Abstract
The Src homology 3 (SH3) motif is found in numerous signal transduction proteins involved in cellular growth and differentiation. We have purified and cloned a novel protein, DEF-1 (differentiation-enhancing factor), from bovine brain by using a Src SH3 affinity column. Ectopic expression of DEF-1 in fibroblasts resulted in the differentiation of a significant fraction of the culture into adipocytes. This phenotype appears to be related to the induction of the transcription factor peroxisome proliferator-activated receptor γ (PPARγ), since DEF-1 NIH 3T3 cells demonstrated augmented levels of PPARγ mRNA and, when treated with activating PPARγ ligands, efficient induction of differentiation. Further evidence for a role for DEF-1 in adipogenesis was provided by heightened expression of DEF-1 mRNA in adipose tissue isolated from obese and diabetes mice compared to that in tissue isolated from wild-type mice. However, DEF-1 mRNA was detected in multiple tissues, suggesting that the signal transduction pathway(s) in which DEF-1 is involved is not limited to adipogenesis. These results suggest that DEF-1 is an important component of a signal transduction process that is involved in the differentiation of fibroblasts and possibly of other types of cells.
Src homology 3 (SH3) domains are found in numerous signal transduction proteins, including the Src family of protein tyrosine kinases. In Src family members, SH3 domains are believed to function in the control of subcellular localization and the regulation of kinase activity and as sites of interaction with other signal transduction proteins (2, 10, 12, 43, 44). Proteins that are known to interact with an SH3 domain typically have a PXXP consensus sequence (P = proline, X = any amino acid) that is believed to adopt a polyproline type II helix conformation (68). Residues adjacent to the prolines also form contacts with the SH3 structure, and these interactions determine the binding specificity between a protein and a particular SH3. For example, the arginine in RPLPXXP forms a salt bridge with aspartate 99 of pp60c-src. However, the C-terminal arginine in the sequence AFAPPLPRR contacts the identical aspartate in pp60c-src, indicating that proteins may interact with SH3 domains in either a plus or minus orientation (termed class I and class II binding, respectively [35, 68]).
SH3 consensus binding sequences have been useful for identifying proteins that are potential targets for interactions with an SH3-containing protein. For example, a proline-rich region in mSOS suggested a mechanism for its association with an SH3 domain of GRB-2 (9). Novel SH3 binding proteins have been isolated by a number of strategies, including affinity chromatography, expression library screening, and yeast two-hybrid screening (13, 20, 25, 27, 47, 55). However, the signal transduction pathways in which many of these SH3 binding proteins are involved have not been delineated.
Signal transduction proteins involved in the differentiation of fibroblasts into adipocytes have been evaluated by using various tissue culture cell lines as model systems. For example, members of the C/EBP and peroxisome proliferator-activated receptor (PPAR) families of transcription factors have been determined to have the potential to induce adipogenesis in NIH 3T3 and 3T3-L1 cells under the appropriate conditions (6, 11, 37, 54). Although numerous studies have confirmed the importance of these transcription factors for the promotion of fibroblastic differentiation, the intracellular signaling involved in the regulation of these factors and others involved in adipogenesis has not been defined completely.
Several proteins that potentially influence the expression and activity of transcription factors involved in adipogenesis are members of the insulin and leptin receptor signal transduction pathways (34, 36). For example, insulin treatment of 3T3-L1 cells results in the phosphorylation of mitogen-activated protein kinase, p90rsk, and c-raf (46). In turn, expression of activated alleles of raf and ras induces adipogenesis in cultured preadipocytes (3, 46). The genes for leptin and its receptor are mutated in the mouse models of obesity, obese (ob), and diabetes (db), respectively (58). The leptin receptor is a member of the class I cytokine receptor family and, consequently, signals through the activation of Jak kinases and the STAT family of transcription factors (4, 22, 42, 51, 56, 59). STAT signaling has been shown to be defective in db/db mice, and presumably, particular STATs control the transcription of genes involved in antiobesity (14, 23). Therefore, there are likely numerous proteins that potentially influence adipogenesis and obesity.
This study describes the purification and characterization of a novel Src SH3 binding protein, DEF-1 (differentiation-enhancing factor). Although DEF-1 was partially purified with a domain from a proto-oncoprotein, its ectopic expression in fibroblasts promotes adipogenesis instead of oncogenic transformation. How DEF-1 promotes this effect is mechanistically unclear, although we have determined that expression of the C terminus of DEF-1 is sufficient for this phenotype. These results, along with the ubiquitous expression pattern of DEF-1, suggest that DEF-1 is an important signal transduction protein involved in the differentiation of fibroblasts and possibly of other cell types.
MATERIALS AND METHODS
GST constructs.
The Src SH3 and Src SH3SH2 affinity columns were constructed by cloning the avian Src SH3 or Src SH3SH2 domains (amino acids 88 to 136 and 88 to 240 of chicken c-Src, respectively) into the plasmid vector pGEX-2T (Pharmacia) by standard PCR techniques. The resulting glutathione S- transferase (GST) Src SH3 domain fusion protein was secured to glutathione-coupled Sepharose beads. Lck SH3 was constructed in a similar fashion, with a murine c-lck gene as the initial template. The GST-DEF-1 C1 (amino acids 777 to 926) and C2+3 (amino acids 928 to 1129) constructs were made by cloning in the appropriate blunt-ended, BglII fragment of bovine DEF-1 into the SmaI site of pGEX-2T (Pharmacia). GST-DEF-1 C2 and C3 comprised amino acids 934 to 1036 and 1074 to 1129 of bovine DEF-1, respectively, and were made by standard PCR techniques and cloned into pGEX-2T.
Protein purification.
Calf brain lysates were made by homogenization in the presence of hypotonic lysis buffer (0.25 M sucrose, 20 mM Tris [pH 8.0], 1 mM EDTA, 1 mM β-mercaptoethanol, 2 mM phenylmethylsulfonyl fluoride [PMSF]) and passed over the respective columns. Each column was washed once in Nonidet P-40 (NP-40) lysis buffer (1% NP-40, Tris [pH 8.0], 137 mM NaCl, 1 mM EDTA, 10% glycerol, 2 mM PMSF, 0.1 TIU of aprotinin/ml), twice in 0.5 M LiCl-20 mM Tris (pH 8.0), and once with phosphate-buffered saline (PBS). Samples were eluted with 10 mM glutathione in 120 mM NaCl-100 mM Tris (pH 8.0) and passed over an ATP-agarose column (Sigma) or eluted with sodium dodecyl sulfate (SDS) sample buffer and loaded onto an SDS–10% polyacrylamide gel. Samples passed over the ATP-agarose column were washed twice with PBS, eluted with SDS sample buffer, and electrophoresed on an SDS–5% polyacrylamide gel. The gel was electroblotted with polyvinylidene difluoride membrane (Bio-Rad) in CAPS buffer [10 mM 3-(cyclohexylamino)-1-propanesulfonic acid, 10% methanol], and the band corresponding to DEF-1 was excised. The starting material for the large-scale purification of DEF-1 was one-half of a calf brain (approximately 250 g). Following in situ digestion with trypsin (17), the resulting peptide mixture was separated by microbe high-pressure liquid chromatography (HPLC) with a Zorbax C18 1.0- by 150-mm reverse-phase column on a Hewlett-Packard 1090 HPLC/1040 diode array detector. Optimum fractions from the chromatogram were chosen based on differential UV absorbance at 205, 277, and 292 nm, peak symmetry, and resolution. Peaks were further screened for length and homogeneity by matrix-assisted laser desorption time-of-flight mass spectrometry on a Finnigan Lasermat 200 (Hemel, England), and selected fractions underwent automated Edman degradation on a Perkin-Elmer/Applied Biosystems (Foster City, Calif.) model 494A or 477A. Details of strategies for the selection of peptide fractions and their microsequencing have been previously described (31).
cDNA cloning.
cDNA cloning with degenerate primers by PCR was performed essentially as described previously (32). Bovine brain RNA was reverse transcribed with the downstream primer 5′ RTCRTTNGTRTCYTC 3′. The cDNA from this reaction was used in a PCR with the same downstream primer and 5′ CAYGTICARAAYGARGARAA 3′ as the upstream primer. This reaction was used as a template for a subsequent PCR with the nested upstream primer, 5′ GARGARAAYTAYGCICARGT 3′, and the downstream primer. The product from this reaction was sequenced and subsequently determined to encode amino acids 92 to 384 of DEF-1. This PCR product was used to screen a bovine brain randomly primed cDNA library in the vector λZapII (Stratagene) obtained from Akio Yamakawa. This resulted in six unique clones, five of which contained DEF-1 coding sequences. The sixth appears to be a related gene (data not shown). A segment of one clone was used to rescreen the library, resulting in three novel DEF-1 clones including the remainder of the coding sequence. The DEF-1 cDNA (comprised of clones S9 and R27), with the hemagglutinin (HA) tag MVYPYDVPDYAG at the N terminus, was cloned into the expression vector pLNSL7 and transfected into ψ2 cells to obtain infectious retroviral supernatants (38). pLNSL7/DEF-1 was digested with BglII and blunt ended with Klenow enzyme, and the vector–DEF-1 backbone was religated to make the DEF-1–Bgl construct.
Immunoprecipitations and GST pull-downs.
Lysates made with NP-40 lysis buffer from NIH 3T3 cells expressing pLNSL7 alone (vector) or HA-tagged DEF-1 (DEF-1) were passed over the noted columns and washed as described above. Bound proteins were immunoblotted with the anti-HA antibody, 12CA5 (Babco). pp60c-src was detected by using the monoclonal antibody 327, a gift from J. Brugge. GST preparations were prepared as described above and quantitated by staining with Coomassie blue. Equal amounts of GST fusion proteins were used for each pull-down experiment. All immunoprecipitates and GST pull-downs were washed once with NP-40 lysis buffer, twice with 0.5 M LiCl–20 mM Tris (pH 8.0), and once with PBS. Anti-DEF-1 antibody polyclonal serum was prepared by injecting rabbits with amino acids 928 to 1129 of DEF-1 (prepared by proteolysis of a GST fusion protein). The preimmune serum used was obtained from this rabbit.
Cell treatments.
BALB/c 3T3, NIH 3T3, and 3T3 F442A cells were infected with the vector or DEF-1 retroviral supernatants and selected with 400 μg of G418 per ml. Only pools of cells derived from more than ∼1,000 infected cells were assayed. Upon confluence, the derivative NIH 3T3 cells were cultured in 10% fetal calf serum (FCS)–Dulbecco modified Eagle medium (DMEM) (Gibco/BRL) and supplemented with combinations of 1 μM dexamethasone (Sigma), 5 μM insulin (Sigma), and 10 μM pioglitazone, as indicated previously (61). 3T3 F442A derivative cell lines were passaged in 10% calf serum–DMEM and assayed with 10% FCS-DMEM with 10 nM pioglitazone. For all assays, the medium was changed every other day. After 9 days, the cells were fixed, stained with Oil-Red-O (39), and photographed by using a 20× objective or harvested for RNA.
Northern blot analysis.
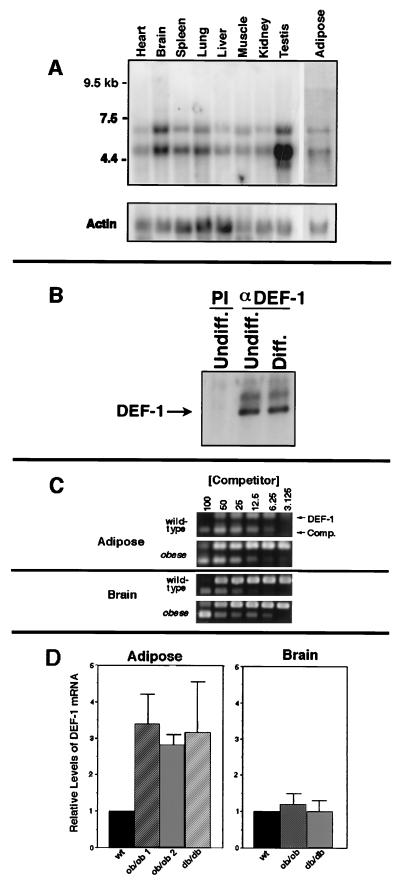
Twenty micrograms of total RNA (Fig. 4C) or poly(A)+ RNA purified from 50 μg of total RNA (Invitrogen; Fig. 5) isolated from the cells treated as described above was probed with an aP2 or PPARγ cDNA, respectively (61). Quantities of RNA for Fig. 5 were normalized with a 36B4 probe (30). The full-length DEF-1 cDNA was used to probe a mouse multiple-tissue Northern blot (Clontech) and 5 μg of poly(A)+ mRNA isolated from mouse adipose tissue. A β-actin probe was subsequently used to normalize mRNA loading.
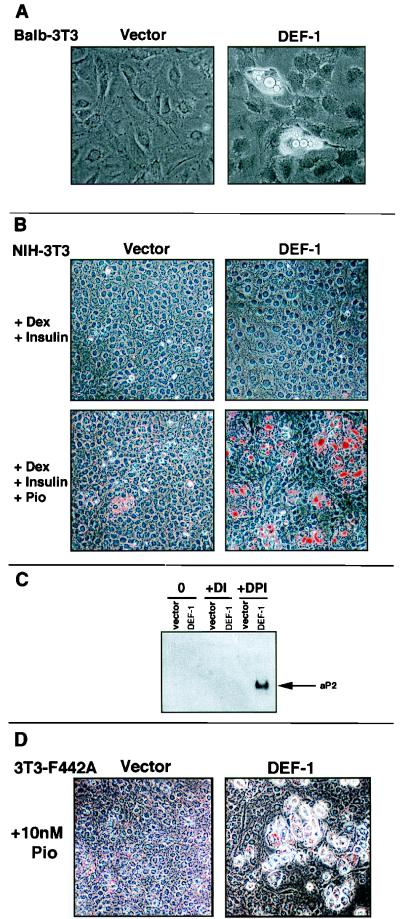
FIG. 4.
DEF-1 induces fibroblastic differentiation. (A) Stably infected DEF-1 BALB/c 3T3 cells were grown to confluence and photographed with a 20× objective after noticeable lipid accumulation. (B) NIH 3T3 cells stably infected with DEF-1 or the expression vector alone were grown to confluence with 10% FCS-DMEM and supplemented with combinations of dexamethasone (Dex), insulin, and pioglitazone (Pio) as indicated. After notable lipid accumulation (9 days), the cells were fixed and lipid droplets were identified by staining with Oil-Red-O. (C) Twenty micrograms of total RNA isolated from the cells treated as shown in panel B was probed with an aP2 cDNA (D, dexamethasone; P, pioglitazone; I, insulin; 0, no treatment) (61). (D) 3T3 F442A cells ectopically expressing DEF-1 or the expression vector alone were grown to confluence and supplemented with 10 nM pioglitazone. After 9 days, the cells were stained with Oil-Red-O and photographed.
FIG. 5.
Expression of PPARγ in DEF-1 cells. RNA isolated from NIH 3T3 cells expressing the vector alone (vector) or DEF-1 (DEF-1) treated as in Fig. 4B was probed with a PPARγ cDNA (60). Quantities of RNA were standardized with a 36B4 probe (30).
Competitive PCR analyses.
A DNA competitor was constructed with primers 5′ TCGTTTTCGGATGTGACGGCTGAGGTTCATCGCCGAGACCA 3′ and 5′ ATTGTGGCTCAGACCCTGGA 3′ and murine DEF-1 (21) as a template. This PCR product represented the 5′ end of murine DEF-1 with an internal 88-nucleotide deletion. Five micrograms of RNA from each tissue sample was reverse transcribed by using reverse transcriptase and the primer 5′ AACAAGGAATCCAAGGTGAAGA 3′ according to the protocol of the manufacturer (Promega). Equivalent quantities of RNA were confirmed by Northern blot analysis of 5 μg of RNA from each sample by using 36B4 as a probe (data not shown), and quantitation of the signal was performed on a PhosphorImager (Molecular Dynamics). PCRs were performed with equal amounts of template and 5′ TCGTTTTCGGATGTGACGGCTGAG 3′ (containing 5′ noncoding sequence of murine DEF-1) and 5′ ATTGTGGCTCAGACCCTGGA 3′ as primers, with decreasing amounts of competitor (starting at 0.1 attomole). Equal amounts of the PCR mixtures were loaded on an agarose gel, and band intensities were determined by using an AlphaImager 2000 version 3.3 analysis system (Alpha Innotech Corporation). The log ([DEF-1]/[competitor]) versus log [competitor added] was plotted, and the value of [DEF-1] = [competitor] was calculated (Clontech). The fold induction versus that of the wild type was then determined and plotted. All values represent the average of three experiments. Adipose and brain tissue samples were run at separate times and thus cannot be directly compared.
Nucleotide sequence accession number.
The sequence of bovine DEF-1 has been assigned GenBank accession no. AF112886.
RESULTS
Purification and cloning of DEF-1.
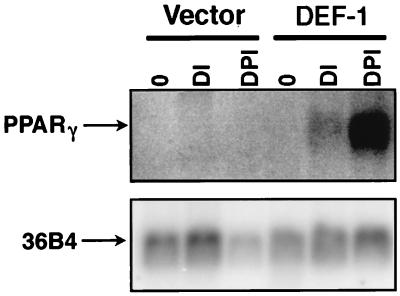
To identify novel Src SH3 binding proteins, we undertook an analysis of proteins isolated from bovine brain extracts that bound to a GST Src SH3 (Src SH3) affinity column. Resolution of the associated proteins by SDS-polyacrylamide gel electrophoresis showed several species that bound to the Src SH3 column but not the GST beads alone (Fig. 1A). Included was a prominent band of approximately 100 kDa, which was subsequently identified as dynamin (data not shown; 24). Because dynamin also has affinity for ATP agarose (50), we determined the ability of the Src SH3-associated proteins to bind to an ATP affinity matrix. This led to the identification of a small number of proteins that bound to both affinity columns, including a protein of approximately 140 kDa (DEF-1; see below) which showed high abundance and good separation relative to the other proteins (Fig. 1B). Therefore, our efforts focused on purifying DEF-1 in a quantity sufficient for amino acid sequencing.
FIG. 1.
Purification of DEF-1 protein. (A) Calf brain lysates were passed over the GST, Src SH3, or Src SH3SH2 affinity column, and associated proteins were electrophoresed on a 10% polyacrylamide gel and visualized by silver stain. Molecular size markers in kilodaltons are indicated on the left. (B) Bound proteins from panel A were eluted with free glutathione and passed over an ATP-agarose column. The associated proteins were electrophoresed on a 5% polyacrylamide gel and visualized by silver staining.
A large-scale protein purification of DEF-1 was performed, resulting in approximately 20 μg of the purified protein. Degenerate oligonucleotides were designed based on the resultant amino acid sequence of six tryptic peptides and were used as primers in a series of nested PCRs, with bovine brain mRNA as the initial template. One positive PCR product was used to screen a randomly primed, bovine brain cDNA library. Positive clones were used to isolate eight overlapping clones that resulted in approximately 5,300 bp of contiguous sequence. The composite sequence contained an open reading frame encoding a protein of 1,129 amino acids (Fig. 2A). All six peptides sequenced were found in the predicted translation product. Comparison of the amino acid sequence with those in the database indicated several motifs, including a zinc finger closely related to the zinc finger found in ARF1 GTPase-activating protein (62), three ankyrin repeats (41), a pleckstrin homology domain (15), a putative lipid binding motif named C2 (19, 57), and an SH3 domain (16). In addition, several proline-rich motifs, including multiple Src SH3 consensus binding sequences, were noted (1, 48, 52, 63). No previously described motifs that would account for DEF-1’s affinity for ATP agarose were apparent.
FIG. 2.
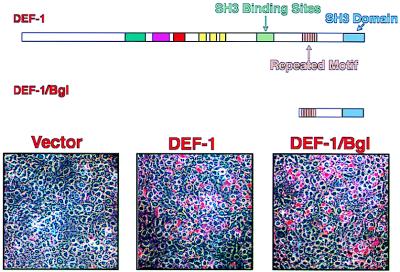
Sequence of DEF-1. (A) Eight unique, overlapping clones were used to obtain the DEF-1 composite sequence. One clone (R19) lacked the sequence SRR at amino acids 304 to 306. The sequence from other clones and an apparent human DEF-1 partial sequence in the EST database suggested that the SRR sequence is an alternative exon (data not shown). The number of the last amino acid in a line is noted on the right. Key: overline, peptide sequenced; underline, putative alternative exon; aqua, pleckstrin homology domain (amino acids 326 to 419); purple, zinc finger (amino acids 457 to 480); red, C2 domain (amino acids 498 to 557); yellow, ankyrin-related motifs (amino acids 604 to 623, 640 to 659, 673 to 692); green, SH3 consensus binding sequences (amino acids 791 to 800, 803 to 809, 828 to 835, 895 to 901, 993 to 999); brown, proline-rich repeat (amino acids 934 to 1001); blue, SH3 domain (amino acids 1074 to 1123). (B) Putative structure of proline-rich motif in DEF-1. The sequence of amino acids 976 to 1001 is drawn in a left-handed poly-proline type II helix. Standard amino acid abbreviations are used.
In addition to the readily identifiable motifs described above, an unusual proline-rich stretch located between the SH3 domain and the predicted SH3 binding sites in DEF-1 was noted (amino acids 934 to 1001). This region can be subdivided into six tandem repeats centered on the consensus sequence GDLPPKP. Although this motif has the PXXP consensus found in SH3 binding proteins, it would not be predicted to form a high affinity interaction with Src SH3, since it lacks a basic amino acid residue at the proper position (with the exception of the last repeat [48]). However, the preponderance of prolines in this repeat suggests that this region forms a polyproline type II helix (64). On the basis of this assumption, the four C-terminal repeats form a trigonal prism with an acidic edge, a basic edge, and an uncharged edge (with the exception noted above; Fig. 2B). The two longer repeats (amino acids 934 to 965) have a similar pattern, yet the relative charge rotates between the repeats (data not shown).
DEF-1 is a Src SH3 binding protein.
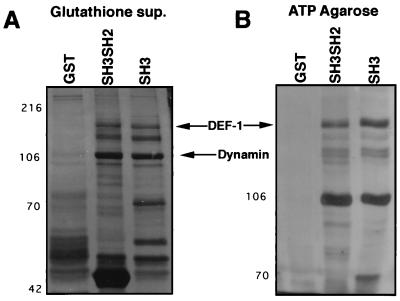
To confirm that the DEF-1 cDNA encoded a Src SH3 binding protein, the full-length DEF-1 coding sequence, fused with an HA tag at the amino terminus, was expressed in NIH 3T3 cells. Lysates from the subsequent drug selected, DEF-1-expressing cells were passed over a Src SH3 column and probed with an anti-HA antibody. The protein produced by the DEF-1 cDNA associated with the Src SH3 beads (Fig. 3A), strongly suggesting that it encodes the protein detected in Fig. 1A. DEF-1 also associated with the SH3 domain of the Src-related protein, lck, indicating that DEF-1 does not exclusively bind Src SH3.
FIG. 3.
DEF-1 is an SH3 binding protein. (A) Lysates made with NP-40 lysis buffer from NIH 3T3 cells expressing pLNSL7 alone (pLN) (38) or pLNSL7/HA-tagged DEF-1 (pLN/DEF-1 HA) were passed over the noted columns. Bound proteins were immunoblotted with an anti-HA antibody. (B) Bovine brain (brain extract) or Sf9 cells infected with a pp60c-src baculovirus (Bv Src) was lysed with NP-40 lysis buffer and passed over a column consisting of GST alone, GST-DEF-1-C1 (amino acids 777 to 926; SH3 binding sites), or GST-DEF-1-C2+3 (amino acids 928 to 1129; proline-rich repeat plus SH3 domain). Bound proteins were immunoblotted with an anti-pp60c-src antibody. (C) Bovine brain extracts were passed over a GST-DEF-1-C1, C2 (amino acids 934 to 1036; proline-rich repeat) or C3 column (amino acids 1074 to 1129; SH3 domain), and bound proteins were blotted for DEF-1 by using an anti-DEF-1 rabbit polyclonal serum. The brain lysate was immunoprecipitated with preimmune (pre-imm.) or anti-DEF-1 serum (αDEF-1) or passed over a column of GST alone (GST) as a control.
DEF-1 copurified with dynamin, a protein known to associate with numerous SH3 domains and ATP agarose (24, 50). Therefore, the interaction between DEF-1 and Src SH3 may have been dependent upon an intermediary such as dynamin. To provide evidence that DEF-1 associated with Src SH3 directly, two GST fusion proteins spanning regions of DEF-1 that had SH3 consensus binding sequences were constructed and named C1 (amino acids 777 to 926 of DEF-1) and C2+3 (amino acids 928 to 1129 of DEF-1). Lysates made from bovine brain or insect cells infected with a baculovirus expressing pp60c-src were passed over the respective columns, and after being washed, proteins eluted from the beads were immunoblotted with an anti-pp60c-src antibody. pp60c-src from either lysate associated efficiently with the C1 column (Fig. 3B). Although proteins found in both lysates may have acted as intermediates between pp60c-src and the column, the results in Fig. 3A and B are most easily explained by a direct interaction between amino acids 777 and 926 of DEF-1 and the SH3 domain in pp60c-src. Even though the C2+3 column contains a consensus Src SH3 binding site (amino acids 993 to 999), no interaction with pp60c-src was detected. However, it is not clear if an intramolecular or intermolecular interaction between the SH3 binding motif and the DEF-1 SH3 domain in this construct might interfere with Src SH3 binding.
To determine if the SH3 domain of DEF-1 could associate with full-length DEF-1, brain lysates were passed over a column comprised of GST fused with the DEF-1 SH3 domain (C3, amino acids 1074 to 1129), C1, or C2 (the proline-rich repeats; amino acids 934 to 1036). The C3 column bound DEF-1 as determined by immunoblotting with an anti-DEF-1 antibody. Presumably, this interaction occurs at one of the SH3 consensus binding sites denoted in Fig. 2A. However, even though the C1 column was capable of associating with pp60c-src (presumably through the SH3 domain of pp60c-src; Fig. 3B), no full-length DEF-1 protein bound to this column. This result suggests that the SH3 domain of full-length DEF-1 may be tightly associated with a protein with an SH3 binding site that may be DEF-1 itself.
DEF-1 induces adipogenesis in fibroblastic cell lines.
Numerous SH3 binding proteins have been cloned, yet their respective cellular functions have not been determined. Since DEF-1 was isolated by using a domain from the proto-oncoprotein pp60c-src, we introduced DEF-1 into BALB/c 3T3 cells by retroviral infection to ascertain if DEF-1 was capable of inducing cellular transformation. DEF-1-expressing cells initially had the same morphology as cells infected with the expression vector alone (data not shown). However, after approximately 2 weeks at confluence, a small number of DEF-1 BALB/c 3T3 cells formed shiny vacuoles that were suggestive of lipid droplets (Fig. 4A). This unexpected result implied that DEF-1 enhanced the potential of fibroblasts to differentiate into adipocytes.
The formation of lipid droplets in the DEF-1 BALB/c 3T3 cells encouraged us to study the role of DEF-1 in adipogenesis, using NIH 3T3 cells as a model system (11). A selected pool of NIH 3T3 cells infected with the DEF-1 retrovirus (DEF-1 NIH 3T3) kept at confluence in 10% FCS-DMEM demonstrated no visible signs of adipogenesis (data not shown). However, parallel cultures supplemented with factors that have been previously shown to enhance differentiation in preadipocytic cell lines, particularly dexamethasone, insulin, and the thiazolidinedione pioglitazone, demonstrated considerable levels of lipid accumulation compared to the level accumulated with the vector alone (Fig. 4B) (7, 18, 28). Northern blot analysis with the adipocyte-specific marker aP2 confirmed that the cultures of treated DEF-1 NIH 3T3 cells that presented lipid droplets underwent adipogenesis (Fig. 4C) (53, 60).
In addition to NIH 3T3 and BALB/c-3T3 cells, other cell lines have been used as model systems to study adipogenesis in culture (reviewed in reference 11). We have used the DEF-1 retroviral supernatants to ectopically express DEF-1 in several of these lines in an attempt to find DEF-1-associated phenotypes which would advance our understanding of how DEF-1 promotes adipogenesis in the assay described above. 3T3 F442A cells have been used extensively to study adipogenic differentiation because well-defined protocols that induce robust and exclusive differentiation into adipocytes have been developed. For example, 3T3 F442A cells have been shown to undergo adipocytic differentiation when they are treated with high levels of insulin or pioglitazone (49). 3T3 F442A cells stably expressing DEF-1 (DEF-1 F442A) showed no evidence of adipogenesis when they were kept at confluence in 10% FCS-DMEM. Supplementation of DEF-1 F442A cultures with pioglitazone demonstrated more extensive differentiation than that observed with the cells expressing the vector alone. The difference between DEF-1 F442A and control cells was most noticeable when the amount of pioglitazone was lowered to a limiting concentration of 10 nM (Fig. 4D and data not shown).
DEF-1 NIH 3T3 cells express augmented levels of PPARγ.
The adipogenic activity seen in the DEF-1 NIH 3T3 and DEF-1 F442A cells was dependent upon the presence of pioglitazone, which is a potent and specific stimulator of the nuclear receptor PPARγ (33). NIH 3T3 cells normally demonstrate no discernible phenotypic changes during pioglitazone treatment, presumably due to low levels of PPARγ expression (61). However, ectopic expression of PPARγ in NIH 3T3 cells followed by treatment with PPARγ-activating ligands has been shown to be sufficient to promote conspicuous adipogenesis (18, 29).
While assaying for the expression of adipocytic markers in DEF-1 NIH 3T3 cells, we noted elevated levels of PPARγ mRNA in cells that had been treated with the complete differentiation cocktail (Fig. 5). Since PPARγ levels increase during adipogenesis, this result could imply that DEF-1 promotes PPARγ expression or that augmented PPARγ levels are the result of DEF-1-induced fibroblastic differentiation (60). Notably, the culture of DEF-1 NIH 3T3 cells supplemented only with dexamethasone and insulin demonstrated increased levels of PPARγ mRNA compared to those in control cells (Fig. 5). This suggests that heightened expression of DEF-1 in NIH 3T3 cells synergizes with the effects of dexamethasone and insulin treatment to increase PPARγ levels. The further supplementation of pioglitazone activates the augmented levels of PPARγ, resulting in the adipogenic phenotype observed in Fig. 4B and C. Similarly, the observed increase in the sensitivity of DEF-1 F442A cells to pioglitazone is consistent with the notion that DEF-1 expression is modulating PPARγ levels in these cells (Fig. 4D).
Expression of DEF-1 in tissues from wild-type mice and mouse models of adipogenesis and obesity.
How DEF-1 operates as a signal transduction protein to promote adipogenesis is unclear at present. Although we have found that DEF-1 induces fibroblastic differentiation under certain conditions, DEF-1 is also expressed in every tissue and cell line that has been tested to date (Fig. 6A and data not shown). Therefore, we postulate that DEF-1 signal transduction activity is not limited to adipogenesis.
FIG. 6.
DEF-1 expression in various tissues and adipose tissue from mouse models of obesity. (A) The full-length DEF-1 cDNA was used to probe a mouse multiple-tissue Northern blot (Clontech), and 5 μg of poly(A)+ mRNA was isolated from mouse adipose tissue. The lower band appears to be smaller than the DEF-1 composite cDNA isolated and, therefore, is believed to be a DEF-1-related mRNA (data not shown). A β-actin probe was subsequently used to normalize mRNA loading. (B) Lysates from 3T3 F442A cells before (Undiff.) and after (Diff.) differentiation with 1 μM pioglitazone were immunoprecipitated with preimmune (PI) or anti-DEF-1 antiserum (αDEF-1). The immunoprecipitates were immunoblotted with the anti-DEF-1 antibody. (C) An example of the data obtained from a competitive PCR analysis used to evaluate the levels of DEF-1 mRNA in adipose and brain tissues isolated from wild-type, ob/ob, and db/db mice. The amount of competitor added that resulted in equivalent DEF-1 and competitor signal intensities was determined. (D) The average fold increase of DEF-1 mRNA levels in adipose tissue isolated from two ob/ob mice and one db/db mouse relative to that of a wild-type standard was determined as described in panel C. A similar analysis was performed with RNA from brain tissue. All values represent the average from three separate trials, and the error bars refer to average deviations.
The expression of numerous genes is altered during adipocytic development (11). These genes include PPARγ, which as noted above, is induced to high levels of expression during late stages of adipocyte differentiation (61). To determine if the levels of DEF-1 are modified during adipogenesis, we analyzed the amount of DEF-1 protein in 3T3 F442A cells before and after differentiation with pioglitazone. There was no apparent alteration in the levels of DEF-1 after differentiation of these cells (Fig. 6B).
In contrast to the pattern of DEF-1 expression in 3T3 F442A differentiation, two mouse models of obesity have provided additional evidence that DEF-1 expression levels correlate with enhanced adipogenesis in vivo. Adipose tissue from ob/ob and db/db mice show heightened levels of DEF-1 mRNA compared to those in wild-type mice (Fig. 6C and D). This relative increase was not seen in RNA isolated from brain tissue from the same mice. The augmentation of DEF-1 expression in ob/ob and db/db adipose tissue over that in wild type was approximately equal to the increase in NIH 3T3 cells due to ectopic expression of DEF-1 (data not shown).
The C terminus of DEF-1 is sufficient to induce adipogenesis.
We have undertaken a deletion analysis of DEF-1 to delineate the domains involved in the adipogenic differentiation described above. A construct involving a BglII digest resulted in the fusion of the first three amino acids of DEF-1 to the C-terminal 204 (DEF-1/Bgl). When assayed for the ability to induce adipogenesis in NIH 3T3 cells in a manner identical to that described above, the truncated DEF-1 mutant was shown to be sufficient to promote adipogenesis to levels equal to or greater than those achieved with the full-length construct (Fig. 7). Since this region of DEF-1 contains the proline-rich repeat and the SH3 domain (Fig. 2A), this result strongly suggests that proteins that interact with the SH3 domain of DEF-1 are involved in the adipogenic phenotype described above.
FIG. 7.
The C terminus of DEF-1 promotes adipogenesis. NIH 3T3 cells stably expressing DEF-1–Bgl were assayed as described in the legend for Fig. 4B in the presence of dexamethasone, insulin, and pioglitazone.
DISCUSSION
We have described a novel signal transduction molecule, DEF-1, whose overexpression in fibroblasts participates in augmentation of PPARγ levels and induction of cellular differentiation. Heightened DEF-1 expression was also found in the adipose tissue from particular mouse models of obesity, which seems to correlate with the phenotype observed by DEF-1 ectopic expression in fibroblasts. The adipogenic effect mapped to the C terminus of DEF-1, implying that key molecules involved in this response interact with this region of DEF-1.
DEF-1 has several motifs, suggesting that it interacts with other proteins to achieve its biological effects and, therefore, may act as a scaffolding protein (Fig. 2A) (45). These proteins may include a GTPase, since DEF-1 has been demonstrated to have GTPase-activating properties (5). The purification of DEF-1 described above involved a Src SH3 affinity column, which implies that DEF-1 is potentially involved in pp60c-src signal transduction. Although we have mapped a pp60c-src binding site to a region of DEF-1 containing Src SH3 consensus binding sequences (Fig. 3B), we have not been able to demonstrate an interaction between full-length DEF-1 and pp60c-src in vivo at physiological levels of expression of the two proteins by any of several different experimental approaches. For example, immunoprecipitates of DEF-1 from brain lysate (where both pp60c-src and DEF-1 are abundant) or cultured cells stably expressing an activated Src allele show no pp60c-src by immunoblotting, and these immunoprecipitates of DEF-1 do not have a kinase activity that comigrates with pp60c-src (data not shown). At least one potential impediment to an in vivo interaction between DEF-1 and pp60c-src may derive from their divergent subcellular locations. Whereas pp60c-src is localized to the plasma membrane, results from experiments involving immunofluorescence, localization of a fusion of DEF-1 and green fluorescent protein, and the purification of DEF-1 with a hypotonic lysis buffer all indicate that most, if not all, DEF-1 is present in the cytosol (Fig. 1) (5, 40). This is in spite of the fact that DEF-1 has multiple motifs that have been found in proteins that associate at least transiently with membranes or lipids (Fig. 2A) (15, 19, 41, 57). These domains within DEF-1 suggest that a hypothetical stimulus could result in the migration of DEF-1 to the plasma membrane, but to date we have not been able to identify conditions under which this occurs. A potential interaction with pp60c-src could be regulated by such a stimulus. In keeping with this idea, Brown et al. have observed an interaction between DEF-1 and an activated pp60c-src allele but not pp60c-src upon transient coexpression (5). Alternatively, DEF-1 may be a target for a different SH3-containing protein in vivo. Therefore, our data presently do not support a role for pp60c-src in adipogenesis.
The potential association between DEF-1 and SH3 domain-containing proteins or SH3 binding proteins may be regulated at steps other than subcellular localization. A GST fusion protein containing several SH3 binding consensus sequences found in DEF-1 demonstrated affinity to pp60c-src but not to DEF-1 (Fig. 3C). This may indicate an intramolecular interaction between the SH3 domain of DEF-1 with one of the potential SH3 binding sites or dimerization of two DEF-1 molecules. Thus, efforts to define the mechanism by which the DEF-1–Bgl construct enhances adipogenesis may be complicated by a potential interaction with the endogenous DEF-1 that would serve to alter the activity of the full-length molecule.
The effect of DEF-1 on PPARγ expression in NIH 3T3 cells was apparent only after the cultures were supplemented with dexamethasone and insulin (Fig. 5). How might the cellular effects brought upon by these chemicals interplay with ectopic expression of DEF-1 to induce adipogenesis? Dexamethasone and insulin have been shown to induce or maintain the expression of particular members of the PPAR and C/EBP families of transcription factors which have fundamental roles in adipogenesis (37, 54). For example, dexamethasone has been shown to induce the expression of C/EBPδ (65, 67). However, currently undefined factors that enhance adipogenesis and are affected by dexamethasone treatment have been suggested previously (65). The changes imparted by dexamethasone and insulin in DEF-1 NIH 3T3 cells that contribute to PPARγ expression may already be present in 3T3 F442A cells, since DEF-1 F442A cells showed heightened sensitivity to pioglitazone in the absence of this treatment.
Increased expression of DEF-1 promoted adipogenesis in cultured fibroblast cell lines and was also detected in adipose tissue of ob/ob and db/db mice. How these two observations might be mechanistically linked is unclear at present. The obese phenotype seen in ob/ob and db/db mice is due (at least partially) to a defect in leptin signaling in the hypothalmus, resulting in the loss of appetite suppression (26, 58). Therefore, leptin may affect DEF-1 expression levels in these mouse models of obesity by a mechanism that does not directly target fat tissue.
The ubiquitous expression pattern of DEF-1 implies that DEF-1 function is not restricted to adipogenesis (Fig. 6A). Moreover, amino acid sequences of cDNAs corresponding to DEF-1 homologues reveal that DEF-1 has been extremely well conserved between zebrafish, mice, rats, cows, and humans, which argues that DEF-1 may be a signal transduction component in all vertebrates (5, 8, 66). Whereas the phenotype of DEF-1 ectopic expression has been fibroblastic differentiation, this may be due to the ability of high levels of DEF-1 (or DEF-1 mutants) to bind a subset of signal transduction proteins, resulting in the formation of an inactive signaling complex. Therefore, ectopic expression of DEF-1 may be acting by a dominant negative mechanism. Determination of the proteins that interact with DEF-1 should elucidate the signal transduction pathways where DEF-1 participates and may provide new insights into cellular signal transduction.
ACKNOWLEDGMENTS
We are indebted to A. Gashler, J. Chan, I. Aksoy, C. Furman, W. Haser, and A. Yamakawa for advice, helpful discussions, and contributions of unpublished data. We are grateful to W. S. Lane, R. Robinson, V. Bailey, J. Neveu, T. Addona, and E. Spooner of the Harvard Microchemistry Facility for their expertise in the HPLC, mass spectrometry, and peptide sequencing. We also thank the members of the Dana-Farber core facility for performing all the DNA sequencing described.
This work was supported by an NIH postdoctoral fellowship to F.J.K. (CA09134) and NIH grants to B.M.S. (R37DK31405) and T.M.R. (CA43803).
REFERENCES
- 1.Alexandropoulos K, Cheng G, Baltimore D. Proline-rich sequences that bind to Src homology 3 domains with individual specificities. Proc Natl Acad Sci USA. 1995;92:3110–3114. doi: 10.1073/pnas.92.8.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar-Sagi D, Rotin D, Batzer A, Mandiyan V, Schlessinger J. SH3 domains direct cellular localization of signaling molecules. Cell. 1993;74:83–91. doi: 10.1016/0092-8674(93)90296-3. [DOI] [PubMed] [Google Scholar]
- 3.Benito M, Porras A, Nebreda A R, Santos E. Differentiation of 3T3-L1 fibroblasts to adipocytes induced by transfection of ras oncogenes. Science. 1991;253:565–568. doi: 10.1126/science.1857988. [DOI] [PubMed] [Google Scholar]
- 4.Bjorbaek C, Uotan S, da Silva B, Flier J S. Divergent signaling capacities of the long and short isoforms of the leptin receptor. J Biol Chem. 1997;272:32686–32695. doi: 10.1074/jbc.272.51.32686. [DOI] [PubMed] [Google Scholar]
- 5.Brown M T, Andrade J, Radhakrishna H, Donaldson J G, Cooper J A, Randazzo P A. ASAP1, a phospholipid-dependent Arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol Cell Biol. 1998;18:7038–7051. doi: 10.1128/mcb.18.12.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brun R P, Tontonoz P, Forman B M, Ellis R, Chen J, Evans R M, Spiegelman B M. Differential activation of adipogenesis by multiple PPAR isoforms. Genes Dev. 1996;10:974–984. doi: 10.1101/gad.10.8.974. [DOI] [PubMed] [Google Scholar]
- 7.Cao Z, Umek R M, McKnight S L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 8.Chan, J., I. Aksov, and C. Furman. Personal communication.
- 9.Chardin P, Camonis J H, Gale N W, van Aelst L, Schlessinger J, Wigler M H, Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 10.Cohen G B, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 11.Cornelius P, MacDougald O A, Lane M D. Regulation of adipocyte development. Annu Rev Nutr. 1994;14:99–129. doi: 10.1146/annurev.nu.14.070194.000531. [DOI] [PubMed] [Google Scholar]
- 12.Courtneidge S A. Protein tyrosine kinases, with emphasis on the Src family. Semin Cancer Biol. 1994;5:239–246. [PubMed] [Google Scholar]
- 13.Dai Z, Pendergast A M. Abi-2, a novel SH3-containing protein interacts with the c-Abl tyrosine kinase and modulates c-Abl transforming activity. Genes Dev. 1995;9:2569–2582. doi: 10.1101/gad.9.21.2569. [DOI] [PubMed] [Google Scholar]
- 14.Darnell J E., Jr Reflections on STAT3, STAT5, and STAT6 as fat STATs. Proc Natl Acad Sci USA. 1996;93:6221–6224. doi: 10.1073/pnas.93.13.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Downing A K, Driscoll P C, Gout I, Salim K, Zvelebil M J, Waterfield M D. Three-dimensional solution structure of the pleckstrin homology domain from dynamin. Curr Biol. 1994;4:884–891. doi: 10.1016/s0960-9822(00)00197-4. [DOI] [PubMed] [Google Scholar]
- 16.Feng S, Kasahara C, Rickles R J, Schreiber S L. Specific interactions outside the proline-rich core of two classes of Src homology 3 ligand. Proc Natl Acad Sci USA. 1995;92:12408–12415. doi: 10.1073/pnas.92.26.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez J, Andrews L, Mische S M. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal Biochem. 1994;218:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- 18.Forman B M, Tontonoz P, Chen J, Brun R P, Spiegelman B M, Evans R M. 15-Deoxy-Δ 12,14-prostaglandin J2 is a ligand for the adiptocyte determination factor PPARγ. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda M, Aruga J, Niinobe M, Aimoto S, Mikoshiba K. Inositol-1,3,4,5-tetrakisphosphate binding to C2B domain of IP4BP/synaptotagmin II. J Biol Chem. 1994;269:29206–29211. [PubMed] [Google Scholar]
- 20.Fumagalli S, Totty N F, Hsuan J J, Courtneidge S A. A target for Src in mitosis. Nature. 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- 21.Furman, C. Unpublished data.
- 22.Ghilardi N, Skoda R C. The leptin receptor activates janus kinase 2 and signals for proliferation in a factor-dependent cell line. Mol Endocrinol. 1997;11:393–399. doi: 10.1210/mend.11.4.9907. [DOI] [PubMed] [Google Scholar]
- 23.Ghilardi N, Ziegler S, Wiestner A, Stoffel R, Heim M H, Skoda R C. Defective STAT signaling by the leptin receptor in diabetic mice. Proc Natl Acad Sci USA. 1996;39:6231–6235. doi: 10.1073/pnas.93.13.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gout I, Dhand R, Hiles I D, Fry M J, Panayotou G, Das P, Truong O, Totty N F, Hsuan J, Booker G W, et al. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- 25.Hasegawa H, Kiyokawa E, Tanaka S, Nagashima K, Gotoh N, Shibuya M, Kurata T, Matsuda M. DOCK180, a major CRK-binding protein, alters cell morphology upon translocation to the cell membrane. Mol Cell Biol. 1996;16:1770–1776. doi: 10.1128/mcb.16.4.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang C S, Loftus T M, Mandrup S, Lane M D. Adipocyte differentiation and leptin expression. Annu Rev Cell Dev Biol. 1997;13:231–259. doi: 10.1146/annurev.cellbio.13.1.231. [DOI] [PubMed] [Google Scholar]
- 27.Kitamura T, Kitamura Y, Yonezawa K, Totty N F, Gout I, Hara K, Waterfield M D, Sakaue M, Ogawa W, Kasuga M. Molecular cloning of p125Nap1, a protein that associates with an SH3 domain of Nck. Biochem Biophys Res Commun. 1996;219:509–514. doi: 10.1006/bbrc.1996.0264. [DOI] [PubMed] [Google Scholar]
- 28.Kletzien R F, Clarke S D, Ulrich R G. Enhancement of adipocyte differentiation by an insulin-sensitizing agent. Mol Pharmacol. 1992;41:393–398. [PubMed] [Google Scholar]
- 29.Kliewer S A, Forman B M, Blumberg B, Ong E S, Borgmeyer U, Mangelsdorf D J, Umesono K, Evans R M. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laborda J. 36B4 cDNA used as an estradiol-independent mRNA control is the cDNA for human acidic ribosomal phosphoprotein PO. Nucleic Acids Res. 1991;19:3998. doi: 10.1093/nar/19.14.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lane W S, Galat A, Harding M W, Schreiber S L. Complete amino acid sequence of the FK506 and rapamycin binding protein, FKBP, isolated from calf thymus. J Protein Chem. 1991;10:151–160. doi: 10.1007/BF01024778. [DOI] [PubMed] [Google Scholar]
- 32.Lee C C, Caskey C T. cDNA cloning using degenerate primers. In: Innis M A, Gelfad D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 46–53. [Google Scholar]
- 33.Lehmann J M, Moore L B, Smith-Oliver T A, Wilkison W O, Willson T M, Kliewer S A. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPAR gamma) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 34.Leibel R L, Chung W K, Chua S C., Jr The molecular genetics of rodent single gene obesities. J Biol Chem. 1997;272:31937–31940. doi: 10.1074/jbc.272.51.31937. [DOI] [PubMed] [Google Scholar]
- 35.Lim W A, Richards F M, Fox R O. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature. 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 36.MacDougald O A, Lane M D. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- 37.Mandrup, S., and M. D. Lane. Regulating adipogenesis. J. Biol. Chem. 272:5367–5370. [DOI] [PubMed]
- 38.Marth J D, Lewis D B, Cooke M P, Mellins E D, Gearn M E, Samelson L E, Wilson C B, Miller A D, Perlmutter R M. Lymphocyte activation provokes modification of a lymphocyte-specific protein tyrosine kinase (p56lck) J Immunol. 1989;142:2430–2437. [PubMed] [Google Scholar]
- 39.McKinney B, Riley M. An orecin-oil red O stain for concomitant demonstration of elastic tissue and lipid. Stain Technol. 1967;42:245–258. doi: 10.3109/10520296709115018. [DOI] [PubMed] [Google Scholar]
- 40.McNamee, H., and I. Aksoy. Personal communication.
- 41.Michaely P, Bennett V. The membrane-binding domain of ankyrin contains four independently folded subdomains, each comprised of six ankyrin repeats. J Biol Chem. 1993;268:22703–22709. [PubMed] [Google Scholar]
- 42.Miller R J, Bell G I. JAK/STAT eats the fat. Trends Neurosci. 1996;19:159–161. doi: 10.1016/s0166-2236(96)30006-4. [DOI] [PubMed] [Google Scholar]
- 43.Pawson, T. 1988. Non-catalytic domains of cytoplasmic protein-tyrosine kinases: regulatory elements in signal transduction. Oncogene 491–495. [PubMed]
- 44.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 45.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 46.Porras A, Muszynski K, Rapp U R, Santos E. Dissociation between activation of Raf-1 kinase and the 42-kDa mitogen-activated protein kinase/90-kDa S6 kinase (MAPK/RSK) cascade in the insulin/Ras pathway of adipocytic differentiation of 3T3 L1 cells. J Biol Chem. 1994;269:12741–12748. [PubMed] [Google Scholar]
- 47.Richard S, Yu D, Blumer K J, Hausladen D, Olszowy M W, Connelly P A, Shaw A S. Association of p62, a multifunctional SH2- and SH3-domain-binding protein, with src family tyrosine kinases, Grb2, and phospholipase Cγ-1. Mol Cell Biol. 1995;15:186–197. doi: 10.1128/mcb.15.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rickles R J, Botfield M C, Zhou X M, Henry P A, Brugge J S, Zoller M J. Phage display selection of ligand residues important for Src homology 3 domain binding specificity. Proc Natl Acad Sci USA. 1995;92:10909–10913. doi: 10.1073/pnas.92.24.10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandouk T, Reda D, Hofmann C. Antidiabetic agent pioglitazone enhances adipocyte differentiation of 3T3-F442A cells. Am J Physiol. 1993;264:C1600–C1608. doi: 10.1152/ajpcell.1993.264.6.C1600. [DOI] [PubMed] [Google Scholar]
- 50.Scaife R, Margolis R L. Biochemical and immunochemical analysis of rat brain dynamin interaction with microtubules and organelles in vivo and in vitro. J Cell Biol. 1990;111:3023–3033. doi: 10.1083/jcb.111.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 52.Sparks A B, Adey N B, Quilliam L A, Thorn J M, Kay B K. Screening phage-displayed random peptide libraries for SH3 ligands. Methods Enzymol. 1995;255:498–509. doi: 10.1016/s0076-6879(95)55052-6. [DOI] [PubMed] [Google Scholar]
- 53.Spiegelman B M, Choy L, Hotamisligil G S, Graves R A, Tontonoz P. Regulation of adipocyte gene expression in differentiation and syndromes of obesity/diabetes. J Biol Chem. 1993;268:6823–6826. [PubMed] [Google Scholar]
- 54.Spiegelman B M, Flier J S. Adipogenesis and obesity: rounding out the big picture. Cell. 1996;87:377–389. doi: 10.1016/s0092-8674(00)81359-8. [DOI] [PubMed] [Google Scholar]
- 55.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 56.Tanabe K, Okuya S, Tanizawa Y, Matsutani A, Oka Y. Leptin induces proliferation of pancreatic beta cell line MIN6 through activation of mitogen-activated protein kinase. Biochem Biophys Res Commun. 1997;241:765–768. doi: 10.1006/bbrc.1997.7894. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka K, Imajoh-Ohmi S, Sawada T, Shirai R, Hashimoto Y, Iwasaki S, Kaibuchi K, Kanaho Y, Shirai T, Terada Y, Kimura K, Nagata S, Fukui Y. A target of phosphatidylinositol 3,4,5-triphosphate with a zinc finger motif similar to that of the ADP-ribosylation-factor GTPase-activating protein and two pleckstrin homology domains. Eur J Biochem. 1997;245:512–519. doi: 10.1111/j.1432-1033.1997.00512.x. [DOI] [PubMed] [Google Scholar]
- 58.Tartaglia L A. The leptin receptor. J Biol Chem. 1997;272:32686–32695. doi: 10.1074/jbc.272.51.32686. [DOI] [PubMed] [Google Scholar]
- 59.Tartaglia L A, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards G J, Campfield L A, Clark F T, Deeds J, et al. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83:1263–1271. doi: 10.1016/0092-8674(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 60.Tontonoz P, Hu E, Graves R A, Budavari A I, Spiegelman B M. mPPARγ2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 61.Tontonoz P, Hu E, Spiegelman B M. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 62.Trainor C D, Evans T, Felsenfeld G, Boguski M S. Structure and evolution of a human erythroid transcription factor. Nature. 1990;343:92–96. doi: 10.1038/343092a0. [DOI] [PubMed] [Google Scholar]
- 63.Weng Z, Rickles R J, Feng S, Richard S, Shaw A S, Schreiber S L, Brugge J S. Structure-function analysis of SH3 domains: SH3 binding specificity altered by single amino acid substitutions. Mol Cell Biol. 1995;15:5627–5634. doi: 10.1128/mcb.15.10.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williamson M P. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu Z, Bucher N L R, Farmer S R. Induction of peroxisome proliferator-activated receptor γ during the conversion of 3T3 fibroblasts into adipocytes is mediated by C/EBPβ, C/EBPδ, and glucocorticoids. Mol Cell Biol. 1996;16:4128–4136. doi: 10.1128/mcb.16.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamabhai M, Kay B K. Examining the specificity of Src homology 3 domain—ligand interactions with alkaline phosphatase fusion proteins. Anal Biochem. 1997;247:143–151. doi: 10.1006/abio.1997.2040. [DOI] [PubMed] [Google Scholar]
- 67.Yeh W C, Cao Z, Classon M, McKnight S L. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- 68.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]