Abstract
This study led to the discovery of three entomopathogenic fungi associated with Kuwanaspis howardi, a scale insect on Phyllostachys heteroclada (fishscale bamboo) and Pleioblastus amarus (bitter bamboo) in China. Two of these species belong to Podonectria: P. kuwanaspidis X.L. Xu & C.L. Yang sp. nov. and P. novae-zelandiae Dingley. The new species P. kuwanaspidis has wider and thicker setae, longer and wider asci, longer ascospores, and more septa as compared with similar Podonectria species. The morphs of extant species P. novae-zelandiae is confirmed based on sexual and asexual morphologies. Maximum likelihood and Bayesian inference analyses of ITS, LSU, SSU, tef1-α, and rpb2 sequence data provide further evidence for the validity of the two species and their placement in Podonectriaceae (Pleosporales). The second new species, Microcera kuwanaspidis X.L. Xu & C.L. Yang sp. nov., is established based on DNA sequence data from ITS, LSU, SSU, tef1-α, rpb1, rpb2, acl1, act, cmdA, and his3 gene regions, and it is characterized by morphological differences in septum numbers and single conidial mass.
Keywords: 2 new taxa, bamboo, entomopathogens, Nectriaceae, Podonectriaceae, scale insect
1. Introduction
Podonectria was introduced by Petch [1] to accommodate species of Ophionectria, which are parasitic on scale insects and have thick-walled asci, long, multiseptate ascospores, and a tetracrium-like conidial stage. Ten species are listed in Index Fungorum [2]. The type species, Podonectria coccicola (Ellis and Everh.) Petch was transferred from Ophionectria coccicola (Ellis & Everh.) Berl. & Voglino and is associated with the scale insects Aonidiella aurantia (Maskell), Aspidiotus perniciosus (Comstock), Chrysomphalus aonidum (Linnaeus), Lepidosaphes beckii (Newman), L. gloverii (Packard), Leucapsis sp., Parlatoria pergandii Comstock, P. ziziphi Lucas, and Unaspis citri (Comstock) which are mainly found on Rutaceae [1,3,4,5]. Puttemansia aurantii (Henn.) Höhn, which was initially found from the type specimen of the asexual morph Tetracrium aurantii Henn. associated with scale insect Parlatoria ziziphi on Citrus aurantium L., was also transferred to Podonectria as P. aurantii (Henn.) Petch [1]. A new species collected from Lepidosaphes sp. on Citrus nobilis Lour. was named as Podonectria echinata [1]. Additionally, two new species, P. gahnia Dingley and P. novae-zelandiae Dingley, were reported by Dingley (1954) from scale insects in New Zealand [3], followed by a new fungus P. tenuispora Dennis collected from Lepidosaphes ulmi (Linnaeus) on Calluna vulgaris (L.) Hull [6]. Subsequently, Rossman transferred Ophionectria coccorum Petch, associated with Fiorinia juniperi Kuwana, and Lasiosphaeria larvaespora Cooke & Massee on an undetermined scale insect to Podonectria, viz. P. coccorum (Petch) Rossman and P. larvaespora (Cooke & Massee) Rossman [7]. The species Trichonectria bambusicola Rehm was referred as P. bambusicola (Rehm) Piroz. on account of scolecosporous ascospores and tetracrium-like conidia by Pirozynski [8]. However, Podonectria bambusicola was excluded because of its occurrence on living leaves of bamboo rather than scale insects and remained an unclassified loculoascomycete [4]. Rossman published a monograph on Podonectria and accepted eight species [4]. An examination of the type specimen of T. bambusicola further revealed that this was a synonym of Uredinophila erinaceae (Rehm) Rossman [9]. The genus Podonectria was characterized by fleshy, white to brown, uninoculated ascomata with bitunicate asci and long, multiseptated ascospores associated with scale insects [4]. Spatafora et al. [10] transferred the previously reported species Podonectria cicadellidicola Kobayasi & Shimizu and P. citrina Kobayasi & Shimizu to Ophiocordyceps supported by the previous phylogenetic analyses presented in Quandt et al. [11]. Yang et al. [12] found P. sichuanensis C.L. Yang & X.L. Xu parasitic around the ascomata of Neostagonosporella sichuanensis C.L. Yang, X.L. Xu & K.D. Hyde on Phyllostachys heteroclada Oliv.
Microcera (Nectriaceae, Hypocreales), typified by Microcera coccophila Desm. and known as the “red-headed fungus”, is mostly parasitic on scale insects with fusarium-like asexual morphs. The genus has been considered as a synonym in major taxonomic revisions of Fusarium Link [13,14,15,16,17]. A multilocus phylogenetic approach was subsequently applied to identify species in the fusarium-like clade since morphological identification was difficult [18,19,20]. Gräfenhan et al. [18] resurrected Microcera based on DNA sequence data and accepted four Microcera species, viz. M. coccophila, M. diploa (Berk. & M.A. Curtis) Gräfenhan & Seifert, M. rubra Gräfenhan & Seifert, M. larvarum (Fuckel) Gräfenhan, Seifert & Schroers. Lombard et al. [19] further investigated phylogenetic relationships of Microcera based on DNA sequence data and reported that it constitutes a lineage distantly related to Fusarium but closely related to Fusicolla and Macroconia.
Armored scale insects (Hemiptera: Coccomorpha: Diaspididae) are major economic pests on agriculture and forestry plants, especially on fruit trees and vegetables. Diaspididae is the largest family of scale insects with 421 accepted genera and four subfamilies recognized, viz. Ancepaspidinae Borchsenius, Furcaspidinae Balachowsky, Diaspidinae Targioni Tozzetti, and Aspidiotinae Westwood. by Normark et al. [21]. The grass-feeding species, Kuwanaspis MacGillivray, which is classified into subtribe Fioriniina Targioni Tozzetti under tribe Diaspidini Targioni Tozzetti within subfamilies Diaspidinae, are harmful to bamboo [22,23]. During our investigations of microfungi associated with bamboo in Sichuan Province, two Podonectria species and a Microcera species were isolated in association with the armored scale insect Kuwanaspis howardi (Cooley) on native bamboo plants Phyllostachys heteroclada and Pleioblastus amarus (Keng) Keng. Morphological characteristics coupled with phylogenetic analyses of the combined ITS, LSU, SSU, tef1-α, and rpb2 sequence dataset support the validity of the P. kuwanaspidis X.L. Xu & C.L. Yang sp. nov. and P. novae-zelandiae Dingley and their placement in Podonectriaceae, Pleosporales. The fusarium-like species Microcera kuwanaspidis is distinguished from similar species based on the sequences’ differences, mainly in the tef1-α, acl1, act, cmdA, rpb1, and his3 regions. This is the first record of these taxa associated with scale insects in China. The taxa are compared with allied species, and comprehensive descriptions and micrographs are provided.
2. Materials and Methods
2.1. Specimen Collection and Morphological Study
During spring to autumn from 2018 to 2021, the specimens were collected from the bamboo forests located in Ya’an City and a neighboring county (Sichuan Province, China), where the environment is characterized by river valley terraces and intermountain basins and a subtropical monsoon humid climate with abundant natural resources, and it is the transition zone from Qinghai-Tibet Plateau to Chengdu Plain. Specimens documented with host, locality, time, and distribution of taxa were returned to the laboratory in suitable containers separately with the collection detail tag, and the substrate with fruiting bodies was checked following the methods described in Senanayake et al. [24]. The fungi were isolated into pure culture using single conidium obtained from sporodochia and single ascospore from ascomata parasitic on Kuwanaspis howardi following the isolation via spore suspension detailed in Chomnunti et al. [25]. The spore suspension was sucked into a Pasteur pipette, small drops were placed on isolation media (potato dextrose agar, PDA) in an incubator (20 °C). Then the plates were examined for single germinated spores under a dissecting microscope, and germinating spores were transferred separately to at least three new PDA plates. After incubation on PDA plates at 20 °C for 20 to 40 days depending on the growth rate, colonies were examined for their diameter, shape, and appearance. Ascomata and sporodochia were observed and photographed using a dissecting microscope NVT-GG (Shanghai Advanced Photoelectric Technology Co. Ltd., Shanghai, China) fitted with a VS-800C micro-digital camera (Shenzhen Weishen Times Technology Co. Ltd., Shenzhen, China). Dimensions of asci, ascospores, pseudoparaphyses, hairs, ascomata wall, conidia, conidiophores, and numbers of septa were based on field samples and were photographed using an Olympus BX43 compound microscope fitted with an Olympus DP22 digital camera in association with ACDSee v3.1 software. Measurements were made using Tarosoft® Image Frame Work v.0.9.7 (Tarosoft (R), Nontha Buri, Thailand). Lactophenol cotton blue reagent was used to observe the number of septa. The gelatinous appendage was observed in Black Indian ink. The type specimens were deposited at the Herbarium of Sichuan Agricultural University, Chengdu, China (SICAU). The ex-type cultures were deposited at the Culture Collection in Sichuan Agricultural University (SICAUCC), and MycoBank numbers are registered (http://www.MycoBank.org, accessed on 10 January 2021).
2.2. DNA Extraction, Amplification and Sequencing
Total genomic DNA was extracted from mycelia grown on PDA at 20 °C for 30 days, using the Plant Genomic DNA extraction kit (Tiangen, China). The internal transcribed spacer (ITS), the partial large subunit nuclear rDNA (LSU), the partial small subunit nuclear rDNA (SSU), translation elongation factor 1-alpha (tef1-α), the RNA polymerase II second largest subunit (rpb2), the large subunit of the ATP citrate lyase (acl1), the RNA polymerase II largest subunit (rpb1), β-tubulin (tub2), histone H3 (his3), translation elongation factor 1-alpha (tef1-α), calmodulin (cmdA), and actin (act) regions were amplified with primer pairs ITS5/ITS4 [26], LR0R/LR5 [27], NS1/NS4 [26], EF1-983F/EF1-2218R [28], fRPB2-5F/fRPB2-7cR [29], acl1-230up/acl1-1220low, RPB1-Ac/RPB1-Cr, T1/CYLTUB1R, CYLH3F/CYLH3R, EF1-728F/EF2, CAL-228F/CAL2Rd, and ACT-512F/ACT-1Rd [19], respectively.
Polymerase chain reaction (PCR) was performed in 25 μL reaction mixture containing 22 μL Master Mix (Beijing TsingKe Biotech Co., Ltd., Beijing, China), 1 μL DNA template, 1 μL each primer (10 μM). The amplification reactions were performed as described by Gräfenhan et al. [18], Lombard et al. [19], Dai et al. [30], and Wanasinghe et al. [31]. PCR products were sequenced at TsingKe Biological Technology Co., Ltd., Chengdu, China. The newly generated sequences were deposited in GenBank.
2.3. Phylogenetic Analyses
To infer relationships of our Podonectria taxa, a combined ITS, LSU, SSU, tef1-α, and rpb2 sequences dataset was used to construct the phylogenetic tree. For Microcera taxa, a combined ITS, LSU, tef1-α, rpb1, rpb2, acl1, act, tub2, cmdA, and his3 sequences dataset was used. Taxa used for phylogenetic analyses were selected based on BLAST searches and recent publications (Table 1 and Table 2). DNA alignments were performed using MAFFT v.7.429 online service [32], and ambiguous regions were excluded using BioEdit version 7.0.5.3 [33]. Phylogenetic trees were inferred with maximum likelihood (ML) and Bayesian inference (BI), according to the details described in Xu et al. [34]. The finalized alignments and trees were deposited in TreeBASE (http://www.treebase.org, accessed on 10 January 2021), submission ID: 27547 and 27549, respectively.
Table 1.
GenBank accession numbers of strains in Pleosporales and Tubeufiales used for the phylogenetic analyses of Podonectria.
| Species | Strain/Voucher No. | GenBank Accession Numbers | References | ||||
|---|---|---|---|---|---|---|---|
| ITS | LSU | SSU | tef1-α | rpb2 | |||
| Alternaria alternata | CBS 916.96 T | AF347031 | DQ678082 | KC584507 | KC584634 | KC584375 | [35] |
| Alternaria aconidiophora | CBS 145419 T | LR133931 | – | – | LR133968 | LR133967 | [36] |
| Alternaria dactylidicola | MFLUCC 15-0466 T | KY703616 | KY703617 | KY703618 | – | KY750720 | [37] |
| Alloleptosphaeria clematidis | MFLUCC 17-2071 T | MT310604 | MT214557 | MT226674 | MT394736 | MT394685 | [38] |
| Astragalicola amorpha | CBS 142999 T | MF795753 | MF795753 | – | MF795842 | MF795795 | [39] |
| Astragalicola vasilyevae | MFLUCC 17-0832 T | MG828870 | MG828986 | MG829098 | MG829193 | MG829248 | [40] |
| Bambusicola bambusae | MFLUCC 11-0614 T | JX442031 | JX442035 | JX442039 | KP761722 | KP761718 | [41,42] |
| Bambusicola irregularispora | MFLUCC 11-0437 T | JX442032 | JX442036 | JX442040 | KP761723 | KP761719 | [41,42] |
| Bambusicola massarinia | MFLUCC 11-0389 T | JX442033 | JX442037 | JX442041 | KP761725 | KP761716 | [41,42] |
| Bambusicola splendida | MFLUCC 11-0439 T | JX442034 | JX442038 | JX442042 | KP761726 | KP761717 | [41,42] |
| Bambusicola didymospora | MFLUCC 10-0557 T | KU940116 | KU863105 | KU872110 | KU940188 | KU940163 | [30] |
| Bambusicola pustulata | MFLUCC 15-0190 T | KU940118 | KU863107 | KU872112 | KU940190 | KU940165 | [30] |
| Bambusicola thailandica | MFLUCC 11-0147 T | KU940119 | KU863108 | KU872113 | KU940191 | KU940166 | [30] |
| Bambusicola triseptatispora | MFLUCC 11-0166 T | KU940120 | KU863109 | – | – | KU940167 | [30] |
| Bambusicola dimorpha | MFLUCC 13-0282 T | KY026582 | KY000661 | KY038354 | – | KY056663 | [37] |
| Bambusicola loculata | MFLUCC 13-0856 T | KP761732 | KP761729 | KP761735 | KP761724 | KP761715 | [42] |
| Bambusicola sichuanensis | SICAUCC 16-0002 T | MK253473 | MK253532 | MK253528 | MK262828 | MK262830 | [43] |
| Bambusicola subthailandica | SICAU 16-0005 T | MK253474 | MK253533 | MK253529 | MK262829 | MK262831 | [43] |
| Boeremia coffeae | CBS 109183 | GU237748 | GU237943 | – | KY484678 | KT389566 | [44] |
| Boeremia rhapontica | CBS 113651 T | KY484662 | – | – | KY484713 | – | [44] |
| Boeremia opuli | CGMCC 3.18354 T | KY742045 | KY742199 | – | – | KY742133 | [44] |
| Boeremia linicola | CBS 116.76 T | GU237754 | GU237938 | – | KY484705 | KT389574 | [44] |
| Boeremia populi | CBS 100167 T | GU237707 | GU237939 | – | KY484706 | – | [44] |
| Coniothyrium telephii | UTHSC DI16-189 | LT796830 | LN907332 | – | – | LT796990 | [45] |
| Coniothyrium chiangmaiense | MFLUCC 16-0891 T | KY568987 | KY550384 | KY550385 | – | KY607015 | [37] |
| Coniothyrium sidae | CBS 135108 T | KF251149 | KF251653 | – | KF253109 | KF252158 | [46] |
| Cucurbitaria berberidis | CBS 363.93 | JF740191 | GQ387606 | – | – | – | [47] |
| Decorospora gaudefroyi | CBS 332.63 | AF394541 | – | AF394542 | – | – | [48] |
| Didymella poaceicola | MFLUCC 13-0212 T | KX965726 | KX954395 | – | – | KX898364 | [37] |
| Dothidotthia robiniae | MFLUCC 16-1175 T | MK751727 | MK751817 | MK751762 | MK908017 | MK920237 | [49] |
| Epicoccum thailandicum | MFLUCC 16-0892 T | KY703619 | KY703620 | – | – | – | [37] |
| Epicoccum poaceicola | MFLUCC 15-0448 T | KX965727 | KX954396 | – | – | KX898365 | [37] |
| Leptosphaeria cichorium | MFLUCC 14-1063 T | KT454720 | KT454712 | KT454728 | – | – | [50] |
| Nothophoma chromolaenae | MFLUCC 17-1443 T | MT214364 | MT214458 | MT214410 | – | – | [51] |
| Ophiosimulans tanaceti | MFLUCC 14-0525 T | KU738890 | KU738891 | KU738892 | MG520910 | – | [52,53] |
| Palmiascoma gregariascomum | MFLUCC 11-0175 T | KP744452 | KP744495 | KP753958 | – | KP998466 | [54] |
| Parafenestella austriaca | CBS 145262 T | MK356304 | MK356304 | – | MK357576 | MK357532 | [55] |
| Parafenestella alpina | CBS 145263 T | MK356302 | MK356302 | – | MK357574 | MK357530 | [55] |
| Paraophiobolus plantaginis | MFLUCC 17-0245 T | KY797641 | KY815010 | KY815012 | MG520913 | – | [53] |
| Phaeosphaeria ampeli | MFLUCC 18-1641 T | MK503797 | MK503808 | MK503814 | MK503802 | – | [56] |
| Podonectria coccicola | DAR 81026 | KU587798 | KU519419 | – | – | – | [5] |
| Podonectria coccicola | PUcS15 | KU720533 | KU519420 | – | – | – | [5] |
| Podonectria novae-zelandiae | PUcS14 | KU720535 | KU559551 | – | – | – | [5] |
| Podonectria novae-zelandiae | PUcS13 | KU720538 | KU559548 | – | – | – | [5] |
| Podonectria novae-zelandiae | PUcS12 | KU720537 | KU529802 | – | – | – | [5] |
| Podonectria novae-zelandiae | PUcS11 | KU720536 | KU568479 | – | – | – | [5] |
| Podonectria sichuanensis | SICAU 16-0003 T | MK305903 | MK296471 | MK296467 | MK313852 | MK313855 | [12] |
| Podonectria sichuanensis | SICAUCC 21-0001 | MW484988 | MW462899 | MW462891 | MW462111 | MW462118 | This study |
| Podonectria kuwanaspidis | SICAUCC 21-0002 T | MW484989 | MW462900 | MW462892 | MW462112 | MW462119 | This study |
| Podonectria kuwanaspidis | SICAUCC 21-0003 | MW484990 | MW462901 | MW462893 | MW462113 | MW462120 | This study |
| Podonectria novae-zelandiae | SICAUCC 21-0004 | MW484991 | MW462902 | MW462894 | MW462114 | MW462121 | This study |
| Podonectria novae-zelandiae | SICAUCC 21-0005 | MW484992 | MW462903 | MW462895 | MW462115 | MW462122 | This study |
| Podonectria kuwanaspidis | SICAUCC 21-0007 | MW484994 | MW462905 | MW462897 | MW462116 | MW462123 | This study |
| Pseudoophiobolus galii | MFLUCC 17-2257 T | MG520947 | MG520967 | MG520989 | MG520926 | – | [53] |
| Pseudopyrenochaeta lycopersici | CBS 306.65 T | AY649587 | EU754205 | – | – | LT717680 | [57] |
| Pseudopyrenochaeta terrestris | CBS 282.72 T | LT623228 | LT623216 | – | – | LT623287 | [57] |
| Sclerenchymomyces clematidis | MFLUCC 17-2180 T | MT310605 | MT214558 | MT226675 | MT394737 | MT394686 | [38] |
| Seltsamia ulmi | CBS 143002 T | MF795794 | MF795794 | MF795794 | MF795882 | MF795836 | [39] |
| Thyrostroma lycii | MFLUCC 16-1170 T | MK751734 | MK751824 | MK751769 | MK908024 | MK920241 | [49] |
| Thyrostroma robiniae | MFLUCC 18-1191 T | MK751735 | MK751825 | MK751770 | MK908025 | MK920242 | [49] |
| Tubeufia javanica | MFLUCC 12-0545 T | KJ880034 | KJ880036 | KJ880035 | KJ880037 | – | [58] |
| Tubeufia chiangmaiensis | MFLUCC 11-0514 T | KF301530 | KF301538 | KF301543 | KF301557 | – | [58] |
Notes: The superscript T represents ex-type or ex-epitype isolates. “–” means that the sequence is missing or unavailable. New sequences are listed in bold. Abbreviations. CBS: Centraalbureau voor Schimmelcultures, Utrecht, Netherlands; CGMCC: China General Microbiological Culture Collection Center; DAR: New South Wales Plant Pathology Herbarium, Orange Agricultural Institute, Orange, NSW, Australia; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; PUcS: unspecified; UTHSC: Fungus Testing Laboratory of the University of Texas Health Science Center at San Antonio, San Antonio, TX, USA; SICAUCC: Sichuan Agricultural University Culture Collection, Sichuan, China; SICAU: Herbarium of Sichuan Agricultural University, Sichuan, China.
Table 2.
GenBank accession numbers of strains in Nectriaceae used for the phylogenetic analyses of Microcera.
| Species | Strain/Voucher No. | GenBank Accession No. | References | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| acl1 | act | cmdA | his3 | ITS | LSU | rpb1 | rpb2 | tef1-α | tub2 | |||
| Cosmospora coccinea | CBS 341.70 T | HQ897913 | KM231221 | KM231398 | KM231550 | HQ897827 | KM231692 | KM232242 | HQ897777 | KM231947 | KM232086 | [18,19] |
| Cosmospora cymosa | CBS 762.69 T | HQ897914 | KM231222 | KM231399 | KM231551 | HQ897828 | KM231693 | KM232243 | HQ897778 | KM231948 | KM232087 | [18,19] |
| Dialonectria episphaeria | CBS 125494 = TG 2006-11 | HQ897892 | KM231227 | KM231404 | KM231556 | HQ897811 | KM231697 | KM232248 | HQ897756 | KM231953 | KM232092 | [18,19] |
| Dialonectria ullevolea | CBS 125493 = TG 2007-56 | HQ897918 | KM231226 | KM231403 | KM231555 | KM231821 | KM231696 | KM232247 | HQ897782 | KM231952 | KM232091 | [18,19] |
| Fusicolla acetilerea | BBA 63789 T = IMI 181488 = NRRL20827 | KM231065 | – | – | – | HQ897790 | U88108 | – | HQ897701 | – | – | [18] |
| Fusicolla aquaeductuum | CBS 837.85 = BBA 64559 = NRRL 20865 | KM231067 | - | KM231406 | - | KM231823 | KM231699 | KM232250 | HQ897744 | KM231955 | KM232094 | [19] |
| Fusicolla epistroma | BBA 62201 T = IMI 85601 = NRRL 20439 | KM231069 | – | – | – | – | AF228352 | – | HQ897765 | – | – | [18] |
| Fusicolla matuoi | CBS 581.78 = ATCC 18694 = MAFF 238445 = NRRL 20427 | KM231070 | KM231228 | KM231405 | KM231557 | KM231822 | KM231698 | KM232249 | HQ897720 | KM231954 | KM232093 | [18,19] |
| Macroconia papilionacearum | CBS 125495 | HQ897912 | KM231233 | KM231411 | KM231561 | HQ897826 | KM231704 | KM232254 | HQ897776 | KM231958 | KM232096 | [18,19] |
| Macroconia leptosphaeriae | CBS 717.74 | KM231062 | KM231236 | KM231414 | KM231564 | KM231827 | KM231707 | KM232257 | KM232390 | JF735695 | KM232099 | [18,19] |
| Macroconia leptosphaeriae | CBS 100001 = CBS H-6030 | KM231063 | KM231234 | KM231412 | KM231562 | HQ897810 | KM231705 | KM232255 | HQ897755 | KM231959 | KM232097 | [18,19] |
| Microcera coccophila | CBS 310.34 = NRRL 13962 | HQ897843 | KM231232 | KM231410 | KM231560 | HQ897794 | KM231703 | – | HQ897705 | JF740692 | – | [18,19,59] |
| Microcera diploa | CBS 735.79 = BBA 62173 = NRRL 13966 | HQ897899 | – | – | – | HQ897817 | – | – | HQ897763 | – | – | [18] |
| Microcera kuwanaspidis | SICAUCC 21-0006 T | MW462125 | MW462126 | MW462127 | MW462128 | MW484993 | MW462905 | MW462129 | MW462124 | MW462117 | MW462130 | This study |
| Microcera kuwanaspidis | SICAUCC 21-0009 | MZ044037 | MZ044038 | MZ044039 | MZ044040 | MZ029437 | MZ029436 | MZ044041 | MZ044036 | MZ044035 | MZ044042 | This study |
| Microcera larvarum | CBS 169.30 | HQ897855 | – | – | EU860049 | EU860064 | EU860064 | – | HQ897717 | – | EU860025 | [18,60] |
| Microcera larvarum | CBS 738.79 = BBA 62239 = MUCL 19033 = NRRL 20473 | KM231060 | KM231230 | KM231408 | KM231559 | KM231825 | KM231701 | KM232252 | KM232387 | KM231957 | EU860026 | [19,60] |
| Microcera larvarum | A.R. 4580 = CBS 133964 | – | – | – | – | KC291751 | KC291759 | KC291894 | – | KC291832 | KC291935 | [61] |
| Microcera rubra | CBS 638.76 T = BBA 62460 = NRRL 20475 | HQ897903 | KM231231 | KM231409 | EU860050 | HQ897820 | KM231702 | KM232253 | HQ897767 | JF740696 | EU860018 | [18,19,60] |
| Pseudocosmospora rogersonii | CBS 133981 T = G.J.S. 90-56 | – | – | – | – | KC291729 | KC291780 | KC291878 | – | KC291852 | KC291915 | [61] |
| Pseudocosmospora eutypellae | CBS 133966 T = A.R. 4562 | – | – | – | – | KC291721 | KC291757 | KC291871 | – | KC291830 | KC291912 | [61] |
| Pseudocosmospora eutypae | C.H. 11-01 = CBS 133961 T | – | – | – | – | KC291735 | KC291766 | KC291884 | – | KC291837 | KC291925 | [61] |
| Tilachlidium brachiatum | CBS 505.67 | KM231076 | KM231249 | KM231436 | – | KM231839 | KM231720 | KM232272 | KM232415 | KM231976 | KM232110 | [19] |
| Tilachlidium brachiatum | CBS 363.97 | KM231077 | KM231248 | KM231435 | KM231583 | KM231838 | KM231719 | KM232271 | KM232414 | KM231975 | KM232109 | [19] |
Notes: superscript T represents ex-type or ex-epitype isolates. “–” means that the sequence is missing or unavailable. New sequences are listed in bold. Abbreviations. A.R.: Amy Y. Rossman, USDA-ARS, MD, USA; ATCC: American Type Culture Collection, U.S.A.; BBA: Julius Kühn-Institute, Institute for Epidemiology and Pathogen Diagnostics, Berlin and Braunschweig, Germany; CBS: Centraalbureau voor Schimmelcultures, Utrecht, Netherlands; C.H.: Cesar S. Herrera, University of Maryland, MD, USA; G.J.S.: Gary J. Samuels, USDA-ARS, MD, USA; IMI: International Mycological Institute, CABI-Bioscience, Egham, UK; MAFF: MAFF Genebank, National Institute of Agrobiological Sciences, Ibaraki, Japan; MUCL: Mycothèque de I’Université Catholique de Louvain, Belgium; NRRL: Agricultural Research Service Culture Collection, USA; TG: T. Gräfenhan collection.
3. Results
3.1. Phylogenetic Analyses
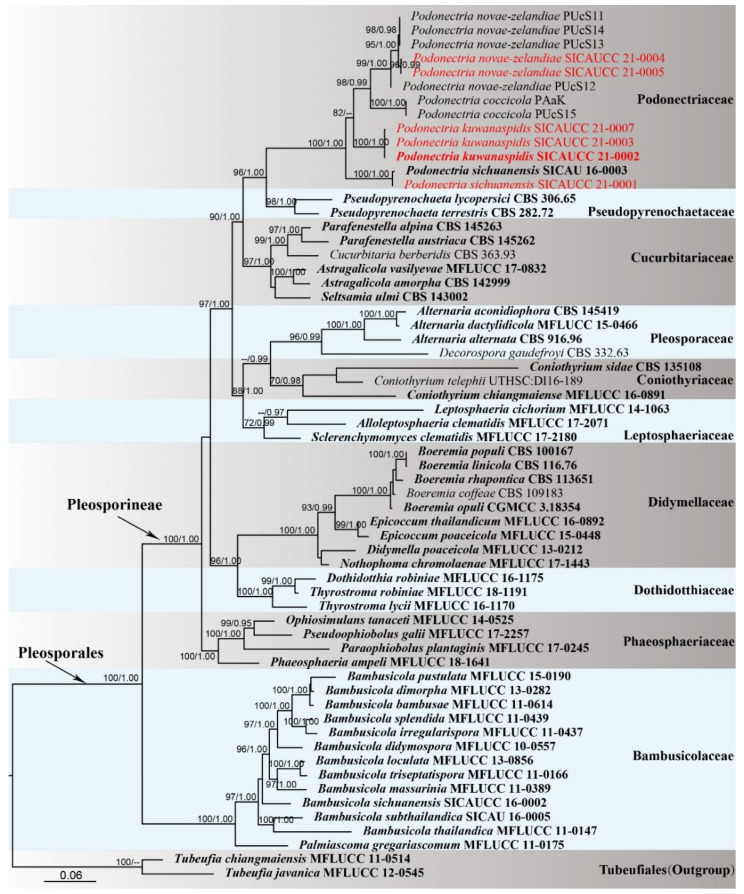
Phylogenetic analyses of a combined five-gene dataset (ITS, LSU, SSU, tef1-α, rpb2) comprised 62 taxa, and the tree is rooted with Tubeufia javanica Penz. & Sacc. (MFLUCC 12-0545) and T. chiangmaiensis Boonmee & K.D. Hyde (MFLUCC 11-0514) (Tubeufiaceae, Tubeufiales). The alignment contained 5721 characters (LSU = 1046, ITS = 821, SSU = 1176, tef1-α = 1507, rpb2 = 1171), including gaps. The best scoring RAxML tree with a final likelihood value of −40,064.587233 is presented. The matrix had 2539 distinct alignment patterns, with 46.29% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.244598, C = 0.248213, G = 0.265992, T = 0.241197, with substitution rates AC = 1.565487, AG = 3.743698, AT = 1.774643, CG = 1.114196, CT = 7.131582, GT = 1.000000. The gamma distribution shape parameter α = 0.240760, and the Tree-Length = 3.685780.
Phylogenetic trees generated from ML and BI analyses were similar in overall topologies. Phylogeny from the combined sequence data analysis indicates that all the Pleosporalean families are monophyletic with strong bootstrap support values (Figure 1). Three species grouped with taxa in Podonectria with 100% ML and 1.00 BYPP support. A species (SICAUCC 21-0004, SICAUCC 21-0005) clustered with P. novae-zelandiae in a clade with 99% ML and 1.00 BYPP statistical support. Our novel species P. kuwanaspidis constitutes a moderately supported independent lineage (82% ML/-- BYPP statistical support) between P. novae-zelandiae and P. coccicola.
Figure 1.
Phylogram generated from RAxML analysis based on ITS, LSU, SSU, tef1-α, and rpb2 Scheme 70. and Bayesian posterior probabilities (BYPP, right) equal to or greater than 0.95 are indicated at the nodes respectively. The sequences from ex-type strains are in bold. The newly generated sequence is in red.
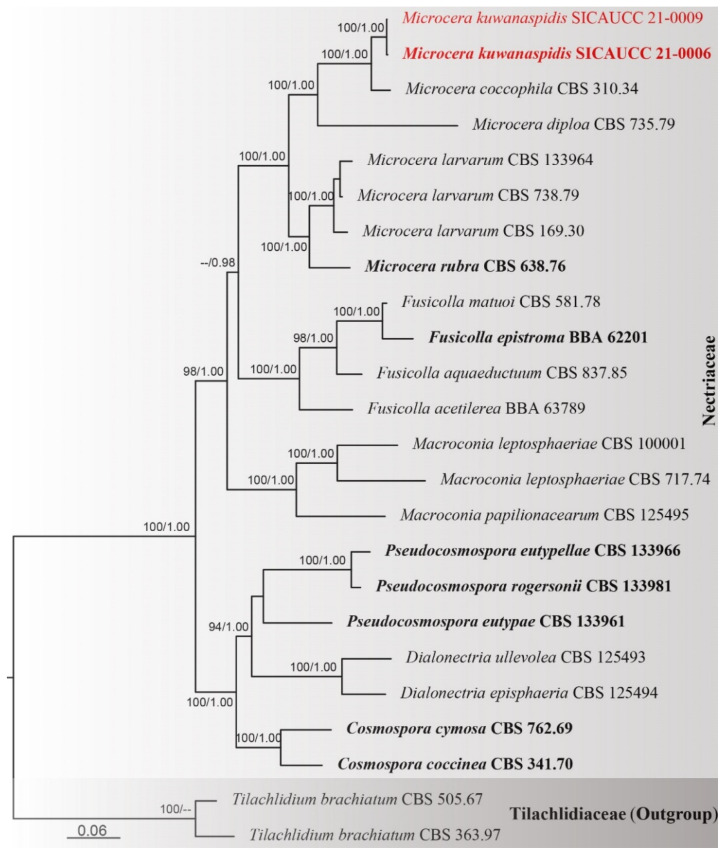
DNA sequences of four known species of Microcera and our new taxon, M. kuwanaspidis, were used in the analyses. The combined dataset comprised 24 taxa within Nectriaceae and two outgroup taxa in Tilachlidiaceae (Table 2). The alignment contained 7447 characters (ITS = 638, LSU = 831, acl1 = 1041, act = 673, cmdA = 778, his3 = 530, rpb1 = 741, rpb2 = 874, tef1-α = 631, tub2 = 710), including gaps. The tree is rooted with Tilachlidium brachiatum (Batsch) Petch (CBS 363.97, CBS 505.67). The best scoring RAxML tree with a final likelihood value of −50,074.064664 is presented. The matrix had 3327 distinct alignment patterns, with 28.65% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.233349, C = 0.272026, G = 0.255053, T = 0.239571, with substitution rates AC = 1.285610, AG = 3.696293, AT = 1.292257, CG = 0.967004, CT = 6.115076, GT = 1.000000. The gamma distribution shape parameter α = 0.261457, and the Tree-Length = 2.372705. In the concatenated phylogenetic analyses of ML and BI, all species of Microcera analyzed clustered in a well-supported clade (ML = 100%, BYPP = 1.00) with a close affinity to Fusicolla and Macroconia (Figure 2). Microcera kuwanaspidis is related to M. coccophila in a subclade with 100% ML and 1.00 BYPP statistical support.
Figure 2.
Phylogram generated from RAxML analysis based on combined ITS, LSU, tef1-α, rpb1, rpb2, acl1, act, tub2, cmdA, and his3 sequence data of Microcera isolates. Bootstrap support values for maximum likelihood (ML, left) higher than 70% and Bayesian posterior probabilities (BYPP, right) equal to or greater than 0.95 are indicated at the nodes respectively. The sequences from ex-type strains are in bold. The newly generated sequence is in red.
3.2. Taxonomy
Podonectriaceae H.T. Dao & Rossman, Mycological Progress 15(5): 47 (2016) amended.
MycoBank number: MB 815827
Type genus: Podonectria Petch, Trans. Br. mycol. Soc. 7(3): 146 (1921).
Parasitic fungus on scale insects, other fungi, or substrates previously colonized by other fungi. Sexual morph: Stromata byssoid, well-developed or scant, white to brown or dark-brown. Ascomata solitary or aggregated, superficial on or immersed in the stroma, globose to subglobose, obpyriform or ovoid, cream white to light yellow, or brown to dark brown, covered with hairs or absent. The hamathecium comprises numerous reticulate, filiform, septate, branched, pseudoparaphyses. Asci 8-spored, bitunicate, long clavate to cylindric. Ascospores long clavate to long cylindric, or vermiform, multiseptate. Asexual morph: Tetracrium-like. Sporodochia formed directly on cushion-shaped, white, orange, or brown, and hard stroma. Conidiophores moniliform. Conidia usually 1–4 “arms”, narrowed toward the apex, joined at the basal cell, multiseptate.
Notes: The family Podonectriaceae was introduced to accommodate Podonectria by Dao et al. [5], in which descriptions of conidia, ascomata, asci, and ascospores were lacking. Here we emend those descriptions and the habitats of Podonectriaceae with the inclusion of fungi or substrates previously colonized by other fungi and not only scale insects [4,5,8,12]. This broadens the taxonomic concept of Podonectria, which is further supported by molecular analyses in this study.
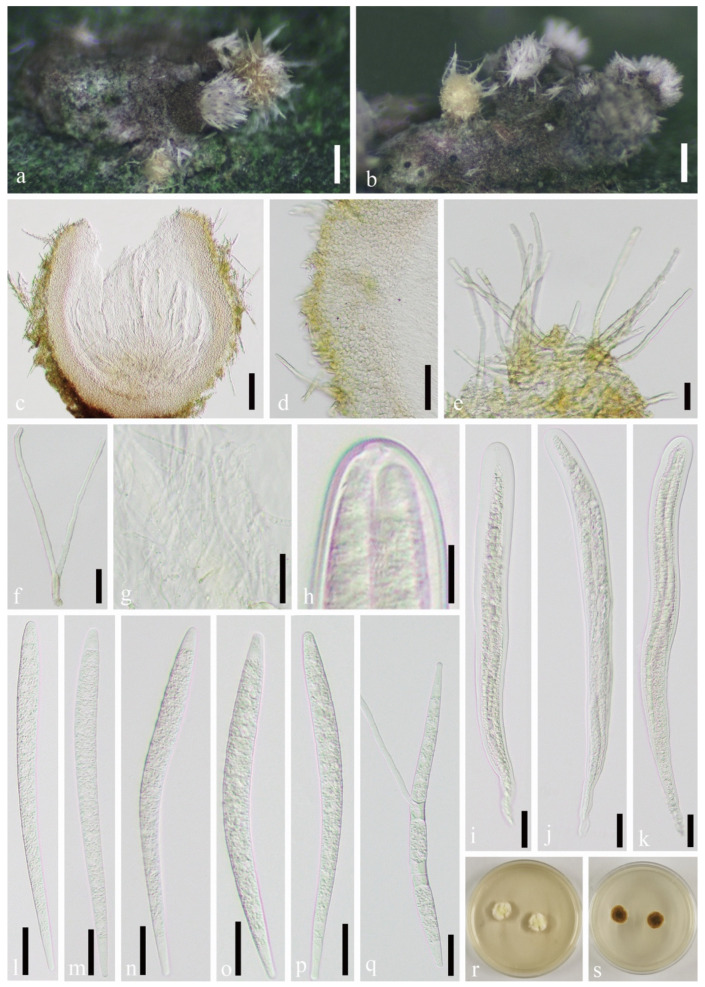
Podonectria novae-zelandiae Dingley, Trans. & Proc. Roy. Soc. N.Z. 81: 496 (1954) (Figure 3 and Figure 4).
Figure 3.
Podonectria novae-zelandiae (SICAU 21-0005). (a,b) Ascomata and sporodochia on host substrate. (c) Section through ascoma. (d) Peridium. (e,f) Hairs covering on ascoma. (g) Pseudoparaphyses. (h) Ocular chamber. (i–k) Asci. (l–p) Ascospores. (q) Germinated ascospores. (r,s) Colonies on PDA after 18 days. Scale bars: (a,b) 200 μm, (c) 100 μm, (d) 50 μm, (e–g) 20 μm, (h) 10 μm, (i–q) 20 μm.
Figure 4.
Podonectria novae-zelandiae (SICAU 21-0004). (a–c) Sporodochia and ascomata on host substrate. (d–f) Immature conidia. (g–j) Mature conidia. (k,l) Conidiophores. (m,n) Colonies on PDA after 20 days and 60 days. (o) Germinated conidium. Scale bars: (a–c) 500 μm, (d–k) 20 μm, (l,o) 10 μm.
MycoBank number: MB 304079
Habitat associated with scale insects Kuwanaspis howardi on Phyllostachys heteroclada. Sexual morph: Stromata byssoid, brown, well-developed, and covered the scale insects. Ascomata solitary, rarely aggregated, superficial on the byssoid stroma, concomitant with sporodochia, light yellow, covered with long hairs, 150–415 μm high ( = 240 μm, n = 20), 100–350 μm wide (without hairs) ( = 192 μm, n = 30). Hairs 60–280 μm long, multiseptate, 3–6.5 μm wide, straight, or curved, abundant, hyaline, slightly narrowed toward the apex, 1–2.5 μm thick-walled (n = 30). Peridium 60–100 μm thick, usually wider at the base, composed of hyaline suborbicular cells forming textura angularis, the cells measuring 5.5–12 × 4.5–10 μm ( = 8.9 × 7.0 μm, n = 20). Hamathecium 1.5–3 μm diameter ( = 2.3 μm, n = 30), 1 μm diameter at the apex, longer than the asci, numerous, filiform, curved, septate, branched pseudoparaphyses. Asci 220–340 × 18–26 μm ( = 267 × 21 μm, n = 20), 8-spored, bitunicate, cylindrical, straight, or curved, rounded at apex. Ascospores 100–160 × 7–10 μm ( = 138 × 9 μm, n = 30), fasciculate, parallel, long-clavate, rounded at ends, multiseptate, 10–22 septa with slight constriction, curved, hyaline, smooth. Asexual morph: Stromata hard, white to grey-brown, cushion-shaped, formed directly on host scales with 1–4 sporodochia. Sporodochia erupted, white, yellowish to grey-brown, scattered or aggregated. Conidiophores inconspicuous, short, 1–2 celled, the cells 3–7 × 4–10 μm ( = 5.0 × 7.5 μm, n = 30), usually globose, subglobose, or shortly cylindrical, attached with 1–2 conidiogenous cells. Conidiogenous cells 3–7 × 4–11 μm ( = 7.3 × 6.5 μm, n = 30), globose or ellipsoidal. Conidia usually with two and three “arms”, occasionally one and four “arms”, each “arm” varies in length and slightly divergent, 85–163 μm long ( = 117 μm, n = 70), 7–11 μm wide ( = 9 μm, n = 70) with 11–25 septa, mature conidium tapering toward the acute apex. All “arms” of single conidium joined at a basal oval or irregular cell, measuring 4–7 × 5–10 μm ( = 5.3 × 7.1 μm, n = 40).
Material examined: CHINA, Sichuan Province, Ya’an City, Lushan County (102°55′58.13″ E, 30°15′24.07″ N, Alt. 1116 m), on scale insect Kuwanaspis howardi, 10 June 2020, Xiu-lan Xu, XXL202006006 (SICAU 21-0005), living culture SICAUCC 21-0005; ibid. XXL202006005 (SICAU 21-0004), living culture SICAUCC 21-0004.
Culture characters: Conidia germinate on PDA within 12 h, and the cultures grow slowly on PDA. Colonies reach 2 cm in diameter after 25 days. Colonies from single conidia are flocculent and hard, with irregular margins. The mycelium is creamy white to light lemon yellow starting at the center but gradually becoming brown to dark brown after 20 days. Aerial hyphae cluster and raise straightly, measuring 2–3 μm diam. Conidia develop on small, sparsely distributed mycelial clumps after two months. Conidiophores moniliform, branched, multi-celled, and longer than those in nature. Conidia commonly have three “arms”, occasionally two and four “arms”, rarely one and five “arms”, each “arm” with 17–22 septa, measuring 115–145 μm long, 6.5–10 μm wide ( = 128 × 7.9 μm, n = 30). Ascospores germinate on PDA within 12 h, and the cultures grow slowly on PDA. Colonies reach 1 cm in diameter after 20 days. Colonies from single ascospores are cottony, with regular margin; the mycelium is creamy white to yellow; and the back of colonies is brown, with concentric rings.
Notes: Here, we follow the recommendation of Rossman et al. [62] by adopting Podonectria over Tetracrium. The asexual morph of P. novae-zelandiae was reported by Dao et al. [5], and was supported with morphology and molecular data. Our observations agree with the descriptions provided by Rossman [4] and Dao et al. [5]. Nucleotide comparison of ITS and LSU (SICAUCC 21-0005) reveals high similarity to P. novae-zelandiae (isolate PUcS13, similarities = 473/476 (99%), 0 gaps (0%); similarities = 517/518 (99%), 0 gaps (0%), respectively) in Dao et al. [5]; however, the latter lack SSU, tef1-α, and rpb2 sequences for further comparisons. The conidia produced here in culture were similar to those on scale insects in the field.
Podonectria kuwanaspidis X.L. Xu & C.L. Yang sp. nov. (Figure 5)
Figure 5.
Podonectria kuwanaspidis (SICAU 21-0002, holotype). (a) Ascomata on or around the scalehost (red arrow). (b) Aggregated ascomata. (c) Solitary ascomata. (d) Section through ascoma. (e) Peridium. (f) Hairs covering ascoma. (g) Pseudoparaphyses. (h–k) Asci. (l–n,q–u) Ascospores. (o,p) Colonies on PDA after 25 days and 50 days. (v,w) Germinated ascospores. Scale bars: (a) 500 μm, (b,c) 200 μm, (d) 100 μm, (e) 50 μm, (f–n) 20 μm, (q–w) 20 μm.
MycoBank number: MB 838465
Etymology: In reference to the generic name for the associated scale insect.
Holotype: SICAU 21-0002.
Habitat associated with scale insects Kuwanaspis howardi on Phyllostachys heteroclada. Sexual morph: Stromata byssoid, white or brown, well-developed and covering the scale insects, or forming a thin, white, and byssoid layer, which spreads out from the scale over the stem. Ascomata solitary to aggregated, superficial on byssoid stroma around the scale hosts, or extending far beyond the scale byssoid stroma, globose to subglobose, creamy white to dirty white, covered with hairs, 200–590 μm high ( = 424 μm, n = 20), 140–600 μm wide ( = 346 μm, n = 70). Hairs 30–120 μm long, 0–4 septa, 8–16 μm wide at the base, 2–7 μm wide at the apex, abundant, hyaline, distinctly narrowed toward the apex, 2–4.5 μm thick-walled (n = 40). Ostiole 40–100 μm wide. Peridium 40–170 μm thick ( = 72 μm, n = 30), usually wider at the base, composed of hyaline elongated cells forming textura prismatica to textura angularis, becoming globose toward outside, the cells measuring 6.5–15× 8–20 μm ( = 10 × 14 μm, n = 30) μm. Hamathecium 1.5–3.5 μm diameter ( = 2.2 μm, n = 40) at the base, 1 μm diameter at the apex, longer than the asci, numerous, filiform, curved, septate, branched pseudoparaphyses. Asci 185–250 × 15–25 μm ( = 219 × 20 μm, n = 40), 8-spored, bitunicate, cylindrical, straight or curved, rounded at apex, shortly pedicellate. Ascospores 150–240 × 5–7 μm ( = 199 × 5.8 μm, n = 40), fasciculate, parallel or spiral, broadly filiform, cylindrical to long fusiform, elongate, rounded at ends, multiseptate, 16–31 septa without constriction, usually tapering toward the base, curved, hyaline, smooth. Asexual morph: Undetermined.
Material examined: CHINA, Sichuan Province, Ya’an City, Lushan County (102°55′58.13″ E, 30°15′24.07″ N, Alt. 1116 m), on scale insect Kuwanaspis howardi, 10 June 2020, Xiu-lan Xu, XXL202006002 (SICAU 21-0002, holotype), ex-type culture, SICAUCC 21-0002; ibid. XXL202006003 (SICAU 21-0003, paratype), living culture SICAUCC 21-0003. ibid. Yucheng District, Kongping Township (103°2′59.87″ E, 29°50′8.56″ N, Alt. 1133 m), on scale insect Kuwanaspis howardi, 19 September 2018, Xiu-lan Xu, YCL201810014 (SICAU 21-0007, paratype), living culture SICAUCC 21-0007.
Culture characters: Ascospores germinating on PDA within 12 h, and the cultures grow slowly on PDA. Colonies reach 2 cm in diameter after 25 days. Colonies from single ascospores are cottony, cling to the medium, with regular margin; the mycelium is creamy white to pale yellow but gradually becomes pale brown after 30 days.
Notes: This new taxon resembles species of Podonectria, in having superficial, bright, or lightly colored fruiting bodies and hairs obscuring the outer wall of ascoma. Morphologically Podonectria kuwanaspidis is comparable with P. novae-zelandiae. It has shorter (30–120 vs. 60–280 μm) and thicker-walled hairs (2–4.5 vs.1–2.5 μm), longer and narrower ascospores (150–240 × 5–7 vs. 100–160 × 7–10 μm). The ITS base-pair comparison between Podonectria kuwanaspidis (SICAUCC 21-0002) and phylogenetically affiliated P. sichuanensis (SICAU 16-0003) reveals 15% (including 20 gaps, 4%) nucleotide differences; the nucleotide differences in the SSU, LSU, tef1-α, and rpb2 region between them are 1% (0 gaps, 0%), 3% (5 gaps, 0%), 4% (0 gaps, 0%), and 10% (0 gaps, 0%), respectively. Hence, we describe our collection as a new species in Podonectria, as recommended by Jeewon and Hyde [63].
Nectriaceae Tul. & C. Tul., Select. fung. carpol. (Paris) 3:3 (1865)
Microcera Desm., Annls Sci. Nat., Bot., sér. 3 10:359 (1848)
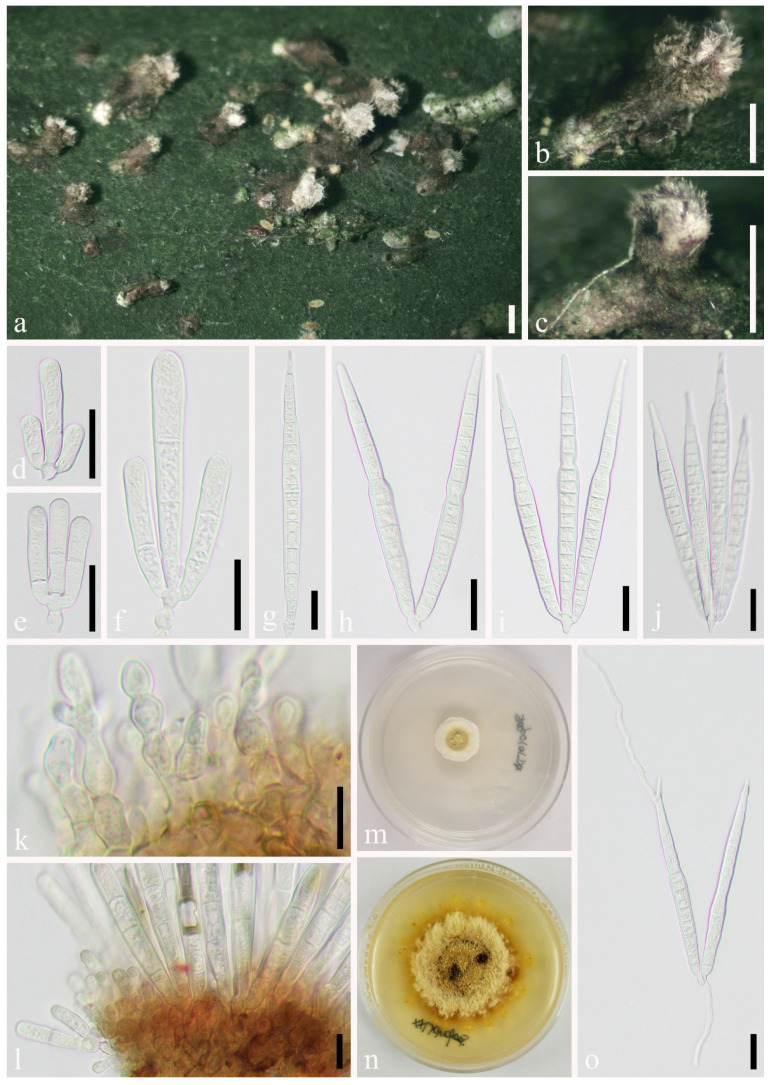
Microcera kuwanaspidis X.L. Xu & C.L. Yang sp. nov. (Figure 6)
Figure 6.
Microcera kuwanaspidis (SICAU 21-0006, holotype). (a,b) Stromata and sporodochia on host substrate. (c–e) Conidiophore with developing macroconidia. (f) Germinated conidium. (g–l) Macroconidia. (m) Colonies on PDA after 12 days. Scale bars: (a,b) 200 μm, (c–e) 50 μm, (f–l) 20 μm.
MycoBank number: MB 838464
Etymology: In reference to the generic name for the associated scale insect.
Holotype: SICAU 21-0006.
Habitat associated with scale insects Kuwanaspis howardi on bamboo. Sexual morph: Undetermined. Asexual morph: Stromata 500–690 μm long, 410–600 μm wide ( = 614 × 524 μm, n = 20), ellipsoid, orange-red, completely covering a single scale insect, or absent. Sporodochia 190–280 μm long, 150–300 μm wide ( = 240 × 227 μm, n = 20), formed singly on the margin of the stroma, or rarely in groups of one to three on the margin of the scale covers. Conidiophores with developing macroconidia form a pink upright mass. Macroconidia (80–)95–120 μm long × 6.5–8.5 ( = 107 × 7.3 μm, n = 20) μm wide, hyaline, cylindrical, slightly curved, slender toward each end, 3–8 septate, mostly 5–6–7 septate, difficult to distinguish apical cell and basal cell. Microconidia and chlamydospores were not observed.
Material examined: CHINA, Sichuan Province, Ya’an City, Lushan County (102°55′58.13″ E, 30°15′24.07″ N, Alt. 1116 m), on scale insect Kuwanaspis howardi on Phyllostachys heteroclada, 10 June 2020, Xiu-lan Xu, XXL202006007 (SICAU 21-0006, holotype), ex-type culture SICAUCC 21-0006, additional GenBank Number: SSU = MW462896; CHINA, Sichuan Province, Meishan City, Hongya County (103°14′2.64″ E, 29°41′53.07″ N, Alt. 538 m), on scale insect Kuwanaspis howardi on Pleioblastus amarus, 9 March 2021, Chun-lin Yang, YCL202103001 (SICAU 21-0009, paratype), living culture SICAUCC 21-0009, additional GenBank Number: SSU = MZ029435.
Culture characters: Colonies from a single macroconidium on PDA grow slowly and reach approximately 2.2 cm in diameter after 12 days at 25 °C, circular, flat, whitish to bright orange with white mycelium on the surface forming concentric circles, and the back of colonies is bright orange.
Notes: Distinguished from the red-headed fungus Microcera coccophila [18,64], in which the sporodochium is usually formed in groups on margin of dead scale or their covers accompanied with perithecia surround the edge of scale covers. However, this new species has distinct stroma covering the host, with a single sporodochium at the edge and without perithecia being discovered. Furthermore, although they are similar in size, Microcera kuwanaspidis is different from M. coccophila in numbers of septa (3–8 vs. 7–9). Microcera kuwanaspidis clusters with M. coccophila (CBS 310.34) with 100% ML and 1.00 BYPP support; however, striking base-pair differences are noted, viz. 1% (0 gaps, 0%), 1% (0 gaps, 0%), 1% (0 gaps, 0%), 13% (23 gaps, 4%), 4% (0 gaps, 0%), 3% (0 gaps, 0%), 5% (3 gaps, 0%), and 9% (6 gaps, 1%) in the ITS, LSU, rpb2, tef1-α, acl1, act, cmdA, and his3 DNA sequence data, respectively. According to the guidelines of Jeewon and Hyde [63], our collection is proposed as a new species.
4. Discussion
Mycologists have questioned the exact familial placement of Podonectria since the beginning of its establishment. Dingley [3] placed the genus in Clavicipitaceae (Hypocreales). Rossman transferred it into the Pleosporaceae (Pleosporales) due to its bitunicate asci rather than the unitunicate asci found in Hypocreales [4,7]. Barr transferred Podonectria to Tubeufiaceae [65], which was erected [66] to accommodate pleosporaceous taxa that are typically hyper saprobic on other fungi or substrates previously colonized by other fungi, hyperparasitic on foliicolous fungi, parasitic on scale insects, or occasionally parasitic on living leaves. This treatment was followed by subsequent authors [9,67,68,69]. However, Tubeufiaceae, which was comprehensively reviewed by Boonmee et al. [70], was accommodated in a new order, Tubeufiales [58]. This placement was followed by Wijayawardene et al. [71,72] and Hongsanan et al. [73]. However, Dao et al. [5] proposed Podonectriaceae, a new family in Pleosporales, to accommodate this genus, which was confirmed by ITS and LSU data. This placement was supported by Yang et al. [12], in which Podonectria sichuanensis was identified based on morphological characteristics and phylogenetic analyses. Based on the phylogenetic results of combined ITS, LSU, SSU, tef1-α, and rpb2 data in this current study, we confirm Podonectriaceae as an accepted family in the suborder Pleosporineae [49]. Podonectriaceae is phylogenetically closely related to Pseudopyrenochaetaceae that has been established to accommodate two species, viz. Pseudopyrenochaeta lycopersici and P. terrestris [57]. However, the two families are morphologically distinct. Pseudopyrenochaetaceae has pycnidial conidiomata, filiform conidiophores, and aseptate, cylindrical to allantoid conidia, whereas Podonectriaceae comprises sporodochial conidiomata, moniliform or inconspicuous conidiophores, and 1–4 armed, multiseptated conidia. In addition, the coelomycete genera Tetranacrium that has septate tetraradiate conidia [74,75] was documented as the anamorph associated with Podonectria gahnia according to substrate observation [4]. However, the association is somewhat confused, as it lacks further phylogenetic investigations and taxonomic studies. Identical molecular sequences of Podonectria novae-zelandiae in our study confirmed the link between the sexual morphs and asexual morphs in Tetracrium. Podonectria was reported to be associated with scale insects on various hosts in previous studies [1,4,5,58]. In this paper, we isolated Podonectria sichuanensis (SICAUCC 21-0001) on the ascomata of Neostagonosporella sichuanensis in our sampling site and confirm that the Podonectria species are not only parasitic on scale insects but also on other fungi or substrates previously colonized by other fungi [12]. According to published studies, most species of Podonectria are associated with armored scale insects, in addition to being associated with the mostly reported hosts Citrus aurantium L. and C. nobilis Lour. (Rutaceae) [1,4] and the known host plants associated with Podonectria are Calluna vulgaris Salisb. (Ericaceae), Gahnia setifolia (A. Rich) Hook.f., G. xanthocarpa (Hook.f.) Hook. f. (Cyperaceae), Juniperus bermudiana L. (Cupressaceae), Olearia rani Druce (Asteraceae), Phyllostachys heteroclada (Poaceae) and Podocarpus ferrugineus G. Benn. ex D. Don (Podocarpaceae) [3,4,5,12].
Gräfenhan et al. [18] reported an association of Microcera to Fusarium, Cladosterigma Pat., Mycogloea L.S. Olive, Tetracrium Henn., and accepted four species in Microcera. Nowadays, taxonomic concepts based on multi-gene phylogenetic inference have provided a deeper understanding of phylogenetic relationships than those based on individual gene regions [76,77,78,79]. Recently, combined ITS-LSU-tef1-α-acl1-act-cmdA-his3-rpb1-rpb2-tub2 datasets were used to clarify intraspecific and intergeneric relationships within Nectriaceae [19], and combined ITS-LSU-tef1-α-cmdA-rpb2-tub2 datasets were similarly used for Hypocreales [80]. In this paper, Microcera kuwanaspidis can be distinguished from M. coccophila and is established as new species on account of base-pair differences, especially in the tef1-α (13%), acl1 (4%), act (3%), cmdA (5%), and his3 (9%). The Microcera species have been mostly reported associated with armored scale insects on citrus (Rutaceae), viz. Aonidiella aurantii, A. citrina, Lepidosaphes beckii, Unaspis citri, and Quadraspidiotus perniciosus on Pyrus communis, Prunus domestica and P. cerasus (Rosaceae), as well associated with nut scale Eulecanium tiliae (Hemiptera: Coccidae) on Salix sp. (Salicaceae) and Fraxinus excelsior (Oleaceae), and an unknown scale insect on Broussonetia kazinoki × B. papyrifera (Moraceae), Laurus nobilis (Lauraceae), Citrus maxima (Rutaceae), and apple trees [18,60,64].
In China, the entomopathogenic fungi associated with scale insects was mainly focused on commercial Citrus plants in the 1990s. Verticillium lecanii (Zimm) Viegas is the most common fungus that is parasitic on scale insects on Citrus since its discovery from Guizhou Province in 1982 [81]. Subsequently, Aschersonia duplex Berk., Beauveria bassiana (Bals.-Criv.) Vuill., Fusarium juruanum Henn., F. moniliforme Sheld., Microcera coccophila, Nigrospora sphaerica (Sacc) Mason, and Podonectria coccicola have also been reported to be associated with the scale insects on citrus [82,83,84]. Microcera and Podonectria were commonly encountered on scale insects within tree canopies and occurred throughout the year but were more noticeable under wet and humid conditions [5,64,85,86], consistent with the observations in this study. Presently, Microcera coccophila and Podonectria coccicola have been the most commonly and worldwide recorded species on scale insects, especially on orange trees [1,4,7,85,86,87,88,89]. This paper provides new records for three entomopathogenic fungi, Podonectria kuwanaspidis, P. novae-zelandiae, and Microcera kuwanaspidis on armored insect scale from bamboo in China. According to the field observation from 2015 to 2020, the three species are commonly associated with Kuwanaspis howardi on native bamboo, especially on Phyllostachys heteroclada, and they effectively cause the scale insect hosts to be infected, which ultimately results in death. As documented by Rossman [4] and Dao et al. [64], the role of entomopathogens in the biological control of destructive scale insects on citrus trees was usually controlled by chemical sprays. These entomopathogenic fungi should be further screened to assess their potential for commercial development as biological control agents.
Acknowledgments
Xiulan Xu acknowledges the Sichuan Agricultural University for providing laboratory facilities, and Konstanze Bensch is thanked for the nomenclatural correction of the name. D.W. would like to thank the CAS President’s International Fellowship Initiative (No. 2021FYB0005) and Postdoctoral Fund from the Human Resources and Social Security Bureau of Yunnan Province.
Author Contributions
X.-L.X. and C.-L.Y.: conceptualization. X.-L.X.: data curation. X.-L.X. and C.-L.Y.: formal analysis, methodology, and writing—original draft. X.-L.X. and Q.-G.X.: funding acquisition. X.-L.X., C.-L.Y., Q.Z. and Y.-C.L.: investigation. Q.-G.X. and Y.-G.L.: project administration. C.-L.Y.: supervision. C.-L.Y., R.J., S.S.N.M., D.N.W. and K.D.H.: writing—review and editing. All authors contributed to the article and approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Chengdu Science and Technology Bureau (2019040509).
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets presented in this study can be found in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/), MycoBank (http://www.MycoBank.org) and TreeBASE (http://www.treebase.org) (all accessed on 18 July 2021).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Petch T. Studies in entomogenous fungi. The Nectriae parasitic on scale insects. Trans. Brit. Myc. Socy. 1921;7:133–167. doi: 10.1016/S0007-1536(21)80018-2. [DOI] [Google Scholar]
- 2.Index Fungorum Database. [(accessed on 15 April 2021)]; Available online: http://www.indexfungorum.org/
- 3.Dingley J.M. The Hypocreales of new zealand VI. The genera Hypocrella, Barya, Claviceps and Podonectria. Trans. R. Soc. N. Z. 1954;81:489–499. [Google Scholar]
- 4.Rossman A.Y. Podonectria, a genus in the Pleosporales on scale insects. Mycotaxon. 1978;7:163–182. [Google Scholar]
- 5.Dao H.T., Beattie G.A.C., Rossman A.Y., Burgess L.W., Holford P. Four putative entomopathogenic fungi of armoured scale insects on Citrus in Australia. Mycol. Prog. 2016;15:47. doi: 10.1007/s11557-016-1188-6. [DOI] [Google Scholar]
- 6.Dennis R.W.G. New British fungi. Kew Bull. 1957;12:399–404. doi: 10.2307/4113704. [DOI] [Google Scholar]
- 7.Rossman A.Y. The genus Ophionectria (Euascomycetes, Hypocreales) Mycologia. 1977;69:355–391. doi: 10.1080/00275514.1977.12020067. [DOI] [Google Scholar]
- 8.Pirozynski K.A. Notes on Hyperparasitic Sphaeriales, Hypocreales and ‘Hypocreoid Dothideales’. Kew Bull. 1977;31:595–610. doi: 10.2307/4119409. [DOI] [Google Scholar]
- 9.Rossman A.Y. The Tubeufiaceae and Similar Loculoascomycetes. C.A.B. International; Farnham Royal, UK: 1987. p. 43. [Google Scholar]
- 10.Spatafora J.W., Quandt C.A., Kepler R.M., Sung G.H., Shrestha B., Hywel-Jones N.L., Luangsa-ard J.J. New 1F1N species Combinations in Ophiocordycipitaceae (Hypocreales) IMA Fungus. 2015;6:357–362. doi: 10.5598/imafungus.2015.06.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quandt C.A., Kepler R.M., Gams W., Araújo J.P.M., Ban S., Evans H.C., Hughes D., Humber R., Hywel-Jones N., Li Z.Z., et al. Phylogenetic-based nomenclatural proposals for Ophiocordycipitaceae (Hypocreales) with new combinations in Tolypocladium. IMA Fungus. 2014;5:121–134. doi: 10.5598/imafungus.2014.05.01.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang C.L., Xu X.L., Liu Y.G. Podonectria sichuanensis, a potentially mycopathogenic fungus from Sichuan Province in China. Phytotaxa. 2019;402:219–231. doi: 10.11646/phytotaxa.402.5.1. [DOI] [Google Scholar]
- 13.Wollenweber H.W., Reinking O.A. Die fusarien, ihre Beschreibung, schadwirkung und bekämpfung. Paul Parey; Berlin, Germany: 1935. pp. 1–355. [Google Scholar]
- 14.Booth C. The Genus Fusarium. Commonwealth Mycological Institute; Kew, UK: 1971. pp. 1–237. [Google Scholar]
- 15.Gerlach W., Nirenberg H. Mitteilungen Aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft. Volume 209. Kommissionsverlag Paul Parey; Berlin, Germany: 1982. The genus Fusarium: A pictorial atlas; pp. 1–406. [Google Scholar]
- 16.Nelson P.E., Toussoun T.A., Marasas W.F.O. Fusarium Species: An Illustrated Manual for Identification. Pennsylvania State University Press; University Park, PA, USA: 1983. pp. 1–193. [Google Scholar]
- 17.Leslie J.F., Summerell B.A. The Fusarium Laboratory Manual. 1st ed. Blackwell Publishing; Ames, IA, USA: 2006. pp. 1–369. [Google Scholar]
- 18.Gräfenhan T., Schroers H.J., Nirenberg H.I., Seifert K.A. An overview of the taxonomy, phylogeny, and typification of nectriaceous fungi in Cosmospora, Acremonium, Fusarium, Stilbella, and Volutella. Stud. Mycol. 2011;68:79–113. doi: 10.3114/sim.2011.68.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombard L., Merwe N.A., Groenewald J.Z., Crous P.W. Generic concepts in Nectriaceae. Stud. Mycol. 2015;80:189–245. doi: 10.1016/j.simyco.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Triest D., Cremer K.D., Piérard D., Hendrickx M. Unique phylogenetic lineage found in the Fusarium-like clade after re-examining BCCM/IHEM fungal culture collection material. Mycobiology. 2016;44:121–130. doi: 10.5941/MYCO.2016.44.3.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Normark B.B., Okusu A., Morse G.E., Peterson D.A., Itioka T., Schneider S.A. Phylogeny and classification of armored scale insects (Hemiptera: Coccomorpha: Diaspididae) Zootaxa. 2019;4616:1–98. doi: 10.11646/zootaxa.4616.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Fang Z.G., Liu J. Biology observation and control on the Kuwanaspis howardi. J. Bamboo Res. 2000;19:78–80. [Google Scholar]
- 23.Malumphy C., Salisbury A. First incursion in Europe of bamboo white scale Kuwanaspis howardi (Hemiptera: Diaspididae), with a review of Kuwanaspis species detected in Britain. Br. J. Entomol. Nat. Hist. 2016;29:97–103. [Google Scholar]
- 24.Senanayake I.C., Rathnayaka A.R., Marasinghe D.S., Calabon M.S., Gentekaki E., Lee H.B., Hurdeal V.G., Pem D., Dissanayake L.S., Wijesinghe S.N., et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere. 2020;11:2678–2754. doi: 10.5943/mycosphere/11/1/20. [DOI] [Google Scholar]
- 25.Chomnunti P., Hongsanan S., Hudson B.A., Tian Q., Peršoh D., Dhami M.K., Alias A.S., Xu J.C., Liu X.Z., Stadler M., et al. The sooty moulds. Fungal Divers. 2014;66:1–36. doi: 10.1007/s13225-014-0278-5. [DOI] [Google Scholar]
- 26.White T.J., Bruns T., Lee S., Taylor J. Amplifcation and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfaud D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Diego, CA, USA: 1990. pp. 315–322. [DOI] [Google Scholar]
- 27.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rehner S.A., Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among Ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 30.Dai D.Q., Phookamsak R., Wijayawardene N.N., Li W.J., Bhat D.J., Xu J.C., Taylor J.E., Hyde K.D., Chukeatirote E. Bambusicolous fungi. Fungal Divers. 2016;82:1–105. doi: 10.1007/s13225-016-0367-8. [DOI] [Google Scholar]
- 31.Wanasinghe D.N., Mortimer P.E., Xu J. Insight into the systematics of microfungi colonizing dead woody twigs of Dodonaea viscosa in Honghe (China) J. Fungi. 2021;7:180. doi: 10.3390/jof7030180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 34.Xu X.L., Yang C.L., Jeewon R., Wanasinghe D.N., Liu Y.G., Xiao Q.G. Morpho-molecular diversity of Linocarpaceae (Chaetosphaeriales): Claviformispora gen. nov. from decaying branches of Phyllostachys heteroclada. MycoKeys. 2020;70:1–17. doi: 10.3897/mycokeys.70.54231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woudenberg J.H.C., Groenewald J.Z., Binder M., Crous P.W. Alternaria redefined. Stud. Mycol. 2013;75:171–212. doi: 10.3114/sim0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marin-Felix Y., Hernández-Restrepo M., Iturrieta-González I., García D., Gené J., Groenewald J.Z., Cai L., Chen Q., Quaedvlieg W., Schumacher R.K., et al. Genera of phytopathogenic fungi: GOPHY 3. Stud. Mycol. 2019;94:1–124. doi: 10.1016/j.simyco.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thambugala K.M., Wanasinghe D.N., Phillips A.J.L., Camporesi E., Bulgakov T.S., Phukhamsakda C., Ariyawansa H.A., Goonasekara I.D., Phookamsak R., Dissanayake A., et al. Mycosphere notes 1-50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere. 2017;8:697–796. doi: 10.5943/mycosphere/8/4/13. [DOI] [Google Scholar]
- 38.Phukhamsakda C., McKenzie E.H.C., Phillips A.J.L., Jones E.B.G., Bhat D.J., Marc S., Bhunjun C.S., Wanasinghe D.N., Thongbai B., Camporesi E., et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020;102:1–203. doi: 10.1007/s13225-020-00448-4. [DOI] [Google Scholar]
- 39.Jaklitsch W.M., Checa J., Blanco M.N., Olariaga I., Tello S., Voglmayr H. A preliminary account of the Cucurbitariaceae. Stud. Mycol. 2018;90:71–118. doi: 10.1016/j.simyco.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wanasinghe D.N., Phukhamsakda C., Hyde K.D., Jeewon R., Lee H.B., Jones E.B.G., Tibpromma S., Tennakoon D.S., Dissanayake A.J., Jayasiri S.C., et al. Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018;89:1–236. doi: 10.1007/s13225-018-0395-7. [DOI] [Google Scholar]
- 41.Dai D.Q., Bhat D.J., Liu J.K., Chukeatirote E., Zhao R.L., Hyde K.D. Bambusicola, a new genus from bamboo with asexual and sexual morphs. Cryptogamie Mycol. 2012;33:363–379. doi: 10.7872/crym.v33.iss3.2012.363. [DOI] [Google Scholar]
- 42.Dai D.Q., Bahkali A.H., Li W.J., Bhat D.J., Zhao R.L., Hyde K.D. Bambusicola loculata sp. nov. (Bambusicolaceae) from bamboo. Phytotaxa. 2015;213:122–130. doi: 10.11646/phytotaxa.213.2.5. [DOI] [Google Scholar]
- 43.Yang C.L., Xu X.L., Liu Y.G. Two new species of Bambusicola (Bambusicolaceae, Pleosporales) on Phyllostachys heteroclada from Sichuan, China. Nova Hedwig. 2019;108:527–545. doi: 10.1127/nova_hedwigia/2019/0526. [DOI] [Google Scholar]
- 44.Jayawardena R.S., Hyde K.D., Jeewon R., Ghobad-Nejhad M., Wanasinghe D.N., Liu N.G., Phillips A.J.L., Oliveira-Filho J.R.C., da Silva G.A., Gibertoni T.B., et al. One stop shop II: Taxonomic update with molecular phylogeny for important phytopathogenic genera: 26–50. Fungal Divers. 2019;94:41–129. doi: 10.1007/s13225-019-00418-5. [DOI] [Google Scholar]
- 45.Valenzuela-Lopez N., Sutton D.A., Cano-Lira J.F., Paredes K., Wiederhold N., Guarro J., Stchigel A.M. Coelomycetous fungi in the clinical setting: Morphological convergence and cryptic diversity. J. Clin. Microbiol. 2016;55:552–567. doi: 10.1128/JCM.02221-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quaedvlieg W., Verkley G.J.M., Shin H.D., Barreto R.W., Alfenas A.C., Swart W.J., Groenewald J.Z., Crous P.W. Sizing up Septoria. Stud. Mycol. 2013;75:307–390. doi: 10.3114/sim0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Gruyter J., Woudenberg J.H.C., Aveskamp M.M., Verkley G.J.M., Groenewald J.Z., Crous P.W. Redisposition of phoma-like anamorphs in Pleosporales. Stud. Mycol. 2013;75:1–36. doi: 10.3114/sim0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inderbitzin P., Kohlmeyer J., Volkmann-Kohlmeyer B., Berbee M.L. Decorospora, a new genus for the marine ascomycete Pleospora gaudefroyi. Mycologia. 2002;94:651–659. doi: 10.1080/15572536.2003.11833193. [DOI] [PubMed] [Google Scholar]
- 49.Senwanna C., Wanasinghe D.N., Bulgakov T.S., Wang Y., Bhat D.J., Tang A.M.C., Mortimer P.E., Xu J., Hyde K.D., Phookamsak R. Towards a natural classification of Dothidotthia and Thyrostroma in Dothidotthiaceae (Pleosporineae, Pleosporales) Mycosphere. 2019;10:701–738. doi: 10.5943/mycosphere/10/1/15. [DOI] [Google Scholar]
- 50.Ariyawansa H.A., Phukhamsakda C., Thambugala K.M., Bulgakov T.S., Wanasinghe D.N., Perera R.H., Mapook A., Camporesi E., Kang J.C., Jones E.B.G., et al. Revision and phylogeny of Leptosphaeriaceae. Fungal Divers. 2015;74:19–51. doi: 10.1007/s13225-015-0349-2. [DOI] [Google Scholar]
- 51.Mapook A., Hyde K.D., McKenzie E.H.C., Jones E.B.G., Bhat D.J., Jeewon R., Stadler M., Samarakoon M.C., Malaithong M., Tanunchai B., et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed) Fungal Divers. 2020;101:1–175. doi: 10.1007/s13225-020-00444-8. [DOI] [Google Scholar]
- 52.Tibpromma S., Liu J.K., Promputtha I., Camporesi E., Bhakali A.H., Hyde K.D., Boonmee S. Ophiosimulans tanaceti gen. et sp. nov. (Phaeosphaeriaceae) on Tanacetum sp. (Asteraceae) from Italy. Mycol. Prog. 2016;15:46. doi: 10.1007/s11557-016-1187-7. [DOI] [Google Scholar]
- 53.Phookamsak R., Wanasinghe D.N., Hongsanan S., Phukhamsakda C., Huang S.K., Tennakoon D.S., Norphanphoun C., Camporesi E., Bulgakov T.S., Promputtha I., et al. Towards a natural classification of Ophiobolus and ophiobolus-like taxa; introducing three novel genera Ophiobolopsis, Paraophiobolus and Pseudoophiobolus in Phaeosphaeriaceae (Pleosporales) Fungal Divers. 2017;87:299–339. doi: 10.1007/s13225-017-0393-1. [DOI] [Google Scholar]
- 54.Liu J.K., Hyde K.D., Jones E.B.G., Ariyawansa H.A., Bhat D.J., Boonmee S., Maharachchikumbura S.S.N., McKenzie E.H.C., Phookamsak R., Phukhamsakda C., et al. Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015;72:1–197. doi: 10.1007/s13225-015-0324-y. [DOI] [Google Scholar]
- 55.Jaklitsch W.M., Voglmayr H. Fenestelloid clades of the Cucurbitariaceae. Persoonia. 2020;44:1–40. doi: 10.3767/persoonia.2020.44.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tennakoon D., Jeewon R., Gentekaki E., Kuo C.H., Hyde K.D. Multi-gene phylogeny and morphotaxonomy of Phaeosphaeria ampeli sp. nov. from Ficus ampelas and a new record of P. musae from Roystonea regia. Phytotaxa. 2019;406:111–128. doi: 10.11646/phytotaxa.406.2.3. [DOI] [Google Scholar]
- 57.Valenzuela-Lopez N., Cano-Lira J.F., Guarro J., Sutton D.A., Wiederhold N., Crous P.W., Stchigel A.M. Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Stud. Mycol. 2018;90:1–69. doi: 10.1016/j.simyco.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boonmee S., Rossman A.Y., Liu J.K., Li W.J., Dai D.Q., Bhat J.D., Jones E.B.G., McKenzie E.H.C., Xu J.C., Hyde K.D. Tubeufiales, ord. nov., integrating sexual and asexual generic names. Fungal Divers. 2014;68:239–298. doi: 10.1007/s13225-014-0304-7. [DOI] [Google Scholar]
- 59.O’Donnell K., Humber R.A., Geiser D.M., Kang S., Park B., Robert V.A.R.G., Crous P.W., Johnston P.R., Aoki T., Rooney A.P., et al. Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia. 2012;104:427–445. doi: 10.3852/11-179. [DOI] [PubMed] [Google Scholar]
- 60.Bills G.F., Platas G., Overy D.P., Collado J., Fillola A., Jiménez M.R., Martín J., del Val A.G., Vicente F., Tormo J.R., et al. Discovery of the parnafungins, antifungal metabolites that inhibit mRNA polyadenylation, from the Fusarium larvarum complex and other Hypocrealean fungi. Mycologia. 2009;101:449–472. doi: 10.3852/08-163. [DOI] [PubMed] [Google Scholar]
- 61.Herrera C.S., Rossman A.Y., Samuels G.J., Chaverri P. Pseudocosmospora, a new genus to accommodate Cosmospora vilior and related species. Mycologia. 2013;105:1287–1305. doi: 10.3852/12-395. [DOI] [PubMed] [Google Scholar]
- 62.Rossman A.Y., Crous P.W., Hyde K.D., Hawksworth D.L., Aptroot A., Bezerra J.L., Bhat J.D., Boehm E., Braun U., Boonmee S., et al. Recommended names for pleomorphic genera in Dothideomycetes. IMA Fungus. 2015;6:507–523. doi: 10.5598/imafungus.2015.06.02.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jeewon R., Hyde K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere. 2016;7:1669–1677. doi: 10.5943/mycosphere/7/11/4. [DOI] [Google Scholar]
- 64.Dao H.T., Beattie G.A.C., Rossman A.Y., Burgess L.W., Holford P. Systematics and biology of two species of Microcera associated with armoured scales on citrus in Australia. Mycol. Prog. 2015;14:17. doi: 10.1007/s11557-015-1044-0. [DOI] [Google Scholar]
- 65.Barr M.E. On the family Tubeufiaceae (Pleosporales) Mycotaxon. 1980;12:137–167. [Google Scholar]
- 66.Barr M.E. A classification of Loculoascomycetes. Mycologia. 1979;71:935–957. doi: 10.1080/00275514.1979.12021099. [DOI] [Google Scholar]
- 67.Kodsueb R., Jeewon R., Vijaykrishna D., McKenzie E.H.C., Lumyong P., Lumyong S., Hyde K.D. Systematic revision of Tubeufiaceae based on morphological and molecular data. Fungal Divers. 2006;21:105–130. [Google Scholar]
- 68.Lumbsch H.T., Huhndorf S.M. Myconet Volume 14. Part One. Outline of Ascomycota—2009. Part Two. Notes on Ascomycete Systematics. Nos. 4751–5113. Fieldiana Life Earth Sci. 2010;1:1–64. doi: 10.3158/1557.1. [DOI] [Google Scholar]
- 69.Hyde K.D., Jones E.B.G., Liu J.K., Ariyawansa H., Boehm E., Boonmee S., Braun U., Chomnunti P., Crous P.W., Dai D.Q., et al. Families of Dothideomycetes. Fungal Divers. 2013;63:1–313. doi: 10.1007/s13225-013-0263-4. [DOI] [Google Scholar]
- 70.Boonmee S., Zhang Y., Comment P., Chukeatirote E., Tsui C.K., Bahkali A.H., Hyde K.D. Revision of lignicolous Tubeufiaceae based on morphological reexamination and phylogenetic analysis. Fungal Divers. 2011;51:51–63. doi: 10.1007/s13225-011-0147-4. [DOI] [Google Scholar]
- 71.Wijayawardene N.N., Hyde K.D., Lumbsch H.T., Liu J.K., Maharachchikumbura S.S.N., Ekanayaka A.H., Tian Q., Phookamsak R. Outline of ascomycota: 2017. Fungal Divers. 2018;88:167–263. doi: 10.1007/s13225-018-0394-8. [DOI] [Google Scholar]
- 72.Wijayawardene N.N., Hyde K.D., Al-Ani L.K.T., Tedersoo L., Haelewaters D., Rajeshkumar K.C., Zhao R.L., Aptroot A., Leontyev D.V., Saxena R.K., et al. Outline of fungi and fungus-like taxa. Mycosphere. 2020;11:1060–1456. doi: 10.5943/mycosphere/11/1/8. [DOI] [Google Scholar]
- 73.Hongsanan S., Hyde K.D., Phookamsak R., Wanasinghe D.N., McKenzie E.H.C., Sarma V.V., Lücking R., Boonmee S., Bhat J.D., Liu N.G., et al. Refined families of Dothideomycetes: Orders and families incertae sedis in Dothideomycetes. Fungal Divers. 2020;105:17–318. doi: 10.1007/s13225-020-00462-6. [DOI] [Google Scholar]
- 74.Punithalingam E. Nuclei, micronuclei and appendages in tri- and tetraradiate conidia of Cornutispora and four other coelomycete genera. Mycol. Res. 2003;107:917–948. doi: 10.1017/S0953756203008037. [DOI] [PubMed] [Google Scholar]
- 75.Hudson H.J., Sutton B.C. Trisulcosporium and Tetranacrium, two new genera of fungi imperfecti. Trans. Brit. Mycol. Soc. 1964;47:197–203. doi: 10.1016/S0007-1536(64)80053-X. [DOI] [Google Scholar]
- 76.Sung G.H., Sung J.M., Hywei-Jones M.L., Spatafora J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007;44:1204–1223. doi: 10.1016/j.ympev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 77.Lombard L., Crous P.W. Phylogeny and taxonomy of the genus Gliocladiopsis. Persoonia. 2012;28:25–33. doi: 10.3767/003158512X635056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lücking R., Aime M.C., Robbertse B., Miller A.N., Ariyawansa H.A., Aoki T., Cardinali G., Crous P.W., Druzhinina I.S., Geiser D.M., et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding. IMA Fungus. 2020;11:14. doi: 10.1186/s43008-020-00033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei D.P., Wanasinghe D.N., Xu J.C., To-anun C., Mortimer P.E., Hyde K.D., Elgorban A.M., Madawala S., Suwannarach N., Karunarathna S.C., et al. Three novel entomopathogenic fungi from China and Thailand. Front. Microbiol. 2021;11:608991. doi: 10.3389/fmicb.2020.608991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hyde K.D., Norphanphoun C., Maharachchikumbura S.S.N., Bhat D.J., Jones E.B.G., Bundhun D., Chen Y.J., Bao D.F., Boonmee S., Calabon M.S., et al. Refined families of Sordariomycetes. Mycosphere. 2020;11:305–1059. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- 81.Fu L.S. Pathogenic microorganisms on citrus insect scale. Hubei Plant Prot. 1994;4:30. [Google Scholar]
- 82.Gao R.X., Ouyang Z.A. A preliminary investigation on parasitic fungi on citrus pests. Microbiol. China. 1981;8:57–58. [Google Scholar]
- 83.Wang H. Entomogenous fungi of scale insects and mealworms and their applications. J. Sichuan For. Sci. Technol. 1999;20:62–65. doi: 10.16779/j.cnki.1003-5508.1999.03.021. [DOI] [Google Scholar]
- 84.Dong Z.Y., Luo M. Isolation and identification of a parasitic fungus of citrus scale insects. Mod. Agric. Sci. Technol. 2012;16:163–164. [Google Scholar]
- 85.Hely P.C., Pasfield G., Gellatley J.G. Insect Pests of Fruit and Vegetable in NSW. Department of Agriculture, New South Wales; Sydney, Australia: 1982. pp. 1–312. [Google Scholar]
- 86.Smith D., Beattie G.A.C., Broadley R. Citrus Pests and Their Natural Enemies: Integrated Pest Management in Australia. 1st ed. Department of Primary Industries; Brisbane, Australia: 1997. [(accessed on 10 April 2021)]. pp. 1–263. Available online: http://hdl.handle.net/10462/pdf/9446. [Google Scholar]
- 87.Petch T. Fungi parasitic on scale insects. Trans. Br. Mycol. Soc. 1921;7:18–40. doi: 10.1016/S0007-1536(21)80005-4. [DOI] [Google Scholar]
- 88.Tyson J.L., Henderson R.C., Fullerton R.A., Jamieson L.E., Froud K.J. Distribution and new host records for Cosmospora aurantiicola and Cosmospora flammea: Entomopathogens of Diaspididae in New Zealand. N. Z. Plant Prot. 2005;58:283–287. doi: 10.30843/nzpp.2005.58.4264. [DOI] [Google Scholar]
- 89.Dao H.T. Ph.D. Thesis. University of Western Sydney; Penrith, Australia: 2012. Ecology of Red Scale (Aonidiella aurantii (Maskell) [Hemiptera: Sternorrhyncha: Diaspididae]) in Citrus Orchards on the Central Coast of New South Wales. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/), MycoBank (http://www.MycoBank.org) and TreeBASE (http://www.treebase.org) (all accessed on 18 July 2021).