Abstract
Staufen (Stau) is a double-stranded RNA (dsRNA)-binding protein involved in mRNA transport and localization in Drosophila. To understand the molecular mechanisms of mRNA transport in mammals, we cloned human (hStau) and mouse (mStau) staufen cDNAs. In humans, four transcripts arise by differential splicing of the Stau gene and code for two proteins with different N-terminal extremities. In vitro, hStau and mStau bind dsRNA via each of two full-length dsRNA-binding domains and tubulin via a region similar to the microtubule-binding domain of MAP-1B, suggesting that Stau cross-links cytoskeletal and RNA components. Immunofluorescent double labeling of transfected mammalian cells revealed that Stau is localized to the rough endoplasmic reticulum (RER), implicating this RNA-binding protein in mRNA targeting to the RER, perhaps via a multistep process involving microtubules. These results are the first demonstration of the association of an RNA-binding protein in addition to ribosomal proteins, with the RER, implicating this class of proteins in the transport of RNA to its site of translation.
It is now believed that the cytoskeleton is widely used to transport mRNAs between their transcription and processing sites in the nucleus and their translation and degradation sites in the cytoplasm (3, 42, 44). One consequence of the interaction between mRNAs and the cytoskeleton is to promote differential localization and/or transport of mRNAs in subcellular compartments. Indeed, examples of mRNA targeting have been observed in both germinal and somatic cells throughout the animal kingdom (51, 55, 63). The universal use of this mechanism is also apparent when we consider the nature of the proteins which are coded by the transported mRNAs; asymmetric localization involving mRNAs coding for cytosolic, secreted, membrane-associated, or cytoskeletal proteins have all been reported. Localization of mRNAs in the cytoplasm is now considered an essential step in the regulation of gene expression and an efficient way to unevenly distribute proteins in polarized cells. In general, it is believed that mRNA localization is used to determine and/or regulate local sites of translation (46, 51, 55). Indeed, ribosomes and many translational cofactors were found in association with the cytoskeletal elements, preventing both mRNAs and translation factors from being diluted by the cellular fluid (44). Transport and local translation of specific mRNAs have been shown to play an important role in processes such as learning and memory (38), synaptic transmission (9, 22, 26, 51, 61), axis formation during development (reviewed in reference 55), cell motility (30), and asymmetric cell division (7, 36, 37, 56).
The mechanisms underlying mRNA localization are not yet fully understood, mainly because of the lack of information on the principal constituents of the ribonucleoprotein (RNP) complexes involved in this process. Nevertheless, it is known to involve both cis-acting signals in mRNA and trans-acting RNA-binding proteins which bind to this signal (55). The signals that allow mRNAs to be recognized as targets for transport and then to be localized have been mapped within their 3′ untranslated regions (UTRs) (55, 63). In contrast, the nature of the RNA-binding proteins is still obscure. Recently, a 68-kDa protein which binds the β-actin mRNA zipcode localization domain was isolated and its transcript was cloned from chicken cDNA libraries (47). This protein, which binds to microfilaments, contains RNA-binding domains (RBDs) which share strong sequence similarities with the RNP and KH motifs. In addition, 69- and 78 kDa proteins in Xenopus laevis oocyte extracts have been shown to bind to the localization signal of Vg1 mRNA (12, 50). While the 69-kDa protein was shown to bind microtubules (15), the 78-kDa Vera protein colocalized with a subdomain of the smooth endoplasmic reticulum (SER) (12). Surprisingly, molecular cloning of the two proteins revealed that they are identical and are similar to the chicken zipcode-binding protein (13, 23).
Genetic and molecular studies have shown that the activity of the staufen gene product in Drosophila is necessary for the proper localization of bicoid and oskar mRNAs to the anterior and posterior cytoplasm of oocytes, respectively, and of prospero mRNA in neuroblasts (7, 16, 28, 36, 52, 53). Staufen (Stau), a member of the double-stranded RNA (dsRNA)-binding protein family, contains (i) three copies of a domain consisting of a 65- to 68-amino-acid consensus sequence which is required to bind RNAs having double-stranded secondary structures and (ii) two copies of a short domain which retains the last 21 amino acids at the C-terminal end of the complete motif (53, 54). In vitro, it has been demonstrated that Stau binds directly to bicoid and prospero mRNAs (36, 54). However, since Stau seems to bind to any dsRNA in vitro, it is not clear whether it binds directly to these RNAs in vivo or needs cellular cofactors which make up part of a larger RNP complex to localize each mRNA. Many experiments have demonstrated that the localization of oskar, prospero, and bicoid mRNAs occurs through a multistep mechanism of active transport that is dependent on elements of the cytoskeleton (7, 17, 45, 55, 58).
To understand the mechanisms of mRNA transport in mammals and determine the nature of both the RNAs and proteins in the RNA-protein complexes, we began the cloning of the human and mouse staufen (hStau and mStau) cDNAs and the characterization of their encoded proteins. Recently, we showed by both Southern blot analysis of human DNA and fluorescent in situ hybridization on human chromosomes in metaphase that the human gene is present as a single copy in the human genome and is localized in the middle of the long arm of chromosome 20 (11). We now report the sequence of the hStau and mStau and show that the transcript is found in all tested tissues. We further demonstrate that Stau binds both dsRNA and tubulin in vitro via specific binding domains. Stau is also shown to be present in the cytoplasm in association with the rough endoplasmic reticulum (RER), implicating this protein in the targeting of RNA to its site of translation.
MATERIALS AND METHODS
Molecular cloning and sequencing of the cDNAs.
To clone an hStau homologue, we searched the GenBank database with Drosophila dsRBD sequences to find consensus sequences and eventually design degenerate oligonucleotide primers for reverse transcription (RT)-PCR. However, searching in the expressed sequence tag database identified a partial sequence, clone HFBDQ83 (GenBank accession no. T06248), with high homology to the Drosophila sequence. This clone was purchased from the American Type Culture Collection and used as a probe to screen both human brain (Clontech) and fetal total mouse (a generous gift from A. Royal) cDNA libraries as described previously (62). DNA from the isolated λgt10 clones was subcloned into a pBluescript vector (Stratagene). Double-stranded DNA (dsDNA) was sequenced by the dideoxynucleotide method according to Sequenase protocols (United States Biochemical Corp.).
Construction of fusion proteins.
The 1.2-kbp BamHI fragment of the human HFBDQ83 cDNA was subcloned in frame in either pQE32 (Qiagen) or pMAL-c (New England Biolabs), thus generating the protein fused to a hexahistidine tag or to the maltose-binding protein (MBP), respectively. The protein was expressed in bacteria by induction with isopropyl-β-d-thiogalactopyranoside (IPTG) as recommended by the manufacturer.
Full-length and internal fragments of the mStau protein were PCR amplified and cloned into pMal-c to produce MBP fusion proteins. For the expression of the internal domains, which do not contain an endogenous stop codon, the PCR fragments were cloned in a modified pMal-c vector (pMal-stop) in which stop codons were introduced at the HindIII site, by the ligation of the annealed complementary oligonucleotides 5′-AGCTTAATTAGCTGAC-3′ and 5′-AGCTGTCAGCTAATTA-3′. The MBP-mStau fusion protein, containing the full-length mStau sequence, was generated by PCR amplification with Vent DNA polymerase (New England BioLabs), using the primer pair 5′-CCTGGATCCGAAAGTATAGCTTCTACCATTG-3′ plus 5′-TACATAAGCTTCTAGATGGCCAGAAAAGGTTCAGCA-3′. The resulting 1,562-bp fragment was digested with HindIII and BamHI and ligated in the pMal-c vector. The C-terminal fragment (mStau-C) was amplified with the primer pair 5′-GGATGAATCCTATTAGTAGACTTGCAC-3′ plus 5′-TACATAAGCTTCTAGATGGCCAGAAAAGGTTCAGCA-3′, digested with HindIII, and cloned in the EagI* and HindIII sites of pMal-c. EagI* was created by filling in the cohesive ends of EagI-digested pMal-c vector, using the Klenow fragment of DNA polymerase I. This fusion vector was then digested with SacI and EcoRI, and the resulting fragment was subcloned in the pMal-stop vector to generate the mStau-RBD3 construct. The mStau-tubulin-binding domain (TBD) construct was prepared by PCR using the primer pair 5′-GCTCTAGATTCAAAGTTCCCCAGGCGCAG-3′ plus 5′-TTAAGCTTCTCAGAGGGTCTAGTGCGAG-3′; the product was digested with XbaI and HindIII and cloned in the pMal-stop vector. mStau-RBD2 and mStau-RBD1 were constructed by first amplifying a fragment using the primer pair 5′-CAATGTATAAGCCCGTGGACCC-3′ and 5′-AAAAAGCTTGTGCAAGTCTACTAATAGGATTCATCC-3′. The resulting product was digested with HindIII and cloned in the EagI* and HindIII sites of the pMal-stop vector. This vector was then used to purify the 398-bp PstI and HindIII fragment, which was subcloned in the pMAL-stop vector to generate the mStau-RBD2 construct. In the same way, the mStau-RBD1 vector was obtained by digestion with SmaI and StuI, followed by recircularization of the digestion product using T4 DNA ligase. The mStau-RBD4 was PCR amplified by using the primer pair 5′-ATAGCCCGAGAGTTGTTG-3′ plus 5′-TACATAAGCTTCTAGATGGCCAGAAAAGGTTCAGCA-3′. The resulting fragment was digested with HindIII and ligated in the pMal-stop vector at the StuI and HindIII sites. All MBP-Stau fusion plasmids were transformed in BL-21 Escherichia coli. The fusion proteins were obtained after induction with 1 mM IPTG for 2 to 3 h. Cells were lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer for immediate use, or frozen at −80°C for storage.
Antibody production and Western blotting.
For the production of antibodies, a large amount of the His-hStau fusion protein was purified on Ni-nitrilotriacetic acid (NTA) resin (Qiagen) as recommended by the manufacturer and injected into rabbits as done previously (2). For Western blotting, cells were lysed in 1% n-octylglucoside–1 mM phenylmethylsulfonyl sulfoxide–aprotinin (1 μg/ml)–pepstatin A (1 μg/ml) in phosphate-buffered saline (PBS). Protein extracts were quantified by the Bradford method (Bio-Rad), and similar amounts of proteins were separated on SDS–10% polyacrylamide gels and transferred onto nitrocellulose membranes. Membranes were blocked for 30 min in TBS (Tris-buffered saline; 10 mM Tris [pH 8.0], 150 mM NaCl)–5% dry milk and incubated with primary antibodies in TBS–0.05% Tween for 1 h at room temperature. Detection was accomplished by incubating the blots with peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) antibodies (Dimension Labs) followed by Supersignal Substrate (Pierce) as recommended by the manufacturer.
RNA-binding assay.
Bacterial extracts from IPTG-induced cultures were separated on SDS–10% polyacrylamide gels and the proteins were transferred onto nitrocellulose membranes. Membranes were incubated in the presence of 32P-labeled RNA probes in 50 mM NaCl–10 mM MgCl2–10 mM HEPES (pH 8.0)–0.1 mM EDTA–1 mM dithiothreitol–0.25% milk for 2 h at room temperature, washed in the same buffer for 30 min, and exposed for autoradiography. For competition assays, a 100- to 1,000-fold excess of cold homopolymers (Pharmacia) was added to the hybridization mixture along with the labeled probe. The 3′ UTR of bicoid cDNA (positions 4016 to 4972), which was PCR amplified from Drosophila genomic DNA and subcloned in the pBluescript vector, was transcribed by using T7 RNA polymerase in the presence of [α-32P]CTP. Synthetic RNAs (Pharmacia) were labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP. The specific activities of the bicoid and synthetic RNA probes were 1.4 × 106 and 0.5 × 106 cpm/μg, respectively.
For RNA-binding assay in solution, dilutions of the purified human His-Stau fusion protein were incubated with in vitro-labeled bicoid RNA (3′ UTR), poly(rI)-poly(rC), poly(rI), or poly(rC) (20,000 cpm; specific activity, 106 cpm/μg) in 10 mM MgCl2–50 mM NaCl–0.1 mM EDTA–1 mM dithiothreitol–10 mM HEPES (pH 8.0) for 30 min at room temperature. The RNA-protein complexes were then filtered through a nitrocellulose membrane (0.45-μm pore size), washed, and counted. Analyses were done with the Graph Pad PRISM (version 2.01) software.
Tubulin-binding assay.
Bacterial extracts from IPTG-induced cultures were separated on SDS–10% polyacrylamide gels, and the MBP-tagged proteins were transferred onto nitrocellulose membranes. Membranes were incubated in TBS–1% Tween 20 for 45 min prior to an overnight overlay with tubulin (7 μg/ml; Sigma) in TBS–0.2% Tween 20. Blots were washed several times in TBS–0.2% Tween 20 and then incubated with a mixture of mouse monoclonal anti-α- and anti-β-tubulin antibodies (ICN). Bound antibodies were detected with secondary peroxidase-conjugated anti-mouse IgG antibodies (Dimension Labs) and Supersignal substrate (Pierce) as stated previously. Separate assays were performed with actin and antiactin antibodies (both from Sigma).
Immunofluorescence.
HStau-hemagglutinin (HA) and hStau-green fluorescent protein (GFP) were constructed by PCR amplification of the full-length cDNA, using the primer pair 5′-TACATGTCGACTTCCTGCCA/GGGCTGCGGG-3′ plus 5′-TACAATCTAGATTATCAGCGGCCGCACCTCCCACACACAGACAT-3′. The 3′ primer was synthesized with a NotI site just upstream from the stop codon allowing ligation of a NotI cassette containing either three copies of the HA tag or the GFP sequence. The resulting fragment was cloned in pBluescript following digestion with SalI and XbaI. The KpnI/XbaI fragment was then subcloned in the pCDNA3/RSV vector (25), and a NotI cassette was introduced at the NotI site. For the TBD-GFP fusion protein, the TBD was PCR amplified with oligonucleotides on each side of this region (5′-TACATAAGCTTAAGCCACCATGGTCAAAGTTCCCCAGGCGC-3′ and 5′-TACAATCTAGAGCGGCCGCGCTCAGAGGGTCTAGTGCGAG-3′). The sense primer contained an ATG initiation codon and the Kozak consensus sequence upstream from the TBD sequence. The antisense primer contained a Not1 site just upstream from a stop codon. The resulting fragment was digested with HindIII and XbaI and cloned into the pCDNA3/RSV vector. The GFP NotI cassette was then introduced at the NotI site.
Mammalian cells were transiently transfected with the cDNAs by the calcium phosphate precipitation technique, fixed in 4% paraformaldehyde in PBS for 25 min at room temperature, and permeabilized with 0.3% Triton X-100 in PBS containing 0.1% bovine serum albumin (BSA). The cells were then blocked with 1% BSA in PBS–0.3% Triton X-100 and incubated with mouse anti-HA, rabbit anticalreticulin, or rabbit anticalnexin antibodies for 1 h at room temperature, as indicated. Cells were washed in permeabilization buffer and incubated with fluorescein-conjugated or Texas red-conjugated species-specific secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, Pa.) in blocking buffer for 1 h. GFP and GFP fusion proteins were detected by autofluorescence. Mounting was done in ImmunoFluor mounting medium (ICN). For the analysis of cytoskeleton-associated proteins, transfected cells were first extracted in 0.3% Triton X-100–130 mM HEPES (pH 6.8)–10 mM EGTA–20 mM MgSO4 for 5 min at 4°C as previously described (10). They were then fixed in 4% paraformaldehyde in PBS and processed for immunofluorescence as described above. Cells were visualized by immunofluorescence, using the 63× planApochromat objective of a Zeiss Axioskop fluorescence microscope.
Confocal microscopy was performed with the 60× Nikon Plan Apochromat objective of a dual-channel Bio-Rad 600 laser scanning confocal microscope equipped with a krypton-argon laser and the corresponding dichroid reflectors to distinguish fluorescein and Texas red labeling. No overlap was observed between the fluorescein and Texas red channels. Confocal images were printed with a Polaroid TX1500 video printer.
Nucleotide sequence GenBank accession numbers.
The human and mouse sequences were deposited in the GenBank database under accession no. AF061938, AF061939, AF061940, and AF061941 (human) and AF061942 (mouse).
RESULTS
Molecular cloning of mammalian staufen cDNAs.
To understand the mechanism of mRNA transport in mammalian cells, we cloned the human and mouse staufen homologues. Thirteen overlapping human cDNAs, ranging in size between 0.8 and 2.5 kb, were isolated from a human central nervous system cDNA library, using the expressed sequence tag HFBDQ83 cDNA as a probe (Fig. 1). Purified human HeLa cell poly(A)+ RNAs were also reverse transcribed and PCR amplified, using different 5′ RACE (rapid amplification of 5′ cDNA ends) protocols, allowing us to clone the 5′ end of the transcript. Two different cDNAs of 3,217 and 3,506 nucleotides were identified from overlapping clones (see below). One of the human cDNAs was then used to screen a fetal total mouse cDNA library under low-stringency conditions, which led to the isolation of a full-length cDNA (mStau). The human and mouse proteins are 90% identical (98% similarity).
FIG. 1.
Amino acid sequences of the hStau cDNAs. Shown is alignment of the cDNAs and PCR fragments with the translation of the putative protein sequences. Numbers refer to the sequence of the short cDNA. Positions of the four consensus dsRBDs (RBD1 to RBD4) and of the TBD are indicated between brackets above the sequence.
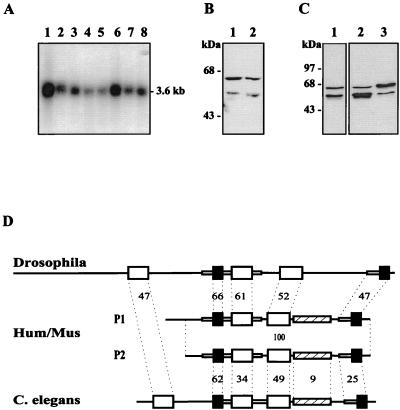
Hybridization of a human multiple-tissue Northern blot with a human cDNA reveals that hStau mRNA is found in every tested tissue (Fig. 2A), unlike the Drosophila staufen gene, which is exclusively expressed in oocytes and in the central nervous system at the larval stage (53). The size of the cDNAs is close to that of the transcripts, which migrate on a Northern blot as an unresolved large band of around 3.6 kb.
FIG. 2.
Characterization of the hStau mRNA and proteins. (A) Northern blot analysis of hStau expression in human tissues. A human multiple-tissue Northern blot (Clontech) was hybridized with the 1.2-kbp BamHI fragment of hStau cDNA. Lane 1, brain; lane 2, pancreas; lane 3, heart; lane 4, skeletal muscles; lane 5, liver; lane 6, placenta; lane 7, lung; lane 8, kidney. (B) Western blot experiment with anti-hStau antibodies. Lane 1, HeLa cell extracts; lane 2, HEK 293 cell extracts. (C) HEK 293 cells were transfected with cDNAs coding for either the short (lane 2) or the long (lane 3) hStau isoform, lysed, and analyzed by Western blotting using the anti-hStau antibodies. Mock-transfected cells are shown in lane 1. (D) Schematic representation of the Drosophila (accession no. M69111), human and mouse (Hum/Mus), and C. elegans (accession no. U67949) Stau proteins. The human protein P1 has an insertion of 81 amino acids at its N-terminal extremity compared to protein P2. Large open and black boxes represent the full-length and short dsRBDs, respectively. Small boxes and lines are regions of high and low sequence similarity, respectively. The hatched boxes indicate the position of the region which is similar to the MAP1B microtubule-binding domain. The percentage of identity between the domains of the human and invertebrate proteins is indicated.
A differential splicing event generates two hStau proteins.
Characterization of the human cDNAs revealed the presence of two types of transcripts which differ only by an insertion of 289 bp at position 324 (T2 and T3 in Fig. 1). To confirm this result and determine the relative expression of the two classes of transcripts, we used RT-PCR to amplify the region of the transcript which overlaps the site of insertion. Unexpectedly, four different fragments were amplified. Cloning and sequencing of the fragments revealed that two correspond to the cDNA sequences (Fig. 1). Compared to the smallest cloned cDNA sequence (T2), a third fragment (T4) has an insertion of 132 bp at position 249, which corresponds to an Alu Sq sequence in an inverted orientation, while the other one (T1) has a deletion of 75 bp between positions 249 and 324. Within a single tissue, the four bands are not expressed at the same level, T2 being the most abundant. However, from one tissue to the other, the relative ratios of the four bands are roughly the same (not shown).
Translation of the cDNAs suggests that three of the four transcripts (T1, T2, and T4) may give rise to a protein of 55 kDa. Interestingly, the DNA insertion in transcript T3 introduces an ATG initiation codon upstream from the first one found in the other transcripts (Fig. 1). This finding suggests that a second putative protein of 63 kDa, exhibiting an 81-amino-acid extension at its N-terminal extremity compared to the other protein, may be translated. Using anti-hStau antibodies in Western blot experiments, we observed two protein bands of around 63 and 55 kDa in human cell extracts (Fig. 2B). To determine whether our cDNAs could account for the presence of the two proteins, we subcloned the T2 and T3 transcripts in an expression vector and expressed them in mammalian cells. As seen in Fig. 2C, each cDNA gives rise to a single overexpressed protein which perfectly comigrates with one of the endogenous proteins. Altogether, these results demonstrate that the hstaufen gene produces four different transcripts and that the transcripts code for two highly homologous proteins which differ in their N-terminal extremities.
Comparison of the mammalian and Drosophila Stau proteins.
The amino acid sequences of the mammalian proteins are similar to that of the Drosophila Stau protein and of the product of an uncharacterized open reading frame on the X chromosome of Caenorhabditis elegans (Fig. 2D). The overall structures and relative positions of the full-length and short RBDs are well conserved, and high sequence identity is found between corresponding dsRBDs. This is highly significant since an alignment of the domains found in the members of the dsRNA-binding protein family shows an average of only 29% amino acid identity to one another (54). In addition, domains 1 and 4 in the human sequence, which are short domains compared to the consensus, are nevertheless highly similar to the corresponding fly sequences, even in the region that extends far beyond the N-terminal side of the consensus sequence, suggesting that they must play an essential role in Stau function.
Mammalian Stau does not contain the first dsRBD or the long N-terminal sequence of the Drosophila protein which was shown to bind to oskar protein (5). In addition, a putative TBD located between the third and fourth dsRBDs of mammalian Stau is not found in the Drosophila protein, at least at the amino acid level. This region contains a stretch of 91 amino acids which show 25% amino acid identity (66% similarity) to a microtubule-binding domain of microtubule-associated protein 1B (MAP1B) (65). It is meaningful that the sequence similarity covers the full microtubule-binding domain of MAP1B and that it is restricted to this domain. Putative nuclear localization signals are also present.
hStau and mStau bind dsRNAs.
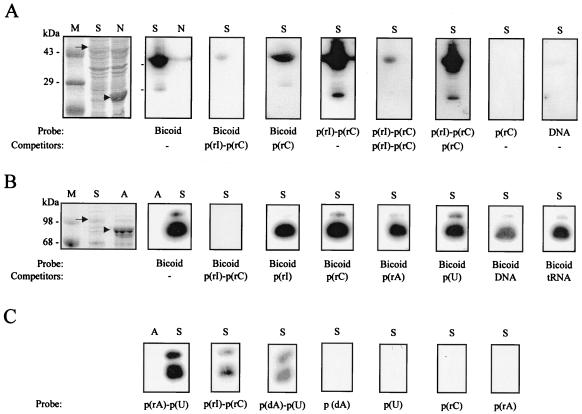
As seen in Fig. 2D, mammalian Stau proteins contain multiple dsRBDs. To determine whether Stau binds RNAs, we used two bacterially expressed fusion proteins, His-hStau and MBP-mStau, in an RNA-binding assay. The fusion proteins were probed with in vitro-labeled bicoid mRNA, which is known to adopt an extensive secondary structure and to strongly bind to Drosophila Stau protein, both in vivo and in vitro (18, 54). Both fusion proteins strongly bind this RNA. The binding is competed by a 100-fold excess of cold poly(rI)-poly(rC) but not by a 1,000-fold excess of poly(rI), poly(rC), poly(rA), or poly(U) or by tRNA or dsDNA (Fig. 3A and B), suggesting that mammalian Stau recognizes double-stranded structures in the RNA rather than a sequence-specific region. Both fusion proteins also directly bind labeled dsRNAs and RNA-DNA hybrids but not single-stranded RNA or DNA homopolymers (Fig. 3A and C). As controls, bacterial extracts containing overexpressed His-neutral endopeptidase or MBP-aminopeptidase fusion proteins were also included on each blot; they did not bind any of these nucleic acids. We also tested other in vitro-labeled RNAs such as those coding for tubulin, neuropeptides from Aplysia, and nuclear RNP B. All of these RNAs bind to Stau in vitro, as reported previously for other members of the dsRNA-binding protein family. This finding demonstrates that both hStau and mStau, regardless of the protein to which they are fused, are able to bind dsRNAs. However, there is no sequence specificity, as reported for other members of the dsRNA-binding protein family (21, 54, 55).
FIG. 3.
RNA-binding assay. Bacterially expressed His-hStau (A, lanes S) and His-neutral endopeptidase (A, lane N) fusion proteins or bacterially expressed MBP-mStau (B and C, lanes S) or MBP-aminopeptidase (B and C, lanes A) fusion proteins were electrophoresed on a polyacrylamide gel, transferred to nitrocellulose, and incubated with 32P-labeled nucleic acids, in the presence or absence of cold competitors, as indicated below each gel. After extensive washing, binding was detected by autoradiography. A representative Coomassie blue staining of the blots is shown on the left (A and B). Arrows, positions of overexpressed Stau; arrowheads, positions of overexpressed control proteins. Lanes M, molecular weight markers.
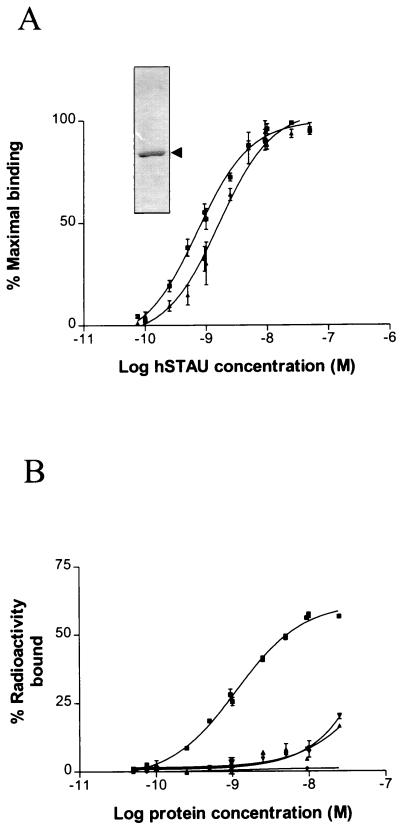
Filter binding assays, using Ni-NTA-purified His-hStau (inset), were used to determine the binding affinity of Stau (Fig. 4). High-affinity binding, with a Kd of about 10−9 M, was observed when the 3′ UTR of bicoid or double-stranded RNA was used as a probe (Fig. 4A). The resulting sigmoidal curves suggest that Stau cooperatively binds dsRNAs. In contrast, only low-affinity binding was observed with single-stranded RNAs, confirming that Stau specifically binds dsRNAs (Fig. 4B).
FIG. 4.
RNA-binding assay in solution. Dilutions of the purified His-hStau fusion protein were incubated with fixed amounts of labeled RNAs, and the RNA-protein complexes were filtered through nitrocellulose membranes. (A) RNA-binding affinity to dsRNAs. Triangles, 3′ UTR of bicoid RNA; squares, poly(rI)-poly(rC). The results are presented as a percentage of maximal retained probe and are the averages of three independent experiments done in duplicate. Inset, Coomassie blue staining of Ni-NTA-purified His/hStau after separation by SDS-PAGE. (B) RNA-binding affinity to RNAs. Squares, poly(rI)-poly(rC); triangles, poly(rI); inverted triangles, poly(rC); diamonds, BSA with poly(rI)-poly(rC), used as a control. The results are presented as a percentage of bound radioactivity and represent a single experiment done in duplicate. The same results were obtained in two other independent experiments.
hStau and mStau bind tubulin in vitro.
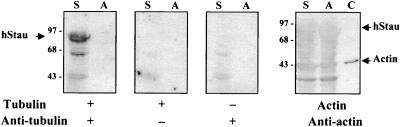
As described above, Stau contains a region which is similar to the microtubule-binding domain of MAP1B. To determine whether mammalian Stau can bind tubulin, bacterially expressed MBP-Stau fusion proteins were used in a tubulin-binding assay. As shown in Fig. 5, hStau binds tubulin in vitro. As a control, the bacterially expressed MBP-aminopeptidase fusion protein was also included on the blot; it did not show any tubulin-binding capability. Under the same conditions, hStau cannot bind actin (Fig. 5), which suggests that the binding of tubulin to Stau is specific. The same results were obtained with the MBP-mStau fusion protein (Fig. 6B, lane 1). Binding to mRNAs and microtubules are two of the characteristics expected of localizing proteins, making hStau and mStau very good candidates for mRNA transport and localization in mammals.
FIG. 5.
Tubulin-binding assay. Bacterially expressed MBP-hStau (lanes S) or MBP-aminopeptidase (lanes A) fusion proteins were electrophoresed on SDS-polyacrylamide gels, transferred to nitrocellulose, and incubated with tubulin or actin. After extensive washing, tubulin and actin were detected with monoclonal antitubulin and antiactin antibodies, respectively. As controls, the same experiments were performed in the absence of either tubulin or antitubulin antibodies. Purified actin was also loaded on the gel as a control (lane C). Sizes are indicated in kilodaltons.
FIG. 6.
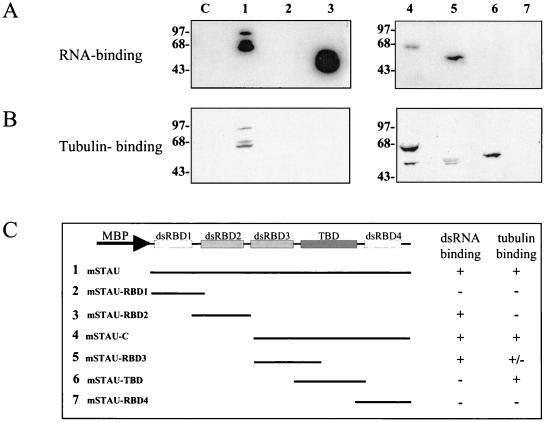
Molecular mapping of the dsRBD and TBD. Bacterially expressed MBP-mStau (lanes 1), MBP-mStau deletion mutants (lanes 2 to 7), or MBP-aminopeptidase (lanes C) fusion proteins were electrophoresed on a polyacrylamide gel, transferred to nitrocellulose, incubated with either 32P-labeled 3′-UTR bicoid RNA (A) or tubulin and antitubulin antibodies (B), and revealed as described above. (C) Schematic representation of the mutant proteins. Their RNA- and tubulin-binding responses are indicated.
Molecular mapping of the RBD and TBD.
To determine which Stau domain(s) is involved in RNA and/or tubulin binding, the MBP-mStau fusion protein was used to construct a series of deletion mutants (Fig. 6). The production and relative abundance of each fusion protein was first verified by Western blotting (not shown). Using the RNA-binding assay, we demonstrated that both of the full-size dsRBDs (dsRBD2 and dsRBD3) are independently sufficient to bind bicoid RNA (Fig. 6A). In contrast, the two short domains (dsRBD1 and dsRBD4) were unable to bind dsRNA in this assay. We also demonstrated that the C-terminal half of mStau is able to bind tubulin (Fig. 6B, lane 4). More specifically, the region which is similar to the MAP1B microtubule-binding domain is sufficient to bind tubulin (Fig. 6B, lane 6). These experiments confirm that the regions that we identified by sequence comparison as putative dsRBDs and TBDs are biochemically functional.
Stau is associated with the detergent-insoluble fraction in vivo.
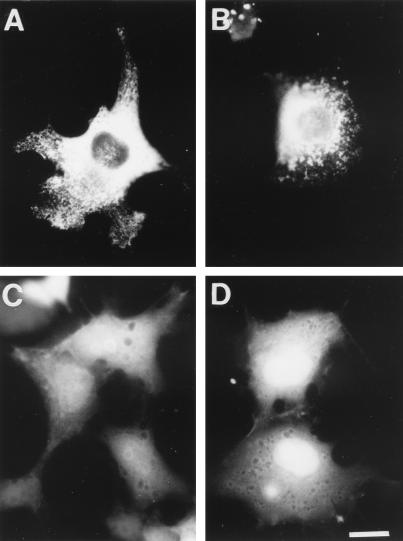
We next addressed the cellular distribution and cytoskeletal association of the two hStau proteins in vivo. To do so, we fused GFP or an HA tag to the 63- and 55-kDa hStau isoforms, respectively. Using confocal microscopy, we first showed that the two fusion proteins colocalize when coexpressed in mammalian cells (not shown). Then, we showed that they are nonhomogeneously distributed throughout the cytoplasm and label numerous vesicular and tubular structures which concentrate in the perinuclear region (Fig. 7A). Minimal staining was found in the nucleus. When the cells were treated with Triton X-100 prior to fixing, allowing soluble proteins to be separated from the cytoskeleton and cytoskeleton-associated proteins (44), the tubulovesicular labeling was still present, demonstrating that hStau is associated with the detergent-insoluble material in vivo (Fig. 7B). Labeled structures were also present in cell processes, suggesting that Stau may target mRNAs to peripheral ER elements. The same results were obtained following expression of the GFP-mStau protein (not shown). The association between hStau and the cytoskeletal-associated material was confirmed by in vitro cell fractionation in the presence of Triton X-100. In this assay, hStau partitioned mainly in the cytoskeleton-associated fractions, although a significant fraction was found in a soluble form, as judged by Western blotting (not shown).
FIG. 7.
Subcellular localization of the hStau-GFP fusion proteins. COS7 cells were transfected with cDNAs coding for either the hStau-GFP (A and B) or TBD-GFP (C) fusion protein, or GFP alone (D). Untreated (A, C, and D) or Triton X-100-treated (B) cells were fixed and visualized by autofluorescence. Bar = 20 μm.
To determine whether the tubulin-binding domain identified in vitro is truly involved in this function in vivo, we transfected mammalian cells with a cDNA coding for a fusion protein in which the minimal TBD was fused to GFP. In contrast to the full-length protein, the TBD-GFP fusion protein is randomly distributed in the cytoplasmic and nuclear domains of the cells (Fig. 7C), as is the GFP protein used as a control (Fig. 7D). This staining was completely extracted by the Triton X-100 treatment (not shown), suggesting that the minimal TBD found in vitro is not sufficient to render the protein insoluble and form a stable association with the microtubule network and/or the cytoskeleton-associated material.
Stau localizes to the RER in vivo.
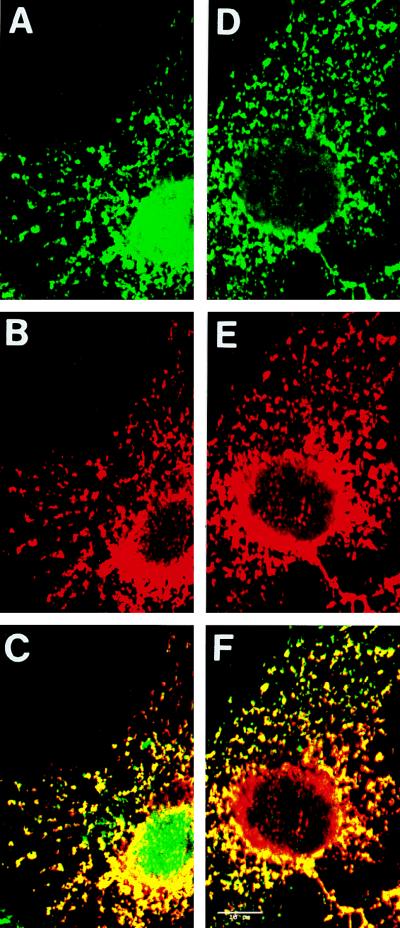
Interestingly, the pattern of localization of Stau resembles that of the ER. To test a putative localization of Stau to the ER, we transfected mammalian cells with a cDNA coding for a fusion protein in which a HA tag was introduced at the C-terminal end of the short hStau protein. We then double labeled transfected cells with anti-HA, to recognize hStau, and with anticalreticulin or anticalnexin, two markers of the ER. Using a confocal microscope, we showed that hStau completely colocalizes with anticalreticulin, although HA-staining appears to be absent in some parts of the ER, in particular around the nucleus (Fig. 8A to C). To confirm these results, we examined the colocalization of Stau and calnexin, a specific marker for the RER (24) (Fig. 8D to F). The patterns of staining obtained with anti-hStau and anticalnexin were identical, demonstrating that hStau colocalizes exclusively with the RER.
FIG. 8.
Colocalization of hStau with markers of the RER by confocal microscopy. A cDNA coding for an hStau-HA fusion protein was transfected into COS7 cells. Triton X-100-treated cells were fixed and double labeled with anti-HA (B) and anticalreticulin (A) or with anti-HA (E) and anticalnexin (D). Anti-HA was detected with Texas red-coupled anti-mouse IgG antibodies, using the Texas Red channel; anticalreticulin and anticalnexin were detected with fluorescein-conjugated anti-rabbit IgG antibodies, using the fluorescein channel. Panels C and F are superpositions of panels A plus B and D plus E, respectively. No overlap was observed between the fluorescein and Texas red channels. Bar = 10 μm.
DISCUSSION
The transport and localization of specific mRNAs have important functions in cell physiology. For example, mRNA targeting plays a key role in the formation of cytoskeletal filaments and in the establishment of morphogenetic gradients (55). However, the nature of the RNP complexes as well as the mechanisms involved in these processes are still largely uncharacterized. In this paper, we describe a novel RNA-binding protein which localizes to the RER in mammalian cells. Although its precise role is still unclear, its biochemical and molecular properties strongly suggest that it is involved in mRNA transport and/or localization. Consistent with such a role, we recently demonstrated that hStau is involved in human immunodeficiency virus type 1 genomic RNA encapsidation (41). Mammalian Stau was also found in the dendrites of rat hippocampal neurons in culture, but not in the axons, and colocalizes with RNPs known to contain mRNA, ribosomes, and translation factors, suggesting a role for Stau in the polarized transport and localization of mRNAs in mammalian neurons (27).
A differential splicing event generates four hStau transcripts.
The significance of the four different classes of transcripts is unknown. They arise by differential splicing since each of the inserts that appears in the 5′ end of the cDNAs is flanked by large introns containing typical consensus splicing sequences (6). They are observed in all the tissues tested by RT-PCR, suggesting that they are not cloning artifacts. The multiplicity of the transcripts and the short size of the alternatively spliced exons may explain the fact that a single but diffuse band is visible on the Northern blot, despite the presence of four transcripts. This alternative splicing, which changes the 5′ UTR of the transcripts, could represent a mechanism by which translation is regulated, although the presence of similar relative levels of the four transcripts in each tissue argues against this possibility. Alternatively, these different classes of transcripts may result from aberrant or incomplete splicing events. In fact, an unusually large number of cDNA clones containing intron sequences were isolated, and this may indicate that the splicing of the premature transcripts is a slow process. The presence of an Alu sequence in the 5′ UTR may arise by the activation of a resident intronic Alu element as an exon, as reported previously (35). Many examples of recruitment, via splicing or intron sliding, of a segment of a resident Alu element into an mRNA have been reported. It is thought that the presence of a polypyrimidine tract which is the complement of the A-rich tail of the element when inserted in the reverse orientation contributes to the creation of the splicing acceptor site. A point mutation, downstream from this site, may generate the splicing donor site, as reported previously. The presence of an old Alu subfamily sequence and of only the right subunit segment of the Alu element is consistent with this interpretation.
Structure and function of Stau.
We observed that mammalian Stau, like all members of the dsRNA-binding protein family (55), can bind any dsRNA or RNAs forming double-stranded structures in vitro, regardless of its primary sequence, as well as RNA-DNA hybrids. The latter adopt a conformation that is more closely related to that of dsRNA than dsDNA, which probably explains why they can bind to Stau. The fact that the full-length Stau protein, as observed with single dsRBD, binds to any dsRNA in vitro suggests that the correspondence between the position of the dsRBDs, and the arrangement of double-stranded stems in the folded RNAs may not be sufficient for specificity; posttranslational modifications and/or essential cofactors capable of forming complex RNP structures along with mRNA molecules could be necessary to discriminate between different RNA secondary structures. Packaging of mRNAs into RNP complexes (1, 18, 20, 32), the intermolecular dimerization of the localization signal of bicoid mRNA (19), and the involvement of untranslatable hnRNAs in mRNA transport (31, 59, 60) are consistent with this interpretation. Until now, specific mRNA-Stau interactions were shown in vivo only after injection of different RNAs into Drosophila embryos, but the mechanisms underlying the specificity are not known (18). Since specific RNA binding cannot be obtained in vitro, it precludes the use of classic techniques to isolate and identify relevant RNAs which would bind staufen in vivo. Cross-linking of mRNA to Stau in vivo and isolation of the resulting complexes will be necessary to identify the nature of bound RNAs.
Regardless of their limitations, the in vitro assays did allow us to map the molecular determinants which are necessary and sufficient to bind RNAs. The presence of two functional domains in the mammalian Stau contrasts with what has been reported for other members of the dsRNA-binding protein family, which contain multiple full-length dsRBDs but only one that is biochemically functional (21, 34, 39, 48). Interestingly, full-length dsRBDs incapable to bind dsRNA by themselves can do so when joined to another inactive full-length domain, suggesting that multiple domains present in a given protein exhibit cooperative binding effect (34, 48). Whether the two mStau dsRBDs exhibit similar or different affinities is not yet clear.
We mapped the TBD to a region which is similar to a microtubule-binding domain of MAP1B. Although this region can efficiently bind tubulin in vitro, it is not sufficient to bring a TBD-GFP fusion protein to the microtubule network. Binding of Stau to microtubules in vivo may involve more than one molecular determinant or the proper localization and folding of the TBD in the full-length protein. Indeed, in our in vitro assay, the fusion protein which contains the C-terminal region in addition to the TBD binds tubulin more efficiently than does the TBD alone, suggesting that this region may be necessary for binding to microtubules in vivo. Interestingly, the corresponding region of the Drosophila Stau protein was shown to bind inscuteable (36), a protein with ankyrin domains which is believed to associate with the cytoskeleton (33), suggesting that corresponding regions of the mammalian and Drosophila proteins may have functional similarities.
Alternatively, binding may be weak and/or transitory in vivo, for example during the early steps of mRNA recruitment, during mRNA transport, and/or at mitosis, as found in Drosophila (18, 45, 55). These steps may be difficult to observe by immunofluorescence in some cell lines (18) and/or be masked by the anchoring of the protein to the RER. A similar conclusion was reached when binding of MAP1B to the microtubule network was studied (65), suggesting that weak binding to the cytoskeleton may be a characteristic of proteins containing this type of TBD. These steps may nevertheless be necessary to allow the efficient and flexible transport of RNA along the cytoskeleton. Interestingly, the immunoelectron microscopic observation of dendrites of hippocampal neurons in culture showed the presence of abundant gold particles close to microtubules, strongly arguing in favor of a Stau-microtubule association in these cells (27). In Drosophila, there is no evidence that Stau directly binds to the microtubule network, although Stau-dependent mRNA transport was shown to rely on this structure (45, 55).
Our studies demonstrate that Stau is anchored to the RER and that the putative TBD is not involved in this function. Indeed, preliminary results suggest that the binding of Stau to the RER is carried out by one of the RBDs (40). Similar domains in other members of the dsRNA-binding proteins were previously shown to be involved in protein dimerization and/or in protein-protein interactions (4, 8). This finding also suggests that different Stau molecular determinants are necessary for binding to tubulin and anchoring to the RER. This is consistent with previous findings demonstrating that in Xenopus and Drosophila, mRNA localization was likely to occur via successive steps involving different elements of the cytoskeleton and overlapping molecular determinants (55).
Localization of Stau to the RER.
When expressed in mammalian cells, Stau isoforms show a tubulovesicular pattern of localization which is found more abundantly in the perinuclear region. Besides ribosomal proteins, Stau is the first RNA-binding protein shown to be associated with the RER in mammals. No signal peptide or putative hydrophobic transmembrane domains are present in either the long or short Stau proteins, indicating that they are cytosolic proteins and not residents of the RER and that their association to the RER is likely to reflect their mRNA transport function. Two recent papers also suggest that mRNA transport may be linked to the ER or ER-like structures. In Xenopus oocytes, Vera, a Vg1 mRNA-binding protein, was shown to cosediment with TRAPα, a protein associated with the protein translocation machinery of the ER. However, in contrast to Stau, Vera-Vg1 complexes were found associated only with a small subdomain of the ER, which was of the smooth variety (12). Similarly, in Drosophila, at least some steps in mRNA transport in nurse cells and oocytes seem to occur within ER-like cisternae (64). As observed for the Vg1 mRNA-SER interaction in Xenopus, this structure seems to exclude most ribosomes, suggesting that translation is not the major function of these associations.
hStau and mStau represent new members of a large family of proteins involved in the transport and/or localization of mRNAs to different subcellular compartments and/or organelles. Stau, the TAR RNA-binding protein and X. laevis homologue Xlrbpa, and Spnr were shown to colocalize with the RER (this paper), with ribosomes and hnRNPs (14), and with the microtubular array of spermatids (49), respectively. Our results strongly suggest that Stau-mRNA RNP complexes are transported along the microtubule network and then anchored to the RER. It is well known that the ER is associated with the microtubule cytoskeleton (57). Therefore, a transient interaction between microtubules and Stau may facilitate the localization of Stau and the targeting of mRNA to the RER. One of the roles of Stau might be to transport and localize specific mRNAs to the RER, such as those coding for secreted or membrane proteins which have to be translocated to the RER. This would bring them in proximity to the signal recognition particles and RER, thus facilitating translation and translocation. The presence of Stau in cell processes, in association with ER structures, may represent a first clue to understanding the role of many mRNAs which were found to be localized in neuronal processes (51). Stau may facilitate the transport of mRNAs to cell processes to ensure efficient local translation and translocation. In addition, the presence of multiple Stau-like proteins in mammals creates the possibility that different members of the family can target subclasses of mRNAs to different subdomains of the ER. This phenomenon has been described before and is thought to be the first step in the differential targeting of proteins in polarized cells (43).
We do not exclude the possibility that Stau plays additional roles in mammals; Stau may first be linked to the RER for storage, and then a subset of molecules may be recruited by specific mRNAs and/or cofactors to form RNP complexes that will be transported along microtubules toward their final destination. The presence of large amounts of Stau in the perinuclear region, which could be awaiting the nucleocytoplasmic transport of mRNAs, is consistent with this possibility. The presence of a putative nuclear localization signal even suggests that Stau transits through the nucleus before being localized in the cytoplasm and plays a role in mRNA or rRNA export. Alternatively, Stau may play key roles in the translational regulation of localized mRNAs, as is the case for Drosophila Stau, which is essential for the translation of oskar mRNA, once it is localized at the posterior pole (29). Indeed, since polysomes and ER-bound ribosomes are not extracted by Triton X-100, it is possible that Stau is associated with the RER via ribosomes and/or mRNAs. Characterization of mRNAs and putative cofactors which bind to staufen will be necessary to understand the process.
In vertebrates, the mechanisms which underly the transport of mRNAs have not yet been deciphered. Characterization of the RNAs and proteins involved in transport and localization is particularly important since understanding the mechanisms responsible for the transport of mRNAs is fundamental for learning more on the development of polarity in cells, both during mammalian development and in somatic cells, at a time where RNA-based gene therapy is being considered as a possible approach to cure different disorders.
ACKNOWLEDGMENTS
We thank Luis Rokeach (Department of Biochemistry, University of Montreal) for the anticalreticulin antibodies, John Bergeron (McGill University) for the anticalnexin antibodies, and André Royal (Department of Pathology and Cell Biology) for the mouse cDNA library. We thank Luis Rokeach and Lea Brakier-Gingras for comments and discussion and Michael Kiebler for sharing unpublished information.
The first two authors contributed equally to this work.
This work was supported by a Natural Sciences and Engineering Research Council of Canada grant to L.D., a National Health Research Development Program grant to L.D., and a Medical Research Council of Canada grant to I.R.N.
REFERENCES
- 1.Ainger K, Avossa D, Morgan F, Hill S J, Barry C, Barbarese E, Carson J H. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aloyz R S, DesGroseillers L. Processing of the L5-67 precursor peptide and characterization of LUQIN in the central nervous system of Aplysia californica. Peptides. 1995;16:331–338. doi: 10.1016/0196-9781(94)00140-5. [DOI] [PubMed] [Google Scholar]
- 3.Bassell G, Singer R H. mRNA and cytoskeletal filaments. Curr Opin Cell Biol. 1997;9:109–115. doi: 10.1016/s0955-0674(97)80159-7. [DOI] [PubMed] [Google Scholar]
- 4.Benkirane M, Neuveut C, Chun R F, Smith S M, Samuel C E, Gatignol A, Jeang K-T. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16:611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitwieser W, Markussen F-H, Horstmann H, Ephrussi A. Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev. 1996;10:2179–2188. doi: 10.1101/gad.10.17.2179. [DOI] [PubMed] [Google Scholar]
- 6.Brizard, F., and L. DesGroseillers. Submitted for publication.
- 7.Broadus J, Fuerstenberg S, Doe C Q. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;391:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- 8.Cosentino G P, Venkatesan S, Serluca F C, Green S, Mathews M B, Sonenberg N. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein from homo- and heterodimers in vivo. Proc Natl Acad Sci USA. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crino P B, Eberwine J. Molecular characterization of the dendritic growth cone: regulated mRNA transport and local protein synthesis. Neuron. 1996;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- 10.Davis L, Banker G A, Steward O. Selective dendritic transport of RNA in hippocampal neurons in culture. Nature. 1987;330:477–479. doi: 10.1038/330477a0. [DOI] [PubMed] [Google Scholar]
- 11.DesGroseillers L, Lemieux N. Localization of a human double-stranded RNA-binding protein gene (STAU) to band 20q13.1 by fluorescence in situ hybridization. Genomics. 1996;36:527–529. doi: 10.1006/geno.1996.0499. [DOI] [PubMed] [Google Scholar]
- 12.Deshler J O, Highett M I, Schnapp B J. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- 13.Deshler J O, Highett M I, Abramson T, Schnapp B J. A highly conserved RNA-binding protein for cytoplasmic localization in vertebrates. Curr Biol. 1998;8:489–496. doi: 10.1016/s0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- 14.Eckmann C R, Jantsch M F. Xlrbpa, a double-stranded RNA-binding protein associated with ribosomes and heterogeneous nuclear RNPs. J Cell Biol. 1997;138:239–253. doi: 10.1083/jcb.138.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elisha Z, Havin L, Ringel I, Yisraeli J K. Vgl RNA binding protein mediates the association of Vgl RNA with microtubules in Xenopus oocytes. EMBO J. 1995;14:5109–5114. doi: 10.1002/j.1460-2075.1995.tb00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ephrussi A, Dickinson L K, Lehamnn R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 17.Erdelyi M, Michon A M, Guichet A, Glotzer J B, Ephrussi A. Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature. 1995;377:524–527. doi: 10.1038/377524a0. [DOI] [PubMed] [Google Scholar]
- 18.Ferrandon D, Elphick L, Nüsslein-Volhard C, St Johnston D. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 19.Ferrandon D, Koch I, Westhof E, Nüsslein-Volhard C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′UTR-staufen ribonucleoprotein particles. EMBO J. 1997;16:1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forristall C, Pondel M, King M L. Patterns of localization and cytoskeletal association of two vegetally localized RNAs, Vgl and Xcat-2. Development. 1995;121:201–208. doi: 10.1242/dev.121.1.201. [DOI] [PubMed] [Google Scholar]
- 21.Gatignol A, Buckler C, Jeang K-T. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila Staufen. Mol Cell Biol. 1993;13:2193–2202. doi: 10.1128/mcb.13.4.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gazzaley A H, Benson D L, Huntley G W, Morrison J H. Differential subcellular regulation of NMDAR1 protein and mRNA in dendrites of dendate gyrus granule cells after perforant path transection. J Neurosci. 1997;17:2006–2017. doi: 10.1523/JNEUROSCI.17-06-02006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Havin L, Git A, Elisha Z, Obeerman F, Yaniv K, Schwartz S P, Standart N, Yisraeli J K. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 1998;12:1593–1598. doi: 10.1101/gad.12.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochstenback F, David V, Watkins S, Brenner M B. Endoplasmic reticulum resident protein of 90 kilodaltons associates with the T- and B-cell antigen receptors and major histocompatibility complex antigens during assembly. Proc Natl Acad Sci USA. 1992;89:4734–4738. doi: 10.1073/pnas.89.10.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jockers R, Da Silva A, Strosberg A D, Bouvier M, Marullo S. New molecular and structural determinants involved in β2-adrenergic receptor desensitization and sequestration. J Biol Chem. 1996;271:9355–9362. doi: 10.1074/jbc.271.16.9355. [DOI] [PubMed] [Google Scholar]
- 26.Kang H, Schuman E M. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 27.Kiebler, M. Personal communication.
- 28.Kim-Ha J, Smith J L, Macdonald P M. Oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell. 1991;66:23–35. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- 29.Kim-Ha J, Kerr K, Macdonald P M. Translational regulation of oskar mRNA by Bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 30.Kislauskis E H, Zhu X, Singer R H. β-Actin messenger RNA localization and protein synthesis augment cell motility. J Cell Biol. 1997;136:1263–1270. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kloc M, Etkin L D. Delocalization of Vg1 mRNA from the vegetal cortex in Xenopus oocytes after destruction of Xlsirt RNA. Science. 1994;265:1101–1103. doi: 10.1126/science.7520603. [DOI] [PubMed] [Google Scholar]
- 32.Knowles R B, Sabry J H, Martone M E, Deerinck T J, Ellisman M H, Bassell G J, Kosik K S. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraut R, Campos-Ortega J A. Inscuteable, a neural precursor gene of Drosophila encodes a candidate for a cytoskeletal adaptor protein. Dev Biol. 1996;174:66–81. doi: 10.1006/dbio.1996.0052. [DOI] [PubMed] [Google Scholar]
- 34.Krovat B C, Jantsch M F. Comparative mutational analysis of the double-stranded RNA binding domains of Xenopus laevis RNA-binding protein. A. J Biol Chem. 1996;271:28112–28119. doi: 10.1074/jbc.271.45.28112. [DOI] [PubMed] [Google Scholar]
- 35.Labuda D, Zietkiewicz E, Mitchell G A. Alu elements as a source of genomic variation: deleterious effects and evolutionary novelties. In: Maraia R J, editor. The impact of short interspersed elements (SINEs) on the host genome. R. G. Austin, Tex: Landes Company; 1995. pp. 1–24. [Google Scholar]
- 36.Li P, Yang X, Wasser M, Cai Y, Chia W. Inscuteable and staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell. 1997;90:437–447. doi: 10.1016/s0092-8674(00)80504-8. [DOI] [PubMed] [Google Scholar]
- 37.Long R M, Singer R H, Meng X, Gonzalez I, Nasmyth K, Jansen R-P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;177:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 38.Martin K C, Casadio A, Zhu H, Yaping E, Rose J C, Chen M, Bailey C H, Kandel E R. Synapse-specific, long-term facilitation of Aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 39.McCormack S J, Ortega L G, Doohan J P, Samuel C E. Mechanism of interferon action: motif 1 of the interferon-induced, RNA-dependent protein kinase (PKR) is sufficient to mediate RNA-binding activity. Virology. 1994;198:92–99. doi: 10.1006/viro.1994.1011. [DOI] [PubMed] [Google Scholar]
- 40.Ming, L., and L. DesGroseillers. Unpublished data.
- 41.Mouland, A. J., J. Mercier, L. Wickham, L. DesGroseillers, and E. A. Cohen. Submitted for publication.
- 42.Nakielny S, Fischer U, Michael W M, Dreyfuss G. RNA transport. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- 43.Okita T W, Li X, Roberts M W. Targeting of mRNAs to domains of the endoplasmic reticulum. Trends Biochem Sci. 1994;4:91–96. doi: 10.1016/0962-8924(94)90181-3. [DOI] [PubMed] [Google Scholar]
- 44.Pachter J S. Association of mRNA with the cytoskeletal framework: its role in the regulation of gene expression. Crit Rev Eukaryotic Gene Expr. 1992;2:1–18. [PubMed] [Google Scholar]
- 45.Pokrywka N J, Stephenson E C. Microtubules are a general component of mRNA localization systems in Drosophila oocytes. Dev Biol. 1995;167:363–370. doi: 10.1006/dbio.1995.1030. [DOI] [PubMed] [Google Scholar]
- 46.Rings EHHM, Büller H A, Neele A M, Dekker J. Protein sorting versus messenger RNA sorting? Eur J Cell Biol. 1994;63:161–171. [PubMed] [Google Scholar]
- 47.Ross A F, Oleynikov Y, Kisllauskis E H, Taneja K L, Singer R H. Characterization of a β-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmedt C, Green S R, Manche L, Taylor D R, Ma Y, Mathews M B. Functional characterization of the RNA-binding domain and motif of the double-stranded RNA-dependent protein kinase DAI (PKR) J Mol Biol. 1995;249:29–44. doi: 10.1006/jmbi.1995.0278. [DOI] [PubMed] [Google Scholar]
- 49.Schumacher J M, Lee K, Edelhoff S, Braun R E. Spnr, a murine RNA-binding protein that is localized to cytoplasmic microtubules. J Cell Biol. 1995;129:1023–1032. doi: 10.1083/jcb.129.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz S P, Aisenthal L, Elisha Z, Oberman F, Yisraeli J K. A 69-kDa RNA-binding protein from Xenopus oocytes recognizes a common motif in two vegetally localized maternal mRNAs. Proc Natl Acad Sci USA. 1992;89:11895–11899. doi: 10.1073/pnas.89.24.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steward O. mRNA localization in neurons: a multipurpose mechanism? Neuron. 1997;18:9–12. doi: 10.1016/s0896-6273(01)80041-6. [DOI] [PubMed] [Google Scholar]
- 52.St. Johnston D, Driever W, Berleth T, Richstein S, Nüsslein-Volhard C. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Dev Suppl. 1989;107:13–19. doi: 10.1242/dev.107.Supplement.13. [DOI] [PubMed] [Google Scholar]
- 53.St Johnston D, Beuchle D, Nüsslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- 54.St Johnston D, Brown N H, Gall J G, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.St Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 56.Takizawa P A, Sil A, Swedlow J R, Herskowitz I, Vale R D. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 57.Terasaki M, Chen L B, Fujiwara K. Microtubules and the endoplasmic reticulum are highly interdependent structures. J Cell Biol. 1986;103:1557–1568. doi: 10.1083/jcb.103.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tetzlaff M T, Jäckle H, Pankratz M J. Lack of Drosophila cytoskeletal tropomyosin affects head morphogenesis and the accumulation of oskar mRNA required for germ cell formation. EMBO J. 1996;15:1247–1254. [PMC free article] [PubMed] [Google Scholar]
- 59.Tiedge H, Fremeau R T, Jr, Weinstock P H, Arancio O, Brosius J. Dendritic localization of neural BC1 RNA. Proc Natl Acad Sci USA. 1991;88:2093–2097. doi: 10.1073/pnas.88.6.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiedge H, Zhou A, Thorn N A, Brosius J. Transport of BC1 RNA in hypothalamo-neurohypophyseal axons. J Neurosci. 1993;13:4214–4219. doi: 10.1523/JNEUROSCI.13-10-04214.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci. 1997;17:9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wickham L, DesGroseillers L. A bradykinin-like neuropeptide precursor gene is expressed in neuron L5 of Aplysia californica. DNA Cell Biol. 1991;10:249–258. doi: 10.1089/dna.1991.10.249. [DOI] [PubMed] [Google Scholar]
- 63.Wilhelm J E, Vale R D. RNA on the move: the mRNA localization pathway. J Cell Biol. 1993;123:269–274. doi: 10.1083/jcb.123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilsch-Bräuninger M, Schwarz H, Nüsslein-Volhard C. A sponge-like structure involved in the association and transport of maternal products during Drosophila oogenesis. J Cell Biol. 1997;139:817–829. doi: 10.1083/jcb.139.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zauner W, Kratz J, Staunton J, Feick P, Wiche G. Identification of two distinct microtubule binding domains on recombinant rat MAP1B. Eur J Cell Biol. 1992;57:66–74. [PubMed] [Google Scholar]