC risk. In the Dai et al. study [57], in which the SNP was associated with decreased BC risk, the ethnicity of the population studied was mainly Han. Zhang et al. found no association between the SNP and BC risk in a population from Zhejiang province, where the ethnic groups include Han, She, Hu, and 49 minority ethnic groups. All of the authors who studied rs11614913 used cases with BC without specifying whether the cancer was familial, sporadic, or early-onset or had other notable characteristics. Consequently, the heterogeneity of the types of BC included in the samples could provoke discrepancies in the results. It is likely that divergent results from Iranian studies are fundamentally due to variations in ethnicity, as this country includes Persians (the main ethnic group), Azeris, Kurds, Lurs, Turkmens, and Baloch, and others.
The discrepancies observed for miRNA-499 rs3746444:T > C and miRNA-27a rs895819:A > G from studies conducted in China and Iran can be explained by the same reasons discussed for miRNA-146a and miRNA-196a2.
It is clear that more studies in Western populations are needed. In South America, only two countries, Chile and Brazil, have performed such studies. This situation underrepresents Western populations. Another issue that all of these studies classified Asians as a single population group despite the fact that Asia is extremely diverse. It was recently reported that the continent has at least ten ancestral lineages, while areas such as northern Europe have only one [111]. In this review, we included studies from China, Iran, India, Saudi Arabia, Israel, and Vietnam, countries with very different ethnicities and genetic profiles. Unfortunately, these differences are not considered in most population-based analyses. The GenomeAsia100k consortium has addressed this problem, noting that underrepresentation of non-Europeans in genetic studies has limited the diversity of individuals in genomic datasets. As a result, many findings have limited medical relevance for a large proportion of the world’s population [112]. The need for more specific population-based studies is clear, with Asian populations separated into more homogenous groups.
In a clinical context, molecular information regarding breast cancer has become highly relevant. The World Health Organization emphasizes that early diagnosis of BC is critical for optimizing outcomes and survival [113]. Unfortunately, the available molecular diagnostic methods may pose limitations. Therefore, miRNAs have emerged as possible diagnostic and prognostic biomarkers. miRNAs also have a potential role in personalized therapy [114]. Srinivasan et al. 2016, has reported that SNPs are more precise genetic determinants than family history; furthermore, SNP genotyping can be performed without the need for invasive techniques [115].
Conclusions
This review examined the sometimes-conflicting results available in the international literature regarding the impact of miRNA polymorphisms on BC risk. We can conclude that: (a) The greatest proportion of studies on this topic have been carried out in Asia (65.0 %), while only one such study has been performed in Africa (1.8 %). In South America, studies have only been conducted in Chile (50 %) and Brazil (50 %), and in Oceania, studies have only been carried out only in Australia; (b) Association studies have been performed for 67 SNPs, located in 53 miRNAs; (c) 146a, 196a2, 499, 27a, and 423 are the most-studied miRNAs, with each included in at least 6 studies; (d) Most of the studies had small samples, possibly limiting the precision of the results and the power to draw conclusions; and (e) This review demonstrates that the effect of a specific SNP on BC risk varies according to the ethnicity the population. It is crucial that comprehensive evaluations be performed in larger cohorts, stratified by ethnicity and histological subtype, to better define the associations between miRNA polymorphisms and BC risk.
Authors' contributions
Conceptualization, SM-P and LJ; Funding acquisition, LJ; Investigation, SM-P and TA; Methodology, SM-P and EM; Project administration, SM-P and LJ; Resources, LJ; Visualization, SM-P and TA; Writing—original draft, TA; Writing—review and editing, SM-P, TA, EM and LJ. All authors read and approved the final manuscript.
Funding
This research was funded by Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT), Grant Number 1200049.
Availability of data and materials
All data are shown within the manuscript.
Code availability
Not applicable.
Declarations
Ethics approval and informed consent
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Trinidad Arancibia and Sebastian Morales-Pison are considered first authors
Contributor Information
Trinidad Arancibia, Email: triniar17@gmail.com.
Sebastian Morales-Pison, Email: seba.morales.p@gmail.com.
Edio Maldonado, Email: emaldona@med.uchile.cl.
Lilian Jara, Email: ljara@med.uchile.cl, Email: ljara@uchile.cl.
References
- 1.Ciriello G, Sinha R, Hoadley KA, Jacobsen AS, Reva B, Perou CM, Sander C, Schultz N. The molecular diversity of Luminal A breast tumors. Breast Cancer Res Treat. 2013;141:409–20. doi: 10.1007/s10549-013-2699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cecilio AP, Takakura ET, Jumes JJ, Dos Santos JW, Herrera AC, Victorino VJ, Panis C. Breast cancer in Brazil: epidemiology and treatment challenges. Breast Cancer. 2015;7:43–9. doi: 10.2147/BCTT.S50361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Couch FJ, Hart SN, Sharma P, Toland AE, Wang X, Miron P, Olson JE, Godwin AK, Pankratz VS, Olswold C, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol. 2015;33:304–11. doi: 10.1200/JCO.2014.57.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–89. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai RP, Weng Y, Su LL, Jin MJ, Xu ZP, Lu LQ, Chen GD. Association of a pre-miR-27a polymorphism with cancer risk: an updated meta-analysis. Asian Pac J Cancer Prevention APJCP. 2014;15:10107–14. doi: 10.7314/apjcp.2014.15.23.10107. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 8.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 9.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Erson AE, Petty EM. MicroRNAs in development and disease. Clin Genet. 2008;74:296–306. doi: 10.1111/j.1399-0004.2008.01076.x. [DOI] [PubMed] [Google Scholar]
- 11.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–7. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–24. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 15.Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer–new paradigms in molecular oncology. Curr Opin Cell Biol. 2009;21:470–9. doi: 10.1016/j.ceb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Iorio MV, Casalini P, Piovan C, Braccioli L, Tagliabue E. Breast cancer and microRNAs: therapeutic impact. Breast. 2011;20(Suppl 3):63–70. doi: 10.1016/S0960-9776(11)70297-1. [DOI] [PubMed] [Google Scholar]
- 17.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 18.Kalemi TG, Lambropoulos AF, Gueorguiev M, Chrisafi S, Papazisis KT, Kotsis A. The association of p53 mutations and p53 codon 72, Her 2 codon 655 and MTHFR C677T polymorphisms with breast cancer in Northern Greece. Cancer Lett. 2005;222:57–65. doi: 10.1016/j.canlet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 19.Chen QH, Wang QB, Zhang B. Ethnicity modifies the association between functional microRNA polymorphisms and breast cancer risk: a HuGE meta-analysis. Tumour Biol. 2014;35:529–43. doi: 10.1007/s13277-013-1074-7. [DOI] [PubMed] [Google Scholar]
- 20.O’Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res: BCR. 2010;12:201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrone I, Bernardo PS, Dos Santos EC, Abdelhay E. MTHFR C677T and A1298C polymorphisms in breast cancer, gliomas and gastric cancer: a review. Genes. 2021 doi: 10.3390/genes12040587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Claus EB, Risch N, Thompson WD. Genetic analysis of breast cancer in the cancer and steroid hormone study. Am J Hum Genet. 1991;48:232–42. [PMC free article] [PubMed] [Google Scholar]
- 23.Angeli D, Salvi S, Tedaldi G. Genetic predisposition to breast and ovarian cancers: how many and which genes to test? Int J Mol Sci. 2020 doi: 10.3390/ijms21031128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Penrose LS, Mackenzie HJ, Karn MN. A genetical study of human mammary cancer. Ann Eugenics. 1948;14 pt:234–266. 10.1111/j.1469-1809.1947.tb02399.x. [DOI] [PubMed]
- 25.Anderson DE. A genetic study of human breast cancer. J Natl Cancer Inst. 1972;48:1029–34. [PubMed] [Google Scholar]
- 26.Bain C, Speizer FE, Rosner B, Belanger C, Hennekens CH. Family history of breast cancer as a risk indicator for the disease. Am J Epidemiol. 1980;111:301–8. doi: 10.1093/oxfordjournals.aje.a112901. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz AG, King MC, Belle SH, Satariano WA, Swanson GM. Risk of breast cancer to relatives of young breast cancer patients. J Natl Cancer Inst. 1985;75:665–8. [PubMed] [Google Scholar]
- 28.Newman B, Austin MA, Lee M, King MC. Inheritance of human breast cancer: evidence for autosomal dominant transmission in high-risk families. Proc Natl Acad Sci USA. 1988;85:3044–8. doi: 10.1073/pnas.85.9.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 30.Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, Collins N, Nguyen K, Seal S, Tran T, Averill D, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265:2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 31.Tavtigian SV, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, Merajver S, Thorlacius S, Offit K, Stoppa-Lyonnet D, et al. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet. 1996;12:333–7. doi: 10.1038/ng0396-333. [DOI] [PubMed] [Google Scholar]
- 32.Oldenburg RA, Meijers-Heijboer H, Cornelisse CJ, Devilee P. Genetic susceptibility for breast cancer: how many more genes to be found? Crit Rev Oncol/Hematol. 2007;63:125–149. doi: 10.1016/j.critrevonc.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Turnbull C, Rahman N. Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genom Hum Genet. 2008;9:321–45. doi: 10.1146/annurev.genom.9.081307.164339. [DOI] [PubMed] [Google Scholar]
- 34.Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40:17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- 35.Prevalence penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Anglian Breast Cancer Study Group. Br J Cancer. 2000;83:1301–8. doi: 10.1054/bjoc.2000.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith P, McGuffog L, Easton DF, Mann GJ, Pupo GM, Newman B, Chenevix-Trench G, kConFab I, Szabo C, Southey M, et al. A genome wide linkage search for breast cancer susceptibility genes. Genes Chromosom Cancer. 2006;45:646–55. doi: 10.1002/gcc.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–33. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srivastava K, Srivastava A. Comprehensive review of genetic association studies and meta-analyses on miRNA polymorphisms and cancer risk. PloS One. 2012;7:e50966. doi: 10.1371/journal.pone.0050966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou K, Liu M, Cao Y. New Insight into microRNA Functions in Cancer: Oncogene-microRNA-Tumor Suppressor Gene Network. Front Mol Biosci. 2017;4:46. doi: 10.3389/fmolb.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Organization WH, Cancer. https://www.who.int/news-room/fact-sheets/detail/cancer . Accessed on May.
- 42.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res: MCR. 2003;1:882–91. [PubMed] [Google Scholar]
- 44.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 45.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 46.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 48.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V, Borbone E, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26:7590–5. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 51.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–76. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 53.Khalife H, Skafi N, Fayyad-Kazan M, Badran B. MicroRNAs in breast cancer: new maestros defining the melody. Cancer Genet. 2020;246–247:18–40. doi: 10.1016/j.cancergen.2020.08.005. [DOI] [PubMed] [Google Scholar]
- 54.Belaiba F, Medimegh I, Ammar M, Jemni F, Mezlini A, Romdhane KB, Cherni L, Benammar Elgaaied A. Expression and polymorphism of micro-RNA according to body mass index and breast cancer presentation in Tunisian patients. J Leukoc Biol. 2019;105:317–27. doi: 10.1002/JLB.3VMA0618-218R. [DOI] [PubMed] [Google Scholar]
- 55.Qi P, Wang L, Zhou B, Yao WJ, Xu S, Zhou Y, Xie ZB. Associations of miRNA polymorphisms and expression levels with breast cancer risk in the Chinese population. Genetics molecular research: GMR. 2015;14:6289–96. doi: 10.4238/2015.June.11.2. [DOI] [PubMed] [Google Scholar]
- 56.He B, Pan Y, Xu Y, Deng Q, Sun H, Gao T, Wang S. Associations of polymorphisms in microRNAs with female breast cancer risk in Chinese population. Tumour Biol. 2015;36:4575–82. doi: 10.1007/s13277-015-3102-2. [DOI] [PubMed] [Google Scholar]
- 57.Dai ZM, Kang HF, Zhang WG, Li HB, Zhang SQ, Ma XB, Lin S, Wang M, Feng YJ, Liu K, et al. The Associations of Single Nucleotide Polymorphisms in miR196a2, miR-499, and miR-608 With Breast Cancer Susceptibility: A STROBE-Compliant Observational Study. Medicine. 2016;95:e2826. doi: 10.1097/MD.0000000000002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang N, Huo Q, Wang X, Chen X, Long L, Jiang L, Ma T, Yang Q. A genetic variant in pre-miR-27a is associated with a reduced breast cancer risk in younger Chinese population. Gene. 2013;529:125–30. doi: 10.1016/j.gene.2013.07.041. [DOI] [PubMed] [Google Scholar]
- 59.Hu Z, Liang J, Wang Z, Tian T, Zhou X, Chen J, Miao R, Wang Y, Wang X, Shen H. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30:79–84. doi: 10.1002/humu.20837. [DOI] [PubMed] [Google Scholar]
- 60.Zhang M, Jin M, Yu Y, Zhang S, Wu Y, Liu H, Liu H, Chen B, Li Q, Ma X, et al. Associations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese population. Eur J Cancer Care. 2012;21:274–80. doi: 10.1111/j.1365-2354.2011.01308.x. [DOI] [PubMed] [Google Scholar]
- 61.Ma F, Zhang P, Lin D, Yu D, Yuan P, Wang J, Fan Y, Xu B. There is no association between microRNA gene polymorphisms and risk of triple negative breast cancer in a Chinese Han population. PloS One. 2013;8:e60195. doi: 10.1371/journal.pone.0060195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao H, Gao A, Zhang Z, Tian R, Luo A, Li M, Zhao D, Fu L, Fu L, Dong JT, et al. Genetic analysis and preliminary function study of miR-423 in breast cancer. Tumour Biol. 2015;36:4763–71. doi: 10.1007/s13277-015-3126-7. [DOI] [PubMed] [Google Scholar]
- 63.Huang AJ, Yu KD, Li J, Fan L, Shao ZM. Polymorphism rs4919510:C > G in mature sequence of human microRNA-608 contributes to the risk of HER2-positive breast cancer but not other subtypes. PloS One. 2012;7:e35252. doi: 10.1371/journal.pone.0035252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun R, Gong J, Li J, Ruan Z, Yang X, Zheng Y, Qing L, He X, Jiang J, Peng Y, et al. A genetic variant rs13293512 in the promoter of let-7 is associated with an increased risk of breast cancer in Chinese women. Biosci Rep. 2019 doi: 10.1042/BSR20182079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J, Qin Z, Jiang Y, Wang Y, He Y, Dai J, Jin G, Ma H, Hu Z, Yin Y, et al. Genetic variations in the flanking regions of miR-101-2 are associated with increased risk of breast cancer. PloS One. 2014;9:e86319. doi: 10.1371/journal.pone.0086319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou J, Wang L, Liu S, Zhou W, Jiang Y, Du J, Dai J, Jin G, Ma H, Hu Z, et al. Genetic variations in miR-30 family member regulatory regions are associated with breast cancer risk in a Chinese population. Biomed Res Int. 2020;2020:8781348. doi: 10.1155/2020/8781348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bansal C, Sharma KL, Misra S, Srivastava AN, Mittal B, Singh US. Common genetic variants in pre-microRNAs and risk of breast cancer in the North Indian population. Ecancermedicalscience. 2014;8:473. doi: 10.3332/ecancer.2014.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bodal VK, Sangwan S, Bal MS, Kaur M, Sharma S, Kaur B. Association between Microrna 146a and Microrna 196a2 Genes Polymorphism and Breast Cancer Risk in North Indian Women. Asian Pac J Cancer Prev APJCP. 2017;18:2345–8. doi: 10.22034/APJCP.2017.18.9.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mashayekhi S, Saeidi Saedi H, Salehi Z, Soltanipour S, Mirzajani E. Effects of miR-27a, miR-196a2 and miR-146a polymorphisms on the risk of breast cancer. Br J Biomed Sci. 2018;75:76–81. doi: 10.1080/09674845.2017.1399572. [DOI] [PubMed] [Google Scholar]
- 70.Omrani M, Hashemi M, Eskandari-Nasab E, Hasani SS, Mashhadi MA, Arbabi F, Taheri M. hsa-mir-499 rs3746444 gene polymorphism is associated with susceptibility to breast cancer in an Iranian population. Biomark Med. 2014;8:259–67. doi: 10.2217/bmm.13.118. [DOI] [PubMed] [Google Scholar]
- 71.Doulah A, Salehzadeh A, Mojarrad M. Association of single nucleotide polymorphisms in miR-499 and miR-196a with susceptibility to breast cancer. Trop J Pharm Res. 2018;17:319–23. doi: 10.4314/tjpr.v17i2.17. [DOI] [Google Scholar]
- 72.Nejati-Azar A, Alivand MR. miRNA 196a2 (rs11614913) & 146a (rs2910164) polymorphisms & breast cancer risk for women in an Iranian population. Personal Med. 2018;14:279–89. doi: 10.2217/pme-2017-0088. [DOI] [PubMed] [Google Scholar]
- 73.Naderi N, Peymani M, Ghaedi K. The protective role of rs56103835 against breast cancer onset in the Iranian population. Mol Genet Genom Med. 2019;7:e540. doi: 10.1002/mgg3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Danesh H, Hashemi M, Bizhani F, Hashemi SM, Bahari G. Association study of miR-100, miR-124-1, miR-218-2, miR-301b, miR-605, and miR-4293 polymorphisms and the risk of breast cancer in a sample of Iranian population. Gene. 2018;647:73–8. doi: 10.1016/j.gene.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 75.Kazemi A, Vallian S. Significant Association of miR-605 rs2043556 with Susceptibility to Breast Cancer. MicroRNA. 2020;9:133–41. doi: 10.2174/2211536608666190926155149. [DOI] [PubMed] [Google Scholar]
- 76.Parchami Barjui S, Reiisi S, Ebrahimi SO, Shekari B. Study of correlation between genetic variants in three microRNA genes (hsa-miR-146a, hsa-miR-502 binding site, hsa-miR-27a) and breast cancer risk. Curr Res Transl Med. 2017;65:141–7. doi: 10.1016/j.retram.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 77.Afsharzadeh SM, Ardebili SMM, Seyedi SM, Fathi NK, Mojarrad M. Association between rs11614913, rs3746444, rs2910164 and occurrence of breast cancer in Iranian population. Meta Gene. 2017;11:20–5. doi: 10.1016/j.mgene.2016.11.004. [DOI] [Google Scholar]
- 78.Eslami-S Z, Tahmaseb M, Ghaderi A. The investigation of miR-196a2 rs11614913 with breast cancer susceptibility in south of IRAN. Meta Gene. 2018;17:43–7. doi: 10.1016/j.mgene.2018.04.007. [DOI] [Google Scholar]
- 79.Kabirizadeh S, Azadeh M, Mirhosseini M, Ghaedi K, Tanha HM. The SNP rs3746444 within mir-499a is associated with breast cancer risk in Iranian population. J Cell Immunotherapy. 2016;2:95–7. doi: 10.1016/j.jocit.2016.08.003. [DOI] [Google Scholar]
- 80.Hashemi M, Sanaei S, Rezaei M, Bahari G, Hashemi SM, Mashhadi MA, Taheri M, Ghavami S. miR-608 rs4919510 C > G polymorphism decreased the risk of breast cancer in an Iranian subpopulation. Exp Oncol. 2016;38:57–9. doi: 10.31768/2312-8852.2016.38(1):57-59. [DOI] [PubMed] [Google Scholar]
- 81.Bahreini F, Ramezani S, Shahangian SS, Salehi Z, Mashayekhi F. miR-559 polymorphism rs58450758 is linked to breast cancer. Br J Biomed Sci. 2020;77:29–34. doi: 10.1080/09674845.2019.1683309. [DOI] [PubMed] [Google Scholar]
- 82.Meshkat M, Mesrian Tanha H, Ghaedi K, Meshkat M. Association of a potential functional mir-520f rs75598818 G > A polymorphism with breast cancer. Journal of genetics. 2018;97:1307–13. doi: 10.1007/s12041-018-1028-3. [DOI] [PubMed] [Google Scholar]
- 83.Meshkat M, Tanha HM, Naeini MM, Ghaedi K, Sanati MH, Meshkat M, Bagheri F. Functional SNP in stem of mir-146a affects Her2 status and breast cancer survival. Cancer Biomark A. 2016;17:213–22. doi: 10.3233/CBM-160633. [DOI] [PubMed] [Google Scholar]
- 84.Sanaei S, Hashemi M, Rezaei M, Hashemi SM, Bahari G, Ghavami S. Evaluation of the pri-miR-34b/c rs4938723 polymorphism and its association with breast cancer risk. Biomed Rep. 2016;5:125–9. doi: 10.3892/br.2016.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Siasi E, Solimani M. Associations of Single Nucleotide Polymorphism in miR-146a Gene with Susceptibility to Breast Cancer in the Iranian Female. Asian Pac J Cancer Prevent APJCP. 2020;21:1585–93. doi: 10.31557/APJCP.2020.21.6.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kontorovich T, Levy A, Korostishevsky M, Nir U, Friedman E. Single nucleotide polymorphisms in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high-risk women. Int J Cancer. 2010;127:589–97. doi: 10.1002/ijc.25065. [DOI] [PubMed] [Google Scholar]
- 87.Alshatwi AA, Shafi G, Hasan TN, Syed NA, Al-Hazzani AA, Alsaif MA, Alsaif AA. Differential expression profile and genetic variants of microRNAs sequences in breast cancer patients. PloS One. 2012;7:e30049. doi: 10.1371/journal.pone.0030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mir R, Al Balawi IA, Abu-Duhier FM. Role of microRNA-423 gene variation in women at higher risk of breast cancer in Tabuk of Saudi Arabia. Indian J Public Health Res Dev. 2019. 10.37506/ijphrd.v10i2.7675.
- 89.Nguyen PBH, Tran MTH, Nguyen TTN, Nguyen HT. The relationship between SNP rs895819 (A > G) on miRNA-27a and the breast cancer in the Vietnamese population. Sci Technol Dev J. 2016;19:39–49. doi: 10.32508/stdj.v19i4.637. [DOI] [Google Scholar]
- 90.Minh TTH, Thanh NTN, Van Thiep T, Hue NT. Association between single nucleotide polymorphism Rs11614913 (C > T) on Mir-196a2 and breast cancer in Vietnamese population. Int Conf Dev Biomed Eng Vietnam 2017;381–386. 10.1007/978-981-10-4361-1_64.
- 91.Ahmad M, Ahmad S, Rahman B, Haq TU, Jalil F, Shah AA. Association of MIR146A rs2910164 variation with a predisposition to sporadic breast cancer in a Pakistani cohort. Ann Hum Genet. 2019;83:325–30. doi: 10.1111/ahg.12316. [DOI] [PubMed] [Google Scholar]
- 92.Garcia AI, Cox DG, Barjhoux L, Verny-Pierre C, Barnes D, Gemo Study C, Antoniou AC, Stoppa-Lyonnet D, Sinilnikova OM, Mazoyer S. The rs2910164:G > C SNP in the MIR146A gene is not associated with breast cancer risk in BRCA1 and BRCA2 mutation carriers. Hum Mutat. 2011;32:1004–7. doi: 10.1002/humu.21539. [DOI] [PubMed] [Google Scholar]
- 93.Yang R, Schlehe B, Hemminki K, Sutter C, Bugert P, Wappenschmidt B, Volkmann J, Varon R, Weber BH, Niederacher D, et al. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res Treat. 2010;121:693–702. doi: 10.1007/s10549-009-0633-5. [DOI] [PubMed] [Google Scholar]
- 94.Catucci I, Yang R, Verderio P, Pizzamiglio S, Heesen L, Hemminki K, Sutter C, Wappenschmidt B, Dick M, Arnold N, et al. Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum Mutat. 2010;31:E1052–7. doi: 10.1002/humu.21141. [DOI] [PubMed] [Google Scholar]
- 95.Yang R, Dick M, Marme F, Schneeweiss A, Langheinz A, Hemminki K, Sutter C, Bugert P, Wappenschmidt B, Varon R, et al. Genetic variants within miR-126 and miR-335 are not associated with breast cancer risk. Breast Cancer Res Treat. 2011;127:549–54. doi: 10.1007/s10549-010-1244-x. [DOI] [PubMed] [Google Scholar]
- 96.McVeigh TP, Mulligan RJ, McVeigh UM, Owens PW, Miller N, Bell M, Sebag F, Guerin C, Quill DS, Weidhaas JB, et al. Investigating the association of rs2910164 with cancer predisposition in an Irish cohort. Endocr Connect. 2017;6:614–24. doi: 10.1530/EC-17-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Catucci I, Verderio P, Pizzamiglio S, Bernard L, Dall’olio V, Sardella D, Ravagnani F, Galastri L, Barile M, Peissel B, et al. The SNP rs895819 in miR-27a is not associated with familial breast cancer risk in Italians. Breast Cancer Res Treat. 2012;133:805–7. doi: 10.1007/s10549-012-2011-y. [DOI] [PubMed] [Google Scholar]
- 98.Pastrello C, Polesel J, Della Puppa L, Viel A, Maestro R. Association between hsa-mir-146a genotype and tumor age-of-onset in BRCA1/BRCA2-negative familial breast and ovarian cancer patients. Carcinogenesis. 2010;31:2124–6. doi: 10.1093/carcin/bgq184. [DOI] [PubMed] [Google Scholar]
- 99.Esteban Cardenosa E, de Juan Jimenez I, Palanca Suela S, Chirivella Gonzalez I, Segura Huerta A, Santaballa Beltran A, El CasalsBusto M, Barragan Gonzalez E, Fuster Lluch O, Bermudez Edo J, et al. Low penetrance alleles as risk modifiers in familial and sporadic breast cancer. Fam Cancer. 2012;11:629–36. doi: 10.1007/s10689-012-9563-1. [DOI] [PubMed] [Google Scholar]
- 100.Hoffman AE, Zheng T, Yi C, Leaderer D, Weidhaas J, Slack F, Zhang Y, Paranjape T, Zhu Y. microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970–7. doi: 10.1158/0008-5472.CAN-09-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yao S, Graham K, Shen J, Campbell LE, Singh P, Zirpoli G, Roberts M, Ciupak G, Davis W, Hwang H, et al. Genetic variants in microRNAs and breast cancer risk in African American and European American women. Breast Cancer Res Treat. 2013;141:447–59. doi: 10.1007/s10549-013-2698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bensen JT, Tse CK, Nyante SJ, Barnholtz-Sloan JS, Cole SR, Millikan RC. Association of germline microRNA SNPs in pre-miRNA flanking region and breast cancer risk and survival: the Carolina Breast Cancer Study. Cancer Causes Control: CCC. 2013;24:1099–109. doi: 10.1007/s10552-013-0187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chacon-Cortes D, Smith RA, Haupt LM, Lea RA, Youl PH, Griffiths LR. Genetic association analysis of miRNA SNPs implicates MIR145 in breast cancer susceptibility. BMC Med Genet. 2015;16:107. doi: 10.1186/s12881-015-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smith RA, Jedlinski DJ, Gabrovska PN, Weinstein SR, Haupt L, Griffiths L.R. A genetic variant located in miR-423 is associated with reduced breast cancer risk. Cancer Genomics Proteom. 2012;9:115–8. [PubMed] [Google Scholar]
- 105.Jedlinski DJ, Gabrovska PN, Weinstein SR, Smith RA, Griffiths LR. Single nucleotide polymorphism in hsa-mir-196a-2 and breast cancer risk: a case control study. Twin Res Hum Genet. 2011;14:417–21. doi: 10.1375/twin.14.5.417. [DOI] [PubMed] [Google Scholar]
- 106.Morales S, Gulppi F, Gonzalez-Hormazabal P, Fernandez-Ramires R, Bravo T, Reyes JM, Gomez F, Waugh E, Jara L. Association of single nucleotide polymorphisms in pre-miR-27a, pre-miR-196a2, pre-miR-423, miR-608 and pre-miR-618 with breast cancer susceptibility in a South American population. BMC Genet. 2016;17:109. 10.1186/s12863-016-0415-0. [DOI] [PMC free article] [PubMed]
- 107.Morales S, De Mayo T, Gulppi FA, Gonzalez-Hormazabal P, Carrasco V, Reyes JM, Gomez F, Waugh E, Jara L. Genetic variants in pre-miR-146a, pre-miR-499, pre-miR-125a, pre-miR-605, and pri-miR-182 are associated with breast cancer susceptibility in a South American population. Genes. 2018. 10.3390/genes9090427. [DOI] [PMC free article] [PubMed]
- 108.Linhares JJ, Azevedo M Jr, Siufi AA, de Carvalho CV, Wolgien Mdel C, Noronha EC, Bonetti TC, da Silva. I.D. Evaluation of single nucleotide polymorphisms in microRNAs (hsa-miR-196a2 rs11614913 C/T) from Brazilian women with breast cancer. BMC Med Genet. 2012;13:119. 10.1186/1471-2350-13-119. [DOI] [PMC free article] [PubMed]
- 109.Brincas HM, Augusto DG, Mathias C, Cavalli IJ, Lima RS, Kuroda F, Urban CA, Gradia DF, de Oliveira J, de Almeida RC, et al. A genetic variant in microRNA-146a is associated with sporadic breast cancer in a Southern Brazilian Population. Genet Mol Biol. 2020;42:e20190278. doi: 10.1590/1678-4685-GMB-2019-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fejerman L, Hu D, Huntsman S, John EM, Stern MC, Haiman CA, Perez-Stable EJ, Ziv E. Genetic ancestry and risk of mortality among U.S. Latinas with breast cancer. Cancer Res. 2013;73:7243–53. doi: 10.1158/0008-5472.CAN-13-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.University NT. Asia-wide genome mapping project reveals insights into Asian ancestry and genetic diversity. https://www.sciencedaily.com/releases/2019/12/191204145813.htm. Accessed 28 Aug.
- 112.GenomeAsia KC. The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature. 2019;576:106–11. doi: 10.1038/s41586-019-1793-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Organization WH. Breast cancer. https://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en/. Accessed 28 Aug.
- 114.Bertoli G, Cava C, Castiglioni I, MicroRNAs New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics. 2015;5:1122–43. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Srinivasan S, Clements JA, Batra J. Single nucleotide polymorphisms in clinics: fantasy or reality for cancer? Crit Rev Clin Lab Sci. 2016;53:29–39. doi: 10.3109/10408363.2015.1075469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are shown within the manuscript.
Not applicable.