Abstract
The outbreak of the Coronavirus disease 2019 (COVID-19) caused the death of a large number of people and declared as a pandemic by the World Health Organization. Millions of people are infected by this virus and are still getting infected every day. As the cost and required time of conventional Reverse Transcription Polymerase Chain Reaction (RT-PCR) tests to detect COVID-19 is uneconomical and excessive, researchers are trying to use medical images such as X-ray and Computed Tomography (CT) images to detect this disease with the help of Artificial Intelligence (AI)-based systems, to assist in automating the scanning procedure. In this paper, we reviewed some of these newly emerging AI-based models that can detect COVID-19 from X-ray or CT of lung images. We collected information about available research resources and inspected a total of 80 papers till June 20, 2020. We explored and analyzed data sets, preprocessing techniques, segmentation methods, feature extraction, classification, and experimental results which can be helpful for finding future research directions in the domain of automatic diagnosis of COVID-19 disease using AI-based frameworks. It is also reflected that there is a scarcity of annotated medical images/data sets of COVID-19 affected people, which requires enhancing, segmentation in preprocessing, and domain adaptation in transfer learning for a model, producing an optimal result in model performance. This survey can be the starting point for a novice/beginner level researcher to work on COVID-19 classification.
Keywords: COVID-19, Deep learning, Medical image, Survey, AI, CT scan, X-ray
Introduction
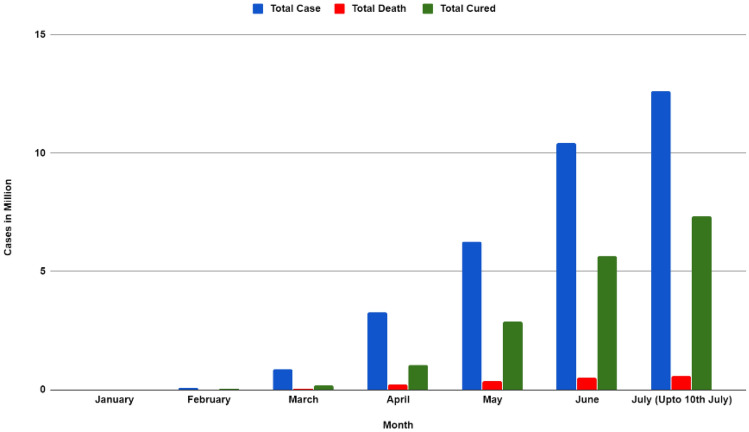
Coronavirus Disease 2019 (COVID-19) is an infectious disease that started to proliferate from Wuhan China, in December 2019 [1]. Within a short period of time, this disease is ravaged every corner of the world and the World Health Organization declared this disease as a pandemic on 11 March 2020 [2]. This disease is caused by the strain of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). In July 2020, cases reached almost 12 million worldwide, and death due to this disease kept rising day by day, and the death toll is 562,039 [3]. From the Worldometers data, the total deaths and total cures (based on month) is illustrated in Fig. 1 [3]. Observing the statistics and properties of COVID-19 it can be asserted that this life-threatening virus can unfurl from individual to individual via cough, sneezing, or even close contact. As a result, it has become important to detect the affected people earlier and isolate them to stop further spreading of this virus.
Fig. 1.
Total case, total death, and total cured (by month) from worldometer [4]
RT-PCR is a procedure of assembling samples from a region of a person’s body, where the coronavirus is most likely to congregate, such as a person’s nose or throat. Then this sample goes through a process called extraction, which separates the genetic data from any virus that may exist. A particular chemical along with a PCR machine (thermal cycler) is applied, which originates a reaction that creates millions of copies of SARS-CoV-2 virus’s genome. A beam of light is produced by one of the chemicals if SARS-COV-2 is present in the sample. The beam of light is traced by the PCR machine which indicates a positive test result for the existence of the coronavirus.
Though RT-PCR can distinctly identify coronavirus disease, it has a high false-negative rate, where the model predicts the result as negative but actually, it is positive (false-negative). Furthermore, in many regions of the world RT-PCR’s availability is limited. Hence, medical images such as Computer Tomography (CT) and X-ray images can be the next best alternative to detect this virus as most of the medical or hospital commonly have this apparatus to generate images. Also, CT or X-ray images are readily available, where there is no RT-PCR. Moreover, RT-PCR is expensive and consumes a considerable amount of time for the identification. In addition, proper training is required for the health workers to collect samples for PCR, whereas it is relatively easy to handle and produce CT and X-ray images.
To work on these medical images, deep learning methods are the most conventional and might be the only direction. Deep Learning is an emerging field that could play a significant role in the detection of COVID-19 in the future. Till now researchers have used machine learning/deep learning models to detect COVID-19 using medical images such as X-ray or CT images and obtained promising results. Many researchers also used transfer learning, attention mechanism [5], and Gradient-weighted Class Activation Mapping (Grad-CAM) [6] to make their results more accurate. Shi et al. [7] and Ilyas et al. [8] discussed some artificial intelligence-based models for diagnosis of COVID-19. In addition, Ulhaq et al. [9] reviewed some papers that worked on diagnosis, prevention, control, treatment, and clinical management of COVID-19. Besides, Ismael et al. [10] approached different types of Machine Learning and Deep Learning techniques COVID-19 detection working on X-ray images. Furthermore, a majority voting-based enseble classifier technique is employed by Chandra et al. [11]. However, as time goes by researchers are finding advanced and improved architectures for the diagnosis of COVID-19. In this paper, we have tried to review these new methods alongside with the basic structures of the earlier COVID-19 classification models. This survey will cover the research papers that are published or in pre-print format. Although it is not the most favorable approach due to the likelihood of below standard and research without peer-review, we intend to share all proposals and information in a single place while giving importance to the automatic diagnosis of COVID-19 in X-ray and CT images of lungs.
The fundamental aim of this paper is to systematically summarize the workflow of the existing researches, accumulate all the different sources of data sets of lung CT and X-ray images, sum up the frequently used methods to automatically diagnose COVID-19 using medical images so that a novice researcher can analyze previous works and find a better solution. We oriented our paper as follows:
First, the Data set source and different types of images used in the papers are described in “COVID-19 Dataset and Resouce Description”.
Second, the methodology where data preprocessing and augmentation techniques, feature extraction methods, classification, segmentation, and evaluation that researchers obtained are charactized in “Methodologies”.
Finally, a discussion is made to aid the new researcher to find future works in detecting COVID-19.
COVID-19 Data Set and Resouce Description
The diagnosis of any disease is like the light at the end of the tunnel. In the case of the COVID-19 pandemic, the importance of earlier diagnosis and detecting the disease is beyond measure. The initial focus must be on the data by which we need to efficiently train a model. This data will help Machine Learning (ML) or Deep Learning (DL) algorithms to diagnose COVID-19 cases. Due to the disadvantages of RT-PCR, researchers adopted an alternative method which is the use of Artificial Intelligence on chest CT or X-ray images to diagnose COVID-19. Fundamentally, a chest CT image is an image taken using the computed tomography (CT) scan procedure, where X-ray images are captured from different angles and compiled to form a single image. A depiction of the CT images (COVID-19 infected and Normal) is illustrated in Fig. 2.
Fig. 2.
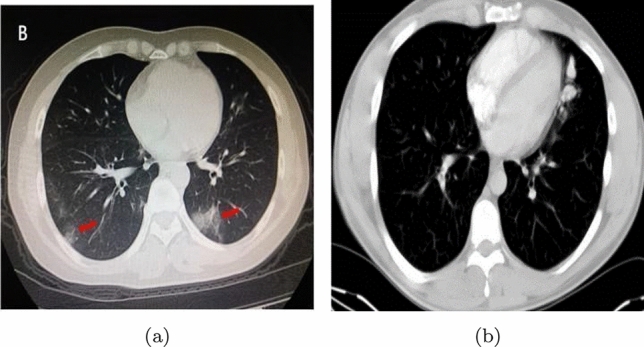
Lung CT-scan images a COVID-19 affected, b normal
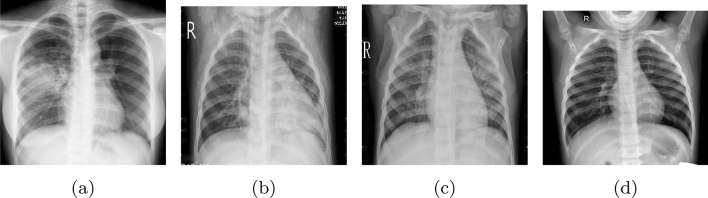
Although a CT scan consumes less time to demonstrate, it is fairly expensive. As a result, many researchers adopted X-ray images instead of CT images to develop a COVID-19 detection model. A chest X-ray is a procedure of using X-rays to generate images of the chest. In addition, it is relatively economical and convenient to maintain. X-ray images of different people with COVID-19, viral pneumonia, bacterial pneumonia, and a person without any disease (normal) are shown in Fig. 3. Furthermore, in this section, an overview of the data set sources used in the existing papers is characterized and data sets of both CT and X-ray images are illustrated and covered in this section.
Fig. 3.
X-ray images a COVID-19, b viral pneumonia, c bacterial pneumonia, d normal from COVID19-XRay-data set
Data Set and Its Sources
Nowadays, the exchange of information between researchers and physicians creates difficulties due to the lockdown phase. Hence, massive COVID-19 data are out of reach or difficult to find for many researchers. As a deep learning architecture needs a considerable number of images to learn a model appropriately and efficiently, the existing COVID-19 automation researches are still in preliminary stages. However, some COVID-19 data sets are proposed and employed by the researchers which show exceptional results in detecting the COVID-19 affected lungs. To corroborate a beginner researcher, we have accumulated the abstract information of the data sets and their sources. A list of the data set sources from February 2020 to June 2020 is embellished in Table 1. In the following, we will cover both CT and X-ray images and their fundamental attributes.
Table 1.
Summary of different data sources used in the papers
Some of the most popular data sets were collected from the following hospitals. Xu et al. [3] collected their data set from First Affiliated Hospital of Zhejiang University, the No. 6 People’s Hospital of Wenzhou, and the No. 1 People’s Hospital of Wenling. Song et al. [64] collected their data sets from three hospitals—Renmin Hospital of Wuhan University, and two affiliated hospitals (the Third Affiliated Hospital and Sun Yat-Sen Memorial Hospital) of the Sun Yat-sen University in Guangzhou. Chen et al. [73] built their data from the Renmin Hospital of Wuhan University (Wuhan, Hubei province, China). Shi et al. [75] built their data set from three hospitals Tongji Hospital of Huazhong University of Science and Technology, Shanghai Public Health Clinical Center of Fudan University, and China–Japan Union Hospital of Jilin University. Selvaraju et al. [6] used five different hospitals data to build their data set, including Beijing Tsinghua Changgung Hospital, Wuhan No. 7 Hospital, Zhongnan Hospital of Wuhan University, Tianyou Hospital Affiliated to Wuhan University of Science and Technology, and Wuhan’s Leishenshan Hospital. Zheng et al. [62] took data from Union Hospital, Tongji Medical College, Huazhong University of Science and Technology.
CT Image Sources
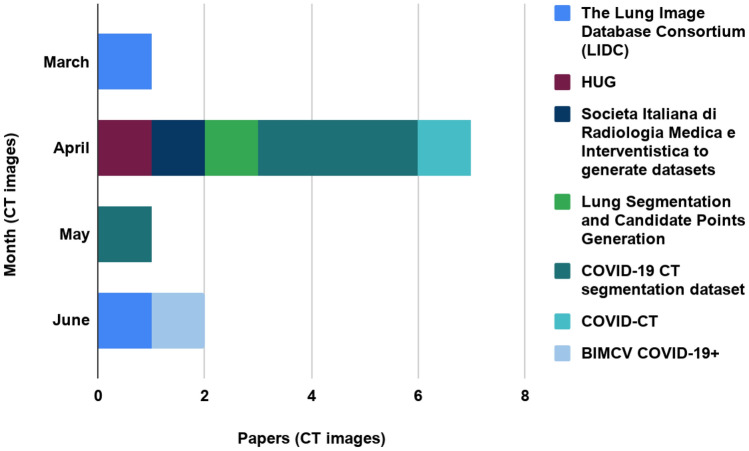
As CT images are said to be detailed than X-ray images, the diagnosis of COVID-19 and developing a model becomes more convenient by employing the CT-scan images. For CT images-based works, four papers used the COVID-19 CT segmentation data set to develop a classification architecture. This data set contains hundred axial CT images from forty patients [17–20]. Chen et al. [17] and Qiu et al. [19] achieved 89% and 83.62% accuracy, respectively, using this data set. Furthermore, two authors adopted the Lung Image Database Consortium (LIDC) data set and accomplished an accuracy above 90% [12, 13]. Besides these, some authors used Societa Italiana di Radiologia Medica e Interventistica to generate data sets [15], Lung Segmentation, and Candidate Points Generation [16], COVID-CT [21], and HUG data set [14] for their purpose. A representation of these data set sources is characterized in Table 1 and depicted in Fig. 4 (based on months). From the table, we can infer that the COVID-19 CT segmentation data set was used mostly in April 2020 and Lung Image Database Consortium (LIDC) data set was used in March 2020 and June 2020 [17–20]. Some researchers also used other lung disease images apart from these mostly used data sets. Nevertheless, they collected CT images from different hospitals to build these data sets.
Fig. 4.
Bar chart showing seven publicly available CT data sets used from March, 2020 to June, 2020
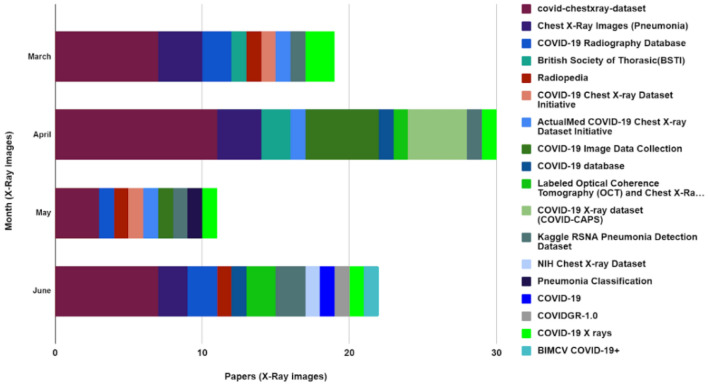
X-ray Image Sources
X-ray image data set is more available than the CT images as the cost of capturing an X-ray image is considerably more economical than a CT image. Studying the existing literature, most of the authors used the COVID-chest X-ray data set [32, 42, 47, 63]. Moreover, Kaggle RSNA Pneumonia Detection Data set [35, 45, 50], COVID-19 database [33, 52, 54], Chest X-ray Images (Pneumonia) is adopted to evaluate their model [27, 46, 59]. These are the most common data set for Chest X-ray-based COVID-19 research (Table 1). However, these data sets contain a limited number of COVID-19 infected lung images which is not efficient to train a deep learning model as the model can overfit the data. For this purpose, most of the researchers utilized different preprocessing techniques to increase the data set size, one of them is data augmentation. Furthermore, the existing works are trained on a hybrid data set combining the COVID-19 data set and normal lung images from another repository. For X-ray-based works, Al-antari et al. [44] used COVID-19 Radiography Database for alternative lung diseases. An illustration of the eighteen X-ray data set usage is depicted in Fig. 5. From there it can be noticed that the COVID-chest x-ray data set was used by most of the authors followed by Kaggle’s Chest X-ray images (Pneumonia) which was used mostly in March 2020, April 2020 and June 2020. Some papers also used both CT and X-ray images from the COVID-19 X-rays and BIMCV COVID-19+ data sets. From both Figs. 4 and 5, it can be observed that BIMCV COVID-19+ emerged in June 2020 in terms of developing a COVID-19 classification model.
Fig. 5.
Bar chart showing 18 publicly available X-ray data sets used from March, 2020 to June, 2020
Types and Properties of Images in the Data Set
Diseases such as Pneumonia, Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS), Influenza and Tuberculosis affect the lungs such as COVID-19 which can lead to misclassification of X-ray and CT images. To avoid this problem, researchers have adapted their data set to have images of diseases affecting similar regions as COVID-19. Moreover, it is important to correctly distinguish COVID-19 patients from people who do not have COVID-19. For this purpose, the authors also used normal lung images collected from healthy people. These data sets are developed by combining COVID-19 images, other lung disease images such as Viral pneumonia [3, 22, 23, 28, 49], Bacterial pneumonia [22, 23, 64–67], fungal pneumonia [68], SARS [60, 69–71], MERS [70], Influenza [13], Tuberculosis [65, 71, 72] and images of healthy people. The distribution of different types of lung disease or normal images and the number of CT images used by papers are illustrated in Table 2. There ‘Not specified (NS)’ indicates that papers used that type of image but did not state the number explicitly and ‘N/A’ indicates those types of images were not used.
Table 2.
Summary of different type of lung disease and normal (healthy patients) CT images used by papers
| Paper | Normal | Pneumonia | COVID-19 | Total images |
|---|---|---|---|---|
| [3] | 175 | 224 | 219 | 618 |
| [64] | 86 | 100 | 88 | 274 |
| [73] | N/A | 14,469 | 20,886 | 35,355 |
| [74] | 1325 | 1735 | 1296 | 4356 |
| [75] | N/A | 1027 | 1658 | 2685 |
| [76] | 91 | NS | 877 | 1418 |
| [12] | 100 | N/A | 106 | 206 |
| [68] | N/A | 40 | 1266 | 1306 |
| [65] | 28,873 | 30,345 | 14,944 | 89,628 |
| [31] | 153 | N/A | 203 | 356 |
| [14] | 1126 | N/A | 938 | 2064 |
| [15] | 6000 | N/A | 6000 | 12,000 |
| [16] | 247 | N/A | 178 | 425 |
| [77] | N/A | N/A | 150 | 150 |
| [78] | 75,541 | N/A | 64,771 | 140,312 |
| [79] | 397 | N/A | 349 | 746 |
| [80] | 195 | N/A | 275 | 470 |
| [21] | 339 | N/A | 391 | 730 |
| [18] | N/A | N/A | 373 | 373 |
| [81] | 495 | N/A | 449 | 944 |
| [19] | N/A | N/A | 100 | 100 |
| [82] | NS | 740 | 325 | 1065 |
| [56] | 344 | N/A | 439 | 783 |
| [83] | N/A | 1027 | 1495 | 2522 |
| [84] | 397 | N/A | 349 | 829 |
| [13] | 3308 | 2296 | 2228 | 7832 |
| [85] | 463 | N/A | 349 | 812 |
| Total | 120,438 | 50,268 | 121,700 | 308,359 |
Furthermore, the number of different types of CT images used in the papers is presented in Fig. 6. From there it can be seen that the number of COVID-19 CT images used for classification is 121,700. A total number of normal CT Images, Pneumonia CT Images, and Other lung disease CT images are 120,438, 50,268, and 15,999, respectively.
Fig. 6.
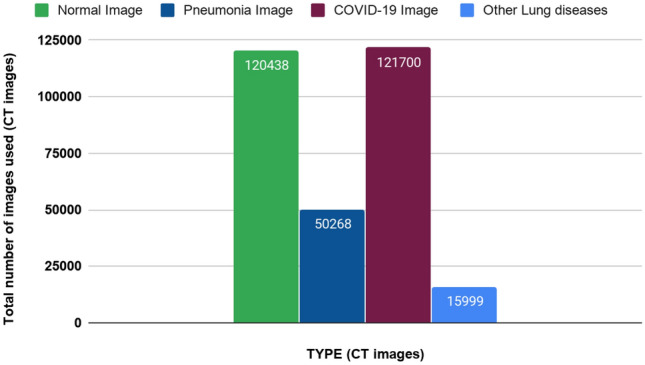
Total number of CT images of different diseases and normal CT images used from February 2020 to June 2020
In addition, the distribution of different types of X-ray images is depicted in Table 3, where the total number of different images used in fifty research works from February to June is represented. In Table 2, the distribution of different types of images was shown for thirty papers which were also from February 2020 to June 2020. Moreover, ‘Not specified (NS)’ and ‘N/A’ are used in Table 3 with the same purpose as it did in Table 2.
Table 3.
Summary of different type of lung disease and normal (healthy patients) X-ray images used by papers
| Paper | Normal | Pneumonia | COVID-19 | Total images |
|---|---|---|---|---|
| [63] | 25 | N/A | 25 | 50 |
| [27] | 50 | N/A | 50 | 100 |
| [28] | 504 | 700 | 224 | 1428 |
| [29] | N/A | 1431 | 100 | 1531 |
| [22] | 1583 | 4290 | 68 | 5941 |
| [49] | 1579 | 1485 | 423 | 3497 |
| [30] | 15 | N/A | 25 | 40 |
| [31] | 85 | N/A | 85 | 170 |
| [50] | N/A | 5551 | 358 | 5909 |
| [54] | 3450 | N/A | 455 | 3905 |
| [86] | 8066 | 5538 | 259 | 13,863 |
| [32] | 207 | N/A | 207 | 414 |
| [33] | N/A | 320 | 135 | 455 |
| [87] | 179 | 179 | 179 | 537 |
| [34] | 310 | 654 | 284 | 1248 |
| [66] | 7595 | 8792 | 313 | 16700 |
| [58] | N/A | N/A | 120 | 120 |
| [35] | 8851 | 6054 | 180 | 15085 |
| [88] | 2400 | 2600 | 536 | 5536 |
| [23] | 127 | 127 | 127 | 381 |
| [59] | 137 | N/A | 137 | 274 |
| [36] | 350 | 322 | 225 | 947 |
| [37] | 8066 | 5521 | 183 | 13,770 |
| [38] | 44,993 | 14,777 | 167 | 59,937 |
| [39] | 9039 | 2306 | 318 | 11,663 |
| [89] | 8851 | 6045 | 215 | 15,111 |
| [40] | 1591 | 4706 | 105 | 6402 |
| [41] | 191 | 131 | 180 | 502 |
| [53] | 1000 | 54 | 90 | 1144 |
| [90] | N/A | N/A | 462 | 462 |
| [51] | 8066 | 5551 | 358 | 13,975 |
| [69] | 80 | N/A | 105 | 196 |
| [24] | 1000 | 1000 | 239 | 2239 |
| [70] | N/A | N/A | 423 | 710 |
| [91] | 1579 | 2760 | 462 | 4801 |
| [42] | 15,000 | 15,000 | 99 | 30,099 |
| [43] | 8851 | 6045 | 386 | 15,282 |
| [71] | 1126 | NS | 1050 | 114,742 |
| [72] | 388 | 1000 | 500 | 2191 |
| [92] | 2880 | 5179 | 415 | 8474 |
| [25] | 1000 | 2000 | 309 | 3309 |
| [48] | 25 | N/A | 25 | 50 |
| [44] | N/A | 120 | 326 | 1312 |
| [60] | 3520 | 500 | 250 | 4281 |
| [67] | N/A | 1583 | 305 | 1888 |
| [52] | 13410 | 13,450 | 8640 | 35,500 |
| [45] | 554 | 554 | 154 | 1262 |
| [46] | 505 | 512 | 236 | 1253 |
| [61] | 377 | N/A | 377 | 754 |
| [47] | 668 | 619 | 132 | 1419 |
| Total | 168,223 | 127,456 | 21,062 | 430,679 |
A depiction of the total number of COVID-19, normal, Pneumonia, and other lung disease X-ray images used by papers is shown in Fig. 7. From the figure, it can be seen that 50 papers used 21,062 COVID-19 images, 168,223 normal images, 127,456 Pneumonia images, and 114,094 other lung disease images were used in total. It can be said that more CT images of COVID-19 were used than COVID-19 X-ray images by comparing Figs. 6 and 7.
Fig. 7.

Total number of X-ray images for different disease and normal patients used from February, 2020 to June, 2020
Methodologies
After data collection, several preeminent steps must be followed to diagnose COVID-19, hence this section depicts different techniques employed by different papers. First, preprocessing techniques along with their characteristics and properties is described. Second, feature extraction methods are thoroughly discussed. After that, segmentation methods and classification techniques are reviewed. Finally, the results obtained in the existing studied papers are briefly described. The workflow of diagnosing COVID-19 from X-ray images demonstrated in Fig. 8.
Fig. 8.
Fundamental architecture of diagnosing COVID-19 from X-ray images
Preprocessing Techniques
There is a high chance that a COVID-19 data set is built with some obscure, duplicate, blur, etc. images that degrade the performance of a model. Hence, it is necessary and mandatory to perform preprocessing techniques on redundant images. Various types of preprocessing techniques can be carried out based on the difficulties of the data set. One of the major problems of deep learning is overfitting. To minimize the effect of overfitting data augmentation is used in the pre-processing stage. Resizing, scaling, cropping, flipping, rotating are the most employed data augmentation techniques. Some of these data augmentation techniques are discussed below:
Resizing is necessary, because the images are not always within the same estimate which postures an issue, whereas preparing the model. To generalize the data set all the images are resized into a fixed dimension such as 224 224 or 299 299.
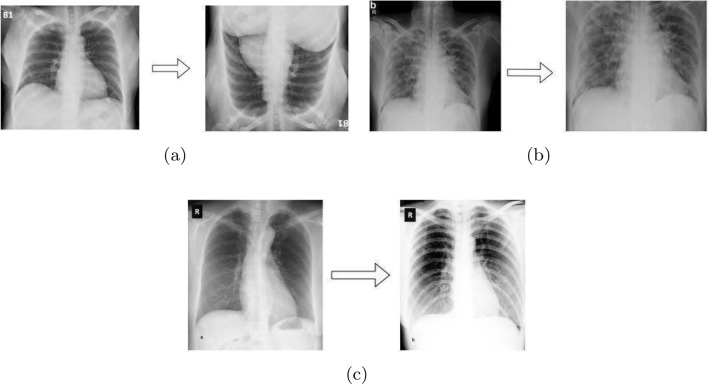
Flipping or Rotating is done to increase the sample size of the data sets. Mainly horizontal and vertical flipping is used to do this as depicted in Fig. 9a.
Scaling or Cropping is the next most used augmentation technique is scaling or cropping. All the portions of the images are not necessary to use. Therefore, to reduce the redundancy researchers used the cropping method as illustrated in Fig. 9b.
Brightness or Intensity adjusting is mandatory to increase or reduce the brightness of the images. An example is shown in Fig. 9c.
As the COVID-19 data set is built with an insufficient number of COVID infected images, Generative Adversarial Networks (GAN) can be employed to generate COVID affected lung images which can be a path to avoid overfitting or data insufficiency. GAN is an unsupervised learning process structured on generative modeling embedded with deep learning architectures. It finds the patterns, similarities in the input data sets and generates new data which is similar to the input data set. GAN [93] increases the sample size in the data set but the quality of the samples is not guaranteed.
Fig. 9.
Some examples of applying Pre-processing Techniques [a flipping by , b cropping, and c adjusting brightness]
A representation of the papers—applying augmentation techniques on their model is characterized in Table 4 and the percentage usage of these augmentation techniques is depicted in Fig. 10. From there it can be seen that resize and flipping has the highest percentage of 27.9% and 27.0%, respectively. Scaling or Cropping, Contrast Adjusting, Brightness Adjusting, and GAN is 22.1%, 12.3%, 7.4%, and 3.3%, respectively. Besides these techniques, some authors used various traditional image preprocessing techniques such as Histogram Equalization [70], Adaptive Winner Filter [80], Affine Transformation [29, 40], Histogram Enhancement [40], Color Jittering [29].
Table 4.
Summary of the preprocessing and augmentation methods used by the papers
| Paper | Resize | Flipping or rotating | Scaling or cropping | Contrast adjusting | Brightness or intensity adjusting | GAN |
|---|---|---|---|---|---|---|
| [3] | ||||||
| [14] | ||||||
| [15] | ||||||
| [17] | ||||||
| [18] | ||||||
| [17] | ||||||
| [21] | ||||||
| [22] | ||||||
| [21] | ||||||
| [22] | ||||||
| [25] | ||||||
| [27] | ||||||
| [28] | ||||||
| [29] | ||||||
| [30] | ||||||
| [31] | ||||||
| [32] | ||||||
| [31] | ||||||
| [34] | ||||||
| [35] | ||||||
| [35] | ||||||
| [37] | ||||||
| [41] | ||||||
| [42] | ||||||
| [43] | ||||||
| [42] | ||||||
| [46] | ||||||
| [48] | ||||||
| [49] | ||||||
| [50] | ||||||
| [49] | ||||||
| [52] | ||||||
| [51] | ||||||
| [54] | ||||||
| [54] | ||||||
| [59] | ||||||
| [60] | ||||||
| [60] | ||||||
| [63] | ||||||
| [62] | ||||||
| [66] | ||||||
| [67] | ||||||
| [68] | ||||||
| [68] | ||||||
| [69] | ||||||
| [72] | ||||||
| [71] | ||||||
| [74] | ||||||
| [77] | ||||||
| [76] | ||||||
| [77] | ||||||
| [78] | ||||||
| [80] | ||||||
| [82] | ||||||
| [84] | ||||||
| [85] | ||||||
| [86] | ||||||
| [87] | ||||||
| [88] | ||||||
| [89] | ||||||
| [92] | ||||||
| [92] |
Fig. 10.
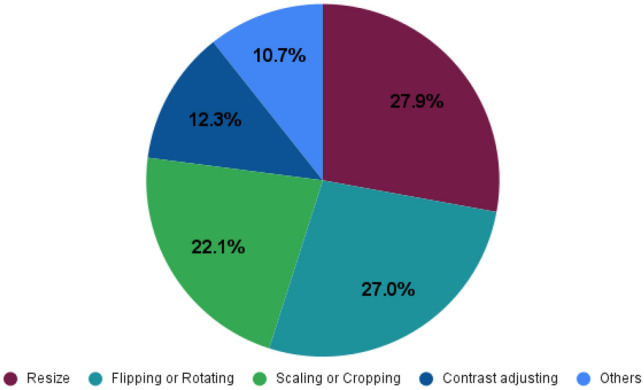
Pie chart illustrates the augmentation techniques used by different papers (Here the percentage of usage of six different augmentation techniques is shown)
Segmentation
It is necessary to train a model with the most significant features as unnecessary features or image region discredit the performance of the model. Therefore, extracting the Region of Interest (ROI) is the preeminent task before the training stage. For that purpose, segmentation comes into the hand as it can segregate the irrelevant and unnecessary regions of an image. In digital image processing and computer vision, image segmentation is defined as the technique of partitioning a digital image into different segments based on some pre-defined criteria, where a segment delineates as a set of pixels. Like other areas of medical image processing, segmentation boosts the effectiveness of COVID-19 detection by finding the ROI such as the lung region. Areas of the image that are redundant and not related to the significant feature area (out of the lung) could meddle the model performance. Using segmentation methods, only ROI areas are preserved which reduces this adverse effect of considering the out of the boundary features. Segmentation can be carried out manually by radiologists, but it takes a substantial amount of time. Several open-source automatic segmentation methods, such as region-based, edge-based, clustering, etc., are feasible to adopt. In the following, we will try to describe the prominent segmentation architecture and their properties.
The U-Net architecture is built with the help of Convolutional Neural Network (CNN) and it is modified such that it can achieve better segmentation in the domain of medical imaging [55]. The main advantage of U-Net is that the location information from the downsampling path and the contextual information in the upsampling path are combined to get general information—containing context and localization, which is the key to predicting a better segmentation map. U-Net-based strategies were utilized in [12–14, 17, 18, 38, 40, 61, 62, 66, 73, 74, 76, 77, 80, 81, 94, 95] for efficient and programmed lung segmentation extracting the lung region as the ROI.
For CT images, to keep contextual information between slices some researchers applied 3D versions of U-Net for lung segmentation named 3D U-Net ([3, 76]). Due to the low contrast at the infected areas in CT images and because of a large variety of both body shape, position over diverse patients, finding the infected areas from the chest CT scans was very challenging. Considering this issue, Narin et al. [27] developed a deep learning-based network named VB-Net. It is a modified 3D convolutional neural network based on V-Net [96]. In some other existing works, this segmentation method is adopted which alleviates the performance of the model [75, 83]. SegNet is also an efficient architecture for pixelwise denotation segmentation [97].
Segmentation methods, such as U-Net, Dense-Net, NABLA-N, SegNet, DeepLab, etc., were also used for the segmentation of lung images in different papers. The different segmentation methods used by different papers are illustrated in Table 5 and the number of papers in which a specific segmentation method is used is shown by a bar chart in Fig. 11.
Table 5.
Summary of different segmentation methods used in COVID-19 detection
Fig. 11.
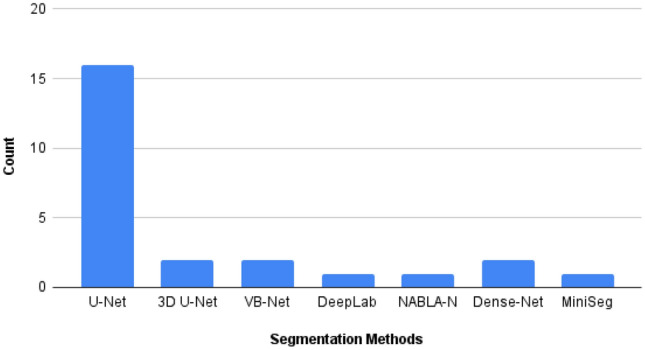
Bar chart showing number of times different segmentation models used in different papers
Feature Extraction Methods
Feature extraction is an essential step for classification as the extracted features provide useful characteristics of the images. For image feature extraction, Deep Neural Networks have extraordinary capabilities to extract the important features from a large-scale data set. As a result, these are used extensively in computer vision algorithms and CNN which is also known as ConvNet. In the following, some of the feature extraction models are briefly described.
Convolutional Neural Network (CNN)
In visual imagery fields, CNN architectures are mostly employed and adopted methods [100]. A CNN architecture is built with various types of network layer—pooling layer, convolutional layer, flatten, etc. corroborating the development and performance of a model.
Convolution layer is the core building block of a CNN. The layer’s parameters are made up of a set of discoverable kernels or filters which have a little responsive field but enlarge through the full input volume. Non-linear layer is the layer, where the change of the output is not proportional to the change of the input. This layer uses activation functions to convey non-linearity to data by adding after each convolution layer. Used activation functions can be Rectified Linear Unit (ReLU) [101], Tanh, etc.
Pooling layer is another important part of CNN architecture, where it is used to downsize the matrix. Pooling can be done in several methods: Max Pooling, Min Pooling, Average Pooling, and Mean Pooling. Fully connected layer is the layer, where every Neuron of a layer is connected with every other neuron of another layer. Traditional Multilayer Perceptron neural networks (MLP) and this layer have common principles.
Existing Pre-trained CNN Models
Most of the COVID-19 diagnosis architectures used various pre-trained CNN models. A representation of the usage of this pre-trained model is shown in Table 6 (CT images) and Table 7 (X-ray images). To work with CT images, Residual Network (ResNet) [102], Densely Connected Convolutional Network (DenseNet) [103], Visual Geometry Group (VGG) [104], SqueezeNet [49] architecture are the most adopted pre-trained architectures by researchers (Table 6) and ResNet [102], DenseNet [103], VGG [104], Inception [105] [106], InceptionResNet [107] models are employed for X-ray images (Table 7). Some of the most used existing pre-trained CNN models are described in the following.
Table 6.
Summary of image feature extraction methods used by different papers for CT images
Table 7.
Summary of image feature extraction methods used by different papers for X-ray images
| CNN | Papers | Count |
|---|---|---|
| ResNet | [22–24, 27, 29, 33, 35, 37, 39–43, 46, 49, 50, 57, 59, 61, 63, 69–72, 86, 88, 89, 91] | 27 |
| DenseNet | [23, 25, 32, 38, 40, 42, 46, 49, 59, 63, 66, 70, 72, 86, 87, 89, 91] | 17 |
| VGG | [23, 24, 28, 33, 37, 48, 49, 59, 60, 63, 66, 71, 72, 86, 92] | 15 |
| Inception | [23, 27, 28, 41, 46, 49, 52, 59, 63, 66, 70, 91] | 12 |
| InceptionResNet | [23, 27, 28, 42, 46, 59, 63, 66] | 8 |
| Xception | [23, 28, 34, 35, 47, 59, 63, 66] | 8 |
| AlexNet | [23, 31, 45, 57, 71, 72] | 6 |
| SqueezeNet | [49, 57, 70, 87, 88] | 5 |
| GoogLeNet | [23, 24, 57, 71] | 4 |
| NASnet | [25, 42, 59] | 3 |
| ShuffleNet | [23, 87] | 2 |
| EfficientNet | [37] | 1 |
| Simple CNN | [31] | 1 |
ResNet [102] is a CNN architecture, designed to enable hundreds or thousands of convolutional layers. While previous CNN architectures had a drop off in the effectiveness of additional layers, ResNet can efficiently add a large number of layers leads to strong performance as an outcome of the model. ResNet is convenient and efficient for data-driven approaches. It has different variants, such as ResNet18, ResNet169, ResNet50, ResNet152, etc. focusing on distinct perspectives. Moreover, studying the works we can infer that, ResNet is the most used architecture for both CT and X-ray- based COVID-19 research. Fourteen papers that have used ResNet in their proposed models for CT image-based works are shown in Table 6 and 27 papers are used for X-ray-based works are represented in Table 7.
DenseNet [103] is one of the current neural networks for visual object recognition. It is quite similar to the architecture ResNet but has some fundamental differences. This model ensures maximum information flow between the layers in the network that helps to extract the optimal features. By matching feature map size all over the network, the authors connected all the layers directly to all of their subsequent layers—a Densely Connected Neural Network, or simply known as DenseNet. DenseNet made strides in the data stream between layers by proposing these distinctive network designs. Unlike many other networks such as ResNet, DenseNets do not sum the output feature maps of the layer with the incoming feature maps but concatenate them. This architecture has different types of variants (DenseNet101, DenseNet169, DenseNet201, etc.) and it has an input shape of . In Table 6 (CT image), DenseNet architecture is used by four papers, and from Table 7 (X-ray image), it is used by seventeen papers.
VGG [104] is another important CNN architecture for the purpose of feature extraction. VGG Network consists of 16 or 19 convolutional layers and is very convenient to demonstrate because of its very uniform architecture . In our survey, we studied four papers who work with VGG for COVID-19 detection purposes to get the features from CT images that are illustrated in Table 6, and fifteen papers from X-ray-based works are shown in Table 7.
Inception [105, 106] is a transfer learning-based method consists of two segments: feature extraction from images with the help of CNN and classification with softmax and fully connected layers . Various versions of Inception architectures are used in the medical imaging field. Among these, InceptionV1, InceptionV2, InceptionV3, and InceptionV4 are the prominent ones with an input image shape of 299 x 299. Twelve papers used an Inception-based model for X-ray-based classification of COVID-19 given in Table 6 and only one paper [82] for CT images used this model to classify COVID-19 disease.
InceptionResnet [107] is the similar architecture as InceptionV4 . InceptionResNetv1 is a half breed Initiation adaptation that encompasses a similar computational fetched to Inceptionv3. InceptionResNetV2 is a convolutional neural arrangement that is prepared on more than a million pictures from the ImageNet [108] database. The arrangement is 164 layers profound and can classify images into 1000 distinct categories. Eight papers are given in Table 7 utilized this strategy for X-ray pictures including extraction.
The number of different CNN models used for CT Images is shown in Fig. 12 (based on month). For feature extraction from CT images, researchers used various types of CNN models from which ResNet is the most used architecture within these 5 months. In February 2020 three types of CNN models are used, two papers used ResNet, one paper used DenseNet, and another paper used VGG. During March 2020 four papers used ResNet, and AlexNet and FCN-8s used once. Whereas in April 2020 ResNet was used four times, three papers used VGG and SqueezeNet, CrNet, EfficientNet, GoogLeNet, and Inception are used once. Moreover, in May 2020 ResNet is used twice and SqueezeNet is used once, and finally, in June 2020 DenseNet and ResNet are used by one and two papers, respectively.
Fig. 12.
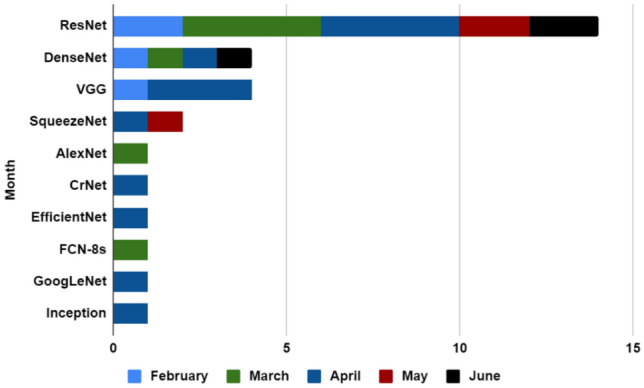
Bar chart for describing used CNN for CT images (by month)
Different CNN models and the number of times of its usage per month is shown for X-ray images in Fig. 13, where ResNet is one of the most used models for feature extraction. In our survey, during March 2020, ResNet is used six times, DenseNet two times, VGG three times, Inception four times, InceptionResNet three times, Xception two times, and AlexNet, SqueezeNet one times each. In April 2020 ResNet is used ten times, DenseNet eight times, VGG six times, Inception three times, InceptionResNet three times and Xception five times. AlexNet, GoogLeNet, and ShuffleNet are both used twice. SqueezeNet, Inception, and InceptionResNet are used three times each. NASnet and EfficientNet are used once each time. During May 2020 ResNet is used five times, DenseNet and Inception are used two times each, VGG, SqueezeNet, and GoogLeNet three of them are used only once. Finally, in June 2020 ResNet is used seven times, DenseNet and VGG both are used five times each. Inception, InceptionResNet, Xception, AlexNet, GoogLeNet, and NASnet are used three, two, one, three, one, and two times, respectively.
Fig. 13.
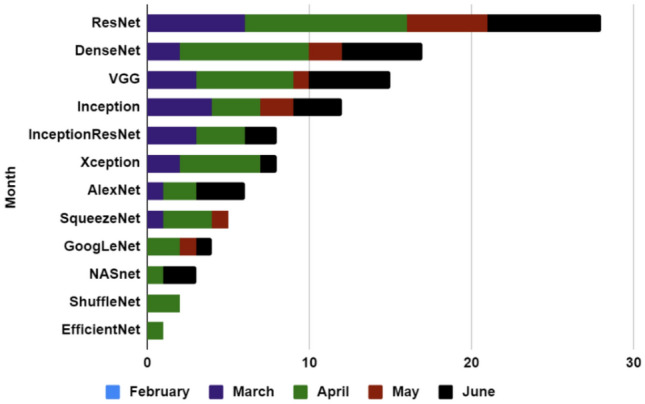
Bar chart describing the use of different CNN models for X-ray images (by month)
Specialized CNN Methods for COVID-19
Some researchers developed several architectures especially for COVID-19 detection with the backbone of basic CNN. These architectures have additional capabilities to classify images into multiple classes such as COVID-19, Viral pneumonia, Bacterial Pneumonia, and Normal case. Because in the primary stage, these models are trained on ImageNet, and then it is trained on various lung diseases CT or X-ray images. In the following, a brief discussion on the ensemble or specialized CNN methods to detect COVID-19 is described.
COVID-19 Detection Neural Network (COVNet) architecture was introduced by Li et al. [74] which is a 3D deep learning architecture to detect COVID-19. This architecture can extract both 2D local and 3D global illustrative features. The COVNet architecture is built with a ResNet architecture as the base model. A max-pooling operation is used for the feature extraction which is carried out for all slices of an image. Moreover, the feature map is connected with a fully connected layer and the author used a softmax activation function for the probability score to accurately classify multiple class (COVID-19, Community-Acquired Pneumonia (CAP), and non-pneumonia).
COVID-Net architecture is specially adapted for COVID-19 detection from chest X-ray images [51]. It has high architectural diversity and selective long-range connectivity. The massive use of a projection–expansion–projection design pattern in the COVID-Net architecture is also observed for the classification. COVID-Net architecture is incorporated into a heterogeneous association of convolution layers. The proposed COVID-Net is pre-trained on the ImageNet data set and then applied to the COVIDx data set. Applying this architecture, they achieved accuracy about 93.3% on the COVIDx data set.
ChexNet is originally a DenseNet-121 type of deep network which is trained on Chest X-ray images introduced by the paper [91]. Therefore, this architecture has been specially designed to diagnose COVID-19.1024-D feature vectors are extracted for the compact classifiers in ChexNet. They used the Softmax activation function to classify COVID-19, Normal, Viral Pneumonia, and Bacterial Pneumonia. The number of trainable parameters in this model is 6955,906.
COVID-CAPS is a capsule-based network architecture invented by Afshar et al. [58]. This model consists of four convolutional layers and three capsule layers. The primary layer is a convolutional layer, then batch-normalization is attached. The second layer is also a convolutional layer, followed by a pooling layer. Correspondingly, the third and fourth layers are convolutional, and the fourth layer is reshaped as the first capsule layer. Three Capsule layers are embedded in the COVID-CAPS to perform the routing. The last Capsule layer contains the classification parameters of the two classes of positive and negative COVID-19. The trainable parameters are 295,488 for this model and achieved 98.3%.
Detail-Oriented Capsule Networks (DECAPS) architecture is introduced by Mobiny et al. [21] which uses a ResNet with three residual blocks. This architecture is trained in CT images. This model obtained an area under the curve (AUC) of 98%. Besides these, some papers adopted different types of ensemble approaches such as Details Relation Extraction neural network (DRE-Net) [64]—ResNet-50 on Feature Pyramid Network [FPN] for extracting top K details from each image. Furthermore, an attention module is combined to learn the importance of every detail. In the training stage, [75] and [13] employed the Least Absolute Shrinkage and Selection Operator (LASSO) to traverse the optimal subset of clinical–radiological features for classification. Moreover, GLCM, HOG, and LBP are used by Sethy et al. [23]. In addition, Gozes et al. [12] used commercial off-the-shelf software that detects nodules and small opacities within a 3D lung volume and subsystem.
Besides some authors applied transfer learning approach [66, 86, 88] with the basic CNN models for better results. Basically, transfer learning is a technique for foretelling modeling on a different but somehow the same problem that can then be reused partially or fully to expedite the training and develop the performance of a model on the problem. In deep learning, transfer learning [109] means regenerating the weights in one or more layers from a pre-trained network architecture in a new model and either keeping the weights fixed, fine-tuning them, or adapting the weights entirely when training the model.
Interpretability
Fundamentally, a learning model consists of algorithms that try to learn patterns and relationships from the data source. To make the results obtained from machines interpretable, researchers use different techniques, such as Class Activation Mapping (CAM), Gradient-weighted Class Activation Mapping (Grad-CAM) based on a heatmap, Local Interpretable Model-agnostic Explanations (LIME) [110], and SHapley Additive exPlanations (SHAP) [111]. CAM is a method that creates heatmaps to show the important portions from the images, especially which regions are essential in terms of the Neural Network. CAM has various versions, such as Score CAM and Grad-CAM. The heatmap generated by CAM is a visualization that can be interpreted as where in the image the neural net is searching to make its decision. LIME tries to interpret models to guess the predictions of the predictive model in specific regions. LIME discovers the set of super pixels with the most coherent connection with the prediction label. It creates explanations by generating another data set of random disturbance by turning on and off a part of the super-pixels in the image. The aim of SHAP is to describe the forecast of a feature vector by calculating the contribution of distinct feature to the forecast. This is very important in image classification and object localization problems.
In our survey, there are few papers that utilized CAM [112] and few papers [12, 14, 22, 36, 38–42, 47, 56, 66, 67, 74, 86] utilized Grad-CAM with heatmap for better understanding of the region it is centering on. At the same time, heatmaps can also provide the radiologists with more useful information and further help them. In Papers [113, 114] LIME is used as one of the interpretable techniques to explain the outcome of different machine learning models for COVID-19 images. SHAP is used to visualize feature importance in [113, 115].
Classification
Almost all of the COVID-19 diagnosis models use Convolutional Neural Network [96] as a feature extractor and as a classifier, it uses softmax or sigmoid. Some authors also attempted to amplify CNN with a sigmoid layer. The authors of [45] merged CNN with the softmax layer along with the SVM classifier [116]. Kassani et al. [59] used CNN with softmax layer along a decision tree, random forest, XGBoost [117], AdaBoost [118], Bagging Classifier [119] and LightGBM [120]. Furthermore, the authors of Ahishali et al. [91] also merged CNN with KNN, support estimator network, and SVM classifier. Nonetheless, these models need a large amount of data for training which is in shortage of COVID-19 images.
Essentially there are two ways of classifying COVID-19 images, Binary Classification, and Multiclass classification. In Binary Classification authors tried to separate COVID-19 and non-COVID-19 patients, but this technique is very inaccurate as other types of lung diseases (viral pneumonia, viral pneumonia, bacterial pneumonia, and Community-Acquired Pneumonia) can be classified as COVID-19. For that reason, many authors differentiate COVID-19, viral pneumonia, bacterial pneumonia, community-Acquired Pneumonia, and normal images by classifying them using a softmax classifier. In terms of accuracy of detecting COVID-19 images, multiclass classifiers performed better than binary classifiers. A summary of different classification techniques used by different papers is illustrated in Tables 8 and 9.
Table 8.
Summary of classification methods used by different papers both for CT and X-ray images
| Classification methods | Papers | Count |
|---|---|---|
| Binary classification | [12, 15, 16, 21, 23, 25, 27, 28, 31–34, 38, 41, 42, 44, 46–49, 56–59, 61, 62, 68, 71, 72, 75, 77–79, 81–85, 87, 88, 90–92] | 43 |
| Multiclass classification | [3, 13, 14, 16, 17, 22, 24, 28, 29, 35–37, 39, 40, 42, 45, 49–54, 60, 63–66, 69, 70, 73, 74, 76, 79, 81, 84, 86, 89] | 37 |
Table 9.
Summary of classification methods used by different papers both for CT and X-ray images (monthwise)
| Months | Binary | Count | Multiclass | Count |
|---|---|---|---|---|
| February, 2020 | – | 0 | [3, 64] | 2 |
| March, 2020 | , [12, 27, 28, 31, 49, 62, 68] | 8 | [22, 28, 29, 49, 50, 63, 65, 73, 74, 76] | 10 |
| April, 2020 | [15, 16, 21, 23, 32–34, 38, 57–59, 77–79, 81, 82, 87, 88] | 19 | [14, 16, 17, 35, 54, 66, 79, 81, 86] [36, 37, 39, 89] | 13 |
| May, 2020 | [41, 83, 84, 90] | 4 | [24, 40, 51, 53, 69, 70, 84] | 7 |
| June, 2020 | [25, 42, 44, 46–48, 61, 71, 72, 85, 91, 92] | 12 | [13, 42, 45, 52, 60] | 5 |
Some authors also tried to detect COVID-19 in several stages. In the beginning, the authors separated normal and pneumonia images. After that, they classify COVID-19 by filtered pneumonia images. Several-stage classification helps the models to memorize various leveled connections. In paper [38, 40], authors used several-stage classification rather than an end to end method to detect COVID-19 which outperforms several the end to end techniques. On the flip side, the performance of multiclass classification relies on data sets. If there is a shortage of data set, the model cannot become familiar with the various leveled connections between classifications such as Pneumonia to Viral Pneumonia to COVID-19.
Experimental Results of the Papers
Researchers used different evaluation metrics to analyze their COVID-19 model’s performance. Among them, the most popular and used metrics for detecting COVID-19 are Accuracy, Precision, Recall/Sensitivity, F1 Score, Specificity, and Area Under Curve (AUC). In our work, we tried to record the performance with these metrics from all the papers which is represented in Table 10 for CT and in Table 11 for X-ray images. In addition, we have given the number of COVID-19 images from the total images used for training, testing, and validation purpose. Some papers explicitly stated the train-test split of COVID-19 images and for some papers, we calculated the split according to the ratio that is provided in the paper. Even so, for some papers, it is not clearly stated how they distributed their data set [16, 17, 20, 23, 75]. In addition, some papers explicitly stated the use of data for validation [27, 32, 33, 38, 40, 41, 44, 49, 52, 56, 63–65, 68, 70, 74, 81, 82, 84, 87, 89].
Table 10.
Summary of result evaluation for CT images
| Publication | Image | Training | Testing+ validation | Result (%) | Citation |
|---|---|---|---|---|---|
| February 21, 2020 [3] | 618 | 189 | 30+ N/A | Accuracy:86.70 | 117 |
| February 25, 2020 [64] | 274 | 79 | 25 + 9 | Accuracy:94.00 AUC:91.00 | 22 |
| March 01, 2020 [73] | 46096 | 691 | 636 + N/A | Accuracy:98.85Sensitivity:94.34Specificity:99.16 | 37 |
| March 19, 2020 [74] | 4356 | 1048 | 131 + 117 | Sensitivity:90.00 Specificity:96.16 AUC:96.00 | 172 |
| March 22, 2020 [75] | 2685 | NS | NS + NS | Accuracy:87.90 Sensitivity:90.70 Specificity:83.30AUC:94.20 | 24 |
| March 23, 2020 [76] | 1418 | 723 | 154 + N/A | Sensitivity:97.4 Specificity:92.2 AUC:99.10 | 13 |
| March 24, 2020[12] | 206 | 50 | 56 + N/A | Sensitivity:98.20 Specificity:92.20 AUC:99.60 | 87 |
| March 26, 2020 [68] | 5372 | 1266 | 102 + 92 | Accuracy:85.00 Sensitivity:79.35 Specificity:71.43 AUC:86.00 | 8 |
| March 26, 2020[62] | 630 | 289 | 76+ N/A | Accuracy:90.10Sensitivity:90.70 Specificity:91.10 | 35 |
| March 30, 2020[65] | 89628 | 7543 | 4887 + 2514 | Accuracy:98.80 Sensitivity: 98.20 Specificity: 98.90 | 2 |
| March 31, 2020[31] | 361 | 339 | 17+ N/A | Accuracy:94.10Sensitivity:90.00 Specificity:100.00 | 11 |
| April 06, 2020[14] | 2064 | 829 | 109 + N/A | Sensitivity:94.00 Specificity:98.00 AUC:99.40 | 11 |
| April 07, 2020 [15] | 12000 | 4500 | 1500 + N/A | Accuracy:98.93 Sensitivity:97.60 Specificity:97.63 | 5 |
| April 10, 2020 [16] | 420 | 375 | 45 + N/A | Accuracy:99.56 | 2 |
| April 12, 2020 [17] | 110 | NS | NS + 10-Fold | Accuracy:89.00 | 7 |
| April 14, 2020 [77] | 360 | 120 | 30 + N/A | Accuracy:89.20 Sensitivity:88.60 Specificity:87.60 AUC:92.30 | 4 |
| April 15, 2020 [78] | 79396 | 2794 | 64711 + N/A | Sensitivity:95.00 Specificity: 93.00 | 11 |
| April 17, 2020 [79] | 746 | 191 | 98 + 60 | Accuracy:83.00 AUC:87.00 | 4 |
| April 17, 2020 [80] | 470 | 165 | 110 + N/A | Accuracy:93.65 Sensitivity:94.25 Specificity:92.79 | 1 |
| April 17, 2020 [21] | 746 | 286 | 105 + N/A | Accuracy:87.60 Specificity:85.20 AUC:96.10 | 2 |
| April 21, 2020 [18] | 829 | 298 | 75 + NS | Sensitivity:86.70 Specificity:99.30 | 1 |
| April 21, 2020 [81] | 1044 | 349 | 50 + 50 | Accuracy:86.00 Sensitivity:94.00 Specificity:94.00 AUC:93.00 | 3 |
| April 21, 2020 [19] | 100 | 60 | 40 + N/A | Accuracy:83.62 Sensitivity:97.42 | 3 |
| April 24, 2020 [82] | 1065 | 340 | N/A + 290 | Accuracy:89.50 Sensitivity: 87.00 Specificity:88.00 | 77 |
| April 24, 2020 [56] | 783 | 251 | 108 + 80 | Accuracy:83.00Sensitivity:85.00 Specificity:81.73 | 2 |
| May 07, 2020 [83] | 2522 | 1196 | 299 +5 fold | Accuracy:97.79 Sensitivity:93.05 Specificity:89.95 AUC:96.35 | 1 |
| May 21, 2020 [20] | 130 | NS | NS + NS | Sensitivity:72.50Specificity:96.00 | 10 |
| May 28, 2020 [84] | 746 | 178 | 95 + 76 | Accuracy:99.40 Sensitivity:100.00 Specificity:98.60 AUC:99.65 | 0 |
| June 02, 2020 [13] | 4260 | 751 | NS + NS | Sensitivity:90.19 Specificity:95.76 AUC:97.17 | 12 |
| June 17, 2020 [85] | 812 | 349 | N/A + N/A | Accuracy:79.50AUC:90.10 | 35 |
| Total = 30 | Average Accuracy: 90.69Sensitivity:91.48 Specificity:92.26 AUC: 94.46 |
Table 11.
Summary of result evaluation for X-ray images
| Publication | Image | Training testing+ | Validation | Result (%) | Citation |
|---|---|---|---|---|---|
| March 24, 2020 [63] | 50 | 10 | 5+ 10 | Accuracy: 90.00 Sensitivity:100.00 Precision:100.00 F1 Score: 91.0 AUC:90.00 | 38 |
| March 24,2020 [27] | 100 | 40 | 10+ 8 | Accuracy:98.00 Sensitivity:96.00 Specificity:100.00 Precision:100.00 F1Score:98.00 | 93 |
| March 25, 2020 [28] | 1427 | 202 | 22 + N/A | Accuracy:96.78 Sensitivity:98.66 Specificity:96.46 | 86 |
| March 27, 2020 [29] | 1531 | 70 | 30 + N/A | Accuracy:96.00 AUC:95.18 | 35 |
| March 27, 2020 [22] | 5941 | 54 | 14+ N/A | Accuracy:89.82 | 19 |
| March 29, 2020 [49] | 3487 | 304 | 85 +34 | Accuracy:99.94 | 25 |
| March 30, 2020 [30] | 40 | N/A | N/A + N/A | Accuracy:97.48 Sensitivity: 95.27 Specificity:99.70 | 5 |
| March 31, 2020[31] | 170 | 120 | 50+ N/A | Accuracy:98.00 Sensitivity:100.00 Specificity:96.00 | 11 |
| March 31, 2020[50] | 13975 | N/A | N/A + N/A | Accuracy:96.23 Precision:100.00 F1 Score:100.00 | 25 |
| April 01, 2020 [54] | 3905 | 409 | 46 + N/A | Accuracy:99.18 Sensitivity:97.36 Specificity:99.42 | 9 |
| April 02, 2020 [57] | 5863 | N/A | N/A + N/A | Accuracy:99.00 Precision:98.97 Sensitivity:98.97 F1 Score:98.97 | 9 |
| April 09, 2020 [86] | 16995 | 181 | 78 + N/A | Accuracy:91.60 Sensitivity:92.45 Specificity:96.12 | 6 |
| April 09, 2020 [32] | 414 | 146 | 31 + 30 | Accuracy:98.00 | 1 |
| April 10, 2020 [16] | 5216 | NS | NS + NS | Accuracy:94.52 | 2 |
| April 13, 2020 [33] | 455 | 102 | 33 + 36 | Accuracy:91.24 AUC:94.00 | 12 |
| April 13, 2020 [87] | 537 | 125 | 36 + 18 | Accuracy:93.5 AUC:94.00 | 6 |
| April 14, 2020 [34] | 1300 | N/A | N/A + N/A | Accuracy:89.60 Precision:93.00 Sensitivity:98.20 | 8 |
| April 16, 2020 [66] | 16700 | 286 | 27 + N/A | Accuracy:99.01 AUC:99.72 | 4 |
| April 16, 2020[58] | 864 | 70 | 50 + N/A | Accuracy:95.70 Sensitivity:90.00 Specificity:95.80 AUC:97.00 | 19 |
| April 17,2020 [35] | 15085 | 149 | 31+ N/A | Accuracy:99.56 Sensitivity:80.53 | 3 |
| April 20,2020 [88] | 5071 | 31 | 40 + N/A | Sensitivity:97.50 Specificity:95.00 AUC:99.60 | 8 |
| April 22,2020 [23] | 381 | NS | NS + NS | Accuracy:95.33 Sensitivity:95.33 F1 Score:95.34 | 43 |
| April 22,2020 [59] | 274 | 137 | NS + N/A | Accuracy:99.00 | 3 |
| April 24,2020 [36] | 109203 | 180 | 45 + N/A | Accuracy:95.30 | 1 |
| April 28,2020 [37] | 13800 | 152 | 31 + N/A | Accuracy:93.90 | 5 |
| April 30,2020 [38] | 59937 | 89 | 35 + 43 | AUC:88.04 | 2 |
| April 30,2020 [39] | 11663 | 258 | 60 + N/A | Precision:98.15 Sensitivity:88.33 AUC:98.50F1 Score:92.98 | 1 |
| April 30,2020 [89] | 15111 | 175 | 20 +20 | Accuracy:89.40 | 0 |
| May 01,2020 [40] | 6297 | 105 | 10 + 10 | Accuracy:97.10 | 1 |
| May 05,2020 [41] | 502 | 126 | 36 + 18 | Accuracy:88.90 Specificity:96.40 F1 Score:84.40 | 9 |
| May 06,2020 [53] | 1144 | 63 | 27 + N/A | F1 Score: 89.60 | 8 |
| May 08,2020 [90] | 6286 | 370 | 92+ 5-fold | Accuracy:95.90 Sensitivity:98.50 Specificity:95.70 | 3 |
| May 11,2020 [51] | 13975 | 258 | 100 + N/A | Accuracy:93.30 Sensitivity:91.00 F1 Score:98.90 | 128 |
| May 17,2020 [69] | 196 | 74 | 31 + N/A | Accuracy:95.12 | 18 |
| May 21,2020 [24] | 2239 | 191 | 48 + N/A | Accuracy:97.01 | 4 |
| May 23,2020 [70] | 701 | 270 | 85 + 68 | Accuracy:97.73 | 0 |
| June 07,20[91] | 5824 | 4659 | 1165 + N/A | Accuracy:99.49 Sensitivity:99.43 Specificity:99.81 | 0 |
| June 18,2020[42] | 30099 | 88 | 11 + N/A | Accuracy:98.00 AUC:99.00 | 2 |
| June 16,2020 [43] | 15282 | 286 | 100 + N/A | Accuracy:98.06 | 0 |
| June 23,2020 [71] | 51960 | 736 | 314 + N/A | Accuracy:97.54 Sensitivity:97.88 Specificity:97.15 | 0 |
| June 23,2020 [72] | 2071 | 350 | 150 + N/A | Accuracy:98.90 | 0 |
| June 11, 2020 [92] | 8474 | 7626 | 848 + N/A | Accuracy:98.60 | 0 |
| June 18, 2020 [25] | 3300 | 247 | 62 + N/A | Sensitivity:100.00 Specificity:99.50 | 0 |
| May June [48] | 50 | 20 | 5 + N/A | Accuracy:91.00 Sensitivity:100.00 Specificity:80.00 | 0 |
| June 19,2020 [44] | 1312 | 228 | 65 + 33 | Accuracy:97.40 Sensitivity:85.15 Specificity:99.05 | 0 |
| June 09,2020 [60] | 6523 | N/A | N/A + N/A | Accuracy:98.00 Sensitivity:96.00 Specificity:98.00 | 0 |
| June 20,2020 [67] | 305 | N/A | N/A + N/A | Accuracy:97.40 | 0 |
| June 18,2020 [52] | 35500 | 7340 | 7340 + 500 | Accuracy:98.00 | 0 |
| June 09,2020 [45] | 1262 | 100 | 54 + N/A | Sensitivity:90.74 Specificity:95.57 | 1 |
| June 10,2020 [46] | 1302 | 142 | 94 + N/A | Precision:88.90 Sensitivity: 85.10 | 0 |
| June 02,2020 [61] | 754 | 302 | 75 + 5 fold | Accuracy:97.37 | 0 |
| June 08,2020 [47] | 1419 | 106 | 26+ k fold | Accuracy:98.94 Sensitivity:96.00 | 0 |
| Total = 52 | Average Accuracy:96.00 Sensitivity:91.09 Specificity:96.45 AUC:95.50 |
A summary of the results obtained by the studied models using CT images is illustrated in Table 10. These papers with their Accuracy, AUC, Sensitivity, and Specificity are given along with their distribution of COVID-19 images in training, testing, and validation set. It can also be observed that CT image-based models gained a minimum accuracy of 79.50% for the paper [85] and maximum accuracy of 99.56% for the paper [16].
A summary of the results obtained by the studied models using X-ray images is illustrated in Table 11. Papers with their Accuracy, AUC, Sensitivity, and Specificity are given along with their distribution of COVID-19 images in training, testing, and validation set. It can also be seen that X-ray image-based models gained a minimum accuracy of 89.82% for the paper [22] and maximum accuracy of 99.94% for the paper [16]. For both Tables 10 and 11, the publication date, the total number of images used by the respective papers is provided. In addition, cited by (Number of papers) indicates the total number of papers that have cited the specific paper up to July 10, 2020.
After analyzing all the papers it can be seen that most models can not accurately distinguish between Pneumonia and COVID-19 from the medical images. All the papers mentioned only focused on medical images but did not consider features, such as initial symptoms, travel history, laboratory assessment, contact history, and distinction between severe and mild COVID-19 [3, 68, 75, 86]. Most papers worked on the type of data sets that are not balanced containing more COVID-19 negative images, hence paper [58] suggested the use of modified loss function to tackle imbalanced data.
A single CNN network is not able to obtain higher dimensional fraternity features that are the decider for the classification. Whereas, modern pre-trained CNN models were fused to obtain higher dimensional fusion features which overcome the problem of insufficient features from a single CNN network model [15]. Due to this most authors used pre-trained models instead of single CNN models to detect COVID-19 from medical images.The main advantage of using pre-trained models such as Inception, ResNet and DenseNet is that they all have strong power of details extraction, but the problem with these models is that they fallaciously focus on some image edges, corners and other image areas that are not related to COVID-19 as these models are pre-trained with non-medical images [79]. Another drawback of using CNN-based models is that these models work like a black box giving no intuitions into the important image features. These methods lack transparency and interpretability [74, 86]. Moreover, most pre-trained models require a lot of time to train due to the immense number of parameters, but Polsinelli et al. [56] used SqueezeNet to solve this problem , a light weight model that reaches similar accuracy to modern CNN models.
Comparing Tables 10 and 11, it can be said that the X-ray image-based models performed better than the CT image-based models. The average Accuracy, Sensitivity, Specificity, and AUC for CT Image-based models are 90.69 %, 91.48%, 92.26%, and 94.46%, respectively, and for X-ray-based models are 96.00%, 91.09%, 96.45%, and 95.50%, respectively.
Conclusion
As COVID-19 is spreading worldwide expeditiously, accurate and faster detection of the disease has become the most essential objective to defend this outbreak. In this article, we tried to present a comprehensive survey of AI-empowered methods that use medical images to combat this pandemic. The fundamental purpose of this survey is to represent the current information so that researchers understand and aware of the up-to-date knowledge and build a model that can accurately detect COVID-19 disease at an economical cost and relatively faster in time. We surveyed a total of 80 COVID-19 diagnosis architectures among which 28 are using CT images, 50 are using X-ray images and 2 are using both CT and X-ray images. Till now none of these models are proved to be as reliable to replace RT-PCR tests and still, researchers are trying to improve these techniques. From our survey, it is noticeable that the X-ray image data set is more widely available than the CT Image data set as a CT scan procedure is costlier and more time-consuming than an X-ray. Therefore, most of the researchers utilized Chest X-ray images for diagnosing COVID-19. After analyzing the existing research works in this domain, we find out that there exists a shortage of annotated medical images of COVID-19 affected people. Enriching quality annotated medical images of COVID-19 affected people can play a significant role to boost up the performance of the mentioned data-hungry models. Furthermore, we remarked that using segmentation as preprocessing has an extensive impact on model performance. We also observed that domain adoption in transfer learning is the widely used technique which gives a promising result. Furthermore, many researchers used Gradient-weighted Class Activation Mapping (Grad-CAM) with heatmap to interpret the performance of the model. Though this survey paper cannot claim to be an in-depth think about those studies, it presents a practical outlook and shows a valid comparison of the researches in this field over these months which can be the conductor for the researcher to find future direction.
Funding
This study was not funded by any funding agency or anybody.
Declarations
Conflict of Interest
All of the authors declare that he/she has no conflict of interest.
Availability of Data and Materials
All the data are collected and cited from published sources.
Code Availability
This a survey paper. We have not conduct any experimental research.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of the topical collection “Computer Aided Methods to Combat COVID-19 Pandemic” guest edited by David Clifton, Matthew Brown, Yuan-Ting Zhang and Tapabrata Chakraborty.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, Xie C, Ma K, Shang K, Wang W et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed]
- 2.Who director-general’s opening remarks at the media briefing on COVID-19—11 march 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020. Accessed 11 Mar 2020.
- 3.Xu X, Jiang X, Ma C, Du P, Li X, Lv S, Yu L, Ni Q, Chen Y, Su J et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020. [DOI] [PMC free article] [PubMed]
- 4.Worldometer: Coronavirus cases. https://www.worldometers.info/coronavirus/. Accessed 11 Mar 2020.
- 5.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need. Adv Neural Inf Process Syst. 2017;5998–6008.
- 6.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-cam: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision, 2017, pp. 618–626.
- 7.Shi F, Wang J, Shi J, Wu Z, Wang Q, Tang Z, He K, Shi Y, Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev Biomed Eng. 2020. [DOI] [PubMed]
- 8.Ilyas M, Rehman H, Naït-Ali A. Detection of COVID-19 from chest x-ray images using artificial intelligence: an early review. arXiv preprint. 2020. arXiv:2004.05436.
- 9.Ulhaq A, Khan A, Gomes D, Pau M. Computer vision for COVID-19 control: a survey. arXiv preprint. 2020. arXiv:2004.09420. [DOI] [PMC free article] [PubMed]
- 10.Ismael AM, Şengür A. Deep learning approaches for COVID-19 detection based on chest x-ray images. Expert Syst Appl. 2021;164:114054. doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandra TB, Verma K, Singh BK, Jain D, Netam SS. Coronavirus disease (COVID-19) detection in chest x-ray images using majority voting based classifier ensemble. Expert Syst Appl. 2021;165:113909. doi: 10.1016/j.eswa.2020.113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gozes O, Frid-Adar M, Greenspan H, Browning PD, Zhang H, Ji W, Bernheim A, Siegel E. Rapid ai development cycle for the coronavirus (COVID-19) pandemic: initial results for automated detection and patient monitoring using deep learning CT image analysis. arXiv preprint. 2020. arXiv:2003.05037.
- 13.Jin C, Chen W, Cao Y, Xu Z, Zhang X, Deng L, Zheng C, Zhou J, Shi H, Feng J. Development and evaluation of an AI system for COVID-19 diagnosis. medRxiv. 2020 doi: 10.1101/2020.03.20.20039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gozes O, Frid-Adar M, Sagie N, Zhang H, Ji W, Greenspan H. Coronavirus detection and analysis on chest ct with deep learning. arXiv preprint. 2020. arXiv:2004.02640.
- 15.Ozkaya U, Ozturk S, Barstugan M. Coronavirus (COVID-19) classification using deep features fusion and ranking technique. arXiv preprint. 2020. arXiv:2004.03698.
- 16.Alom MZ, Rahman M, Nasrin MS, Taha, TM, Asari VK. Covid\_mtnet: Covid-19 detection with multi-task deep learning approaches. arXiv preprint. 2020. arXiv:2004.03747.
- 17.Chen X, Yao L, Zhang Y. Residual attention u-net for automated multi-class segmentation of COVID-19 chest CT images. arXiv preprint. 2020. arXiv:2004.05645.
- 18.Zhou T, Canu S, Ruan S. An automatic COVID-19 CT segmentation based on u-net with attention mechanism. arXiv preprint. 2020. arXiv:2004.06673. [DOI] [PMC free article] [PubMed]
- 19.Qiu Y, Liu Y, Xu J. Miniseg: An extremely minimum network for efficient COVID-19 segmentation. arXiv preprint. 2020. arXiv:2004.09750. [DOI] [PubMed]
- 20.Fan D-P, Zhou T, Ji G-P, Zhou Y, Chen G, Fu H, Shen J, Shao L. Inf-net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans Med Imaging. 2020. [DOI] [PubMed]
- 21.Mobiny A, Cicalese PA, Zare S, Yuan P, Abavisani M, Wu CC, Ahuja J, de Groot PM, Van Nguyen H. Radiologist-level COVID-19 detection using CT scans with detail-oriented capsule networks. arXiv preprint. 2020. arXiv:2004.07407.
- 22.Ghoshal B, Tucker A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv preprint. 2020. arXiv:2003.10769.
- 23.Sethy PK, Behera SK. Detection of coronavirus disease (COVID-19) based on deep features. Preprints. 2020;2020030300:2020. [Google Scholar]
- 24.Li T, Han Z, Wei B, Zheng Y, Hong Y, Cong J. Robust screening of COVID-19 from chest x-ray via discriminative cost-sensitive learning. arXiv preprint. 2020. arXiv:2004.12592.
- 25.Boudrioua MS. COVID-19 detection from chest x-ray images using cnns models: further evidence from deep transfer learning. Available at SSRN 3630150, 2020.
- 26.de la Iglesia Vayá M, Saborit JM, Montell JA, Pertusa A, Bustos A, Cazorla M, Galant J, Barber X, Orozco-Beltrán D, García-García F, et al. Bimcv COVID-19+: a large annotated dataset of RX and CT images from COVID-19 patients. arXiv preprint. 2020. arXiv:2006.01174.
- 27.Narin A, Kaya C, Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using x-ray images and deep convolutional neural networks. arXiv preprint. 2020. arXiv:2003.10849. [DOI] [PMC free article] [PubMed]
- 28.Apostolopoulos ID and Mpesiana TA. COVID-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;1. [DOI] [PMC free article] [PubMed]
- 29.Zhang J, Xie Y, Li Y, Shen C, Xia Y. COVID-19 screening on chest x-ray images using deep learning based anomaly detection. arXiv preprint. 2020. arXiv:2003.12338.
- 30.Hassanien AE, Mahdy LN, Ezzat KA, Elmousalami HH, and Ella HA. Automatic x-ray COVID-19 lung image classification system based on multi-level thresholding and support vector machine. medRxiv. 2020.
- 31.Maghdid HS, Asaad AT, Ghafoor KZ, Sadiq AS, and Khan MK. Diagnosing COVID-19 pneumonia from x-ray and CT images using deep learning and transfer learning algorithms. arXiv preprint. 2020. arXiv:2004.00038.
- 32.Ezzat D, Ella HA, et al. Gsa-densenet121-COVID-19: a hybrid deep learning architecture for the diagnosis of COVID-19 disease based on gravitational search optimization algorithm. arXiv preprint. 2020. arXiv:2004.05084. [DOI] [PMC free article] [PubMed]
- 33.Hall LO, Paul R, Goldgof DB, Goldgof GM. Finding COVID-19 from chest x-rays using deep learning on a small dataset. arXiv preprint. 2020. arXiv:2004.02060.
- 34.Khan AI, Shah JL, and Bhat MM. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Programs Biomed.2020;p. 105581. [DOI] [PMC free article] [PubMed]
- 35.Rahimzadeh M and Attar A. A new modified deep convolutional neural network for detecting COVID-19 from x-ray images. arXiv preprint. 2020. arXiv:2004.08052. [DOI] [PMC free article] [PubMed]
- 36.Basu S and Mitra S. Deep learning for screening COVID-19 using chest x-ray images. arXiv preprint. 2020. arXiv:2004.10507.
- 37.Luz EJS, Silva PL, Silva R, Silva L, Moreira G, and Menotti D. Towards an effective and efficient deep learning model for COVID-19 patterns detection in x-ray images. CoRR. 2020.
- 38.Yeh C-F, Cheng H-T, Wei A, Liu K-C, Ko M-C, Kuo P-C, Chen R-J, Lee P-C, Chuang J-H, Chen C-M, et al. A cascaded learning strategy for robust COVID-19 pneumonia chest x-ray screening. arXiv preprint. 2020. arXiv:2004.12786.
- 39.Zhang Y, Niu S, Qiu Z, Wei Y, Zhao P, Yao J, Huang J, Wu Q, Tan M. Covid-da: Deep domain adaptation from typical pneumonia to COVID-19. arXiv preprint. 2020. arXiv:2005.01577.
- 40.Lv D, Qi W, Li Y, Sun L, Wang Y. A cascade network for detecting COVID-19 using chest x-rays. arXiv preprint. 2020. arXiv:2005.01468.
- 41.Oh Y, Park S, and Ye JC. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans Med Imaging. 2020. [DOI] [PubMed]
- 42.Punn NS and Agarwal S. Automated diagnosis of COVID-19 with limited posteroanterior chest x-ray images using fine-tuned deep neural networks. arXiv preprint. 2020. arXiv:2004.11676. [DOI] [PMC free article] [PubMed]
- 43.Liu B, Yan B, Zhou Y, Yang Y, and Zhang Y. Experiments of federated learning for COVID-19 chest x-ray images. arXiv preprint. 2020. arXiv:2007.05592.
- 44.Al-antari MA, Hua C-H, Lee S. Fast deep learning computer-aided diagnosis against the novel COVID-19 pandemic from digital chest x-ray images. 2020. [DOI] [PMC free article] [PubMed]
- 45.Salih SQ, Abdulla HK, Ahmed ZS, Surameery NMS, and Rashid RD. Modified alexnet convolution neural network for COVID-19 detection using chest x-ray images. Kurdistan J Appl Res. 2020;119–130.
- 46.Chatterjee S, Saad F, Sarasaen C, Ghosh S, Khatun R, Radeva P, Rose G, Stober S, Speck O, and Nürnberger A. Exploration of interpretability techniques for deep COVID-19 classification using chest x-ray images. arXiv preprint. 2020. arXiv:2006.02570. [DOI] [PMC free article] [PubMed]
- 47.Singh KK, Siddhartha M, Singh A. Diagnosis of coronavirus disease (COVID-19) from chest x-ray images using modified xceptionnet. Romanian J Inf Sci Technol. 2020;23:S91–S105. [Google Scholar]
- 48.Manapure P, Likhar K, and Kosare H. Detecting COVID-19 in x-ray images with Keras, tensor flow, and deep learning. Assessment. 2(3).
- 49.Chowdhury ME, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Al-Emadi N, et al. Can AI help in screening viral and COVID-19 pneumonia? arXiv preprint. 2020. arXiv:2003.13145.
- 50.Farooq M and Hafeez A. COVID-resnet: a deep learning framework for screening of covid19 from radiographs. arXiv preprint. 2020. arXiv:2003.14395.
- 51.Wang L and Wong A. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images. arXiv preprint. 2020. arXiv:2003.09871. [DOI] [PMC free article] [PubMed]
- 52.Asif S, Wenhui Y, Jin H, Tao Y, Jinhai S. Classification of COVID-19 from chest x-ray images using deep convolutional neural networks. medRxiv. 2020.
- 53.Pereira RM, Bertolini D, Teixeira LO, Silla CN Jr, Costa YM. COVID-19 identification in chest x-ray images on flat and hierarchical classification scenarios. Comput Methods Programs Biomed.. 2020. p. 105532. [DOI] [PMC free article] [PubMed]
- 54.Apostolopoulos ID, Aznaouridis SI, Tzani MA. Extracting possibly representative COVID-19 biomarkers from x-ray images with deep learning approach and image data related to pulmonary diseases. J Med Biol Eng. 2020. 1. [DOI] [PMC free article] [PubMed]
- 55.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention. Springer, 2015, pp. 234–241.
- 56.Polsinelli M, Cinque L and Placidi G. A light cnn for detecting COVID-19 from CT scans of the chest. arXiv preprint. 2020. arXiv:2004.12837. [DOI] [PMC free article] [PubMed]
- 57.Khalifa NEM, Taha MHN, Hassanien AE and Elghamrawy S. Detection of coronavirus (COVID-19) associated pneumonia based on generative adversarial networks and a fine-tuned deep transfer learning model using chest x-ray dataset. arXiv preprint. 2020. arXiv:2004.01184.
- 58.Afshar P, Heidarian S, Naderkhani F, Oikonomou A, Plataniotis KN and Mohammadi A. Covid-caps: a capsule network-based framework for identification of COVID-19 cases from x-ray images. arXiv preprint. 2020. arXiv:2004.02696. [DOI] [PMC free article] [PubMed]
- 59.Kassani SH, Kassasni PH, Wesolowski MJ, Schneider KA and Deters R. Automatic detection of coronavirus disease (COVID-19) in x-ray and CT images: a machine learning-based approach. arXiv preprint. 2020. arXiv:2004.10641. [DOI] [PMC free article] [PubMed]
- 60.Brunese L, Mercaldo F, Reginelli A and Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from x-rays. Comput Methods Programs Biomed. 2020; p. 105608. [DOI] [PMC free article] [PubMed]
- 61.Tabik S, Gómez-Ríos A, Martín-Rodríguez J, Sevillano-García I, Rey-Area M, Charte D, Guirado E, Suárez J, Luengo J, Valero-González M, et al. Covidgr dataset and COVID-sdnet methodology for predicting covid-19 based on chest x-ray images. arXiv preprint. 2020. arXiv:2006.01409. [DOI] [PMC free article] [PubMed]
- 62.Zheng C, Deng X, Fu Q, Zhou Q, Feng J, Ma H, Liu W and Wang X. Deep learning-based detection for COVID-19 from chest CT using weak label. medRxiv. 2020.
- 63.Hemdan EE-D, Shouman MA and Karar ME. Covidx-net: a framework of deep learning classifiers to diagnose COVID-19 in x-ray images. 2020. arXiv preprintarXiv:2003.11055.
- 64.Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, Chen J, Zhao H, Jie Y, Wang R, et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 65.Fu M, Yi S-L, Zeng Y, Ye F, Li Y, Dong X, Ren Y-D, Luo L, Pan J-S and Zhang Q. Deep learning-based recognizing COVID-19 and other common infectious diseases of the lung by chest ct scan images. medRxiv. 2020.
- 66.Rajaraman S, Siegelman J, Alderson PO, Folio LS, Folio LR and Antani SK. Iteratively pruned deep learning ensembles for COVID-19 detection in chest x-rays. arXiv preprint. 2020. arXiv:2004.08379. [DOI] [PMC free article] [PubMed]
- 67.Mahmud T, Rahman MA and Fattah SA. Covxnet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest x-ray images with transferable multi-receptive feature optimization. Comput Biol Med. 2020;p. 103869. [DOI] [PMC free article] [PubMed]
- 68.Wang S, Zha Y, Li W, Wu Q, Li X, Niu M, Wang M, Qiu X, Li H, Yu H, et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur Respir J. 2020. [DOI] [PMC free article] [PubMed]
- 69.Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID-19 in chest x-ray images using detrac deep convolutional neural network. arXiv preprint. 2020. arXiv:2003.13815. [DOI] [PMC free article] [PubMed]
- 70.Tahir A, Qiblawey Y, Khandakar A, Rahman T, Khurshid U, Musharavati F, Kiranyaz S, and Chowdhury ME. Coronavirus: comparing COVID-19, sars and mers in the eyes of AI. arXiv preprint. 2020. arXiv:2005.11524.
- 71.Abbas A, Abdelsamea MM and Gaber M. 4s-dt: self supervised super sample decomposition for transfer learning with application to COVID-19 detection. arXiv preprint. 2020. arXiv:2007.11450. [DOI] [PMC free article] [PubMed]
- 72.Shelke A, Inamdar M, Shah V, Tiwari A, Hussain A, Chafekar T and Mehendale N. Chest x-ray classification using deep learning for automated COVID-19 screening. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 73.Chen J, Wu L, Zhang J, Zhang L, Gong D, Zhao Y, Hu S, Wang Y, Hu X, Zheng B, et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. MedRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 74.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020. [DOI] [PMC free article] [PubMed]
- 75.Shi F, Xia L, Shan F, Wu D, Wei Y, Yuan H, Jiang H, Gao Y, Sui H and Shen D. Large-scale screening of COVID-19 from community acquired pneumonia using infection size-aware classification. arXiv preprint. 2020. arXiv:2003.09860. [DOI] [PubMed]
- 76.Jin S, Wang B, Xu H, Luo C, Wei L, Zhao W, Hou X, Ma W, Xu Z, Zheng Z, et al. Ai-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 77.Hu S, Gao Y, Niu Z, Jiang Y, Li L, Xiao X, Wang M, Fang EF, Menpes-Smith W, Xia J, et al. Weakly supervised deep learning for COVID-19 infection detection and classification from CT images. IEEE Access. 2020;8:118869–118883. doi: 10.1109/ACCESS.2020.3005510. [DOI] [Google Scholar]
- 78.Wu Y-H, Gao S-H, Mei J, Xu J, Fan D-P, Zhao C-W and Cheng M-M. Jcs: an explainable COVID-19 diagnosis system by joint classification and segmentation. arXiv preprint. 2020. arXiv:2004.07054. [DOI] [PubMed]
- 79.He X, Yang X, Zhang S, Zhao J, Zhang Y, Xing E and Xie P. Sample-efficient deep learning for COVID-19 diagnosis based on CT scans. medRxiv. 2020.
- 80.Al-Karawi D, Al-Zaidi S, Polus N and Jassim S. Machine learning analysis of chest CT scan images as a complementary digital test of coronavirus (COVID-19) patients. medRxiv. 2020.
- 81.Amyar A, Modzelewski R and Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19: classification and segmentation. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 82.Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X, et al. A deep learning algorithm using CT images to screen for corona virus disease (COVID-19). MedRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 83.Sun L, Mo Z, Yan F, Xia L, Shan F, Ding Z, Shao W, Shi F, Yuan H, Jiang H, et al. Adaptive feature selection guided deep forest for COVID-19 classification with chest CT. arXiv preprint. 2020. arXiv:2005.03264. [DOI] [PMC free article] [PubMed]
- 84.Ahuja S, Panigrahi BK, Dey N, Rajinikanth V and Gandhi TK. Deep transfer learning-based automated detection of COVID-19 from lung CT scan slices. 2020. [DOI] [PMC free article] [PubMed]
- 85.Zhao J, Zhang Y, He X and Xie P. Covid-CT-dataset: a CT scan dataset about COVID-19. arXiv preprint. arXiv:2003.13865. 2020.
- 86.Karim M, Döhmen T, Rebholz-Schuhmann D, Decker S, Cochez M, Beyan O, et al. Deepcovidexplainer: explainable COVID-19 predictions based on chest x-ray images. 2020. arXiv preprint arXiv:2004.04582.
- 87.Li X and Zhu D. Covid-xpert: an AI powered population screening of COVID-19 cases using chest radiography images. arXiv preprint. 2020. arXiv:2004.03042.
- 88.Minaee S, Kafieh R, Sonka M, Yazdani S, Soufi GJ. Deep-COVID: predicting COVID-19 from chest x-ray images using deep transfer learning. arXiv preprint. 2020. arXiv:2004.09363. [DOI] [PMC free article] [PubMed]
- 89.Goodwin BD, Jaskolski C, Zhong C, Asmani H. Intra-model variability in COVID-19 classification using chest x-ray images. arXiv preprint. 2020. arXiv:2005.02167.
- 90.Yamac M, Ahishali M, Degerli A, Kiranyaz S, Chowdhury ME and Gabbouj M. Convolutional sparse support estimator based COVID-19 recognition from x-ray images. arXiv preprint. 2020. arXiv:2005.04014. [DOI] [PMC free article] [PubMed]
- 91.Ahishali M, Degerli A, Yamac M, Kiranyaz S, Chowdhury ME, Hameed K, Hamid T, Mazhar R and Gabbouj M. A comparative study on early detection of COVID-19 from chest x-ray images. arXiv preprint. 2020. arXiv:2006.05332.
- 92.Heidari M, Mirniaharikandehei S, Khuzani AZ, Danala G, Qiu Y, Zheng B. Improving performance of CNN to predict likelihood of COVID-19 using chest x-ray images with preprocessing algorithms. arXiv preprint. 2020. arXiv:2006.12229. [DOI] [PMC free article] [PubMed]
- 93.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A and Bengio Y. Generative adversarial nets. Adv Neural Inf Process Syst. 2014;pp. 2672–2680.
- 94.Seum A, Raj AH, Sakib S and Hossain T. A comparative study of cnn transfer learning classification algorithms with segmentation for COVID-19 detection from CT scan images. In 2020 11th international conference on electrical and computer engineering (ICECE). IEEE. 2020, pp. 234–237.
- 95.Ahmed S, Hossain T, Hoque OB, Sarker S, Rahman S, Shah FM. Automated COVID-19 detection from chest x-ray images: a high-resolution network (hrnet) approach. SN Comput Sci. 2021;2(4):1–17. doi: 10.1007/s42979-021-00690-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Milletari F, Navab N and Ahmadi S. V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV). 2016, pp. 565–571.
- 97.Badrinarayanan V, Kendall A, Cipolla R. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(12):2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 98.Ramesh V, Rister B and Rubin DL. Covid-19 lung lesion segmentation using a sparsely supervised mask R-CNN on chest x-rays automatically computed from volumetric CTS. arXiv preprint. 2021. arXiv:2105.08147.
- 99.S. Walvekar and S. Shinde, “Efficient medical image segmentation of covid-19 chest ct images based on deep learning techniques,” in 2021 International Conference on Emerging Smart Computing and Informatics (ESCI). IEEE, 2021, pp. 203–206.
- 100.LeCun Y, Haffner P, Bottou L and Bengio Y. Object recognition with gradient-based learning. In: Shape, contour and grouping in computer vision. Springer, 1999, pp. 319–345.
- 101.Nair V and Hinton GE. Rectified linear units improve restricted boltzmann machines. ICML. 2010.
- 102.He K, Zhang X, Ren S and Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778.
- 103.Huang G, Liu Z, Van Der Maaten V and Weinberger KQ. Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 4700–4708.
- 104.K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” arXiv preprintarXiv:1409.1556, 2014.
- 105.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V and Rabinovich A. Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp. 1–9.
- 106.Szegedy C, Vanhoucke V, Ioffe S, Shlens J and Wojna Z. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 2818–2826.
- 107.Szegedy C, Ioffe S, Vanhoucke V and Alemi A. Inception-v4, inception-resnet and the impact of residual connections on learning. arXiv preprint. 2016. arXiv:1602.07261.
- 108.Deng J, Dong W, Socher R, Li L-J, Li K and Fei-Fei L. Imagenet: a large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition. IEEE, 2009, pp. 248–255.
- 109.Brownlee J. How to improve performance with transfer learning for deep learning neural networks. 2020. https://machinelearningmastery.com/how-to-improve-performance-with-transfer-learning-for-deep-learning-neural-networks/. Accessed 11 Mar 2020.
- 110.Ribeiro MT, Singh S, Guestrin C. “Why should i trust you?” explaining the predictions of any classifier. In: Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining, 2016, pp. 1135–1144.
- 111.Lundberg SM and Lee S-I. A unified approach to interpreting model predictions. In: Proceedings of the 31st international conference on neural information processing systems, 2017, pp. 4768–4777.
- 112.Zhou B, Khosla A, Lapedriza A, Oliva A and Torralba A. Learning deep features for discriminative localization. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 2921–2929.
- 113.Wu H, Ruan W, Wang J, Zheng D, Liu B, Geng Y, Chai X, Chen J, Li K, Li S, et al. Interpretable machine learning for COVID-19: an empirical study on severity prediction task. IEEE Trans Artif Intell. 2021. [DOI] [PMC free article] [PubMed]
- 114.Ahsan MM, Gupta KD, Islam MM, Sen S, Rahman M, Shakhawat Hossain M, et al. COVID-19 symptoms detection based on nasnetmobile with explainable AI using various imaging modalities. Mach Learn Knowl Extract. 2020;2(4):490–504. doi: 10.3390/make2040027. [DOI] [Google Scholar]
- 115.Alorf A. The practicality of deep learning algorithms in COVID-19 detection: application to chest x-ray images. Algorithms. 2021;14(6):183. doi: 10.3390/a14060183. [DOI] [Google Scholar]
- 116.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273–297. [Google Scholar]
- 117.Chen T and Guestrin C. Xgboost: a scalable tree boosting system. In: Proceedings of the 22nd ACM sigkdd international conference on knowledge discovery and data mining, 2016, pp. 785–794.
- 118.Schapire R. A brief introduction to boosting. ijcai’99: Proc. of the sixteenth international joint conference on artificial intelligence (pp. 1401–1406). 1999.
- 119.Breiman L. Bagging predictors. Mach Learn. 1996;24(2):123–140. [Google Scholar]
- 120.Ke G, Meng Q, Finley T, Wang T, Chen W, Ma W, Ye Q and Liu T-Y. Lightgbm: a highly efficient gradient boosting decision tree. Adv Neural Inf Process Syst. 2017;3146–3154.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data are collected and cited from published sources.