Abstract
In the course of a two-hybrid screen with the NS1 protein of influenza virus, a human clone capable of coding for a protein with high homology to the Staufen protein from Drosophila melanogaster (dmStaufen) was identified. With these sequences used as a probe, cDNAs were isolated from a λ cDNA library. The encoded protein (hStaufen-like) contained four double-stranded RNA (dsRNA)-binding domains with 55% similarity and 38% identity to those of dmStaufen, including identity at all residues involved in RNA binding. A recombinant protein containing all dsRNA-binding domains was expressed in Escherichia coli as a His-tagged polypeptide. It showed dsRNA binding activity in vitro, with an apparent Kd of 10−9 M. Using a specific antibody, we detected in human cells a major form of the hStaufen-like protein with an apparent molecular mass of 60 to 65 kDa. The intracellular localization of hStaufen-like protein was investigated by immunofluorescence using a series of markers for the cell compartments. Colocalization was observed with the rough endoplasmic reticulum but not with endosomes, cytoskeleton, or Golgi apparatus. Furthermore, sedimentation analyses indicated that hStaufen-like protein associates with polysomes. These results are discussed in relation to the possible functions of the protein.
The establishment and maintenance of asymmetries in certain cells implies the localized expression of many proteins, a property often enhanced by the localization of the corresponding mRNAs (for reviews, see references 2 and 47). The relevance of mRNA localization at precise sites of the cell in the definition of polarity of developing embryos has been documented in both Drosophila melanogaster and Xenopus laevis. Thus, the positions of gurken and oskar mRNAs in the fly oocyte define its dorsoventral axis (36) and the location of the pole plasm at the posterior pole (15), respectively. Likewise, the localization of the bicoid and nanos mRNAs at the anterior and posterior poles of the embryo, respectively, leads to the generation of two opposing gradients of their protein products and ultimately to the definition of the head, thorax, and abdomen of the embryo (50). In the case of X. laevis, several mRNAs, such as Xcat2, Xcat3, and Xlsirt, are directed to the germ plasm (34), while others, such as Vg1 and Xwnt11, accumulate at the vegetal cortex (30). By analogy to the D. melanogaster genes, these mRNAs are thought to play a role in defining patterns in the X. laevis embryo. In fact, a region of the Xcat2 protein shows sequence homology to the zinc finger domain of nanos protein (34).
The specific localization of certain mRNAs to precise sites within the cell is not an exclusive property of germ cells or developing embryos. A number of observations indicate that some of the mRNAs of somatic cells are also localized at different sites within the cell. Thus, myelin-binding protein is translated in oligodendrocytes from free ribosomes on localized mRNA (51), and the microtubule-associated protein MAP2 is translated preferentially in dendrites of the neurons (23). Likewise, different actin protein isoforms are translated from their mRNAs at distinct sites of myoblasts (27).
In some cases, the intracellular localization of mRNAs involves cis-acting signals consisting of distinct secondary structures of their untranslated regions (UTRs) (reviewed in reference 47). These cis signals are thought to mediate localization by interaction with RNA-binding proteins, the best studied of which is the Staufen protein of D. melanogaster (dmStaufen) (48; reviewed in reference 50). dmStaufen protein is the product of a maternal mRNA of D. melanogaster that is involved in the accumulation of bicoid and oskar mRNAs at the anterior pole of the embryo and the posterior pole of the oocyte, respectively (48). It contains double-stranded RNA (dsRNA)-binding domains that associate with bicoid mRNA through a precise secondary structure in its 3′ UTR (18).
Our group has been studying the influenza virus nonstructural protein NS1, an RNA-binding protein (26, 33, 42) that may be involved in several regulatory processes during viral infection (reviewed in reference 37). These include the modulation of pre-mRNA splicing (20, 21, 32), the retention of poly(A)-containing RNA in the nucleus (20, 42), and the stimulation of viral mRNA translation (10, 14). These properties of NS1 protein seem related to particular interactions with certain cellular or viral RNA molecules and may also involve interaction with specific cellular factors (26, 40, 42, 43). In the course of a genetic screen to detect candidate cellular proteins that may pertain to these biochemical effects, we identified a human gene with homology to the dmStaufen gene. The encoded protein (hStaufen-like) was localized by immunofluorescence to the rough endoplasmic reticulum in cultured cells, was associated with polysomes, and behaved like an RNA-binding protein.
MATERIALS AND METHODS
Biological materials.
The COS-1 cell line (25), kindly provided by Y. Gluzman, and the HeLa cell line, purchased from the American Type Culture Collection, were cultured as described previously (39). Saccharomyces cerevisiae HF7c (MATa his3 GAL1-HIS3 GAL4-lacZ trp1 leu2) was obtained from Clontech and used for two-hybrid screening. The vaccinia virus recombinant vTF7-3 (22) was kindly provided by B. Moss. Plasmids pGBT9 and pGAD424, used for interaction tests in the two-hybrid screen, as well as plasmids pVA3, pTD1, and pCL1, used as internal controls, were obtained from Clontech. pRSET expression vectors were obtained from Invitrogen. Plasmid pGEM1-Bcd (4) containing the bicoid cDNA cloned into pGEM vector was provided by A. Ephrussi. Plasmid pArII contained the rDNA from Artemia salina and was provided by J. Renard. The monoclonal antibody specific for the T7 tag present in the pRSET vectors was purchased from Novagen. The following monoclonal antibodies were used to detect cytological markers: 18A4 (28), specific for ribophorin I, kindly provided by F. Becker; antibodies specific for Bip (kindly provided by S. Fuller) and tubulin (Amersham); anti-lamp2 (Developmental Studies Hybridoma Bank, University of Iowa), specific for late endosomes, kindly provided by J. Krijnse-Locker; and anti-mannose II (mannII), specific for the Golgi apparatus, provided by T. Nilsson. An antibody specific for ribosomal P proteins was kindly donated by J. P. García-Ballesta.
Protein expression and purification.
The thSTL DNA fragment of clone C was transferred in frame to pRSET vector to generate plasmid pRSTL, in which the thSTL protein was fused to a His tag and the T7 antigenic tag. The recombinant pRSTL plasmid was transformed into Escherichia coli BL21(DE3)(pLysS), the expression of His-thSTL was induced by isopropyl-β-d-thiogalactopyranoside, and the recombinant protein was purified by chromatography on Ni-nitrilotriacetic acid (NTA)-agarose as described previously (33). In addition, the hStaufen-like cDNA sequence encoding the dsRNA-binding domains was also fused in frame to pRSET vector to generate plasmid pRHST. This plasmid was transformed into E. coli BL21(DE3)(pLysS), and the His-HST protein was expressed and purified by chromatography on Ni-NTA-agarose as described above. For immunofluorescence studies, the thSTL cDNA, fused to the sequence of the tags indicated above, was transferred to vector pCMV (Clontech) to generate plasmid pCMV-STL.
Two-hybrid screen.
The NS1 cDNA from plasmid pSVa232NS1 (20) was transferred to vector pGBT9, and the resulting plasmid (pGBT-NS1) was used to screen a human kidney cDNA fusion library cloned into pGAD vector. With 10 mM 3-aminotriazole, the recombinant plasmid pGBT-NS1 alone did not induce growth in histidine-free medium. The procedures for library amplification, yeast cell transformation, screening for growth in the absence of histidine, and β-galactosidase activity were those recommended in the Matchmaker protocol (Clontech). Rescue of positive pGAD plasmids was done by transformation into E. coli MH4 (Leu−) cells and selection in M9 plates lacking leucine. cDNA clones corresponding to the thSTL insert were obtained from a HeLa cDNA library constructed in λgt10 (Clontech), using standard procedures (45). Sequencing was carried out in a Perkin-Elmer 373 automatic sequencer, using specific oligonucleotide primers.
Transfections.
Transfection of HeLa cells with expression plasmids driven by polymerase II was done with cationic liposomes (44) as described elsewhere (33). When expression vectors driven by the T7 promoter were used COS-1 cells were previously infected with vaccinia virus vTF7-3 and then transfected as indicated above.
Probe labeling and RNA analyses.
Labeling of cDNA was carried out by random priming using a Stratagene kit. Riboprobe synthesis and poly(A)+ RNA isolation from cultured cells were done as described elsewhere (41). Northern hybridization was performed by using cDNA probes and standard conditions (45). Dot blot hybridization was carried out as described elsewhere (33). To generate a 3′ UTR probe from bicoid, plasmid pGEM1-Bcd was digested with SacI, treated with mung bean nuclease, and further digested with EcoRV. After religation and digestion with EcoRI, the resulting DNA was used for transcription with Sp6 RNA polymerase. To prepare an unrelated probe containing highly structured dsRNA regions, we used plasmid pNSZ (41), which generates a 240-nucleotide RNA upon transcription with T7 RNA polymerase. As a nonstructured probe, we transcribed with T7 RNA polymerase the synthetic oligonucleotide A40CCTATAGTGAGTCGTATTAACC annealed to a T7 promoter-complementary oligonucleotide (GGTTAATACGACTCACTATAGG), using the conditions described by Seong and Brownlee (46). To test RNA-protein interaction, various amounts of His-HST protein were mixed with a fixed quantity of labeled probe (10,000 cpm; specific activity, 108 cpm/μg), incubated for 20 min at room temperature in a buffer containing 100 mM KCl, 5 mM MgCl2, and 50 mM Tris-HCl (pH 7.5), and filtered through a nitrocellulose filter in a dot blot apparatus. The proportion of probe retained was determined with a phosphorimager.
Cell fractionations and polysome analysis.
Polysomes were isolated as described previously (38). In brief, the cell culture was treated with cycloheximide (100 μg/ml) for 15 min before cell harvesting. The cytoplasmic fraction was obtained by cell lysis in isotonic buffer (150 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl [pH 8.5]) containing 0.5% Nonidet P-40, centrifuged for 10 min at 10,000 × g and 4°C, and finally centrifuged on a 7 to 47% sucrose gradient in isotonic buffer for 2 h at 40,000 rpm and 2°C in a SW41 rotor. For EDTA treatment, the cytoplasmic fraction and sucrose gradients were adjusted to 25 mM EDTA. Fractions were collected and analyzed as indicated below. As a negative control, polysomes were disrupted by incubation of the cell cultures with puromycin (100 μg/ml), instead of cycloheximide, for 60 min before cell harvesting.
Immunological techniques.
Immunological detection of hStaufen-like protein was carried out with a rabbit antiserum prepared by immunization with purified His-STL protein. We used as controls both preimmune serum and the immune serum depleted of specific reactivity by incubation with purified His-HST protein bound to Ni2+-NTA resin.
For Western blot analysis, cell extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Immobilon filters, and the membranes were saturated with 3% bovine serum albumin (BSA) for 1 h at room temperature. The filters were incubated with a 1:400 to 1:2,000 dilutions of the anti-STL or control serum for 1 h at room temperature. After being washed twice for 30 min with phosphate-buffered saline (PBS) containing 0.25% Tween 20, the filters were incubated with a 1:10,000 dilution goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase. Finally, the filters were washed two times for 30 min as described above and developed by enhanced chemiluminescence.
Immunofluorescence analysis was performed as follows. HeLa cell cultures were washed with PBS, fixed for 20 min in 4% paraformaldehyde, and washed with PBS. After treatment with 50 mM NH4Cl for 20 min, cells were permeabilized for 10 min with 0.1% Triton X-100 and blocked with 10% BSA in PBS. The cells were incubated with anti-STL rabbit or control serum (at a 1:200 dilution in PBS–1% BSA) or the appropriate dilutions of the antibodies specific for the various intracellular markers (1:100 for monoclonal anti-ribophorin I, 1:50 for monoclonal anti-lamp2, 1:1,000 for monoclonal antibodies specific for Bip and tubulin, and 1:400 for monoclonal anti-T7 tag) for 1 h at room temperature. After washing with PBS–1% BSA, the bound antibodies were revealed with fluorescein isothiocyanate-conjugated rabbit anti-mouse antibody (1:200 dilution) or Texas red-conjugated goat anti-rabbit antibody (1:200 dilution) by incubation for 1 h at room temperature. Finally, the cells were washed with PBS and the preparations were mounted with Mowiol. Images were obtained with a Zeiss Axiophot fluorescence microscope equipped with a high-resolution charge-coupled device (CCD) camera. For double-fluorescence experiments, the images were superimposed with Adobe Photoshop.
RESULTS
Identification of hStaufen-like by interaction with influenza virus NS1 protein.
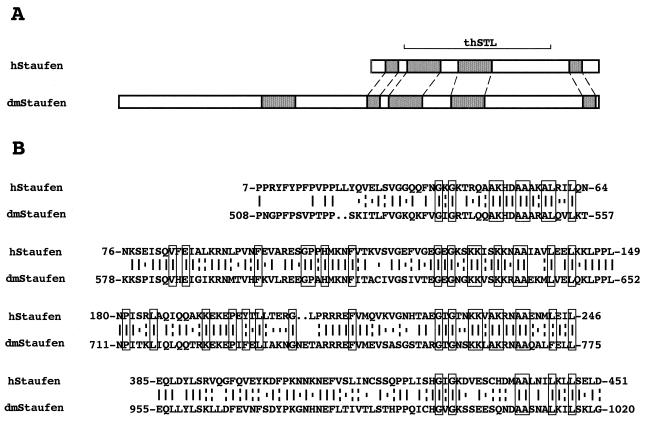
We carried out a two-hybrid screen in yeast with NS1 protein as a bait. Under the conditions used, transformation of S. cerevisiae with plasmid pGBT-NS1 did not stimulate growth of the cells in the absence of histidine (data not shown). Cotransformation with a human kidney cDNA fusion library constructed in plasmid pGAD led to the growth of about 1,000 independent clones after screening of 2 million colonies. Only a few of them were positive in the β-Galactosidase in situ assay, and these were further tested after isolation of the plasmids and retransformation. One of the clones (clone C) was confirmed as positive and fulfilled all controls in the two-hybrid interaction protocol (data not shown). Clone C was analyzed by restriction assay and partial sequence. It contained an insert of about 1 kb (thSTL) with an open reading frame capable of encoding a polypeptide with homology to the Staufen protein of D. melanogaster (48) (Fig. 1A). In view of this homology, the thSTL insert present in clone C was used to screen a standard human cDNA library. Several λ clones were identified, and the cDNA inserts were sequenced. Although the complete sequence of the gene is not yet available, about 3.0 kb have been characterized to date. This portion of the cDNA contains a 471-amino-acid open reading frame and a long 3′ UTR. The predicted protein sequence is highly homologous to the C-terminal half of the dmStaufen protein, with 38% sequence identity and 55% sequence similarity (Fig. 1) but shows much less homology to other members of the Staufen family of dsRNA-binding proteins. It is remarkable that the Staufen dsRNA-binding domains, including all the positions relevant for RNA binding, are very conserved (7). The locations of these RNA-binding domains along hStaufen-like protein reflect the situation found for the corresponding domains in dmStaufen protein (Fig. 1A). These results suggest that the gene identified as an NS1-interacting clone is the human sequence homologue of the dmStaufen gene.
FIG. 1.
Identification of the human sequence homologue of the dmStaufen gene. (A) Structure of hStaufen-like protein, including the four RNA-binding domains (hatched boxes) and the thSTL protein fragment encoded in clone C, aligned with the structure of dmStaufen protein. (B) Comparison of the protein sequences of dmStaufen and the protein predicted from λ clones obtained by using the thSTL insert present in clone C. Boxed residues show the positions conserved among dsRNA-binding domains present in protein members of the dmStaufen family (49).
The hStaufen-like gene is expressed in human cell lines and organs.
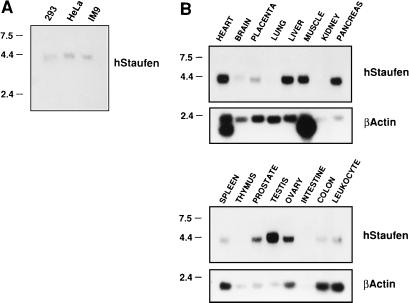
To ascertain whether the cDNA identified in the two-hybrid screen indeed corresponds to a gene expressed in human cells, we carried out Northern analyses using poly(A)+ RNA from human cell lines and organs. Cytoplasmic poly(A)+ RNA was isolated from three human cell lines (293, HeLa, and IM9) and assayed by Northern blotting. In addition, we tested premade blots generated with RNA from a variety of human organs (Clontech). The results are presented in Fig. 2. A single hybridization band was apparent, both in the three cell lines tested (Fig. 2A) and in several of the organs assayed (Fig. 2B). The size of the transcript detected was around 4 to 4.5 kb, and some variation in mobility was observed among the transcripts derived from various organs. The hStaufen-like mRNA was most abundant in the heart, liver, muscle, testis, and ovary (Fig. 2B).
FIG. 2.
Characterization of hStaufen-like mRNA by Northern blot hybridization. Poly(A)+ RNA from human cell lines (A) or human organs (B) was separated by denaturing agarose gel electrophoresis and probed with a thSTL-specific probe or a β-actin probe as described in Materials and Methods. Size of molecular weight markers are indicated in kilobases to the left.
Characterization of the hStaufen-like protein.
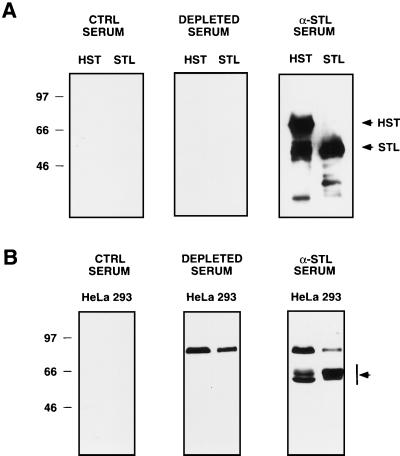
To characterize the hStaufen-like protein, we used a monospecific antiserum generated by immunization of rabbits with the thSTL recombinant protein encoded in clone C (see Materials and Methods and Fig. 1A). The specificity of the anti-STL serum was ascertained by Western blot analysis of extracts obtained from COS-1 cells transfected with plasmids expressing the recombinant His-HST or His-STL protein. Specific reactive bands of the expected sizes were clearly detectable in the extracts of plasmid-transfected but not mock-transfected cells (data not shown). Using purified His-STL and His-HST proteins, we checked that the preimmune serum detected no specific reactivity and that the signal was lost when an anti-STL serum depleted with purified His-HST protein bound to Ni2+-NTA resin was used (Fig. 3A). The availability of the specific anti-STL serum allowed us to search for the endogenous hStaufen-like protein by Western blotting of total human cell extracts. Two reactive bands corresponding to molecular masses of 60 to 65 kDa, as well as a minor band of about 90 kDa, were detected. Depletion of the anti-STL serum in a column containing purified His-HST protein abrogated detection of the 60- to 65-kDa bands but not the minor band of 90 kDa, indicating that the former correspond to hStaufen-like protein. No reactivity with the preimmune serum was detected (Fig. 3B). In extracts of rat hippocampal neurons, a protein band of 65 kDa was also recognized by the anti-STL serum, as well as by two sera raised against a peptide corresponding to the Staufen cDNA sequence (28a).
FIG. 3.
Characterization of hStaufen-like protein by Western blotting. (A) Purified His-STL and His-HST proteins were analyzed by Western blotting as indicated in Materials and Methods, using anti-STL serum, preimmune serum, or anti-STL serum depleted with purified His-HST protein bound to Ni2+-NTA resin. (B) Total extracts from HeLa or 293 cells were analyzed as indicated above. Sizes of molecular weight markers are indicated in kilodaltons to the left. Arrows indicate the relevant protein bands.
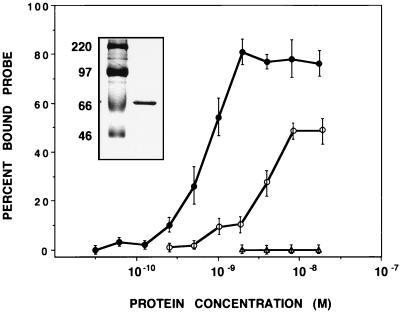
The sequence homology of hStaufen-like and dmStaufen proteins suggested that the former should behave as an RNA-binding protein. To test this prediction, we used plasmid pRHST, which contained the cDNA sequence corresponding to the RNA-binding domains of the hStaufen-like protein (Fig. 1B) subcloned into pRSET vector. This allowed expression of His-HST in E. coli and its purification in a Ni2+-NTA resin. The purified recombinant protein (Fig. 4, inset) was used for in vitro binding assays with a probe containing the 3′ UTR of bicoid mRNA. High-affinity interaction was detected (Fig. 4), with an apparent Kd of approximately 10−9 M. Similar interaction was observed with another highly structured RNA probe derived from an influenza virus-chloramphenicol acetyltransferase chimeric gene (NSZ probe) (41), but the affinity for binding to a single-stranded probe [polyU)] was less (Fig. 4).
FIG. 4.
RNA-binding properties of hStaufen-like protein. Fixed amounts of labeled RNA probe (10,000 cpm) were incubated with increasing amounts of purified His-HST protein. The protein-bound probe was determined by filtration on a nitrocellulose filter. The results are presented as percentage of maximal retained probe and are averages and standard deviations of two to four independent experiments. The insert shows the purified His-HST protein used in the RNA-binding assays as analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue staining. Closed circles, His-HST protein/bicoid probe; open circles, His-HST protein/poly(U) probe; triangles, BSA/bicoid probe.
hStaufen-like protein localizes to the endoplasmic reticulum.
We next studied the intracellular localization of hStaufen-like protein. To this end we used the anti-STL serum; its specificity at the cytological level was verified first. Cultures of HeLa cells were transfected with plasmid pCMV-STL, which encodes a His-tagged thSTL protein. The fixed cells were analyzed by immunofluorescence using an anti-T7 tag antibody or the anti-STL antiserum. The results are presented in Fig. 5. The staining patterns obtained with both reagents were essentially identical (Fig. 5A to C), confirming that the anti-STL serum specifically recognizes the thSTL protein. We observed a more intense pattern of staining next to the nucleus but also irradiating toward the cell periphery. The use of the mild detergent saponin before fixation did not result in a great loss of staining (data not shown), suggesting the interaction of thSTL to membrane or cytoskelton constitutents. As expected, depletion of the serum with purified His-HST protein bound to Ni2+-NTA resin diminished the signal to a large extent (Fig. 5D to F) and the preimmune serum revealed no specific staining pattern (data not shown).
FIG. 5.
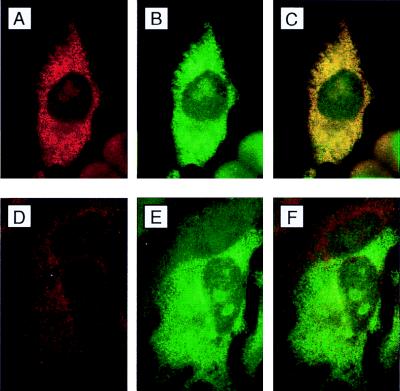
Immunofluorescence analysis of hStaufen-like in transfected HeLa cells. (A and B) Cultures of HeLa cells transfected with plasmid pCMV-STL, fixed, and processed for immunofluorescence as indicated in Materials and Methods, using anti-STL serum (A) and anti-T7 tag monoclonal antibody (B). (C) Overlay of the preceding images. (D) Anti-STL serum depleted with purified His-HST protein bound to Ni2+-NTA resin. (E) Anti-T7 tag monoclonal antibody. (F) Overlay of the preceding images. In these analyses, staining with anti-STL serum and with anti-STL serum depleted with purified His-HST protein were carried out under identical conditions, including dilutions of the sera and exposure times.
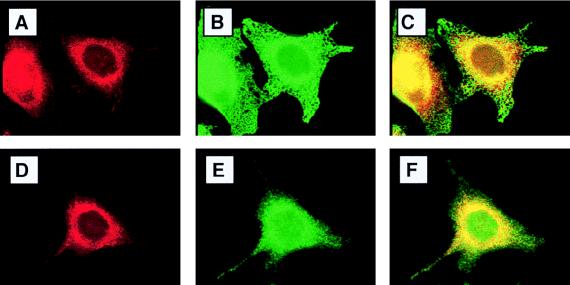
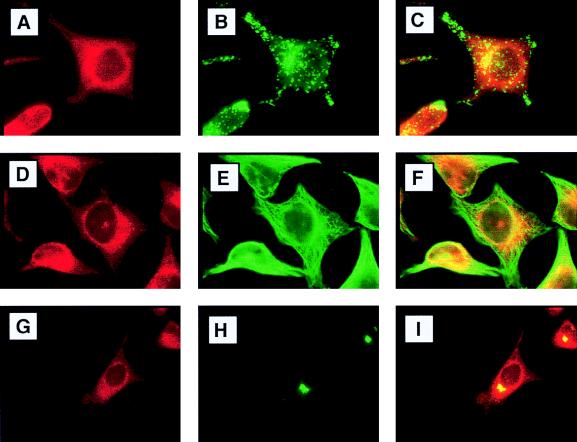
To determine the precise localization of endogenous hStaufen-like protein, we carried out double-immunofluorescence experiments using a variety of cellular markers (Fig. 6 and 7). Partial colocalization of hStaufen-like protein with the endoplasmic reticulum (Fig. 6 to C; Bip marker) as well as a more precise colocalization with the rough endoplasmic reticulum (Fig. 6D to F; ribophorin marker) were observed. In contrast, no colocalization was detected with markers of either the endocytic pathway (Fig. 7A to C; lamp2 marker) or the cytoskeleton (Fig. 7D to F; tubulin marker). The partial overlapping of hStaufen-like staining and the Golgi apparatus (Fig. 7G to I; mannII marker) does not allow us to exclude its presence in this organelle, but the hStaufen-like staining pattern did not correspond to a typical Golgi distribution. Therefore, we conclude from the cytological data that the hStaufen-like protein is associated with the rough endoplasmic reticulum.
FIG. 6.
Colocalization of hStaufen-like with rough endoplasmic reticulum. (A and B) Cultures of HeLa cells fixed and processed for immunofluorescence as indicated in Materials and Methods and the legend to Fig. 5, using anti-STL serum (A) and anti-Bip monoclonal antibody (B). (C) Overlay of the preceding images. (D) Anti-STL serum. (E) Anti-ribophorin I monoclonal antibody. (F) Overlay of the preceding images.
FIG. 7.
Double-immunofluorescence studies with hStaufen-like and cellular markers. (A and B) cultures of HeLa cells fixed and processed for immunofluorescence as indicated in Materials and Methods and the legend to Fig. 5, using anti-STL serum (A) and anti-lamp2 monoclonal antibody (B). (C) Overlay of the preceding images. (D) Anti-STL serum. (E) Antitubulin monoclonal antibody. (F) Overlay of the preceding images. (G) Anti-STL serum. (H) Anti-mannII monoclonal antibody. (I) Overlay of the preceding images.
The hStaufen-like protein is associated with polysomes.
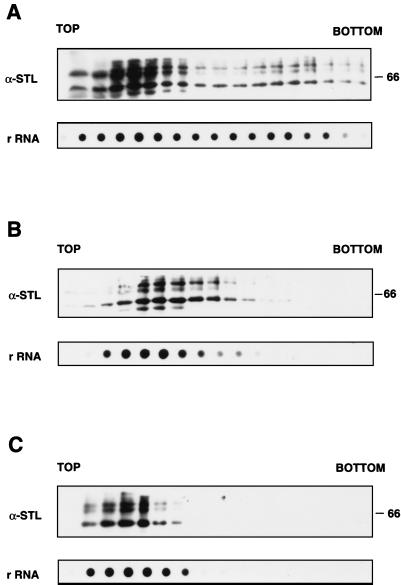
In view of the presence of hStaufen-like in the rough endoplasmic reticulum fraction, we examined whether it is associated with polysomes. Cultures of HeLa cells were fractionated by lysis with Nonidet P-40, and the polysomes were isolated from the cytoplasmic fraction by sedimentation on sucrose gradients as indicated in Materials and Methods. Each fraction was tested for the presence of hStaufen-like protein, using anti-STL serum, and for the presence of ribosomes, using an rDNA probe or an anti-P-protein serum (data not shown). The results are presented in Fig. 8. hStaufen-like protein was not present as a soluble protein; rather, it cosedimented with the ribosomes and the polysomes (Fig. 8A). To verify that the fast-sedimenting forms in the gradient corresponded to polysomes, the cytoplasmic fraction was treated with EDTA before centrifugation. Under these conditions, the ribosomal subunits should dissociate from the mRNAs and sediment more slowly; indeed, we observed the accumulation of rRNA marker in slow-sedimenting forms (Fig. 8C). This mobility shift was also apparent for hStaufen-like protein, which under these conditions was not present in the bottom fractions of the gradient and accumulated with the ribosomal marker (Fig. 8C). Alternatively, the cell cultures were incubated with puromycin before preparation of cytoplasmic extracts (Fig. 8B). As with EDTA treatment, the dissociation of polysomes, shown by the disappearance of fast-sedimenting ribosomes, correlated with a parallel loss of hStaufen-like from the bottom fractions of the gradient (Fig. 8B). These results indicate that the hStaufen-like protein is present in the cell in association with the polysome complexes.
FIG. 8.
Association of hStaufen-like protein with polysomes. Soluble extracts of HeLa cells were centrifuged in a sucrose gradient as described in Materials and Methods. (A) Untreated extracts. The fractions were analyzed by Western blotting using anti-STL serum. In addition, aliquots of each fraction were used to isolate RNA to carry out dot blot hybridization with an rDNA probe as indicated in Materials and Methods. (B) Extracts from cultures treated with puromycin. The cultures were treated with puromycin (100 μg/ml) for 60 min prior to preparation of cytoplasmic extracts. (C) Extracts treated with EDTA. The extracts prepared as described above were treated with 25 mM EDTA and separated as indicated except that the sucrose gradient was adjusted to 25 mM EDTA. The fractions were analyzed as described for panel A. Sizes of molecular weight markers are indicated in kilodaltons to the right.
DISCUSSION
In the course of a two-hybrid screen with influenza virus NS1 protein as a bait, we identified a human cDNA (thSTL) with high sequence homology to dmStaufen protein. The encoded protein was shown to colocalize in vivo and to interact in vitro with NS1 protein by coimmunoprecipitation (19a). The possible significance of this interaction for influenza virus infection is under investigation.
Characterization of hStaufen-like protein.
In view of the high homology between the RNA-binding regions of the protein encoded in the thSTL clone and those of dmStaufen protein, longer cDNA clones were identified and sequenced. The predicted protein showed strong conservation of four of the dsRNA-binding domains present in dmStaufen protein (Fig. 1B), although no such a conservation was observed in comparison to other members of the dsRNA-binding protein family (49). We therefore concluded that the cDNA obtained probably corresponds to a human homologue of dmStaufen.
Previous reports in the literature did not provide much information about possible human homologues of the dmStaufen protein. The use of a cDNA probe for in situ hybridization on mitotic spreads indicated a single-copy gene located at band 20q13.1 (11). We took advantage of the isolation of cDNAs corresponding to the thSTL clone to investigate the pattern of expression of hStaufen-like mRNAs and to study the RNA-binding properties and the intracellular localization of hStaufen-like protein.
The Northern analysis clearly indicated differences in the expression level of the gene in several human organs. Consistent with the function assigned for dmStaufen protein in D. melanogaster early development, we found abundant expression of hStaufen-like gene in both human testis and ovary (Fig. 2B). In addition, several organs in which muscular tissue is present proved positive for hStaufen-like expression. In contrast, we could detect only low-level hStaufen-like gene expression in the brain.
The characterization of hStaufen-like protein in cultured human cells included the evaluation of its apparent molecular weight by Western blotting. We observed prominent bands, with mobilities corresponding to about 60 to 65 kDa, in HeLa and 293 cells (Fig. 3B), as well as in IM9 lymphocytes or human neuroblastoma cells (data not shown). hStaufen-like protein might have been present in more than one isoform; indeed, small differences were observed in the mobilities of the mRNAs detected in various organs (Fig. 2B). The specificity of this signal was corroborated by its absence in Western blotting experiments using a hStaufen-like serum that had been depleted with a Ni2+-NTA resin containing purified His-HST protein (Fig. 3B). The detection of a 65-kDa protein band with identical mobility using sera raised against a Staufen peptide (28a) indicates that the 60- to 65-kDa protein bands detected are indeed the products of hStaufen-like gene and not cross-reacting proteins. The significance of additional signals observed with mobilities corresponding to about 90 kDa is not clear at present.
Next, the RNA-binding properties of the hStaufen-like protein were studied by means of purified His-HST protein, the portion of the polypeptide that contains four dsRNA-binding domains homologous to dmStaufen. High-affinity binding to the 3′ UTR bicoid probe could be observed and an apparent Kd of about 10−9 M was determined, but its affinity of binding to poly(U) was smaller (Fig. 4), indicating that hStaufen-like is a dsRNA-binding protein, as inferred from its amino acid sequence. The high-affinity binding of hStaufen-like to the bicoid probe cannot be interpreted as demonstrating that an mRNA of this sequence is the physiological target of hStaufen-like in human cells, since other dsRNA probes showed similar affinities of binding (data not shown). The identification of the relevant RNAs that interact with hStaufen-like in vivo represents a fundamental research objective for the future.
The characterization of hStaufen-like protein also included the study of its intracellular localization. Indirect immunofluorescence with anti-STL serum showed a granular cytoplasmic staining, more intense around the nucleus (Fig. 6). Double-immunofluorescence experiments demonstrated that it associates mainly with the rough endoplasmic reticulum (Fig. 6). Within the limits of detection of this technique, the endogenous hStaufen-like protein did not colocalize with a variety of markers representative of other cell compartments such as endosomes, cytoskeleton, or Golgi apparatus and could not be detected in the nucleus (Fig. 7), although some nucleolar staining was apparent when the thSTL protein fragment was expressed by transfection (Fig. 5). Analyses of extracts from HeLa cells indicated that hStaufen-like protein cosedimented with ribosomes and polysomes, and its association with the latter was confirmed by the change in sedimentation properties observed upon in vivo disruption of polysomes by puromycin treatment (Fig. 8B) or their in vitro dissociation by EDTA treatment (Fig. 8C). Such a protein distribution is reminiscent of the localization of the protein kinase PKR, a fraction of which is associated to ribosomes and polysomes (53).
As a whole, the results presented in this report support the notion that the human homologue of dmStaufen protein is a dsRNA-binding protein and that it may be related to the localization and/or the translation of cellular mRNA.
Possible functions of hStaufen-like in mRNA localization.
The characterization of hStaufen-like reported here provides only circumstantial evidence about its function in human cells. The role of dmStaufen protein in the early stages of D. melanogaster development is well established. It binds specifically, but not necessarily directly to, some mRNAs, such as oskar mRNA or bicoid mRNA, and allows their localization at opposite poles in the oocyte and early embryo, respectively (48). This precise localization is required to express the oskar and bicoid gene products only at those sites. The expression of oskar induces the formation of the pole plasm, which will determine the posterior pole of the embryo. The importance of its localization is stressed by the phenotype of mutants in which its expression is produced ectopically (16). On the other hand, the localized translation of bicoid mRNA allows the formation of an anterior-posterior gradient of bicoid protein, opposite that formed by nanos protein, that determines the formation of the head and thorax (50). The transport and retention of bicoid mRNA at the anterior pole is mediated by the formation of large ribonucleoprotein complexes that include dmStaufen protein and require the presence of specific sequences located at the 3′ UTR of the mRNA (18). These large complexes involve not only RNA-protein interactions but also intermolecular RNA-RNA interactions (19). In this context, it should be mentioned that despite the high-affinity interaction observed between hStaufen-like protein and the bicoid 3′ UTR probe in solution (Fig. 4), we detected no retention of the same probe by His-HST protein immobilized on a Ni2+-NTA resin (data not shown). The large dmStaufen-bicoid mRNA particles seem to move along microtubule bundles to the anterior pole (18), although localization of oskar mRNA also requires tropomyosin (17). In the case of hStaufen-like, the expression of the gene in ovaries and testis is compatible with a role in development, but so far we have no direct evidence to support such a possibility.
Recent evidence indicates that dmStaufen also plays a role in somatic cells. Thus, dmStaufen protein interacts with prospero mRNA by binding to its 3′ UTR and mediates, in cooperation with inscuteable, its accumulation its accumulation in the basal area of neuroblasts (5, 6, 31; reviewed in reference 8), leading to an asymmetrical distribution of this mRNA in the progeny cells. Such specific localization of mRNAs is reminiscent of similar phenomena described for specialized mammalian cells. Thus, specific intracellular localization of the mRNAs encoding different isoforms of actin, myelin-binding protein, and creatine kinase has been described (27, 51, 52). Furthermore, this specific localization can operate on microinjected myelin-binding protein mRNA (1). Recently, a protein factor present in certain stages of X. laevis has been implicated in the localization of Vg1 mRNA (12, 35). It is noteworthy that this so called Vera protein accumulates in the rough endoplasmic reticulum (12) as we describe for hStaufen-like protein. It is tempting to speculate that the hStaufen-like protein characterized in this report might also play a role in these localization processes in somatic cells. Thus, a similar rat Staufen-like homologue accumulates in dendrites but not in the axons of neurons in culture (28a).
Is hStaufen-like involved in regulation of mRNA translation?
The dmStaufen protein belongs to a family of proteins that share a number of dsRNA-binding domains and include, among others, PKR, TAR RNA-binding protein (TRBP), and its homologue in X. laevis, Xlrbpa (13, 24, 49). PKR is a protein kinase that can be activated by interaction with dsRNA and specifically phosphorylates the α subunit of protein synthesis initiation factor eIF2. This modification abolishes its recycling and therefore inhibits protein synthesis. On the other hand, TRBP is essential for human immunodeficiency virus replication, although its function in the uninfected cell may be related to efficient gene expression. Thus, it has been shown that TRBP interacts with PKR in a manner that may be RNA independent (3, 9) and inhibits its activity. It is conceivable that TRBP blocks PKR activation by binding to dsRNA regions in the mRNAs, therefore avoiding their interaction to the dsRNA-binding domain of PKR, but it is also possible that the observed inhibition is exerted by directly blocking PKR through protein-protein interaction. Since both PKR and TRBP, as well as Xlrbpa, are found associated to the ribosome (13, 53), TRBP could be involved in establishing an appropriate milieu for mRNA translation by local inhibition of the PKR protein bound to the ribosome. In this regard, it should be mentioned that these proteins do not show sequence specificity for RNA binding and therefore could act as general cellular factors. In contrast, dmStaufen has been shown to colocalize specifically with certain mRNA targets (8, 47). We presume that hStaufen-like would show preferential binding to specific mRNAs in human cells. Since it is associated with polysomes, it is conceivable that it plays a dual role: (i) positioning specific mRNAs at given sites in the cell and (ii) stimulating their translation at the site. These roles are consistent with the activity of dmStaufen in mRNA localization (47), with the localization of the rat homologue of hStaufen-like in dendrites of hippocampal neurons (28a), and with the diminished translation of oskar mRNA in D. melanogaster Staufen mutants lacking bruno activity (29).
ACKNOWLEDGMENTS
We are indebted to A. Nieto, M. Kiebler, I. Mattaj, J. A. Melero, and T. Zürcher for critical comments on the manuscript. We thank F. Becker, A. Ephrussi, J. P. García-Ballesta, J. Krijnse-Locker, B. Moss, and J. Renard for providing biological materials. The technical assistance of J. Fernández is gratefully acknowledged.
P.F. and R.M.M. were fellows from Programa Nacional de Formación de Personal Investigador. This work was supported by Programa Sectorial de Promoción General del Conocimiento (grant PB94-1542).
The first two authors contributed equally to this work.
REFERENCES
- 1.Ainger K, Avossa D, Morgan F, Hill S J, Barry C, Barbarese E, Carson J H. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassell G, Singer R H. mRNA and cytoskeletal filaments. Curr Opin Cell Biol. 1997;9:109–115. doi: 10.1016/s0955-0674(97)80159-7. [DOI] [PubMed] [Google Scholar]
- 3.Benkirane M, Neuveut C, Chun R F, Smith S M, Samuel C E, Gatignol A, Jeang K T. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16:611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nüsslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. Cell. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broadus J, Doe C Q. Extrinsic cues, intrinsic cues and microfilaments regulate asymmetric protein localization in Drosophila neuroblasts. Curr Biol. 1997;7:827–835. doi: 10.1016/s0960-9822(06)00370-8. [DOI] [PubMed] [Google Scholar]
- 6.Broadus J, Fuerstenberg S, Doe C Q. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;391:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- 7.Bycroft M, Grunert S, Murzin A G, Proctor M, St Johnston D. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J. 1995;14:3563–3571. doi: 10.1002/j.1460-2075.1995.tb07362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campos-Ortega J A. Dynastic intrincacies of neuroblast division. Curr Biol. 1997;7:R726–R728. doi: 10.1016/s0960-9822(06)00367-8. [DOI] [PubMed] [Google Scholar]
- 9.Cosentino G P, Venkatesan S, Serluca F C, Green S R, Mathews M B, Sonenberg N. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc Natl Acad Sci USA. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Luna S, Fortes P, Beloso A, Ortín J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DesGroseillers L, Lemieux N. Localization of a human double-stranded RNA-binding protein gene (STAU) to band 20q13.1 by fluorescence in situ hybridization. Genomics. 1996;36:527–529. doi: 10.1006/geno.1996.0499. [DOI] [PubMed] [Google Scholar]
- 12.Deshler J O, Highett M I, Schnapp B J. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- 13.Eckmann C R, Jantsch M F. Xlrbpa, a double-stranded RNA-binding protein associated with ribosomes and heterogeneous nuclear RNPs. J Cell Biol. 1997;138:239–253. doi: 10.1083/jcb.138.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enami K, Sato T A, Nakada S, Enami M. Influenza virus NS1 protein stimulates translation of the M1 protein. J Virol. 1994;68:1432–1437. doi: 10.1128/jvi.68.3.1432-1437.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ephrussi A, Dickinson L K, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 16.Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- 17.Erdelyi M, Michon A M, Guichet A, Glotzer J B, Ephrussi A. Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature. 1995;377:524–527. doi: 10.1038/377524a0. [DOI] [PubMed] [Google Scholar]
- 18.Ferrandon D, Elphick L, Nusslein Volhard C, St Johnston D. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 19.Ferrandon D, Koch I, Westhof E, Nusslein V C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-Staufen ribonucleoprotein particles. EMBO J. 1997;16:1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Fortes, P. Unpublished data.
- 20.Fortes P, Beloso A, Ortín J. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks RNA nucleocytoplasmic transport. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fortes P, Lamond A I, Ortín J. Influenza virus NS1 protein alters the subnuclear localization of cellular splicing components. J Gen Virol. 1995;76:1001–1007. doi: 10.1099/0022-1317-76-4-1001. [DOI] [PubMed] [Google Scholar]
- 22.Fuerst T R, Earl P L, Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garner C C, Tucker R P, Matus A. Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature. 1988;336:674–677. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 24.Gatignol A, Buckler C, Jeang K T. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila Staufen. Mol Cell Biol. 1993;13:2193–2202. doi: 10.1128/mcb.13.4.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gluzman Y. SV40 transformed simian cells support the replication or early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 26.Hatada E, Saito S, Okishio N, Fukuda R. Binding of the influenza virus NS1 protein to model genome RNAs. J Gen Virol. 1997;78:1059–1063. doi: 10.1099/0022-1317-78-5-1059. [DOI] [PubMed] [Google Scholar]
- 27.Hill M A, Gunning P. Beta and gamma actin mRNAs are differentially located within myoblasts. J Cell Biol. 1993;122:825–832. doi: 10.1083/jcb.122.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hortsch M, Avossa D, Meyer D I. Characterization of secretory protein translocation: ribosome-membrane interaction in endoplasmic reticulum. J Cell Biol. 1986;103:241–253. doi: 10.1083/jcb.103.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Kiebler, M. A., I. Hemraj, P. Verkade, M. Köhrmann, P. Fortes, R. M. Marión, J. Ortín, and C. Dotti. The mammalian Staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J. Neurosci., in press. [DOI] [PMC free article] [PubMed]
- 29.Kim-Ha J, Kerr K, MacDonald P M. Translational regulation of oskar mRNA by Bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 30.Ku M, Melton D A. Xwnt-11: a maternally expressed Xenopus wnt gene. Developments. 1993;119:1161–1173. doi: 10.1242/dev.119.4.1161. [DOI] [PubMed] [Google Scholar]
- 31.Li P, Yang X, Wasser M, Cai Y, Chia W. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell. 1997;90:437–447. doi: 10.1016/s0092-8674(00)80504-8. [DOI] [PubMed] [Google Scholar]
- 32.Lu Y, Qian X Y, Krug R M. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 1994;8:1817–1828. doi: 10.1101/gad.8.15.1817. [DOI] [PubMed] [Google Scholar]
- 33.Marión R M, Aragón T, Beloso A, Nieto A, Ortín J. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 1997;25:4271–4277. doi: 10.1093/nar/25.21.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosquera L, Forristall C, Zhou Y, King M L. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development. 1993;117:377–386. doi: 10.1242/dev.117.1.377. [DOI] [PubMed] [Google Scholar]
- 35.Mowry K L. Complex formation between stage-specific oocyte factors and a Xenopus mRNA element. Proc Natl Acad Sci USA. 1996;93:14608–14613. doi: 10.1073/pnas.93.25.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuman S F, Schupbach T. Dorsoventral axis formation in Drosophila depends on the correct dosage of the gene gurken. Development. 1994;120:2457–2463. doi: 10.1242/dev.120.9.2457. [DOI] [PubMed] [Google Scholar]
- 37.Ortín J. Multiple levels of post-transcriptional regulation of Influenza virus gene expression. Semin Virol. 1998;3:335–342. [Google Scholar]
- 38.Ortín J, Doerfler W. Transcription of the genome of adenovirus type 12: I. Viral mRNA in abortively infected and transformed cells. J Virol. 1975;15:27–35. doi: 10.1128/jvi.15.1.27-35.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortín J, Nájera R, López C, Dávila M, Domingo E. Genetic variability of Hong Kong (H3N2) influenza viruses: spontaneous mutations and their location in the viral genome. Gene. 1980;11:319–331. doi: 10.1016/0378-1119(80)90072-4. [DOI] [PubMed] [Google Scholar]
- 40.Park Y W, Katze M G. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J Biol Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 41.Perales B, Ortín J. The influenza A virus PB2 polymerase subunit is required for the replication of viral RNA. J Virol. 1997;71:1381–1385. doi: 10.1128/jvi.71.2.1381-1385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu Y, Krug R M. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A) J Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu Y, Nemeroff M, Krug R M. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA. 1995;1:304–316. [PMC free article] [PubMed] [Google Scholar]
- 44.Rose J K, Buonocore L, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Seong B L, Brownlee G G. A new method for reconstituting influenza polymerase and RNA in vitro: a study of the promoter elements for cRNA and vRNA synthesis in vitro and viral rescue in vivo. Virology. 1992;186:247–260. doi: 10.1016/0042-6822(92)90079-5. [DOI] [PubMed] [Google Scholar]
- 47.St Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 48.St Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- 49.St Johnston D, Brown N H, Gall J G, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.St Johnston D, Nusslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68:201–19. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- 51.Trapp B D, Moench T, Pulley M, Barbosa E, Tennekoon G, Griffin J. Spatial segregation of mRNA encoding myelin-specific proteins. Proc Natl Acad Sci USA. 1987;84:7773–7777. doi: 10.1073/pnas.84.21.7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson I A, Brindle K M, Fulton A M. Differential localization of the mRNA of the M and B isoforms of creatine kinase in myoblasts. Biochem J. 1995;308:599–605. doi: 10.1042/bj3080599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu S, Romano P R, Wek R C. Ribosome targeting of PKR is mediated by two double-stranded RNA-binding domains and facilitates in vivo phosphorylation of eukaryotic initiation factor-2. J Biol Chem. 1997;272:14434–14441. doi: 10.1074/jbc.272.22.14434. [DOI] [PubMed] [Google Scholar]