Abstract
Sand flies transmit Leishmania infantum, which is responsible for causing leishmaniasis, as well as many phleboviruses, including the human pathogenic Toscana virus. We screened sand flies collected from a single site between 2017 and 2020 for the presence of both phleboviruses and Leishmania. The sand flies were sampled with attractive carbon dioxide traps and CDC light traps between May and October. We collected more than 50,000 sand flies; 2826 were identified at the species level as Phlebotomus perfiliewi (98%) or Phlebotomus perniciosus (2%). A total of 16,789 sand flies were tested in 355 pools, and phleboviruses were found in 61 pools (6 Toscana virus positive pools, 2 Corfou virus positive pools, 42 Fermo virus positive pools, and 7 Ponticelli virus positive pools, and 4 unidentified phlebovirus positive pools). Leishmania was found in 75 pools and both microorganisms were detected in 16 pools. We isolated nine phleboviruses from another 2960 sand flies (five Ponticelli viruses and for Fermo viruses), not tested for Leishmania; the complete genome of a Fermo virus isolate was sequenced. The simultaneous detection in space and time of the Fermo virus and L. infantum is evidence that supports the co-circulation of both microorganisms in the same location and partial overlap of their cycles. A detailed characterization of the epidemiology of these microorganisms will support measures to limit their transmission.
Keywords: sand fly, phlebotomus perfiliewi, phlebotomus perniciosus, leishmania infantum, phlebovirus, toscana virus, fermo virus, corfou virus, ponticelli I virus, ponticelli II virus, ponticelli III virus
1. Introduction
Phleboviruses (family: Phenuiviridae, genus: Phlebovirus) are ssRNA viruses characterized by a tri-segmented genome. The L segment (6.4 kb) codes the RNA-dependent RNA polymerase (RdRp), the M segment (3.2 kb) encodes for a polyprotein that is the origin of several proteins, and the S segment (1.7 kb) encodes for two proteins with an ambisense strategy (https://viralzone.expasy.org/252 accessed on 10 May 2021). Species in the genus were previously defined by the degree of serological cross-reactivity, but the frequent isolation of new viruses and the reassorting capacities of these viruses suggested a need to change the classification standards [1]. More than 60 phleboviral species have now been defined using a threshold of 95% identity in the amino acid sequences of the RdRp [2].
Some sand fly-borne phleboviruses cause disease in humans. One example, Toscana virus (TOSV), is the etiological agent of neuroinvasive diseases including meningitis and meningoencephalitis. The TOSV was first isolated in 1971 [3] and later recognized as an agent of meningitis in humans [4]. This virus has been detected in large parts of many Mediterranean countries, from Spain to Turkey [5]. Previously unknown phleboviruses have been detected in sand flies, but little is known about their pathogenic potential, ecological cycles, mechanisms of persistence, or presence in vertebrate reservoirs.
Several of these viruses had been detected in Northern Italy. In addition to TOSV, three phleboviruses—Ponticelli I virus, Ponticelli II virus, and Ponticelli III virus—were isolated [6]. These three viruses differ in their M segments, likely as a result of reassortant events. In the same area, Fermo and Corfou viruses, in addition to phleboviruses possessing sequences with no known among previously described strains, were found in sand flies [7].
Among the various sand fly-borne pathogens, the intracellular protozoa of the Leishmania genus (family: Trypanosomatidae; order: Kinetoplastida) contribute significantly to human diseases in Italy and elsewhere. The species circulating in the Mediterranean region include L. donovani and L. infantum, which cause visceral leishmaniasis (VL), and L. tropica and L. major, which, together with dermotropic L. infantum strains, cause cutaneous leishmaniasis (CL). The estimated annual incidence of leishmaniasis in this area is 1200–2000 cases of VL and 239,500–393,600 cases of CL [8].
Leishmania infantum is almost the only causal agent of leishmaniasis in Italy, where it causes VL and CL in humans, as well as canine leishmaniasis (canL) in dogs. Dogs are regarded as the main source of Leishmania infection in many epidemiological settings, but other domestic and wild animal species have also been implicated as reservoirs [9].
VL and CL are endemic to the hilly part of Emilia-Romagna. The first outbreak of 60 cases of VL was recorded in 1971–1972 [10], and a recrudescence of human leishmaniasis was reported to have occurred in 2012–2013, with more than 30 cases of VL [11,12]. Recent epidemiological studies based on molecular typing suggest that dogs cannot be the main reservoir for human leishmaniasis in this area because the strains involved in VL cases are only distantly related to canine Leishmania strains compared to other strains [13,14].
Phleboviruses and Leishmania share the same principal vectors: sand flies. Sand flies are tiny hematophagous insects that often live close to domestic animals and humans. Only females bite, typically from dusk to dawn. They are poor fliers, flying silently for short distances, and they have activity peaks during summertime in non-tropical regions; sand flies are widely distributed in Mediterranean countries. The two species that are most abundant in the natural and rural environments of the Emilia-Romagna region are Ph. perniciosus and Ph. perfiliewi; both species are proven vectors of Leishmania infantum and TOSV in Italy [15].
In this work, we report the contemporaneous detection of diverse phleboviruses and Leishmania parasites in sand flies collected at a site in the Valsamoggia municipality (BO) in the Emilia-Romagna region, Italy from 2017 to 2020.
2. Materials and Methods
2.1. Sampled Site
Between 2017 and 2020, we sampled a single site that had already been surveyed, beginning in 2012 (44°17′15″ N, 11°41′46″ E) [16]. This site was located in the hilly area of the Emilia-Romagna region (altitude 196 m) in the garden of an uninhabited building. The surroundings were characterized by a typical hilly landscape with cultivated fields and hedges interspersed by plots of woodlands, which were prevalent on the hilltops.
2.2. Sand Fly Sampling and Identification
Sampling was performed fortnightly at night in the period from May to October. We used two models of traps: traps baited with dry ice, to produce carbon dioxide (CO2), and standard miniature CDC light traps. One carbon dioxide trap was used at the same site for every sampling and was used as an estimate of the abundance of sand flies. Other traps were employed in variable numbers, from one to nine, to maximize the number of insects collected, keeping a minimum distance of 20 m between them to avoid reciprocal interference (Table S1). A subset of the captured sand flies was clarified in chlorolactophenol and morphologically identified using a light microscope using specific morphological keys [17,18]. We grouped other sub-samples of sand flies in pools, which were submitted to biomolecular analysis for phlebovirus and Leishmania detection or utilized for virus isolation in cell cultures.
Weather data from the closest climatic stations were retrieved with the web app of the Regional Environmental Protection Agency (www.arpae.it accessed on 10 April 2021), Dext3r (https://simc.arpae.it/dext3r/ accessed on 10 April 2021). The average of the precipitation recorded at four stations (Guiglia, Monte San Pietro, Bazzano, Savignano sul Panaro) was used, and average temperatures were recorded at the Guiglia station.
2.3. Pathogen Detection and Sequencing
We examined pools of 50 females for biomolecular analysis. The pools were ground, and the genetic material was extracted with an automated extractor (BioSprint 96, Qiagen, Germany) and retro-transcribed. We searched phleboviruses with PCR (henceforth pan-phlebo-PCR), targeting a 370-nucleotide region of the S segment [19]. The amplicons obtained with the pan-phlebo-PCR were sequenced, and the sequences were used to identify detected viruses by BLAST analysis in the GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi accessed on 10 April 2021). We detected Leishmania parasites with real-time PCR targeting the kinetoplast DNA [20]. The minimum infection rates (MIR) of Leishmania were calculated for every sample, assuming the presence of at least one Leishmania-positive sand fly per positive pool. Primers of the PCRs are reported in Table S2.
A Fermo virus isolate was sequenced on a MiSeq Instrument (Illumina Inc., San Diego, CA, USA) as described elsewhere [6]. The sequences obtained were aligned with homologous sequences that were available in Gen Bank using MAFFT [21], and they were used to obtain a neighbor-joining tree and maximum likelihood trees with the IQ-Tree software [22].
2.4. Virus Isolation
A sub-sample of the females was grouped in 25-specimen pools on a surface kept at −80 °C, and tested for isolation as quickly as possible, as reported previously [6]. The insects were ground in a glass potter in minimal essential medium supplemented with penicillin and streptomycin and were centrifugated at 3000× g for 15 min. An aliquot of these homogenates was submitted to pan-phlebo-PCR. The samples were inoculated in a confluent monolayer of VERO cells (African green monkey kidney cells) at passage 170 (cell culture collection of IZSLER, code BSCL86) and incubated at 37 °C with 5% CO2. We observed the cultures daily for 7 days to detect and characterize cytopathic effect (CPE). Then, we sub-cultured the cryolysates twice into fresh monolayers. Pools that were utilized for virus isolation were not tested for the presence of Leishmania.
3. Results
3.1. Sampled Sand Flies
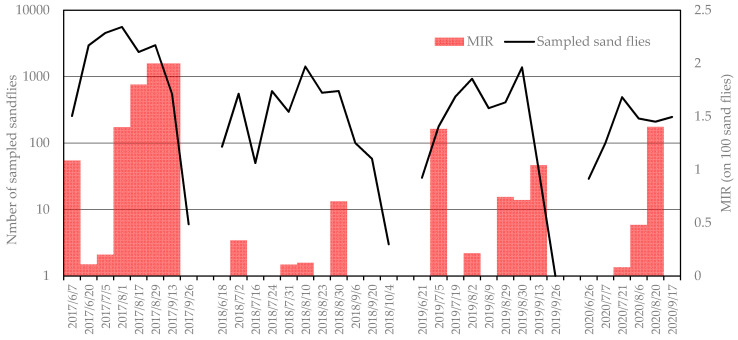
We collected 50,510 sand flies during the study (2017–2020), and morphologically identified 2826 specimens: 2622 were identified as Ph. perfiliewi (98%) and 45 were identified as Ph. perniciosus (2%) (Table 1). The number of sand flies collected in CO2 traps revealed the great abundance of sand flies in 2017 compared to the other years (Figure 1), with a record of 5600 insects in one trap in August. The averages of the temperatures and monthly precipitation recorded in the climatic stations near the sampling site are reported in Figure S1. Interestingly, the 2017 season, which was characterized by a large number of sandflies, had a drier spring than the other years; the lowest precipitation was between March and June (Figure 2, Table S3).
Table 1.
Collection, testing, and identification of sand flies during the years of surveillance.
| 2017 | 2018 | 2019 | 2020 | Total | ||
|---|---|---|---|---|---|---|
| Collected sand flies | 25,966 | 7696 | 12,559 | 4289 | 50,510 | |
| Identified | Ph. perfiliewi | 768 | 1123 | 427 | 453 | 2771 |
| Ph perniciosus | 7 | 22 | 13 | 13 | 55 | |
| Tested | Leishmania/Pan-phlebo PCR | 5443 | 3363 | 5893 | 2090 | 16,789 |
| Isolation in cell cultures | 2460 | 500 | 2960 |
Figure 1.
Number of sand flies sampled per night per trap (logarithmic scale) with reference to the Leishmania minimum infection rates per 100 specimens.
Figure 2.
Monthly precipitation (histograms) and average temperature (black circles) from March to July at the sampled sites during the period of surveillance.
3.2. Molecular Analyses
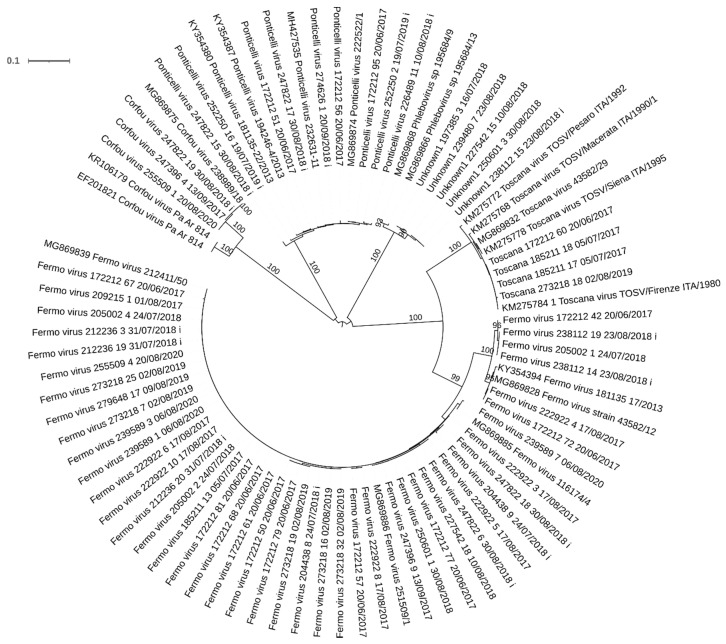
We tested 16,789 sandflies (Table 1)—grouped in 355 pools—using PCR. Of these, 75 tested positive for Leishmania through the PCR, and 61 produced a sequence ascribable to a phlebovirus: 6 Toscana viruses, 42 Fermo viruses, 7 Ponticelli viruses, 2 Corfou viruses, and 4 of a putative phlebovirus that had been detected in the surveyed area (Table 2). The partial sequence of the putative phlebovirus was distantly related to other phleboviruses detected in Mediterranean countries, but it showed the highest amino acid identity with the South American phleboviruses Anhanga virus (67.5% GB: NC_033837). The neighbor joining tree obtained with these sequences and the previously deposited sequence of the same virus are shown in Figure 3 (Jukes–Cantor model, bootstrap resampling of 1000). Both Leishmania and phleboviruses were detected in 16 pools (14 Fermo viruses, 1 Ponticelli virus, 1 Corfou virus) (Table 2). Using only data concerning the more detected virus, the Fermo virus, on the 34 days of sampling, we detected both Fermo virus and Leishmania parasite in 15 days, while we detect only Fermo virus in 2 days and only Leishmania in 3 days. Considering the days of sampling, the detection of both Fermo virus and Leishmania was significantly related, with an odds ratio of 8 (95% confidence, 1.74–36.71, p < 0.01).
Table 2.
Leishmania spp. and phleboviruses detected in double-tested pools by year of surveillance. Phlebovirus-positive pools that also tested positive for Leishmania spp. are shown in parentheses.
| 2017 | 2018 | 2019 | 2020 | Total | |
|---|---|---|---|---|---|
| Tested pools | 110 | 73 | 126 | 46 | 355 |
| Leishmania | 42 | 5 | 22 | 6 | 75 |
| Toscana virus | 4 | 2 | 6 | ||
| Fermo virus | 23 (10) | 5 (1) | 8 (2) | 6 (1) | 42 (14) |
| Ponticelli virus | 4 | 2 (1) | 1 | 7 (1) | |
| Corfou virus | 1 (1) | 1 | 2 (1) | ||
| Unknown1 | 4 | 4 |
Figure 3.
Neighbor joining tree of partial sequences of the S segments obtained from the field samples with reference to the strain collection day and utilization for isolation (i); the selected homologous sequences are available in GenBank (by referencing the accession number), bootstrap supports >90% showed near the branch.
The MIRs of Leishmania recorded in different seasons are shown in Figure 1. The 2017 season, which was characterized by a great abundance of sand flies, showed more intense Leishmania circulation (Figure 1), and the proportion of Leishmania-positive pools in 2017 differed significantly from the proportion in the other years (42/110 in 2017 vs. 37/255 in 2018–2020, Pearson χ2, p < 0.001).
3.3. Virus Isolation
We attempted to isolate virus from 120 pools—100 in 2018 (collected from 18 June to 20 September) and 20 in 2019 (collected on 19 July)—for a total of 2960 sand flies (Table 1). We detected phleboviruses in 20 of these pools (10 Fermo viruses, 7 Ponticelli viruses, 1 Corfou virus, and 1 virus referred to as Unknown1). Cytopathic effects—from moderate to evident—were observed in nine cultures, which were all sampled in 2018. Virus isolation was then confirmed by testing the cryolysates by pan-phlebo PCR, confirming the isolation of four strains of Fermo virus and five strains of Ponticelli virus (Table 3). Interestingly, one of these Ponticelli virus strains was isolated from a homogenate that tested negative in the pan-phlebo PCR and another that was isolated from a homogenate in which a Fermo virus sequence was detected. We submitted the isolated Fermo virus (strain 212236-3) for next generation sequencing and obtained the complete genome, which was then deposited in ENA (European Nucleotide Archive) with the accession numbers OU230765 (L segment), OU230766 (M segment) OU230767 (S segment), to make available the first complete genome sequence of that virus. The amino acid RdRp sequence of this virus showed an identity of 92.1% with the closest deposited virus, the Tehran virus (GB JF939846), which was isolated from Ph. perfiliewi in Iran in 1976. The maximum likelihood tree obtained with the Fermo RdRp amino acid sequence is shown in Figure 4 (LG + F + R3 model selected by IQ-Tree software, ultrafast bootstrap approximation of 1000 iterations). We were able to confirm a high identity between the complete genome nucleotide sequences and the partial Fermo virus sequences deposited in GenBank, which was ascribable to the L segments (between 92.1% and 95.2%) or the S segments (between 91.7% and 99.7%). High identity was also recorded with respect to partial Balkan virus sequences detected in sand flies in Balkan countries in 2015 [23]. More precisely, an identity of between 86.9% and 89.1% was found for the partial L segments, and an identity of from 88.5% to 89.5% was found for the partial S segments.
Table 3.
Results of the analysis of the sand fly pools (25 specimens) with successful isolation.
| Data | Pool Code | Homogenate PCR | CPE 1 | Cryolysate PCR |
|---|---|---|---|---|
| 24/07/2018 | 204438-8 | Fermo virus | ++ | Fermo virus |
| 24/07/2018 | 204438-9 | Fermo virus | + | Fermo virus |
| 31/07/2018 | 212236-3 | Fermo virus | ++ | Fermo virus |
| 31/07/2018 | 212236-20 | Fermo virus | + | Fermo virus |
| 10/08/2018 | 226489-11 | Ponticelli virus | ++ | Ponticelli virus |
| 30/08/2018 | 247822-15 | Ponticelli virus | ++ | Ponticelli virus |
| 30/08/2018 | 247822-17 | Ponticelli virus | ++ | Ponticelli virus |
| 30/08/2018 | 247822-18 | Fermo virus | ++ | Ponticelli virus |
| 06/09/2018 | 256303-2 | Neg. | ++ | Ponticelli virus |
1 Cytopathic effect.
Figure 4.
Maximum likelihood tree obtained using the amino acid sequences of the RpRd of complete sequences of the Fermo virus and homologous sequences of sand fly-borne phleboviruses detected in Mediterranean and Middle Eastern countries. GenBank accession numbers reported as reference, bootstrap supports >90% showed near the branch.
4. Discussion
Sand flies were abundant at this study site, as demonstrated by the 5600 specimens collected in one trap in one night in 2017. This abundance was mainly sustained by the Ph. perfiliewi species, which was much more abundant than Ph. perniciosus at the same site, as previously recorded in the neighboring area [16,24]. The abundance of sand flies differed by year, and we hypothesize that this was influenced by weather conditions; in particular, there was a correlation between abundance and low precipitation in the spring months. A similar relation has been shown for Lutzomyia species in South America [25,26]. As already noted in this area of Italy, the abundance of sand flies directly affected the circulation of Toscana virus [27]. Similarly, the recrudescence of human Leishmaniasis was recorded in the Emilia-Romagna region after seasons with abundant captures of sand flies, such as in 2013–2014 [6,28]. Interestingly, a relationship between drought conditions and the 1970–1971 outbreak was already hypothesized by Pampiglione [10]. If this relationship can be precisely characterized, the definition of factors that influence the number of sand flies will also help in assessing the intensity of pathogen circulation and may, perhaps, be useful in predicting increases of infections.
We recorded the presence of at least four different phleboviruses at the same site during the four seasons of surveillance, similarly to the results obtained in the geographical area around the site [7]. In addition to the well-known TOSV, we confirm the presence of Corfou virus, which was first isolated in Greece in 1981 [29], and of Fermo virus, which was first isolated in the Marche region of Italy in 2012 [30], about 250 km from our site. In addition, amplicons referable to Ponticelli virus were generated, although we did not type them further, as more information on the M segment sequence was needed. We also sequenced amplicons that had already been generated from other sites (defined as Unknown1) and that were ascribable to a putative phlebovirus that has not been isolated; this phlebovirus has been detected since 2013 as far as about 80 km from this site in Emilia-Romagna [6]. The detection of these viruses through a non-virus species-specific PCR, with an intrinsic limit on the sensitivity, suggests that they had a greater presence. Among them, TOSV has the greatest relevance to health because of its capacity to cause neuroinvasive infections. The other phleboviruses detected have not been clearly associated with diseases in humans, though serological reactions to Ponticelli II virus were observed in sick persons in the neighboring Lombardia region [31]. Interestingly, only with this virus have serological reactions been detected, while the other two reassortant viruses—which had different M segments (Ponticelli I virus and Ponticelli III virus)—did not show any reactivity. This highlights the importance of the M segment in defining the ability of these viruses to infect vertebrates, and the ability to reassort this segment is likely an important factor in determining the evolution and pathogenicity of phleboviruses, similar to that observed with viruses in the Orthobunyavirus genus [32,33]. The close proximity of these phleboviruses detected at the same site likely offers them the opportunity to reassort among themselves.
The isolates obtained confirmed the major adaptability of the Ponticelli viruses to cell cultures, highlighting their ability to displace other viruses that are present in an insect pool, as observed with the isolation of one Ponticelli virus from a pool that was positive for Fermo virus. Fermo virus also caused a weak CPE, suggesting a minor adaptability to cell culture; despite this, we were able to isolate and make available the complete genome of Fermo virus. Its relationship with the closest Tehran virus confirmed that Fermo virus represents a new phlebovirus species according to the 95% threshold in the RdRp sequence. The virus is closely related to Balkan viruses; however, in the absence of a complete sequence of the L segment, it is not possible to draw firm conclusions on their relation. The possible presence of a vertebrate host of this virus and its potential pathogenicity require additional studies.
Leishmaniasis is a well-known human health threat, and the surveyed site is located in an area in which the recrudescence of VL caused by L. infantum has been recorded since 2012 [11,28]. Moreover, a screening of blood donors performed in 2014–2015 using serological methods resulted in detection of asymptomatic infections in 12.5% of the tested individuals, indicating an important cumulative exposure to the parasite in the same area [34]. The high rate of infection recorded in this study attests to the active circulation of these parasites in sand flies. However, the reasons for the recent upsurge in VL cases in Emilia-Romagna are unknown. The seroprevalence of L. infantum in the canine population in the Emilia-Romagna region is low when compared to the CanL seroprevalence in other endemic areas of Italy, and it has shown a decreasing trend over time [13]. It is noteworthy that previous molecular studies showed the circulation of two genetically distinct populations of L. infantum in the Emilia-Romagna region—one affecting dogs and the other circulating in Ph. perfiliewi and humans [13,14,16]. This suggests the existence of two overlapping transmission cycles that probably involve different reservoirs.
The co-circulation of L. infantum and phleboviruses has already been described in the Mediterranean basin [35,36,37,38]; we confirmed this phenomenon by detecting the contemporary presence of both in the surveyed sites. We were also able to assess the correlation between the most commonly detected phlebovirus (Fermo virus) and Leishmania, and we found a significant relationship in the co-detection of both on particular days of sampling. The simultaneous detection in space and time of the Fermo virus and Leishmania is an evidence of the cycle similarity between the two microorganisms and strongly suggests an overlap between their cycles. An obvious point of convergence was the sharing of the vector, Ph. perfiliewi, which was the most abundant sand fly species recorded and the main vector of L. infantum in this area [16]. Interestingly, this species was also incriminated as one of the main vectors for TOSV. It will be of interest to determine whether these two microorganisms share other epidemiological characteristics, such as reservoirs. Leishmania sampled in that area showed a unique epidemiology, which strongly suggests the presence of a reservoir that has not yet been determined. Additional studies are required to determine if this statement can be extended to other protozoa and Fermo viruses. The possibility of its similarity to the ecology of phleboviruses can help in clarifying the cycles of both.
5. Conclusions
Sand flies can host a variety of microorganisms, including viruses and parasites, in strict spatial and temporal coexistence. The definition of the cycles of these microorganisms and the possible interactions between them could lead to better understanding of this complex picture. The potential ability to increase the infective capacity of Leishmania through coinfection with phleboviruses also has been suggested [39]. The cycles of sand fly-transmitted pathogens are often enigmatic because it can involve different species of sand fly and different reservoirs in different areas. The advances in the cycle characterization of one of these microorganisms can help in clarifying the cycle of another. An important experimental effort must be made to define the cycle of these microorganisms, which can lead to the assessment of the risks linked to their presence and development of control measures necessary for limiting their transmission.
Acknowledgments
The authors acknowledge Charles H. Calisher for the suggestions and the helpful editing of the paper.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/v13081660/s1: Figure S1: Average temperature (running mean of 15 days) and monthly precipitation in the sampled site during the period of surveillance; Table S1: Sampled, tested, and identified sand flies for every day of sampling; Table S2: PCR primers used in this work; Table S3: Details of the average temperature and monthly precipitation recorded during the survey.
Author Contributions
Conceptualization, M.C. and M.D.; methodology, P.B., C.C., E.C. (Elena Carra), R.T. and D.L.; formal analysis, M.C.; field sampling G.R. (Giuseppe Romeo) and E.C. (Emanuele Callegari); data curation, M.C. and G.R. (Giuseppe Romeo); writing—original draft preparation, M.C.; writing—review and editing, M.C. and G.R. (Gianluca Rugna); funding acquisition, M.D. and D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministero della Salute PRC “Isolation of phleboviruses from sand flies and evaluation of their diffusion in humans and domestic animals” code E89I17000230001, internal code IZSLER 2017/013 RC.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Fermo virus sequences are available at ENA (https://www.ebi.ac.uk/ena/browser/home accessed on 10 April 2021), other data are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Palacios G., Tesh R., Travassos da Rosa A., Savji N., Sze W., Jain K., Serge R., Guzman H., Guevara C., Nunes M.R., et al. Characterization of the Candiru antigenic complex (Bunyaviridae: Phlebovirus), a highly diverse and reassorting group of viruses affecting humans in tropical America. J. Virol. 2011;85:3811–3820. doi: 10.1128/JVI.02275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhn J.H., Adkins S., Alioto D., Alkhovsky S.V., Amarasingh G.K., Anthony S.J., Avšič-Županc T., Ayllón M.A., Bahl J., Balkema-Buschmann A., et al. 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2020;165:3023–3072. doi: 10.1007/s00705-020-04731-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verani P., Ciufolini M.G., Nicoletti L., Balducci M., Sabatinelli G., Coluzzi M., Paci P., Amaducci L. Ecological and epidemiological studies of Toscana virus, an arbovirus isolated from Phlebotomus. Ann. Ist. Super. Sanita. 1982;18:397–399. [PubMed] [Google Scholar]
- 4.Calisher C.H., Weinberg A.N., Muth D.J., Lazuick J.S. Toscana virus infection in United States citizen returning from Italy. Lancet. 1987;1:165–166. doi: 10.1016/S0140-6736(87)92005-8. [DOI] [PubMed] [Google Scholar]
- 5.Moriconi M., Rugna G., Calzolari M., Bellini R., Albieri A., Angelini P., Cagarelli R., Landini M.P., Charrel R.N., Varani S. Phlebotomine sand fly-borne pathogens in the Mediterranean Basin: Human leishmaniasis and phlebovirus infections. PLoS Negl. Trop. Dis. 2017;11:e0005660. doi: 10.1371/journal.pntd.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calzolari M., Chiapponi C., Bellini R., Bonilauri P., Lelli D., Moreno A., Barbieri I., Pongolini S., Lavazza A., Dottori M. Isolation of three novel reassortant phleboviruses, Ponticelli I, II, III, and of Toscana virus from field-collected sand flies in Italy. Parasites Vectors. 2018;11:84. doi: 10.1186/s13071-018-2668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calzolari M., Ferrarini G., Bonilauri P., Lelli D., Chiapponi C., Bellini R., Dottori M. Co-circulation of eight different phleboviruses in sand flies collected in the Northern Apennine Mountains (Italy) Infect. Genet. Evol. 2018;64:131–134. doi: 10.1016/j.meegid.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Alvar J., Vélez I.D., Bern C., Herrero M., Desjeux P., Cano J., Jannin J., den Boer M., WHO Leishmaniasis Control Team Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE. 2012;7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardoso L., Schallig H., Persichetti M.F., Pennisi M.G. New Epidemiological Aspects of Animal Leishmaniosis in Europe: The Role of Vertebrate Hosts Other Than Dogs. Pathogens. 2021;10:307. doi: 10.3390/pathogens10030307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pampiglione S., La Placa M., Schlick G. Studies on mediterranean Leishmaniasis. I. An outbreak of visceral leishmaniasis in Northern Italy. Trans. R. Soc. Trop. Med. Hyg. 1974;68:349–359. doi: 10.1016/0035-9203(74)90148-5. [DOI] [PubMed] [Google Scholar]
- 11.Varani S., Cagarelli R., Melchionda F., Attard L., Salvadori C., Finarelli A.C., Gentilomi G.A., Tigani R., Rangoni R., Todeschini R., et al. Ongoing outbreak of visceral leishmaniasis in Bologna Province, Italy, November 2012 to May 2013. Eurosurveillance. 2013;18:20530. doi: 10.2807/1560-7917.ES2013.18.29.20530. [DOI] [PubMed] [Google Scholar]
- 12.Franceschini E., Puzzolante C., Menozzi M., Rossi L., Bedini A., Orlando G., Gennari W., Meacci M., Rugna G., Carra E., et al. Clinical and Microbiological Characteristics of Visceral Leishmaniasis Outbreak in a Northern Italian Nonendemic Area: A Retrospective Observational Study. BioMed Res. Int. 2016;2016:6481028. doi: 10.1155/2016/6481028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rugna G., Carra E., Corpus F., Calzolari M., Salvatore D., Bellini R., Di Francesco A., Franceschini E., Bruno A., Poglayen G., et al. Distinct Leishmania infantum Strains Circulate in Humans and Dogs in the Emilia-Romagna Region, Northeastern Italy. Vector Borne Zoonotic Dis. 2017;17:409–415. doi: 10.1089/vbz.2016.2052. [DOI] [PubMed] [Google Scholar]
- 14.Rugna G., Carra E., Bergamini F., Calzolari M., Salvatore D., Corpus F., Gennari W., Baldelli R., Fabbi M., Natalini S., et al. Multilocus microsatellite typing (MLMT) reveals host-related population structure in Leishmania infantum from northeastern Italy. PLoS Negl. Trop. Dis. 2018;12:e0006595. doi: 10.1371/journal.pntd.0006595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maroli M., Feliciangeli M.D., Bichaud L., Charrel R.N., Gradoni L. Phlebotomine sandflies and the spreading of leishmaniases and other diseases of public health concern. Med. Vet. Entomol. 2013;27:123–147. doi: 10.1111/j.1365-2915.2012.01034.x. [DOI] [PubMed] [Google Scholar]
- 16.Calzolari M., Carra E., Rugna G., Bonilauri P., Bergamini F., Bellini R., Varani S., Dottori M. Isolation and Molecular Typing of Leishmania infantum from Phlebotomus perfiliewi in a Re-Emerging Focus of Leishmaniasis, Northeastern Italy. Microorganisms. 2019;7:644. doi: 10.3390/microorganisms7120644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romi R., Khoury C., Bigliocchi F., Maroli M. Fact Sheet on Mites and Insects of Medical Importance. Schede Guida su Acari e Insetti di Interesse Sanitario. Istituto Superiore di Sanità; Rome, Italy: 1994. [Google Scholar]
- 18.Dantas-Torres F., Tarallo V.D., Otranto D. Morphological keys for the identification of Italian phlebotomine sand flies (Diptera: Psychodidae: Phlebotominae) Parasites Vectors. 2014;7:479. doi: 10.1186/s13071-014-0479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert A.J., Lanciotti R.S. Consensus amplification and novel multiplex sequencing method for S segment species identification of 47 viruses of the Orthobunyavirus, Phlebovirus, and Nairovirus genera of the family Bunyaviridae. J. Clin. Microbiol. 2009;47:2398–2404. doi: 10.1128/JCM.00182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galletti E., Bonilauri P., Bardasi L., Fontana M.C., Ramini M., Renzi M., Dosa G., Merialdi G. Development of a minor groove binding probe based real-time PCR for the diagnosis and quantification of Leishmania infantum in dog specimens. Res. Vet. Sci. 2011;91:243–245. doi: 10.1016/j.rvsc.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayhan N., Alten B., Ivovic V., Dvořák V., Martinkovic F., Omeragic J., Stefanovska J., Petric D., Vaselek S., Baymak D., et al. Direct evidence for an expanded circulation area of the recently identified Balkan virus (Sandfly fever Naples virus species) in several countries of the Balkan archipelago. Parasit. Vectors. 2017;10:402. doi: 10.1186/s13071-017-2334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corradetti A. Phlebotomus and leishmaniasis in North-Central Italy (Apennine Region) Sci. Rep. Ist. Super. Sanità. 1962;2:103–109. [Google Scholar]
- 25.Vásquez Trujillo A., González Reina A.E., Góngora Orjuela A., Prieto Suárez E., Palomares J.E., Buitrago Alvarez L.S. Seasonal variation and natural infection of Lutzomyia antunesi (Diptera: Psychodidae: Phlebotominae), an endemic species in the Orinoquia region of Colombia. Mem. Inst. Oswaldo Cruz. 2013;108:463–469. doi: 10.1590/S0074-0276108042013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duque P.L., Arrivillaga-Henríquez J., Enríquez S., Ron-Garrid L., Benítez W., Navarro J.C. Spatial-Temporal Analysis of Lutzomyia trapidoi and Lutzomyia reburra (Diptera: Phlebotominae), in Rural Tourist Locations, Biosphere Reserve and Leishmaniasis Endemic Area, Ecuador. J. Med. Entomol. 2020;57:1905–1912. doi: 10.1093/jme/tjaa102. [DOI] [PubMed] [Google Scholar]
- 27.Morini S., Calzolari M., Rossini G., Pascarelli N., Porcellini A., Randi V., Re M.C., Albieri A., Bonilaur P., Bellini R., et al. Detection of Specific Antibodies against Toscana Virus among Blood Donors in Northeastern Italy and Correlation with Sand Fly Abundance in 2014. Microorganisms. 2020;8:145. doi: 10.3390/microorganisms8020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattivi A., Massimiliani E., Cagarell R., Albieri A. Leishmaniosi In Emilia-Romagna Aggiornamento epidemiologico 1999–2015. Report of Emilia-Romagna Region 2016. [(accessed on 10 May 2021)]; Available online: https://salute.regione.emilia-romagna.it/normativa-e-documentazione/rapporti/malattie-infettive.
- 29.Rodhain F., Madulo-Leblond G., Hannoun C., Tesh R.B. Le virus corfou: Un nouveau Phlebovirusisolé de phlébotomes en Grèce. Ann. L’institut Pasteur Virol. 1985;136:161–166. doi: 10.1016/S0769-2617(85)80042-3. [DOI] [Google Scholar]
- 30.Remoli M.E., Fortuna C., Marchi A., Bucci P., Argentini C., Bongiorno G., Maroli M., Gradoni L., Gramiccia M., Ciufolini M.G. Viral isolates of a novel putative phlebovirus in the Marche Region of Italy. Am. J. Trop. Med. Hyg. 2014;90:760–763. doi: 10.4269/ajtmh.13-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Percivalle E., Cassaniti I., Calzolari M., Lelli D., Baldanti F. Thirteen Years of Phleboviruses Circulation in Lombardy, a Northern Italy Region. Viruses. 2021;13:209. doi: 10.3390/v13020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Briese T., Bird B., Kapoor V., Nichol S.T., Lipkin W.I. Batai and Ngari viruses: M segment reassortment and association with severe febrile disease outbreaks in East Africa. J. Virol. 2006;80:5627–5630. doi: 10.1128/JVI.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerrard S.R., Li L., Barrett A.D., Nichol S.T. Ngari virus is a Bunyamwera virus reassortant that can be associated with large outbreaks of hemorrhagic fever in Africa. J. Virol. 2004;78:8922–8926. doi: 10.1128/JVI.78.16.8922-8926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortalli M., De Pascali A.M., Longo S., Pascarelli N., Porcellini A., Ruggeri D., Randi V., Procopio A., Re M.C., Varani S. Asymptomatic Leishmania infantum infection in blood donors living in an endemic area northeastern Italy. J. Infect. 2020;80:116–120. doi: 10.1016/j.jinf.2019.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Bichaud L., Souris M., Mary C., Ninove L., Thirion L., Piarroux R.P., Piarroux R., De Lamballerie X., Charrel R.N. Epidemiologic relationship between Toscana virus infection and Leishmania infantum due to common exposure to Phlebotomus perniciosus sandfly vector. PLoS Negl. Trop. Dis. 2011;5:e1328. doi: 10.1371/journal.pntd.0001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faucher B., Bichaud L., Charrel R., Mary C., Izri A., de Lamballerie X., Piarroux R. Presence of sandflies infected with Leishmania infantum and Massilia virus in the Marseille urban area. Clin. Microbiol. Infect. 2014;20:340–343. doi: 10.1111/1469-0691.12404. [DOI] [PubMed] [Google Scholar]
- 37.Ergunay K., Kasap O.E., Orsten S., Oter K., Gunay F., Yoldar A.Z., Dincer E., Alten B., Ozkul A. Phlebovirus and Leishmania detection in sandflies from eastern Thrace and northern Cyprus. Parasit. Vectors. 2014;7:575. doi: 10.1186/s13071-014-0575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fares W., Dachraoui K., Barhoumi W., Cherni S., Chelbi I., Zhioua E. Co-circulation of Toscana virus and Leishmania infantum in a focus of zoonotic visceral leishmaniasis from Central Tunisia. Acta Trop. 2020;204:105342. doi: 10.1016/j.actatropica.2020.105342. [DOI] [PubMed] [Google Scholar]
- 39.Rath C.T., Schnellrath L.C., Damaso C.R., de Arruda L.B., Vasconcelos P.F.D.C., Gomes C., Laurenti M.D., Calegari T.C., Vivarini Á.C., Fasel N., et al. Amazonian Phlebovirus (Bunyaviridae) potentiates the infection of Leishmania (Leishmania) amazonensis: Role of the PKR/IFN1/IL-10 axis. PLoS Negl. Trop. Dis. 2019;13:e0007500. doi: 10.1371/journal.pntd.0007500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fermo virus sequences are available at ENA (https://www.ebi.ac.uk/ena/browser/home accessed on 10 April 2021), other data are available on request from the corresponding author.