Abstract
We collected 92 isolates belonging to the genus Bacillus from the sake brewing process at Shiraki Tsunesuke Sake Brewery in Gifu, Japan to determine whether there is strain specificity at individual sake breweries. After distributing the isolates into seven groups, we observed that at least two groups (68 isolates) were kuratsuki bacteria at Shiraki Tsunesuke Sake Brewery. The kuratsuki Bacillus isolates were collected from different samples at the early and late stages of sake brewing in 2021 and 2019, respectively. These results showed that kuratsuki Bacillus entered the sake brewing process at this location. These kuratsuki Bacillus isolates had a high ethanol tolerance. Our previous paper showed the existence of kuratsuki Kocuria at Narimasa Sake Brewery in Toyama, Japan, but this study demonstrated that it is not found at Shiraki Tsunesuke Sake Brewery. Therefore, each sake brewery has specific kuratsuki bacterial strains, which are isolated with high frequency and contribute a specific flavor or taste to each sake brewery.
Keywords: Bacillus, bacteria, environment, ethanol tolerance, kuratsuki, PCR, sake brewery
1. Introduction
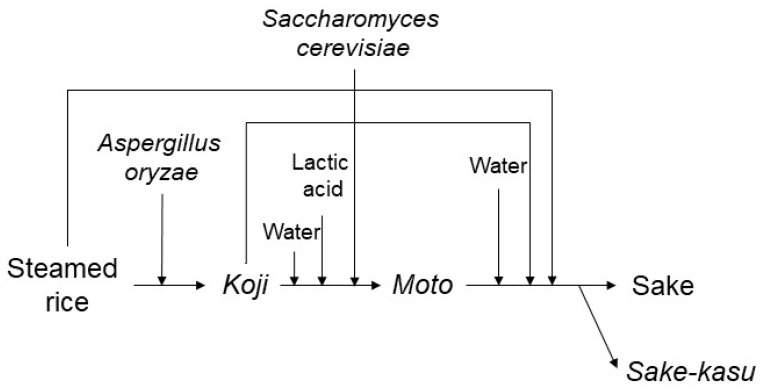
Sake is a traditional Japanese alcoholic drink, which has been brewed for over 1300 years [1]. At present, ~1400 sake breweries are widely distributed throughout Japan. Aspergillus oryzae is a fungus that converts rice starch into sugars, and then another fungus (yeast), Saccharomyces cerevisiae, converts the sugars into ethanol during sake brewing (Figure 1). Sake brewing is not performed aseptically, so bacterial flora analysis based on DNA sequence comparison in sake can detect several bacterial DNA [2,3,4,5,6], indicating that the bacteria may enter the production process by chance and briefly grow. The research question was whether all bacteria entered by chance or not. Thus, are there microorganisms that inevitably enter during the sake production process?
Figure 1.
Sake production scheme. Koji is a mixture of steamed rice and Aspergillus oryzae. Moto is an ethanol fermentation starter. Sake and sake-kasu are separated by filtration.
Lactic acid treatment is performed before the addition of S. cerevisiae for the effective ethanol fermentation (Figure 1). Kuratsuki (living in the sake brewery) lactic acid bacteria have been used for preventing the growth of other bacteria, which can lead to sake spoilage [3]. In the traditional method of producing fermentation starter, the kuratsuki lactic acid bacteria grow and produce lactic acid [3]. The lactic acid bacteria interact with other microorganisms in sake [7,8], but are killed by self-produced lactic acid or ethanol produced by S. cerevisiae. In contrast, some highly ethanol-tolerant lactic acid bacteria, for example Lactobacillus fructivorans, spoil sake [9,10,11], demonstrating that lactic acid bacteria can have a positive and a negative effect on sake brewing.
To elucidate whether kuratsuki bacteria other than lactic acid bacteria exist during the sake brewing process, 46 bacterial isolates were collected at the early stage of the sake brewing process from Narimasa Sake Brewery in Toyama, Japan [6,12]. Among the isolates, 23 had similar sequences to the genera Kocuria, 12 to Staphylococcus, 6 to Bacillus, 2 to Leifsonia, 2 to Microbacterium, and 1 to Enterococcus, inferred from the partial 16S rRNA gene sequences [6,12]. All isolates were not lactic acid bacteria. We showed that the Kocuria isolates have inhabited Narimasa Sake Brewery and are kuratsuki bacteria [12,13]. Kocuria does not belong to the phylum Firmicutes but Actinobacteria [14]. This was the first report of actinomycete living at a sake brewery. Recently, we showed that the kuratsuki bacteria in the Narimasa Sake Brewery interact with S. cerevisiae and affect flavor of the sake products [15]. The bacterial flora affects the chemical components in sake [16,17].
Based on the DNA sequence data in sake, Kocuria is specific to Narimasa Sake Brewery [6]. However, it was uncertain whether Kocuria is not isolated from other sake breweries, so we elucidated the same in this study. In 2019, six bacterial isolates were collected from the sample at the late stage of the sake brewing process of Shiraki Tsunesuke Sake Brewery in Gifu, Japan. In this study, we isolated bacteria during the early stage of the sake brewing processes of this sake brewery, compared them, and then discussed them.
2. Materials and Methods
2.1. Bacterial Isolation
Bacterial isolation was performed for four different sampling date samples from Shiraki Tsunesuke Sake Brewery (Table 1). Three different media, Difco Marine Broth 2216 (DMB) (BD, Franklin Lakes, NJ, USA), LB (10 g/L of tryptone, 10 g/L of NaCl, 5 g/L of yeast extract), and TGY (5 g/L of tryptone, 3 g/L of yeast extract, 1 g/L of glucose), containing 25 μg/mL of cycloheximide, were used [6]. Each sample (30 μL) was added to each medium (20 mL) and cultured at 15 °C and 30 °C for 72 h. Sake brewing was performed at 15 °C. Then, a single colony was chosen on the TGY agar plate.
Table 1.
Bacillus isolates from the sake brewing process in Shiraki Tsunesuke Sake Brewery.
| Isolate | Sample Number | Sampling Date | Process Stage | Medium |
|---|---|---|---|---|
| LB2 | Sample 1 | 30 January 2019 | Late | LB |
| LB3 | Sample 1 | 30 January 2019 | Late | LB |
| LBjs1 | Sample 1 | 30 January 2019 | Late | LB |
| MB1 | Sample 1 | 30 January 2019 | Late | DMB |
| MBjs1 | Sample 1 | 30 January 2019 | Late | DMB |
| TGY2 | Sample 1 | 30 January 2019 | Late | TGY |
| A1 | Sample 2 | 26 January 2021 | Early | DMB |
| A2 | Sample 2 | 26 January 2021 | Early | DMB |
| A3 | Sample 2 | 26 January 2021 | Early | LB |
| A4 | Sample 2 | 26 January 2021 | Early | LB |
| A5 | Sample 2 | 26 January 2021 | Early | LB |
| A6 | Sample 2 | 26 January 2021 | Early | TGY |
| A7 | Sample 2 | 26 January 2021 | Early | TGY |
| A8 | Sample 2 | 26 January 2021 | Early | TGY |
| A9 | Sample 2 | 26 January 2021 | Early | DMB |
| A10 | Sample 2 | 26 January 2021 | Early | TGY |
| A11 | Sample 2 | 26 January 2021 | Early | TGY |
| A12 | Sample 2 | 26 January 2021 | Early | TGY |
| A13 | Sample 2 | 26 January 2021 | Early | LB |
| A14 | Sample 2 | 26 January 2021 | Early | LB |
| a1 | Sample 2 | 26 January 2021 | Early | LB |
| a2 | Sample 2 | 26 January 2021 | Early | LB |
| a3 | Sample 2 | 26 January 2021 | Early | LB |
| a4 | Sample 2 | 26 January 2021 | Early | LB |
| a5 | Sample 2 | 26 January 2021 | Early | LB |
| a6 | Sample 2 | 26 January 2021 | Early | DMB |
| a7 | Sample 2 | 26 January 2021 | Early | DMB |
| a8 | Sample 2 | 26 January 2021 | Early | DMB |
| a9 | Sample 2 | 26 January 2021 | Early | DMB |
| a10 | Sample 2 | 26 January 2021 | Early | DMB |
| a11 | Sample 2 | 26 January 2021 | Early | TGY |
| a12 | Sample 2 | 26 January 2021 | Early | TGY |
| a13 | Sample 2 | 26 January 2021 | Early | TGY |
| a14 | Sample 2 | 26 January 2021 | Early | TGY |
| B1 | Sample 3 | 28 January 2021 | Early | LB |
| B2 | Sample 3 | 28 January 2021 | Early | LB |
| B3 | Sample 3 | 28 January 2021 | Early | LB |
| B4 | Sample 3 | 28 January 2021 | Early | LB |
| B5 | Sample 3 | 28 January 2021 | Early | LB |
| B6 | Sample 3 | 28 January 2021 | Early | LB |
| B7 | Sample 3 | 28 January 2021 | Early | LB |
| B8 | Sample 3 | 28 January 2021 | Early | TGY |
| B9 | Sample 3 | 28 January 2021 | Early | TGY |
| B10 | Sample 3 | 28 January 2021 | Early | TGY |
| B11 | Sample 3 | 28 January 2021 | Early | TGY |
| B12 | Sample 3 | 28 January 2021 | Early | TGY |
| B13 | Sample 3 | 28 January 2021 | Early | TGY |
| B14 | Sample 3 | 28 January 2021 | Early | DMB |
| B15 | Sample 3 | 28 January 2021 | Early | DMB |
| B16 | Sample 3 | 28 January 2021 | Early | DMB |
| B17 | Sample 3 | 28 January 2021 | Early | DMB |
| B18 | Sample 3 | 28 January 2021 | Early | DMB |
| B19 | Sample 3 | 28 January 2021 | Early | DMB |
| b1 | Sample 3 | 28 January 2021 | Early | LB |
| b2 | Sample 3 | 28 January 2021 | Early | LB |
| b3 | Sample 3 | 28 January 2021 | Early | LB |
| b4 | Sample 3 | 28 January 2021 | Early | DMB |
| b5 | Sample 3 | 28 January 2021 | Early | DMB |
| b6 | Sample 3 | 28 January 2021 | Early | DMB |
| b7 | Sample 3 | 28 January 2021 | Early | DMB |
| b8 | Sample 3 | 28 January 2021 | Early | TGY |
| b9 | Sample 3 | 28 January 2021 | Early | TGY |
| b10 | Sample 3 | 28 January 2021 | Early | TGY |
| b11 | Sample 3 | 28 January 2021 | Early | TGY |
| C1 | Sample 4 | 9 February 2021 | Early | LB |
| C2 | Sample 4 | 9 February 2021 | Early | LB |
| C3 | Sample 4 | 9 February 2021 | Early | LB |
| C4 | Sample 4 | 9 February 2021 | Early | LB |
| C5 | Sample 4 | 9 February 2021 | Early | DMB |
| C6 | Sample 4 | 9 February 2021 | Early | DMB |
| C7 | Sample 4 | 9 February 2021 | Early | DMB |
| C8 | Sample 4 | 9 February 2021 | Early | DMB |
| C9 | Sample 4 | 9 February 2021 | Early | TGY |
| C10 | Sample 4 | 9 February 2021 | Early | TGY |
| C11 | Sample 4 | 9 February 2021 | Early | TGY |
| C12 | Sample 4 | 9 February 2021 | Early | TGY |
| C13 | Sample 4 | 9 February 2021 | Early | TGY |
| C14 | Sample 4 | 9 February 2021 | Early | TGY |
| c1 | Sample 4 | 9 February 2021 | Early | LB |
| c2 | Sample 4 | 9 February 2021 | Early | LB |
| c3 | Sample 4 | 9 February 2021 | Early | LB |
| c4 | Sample 4 | 9 February 2021 | Early | LB |
| c5 | Sample 4 | 9 February 2021 | Early | DMB |
| c6 | Sample 4 | 9 February 2021 | Early | DMB |
| c7 | Sample 4 | 9 February 2021 | Early | DMB |
| c8 | Sample 4 | 9 February 2021 | Early | DMB |
| c9 | Sample 4 | 9 February 2021 | Early | DMB |
| c10 | Sample 4 | 9 February 2021 | Early | DMB |
| c11 | Sample 4 | 9 February 2021 | Early | TGY |
| c12 | Sample 4 | 9 February 2021 | Early | TGY |
| c13 | Sample 4 | 9 February 2021 | Early | TGY |
| c14 | Sample 4 | 9 February 2021 | Early | TGY |
2.2. Colony PCR
The V3–V4 region of the 16S rRNA gene from each single colony was amplified using region-specific PCR primers [12], designed by modification of a method by Illumina, San Diego, USA. The PCR products were sequenced using the primer 5′-TGTATAAGAGACAGGACTAC-3′.
2.3. Phylogenetic Analysis
Multiple alignment (Supplementary File S1) was constructed using MEGA X [18] for 92 DNA sequences of the bacterial isolates from Shiraki Tsunesuke Sake Brewery, 6 DNA sequences of the Bacillus isolates from Narimasa Sake Brewery [6,12], and 10 DNA sequences of the known Bacillus species from DNA database (Table 2). A neighbor-joining tree was constructed with 1000 bootstrap replicates using MEGA X [18]. The evolutionary distances were computed using the maximum composite likelihood method [19], and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated (complete deletion option). There were a total of 392 positions in the final dataset.
Table 2.
Bacillus species used in the 16S rRNA sequence comparison.
| Species | Strain | Sequence ID |
|---|---|---|
| Bacillus aryabhattai | B8W22 | NR_115953.1 |
| B. cereus | ATCC 14579 | NR_074540.1 |
| B. endophyticus | 2DT | MK554520.1 |
| B. flexus | IFO 15715 | NR_024691.1 |
| B. megaterium | ATCC 14581 | AB271751.1 |
| B. pumilus | ATCC 7061 | NR_043242.1 |
| B. safensis | FO-36b | NR_041794.1 |
| B. subtilis | IAM 12118 | NR_112116.1 |
| B. velezensis | FZB42 | NR_075005.2 |
| B. zhangzhouensis | MCCC 1A08372 | NR_148786.1 |
2.4. Ethanol Resistance Test
The isolates a12 and C3 were statically cultured in TGY including 20% ethanol at 30 °C. Each sample (100 μL) was spread on a TGY agar to count the number of colonies.
2.5. Statistical Test
A chi-square test was performed between isolated numbers from the cultures at 15 °C and 30 °C using the software R (http://www.R-project.org/).
3. Results
3.1. All Isolates from Shiraki Tsunesuke Sake Brewery belong to the Genus Bacillus
We obtained 6 DNA sequences from the samples at the late stage of the sake brewing process in 2019, 28 at the early stage on 26 January 2021, 30 at the early stage on 28 January 2021, and 28 at the early stage on 9 February 2021 (Table 1). Using the basic local alignment search on National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/), all 92 DNA sequences of bacterial isolates from Shiraki Tsunesuke Sake Brewery were found to be DNA sequences of the genus Bacillus. All partial 16S rRNA gene sequences are shown in Supplementary File S1.
3.2. Bacillus Isolates Distributed to Seven Different Groups
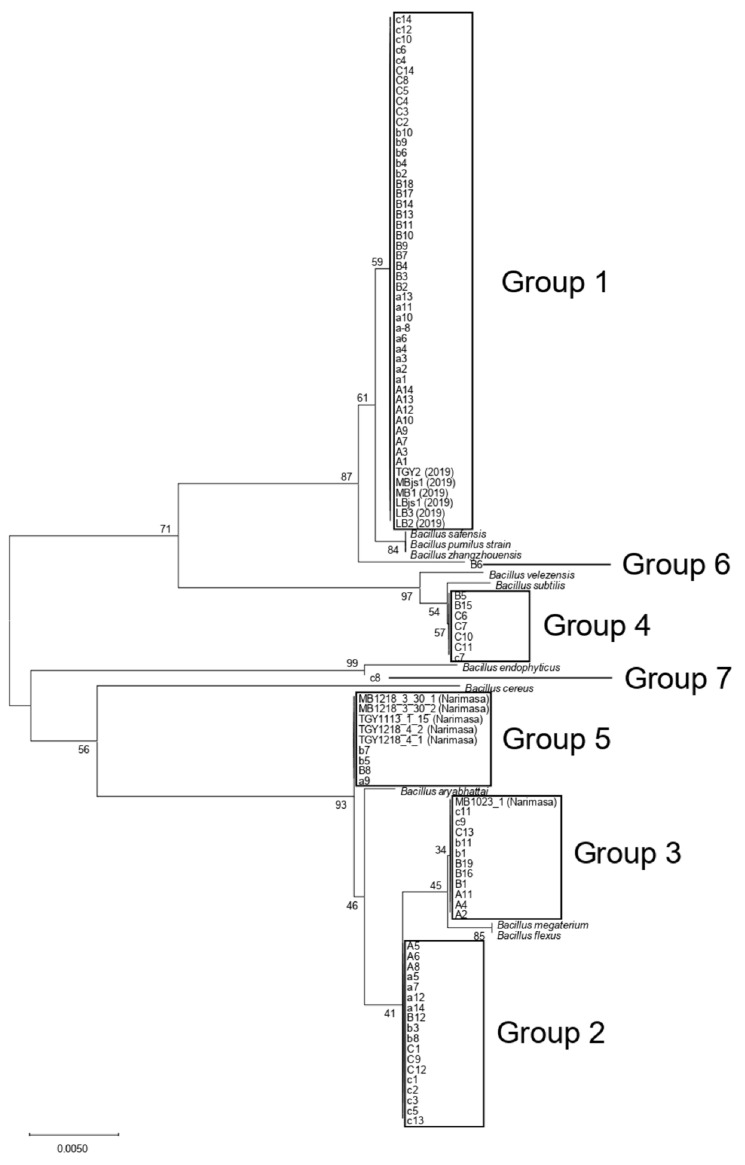
Phylogenetic analysis showed that the bacterial DNA sequences from Shiraki Tsunesuke Sake Brewery could be distributed into seven groups (Figure 2). The groups were numbered 1–7, with Group 1 being the largest and Groups 6 and 7 being the smallest. Group 1 consisted of 50 sequences, including all six DNA sequences from the samples in 2019 (Figure 2). This group consisted of DNA sequences from three different samples at the early stage of the sake brewing process and contained no sequences from Narimasa Sake Brewery. Thus, this largest group was specific to Shiraki Tsunesuke Sake Brewery. Group 2 consisted of 18 sequences from three different samples, with no sequences from Narimasa Sake Brewery (Figure 2). The isolates of Groups 1 and 2 were obtained from both cultures at 15 °C and 30 °C (Table 3). The isolation frequency bias was not detected (p > 0.05, chi-square test) between different sampling dates of cultures at 15 °C and 30 °C in each group and between different groups of cultures at 15 °C and 30 °C. An ethanol resistance test showed that isolate C1 belonging to Group 1 and isolate a12 belonging to Group 2 had an ethanol resistance to 20% ethanol (Figure 3).
Figure 2.
Phylogenetic relationships among 16S rRNA gene sequences of the 92 isolates from Shiraki Tsunesuke Sake Brewery, 6 isolates from Narimasa Sake Brewery, and 10 known species of Bacillus. A neighbor-joining tree was constructed with 1000 bootstrap replicates using MEGA X [18]. The evolutionary distances were computed using the maximum composite likelihood method [19], and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated (complete deletion option). There were total 392 positions in the final dataset.
Table 3.
Number of isolates in each group.
| Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | Group 6 | Group 7 | |
|---|---|---|---|---|---|---|---|
| From sample 2019.1.30 | 6 | 0 | 0 | 0 | 0 | 0 | 0 |
| From sample 2021.1.26; Culture at 30 °C |
8 | 3 | 3 | 0 | 0 | 0 | 0 |
| From sample 2021.1.26; Culture at 15 °C |
9 | 4 | 0 | 0 | 1 | 0 | 0 |
| From sample 2021.1.28; Culture at 30 °C |
11 | 1 | 3 | 2 | 1 | 1 | 0 |
| From sample 2021.1.28; Culture at 15 °C |
5 | 2 | 2 | 0 | 2 | 0 | 0 |
| From sample 2021.2.9; Culture at 30 °C |
6 | 3 | 1 | 4 | 0 | 0 | 0 |
| From sample 2021.2.9; Culture at 15 °C |
5 | 5 | 2 | 1 | 0 | 0 | 1 |
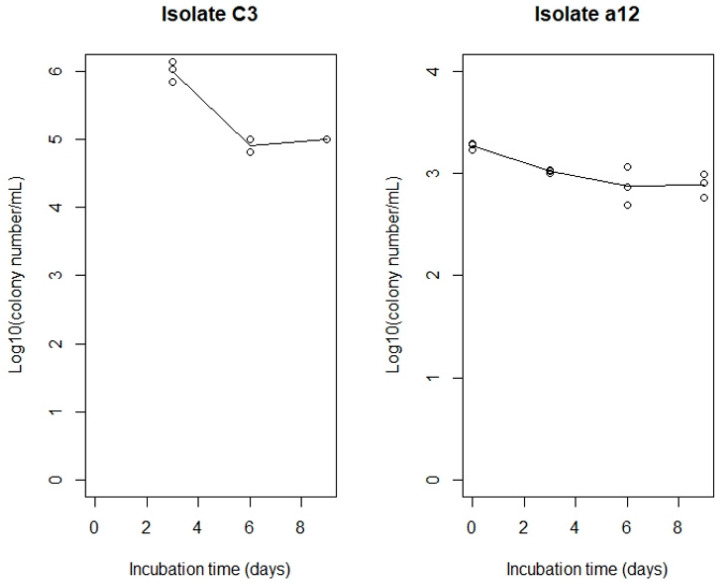
Figure 3.
Growth of kuratsuki Bacillus isolates in TGY including 20% ethanol. The isolates C3 and a12 belonging to Groups 1 and 2, respectively, were statically cultured in TGY including 20% ethanol at 30 °C.
Group 3 consisted of 11 sequences from three different samples (Figure 2). This group included one sequence from Narimasa Sake Brewery, indicating that it was not specific to Shiraki Tsunesuke Sake Brewery. Group 4 consisted of seven sequences from two different samples, with no sequences from Narimasa Sake Brewery (Figure 2). Group 5 consisted of four sequences from two different samples (Figure 2). This group included five sequences from Narimasa Sake Brewery, indicating that this group was not specific to Shiraki Tsunesuke Sake Brewery. In addition, Groups 6 and 7 were located alone (Figure 2).
Group 1 clustered with B. safensis, B. pumilus, B. zhangzhouensis, and isolate B6 (Group 6) with 87% support of bootstrap (Figure 2). Groups 2, 3, and 5 were clustered with B. aryabhattai, B. flexus, and B. megaterium with 93% support of bootstrap (Figure 2). Group 4 was clustered with B. subtilis and B. velezensis with 97% support of bootstrap (Figure 2). Group 7 was clustered with B. endophyticus with 99% support of bootstrap (Figure 2).
4. Discussion
Although Bacillus is frequently detected in the sake brewery environments [2], it was surprising that all 92 isolates from four different samples at Shiraki Tsunesuke Sake Brewery belong to the genus Bacillus. In Narimasa Sake Brewery, 46 isolates from 6 different samples belong to the 6 different genera: Kocuria (23 isolates), Staphylococcus (12 isolates), Bacillus (6 isolates), Leifsonia (2 isolates), Microbacterium (2 isolates), and Enterococcus (1 isolate) [6,12]. This strongly suggests that different sake breweries have different bacteria compositions entering the brewing process. Narimasa Sake Brewery has kuratsuki Kocuria at the different species level [6,12,13], leading us to hypothesize that Shiraki Tsunesuke Sake Brewery may also have kuratsuki Bacillus at the different species level. Bacillus species have both positive and negative effects on drinks and foods [20,21,22,23,24].
We believe that the Bacillus isolates of Groups 1 and 2 are kuratsuiki bacteria in Shiraki Tsunesuke Sake Brewery because these Bacillus isolates are specific to this sake brewery. In particular, the isolates of Group 1 are kuratsuki bacteria because this group includes the Bacillus strains that were isolated in 2019 at late sake production process, which has a high ethanol concentration (>15% of ethanol). Considering that the same Bacillus isolates were collected from the different samples with different sampling year, these isolates may have inhabited the Shiraki Tsunesuke Sake Brewery and entered the annual sake brewing process. The Bacillus isolates of Group 1 have a high ethanol tolerance and survive at the late stage of the sake brewing process. In contrast, Group 2 does not include the Bacillus isolates in 2019. This suggested that the Bacillus isolates of Group 2 might be less ethanol-tolerant compared with those of Group 1. However, the isolates of Group 2 had a high ethanol tolerance (Figure 3).
Group 3 consists of isolates from different three samples as well as Groups 1 and 2 (Table 3). However, Group 3 includes the isolate MB1023_1 from Narimasa Sake Brewery (Figure 1), indicating that this group is not specific to Shiraki Tsunesuke Sake Brewery. It is possible that different sake breweries have common kuratsuki bacteria. However, kuratsuki bacteria undergo genome modification during the sake brewing to adapt to the brewery environments [12]. For example, although the optimal growth temperature of the genus Kocuria is generally 30–37 °C, the kuratsuki Kocuria isolates in Narimasa Sake Brewery is 15 °C, which is the same temperature during the sake brewing process [12]. Even if the two different sake breweries have a common kuratsuki bacterial genus Bacillus, those genomes should differ at the species or strain level. To elucidate it, the genome sequence analysis is necessary.
Groups 4–7 do not consist of isolates from all of the different three samples (Table 3). In addition, from the viewpoint of detection frequency, the detection frequency of isolates in these groups is so low that these isolates have a low possibility of kuratsuki bacteria in Shiraki Tsunesuke Sake Brewery. Because kuratsuki bacteria have inhabited a sake brewery for a long time and entered every sake brewing process, the detection frequency of kuratsuki bacteria is usually higher than that of other bacteria entered in the sake brewing process.
Groups 3 and 5 consist of isolates from both Narimasa and Shiraki Tsunesuke breweries. It is uncertain if these isolates are also kuratsuki Bacillus. Nevertheless, these isolates may have an affinity to sake brewing. Thus, these Bacillus isolates may influence metabolism of S. cerevisiae and/or other microorganisms. If so, both kuratsuki bacteria and other bacteria may affect flavor and taste specific to each sake brewery. To elucidate how the Bacillus isolates affect chemical components in sake, further research is warranted.
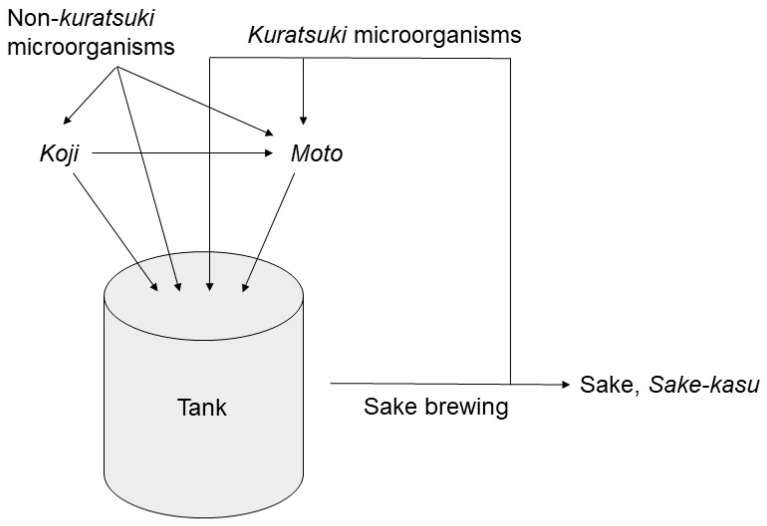
This study shows that different sake breweries have distinct kuratsuki bacteria; Narimasa Sake Brewery has kuratsuki Kocuria and Shiraki Tsunesuke Sake Brewery has kuratsuki Bacillus. The genera Bacillus and Kocuria differ phylogenetically, and these different bacteria are specific to each sake brewery, having evolved in each brewery. This means that kuratsuki bacteria differ from other environmental microorganisms (Figure 4), in that they have a life cycle in the sake brewery and enter the sake brewing process. In contrast, environmental microorganisms enter the sake brewing process by chance. We have a hypothesis that kuratsuki bacteria were derived from the environmental microorganisms and they have adapted to the sake brewing conditions by continuing to enter the sake production process. Kuratsuki bacteria may have a key role in the interaction among microorganisms during the sake production process [13].
Figure 4.
Difference between kuratsuki and non-kuratsuki microorganisms in the sake brewing process. Koji contains Aspergillus oryzae. Moto contains A. oryzae and Saccharomyces cerevisiae. S. cerevisiae in moto produces ethanol during the sake brewing process.
It is important that kuratsuki bacteria do not consist of single lineage but multiple lineages. This strongly suggests that kuratsuki bacteria evolved in sake tank during the sake brewing (Figure 4) and then the bacteria distributed into the environment of the sake brewery. The kuratsuki bacteria have continued to enter and distribute repeatedly for a long time. In each sake brewing, kuratsuki bacteria may be selected from the multiple lineages. Considering the evolution of kuratsuki bacteria, their genome analyses are important. In kuratsuki Kocuria isolates, plasmid and transposon were identified and genome modification between the different isolates were suggested [12]. Kuratsuki yeasts also probably have multiple lineages. However, at present, most sake breweries do not use kuratsuki yeasts but selected sake yeasts (Kyokai yeasts) that are managed by the Brewery Society of Japan. Each Kyokai yeast consists of a single lineage. It is possible that some Kyokai yeasts are not suitable for some sake brewery environments.
5. Conclusions
Microorganisms play an important role in the production of flavor and taste, and influence the sake quality. Although bacteria die at the final stage of sake production, bacteria in sake are alive and grow temporarily during the sake production process. Kuratsuki bacteria are not any contaminated bacteria, which are always entered in sake brewing. To our knowledge, although research on kuratsuki yeasts has been carried out, research on kuratsuki bacteria has not been performed except for lactic acid bacteria. We are changing the characteristics of sake and will produce a unique sake with different tastes and flavors by exchanging the kuratsuki bacteria from different sake breweries during the production process.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9081760/s1, Supplementary File S1. Mega file used in the phylogenetic analysis.
Author Contributions
H.N. designed the experiments. E.K. performed the experiments. K.T. and Y.S. provided the samples. H.N. analyzed the data and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by JSPS KAKENHI Grant Number 21H02109 (to H.N.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in Supplementary File S1.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kitagaki H., Kitamoto K. Breeding research on sake yeasts in Japan: History, recent technological advances, and future perspectives. Ann. Rev. Food Sci. Technol. 2013;4:215–235. doi: 10.1146/annurev-food-030212-182545. [DOI] [PubMed] [Google Scholar]
- 2.Bokulich N.A., Ohta M., Lee M., Mills D.A. Indigenous bacteria and fungi drive traditional kimoto sake fermentations. Appl. Environ. Microbiol. 2014;80:5522–5529. doi: 10.1128/AEM.00663-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koyanagi T., Nakagawa A., Kiyohara M., Matsui H., Tujii A., Barla F., Take H., Katsuyama Y., Tokuda K., Nakamura S., et al. Tracing microbiota changes in yamahai-moto, the traditional Japanese sake starter. Biosci. Biotechnol. Biochem. 2016;80:399–406. doi: 10.1080/09168451.2015.1095067. [DOI] [PubMed] [Google Scholar]
- 4.Terasaki M., Fukuyama A., Takahashi Y., Yamada M., Nishida H. Bacterial DNA detected in Japanese rice wines and the fermentation starters. Curr. Microbiol. 2017;74:1432–1437. doi: 10.1007/s00284-017-1337-4. [DOI] [PubMed] [Google Scholar]
- 5.Terasaki M., Miyagawa S., Yamada M., Nishida H. Detection of bacterial DNA during the process of sake production using sokujo-moto. Curr. Microbiol. 2018;75:874–879. doi: 10.1007/s00284-018-1460-x. [DOI] [PubMed] [Google Scholar]
- 6.Terasaki M., Nishida H. Bacterial DNA diversity among clear and cloudy sakes, and Sake-kasu. Open Bioinform. J. 2020;13:74–82. doi: 10.2174/1875036202013010074. [DOI] [Google Scholar]
- 7.Taniguchi M., Ishiyama Y., Takata T., Nakanishi T., Kaneoke M., Watanabe K., Yanagida F., Chen Y.S., Kouya T., Tanaka T. Growth-inhibition of hiochi bacteria in namazake (raw sake) by bacteriocins from lactic acid bacteria. J. Biosci. Bioeng. 2010;109:570–575. doi: 10.1016/j.jbiosc.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe D., Kumano M., Sugimoto Y., Ito M., Ohashi M., Sunada K., Takahashi T., Yamada T., Takagi H. Metabolic switching of sake yeast by kimoto lactic acid bacteria through the [GAR+] non-genetic element. J. Biosci. Bioeng. 2018;126:624–629. doi: 10.1016/j.jbiosc.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Kitahara K., Kaneko T., Goto O. Taxonomic studies of the hiochi-bacteria, specific saprophytes of sake. I. Isolation and grouping of bacterial strains. J. Gen. Appl. Microbiol. 1957;3:102–110. doi: 10.2323/jgam.3.102. [DOI] [Google Scholar]
- 10.Kitahara K., Kaneko T., Goto O. Taxonomic studies of the hiochi-bacteria, specific saprophytes of sake. II. Identification and classification of hiochi-bacteria. J. Gen. Appl. Microbiol. 1957;3:111–120. doi: 10.2323/jgam.3.111. [DOI] [Google Scholar]
- 11.Suzuki K., Asano S., Iijima K., Kitamoto K. Sake and beer spoilage lactic acid bacteria—A review. J. Inst. Brew. 2008;114:209–223. doi: 10.1002/j.2050-0416.2008.tb00331.x. [DOI] [Google Scholar]
- 12.Terasaki M., Kimura Y., Yamada M., Nishida H. Genomic information of Kocuria isolates from sake brewing process. AIMS Microbiol. 2021;7:114–123. doi: 10.3934/microbiol.2021008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishida H. Sake brewing and bacteria inhabiting sake breweries. Front. Microbiol. 2021;12:602380. doi: 10.3389/fmicb.2021.602380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stackebrandt E., Koch C., Gvozdiak O., Schumann P. Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov., Kytococcus gen. nov., Dermacoccus gen. nov., and Micrococcus Cohn 1872 gen. emend. Int. J. Syst. Evol. Microbiol. 1995;45:682–692. doi: 10.1099/00207713-45-4-682. [DOI] [PubMed] [Google Scholar]
- 15.Terasaki M., Inoue A., Kanamoto E., Yoshida S., Yamada M., Toda H., Nishida H. Co-cultivation of sake yeast and Kocuria isolates from the sake brewing process. FEMS Microbiol. Lett. 2021;368:fnab053. doi: 10.1093/femsle/fnab053. [DOI] [PubMed] [Google Scholar]
- 16.Tsuji A., Kozawa M., Tokuda K., Enomoto T., Koyanagi T. Robust domination of Lactobacillus sakei in microbiota during traditional Japanese sake starter yamahai-moto fermentation and the accompanying changes in metabolites. Curr. Microbiol. 2018;75:1498–1505. doi: 10.1007/s00284-018-1551-8. [DOI] [PubMed] [Google Scholar]
- 17.Akaike M., Miyagawa H., Kimura Y., Terasaki M., Kusaba Y., Kitagaki H., Nishida H. Chemical and bacterial components in sake and sake production process. Curr. Microbiol. 2020;77:632–637. doi: 10.1007/s00284-019-01718-4. [DOI] [PubMed] [Google Scholar]
- 18.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetic analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura K., Nei M., Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granum P.E., Lund T. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 1997;157:223–228. doi: 10.1111/j.1574-6968.1997.tb12776.x. [DOI] [PubMed] [Google Scholar]
- 21.Ter Beek A., Brul S. To kill or not to kill Bacilli: Opportunities for food biotechnology. Curr. Opin. Biotechnol. 2010;21:168–174. doi: 10.1016/j.copbio.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Ito T., Konno M., Shimura Y., Watanabe S., Takahashi H., Hashizume K. Formation of guaiacol by spoilage bacteria from vanillic acid, a product of rice koji cultivation, in Japanese sake brewing. J. Agric. Food Chem. 2016;64:4599–4605. doi: 10.1021/acs.jafc.6b01031. [DOI] [PubMed] [Google Scholar]
- 23.Elshaghabee F.M.F., Rokana N., Gulhane R.D., Sharma C., Panwar H. Bacillus as potential probiotics: Status, concerns, and future perspectives. Front. Microbiol. 2017;8:1490. doi: 10.3389/fmicb.2017.01490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi M., Kita Y., Ninakami R., Mukai N. Growth characteristics of Bacillus cereus in sake and during its manufacture. J. Food Prot. 2021;84:213–219. doi: 10.4315/JFP-20-196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are openly available in Supplementary File S1.