Abstract
Inadequate effectiveness of Indian antivenoms in treating envenomation caused by the Spectacled Cobra/Indian Cobra (Naja naja) in Sri Lanka has been attributed to geographical variations in the venom composition. This study investigated the de novo venom-gland transcriptomics and venom proteomics of the Sri Lankan N. naja (NN-SL) to elucidate its toxin gene diversity and venom variability. The neutralization efficacy of a commonly used Indian antivenom product in Sri Lanka was examined against the lethality induced by NN-SL venom in mice. The transcriptomic study revealed high expression of 22 toxin genes families in NN-SL, constituting 46.55% of total transcript abundance. Three-finger toxins (3FTX) were the most diversely and abundantly expressed (87.54% of toxin gene expression), consistent with the dominance of 3FTX in the venom proteome (72.19% of total venom proteins). The 3FTX were predominantly S-type cytotoxins/cardiotoxins (CTX) and α-neurotoxins of long-chain or short-chain subtypes (α-NTX). CTX and α-NTX are implicated in local tissue necrosis and fatal neuromuscular paralysis, respectively, in envenomation caused by NN-SL. Intra-species variations in the toxin gene sequences and expression levels were apparent between NN-SL and other geographical specimens of N. naja, suggesting potential antigenic diversity that impacts antivenom effectiveness. This was demonstrated by limited potency (0.74 mg venom/ml antivenom) of the Indian polyvalent antivenom (VPAV) in neutralizing the NN-SL venom. A pan-regional antivenom with improved efficacy to treat N. naja envenomation is needed.
Keywords: Indian cobra, venomics, venom-induced toxicity, antivenom, neutralization
1. Introduction
Worldwide, it is estimated that 1.8–2.7 million cases of snakebite envenomation occur each year, with fatalities ranging from around 81,000 to 138,000, and around three times as many continue to suffer permanent complications resulting from amputations, deformity and chronic organ failures [1]. Snakebite envenomation is an important cause of morbidity and mortality that heavily affects impoverished populations in South Asia and Southeast Asia, where the highest incidence of venomous snakebites in the world has been reported [2,3]. The sociodemographic and agriculture-related occupational profile of the population contributes to the increased risk of snake–human encounters in the region, which is a biodiversity hotspot of venomous snakes [4,5].
In South Asia, the major venomous snake species responsible for causing snakebite envenomation are typically referred to as the “Big Four”, which include the spectacled cobra (Naja naja), common krait (Bungarus caeruleus), Russell’s viper (Daboia russelii) and saw-scaled viper (Echis carinatus). The diverse topology and socio-ecological systems in South Asia, however, greatly influence the biogeographical distribution of the snakes and their adaptation to different habitats. This would have modulated the venom phenotype in terms of toxin composition, thus contributing to intra-species variation in the snake venom [6,7,8,9,10,11]. Sri Lanka, which is a tropical island country in the Indian Ocean, is long separated by the Palk Strait and biogeographically distinct from the Indian Peninsula. This has an impact on the management of snakebite envenomation in South Asia, as intra-species venom variation arose from biogeographical factor greatly affects the efficacy of Indian antivenom product used in Sri Lanka [12,13]. Based on recent hospital records and community surveys, over 33,000 snakebite cases and 400 deaths occur in Sri Lanka annually [14]. The Spectacled Cobra (N. naja, also known as Indian Cobra, Binocellate Cobra), which is widely distributed in the island country, is a leading cause of snakebite envenomation, with toxicity manifested as systemic neuromuscular paralysis and local tissue damage [15,16]. As there are no domestic antivenoms produced in Sri Lanka, antivenoms clinically used in the country are imported Indian polyvalent antivenom products, which are raised against the Big Four commonly from a specific region (Tamil Nadu) in India. For decades, the use of Indian antivenom in Sri Lanka has been questioned for low efficacy in treating envenomation caused by the local snake species. Presumably, the discrepancy of antivenom efficacy is due to snake venom variability between Sri Lanka and India [7,12,17,18,19,20]. To warrant safe and effective antivenom use, a robust assessment of imported antivenoms is necessary, and in the long run, developing a domestic antivenom product specific for native venomous snakes in Sri Lanka is the way forward [21,22].
The knowledge of snake venom composition details is essential to elucidate the intra-species venom variability. Current -omics technologies allow high throughput sequencing and identification of snake venom proteins and toxin genes, thereby unveiling the diversity of toxin genes and the dynamic range of venom complexities in many species, including widely distributed venomous snakes such as the cobras. Previous enzymatic, proteomic and transcriptomic studies of Naja species have revealed significant inter- and intra-species variation in venom composition and toxin gene diversity [6,7,23,24,25,26,27,28,29,30]. More recently, the genome of Indian N. naja (Kerala State) was reported, and the venom-gland transcriptomes of six cobra specimens sourced from Kerala in India and Kentucky Reptile Zoo in USA were profiled [31]. The study identified 15 toxin genes in the Indian Cobra N. naja, with three-finger toxins (3FTX) being the most highly expressed. Striking variations were, however, noted in the diversity and expression of the toxin genes across the different specimens even though they were sourced from the same species of the Indian origin. Another recent proteomic study further showed substantial intraspecific venom variation among six N. naja specimens from various geographical locales in India, including the Deccan plateau, Gangetic plain, semi-arid, desert and coastal regions [6]. The variation in venom composition resulted in differential venom toxicity and disparity in the efficacy of antivenom neutralization. It was observed that Indian cobra venom samples with a higher abundance of neurotoxins appeared to be less effectively neutralized by existing Indian antivenom products in the market [6]. In light of the finding, the toxin gene profile and venom proteome of the Sri Lankan N. naja are anticipated to vary markedly from the Indian counterpart, but this has not been investigated in depth. Earlier, there was one proteomic study comparing venom proteomes between the Indian and Sri Lankan N. naja, in which merely six toxin families comprising 26 and 25 proteins, respectively, were identified [7]. Additionally, the study showed that Indian VINS polyvalent antivenom bound less avidly to the Sri Lankan cobra venom toxins than the homologous Indian cobra toxins, supporting that Indian antivenom was generally less effective in treating cobra bite envenomation in Sri Lanka [20].
To elucidate the toxin gene diversity of the Sri Lankan Spectacled Cobra (N. naja), this study investigated the de novo venom-gland transcriptome of the snake applying next-generation sequencing techniques, and the venom proteomics of the snake was studied through a protein decomplexation strategy [32]. The neutralization efficacy of a commonly used Indian antivenom product (VINS polyvalent antivenom) was also assessed against the lethality induced by the venom in vivo. Findings from the study shed light on the variation of snake venom toxins in N. naja, and provide insights into the optimization of snakebite management in Sri Lanka.
2. Results and Discussion
2.1. De Novo RNA Sequencing and Transcriptome Assembly
De novo sequencing on Illumina platforms generated 54,477,234 paired-end raw reads from the venom gland tissue of the Sri Lankan Naja naja (NN-SL). After adapter trimming and quality filtering, a total of 51,595,008 clean reads were obtained. The clean reads were assembled into 106,637 contigs (N50 = 361) by Trinity program and subsequently clustered into 53,595 Unigenes (N50 = 676) (Table 1). Low-frequency transcripts were filtered at FPKM (fragments per kilobase of transcript per million mapped reads) below 1, streamlining the number of transcripts to 50,922 for expressed genes in the venom gland. The genes were further categorized based on their functional annotations: (i) toxin (80 transcripts), with known or putative toxinological role; (ii) non-toxin (24,226 transcripts), with encoded cellular or housekeeping proteins; (iii) unidentified (26,616 transcripts), where the gene identities could not be determined (Supplementary File S1). Among these, the toxin group constituted the most abundantly expressed genes with the highest percentage of total FPKM (46.55% of total gene expression) and redundancy value (5699.14 FPKM/transcript), as compared to the non-toxins (FPKM = 34.33%, redundancy = 13.88 FPKM/transcript) and unidentified (FPKM = 19.12%, redundancy = 7.03 FPKM/transcript) groups. The dominant expression and high redundancy of toxin genes in the venom gland transcriptome of NN-SL were consistent with findings reported for various elapid snakes, including the Hydrophis curtus (48.18% of total FPKM) [33], Ophiophagus hannah (35.3%) [34], Malaysian and Thai Naja kaouthia (41.2% and 82%, respectively) [27], Naja sumatrana (79.2%) [35], Chinese Naja atra and Bungarus multicinctus (70% of expressed sequences tags) [36] and American Micrurus fulvius (45.8%) [37].
Table 1.
Descriptive statistics of RNA-sequencing data for NN-SL venom gland transcriptome.
| Parameter | Sequencing Output |
|---|---|
| Total raw reads | 54,477,234 |
| Total clean reads | 51,595,008 |
| Total clean nucleotides (nt) | 4,643,550,720 |
| Q20 (%) | 98.50 |
| N (%) | 0.01 |
| GC (%) | 44.62 |
| Contigs assembly statistics | 106,637 |
| Total length (nt) | 28,724,569 |
| Mean length (nt) | 269 |
| N50 length (bp) | 361 |
| Unigenes assembly statistics | 53,595 |
| Total length (nt) | 25,919,321 |
| Mean length (nt) | 484 |
| N50 length (bp) | 676 |
Q20 is the proportion of nucleotides with quality value larger than 20; N percentage is the proportion of unknown nucleotides in clean reads; GC percentage is the proportion of guanidine and cytosine nucleotides among the total nucleotides; N50 is the shortest contig length to cover 50% of the transcriptome.
2.2. Venom Gland Transcriptome of NN-SL
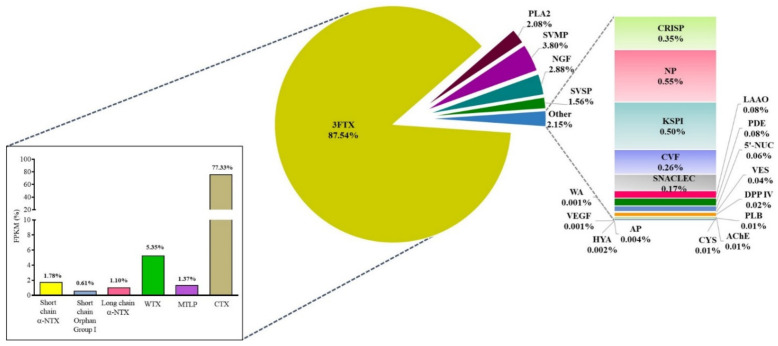
A total of 22 toxin gene families comprising 80 non-redundant transcripts were identified in the venom gland transcriptome of NN-SL (Figure 1). Of these, the three-finger toxins (3FTX) were the most abundantly and diversely expressed (87.54% of total toxin FPKM, 13 transcripts), followed by snake venom metalloproteinase (SVMP, 3.80%, 18 transcripts), nerve growth factor (NGF, 2.88%, 1 transcripts), phospholipase A2 (PLA2, 2.08%, 5 transcripts) and snake venom serine protease (SVSP, 1.56%, 3 transcripts) (Table 2). Other toxin gene families included acetylcholinesterase (AChE), aminopeptidase (AP), cysteine-rich secretory protein (CRISP), cobra venom factor (CVF), cystatin (CYS), dipeptidyl peptidase IV (DPP IV), hyaluronidase (HYA), L-amino acid oxidase (LAAO), natriuretic peptide (NP), Kunitz-type serine protease inhibitor (KSPI), snake C-type lectin (SNACLEC), phosphodiesterase (PDE), 5’-nucleotidase (5’-NUC), vespryn (VES), phospholipase B (PLB), vascular endothelial growth factor (VEGF) and waprin (WA), which were all minimally expressed (<1% of all toxin FPKM) and less diversified (1–4 transcripts each) (Table 2). All toxin transcripts from NN-SL were annotated based on sequence similarity with known toxins of elapid species, while approximately half of the toxin transcripts (mainly 3FTX) were matched to related cobra species (N. atra, N. kaouthia, N. sputatrix, Naja mossambica), implying conserved sequences of 3FTX genes in the genus. A total of 31 toxin transcripts were encoded in full-length sequences with >90% coverage of the gene sequences (Table 2).
Figure 1.
De novo venom-gland transcriptome of Sri Lankan Naja naja: the expression profile of toxin genes in the venom gland of spectacled cobra, Naja naja from Sri Lanka. Percentages indicate the relative abundances of transcripts by total toxin FPKM, sorted according to gene family/subfamily. Abbreviations: 3FTX, three-finger toxins; α-NTX, α-neurotoxin; WTX, weak neurotoxin; MTLP, muscarinic toxin-like protein; CTX, cytotoxin/cardiotoxin; PLA2, phospholipase A2; SVMP, snake venom metalloproteinases; NGF, nerve growth factor; SVSP, snake venom serine protease; CRISP, cysteine-rich secretory protein; NP, natriuretic peptide; KSPI, Kunitz-type serine protease inhibitor; CVF, cobra venom factor; SNACLEC, snake C-type lectins; LAAO, L-amino acid oxidase; PDE, phosphodiesterase; 5’-NUC, 5’-nucleotidase; VES, vespryn; DPP IV, dipeptidylpeptidase IV; PLB, phospholipase B; AChE, acetylcholinesterase; CYS, cystatin; AP, aminopeptidase; HYA, hyaluronidase; VEGF, vascular endothelial growth factor; WA, waprin. FPKM, fragments per kilobase of transcript per million mapped reads.
Table 2.
Overview of toxin gene families and transcript abundances (%) in venom gland transcriptome of NN-SL.
| Gene ID | Protein Family/ Subfamily |
Annotated Accession (NCBI/Uniprot) | Species | Coverage A (%) |
Transcript B Abundance (%) |
|---|---|---|---|---|---|
| Three-finger toxins (3FTX) | 87.54% | ||||
| Short-chain 3FTX: Short α-NTX (SNTX) | 1.78% | ||||
| NN-3FTX01 | Short neurotoxin 2 | P62376 | Hydrophis cyanocinctus | 13–79 | 0.11% |
| NN-3FTX02 * | Cobrotoxin | P60770 | Naja atra | 1–82 (98.80) | 0.69% |
| NN-3FTX03 * | Cobrotoxin-b | P80958 | Naja atra | 1–82 (100) | 0.97% |
| Short-chain 3FTX: Orphan Group I | 0.61% | ||||
| NN-3FTX04 | Neurotoxin-like protein NTL2 | Q9W717 | Naja atra | 1–75 | 0.61% |
| Short-chain 3FTX: Muscarinic toxin-like protein (MTLP) | 1.37% | ||||
| NN-3FTX05 * | Neurotoxin homolog NL1 | Q9DEQ3 | Naja atra | 6–86 (94.19) | 0.96% |
| NN-3FTX06 | Muscarinic toxin-like protein 2 | P82463 | Naja kaouthia | 1–46 | 0.41% |
| NN-3FTX07 | Haditoxin | A8N286 | Ophiophagus hannah | 36–84 | 0.001% |
| Short-chain 3FTX: Cytotoxin/cardiotoxin (CTX) | 77.33% | ||||
| NN-3FTX08 * | Cytotoxin 5 | Q98961 | Naja atra | 1–81 (100) | 77.33% |
| Long-chain 3FTX: Long α-NTX (LNTX) | 1.10% | ||||
| NN-3FTX09 * | Long neurotoxin 7 | O42257 | Naja sputatrix | 12–88 (97.78) | 1.00% |
| NN-3FTX10 | Long neurotoxin 1 | Q8UW29 | Hydrophis hardwickii | 12–91 | 0.01% |
| NN-3FTX11 | Long neurotoxin-like OH-31 | Q53B55 | Ophiophagus hannah | 20–59 | 0.08% |
| NN-3FTX12 | Long neurotoxin-like OH-31 | Q53B55 | Ophiophagus hannah | 5–59 | 0.003% |
| Non-conventional 3FTX: Weak neurotoxin (WTX) | 5.35% | ||||
| NN-3FTX13 | Probable weak neurotoxin NNAM1 | Q9YGI2 | Naja atra | 15–86 | 5.35% |
| Phospholipase A2 (PLA2) | 2.08% | ||||
| NN-PLA01 | Basic phospholipase A2 | P00610 | Hydrophis schistosus | 23–119 | 0.01% |
| NN-PLA02 | Basic phospholipase A2 | P00610 | Hydrophis schistosus | 23–119 | 0.002% |
| NN-PLA03 | Acidic phospholipase A2 57 | Q8UW31 | Hydrophis hardwickii | 51–135 | 0.01% |
| NN-PLA04 * | Acidic phospholipase A2 2 | P00597 | Naja kaouthia | 1–146 (100) | 2.05% |
| NN-PLA05 | Basic phospholipase A2 73 | Q8UW30 | Hydrophis hardwickii | 1–56 | 0.01% |
| Snake venom metalloproteinase (SVMP) | 3.80% | ||||
| NN-SVMP01 | Scutatease-1 | B5KFV7 | Notechis scutatus | 337–388 | 0.04% |
| NN-SVMP02 | Zinc metalloproteinase mocarhagin | Q10749 | Naja mossambica | 502–568 | 0.03% |
| NN-SVMP03 | Zinc metalloproteinase mocarhagin | Q10749 | Naja mossambica | 191–241 | 0.03% |
| NN-SVMP04 | Zinc metalloproteinase mocarhagin | Q10749 | Naja mossambica | 143–195 | 0.03% |
| NN-SVMP05 | Zinc metalloproteinase-disintegrin cobrin | Q9PVK7 | Naja kaouthia | 159–499 | 0.27% |
| NN–SVMP06 | Zinc metalloproteinase-disintegrin cobrin | Q9PVK7 | Naja kaouthia | 159–600 | 0.16% |
| NN-SVMP07 | Zinc metalloproteinase-disintegrin cobrin | Q9PVK7 | Naja kaouthia | 159–600 | 0.13% |
| NN-SVMP08 | Zinc metalloproteinase-disintegrin atrase-B | D6PXE8 | Naja atra | 322–369 | 0.01% |
| NN-SVMP09 | Zinc metalloproteinase-disintegrin atrase-A | D5LMJ3 | Naja atra | 456–607 | 0.10% |
| NN-SVMP10 | Zinc metalloproteinase-disintegrin atrase-A | D5LMJ3 | Naja atra | 456–607 | 0.10% |
| NN-SVMP11 | Zinc metalloproteinase-disintegrin atrase-A | D5LMJ3 | Naja atra | 456–607 | 0.15% |
| NN-SVMP12 | Zinc metalloproteinase-disintegrin atrase-A | D5LMJ3 | Naja atra | 403–607 | 0.13% |
| NN-SVMP13 | Zinc metalloproteinase-disintegrin atrase-A | D5LMJ3 | Naja atra | 403–607 | 0.17% |
| NN-SVMP14 | Zinc metalloproteinase-disintegrin atrase-A | D5LMJ3 | Naja atra | 456–607 | 0.28% |
| NN-SVMP15 | Zinc metalloproteinase-disintegrin atrase-A | D5LMJ3 | Naja atra | 1–229 | 1.25% |
| NN-SVMP16 | Hemorrhagic metalloproteinase-disintegrin kaouthiagin | P82942 | Naja kaouthia | 6–64 | 0.03% |
| NN-SVMP17 | Zinc metalloproteinase-disintegrin-like atragin | D3TTC2 | Naja atra | 527–613 | 0.80% |
| NN-SVMP18 | Hemorrhagic metalloproteinase-disintegrin kaouthiagin | P82942 | Naja kaouthia | 130–201 | 0.07% |
| Cysteine-rich secretory protein (CRISP) | 0.35% | ||||
| NN-CRISP01 * | Natrin-1 | Q7T1K6 | Naja atra | 1–239 (100) | 0.35% |
| Snake venom serine protease (SVSP) | 1.56% | ||||
| NN-SVSP01 * | Serine protease harobin | Q5MCS0 | Hydrophis hardwickii | 1–265 (96.60) | 1.56% |
| NN-SVSP02 | Serine protease HTRA1 | V8NGT0 | Ophiophagus hannah | 79–304 | 0.001% |
| NN-SVSP03 * | Serine protease 23 | V8N8N4 | Ophiophagus hannah | 8–372 (98.12) | 0.001% |
| Cobra venom factor (CVF) | 0.26% | ||||
| NN-CVF01 | Cobra venom factor | Q91132 | Naja kaouthia | 1586–1642 | 0.13% |
| NN-CVF02 * | Cobra venom factor | Q91132 | Naja kaouthia | 1–1592 (96.96) | 0.10% |
| NN-CVF03 | Cobra venom factor | Q91132 | Naja kaouthia | 1197–1592 | 0.02% |
| NN-CVF04 | Cobra venom factor | Q91132 | Naja kaouthia | 1–640 | 0.01% |
| Kunitz-type serine protease inhibitor (KSPI) | 0.50% | ||||
| NN-KSPI01 * | Putative Kunitz-type serine protease inhibitor | B2BS84 | Austrelaps labialis | 1–252 (98.81) | 0.02% |
| NN-KSPI02 * | Kunitz-type protease inhibitor | U3FZD6 | Micrurus fulvius | 1–511 (99.61) | 0.004% |
| NN-KSPI03 | Protease inhibitor 3 | C1IC51 | Walterinnesia aegyptia | 1–67 | 0.17% |
| NN-KSPI04 | Kunitz-type serine protease inhibitor NACI | Q5ZPJ7 | Naja atra | 43–81 | 0.30% |
| Phosphodiesterase (PDE) | 0.08% | ||||
| NN-PDE01 * | Snake venom phosphodiesterase | A0A2D0TC04 | Naja atra | 21–850 (97.65) | 0.08% |
| 5’-Nucleotidase (5’-NUC) | 0.06% | ||||
| NN-NUC01 * | Ecto-5’-nucleotidase | A0A194AS98 | Micrurus tener | 1–569 (99.13) | 0.05% |
| NN-NUC02 * | Ecto-5’-nucleotidase | A0A194AS98 | Micrurus tener | 1–536 (93.38) | 0.008% |
| NN-NUC03 * | Ecto-5’-nucleotidase | A0A194AS98 | Micrurus tener | 1–536 (93.38) | 0.004% |
| NN-NUC04 | 5’ nucleotidase | A6MFL8 | Demansia vestigiata | 134–559 | 0.001% |
| Acetylcholinesterase (AChE) | 0.01% | ||||
| NN-ACHE01 * | Acetylcholinesterase | Q92035 | Bungarus fasciatus | 1–566 (95.55) | 0.004% |
| NN-ACHE02 | Acetylcholinesterase | Q92035 | Bungarus fasciatus | 1–566 (95.55) | 0.002% |
| Hyaluronidase (HYA) | 0.002% | ||||
| NN-HYA01 | Hyaluronidase | V8PHI0 | Ophiophagus hannah | 26–71 | 0.001% |
| NN-HYA02 | Hyaluronidase | A0A2D4JS30 | Micrurus lemniscatus lemniscatus | 89–208 | 0.0005% |
| NN-HYA03 | Hyaluronidase | A0A2D4JS30 | Micrurus lemniscatus lemniscatus | 27–91 | 0.0003% |
| NN-HYA04 * | Hyaluronidase | A0A194APD1 | Micrurus tener | 1–440 (98.43) | 0.0003% |
| Nerve growth factor (NGF) | 2.88% | ||||
| NN-NGF01 * | Venom nerve growth factor 2 | Q5YF89 | Naja sputatrix | 1–241 (100) | 2.88% |
| Vespryn (VES) | 0.04% | ||||
| NN-VES01 * | Ohanin | P83234 | Ophiophagus hannah | 1–181 (100) | 0.03% |
| NN-VES02 * | Ohanin | P83234 | Ophiophagus hannah | 1–181 (100) | 0.02% |
| Vascular endothelial growth factor (VEGF) | 0.001% | ||||
| NN-VEGF01 * | Vascular endothelial growth factor 1 | U3F558 | Micrurus fulvius | 1–142 (95.95) | 0.001% |
| NN-VEGF02 * | Vascular endothelial growth factor 1 | U3F558 | Micrurus fulvius | 1–142 (95.95) | 0.0003% |
| Dipeptidyl peptidase IV (DPP IV) | 0.02% | ||||
| NN-DPP01 | Dipeptidyl peptidase 4 | V8P9G9 | Ophiophagus hannah | 0.02% | 0.02% |
| Phospholipase B (PLB) | 0.01% | ||||
| NN-PLB01 * | Phospholipase-B 81 | F8J2D3 | Drysdalia coronoides | 1–553 (100) | 0.01% |
| NN-PLB02 | Phospholipase B-like | V8NLQ9 | Ophiophagus hannah | 146–196 | 0.001% |
| Snake C-type lectin (SNACLEC) | 0.17% | ||||
| NN-SNAC01 * | C-type lectin BfL-1 | Q90WI8 | Bungarus fasciatus | 1–158 (100) | 0.06% |
| NN-SNAC02 * | C-type lectin BfL-1 | Q90WI8 | Bungarus fasciatus | 1–148 (98.67) | 0.11% |
| NN-SNAC03 | Venom C-type lectin mannose binding isoform 1 variant 3 | D2YVK0 | Tropidechis carinatus | 57–158 | 0.001% |
| Cystatin (CYS) | 0.01% | ||||
| NN-CYS01 | Cystatin | E3P6P4 | Naja kaouthia | 0.01% | 0.01% |
| Aminopeptidase (AP) | 0.004% | ||||
| NN-AP01 * | Aminopeptidase | U3FZS8 | Micrurus fulvis | 1–992 (98.29) | 0.003% |
| NN-AP02 * | Aminopeptidase | U3FZS8 | Micrurus fulvius | 1–992 (98.29) | 0.001% |
| Waprin (WA) | 0.001% | ||||
| NN-WAP01 | Waprin-Phi3 | A7X4M7 | Philodryas olfersii | 33–80 | 0.001% |
| Natriuretic peptide (NP) | 0.55% | ||||
| NN-NP01 | Natriuretic peptide Na-NP | D9IX97 | Naja atra | 34–77 | 0.41% |
| NN-NP02 | Natriuretic peptide Na-NP | D9IX97 | Naja atra | 115–165 | 0.08% |
| NN-NP03 | Natriuretic peptide Na-NP | D9IX97 | Naja atra | 25–80 | 0.06% |
| L-amino acid oxidase (LAAO) | 0.08% | ||||
| NN-LAAO01 | L-amino-acid oxidase | Q4JHE3 | Oxyuranus scutellatus scutellatus | 1–517 | 0.04% |
| NN-LAAO02 | L-amino-acid oxidase | Q4JHE3 | Oxyuranus scutellatus scutellatus | 1–517 | 0.02% |
| NN-LAAO03 | L-amino-acid oxidase | Q4JHE3 | Oxyuranus scutellatus scutellatus | 1–517 | 0.02% |
* Indicates full-length transcripts of NN-SL with >90% coverage to the annotated sequence. A indicates the length of coverage of toxin sequence when compared to the annotated sequence, and the value in bracket refers to the % coverage of full-length transcript of NN-SL. B Transcript abundance (in %) was based on FPKM (fragments per kilobase of transcript per million mapped reads) of toxin genes in NN-SL.
2.3. Venom Proteome of NN-SL
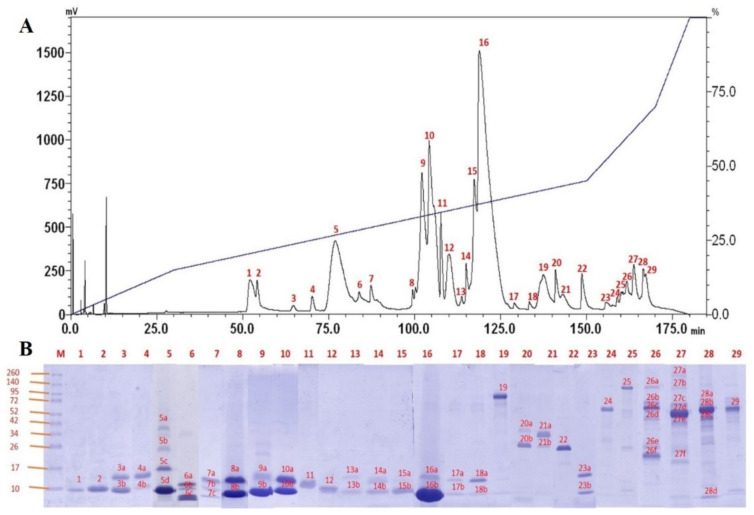
Using C18 reverse-phase HPLC, the NN-SL venom was resolved into 29 protein fractions with increasing hydrophobicity (Figure 2A). The protein fractions were individually subjected to SDS-PAGE under 15% reducing conditions, and a total of 63 heterogeneous protein bands were yielded (Figure 2A,B). The decomplexation profile of NN-SL venom was comparable to that of several Asiatic Naja species reported, studied under similar experimental conditions [8,23,38,39]. In brief, the majority of NN-SL venom proteins were 7–20 kDa and eluted between 50 min and 135 min of reverse-phase chromatography (fractions 1–18), accounting for approximately 88% of total venom composition as estimated by the chromatogram peak areas. Proteins with moderate-to-high molecular weight (>25 kDa) were eluted in the subsequent 40 min and accounting for approximately 12% of total venom proteins.
Figure 2.
(A) C18 reverse-phase HPLC fractionation of Sri Lankan Spectacled Cobra (Naja naja) venom. (B) SDS-PAGE of the HPLC-fractionated venom proteins under 15% reducing conditions. M: Protein ladder with molecular weight 10–260 kDa.
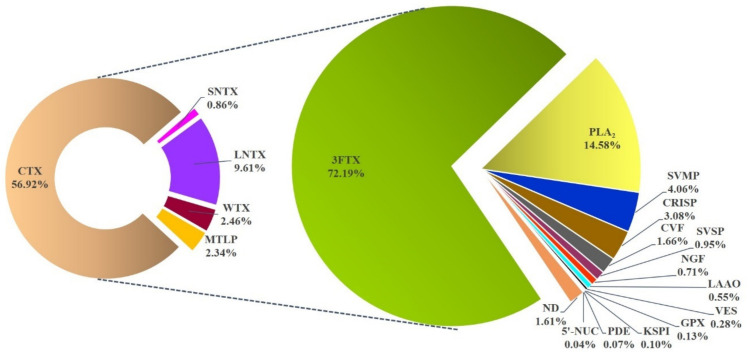
MALDI TOF/TOF and nano-ESI-LCMS/MS analyses of the in-gel digested peptides identified a total of 45 proteins that were clustered into 12 toxin families and a non-toxin protein family in the NN-SL venom proteome (Figure 3). The proteome was dominated by the low molecular weight 3FTX (72.19% of total venom proteins) and PLA2 (14.58%). The remaining ten toxin families collectively constituted less than 12% of total venom proteins, and these were snake venom metalloproteinase (SVMP, 4.06%), cysteine-rich secretory protein (CRISP, 3.08%), cobra venom factor (CVF, 1.66%), snake venom serine protease (SVSP, 0.95%), nerve growth factor (NGF, 0.71%), L-amino acid oxidase (LAAO, 0.55%), vespryn (VES, 0.28%), Kunitz-type serine protease inhibitor (KSPI, 0.10%), phosphodiesterase (PDE, 0.07%), 5’-nucleotidase (5’-NUC, 0.04%) and a non-toxin glutathione peroxidase (GPX, 0.13%) (Table 3). The dominating feature of 3FTX and PLA2 in NN-SL venom proteome follows a somewhat predictable trend as observed in virtually all previously reported venom proteomes of Asiatic cobras, notwithstanding variability in terms of the diversity (subtypes) and quantity (relative abundances) of their toxins [6,7,8,23,24,26,28,30,38,39,40,41,42,43,44].
Figure 3.
Venom proteome of the Sri Lankan Spectacled Cobra (Naja naja), profiled according to toxin families and relative protein abundances (% of total venom proteins). Abbreviations: 3FTX, three-finger toxins; SNTX, short neurotoxin; LNTX, long neurotoxin; WTX, weak neurotoxin; MTLP, muscarinic toxin; CTX, cytotoxin/cardiotoxin; PLA2, phospholipase A2; SVMP, snake venom metalloproteinase; CRISP, cysteine-rich secretory protein; CVF, cobra venom factor; SVSP, snake venom serine protease; NGF, nerve growth factor; LAAO, L-amino acid oxidase; VES, vespryn; GPX, glutathione peroxidase; KSPI, Kunitz-type serine protease inhibitor; PDE, phosphodiesterase; 5‘-NUC, 5’-nucleotidase; ND, not determined/ unidentified.
Table 3.
Overview of NN-SL venom proteins sorted according to families, subtypes and their relative abundance. The identification of tryptic digested peptides was performed using MALDI TOF/TOF and nano-ESI-LCMS/MS analyses.
| Protein Name | Accession Number * | Species * | Relative Abundance |
|---|---|---|---|
| 3FTX | 72.19% | ||
| SNTX | 0.86% | ||
| Short neurotoxin 1 | P01427 | Naja oxiana | 0.86% |
| LNTX | 9.61% | ||
| Alpha-elapitoxin-Nk2a | P01391 | Naja kaouthia | 2.22% |
| Long neurotoxin 1 | P25668 | Naja naja | 0.54% |
| Long neurotoxin 1 | P25671 | Naja kaouthia | 6.60% |
| Long neurotoxin 4 | P25672 | Naja naja | 0.25% |
| MTLP | 2.34% | ||
| Muscarinic toxin-like protein 3 | P82464 | Naja kaouthia | 2.16% |
| Muscarinic toxin-like protein 2 | P82463 | Naja kaouthia | 0.18% |
| WTX | 2.46% | ||
| Weak neurotoxin 7 | P29181 | Naja naja | 0.15% |
| Weak neurotoxin 6 | P29180 | Naja naja | 2.31% |
| CTX | 56.92% | ||
| Cytotoxin 3 | P01446 | Naja kaouthia | 0.06% |
| Cytotoxin 1d/1e | Q98958 | Naja atra | 7.83% |
| Cytotoxin-like basic protein | P62377 | Naja naja | 4.34% |
| Cytotoxin 4 | P60303 | Naja kaouthia | 0.38% |
| Cytotoxin 2c | O93472 | Naja sputatrix | 37.34% |
| Cytotoxin 1 | P01447 | Naja naja | 6.97% |
| PLA2 | 14.58% | ||
| Neutral phospholipase A2 muscarinic inhibitor | Q92084 | Naja sputatrix | 1.88% |
| Acidic phospholipase A2 2 | P15445 | Naja naja | 11.63% |
| Basic phospholipase A2 homolog 1 | P10117 | Laticauda colubrina | 0.54% |
| Acidic phospholipase A2 1 | P00596 | Naja kaouthia | 0.32% |
| Acidic phospholipase A2 2 | P00597 | Naja kaouthia | 0.21% |
| KSPI | 0.10% | ||
| Protease inhibitor 2 | Unigene36921_ESM | Hydrophis schistosus | 0.10% |
| NGF | 0.71% | ||
| Venom nerve growth factor | P01140 | Naja naja | 0.58% |
| Venom nerve growth factor | P61898 | Naja atra | 0.12% |
| VES | 0.28% | ||
| Thaicobrin | P82885 | Naja kaouthia | 0.28% |
| SVMP | 4.06% | ||
| Zinc metalloproteinase-disintegrin-like atrase-A | D5LMJ3 | Naja atra | 3.91% |
| Hemorrhagic metalloproteinase kaouthiagin | P82942 | Naja kaouthia | 0.15% |
| CRISP | 3.08% | ||
| Cysteine-rich venom protein natrin-1 | Q7T1K6 | Naja atra | 3.08% |
| SVSP | 0.95% | ||
| Snake venom serine protease NaSP | A8QL53 | Naja atra | 0.95% |
| CVF | 1.66% | ||
| Cobra venom factor precursor | Q91132 | Naja kaouthia | 1.66% |
| PDE | 0.07% | ||
| Phosphodiesterase | A0A194ARD7 | Micrurus tener | 0.04% |
| Phosphodiesterase 1 | Unigene5869_Naja-sumatrana | Naja sumatrana | 0.03% |
| LAAO | 0.55% | ||
| L-amino acid oxidase | A8QL58 | Naja atra | 0.44% |
| L-amino-acid oxidase precursor | Q4JHE3 | Oxyuranus scutellatus scutellatus | 0.11% |
| 5’-NUC | 0.04% | ||
| Snake venom 5’-nucleotidase | CL3600.Contig1_NS2A | Naja sumatrana | 0.04% |
| GPX | 0.13% | ||
| Glutathione peroxidase | V8P395 | Ophiophagus hannah | 0.13% |
| ND | 1.61% |
* accession number and species annotated based on sequence similarity by BLAST. Abbreviations: 3FTX, three-finger toxins; SNTX, short neurotoxin; LNTX, long neurotoxin; WTX, weak neurotoxin; MTLP, muscarinic toxin; CTX, cytotoxin/cardiotoxin; PLA2, phospholipase A2; SVMP, snake venom metalloproteinase; CRISP, cysteine-rich secretory protein; CVF, cobra venom factor; SVSP, snake venom serine protease; NGF, nerve growth factor; LAAO, L-amino acid oxidase; VES, vespryn; GPX, glutathione peroxidase; KSPI, Kunitz-type serine protease inhibitor; PDE, phosphodiesterase; 5‘-NUC, 5’-nucleotidase; ND, not determined/unidentified.
Comparing between the venom gland transcriptome and venom proteome, we observed a significant correlation between the toxin gene expression (% of all toxin FPKM) and the venom protein abundance (% of total venom proteins) of NN-SL (Spearman correlation coefficient, r = 0.7062, p <0.05) (Supplementary Figure S1). Among the 22 toxin families identified in the venom gland transcriptome, 12 protein families (3FTX, PLA2, SVMP, NGF, SVSP, CRISP, KSPI, CVF, LAAO, PDE, VES, 5’-NUC) were detected at the proteomic level. The toxin genes found only at the transcriptomic level were primarily those with very low expression, including NP, SNACLEC, CYS, AChE, VEGF, DPP IV, PLB, AP, WA and HYA. These proteins might be present at an extremely low amount below the mass spectrometry detection limit, or, the translation of the mRNA had been negatively regulated post-transcriptionally [45].
The data of the mass spectrometry analysis, including peptide sequences, values of spectral mass/charge and relative abundances of proteins identified was provided in Supplementary File S2.
2.4. Three-Finger Toxins (3FTX)
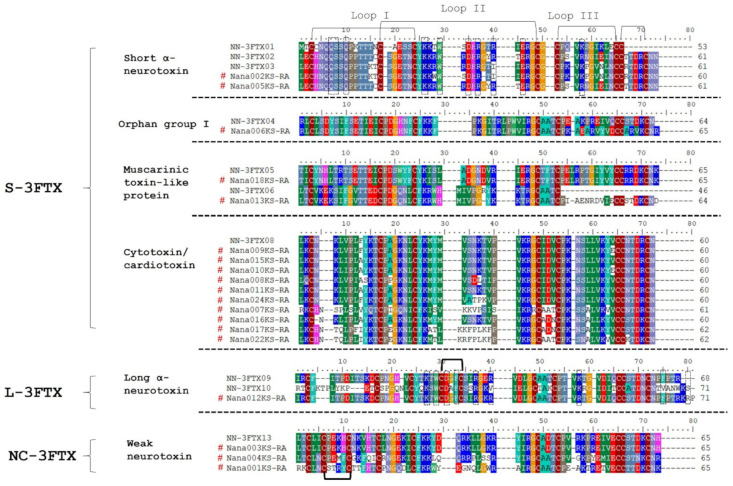
The high expression levels of 3FTX in the transcriptome and proteome of NN-SL support a pivotal role of these toxins in predation and pathogenesis of envenomation. Three-finger toxins are non-enzymatic polypeptides containing 60–74 amino acids with 8–10 conserved cysteinyl residues. The molecule adopts a three-finger-like structure that consists of three antiparallel ß-stranded loops extending from a small hydrophobic core cross-linked by conserved disulfide bridges [46,47]. Of the 13 diverse 3FTX genes expressed in NN-SL, eight belonged to the short-chain 3FTX (S-3FTX, characterized by the presence of four disulfide bonds); these were denoted as NN-3FTX01–08, comprising three short-chain alpha-neurotoxins (SNTX), one cytotoxin/cardiotoxin (CTX), three muscarinic toxin-like proteins (MTLP), and one orphan group I 3FTX (Table 2). The remaining five 3FTX genes were of long-chain 3FTX (L-3FTX, with 5 disulfide bonds), comprising four long-chain alpha-neurotoxins (LNTX) and one non-conventional 3FTX (NC-3FTX) (Table 2). The primary structures of the 3FTX were shown in Figure 4, illustrating the four conserved disulfide bonds between loop I and loop III of SC-3FTX, and the additional fifth disulfide bond present at the N-terminal loop I of NC-CTX and the central loop II of LNTX.
Figure 4.
Multiple sequence alignment of the three-finger toxins (3FTX) transcripts from the venom gland transcriptome of N. naja. The alignment of 3FTX transcripts based on subfamilies: short-chain, long-chain and non-conventional toxins. The 3FTX transcripts obtained in the current study were aligned and compared with the gene sequence from the transcriptome of N. naja specimens sourced from Kerala, India and Kentucky Reptile Zoo, USA. # indicates the gene sequences of 3FTX retrieved from [31]. The black bracket indicates the disulfide bridges located at the first, second and third loop of 3FTX, respectively. The additional bracket denotes the fifth disulfide bridge located at the first and second loops of NC-3FTX and L-3FTX, respectively. Dotted square box in the amino acid sequence of short α-neurotoxins and long α-neurotoxins highlights the critical region of amino acids that bind to nAChR [66].
Of the 13 3FTX genes identified in NN-SL, the CTX transcript NN-3FTX08 was the most abundantly expressed in its venom gland (77.33% of total toxin FPKM). The sequence of NN-3FTX08 was highly similar to Nana009KS-RA, the major CTX gene of N. naja from India with one amino acid substitution at the 52nd residue position (E52 → V52) [31] (Figure 4; note that the position in alignment figure is 64 due to artificial gapping). Both NN-3FTX08 and Nana009KS-RA are S-type CTX based on the presence of S28 residue (Figure 4, position in alignment figure: 35) in the protein, as proposed in the S-type/P-type CTX classification system [48,49]. The P-type CTX, as exemplified by CTX A3 from the Taiwanese Cobra, N. atra, has a more pronounced lipid-binding and membrane pore-forming activities than the S-type CTX [50,51], although the latter also exhibits cytotoxicity in human cell lines [52]. Between NN-3FTX08 and Nana009KS-RA, the substitution of polar, negatively charged glutamic acid (E52) in Nana009KS-RA to hydrophobic valine (V52) (Figure 4, position in alignment figure: 64) in NN-3FTX08 may result in increased hydrophobicity of loop III by strengthening the interaction of tyrosine (Y51, position in alignment figure: 63) with the lipid membrane [53], thereby causing a more pronounced lipolytic activity of the CTX. There is, however, a dearth of study that compares the clinical toxicity of N. naja envenomation between India and Sri Lanka to shed light on the potential difference in cytotoxicity of the venoms. Hence, studies with a focus on the comparative characterization of CTX of N. naja from different locales should be the subject of future research.
The relative protein abundance of CTX present in the NN-SL venom proteome was highly correlated with the CTX gene expression in the venom gland. The high abundance of CTX (56.92% of total venom proteins) is consistent with the development of severe tissue necrosis in envenomation caused by N. naja [13,20]. With a median lethal dose (LD50) of 1–2 ug/g in mice, cobra venom CTX usually do not cause a direct lethal activity [54,55] but are implicated in the local tissue damage and venom ophthalmia [56,57]. Cationic CTX are able to bind to the anionic phospholipid bilayer of cell membrane, thereby disrupting the membrane integrity, forming pore and initiating cell death [48,58,59]. CTX has also been known to elicit a cardiotoxic effect by causing systolic heart arrest in ex vivo experiments [60,61], albeit direct cardiotoxicity is rarely seen in cobra envenomation. From the evolutionary perspective, it has been suggested that the cytotoxic effect of CTX co-evolved with the hooding display behavior and aposematic coloration of cobras for defense purposes [62].
The alpha-neurotoxins are the principal lethal components (with LD50 of 0.05–0.2 mg/kg in mice) in cobra venoms [38,39,54,55,63]. These toxins are post-synaptic neurotoxins that bind to the nicotinic acetylcholine receptors (nAChR) at the neuromuscular junction, thereby interrupting neurotransmission, and rapidly resulting in descending paralysis as manifested clinically in cobra bite envenomation [64,65,66]. Three transcripts of short-chain α-neurotoxin (SNTX, NN-3FTX01-03) and four transcripts of long-chain α-neurotoxin (LNTX, NN-3FTX9-12) were identified in the venom gland transcriptome of NN-SL (Table 2). In comparison to alpha-neurotoxins of the Indian N. naja, the SNTX in NN-SL (NN-3FTX02) was found identical to Nana005KS-RA [31], whilst two amino acid substitutions occurred between NN-3FTX03 and Nana002KS-RA at loop II: W27 and S29 in Nana002KS-RA were substituted by R27 and R29 in NN-3FTX03, respectively, as shown in Figure 4 (positions in alignment figure: 28 and 34). The substitutions involving these polar amino acids (Trp, Ser and Arg) were predicted to be conservative based on the Sorting Intolerant from Tolerant (SIFT) algorithm [67,68], thus unlikely to result in a significant change of the SNTX activity. Meanwhile, NN-3FTX01 was not previously reported in the N. naja specimens sourced from India and Kentucky Reptile Zoo, hence it could be a diverging SNTX form unique to the Sri Lankan cobra population.
The main transcript of LNTX in NN-SL (NN-3FTX09) was homologous to the LNTX transcript Nana012KS-RA of the Indian N. naja, with two amino acid substitutions that occurred at the residue positions of 35 and 49 (from NN-SL to the Indian Cobra of Kerala: E35 → K35 and K49 → R49) (Figure 4, positions in alignment figure: 39 and 58). The substitution of R49 with K49 (Arg and Lys are basic and charged) was conservative while the replacement involving E35 (acidic Glu) and K35 (basic Lys) might result in altered activity, although LNTX were typically the most lethal component in cobra venoms [54,55]. Irrespective of the subtle sequence variation between NN-SL and the Indian Cobra, these alpha-neurotoxins (SNTX and LNTX) bind to similar sites on the nicotinic acetylcholine receptors (nAChR) with high affinity [69]. The SNTX binds to the nAChR six- to seven-times faster and dissociates five- to nine-times faster than LNTX, which is known to cause relatively irreversible block compared to SNTX [70,71]. Toxin neutralization experiments, on the other hand, showed that most antivenoms neutralized SNTX less efficaciously compared to LNTX, implying that the SNTX has a lower immunogenicity than LNTX [54,55]. It should be noted though that the neutralization efficacy of antivenoms against the lethality of cobra venoms is generally low (potency <1 mg/mL in terms of the amount of venom completely neutralized by one mLof antivenom) [72,73], compared to the neutralization of viper and pit viper venoms in which the antivenom potency commonly exceeded 2 mg/mL [74,75,76]. The abundance of alpha-neurotoxins, therefore, appears to be the limiting factor of antivenom neutralization. Proteomic studies of cobra venoms should aim to characterize the quantitative toxin abundance in order to elucidate differences in the venom lethality and neutralization efficacy of antivenom [39].
The venom-gland transcriptomics also unveiled the genes of minor 3FTX including muscarinic toxin-like protein (MTLP), neurotoxin-like protein (Orphan Group I) and weak neurotoxin (WTX) (Table 2). These toxin genes, albeit lowly expressed, were rather conserved venom phenotype between NN-SL and N. naja specimens from different geographical locales [31] (Figure 4). The MTLP-encoding genes in NN-SL (NN-3FTX05 and NN-3FTX06) show sequences identical with those of the Indian Cobra (Nana018KS-RA and Nana013KS-RA). Similarly, the WTX genes in NN-SL (NN-3FTX13) and the Indian Cobra (Nana003KS-RA) were also identically matched by their sequences (Figure 4). The genes encoding Orphan Group I neurotoxin-like protein in NN-SL (NN-3FTX04) were similar to the Indian Cobra (Nana006KS-RA) except with some amino acid differences in loop III and C-terminal region (Figure 4). These 3FTX genes were expressed in low amount into venom proteome, accounting for ~2% of total venom proteins of NN-SL (Figure 3). These minor toxins have been shown to have low toxicity and lack lethal activity [77,78,79].
2.5. Phospholipase A2 (PLA2)
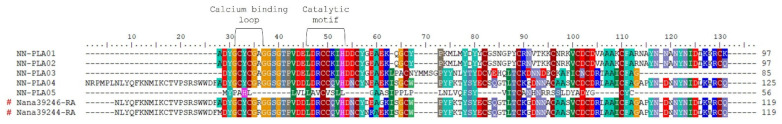
A total of five PLA2 transcripts (NN-PLA01–05, Table 2) were identified in the venom-gland transcriptome of NN-SL. NN-PLA04 was the most abundantly expressed secretory PLA2 transcript, with a sequence highly similar (>90%) to the PLA2 transcripts (Nana39246-RA and Nana39244-RA) from the Indian Cobra of Kerala [31] (Figure 5). NN-PLA04 belongs to the snake venom PLA2 of Group IA family, and its calculated pI (5.22) indicated that it is an acidic PLA2. The PLA2 sequence contains conserved amino acid residues H53, Y57 and D104 as with other svPLA2, and in addition, D49 (Figure 5; positions in alignment figure: 53, 57, 104, 54, respectively) which is critical for its enzymatic activity [80,81]. In the venom proteomics, the protein abundance of PLA2 expressed was relatively higher (~14% of total venom proteins) than the transcription level in the venom gland (~2.1% of toxin FPKM). Low expression levels of PLA2 in snake venom gland transcriptomics have been reported for a number of Asiatic cobras (for instance, N. naja [31], Naja kaouthia [27], Naja sumatrana [35]) and the king cobra, Ophiophagus hannah [34,82], despite showing high protein abundances of PLA2 in their venom proteomes. The discrepancy could be due to a lower rate of PLA2 gene transcription, or a longer half-life of the PLA2 transcripts that allows prolonged protein translation. The protein abundance of PLA2 in NN-SL venom is consistent with that reported in most cobra venom proteomes (ranging from 11–21% of total venom proteins) [7,8,29,38,39], except for those in the Uraeus subgenus (African non-spitting cobras) whose venoms contain no or negligible amount of PLA2 [83,84].
Figure 5.
Comparison of PLA2 transcripts of NN-SL and N. naja specimens from Kerala, India and Kentucky Reptile Zoo, USA. # indicates the gene sequences of PLA2 retrieved from [31]. Top brackets indicate calcium-binding loop and catalytic motif.
The acidic PLA2 in cobra venoms generally have low toxicity (i.v. LD50 >10 mg/kg in mice) [54,55,63,85], but probably serve ancillary functions through synergistic interaction with other venom proteins such as WTX [86], CTX [87] and SVMP [88]. A recent study also demonstrated the synergistic action of upregulated PLA2 and CTX in eliciting pain for defense in three lineages of spitting cobras [89]. While N. naja is a non-spitting cobra, its PLA2 may exhibit similar potentiating activity with the CTX abundantly expressed in its venom, thereby enhancing the local tissue toxicity in envenomation.
2.6. Comparison of Naja naja Venomics and Implication on Antivenom Treatment for Cobra Bite Envenomation
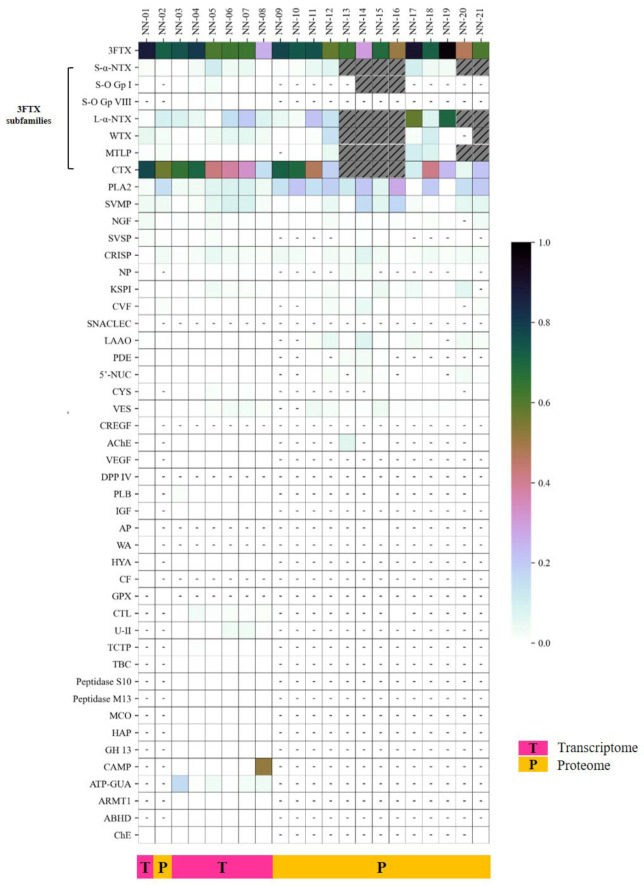
Intra-species variation in snake venom composition is associated with differential venom toxicity and variable antivenom efficacy [23,90,91,92]. Across many studies reported for N. naja venom composition, the venom samples were typically sourced from various locales and assayed with different approaches, and therefore it is challenging for meaningful interpretation of the differential expression of the diverse toxins therein. To address this, we compiled data of N. naja venom proteomes and transcriptomes derived from the current work and previous studies, and the variations were illustrated as shown in Figure 6 for comparison purposes across different specimens, where the quantitative abundances were normalized on a scale of 0–1.0.
Figure 6.
Heatmap of the comparative expression profiles of toxin families derived from the venom-gland transcriptome and venom proteome of N. naja from different geographical locales. The quantitative abundances were normalized on a scale of 0–1.0 for comparison purposes across different specimens, where 1.0 depicts 100% of relative abundance, as reported in respective studies. Unreported/undetected genes or proteins were denoted in dashed lines and unsorted subtypes (mainly 3FTX) were indicated with grey stripes. Details are available in Supplementary File S3. NN-01 and NN-02 represent the relative abundance ratio of toxin families expressed in the transcriptome and proteome of NN-SL (Colombo, Sri Lanka) in the current study, respectively. NN-03 and NN-04 correspond to the relative abundance ratio of toxin families expressed in the venom gland of N. naja specimens from Kerala, India, while NN-05 to NN-08 refers to the samples from the Kentucky Reptile Zoo [31]. NN-09 and NN-10 correspond to the relative abundance ratio of N. naja venom proteins from Sri Lanka and India, respectively [7]. NN-11 corresponds to the relative abundance ratio of N. naja venom proteins from Pakistan (Sindh) [8]. NN-12 corresponds to the relative abundance ratio of N. naja venom proteins from Southern Punjab, Pakistan [44]. NN-13 corresponds to the relative abundance ratio of N. naja venom proteins from West Bengal, India [26]. NN-14 corresponds to the relative abundance ratio of N. naja venom proteins from Tamil Nadu, India [40]. NN-15 and NN-16 correspond to the relative abundance ratio of N. naja venom proteins from Western Maharashtra [93]. NN-17, NN-18 and NN-19 correspond to the relative abundance ratio of N. naja venom proteins from Indian Punjab, Rajasthan and West Bengal [6]. NN-20 corresponds to the relative abundance ratio of N. naja venom proteins from Maharashtra, India [94]. NN-21 corresponds to the relative abundance ratio of N. naja venom proteins from West Bengal, India [30]. Abbreviations: 3FTX, three-finger toxins; S-α-NTX, short chain α-neurotoxin; S-O Gp I, short chain orphan group I, S-O Gp VIII, short chain orphan group VIII; L-α-LNTX, long chain α-neurotoxin; WTX, weak neurotoxin; MTLP, muscarinic toxin-like protein; CTX, cytotoxin/cardiotoxin; PLA2, phospholipase A2; SVMP, snake venom metalloproteinases; NGF, nerve growth factor; SVSP, snake venom serine protease; CRISP, cysteine-rich secretory protein; NP, natriuretic peptide; KSPI, Kunitz-type serine protease inhibitor; CVF, cobra venom factor; SNACLEC, snake C-type lectins; LAAO, L-amino acid oxidase; PDE, phosphodiesterase; 5’-NUC, 5’-nucleotidase; CYS, cystatin; VES, vespryn; CREGF, cysteine-rich with EGF domain-like; AChE, acetylcholinesterase; VEGF, vascular endothelial growth factor; DPP IV, dipeptidylpeptidase IV; PLB, phospholipase B; IGF, insulin-like growth factor; AP, aminopeptidase; WA, waprin; HYA, hyaluronidase; CF, coagulation factor; GPX, glutathione peroxidase; CTL, C-type lectin; U-II, urotensin-II; TCTP, translationally-controlled tumor protein; TBC, Type-B carboxylesterase; MCO, multicopper oxidase; HAP, histidine acid phosphatase; GH 13, glycosyl hydrolase 13; CAMP, cathelicidin antimicrobial peptide; ATP-GUA, ATP: guanido phosphotransferase; ARMT1, ARM-transferase; ABHD, AB hydrolase; ChE, cholinesterase.
We found that a total of 40 protein families with medical and toxinological relevance have been variably reported from the venom-gland transcriptomics and venom proteomics of N. naja specimens sourced from Sri Lanka (NN-01 and NN-02, Colombo region for current work; NN-09, unspecified locality [7]), India (NN-03 and NN-04, Kerala [31]; NN-10, Rajasthan and Gujarat [7]; NN-13, West Bengal [26]; NN-14, Tamil Nadu [40]; NN-15 and NN-16, Maharashtra [93]; NN-17, Punjab; NN-18, Rajasthan; NN-19, West Bengal [6]; NN-20, Maharashtra [94]; NN-21, West Bengal [30]), Pakistan (NN-11, related to Sindh [8]; NN-12, Punjab [44]) and the USA (NN-05 to NN-08, in Kentucky Reptile Zoo, supposedly originated from India or a captive-bred of Indian lineage). Four toxin families, 3FTX, PLA2, SVMP and CRISP, were detected and reported across all 21 specimens of N. naja, while other toxin families were much lowly expressed and not consistently present in all specimens. Of all toxins profiled, the 3FTX were the most highly expressed in all specimens irrespective of geographical origins, except in specimen NN-08 sourced from the Kentucky Reptile Zoo with cathelicidin antimicrobial peptide (CAMP) being the most abundantly expressed [31] (Figure 6). At the subproteome level, the 3FTX subtypes were variably detected. CTX were generally the major 3FTX expressed across all specimens while the content of lethal alpha-neurotoxins varied geographically. The alpha-neurotoxins were relatively higher in expression (abundance ratio of ~0.3–0.7 based on a scale of 0–1.0 per Figure 6) in the majority of N. naja specimens from the northern and eastern parts of the Indian subcontinent including Pakistan (NN-11 and NN-12) [8,44], Indian Punjab (NN-17) and West Bengal (NN-19) [6] compared to specimens from the southern part of South Asia, i.e., Kerala in South West India (NN-03 and NN-04, ~0.08) [31] and Sri Lanka (NN-01 and NN-02, present study; NN-09, [7]) (abundance ratio of < 0.2). The CTX expressions in the southern populations were, however, markedly higher (abundance ratio >0.6) than those in the northern and northeastern parts. With exceptions, specimens from Rajasthan and Gujarat in West India (NN-10 and NN-18) showed a relative higher CTX expression (abundance ratio of ~0.7) in contrast to their lowly expressed alpha-neurotoxins (abundance ratio of ~0.1) [6,7].
The finding, in general, implies a venom phenotypic dichotomy characterized by differential expression of alpha-neurotoxins/cytotoxins in N. naja venom, and a somewhat imperfect distributional pattern of these venom phenotypes along the north–south axis of the Indian subcontinent including Sri Lanka (Figure 7). In brief, the Sri Lankan N. naja, representing the southernmost dispersed population exhibited a venom phenotype characterized by a lower alpha-neurotoxin abundance in exchange for a relatively higher CTX abundance, as opposed to that observed in the northern lineages from Pakistan, Indian Punjab and West Bengal [6,8,44]. Similar patterns of toxin compositional dichotomy within closely related species have been found among evolutionarily distant snake lineages from Elapidae (kraits, sea snakes, coral snakes) [95,96,97,98,99,100], and Viperidae (rattlesnakes, Russell’s vipers, etc.) [18,91,101,102], suggesting that intra-species venom phenotypic dichotomy might be more common than previously thought, and underscoring its impact on the effectiveness of antivenom treatment. The venom divergence within N. naja that follows a biogeographical pattern of north–south distribution implies local adaptation of the species to novel ecological niches. This results in diverging phenotypes that are shaped by the frequency of use and advantage of one toxin class/protein subtype over another as the species distributes southward. The phylogeographical factor alone, however, may not have represented the major driving force of such intra-species venom variation, and other evolutionary paths responsible for generating such venom variability should be studied with more extensive sampling across the biogeographical range of N. naja. Further research should also aim to profile the venom proteome of N. naja populations from biologically significant areas such as Tamil Nadu (from where snake venoms are sourced for the production of most Indian antivenoms), Kerala (for the unique shola ecosystem in the Western Ghats with high biological diversity and endemism), Maharashtra and Rajasthan (for correlation of alpha-neurotoxin/cytotoxin abundances and venom LD50). Earlier, the venom proteome of cobra from Maharashtra was shown to contain as high as 41.8% of neurotoxic 3FTX (alpha-neurotoxins clustered with various atypical and unconventional 3FTX), and very little cytotoxic 3FTX (CTX) at only 5.4% of total venom protein [94]. The venom seems to exhibit atypical toxicity that is highly varied from other cobra species, and is therefore worthy of investigation. Its very high content of neurotoxic 3FTX (reported at 41.8% of protein abundance) apparently did not contribute proportionally to the venom-induced neurotoxicity, judging from its reported i.v. LD50 of 0.73 µg/g which indicates modest lethality for a cobra venom [39]. The abundance of cytotoxic 3FTX reported was exceptionally low (compared to well-established CTX composition in cobra venoms), notwithstanding the fact that cobra bite envenomation in Maharashtra produces local envenoming effect of tissue toxicity [103,104]. On the other hand, N. naja venom from Rajasthan was shown to have extremely low lethality (i.v. LD50 = 2.53 µg/g) [6]. Based on the study, the Rajasthani N. naja venom is perhaps the least lethal amongst all cobra species known, despite having a moderate amount of alpha-neurotoxins at 11.4% (note: reported at 30.1% as “neurotoxic 3FTX”) [6]. This protein abundance of alpha-neurotoxins is at par with N. atra from Zhejiang (China, 11.2%) and N. kaouthia from Vietnam (9.2%) and Guangxi (China, 13.4%), whose LD50 values were established in the range between 0.68 and 0.90 µg/g, in mice [23,43,105]. Clinically, cobra bite in Rajasthan significantly contributed to 53.6% of 138 envenomation cases and was associated with a high mortality of 61.5% in a 2-year hospital-based epidemiological study, where patients were found to manifest neurological deficits with diminished oxygen saturation suggestive of prominent paralysis [106]. The intriguing though somewhat conflicting findings between previously reported venom proteomes, venom toxicity study and clinical manifestation of envenoming warrant a revisitation to resolve the discrepancies.
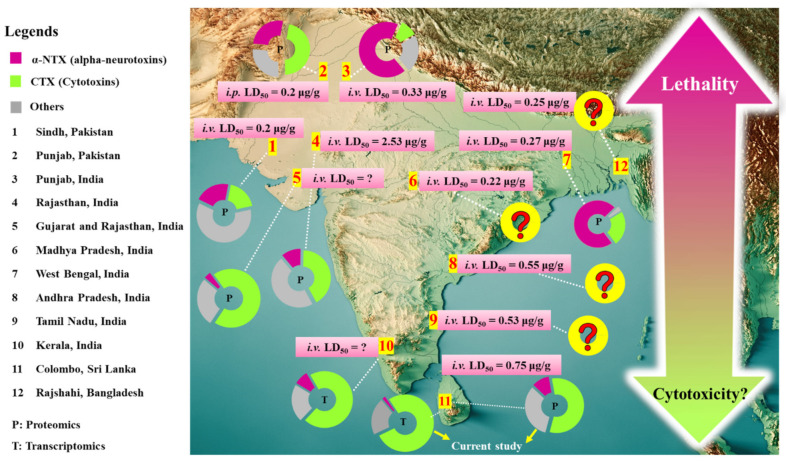
Figure 7.
Comparison of the relative expression levels of alpha-neurotoxins and cytotoxins reported in the venom proteomes and/or venom-gland transcriptomes of Naja naja for snake populations from different localities in South Asia, including Pakistan, India, Sri Lanka (current work) and Bangladesh. Values of intravenous median lethal doses of the venoms were included where applicable, based on available references [6,8,44,107] except for locality #8 (Tamil Nadu, India, unpublished data from authors). Details of toxin abundances sorted were provided in Supplementary File S3.
The comparative venomics to date suggests a high degree of 3FTX evolvability in N. naja, and the consequent variability in toxin expression that bears toxicological correlations with effects attributed to the venom proteins and their neutralization. In mice, the lethal potencies of various cobra venoms have been shown to correlate with the content of alpha-neurotoxins in the venom [39]; here, we observed a similar trend even within the same species of N. naja from different locales. For N. naja of the northern distribution (NN-11: Sindh, Pakistan; NN-12: Punjab, Pakistan; NN-17, Punjab, India; NN-19; West Bengal, India), whose alpha-neurotoxins are more abundant, the venoms elicited rapid neurotoxic effect and high lethality with low median lethal doses (i.v. or i.p. LD50) ranging from 0.2 µg/g to 0.3 µg/g [6,8,44] (Figure 7). Similarly, the venom of N. naja from Rajshahi, Bangladesh has a low i.p. LD50 of 0.25 µg/g [107], although its venom proteomics has not been studied to reveal the quantitative abundance of alpha-neurotoxins in the venom. On the other hand, the venom of NN-SL tested in the present study (representing the southernmost dispersal of N. naja) has an i.v. LD50 of 0.75 µg/g in mice, which is comparable to values previously reported for Sri Lankan cobras (0.67 mg/kg [12]; 1.13 mg/kg [22]) and is consistent with the relatively lower abundance of alpha-neurotoxins in its venom. Meanwhile, the venoms of N. naja from the south (Tamil Nadu) and south-east (Andhra Pradesh) of the Indian subcontinent have i.v. LD50 values of 0.53 µg/g (unpublished) and 0.55 µg/g [6], respectively, suggesting that the venoms likely contain a moderate amount of alpha-neurotoxins that is intermediate between the northern and the Sri Lankan populations. The finding indicates that the lethality of N. naja cobra venom varies with the expression and the abundance of α-NTX in the venom proteome, with an observable shift in potency decreasing as the species distributes southward (Figure 7). A trend of increasing cytotoxins appears to accompany the shift, though clinically local tissue necrosis caused bty cobra bite is common throughout the region.
Consequently, the variable expression of the principal toxins (α-NTX and CTX) is anticipated to influence the effectiveness of antivenom treatment for envenomation caused by N. naja in different geographical areas. This study further investigated the neutralization activity of VPAV, a commonly used Indian polyvalent antivenom in Sri Lanka, against the lethality of NN-SL venom at a challenge lethal dose of 2.5 LD50. The antivenom moderately neutralized the NN-SL venom lethality with a median effective dose (ED50) of 35 µL of antivenom, a dose at which half of the challenged mice survived. The neutralization potency of VPAV against NN-SL venom was determined accordingly as 0.74 mg venom per mLantivenom, and compared with the neutralization activities of Indian antivenoms previously reported for N. naja of various locales (Table 4). The data from the current study fell within the range of potency (Potency = 0.3–0.7 mg/mL) reported for commercially available Indian antivenom products, namely VINS polyvalent antivenom, Bharat antivenom and Premium Serums antivenom, in neutralizing the lethality of cobra venoms from various locales in South Asia [6,8,12,22,54,94]. Based on the neutralization potency, a vial of antivenom reconstituted in 10 mLwater should be able to neutralize approximately 7 mg of the NN-SL cobra venom. Considering an adult cobra snake can deliver more than 50 mg of venom in a bite, it is estimated that approximately 10 vials of antivenom and above would be required for effective treatment. Clinically, neurotoxic envenomation cases caused by cobra bite in Sri Lanka were treated with considerably high doses of Indian antivenoms ranging from 8–30 vials [15,20]. Meanwhile, antivenoms generally have limited effectiveness against the local envenoming effect of cobra bite that causes tissue necrosis [13]. A recent study demonstrated that the Indian antivenoms (VINS and Bharat) were less effective in reversing the local myotoxicity of Sri Lankan N. naja venom tested in an in vitro chick biventer nerve-muscle preparation [17]. In fact, the neutralization effect of antivenom against venom-induced tissue destruction in envenomation caused by most Asiatic cobra bites is known to be weak, as shown in humans clinically and assessed in laboratory animals [56,108,109,110]. Overall, the finding underscores the need for antivenom products with higher efficacy to neutralize the venom-induced toxicity caused by cobra bite envenomation in Sri Lanka. The existing antivenom manufacturing process based in India may be improved by reformulation of immunogen based on the venomics of N. naja from various locales. The principal toxins specific to different locales including Sri Lanka can be incorporated into the immunogen mixture to optimize the immunization of horses during antiserum production [111,112,113].
Table 4.
Lethality and antivenom neutralization of N. naja venoms from different geographical locales.
| Snake Locality | i.v. LD50 (mg/kg) | Antivenom Used | Challenge Dose | ED50 (µL) a | ER50 (mg/mL) b | Neutralization c Potency (mg/mL) | Reference |
|---|---|---|---|---|---|---|---|
| Sri Lanka | 0.75 (0.48–1.18) |
VINS polyvalent antivenom | 2.5 LD50 | 35.00 | 1.23 (1.11–1.37) |
0.74 | Current study |
| Sri Lanka | 1.13 (0.80–1.49) |
VINS polyvalent antivenom | 3.0 LD50 | 96.86 | 0.70 (0.60–0.90) |
0.47 | [22] |
| Instituto Clodomiro Picado polyvalent antivenom | 3.0 LD50 | 195 | 0.40 (0.30–0.50) |
0.23 | |||
| Sri Lanka | 0.67 (0.48–0.99) |
VINS polyvalent antivenom | 5.0 LD50 | # | - | [12] | |
| Bharat polyvalent antivenom | 5.0 LD50 | # | - | ||||
| Maharashtra, West India | 0.73 (0.50–0.88) |
Premium Serums antivenom | 5.0 LD50 | 81.27 | 0.90 (0.74–1.08) |
0.72 | [94] |
| Punjab, North India | 0.33 (0.28–0.38) |
Premium Serums antivenom | 5.0 LD50 | 67.37 | 0.49 | 0.39 | [6] |
| Rajasthan, West India | 2.53 | Premium Serums antivenom | 5.0 LD50 | NE | NE | NE | |
| West Bengal, East India | 0.27 (0.22–0.32) |
Premium Serums antivenom | 5.0 LD50 | 60.45 | 0.45 | 0.36 | |
| Andhra Pradesh, Southeast India | 0.55 (0.45–0.65) |
Premium Serums antivenom | 5.0 LD50 | 54.24 | 1.01 | 0.81 | |
| Madhya Pradesh, Central India | 0.22 (0.18–0.25) |
Premium Serums antivenom | 5.0 LD50 | 60.45 | 0.36 | 0.29 | |
| Pakistan | 0.22 (0.12–0.40) |
VINS polyvalent antivenom | 5.0 LD50 | 32.77 | 0.77 (0.69–0.85) |
0.61 | [8,54] |
Abbreviation: i.v., intravenous; LD50, median lethal dose; ED50, median effective dose; ER50, median effective ratio. a the dose of antivenom (μL) to neutralize the challenge dose of venom (ΧLD50) at which 50% of mice survived; b ratio of the amount of venom (mg) to the volume of the antivenom (ml) at which 50% of mice survived; c the amount of venom (mg) completely neutralized by per unit volume of antivenom (ml). # Median effective dose (ED50) defined differently as the amount of antivenom (μg) per g mice weight (μg/g) to neutralize the challenge dose of venom (ΧLD50), where VINS = 0.77 μg/g (95% C.I.: 0.48–1.05) and Bharat = 1.38 μg/g (0.99–1.63); 95% confidence intervals are indicated in parentheses, unreported data are denoted in dashed line, antivenom not effective toward the challenge dose of venom at a maximum volume of 166 μL is indicated as NE.
3. Conclusions
Incorporating de novo venom-gland transcriptomics and venom proteomics, the present study elucidated the complexity of the Sri Lankan N. naja (NN-SL) venom composition. A total of 22 toxin gene families unique to NN-SL were uncovered, and 12 of these gene families were translated into various protein forms detected in the venom proteome. The 3FTX were the most abundantly expressed, while variation in the 3FTX subtypes, in particular alpha-neurotoxins (α-NTX) and cytotoxins (CTX), correlated with the differential venom toxicity of N. naja from various locales. The NN-SL which represented the southernmost dispersal of N. naja, has a relatively lower content of venom α-NTX and consequently a higher LD50 of its venom, in contrast to the far northern and northeastern populations whose venoms contain more α-NTX and show higher lethality. Despite the venom variability, the Indian polyvalent antivenom moderately neutralized the lethality induced by the Sri Lankan cobra venom. Its neutralization potency was limited, and thus underscoring the need for an improved antivenom product with higher efficacy and broader geographical utility in the region.
4. Materials and Methods
4.1. Snake Venom, Venom Gland Preparation and Antivenom
The cobra, N. naja was an adult snake originated from Colombo, Sri Lanka. The venom was collected by inducing the snake to bite through a container covered with a sterile plastic film, and the venom was lyophilized and stored at −20 °C until use. The snake was rested for four days for the transcription process to continue in its venom glands for four days. This was followed by euthanasia of the snake and quick removal of the venom gland, which was sectioned and preserved in the RNAlater® solution (Ambion, TX, USA). The procedures of venom milking and tissue collections were carried out according to the protocol and guideline approved by the Institutional Animal Use and Care Committee (IACUC) of the University of Malaya, Malaysia (approval number: #2013-11-12/PHAR/R/TCH). Indian VINS polyvalent antivenom (VPAV, batch no: 01AS12041) used in the current study was produced from horse antisera hyperimmunized against the venom of Indian “Big Four”, i.e., Daboia russelii, Naja naja, Bungarus caeruleus and Echis carinatus. The antivenom was reconstituted in 10 mLultrapure water as per manufacturer’s instruction prior to use.
4.2. Chemicals and Materials
All chemicals and materials used were of analytical grade. Ammonium bicarbonate, dithiothreitol (DTT) and iodoacetamide (IAA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). MS grade trypsin protease, Spectra™ Multicolor Broad Range Protein Ladder (Catalog number: 26634, 10 to 260 kDa), and HPLC grade solvents were purchased from Thermo Scientific™ (Pierce™, Rockford, IL, USA). Millipore ZipTip® C18 Pipette Tips were obtained from Merck KGaA (Darmstadt, Germany). Other chemicals and reagents of analytical grade were purchased from Sigma-Aldrich (St. Louis, MO, USA).
4.3. RNA Extraction and Purification
The dissected venom gland tissue was homogenized under sterile conditions in a 1 mLglass homogenizer submerged with TRIzol solution (Invitrogen, Carlsbad, CA, USA). This was followed by the addition of chloroform and treated with RNA-free DNAase I (Thermo Fisher Scientific, Waltham, MA, USA) to separate RNA from cellular debris and residual DNA. The RNA was isolated with isopropyl alcohol and ethanol precipitation. Polyadenylated mRNA was subsequently purified with oligo (dT) magnetic beads according to the manufacturer’s protocol (Illumina, San Diego, CA, USA). The quality of the purified mRNA was subsequently checked using the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany).
Enriched poly(A)+ mRNA isolated from the total venom-gland RNA was used for cDNA construction. The isolated mRNA was fragmented into short fragments, which acted as templates for cDNA synthesis [114]. Random hexamer-primer (N6) was used to synthesis the first-stranded cDNA as input materials, using second strand buffers, dNTPs, RNase H and DNA polymerase I. From these cDNA, a paired-end library was synthesized using the Genomic Sample Prep kit (Illumina, San Diego, CA, USA), according to the manufacturer’s instructions. The cDNA fragments generated were purified with QIAquick PCR extraction kit (Qiagen, Valencia, CA, USA) and dissolved in elution buffer for end repair and the addition pf poly(A) to aid in the subsequent ligation of Illumina adaptors that contain a single thymine (T) base overhang at the 3’ ends. Following the ligation, these cDNA fragments were amplified via polymerase chain reaction (PCR) electrophoresed on a 1.5–2% TAE (Tris-base, acetic acid and EDTA) agarose gel. From the gel, suitable fragments (200–700 bp) were selected as templates for subsequent PCR amplification. Sequencing of the amplified samples library was achieved in a single lane on the Illumina HiSeq™ 2000 platform (Illumina, San Diego, CA, USA) with 100-base-pair, paired-end reads.
4.4. Raw Sequenced Reads and De Novo Transcriptome Assembly
Sequenced data generated from Illumina HiSeq™ 2000 were transformed by base calling into sequence data, called the raw reads and stored in a FASTQ format. Prior to transcriptome assembly, raw reads were filtered to generate clean reads as part of the quality control process in the pre-analysis stage [115]. This involved the removal of (i) adaptor; (ii) reads with >5% of unknown nucleotides or (ii) low-quality reads with > 20% of low-quality bases (determined as base quality <10).
De novo transcriptome assembly was performed using short reads assembly program, Trinity (version 2.0.6) [116,117]. Three independent software modules, i.e., Inchworm, Chrysalis and Butterfly, comprised the Trinity program, were sequentially applied to process the large volumes of RNA-seq reads based on the algorithm of de Bruijn graph construction. The process began by aligning k-mers (k = 25), and sequence reads with a certain length of overlap were joined to form linear contigs. The sequence reads were mapped back onto contigs, and by referring to paired-end reads, contigs from the same transcript, as well as the distances between them were determined. The contigs were then partitioned into clusters, each of which carried a complete set of de Bruijn graphs as a representation of the transcriptional complexity at a given gene or locus. The graphs were independently processed to obtain full-length transcripts for alternatively spliced isoforms and to tease apart transcripts that corresponded to paralogous genes. The clean read Q20 percentage, a point of reference for quality control assessment was obtained as a benchmark for successful de novo assembly of the transcriptome.
4.5. Transcripts Clustering and Functional Annotation
The transcript sequences generated from Trinity (known as Unigenes) were further processed for sequence splicing and the removal of redundancy using TGI clustering tools (TGICL) (version 2.1) to acquire the longest possible length of non-redundant (NR) transcripts. The transcripts were then subjected to family clustering into two classes of transcripts: (a) clusters, with a prefix CL and the cluster ID behind as contig; (b) singletons, ID with a prefix of Unigene. In each cluster, there were several transcripts with sequence similarities among them being >70% while the singletons “Unigenes” lack of overlapping with other fragments at the given stringency. In brief, the value 70% was used to categorize the assembled sequences based on similarity; sequences similar to each other (may or may not be homologous as having >70% similarity) were grouped under a cluster comprising various contigs.
Subsequently, transcript Unigenes were aligned using BLASTx to NCBI non-redundant (NR) protein database, with a cut-off value of E < 10−5. The proteins with the highest rank in the BLASTx results were referred to determine the coding region sequences of the Unigenes, followed by translation into amino acid sequences using standard codon table. Hence, both nucleotide sequences (5′–3′) and amino acid sequences of the Unigene-coding regions were acquired. To remove redundancy from each cluster, the longest sequence in each cluster was chosen as the transcript, and the length of the scaffold was extended based on overlapping sequences using Phrap assembler (release 23.0) (http://www.phrap.org) (accessed on 14 September 2020). The distribution lengths of contigs, scaffolds and Unigenes were calculated, and the N50 length (assembly quality indicator) was set at >500 for success assembly.
4.6. Quantification of Transcript Abundance
Clean reads were aligned to Unigene using Bowtie2 [118]. The transcript abundances were calculated using RNA-seq with expectation maximization (RSEM) software [119]. Fragments per kilobase of exon model per million reads mapped (FPKM) were used to determine the transcript abundance for the identified genes [120]. FPKM is the summation of normalized read counts based on gene length and the total number of mapped reads. The data was obtained using RSEM tool in conjunction with Trinity based on a computational formula:
| (1) |
FPKM is the expression of gene A; B is the number of fragments/reads which are aligned to gene A, N is the total number of fragments/reads that are aligned to all genes; C is the base number in the coding sequence of gene A.
4.7. Categorization of Transcripts
The de novo assembled transcripts were subjected to BLASTx search to obtain the closest resembling sequences from the NR protein database for further classification based on functional annotations. The transcripts were then sifted to remove those with an FPKM value of less than 1, followed by categorization into three groups: “toxins”, “non-toxins” and “unidentified” (Supplementary File S1) [33,35]. “Toxin” transcripts were recruited by toxin-related keyword searches against the annotated transcripts. “Non-toxin” and “unidentified” groups contain transcripts of cellular proteins or house-keeping genes and transcripts that could not be identified, respectively. The redundancy of gene expression was determined by dividing the total FPKM of each group by the total number of transcripts in the respective group of transcripts [27]. In the “toxin” group, the amino acid sequences were used to further validate the toxin identity through BLASTp suite (Basic Local Alignment Search Tool-Protein) in the Uniprot (Universal Protein Resource Knowledgebase) database platform. The transcripts were searched against Serpentes database (taxid: 8570) and validated based on the lowest E-score value with the highest percentage of sequence similarity (updated as of September 2019). The annotation was manually filtered to proteins specific for elapids only.
4.8. Multiple Sequence Alignment
Multiple sequence alignment was conducted using Jalview software (version 2.11.1.4) [121,122] and aligned using MUSCLE (multiple sequence comparison by log-expectation) program [123]. Sequences of related species used in multiple sequence alignment were retrieved from UniprotKB depository (http://www.uniprot.org/) (accessed on 14 September 2020). The selection was based on their relevance to the toxins in comparison to elucidate the similarity and variation as well as conserved regions of the sequences.
4.9. Correlation Analysis
The correlation of the toxin gene expression of each family (from venom-gland transcriptome) and the protein abundance of the corresponding toxin family (from venom proteome) was analyzed by Spearman’s correlation coefficient and constructed using GraphPad Prism 6 (San Diego, CA, USA). The significance of the correlation was estimated by the coefficient of determination (r) at a cutoff value of p < 0.05.
4.10. Heatmap of Differentially Expressed Toxins Genes and Proteins
A heatmap of relative abundance pattern in the differentially expressed toxin genes and venom proteins of N. naja from various geographical locales were generated with the custom scripts of Python version 3.8.5. The dataset was derived from previous proteomic and transcriptomic studies on N. naja from Sri Lanka, India and Pakistan [6,7,8,26,30,31,40,44,93,94] and the current work on NN-SL from Sri Lanka (dataset available in Supplementary File S3). The relative abundances were depicted by color scheme ranging from low (white) to high (black) intensity, based on a scale of 0 to 1.0 where 1.0 represented 100% of relative abundances of transcript or protein, as reported in the respective studies. Unreported or undetected genes and proteins were denoted with dashed lines, and unsorted subtypes (3FTX mainly) were indicated with grey stripes.
4.11. Reverse-Phase High-Performance Liquid Chromatography (RP-HPLC)
Naja naja venom (3 mg) was reconstituted in 200 µL ultrapure water and subjected to C-18 RP-HPLC using Shimadzu LC-20 AD HPLC system (Shimadzu, Kyoto, Japan). The LiChrospher® WP 300 C-18 column (5 µm pore size) was pre-equilibrated with Eluent A (0.1% TFA in water) and eluted with Eluent B (0.1% TFA in ACN) using a linear gradient of 5% B for 10 min, 5–15% B over 20 min, 15–45% B over 120 min and 45–70% B over 20 min. The flow rate was set to 1 ml/min. The fractionation was monitored at 215 nm absorbance, and the resulting peaks were collected, lyophilized and stored at −20 °C prior to use.
4.12. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
The fractions collected from RP-HPLC were subjected to SDS-PAGE according to Laemmli [124]. Approximately 20 µg of venom protein fractions were loaded into the wells, and fractionation was performed with 15% SDS-PAGE under reducing condition at 90 V for 2 h. Thermo Scientific Spectra™ Multicolor Broad Range Protein Ladder (10–260 kDa) was used as calibration standards. The gels were stained with Coomassie Brilliant Blue R-250, and protein bands were visualized and scanned using ImageScanner III (GE Healthcare, Danderyd, Sweden).
4.13. In-Gel Tryptic Digestion and Tandem Mass Spectrometry
Protein bands of interest were excised into small gel pieces (~2 mm), reduced with dithiothreitol, alkylated with iodoacetamide and digested with MS grade Pierce trypsin protease according to the manufacturer’s protocol (Thermo Scientific™ Pierce™, Rockford, IL, USA). Digested peptides were extracted and desalted using Millipore ZipTip® C18 Pipette Tips.
The tryptic digested peptides were then subjected to MALDI TOF/TOF using AB SCIEX TOF/TOF™ 5800 System Proteomic Analyzer (AB SCIEX, Framingham, MA, USA) equipped with neodymium: yttrium-aluminum-garnet laser (laser wavelength was 355 nm). Samples (0.5 µL) were mixed with alpha-cyano-4-hydroxycinnamic acid matrix (0.5 µL) and spotted on OPTI-TOF™ LC/MALDI insert plate (123 mm × 81 mm). For MS mode, peptide mass maps were acquired in positive reflection mode at 1000 laser shots per spectrum at 1 kV with a mass range of 800–4000 m/z. The MS/MS peak detection criteria were a minimum signal-to-noise ratio of 100. The raw mass spectra acquired were exported to AB SCIEX ProteinPilot™ Software and search against a customized database consisting of the non-redundant protein sequence database from NCBI (taxonomy: Serpentes; taxid: 8570) and our in-house transcripts database. MS peak filter mass range 800–4000 m/z was applied. Precursor and fragment mass tolerances were set to 100 ppm and 0.2 Da and allowing one missed cleavage. Oxidation (M) was set as a variable modification, and carbamidomethylation (C) was set as a fixed modification. The protein score intervals (C.I.) above 95% were considered as confident identification.
The unidentified peptide samples from MALDI TOF/TOF were subjected to nano-electrospray ionization MS/MS using Agilent 1200 HPLC-Chip/MS Interface, coupled with Agilent 6550 Accurate-Mass Q-TOF LC/MS system. Samples were loaded in a large capacity chip 300 Å, C18, 160 nl enrichment column and 75 µm × 150 mm analytical column (Agilent part no. G4240-62010) with a flow rate of 4 µL/ min from a capillary pump and 0.4 µL/min from a nano pump of Agilent 1200 series. The injection volume was 2 µL per sample, and the mobile phase was 0.1% formic acid in water (Solution A) and 0.1% formic acid in 100% acetonitrile (Solution B). The gradient applied was: 5–50% B for 11 min, 50–70% B for 4 min and 70% B for 3 min using Agilent 1200 series nano-flow LC pump. Ion polarity was set to positive ionization mode. Drying gas flow was 11 L/min and drying gas temperature was 290 °C. Fragmentor voltage was 175 V and the capillary voltage was set to 1800 V. Spectra was acquired in a MS/MS mode with an MS scan range of 200–3000 m/z and MS/MS scan range of 50–3200 m/z. Precursor charge selection was set as doubly charge state and above with the exclusion of precursors 1221.9906 m/z (z = 1) and 299.2944 (z = 1) set as reference ions. Data were extracted with MH+ mass range between 50 and 3200 Da and processed with Agilent Spectrum Mill MS Proteomics Workbench software packages. Carbamidomethylation of cysteine was set as a single modification. The peptide finger mapping was modified to specifically search against a customized database consisting of the non-redundant protein sequence database from NCBI (taxonomy: Serpentes; taxid: 8570) and our in-house transcripts database. Protein identification were validated with the following filters: protein score >11, peptide score >10 and score peak intensity (SPI) >70%. The result for “Distinct peptide” identification of greater than one is considered significant.
4.14. Estimation of Protein Relative Abundance
The protein relative abundance was estimated according to the method used earlier in proteomic studies [8,23]. In brief, the relative abundance of venom fractions was estimated by peak area under curve using Shimadzu LC Solution Software (Shimadzu, Kyoto, Japan). The fractions that show a homogenous protein band in SDS-PAGE was directly implemented with the relative abundance obtained from peak area under curve (AUC) of chromatogram. For the fractions with various protein bands in SDS-PAGE, the relative abundance of individual bands in a fraction were estimated by gel densitometry using Thermo Scientific™ Pierce™ MyImageAnalysis™ Software (Thermo Fisher Scientific, Waltham, MA, USA). The relative abundance (in percentage) of these individual protein bands was the product of multiplying the peak area under the curve (AUC) with gel densitometry of protein bands. The relative abundances were then accumulated according to the protein family.
4.15. Determination of Venom Lethality and Neutralization by Antivenom
The study was carried out by intravenous administration of various doses of venom (four doses of venom tested) via the caudal vein into the ICR albino mice (20–25 g, n = 4 per dose). The mice were allowed free access to food and water ad libitum. The survival ratio of mice was recorded at 24 h post-injection for the determination of intravenous median lethal dose (i.v., LD50) at which 50% of the tested animal were dead.
For antivenom neutralization, the study was conducted as previously described in [125]. In brief, a challenge dose of venom at 2.5 times LD50 dissolving in saline water was pre-incubated with various dilutions of antivenom (4 dilutions of antivenom tested, 15 μL, 35 μL, 75 μL and 100 μL) at 37 °C for 30 min. The venom–antivenom mixture was then injected intravenously into the mice (20–25 g, n = 4 per dose). The mice were allowed free access to food and water ad libitum. The survival ratio of the mice was recorded at 24-h post-injection for the determination of median effective dose (ED50) of the antivenom against the venom. ED50 was defined as the antivenom dose (µL) that prevented death in 50% of the tested animals.
Both LD50 and ED50 values and 95% C.I. were calculated using the Probit analysis method using Biostat 2009 software [126]. The neutralization efficacy was also expressed as Potency, (P), defined as the amount of venom (mg) completely neutralized per unit volume of antivenom (ml) [127,128].
4.16. Supporting Data
Sequencing data from the de novo venom-gland transcriptomics of Naja naja was deposited in National Centre for Biotechnology Information (NCBI) Sequence Read Archive (http://submit.ncbi.nlm.nih.gov/subs/sra/) (submitted on 28 June 2021) under SRA accession: PRJNA741865). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository (http://www.iprox.org) (accessed on 14 September 2020) [129] with dataset identifier PXD026735 (Subproject ID: IPX0003192001) (submitted on 16 June 2021).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/toxins13080558/s1, Figure S1: Correlation between the venom gland transcriptome and venom proteome of NN-SL, Supplementary File S1: Venom gland transcriptomic of N. naja from Sri Lanka, Supplementary File S2: Mass spectrometry analysis of in-gel tryptic digested peptides of RP-HPLC fractions of Sri Lankan N. naja venom, Supplementary File S3: Comparative expression profile of toxin families derived from the venom gland transcriptome and venom proteome of N. naja from different geographical locales.
Author Contributions
Conceptualization, C.H.T. and K.Y.T.; methodology, C.H.T., K.Y.W. and K.Y.T.; validation, K.Y.T. and C.H.T.; formal analysis, K.Y.W.; investigation, K.Y.W.; resources, C.H.T., N.H.T. and C.A.G.; data curation, C.H.T. and K.Y.T.; writing—original draft preparation, K.Y.W.; writing—review and editing, K.Y.W., C.H.T., K.Y.T., N.H.T. and C.A.G.; visualization, K.Y.T. and C.H.T.; supervision, C.H.T., K.Y.T. and N.H.T.; funding acquisition, C.H.T. and K.Y.T. All authors have read and agreed to the published version of the manuscript.
Funding
Authors acknowledged Fundamental Research Grant (FRGS/1/2019/SKK08/UM/02/2) from the Ministry of Higher Education, Malaysia, and Special Reserch Grant (BKS003-2020) from the University of Malaya, Malaysia.
Institutional Review Board Statement
The protocol for animal experimentation was carried out based on the Council for International Organizations of Medical Sciences (CIOMS) guidelines and was approved by the Institutional Animal Care and Use Committee of the University of Malaya (Ethics clearance number: 2013-11-12/PHAR/R/TCH, approved 11 December 2013).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
Key Contribution
The study unveils comprehensive profiles of de novo venom gland transcriptome and venom proteome of the Spectacled Cobra, Naja naja from Sri Lanka. The intra-species variations in toxin diversity and venom composition were elucidated, providing insights into the pathophysiology and antivenom treatment of cobra bite envenomation in Sri Lanka. The findings enrich the current knowledgebase of snake toxins and indicate the need for effective antivenom with broader geographical utility.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO Snakebite Envenoming. [(accessed on 12 June 2021)]; Available online: https://www.who.int/news-room/fact-sheets/detail/snakebite-envenoming.
- 2.Kasturiratne A., Wickremasinghe A.R., de Silva N., Gunawardena N.K., Pathmeswaran A., Premaratna R., Savioli L., Lalloo D.G., de Silva H.J. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008;5:e218. doi: 10.1371/journal.pmed.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gutiérrez J.M., Calvete J.J., Habib A.G., Harrison R.A., Williams D.J., Warrell D.A. Snakebite envenoming. Nat. Rev. Dis. Primers. 2017;3:17063. doi: 10.1038/nrdp.2017.63. [DOI] [PubMed] [Google Scholar]
- 4.Ralph R., Sharma S.K., Faiz M.A., Ribeiro I., Rijal S., Chappuis F., Kuch U. The timing is right to end snakebite deaths in South Asia. BMJ Clin. Res. Ed. 2019;364:k5317. doi: 10.1136/bmj.k5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longbottom J., Shearer F.M., Devine M., Alcoba G., Chappuis F., Weiss D.J., Ray S.E., Ray N., Warrell D.A., Ruiz de Castaneda R., et al. Vulnerability to snakebite envenoming: A global mapping of hotspots. Lancet. 2018;392:673–684. doi: 10.1016/S0140-6736(18)31224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senji Laxme R.R., Attarde S., Khochare S., Suranse V., Martin G., Casewell N.R., Whitaker R., Sunagar K. Biogeographical venom variation in the Indian spectacled cobra (Naja naja) underscores the pressing need for pan-India efficacious snakebite therapy. PLoS Negl. Trop. Dis. 2021;15:e0009150. doi: 10.1371/journal.pntd.0009150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sintiprungrat K., Watcharatanyatip K., Senevirathne W.D., Chaisuriya P., Chokchaichamnankit D., Srisomsap C., Ratanabanangkoon K. A comparative study of venomics of Naja naja from India and Sri Lanka, clinical manifestations and antivenomics of an Indian polyspecific antivenom. J. Proteom. 2016;132:131–143. doi: 10.1016/j.jprot.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Wong K.Y., Tan C.H., Tan K.Y., Quraishi N.H., Tan N.H. Elucidating the biogeographical variation of the venom of Naja naja (spectacled cobra) from Pakistan through a venom-decomplexing proteomic study. J. Proteom. 2018;175:156–173. doi: 10.1016/j.jprot.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Faisal T., Tan K.Y., Sim S.M., Quraishi N., Tan N.H., Tan C.H. Proteomics, functional characterization and antivenom neutralization of the venom of Pakistani Russell’s viper (Daboia russelii) from the wild. J. Proteom. 2018;183:1–13. doi: 10.1016/j.jprot.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Bhatia S., Vasudevan K. Comparative proteomics of geographically distinct saw-scaled viper (Echis carinatus) venoms from India. Toxicon X. 2020;7:100048. doi: 10.1016/j.toxcx.2020.100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalita B., Patra A., Das A., Mukherjee A.K. Proteomic analysis and immuno-profiling of eastern India Russell’s viper (Daboia russelii) Venom: Correlation between RVV composition and clinical manifestations post RV bite. J. Proteom. Res. 2018;17:2819–2833. doi: 10.1021/acs.jproteome.8b00291. [DOI] [PubMed] [Google Scholar]
- 12.Maduwage K., Silva A., O’Leary M.A., Hodgson W.C., Isbister G.K. Efficacy of Indian polyvalent snake antivenoms against Sri Lankan snake venoms: Lethality studies or clinically focussed in vitro studies. Sci. Rep. 2016;6:26778. doi: 10.1038/srep26778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kularatne S.A., Budagoda B.D., Gawarammana I.B., Kularatne W.K. Epidemiology, clinical profile and management issues of cobra (Naja naja) bites in Sri Lanka: First authenticated case series. Trans. R. Soc. Trop. Med. Hyg. 2009;103:924–930. doi: 10.1016/j.trstmh.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Ediriweera D.S., Kasturiratne A., Pathmeswaran A., Gunawardena N.K., Wijayawickrama B.A., Jayamanne S.F., Isbister G.K., Dawson A., Giorgi E., Diggle P.J., et al. Mapping the risk of snakebite in Sri Lanka—A national survey with geospatial analysis. PLoS Negl. Trop. Dis. 2016;10:e0004813. doi: 10.1371/journal.pntd.0004813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rathnayaka R., Kularathna S.A.M., Sugathapala A.G.H., Pathiraja P., Ranathunga P. A cobra bite—Uncommon bitten site. Sri Lankan J. Anaesthesiol. 2015;23:89. doi: 10.4038/slja.v23i2.8069. [DOI] [Google Scholar]
- 16.Seneviratne U., Dissanayake S. Neurological manifestations of snake bite in Sri Lanka. J. Postgrad. Med. 2002;48 [PubMed] [Google Scholar]
- 17.Madhushani U., Thakshila P., Hodgson W.C., Isbister G.K., Silva A. Effect of Indian Polyvalent Antivenom in the Prevention and Reversal of Local Myotoxicity Induced by Common Cobra (Naja naja) Venom from Sri Lanka In Vitro. Toxins. 2021;13:308. doi: 10.3390/toxins13050308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pla D., Sanz L., Quesada-Bernat S., Villalta M., Baal J., Chowdhury M.A.W., León G., Gutiérrez J.M., Kuch U., Calvete J.J. Phylovenomics of Daboia russelii across the Indian subcontinent. Bioactivities and comparative in vivo neutralization and in vitro third-generation antivenomics of antivenoms against venoms from India, Bangladesh and Sri Lanka. J. Proteom. 2019;207:103443. doi: 10.1016/j.jprot.2019.103443. [DOI] [PubMed] [Google Scholar]
- 19.Kularatne S.A.M., Silva A., Weerakoon K., Maduwage K., Walathara C., Paranagama R., Mendis S. Revisiting Russell’s viper (Daboia russelii) bite in Sri Lanka: Is abdominal pain an early feature of systemic envenoming? PLoS ONE. 2014;9:e90198. doi: 10.1371/journal.pone.0090198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theakston R.D., Phillips R.E., Warrell D.A., Galagedera Y., Abeysekera D.T., Dissanayaka P., de Silva A., Aloysius D.J. Envenoming by the common krait (Bungarus caeruleus) and Sri Lankan cobra (Naja naja naja): Efficacy and complications of therapy with Haffkine antivenom. Trans. R. Soc. Trop. Med. Hyg. 1990;84:301–308. doi: 10.1016/0035-9203(90)90297-R. [DOI] [PubMed] [Google Scholar]
- 21.Keyler D.E., Gawarammana I., Gutierrez J.M., Sellahewa K.H., McWhorter K., Malleappah R. Antivenom for snakebite envenoming in Sri Lanka: The need for geographically specific antivenom and improved efficacy. Toxicon. 2013;69:90–97. doi: 10.1016/j.toxicon.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 22.Villalta M., Sánchez A., Herrera M., Vargas M., Segura Á., Cerdas M., Estrada R., Gawarammana I., Keyler D.E., McWhorter K., et al. Development of a new polyspecific antivenom for snakebite envenoming in Sri Lanka: Analysis of its preclinical efficacy as compared to a currently available antivenom. Toxicon. 2016;122:152–159. doi: 10.1016/j.toxicon.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Tan K.Y., Tan C.H., Fung S.Y., Tan N.H. Venomics, lethality and neutralization of Naja kaouthia (monocled cobra) venoms from three different geographical regions of Southeast Asia. J. Proteom. 2015;120:105–125. doi: 10.1016/j.jprot.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Huang H.W., Liu B.S., Chien K.Y., Chiang L.C., Huang S.Y., Sung W.C., Wu W.G. Cobra venom proteome and glycome determined from individual snakes of Naja atra reveal medically important dynamic range and systematic geographic variation. J. Proteom. 2015;128:92–104. doi: 10.1016/j.jprot.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 25.Deka A., Reza M.A., Faisal Hoque K.M., Deka K., Saha S., Doley R. Comparative analysis of Naja kaouthia venom from North-East India and Bangladesh and its cross reactivity with Indian polyvalent antivenoms. Toxicon. 2019;164:31–43. doi: 10.1016/j.toxicon.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 26.Dutta S., Chanda A., Kalita B., Islam T., Patra A., Mukherjee A.K. Proteomic analysis to unravel the complex venom proteome of eastern India Naja naja: Correlation of venom composition with its biochemical and pharmacological properties. J. Proteom. 2017;156:29–39. doi: 10.1016/j.jprot.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 27.Tan K.Y., Tan C.H., Chanhome L., Tan N.H. Comparative venom gland transcriptomics of Naja kaouthia (monocled cobra) from Malaysia and Thailand: Elucidating geographical venom variation and insights into sequence novelty. PeerJ. 2017;5:e3142. doi: 10.7717/peerj.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C.-C., You C.-H., Wang P.-J., Yu J.-S., Huang G.-J., Liu C.-H., Hsieh W.-C., Lin C.-C. Analysis of the efficacy of Taiwanese freeze-dried neurotoxic antivenom against Naja kaouthia, Naja siamensis and Ophiophagus hannah through proteomics and animal model approaches. PLoS Negl. Trop. Dis. 2017;11:e0006138. doi: 10.1371/journal.pntd.0006138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petras D., Sanz L., Segura Á., Herrera M., Villalta M., Solano D., Vargas M., León G., Warrell D.A., Theakston R.D.G., et al. Snake venomics of African spitting cobras: Toxin composition and assessment of congeneric cross-reactivity of the pan-african EchiTAb-Plus-ICP antivenom by antivenomics and neutralization approaches. J. Proteom. Res. 2011;10:1266–1280. doi: 10.1021/pr101040f. [DOI] [PubMed] [Google Scholar]
- 30.Chanda A., Patra A., Kalita B., Mukherjee A.K. Proteomics analysis to compare the venom composition between Naja naja and Naja kaouthia from the same geographical location of eastern India: Correlation with pathophysiology of envenomation and immunological cross-reactivity towards commercial polyantivenom. Expert Rev. Proteom. 2018;15:949–961. doi: 10.1080/14789450.2018.1538799. [DOI] [PubMed] [Google Scholar]
- 31.Suryamohan K., Krishnankutty S.P., Guillory J., Jevit M., Schröder M.S., Wu M., Kuriakose B., Mathew O.K., Perumal R.C., Koludarov I., et al. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat. Genet. 2020;52:106–117. doi: 10.1038/s41588-019-0559-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan C., Tan K.Y., Tan N. A protein decomplexation strategy in snake venom proteomics. Funct. Proteom. 2019;1871:83–92. doi: 10.1007/978-1-4939-8814-3_5. [DOI] [PubMed] [Google Scholar]
- 33.Tan C.H., Tan K.Y. De novo venom-gland transcriptomics of spine-bellied sea snake (Hydrophis curtus) from Penang, Malaysia–Next-generation sequencing, functional annotation and toxinological correlation. Toxins. 2021;13:127. doi: 10.3390/toxins13020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan C.H., Tan K.Y., Fung S.Y., Tan N.H. Venom-gland transcriptome and venom proteome of the Malaysian king cobra (Ophiophagus hannah) BMC Genom. 2015;16:1–21. doi: 10.1186/s12864-015-1828-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chong H.P., Tan K.Y., Tan N.H., Tan C.H. Exploring the diversity and novelty of toxin genes in Naja sumatrana, the equatorial spitting cobra from Malaysia through de novo venom-gland transcriptomics. Toxins. 2019;11:104. doi: 10.3390/toxins11020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y., Li Y., Lee W., Xu X., Zhang Y., Zhao R., Zhang Y., Wang W. Venom gland transcriptomes of two elapid snakes (Bungarus multicinctus and Naja atra) and evolution of toxin genes. BMC Genom. 2011;12:1. doi: 10.1186/1471-2164-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margres M.J., Aronow K., Loyacano J., Rokyta D.R. The venom-gland transcriptome of the eastern coral snake (Micrurus fulvius) reveals high venom complexity in the intragenomic evolution of venoms. BMC Genom. 2013;14:531. doi: 10.1186/1471-2164-14-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan N.H., Wong K.Y., Tan C.H. Venomics of Naja sputatrix, the Javan spitting cobra: A short neurotoxin-driven venom needing improved antivenom neutralization. J. Proteom. 2017;157:18–32. doi: 10.1016/j.jprot.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 39.Tan C.H., Wong K.Y., Chong H.P., Tan N.H., Tan K.Y. Proteomic insights into short neurotoxin-driven, highly neurotoxic venom of Philippine cobra (Naja philippinensis) and toxicity correlation of cobra envenomation in Asia. J. Proteom. 2019;206:103418. doi: 10.1016/j.jprot.2019.103418. [DOI] [PubMed] [Google Scholar]
- 40.Choudhury M., McCleary R.J.R., Kesherwani M., Kini R.M., Velmurugan D. Comparison of proteomic profiles of the venoms of two of the ‘Big Four’ snakes of India, the Indian cobra (Naja naja) and the common krait (Bungarus caeruleus), and analyses of their toxins. Toxicon. 2017;135:33–42. doi: 10.1016/j.toxicon.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Yap M.K., Fung S.Y., Tan K.Y., Tan N.H. Proteomic characterization of venom of the medically important Southeast Asian Naja sumatrana (Equatorial spitting cobra) Acta Trop. 2014;133:15–25. doi: 10.1016/j.actatropica.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 42.Laustsen A.H., Gutierrez J.M., Lohse B., Rasmussen A.R., Fernandez J., Milbo C., Lomonte B. Snake venomics of monocled cobra (Naja kaouthia) and investigation of human IgG response against venom toxins. Toxicon. 2015;99:23–35. doi: 10.1016/j.toxicon.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Shan L.L., Gao J.F., Zhang Y.X., Shen S.S., He Y., Wang J., Ma X.M., Ji X. Proteomic characterization and comparison of venoms from two elapid snakes (Bungarus multicinctus and Naja atra) from China. J. Proteom. 2016;138:83–94. doi: 10.1016/j.jprot.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 44.Asad M.H.H.B., McCleary R.J.R., Salafutdinov I., Alam F., Shah H.S., Bibi S., Ali A., Khalid S., Hasan S.M.F., Sabatier J.-M., et al. Proteomics study of Southern Punjab Pakistani cobra (Naja naja: Formerly Naja naja karachiensis) venom. Toxicol. Environ. Chem. 2019;101:91–116. doi: 10.1080/02772248.2019.1619743. [DOI] [Google Scholar]
- 45.Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Utkin Y.N. Last decade update for three-finger toxins: Newly emerging structures and biological activities. World J. Biol Chem. 2019;10:17–27. doi: 10.4331/wjbc.v10.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kini R.M., Doley R. Structure, function and evolution of three-finger toxins: Mini proteins with multiple targets. Toxicon. 2010;56:855–867. doi: 10.1016/j.toxicon.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Chien K.Y., Chiang C.M., Hseu Y.C., Vyas A.A., Rule G.S., Wu W. Two distinct types of cardiotoxin as revealed by the structure and activity relationship of their interaction with zwitterionic phospholipid dispersions. J. Biol. Chem. 1994;269:14473–14483. doi: 10.1016/S0021-9258(17)36647-4. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki-Matsubara M., Athauda S.B., Suzuki Y., Matsubara K., Moriyama A. Comparison of the primary structures, cytotoxicities, and affinities to phospholipids of five kinds of cytotoxins from the venom of Indian cobra, Naja naja. Comp. Biochem. Physiol. Toxicol. Pharmacol. CBP. 2016;179:158–164. doi: 10.1016/j.cbpc.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 50.Wu P.-L., Chiu C.-R., Huang W.-N., Wu W.-G. The role of sulfatide lipid domains in the membrane pore-forming activity of cobra cardiotoxin. Biochim. Biophys. Acta (BBA) Biomembr. 2012;1818:1378–1385. doi: 10.1016/j.bbamem.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 51.Forouhar F., Huang W.N., Liu J.H., Chien K.Y., Wu W.G., Hsiao C.D. Structural basis of membrane-induced cardiotoxin A3 oligomerization. J. Biol. Chem. 2003;278:21980–21988. doi: 10.1074/jbc.M208650200. [DOI] [PubMed] [Google Scholar]
- 52.Chong H.P., Tan K.Y., Tan C.H. Cytotoxicity of snake venoms and cytotoxins from two southeast Asian cobras (Naja sumatrana, Naja kaouthia): Exploration of anticancer potential, selectivity, and cell death mechanism. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.583587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konshina A.G., Boldyrev I.A., Omelkov A.V., Utkin Y.N., Efremov R.G. Anionic lipids: Determinants of binding cytotoxins from snake venom on the surface of cell membranes. Acta Nat. 2010;2:88–96. doi: 10.32607/20758251-2010-2-2-88-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong K.Y., Tan C.H., Tan N.H. Venom and purified toxins of the spectacled cobra (Naja naja) from Pakistan: Insights into toxicity and antivenom neutralization. Am. J. Trop. Med. Hyg. 2016;94:1392–1399. doi: 10.4269/ajtmh.15-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan K.Y., Tan C.H., Fung S.Y., Tan N.H. Neutralization of the principal toxins from the venoms of Thai Naja kaouthia and Malaysian Hydrophis schistosus: Insights into toxin-specific neutralization by two different antivenoms. Toxins. 2016;8:86. doi: 10.3390/toxins8040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu C.-C., Chou Y.-S., Chen C.-Y., Liu K.-L., Huang G.-J., Yu J.-S., Wu C.-J., Liaw G.-W., Hsieh C.-H., Chen C.-K. Pathogenesis of local necrosis induced by Naja atra venom: Assessment of the neutralization ability of Taiwanese freeze-dried neurotoxic antivenom in animal models. PLoS Negl. Trop. Dis. 2020;14:e0008054. doi: 10.1371/journal.pntd.0008054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang K.C., Huang Y.K., Chen Y.W., Chen M.H., Tu A.T., Chen Y.C. Venom ophthalmia and ocular complications caused by snake venom. Toxins. 2020;12:567. doi: 10.3390/toxins12090576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Konshina A.G., Boldyrev I.A., Utkin Y.N., Omel’kov A.V., Efremov R.G. Snake Cytotoxins bind to membranes via interactions with phosphatidylserine head groups of lipids. PLoS ONE. 2011;6:e19064. doi: 10.1371/journal.pone.0019064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tjong S.C., Wu P.L., Wang C.M., Huang W.N., Ho N.L., Wu W.G. Role of glycosphingolipid conformational change in membrane pore forming activity of cobra cardiotoxin. Biochemistry. 2007;46:12111–12123. doi: 10.1021/bi700871x. [DOI] [PubMed] [Google Scholar]
- 60.Tönsing L., Potgieter D.J., Louw A.I., Visser L. The binding of snake venom cardiotoxins to heart cell membranes. Biochim. Biophys. Acta. 1983;732:282–288. doi: 10.1016/0005-2736(83)90213-4. [DOI] [PubMed] [Google Scholar]
- 61.Lee C.Y., Chang C.C., Chiu T.H., Chiu P.J.S., Tseng T.C., Lee S.Y. Pharmacological properties of cardiotoxin isolated from Formosan cobra venom. Naunyn-Schmiedebergs Arch. Für Pharmakol. Exp. Pathol. 1968;259:360–374. doi: 10.1007/BF00536909. [DOI] [PubMed] [Google Scholar]
- 62.Panagides N., Jackson T., Ikonomopoulou M., Arbuckle K., Pretzler R., Yang D., Ali S., Koludarov I., Dobson J., Sanker B., et al. How the cobra got its flesh-eating venom: Cytotoxicity as a defensive innovation and its co-evolution with hooding, aposematic marking, and spitting. Toxins. 2017;9:103. doi: 10.3390/toxins9030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lauridsen L.P., Laustsen A.H., Lomonte B., Gutierrez J.M. Exploring the venom of the forest cobra snake: Toxicovenomics and antivenom profiling of Naja melanoleuca. J. Proteom. 2017;150:98–108. doi: 10.1016/j.jprot.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 64.Bourne Y., Talley T.T., Hansen S.B., Taylor P., Marchot P. Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors. EMBO J. 2005;24:1512–1522. doi: 10.1038/sj.emboj.7600620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang C.C., Lee C.Y. Electrophysiological study of neuromuscular blocking action of cobra neurotoxin. Br. J. Pharmacol. Chemother. 1966;28:172–181. doi: 10.1111/j.1476-5381.1966.tb01883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nirthanan S., Gwee M.C.E. Three-finger alpha-neurotoxins and the nicotinic acetylcholine receptor, forty years on. J. Pharmacol. Sci. 2004;94:1–17. doi: 10.1254/jphs.94.1. [DOI] [PubMed] [Google Scholar]
- 67.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nirthanan S., Awal W., Niranjan N.R. Snake α-neurotoxins and the nicotinic acetylcholine receptor. In: Gopalakrishnakone P., Inagaki H., Mukherjee A.K., Rahmy T.R., Vogel C.-W., editors. Snake Venoms. Springer; Dordrecht, The Netherlands: 2015. pp. 1–39. [Google Scholar]
- 70.Silva A., Cristofori-Armstrong B., Rash L.D., Hodgson W.C., Isbister G.K. Defining the role of post-synaptic alpha-neurotoxins in paralysis due to snake envenoming in humans. Cell Mol. Life Sci. 2018;75:4465–4478. doi: 10.1007/s00018-018-2893-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chicheportiche R., Vincent J.P., Kopeyan C., Schweitz H., Lazdunski M. Structure-function relationship in the binding of snake neurotoxins to the torpedo membrane receptor. Biochemistry. 1975;14:2081–2091. doi: 10.1021/bi00681a007. [DOI] [PubMed] [Google Scholar]
- 72.Leong P.K., Tan N.H., Fung S.Y., Sim S.M. Cross neutralisation of Southeast Asian cobra and krait venoms by Indian polyvalent antivenoms. Trans. R. Soc. Trop. Med. Hyg. 2012;106:731–737. doi: 10.1016/j.trstmh.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 73.Leong P.K., Sim S.M., Fung S.Y., Sumana K., Sitprija V., Tan N.H. Cross neutralization of afro-Asian cobra and Asian krait venoms by a thai polyvalent snake antivenom (neuro polyvalent snake antivenom) PLoS Negl. Trop. Dis. 2012;6:e1672. doi: 10.1371/journal.pntd.0001672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leong P.K., Tan C.H., Sim S.M., Fung S.Y., Sumana K., Sitprija V., Tan N.H. Cross neutralization of common southeast Asian viperid venoms by a Thai polyvalent snake antivenom (Hemato polyvalent snake antivenom) Acta Trop. 2014;132:7–14. doi: 10.1016/j.actatropica.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 75.Tan C.H., Leong P.K., Fung S.Y., Sim S.M., Ponnudurai G., Ariaratnam C., Khomvilai S., Sitprija V., Tan N.H. Cross neutralization of Hypnale hypnale (hump-nosed pit viper) venom by polyvalent and monovalent Malayan pit viper antivenoms in vitro and in a rodent model. Acta Trop. 2011;117:119–124. doi: 10.1016/j.actatropica.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 76.Liew J.L., Tan N.H., Tan C.H. Proteomics and preclinical antivenom neutralization of the mangrove pit viper (Trimeresurus purpureomaculatus, Malaysia) and white-lipped pit viper (Trimeresurus albolabris, Thailand) venoms. Acta Trop. 2020;209:105528. doi: 10.1016/j.actatropica.2020.105528. [DOI] [PubMed] [Google Scholar]
- 77.Utkin Y.N., Kukhtina V.V., Kryukova E.V., Chiodini F., Bertrand D., Methfessel C., Tsetlin V.I. Weak toxin from Naja kaouthia is a nontoxic antagonist of alpha 7 and muscle-type nicotinic acetylcholine receptors. J. Biol. Chem. 2001;276:15810–15815. doi: 10.1074/jbc.M100788200. [DOI] [PubMed] [Google Scholar]
- 78.Kukhtina V.V., Weise C., Muranova T.A., Starkov V.G., Franke P., Hucho F., Wnendt S., Gillen C., Tsetlin V.I., Utkin Y.N. Muscarinic toxin-like proteins from cobra venom. Eur. J. Biochem. 2000;267:6784–6789. doi: 10.1046/j.1432-1033.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- 79.Chang L.S., Chou Y.C., Lin S.R., Wu B.N., Lin J., Hong E., Sun Y.J., Hsiao C.D. A novel neurotoxin, cobrotoxin b, from Naja naja atra (Taiwan cobra) venom: Purification, characterization, and gene organization. J. Biochem. 1997;122:1252–1259. doi: 10.1093/oxfordjournals.jbchem.a021889. [DOI] [PubMed] [Google Scholar]
- 80.Pereira M.F., Novello J.C., Cintra A.C.O., Giglio J.R., Landucci E.T., Oliveira B., Marangoni S. The amino acid sequence of Bothropstoxin-II, an Asp-49 myotoxin from Bothrops jararacussu (Jararacucu) venom with low phospholipase A2 activity. J. Protein Chem. 1998;17:381–386. doi: 10.1023/A:1022563401413. [DOI] [PubMed] [Google Scholar]
- 81.van den Bergh C.J., Slotboom A.J., Verheij H.M., de Haas G.H. The role of aspartic acid-49 in the active site of phospholipase A2. A site-specific mutagenesis study of porcine pancreatic phospholipase A2 and the rationale of the enzymatic activity of [lysine49]phospholipase A2 from Agkistrodon piscivorus piscivorus’ venom. Eur. J. Biochem. 1988;176:353–357. doi: 10.1111/j.1432-1033.1988.tb14288.x. [DOI] [PubMed] [Google Scholar]
- 82.Vonk F.J., Casewell N.R., Henkel C.V., Heimberg A.M., Jansen H.J., McCleary R.J., Kerkkamp H.M., Vos R.A., Guerreiro I., Calvete J.J. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA. 2013;110:20651–20656. doi: 10.1073/pnas.1314702110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tan C.H., Wong K.Y., Tan N.H., Ng T.S., Tan K.Y. Distinctive Distribution of Secretory Phospholipases A2 in the Venoms of Afro-Asian Cobras (Subgenus: Naja, Afronaja, Boulengerina and Uraeus) Toxins. 2019;11:116. doi: 10.3390/toxins11020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tan K.Y., Wong K.Y., Tan N.H., Tan C.H. Quantitative proteomics of Naja annulifera (sub-Saharan snouted cobra) venom and neutralization activities of two antivenoms in Africa. Int. J. Biol. Macromol. 2020;158:605–616. doi: 10.1016/j.ijbiomac.2020.04.173. [DOI] [PubMed] [Google Scholar]
- 85.Sudarshan S., Dhananjaya B.L. The antimicrobial activity of an acidic phospholipase A2 (NN-XIa-PLA2) from the venom of Naja naja naja (indian cobra) Appl. Biochem. Biotechnol. 2015;176:2027–2038. doi: 10.1007/s12010-015-1698-8. [DOI] [PubMed] [Google Scholar]
- 86.Mukherjee A.K. Non-covalent interaction of phospholipase A2 (PLA2) and kaouthiotoxin (KTX) from venom of Naja kaouthia exhibits marked synergism to potentiate their cytotoxicity on target cells. J. Venom Res. 2010;1:37–42. [PMC free article] [PubMed] [Google Scholar]
- 87.Gasanov S.E., Alsarraj M.A., Gasanov N.E., Rael E.D. Cobra venom cytotoxin free of phospholipase A2 and its effect on model membranes and T leukemia cells. J. Membr. Biol. 1997;155:133–142. doi: 10.1007/s002329900165. [DOI] [PubMed] [Google Scholar]
- 88.Bustillo S., Garcia-Denegri M.E., Gay C., Van de Velde A.C., Acosta O., Angulo Y., Lomonte B., Gutierrez J.M., Leiva L. Phospholipase A2 enhances the endothelial cell detachment effect of a snake venom metalloproteinase in the absence of catalysis. Chem.-Biol. Interact. 2015;240:30–36. doi: 10.1016/j.cbi.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 89.Kazandjian T.D., Petras D., Robinson S.D., van Thiel J., Greene H.W., Arbuckle K., Barlow A., Carter D.A., Wouters R.M., Whiteley G., et al. Convergent evolution of pain-inducing defensive venom components in spitting cobras. Science. 2021;371:386–390. doi: 10.1126/science.abb9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hia Y.L., Tan K.Y., Tan C.H. Comparative venom proteomics of banded krait (Bungarus fasciatus) from five geographical locales: Correlation of venom lethality, immunoreactivity and antivenom neutralization. Acta Trop. 2020;207:105460. doi: 10.1016/j.actatropica.2020.105460. [DOI] [PubMed] [Google Scholar]
- 91.Tan K.Y., Tan N.H., Tan C.H. Venom proteomics and antivenom neutralization for the Chinese eastern Russell’s viper, Daboia siamensis from Guangxi and Taiwan. Sci. Rep. 2018;8:8545. doi: 10.1038/s41598-018-25955-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Senji Laxme R.R., Khochare S., Attarde S., Suranse V., Iyer A., Casewell N.R., Whitaker R., Martin G., Sunagar K. Biogeographic venom variation in Russell’s viper (Daboia russelii) and the preclinical inefficacy of antivenom therapy in snakebite hotspots. PLoS Negl. Trop. Dis. 2021;15:e0009247. doi: 10.1371/journal.pntd.0009247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chanda A., Kalita B., Patra A., Senevirathne W., Mukherjee A.K. Proteomic analysis and antivenomics study of Western India Naja naja venom: Correlation between venom composition and clinical manifestations of cobra bite in this region. Expert Rev. Proteom. 2019;16:171–184. doi: 10.1080/14789450.2019.1559735. [DOI] [PubMed] [Google Scholar]
- 94.Senji Laxme R.R., Khochare S., de Souza H.F., Ahuja B., Suranse V., Martin G., Whitaker R., Sunagar K. Beyond the ‘big four’: Venom profiling of the medically important yet neglected Indian snakes reveals disturbing antivenom deficiencies. PLoS Negl. Trop. Dis. 2019;13:e0007899. doi: 10.1371/journal.pntd.0007899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oh A.M.F., Tan C.H., Tan K.Y., Quraishi N.H., Tan N.H. Venom proteome of Bungarus sindanus (Sind krait) from Pakistan and in vivo cross-neutralization of toxicity using an Indian polyvalent antivenom. J. Proteom. 2018;193:243–254. doi: 10.1016/j.jprot.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 96.Sanz L., Quesada-Bernat S., Ramos T., Casais-e-Silva L.L., Corrêa-Netto C., Silva-Haad J.J., Sasa M., Lomonte B., Calvete J.J. New insights into the phylogeographic distribution of the 3FTx/PLA2 venom dichotomy across genus Micrurus in South America. J. Proteom. 2019;200:90–101. doi: 10.1016/j.jprot.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 97.Tan C.H., Tan K.Y., Ng T.S., Sim S.M., Tan N.H. Venom proteome of spine-bellied sea snake (Hydrophis curtus) from Penang, Malaysia: Toxicity correlation, immunoprofiling and cross-neutralization by sea snake antivenom. Toxins. 2019;11:3. doi: 10.3390/toxins11010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanz L., Pla D., Pérez A., Rodríguez Y., Zavaleta A., Salas M., Lomonte B., Calvete J.J. Venomic analysis of the poorly studied desert coral snake, Micrurus tschudii tschudii, supports the 3FTx/PLA2 dichotomy across Micrurus venoms. Toxins. 2016;8:178. doi: 10.3390/toxins8060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fernández J., Rey-Súarez P., Pla D., Sanz L., Sasa M., Núñez V., Gutiérrez J., Calvete J., Lomonte B. Proteomic studies on Micrurus (Coral Snakes) venom reveal a dichotomy of phenotypes. Toxicon. 2018;150:319–320. doi: 10.1016/j.toxicon.2018.06.015. [DOI] [Google Scholar]
- 100.Oh A.M.F., Tan K.Y., Tan N.H., Tan C.H. Proteomics and neutralization of Bungarus multicinctus (many-banded krait) venom: Intra-specific comparisons between specimens from China and Taiwan. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2021;247:109063. doi: 10.1016/j.cbpc.2021.109063. [DOI] [PubMed] [Google Scholar]
- 101.Lomonte B., Díaz C., Chaves F., Fernández J., Ruiz M., Salas M., Zavaleta A., Calvete J.J., Sasa M. Comparative characterization of Viperidae snake venoms from Perú reveals two compositional patterns of phospholipase A2 expression. Toxicon X. 2020;7:100044. doi: 10.1016/j.toxcx.2020.100044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zancolli G., Calvete J.J., Cardwell M.D., Greene H.W., Hayes W.K., Hegarty M.J., Herrmann H.-W., Holycross A.T., Lannutti D.I., Mulley J.F., et al. When one phenotype is not enough: Divergent evolutionary trajectories govern venom variation in a widespread rattlesnake species. Proc. R. Soc. B Biol. Sci. 2009;286:20182735. doi: 10.1098/rspb.2018.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Punde D.P. Management of snake-bite in rural Maharashtra: A 10-year experience. Natl. Med. J. India. 2005;18:71–75. [PubMed] [Google Scholar]
- 104.Bawaskar H.S., Bawaskar P.H., Punde D.P., Inamdar M.K., Dongare R.B., Bhoite R.R. Profile of snakebite envenoming in rural Maharashtra, India. J. Assoc. Phys. India. 2008;56:88–95. [PubMed] [Google Scholar]
- 105.Xu N., Zhao H.Y., Yin Y., Shen S.S., Shan L.L., Chen C.X., Zhang Y.X., Gao J.F., Ji X. Combined venomics, antivenomics and venom gland transcriptome analysis of the monocoled cobra (Naja kaouthia) from China. J. Proteom. 2017;159:19–31. doi: 10.1016/j.jprot.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 106.Pandey S., Pandey S. A study of snake bite cases in rural area of south eastern Rajasthan. Int. J. Biol. Med. Res. 2012;3:2348–2351. [Google Scholar]
- 107.Al-Mamun M., Hakim M., Zaman M., Hoque K., Ferdousi Z., Reza M. Histopathological alterations induced by Naja naja crude venom on renal, pulmonary and intestinal tissues of mice model. Br. Biotechnol. J. 2015;6:119–125. doi: 10.9734/BBJ/2015/16188. [DOI] [Google Scholar]
- 108.Wong O.F., Lam T.S., Fung H.T., Choy C.H. Five-year experience with Chinese cobra (Naja atra)-related injuries in two acute hospitals in Hong Kong. Hong Kong Med. J. 2010;16:36–43. [PubMed] [Google Scholar]
- 109.Mao Y.C., Liu P.Y., Chiang L.C., Lai C.S., Lai K.L., Ho C.H., Wang T.H., Yang C.C. Naja atra snakebite in Taiwan. Clin. Toxicol. 2017;56:1–8. doi: 10.1080/15563650.2017.1366502. [DOI] [PubMed] [Google Scholar]
- 110.Wongtongkam N., Wilde H., Sitthi-Amorn C., Ratanabanangkoon K. A study of thai cobra (Naja kaouthia) bites in Thailand. Mil. Med. 2005;170:336–341. doi: 10.7205/MILMED.170.4.336. [DOI] [PubMed] [Google Scholar]
- 111.Ratanabanangkoon K., Tan K.Y., Pruksaphon K., Klinpayom C., Gutiérrez J.M., Quraishi N.H., Tan C.H. A pan-specific antiserum produced by a novel immunization strategy shows a high spectrum of neutralization against neurotoxic snake venoms. Sci. Rep. 2020;10:11261. doi: 10.1038/s41598-020-66657-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ratanabanangkoon K., Tan K.Y., Eursakun S., Tan C.H., Simsiriwong P., Pamornsakda T., Wiriyarat W., Klinpayom C., Tan N.H. A Simple and novel strategy for the production of a pan-specific antiserum against elapid snakes of Asia. PLoS Negl. Trop. Dis. 2016;10:e0004565. doi: 10.1371/journal.pntd.0004565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ratanabanangkoon K. A quest for a universal plasma-derived antivenom against all elapid neurotoxic snake venoms. Front. Immunol. 2021;12:1363. doi: 10.3389/fimmu.2021.668328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wery M., Descrimes M., Thermes C., Gautheret D., Morillon A. Zinc-mediated RNA fragmentation allows robust transcript reassembly upon whole transcriptome RNA-Seq. Methods. 2013;63:25–31. doi: 10.1016/j.ymeth.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 115.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M.W., Gaffney D.J., Elo L.L., Zhang X., et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li B., Dewey C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 121.Thompson J.D., Muller A., Waterhouse A., Procter J., Barton G.J., Plewniak F., Poch O. MACSIMS: Multiple alignment of complete sequences information management system. BMC Bioinform. 2006;7:318. doi: 10.1186/1471-2105-7-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Edgar R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 125.Wong K.Y., Tan K.Y., Tan N.H., Tan C.H. A neurotoxic snake venom without phospholipase A2: Proteomics and cross-neutralization of the venom from Senegalese cobra, Naja senegalensis (Subgenus: Uraeus) Toxins. 2021;13:60. doi: 10.3390/toxins13010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Finney D.J. Probit Analysis. 2nd ed. Cambridge University Press; New York, NY, USA: 1952. [Google Scholar]
- 127.Morais V., Ifran S., Berasain P., Massaldi H. Antivenoms: Potency or median effective dose, which to use? J. Venom. Anim. Toxins Incl. Trop. Dis. 2010;16:191–193. doi: 10.1590/S1678-91992010000200002. [DOI] [Google Scholar]
- 128.Tan C.H., Tan K.Y. Functional application of snake venom proteomics in in vivo antivenom assessment. In: Wang X., Kuruc M., editors. Functional Proteomics: Methods and Protocols. Springer; New York, NY, USA: 2019. pp. 153–158. [DOI] [PubMed] [Google Scholar]
- 129.Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., Li K., Zhang G., Jin Z., He F., et al. iProX: An integrated proteome resource. Nucleic Acids Res. 2019;47:1211–1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article or supplementary material.