Abstract
Xeroderma pigmentosum variant (XPV) cells are characterized by a cellular defect in the ability to synthesize intact daughter DNA strands on damaged templates. Molecular mechanisms that facilitate replication fork progression on damaged DNA in normal cells are not well defined. In this study, we used single-stranded plasmid molecules containing a single N-2-acetylaminofluorene (AAF) adduct to analyze translesion synthesis (TLS) catalyzed by extracts of either normal or XPV primary skin fibroblasts. In one of the substrates, the single AAF adduct was located at the 3′ end of a run of three guanines that was previously shown to induce deletion of one G by a slippage mechanism. Primer extension reactions performed by normal cellular extracts from four different individuals produced the same distinct pattern of TLS, with over 80% of the products resulting from the elongation of a slipped intermediate and the remaining 20% resulting from a nonslipped intermediate. In contrast, with cellular extracts from five different XPV patients, the TLS reaction was strongly reduced, yielding only low amounts of TLS via the nonslipped intermediate. With our second substrate, in which the AAF adduct was located at the first G in the run, thus preventing slippage from occurring, we confirmed that normal extracts were able to perform TLS 10-fold more efficiently than XPV extracts. These data demonstrate unequivocally that the defect in XPV cells resides in translesion synthesis independently of the slippage process.
Xeroderma pigmentosum (XP) is an autosomal recessive disorder characterized by a genetic predisposition to sunlight-induced skin cancer. Fibroblasts derived from patients with XP are extremely sensitive to the mutagenic effect of UV irradiation (19). The majority of XP patients are deficient in nucleotide excision repair, and there have been dramatic advances in our understanding of the molecular defects in these patients. In contrast, there has been little progress in our understanding of the molecular defect in the XP variant (XPV) group, which comprises a substantial minority (approximately 20%) of XP patients. They have normal levels of nucleotide excision repair and normal sensitivity to the lethal effects of UV irradiation (5) but a marked defect in the ability to synthesize intact daughter DNA strands during DNA replication after some (4, 17, 20, 26) but not all (4, 7) types of carcinogenic damage. The cellular defect in DNA replication in UV-irradiated XPV cells was discovered as long ago as 1975 (17), but because of the normal sensitivity to killing by UV light, the XPV gene has been refractory to cloning. The precise nature of the molecular defect in this important class of XP patients remains one of the major unsolved problems in this area.
The UV hypermutability of XPV cells could be due to an abnormal error-prone mechanism of replication. This hypothesis is supported by results showing that mutation spectra in UV-irradiated XPV cells are distinct from those observed in normal cells (32, 33). In XPV cells, the UV-induced substitutions are mainly transversions (C→A), whereas in normal cells, transitions (C→T) predominate. An altered mutation pattern was also generated by psoralen photoadducts in XPV compared to normal cells (27). To determine whether the process of translesion synthesis (TLS) in XPV extracts differs from that of normal cells, we compared the abilities of cellular extracts from either normal or XPV primary fibroblasts to perform primer elongation past a unique blocking lesion located on a single-stranded circular template. This approach allows TLS to be investigated both quantitatively and qualitatively in the absence of other cellular responses such as repair, recombination, or polymerase strand switching. This assay will greatly facilitate the identification of the XPV gene product(s) and the biochemical features of replication of damaged DNA in human cells.
MATERIALS AND METHODS
Construction of single-stranded plasmid containing a single AAF adduct.
The strategy used to construct double-stranded molecules containing single N-2-acetylaminofluorene (AAF) adducts involved the formation of gapped-duplex molecules (14). A 14-mer oligonucleotide d(ATACCCG1G2G3ACATC) was reacted with N-acetoxy-N-2-acetylaminofluorene under conditions such as to create one adduct per oligonucleotide on average. The crude reaction mixture was subjected to reverse-phase high-pressure liquid chromatography, and the oligonucleotides with a single AAF adduct at G1 or G3 were purified and ligated into the gap to generate plasmid pUC-3G1 or pUC-3G3, respectively. A control undamaged plasmid, pUC-3G0, was constructed with the unreacted control oligonucleotide. The single-stranded vectors (pUC-3G0.ss, pUC-3G1.ss, and pUC-3G3.ss) were produced from the corresponding double-stranded plasmids by selective enzymatic degradation of the nonadducted uracil-containing strand. A detailed description of this procedure using an enzymatic cocktail containing uracil-DNA glycosylase, exonuclease III, and the 3′→5′ exonuclease activity associated with T7 DNA polymerase has been described recently (23).
Cell cultures.
The cell strains used in this study were fibroblast cultures derived from the skin of normal individuals (1BR3, 205BR, 250BR, and 368BR) and XP variants (XP7BR, XP11BR, XP6DU, XP7DU, and XP30R0). The XP variants were all defective in postreplication repair of UV damage as shown by sucrose density gradient analysis of newly synthesized DNA in UV-irradiated cells (reference 17 [XP30R0] and our unpublished results [other cell strains]). XP11BR cells are derived from the patient described in reference 3. All cell strains were grown in Dulbecco’s modified Eagle’s medium (Sigma) supplemented with 15% fetal calf serum (Eurobio) and 50 μg of gentamicin (Sigma) per ml.
Preparation of cell extracts.
Cell extracts (100 μl) were obtained from 107 exponentially growing cells essentially as described previously (13). The cells were washed twice with phosphate-buffered saline (PBS). trypsin (0.25% in PBS) was added to the plates, which were then incubated at 37°C until cells rounded up and were almost ready to detach. Excess buffer was removed, and the cells were collected by agitation in the culture medium. The cells were pelleted by centrifugation (1,000 × g for 5 min) and washed once in the culture medium and twice in PBS. The cell pellet was resuspended in 4 volumes of ice-cold hypotonic buffer (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol [DTT]) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride and 5 μg each of leupeptin, chymostatin, and aprotinin per ml). The cells were allowed to swell on ice for 30 min at 4°C and disrupted with 20 strokes of a tight-fitting pestle in a Dounce homogenizer. Cell disruption and integrity of the nuclei were examined under light microscopy by exclusion of trypan blue. Nuclei were harvested by centrifugation for 10 min at 3,000 × g at 4°C, and cytosolic supernatants were kept on ice. After 1 h of extraction at 0°C in hypotonic buffer containing 350 mM NaCl, the nuclear extracts were centrifuged at 10,000 × g for 10 min. Cytosolic and nuclear extracts were mixed, and the proteins were precipitated by the addition of ammonium sulfate (0.33 g/ml) and gentle stirring for 1 h at 4°C. The precipitates were collected by centrifugation (45 min at 10,000 × g), resuspended in dialysis buffer (100 mM potassium glutamate, 30 mM HEPES [pH 7.5], 1 mM DTT, 10% glycerol), and dialyzed for 2 h at 4°C. The extracts were clarified by centrifugation for 10 min at 10,000 × g and stored at −80°C. The protein concentration of extracts is typically between 5 and 15 mg/ml as measured by the Bradford protein assay (Bio-Rad) using bovine serum albumin as the standard.
In vitro primer extension assays.
A primer (24-mer oligonucleotide) was phosphorylated with T4 polynucleotide kinase (New England Biolabs), using 50 pmol of [γ-32P]ATP (3,000 Ci/mmol; Amersham). After purification by electrophoresis on a 20% polyacrylamide–7 M urea denaturing gel, the primer (twofold molar excess) was annealed to single-stranded DNA in a buffer containing 60 mM HEPES (pH 7.5) and 20 mM MgCl2. The mixture was incubated at 50°C for 15 min with Escherichia coli SSB (Pharmacia). For primer extension assays, the reaction mixture (6.25 μl) containing 10 fmol of SSB-coated primed DNA, and whole-cell extract was incubated at 37°C in 50 mM HEPES-KOH (pH 7.8)–7 mM MgCl2–1 mM DTT–4 mM ATP–500 μM deoxynucleoside triphosphates (dNTPs)–200 μM each UTP, CTP, and GTP–40 mM creatine phosphate–100 μg of creatine kinase per ml. The reaction was stopped by adding an equal volume of proteinase K (4 mg/ml)-sodium dodecyl sulfate (2%) and incubated for 30 min at 37°C. The samples were precipitated in the presence of 1 M ammonium acetate and 50% isopropanol. Replication products were digested with restriction enzymes EcoRI, PvuII, and SmaI in buffers recommended by the manufacturers and analyzed by electrophoresis on a polyacrylamide–7 M urea denaturing gel.
RESULTS
Design of the damaged single-stranded DNA substrate.
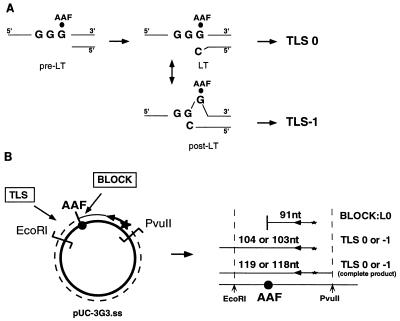
We used AAF as a model DNA-damaging agent to analyze the ability of cell extracts to carry out TLS. AAF adducts at the C8 position of guanine (dGua-C8-AAF) are severe blocks to in vitro DNA synthesis by purified prokaryotic and eukaryotic DNA polymerases (2, 21, 22). TLS past AAF adducts in both double-stranded (30, 31) and forked single-stranded (13) DNA templates has been observed to some extent in cell extracts. This suggests that extracts accurately mimic at least some of the mechanisms that rescue a blocked replication fork in vivo. In the present study, single-stranded plasmids containing single AAF adducts at G1 or G3 in a run of three guanines (5′-G1G2G3-3′) were constructed to analyze TLS. In E. coli, when located at G3 in the run (5′-GGGAAF-3′), an AAF adduct induces −1 frameshift mutations at least 100-fold more efficiently than when located at G1 (5′-GAAFGG-3′) (15). Indeed, a G3 adduct can trigger a primer-template misalignment event yielding a slipped intermediate that is in equilibrium with its nonslipped counterpart, while such an event is not favored for a G1 adduct (Fig. 1A). Cellular extracts from either normal or XPV primary fibroblasts were tested for their abilities to elongate a 32P-end-labeled oligonucleotide (24-mer) annealed to the single-stranded circular template, at a distance of 91 (pUC-3G3.ss) or 93 (pUC-3G1.ss) nucleotides from the lesion site. TLS was analyzed on sequencing gels following PvuII and EcoRI restriction digestion of the DNA products (Fig. 1B). For both substrates, TLS resulting from the elongation of nonslipped (TLS0) and slipped (TLS−1 [−1 frameshift event]) intermediates will yield fragments with lengths of 104 and 103 nucleotides, respectively.
FIG. 1.
AAF-induced −1 frameshift pathway. The primer terminus when located opposite the lesion site is designated the lesion terminus (LT). The LT can isomerize via a slippage mechanism into the slipped intermediate, which has a paired primer terminus (post-LT). Elongation from the nonslipped and slipped intermediates lead to TLS0 and TLS−1 products, respectively. (B) Diagram of the modified template, pUC-3G3.ss. A single AAF adduct is located with the recognition site of SmaI. The positions of PvuII and EcoRI restriction sites and lengths of the strands produced upon elongation of the labeled primer are indicated. L0, fragment elongated up the lesion site; nt, nucleotides.
Primer extension using the unmodified substrate.
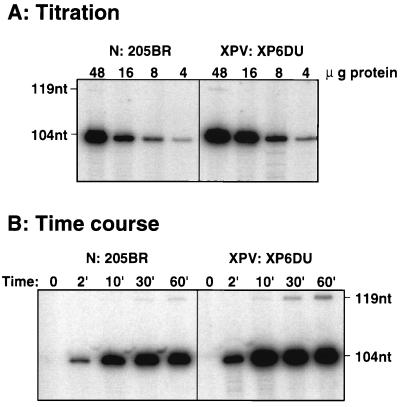
The replication activities of extracts from normal (205BR) and XPV (XP6DU) cells were tested by using a primed unmodified single-stranded template (pUC-3G0.ss). For both extracts, product yield increased with increasing amounts of protein (Fig. 2A). In a typical time course experiment, radiolabeled 104-nucleotide products were detected after a 2-min reaction and reached a plateau at 10 min. Completion of DNA synthesis around the whole template followed by subsequent ligation was observed by the appearance of a band at 119 nucleotides (Fig. 2B). Quantitative analysis of the results indicates that after a 1-h incubation with 48 μg of either extract, about 20% of the primers were elongated. All of these data indicate that XPV cell extracts have replication activity similar to that of normal cells on a lesion-free template, consistent with the normal DNA replication observed in undamaged XPV cells in vivo (17).
FIG. 2.
DNA synthesis catalyzed by normal (N: 205BR) or XPV (XPV: XP6DU) extracts, using the unmodified single-stranded template (pUC-3G0.ss). (A) Titration of DNA synthesis activity after 1 h of incubation. (B) Time course of DNA synthesis catalyzed by 48 μg of normal and XPV proteins. DNA products obtained after 1 h of incubation with pUC-3G0.ss were cleaved with enzymes PvuII and EcoRI and subjected to electrophoresis on an 8% polyacrylamide–7 M urea denaturing gel. nt, nucleotides.
Differential TLS catalyzed by extracts from normal or XPV extracts.
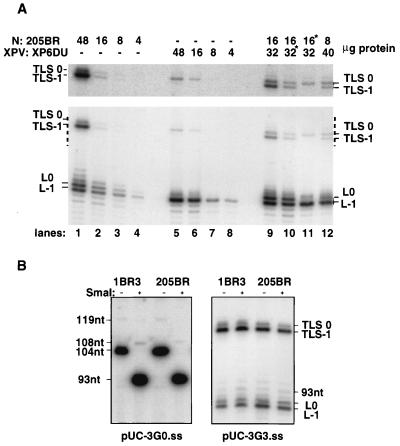
TLS can be described in terms of the different replication intermediates that are involved. The replication intermediate in which the last nucleotide of the primer is located opposite the lesion in the template will be referred to as a lesion terminus (LT). All replication intermediates preceding and succeeding this step will be referred to as prelesion termini (pre-LT) and postlesion termini (post-LT), respectively. Given these definitions, TLS can be viewed as a succession of at least two reactions (pre-LT→LT→post-LT). The progression from one step to the next normally requires a DNA synthesis step. A marked difference between normal and XPV extracts was observed for TLS past the AAF adduct at the G3 position (Fig. 3A). Despite the presence of the lesion on the modified substrate (pUC-3G.3ss), normal extracts (205BR) were able to catalyze TLS efficiently as 40 to 60% of the elongated primers were extended past the adduct site upon incubation with 48 μg of normal extract proteins. Both nonslipped (TLS0) and slipped (TLS−1) elongation products were seen (Fig. 3A, lanes 1 to 4), the latter constituting over 80% of the TLS products formed. This finding indicates that the replication complex is able to elongate the slipped LT more efficiently than the corresponding nonslipped intermediate. The slipped intermediate exhibits a normal G · C base pair at its terminus and therefore mimics a post-LT (Fig. 1A). A distinct feature of the slipped TLS reaction is that the formation of the slipped intermediate occurs by isomerization of the LT in the absence of a DNA synthesis step. As a consequence, the slipped TLS reaction is unique in the sense that the otherwise critical step converting the LT to a post-LT occurs in the absence of DNA synthesis. With normal cell extracts, a distinct pattern of bands is also observed near the adduct site, as elongation products blocked both one nucleotide before the lesion site (L−1) and opposite the lesion (L0) are seen.
FIG. 3.
TLS by extracts from normal or XPV cells, using the pUC-3G3.ss substrate. (A) Analysis of TLS catalyzed by various amounts of either normal (N: 205BR; lanes 1 to 4) or XPV (XPV: XP6DU; lanes 5 to 8) cell extracts. In lanes 9 to 12, different amounts of normal and XPV extracts were mixed. Asterisks indicate that the proteins were heated at 50°C for 10 min before the assay. DNA products obtained after 1 h of incubation with pUC-3G3.ss were cleaved with enzymes PvuII and EcoRI and subjected to electrophoresis on an 8% polyacrylamide–7 M urea denaturing gel. L−1 and L0 are products generated if synthesis is blocked one nucleotide before and opposite the lesion, respectively. TLS0 and TLS−1 are products from TLS via nonslipped and slipped intermediates. Overexposure (fivefold) of the portion of the gel indicated by dashes is shown at the top. (B) Resistance to SmaI digestion of TLS0 products generated by 30 μg of normal (1BR3 and 205BR) cell extracts. DNA products obtained after 1 h of incubation with pUC-3G0.ss or pUC-3G3.ss were cleaved with enzymes PvuII and EcoRI. Half of the DNA samples were further digested by SmaI as indicated and subjected to electrophoresis on an 8% polyacrylamide–7 M urea denaturing gel. nt, nucleotides.
In marked contrast, comparable reactions with an XPV extract (lanes 5 to 8) resulted in the accumulation of replication products blocked one nucleotide before the lesion (position L−1) and of low amounts of nonslipped elongation product (TLS0). The slipped elongation product (TLS−1) was barely detectable. Therefore, bands L0 and TLS−1 appear to be diagnostic for normal cells (which we will refer to as the normal cell TLS pattern), as they are absent in all five XPV extracts tested (Fig. 3A and 4). It should be stressed that failure of XP variants to effect TLS via the slipped intermediate was obtained even under the forcing conditions used, such as long incubation periods (1 h) and high dNTP concentrations (500 μM dNTPs). These conditions increased the overall efficiency of TLS in normal extracts but did not modify the relative ratio of TLS0 to TLS−1 (data not shown). In addition, neither the passage number of the cells nor the position of the primer with respect to the lesion site altered the reaction (data not shown). Our results were confirmed in a blind study in which coded samples of two normal and two XPV cell strains, provided by A. R. Lehmann, were correctly identified.
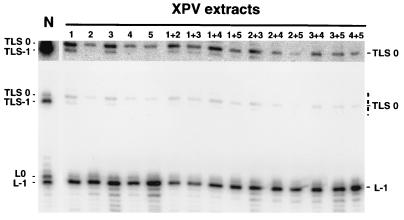
FIG. 4.
Absence of complementation between extracts from five different XPV individuals. XPV extracts (15 μg of each) were mixed together as indicated and incubated for 1 h with pUC-3G3.ss. In parallel, 30 μg of normal (N; 205BR) or XPV extracts was incubated with pUC-3G3.ss. DNA products were cleaved with enzymes PvuII and EcoRI and subjected to electrophoresis on an 8% polyacrylamide–7 M urea denaturing gel. Overexposure (13-fold) of the portion of the gel indicated by dashes is shown at the top. XPV extracts: 1, XP11BR; 2, XP30R0; 3, XP7BR; 4, XP6DU; 5, XP7DU. All of these cell strains have the cellular defect in DNA synthesis after UV irradiation (unpublished data and reference 17) that is diagnostic for XP variants.
It was important to confirm that the TLS products actually resulted from synthesis past the AAF lesion, rather than on contaminating undamaged substrate. The AAF lesion is situated in a sequence of DNA which, in the absence of the lesion, is a substrate for cleavage by the restriction enzyme SmaI. To provide direct evidence that DNA synthesis had actually occurred past the lesion, we tested the resistance of the TLS0 product DNA to cleavage by SmaI. With pUC-3G0.ss, incision at the recognition site converted the labeled major fragment of 104 nucleotides and minor fragment of 119 nucleotides (Fig. 1B) to smaller bands of 93 and 108 nucleotides, respectively (Fig. 3B). In contrast, TLS0 elongation products obtained with damaged template were resistant to cleavage, indicating that the adduct was indeed present in the double-stranded DNA obtained after replication in vitro (Fig. 3B).
Increasing amounts of normal extract stimulated TLS (Fig. 3A, lanes 1 to 4). Generally, optimal activity was obtained with 30 to 50 μg of protein per 10 ng of DNA, and at a higher protein concentration, TLS declined (data not shown). Addition of XPV extracts to limiting amounts of normal extracts (8 and 16 μg) did not inhibit TLS; on the contrary, increasing amounts of products at position L0 and TLS−1 were detectable when the two extracts were combined (Fig. 3A, lanes 9 to 12). This observation demonstrating the absence of a dominant negative factor in XPV extracts is consistent with the recessive mode of transmission of the XPV defect. The addition of 32 μg of XPV extract to a limiting amount (16 μg) of normal cell extract stimulates the normal cell TLS pattern (i.e., presence of bands TLS-1 and L0) compared to the replication assay where the same quantity of heat-inactivated XPV extract is added to limiting quantities of normal cell extract (Fig. 3A, lanes 9 and 10). This finding demonstrates that it is not the XPV factor that limits the TLS reaction with 16 μg of normal cell extracts since XPV extracts can stimulate the normal cell TLS pattern under these conditions. On the other hand, heat inactivation of the normal cell extract (lane 11) yields the TLS pattern typical for XPV cells (i.e., absence of both TLS−1 and L0 bands).
Complementation studies.
The assay described above provides a complementation approach to isolate proteins from normal extracts that would allow XPV extracts to produce TLS−1 efficiently with pUC-3G3.ss as a template. As a first step, we tested complementation between extracts from five different XPV individuals (Fig. 4). Complementation was not observed upon mixing equal amounts of any of the five extracts, suggesting that these five XPV patients are mutated in the same gene.
Analysis of TLS by using a substrate where the possibility of slippage is reduced.
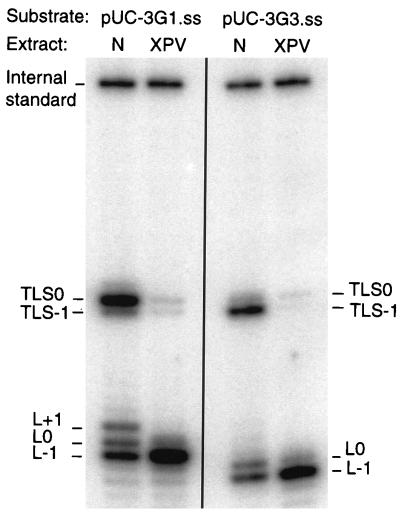
The data obtained with the pUC-3G3.ss substrate indicate that XPV extracts are less able than normal extracts to elongate a primer past an AAF adduct. However, this defect could be specific for the elongation of the post-LT generated by slippage (Fig. 1). Another possibility could be that a misincorporation at this site prevents slippage from occurring. To further investigate these points, we analyzed TLS by using a substrate where the slippage process is minimized because the AAF adduct is located on the first G of the run (pUC-3G1.ss). We previously showed (15) that at this position, the frequency of induced −1 frameshift mutation was reduced 100-fold in vivo in E. coli. Interestingly, cellular extracts from normal cells were able to perform TLS with pUC-3G1.ss as efficiently as with pUC-3G3.ss (Fig. 5). In agreement with the result obtained with E. coli, TLS using substrate pUC-3G1.ss resulted mainly in normal elongation products (TLS0). The TLS−1 products could consist of either targeted deletions (-G within the run of guanine residues) or semitargeted deletions (-C in the 5′ flanking repetitive cytosine sequence) as found in E. coli (15). In the vicinity of the lesion site, the presence of three bands at L−1, L0, and L+1 revealed that the adduct hindered the progression of DNA synthesis. Obviously, the absence of band L+1 when pUC-3G3.ss was used as the template indicates that this step was obviated by the slippage event. Irrespective of the site of the lesion (G1 or G3), the efficiency of TLS was highly reduced with XPV extracts.
FIG. 5.
TLS past and AAF adduct by extracts from normal or XPV cells. Analysis of TLS catalyzed by 30 μg of either normal (N; 205BR) or XPV (XP6DU) cell extracts. DNA products obtained after 1 h of incubation with pUC-3G1.ss or pUC-3G3.ss were cleaved with enzymes PvuII and EcoRI and subjected to electrophoresis on an 10% polyacrylamide–7 M urea denaturing gel. L−1, L0, and L+1 are products generated if synthesis is blocked one nucleotide before, opposite, and one nucleotide after the lesion, respectively. TLS0 and TLS−1 are products from TLS via nonslipped and slipped intermediates. As an internal standard, an identical amount of a 120-nucleotide fragment (end labeled with [γ-32P]ATP by T4 polynucleotide kinase) was added to each reaction at the end of the replication step.
The high amount of TLS0 obtained in normal extracts with the pUC-3G1.ss substrate indicates that efficient TLS can occur independently of the slippage process. In XPV extracts, the efficiency of TLS was reduced 10-fold, providing evidence that the nonslipped TLS product formed with normal cell extracts upon incubation with pUC-3G1.ss arises by a mechanism that required the XPV factor, similarly to the slipped TLS product formed with pUC-3G3.ss.
DISCUSSION
Complete replication past site-specific UV-induced lesions (6, 29) or AAF adducts (30, 31) in double-stranded DNA carrying the simian virus 40 origin of replication has been observed in HeLa cell extracts, in the presence of T antigen. Recently, using a similar approach, three groups (8, 9, 28) demonstrated that in contrast to normal cell extracts, extracts from XPV cells were completely (8) or partially (9, 28) deficient in the bypass of a single cis-syn thymine dimer. In these studies, impaired replication fork bypass observed in XPV extracts could be due to a defect either in mechanisms broadly referred to as postreplication repair (such as polymerase template switching or recombinational strand transfer) or in a simple TLS reaction. However, in the present work, using a single-stranded template with a single lesion, we show directly and unequivocally that it is TLS that is impaired in XPV extracts. This result highlights the biological importance of a factor(s) involved in TLS in human cells, since a defect in this pathway leads to enhanced mutagenesis and to sunlight-induced skin cancer.
TLS in normal cell extracts.
In normal cell extracts, the slow process of formation of complete TLS products past the AAF adduct and the pattern of bands seen in the vicinity of the adduct site show that the replication apparatus stalls at the lesion. This finding suggests that the conformational change induced by AAF adducts in the template DNA (rotation of the guanine moiety from the anti to the syn conformation [for a review, see reference 12]) hinders this replication step. It is thought that modification of the replication multiprotein apparatus (such as ubiquitination by the Rad6-Rad18 heterodimer in yeast), release and/or recruitment of accessory proteins, and replacement of the DNA polymerase are necessary to overcome the block in vivo (16). Interestingly, in Saccharomyces cerevisiae, TLS past an AAF adduct in the three-G sequence context requires the Rev3 (1) and to a large extent the Rev1 protein (1a). Rev3p together with Rev7p forms a heterodimeric DNA polymerase designated Polζ, dedicated to replication on damaged DNA (25). Rev1p has weak homology with the E. coli UmuC protein and has a deoxycytidyltransferase activity (24). Human homologs of Rev1 and Rev3 (hsREV3) have been identified (10, 16). In view of the involvement of these proteins in TLS past AAF adducts in yeast, it is tempting to speculate that they may also participate in TLS past AAF adducts in our assay. This raises the possibility that homologs of the Rev1, Rev3, and Rev7 protein are products of the XPV gene(s). In a study to be presented in detail elsewhere, using sequence and segregation analysis, we have been able to exclude the hsREV3 as the gene defective in XPV patients (4a). Recent work demonstrated that the great majority of UV-induced mutations were abolished in cultured human cells expressing an hsREV3 antisense RNA fragment (10), supporting a mechanism of Polζ-dependent translesion replication of UV photoproducts in human cells. However, since UV-irradiated XPV cells are hypermutable, the XPV defect cannot be explained simply by an abrogation of TLS via Polζ, but rather involves subtle alteration of this specific pathway leading to enhanced UV-induced mutagenesis. On the other hand, Polδ (11) was also shown to be involved in UV-induced mutagenesis in yeast. Thus, the XPV factor could be a component of the normal replication machinery.
TLS in XPV extracts.
TLS must overcome two problems: first the insertion of a nucleotide opposite the residue containing the adduct (insertion) and second, following insertion, extension of the improperly base-paired terminus (elongation). From the present work, it appears that the primary defect in XPV extracts residues in their reduced ability to incorporate (or stably maintain) a nucleotide in front of the AAF adduct (absence of band L0). This defect observed in vitro reflects the in vivo situation, in which replication blocks occur at the sites of damage in UV-irradiated XPV cells (4, 17, 26).
The qualitative difference between the TLS patterns produced in normal extracts and in XPV extracts suggests that different pathways of TLS coexist in mammalian cells. In the absence of a functional XPV factor, an alternative, minor TLS pathway yields the small amount of products specific for XPV extracts (i.e., relatively high amount of TLS−1 with pUC-3G1.ss and absence of TLS−1 with pUC-3G3.ss). The enhancement by caffeine of the defect in replication on damaged templates in XPV cells (17, 18) as well as the distinct UV- and psoralen-induced mutation spectrum observed in these cells compared to normal cells (27, 32, 33) might be hallmarks of this residual TLS pathway. Hypermutability of XPV cells after UV irradiation can thus be explained by the use of this specific pathway of TLS, less efficient but more error prone than the major pathway observed in normal extracts. To confirm this hypothesis and to get a better understanding of XPV pathogenesis, we are currently extending our analysis to several different photoproducts.
The assay described in this paper provides a unique opportunity to distinguish different pathways of TLS in eukaryotic cell extracts. It provides compelling evidence that XPV cells are defective in a major TLS pathway. This assay should greatly facilitate attempts to clone the XPV gene and to identify proteins involved in bypass of lesions in eukaryotes.
ACKNOWLEDGMENTS
We are grateful to Heather Fawcett for cell culture and to Marc Bichara, Jenny Lees, Rita Napolitano, and Elaine Taylor for helpful critical comments.
This work was supported in part by grant 9616 from the Association pour la Recherche contre le Cancer (Villejuif, France).
REFERENCES
- 1.Baynton K, Bresson-Roy A, Fuchs R P P. Analysis of damage tolerance pathways in Saccharomyces cerevisiae: a requirement for Rev3 DNA polymerase in translesion synthesis. Mol Cell Biol. 1998;18:960–966. doi: 10.1128/mcb.18.2.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Baynton, K. Personal communication.
- 2.Belguise-Valladier P, Maki H, Sekiguchi M, Fuchs R P P. Effect of single DNA lesions on in vitro replication with DNA polymerase III holoenzyme. J Mol Biol. 1994;236:151–164. doi: 10.1006/jmbi.1994.1125. [DOI] [PubMed] [Google Scholar]
- 3.Berth-Jones J, Cole J, Lehmann A R, Arlett C F, Graham-Brown R A C. Xeroderma pigmentosum variant: 5 years of tumor suppression by etretinate. J R Soc Med. 1993;86:355–356. doi: 10.1177/014107689308600615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer J C, Kaufmann W K, Brylawski B P, Cordeiro-Stone M. Defective postreplication repair in xeroderma pigmentosum variant fibroblasts. Cancer Res. 1990;50:2593–2598. [PubMed] [Google Scholar]
- 4a.Broughton, B. C., and A. R. Lehmann. Unpublished data.
- 5.Burk P G, Lutzner M A, Clarke D D, Robbins J H. Ultraviolet-stimulated thymidine incorporation in xeroderma pigmentosum lymphocytes. J Lab Clin Med. 1971;77:759–761. [PubMed] [Google Scholar]
- 6.Carty M P, Lawrence C W, Dixon K. Complete replication of plasmid DNA containing a single UV-induced lesion in human cell extracts. J Biol Chem. 1996;271:9637–9647. doi: 10.1074/jbc.271.16.9637. [DOI] [PubMed] [Google Scholar]
- 7.Cordeiro-Stone M, Boyer J C, Smith B A, Kaufmann W K. Xeroderma pigmentosum variant and normal fibroblasts show the same response to the inhibition of DNA replication by benzo[a]pyrene-diol-epoxide-I. Carcinogenesis. 1986;7:1783–1786. doi: 10.1093/carcin/7.10.1783. [DOI] [PubMed] [Google Scholar]
- 8.Cordeiro-Stone M, Zaritskaya L S, Price L K, Kaufmann W K. Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J Biol Chem. 1997;272:13945–13954. doi: 10.1074/jbc.272.21.13945. [DOI] [PubMed] [Google Scholar]
- 9.Ensch-Simon I, Burgers P M, Taylor J-S. Bypass of a site-specific Cis-Syn dimer in an SV40 vector during in vitro replication by HeLa and XPV cell-free extracts. Biochemistry. 1998;37:8218–8226. doi: 10.1021/bi972460j. [DOI] [PubMed] [Google Scholar]
- 10.Gibbs P E, Glenn McGregor W, Maher V M, Nisson P, Lawrence C W. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase ζ. Proc Natl Acad Sci USA. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giot L, Chanet R, Simon M, Facca C, Faye G. Involvement of the yeast DNA polymerase δ in DNA repair in vivo. Genetics. 1997;146:1239–1251. doi: 10.1093/genetics/146.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann G, Fuchs R P P. Mechanisms of frameshift mutations: insight from aromatic amines. Chem Res Toxicol. 1997;10:347–359. doi: 10.1021/tx960128n. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann J S, Pillaire M J, Lesca C, Burnouf D, Fuchs R P P, Defais M, Villani G. Fork-like DNA templates support bypass replication of lesions that block DNA synthesis on single-stranded templates. Proc Natl Acad Sci USA. 1996;93:13766–13769. doi: 10.1073/pnas.93.24.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koehl P, Burnouf D, Fuchs R P P. Construction of plasmids containing a unique acetylaminofluorene adduct located within a mutation hot spot. J Mol Biol. 1989;207:355–364. doi: 10.1016/0022-2836(89)90259-3. [DOI] [PubMed] [Google Scholar]
- 15.Lambert I B, Napolitano R L, Fuchs R P P. Carcinogen-induced frameshift mutagenesis in repetitive sequences. Proc Natl Acad Sci USA. 1992;89:1310–1314. doi: 10.1073/pnas.89.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence C W, Hinkle D C. DNA polymerase ζ and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv. 1997;28:21–31. [PubMed] [Google Scholar]
- 17.Lehmann A R, Kirk-Bell S, Arlett C F, Paterson M C, Lohman P H M, de Weerd-Kastelein E A, Bootsma D. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc Natl Acad Sci USA. 1975;72:219–233. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehmann A R. The relationship between pyrimidine dimers and replicating DNA in UV-irradiated human fibroblasts. Nucleic Acids Res. 1979;7:1901–1912. doi: 10.1093/nar/7.7.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maher V M, Ouellette L M, Curren R D, McCormick J J. Frequency of ultraviolet light-induced mutations is higher in xeroderma variant cells than in normal human cells. Nature. 1976;261:593–595. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- 20.Misra R R, Vos J-M H. Defective replication of psoralen adducts detected at the gene-specific level in xeroderma pigmentosum variant cells. Mol Cell Biol. 1993;13:1002–1012. doi: 10.1128/mcb.13.2.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore P D, Bose K K, Rabkin S D, Strauss B S. Sites of termination of in vitro DNA synthesis on ultraviolet and N-acetylaminofluorene treated ΦX 174 templates by prokaryotic and eukaryotic polymerases. Proc Natl Acad Sci USA. 1981;78:110–114. doi: 10.1073/pnas.78.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mozzherin D J, Shibutani S, Tan C K, Downey K M, Fisher P A. Proliferating cell nuclear antigen promotes DNA synthesis past template lesions by mammalian DNA polymerase δ. Proc Natl Acad Sci USA. 1997;94:6126–6131. doi: 10.1073/pnas.94.12.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Napolitano R L, Fuchs R P P. New strategy for the construction of single-stranded plasmids with single mutagenic lesions. Chem Res Toxicol. 1997;10:667–671. doi: 10.1021/tx970018w. [DOI] [PubMed] [Google Scholar]
- 24.Nelson J R, Lawrence C W, Hinkle D C. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 25.Nelson J R, Lawrence C W, Hinkle D C. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 26.Park S D, Cleaver J E. Postreplication repair: question of its definition and possible alteration in xeroderma pigmentosum cell strains. Proc Natl Acad Sci USA. 1979;76:3927–3931. doi: 10.1073/pnas.76.8.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raha M, Wang G, Seidman M M, Glazer P M. Mutagenesis by third-strand-directed psoralen adducts in repair-deficient human cells: high frequency and altered spectrum in a xeroderma pigmentosum variant. Proc Natl Acad Sci USA. 1996;93:2941–2946. doi: 10.1073/pnas.93.7.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svoboda D L, Briley L P, Vos J-M H. Defective bypass replication of a leading strand cyclobutane thymine dimer in xeroderma pigmentosum variant cell extracts. Cancer Res. 1998;58:2445–2448. [PubMed] [Google Scholar]
- 29.Svoboda D L, Vos J-M H. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: fork uncoupling or gap formation. Proc Natl Acad Sci USA. 1995;92:11975–11979. doi: 10.1073/pnas.92.26.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas D C, Veaute X, Fuchs R P P, Kunkel T A. Frequency and fidelity of translesion synthesis of site-specific N-2-acetylaminofluorene adducts during DNA replication in a human cell extract. J Biol Chem. 1995;270:21226–21233. doi: 10.1074/jbc.270.36.21226. [DOI] [PubMed] [Google Scholar]
- 31.Thomas D C, Veaute X, Kunkel T A, Fuchs R P P. Mutagenic replication in human cell extracts of DNA containing site-specific N-2-acetylaminofluorene adducts. Proc Natl Acad Sci USA. 1994;91:7752–7756. doi: 10.1073/pnas.91.16.7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y C, Maher V M, McCormick J J. Xeroderma pigmentosum variant cells are less likely than normal cells to incorporate dAMP opposite photoproducts during replication of UV-irradiated plasmids. Proc Natl Acad Sci USA. 1991;88:7810–7814. doi: 10.1073/pnas.88.17.7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y C, Maher V M, Mitchell D L, McCormick J J. Evidence from mutation spectra that the UV hypermutability of xeroderma pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol Cell Biol. 1993;13:4276–4283. doi: 10.1128/mcb.13.7.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]