Abstract
Down-regulation of the Drosophila ribosomal protein S21 gene (rpS21) causes a dominant weak Minute phenotype and recessively produces massive hyperplasia of the hematopoietic organs and moderate overgrowth of the imaginal discs during larval development. Here, we show that the S21 protein (RpS21) is bound to native 40S ribosomal subunits in a salt-labile association and is absent from polysomes, indicating that it acts as a translation initiation factor rather than as a core ribosomal protein. RpS21 can interact strongly with P40, a ribosomal peripheral protein encoded by the stubarista (sta) gene. Genetic studies reveal that P40 underexpression drastically enhances imaginal disc overgrowth in rpS21-deficient larvae, whereas viable combinations between rpS21 and sta affect the morphology of bristles, antennae, and aristae. These data demonstrate a strong interaction between components of the translation machinery and showed that their underexpression impairs the control of cell proliferation in both hematopoietic organs and imaginal discs.
In Drosophila melanogaster more than 50 genes controlling cell proliferation have been identified by using mutations (25, 45). Mutations in these genes act as recessive determinants of tissue overgrowth and are classified as tumor suppressor genes. The Drosophila tissues susceptible to becoming overgrown derive from cells which divide actively during embryonic and larval development and include organs such as the larval brain hemispheres, the imaginal discs that will form the adult hypoderm and cuticle, the hematopoietic organs, and the germ line (25). With the exception of tumors in the germ line which result only in adult sterility, overgrowth in other tissues is accompanied by developmental arrest at the larval-to-pupal transition phase. As a consequence of the developmental arrest, the larval life of the mutant animals is extended over several days and the tumorous organs can reach a considerable mass that is readily observed upon dissection.
Mutations in more than 25 genes were found to cause overgrowth of the hematopoietic organs (25, 45, 75), which consist of five to seven pairs of glandular structures located along the dorsal heart vessel behind the brain hemispheres and which produce hemocytes by a stem-cell mechanism. In wild-type larvae the hemocytes are released into the hemolymph at the end of the third larval instar (55, 64). By contrast, in homozygous l(2)k168-14 larvae, the proliferating hemocytes remain mainly in the hematopoietic organs, which become massively enlarged (72). These organs retain a globular and compact structure and can reach up to 50-fold their normal size. Although the tumorous organs are filled with partially differentiated hemocytes, these cells are unable to form melanotic masses as is usually the case in other mutations producing overgrowth of the hematopoietic organs (25, 75).
We have cloned and sequenced the gene mutated in l(2)k168-14 and found that it encodes the ribosomal protein S21, which has been previously identified in species as diverse as rats (31), yeast cells (66), humans (8), and rice (48). We show that the l(2)k168-14 mutation produces a dominant weak Minute phenotype similar to the phenotype produced by mutations in other ribosomal protein genes, encoding the ribosomal proteins 49 (or L32) (39), S2 (15), S3 (1), S5 (44), S6 (58, 65, 76), S13 (56), L9 (61), L14 (57), and L19 (28). Furthermore, our analysis revealed that the ribosomal protein S21 (RpS21) is essentially associated with the native 40S ribosomal subunits and absent from polysomes, indicating that this protein acts presumably as an initiation factor rather than as a core ribosomal protein. Following the recent finding that mutations in another Drosophila gene encoding the ribosomal protein S6 can also produce tumorous growth in the hematopoietic organs (65, 76), our studies confirm that, in addition to their function in protein synthesis, ribosomal and ribosome-associated proteins may play a role in the regulation of cell proliferation.
Although no ribosomal gene has yet been assigned to any known inherited cancer susceptibility locus in humans, the divergent expression of ribosomal protein transcripts has been reported in a series of human transformed cells. The expression of numerous transcripts encoding ribosomal proteins was found to be enhanced in colon carcinomas and squamous carcinomas. The identified sequences include the ribosomal proteins L31 (14); P0, S3, S6, S8, and S12 (53); S2 (13); S19 (38); L18 (5); ubiquitin-S27a (79); L19 (29); P2 (62); and L37 (5).
By contrast, the expression of the transcripts encoding the QM associated ribosomal protein (21, 41) and the S29 ribosomal protein (37) is down-regulated in Wilms’ tumor and colon carcinoma, respectively. Evidence for growth suppression has been obtained for S29 by transfecting human rpS29 cDNA into mouse v-Ki-ras-transformed NIH 3T3 cells (37). These studies showed that rpS29 alone induces flat revertants at low frequency but that it significantly enhances the potential for suppression of transformation by Krev-1, which antagonizes an activated ras oncogene (35).
Although no direct functional growth suppression has been shown for QM, which shares 99% identity with the recently identified rat ribosomal protein L10 (12), this protein was found to be particularly elevated in tissues undergoing rapid proliferation (20, 22, 33). Of particular interest are the recent findings that the yeast homolog of QM, designated as Qsr1p, acts as a peripheral ribosomal protein of the 60S ribosomal subunit and is required for ribosomal joining (19, 21, 36), suggesting a regulatory function in the rates of growth and protein synthesis.
Finally, by using the yeast two-hybrid system we have shown that RpS21 can interact strongly with the ribosome-associated protein P40, encoded by the stubarista (sta) gene (46). Further support in favor of a functional interaction between RpS21 and P40 was obtained from genetic and biophysical studies showing that (i) heterozygous combinations between different sta alleles and l(2)k168-14 show enhancement of the antennal and bristle phenotypes, (ii) underexpression of P40 enhances the tumor phenotype in l(2)k168-14 larvae, and (iii) bacterially expressed RpS21 and P40 can bind in an in vitro assay. With this approach, we have identified two ribosomal protein genes whose functions regulate protein synthesis and cell growth and in which defects may lead to tissue-specific tumors in D. melanogaster.
MATERIALS AND METHODS
Drosophila stocks and yeast strains.
All fly stocks were maintained on standard Drosophila medium at room temperature (2); all crosses were done at 25°C. The genetic nomenclature is as outlined by Lindsley and Zimm (40). Stocks were kindly provided by the European Drosophila Stock Center, Umeå, Sweden, and by the Bloomington Drosophila Stock Center.
The l(2)k168-14 mutant was recovered from a large-scale P-element mutagenesis screen by using the y w P-lacW procedure as described in Török et al. (70). We constructed the stock y sta2/Binsn; l(2)k168-14/P[y+] CyO to facilitate the selection by the y marker mutation of the y sta2/Y; l(2)k168-14/l(2)k168-14 double-mutant larvae.
The YRG-2 yeast strain (MATα ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3 gal4-542 gal80-538 LYS2::UASGAL1-TATAGAL1-HIS3 URA3::UASGAL4-17mer(3x)-TATACYC1-LacZ) (Stratagene, La Jolla, Calif.) was used in the yeast two-hybrid procedure.
Examination of mutant larval phenotypes.
Larval phenotypes were examined as described previously (71). In short, eggs were collected for 24 h, and larvae were washed out of the medium on days 7 to 8 after egg collection. By this time, the majority of the normal siblings have pupariated, and slowly growing mutant larvae were enriched in the medium. As the mutant l(2)k168-14 stocks were maintained in a homozygous y w background over a CyO balancer marked with a y+ transgene (P[y+] CyO), homozygous mutant larvae were selected by their yellow mouth hooks. Mutant larvae were transferred to fresh vials and left in a humidified atmosphere until dissection. Hemocyte counts were determined according to the method of Szabad and Bryant (66a).
Isolation of viable and lethal revertants.
The isolation of viable and lethal white-eyed revertants was carried out as previously described (71, 72).
Cytogenetic mapping.
Cytogenetic mapping by in situ hybridization to salivary gland polytene chromosomes was performed essentially as described in De Frutos et al. (17), using digoxigenin-labeled pUC18 DNA probe (Boehringer GmbH, Mannheim, Germany).
Germ line clone analysis.
The possible function of rpS21 in the ovary was tested by using the dominant female sterile technique (74). Mitotic recombination was induced in oogonial cells of 65- to 75-h-old larvae heterozygous for the Fs(2)1 and l(2)k168/14 mutations by X-irradiating them with 1,200 rads.
Nucleic acid procedures.
Basic DNA manipulations, unless otherwise indicated, were performed according to standard protocols (59). Plasmid rescue, genomic and cDNA library screening, RNA purification and Northern blotting were done as previously described (71). Sequencing was carried out with double-stranded DNA templates and oligodeoxynucleotide primers by using the USB sequencing kit (U.S. Biochemical Corp., Cleveland, Ohio).
Immunochemical procedures.
To raise antibodies against the RpS21 protein, synthetic peptides corresponding to its NH2- and COOH-terminal ends with the sequences MENDAGENVDLYVPRKCSASNRIC and CRMGESDDCIVRLAKKDGIITKNF, respectively, were conjugated to maleimide-activated keyhole limpet hemocyanin (Pierce Chemical Co., Rockford, Ill.) through the terminal cysteine of each peptide, and the resulting conjugates were used to immunize rabbits. Anti-peptide antibodies were affinity purified by column chromatography in two steps as described by Harlow and Lane (27) by using protein A-agarose (Boehringer GmbH) and then the immunizing peptide coupled to Sulfo-Link gel (Pierce Chemical Co.).
To prepare antibodies against the P40 ribosomal associated protein, the whole protein coding sequence of the p40 cDNA was first PCR amplified and then cloned into pGEX-4T-1 expression vector (Pharmacia LKB Biotechnology, Uppsala, Sweden). The growth and induction were performed in the BL-21 Escherichia coli strain at 25°C with 400 μM IPTG (isopropyl-β-d-thiogalactopyranoside). The P40 fusion protein, designated GST-P40, was extracted with sonication in extraction buffer (10 mM Tris-HCl, pH 8.0; 150 mM NaCl; 1 mM EDTA; 5 mM dithiothreitol [DTT], 2% Triton X-100; 1.5% Sarkosyl). The GST-P40 protein was affinity purified on glutathione-Sepharose 4B (Pharmacia LKB Biotechnology), eluted with 10 mM glutathione and 50 mM Tris-HCl at pH 8.0, and used to immunize rabbits. Anti-P40 antibodies were purified from the serum in three steps. First, total immunoglobulins were separated on a protein A-agarose column. Second, the immunoglobulins were depleted of glutathione S-transferase (GST)-reacting antibodies by passage through a GST-Sepharose 4B column. Third, the purified anti-P40 antibodies were recovered on a GST-P40 Sepharose 4B column. Western blotting and immunoprecipitation were done as indicated previously (71).
P-element transformation and rescue.
The 2.8-kb BamHI-XhoI fragment overlapping the P-lacW insertion site in l(2)k168-14 and containing the rpS21 gene sequence was cloned into the pCaSpeR4 transformation vector of Pirrotta (52), which carries the white+ (w+) eye color gene as a marker. This construct was coinjected with the wing-clipped helper P-element (wc Δ2-3) into w1118 homozygous embryos prior to the cellular blastoderm stage. Emerging adult flies were individually crossed back to w1118, and progeny with pigmented (red) eyes were crossed with y w; Bc Gla/CyO flies. Their progeny were further pair-crossed to isolate lines with insertion of the P-transposon containing wild-type copies of the white and rpS21 genes, designated as P[w+ rpS21+], on either the second or the third chromosome. A third-chromosome insertion line (y w; Bc Gla/CyO; P[w+ rpS21+]) was crossed with the original mutant line y w, l(2)k168-14/CyO, or with lethal revertant lines obtained by imprecise excision of the P-lacW transposon: l(2)k168-14R7 and l(2)k168-14R8, respectively. Progeny with curled wings and normal (Gla+) eyes were further pair-crossed, and their progeny were scored for non-curly homozygous l(2)k168-14 flies (as well as l(2)k168-14R7 or l(2)k168-14R12 flies) rescued by the insertion of the P[w+ rpS21+] transposon on chromosome 3.
Yeast two-hybrid screen.
The experiments were performed according to the protocol of the Hybri-Zap two-hybrid system as described by the manufacturer (Stratagene). In short, the protein-coding sequence of the rpS21 cDNA was PCR amplified and ligated in frame to the binding domain of the pBD(GAL4) vector. An in-frame clone was selected by DNA sequencing. The pBD(GAL4)-rpS21-transformed YRG-2 yeast cells were retransformed with DNA from Hybri-ZAP cDNA libraries made of either Drosophila ovarian or embryonic poly(A)+ RNA. Then, 3 × 106 to 5 × 106 recombinant clones harboring both plasmids with binding and activation domains and showing growth on Leu− Trp− SD plates were further tested on Leu− Trp− His− SD plates. A total of 26 yeast clones were able to grow on Leu− Trp− His− SD plates. Of these clones, 18 proved to be positive in a β-galactosidase test. The specificity of the interactions was verified by backtransforming the constructs containing the binding and activation domains and appropriate control plasmids in the different pairwise combinations into fresh YRG-2 cells. Plasmid DNA of the positively growing yeast cells was transformed into XL-1 Blue-competent E. coli cells, and the plasmid containing the activation domain was isolated and sequenced.
In vitro binding assay.
Bacterially expressed GST and GST-P40 proteins were purified in large amounts as described above. In vitro translation of RpS21 proteins was performed with the Promega TNT Coupled reticulocyte lysate system and [35S]methionine (Amersham). Affinity-purified GST and GST-P40 fusion proteins were bound to glutathione-Sepharose 4B beads and washed three times with 10 volumes of binding buffer including 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 0.05% sarcosyl, 1 mM DTT, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride, and 1 μg of leupeptin and 0.5 μg of aprotinin per ml to remove the excess of nonbound proteins. 35S-labeled RpS21 proteins were mixed with the beads bound to GST or GST-P40 and incubated at room temperature for 2 h. Beads were washed three times with 10 volumes of binding buffer; this treatment was followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Immunohistochemistry.
Brain-disc complexes including the hematopoietic organs and ring gland of Oregon-R wild-type and l(2)k168-14 third-instar larvae were dissected in phosphate-buffered saline (PBS), fixed in 4% formaldehyde in PBS for 20 min at room temperature, and extensively washed with PBS containing 0.1% Triton X-100 (PBT). The brain-disc complexes were then blocked overnight in PBT containing 5% normal goat serum and 1% bovine serum albumin. The blocking solution was removed, and the brain complexes were incubated for 20 min at room temperature with 1 μg of fluorescein isothiocyanate FITC-conjugated phalloidin (Sigma Chemical Co., St. Louis, Mo.). After being washed with PBS, the brain complexes were treated for 2 h at room temperature with RNase A (400 μg/ml in PBS), washed with PBS, stained for 2 h at room temperature with 5 μg of propidium iodine per ml, washed overnight at 4°C in PBS, mounted in Vectashield embedding medium (Vector Laboratories, Burlingame, Calif.), and examined under a Zeiss confocal laser scanning microscope (Carl Zeiss Jena GmbH, Jena, Germany).
Scanning electron microscopy.
Next, 2-day-old flies were fixed overnight at 4°C in 2% glutaraldehyde–0.1% Tween 20, washed twice with PBS, and dehydrated through a graded ethanol series (30, 50, 70, 95, and 100%) with a 30-min incubation at each step. The flies were then equilibrated with ethanol-acetone mixtures (3:1, 1:1, and 1:3) with a 30-min incubation at each step. Critical-point drying was replaced by incubating the flies for 30 min in hexamethylenedisilazane (Sigma Chemical Co.)-acetone (1:1) and 30 min again in 100% hexamethylenedisilazane (9a). The flies were dried on a filter paper, mounted onto scanning electron microscopy (SEM) stubs, and sputter coated with a 2-nm-thick gold coat. Samples were viewed and photographed on a Philips SEM 505 instrument.
Preparation of ribosomes.
Postnuclear extracts from overnight collections of Drosophila embryos or Schneider cells grown at 25°C were centrifuged on 15 to 40% linear sucrose density gradients in 0.08 to 0.5 M KCl, 0.002 M MgCl2, and 0.05 M Tris-HCl (pH 7.4) loaded over a 1-ml 50% sucrose layer made in the same buffer and centrifuged in a Spinco SW41 rotor (Beckman Instruments, Spinco Div., Palo Alto, Calif.). For the analysis of the polysome profile, centrifugation was carried out at 4°C for 5 h at 23,000 rpm. For the analysis of the 80S ribosomes and native subunits, the centrifugation was performed at 4°C for 18 h at 23,000 rpm. The gradients were collected in fractions of ∼0.4 ml, and the absorbance profile was measured at 254 nm. Proteins contained in the sucrose gradient fractions were precipitated by the addition of 3 volumes of ethanol in the presence of 0.2 M NaCl, resuspended in 40 μl of SDS sample buffer, and resolved by SDS-PAGE. Western blots and immunodetections were performed essentially as previously described (71).
RESULTS
Slow rate of growth of the homozygous l(2)168-14 larvae with hyperplasia of the hematopoietic organs.
The l(2)k168-14 mutation was isolated during a large-scale P-element mutagenesis screening designed to identify genes in control of cell growth and proliferation (72). In the l(2)k168-14 mutant line the P-lacW transposon was found to be integrated into the 23B region of the second chromosome. Compared to wild-type or heterozygous animals, the development of homozygous l(2)k168-14 animals is considerably delayed and, when the heterozygous sibling imagos emerge from the pupal case, the homozygous mutant larvae are the size of young third-instar larvae. These larvae were found to reach a nearly mature size 10 to 15 days after egg laying and to remain as third-instar larvae for up to 3 weeks. Thereafter, they undertake puparium formation, albeit with incomplete eversion of the leg and wing imaginal discs. Similarly, eversion of the anterior spiracles, as well as the tanning of the larval cuticle, is incomplete. In addition, in about one-third of the larvae we found that the imaginal discs were slightly larger than normal (Table 1). Although the retraction of the epidermis takes place and leaves gas bubbles on both extremities of the pupal case, no further development occurs as judged by the absence of head eversion, which normally takes place 12 h after puparium formation.
TABLE 1.
Characterization of l(2)k168-14 and stubarista lethal phenotypes
| Genotype | Phenotype |
|---|---|
| l(2)k168-14/l(2)k168-14 | Lethality at larval-to-pupal transition phase, slow larval growth, extended larval life, overgrown hematopoietic organs, rare overgrown imaginal discs, few larvae forming a puparium |
| sta2/sta2 | Lethality during third larval instar with slow growth, overgrown hematopoietic organs, small size imaginal discs |
| sta2/sta2; l(2)k168-14/l(2)k168-14 or sta2/Y; l(2)k168-14/l(2)k168-14 | Early larval lethality during first or second larval instar |
| sta2/+; l(2)k168-14/l(2)k168-14 | Lethality at larval-to-pupal transition phase, almost normal larval growth with extended larval life, overgrown hematopoietic organs, high proportion of overgrown imaginal discs, numerous larvae form a puparium and then die |
| sta2/sta2; l(2)k168-14/+ or sta2/Y; l(2)k168-14/+ | Early larval lethality during first or second larval instar |
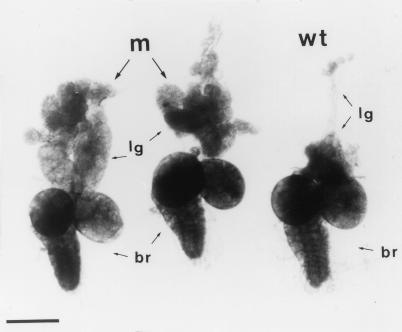
Examination of aged mutant larvae revealed that the hematopoietic organs, which in wild-type larvae consist of four to seven small paired lobes arrayed along the dorsal aorta, are considerably enlarged, with hyperplasia occurring maximally in the first pair of lobes (Fig. 1) and gradually decreasing in the more posterior lobes. The hypertrophied organs, which can reach up to 50-fold their normal size, retained a globular and compact structure. No melanotic masses could be detected in aged mutant larvae in either the hematopoietic organs or within the body cavity. Furthermore, in l(2)k168-14 homozygous larvae the number of circulating hemocytes was strongly reduced compared to wild-type wandering third-instar larvae (0.7 × 104 cells/μl instead of 2.7 × 104 cells/μl), showing that the release of hemocytes is blocked in the mutant animals. Furthermore, examination by confocal microscopy (Fig. 2) revealed that the mutant hematopoietic organs consist predominantly of compact masses (arrowheads) of relatively small cells and a few nodules of less densely packed cells (arrows), resembling morphologically the wild-type hemocytes. The small size of the hemocytes and their compactness are characteristics of hyperplastic tissues. In conjunction with the reduced number of circulating hemocytes, these data suggest that the l(2)k168-14 hematopoietic cell lineage is affected at a relatively early stage of differentiation.
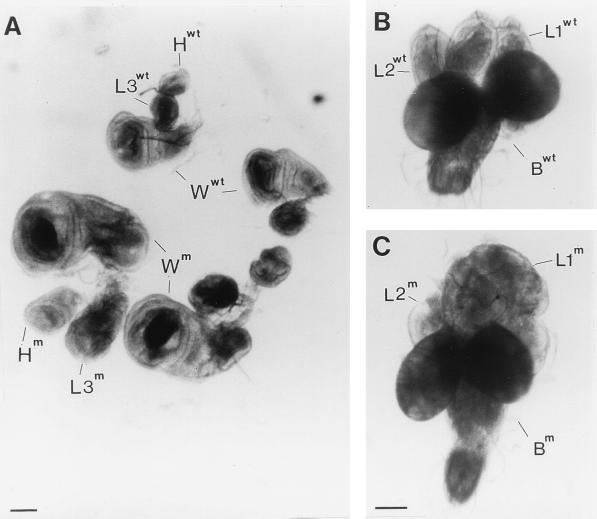
FIG. 1.
Hematopoietic organs, lymph glands (lg), and brains (br) dissected from l(2)k168-14 (m) and Oregon-R wild-type (wt) third-instar larvae. Bar = 200 μm.
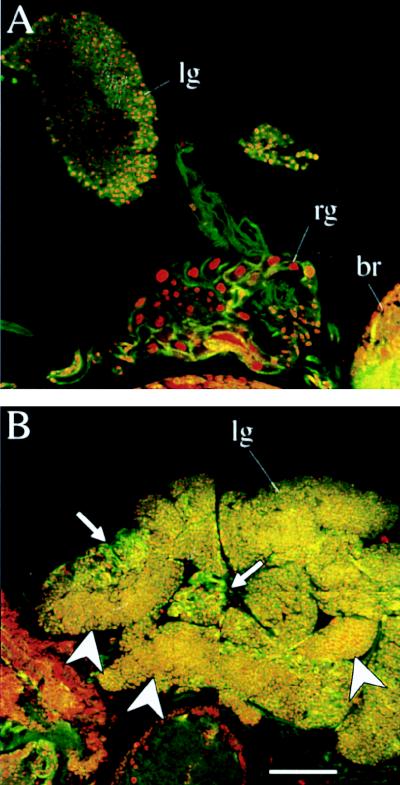
FIG. 2.
Whole-mount hematopoietic organs of (A) wild-type and (B) l(2)k168-14 larvae double stained for F-actin and DNA with FITC-conjugated phalloidin and propidium iodide, respectively. Confocal laser scan images were collected on separate channels simultaneously and processed to show F-actin as green and DNA as red. The yellow color represents coincidental staining and is more intense over relatively small-sized cells. In the l(2)k168-14 overgrown hematopoietic organs, arrows indicate areas with cells retaining a wild-type morphology, whereas arrowheads indicate areas with compacted cells of relatively small size. The positions of hematopoietic organs or lymph glands (lg), ring gland (rg), and brain (br) are indicated. Bar = 50 μm.
Dominant Minute phenotype.
Examination of heterozygous l(2)k168-14/+ animals revealed a characteristic Minute phenotype consisting of short slender bristles (∼20% reduction in length), small body (∼12 to 20% reduction in body width and length), and a 2-day delay in eclosion at 25°C owing to the delay in puparium formation (Fig. 3A and B). To determine whether we can obtain a stronger Minute phenotype, we have mobilized the P-lacW transposon at position 23A and selected w revertant lines. Among these lines we found that the Minute phenotype is enhanced in the R12 line whose heterozygous adults exhibit shorter bristles (∼40% reduction in length), a reduced body size, and a 4-day delay in eclosion (Fig. 3C) and whose homozygous animals die during the second and early third larval instars. Moreover, on both sides of the scutum of l(2)k168-14/+ and l(2)k168-14R12/+ flies, we observed a partial cleft extending anteriorly from the scutoscutellar suture, which is more pronounced in l(2)k168-14R12/+ flies. These findings suggest (i) that the imprecise excision of the P-lacW transposon in the R12 line has produced a chromosomal deletion removing part or all of the gene affected by the P-lacW insert in l(2)k168-14 and (ii) that the original l(2)k168-14 line corresponds to a hypomorphic allele, whereas the R12 line represents a stronger allele. As revealed by molecular investigations (vide infra), the R12 allele is characterized by a chromosomal deletion which removed part of the 5′ UTR as well as the upstream control and promoter region.
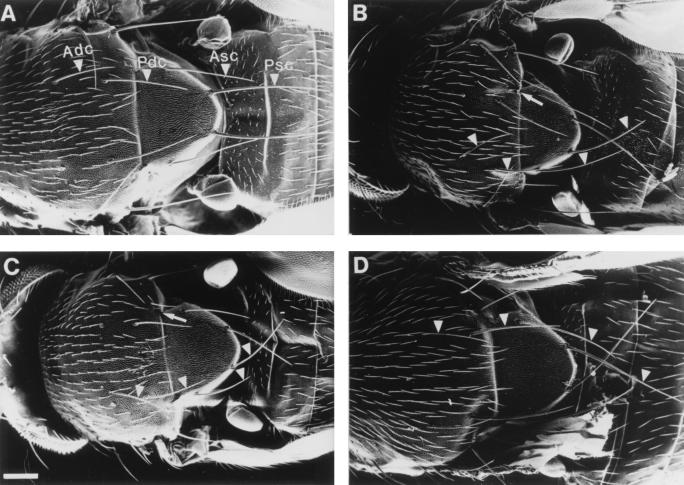
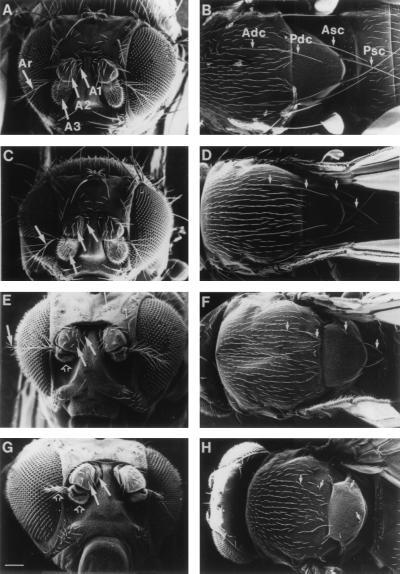
FIG. 3.
The Minute phenotype in different l(2)k168-14 variants. SEM images of the scutum and scutellum of (A) wild-type Oregon-R, (B) l(2)k168-14/+, (C) l(2)k168-14R12/+, and (D) l(2)k168-14R12/+; P-[rpS21+]/+. Compared to the bristles of wild-type animals, the size of the bristles is reduced by ∼20% in l(2)k168-14/+ and by ∼40% in l(2)k168-14R12/+ flies. In addition, a partial cleft, indicated by an arrow, extends anteriorly from the scutoscutellar suture on both sides of the scutum of l(2)k168-14 and l(2)k168-14R12/+ flies. The nomenclature of the bristles indicated by arrowheads: Adc, anterior dorsocentral bristle; Asc, anterior scutellar bristle; Pdc, posterior dorsocentral bristle; Psc, posterior scutellar bristle. Bar = 100 μm.
Cloning and sequencing of the S21 ribosomal protein gene.
A genomic DNA fragment flanking the P-insert in l(2)k168-14 was isolated by plasmid rescue (70) and used to screen a Drosophila Oregon-R wild-type genomic library. As shown in Fig. 4A, a chromosomal segment of ∼28-kb genomic DNA and encompassing the P-insertion site was mapped and investigated by reverse Southern hybridization for the presence of reiterated sequences (data not shown). The detection of an ∼5-kb repetitive fragment located on the left side of the P-insert at a distance of ∼1 kb prompted us to further isolate clones from a Canton-S genomic library. Alignment of the Oregon-R and Canton-S genomic maps revealed the occurrence of an additional 4.5-kb DNA segment in the Oregon-R genome corresponding to the reiterated sequence. This repetitive element was not further analyzed. No further repeated sequence could be detected in the isolated genomic DNA segments.
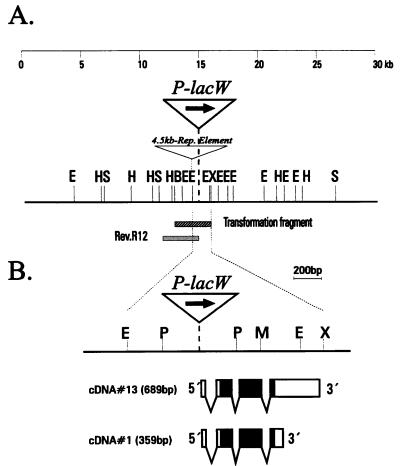
FIG. 4.
Map of the chromosomal locus around the P-lacW insert of l(2)k168-14 in the region 23B in D. melanogaster. (A) A composite map of ∼30 kb of DNA from the rpS21 region is shown with the coordinate scale above the map. Coordinate 0 is arbitrarily chosen at the left end of the cloned Drosophila DNA segment. The exact location of the P-lacW insertion in l(2)k168-14 was mapped by DNA sequencing, as indicated in Fig. 3. An unknown repetitive element with a size of ∼4.5 kb was found in genomic DNA cloned derived from Oregon-R flies but absent in DNA from Canton-S flies. The genomic fragment used to generate transgenic flies is indicated below the map as a shaded box (transformation fragment), as well as the interstitial deletion Rev.R12 which has been induced by imprecise excision of the P-lacW element in l(2)k168-14. (B) Enlargement of the map around the site of the P-lacW insertion with localization of the two size classes of transcripts encoded by the rpS21 gene, as determined from sequence analysis of cDNA 13 and cDNA 1, containing a 689- and a 359-nucleotide long transcript, respectively. The coding sequences are indicated by solid boxes, and the noncoding transcribed regions are marked by open boxes. Restriction sites: BamHI (B), EcoRI (E), HindIII (H), PstI (P), SalI (S), SmaI (M), and XhoI (X).
A transcript map for a region covering ∼14 kb around the P-lacW insertion site was generated by probing Northern blots made of poly(A)+ RNA extracted from embryos, larvae, and adult flies with a series of genomic fragments. Only one small (∼0.4-kb) poly(A)+ transcript was detected on the proximal side of the P-lacW insertion site, and this transcript was found to exhibit a relatively uniform expression throughout development. On the distal side of the P-insert no transcript could be identified by Northern blotting between the insertion site and the reiterated sequence present in the Oregon-R genomic DNA. These results suggest that the P-lacW element responsible for the l(2)k168-14 lethality is inserted within or in the immediate vicinity of the transcription unit encoding the 0.4-kb poly(A)+ RNA.
To determine the organization of this transcription unit, embryonic cDNA libraries made with poly(A)+ RNA extracted from either 0- to 9-h-old or 0- to 16-h-old embryos were screened with a 3.9-kb HindIII-XhoI DNA fragment from the Canton-S genome, as shown in Fig. 4A. From this, 14 independent cDNA clones were recovered and assigned to two different size classes of transcripts of 0.4 and 0.7 kb, respectively. Determination of the nucleotide sequence of the genomic DNA and cDNAs showed that both classes of transcripts belong to the same transcription unit, with nearly identical 5′ ends, but exhibit different lengths in their 3′ ends, as indicated in Fig. 4B. The 0.7-kb transcripts were found to contain a 3′ extension of 326 nucleotides compared to the 0.4-kb transcripts (Fig. 5). Alignment of the sequence of the 0.4-kb class of cDNAs (12 clones) with the genomic DNA sequence revealed four exons of 41 (or larger), 60, 191, and 75 nucleotides separated by three relatively small introns of 71, 71, and 63 nucleotides. The last exon of the 0.7-kb class of cDNAs (two clones) is larger, with 400 nucleotides. A long exposure of a Northern blot of poly(A)+ RNA hybridized with a genomic fragment covering the transcribed region showed that the 0.7-kb transcripts are relatively rare, with an abundance less than 5% of the 0.4-kb class of transcripts (data not shown). Therefore, the finding of only two cDNA clones from the 0.7-kb transcripts among the 14 isolated clones reflects the relative abundance of this class of transcripts.
FIG. 5.
Nucleotide sequence of the rpS21 gene and the predicted amino acid sequence of its product. Introns and untranscribed sequences are shown in lowercase letters; exons are shown in uppercase letters. The first nucleotides of the cDNAs 1 and 13 are indicated by an arrow with an asterisk, and the last nucleotides of these cDNAs are indicated by an arrow with an open circle. The insertion of the P-lacW transposon in l(2)k168-14 is marked by a diamond. Arrowheads indicate transcription initiation sites as determined by 5′-end amplification of S21 cDNAs by using an ovarian cDNA library as a template, with the major initiation site located at −68. The primer sequence used for the amplification is double underlined. The putative polyadenylation site is underlined. The GenBank/EMBL/DDBJ accession number for the genomic and amino acid sequences of Drosophila rpS21 is AJ009557.
To further ascertain the position of the P-insertion site relative to the initiation site of the 0.4-kb transcript, the 5′ extremity of this transcript was amplified from a cDNA library made with poly(A)+ RNA extracted from ovaries of Oregon-R adult flies in λ Hybri-ZAPII vector by using a 5-AD PCR primer from the GAL-4 activation domain and a primer located at the junction between the third and fourth exons of the rpS21 transcript (3′-TGCCGTAGTAATGGTTCTTGAAGATT), as indicated in Fig. 5. The amplified fragments were subcloned in CR-ScriptAMP/SK+ plasmid (Stratagene). The 5′-terminal sequence of the insert in 13 independent clones was determined, revealing that the 5′ ends of three cDNAs were located upstream from the P-insertion site. This finding indicates that the P-lacW transposon is inserted at the beginning of the 5′ untranslated region of the 0.4-kb transcript. This was further confirmed by primer extension analysis of embryonic poly(A)+ RNA, which showed that a series of extended fragments initiate upstream from the P-insertion site, as indicated in Fig. 5. As in the case of other genes encoding ribosomal proteins and protein synthesis elongation factors (32), the region upstream from the presumptive transcription initiation site lacks TATA or CAAT motifs. The absence of such motifs may explain the apparent scattered initiation of rpS21 transcription. However, it should be noted that, downstream of the major transcription initiation sites and the insertion site of the P-lacW transposon, there is a 16-pyrimidine tract (nucleotide positions −10 to −25 in Fig. 5) highly reminiscent of the TOP sequences found around the transcription start of insect and vertebrate ribosomal protein genes (47).
The cDNA sequence displays an open reading frame of 83 codons initiated by an ATG present in the second exon (Fig. 5). This open reading frame encodes a protein with a predicted molecular mass of 9,275 Da and is preceded by a sequence which conforms to the Drosophila translation-start consensus sequence ANN (C/A) A (A/C) (A/C) ATGN (10). A canonical poly(A) addition site, AATAAA, is located 26 nucleotides from the start of the poly(A) tract in the 0.4-kb class of transcripts. No such site could be identified at the 3′ end of the sequence encoding the 0.7-kb class of transcripts.
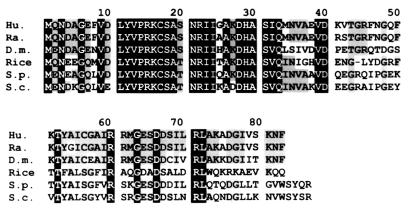
Analysis of the encoded sequence revealed strong homology between the deduced protein sequence and the S21 ribosomal protein of different organisms. As shown in Fig. 6, all identified RpS21 homologues are made of 83-amino-acid residues, except for the two yeast proteins, which contain four additional residues. Amino acid identity is particularly high in the N-terminal region of RpS21 and is well conserved between distantly related species. Drosophila RpS21 shows 73% identity with both the human and rat proteins (8, 31); 60 and 53% identity with the RpS25 protein of the fission yeast Schizosaccharomyces pombe and the baker’s yeast Saccharomyces cerevisiae, respectively (31, 66); and 51% identity with rice RpS21 (48). The N-terminal moiety of RpS21 contains several potential phosphorylation sites at position S18 for cAMP-cGMP dependent protein kinase, position S35 for casein kinase II, and positions S20 and T43 for protein kinase C which, with the exception of the T43 site, are conserved in all other species so far examined.
FIG. 6.
Amino acid alignment of six RpS21 proteins. The species names and the respective GenBank/EMBL/DDBJ and Swissprot accession numbers are given as follows: Hu, human (numbers L04483 and P35265); Ra, rat (X79059 and P05765); Rice (D12633/P35687); D.m., D. melanogaster; S.p., the fission yeast Schizosaccharomyces pombe (—/P05764); and S.c., the baker’s yeast Saccharomyces cerevisiae (X07811 and P05760). Sequence identity of the Drosophila RpS21 reaches 70% with both human and rat proteins, 60% with the fission yeast protein, 53% with the baker’s yeast protein, and 51% with the rice protein. Invariant amino acids are boxed in black, and amino acids conserved in at least 50% of the proteins are boxed in grey.
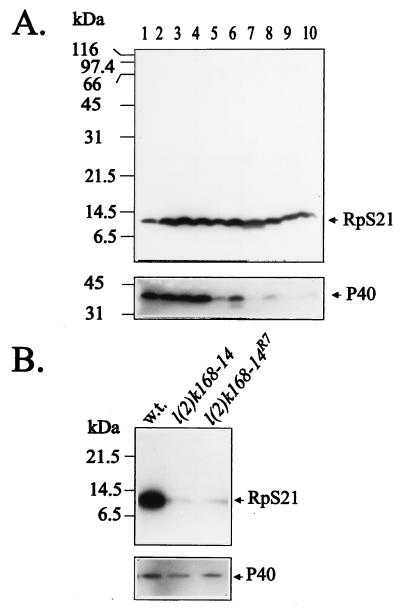
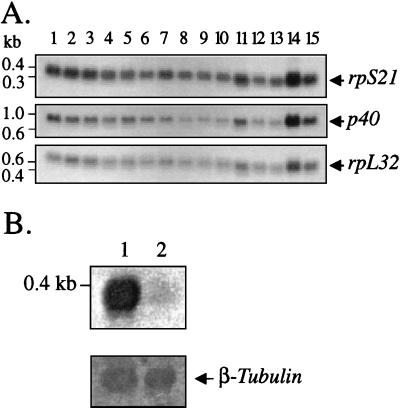
To show that the isolated sequence encodes RpS21, a cDNA corresponding to the 0.4-kb class of transcripts was translated in vitro in a coupled transcription-translation reticulocyte system by using T7 or T3 RNA polymerase. The reaction driven by the T7 RNA polymerase was found to produce a single [35S]methionine-labeled polypeptide with an apparent molecular mass of ∼10 kDa corresponding to the calculated molecular weight of RpS21 (see Fig. 11, lane 3). Moreover, the 35S-labeled polypeptide was specifically precipitated with antibodies raised against synthetic peptides corresponding to either the NH2- or the COOH-terminal ends of the conceptual translation product of the rpS21 gene. Immunoblot analysis revealed that RpS21 is relatively uniformly expressed at all developmental stages (Fig. 8A), reflecting the abundance of rpS21 transcripts during development (Fig. 7A) which appears to be three- to fourfold greater than the abundance of the transcripts encoding the ribosomal proteins RpL32 (formerly Rp49) and P40 with sizes of 0.6 and 1.0 kb, respectively. Further Northern blot analysis showed that the level of rpS21 transcription (Fig. 7B) is strongly reduced in l(2)k168-14 larvae, and Western blot studies confirmed that the level of RpS21 protein expression (Fig. 8B) is similarly lowered in l(2)k168-14 larvae and in l(2)k168-14R7 larvae. The l(2)k168-14R7 allele is characterized by an internal deletion of the P-lacW transposon that has left 48 nucleotides of both 3′ and 5′ ends flanked by the 8-bp duplication of genomic sequence that is characteristic of P-element insertions (49a). This allele displays a phenotype similar to that of the original l(2)k168-14 mutation, indicating that the insertion of ∼50 nucleotides can produce an effect identical to that caused by the insertion of a 10.2-kb P-lacW transposon.
FIG. 11.
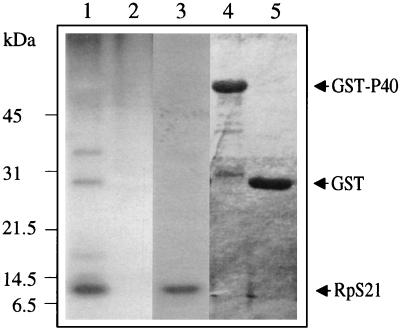
Binding of RpS21 with P40 in vitro. The reticulolysate translation system was used to synthesize RpS21 in the presence of [35S]methionine. GST-P40 and GST proteins bound to glutathione-Sepharose 4B beads were incubated with labeled RpS21, and the beads were extensively washed. 35S-labeled RpS21 proteins bound to GST-P40 (lane 1) or GST proteins (lane 2) were analyzed by SDS-PAGE and detected by autoradiography. Lane 3 shows input 35S-labeled RpS21 proteins. Lanes 4 and 5 show Coomassie blue staining of lanes 1 and 2 showing the loaded GST-P40 and GST proteins, respectively.
FIG. 8.
Expression of RpS21 and P40 throughout development in wild-type animals and in various genotypes. (A) Developmental profile of RpS21 and P40 expression. Immunodetection of RpS21 and P40 in extracts of ovaries of 3-day-old females (lane 1); embryonic stages of 0 to 3, 0 to 6, 6 to 9, 9 to 13, and 13 to 24 h after egg laying (lanes 2 to 6 respectively); third-instar larvae (lane 7); pupae (lane 8); and adult females (lane 9) and males (lane 10). The same Western blot was successively probed for RpS21 (above) and P40 (below), respectively. (B) Expression of RpS21 and P40 in proteins extracted from wild-type, homozygous l(2)k168-14, or homozygous l(2)k168-14R7 third-instar larvae shows that the level of RpS21 is greatly reduced in mutant larvae, whereas the expression of P40 is similar in wild-type and mutant larvae.
FIG. 7.
Abundance of the rpS21 and sta transcripts during development. (A) A developmental Northern blot was successively probed with 32P-labeled rpS21, sta, and rpL32 cDNAs. Embryonic stages (in hours): 1 (0 to 3), 2 (3 to 6), 3 (6 to 9), 4 (9 to 12), 5 (11 to 14), 6 (14 to 17), 7 (17 to 20), 8 (18 to 21), and 9 (21 to 24). Larval stages, 10 (first), 11 (second), and 12 (third); pupal stage, 13; adult stages: 14 (females) and 15 (males). rpL32 cDNA probe was used as a control for loading. (B) rpS21 expression is strongly reduced in l(2)k168-14 10- to 15-day-old third-instar larvae (lane 1) compared to Oregon-R wild-type larvae (lane 1). The amount of poly(A)+ RNA in each sample was equilibrated by using a β-tubulin probe as a control for loading.
A single copy of a 2.8-kb BamHI-XhoI genomic fragment containing the rpS21 gene with an ∼1-kb upstream sequence was found to restore the full development and viability of homozygous l(2)k168-14 animals. These results confirm that the P-insert in l(2)k168-14 affects only the expression of the rpS21 gene. In addition, the transgene could abolish the Minute phenotype of l(2)k168-14/+ flies, as well as the stronger Minute phenotype associated with Df(2L)k168-14R12 (Fig. 3D) which consists of a deletion removing ∼2.5-kb genomic DNA upstream from the rpS21 gene and was generated by mobilization of the P-lacW transposon present in l(2)k168-14. However, the lethality associated with Df(2L)k168-14R12 could be rescued only by two copies of the rpS21+ transgene. This requirement indicates that the R12 deletion, which has removed the promoter and upstream control elements of the rpS21 gene without affecting any other essential genes, behaves like a strong hypomorph or null allele.
RpS21 interacts with P40 encoded by the stubarista gene.
The sequence analysis provides evidence that the affected gene encodes a component of the 40S ribosomal subunit. However, the viability of the tumorous hematopoietic cells suggests that RpS21 may play an accessory role in the protein synthesis machinery rather than constituting a structural component of the ribosome. To investigate the nature of the association of RpS21 with ribosomes, we have undertaken a search for proteins interacting with RpS21 and determined the distribution of RpS21 within ribosomal particles, including polysomes, monosomes, and native ribosomal subunits.
To determine with which protein(s) RpS21 can interact, we carried out a yeast two-hybrid search (6) with full-length RpS21 as a bait and found 18 positive clones among 5 × 106 double-transformed yeast colonies. These clones consisted of double transformants which grew in medium lacking His, Leu, and Trp and were expressing β-galactosidase. Determination of the nucleotide sequence of the cDNA insert in the vector containing the GAL4 activation domain revealed that the 18 clones contain a full coding sequence for the ribosome-associated P40 protein (or P40) linked through a short in-frame sequence (6 to 10 codons) originating from the 5′ untranslated region of the P40 transcript to the GAL4 activation domain. P40 was previously identified as a protein associated with polysomes (3), presumably bound to the 40S ribosomal subunit (46), and was found to be particularly abundant in human colon carcinoma cells (82). In D. melanogaster, P40 is encoded by the stubarista gene (sta), and the viable sta1 mutation displays Minute-like characteristics with malformed antennae, short and thin bristles, and female sterility (46). To further confirm the interaction between P40 and RpS21, we performed a complementary yeast two-hybrid screening in which P40 was used as a bait. With this construct we screened 2 × 106 yeast recombinant colonies and found 16 positives clones, one of which was found to encode RpS21. Transformation of fusion constructs of RpS21 and P40 with both the GAL4 DNA-binding domain (pBD-GAL4) and the GAL4 activation domain (pAD-GAL4) into fresh yeast cells confirmed that only the double transformants were able to grow on medium lacking His, Leu, and Trp and were expressing β-galactosidase. These assays further established that the two proteins interact directly. Moreover, we mapped by deletion analysis the region in p40 which is involved in the RpS21-P40 interaction. This analysis showed that a centrally located area in P40 (amino acid residues 55 to 230) is important for interaction with RpS21.
Hyperplasia of the hematopoietic organs in lethal stubarista third-instar larvae.
The finding of a strong interaction between RpS21 and P40 in the yeast two-hybrid system prompted us to investigate the phenotype of the zygotic lethal sta mutant. Two categories of sta mutations have been described. The first category is characterized by chromosomal rearrangements producing weak hypomorphs, such as sta1. This viable mutation was induced by X rays and is a combination of the deletion of the 1E1-2 2B3-4 region of the X chromosome, causing the deficiency Df(1)sta and the translocation of the deleted region onto the third chromosome at 89B21-C4, resulting in the transposition Tp(1;3)sta (7). Since Tp(1;3)sta can be separated from the translocation without impairing viability, it is designated Dp(1;3)sta. Homozygous sta1 mutations give rise to viable adults displaying shortened antennae, aristae with thickened and irregular branches, and short and sparse bristles (50, 46). The second category of sta mutations, like the sta2 allele, is defined by recessive lethality without cytologically detectable rearrangements. The sta2 mutation was induced with ethyl methanesulfonate and produces zygotic lethality. Although the stubarista gene has been molecularly characterized (46), no information is currently available on the lesions affecting the sta alleles except for genetic data indicating that sta1 and sta2 alleles retain partial sta+ activity and that sta1 is a much weaker hypomorph than sta2 (46).
Our analysis of the lethal phase of sta2 showed that homozygous larvae can reach the third larval stage, albeit with a considerable delay in their development. sta2 third-instar larvae tend to die early when they are left in the food but when they are placed in a humid chamber they can survive as relatively underdeveloped third-instar larvae for up to 2 weeks after the heterozygous siblings have emerged from the pupal case, and they gradually die as larvae without forming a puparium. In aged mutant larvae, as in l(2)k168-14 larvae, the hematopoietic organs are considerably enlarged, with the maximal hyperplasia occurring in the first lobes (data not shown). Moreover, the sta2 overgrown organs remained globular and compact, and no melanotic masses could be detected in either the hematopoietic organs or in other parts of the larval body.
Enhancement of the l(2)k168-14 tumor phenotype by underexpression of P40.
Our hypothesis was that a decrease in P40 expression should enhance or suppress the tumorous phenotype associated with the absence of RpS21 expression. In order to test for genetic interaction between l(2)k168-14 and stubarista, we used first the strongest available allele sta2 and tested a series of genetic combinations. In these experiments the lethal phase and the structure of the internal organs in genetically marked larvae were determined, as indicated in Tables 1 and 2. Although hemizygous sta2/Y; l(2)k168-14/l(2)k168-14 animals die as first- or early-second-instar larvae, we found that homozygous l(2)k168-14 female third-instar larvae which are heterozygous for sta2 are larger in size and display considerably larger overgrown imaginal discs (Fig. 9). In particular, the first leg discs tend to fuse together (Table 2 and Fig. 9B and C), and the hematopoietic organs are larger than in corresponding homozygous l(2)k168-14 third-instar larvae with two sta+ copies. The finding that the tumor phenotype is enhanced when P40 expression is reduced in rpS21-deficient larvae indicates a direct genetic interaction between stubarista and l(2)k168-14. Furthermore, the reduction in the expression of P40 was found to exert also a behavioral effect, as revealed by the observation that sta2/+; l(2)k168-14/l(2)k168-14 third-instar larvae are more active than homozygous l(2)k168-14 third-instar larvae.
TABLE 2.
Phenotype of internal organs in l(2)k168-14 and stubarista larvaea
| Larval genotype | Age (days) at dissection | No. of dissected larvae | Avg larval size | Avg organ size
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain
|
Imaginal discs
|
Hematopoietic organs | ||||||||||
| Hemi-spheres | Ventral ganglion | W | L1 | L2 | L3 | E/A | H | |||||
| y sta2/Y | 13–21 | 11 | 2.0 | 2.0 | 3.3 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 3.8 |
| y w/y w; l(2)k168-14/l(2)k168-14 or y w/Y; l(2)k168-14/l(2)k168-14 | 13–19 | 25 | 2.8 | 2.8 | 3.4 | 2.6 | 2.9 | 2.9 | 2.5 | 2.8 | 2.4 | 4.4 |
| y sta2/y w; l(2)k168-14/l(2)k168-14 | 13–22 | 31 | 3.0 | 3.1 | 3.8 | 3.2 | 3.6 | 3.6 | 3.1 | 3.4 | 3.1 | 4.8 |
The sizes of the larval body and the organs dissected from mutant larvae were compared with those of mature third-instar Oregon-R larvae and estimated according to an arbitrary scale: 1.0, strongly reduced size (≤50%); 2.0, reduced size (50 to 90%); 3.0, normal size (90 to 110%); 4.0, increased size (110 to 150%); and 5.0, strongly increased size (≥150%). The average values are indicated. Abbreviations: E/A, eye-antenna disc; H, haltere disc; L1, L2, and L3, first, second, and third pairs of leg discs, respectively; W, wing disc.
FIG. 9.
Enhanced overgrowth of l(2)k168-14 imaginal discs by underexpression of P40. (A) Wing (W), halter (H), and third leg (L3) imaginal discs dissected from Oregon-R wild-type (wt) and sta2/+; l(2)k168-14/l(2)k168-14 (m) third-instar larvae. (B and C) Brain hemispheres (B) and associated first-leg (L1) and second-leg (L2) imaginal discs of the same larvae as in panel A. As shown in panel C, the overgrown first pair of leg discs of the sta2/+; l(2)k168-14/l(2)k168-14 third-instar larva tends to fuse together. Note that the ventral ganglia of both wild-type and mutant brains were damaged during dissection. Bar = 100 μm.
Dosage interaction between l(2)k168-14 and stubarista.
Since homozygous and hemizygous sta1 animals exhibit shorter scutellar and thoracic bristles and a reduced body size (46), we tested whether the weak Minute phenotype in l(2)k168-14/+ animals can be enhanced by reduced expression of P40. For this purpose, we examined a series of viable combinations of l(2)k168-14 and stubarista alleles. As indicated in Table 3, we found that sta mutations behave as recessive enhancers of l(2)k168-14, causing a marked reduction in the length and width of the scutellar and dorsothoracal bristles and a reduction in size of the antennae and aristae. The antennal and bristle phenotypes of Df(1)sta; l(2)k168-14/+; Dp(1; 3)sta/+ males and females (Fig. 10) are more severe than the corresponding phenotypes of Df(1)sta/sta2; l(2)k168-14/+; Dp(1;3)sta/+ which are in turn more severe than the phenotypes of sta2; l(2)k168-14/+; Dp(1;3)sta/+. Thus, these phenotypes reflect the degree of loss of function of the sta alleles, with Df(1)sta being a null allele, sta2 being a strong hypomorph, and Df(1)sta; Dp(1;3)sta being a weak hypomorph. The phenotypic characteristics of lethal and viable combinations between l(2)k168-14 and stubarista, together with the molecular interaction detected in the yeast two-hybrid system, indicate a strong interaction between RpS21 and P40.
TABLE 3.
Comparison of bristle, antenna, and arista phenotypes in viable combinations of l(2)k168-14 and stubarista
| Combina-tion no. | Genotype | Phenotypea
|
||
|---|---|---|---|---|
| Bristles | Antenna | Arista | ||
| 1 | Wild type | + | + | + |
| 2 | l(2)k168-14/+ | Weak Minute | + | + |
| 3 | l(2)k168-14/+; Dp(1;3)sta/+ | Weak Minute (like combination 2) | + | + |
| 4 | sta2; Dp(1;3)sta/+ | Shorter and thinner than in combination 3 | Fifth segment enlarged | Branches shorter and axis proximally thicker than in wild type |
| 5 | Df(1)sta/sta2; Dp(1;3)sta/+ | Shorter and thinner than in combination 4 | Third segment hairless and smaller than in combination 4 | Similar to combination 4 |
| 6 | Df(1)sta; Dp(1;3)sta/+ | Shorter and thinner than in combination 5 | Third segment smaller (0.5×) than in combination 5 | As in combination 5, albeit with a shorter axis |
| 7 | sta2; l(2)k168-14/+; Dp(1;3)sta/+ | Shorter and thinner than in combination 5 | Third segment smaller than in combination 5, with a slight protrusion towards arista | As in combination 5, but slightly shorter (albeit still longer than in combination 6) |
| 8 | Df(1)sta/sta2; l(2)k168-14/+; Dp(1;3)sta/+ | Similar to combination 7 | Third segment similar to combination 7, but even smaller | Similar to combination 7 |
| 9 | Df(1)sta; l(2)k168-14/+; Dp(1;3)sta/+ | Shorter and thinner than in combination 8 | Smaller than in combination 8, with a marked protrusion towards the arista | Axis shorter (0.5×) and thicker than in combination 8, with an equal number of shorter branches |
With respect to the length and width of the scutellar and dorsothoracal bristles, as well as the size of the antennae, the various genetic combinations between l(2)k168-14 and the sta alleles can be organized in the following phenotypic series, ranging from normal (combination 1) to the most severe size reduction, as observed in combination 9. For the length and width of the bristles, the combinations were as follows: 1 > (2 = 3) > 4 > 5 > 7 > 8 > 6 > 9. For the sizes of the antennae, the combinations were as follows: (1 = 2 = 3) > 4 > 5 > 7 > 8 > 6 > 9. In addition, all of these combinations were found to be fertile.
FIG. 10.
Dosage interaction between l(2)k168-14 and stubarista. SEM images of the antennal and bristle phenotypes of (A and B) wild-type Oregon-R, (C and D) l(2)k168-14/+, (E and F) sta1/Y (or sta1/sta1), and (G and H) sta1/Y; l(2)k168-14/+. In panels A, C, E, and G the arrows A1, A2, A3, and Ar indicate the first, second, and third segments of the antenna and the arista, respectively. In panels B, D, F, and H the arrows Adc, Asc, Pdc, and Psc point out the thoracic and scutellar bristles as indicated in Fig. 2. In panels E and G, the open arrows point to the particularly smaller A3 and Ar segments observed in these genetic combinations. The reduction in the size of the bristles, antennal segments, and arista in the different viable combinations between l(2)k168-14 and sta are detailed in Table 3. sta1 corresponds to Tp(1;3)1E1-2; 2B3-4; 89B21-C4, which can be also described as Df(1)sta/+ (or Df(1)sta/Y); Dp(1;3)sta/+. Bar = 100 μm.
Biophysical interaction between RpS21 and P40.
To further show a direct interaction between RpS21 and P40, we performed an in vitro binding assay. The rpS21 cDNA was expressed in the in vitro transcription-translation reticulocyte lysate system in the presence of [35S]methionine (Fig. 11, lane 3), producing an ∼10-kb polypeptide. To examine binding with P40, the GST-P40 fusion protein and the GST protein, produced in E. coli and purified on glutathione-Sepharose 4B beads (Fig. 11, lanes 4 and 5) were incubated with 35S-labeled RpS21 proteins. The beads were then washed, and bound labeled proteins were analyzed by SDS-PAGE and autoradiography (Fig. 11, lanes 1 and 2). Radiolabeled RpS21 was detected to be bound to GST-P40 but not to GST alone. These results confirm a direct biophysical interaction between RpS21 and P40.
RpS21 is associated with the native 40S ribosomal subunit.
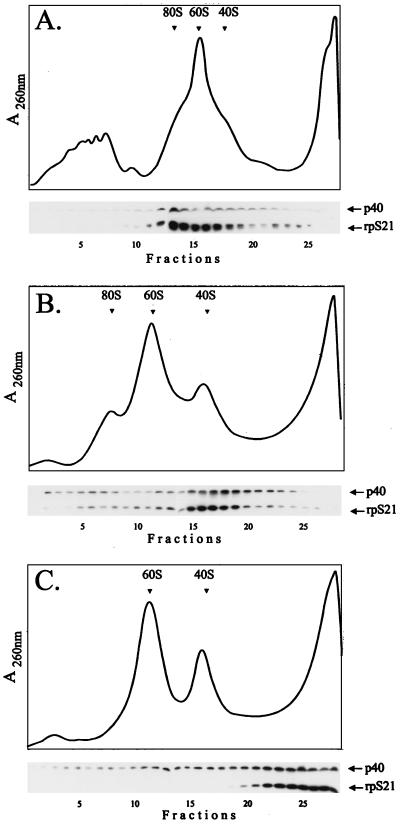
To establish the pattern of association of RpS21 and P40 with the different categories of ribosomal particles, we prepared postnuclear cytoplasmic extracts from Drosophila embryos or Drosophila Schneider S2 cells grown at 25°C. Furthermore, to determine whether the association of these proteins with the ribosomal particles is salt labile, the extracts were centrifuged under different salt conditions. Proteins from fractions across the gradients were analyzed by gel electrophoresis, and both RpS21 and P40 were detected by immunoblotting by using specific polyclonal antibodies raised against the carboxyl end of RpS21 or the GST-P40 fusion protein, respectively. As shown in Fig. 12, this analysis revealed that at a low salt concentration (80 mM KCl), P40 is evenly distributed among ribosomal particles including polysomes, monomeric 80S ribosomes, and native ribosomal subunits. By contrast, RpS21 is found only in association with monomeric 80S ribosomes and native ribosomal subunits, presumably the native 40S subunit. Longer centrifugation in moderate salt concentration (300 mM KCl) showed that RpS21 is predominantly associated with the native 40S ribosomal subunits which are also enriched in P40. When the same extracts are centrifuged at a higher salt concentration (500 mM KCl), RpS21 is fully dissociated from the ribosomal particles and recovered at the top of the gradient. By contrast, although a large fraction of P40 is released from the ribosomes, a significant portion of this protein remains associated with polysomes and ribosomes. Treatment of the cytoplasmic extracts with RNase had no effect on the pattern of association of RpS21 and P40 with ribosomes, whereas treatment with 20 mM EDTA resulted in the release of both RpS21 and P40. These results indicate that RpS21 is associated with the native 40S ribosomal subunit and specifically interacts with P40, another ribosome-associated protein.
FIG. 12.
Effect of salt on the distribution of RpS21 and P40 among ribosomal particles. Extracts from Drosophila embryos collected overnight were fractionated on a 15 to 40% sucrose gradient containing 5 mM MgCl2 and either 80 mM (A), 300 mM (B), or 500 mM (C) KCl. The extracts were centrifuged at 23,000 rpm in an SW41 Beckmann rotor for either 5 h in order to obtain a complete profile of the distribution of ribosomal particles including polysome, 80S ribosomes, and native ribosomal subunits (A) or 18 h in order to improve the separation of the ribosomal subunits (B and C). Furthermore, dot blot analysis was performed on each fraction to determine the distribution of the 18S ribosomal RNA. The 40S, 60S, and 80S peaks are marked in the absorbance (A254) profile. Immunoblots of gradients fractions are aligned below the tracing to show the corresponding distributions of the ribosomal S21 and P40 proteins.
DISCUSSION
Here we show that the ribosome-associated proteins RpS21 and P40 can specifically interact and that the inactivation of the genes encoding these proteins leads to two phenotypes, depending upon their dosage. A reduction of approximately one-half of their expression produces a dominant viable Minute phenotype, including delayed development, short slender bristles, and small body size, whereas a nearly complete absence of RpS21 or P40 results in larval lethality with marked hyperplasia of the hematopoietic organs. However, the recessive phenotypes differ between l(2)k168-14 and sta2 larvae, in particular by the greater viability and longevity of the l(2)k168-14 larvae and a moderate hyperplasia of their imaginal discs, indicating that both proteins may exert specific functions in addition to their participation in the protein synthesis machinery.
Whether the cause of tissue overgrowth in l(2)k168-14 and sta2 larvae results from a direct involvement of RpS21 and P40 in the control of cell proliferation or from a secondary effect due to a defect in protein synthesis remains an open question. If tissue overgrowth results from a defect in protein synthesis, this defect should, however, be extremely selective since homozygous mutations in genes encoding other ribosomal or ribosome-associated proteins, with the exception of mutations in the rpS6 gene (65, 75, 76), cause no excessive cell proliferation (1, 15, 28, 39, 44, 56, 57, 61).
The dominant Minute and recessive lethal tumorous phenotypes are highly reminiscent of the abnormalities observed in mutations affecting the ribosomal protein rpS6 gene, which give rise to a weak viable Minute phenotype in heterozygous females (58) and to lethality with hyperplastic hematopoietic organs in hemizygous larvae (65, 75, 76). Furthermore, as with rpS6 (76), mosaic studies revealed that rpS21 is required for egg development in the ovary and cell viability in developing imaginal discs (data not shown). However, in contrast to rpS21 inactivation, a nearly complete absence of rpS6 expression results in an increased number of circulating plasmatocytes and lamellocytes which accumulate in various locations of the hemocoel and give rise to massive melanotic pseudotumors of self-encapsulated lamellocytes (65, 76). In RpS21- or P40-deficient larvae, no trace of melanization could be detected in the hemocoel or within the organs, even after a prolonged extension of larval life. Moreover, the hemolymph of RpS21-deficient larvae is strongly depleted of hemocytes. These features suggest that the differentiation of the hemocyte cell lineage is blocked at an earlier stage in RpS21- or P40-deficient larvae than in rpS6 mutant larvae, suggesting that these genes may control different stages of hematopoiesis in Drosophila spp. The block in hemocyte differentiation may result either from a defect in a nonribosomal function of these proteins or from a failure in the spatiotemporally coordinated translation of specific factors that control the pathway of hemocyte differentiation.
The current explanation proposed to account for the Minute phenotype is haploinsufficiency for a component of the protein synthesis machinery (24) reducing the overall rate of protein synthesis, which produces a lower mitotic rate compared to wild-type cells and a reduction in the rate of bristle synthesis. Molecular analysis of more than 40 ribosomal protein genes has revealed that, in addition to rpS21, rpS6, and p40, mutations in eight other ribosomal protein genes, including rp49 (or rpL32) (39), rpS2 (15), rpS3 (1), rpS5 (44), rpS13 (56), rpL9 (61), rpL14 (57), rpL19 (28), give rise to Minute phenotype. Furthermore, the Minute phenotype is also produced by mutations in the bobbed and mini loci, which are partial deletions of rRNA genes (54, 73), and by mutations in the suppressor of forked locus which encodes a protein thought to be involved in RNA stability (47a). Thus, the available evidence indicates that, in general, partial impairment of the protein synthesis machinery delays larval development and produces a dominant Minute phenotype.
Increasing evidence suggests that, in addition to their role in protein synthesis, some ribosomal proteins are involved in specific regulatory steps during normal development or else display enzymatic activities required in distinct cellular functions. For instance, a P-element insertion in the promoter region of the string of pearls (sop) gene, which encodes the S2 ribosomal protein, causes recessive female sterility with a block in oogenesis at the end of previtellogenesis (15). The Drosophila S3 ribosomal protein, which forms part of the domain on the ribosome where translation is initiated (9), contains three distinct enzymatic activities which were biochemically assayed: (i) an apurinic-apyrimidinic (AP) lyase activity, cleaving phosphodiester bonds via a β,δ-elimination reaction (78); (ii) an N-glycosylase activity, cleaving DNA containing 8-oxoguanine residues (18, 80); and (iii) a DNA deoxyribophosphodiesterase activity (60). These findings indicate that RpS3 can function in several steps of the DNA excision-repair pathway. Consistent with its multifunctionality, the Drosophila RpS3 protein is not only present in ribosomes but is also tightly associated with the nuclear matrix (78). The DNA cleavage activity of RpS3 suggests that this protein may be involved in the repair of oxidative and ionizing-radiation-induced DNA damage. Consistent with this idea, the human S3 ribosomal protein was identified as the AP endonuclease III missing or altered in group-D patients with xeroderma pigmentosum (34). However, although the endonuclease activity associated with RpS3 is lacking in xeroderma pigmentosum group D cells, the protein is clearly present in these cells in association with ribosomes suggesting that it may require association with specific proteins for endonuclease activity. Similarly, another Drosophila ribosomal protein, P0, displays endonuclease activity and is also found in association with the nuclear matrix (81). Moreover, in human cells, expression of the ribosomal P0 protein is induced by functional alkylating agents that cause DNA damage (26). RpS3 is also overexpressed in colorectal cancer cells (53), supporting the idea that both RpS3 and P0 may be involved in DNA damage processing, particularly in drug-resistant cells treated with DNA cross-linking agents.
Among ribosomal proteins, the S6 protein has attracted particular attention because it is phosphorylated in vivo in response to various mitogens. Phosphorylation of RpS6 was found to be a critical event in the initiation of cell growth and proliferation and to be a ubiquitous response when cells are induced to reenter the cell cycle (for a review, see reference 23). The higher rate of protein synthesis resulting from RpS6 phosphorylation can be attributed to a selective advantage of RpS6-phosphorylated 40S subunits to enter into polysomes and to a modification in the ability of the 40S ribosomal subunit to recognize specific mRNAs (51). Recent studies have revealed that the mechanism underlying this selectivity involves the presence of an oligopyrimidine tract in the 5′ UTR of the target mRNAs, which are designated TOP mRNAs (32, 47, 67). This class of transcripts includes mRNAs for ribosomal proteins and other components of the translation machinery, such as elongation factors. Interestingly, our analysis of the 5′ UTR of the rpS21 mRNA revealed the occurrence of a pyrimidine tract located at or in the vicinity of the 5′ extremity of the rpS21 transcripts, indicating that their translation may be regulated by a mechanism similar to that shown for transcripts encoding other ribosomal proteins. The involvement of RpS6 in the selection of mRNAs is further sustained by structural analyses of the 40S ribosomal subunit, which showed that RpS6 maps on the small head region on the inner side of the beak, in a position juxtaposed to the larger 60S subunit in an area implicated in mRNA binding (49, 69).
A common feature of all ribosomal proteins involved in the regulation of either Drosophila or human cell proliferation is their involvement in different steps of translation initiation, as defined by the process through which a 40S ribosomal subunit associates with the correct AUG codon, binds an initiator tRNA, and joins a 60S ribosomal subunit to form an actively translating ribosome. As part of this process, the S6 ribosomal protein plays a role in the entry of the 40S ribosomal subunit into polysomes (51). The QM or L10 protein is involved in joining of the 40S with the 60S ribosomal subunit (21). Although the precise molecular function of RpS21 remains yet to be determined, this protein is associated with the surface of mammalian 40S ribosomal subunits (42) and our work shows that it is associated with native 40S ribosomal subunits and absent from polysomes, indicating a role in translation initiation. Furthermore, immunostaining and cell fractionation procedures revealed no detectable amount of RpS21 in the nucleus (data not shown), suggesting no function in the maturation or transport of the 40S ribosomal subunit from the nucleolus to the cytoplasm. A relevant function of P40 in translation initiation can be inferred from its homology with the prokaryotic ribosomal protein, S2, of E. coli (16, 46). The S2 protein, like P40, is associated with the small ribosomal subunit at its surface, where it contributes to stabilizing conformation (43), and is involved in tRNA binding (63, 68). Consistent with these results, our analysis reveals that Drosophila P40 is preferentially bound to native 40S subunits. Furthermore, in Hydra spp. the intracellular localization of P40 varies according to the stage of the cell cycle. In dividing cells P40 is diffusely distributed throughout the cytoplasm, but in nondividing cells it appears to be associated with the cytoskeleton (33a), a finding consistent with studies showing an association between polysomes and the cytoskeleton (11, 30). However, by carrying out a yeast two-hybrid search with P40 as a bait, we found that P40 binds to other ribosomal proteins, including S21 and S17, and novel proteins with no assigned function (data not shown), indicating that the major structure with which P40 is associated is the ribosome.
In conclusion, the present work shows that two ribosome-associated proteins, which are presumably involved in one or several steps of translation, contribute to the regulation of cell proliferation in the hematopoietic organs and the imaginal discs of Drosophila larvae. Although reduced expression of RpS21 or P40 impairs protein synthesis and decreases the rate of cell growth, as revealed by the Minute phenotype, a nearly complete absence of these proteins leads to excessive cell proliferation in specific tissues. However, tissue overgrowth becomes apparent only in aged larvae, indicating that the rate of protein synthesis is considerably decreased but sufficient to maintain cell proliferation. A knowledge of the interaction between RpS21 and P40 and their function during protein synthesis should provide a better understanding of the regulatory mechanism of cell growth and tumorigenesis.
ACKNOWLEDGMENTS
We thank Andrew Lambertsson for his help in the preliminary characterization of the Minute phenotype.
This work was supported by grants of the Deutsche Forschungsgemeindschaft (436UNG113/81/0) within the framework of a German-Hungarian program of scientific cooperation, the European Commission Biomed Programme (BMH1-CT94-1572) and Biotechnology Programme (BIO4-CT95-0202), a grant given to I.K. by the Hungarian Scientific Research Fund (OTKA T166/93), and an International Research Scholar’s award from the Howard Hughes Medical Institute given to I.K. and B.M.M.
REFERENCES
- 1.Andersson S, Saebøe-Larssen S, Lambertsson A, Merriam J, Jacobs-Lorena M. A Drosophila third chromosome Minute locus encodes a ribosomal protein. Genetics. 1994;137:513–520. doi: 10.1093/genetics/137.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner M. Drosophila: a laboratory handbook. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 3.Auth D, Brawerman G. A 33-kDa polypeptide with homology to the laminin receptor: component of translation machinery. Proc Natl Acad Sci USA. 1992;89:4368–4372. doi: 10.1073/pnas.89.10.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnard G F, Staniunas R J, Mori M, Puder M, Jessup M J, Steele G D, Chen L B. Gastric and hepatocellular carcinomas do not overexpress the same ribosomal protein messenger RNAs as colonic carcinoma. Cancer Res. 1993;53:4048–4052. [PubMed] [Google Scholar]
- 5.Barnard G F, Staniunas R J, Puder M, Steele G D, Chen L B. Human ribosomal protein L37 has motifs predicting serine/threonine phosphorylation and a zinc-finger domain. Biochim Biophys Acta. 1994;1218:425–428. doi: 10.1016/0167-4781(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 6.Bartel P, Chien C T, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. BioTechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 7.Belyaeva E S, Aizenzon M G, Semeshin V F, Kiss I I, Koczka K, Baritcheva E M, Gorelova T V, Zhimulev I F. Cytogenetic analysis of the 2B3-4-2B11 region of the X-chromosome of Drosophila melanogaster. I. Cytology of the region and mutant complementation groups. Chromosoma. 1980;81:281–306. doi: 10.1007/BF00285954. [DOI] [PubMed] [Google Scholar]
- 8.Bhat K S, Morrison S G. Primary structure of human ribosomal protein S21. Nucleic Acids Res. 1993;21:2939. doi: 10.1093/nar/21.12.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bommer U A, Lutsch G, Stahl J, Bielka H. Eukaryotic initiation factors eIF-2 and eIF-3: interactions, structure and localization in ribosomal initiation complexes. Biochemie. 1991;73:1007–1019. doi: 10.1016/0300-9084(91)90142-n. [DOI] [PubMed] [Google Scholar]
- 9a.Bray D F, Bagu J, Koegler P. Comparison of hexamethyldisilazane (HMDS), Peldri II, and critical-point drying methods for scanning electron microscopy of biological specimens. Microsc Res Tech. 1993;15:489–495. doi: 10.1002/jemt.1070260603. [DOI] [PubMed] [Google Scholar]
- 10.Cavener D R. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cervera M, Dreyfuss G, Penman S. Messenger RNA is translated when associated with the cytoskeletal framework from normal and VSV-infected HeLa cells. Cell. 1981;23:113–120. doi: 10.1016/0092-8674(81)90276-2. [DOI] [PubMed] [Google Scholar]
- 12.Chan Y-L, Diaz J-J, Deneroy L, Madjar J-J, Wool I G. The primary structure of rat ribosomal protein L10: relationship to a Jun-binding protein and to a putative Wilms’ tumor suppressor. Biochem Biophys Res Commun. 1996;225:952–956. doi: 10.1006/bbrc.1996.1277. [DOI] [PubMed] [Google Scholar]
- 13.Chiao P J, Shin D M, Sacks P G, Hong W K, Tainsky M A. Elevated expression of the ribosomal protein S2 gene in human tumors. Mol Carcinog. 1992;5:219–231. doi: 10.1002/mc.2940050309. [DOI] [PubMed] [Google Scholar]
- 14.Chester K A, Robson L, Begent R H J, Talbot I C, Pringle J H, Primrose L, Macpherson A J S, Boxer G, Southall, P. P, Malcolm A D B. Identification of a human ribosomal protein mRNA with increased expression in colorectal tumours. Biochim Biophys Acta. 1989;1009:297–300. doi: 10.1016/0167-4781(89)90119-x. [DOI] [PubMed] [Google Scholar]
- 15.Cramton S E, Laski F A. string of pearls encodes Drosophila ribosomal protein S2, has Minute-like characteristics, and is required during oogenesis. Genetics. 1994;137:1039–1048. doi: 10.1093/genetics/137.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis S C, Tzagoloff A, Ellis S R. Characterization of a yeast mitochondrial ribosomal protein structurally related to the mammalian 67-kDa high-affinity laminin receptor. J Biol Chem. 1992;267:5508–5514. [PubMed] [Google Scholar]
- 17.De Frutos R, Kimura K, Peterson K. In situ hybridization of Drosophila polytene chromosomes with digoxigenin-dUTP labelled probes. Methods Mol Cell Biol. 1990;2:32–36. [Google Scholar]
- 18.Deutsch W A, Yacoub A, Jaruga P, Zastawny T H, Dizdaroglu M. Characterization and mechanism of action of ribosomal protein S3 DNA glycosylase activity for the removal of oxidatively damaged DNA bases. J Biol Chem. 1997;272:32857–32860. doi: 10.1074/jbc.272.52.32857. [DOI] [PubMed] [Google Scholar]
- 19.Dick F A, Karamamou S, Trumpower B L. QSR1, an essential yeast gene with a genetic relationship to a subunit of the mitochondrial cytochrome bc1 complex codes for a 60S ribosomal subunit protein. J Biol Chem. 1997;272:13373–13379. doi: 10.1074/jbc.272.20.13372. [DOI] [PubMed] [Google Scholar]
- 20.Dowdy S F, Lai K M, Weissman B E, Matsui Y, Hogan B L, Stanbridge E J. The isolation and characterization of a novel cDNA demonstrating an altered mRNA level in nontumorigenic Wilms’ microcell hybrid cell. Nucleic Acids Res. 1991;19:5763–5769. doi: 10.1093/nar/19.20.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisinger D F P, Dick F A, Trumpower B L. Qsr1p, a 60S ribosomal subunit protein is required for joining of 40S and 60S subunits. Mol Cell Biol. 1997;17:5136–5145. doi: 10.1128/mcb.17.9.5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisinger D F P, Jiang H P, Serrero G. A novel mouse gene highly conserved throughout evolution: regulation in adipocyte differentiation and in tumorigenic lines. Biochem Biophys Res Commun. 1993;196:1227–1232. doi: 10.1006/bbrc.1993.2383. [DOI] [PubMed] [Google Scholar]
- 23.Ferrari S, Thomas G. S6 phosphorylation and the p70s6k/p85s6k. CRC Crit Rev Biochem Mol Biol. 1994;29:385–413. doi: 10.3109/10409239409083485. [DOI] [PubMed] [Google Scholar]
- 24.Fisher E M C, Beer-Romero P, Brown L G, Ridley A, McNell J A, Lawrence J B, Willard H F, Bieber F R, Page D C. Homologous ribosomal protein genes on the X and Y chromosomes: escape from X inactivation and possible implications for Turner syndrome. Cell. 1990;63:1205–1218. doi: 10.1016/0092-8674(90)90416-c. [DOI] [PubMed] [Google Scholar]
- 25.Gateff E, Mechler B M. Tumour suppressor genes of Drosophila melanogaster. Crit Rev Oncog. 1989;1:221–245. [PubMed] [Google Scholar]
- 26.Grabowski D T, Pieper R O, Futscher B W, Deutsch W A, Erikson L C, Kelley M R. Expression of ribosomal phosphoprotein P0 is induced by antitumor agents and increased in Mer-human tumor cell lines. Carcinogenesis. 1992;13:259–263. doi: 10.1093/carcin/13.2.259. [DOI] [PubMed] [Google Scholar]
- 27.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 28.Hart K, Klein T, Wilcox M. A Minute encoding a ribosomal protein enhances wing morphogenesis mutants. Mech Dev. 1993;43:101–110. doi: 10.1016/0925-4773(93)90028-v. [DOI] [PubMed] [Google Scholar]
- 29.Henry J L, Coggin D L, King C R. High-level expression of the ribosomal protein L19 in human breast tumors that overexpress erbB-2. Cancer Res. 1993;53:1403–1408. [PubMed] [Google Scholar]
- 30.Howe J G, Hershey J W B. Translation initiation factor and ribosome association with the cytoskeletal framework fraction from HeLa cells. Cell. 1984;37:85–93. doi: 10.1016/0092-8674(84)90303-9. [DOI] [PubMed] [Google Scholar]
- 31.Itoh T, Otaka E, Matsui K A. Primary structures of ribosomal YS25 from Saccharomyces cerevisiae and its counterparts from Schizosaccharomyces pombe and rat liver. Biochemistry. 1985;24:7418–7423. doi: 10.1021/bi00346a058. [DOI] [PubMed] [Google Scholar]
- 32.Jefferies H B J, Reinhard C, Kozma S C, Thomas G. Rapamycin selectively represses translation of the “polypyrimine tract” mRNA family. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaneko K, Kobayashi H, Onodera O, Miyatake T, Tsuji S. Genomic organization of a cDNA (QM) demonstrating an altered mRNA level in nontumorigenic Wilms’ microcell hybrid cells and its localization to Xq28. Hum Mol Genet. 1992;1:529–533. doi: 10.1093/hmg/1.7.529. [DOI] [PubMed] [Google Scholar]
- 33a.Keppel E, Schaller H C. A 33-kDa protein with sequence homology to the “laminin binding protein” is associated with the cytoskeleton in hydra and in mammalian cells. J Cell Sci. 1991;100:789–797. doi: 10.1242/jcs.100.4.789. [DOI] [PubMed] [Google Scholar]
- 34.Kim J, Chubatsu L S, Admon A, Stahl J, Fellous R, Linn S. Implication of mammalian ribosomal protein S3 in the processing of DNA damage. J Biol Chem. 1995;270:13620–13629. doi: 10.1074/jbc.270.23.13620. [DOI] [PubMed] [Google Scholar]
- 35.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. A ras-related gene with transformation suppressor activity. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 36.Koller H T, Klade T, Ellinger A, Breitenbach M. The yeast growth control gene GRC5 is highly homologous to the mammalian putative tumor suppressor gene QM. Yeast. 1996;12:53–65. doi: 10.1002/(SICI)1097-0061(199601)12:1%3C53::AID-YEA886%3E3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 37.Kondoh N, Noda M, Fisher R J, Schweinfest C W, Papas T S, Kondoh A, Samuel K P, Oikawa T. The S29 ribosomal protein increases tumor suppressor activity of Krev-1 gene on v-K ras-transformed NIH3T3 cells. Biochim Biophys Acta. 1996;1313:41–46. doi: 10.1016/0167-4889(96)00052-3. [DOI] [PubMed] [Google Scholar]
- 38.Kondoh N, Schweinfest C W, Henderson K W, Papas T S. Differential expression of S19 ribosomal protein, laminin-binding protein, and human lymphocyte antigen class I messenger RNAs associated with colon carcinoma progression and differentiation. Cancer Res. 1992;52:791–796. [PubMed] [Google Scholar]
- 39.Kongsuwan K, Qiang Y, Vincent A, Frisardi M C, Rosbash M. A Drosophila Minute gene encodes a ribosomal protein. Nature. 1985;317:555–558. doi: 10.1038/317555a0. [DOI] [PubMed] [Google Scholar]
- 40.Lindsley D L, Zimm G G. The genome of Drosophila melanogaster. San Diego, Calif: Academic Press, Inc.; 1992. [Google Scholar]
- 41.Loftus T M, Nguyen Y E, Stanbridge E J. The QM protein associates with ribosomes in the rough endoplasmic reticulum. Biochemistry. 1997;36:8224–8230. doi: 10.1021/bi970288d. [DOI] [PubMed] [Google Scholar]
- 42.Lutsch G, Noll F, Theise H, Enzmann G, Bielka H. Localization of proteins S1, S2, S16 and S23 on the surface of small subunits of rat liver ribosomes by immune electron microscopy. Mol Gen Genet. 1979;176:281–291. doi: 10.1007/BF00273223. [DOI] [PubMed] [Google Scholar]
- 43.Marion M-J, Marion C. Ribosomal proteins S2, S6, S10, S14, S15, and S25 are localized on the surface of mammalian 40S subunits and stabilized their conformation. FEBS Lett. 1988;232:281–285. doi: 10.1016/0014-5793(88)80753-1. [DOI] [PubMed] [Google Scholar]
- 44.McKim K S, Dahmus J B, Hawley R S. Cloning of the Drosophila melanogaster meiotic recombination gene mei-218: a genetic and molecular analysis of interval 15E. Genetics. 1996;144:215–228. doi: 10.1093/genetics/144.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mechler B M. Genes in control of cell proliferation and tumorigenesis in Drosophila. In: Gordon S, editor. The legacy of cell fusion. Oxford, England: Oxford University Press; 1994. pp. 183–198. [Google Scholar]
- 46.Melnick M B, Noll E, Perrimon N. The Drosophila stubarista phenotype is associated with a dosage effect of the putative ribosome-associated protein D-p40 on spineless. Genetics. 1993;135:553–564. doi: 10.1093/genetics/135.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyuhas O, Avni D, Shama S. Translational control of ribosomal protein mRNAs in eukaryotes. In: Hershey J W B, Mathews M, Sonenberg S, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 363–388. [Google Scholar]
- 47a.Mitchelson A, Simonelig M, Williams C J, O’Hare K. Homology with Saccharomyces cerevisiae RNA14 suggests that phenotypic suppression in Drosophila melanogaster by suppressor of forked occurs at the level of RNA stability. Genes Dev. 1993;7:241–249. doi: 10.1101/gad.7.2.241. [DOI] [PubMed] [Google Scholar]
- 48.Nishi R, Hashimoto H, Uchimiya H, Kato A. The primary structure of two proteins from the small ribosomal subunit of rice. Biochim Biophys Acta. 1993;1216:113–114. doi: 10.1016/0167-4781(93)90044-e. [DOI] [PubMed] [Google Scholar]
- 49.Nygärd O, Nilsson L. Translational dynamics. Interactions between the translation factors, tRNA and ribosomes during eukaryotic protein synthesis. Eur J Biochem. 1990;191:1–17. doi: 10.1111/j.1432-1033.1990.tb19087.x. [DOI] [PubMed] [Google Scholar]
- 49a.O’Hare K, Rubin G M. Structure of P-transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983;34:25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- 50.Oliver C P. New mutants report. Drosophila Infor Service. 1935;4:15. [Google Scholar]
- 51.Palen E, Traugh J A. Phosphorylation of ribosomal protein S6 by cAMP-dependent protein kinase and mitogen-stimulated S6 kinase differently alters translation of globin mRNA. J Biol Chem. 1987;262:3528–3523. [PubMed] [Google Scholar]
- 52.Pirrotta V. Vectors for P-element transformation in Drosophila. In: Rodriguez R L, Renhardt D T, editors. Vectors: a survey of molecular cloning vectors and their uses. London, England: Butterworths; 1988. pp. 437–456. [Google Scholar]
- 53.Pogue-Geile K, Geiser J R, Shu M, Miller C, Wool I G, Meisler A I, Pipas J M. Ribosomal protein genes are overexpressed in colorectal cancer: isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol Cell Biol. 1991;11:3842–3849. doi: 10.1128/mcb.11.8.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritossa F. The bobbed locus. In: Ashburner M, Novitski E, editors. The genetics and biology of Drosophila. 1B. London, England: Academic Press, Inc.; 1976. pp. 801–846. [Google Scholar]
- 55.Rizki T M. The circulatory system and associated cells and tissues. In: Ashburner M, Wright T R F, editors. The genetics and biology of Drosophila. 2B. London, England: Academic Press, Inc.; 1978. pp. 397–452. [Google Scholar]
- 56.Saebøe-Larssen S, Lambertsson A. A novel Drosophila Minute locus encodes ribosomal protein S13. Genetics. 1996;143:877–885. doi: 10.1093/genetics/143.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saebøe-Larssen S, Urbanczyk-Mohebi B, Lambertsson A. The Drosophila ribosomal protein L14-encoding gene, identified by a novel Minute mutation in a dense cluster of previously undescribed genes in cytogenetic region 66D. Mol Gen Genet. 1997;225:141–151. doi: 10.1007/s004380050482. [DOI] [PubMed] [Google Scholar]
- 58.Saebøe-Larssen S, Lyamouri M, Merriam J, Oksvold M P, Lambertsson A. Ribosomal protein insufficiency and the Minute syndrome in Drosophila: a dose-response relationship. Genetics. 1998;148:1215–1224. doi: 10.1093/genetics/148.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 60.Sandigursky M, Yacoub A, Kelley M R, Deutsch W A, Franklin W A. The Drosophila ribosomal protein S3 contains a DNA deoxyribophosphodiesterase (dRpase) activity. J Biol Chem. 1997;272:17480–17484. doi: 10.1074/jbc.272.28.17480. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt A, Hollmann M, Schäfer U. A newly identified Minute locus, M(2)32D, encodes the ribosomal protein L9 in Drosophila melanogaster. Mol Gen Genet. 1996;251:381–387. doi: 10.1007/BF02172530. [DOI] [PubMed] [Google Scholar]
- 62.Sharp M G F, Adams S M, Elvin P, Walker R A, Brammar W J, Varley J M. A sequence previously identified as metastasis-related encodes an acidic ribosomal phosphoprotein, P2. Br J Cancer. 1990;61:83–88. doi: 10.1038/bjc.1990.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shimuzu M, Craven G R. Chemical inactivation of Escherichia coli 30-S ribosomes by iodination. Eur J Biochem. 1976;61:307–315. doi: 10.1111/j.1432-1033.1976.tb10023.x. [DOI] [PubMed] [Google Scholar]
- 64.Shrestha R, Gateff E. Ultrastructure and cytochemistry of the cell types in the larval hematopoietic organs and hemolymph of Drosophila melanogaster. Dev Growth Differ. 1982;24:65–82. doi: 10.1111/j.1440-169X.1982.00065.x. [DOI] [PubMed] [Google Scholar]
- 65.Stewart M J, Denell R. Mutations in the Drosophila gene encoding ribosomal protein S6 cause tissue overgrowth. Mol Cell Biol. 1993;13:2524–2535. doi: 10.1128/mcb.13.4.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzuki K, Otoka E. Cloning and nucleotide sequence of the gene encoding yeast ribosomal protein S25. Nucleic Acids Res. 1988;16:6223. doi: 10.1093/nar/16.13.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66a.Szabad J, Bryant P J. The mode of action of discless mutations in Drosophila melanogaster. Dev Biol. 1982;93:240–256. doi: 10.1016/0012-1606(82)90256-1. [DOI] [PubMed] [Google Scholar]
- 67.Terada N, Patel H R, Takase K, Kohno K, Nairn A C, Gelfand E W. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc Natl Acad Sci USA. 1994;91:11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas G, Sweeney R, Chang C, Noller H F. Identification of proteins functionally altered by chemical modification of the transfer RNA and polyuridilic acid binding sites of 30S ribosomal subunits. J Mol Biol. 1975;95:91–102. doi: 10.1016/0022-2836(75)90338-1. [DOI] [PubMed] [Google Scholar]
- 69.Tolan D R, Traut R R. Protein topography of the 40S ribosomal subunit from rabbit reticulocytes shown by cross-linking with 2-iminothiolane. J Biol Chem. 1981;256:10129–10136. [PubMed] [Google Scholar]
- 70.Török I, Hartenstein K, Kalmes A, Schmitt R, Strand D, Mechler B M. The l(2)gl homologue of Drosophila pseudoobscura suppresses tumorigenicity in transgenic Drosophila melanogaster. Oncogene. 1993;8:1537–1549. [PubMed] [Google Scholar]
- 71.Török I, Strand D, Schmitt R, Tick G, Török T, Kiss I, Mechler B M. The overgrown hematopoietic organs-31 tumor suppressor gene of Drosophila encodes an importin-like protein accumulating in the nucleus at the onset of mitosis. J Cell Biol. 1995;129:1473–1489. doi: 10.1083/jcb.129.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Török T, Tick G, Alvarado M, Kiss I. P-lacW insertional mutagenesis on the second chromosome of Drosophila melanogaster: isolation of lethals with different overgrowth phenotypes. Genetics. 1993;135:71–80. doi: 10.1093/genetics/135.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tschudi C, Pirrotta V, Junakovic N. Rearrangements of the 5S RNA gene cluster of Drosophila melanogaster associated with insertion of a B104 element. EMBO J. 1982;1:977–985. doi: 10.1002/j.1460-2075.1982.tb01281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tron T, Yang M, Dick F A, Schmitt M E, Trumpower B L. QSR1, an essential yeast gene with a genetic relationship to a subunit of the mitochondrial cytochrome bc1 complex is homologous to a gene implicated in eukaryotic cell differentiation. J Biol Chem. 1995;270:9961–9970. doi: 10.1074/jbc.270.17.9961. [DOI] [PubMed] [Google Scholar]
- 75.Watson K L, Johnson T K, Denell R E. Lethal(1)aberrant immune response mutations leading to melanotic tumor formation in Drosophila melanogaster. Dev Genet. 1991;12:173–187. doi: 10.1002/dvg.1020120302. [DOI] [PubMed] [Google Scholar]
- 76.Watson K L, Konrad K D, Woods D F, Bryant P J. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc Natl Acad Sci USA. 1992;89:11302–11306. doi: 10.1073/pnas.89.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wieschaus E. A combined genetic and mosaic approach to the study of oogenesis in Drosophila. In: Siddiqui O, Babu P, Hall L M, Hall J C, editors. Development and neurobiology of Drosophila. Basic Life Sciences. Vol. 16. New York, N.Y: Plenum, Inc.; 1980. pp. 85–94. [DOI] [PubMed] [Google Scholar]
- 78.Wilson D M, Deutsch W A, Kelley M R. Drosophila ribosomal protein S3 contains an activity that cleaves DNA at apurinic/apyrimidic sites. J Biol Chem. 1994;269:25359–25364. [PubMed] [Google Scholar]
- 79.Wong J M, Mafune K-I, Yow H, Rivers E N, Ravikumar T S, Steele G D, Chen L B. Ubiquitin-ribosomal protein S27a gene overexpressed in human colorectal carcinoma is an early growth response gene. Cancer Res. 1993;53:1916–1920. [PubMed] [Google Scholar]
- 80.Yacoub A, Augeri L, Kelley M R, Doetsch P W, Deutsch W A. A Drosophila ribosomal protein contains 8-oxoguanine and abasic site DNA repair activities. EMBO J. 1996;15:2306–2312. [PMC free article] [PubMed] [Google Scholar]
- 81.Yacoub A, Kelley M R, Deutsch W A. Drosophila ribosomal protein P0 contains apurinic/apyrimidinic endonuclease activity. Nucleic Acids Res. 1996;24:4298–4303. doi: 10.1093/nar/24.21.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yow H K, Wong J M, Chen H S, Lee C G, Davis S, Steele G D, Chen L B. Increased mRNA expression of a laminin-binding protein in human carcinoma: complete sequence of a full-length cDNA encoding the protein. Proc Natl Acad Sci USA. 1988;85:6394–6398. doi: 10.1073/pnas.85.17.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]