Abstract
Pax family transcription factors bind DNA through the paired domain. This domain, which is comprised of two helix-turn-helix motifs and a β-hairpin structure, is a target of mutations in congenital disorders of mice and humans. Previously, we showed that Pax-5 (B-cell-specific activator protein) recruits proteins of the Ets proto-oncogene family to bind a composite DNA site that is essential for efficient transcription of the early-B-cell-specific mb-1 promoter. Here, evidence is provided for specific interactions between Ets-1 and the amino-terminal subdomains of Pax proteins. By tethering deletion fragments of Pax-5 to a heterologous DNA-binding domain, we show that 73 amino acids (amino acids 12 to 84) of its amino-terminal subdomain can recruit the ETS domain of Ets-1 to bind the composite site. Furthermore, an amino acid (Gln22) within the highly conserved β-hairpin motif of Pax-5 is essential for efficient recruitment of Ets-1. The ability to recruit Ets proteins to bind DNA is a shared property of Pax proteins, as demonstrated by cooperative DNA binding of Ets-1 with sequences derived from the paired domains of Pax-2 and Pax-3. The strict conservation of sequences required for recruitment of Ets proteins suggests that Pax-Ets interactions are important for regulating transcription in diverse tissues during cellular differentiation.
The highly conserved Pax family of transcriptional regulators is important for the control of gene expression during cellular differentiation and diversification in species ranging from Drosophila to jellyfish to humans (13). Naturally occurring mutations in mice and humans have identified Pax proteins as important for the control of cellular differentiation and organogenesis. For example, mutations in the pax-1 (undulated) or pax-3 (splotch) gene result in the impaired development of the vertebral column or neural crest cell-derived tissues, respectively, in mutant mice (10, 15). Mutations in pax-6 genes in mice (Small eye) cause a loss of the eye lens placode, changes in the forebrain, and impaired pancreas function (27, 39), and mutation of the pax-6 gene in humans results in aniridia (44). Mutations in human pax-3 genes have been linked to Waardenburg syndrome type I (WS-I) (4, 42, 43) WS-III (28) and to craniofacial-deafness-hand syndrome (2). In other studies, the lack of Pax-5 (B-cell-specific activator protein) in pax-5−/−knockout mice caused the developmental arrest of B lymphocytes at an early stage of differentiation and altered morphogenesis of the midbrain (47). In Drosophila, the eyeless mutation defined an evolutionarily conserved requirement for this Pax-6 homolog for normal eye development, and overexpression of the Eyeless protein directed the formation of ectopic eyes in transgenic flies (38).
The defining feature of the Pax family is the paired domain (or paired box), a 128-amino-acid DNA-binding motif comprising two distinct subdomains that function together to coordinately recognize specific DNA sequences. X-ray crystallographic analysis of the paired domain of the Drosophila Paired protein (which regulates expression of the even-skipped gene) identified amino-terminal and carboxy-terminal subdomains, as well as a linker region that interacts directly with DNA (53). The two subdomains, which do not contact one another, are each comprised of three α-helices that assemble into helix-turn-helix motifs typical of homeodomains and Hin recombinase. However, side chains of the recognition helix (α3) within the amino-terminal subdomain dock into the major groove of DNA in a manner that is more reminiscent of the interaction of λ repressor with its operator DNA. Although the carboxy-terminal subdomain was not bound to DNA in the crystal structure of the Drosophila Paired domain, it is clear that it resembles conformationally the amino-terminal subdomain. Biochemical studies indicate that the carboxy-terminal subdomain of other Pax proteins, e.g., Pax-5, directly contacts DNA on other binding sites (12). In addition to connecting the amino- and carboxy-terminal subdomains, DNA binding is further facilitated by residues within the linker that make significant contacts with the phosphodiester backbone along the minor groove. DNA binding is also assisted by a β-hairpin and type II β-turn motif in the amino-terminal subdomain that precede the helical region. The β-hairpin is formed by two small antiparallel β-strands linked by a type I β-turn. One of the most notable features of the crystal structure was the novel use of the type II β-turn by Paired for base-specific minor groove recognition and of the β-hairpin for contacts to the sugar-phosphate backbone. A number of developmental anomalies result from missense mutations that are in or near the β-hairpin and β-turn motifs of paired domains, suggesting their importance for Pax protein function. In addition to protein-DNA contacts mentioned above, side chain interactions between various parts of the paired domain, e.g., between the β-turn motif and the linker, were identified.
The recognition of specific DNA sequences by paired domains reflects the presence of two functional DNA-binding motifs. In general, paired domains recognize two half-sites separated by approximately one turn of the DNA helix (12, 17). These observations are consistent with DNA recognition by both of the two subdomains within the major groove on one face of the double helix. However, other sites are efficiently bound by paired domain polypeptides lacking the carboxy-terminal subdomain, suggesting that Pax proteins can interact with DNA in multiple ways (12). Another important feature of paired domain-DNA interactions is their relatively relaxed nucleotide sequence specificity. The various Pax proteins exhibit preferences for binding different sets of nucleotide sequences that can be very degenerate. In this regard, it has been estimated that a binding site for Pax-5 occurs every 1 kb or so throughout the mouse genome (9). To account for the regulation of specific genes by Pax-5, other mechanisms, including interactions with partner proteins, have been proposed to increase the specificity of Pax-5 (and other Pax proteins) for their targets in vivo.
As one mechanism that enhances the target gene specificity of Pax proteins, we have described ternary complexes (B-cell-specific ternary complexes [BTCs]) comprised of Pax-5, Ets proto-oncogene family proteins, and specific DNA (20). Pax-5 is expressed in a restricted fashion in early B cells prior to terminal differentiation to the plasma cell stage (1). Pax-5 can recruit at least three Ets proteins, Fli-1, Ets-1, and GABPα (together with the ankyrin-repeat protein GABPβ), to bind a functionally important composite site in the early B-cell-specific mb-1 (immunoglobulin alpha-chain [Igα]) promoter in vitro. A role for this site for mb-1 transcription in vivo was directly confirmed by mutations in either the Pax-5 or Ets binding sites that similarly reduce mb-1 promoter function in transfected B cells. This observation is supported by the 10-fold downregulation of mb-1 expression in pax-5−/− knockout mice (35). The recruitment of Ets proteins by Pax-5 has a number of novel features. First, although the sequence recognized by Ets proteins in the promoter (5′CCGGAG) resembles the consensus site for Ets-1 DNA binding (5′CCGGAA/T [19, 26, 36, 52]), the promoter sequence is not bound detectably by most Ets proteins in the absence of Pa ax-5. Intriguingly, the last base in the core Ets site in the mb-1 promoter (5′CCGGAG) is also an important DNA contact for Pax-5. Second, ternary complex assembly requires only the paired and ETS DNA-binding domains. Third, recruitment by Pax-5 is dependent on an aspartic acid (Asp398 in Ets-1) that follows immediately the DNA recognition α-helix (α3) of the ETS domain. Together, these data suggest that Pax-5 and Ets bind the mb-1 promoter in close association and are likely to directly contact each other.
Here, we show that the β-hairpin, which is predicted by homology to form in Pax-5, is required both for binding of mb-1 promoter DNA and for efficient recruitment of Ets proteins. Our results suggest that the very high degree of sequence conservation of this motif in different Pax proteins and across species reflects its functional roles for DNA recognition and protein-protein interactions. Additional evidence supporting this hypothesis is provided by the recruitment of Ets-1 by Pax-2, or by amino-terminal sequences from Pax-3, which represents a distinct subfamily of Pax proteins. Together, these results support the hypothesis that interactions with other factors increase the specificity of Pax DNA binding in vivo, and they implicate Ets proteins as evolutionarily conserved partners of Pax proteins for regulating transcription.
MATERIALS AND METHODS
Model for Pax-Ets interactions.
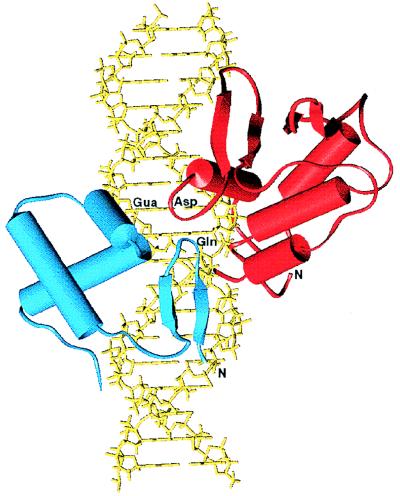
Figure 2 was generated with the graphic program SETOR (18) to represent (i) the nuclear magnetic resonance (NMR)-derived structure of the murine Ets-1 ETS domain and flanking carboxy-terminal α-helix (14, 51) (Brookhaven Protein Data Bank [PDB] code, 1ETC) docked on idealized B-DNA in an orientation similar to that observed in the crystal structure of the PU.1 ETS domain-DNA complex (30) (PDB code; 1PUE) and (ii) the amino-terminal subdomain of Drosophila Paired (53) (PDB) code, 1PDN) docked by alignment of the mb-1 promoter and optimized Paired binding site sequences.
FIG. 2.
A model of the Pax-5 and Ets-1 DNA-binding domains bound to a portion of the mb-1 promoter sequence. The murine Ets-1 ETS domain and flanking carboxy-terminal α-helix (red) and amino-terminal subdomain of Drosophila Paired (blue) are shown docked on idealized B-DNA (yellow). The guanosine common to the Pax-5 and Ets-1 binding sites in mb-1 is indicated as Gua. Positions of the aspartic acid (Asp) following the recognition helix in Ets-1 and the glutamine (Gln) in the β-hairpin of Pax-5 are indicated. These residues are identified as playing a key role in ternary complex formation. The amino termini of the two protein fragments are identified by N.
Plasmids and in vitro mutagenesis.
All PCR amplifications were performed with Pfu DNA polymerase (Stratagene, La Jolla, Calif.). For expression of human Pax-5 (hPax-5) in Escherichia coli, plasmid pbPax-5(1–149) was constructed by amplification of DNA encoding the first 149 amino acids of human Pax-5 from the pBLKS+-λP5 template plasmid (kindly provided by Peter Gruss), using oligonucleotides 5′-TGGATTTAGAGAAAAATTATCCG (5′ sense) and 5′-GGCGGCAAGCTTATTGGTTGGGTGGCTGCT (3′ antisense), and ligating the fragment into the blunted (Klenow fragment) NdeI site of plasmid pET-11a (Novagen, Madison, Wis.). DNA segments encoding other truncated hPax-5 polypeptides were generated by using PCR amplification of pbPax5(1–149) with the 5′ sense primer described above and downstream primers as follows: 1–107, 5′-AAGCTTTCAGGGATTTTGGCGTTTATATTCAGCG; 1–93, 5′-TCAGGGTGTGGCGACCTTTGGTTTGGA; 1–89, 5′-TCACTTTGGTTTGGATCCTCCAAT; and 1–84, 5′-TCATCCAATTACCCCAGGCTTGAT. Resulting inserts were ligated into pET-11a prepared as described above.
PCR mutagenesis was performed as described previously (20). For Pax-5 mutants, pbPax-5(1–149) was used as the template. Fragments were generated from two pairs of primers for each mutation, using PCR for the Q22A construct, 5′ sense primer and 5′-CCCAAGAGCATTCACTCCTCCATGTCCTG (hPax-5 Q22A, antisense), plus 5′-GTGAATGCTCTTGGGGGGGTTTTTGTGAA (hPax-5 Q22A, sense) and 3′ antisense primer. Amplified fragments were purified and combined in subsequent PCRs using the 5′ sense and 3′ antisense oligonucleotides described above. Resulting fragments were ligated into the NdeI site of pET-11a. All constructs were sequenced by using an ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems, Foster City, Calif.).
Lymphoid enhancer factor 1 (LEF-1) high-mobility-group (HMG) domain–Pax-5 fusion proteins were generated by amplifying the HMG domain (amino acids 296 to 381) of murine LEF-1 (the kind gift of Rudolf Grosschedl), using oligonucleotides 5′-TGCATATTAAGAAGCCTCTGAATGCT (LEF-1 5′ sense) and 5′-TCAAAGCTTCTCTCTCTTCCTCTTCTTC. The fragment was inserted into a pET-11a vector (in which the downstream HindIII site was destroyed by filling in and religating to make pET-11H−) to make pET-LEF-1-HMG. A double-stranded oligonucleotide encoding the (Gly3Ser/Thr)4 linker was assembled by annealing 5′-AGCTTGGTGGCGGTAGCGGCGGTGGCACCGGCGGTGGCAGCGGTGGCGGTACCTG) and 5′-AGCTCAGGTACCGCCACCGCTGCCACCGCCGGTGCCACCGCCGCTACCGCCACCA and ligating into the HindIII site of pET-LEF-1-HMG. The resulting pET-LEF-1-linker construct was digested with Asp718 and blunted for ligation with Pax-5 fragments. Oligonucleotides used to generate Pax-5 segments to make LEF-1–Pax-5 hybrid proteins were identical to those used for preparation of the Pax-5 truncations, except that the upstream sense primer was 5′-CAGCAGGACAGGACATGGAGGAGTG (from Thr12). All constructs were sequenced as described above.
Plasmids with Pax-2 or Pax-3 cDNA were the generous gifts of P. Gruss. For production of Pax-2(1–148) protein in reticulocyte lysates, a segment encoding the paired domain was generated by PCR using primers 5′ CTCACCATGGATATGCACTGCAAAGCAGA and 5′-ATCATGGGTGGAAAGGCTGCTGAACTTT along with pC31A(murine Pax-2) DNA. The amplified Pax-2 fragment was ligated into the filled HindIII site of pBluescript KS+ (Stratagene) to make BSPax-2(1–148). Pax-3 and Pax-6 sequences were ligated into the Asp718 site of pET-LEF-LK. The murine Pax-3 insert was generated by using Pax3 cDNA pBH3.2 as a template and primers 5′-CACCCCTCTTGGCCAGGGCCGAGT (Pax-3 sense) and 5′-CTACTTGGGTTTGCTGCCGCCGATGGC (Pax-3 antisense). The resulting plasmid was termed pLEF-LK-P3. Fragments for generation of the Q40A mutation in Pax-3 pLEF-LK-P3 were generated by using the pLEF-LK-P3 template, Pax-3 Q40A sense primer 5′-5CACCCCTCTTGGCCAGGGCCGAGTCAACGCGCTCGGAGGAGTATTT, and the Pax-3 antisense primer described above. The resulting fragment was ligated into pET-LEF-LK to make pLEF-LK-P3Q40A.
BSEts-1(333–440) was made by inserting the AccI restriction fragment including murine Ets-1 carboxy-terminal sequences from SK-c-ets-1.6 into BSEts-1(333–420) (20) linearized with AccI.
Production of recombinant proteins.
Pax and LEF-1-Pax hybrid proteins were overexpressed in E. coli BL21(λDE3)pLysS (Novagen). Expression was significantly increased by including the plasmid dnaY (21) (generously provided by Peter Love) for coexpression of E. coli Arg tRNAs. Single colonies were picked from plates of freshly transformed bacteria and inoculated into 50-ml cultures of Luria broth supplemented with carbenicillin (250 to 500 μg/ml), chloramphenicol (34 μg/ml), and kanamycin (50 μg/ml). Bacteria were grown at 37°C to an optical density at 600 nm of 0.4 and induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to 1 mM, and growth was continued for 2 h. Bacteria were harvested by centrifugation, and pellets were stored at −80°C. Pellets were resuspended in ice-cold buffer Z (25 mM HEPES [pH 7.7], 100 mM KCl, 12.5 mM MgCl2, 20% glycerol, 0.1% nonidet P-40, 1 mM dithiothreitol) supplemented with aprotinin (2 μg/ml), phenylmethylsulfonyl fluoride (100 μg/ml), leupeptin (2 μg/ml), and pepstatin A (1 μg/ml). Pellets were sonicated 10 s and centrifuged for 20 min in a Sorvall SS34 rotor at 12,000 rpm to remove bacterial debris. Protein concentrations of supernatants were determined by the Bradford assay (Bio-Rad, Hercules, Calif.). Relative protein expression was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. Protein concentrations were normalized relative to wild-type Pax-5(1–149); 1 to 5 μg of total bacterial lysate proteins was used in each binding assay.
To generate Pax-2 or Ets-1 DNA-binding domain protein, plasmid BSPax-2(1–148) or BSEts-1(333–440) was linearized and transcribed with T7 RNA polymerase, and RNAs were purified and translated as reported previously (20).
DNA probes and EMSA.
Annealing, labeling of DNA probes, and electrophoretic mobility shift assay (EMSA) were performed as described previously (20). The wild-type mb-1 promoter probe (mb-1 probe) was assembled by using 5′-TCGAAGGGCCACTGGAGCCCATCTCCGGCACGGC and 5′-TCGAGCCGTGCCGGAGATGGGCTCCAGTGGCCCT. Oligonucleotides comprising the various probes were as follows: mut 1 probe, 5′-TCGAAGGGCCACTGGAGCCCATCTAAGGCACGGC and 5′-TCGAGCCGTGCCTTAGATGGGCTCCAGTGGCCCT; mut 2 probe, 5′-TCGAAGGGCAAATTGAGCCCATCTCCGGCACGGC and 5′-TCGAGCCGTGCCGGAGATGGGCTCAATTTGCCCT; and G→A probe, 5′ TCGAAGGGCCACTGGAGCCCATTTCCGGCACGGC and 5′-TCGAGCCGTGCCGGAAATGGGCTCCAGTGGCCCT. The Sγ2a probe was assembled by using 5′-TCGAGATCAGAATTGTGAAGCGTGACCATAGAAA and 5′-TCGATTTCTATGGTCACGCTTCACAATTCTGATC. LEF-1–Pax-5–Ets (LPE) phasing probes were made by annealing each of the following oligonucleotides with 5′GCCGTGCCGGAGATGGGCTCCA and extending the gap with Klenow enzyme, [α-32P]dCTP, and unlabeled dATP, dCTP, and TTP: 5′GACACCCTTTGAAGCTTCTGGAGCCCATCTCCGGCACGGC (+1), 5′GACACCCTTTGAAGCTCTGGAGCCCATCTCCGGCACGGC (0), 5′GACACCCTTTGAAGTCTGGAGCCCATCTCCGGCACGGC (−1), 5′GACACCCTTTGAAGCTGGAGCCCATCTCCGGCACGGC (−2), and 5′GACACCCTTTGAAGTGGAGCCCATCTCCGGCACGGC (−3). The LPE (−1) mut 1 probe was made by annealing 5′GCCGTGCCTTAGATGGGCTCCA with 5′GACACCCTTTGAAGTCTGGAGCCCATCTAAGGCACGGC and filling in with Klenow enzyme as described for the wild-type LPE probes. Relative levels of DNA binding were estimated by using a Molecular Dynamics (Sunnyvale, Calif.) PhosphorImager.
RESULTS
Requirements for Pax-5–Ets-1 ternary complex assembly.
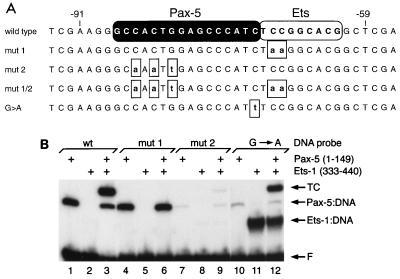
In our previous study (20), we defined DNA sequences required for the assembly of ternary complexes comprised of full-length Pax-5 and Ets proteins together with mb-1 promoter DNA. Although we identified residues in the ETS domain required for interaction with Pax-5, we did not identify amino acids within the Pax-5 paired domain that participate in the recruitment of Ets proteins. To begin to address this question, we first confirmed that BTCs assembled in vitro by using recombinant DNA-binding domains exhibit the cooperative DNA binding observed with full-length proteins. Polypeptides comprising the paired domain of Pax-5 (amino acids 1 to 149, synthesized in E. coli) and ETS domain of murine Ets-1 (amino acids 333 to 440, translated in vitro by using rabbit reticulocyte lysates), which together comprise the BTC2 complex identified previously, were used in our studies. In an EMSA with 32P-labeled probe DNA comprising wild-type mb-1 promoter sequences, Pax-5(1–149) bound the promoter probe in the absence of the Ets-1 ETS domain (Fig. 1B, lane 1), while ETS domain binding by itself was detected only weakly (lane 2). When combined, the two DNA-binding domains efficiently assembled ternary complexes that include both polypeptides (lane 3). Assembly of the complex requires an intact Ets core nucleotide sequence, because mutation of this sequence (5′CCGGAG to 5′CCttAG) in the mut 1 probe prevents ternary complex formation (lanes 4 to 6). Mutation of three bases in the mut 2 probe (lanes 7 to 9) results in greatly reduced but detectable levels of DNA binding by Pax-5(1–149) by itself (less than 2% of binding to the wild-type probe), supporting the previous observation that the substitutions delete important base contacts for this protein (as determined by methylation interference [20]). Although Pax-5 binding to the mut 2 probe is greatly diminished, a low level of ternary complex was observed with the addition of Ets-1(333–440). Thus, the recruitment of Ets proteins does not require DNA contacts for Pax-5 that are mutated in the mut 2 probe. In contrast, a single base mutation within the Ets recognition sequence to form a consensus core sequence (CCGGAG to CCGGAa on the antisense strand) greatly reduces Pax-5 binding by itself (to 5% of wild-type binding [lane 10]) and allows Ets-1(333–440) to bind efficiently in the absence of Pax-5 (increases by >100-fold [lane 11]). This finding confirms the importance of the last base at the wild-type G position to the binding of Pax-5 and Ets-1. Combining the two polypeptides results in a level of ternary complex formation similar to that observed with the wild-type promoter probe (lane 12). We conclude that, depending on the nucleotide sequence of the labeled probe, the Pax-5 paired domain can recruit the ETS domain of Ets-1, or in reciprocal fashion, the ETS domain can recruit the paired domain. These data suggest that interactions between the two proteins contribute to ternary complex assembly.
FIG. 1.
The Pax-5 paired domain (residues 1 to 149) and the ETS domain of Ets-1 (residues 333 to 440) exhibit cooperative binding to the wild type mb-1 promoter. (A) Double-stranded oligonucleotide probe sequences used in this study. Only the sense sequence is shown, numbered relative to +1 of the wild-type mb-1 promoter (45). Nucleotides contacted by Pax-5 and those contacted by Ets-1, as estimated by methylation interference and footprinting studies (20, 24a), are highlighted at the top. Boxed lowercase letters represent mutations. (B) Relative DNA binding by Pax-5 and ETS DNA-binding domains. DNA probes and inclusion of proteins are indicated at the top. wt, wild type; TC, ternary complexes; F, free probe.
We hypothesized that Pax-5 and Ets-1 physically interact upon binding DNA, but our results did not distinguish between various potential mechanisms for assembling the complex. Although it is likely that Ets-1 binds the promoter site in much the same manner that Ets-1 binds closely related nucleotide sequences (51), insufficient information precluded making similar assumptions for Pax-5. For example, the structural features of the Pax-5 paired domain on the mb-1 promoter, including whether the amino-terminal or carboxy-terminal subdomain of the paired domain is oriented toward the adjacently bound ETS domain in the ternary complex, are not known. In this regard, a preliminary experiment suggested that the amino-terminal subdomain of Pax-5 is oriented toward the ETS domain (data not shown): a mutation in the putative recognition α-helix of the carboxy-terminal subdomain (α6) greatly decreased the binding of Pax-5 to the wild-type promoter probe (to 1 to 2% of the wild-type level), but levels of binding of wild-type or α6 mutant Pax-5 to the mut 2 probe were nearly equivalent. These results suggested that the bases changed in the mut 2 probe are recognized by the carboxy-terminal subdomain and that the amino-terminal subdomain binds DNA nearest the Ets recognition site.
A structural model for Pax-Ets interactions.
To make predictions concerning the interaction of Pax-5 with Ets-1 on specific DNA, we constructed a model using previously determined X-ray crystallographic and NMR structures of several related proteins bound to their cognate sites. Although structural determinations were not available for Pax-5 itself, a crystallographic analysis of the highly homologous paired domain (77% identity) of Drosophila Paired bound to an optimal binding site has been reported (53). In this crystal structure, the amino-terminal subdomain and linker of the paired domain contact an optimized site, while the carboxy-terminal subdomain does not interact with DNA. Preliminary data suggested that the amino-terminal subdomain of Pax-5 is oriented toward the Ets binding site (see above). Therefore, in our model (Fig. 2), only the amino-terminal subdomain, including the β-hairpin, β-turn, and linker, were docked onto an ideal B-form DNA corresponding to the mb-1 sequence. For the ETS domain, the recently determined structures of the Ets-1 and PU.1 DNA-binding domains bound to their recognition sites were readily appropriated for the ternary complex model (14, 30, 51). Relative positioning and orientation of the ETS domain was suggested by the obvious relationship between the mb-1 and consensus Ets-1 binding sites (5′CCGGAG and 5′CCGGA[A/T], respectively). Positioning of the amino-terminal subdomain of Paired was based on the assumption that the G common to the Ets-1 and Pax-5 sites (5′CCGGAG) is, by analogy with Paired DNA binding, contacted by the first residue of its DNA recognition α-helix (α3). Small distortions of the DNA observed in the structures of the Paired-DNA and ETS domain-DNA complexes were neglected for this modeling.
The model suggests that, indeed, Pax and Ets proteins can bind the mb-1 promoter in very close proximity to one another. Furthermore, the model is consistent with previous studies indicating that Asp398 of Ets-1 is important for Ets-Pax interactions and suggests that this amino acid may contact the paired domain. If this view is correct, recruitment of Ets proteins would be a function of the amino-terminal subdomain alone, with the linker of the paired domain functioning to stabilize the conformation of the amino-terminal subdomain, or binding of the domain to DNA. As one caveat, our data show that the carboxy-terminal subdomain is required for high-affinity binding of the mb-1 promoter by Pax-5, and the model cannot address the role of this subdomain. However, the model suggests that the contribution of the carboxy-terminal subdomain in the ternary complex may be the stabilization of Pax-5 binding through additional contacts with DNA, not through contacts with the ETS domain.
Amino acid sequences required for Ets recruitment by Pax-5.
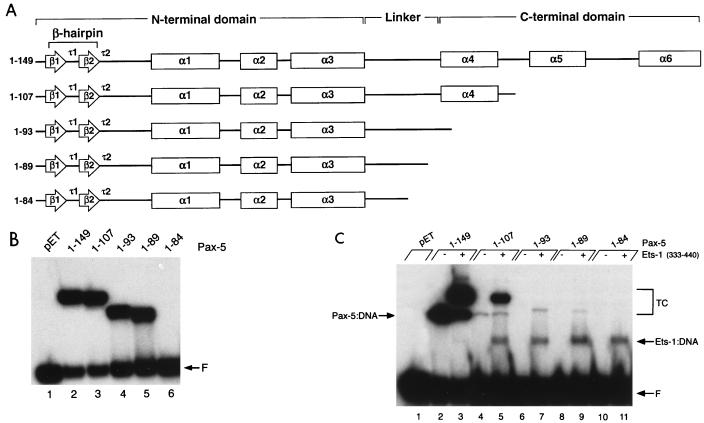
Our model suggests that the amino-terminal subdomain of Pax-5 should include sequences for recruiting Ets proteins to bind DNA. To test this hypothesis, we prepared a set of progressively truncated paired domain polypeptides (Fig. 3A) expressed in E. coli. To examine the relative abilities of these polypeptides to bind DNA, we used probes derived either from the switch region promoter of the Igγ2a heavy-chain gene (Sγ2a) or from the mb-1 promoter in the absence or presence of the Ets-1 ETS domain. With the exception of the shortest polypeptide tested (amino acids 1 to 84), each of the truncated Pax-5 polypeptides bound the Sγ2a probe in the absence of Ets-1 (Fig. 3B), suggesting that this sequence is bound by the amino-terminal subdomain and linker alone. In contrast, relative to binding of the full-length paired domain (Fig. 3C, lane 2), a very large decrease in binding to the mb-1 promoter was observed following deletion of sequences including helices α5 and α6 (lane 4), consistent with recognition of the mb-1 promoter by both of the two subdomains. Addition of the Ets-1 ETS domain still resulted in cooperative assembly of ternary complexes with the truncated Pax-5 polypeptide (lane 5). Binding of probe DNA by the ETS domain alone, which was included at a limiting concentration in this experiment, was detected due to the extended autoradiography necessary for detection of weak ternary complex formation. With further truncation, binding of the Pax-5 paired domain in the absence of Ets-1 was not detected (lanes 6, 8, and 10). With the addition of the ETS domain, a very low level of ternary complex was observed for either Pax-5(1–93) or Pax-5(1–89) but not Pax-5(1–84) (lanes 7, 9, or 11, respectively). These data suggest that Pax-5(1–89) comprises minimal sequences for interactions with the ETS domain on the mb-1 promoter. However, the lack of detection of ternary complexes with Pax-5(1–84) could be due either to deletion of sequences that are required for recruitment or to the general impairment of DNA binding by the truncation of Pax-5.
FIG. 3.
DNA binding by truncated Pax-5 polypeptides. (A) Schematic representation of truncated polypeptides used for panels B and C. The secondary structure of Pax-5 was predicted from the crystal structure of Drosophila Paired (53). Arrows are regions of β-sheet, boxes are α-helices, and τ1 and τ2 are turns. The β-hairpin is formed by β1-τ1-β2, and the type II turn (β-turn) is τ2. Relative to the diagram, Pax-5(1–149) includes six additional carboxy-terminal amino acids. (B) Control EMSA showing binding of truncated Pax-5 polypeptides to the Sγ2a probe. The lysate in lane pET was prepared from E. coli transformed with empty pET-11a vector. The gel was exposed to X-ray film for 15 h. F, free probe. (C) EMSA showing binding of truncated Pax-5 polypeptides to the mb-1 probe with or without added Ets-1 ETS domain. The concentration of Pax-5(1–149) used in lanes 2 and 3 is approximately one-fourth that of other Pax-5 polypeptides used in this experiment. Binding of the Ets-1 ETS domain by itself was detected due to the extended period of autoradiography (3 days) necessary for detection of weak ternary complex formation. Lane pET is as in panel B. TC, ternary complexes; F, free probe.
As shown in Fig. 3, our studies of Pax-5–Ets interactions were dependent on the detection of DNA binding. To further address requirements for Pax-5–Ets interactions, we sought to reduce the dependence of our studies on the affinity of Pax-5 for DNA. To achieve this end, we tethered the amino-terminal subdomain of Pax-5 to a heterologous DNA-binding domain derived from LEF-1, which encodes an HMG domain protein that binds as a monomer to a short nucleotide sequence with high affinity (KD ≃ 1 nM) (22). The HMG domain (85 amino acids) was placed amino terminal to Pax-5 in the hybrid proteins because the crystallographic structure of Paired suggested that its amino terminus (and therefore that of Pax-5) points back toward sequences recognized by its carboxy-terminal subdomain (53). With this configuration, we reasoned that replacement of sequences recognized by the carboxy-terminal subdomain of Pax-5 with a LEF-1 binding site would allow for simultaneous DNA binding by both domains of a LEF-1–Pax-5 hybrid protein. Moreover, this configuration is predicted to maintain the orientation of Pax sequences in the hybrid protein relative to an ETS domain bound at the adjacent Ets binding site, which should allow for progressive carboxy-terminal deletions in paired domain sequences without otherwise changing the hybrid polypeptides. A linker consisting of four repeats of Gly3Ser/Thr was inserted between LEF-1 and Pax-5 sequences to act as a flexible tether. As a potential complication to these experiments, it has been reported that LEF-1 induces a very sharp bend in bound DNA (23, 33). However, effects of the bend upon an LEF-1–Pax-5–Ets ternary complex should be minimal, because the interaction between Pax and Ets sequences would take place entirely on one side of the DNA bend.
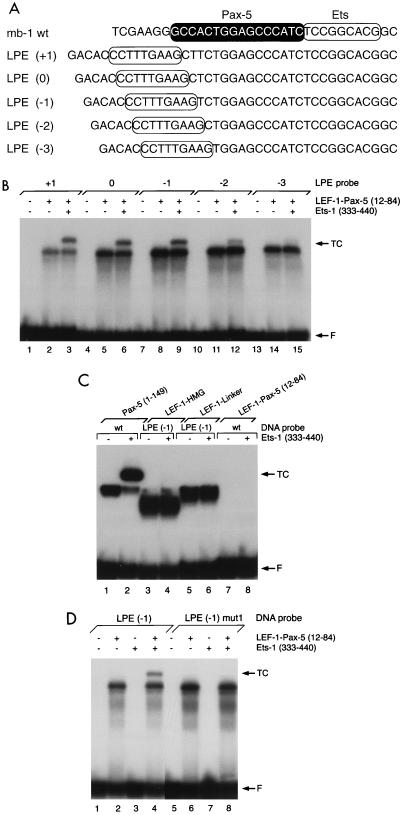
To examine DNA binding by LEF-1–Pax-5 hybrid proteins, we used EMSA and five probes (LEF-1–Pax–Ets or LPE probes) that vary the position of the LEF-1 binding site through one-half turn of the double helix relative to the downstream Pax-5 and Ets recognition sequences (Fig. 4A). A hybrid protein comprising the HMG domain, linker, and Pax-5(12–84) was synthesized in E. coli and demonstrated to have the appropriate molecular mass when analyzed by SDS-PAGE (data not shown). Amino acids 1 to 11, which precede sheet β1, are not included in the paired domain and are dispensable for DNA binding (12, 51a). Probe DNAs were labeled with 32P and incubated with LEF-1–Pax-5(12–84) before analysis by EMSA. By itself, the hybrid protein bound each of the five helically phased LPE probes (Fig. 4B, lanes 2, 5, 8, 11, and 14). Variations in levels of binding by the five probes were observed, suggesting that optimal spacing between the LEF-1 and Pax-5 recognition sites in a subset of the probes allows for simultaneous DNA contacts by both the LEF-1 HMG and Pax-5 domains. This ability would suggest that paired domain sequences in the hybrid protein can dock with DNA in a manner that approximates complexes assembled with the intact paired domain. In support of this conclusion, the LEF-1-Pax(12–84) polypeptide recruited the Ets-1 ETS domain to bind three of the five LPE probes (lanes 3, 6, and 9) similarly and less well to bind probes with fewer nucleotides between the LEF-1 and Ets binding sites (lanes 12 and 15). This result is particularly striking because Pax-5(1–84) did not bind either the Sγ2a (Fig. 3B) or mb-1 probe and was not recruited detectably by the Ets-1 ETS domain (Fig. 3C). Similar results were obtained with a second hybrid polypeptide, LEF-1–Pax-5(1–89) (data not shown). Our studies suggest that Pax-5(12–84), although not sufficient for DNA binding by itself, includes minimal sequences for recruiting Ets-1 to bind the mb-1 promoter.
FIG. 4.
DNA binding and recruitment of the Ets-1 ETS domain by LEF-1–Pax-5 amino-terminal subdomain hybrid polypeptides. (A) DNA probes used in these assays. Pax-5 and Ets-1 binding sites in the wild-type (wt) probe are highlighted as in Fig. 1A. LEF-1 recognition sequences are circled. LPE (0) was the sequence predicted by molecular modeling to be optimal for LEF-1–Pax-5 binding. (B) EMSA of LEF-1–Pax-5(12–84) binding to the five LPE probes. TC, ternary complexes; F, free probe. (C) Control experiments. EMSA of LEF-1–HMG (residues 296 to 381 of murine LEF-1), LEF-1–linker [the HMG domain of LEF-1 and the (Gly3Ser/Thr)4 linker], or LEF-1–Pax-5(12–84) binding to the wild-type mb-1 or LPE (−1) probe. Ternary complexes are detected only with Pax-5, Ets-1, and the wild-type mb-1 probe. F, free probe. (D) Ternary complex assembly with the hybrid protein requires the Ets-1 binding site. EMSA was performed with the wild-type or mut 1 (Fig. 1A) version of the LPE (−1) probe together with the LEF-1–Pax-5(12–84) protein and Ets-1 ETS domain. TC, ternary complexes; F, free probe. The gels in panels B to D were exposed to X-ray film for 12 to 15 hours.
It was important to show that recruitment of Ets-1 by LEF-1–Pax-5 hybrid proteins mimics aspects of Ets recruitment by intact Pax-5 on the mb-1 promoter. Binding of either the HMG domain of LEF-1 or the LEF-1-linker polypeptide alone was readily detected with the LPE (−1) probe (Fig. 4C, lanes 3 and 5). Neither of these polypeptides recruited Ets proteins to bind the probe (lanes 4 and 6). Consistent with experiments with Pax-5(1–84), the LEF-1–Pax-5(12–84) polypeptide did not bind detectably to the wild-type mb-1 promoter probe without or with added Ets-1 (lanes 7 and 8). Recruitment of Ets proteins by the hybrid polypeptide also requires an intact Ets binding site in the LPE (−1) probe, as shown by the lack of Ets-1 recruitment when the Ets core sequence is mutated (Fig. 4D, lane 8), and Asp398 in the ETS domain, which is essential for efficient ternary complex assembly (data not shown). Therefore, Ets-1 interacts with LEF-1-Pax-5(12–84) and with intact Pax-5 in very similar manners.
To further localize sequences in Pax-5 that are required for ternary complex assembly, we introduced additional deletions in the Pax-5 segment of the hybrid protein and tested their binding to the LPE (−1) probe by EMSA. An additional truncation removed five amino acids of the linker region. The hybrid polypeptide, LEF-1–Pax-5(12–79), bound the LPE (−1) probe but did not recruit Ets-1 detectably (data not shown). This result is consistent with the observation that intramolecular interactions within the amino-terminal subdomain and linker region stabilize its overall structure (53).
The paired domain β-hairpin is a highly conserved protein-DNA and protein-protein interaction motif.
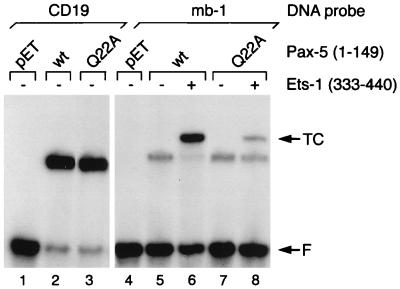
In our previous analysis, we showed that an aspartic acid (Asp398) carboxy terminal to the major DNA recognition α-helix in Ets-1 (α3) is required for its recruitment by Pax-5 (20). Our model for Pax-5–Ets-1 interactions suggests that sequences within the paired domain nearest the aspartic acid of Ets-1 include the β-hairpin motif (Fig. 2). Mutations in the β-hairpin and adjacent β-turn have profound effects on the function of Pax proteins in vivo (5, 43, 48). Previously, X-ray crystallographic analysis showed that a glutamine located within the turn (τ1) of the β-hairpin of Paired (corresponding to Gln22 of Pax-5) interacts with one strand of the sugar-phosphate backbone of DNA (53). In our model, this amino acid is also positioned close to Asp398 in the Ets-1 ETS domain (Fig. 2B). Therefore, we used oligonucleotide-directed mutagenesis to convert the glutamine to alanine in the context of the intact paired domain of Pax-5(1–149). This substitution is not expected to affect the structure of the β-hairpin. Wild-type and Q22A polypeptides were expressed in E. coli at similar levels and were used in an EMSA with labeled DNA probes comprising binding sites from the CD19 or mb-1 promoter (Fig. 5). DNA binding by the Q22A polypeptide to either the CD19 or mb-1 probe was equivalent to binding by the wild-type polypeptide (lanes 3 versus 2 and 7 versus 5). However, although Gln22 is not critical for DNA binding by Pax-5, the mutation decreased recruitment of the Ets-1 ETS domain to one-fourth (determined by phosphorimaging) of that observed with the wild-type paired domain (lane 8 versus 6). These data indicate that Gln22 plays an important role in the recruitment of Ets-1 by Pax-5. However, the residual recruitment of Ets proteins observed with the Q22A mutant suggests that other amino acids in Pax-5 may participate in Pax-5–Ets-1 interactions.
FIG. 5.
Gln22 of Pax-5 is required for efficient assembly of ternary complexes with Ets-1. EMSA was performed with wild-type (wt) or mutated Pax-5(1–149) polypeptide and either the CD19 or mb-1 DNA probe, as indicated. Lysates in lanes pET were prepared from E. coli transformed with empty pET-11a vector. TC, ternary complexes; F, free probe.
Interactions of other Pax proteins with Ets-1.
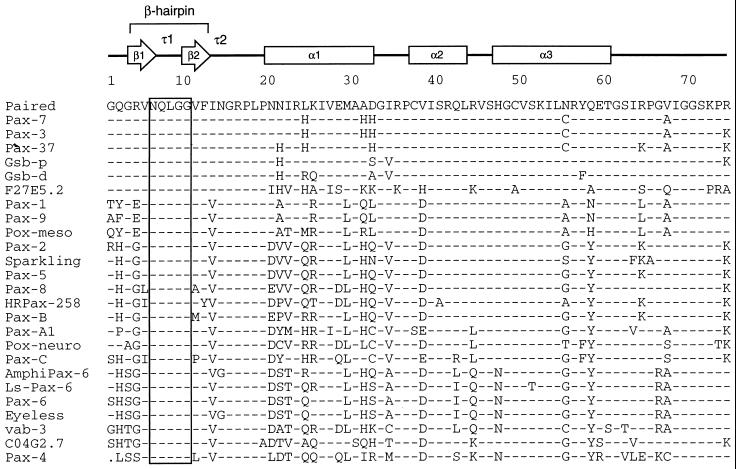
Our studies confirmed that the β-hairpin is a likely candidate for an Ets interaction motif. Provocatively, alignment of a large number of Pax protein sequences shows that a stretch of five amino acids from the β-hairpin (including Gln22 of Pax-5) is absolutely conserved (Fig. 6). The comparison includes sequences of the nine Pax proteins identified in mice and humans along with sequences from Drosophila, Caenorhabditis elegans, Aromphioxus, ribbonworms, jellyfish, and coral polyps. The strict conservation of the sequence and, by inference, structure of the β-hairpin between Pax proteins of animals as divergent as humans and jellyfish suggests that the function(s) of these structures evolved prior to the extensive duplication and diversification of pax genes at the Cambrian radiation of species (3). Indeed, the sequence has been conserved as well as or better than Pax sequences involved in recognition of the major groove (α3).
FIG. 6.
The glutamine required for efficient recruitment of Ets proteins (and four flanking amino acids) has been perfectly conserved throughout the evolution of Pax proteins. Amino acid sequences comprising the amino-terminal subdomains and linker regions of Pax proteins were aligned for comparison as shown. Secondary structure of Drosophila Paired is shown with arrows indicating β-strands, boxes indicating α-helical regions, and τ1 and τ2 indicating turns identified in the crystal structure of Paired (53). The β-hairpin is formed by strands β1 and β2 and the type I β-turn τ1, while τ2 is a type II β-turn. Shaded vertical bar indicates the completely conserved region of the β-hairpin, including the glutamine residue analyzed in this report. Sequences were derived from Homo sapiens Pax-1 (EMBL/GenBank accession no. P15863), Pax-3 (P23760), Pax-5 (M96944), Pax-6 (M77844), Pax-7 (Z35141), Pax-8 (L19606), and Pax-9 (S36115); Mus musculus Pax-2 (280984) and Pax-4 (P32115); Branchiostoma floridae (amphioxus) AmphiPax-6 (AJ223440); Halocynthia roretzi (ascidian) Pax-37 (D84254) and HRPax-258 (AB006675); Lineus sanguineus (ribbonworm) Ls-Pax-6 (X95594); Acropora millepora (coral) Pax-C (AF053459); Chrysaora quinquecirrha (sea nettle) Pax-A1 (U96195) and Pax-B (U96197); Drosophila melanogaster Paired (P06601), Gooseberry proximal (Gsb-p; P09083), Gooseberry distal (Gsb-d; P09082), Sparkling (AF010256), Eyeless (X79492), Pox-meso (P23757), and Pox-neuro (P23758); and C. elegans Pax homologs C04G2.7 (Z70718) and F27E5.2 (Z48582) and Pax-6 homolog vab-3 (U31537). The dot in the Pax-4 sequence represents a gap in the alignment.
The high degree of sequence conservation of the β-hairpin suggests that recruitment of Ets proteins may be a common function of the paired domain. To address this hypothesis, we translated other Pax paired domains in vitro, using reticulocyte lysates, and examined their ability to bind the mb-1 probe and recruit the Ets-1 ETS domain. Of those tested, only Pax-2(1–148) bound the probe efficiently by itself (Fig. 7A, lane 2), and the Pax-2 paired domain recruited Ets-1 (lane 4). This result is not surprising, considering that the sequences of the Pax-2 and Pax-5 paired domains exhibit only one and three amino acid differences between their amino-terminal and carboxy-terminal subdomains. In contrast, DNA binding by the paired domain of Pax-3 was detected only very weakly, and binding of Pax-6 was not observed (data not shown). The lack of DNA binding by these proteins is due to their divergent DNA-binding specificities, which are significantly different from those of the Pax-2/5/8 subfamily.
FIG. 7.
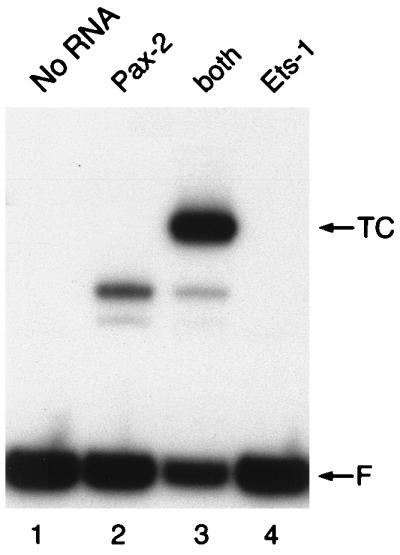
Recruitment of the Ets-1 ETS domain by Pax-2. (A) The paired domain of Pax-2 (residues 1 to 148) binds the mb-1 probe and recruits Ets-1. EMSA was performed with the mb-1 probe. Binding assays were performed with rabbit reticulocyte lysates programmed with synthetic RNA encoding the Pax-2 paired or Ets-1 ETS domain or with a translation without added RNA (No RNA). Both, Pax-2 and Ets-1 were both added after their separate translation; TC, ternary complexes; F, free probe.
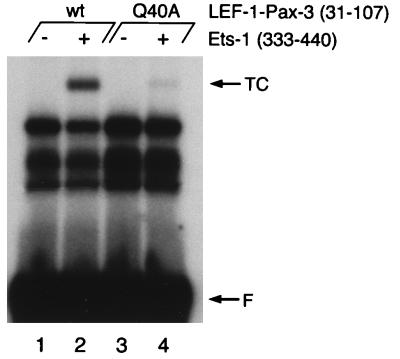
We have shown that polypeptides comprising the amino-terminal subdomain of Pax-5 can recruit Ets proteins to bind the mb-1 promoter when tethered to a heterologous DNA-binding domain. As observed with the amino-terminal subdomain of Pax-5, the lack of detectable binding of the paired domains of Pax-3 or Pax-6 to the mb-1 promoter is likely due to inadequate contacts with the DNA probe. Therefore, we reasoned that the amino-terminal subdomains of these proteins may functionally substitute for Pax-5 sequences in the context of LEF-1–Pax-5 hybrid polypeptides. We tethered the amino-terminal subdomains of Pax-3(31–107) to the LEF-1-linker polypeptide and expressed the hybrid protein in E. coli. As observed with LEF-1–Pax-5(12–84), the hybrid polypeptides bound the LPE(−1) probe (Fig. 8, lanes 1 and 2). Moreover, the hybrid polypeptide recruited the Ets-1 ETS domain to bind the probe (lanes 2). Recruitment of ETS domains by LEF-1–Pax-3(31–107) likely involved the same mechanism as observed for Pax-5, because binding was similarly decreased by a Gln-to-Ala mutation (Q40A; homologous to Q22A in Pax-5). We also observed recruitment of the ETS domain by the amino-terminal subdomain of Pax-6 (data not shown). We conclude that recruitment of Ets proteins is a capability of many, if not all, Pax proteins, and is due to the β-hairpin.
FIG. 8.
DNA binding and recruitment of Ets-1 by LEF-1–Pax-3(31–107) fusion protein. EMSA was performed with the LPE (−1) probe. Faster-migrating bands are likely due to limited proteolysis during preparation of proteins from E. coli. TC, ternary complexes; F, free probe; wt, wild type.
DISCUSSION
Partnerships between transcription factors form the basis of the combinatorial control of gene expression in eukaryotic cells (reviewed in reference 24). Cooperative interactions between factors enhance the specificity of DNA binding by increasing their relative affinity for sites comprised of recognition sequences for each partner. Two major types of protein-protein interactions contribute to combinatorial gene regulation. First, the assembly of stably associated multimers in solution (e.g., to form homo- or heterodimers) is a common mechanism that increases the affinity and specificity of proteins that do not fold and/or bind DNA as monomers (e.g., assembly of AP-1 by Fos and Jun). In the other general case, partner transcription factors that recognize adjacent promoter sites assemble into multimeric complexes via protein-protein and protein-DNA interactions. Assembly of these complexes may also be dependent on protein-induced changes in the conformation of DNA, e.g., by bending. Interactions with partner proteins are often obligatory for specific binding to one set of sites but not to others. Although interactions of this type are quite common, the structural basis for complex formation of this type has only been determined for a small number of examples (e.g., AP-1–NFAT [11]). Therefore, it is significant that our studies identify specific amino acids involved in the interaction of Pax-5 and Ets-1 and thereby support the basic aspects of the model presented in Fig. 2. Further confirmation of this model will require direct structural studies of the Pax-5–Ets-1–DNA ternary complex by NMR spectroscopic or X-ray crystallographic methods.
Our studies increase the number of possible combinations of Pax and Ets proteins. Previously, we showed that Pax-5 can assemble complexes in vitro with at least five Ets proteins (20). In this report, we show that in addition to Pax-5, other Pax proteins including Pax-2 and Pax-3 (and tentatively Pax-6) can recruit Ets proteins to bind DNA through interactions with the β-hairpin of their paired domains. Because all Pax proteins have the potential to form similar β-hairpins, we hypothesize that interactions between Pax and Ets proteins are a common function that contributes to the combinatorial regulation of gene expression in organisms that express these factors.
Studies suggest that interactions with Ets, and potentially other proteins, are an important feature of Pax functions in vivo. Mutation of either the Pax-5 or Ets binding sites in the mb-1 promoter results in a similar decrease in promoter function in transfected mb-1-expressing cells (20). In support of these data, targeted deletion of genes encoding Pax-5 in mice resulted in the reduction of mb-1 expression to 1/10 of that of wild-type mice (35). Moreover, in an elegant study, pax-5−/−pre-B cells were expanded ex vivo with interleukin-7 and infected with a retrovirus for expression of a Pax-5–estrogen receptor hybrid protein (34). Upon treatment with 17-β-estradiol, the cells upregulated expression of the CD19, N-myc, mb-1, and LEF-1 genes. Intriguingly, the transcriptional activation domain of Pax-5 was not required for the upregulation of mb-1 transcription. The authors concluded that the role of Pax-5 for mb-1 activation likely involves its ability to recruit partner proteins to bind the mb-1 promoter, which in turn leads to promoter activation. The activation domain was also not required for upregulation of the LEF-1 gene.
Requirements for ternary complex assembly.
Our studies revealed new information concerning DNA binding by Pax-5. Pax-5 exhibits multiple modes of DNA binding on different nucleotide sequences. A number of sites, including those in the Ig Sγ2a and Iɛ promoters, as well as a site identified in the p53 gene, are bound equally well by the intact paired domain of Pax-5 or a truncated polypeptide that lacks the putative recognition α-helix (α6) of the carboxy-terminal subdomain (this study and reference 51a). In contrast, both subdomains of the paired domain are required to bind other sites, including those in the sea urchin histone H2A-2.2 and murine mb-1 promoters as well as the murine Ig 3′ Cα enhancer (12, 51a). Truncation of Pax-5 to amino acids 1 to 107 resulted in nearly 2-orders-of-magnitude-lower binding to the wild-type mb-1 promoter site. Similar results were obtained for the binding of the intact paired domain to the mut 2 probe, which includes mutations in putative recognition sequences for the carboxy-terminal subdomain. Evidence was also obtained for a role for the linker region in binding the mb-1 probe, because significantly lower binding was obtained with truncated polypeptides that lacked this region.
Although we cannot rule out a role for the carboxy-terminal subdomain of Pax-5 for recruitment of Ets-1, this recruitment appears to be largely a function of its amino-terminal subdomain. Amino acids 12 to 84 of Pax-5 can recruit the ETS domain, but only when tethered to a heterologous DNA-binding domain bound to DNA. Ternary complexes were not detected when additional amino acids of the linker region were deleted. These data can be interpreted in two ways. Amino acids of the linker region may be involved directly in interactions with Ets proteins; however, our model suggests that the linker is on the opposite side of the paired domain relative to DNA-bound Ets-1. It is more likely that the truncation to amino acid 79 removes amino acids that contact DNA or contribute to the structure of the paired domain such that its affinity for DNA and/or Ets-1 is severely reduced. In the Paired-DNA structure, amino acids within the linker region interact with the DNA backbone and make intramolecular contacts with residues within helix α1 and near the type II β-turn. These contacts may be important for folding of the β-hairpin and β-turn and/or their positioning relative to the minor groove of DNA (53). Physical studies of isolated paired domain polypeptides have shown that they are largely structureless in solution in the absence of DNA (21a) and assume a more structured conformation on DNA (16). In the absence of the linker, it is plausible that the amino-terminal subdomain of Pax-5 will not assume an appropriate conformation for binding DNA and/or for interaction with Ets proteins.
A key amino acid for Ets-1 recruitment is an invariant glutamine residue present in the β-hairpin of all paired domains (Fig. 6). In our model for the ternary complex, this residue is near the region of the ETS domain that comprises the recognition helix and Asp398, suggesting that it plays a direct role in interactions between these two proteins. However, we cannot exclude an indirect structural role for this residue in ternary complex formation. In the Paired-DNA structure, Gln7 of Paired contacts a phosphate of DNA (53). By analogy with Paired, Gln22 of Pax-5 also is likely to lie near DNA in Pax-5–mb-1 promoter complexes and could contact it directly. The observation that Q22A does not alter the affinity of Pax-5 for the mb-1 promoter suggests that this contact, if present, does not contribute significantly to the stability of the Pax-5–DNA complex. This is in agreement with the Paired–DNA structure, which suggests that contacts between the glutamine and a DNA phosphate are likely to be weak and may be dispensable for DNA binding. Mutation of Gln22 to alanine reduces but does not eliminate detection of ternary complexes, indicating that additional contacts between the proteins contribute to ternary complex assembly. The two amino acids required for Pax-5–Ets-1 interactions do not have reciprocal functions, because Pax-5 Q22D did not recruit Ets-1 D398Q to bind DNA (51a). Further structural and biochemical studies will be required to define the precise mechanism for the assembly of the Pax-5–Ets-1 ternary complex.
Interaction with Pax-5 somehow overcomes obstacles to Ets-1 DNA binding by stabilizing both proteins in the ternary complex (25a). First, interaction with Pax-5 allows for the binding of Ets-1 to a suboptimal recognition sequence (5′CCGGAG). Second, as reported previously, autoinhibitory sequences flanking either side of the ETS domain of Ets-1 further decrease its affinity for DNA (25, 31, 36, 49). The fragment of Ets-1 used in this study (amino acids 333 to 440) exhibits high-affinity DNA binding (to most Ets binding sites) due to the absence of inhibitory sequences amino-terminal to the ETS domain (29, 37, 40). Interactions between Pax-5 and Ets proteins requires Asp398 of the ETS domain (20), which is immediately carboxy terminal to its major recognition α-helix (α3). Recently, we determined that either aspartic acid or glutamic acid at this key position can support ternary complex assembly with Pax-5 (19a). We cannot predict the consequences of ternary complex formation on the inhibitory domains but note that the fourth helix of Ets-1(333–440), which corresponds to the carboxy-terminal inhibitory sequence, is positioned adjacent the Pax-5 paired domain in our model (Fig. 2). This raises the intriguing possibility that formation of the Pax-5–Ets-1 ternary complex leads to derepression of Ets-1 DNA binding by protein-protein interactions with residues within its amino- or carboxy-terminal inhibitory domains.
Interactions with other proteins on specific DNA sequences are common among Ets proteins (reviewed in reference 24). The consequences of Ets-1 interactions with Pax-5 include enhanced specificity and stabilization of DNA binding. A well-characterized example of a ternary complex involving Ets proteins is that of GABPα, which interacts with the ankyrin repeats of GABPβ via sequences carboxy-terminal to its ETS domain (7). Interestingly, Pax-5 can assemble complexes with GABPα and GABPβ simultaneously (20), suggesting the presence of two protein-protein interfaces in the GABPα ETS domain. Other Ets proteins possess protein-protein interaction motifs outside their ETS domains. For example, the Ets protein PU.1 recruits the PU.1 interaction partner protein to bind a composite site through a single phosphorylated serine residue that is amino terminal to its ETS domain (8). In Ets proteins of the ternary complex factor subfamily, a specialized α-helical region has evolved for cooperative interactions with the MADS domain protein serum response factor (32). The adaptation of different mechanisms for protein-protein interactions reflects the structural diversity of partners for Ets proteins, e.g., of paired, ankyrin repeat, interferon response factor, or MADS domain proteins.
Importance of the β-hairpin motif.
Evidence for the functional importance of sequences in and around the conserved β-hairpin motif has been gathered from congenital diseases that affect mice and humans. A number of mutations in the mouse Pax-3 locus have been linked to the splotch phenotype, which is characterized by defects in neural crest cell migration and closure of the neural tube. One allele of splotch, termed splotch-delayed (Spd), presents a less severe phenotype due to a glycine-to-arginine substitution (Gly9 in Fig. 6) within the β-hairpin (48). Analysis of DNA binding by Pax-3 (Spd) showed that the mutated polypeptide binds a site in the Drosophila even-skipped promoter only 1/17 as well as does wild-type Pax-3 (46). In humans, autosomal dominant mutations in pax-3 genes have been linked to disorders that exhibit similarities with splotch. WS-I typically presents piebald-like abnormalities of the skin and hair, pigmentary anomalies of the iris, displacement of the inner canthi of the eyes (dystopia canthorum), and sensorineural deafness due to defects of neural crest-derived tissues (reviewed in reference 41). pax-3 mutations in WS-I include missense substitutions, small in-frame deletions, and nonsense, frameshift, and splice junction mutations. In two groups of WS-I patients, phenylalanine (Phe12 in Fig. 6)-to-leucine or proline (Pro17 in Fig. 6)-to-leucine mutations were identified within or just past the second β-turn (4, 43). In other patients, mutation of an asparagine (Asn14 in Fig. 6) to histidine in the β-hairpin was detected in individuals diagnosed with the related Klein-Waardenburg syndrome (WS-III) (28). Mutation of the same amino acid in Pax-3 to lysine was identified as the probable cause of craniofacial-deafness-hand syndrome (2), suggesting that different amino acids (histidine versus lysine) at a single position result in distinct phenotypes. In mice, the undulated mutation results in distortions of the vertebral column and sternum due to a glycine (Gly15 in Fig. 6)-to-serine mutation that occurs at the end of the type II β-turn in Pax-1 (5). It is unclear how these mutations affect the expression of Pax-regulated genes, but it has been suggested that DNA binding affinity and specificity are likely to be perturbed, similar to data obtained with the Pax-3 (Spd) protein.
In conclusion, we speculate that pressures to maintain the β-hairpin and β-turn sequences may have included a requirement(s) for functional interactions with Ets proteins, which have been detected in a similar spectrum of species (reviewed in reference 24). It is interesting that the ability to recruit Ets proteins has been conserved, even though the various Pax proteins (e.g., Pax-2/5 versus Pax-3) have diversified to recognize different sets of nucleotide sequences. Although evidence has accumulated for at least a limited role for Pax-Ets complexes in vivo, the question arises as to whether any of the phenotypes associated with expression of abnormal Pax proteins result from altered interactions with Ets proteins. In addition to the congenital disorders, overexpression of Pax and Pax hybrid proteins resulting from chromosomal translocations has been linked to oncogenesis in multiple tissue types (reviewed in reference 6). It is intriguing to speculate that a role exists for Ets proteins in Pax-mediated oncogenesis, because the Ets family includes many proto-oncogenes and can activate transcription as nuclear effectors of the Ras/mitogen-activated protein kinase signaling pathway (50). Addressing these questions will be important to determine whether Pax-Ets interactions are a point for pharmaceutical intervention in cancer and other diseases.
ACKNOWLEDGMENTS
We thank Larry Borish, Barbara J. Graves, Arthur Gutierrez-Hartmann, and Robert Scheinman for helpful discussions and comments. We thank Klaus Giese, Rudolf Grosschedl, and John Love for supplying reagents and for helpful discussions. We are greatly indebted to Peter Gruss for supplying cDNAs encoding Pax proteins. We also thank Julie Negri for excellent technical support, Leigh Landskroner for illustration, Carolyn Slupsky, Nancy Wilson, and John Kappler for help with figures, and Amy Marrs and Randal Anselment, (Molecular Resource Center at National Jewish Medical and Research Center) for oligonucleotide synthesis and support.
W.W. was supported by National Institutes of Health training grant AI00048, by a generous grant from the Cancer League of Colorado, and by funds from NJMRC. D.F. was supported by NJMRC. S.R.K. received a University of Colorado Cancer Center Summer Student Fellowship supported (in part) by research grants from NCI Cancer Education grant R25 CA49981 and ACS Colorado Division, Brooks Trust. L.N.G. was the recipient of a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. L.P.M. was supported by the National Cancer Institute of Canada with funds from the Canadian Cancer Society. This work was supported by generous awards to J.H. from the American Cancer Society (DB-8309) and National Institutes of Health (R01 AI37574 and P01 AI22295) and by a grant from the Rocky Mountain Chapter of the Arthritis Foundation.
REFERENCES
- 1.Adams B, Dorfler P, Aguzzi A, Kozmik Z, Urbanek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992;6:1589–607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- 2.Asher J H, Jr, Sommer A, Morell R, Friedman T B. Missense mutation in the paired domain of PAX3 causes craniofacial-deafness-hand syndrome. Human Mutat. 1996;7:30–35. doi: 10.1002/(SICI)1098-1004(1996)7:1<30::AID-HUMU4>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 3.Balczarek K A, Lai Z-C, Kumar S. Evolution and functional diversification of the paired box (Pax) DNA-binding domains. Mol Biol Evol. 1997;14:829–842. doi: 10.1093/oxfordjournals.molbev.a025824. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin C T, Hoth C F, Amos J A, daSilva E O, Milunsky A. An exonic mutation in the HuP2 paired domain gene causes Waardenburg’s syndrome. Nature. 1992;355:637–638. doi: 10.1038/355637a0. [DOI] [PubMed] [Google Scholar]
- 5.Balling R, Deutsch U, Gruss P. undulated, a mutation affecting the development of the mouse skeleton, has a point mutation in the paired box of Pax-1. Cell. 1988;55:531–535. doi: 10.1016/0092-8674(88)90039-6. [DOI] [PubMed] [Google Scholar]
- 6.Barr F G. Chromosomal translocations involving paired box transcription factors in human cancer. Int J Biochem Cell Biol. 1997;29:1449–1461. doi: 10.1016/s1357-2725(97)00095-2. [DOI] [PubMed] [Google Scholar]
- 7.Batchelor A H, Piper D E, de la Brousse F C, McKnight S L, Wolberger C L. The structure of GABPα/β: an ETS domain-ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- 8.Brass A L, Kehrli E, Eisenbeis C F, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 9.Busslinger M, Urbánek P. The role of BSAP (Pax-5) in B-cell development. Curr Opin Genet Dev. 1995;5:595–601. doi: 10.1016/0959-437x(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 10.Chalepakis G, Fritsch R, Fickenscher H, Deutsch U, Goulding M, Gruss P. The molecular basis of the undulated/Pax-1 mutation. Cell. 1991;66:873–884. doi: 10.1016/0092-8674(91)90434-z. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Glover J N, Hogan P G, Rao A, Harrison S C. Structure of the DNA-binding domains from NFAT, Fos, and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 12.Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- 13.Dahl E, Koseki H, Balling R. Pax genes and organogenesis. Bioessays. 1997;19:755–765. doi: 10.1002/bies.950190905. [DOI] [PubMed] [Google Scholar]
- 14.Donaldson L W, Petersen J M, Graves B J, McIntosh L P. Solution structure of the ETS domain from murine Ets-1: a winged helix-turn-helix DNA binding motif. EMBO J. 1996;15:125–134. [PMC free article] [PubMed] [Google Scholar]
- 15.Epstein D J, Vekemans M, Gros P. Splotch (Sp2H), a mutation affecting development of the mouse neural tube, shows a deletion within the paired homeodomain of Pax-3. Cell. 1991;57:767–774. doi: 10.1016/0092-8674(91)90071-6. [DOI] [PubMed] [Google Scholar]
- 16.Epstein J A, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 17.Epstein J A, Glaser T, Cai J, Jepeal L, Walton D S, Maas R L. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 18.Evans S. SETOR: hardware lighted three-dimensional solid representation of macromolecules. J Mol Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 19.Fisher R J, Mavrothalassitis G, Kondoh A, Papas T S. High affinity DNA-protein interactions of the cellular ETS1 protein: the determination of the ETS binding motif. Oncogene. 1991;6:2249–2254. [PubMed] [Google Scholar]
- 19a.Fitzsimmons, D. Unpublished data.
- 20.Fitzsimmons D, Hodsdon W, Wheat W, Maira S-M, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- 21.Ford K G, Whitmarsh A J, Hornby D P. Overexpression and purification of eukaryotic transcription factors as glutathione-S-transferase fusions in E. coli. Methods Mol Biol. 1994;30:185–197. doi: 10.1385/0-89603-256-6:185. [DOI] [PubMed] [Google Scholar]
- 21a.Gentile, L. N., and L. P. McIntosh. Unpublished data.
- 22.Giese K, Amsterdam A, Grosschedl R. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 1991;5:2567–2578. doi: 10.1101/gad.5.12b.2567. [DOI] [PubMed] [Google Scholar]
- 23.Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 24.Graves B J, Petersen J M. Specificity within the ets family of transcription factors. Adv Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 24a.Hagman, J. Unpublished data.
- 25.Hagman J, Grosschedl R. An inhibitory carboxyl-terminal domain in Ets-1 and Ets-2 mediates differential binding of ETS family factors to promoter sequences of the mb-1 gene. Proc Natl Acad Sci USA. 1992;89:8889–8893. doi: 10.1073/pnas.89.19.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Hagman, J., and W. Wheat. Unpublished data.
- 26.Hahn S L, Wasylyk B. The oncoprotein v-Ets is less selective in DNA binding than c-Ets-1 due to the C-terminal sequence change. Oncogene. 1994;9:2499–2512. [PubMed] [Google Scholar]
- 27.Hill R E, Favor J, Hogan B L M, Ton C C T, Saunders G F, Hanson I M, Prosser J, Jordan T, Hastie N D, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 28.Hoth C F, Milunsky A, Lipsky N, Sheffer R, Clarren S K, Baldwin C T. Mutations in the paired domain of the human PAX3 gene cause Klein-Waardenburg syndrome (WS-III) as well as Waardenburg syndrome type I (WS-I) Am J Hum Genet. 1993;52:455–462. [PMC free article] [PubMed] [Google Scholar]
- 29.Jonsen M D, Petersen J M, Xu Q-P, Graves B J. Characterization of the cooperative function of inhibitory sequences in Ets-1. Mol Cell Biol. 1996;16:2065–2073. doi: 10.1128/mcb.16.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kodandapani R, Pio F, Ni C-Z, Piccialli G, Klemsz M, McKercher S, Maki R A, Ely K R. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature. 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 31.Lim F, Kraut N, Frampton J, Graf T. DNA binding by c-Ets-1, but not v-Ets, is repressed by an intramolecular mechanism. EMBO J. 1992;11:643–652. doi: 10.1002/j.1460-2075.1992.tb05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ling Y, Lakey J H, Roberts C E, Sharrocks A D. Molecular characterization of the B-box protein-protein interaction motif of the ETS-domain transcription factor Elk-1. EMBO J. 1997;16:2431–2440. doi: 10.1093/emboj/16.9.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love J J, Li X, Case D A, Giese K, Grosschedl R, Wright P E. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 34.Nutt S L, Morrison A M, Dörfler P, Rolink A, Busslinger M. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 1998;17:2319–2333. doi: 10.1093/emboj/17.8.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nutt S L, Urbánek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11:476–491. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- 36.Nye J A, Petersen J M, Gunther C V, Jonsen M D, Graves B J. Interaction of murine Ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 37.Petersen J M, Skalicky J J, Donaldson L W, McIntosh L P, Alber T, Graves B J. Modulation of transcription factor Ets-1 DNA binding: DNA-induced unfolding of an α helix. Science. 1995;269:1866–1869. doi: 10.1126/science.7569926. [DOI] [PubMed] [Google Scholar]
- 38.Quiring R, Walldorf U, Kloter U, Gehring W J. Homology of the eyeless gene of Drosophila to the small eye gene in mice and aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 39.Sander M, Neubüser A, Kalamaras J, Ee H C, Martin G R, German M S. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 40.Skalicky J J, Donaldson L W, Peterson J M, Graves B J, McIntosh L P. Structural coupling of the inhibitory regions flanking the ETS domain of murine Ets-1. Protein Sci. 1996;3:296–309. doi: 10.1002/pro.5560050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spritz R A. Piebaldism, Waardenburg syndrome, and related disorders of melanocyte development. Semin Cutan Med Surg. 1997;16:15–23. doi: 10.1016/s1085-5629(97)80031-4. [DOI] [PubMed] [Google Scholar]
- 42.Tassabehji M, Read A P, Newton V E, Harris R, Balling R, Gruss P, Strachan T. Waardenburg’s syndrome patients have mutations in the human homologue of the Pax-3 paired-box gene. Nature. 1992;355:635–636. doi: 10.1038/355635a0. [DOI] [PubMed] [Google Scholar]
- 43.Tassabehji M, Read A P, Newton V E, Patton M, Gruss P, Harris R, Strachan T. Mutations in the PAX3 gene causing Waardenburg syndrome type 1 and type 2. Nat Genet. 1993;3:26–30. doi: 10.1038/ng0193-26. [DOI] [PubMed] [Google Scholar]
- 44.Ton C C T, Hirvonen H, Miwa H, Weil M M, Monaghan P, Jordan T, van Heyningen V, Hastie N D, Mjiers-Heijboer H, Dreschler M, Royer-Pokora B, Collins F, Swaroop A, Strong L C, Saunders G F. Positional cloning and characterization of a paired-box- and homeobox-containing gene from the aniridia region. Cell. 1991;57:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 45.Travis A, Hagman J, Grosschedl R. Heterogeneously initiated transcription from the pre-B- and B-cell-specific mb-1 promoter: analysis of the requirement for upstream factor-binding sites and initiation site sequences. Mol Cell Biol. 1991;11:5756–5766. doi: 10.1128/mcb.11.11.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Underhill D A, Vogan K J, Gros P. Analysis of the mouse Splotch-delayed mutation indicates that the Pax-3 paired domain can influence homeodomain DNA-binding activity. Proc Natl Acad Sci USA. 1995;92:3692–3696. doi: 10.1073/pnas.92.9.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urbánek P, Wang Z-Q, Fetka I, Wagner E F, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 48.Vogan K J, Epstein D J, Trasler D G, Gros P. The splotch-delayed (Spd) mouse mutant carries a point mutation within the paired box of the pax3 gene. Genomics. 1993;17:364–369. doi: 10.1006/geno.1993.1333. [DOI] [PubMed] [Google Scholar]
- 49.Wasylyk C, Kerckaert J-P, Wasylyk B. A novel modulator domain of Ets transcription factors. Genes Dev. 1992;6:965–974. doi: 10.1101/gad.6.6.965. [DOI] [PubMed] [Google Scholar]
- 50.Wasylyk C, Maira S-M, Sobieszczuk P, Wasylyk B. Reversion of Ras transformed cells by Ets transdominant mutants. Oncogene. 1994;9:3665–3673. [PubMed] [Google Scholar]
- 51.Werner M H, Clore G M, Fisher C L, Fisher R J, Trinh L, Shiloach J, Gronenborn A M. Correction of the NMR structure of the Ets1/DNA complex. J Biomol NMR. 1997;10:317–328. doi: 10.1023/a:1018399711996. [DOI] [PubMed] [Google Scholar]
- 51a.Wheat, W. Unpublished data.
- 52.Woods D B, Ghysdael J, Owen M J. Identification of nucleotide preferences in DNA sequences recognized specifically by c-Ets-1 protein. Nucleic Acids Res. 1992;20:699–704. doi: 10.1093/nar/20.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu W, Rould M A, Jun S, Desplan C, Pabo C O. Crystal structure of a paired domain-DNA complex at 2.5 Å resolution reveals structural basis for Pax developmental mutations. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]