Abstract
The nuclear factor of activated T cells (NFAT) transcription factor is implicated in expression of the cytokine interleukin-2 (IL-2). Binding sites for NFAT are located in the IL-2 promoter. Furthermore, pharmacological studies demonstrate that the drug cyclosporin A inhibits both NFAT activation and IL-2 expression. However, targeted disruption of the NFAT1 and NFAT2 genes in mice does not cause decreased IL-2 secretion. The role of NFAT in IL-2 gene expression is therefore unclear. Here we report the construction of a dominant-negative NFAT mutant (dnNFAT) that selectively inhibits NFAT-mediated gene expression. The inhibitory effect of dnNFAT is mediated by suppression of activation-induced nuclear translocation of NFAT. Expression of dnNFAT in cultured T cells caused inhibition of IL-2 promoter activity and decreased expression of IL-2 protein. Similarly, expression of dnNFAT in transgenic mice also caused decreased IL-2 gene expression. These data demonstrate that NFAT is a critical component of the signaling pathway that regulates IL-2 expression.
The nuclear factor of activated T cells (NFAT) group of proteins were first characterized as transcription factors that bind to the interleukin-2 (IL-2) promoter (13, 22, 40, 42). The NFAT transcription factor consists of two components: a cytoplasmic Rel domain protein (NFAT family member) and a nuclear component consisting of activating protein 1 (AP-1) transcription factors (Fos- and Jun-related proteins) and probably other transcription factors (reviewed in reference 38). Four members of the NFAT group have been identified: NFAT1 (NFATp/NFATc2), NFAT2 (NFATc/NFATc1), NFAT3, and NFAT4 (NFATx/NFATc3) (17, 19, 27, 29, 31). NFAT1 and NFAT2 are expressed predominantly in lymphoid tissues (thymocytes, T cells, B cells, mast cells and NK cells), but NFAT2 is also expressed in muscle cells. NFAT4 is expressed mainly in the thymus and NFAT3 is expressed primarily in nonlymphoid tissues. The major NFAT proteins expressed in peripheral T cells that produce IL-2 correspond to the isoforms NFAT1 and NFAT2.
While NFAT is thought to be important for IL-2 gene expression, recent studies demonstrate that NFAT may also contribute to the expression of other cytokines, including IL-3, IL-4, IL-5, granulocyte-macrophage colony-stimulating factor, and tumor necrosis factor (8, 10, 12, 28, 43, 47, 48). NFAT is also implicated in the regulation of expression of the cell surface molecules Fas ligand and CD40 ligand (25, 50).
NFAT activation requires signals that are initiated by Ca2+ and protein kinase C (PKC) (38). Activators of PKC, such as phorbol 12-myristate 13-acetate (PMA), induce the synthesis of the nuclear component (e.g., Fos and Jun family proteins). A sustained increase in intracellular Ca2+ is required to activate calcineurin, a Ca2+-dependent phosphatase (49). Calcineurin dephosphorylates NFAT proteins and induces their translocation from the cytoplasm to the nucleus (9). The immunosuppressive drugs cyclosporin A and FK506 inhibit calcineurin and, thereby, nuclear translocation of NFAT (14, 37). Nuclear NFAT binds to specific DNA elements and activates transcription. These processes are mediated, in part, by the interaction of NFAT with AP-1 proteins. Some NFAT isoforms may also interact with other transcription factors, including GATA-4 (30).
In T cells, NFAT is activated by the engagement of the antigen-specific T-cell receptor (44). NFAT activity can also be induced in other cell types in response to extracellular stimulation. For example, ligation of surface immunoglobulins or CD40 receptors in combination with IL-4 causes NFAT activation in B cells (20). Stimulation of NK cells by CD16 ligands also causes NFAT activation (1). However, although NFAT is activated in response to these stimuli, the contribution of NFAT-mediated transcription to the expression of cytokine genes and the specific role of NFAT in the immune response remains unclear.
Recent studies designed to examine the role of the NFAT transcription factor have used homologous recombination to prepare mice that are defective in NFAT expression. Mice that are deficient in the expression of NFAT1 and NFAT2 have been reported (18, 24, 35, 41, 51, 53). NFAT1-deficient mice show increased proliferation and dysregulation of IL-4 gene expression (18, 24, 51). In contrast, IL-2 gene expression was not affected in these mice. More recently, it has been reported that mice deficient in NFAT2 show reduced IL-4 production but normal or increased IL-2 secretion (35, 53). These data do not provide evidence for a clear role of NFAT in the regulation of IL-2 gene expression. The potential redundancy of NFAT isoforms and the possible compensation by other NFAT isoforms in these knockout mice complicates the interpretation of the phenotypes that have been reported.
The purpose of this study was to examine the role of NFAT in the expression of IL-2. Since IL-2 expression is either unaffected or enhanced in mice without NFAT1 or NFAT2, we used a different approach to test whether NFAT activity is required for IL-2 gene expression. We report the construction of a mutated NFAT protein that dominantly inhibits NFAT function in vivo. This dominant-negative NFAT protein (dnNFAT) acts as a strong inhibitor of IL-2 expression. Thus, NFAT activity is required for IL-2 expression.
MATERIALS AND METHODS
Cell culture and reagents.
BHK fibroblasts, Jurkat T cells, and COS cells were cultured in minimal essential medium, RPMI 1640, and Dulbecco modified Eagle medium, respectively, supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (Life Technologies Inc.). Ionomycin and PMA were obtained from Sigma.
Plasmids.
Mammalian expression vectors for NFAT and luciferase reporter plasmids (NFAT and IL-2 promoter) were provided by T. Hoey (19). The green fluorescence protein (GFP) expression vector pCMV-GFP was provided by D. Kerr (University of Vermont). The calcineurin expression vector was obtained from T. Soderling (32). The GAL4-luciferase, AP-1–luciferase, NF-κB–luciferase, and pRSV β-galactosidase reporter plasmids and the expression vectors for Flag-tagged NFAT and GAL4-NFAT have been described previously (7, 33, 39). The Rel homology domain of NFAT4 (NFAT4 Rel; amino acids 365 to 708) was subcloned with an NH2-terminal hemagglutinin epitope tag in the expression vector pCDNA3 (Invitrogen Inc.). Deletion and point mutations were constructed by PCR and sequenced with an Applied Biosystems machine.
Luciferase reporter gene assays.
BHK cells were transfected by using Lipofectamine as specified by the manufacturer (Life Technologies Inc.). A full-length NFAT expression vector (0.3 μg) was cotransfected with the NFAT-luciferase reporter plasmid (0.2 μg) and the pRSV β-galactosidase control plasmid (0.2 μg). Various amounts (0.1 to 0.3 μg) of expression vectors for NFAT deletion mutants were cotransfected. Jurkat T cells (5 × 106) were transfected by electroporation (1,080 μF and 250 V; Life Technologies Inc.). Luciferase reporter plasmids (5 μg) and the pRSV β-galactosidase control plasmid (5 μg) were cotransfected together with an NFAT expression vector (1 to 10 μg). The total amount of DNA was adjusted to 20 μg with plasmid pCDNA3. Luciferase activity was measured 48 h after transfection. Unless otherwise indicated, cells were stimulated with 2 μM ionomycin and 100 nM PMA for 16 h prior to harvesting. The data are presented as luciferase activity/β-galactosidase activity (mean ± standard deviation SD [n = 3]).
Immunoblot analysis.
COS cells were transfected with NFAT expression vectors by the Lipofectamine method (Life Technologies Inc.). The cells were harvested in Triton-lysis buffer (20 mM Tris [pH 7.4], 137 mM NaCl, 2 mM EDTA, 1% Triton X-100, 25 mM β-glycerophosphate, 1 mM sodium vanadate, 2 mM sodium pyrophosphate, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml) 48 h after transfection. Cell extracts were separated by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Millipore Inc.). The epitope-tagged NFAT proteins were detected with a monoclonal antibody (MAb) to Flag (Sigma) and enhanced chemiluminescence (Kirkegaard & Perry Laboratories).
Immunofluorescence analysis.
Transfected BHK cells were treated without and with ionomycin (30 min) prior to fixation. Immunofluorescence analysis was performed as described elsewhere (7). NFAT1 was detected with a rabbit polyclonal antibody (1:200; Upstate Biotechnology), and NFAT2 was detected with a mouse MAb (1:200; Affinity Bioreagents). The secondary antibody was either Texas red-conjugated anti-mouse or anti-rabbit immunoglobulin antibody (1:100; Jackson Immunoresearch). Nuclei were visualized with 4,6-diamidino-2-phenylindole (DAPI; Sigma).
IL-2 expression assays.
Jurkat T cells (5 × 106) were transfected with expression vectors for NFAT (20 μg) and GFP (5 μg). GFP-positive and GFP-negative cells were selected by cell sorting (Becton Dickinson) and treated without or with ionomycin (2 μM) and PMA (100 nM) for 20 h. Culture supernatants were collected and IL-2 was assayed with CTL.L cells as described previously (15).
Intracellular staining for IL-2 was performed with reagents from Pharmingen Inc. according to the manufacturer’s protocol. Jurkat T cells were transfected with expression vectors for NFAT and GFP. These cells were incubated for 20 h without and with PMA and ionomycin. Four hours prior to harvesting, the cells were incubated with monensin (2 μM) and subsequently fixed with paraformaldehyde (4%). Rat preimmune antibody (1 μg/ml) was used to block nonspecific binding. Phycoerythrin-conjugated rat anti-human IL-2 antibody was used for staining. The fluorescence intensity of GFP and phycoerythrin was measured by flow cytometry (Becton Dickinson).
Mice.
The DNA fragment encoding Flag-tagged dnNFAT (NFAT3 amino acids 1 to 130) was subcloned downstream of the proximal lck promoter, and transgenic mice were generated as described previously (39). Three expressing positive founders lines were established and backcrossed onto B10.BR/SGSNJ mice (The Jackson Laboratory). Thymocytes isolated from transgenic mouse lines 4 and 8 were used for further studies. Expression of dnNFAT was confirmed by immunoblot analysis of thymocyte lysates using MAb M2, specific to the Flag epitope. After 24 h of activation, IL-2 production by these thymocytes (2 × 106 cells/ml) was determined by the CTL.L assay (15).
RESULTS
Construction of dnNFAT.
Functional studies have identified a transcription activation domain (TAD) in the NH2-terminal region of NFAT (26). Adjacent to the TAD is a conserved NFAT homology region that is similar in all members of the NFAT group of transcription factors (17, 19, 27). This homology region is highly phosphorylated and includes the sites of regulatory phosphorylation that are substrates for the phosphatase calcineurin (38). The COOH-terminal region of NFAT proteins includes a Rel homology domain which mediates DNA binding (21). Studies of other transcription factors indicate that the DNA binding domain can act as a dominant-negative inhibitor by competition for DNA binding (5). This approach to create a dominant-negative transcription factor has not been successful for NFAT (data not shown). The lack of success is caused by the finding that the DNA binding Rel homology domain of NFAT activates NFAT-dependent reporter gene expression (26). This may be accounted for, in part, by the interaction of the Rel homology domain with AP-1 complexes. Indeed, structural analysis indicates that the Rel homology domain of NFAT is sufficient for complex formation with AP-1 on DNA (6, 54).
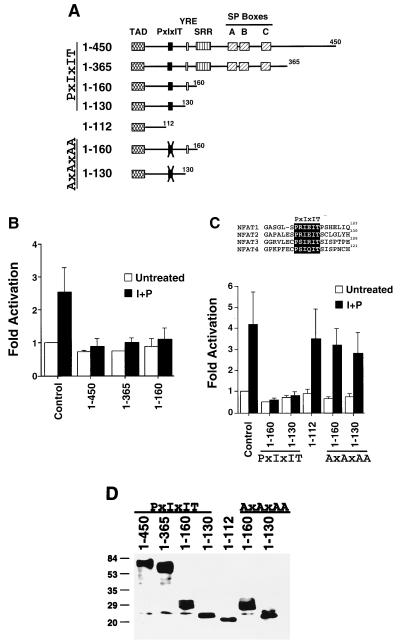
As the DNA binding domain of NFAT does not appear to function as a dominant inhibitor of NFAT function, we examined whether the conserved NH2-terminal NFAT homology domain could interfere with NFAT-mediated transcription. These experiments were performed by expression of a truncated protein encoding the NFAT homology domain of NFAT3 (residues 1 to 450) in BHK cells (Fig. 1A). The transcription activity of NFAT2 was measured in cotransfection assays using an NFAT-luciferase reporter plasmid. This reporter plasmid contains three copies of an NFAT–AP-1 composite element derived from the IL-2 promoter. Treatment with PMA and ionomycin induced NFAT2 transcriptional activity (Fig. 1B). In the absence of NFAT2, transcription activity was not observed in either the absence or the presence of the NH2-terminal NFAT homology domain (data not shown). In contrast, expression of the NH2-terminal NFAT homology domain (residues 1 to 450) inhibited transcription mediated by NFAT2 (Fig. 1B). These data indicated that the NH2-terminal NFAT homology domain interferes with NFAT-mediated transcription.
FIG. 1.
The PxIxIT domain acts as a dominant inhibitor of NFAT transcription activity. (A) Schematic representation of the NH2-terminal region of NFAT transcription factors. The conserved SP boxes (A, B, and C), TAD, PxIxIT motif, YRE motif, and SRR are indicated. Mutation of the PxIxIT motif by replacement of the Pro, Ile, and Thr residues with Ala (AxAxAA) is indicated by a cross. The deletion mutations correspond to the NFAT3 isoform. (B) Expression of the NH2-terminal NFAT homology region inhibits NFAT-mediated transcription activity. Various NFAT3 deletion mutants (residues 1 to 450, 1 to 365, and 1 to 160) were coexpressed with full-length NFAT2 and an NFAT-luciferase reporter plasmid in BHK cells. Luciferase activity was measured in cultures incubated without (Untreated) or with ionomycin (2 μM) and PMA (100 nM) (I+P). The data are presented as fold activation compared to an untreated control. (C) The PxIxIT motif is responsible for the dominant-negative activity of the NH2-terminal NFAT homology region. The effects of NFAT3 deletion mutants (residues 1 to 160, 1 to 130, and 1 to 112) on NFAT2-mediated transcription activity were examined by using an NFAT-luciferase reporter plasmid in BHK cells. The effect of mutation of the PxIxIT motif by replacement of the Pro, Ile, and Thr residues with Ala (AxAxAA) was investigated. Luciferase activity was measured in cultures incubated without (Untreated) or with ionomycin (2 μM) and PMA (100 nM) (I+P). The data are presented as fold activation compared to an untreated control. (D) Epitope-tagged Flag-NFAT3 proteins were expressed in COS cells, and detected by protein immunoblotting of cell lysates with MAb M5, specific to the Flag epitope (Sigma). Sizes are indicated in kilodaltons.
The conserved NH2-terminal NFAT homology domain is formed by distinct subregions (Fig. 1A). These include the Ser-rich region (SRR) and three conserved Ser-Pro repeats (SP boxes A, B, and C). The SP boxes represent major sites of interaction of NFAT with calcineurin in vitro (7), and sites of NFAT phosphorylation in vivo have been identified in the SRR (3, 7, 55). To test whether these conserved subregions (SRR and SP boxes) are required for the inhibitory function of the NFAT homology domain, we generated a series of truncated NFAT proteins (Fig. 1A). Removal of the COOH-terminal portion of the SP boxes (residues 365 to 450) did not affect the inhibitory activity (Fig. 1B), nor did truncation at residue 160, which deletes the SP boxes and the adjacent SRR (Fig. 1B). These data indicate that neither the SRR nor the SP boxes are required for the inhibitory activity of the NH2-terminal NFAT homology region.
The region identified that confers inhibitory transcription activity (residues 1 to 160) includes the TAD, the conserved Pro-Xaa-Ile-Xaa-Ile-Thr (PxIxIT) box (residues 114 to 119), and the Tyr-Arg-Glu (YRE) box (residues 155 to 157) (Fig. 1A). To examine the role of these conserved subregions, we performed further deletion analysis. Truncation at residue 130 removes the YRE box but does not alter the inhibitory activity of the NH2-terminal NFAT homology region (Fig. 1C). In contrast, truncation at residue 112, which deletes the PxIxIT box, abolished the inhibitory activity (Fig. 1C). Control experiments demonstrated that these truncated NFAT proteins were expressed at similar levels (Fig. 1D). Thus, it appears that the conserved PxIxIT box is required for the dominant-negative function of the NH2-terminal NFAT homology region. To test this hypothesis, we replaced the conserved Pro, Ile, and Thr residues in the PxIxIT motif with Ala residues (Fig. 1A, AxAxAA). This mutation eliminated the inhibitory activity of the NH2-terminal NFAT homology region (Fig. 1C). These data indicate that the PxIxIT box mediates the dominant-negative action of the NH2-terminal NFAT homology region.
The PxIxIT box selectively inhibits NFAT transcription activity.
The NH2-terminal NFAT homology domain is conserved in the four members of the NFAT group of transcription factors (38). We therefore reasoned that the dominant-negative action of the PxIxIT box may inhibit transcription activity of all members of this group. To test this hypothesis, we examined the transcription activity of NFAT1, NFAT2, NFAT3, and NFAT4 in a cotransfection assay with an NFAT-luciferase reporter gene (Fig. 2). Transcription activity mediated by each of these NFAT proteins was inhibited by coexpression with the PxIxIT box (NFAT3 residues 1 to 130). In contrast, expression of the Ala-substituted PxIxIT box did not inhibit transcription activity. These data indicate that the PxIxIT box can function as a dominant-negative NFAT mutant that suppresses transcription mediated by NFAT transcription factors.
FIG. 2.
dnNFAT inhibits transcription activity of all four NFAT isoforms. NFAT proteins were expressed in BHK cells together with dnNFAT (NFAT3 amino acids 1 to 130). The effect of mutation of the PxIxIT motif by replacement of the Pro, Ile, and Thr residues with Ala (AxAxAA) was investigated. Cotransfection assays in BHK cells using an NFAT-luciferase reporter plasmid and NFAT1 (A), NFAT2 (B), NFAT3 (C), and NFAT4 (D) were performed. Luciferase activity was measured in cultures treated without (open bar) and with (filled bar) PMA and ionomycin. The data are presented as fold activation compared to an untreated control.
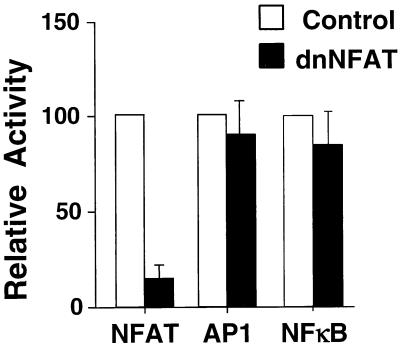
It was possible that dnNFAT could inhibit transcription activity nonspecifically. We therefore examined the effect of dnNFAT on transcription activity mediated by AP-1 and NF-κB. dnNFAT did not inhibit AP-1- or NF-κB-dependent reporter gene expression in cotransfection assays but did cause selective inhibition of NFAT transcription activity (Fig. 3).
FIG. 3.
AP-1 and NF-κB transcription activities are not inhibited by dnNFAT. NFAT, AP-1, and NF-κB transcription activities were measured by using luciferase reporter plasmids cotransfected in Jurkat T cells without (Control) and with dnNFAT. Luciferase activity was measured in cultures incubated with ionomycin (2 μM) and PMA (100 nM). The data are presented as relative percentage activity compared to a control without dnNFAT.
Mechanism of the inhibitory activity of dnNFAT.
Previous studies indicated that both the NH2- and COOH-terminal regions of NFAT proteins can mediate transcription activity (26). The NH2-terminal region of NFAT acts as a strong transactivation domain (Fig. 1A). In addition, the COOH-terminal region, which includes the Rel homology domain, can also mediate transactivation which may be caused, in part, by the association of the NFAT Rel domain with other transcription factors, such as AP-1 (4, 23, 52). To gain insight into the mechanism by which dnNFAT inhibits NFAT transcription activity, we examined the effect of dnNFAT on the transcription activity mediated by the NH2- and COOH-terminal regions of NFAT. Interestingly, while dnNFAT did inhibit transcription activity of full-length NFAT, no significant inhibition of transcription activity mediated by the COOH-terminal region of NFAT was detected (Fig. 4A). The absence of an effect of dnNFAT on transcription mediated by the COOH-terminal region of NFAT suggests that the inhibition of NFAT transcription activity requires the NH2-terminal NFAT homology domain. To test this hypothesis, we fused the NH2-terminal region of NFAT4 (residues 1 to 207) to the GAL4 DNA binding domain and examined the effect of dnNFAT on transcription activation in cotransfection assays with a GAL4-luciferase reporter plasmid. dnNFAT was found to inhibit the transcription activity of this GAL4-NFAT4 fusion protein (Fig. 4B). These data indicate that the NH2-terminal region of NFAT is both necessary and sufficient for the response of the NFAT transcription factor to the inhibitory action of dnNFAT.
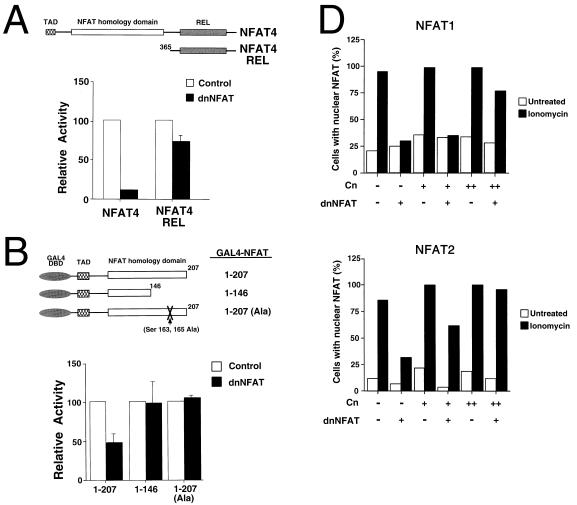
FIG. 4.
Mechanism of dominant inhibitory activity of dnNFAT. (A) Transcription activity mediated by the COOH-terminal region of NFAT is not affected by dnNFAT. Full-length NFAT4 and the NFAT4 COOH-terminal region (NFAT4 Rel; residues 365 to 708) were expressed together with an NFAT-luciferase reporter plasmid in BHK cells without (Control) and with dnNFAT. Luciferase activity was measured in cultures incubated with ionomycin (2 μM) and PMA (100 nM). The data are presented as relative percentage activity compared to a control without dnNFAT. (B) Transcription activity mediated by the NH2-terminal activation domain of NFAT is not affected by dnNFAT. GAL4-NFAT4 fusion proteins were expressed in BHK cells together with a GAL4-luciferase reporter plasmid and dnNFAT. Luciferase activity was measured in cultures incubated with ionomycin (2 μM) and PMA (100 nM). The data are presented as relative percentage activity compared to a control without dnNFAT. The effect of replacement of the phosphorylation sites Ser-163 and Ser-165 with Ala is shown. DBD, DNA binding domain. (C) Regulation of the subcellular distribution of NFAT proteins by dnNFAT. NFAT1 and NFAT2 were coexpressed with dnNFAT in BHK cells. Immunofluorescence analysis was performed on cells treated without or with ionomycin (2 μM, 30 min). NFAT proteins (red) and the nucleus (blue) were visualized. Arrowheads indicate the nuclei of cells expressing transfected proteins. (D) Overexpression of calcineurin opposed the inhibitory effect of dnNFAT. Various amounts of calcineurin expression vector (50 and 100 ng) were coexpressed with dnNFAT in BHK cells. Immunofluorescence analysis was performed to examine the subcellular distribution of NFAT1 and NFAT2 proteins in the absence or presence of ionomycin (2 μM, 30 min). One hundred transfected cells were examined. The percentage of cells with NFAT in the nucleus is presented.
It was possible that dnNFAT interferes with the mechanism of transcription activation mediated by NFAT. However, the results of deletion analysis of the NFAT4 NH2-terminal region in the GAL4 fusion protein assay did not support this hypothesis (Fig. 4B). While dnNFAT inhibited the transcription activity of GAL4-NFAT4 (residues 1 to 207), dnNFAT did not inhibit transcription activity of GAL4-NFAT4 (residues 1 to 146). Since NFAT4 residues 1 to 146 include the NH2-terminal TAD, the absence of inhibition by dnNFAT demonstrates that dnNFAT does not act by directly interfering with transcription activation.
It appears that the inhibitory effect of dnNFAT is not mediated by direct inhibition of transcription activation (Fig. 4B) and is not mediated by regulation of the Rel homology region that binds DNA (Fig. 4A). However, residues 146 to 207 of the NH2-terminal homology region of the target NFAT molecule are required for inhibition by dnNFAT (Fig. 4B). Since this region contributes to the regulated nuclear translocation of NFAT (3, 7), we tested the effect of dnNFAT on Ca2+-stimulated nuclear accumulation of NFAT proteins. Immunofluorescence analysis indicated that NFAT1 is located in the cytosol of unstimulated cells (Fig. 4C). Upon treatment with ionomycin, NFAT1 translocates into the nucleus (Fig. 4C). However, the ionomycin-induced nuclear translocation of NFAT1 was blocked by the expression of dnNFAT (Fig. 4C). Similar inhibitory effects on nuclear translocation of NFAT2 caused by the expression of dnNFAT was observed (Fig. 4C). These data indicate that Ca2+-stimulated nuclear translocation of NFAT transcription factors is inhibited by dnNFAT. This conclusion is consistent with the observation that dnNFAT did not inhibit the transcriptional activity of constitutively nuclear GAL4-NFAT4 (Ala-163, Ala-165) (Fig. 4B).
Previous studies indicated that nuclear translocation of NFAT is mediated, in part, by calcineurin upon sustained increase in intracellular calcium (45, 49). Since dnNFAT blocks nuclear translocation of NFAT, we tested whether overexpression of calcineurin in cells would oppose the inhibitory effect by dnNFAT. We performed immunofluorescence analysis and examined the subcellular distribution of NFAT proteins. Expression of calcineurin did not affect the subcellular distribution of the NFAT proteins in the presence or absence of ionomycin (Fig. 4D). Expression of dnNFAT caused decreased nuclear accumulation of NFAT proteins (Fig. 4D). Overexpression of calcineurin, however, opposed the inhibitory effect of dnNFAT and increased nuclear accumulation of NFAT proteins (Fig. 4D). These data indicate that dnNFAT blocks nuclear translocation of NFAT proteins by interfering with calcineurin.
IL-2 expression is inhibited by dnNFAT.
NFAT was initially characterized as a nuclear transcription factor of activated T cells that binds to the IL-2 promoter (13). However, the contribution of NFAT-mediated transcription to IL-2 expression remains unclear. Recent studies demonstrate that IL-2 production is not decreased in mice lacking NFAT1 or NFAT2 (18, 24, 36, 41, 51, 53). These data may indicate that NFAT is not relevant to IL-2 expression, that the functions of NFAT isoforms are redundant, or that there are compensatory changes in the expression of other NFAT family members in NFAT-deficient mice. The role of NFAT in IL-2 expression therefore remains to be established. To test the involvement of NFAT in IL-2 expression, we examined the effect of dnNFAT.
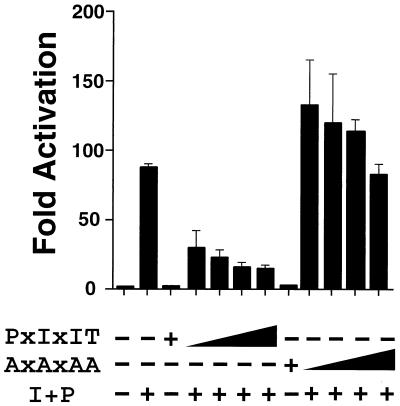
We examined whether dnNFAT inhibited the endogenous NFAT activity in Jurkat T cells in a transfection experiment using an NFAT-luciferase reporter plasmid (Fig. 5). Expression of dnNFAT caused a dose-dependent inhibition of NFAT transcription activity. This inhibition was blocked by the replacement of the conserved Pro, Ile, and Thr residues in the PxIxIT motif with Ala residues. Similar studies were performed with a luciferase reporter plasmid under the control of the IL-2 promoter (Fig. 6A). The dnNFAT (PxIxIT), but not the Ala-substituted mutant (AxAxAA), caused inhibition of IL-2 promoter activity in Jurkat T cells. The inhibition of IL-2 promoter activity caused by dnNFAT was similar to that caused by mutation of an NFAT binding site in the IL-2 promoter (13). These data indicate that NFAT transcription activity is required for IL-2 gene expression.
FIG. 5.
dnNFAT causes a dose-dependent inhibition of NFAT activity in Jurkat T cells. Increasing amounts (1, 3, 6, and 9 μg) of a dnNFAT expression vector were transfected in Jurkat T cells. The transcription activity of endogenous NFAT was detected with an NFAT-luciferase reporter plasmid. The effect of mutation of the PxIxIT motif by replacement of the Pro, Ile, and Thr residues with Ala (AxAxAA) was investigated. Luciferase activity was measured in cultures incubated without (−) or with (+) ionomycin (2 μM) and PMA (100 nM) (I+P). The data are presented as fold activation compared to an untreated control.
FIG. 6.
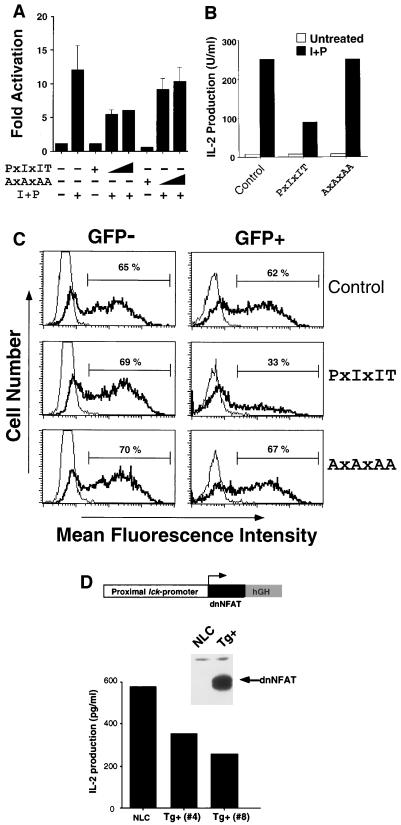
dnNFAT inhibits IL-2 production. (A) The activity of the IL-2 promoter is inhibited by dnNFAT. Jurkat T cells were cotransfected with an IL-2 promoter-luciferase reporter plasmid and various amounts (1 and 10 μg) of dnNFAT expression vector. The effect of mutation of the PxIxIT motif by replacement of the Pro, Ile, and Thr residues with Ala (AxAxAA) was investigated. Luciferase activity was measured in cultures incubated without (−) or with (+) ionomycin (2 μM) and PMA (100 nM) (I+P). The data are presented as fold activation compared to an untreated control. (B) IL-2 secretion is inhibited by dnNFAT. Jurkat T cells were cotransfected with expression vectors for GFP and dnNFAT (either wild-type PxIxIT or mutated AxAxAA). Transfected cells expressing GFP were selected by flow cytometry and treated without (Untreated) or with ionomycin (2 μM) and PMA (100 nM) (I+P), and the amount of IL-2 secreted in the culture medium was measured. (C) IL-2 expression is inhibited by dnNFAT. Jurkat T cells were cotransfected without (Control) and with expression vectors for GFP and dnNFAT (either wild-type PxIxIT or mutated AxAxAA). The cells were treated without (thin line) or with (thick line) ionomycin (2 μM) and PMA (100 nM) (I+P). The intracellular IL-2 and GFP was measured by flow cytometry. IL-2 expression (mean fluorescence intensity) of the transfected GFP positive (+) and untransfected GFP negative (−) cells in each culture is shown. (D) Thymocytes from dnNFAT transgenic mice have reduced IL-2 expression. Thymocytes were isolated from dnNFAT transgenic mice (Tg+) and control nontransgenic littermates (NLC). Expression of dnNFAT was detected by protein immunoblot analysis using MAb M2, specific to the Flag epitope. Cells were stimulated with ionomycin (2 μM) and PMA (100 nM), and the amount of IL-2 secreted in the culture medium was measured. hGH, human growth hormone.
To confirm the results obtained with the IL-2 promoter reporter plasmid, we examined the effect of dnNFAT on IL-2 secretion. Jurkat T cells were cotransfected with expression vectors for GFP and dnNFAT. The transfected cells were selected by cell sorting and treated with PMA and ionomycin (Fig. 6B). Cells transfected with GFP alone demonstrated a large increase in IL-2 secretion following treatment with PMA and ionomycin. In contrast, IL-2 secretion was markedly reduced in cells cotransfected with GFP and dnNFAT. The ability of dnNFAT to reduce IL-2 secretion was eliminated if the conserved residues in the PxIxIT motif were replaced with Ala. These data indicate that dnNFAT inhibits IL-2 production in vivo.
To further confirm that IL-2 production is inhibited by dnNFAT, we directly measured the amount of IL-2 expressed by individual Jurkat T cells by the intracellular cytokine staining procedure (Fig. 6C). Jurkat T cells were transfected with GFP alone, GFP plus dnNFAT (PxIxIT), and GFP plus the mutated dnNFAT (AxAxAA). These cultures were treated without and with PMA plus ionomycin and then examined by flow cytometric analysis of GFP and IL-2. Treatment with PMA plus ionomycin caused similar increases in IL-2 detected in the untransfected (GFP-negative) cells present in each culture. Increased amounts of IL-2 were also detected in the transfected (GFP-positive) cells. Expression of dnNFAT caused a large decrease in IL-2 accumulation. In contrast, the mutated dnNFAT (AxAxAA) caused no change in IL-2 accumulation. These data indicate that dnNFAT inhibits expression of IL-2.
To test the effect of dnNFAT in IL-2 production in primary cells, we generated transgenic mice that express dnNFAT in the thymus using the proximal lck promoter. Immunoblot analysis showed the expression of dnNFAT in the thymus of the positive transgenic mice (Fig. 6D). We isolated thymocytes from control littermates and dnNFAT transgenic mice and measured IL-2 production in response to PMA plus ionomycin. We found that the production of IL-2 was markedly reduced in thymocytes from two different dnNFAT transgenic mouse lines than in those from the negative littermate control mice (Fig. 6D). Together, these data demonstrate that dnNFAT inhibits IL-2 expression not only in a T-cell clone (e.g., Jurkat cells) but also in primary cells. These data therefore provide strong support for the conclusion that NFAT is critically important for IL-2 gene expression.
DISCUSSION
Disruption of the NFAT1 gene in mice has been reported to cause enhanced immune responses (18, 24, 41, 51). The molecular basis for this effect of NFAT1 gene disruption is unclear. The levels of production of IL-2, IL-4, tumor necrosis factor alpha, and gamma interferon by wild-type and NFAT1−/− T cells in response to anti-CD3 MAb or concanavalin A are similar (51). In contrast, NFAT1−/− T cells expressed reduced amounts of IL-4 when treated with concanavalin A in vitro (41). A similar decrease in IL-4 expression was reported in response to the administration of anti-CD3 MAb in vivo, but Th2 cell development and late IL-4 production in vitro were enhanced (18). In a separate study, no differences in early IL-4 gene expression were detected, but the expression of IL-4 was more sustained in NFAT1−/− mice (24). None of these reports demonstrate changes in the expression of IL-2, suggesting either that NFAT1 is not required for IL-2 gene expression or that other members of the NFAT family can compensate for the absence of NFAT1 in these mice.
Mice deficient in the expression of NFAT2 have also been reported (36, 53). Disruption of the NFAT2 gene causes early embryonic death due to impairment of heart development (11, 35). However, the creation of Rag2−/− NFAT2−/− chimeric mice has enabled studies of immune responses. These studies have demonstrated that NFAT2 gene disruption causes impaired Th2 responses with reduced IL-4 production (36, 53). However, the effect on IL-2 production is unclear. The study by Ranger et al. shows increased IL-2 production (36). In contrast, Yoshida et al. detected no differences in IL-2 expression in NFAT2-deficient mice (53). The interpretation of these data is confounded by the observation that disruption of the NFAT2 gene alters the development of T cells in the thymus. Thus, it is possible that the population of T cells present in the spleen or lymph nodes of the Rag2−/− NFAT2−/− chimeric mice does not represent normal T cells.
The failure of the reported gene disruption studies to demonstrate a role for NFAT in IL-2 gene expression may result from functional redundancy or compensatory changes in the knockout mice. It is therefore possible that NFAT contributes to the expression of IL-2 in T cells. Indeed, several lines of evidence that support the contention that NFAT contributes to the regulation of IL-2 gene expression have been reported. First, NFAT binding sites are located in the IL-2 promoter (44). Second, mutational analysis of the distal NFAT binding site present in the IL-2 promoter demonstrates that this DNA element contributes to IL-2 gene expression (13). Third, NFAT activation correlates with IL-2 secretion (22, 44). Fourth, immunosuppressive drugs (e.g., cyclosporin A and FK506) which reduce calcineurin activity inhibit both NFAT-mediated transcription and IL-2 gene expression (14, 16, 34). Together, these data provide strong support for the hypothesis that NFAT contributes to IL-2 secretion. However, the requirement of NFAT binding sites and calcineurin activity for IL-2 expression does not establish that NFAT is necessary for this process. Further studies are therefore required to demonstrate a role for NFAT in IL-2 gene expression.
We have tested the involvement of NFAT in IL-2 expression by using the dnNFAT molecule. The active component of this inhibitor corresponds to the PxIxIT box located in the conserved NH2-terminal homology region of NFAT (Fig. 1). dnNFAT selectively inhibited NFAT transcription activity by interfering with the activation-induced nuclear import of NFAT. These data suggest that the normal function of the PxIxIT box in NFAT contributes to nuclear accumulation. Indeed, deletion of the PxIxIT box inhibits activation-induced nuclear import of NFAT (2, 55). The mechanism of action of dnNFAT is likely to be mediated by interference with the normal function of the conserved PxIxIT box. This function may involve the targeting of NFAT to calcineurin, which is required for NFAT activation. Interestingly, overexpression of calcineurin opposed the inhibitory effect mediated by the PxIxIT box (dnNFAT) (Fig. 4D). In addition, in vitro studies demonstrate that peptides corresponding to the PxIxIT box inhibit the dephosphorylation of NFAT by calcineurin (2). This effect of the PxIxIT peptide is not mediated by inhibition of calcineurin activity. Instead, the PxIxIT peptide prevents the recognition of NFAT as a substrate by calcineurin without altering the ability of calcineurin to dephosphorylate other substrates (2). Thus, in contrast to the immunosuppressive drugs cyclosporin A and FK506, which cause inhibition of all calcineurin signaling functions, the PxIxIT box is a selective inhibitor of NFAT dephosphorylation in vitro. In this study, we demonstrate that expression of the PxIxIT box (dnNFAT) in T cells causes selective inhibition of NFAT transcription activity.
We have used dnNFAT to test the role of NFAT in IL-2 gene expression. Inhibition of NFAT-mediated transcription by dnNFAT resulted in dose-dependent inhibition of IL-2 promoter activity in Jurkat T cells. These data indicate that the NFAT transcription factor is required for normal expression of the IL-2 gene. Moreover, we have shown that IL-2 secretion is markedly inhibited by dnNFAT (Fig. 6). More importantly, we have demonstrated that dnNFAT inhibited IL-2 production in a transgenic animal model (Fig. 6D). Together, our results demonstrate that dnNFAT inhibits the production of IL-2. Thus, the NFAT transcription factor contributes to the regulation of IL-2 gene expression and therefore plays a critical role in the initiation of immune responses.
ACKNOWLEDGMENTS
We thank S. Ghosh, T. Hoey, D. Kerr, and T. Soderling for providing reagents; T. Barrett and M. Sharma for technical assistance; M. McFadden for assistance with flow cytometry; and K. Gemme for administrative assistance.
C.-W. Chow is an Arthritis Foundation fellow. This work was supported in part by grants CA65861 and CA72009 from the National Cancer Institute (R.J.D.) and AI42138 from the National Institutes of Health (M.R.). R.J.D. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Aramburu J, Azzoni L, Rao A, Perussia B. Activation and expression of the nuclear factors of activated T cells, NFATp and NFATc, in human natural killer cells: regulation upon CD16 ligand binding. J Exp Med. 1995;182:801–810. doi: 10.1084/jem.182.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aramburu J, Garcia-Cozar F, Raghavan A, Okamura H, Rao A, Hogan P G. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol Cell. 1998;1:627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 3.Beals C R, Clipstone N A, Ho S N, Crabtree G R. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- 4.Boise L H, Petryniak B, Mao X, June C H, Wang C Y, Lindsten T, Bravo R, Kovary K, Leiden J M, Thompson C B. The NFAT-1 DNA binding complex in activated T cells contains Fra-1 and JunB. Mol Cell Biol. 1993;13:1911–1919. doi: 10.1128/mcb.13.3.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown P H, Kim S H, Wise S C, Sabichi A L, Birrer M J. Dominant-negative mutants of cJun inhibit AP-1 activity through multiple mechanisms and with different potencies. Cell Growth Differ. 1996;7:1013–1021. [PubMed] [Google Scholar]
- 6.Chen L, Glover J N, Hogan P G, Rao A, Harrison S C. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 7.Chow C W, Rincon M, Cavanagh J, Dickens M, Davis R J. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science. 1997;278:1638–1641. doi: 10.1126/science.278.5343.1638. [DOI] [PubMed] [Google Scholar]
- 8.Chuvpilo S, Schomberg C, Gerwig R, Heinfling A, Reeves R, Grummt F, Serfling E. Multiple closely-linked NFAT/octamer and HMG I(Y) binding sites are part of the interleukin-4 promoter. Nucleic Acids Res. 1993;21:5694–5704. doi: 10.1093/nar/21.24.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clipstone N A, Crabtree G R. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- 10.Cockerill P N, Shannon M F, Bert A G, Ryan G R, Vadas M A. The granulocyte-macrophage colony-stimulating factor/interleukin 3 locus is regulated by an inducible cyclosporin A-sensitive enhancer. Proc Natl Acad Sci USA. 1993;90:2466–2470. doi: 10.1073/pnas.90.6.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Pompa J L, Timmerman L A, Takimoto H, Yoshida H, Eila A J, Samper E, Potter J, Wakeham A, Marengere L, Langille B L, Crabtree G R, Mak T W. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature. 1998;392:182–186. doi: 10.1038/32419. [DOI] [PubMed] [Google Scholar]
- 12.Duncliffe K N, Bert A G, Vadas M A, Cockerill P N. A T cell-specific enhancer in the interleukin-3 locus is activated cooperatively by Oct and NFAT elements within a DNase I-hypersensitive site. Immunity. 1997;6:175–185. doi: 10.1016/s1074-7613(00)80424-0. [DOI] [PubMed] [Google Scholar]
- 13.Durand D B, Shaw J-P, Bush M R, Repogla R E, Belagaje R, Crabtree G R. Characterization of antigen receptor response elements within the interleukin-2 enhancer. Mol Cell Biol. 1988;8:1715–1724. doi: 10.1128/mcb.8.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flanagan W M, Corthesy B, Bram R J, Crabtree G R. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. 1991;352:803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- 15.Gillis S, Ferm M N, Smith K A. T cell growth factor: parameters of production and quantative microassay for activity. J Immunol. 1978;120:1109–1113. [PubMed] [Google Scholar]
- 16.Granelli-Piperno A, Nolan P, Inaba K, Steinman R M. The effect of immunosuppressive agents on the induction of nuclear factors that bind to sites on the interleukin 2 promoter. J Exp Med. 1990;172:1869–1872. doi: 10.1084/jem.172.6.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho S N, Thomas D J, Timmerman L A, Li X, Francke U, Crabtree G R. NFATc3, a lymphoid-specific NFATc family member that is calcium-regulated and exhibits distinct DNA binding specificity. J Biol Chem. 1995;270:19898–19907. doi: 10.1074/jbc.270.34.19898. [DOI] [PubMed] [Google Scholar]
- 18.Hodge M R, Ranger A M, de la Brousse C F, Hoey T, Grusby M J, Glimcher L H. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity. 1996;4:397–405. doi: 10.1016/s1074-7613(00)80253-8. [DOI] [PubMed] [Google Scholar]
- 19.Hoey T, Sun Y-L, Williamson K, Xu X. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity. 1995;2:461–472. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 20.Hutchinson L E, McCloskey M A. Fc epsilon RI-mediated induction of nuclear factor of activated T-cells. J Biol Chem. 1995;270:16333–16338. doi: 10.1074/jbc.270.27.16333. [DOI] [PubMed] [Google Scholar]
- 21.Jain J, Burgeon E, Badalian T M, Hogan P G, Rao A. A similar DNA-binding motif in NFAT family proteins and the Rel homology region. J Biol Chem. 1995;270:4138–4145. [PubMed] [Google Scholar]
- 22.Jain J, Loh C, Rao A. Transcriptional regulation of the IL-2 gene. Curr Opin Immunol. 1995;7:333–342. doi: 10.1016/0952-7915(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 23.Jain J, McCaffrey P G, Miner Z, Kerppola T K, Lambert J N, Verdine G L, Curran T, Rao A. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- 24.Kiani A, Viola J P, Lichtman A H, Rao A. Down-regulation of IL-4 gene transcription and control of Th2 cell differentiation by a mechanism involving NFAT1. Immunity. 1997;7:849–860. doi: 10.1016/s1074-7613(00)80403-3. [DOI] [PubMed] [Google Scholar]
- 25.Latinis K M, Carr L L, Peterson E J, Norian L A, Eliason S L, Koretzky G A. Regulation of CD95 (Fas) ligand expression by TCR-mediated signaling events. J Immunol. 1997;158:4602–4611. [PubMed] [Google Scholar]
- 26.Luo C, Burgeon E, Rao A. Mechanisms of transactivation by nuclear factor of activated T cells-1. J Exp Med. 1996;184:141–147. doi: 10.1084/jem.184.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda E S, Naito Y, Tokumitsu H, Campbell D, Saito F, Hannum C, Arai K, Arai N. NFATx, a novel member of the nuclear factor of activated T cells family that is expressed predominantly in the thymus. Mol Cell Biol. 1995;15:2697–2706. doi: 10.1128/mcb.15.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCaffrey P G, Goldfeld A E, Rao A. The role of NFATp in cyclosporinA-sensitive tumore necrosis factor-α gene transcription. J Biol Chem. 1994;269:30445–30450. [PubMed] [Google Scholar]
- 29.McCaffrey P G, Luo C, Kerppola T K, Jain J, Badalian T M, Ho A M, Burgeon E, Lane W S, Lambert J N, Curran T, et al. Isolation of the cyclosporin-sensitive T cell transcription factor NFATp. Science. 1993;262:750–754. doi: 10.1126/science.8235597. [DOI] [PubMed] [Google Scholar]
- 30.Molkentin J D, Lu J-R, Antos C L, Robbins J, Grant S R, Olson E N. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Northrop J P, Ho S N, Chen L, Thomas D J, Timmerman L A, Nolan G P, Admon A, Crabtree G R. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature. 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- 32.Perrino B A, Fong Y L, Bricky D A, Saitoh Y, Ushio Y, Fukunaga K, Miyamoto E, Soderling T R. Characterization of the phosphatase activity of a baculovirus-expressed calcineurin A isoform. J Biol Chem. 1992;267:15965–15969. [PubMed] [Google Scholar]
- 33.Phillips R J, Gustafson S, Ghosh S. Identification of a novel NF-κB p50-related protein in B lymphocytes. Mol Cell Biol. 1996;16:7089–7097. doi: 10.1128/mcb.16.12.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Randak C, Brabletz T, Hergenrother M, Sobotta I, Serfling E. Cyclosporin A suppresses the expression of the interleukin 2 gene by inhibiting the binding of lymphocyte-specific factors to the IL-2 enhancer. EMBO J. 1990;9:2529–2536. doi: 10.1002/j.1460-2075.1990.tb07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranger A M, Grusby M J, Hodge M R, Gravallese E M, de la Brousse C F, Hoey T, Mickanin C, Baldwin H S, Glimcher L H. The transcription factor NF-ATc is essential for cardiac valve formation. Nature. 1998;392:186–190. doi: 10.1038/32426. [DOI] [PubMed] [Google Scholar]
- 36.Ranger A M, Hodge M R, Gravallese E M, Oukka M, Davidson L, Alt F W, de la Brousse F C, Hoey T, Brusby M, Glimcher L H. Delayed lymphoid repopulation with defects in IL-4 driven responses produced by inactivation of NF-ATc. Immunity. 1998;8:125–134. doi: 10.1016/s1074-7613(00)80465-3. [DOI] [PubMed] [Google Scholar]
- 37.Rao A. NFATp, a cyclosporin-sensitive transcription factor implicated in cytokine gene induction. J Leukoc Biol. 1995;57:536–542. doi: 10.1002/jlb.57.4.536. [DOI] [PubMed] [Google Scholar]
- 38.Rao A, Luo C, Hogan P G. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 39.Rincón M, Flavell R A. AP-1 transcriptional activity requires both T-cell receptor-mediated and co-stimulatory signals in primary T lymphocytes. EMBO J. 1994;13:4370–4381. doi: 10.1002/j.1460-2075.1994.tb06757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothenberg E V, Ward S B. A dynamic assembly of diverse transcription factors integrates activation and cell-type information for interleukin 2 gene regulation. Proc Natl Acad Sci USA. 1996;93:9358–9365. doi: 10.1073/pnas.93.18.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuh K, Kneitz B, Heyer J, Siebelt F, Fischer C, Jankevics E, Rude E, Schmitt E, Schimpl A, Serfling E. NF-ATp plays a prominent role in the transcriptional induction of Th2-type lymphokines. Immunol Lett. 1997;57:171–175. doi: 10.1016/s0165-2478(97)00068-0. [DOI] [PubMed] [Google Scholar]
- 42.Serfling E, Avots A, Neumann M. The architecture of the interleukin-2 promoter: a reflection of T lymphocyte activation. Biochim Biophys Acta. 1995;1263:181–200. doi: 10.1016/0167-4781(95)00112-t. [DOI] [PubMed] [Google Scholar]
- 43.Shannon M F, Coles L S, Vadas M A, Cockerill P N. Signals for activation of the GM-CSF promoter and enhancer in T cells. Crit Rev Immunol. 1997;17:301–323. doi: 10.1615/critrevimmunol.v17.i3-4.30. [DOI] [PubMed] [Google Scholar]
- 44.Shaw J P, Utz P J, Durand D B, Toole J J, Emmel E A, Crabtree G R. Identification of a putative regulator of early T cell activation genes. Science. 1988;241:202–205. [PubMed] [Google Scholar]
- 45.Shibasaki F, Price E R, Milan D, McKeon F. Role of kinases and phosphatase calcineurin in the nuclear shuttling of transcription factor NFAT4. Nature. 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- 46.Silver P, Keegan L, Ptashne M. Amino terminus of the yeast GAL4 gene product is sufficient for nuclear localization. Proc Natl Acad Sci USA. 1984;81:5951–5955. doi: 10.1073/pnas.81.19.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stranick K S, Zambas D N, Uss A S, Egan R W, Billah M M, Umland S P. Identification of transcription factor binding sites important in the regulation of the human interleukin-5 gene. J Biol Chem. 1997;272:16453–16465. doi: 10.1074/jbc.272.26.16453. [DOI] [PubMed] [Google Scholar]
- 48.Szabo S J, Gold J S, Murphy T L, Murphy K M. Identification of cis-acting regulatory elements controlling interleukin-4 gene expression in T cells: roles for NF-Y and NF-ATc. Mol Cell Biol. 1993;13:4793–4805. doi: 10.1128/mcb.13.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timmerman L A, Clipstone N A, Ho S N, Northrop J P, Crabtree G R. Rapid shuttling of NF-AT in discrimination of Ca2+ signals and immunosuppression. Nature. 1996;383:837–840. doi: 10.1038/383837a0. [DOI] [PubMed] [Google Scholar]
- 50.Tsysykova A V, Tsitsikov E N, Geha R S. The CD40L promoter contains nuclear factor of activated T cells-binding motifs which require AP-1 binding for activation of transcription. J Biol Chem. 1996;271:3763–3770. doi: 10.1074/jbc.271.7.3763. [DOI] [PubMed] [Google Scholar]
- 51.Xanthoudakis S, Viola J P, Shaw K T, Luo C, Wallace J D, Bozza P T, Luk D C, Curran T, Rao A. An enhanced immune response in mice lacking the transcription factor NFAT1. Science. 1996;272:892–895. doi: 10.1126/science.272.5263.892. [DOI] [PubMed] [Google Scholar]
- 52.Yaseen N R, Maizel A L, Wang F, Sharma S. Comparative analysis of NFAT (nuclear factor of activated T cells) complex in human T and B lymphocytes. J Biol Chem. 1993;268:14285–14293. [PubMed] [Google Scholar]
- 53.Yoshida H, Nishina H, Takimoto H, Marengere L E M, Wakeham A C, Bouchard D, Shahinian A, Bachmann M, Ohashi P S, Penninger J M, Crabtree G R, Mak T W. The transcription factor NF-ATc1 regulates lymphocyte proliferation and Th2 cytokine production. Immunity. 1998;8:115–124. doi: 10.1016/s1074-7613(00)80464-1. [DOI] [PubMed] [Google Scholar]
- 54.Zhou P, Sun L J, Dotsch V, Wagner G, Verdine G L. Solution structure of the core NFATC1/DNA complex. Cell. 1998;92:687–696. doi: 10.1016/s0092-8674(00)81136-8. [DOI] [PubMed] [Google Scholar]
- 55.Zhu J, Shibasaki F, Price R, Guillemot J C, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]