Abstract
GATA transcription factors are a class of transcriptional regulatory proteins that contain a characteristic type-IV zinc finger DNA-binding domain, which play important roles in plant growth and development. The GATA gene family has been characterized in various plant species. However, GATA family genes have not been identified in cucumber. In this study, 26 GATA family genes were identified in cucumber genome, whose physicochemical characteristics, chromosomal distributions, phylogenetic tree, gene structures conserved motifs, cis-regulatory elements in promoters, homologous gene pairs, downstream target genes were analyzed. Tissue expression profiles of cucumber GATA family genes exhibited that 17 GATA genes showed constitutive expression, and some GATA genes showed tissue-specific expression patterns. RNA-seq analysis of green and virescent leaves revealed that seven GATA genes might be involved in the chloroplast development and chlorophyll biosynthesis. Importantly, expression patterns analysis of GATA genes in response to abiotic and biotic stresses indicated that some GATA genes respond to either abiotic stress or biotic stress, some GATA genes such as Csa2G162660, Csa3G017200, Csa3G165640, Csa4G646060, Csa5G622830 and Csa6G312540 were simultaneously functional in resistance to abiotic and biotic stresses. Overall, this study will provide useful information for further analysis of the biological functions of GATA factors in cucumber.
Keywords: cucumber, GATA, gene family, expression pattern, stress response
1. Introduction
GATA transcription factors, a group of transcriptional regulatory proteins, are encoded by small multigene families. GATA protein sequences contain either one or two highly conserved type-IV zinc-finger motifs (C-X2-C-X17–20-C-X2-C) and a DNA-binding domain recognized as the DNA consensus sequence (A/T)GATA(A/G) [1]. The GATA family is widely distributed in eukaryotic organisms including animals, plants and fungi. In animals, GATA factors typically contain two conserved C-X2-C-X17-C-X2-C zinc-finger domains, but only the C-terminal finger is involved in DNA binding, and they have been shown to play critical roles in the processes of development, differentiation and cell proliferation [2]. In plants, the majority of GATA factors contain only a single C-X2-C-X18-C-X2-C or C-X2- C-X20-C-X2-C zinc finger domain, and several GATA factors encode two zinc finger domains. Plant GATA factors play important roles in plant growth and development, biotic and abiotic stresses, secondary metabolism and other biological processes [3]. The fungal GATA factors generally contain a C-X2-C-X17-C-X2-C or C-X2-C-X18-C-X2-C domain, and they have been shown to be involved in the global regulation of nitrogen metabolism, light-regulated photomorphogenesis, circadian regulation and mating-type switching [4,5].
The first plant GATA gene (NTL1) was cloned from Nicotiana tabacum [6]. The GATA family was subsequently identified in a number of plants, such as Oryza sativa [3], Arabidopsis thaliana [3,7], Glycine max [8], Malus domestica [9], Vitis vinifera [10], Solanum lycopersicum [11], Gossypium raimondii, Gossypium arboreum, Gossypium hirsutum [12], Brassica napus [13], Brachypodium distachyon [14], Capsicum annuum [15] and so on. In higher plants, GATA genes are involved in various biological processes. For instance, the Arabidopsis GATA2 (At2g45050) is a key light-signaling transcription factor that mediates photomorphogenesis [16]. GATA factor, Nitrate-inducible, Carbon metabolism-involved (GNC) and Cytokinin-responsive GATA1/GNC-Like (CGA1/GNL) serve important functions in chlorophyll synthesis and potentially regulate carbon and nitrogen metabolism [17]. It was also found that GATA transcription factor PdGNC regulates chloroplast ultrastructure and photosynthesis in poplar [18]. ZIM (At4g24470) is a GATA transcription factor involved in inflorescence and flower development [19]. The GATA transcription factor HANABA TARANU is required in early embryo development of Arabidopsis [20]. GATA factors have also been implicated in the regulation of nitrogen assimilation in plants. GATA motifs have been identified in the regulatory regions of many genes involved in nitrate assimilation such as nitrate reductase (NIA), nitrite reductase (NiR) and glutamine synthetase [21,22]. Furthermore, GATA factors are responsive to hormone signals, such as auxin and gibberellin signals, which regulate the downstream target genes GNC and GNL during plant growth and development; these signals also regulate brassinosteroid, which participates in the regulation of the GATA transcription factor GATA2 during Arabidopsis photomorphogenesis [16,23]. These studies reported that the GATA transcription factors were involved in the regulation of photomorphogenesis, nitrogen metabolism, light-responsive development, chlorophyll biosynthesis, flowering transition and abiotic stress response. However, the biological functions of GATA transcription factor family members remain poorly understood.
Cucumber (Cucumis sativus L.), one of the most economically important vegetable crop species, is the first vegetable crop whose complete genome sequencing project has been finished [24]. Lots of gene families such as WRKY [25], MADS-box [26], NBS [27], bZIP [28], LEA [29], CLE [30] and so on have been reported in cucumber. However, the GATA transcription factors family has not been identified in cucumber. In addition, the functional analysis of GATA transcription factors mainly focused on the abiotic stress such as low nitrogen [8], light [9], cold, drought, salt [11,13] and phytohormones [10,14], but lack of expression patterns analysis of GATA genes in response to biotic stress. Therefore, in this study, we performed the systematic bioinformatics analysis of GATA transcription factors and analyzed the expression profiles of GATA family genes under the abiotic and biotic stresses in cucumber, which provides valuable information and candidate genes for cucumber resistance breeding.
2. Results
2.1. Genome-Wide Identification and Chromosomal Distribution of GATA Family Genes in Cucumber
A total of 26 GATA family members were identified from cucumber genome using HMMER 3.0 software. The physical and chemical properties of 26 cucumber GATA genes and encoded proteins, including coding sequence (CDS) sizes, number of amino acids, molecular weights, protein isoelectric points (pI), instability indexes, aliphatic indexes, grand average of hydropathicity (GRAVY) values, and genomic locations, were analyzed as shown in Table 1. The CDS size of 26 cucumber GATA genes ranged from 420 bp (Csa6G312540) to 1620 bp (Csa3G017200), with the number of amino acids of GATA proteins accordingly ranging from 139 to 539 aa. The molecular weights of 26 GATA proteins ranged from approximately 15.13 to 59.97 kD. The aliphatic indexes of 26 GATA proteins ranged from 32.48 (Csa6G502700) to 77.40 (Csa7G405980). The pI of 26 GATA proteins varied from 4.86 (Csa2G370420) to 9.83 (Csa4G286370). The instability index was greater than 40 for each GATA protein except Csa4G286370 and Csa7G405980, which suggested that most GATA proteins were stable proteins except Csa4G286370 and Csa7G405980. The GRAVY values of all 26 GATA proteins were less than zero, indicating that these proteins were hydrophilic.
Table 1.
Detailed information of 26 predicted GATA proteins in cucumber. CDS, the coding sequence of a gene; pI, protein isoelectric point.
| Gene Name | CDS Size (bp) | Number of Amino Acids (aa) | Molecular Weight (kD) | pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Genomic Location |
|---|---|---|---|---|---|---|---|---|
| Csa1G569090 | 480 | 159 | 17.40 | 9.41 | 43.36 | 63.14 | −0.597 | Chr1:20743377-20744463 |
| Csa1G587970 | 978 | 325 | 35.68 | 8.71 | 57.34 | 64.25 | −0.664 | Chr1:22118620-22120914 |
| Csa2G162660 | 864 | 287 | 31.02 | 6.53 | 67.35 | 62.2 | −0.570 | Chr2:9330657-9332857 |
| Csa2G251490 | 1005 | 334 | 36.47 | 5.86 | 63.14 | 60.42 | −0.580 | Chr2:12381097-12382974 |
| Csa2G370420 | 1059 | 352 | 38.58 | 4.86 | 44.39 | 62.84 | −0.706 | Chr2:18236800-18244086 |
| Csa2G370430 | 855 | 284 | 30.70 | 6.32 | 43.10 | 66.58 | −0.630 | Chr2:18245818-18251412 |
| Csa2G373450 | 1125 | 374 | 40.47 | 5.25 | 60.87 | 62.17 | −0.647 | Chr2:18666831-18668584 |
| Csa3G017200 | 1620 | 539 | 59.97 | 6.49 | 56.47 | 68.72 | −0.632 | Chr3:1732854-1739477 |
| Csa3G165640 | 447 | 148 | 16.11 | 9.71 | 66.68 | 63.92 | −0.785 | Chr3:10937552-10939059 |
| Csa3G457670 | 960 | 319 | 34.84 | 5.67 | 55.15 | 52.07 | −0.579 | Chr3:20795136-20796243 |
| Csa3G843820 | 873 | 290 | 32.23 | 9.38 | 68.64 | 65.24 | −0.662 | Chr3:34100073-34101438 |
| Csa3G895650 | 1056 | 351 | 39.08 | 5.56 | 42.17 | 53.39 | −0.813 | Chr3:38551528-38552834 |
| Csa3G912920 | 1542 | 513 | 57.19 | 6.19 | 58.56 | 68.58 | −0.659 | Chr3:39577751-39583126 |
| Csa4G043890 | 924 | 307 | 34.24 | 5.48 | 75.12 | 63.49 | −0.678 | Chr4:3394536-3395860 |
| Csa4G046650 | 768 | 255 | 27.06 | 9.21 | 49.09 | 50.24 | −0.656 | Chr4:3624324-3626040 |
| Csa4G286370 | 531 | 176 | 19.86 | 9.83 | 37.02 | 47.73 | −1.150 | Chr4:11065609-11066229 |
| Csa4G646060 | 1254 | 417 | 46.03 | 7.67 | 48.48 | 53.12 | −0.953 | Chr4:21924862-21927071 |
| Csa5G622830 | 1068 | 355 | 38.46 | 5.85 | 57.10 | 66.51 | −0.543 | Chr5:24735193-24738505 |
| Csa6G312540 | 420 | 139 | 15.13 | 9.76 | 60.79 | 52.01 | −0.950 | Chr6:14872370-14873463 |
| Csa6G405920 | 984 | 327 | 36.03 | 5.39 | 58.06 | 72.14 | −0.570 | Chr6:18352364-18354766 |
| Csa6G502700 | 645 | 214 | 24.17 | 7.6 | 56.34 | 32.48 | −1.028 | Chr6:25337427-25338242 |
| Csa6G504690 | 807 | 268 | 30.48 | 7.22 | 55.43 | 53.84 | −0.962 | Chr6:25624800-25625808 |
| Csa7G064580 | 1335 | 444 | 48.13 | 6.15 | 46.66 | 75.97 | −0.405 | Chr7:3845510-3853491 |
| Csa7G405980 | 510 | 169 | 18.21 | 9.08 | 38.63 | 77.4 | −0.275 | Chr7:15586933-15588082 |
| Csa7G447800 | 912 | 303 | 33.95 | 6.39 | 44.74 | 64.65 | −0.871 | Chr7:18027186-18032948 |
| Csa7G452960 | 1002 | 333 | 36.32 | 6.26 | 49.71 | 59.19 | −0.608 | Chr7:19135685-19136959 |
Based on the physical positions of GATA genes annotated in the cucumber_ChineseLong_v2 GFF file, the chromosomal locations of 26 GATA genes were marked on the physical map of cucumber. The 26 GATA genes were located on all the seven cucumber chromosomes with different densities (Supplementary Figure S1). Chromosome 3 contained the largest number of GATA genes with six GATA genes. Chromosome 5 contained the lowest number of GATA genes with only one GATA gene.
2.2. Phylogenetic Analysis and Sequence Alignment of GATA Proteins
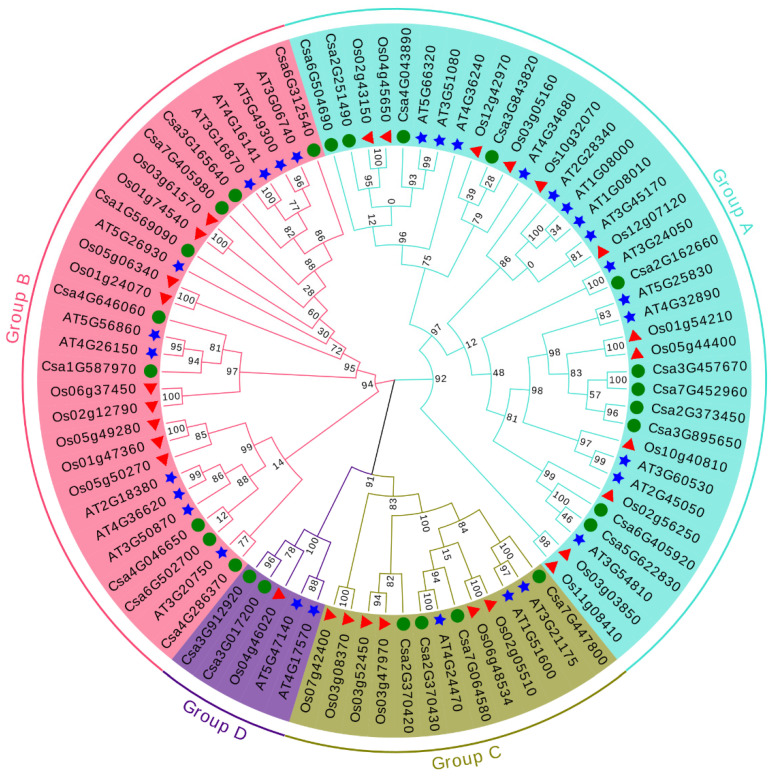
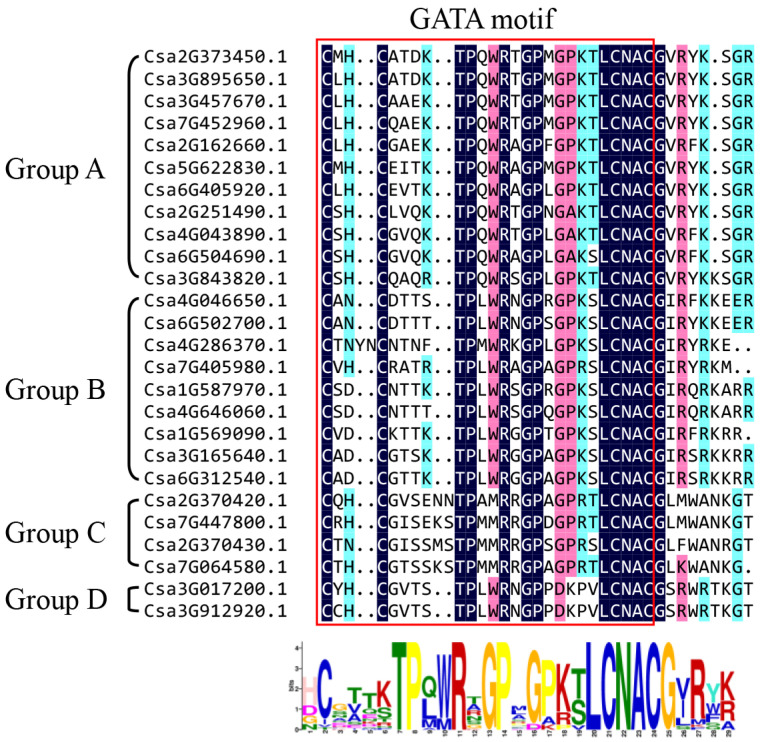
To analyze the phylogenetic relationship of the GATA genes among different species and classify the GATA genes identified in cucumber, a maximum likelihood phylogenetic tree was constructed based on the multiple sequences alignment of 26 cucumber GATA proteins, 30 A. thaliana GATA proteins and 28 rice GATA proteins (Figure 1). According to the classification of Arabidopsis and rice GATA proteins, cucumber GATA family proteins were divided into four groups (A, B, C and D). Among the four classified groups, group A had the largest number of cucumber GATA proteins (11 GATA proteins), accounting for 42.3% of the total cucumber GATA proteins. Group D had the least number of GATA proteins with only two members (8.0%), namely, Csa3G017200 and Csa3G912920. To further analyze the sequence features of the 26 cucumber GATA proteins, their conserved domain sequences were aligned. The multiple sequence alignment revealed that all GATA proteins contained the conserved domain C-X2-C-X18–20-C-X2-C with the exception of Csa4G286370 which possessed two extra amino acids to form C-X4-C-X18-C-X2-C (Figure 2). The characteristics of cucumber GATA domains in each group were generally consistent with previously studied GATA domains in A. thaliana. For example, all GATA members in group C had an insertion of two amino acids. The GATA motifs and conserved amino acid sites in different groups may contribute to the various functions of these GATA proteins.
Figure 1.
The phylogenetic tree of the total GATA proteins from cucumber, Arabidopsis and rice. Phylogenetic relationship of GATA proteins from cucumber (26), Arabidopsis (30) and rice (28) were performed with MEGA 7.0.26 using the maximum likelihood method with 1000 bootstrap replicates. The arcs with different colors represent four major groups of GATA proteins. GATA members of cucumber, Arabidopsis, and rice were represented by green circles, blue stars, and red triangles, respectively. The number represented the bootstrap replicates.
Figure 2.
Alignment of conserved domain sequences from 26 GATA proteins in cucumber. GATA motif and amino acid sites were shown at the top, and sequence identities were shown at the bottom.
2.3. Phylogenetic, Gene Structure and Conserved Motif Analysis of Cucumber GATA Proteins
Analysis of the exon/intron organization of 26 cucumber GATA genes revealed that the numbers of exon in GATA genes varied from 1 (Csa6G504690) to 11 (Csa7G064580). Group A contained the lowest average number of exons per gene, 1.9, while group C had the highest, 8.8. Furthermore, the structural characteristics of GATA genes in the same group were similar but varied among different groups (Figure 3). For example, in group C, each GATA gene contained more than seven exons, while each GATA gene in group B comprised two or three exons. A total of 10 conserved motifs, designated as motifs 1–10, were identified in the 26 cucumber GATA proteins. The amino acid sequences of each conserved motif were shown in Supplementary Table S1. Most GATA proteins in the same group generally contained similar conserved motif compositions (Figure 3). For example, GATA proteins in group A had an average of five conserved motifs, including motif 1, which was annotated as the GATA zinc finger domain according to the Pfam database, and the other motifs 2, 4, 6, 9. In addition to motif 1, all GATA proteins in group C contained conserved motifs 3 and 5, representing CCT and TIFY domains, respectively. All GATA proteins in group D contained motif 7 and motif 10 (representing ASXH and RPN13_C domains, respectively). Taken together, the conserved motif compositions of GATA proteins in the same group were similar but varied among different groups.
Figure 3.
Phylogenetic relationship, gene structure, and conserved motif analysis of cucumber GATA genes. Left: Phylogenetic tree of 26 cucumber GATA proteins. The neighbor-joining phylogenetic tree was constructed using MEGA 7.0.26, with 1000 replicates. Middle: Exon-intron structures of cucumber GATA genes. Orange boxes represent exons, black lines represent introns, and the upstream/downstream regions of GATA genes are represented by green boxes. Right: Conserved motifs of cucumber GATA proteins. Ten conserved motifs are shown in different colored boxes, and the details of the motifs are provided in Table S1.
2.4. Homologous Gene Pairs and Synteny Analysis
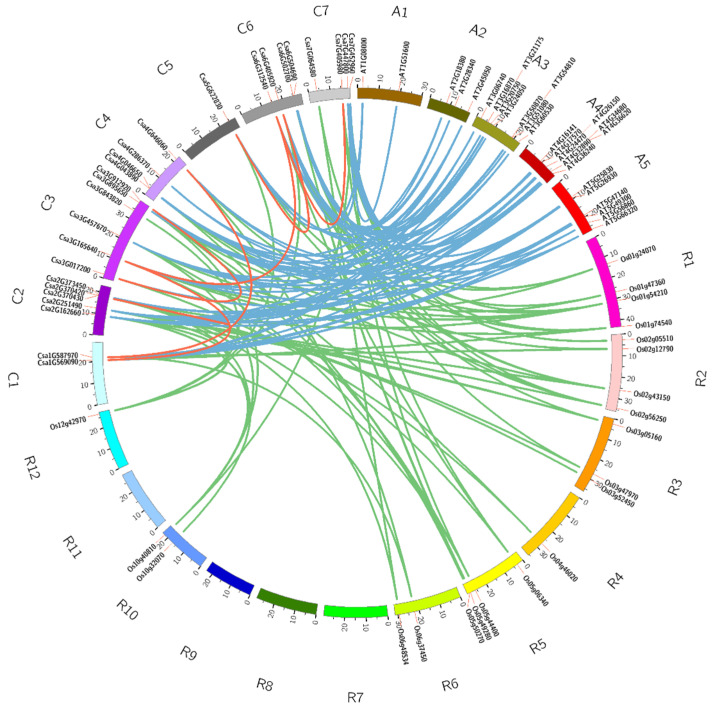
Analysis of cucumber GATA gene duplication events identified seven pairs of putative paralogous genes including one tandem duplication (Csa2G370420/Csa2G370430) and six segmental duplications (Csa1G569090/Csa3G165640, Csa1G587970/Csa4G646060, Csa3G017200/Csa3G912920, Csa3G165640/Csa6G312540, Csa5G622830/Csa6G405920, Csa6G312540/Csa7G405980), which suggest that segmental duplication played a crucial role in the expansion of the GATA gene family in cucumber. The orthologous GATA gene pairs among cucumber, A. thaliana and rice were also investigated in this study. The results indicated that 22 cucumber GATA genes and 28 A. thaliana GATA genes were orthologous gene pairs, which resulted in the 71 syntenic relationships across these two species (Supplementary Table S2). 25 cucumber GATA genes and 21 rice GATA genes were orthologous gene pairs with 59 syntenic relationships (Figure 4 and Supplementary Table S3). Only Csa4G286370 gene in cucumber did not form the syntenic relationship with neither A. thaliana nor rice, which means that Csa4G286370 was conservative in cucumber GATA gene family.
Figure 4.
Syntenic relationships of GATA gene family in cucumber, Arabidopsis and rice. The red lines represent the segmentally duplicated GATA genes in cucumber. The blue lines represent the orthologous relationships of GATA genes between cucumber and Arabidopsis. The green lines represent the orthologous relationships of GATA genes between cucumber and rice.
2.5. Cis-Acting Regulatory Elements in the Promoters of Cucumber GATA Genes
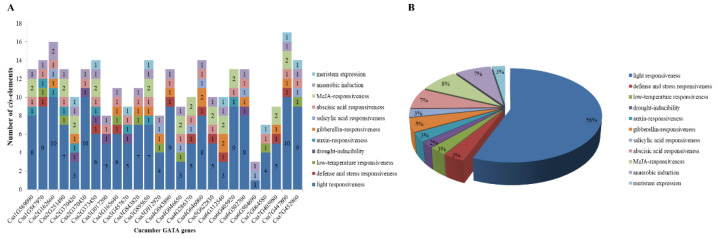
Cis-acting regulatory elements analysis identified 11 main types of cis-regulatory elements in the promoter sequences of cucumber GATA genes. The light-responsiveness cis-regulatory elements accounts for the largest proportion (up to 56%) in the total across the promoters of 26 GATA genes, which contains different kinds of cis-regulatory elements such as ACE, G-box, and MRE. Additionally, the cis-regulatory elements associated with hormone response (including auxin, salicylic acid, gibberellins, abscisic acid, and MeJA), stress response (including drought, low temperature, defense and stress); meristem expression, anaerobic induction were also identified in promoter sequences of the cucumber GATA genes (Figure 5).
Figure 5.
Cis-regulatory elements in the promoters of 26 cucumber GATA genes. (A) The number of various cis-regulatory elements in the promoters of each cucumber GATA gene. (B) The relative proportions of different cis-regulatory elements in the promoters of cucumber GATA genes are indicated by the pie chart. Cis-regulatory elements sharing identical or similar functions are represented by the same color.
2.6. The Downstream Target Genes Analysis of Cucumber GATA Genes
Through the website of Plant Transcriptional Regulatory Map, the target genes analyses of cucumber GATA genes were conducted. The target genes of cucumber GATA family genes were shown in Supplementary Table S4. Only seven cucumber GATA genes including Csa2G162660, Csa2G370430, Csa2G373450, Csa3G165640, Csa3G895650, Csa6G405920 and Csa7G447800 were found to regulate the target genes; no target gene was found for other GATA genes (Figure 6 and Table S4). Among the seven cucumber GATA genes, the gene Csa2G162660 has the largest number of target genes (1910), and the gene Csa3G895650 has the lowest number of target genes (99). These results will be a benefit to the research of transcriptional regulatory network of GATA genes.
Figure 6.
The regulatory interactions network between cucumber GATA genes and their target genes. The genes marked in red are the cucumber GATA genes. The detailed downstream target genes of cucumber GATA genes are shown in the Table S4.
2.7. Tissue Expression Profiles Analysis of Cucumber GATA Genes
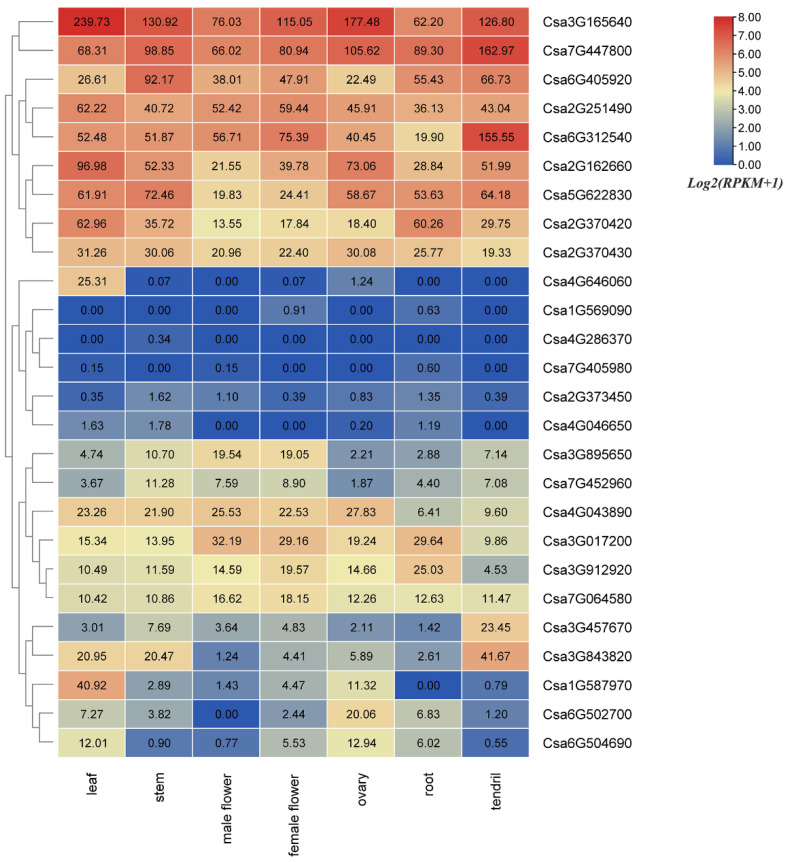
The expression patterns of all 26 cucumber GATA genes were investigated based on public transcriptomic data of different tissues of cucumber, including leaf, stem, male flower, female flower, ovary, root and tendril. Among the 26 GATA genes, 19 GATA genes were expressed in all detected samples (RPKM > 0), and 17 genes showed constitutive expression (RPKM > 1 in all samples). Overall, 34.6% (9/26) of GATA genes were highly expressed in different tissues of cucumber. Of GATA genes, 19.2% (5/26) were low or not expressed in any tissues. Additionally, 23.1% (6/26) of GATA genes were middle expressed in different tissues of cucumber. The other cucumber GATA genes were specially expressed in some tissues, such as Csa1G587970 and Csa4G646060 were highly expressed in leaf, Csa3G457670 and Csa3G843820 were highly expressed in tendril, Csa6G502700 was highly expressed in ovary, Csa6G504690 was highly expressed in leaf and ovary (Figure 7). These results revealed that the expression patterns of cucumber GATA genes were diverse in different tissues.
Figure 7.
Tissue-specific expression of GATA genes in cucumber. The transcriptional levels of GATA genes in seven tissues (leaf, stem, male flower, female flower, ovary, root, and tendril) of cucumber 9930 were investigated based on a public transcriptome data. The heatmap was constructed using the TBtools software, and the RPKM (reads per kilobase per million mapped reads) values of GATA genes were transformed by log2(RPKM+1). The data in the boxes indicate original RPKM values. The red and blue colors represent the higher and lower relative expression levels, respectively.
2.8. Expression Profiles Analysis of Cucumber GATA Genes during Chlorophyll Biosynthesis
To explore the potential functions of cucumber GATA genes in chloroplast development and chlorophyll biosynthesis, RNA-seq analysis of green and virescent true leaves were conducted. As compared with the virescent leaf, most GATA genes were up-regulated in the green leaf. As shown in Figure 8, eight cucumber GATA genes were differentially expressed between green and virescent leaves. Among them, seven cucumber GATA genes including Csa3G165640, Csa5G622830, Csa3G843820, Csa6G405920, Csa6G502700, Csa6G504690 and Csa7G452960 were significantly induced in the green leaf compared with virescent leaf, only one cucumber GATA gene Csa3G017200 was significantly down-regulated in the green leaf compared with virescent leaf. Notably, although the Log2FC(EC_1/104Y_1) value of Csa4G046650 was 2.69, the FPKM value in green and virescent leaves were all lower than five, which would be filtered. Thus, the above results revealed that seven cucumber GATA family genes might be involved in the chloroplast development and chlorophyll biosynthesis.
Figure 8.
Expression profiles of cucumber GATA genes between green and virescent leaves. The fragments per kilobase of transcript per million fragments (FPKM) values of GATA genes were transformed by log2(FPKM+1). The data in the boxes indicated original FPKM values. The red and blue colors represent the higher and lower relative expression levels, respectively. In the right table, differentially expressed genes (DEGs) are highlighted by red (up-regulation) and green (down-regulation). FC represent fold-change. 104Y-1 represent the first true leaf of virescent plant 104Y, 104Y-1-1, 104Y-1-2 and 104Y-1-3 were three biological replications of virescent leaves. EC1-1 represent the first true leaf of green plant EC1, EC1-1-1, EC1-1-2 and EC1-1-3 were three biological replications of green leaves.
2.9. Expression Profiles Analysis of Cucumber GATA Genes under Abiotic Stresses
To understand the expression profiles of cucumber GATA genes under abiotic stresses, the available transcriptomic data were used to analysis the expression levels of cucumber GATA genes in response to the treatments of high temperature and low nitrogen. Under high temperature treatment, six GATA genes were significantly induced/repressed by high temperatures. Among them, the expression levels of Csa2G162660, Csa3G017200, Csa7G064580, Csa4G646060 and Csa3G457670 were significantly up-expressed. The expression levels of these five GATA genes in response to high temperature for 3 h (hours) was higher than that for 6 h, which revealed that these five GATA genes responded quickly to high temperatures. The expression level of Csa5G622830 was significantly down-expressed in resistance to high temperature, which suggested that this GATA gene was repressed by high temperatures. Under the low nitrogen stress, Csa6G312540 and Csa7G452960 were down-regulated in root, Csa2G162660 was down-regulated expressed in leaf, and Csa4G043890 was up-regulated in leaf. Thus, these four cucumber GATA genes were associated with the response to low nitrogen (Figure 9).
Figure 9.
Expression profiles of cucumber GATA genes in response to various abiotic stress treatments including high temperature, low nitrogen and GA. HT = high temperature; HT_0h = heat treatment for 0 h (hours); HT_3h = heat treatment for 3 h; HT_6h = heat treatment for 6 h. CK means control plant; LN means low nitrogen. The FPKM or RFPKM values of GATA genes were transformed by log2(FPKM+1) and log2(RPKM+1). The data in the boxes indicate original FPKM or RPKM values. The red and blue colors represent the higher and lower relative expression levels, respectively.
2.10. Expression Profiles Analysis of Cucumber GATA Genes under Biotic Stresses
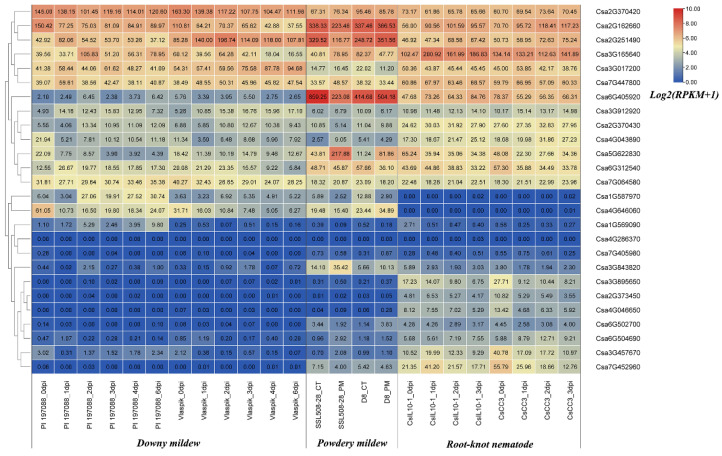
Previous studies only performed the functional analysis of plant GATA genes under abiotic stresses such as cold, salt and drought. In this study, the expression patterns of cucumber GATA genes under biotic stresses including downy mildew, powdery mildew and root-knot nematode were analyzed with the big data of cucumber transcriptome sequencing (Figure 10). After the treatment with downy mildew inoculation, the expression levels of Csa2G162660, Csa4G646060 and Csa5G622830 in both resistant and susceptible cucumber lines were all down-regulated. With the extension of inoculation time in the resistant cucumber line, the expression levels of Csa2G251490 and Csa3G017200 were initially increased and then decreased to the similar expression level as the control plant. While in the susceptible cucumber line, the expression levels of Csa2G251490 and Csa3G017200 were up-regulated after inoculation and then down-regulated to the expression levels that were higher than the expression levels of mock plant. Csa6G312540 was up-regulated after inoculation in the resistant cucumber line and then decreased to the expression level that was higher than the expression level of mock plant; however, Csa6G312540 was up-regulated after inoculation in the susceptible cucumber line and then down-regulated to the expression level that was lower than the expression level of mock plant. Thus, these six cucumber GATA genes were associated with downy mildew resistance in cucumber.
Figure 10.
Expression profiles of cucumber GATA genes in response to various biotic stress treatments including downy mildew, powdery mildew and root-knot nematode. PI 197088 is the downy mildew-resistant cucumber plant; Vlaspik is the downy mildew-susceptible cucumber plant; dpi means the days post inoculation. SSL508-28 is the powdery mildew-resistant cucumber plant; D8 is the powdery mildew-susceptible cucumber plant; CT is the control plant; PM is the abbreviation of powdery mildew. CsIL10_1 is the root-knot nematode resistant cucumber plant, CsCC3 is the root-knot nematode susceptible cucumber plant. The RPKM values of GATA genes were transformed by log2(RPKM+1). The data in the boxes indicated original RPKM values. The red and blue colors represented the higher and lower relative expression levels, respectively.
Under the stress of powdery mildew inoculation, the expression levels of Csa5G622830 were up-regulated in the resistant and susceptible cucumber lines. The expression levels of Csa2G162660, Csa2G251490 and Csa6G405920 were down-regulated in the resistance cucumber line and up-regulated in the susceptible cucumber line. The expression level of Csa3G165640 was up-regulated in the resistance cucumber line and down-regulated in the susceptible cucumber line. The expression levels of Csa3G017200 and Csa6G312540 did not change in the resistant cucumber line; however, they were down-regulated in the susceptible cucumber line. These results indicate that the above seven cucumber GATA genes were related to the powdery mildew resistance in cucumber.
After the treatment of root-knot nematode infection, the expression levels of Csa2G162660 and Csa3G165640 were up-regulated in the resistant and susceptible cucumber plants, and the expression levels in the resistant cucumber were higher than those in the susceptible cucumber. The expression levels of Csa5G622830 were down-regulated in the resistant and susceptible cucumber lines. The expression level of Csa6G405920 was up-regulated in the resistant cucumber and down-regulated in the susceptible cucumber. The results showed that these four cucumber GATA genes responded to the root-knot nematode infection.
3. Discussion
Plant transcription factors, such as WRKY [31], MYB [32], bHLH [33], and zinc-finger [34], play a key role in governing gene regulation that mediates diverse biological processes in plant developmental processes, stress responses, and hormone signaling pathways [35]. GATA proteins are defined as GATA transcription factors due to their specific binding to the consensus sequence (A/T) GATA (A/G), which play important roles in plant growth and development [3]. The GATA gene family has been identified in various plant species, such as Oryza sativa [3], Arabidopsis thaliana [3,7], Glycine max [8], Malus domestica [9], Vitis vinifera [10], Solanum lycopersicum [11], Gossypium raimondii, Gossypium arboreum, Gossypium hirsutum [12], Brassica napus [13], Brachypodium distachyon [14], Capsicum annuum [15] and so on. Cucumber is the first vegetable crop whose whole-genome sequencing has been finished; however, genome-wide identification of GATA gene family in cucumber has not been conducted yet. Therefore, genome-wide characterization and expression analysis of GATA gene family in cucumber will help us understand further GATA gene functions.
In this study, it is the first time to identify and characterize GATA gene family in cucumber using bioinformatics methods. A total of 26 GATA genes were identified and classified into four subfamilies (groups A to D) in cucumber (Figure 1; Figure 3). Consistent with A. thaliana and rice, group A harbored the largest number of GATA genes (Figure 1). The results of phylogenetic tree analysis were, to some extent, consistent with the results of synteny analysis, which means that these GATA homologous gene pairs were more closely related to each other ( Figure 1; Figure 4). The analysis of exon/intron structure and conserved motifs revealed that the GATA genes in each subfamily have special characteristics. A comparison of gene structures indicated that the number of exons/introns and motifs varies between subfamilies, but is similar within each subfamily (Figure 3).
Gene duplication events including tandem, segment and transposition duplications are crucial in genomic rearrangement, which often result in expansion of gene family [36]. The GATA genes in cucumber only contained one tandem duplication and six segmental duplications (Figure 4), indicating that the GATA genes did not undergo the large-scale gene expansion. Most GATA genes in cucumber may involve an early divergence time or be obtained from gene transposition, which is consistent with previous studies demonstrating the absence of recent whole-genome duplication resulting the presence of few tandem in cucumber [24].
Along with the rapid development of high-throughput sequencing technologies, numerous omics studies, especially genome and transcriptome analysis, have been widely conducted. The time of big data has been coming. The big data of cucumber transcriptome sequencing have been validated with the qRT-PCR analysis and peer-reviewed, which could be considered as reliable data. Therefore, the effective utilization of these big data regarding cucumber transcriptome sequencing can not only reduce the research cost, but also facilitate the deep mining of the data of each transcriptome sequence [37,38,39,40]. In this study, the expression profiles of GATA genes were performed based on eight types of public cucumber transcriptome data. The genes Csa4G286370 and Csa7G405980 were not expressed in any tissues (Figure 7), which reveals that the two genes might be non-functional genes or occur transcriptional gene silencing and post-transcriptional gene silencing [41,42]. Some genes were highly expressed in different tissues and some genes were expressed in specific tissues, which showed the functional difference in GATA genes in cucumber. The expression profiles analysis of cucumber GATA genes in green and virescent leaves shows that seven GATA genes might be involved in the chloroplast development and chlorophyll biosynthesis (Figure 8), which is consistent with the functions of Arabidopsis homologous GATA genes GNC and CGA1 [17]. In the soybean and poplar, the GATA genes GmGATA58 and PdGATA19 were also demonstrated to be involved in regulating chlorophyll biosynthesis [18,43].
To comment on the role of cucumber GATA genes in abiotic stresses, we analyzed the expression patterns of these GATA genes in response to high temperature and low nitrogen treatments based on the published cucumber transcriptome sequencing data (Figure 9). In our study, four cucumber GATA genes (Csa6G312540, Csa7G452960, Csa2G162660 and Csa4G043890) were associated with the response to low nitrogen. In a previous study, four soybean GATA genes (GmGATA10/16/24/62) also exhibited different expression levels in both leaves and roots compared with the control under the low nitrogen treatment [8]. Interestingly, several cucumber GATA genes were identified to respond to both kinds of abiotic stresses. For example, under the treatments of high temperature and low nitrogen, the GATA gene Csa2G162660 was simultaneously differentially expressed between the control and treated materials. However, some other GATA genes were only differentially expressed under one type of abiotic stresses, for example, Csa5G622830 was only down-regulated under high temperature treatment, the expression level did not change under low nitrogen treatment. The results revealed that some GATA genes play the general roles under several kinds of abiotic stresses, while some other GATA genes only play roles under one specific abiotic stress.
In addition to the abiotic stresses, the expression patterns of cucumber GATA genes were also observed under the biotic stresses (Figure 10). Expression patterns analysis showed that six, seven, and four cucumber GATA genes responded to downy mildew, powdery mildew, and root-knot nematode infections, respectively. Among them, two GATA genes Csa2G162660 and Csa5G622830 were all differentially expressed between control and treated materials after the infections of downy mildew, powdery mildew and root-knot nematode. Earlier, it had been reported that Csa5G622830 was the candidate gene for the downy and powdery mildew resistance in cucumber from our lab [44]. After the infections of downy mildew and powdery mildew, the expression levels of three GATA genes including Csa2G251490, Csa3G017200 and Csa6G312540 were simultaneously changed between control and treated materials. After the infections of powdery mildew and root-knot nematode, Csa6G312540 and Csa6G405920 were differentially expressed between control and treated materials. The cucumber GATA gene Csa4G646060 was only functional to downy mildew, while not resistant to powdery mildew and root-knot nematode. These results show that some GATA genes such as Csa2G162660 and Csa5G622830 were broad-spectrum resistance, while some GATA genes such as Csa4G646060 were specific resistance. In the previous study, it had been reported that plant GATA transcription were related with some diseases. For instance, 10 Brachypodium distachyon GATA genes responded to invasion of the fungal pathogen Magnaporthe oryzae [14]. grape VdGATA2 enhanced the resistance to powdery mildew [45]. DvGATA was involved in defense to wheat powdery mildew [46]. Arabidopsis GATA23 is the essential gene for gall formation [47]. Wheat TaGATA1 positively modulates host immune response to Rhizoctonia cerealis [48].
Additionally, some GATA genes not only play the roles in response to the biotic stress, but also in response to the abiotic stress; for example, Csa5G622830 were not only functioned after the treatments of downy mildew, powdery mildew and root-knot nematode infections, but also responded to high temperature treatment. Csa2G162660 and Csa6G312540 were not only functioned after the treatments of downy mildew, powdery mildew and root-knot nematode infections, but also responded to low nitrogen stress. The results showed that some GATA genes play important roles in response to abiotic and biotic stresses. In this study, in total, six cucumber GATA genes including Csa2G162660, Csa3G017200, Csa3G165640, Csa4G646060, Csa5G622830 and Csa6G312540 were simultaneously functional in resistance to abiotic and biotic stresses.
4. Materials and Methods
4.1. Identification and Chromosomal Distribution of GATA Genes in Cucumber
To identify all the members of GATA transcription factors in cucumber, the Hidden Markov Model (HMM) file corresponding to GATA zinc finger domain (PF00320) was downloaded from protein family (Pfam) database and used as a query to search all the putative GATA genes in the cucumber genome based on an expected value (E-value) cutoff of 1 × 10−5 in HMMER 3.0 [49]. Subsequently, each of all putative cucumber GATA genes was confirmed in the SMART database (http://smart.embl-heidelberg.de/ (accessed on 12 April 2021)) [50] and the NCBI Conserved Domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (accessed on 12 April 2021)) [51]. The protein sequences of confirmed cucumber GATA transcription factor family members were analyzed with Prosite ExPASy server (http://web.expasy.org/protparam/ (accessed on 13 April 2021)) to predict their physicochemical characteristics. The chromosomal position of each confirmed GATA gene was retrieved from the GFF3 file of ChineseLong_V2 and then visualized on the cucumber chromosomes with TBtools [52].
4.2. Phylogenetic Analysis of GATA Family Genes in Cucumber, Arabidopsis and Rice
Based on the studies of GATA family genes in Arabidopsis and rice [3,7], the GATA zinc finger domain sequences of 30 Arabidopsis thaliana GATA proteins and 28 rice GATA proteins were downloaded, respectively. Multiple alignments of GATA protein sequences of cucumber, Arabidopsis and rice were performed by Muscle in MEGA 7.0.26 [53] with default parameters. Phylogenetic trees were then constructed based on the alignments using the maximum likelihood method with 1000 bootstrap replicates. The parameters were Jones-Taylor-Thornton (JTT), gamma distributed (G) rates, and partial deletion. The trees were visualized and optimized via Evolview (http://www.evolgenius.info/evolview (accessed on 15 April 2021)).
4.3. Gene Structure, Conserved Motif, Promoter Sequence Analyses of Cucumber GATA Genes
The locations of exons, introns and untranslated regions of each cucumber GATA gene were retrieved from GFF3 file of ChineseLong_V2. The conserved motifs in cucumber GATA proteins were determined with MEME server (http://memesuite.org/ (accessed on 15 April 2021)) [54] using the following parameters: maximum number of motifs, 10; minimum motif width, 6; and maximum motif width, 100. Exon/intron structures of cucumber GATA genes and conserved motifs of cucumber GATA proteins were visualized using the software TBtools [52]. Conserved domains sequences of cucumber GATA proteins were analyzed using DNAMAN software (http://en.bio-soft.net/format/DNAMAN.html (accessed on 15 April 2021)). The 1500 bp sequences upstream of the start codon of each cucumber GATA gene was extracted from the cucumber genome sequences and then submitted to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/ (accessed on 15 April 2021)) [55] for cis-regulatory elements prediction. The predicted cis-regulatory elements were classified according to their regulatory functions.
4.4. Detection of Homologous Gene Pairs and Synteny Analysis
The homologous gene pairs and syntenic relations of GATA family genes in cucumber were identified using Multiple Collinearity Scan toolkit (MCScanX) software [56] with default parameters. To predict the gene functions of cucumber GATA genes, the syntenic relationships of the orthologous GATA genes between cucumber and the model plants (Arabidopsis and rice) were examined. The syntenic relationships of GATA genes among cucumber, Arabidopsis and rice were explored using MCScanX software with the default parameters. The homologous genetic relationships of GATA genes among cucumber, Arabidopsis and rice were illustrated with Circos software [57].
4.5. Regulatory Interactions Analysis between GATA Genes and Their Target Genes
The target genes of each GATA gene were retrieved and counted from the total transcription regulatory networks of cucumber downloaded from Plant Transcriptional Regulatory Map (http://plantregmap.cbi.pku.edu.cn/download.php#networks (accessed on 17 April 2021)). The regulatory interactions network between cucumber GATA genes and their target genes was visualized with Cytoscape version 3.7.0 software (http://cytoscape.org/ (accessed on 17 April 2021)) [58].
4.6. Expression Profiles Analysis of Cucumber GATA Genes with Cucumber Transcriptome Sequencing Big Data
The expression data of cucumber GATA genes between the first true leaves of green and virescent plant were obtained from our previous study (PRJNA612596) [59]. The expression data of cucumber GATA genes in different tissue (PRJNA80169) [60] and different biotic stresses including downy mildew resistance (PRJNA285071) [61], powdery mildew resistance (PRJNA321023) [62], root-knot nematode resistance (PRJNA419665) [63] were all obtained from Cucurbit Genomics Database (CuGenDB) (http://cucurbitgenomics.org/rnaseq/home (accessed on 20 April 2021)). The expression data of cucumber GATA genes under high temperature (GSM4565536) [40] and low nitrogen (GSE46678) [64] treatments were downloaded from the NCBI’s GEO (Gene Expression Omnibus) database (https://www.ncbi.nlm.nih.gov/geo/ (accessed on 20 April 2021)). The heatmap of each GATA gene in above experiments were visualized with the TBtools software [52].
5. Conclusions
In this study, it is the first time the GATA gene family in cucumber have been identified and characterized. A total of 26 cucumber GATA genes were obtained and classified into subfamilies A–D after systematic investigations. An overview of the cucumber GATA factor gene family was revealed through the comprehensive investigation of their physicochemical characteristics, chromosomal location, phylogenetic tree, gene structure, conserved motif, cis-regulatory elements in the promoters, homologous gene pairs, synteny, and target genes. Tandem and segmental duplications contributed to the expansion of the GATA gene family, and segmental duplication tended to play the predominant role. A comparative analysis of the GATA factor gene family across cucumber, Arabidopsis, and rice helped us facilitate further gene function analysis of cucumber GATA genes. The expression patterns of the cucumber GATA genes in different cucumber tissues, between green and virescent leaves, and in response to various stresses then showed that these genes may play important roles in cucumber growth and development. Our results also provide useful information by identifying candidate tissue-specific, chlorophyll biosynthesis, abiotic and biotic stresses responsive cucumber GATA genes. This study not only provided a scientific foundation for the comprehensive understanding of the cucumber GATA gene family, but was also helpful for screening more candidate genes and breeding new varieties of cucumber with a high yield and stress resistance.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/plants10081626/s1, Figure S1: The chromosomal distribution of cucumber GATA genes. Table S1: Detail information of the conserved motifs of cucumber GATA proteins. Table S2: The detail syntenic relationships between cucumber and Arabidopsis GATA genes. Table S3: The detail syntenic relationships between cucumber and rice GATA genes. Table S4: The downstream target genes of cucumber GATA family genes.
Author Contributions
C.Y., X.L. and K.Z. conceived and designed the project; K.Z., L.J. and D.Y. conducted the bioinformatics analysis; K.Z., Y.H. and P.W. performed the analysis of cucumber transcriptome sequencing big data; K.Z., L.J. and M.K.N. wrote the paper. All authors reviewed and approved the final manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (32002061), the Talent Foundation of Anhui Science and Technology University (NXYJ202103), and the College Students’ Innovative Entrepreneurial Training Plan Program (202010879108, S202110879222).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lowry J.A., Atchley W.R. Molecular evolution of the GATA family of transcription factors: Conservation within the DNA-binding domain. J. Mol. Evol. 2000;50:103–115. doi: 10.1007/s002399910012. [DOI] [PubMed] [Google Scholar]
- 2.Patient R.K., McGhee J.D. The GATA family (vertebrates and invertebrates) Curr. Opin. Genet. Dev. 2002;12:416–422. doi: 10.1016/S0959-437X(02)00319-2. [DOI] [PubMed] [Google Scholar]
- 3.Reyes J.C., Muro-Pastor M.I., Florencio F.J. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004;134:1718–1732. doi: 10.1104/pp.103.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teakle G.R., Gilmartin P.M. Two forms of type IV zinc-finger motif and their kingdom-specific distribution between the flora, fauna and fungi. Trends Biochem. Sci. 1998;23:100–102. doi: 10.1016/S0968-0004(98)01174-8. [DOI] [PubMed] [Google Scholar]
- 5.Scazzocchio C. The fungal GATA factors. Curr. Opin. Microbiol. 2000;3:126–131. doi: 10.1016/S1369-5274(00)00063-1. [DOI] [PubMed] [Google Scholar]
- 6.Danielvedele F., Caboche M. A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol. Gen. Genet. 1993;240:365–373. doi: 10.1007/BF00280388. [DOI] [PubMed] [Google Scholar]
- 7.Bi Y.M., Zhang Y., Signorelli T., Zhao R., Zhu T., Rothstein S. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 2005;44:680–692. doi: 10.1111/j.1365-313X.2005.02568.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C., Hou Y., Hao Q., Chen H., Chen L., Yuan S., Shan Z., Zhang X., Yang Z., Qiu D., et al. Genome-wide survey of the soybean GATA transcription factor gene family and expression analysis under low nitrogen stress. PLoS ONE. 2015;10:e0125174. doi: 10.1371/journal.pone.0125174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H., Shao H., Li K., Zhang D., Fan S., Li Y., Han M. Genome-wide identification, evolution, and expression analysis of GATA transcription factors in apple (Malus × domestica Borkh.) Gene. 2017;627:460–472. doi: 10.1016/j.gene.2017.06.049. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z., Ren C., Zou L., Wang Y., Li S., Liang Z. Characterization of the GATA gene family in Vitis vinifera: Genome-wide analysis, expression profiles, and involvement in light and phytohormone response. Genome. 2018;61:713–723. doi: 10.1139/gen-2018-0042. [DOI] [PubMed] [Google Scholar]
- 11.Yuan Q., Zhang C., Zhao T., Yao M., Xu X. A Genome-wide analysis of GATA transcription factor family in tomato and analysis of expression patterns. Int. J. Agric. Biol. 2018;20:1274–1282. [Google Scholar]
- 12.Zhang Z., Zou X., Huang Z., Fan S., Qun G., Liu A., Gong J., Li J., Gong W., Shi Y., et al. Genome-wide identification and analysis of the evolution and expression patterns of the GATA transcription factors in three species of Gossypium genus. Gene. 2019;680:72–83. doi: 10.1016/j.gene.2018.09.039. [DOI] [PubMed] [Google Scholar]
- 13.Zhu W., Guo Y., Chen Y., Wu D., Jiang L. Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in Brassica napus. BMC Plant Biol. 2020;20:543. doi: 10.1186/s12870-020-02752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng W., Li W., Song N., Tang Z., Liu J., Wang Y., Pan S., Dai L., Wang B. Genome-wide characterization, evolution, and expression profile analysis of GATA transcription factors in Brachypodium distachyon. Int. J. Mol. Sci. 2021;22:2026. doi: 10.3390/ijms22042026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu C., Li N., Yin Y., Wang F., Gao S., Jiao C., Yao M. Genome-wide identification and function characterization of GATA transcription factors during development and in response to abiotic stresses and hormone treatments in pepper. J. Appl. Genet. 2021;62:265–280. doi: 10.1007/s13353-021-00618-3. [DOI] [PubMed] [Google Scholar]
- 16.Luo X.M., Lin W.H., Zhu S., Zhu J.Y., Sun Y., Fan X.Y., Cheng M., Hao Y., Oh E., Tian M., et al. Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell. 2010;19:872–883. doi: 10.1016/j.devcel.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hudson D., Guevara D., Yaish M.W., Hannam C., Long N., Clarke J.D., Bi Y.M., Rothstein S.J. GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS ONE. 2011;6:e26765. doi: 10.1371/journal.pone.0026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.An Y., Han X., Tang S., Xia X., Yin W. Poplar GATA transcription factor PdGNC is capable of regulating chloroplast ultrastructure, photosynthesis, and vegetative growth in Arabidopsis under varying nitrogen levels. Plant Cell Tissue Organ. 2014;119:313–327. doi: 10.1007/s11240-014-0536-y. [DOI] [Google Scholar]
- 19.Nishii A., Takemura M., Fujita H., Shikata M., Yokota A., Kohchi T. Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci. Biotech. Bioch. 2000;64:1402–1409. doi: 10.1271/bbb.64.1402. [DOI] [PubMed] [Google Scholar]
- 20.Nawy T., Bayer M., Mravec J., Friml J., Birnbaum K.D., Lukowitz W. The GATA factor HANABA TARANU is required to position the proembryo boundary in the early Arabidopsis embryo. Dev. Cell. 2010;19:103–113. doi: 10.1016/j.devcel.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Rastogi R., Bate N.J., Sivasankar S., Rothstein S.J. Footprinting of the spinach nitrite reductase gene promoter reveals the preservation of nitrate regulatory elements between fungi and higher plants. Plant Mol. Biol. 1997;34:465–476. doi: 10.1023/A:1005842812321. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira I.C., Coruzzi G.M. Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in arabidopsis. Plant Physiol. 1999;121:301–309. doi: 10.1104/pp.121.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richter R., Behringer C., Zourelidou M., Schwechheimer C. Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2013;110:13192–13197. doi: 10.1073/pnas.1304250110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang S., Li R., Zhang Z., Li L., Gu X., Fan W., Lucas W.J., Wang X., Xie B., Ni P., et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 25.Ling J., Jiang W., Zhang Y., Yu H., Mao Z., Gu X., Huang S., Xie B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011;12:471. doi: 10.1186/1471-2164-12-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu L., Liu S. Genome-wide analysis of the MADS-box gene family in cucumber. Genome. 2012;55:245–256. doi: 10.1139/g2012-009. [DOI] [PubMed] [Google Scholar]
- 27.Wan H., Yuan W., Bo K., Shen J., Pang X., Chen J. Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genom. 2013;14:109. doi: 10.1186/1471-2164-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baloglu M.C., Eldem V., Hajyzadeh M., Unver T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE. 2014;9:e96014. doi: 10.1371/journal.pone.0096014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altunoglu Y.C., Baloglu P., Yer E.N., Pekol S., Baloglu M.C. Identification and expression analysis of LEA gene family members in cucumber genome. Plant Growth Regul. 2016;80:225–241. doi: 10.1007/s10725-016-0160-4. [DOI] [Google Scholar]
- 30.Qin N., Gao Y., Cheng X., Yang Y., Wu J., Wang J., Li S., Xing G. Genome-wide identification of CLE gene family and their potential roles in bolting and fruit bearing in cucumber (Cucumis sativus L.) BMC Plant Biol. 2021;21:143. doi: 10.1186/s12870-021-02900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Yang T., Lin Z., Gu B., Xing C., Zhao L., Dong H., Gao J., Xie Z., Zhang S., et al. A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol. J. 2019;17:1770–1787. doi: 10.1111/pbi.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Guo C., Ahmad S., Wang Q., Yu J., Liu C., Guo Y. Systematic analysis of MYB family genes in potato and their multiple roles in development and stress responses. Biomolecules. 2019;9:317. doi: 10.3390/biom9080317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geng J., Liu J.H. The transcription factor CsbHLH18 of sweet orange functions in modulation of cold tolerance and homeostasis of reactive oxygen species by regulating the antioxidant gene. J. Exp. Bot. 2018;69:2677–2692. doi: 10.1093/jxb/ery065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Lan H., Shao Q., Wang R., Chen H., Tang H., Zhang H., Huang J. An A20/AN1-type zinc finger protein modulates gibberellins and abscisic acid contents and increases sensitivity to abiotic stress in rice (Oryza sativa L.) J. Exp. Bot. 2016;67:315–326. doi: 10.1093/jxb/erv464. [DOI] [PubMed] [Google Scholar]
- 35.Franco-Zorrilla J.M., Lopez-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cannon S.B., Mitra A., Baumgarten A., Young N.D., May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mou Y., Liu Y., Tian S., Guo Q., Wang C., Wen S. Genome-wide identification and characterization of the OPR gene family in wheat (Triticum aestivum L.) Int. J. Mol. Sci. 2019;20:1914. doi: 10.3390/ijms20081914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen C., Chen X., Han J., Lu W., Ren Z. Genome-wide analysis of the WRKYgene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020;20:443. doi: 10.1186/s12870-020-02625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K., He S., Sui Y., Gao Q., Jia S., Lu X., Jia L. Genome-wide characterization of HSP90 gene family in cucumber and their potential roles in response to abiotic and biotic stresses. Front. Genet. 2021;12:584886. doi: 10.3389/fgene.2021.584886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao P., Qin T., Chen W., Sang X., Zhao Y., Wang H. Genome-wide study of NOT2_3_5 protein subfamily in cotton and their necessity in resistance to verticillium wilt. Int. J. Mol. Sci. 2021;22:5634. doi: 10.3390/ijms22115634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jury J.K., Frayne J., Hall L. Sequence analysis of a variety of primate fertilin alpha genes: Evidence for non-functional genes in the gorilla and man. Mol. Reprod. Dev. Inc. Gamete Res. 1998;51:92–97. doi: 10.1002/(SICI)1098-2795(199809)51:1<92::AID-MRD11>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 42.Vaucheret H., Beclin C., Fagard M. Post-transcriptional gene silencing in plants. J. Cell Sci. 2001;114:3083–3091. doi: 10.1242/jcs.114.17.3083. [DOI] [PubMed] [Google Scholar]
- 43.Zhang C., Huang Y., Xiao Z., Yang H., Hao Q., Yuan S., Chen H., Chen L., Chen S., Zhou X., et al. A GATA transcription factor from soybean (Glycine max) regulates chlorophyll biosynthesis and suppresses growth in the transgenic Arabidopsis thaliana. Plants. 2020;9:1036. doi: 10.3390/plants9081036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang K., Wang X., Zhu W., Qin X., Xu J., Cheng C., Lou Q., Li J., Chen J. Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix introgression line of cucumber were controlled by a co-localized locus. Theor. Appl. Genet. 2018;131:2229–2243. doi: 10.1007/s00122-018-3150-2. [DOI] [PubMed] [Google Scholar]
- 45.Yu Y.H., Bian L., Yu K.K., Yang S.D., Zhang G.H., Guo D.L. Grape (Vitis davidii) VdGATA2 functions as a transcription activator and enhances powdery mildew resistance via the active oxygen species pathway. Sci. Hortic. 2020;267:109327. doi: 10.1016/j.scienta.2020.109327. [DOI] [Google Scholar]
- 46.He H., Zhu S., Jiang Z., Ji Y., Wang F., Zhao R., Bie T. Comparative mapping of powdery mildew resistance gene Pm21 and functional characterization of resistance-related genes in wheat. Theor. Appl. Genet. 2016;129:819–829. doi: 10.1007/s00122-016-2668-4. [DOI] [PubMed] [Google Scholar]
- 47.Olmo R., Cabrera J., Diaz-Manzano F.E., Ruiz-Ferrer V., Barcala M., Ishida T., Garcia A., Andres M.F., Ruiz-Lara S., Verdugo I., et al. Root-knot nematodes induce gall formation by recruiting developmental pathways of post-embryonic organogenesis and regeneration to promote transient pluripotency. New Phytol. 2020;227:200–215. doi: 10.1111/nph.16521. [DOI] [PubMed] [Google Scholar]
- 48.Liu X., Zhu X., Wei X., Lu C., Shen F., Zhang X., Zhang Z. The wheat LLM-domain-containing transcription factor TaGATA1 positively modulates host immune response to Rhizoctonia cerealis. J. Exp. Bot. 2020;71:344–355. doi: 10.1093/jxb/erz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finn R.D., Clements J., Eddy S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Letunic I., Khedkar S., Bork P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021;49:D458–D460. doi: 10.1093/nar/gkaa937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 53.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouze P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T., Jin H., Marler B., Guo H., et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang K., Li Y., Zhu W., Wei Y., Njogu M.K., Lou Q., Li J., Chen J. Fine mapping and transcriptome analysis of virescent leaf gene v-2 in cucumber (Cucumis sativus L.) Front. Plant Sci. 2020;11:570817. doi: 10.3389/fpls.2020.570817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z., Zhang Z., Yan P., Huang S., Fei Z., Lin K. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom. 2011;12:540. doi: 10.1186/1471-2164-12-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burkhardt A., Day B. Transcriptome and small RNAome dynamics during a resistant and susceptible interaction between cucumber and downy mildew. Plant Genome. 2016;9 doi: 10.3835/plantgenome2015.08.0069. [DOI] [PubMed] [Google Scholar]
- 62.Xu Q., Xu X., Shi Y., Qi X., Chen X. Elucidation of the molecular responses of a cucumber segment substitution line carrying Pm5.1 and its recurrent parent triggered by powdery mildew by comparative transcriptome profiling. BMC Genom. 2017;18:21. doi: 10.1186/s12864-016-3438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X., Cheng C., Zhang K., Tian Z., Xu J., Yang S., Lou Q., Li J., Chen J.F. Comparative transcriptomics reveals suppressed expression of genes related to auxin and the cell cycle contributes to the resistance of cucumber against Meloidogyne incognita. BMC Genom. 2018;19:583. doi: 10.1186/s12864-018-4979-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xin M., Wang L., Liu Y., Feng Z., Zhou X., Qin Z. Transcriptome profiling of cucumber genome expression in response to long-term low nitrogen stress. Acta Physiol. Plant. 2017;39:130. doi: 10.1007/s11738-017-2429-2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.