Abstract
Late assembly (L) domains are conserved sequences that are necessary for the late steps of viral replication, acting like cellular adaptors to engage the ESCRT membrane fission machinery that promote virion release. These short sequences, whose mutation or deletion produce the accumulation of immature virions at the plasma membrane, were firstly identified within retroviral Gag precursors, and in a further step, also in structural proteins of many other enveloped RNA viruses including arenaviruses, filoviruses, rhabdoviruses, reoviruses, and paramyxoviruses. Three classes of L domains have been identified thus far (PT/SAP, YPXnL/LXXLF, and PPxY), even if it has recently been suggested that other motifs could act as L domains. Here, we summarize the current state of knowledge of the different types of L domains and their cellular partners in the budding events of RNA viruses, with a particular focus on retroviruses.
Keywords: viral assembly, budding, late assembly domains, retrovirus, RNA enveloped viruses, retroviral gag precursor, structural viral proteins, ubiquitination, ESCRT machinery
1. Introduction
Viruses are ubiquitous obligate intracellular parasites multiplying only within living cells by usurping the cellular machinery of their host to produce progeny virions. They are composed of extremely diverse nucleoprotein entities, classified into DNA or RNA viruses according to the type of nucleic acid constituting their genome. Even though DNA and RNA viruses are likely to infect both prokaryotes and eukaryotes, the two different classes are not found with the same abundance. Indeed, while the prokaryotic virosphere is dominated by DNA viruses (especially with double-stranded DNA genome), eukaryotes are rather found hosting a wide variety of RNA viruses (for a review, see [1,2]). A considerable number of those RNA viruses are responsible for severe human diseases, and thus represent a serious threat to global health. Since pathogenic RNA viruses are widely involved in zoonoses and can quickly adapt to the new host, they are considered as major etiological agents of emerging infectious diseases in humans (for a review, see [3]). Indeed the pandemic caused by influenza A (H1N1) in 1918, which led to more than 50 million deaths worldwide, was followed during this last century by many other epidemics and pandemics caused by RNA viruses, that include human immunodeficiency virus (HIV-1), identified in 1983 as the causative agent of the acquired immunodeficiency syndrome (AIDS), which has caused an estimated 34.7 million deaths according to UNIAIDS [4]; ebolaviruses, causing systemic disease with high lethality rate; and also coronaviruses, associated with severe respiratory illness, such as the severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome coronavirus (MERS). Most recently the SARS coronavirus 2 (SARS-CoV-2) emerged in 2019, having infected more than 178 million people by mid-June 2021 and continuing to spread around the world. In this context, the surveillance and the biological study of RNA viruses are essential tools to control their spread and the diseases they cause.
Viral dissemination relies on an extracellular phase in the virus life cycle. Indeed, the progeny virions formed in infected cells must be released and enter new target cells, therefore overcoming the barriers formed by cellular membranes and cytoskeleton. To this aim, viruses use three main strategies for egress: cell lysis, budding, and exocytosis. Non-enveloped RNA viruses (also termed as “naked viruses”) are generally thought to exit infected cells by lysis, thereby causing disruption of cellular membranes. However, there are some exceptions to this rule, and it has been demonstrated that viruses such as hepatitis A virus, poliovirus, rotavirus, and norovirus can exit cells non-lytically within vesicles, thus enclosing themselves into host-derived membranes [5,6,7]. As this feature is usually considered as the major difference between enveloped and non-enveloped viruses, viruses using this strategy for cellular egress are sometimes defined as “quasi-enveloped” (see Section 7). The budding process is commonly employed by enveloped RNA viruses that derive their envelopes enriched by viral glycoproteins, directly from the host. Budding of many RNA viruses, including retroviruses, orthomyxoviruses, filoviruses, alphaviruses, and rhabdoviruses, occur at the plasma membrane (PM), and in this case, progeny virions are directly released in the extracellular environment. For other enveloped RNA viruses, including coronaviruses, flaviviruses, and bunyaviruses, budding occurs at intracellular membranes into the lumen of organelles such as the endoplasmic reticulum (ER), the Golgi apparatus, the ER–Golgi intermediate compartment (ERGIC), and the endosomes. In this case, virus egress relies on the cellular secretory pathway, and the viral budding process is mechanistically equivalent at the PM and at cellular organelles, since in both cases it implies the deformation of the membrane during the viral particle envelopment process. This event is then followed by membrane fission, thus allowing the release of viral particles and the generation of two distinct cellular and viral membranes. Importantly, membrane fission is achieved by hijacking the endosomal sorting complexes required for transport (ESCRT) pathway (for a review, see [8] and Section 6). These cellular factors comprise three subcomplexes (ESCRT-I, ESCRT-II, ESCRT-III), as well as the ESCRT-III-associated ALG-2-interacting protein X (ALIX) and the ATPase vacuolar protein sorting-associated 4 (VSP4). The early acting ESCRT complexes (ESCRT-I and ESCRT-II) assemble stably within the cytoplasm and can associate with adaptor proteins such as the HRS/STAM complex (also named ESCRT-0) to recruit and activate late-acting ESCRT-III and VSP4 factors at specific membrane sites where the assembling virions and fission events occur (for reviews, see [9,10,11]). Viral assembly and the recruitment of the ESCRT machinery must thus be coordinated in space and time, and it is therefore not surprising that these steps are finely orchestrated by viral structural proteins. Indeed, these multifunctional proteins were found to recruit the ESCRT machinery at viral budding sites via conserved motifs, termed late assembly domains (L domains) that act like cellular adaptor proteins (for a review, see [12]). Historically L domains were first identified within Gag (group-specific antigen) structural precursors of retroviruses (Figure 1) as HIV-1, Rous sarcoma virus (RSV) and equine infectious anemia virus (EIAV), HIV-2, feline immunodeficiency virus (FIV), human T cell leukemia virus type 1 (HTLV-1), murine leukemia virus (MLV), porcine endogenous retrovirus (PERV), human endogenous retrovirus-K (HML-2), Mason–Pfizer monkey virus (M-PMV), and prototypic foamy viruses (PFV) [13,14,15,16,17,18,19,20,21,22,23,24,25]. The different retroviral Gag proteins are ubiquitinated, and importantly, ubiquitin was observed to be linked to the budding machinery (see Section 6) [26,27,28,29].
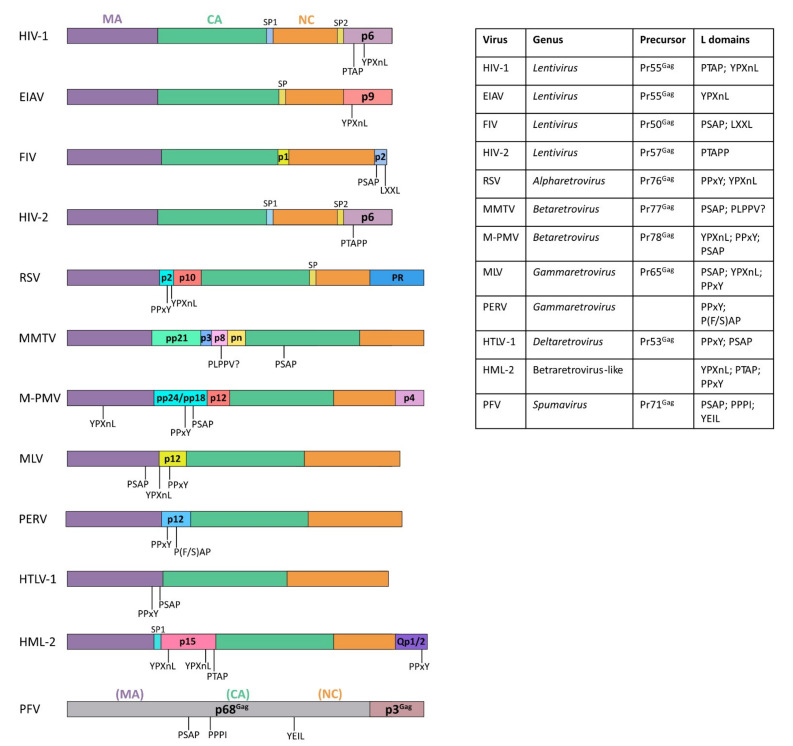
Figure 1.
Retroviral Gag organization domains. All retroviruses display matrix domain (MA, in violet), capsid (CA, in green), and nucleocapsid (NC, in orange). Spacer peptides (SP) and peptide sequences containing L domains are indicated within structural precursors of HIV-1, EIAV, FIV, HIV-2, RSV, MMTV, M-PMV, MLV, PERV, HTLV-I, HML-2, and PFV.
In a further step, L domains were also found in structural proteins of many other RNA enveloped viruses, including arenaviruses, filoviruses, rhabdoviruses, reoviruses, and paramyxoviruses (Figure 2).
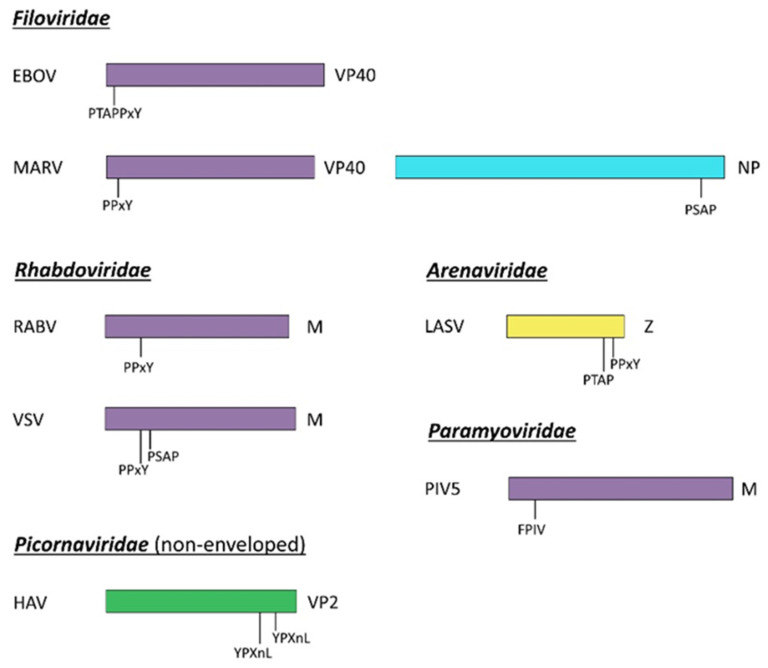
Figure 2.
L domains in the structural proteins of RNA viruses including filoviruses (EBOV and MARV), rhabdoviruses (RABV and VSV), arenaviruses (LASV), paramyoviruses (PIV5), and picornavirus (HAV).
Of note, some enveloped RNA viruses such as influenza A [30,31] were found to not rely on L domains, and thus cellular egress is regulated by ESCRT-independent budding mechanisms, as extensively described in [32].
This review relates the history of L domains discovery and aims to provide a current state of knowledge concerning the function of L domains and their cellular partners involved in the budding events of RNA viruses, with a particular focus on retroviruses. Considering the importance of budding process in the viral life cycle, a better comprehension of molecular mechanisms driving this step would constitute an asset to the development of new antiviral drugs that could be effective against multiple viruses.
2. Identification of L Domains within HIV-1 p6 Domain of Pr55Gag Precursor
All retroviruses express structural Gag polyproteins that drive the assembly and the release of virus-like particles (VLPs), even in the absence of other viral components (for reviews, see [33,34]), and all retroviral Gag precursors contain the three structural domains of matrix (MA), capsid (CA), and nucleocapsid (NC), each of them playing an essential role in the retroviral particle assembly and in budding processes. Gag usually also carry additional domains and spacer peptides such as, for example, the HIV-1 p6 domain, as well as SP1 and SP2 linkers (for reviews, see [34,35,36,37]) (Figure 3).
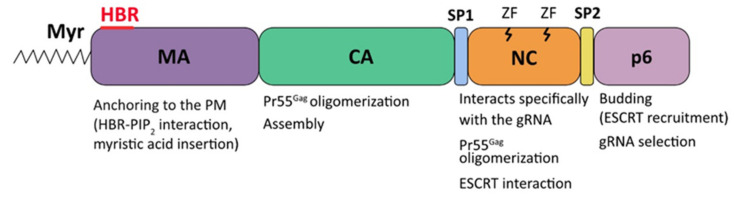
Figure 3.
Schematic representation of the domains in HIV-1 Pr55Gag precursor. The myristoyl moiety is indicated at the N-terminus of the precursor. The MA domain drives the interaction between the Pr55Gag and the PM through a bipartite signal consisting of a HBR domain and the covalently attached myristic acid moiety. The CA domain of Pr55Gag mediates Pr55Gag oligomerization and ensures formation of the core of the mature viral particles, and the NC domain, which contains two zinc-finger motifs, corresponds to the primary binding motif to nucleic acids, and contributes to the Pr55Gag multimerization. The p6 domain contributes to the specificity of Pr55Gag–gRNA interactions and is essential for viral budding. Pr55Gag contains also two spacer peptides which are SP1 and SP2.
Amongst retroviral precursors, the role of the different domains of HIV-1 Gag (also named Pr55Gag) in the encapsidation of gRNA into the viral particles was extensively characterized [34,35,36,37,38,39] (Figure 3). The HIV-1 assembly process occurs at the PM, where about 2000 copies of Pr55Gag accumulate to produce immature particles. The targeting of Pr55Gag polyproteins to lipid rafts at the PM [40] is driven by the MA domain at the N-terminus of the precursor via a bipartite signal. The latter is composed by a myristic acid moiety, and a highly basic region (HBR) located on the surface of MA domain, which forms electrostatic interactions with negatively charged phosphatidylinositol-(4,5)-bisphosphate (PIP2) that are specifically enriched at the inner leaflet of PM [41]. The myristic acid is initially sequestered within the MA domain to prevent unspecific interactions with cellular internal membranes. In a further step, the interactions between the HBR and PIP2 at PM, in parallel with Pr55Gag-Pr55Gag interactions, lead to the exposure of the myristic acid moiety [42,43,44], which is then inserted into the PM, thus allowing the anchoring of the precursor by hydrophobic interactions. The CA domain mediates the oligomerization of Pr55Gag and drives the immature particle assembly process. Then, the SP1 spacer, which is located between CA and NC domains, contributes to Pr55Gag multimerization and acts as a molecular switch to initiate the assembly of immature VLPs [45,46,47,48]. The NC domain contains two zinc-finger motifs (ZFs) and interacts specifically with the dimeric viral RNA genome (gRNA) to ensure its encapsidation within the virions [49,50,51,52,53,54,55], and the SP2 domain that separates the NC and the C-terminus p6 domain plays a role in Pr55Gag processing, even though the deletion of this domain has only a minor effect on viral infectivity, and did not induce major alterations in mature core morphology [56]. Finally, the p6 domain seems to mediate specific interactions occurring between Pr55Gag and gRNA [57] and regulates the budding of viral particles at the PM by recruiting the cellular factors of the ESCRT pathway. Indeed, mutations in this domain completely inhibited the budding of virions from the PM of COS-7 cells [13]. In particular, truncated forms of Pr55Gag lacking the p6 domain were found to be able to assemble into VLP, even though, as shown by electron microscopy, they are not released into extracellular medium and remain attached to the cell membrane by a thin tether [13]. All together, these observations provided the first evidence that retroviruses code for regions specifically required for virus release. Thereafter, extensive mutagenesis analyses of the p6 domain showed that a PTAP motif is crucial for the release of viral particles [58,59]. Subsequently, P(T/S)AP motifs (where the second residue can be either a Ser or a Thr) were also identified within structural proteins of others retroviruses, filoviruses, rhabdoviruses, and arenaviruses (for a list, see [8]), and at least two other conserved motifs (PPxY and YPXnL) displaying similar functions in efficient viral budding were identified within retroviral Gag proteins, as a number of studies have demonstrated their interactions with components of the ESCRT machinery (for a review, see [60]). All these short sequences were thus termed “L domains” to emphasize their role in the late phases of the assembly process during the separation of virions from the cell membrane.
In the specific HIV-1 context, both PTAP and YPXnL motifs within the p6 domain were observed to promote the viral egress by interacting with the ESCRT machinery, as the PTAP domain was found to be bound by the ESCRT-I tumor suppressor gene 101 (TSG101) [61] and the YPXnL domain by the ESCRT-III associated ALIX protein [62] (see Section 6.1 and Section 6.2).
In Pr55Gag, in addition to p6, the NC domain was also found to play a role in viral release as the latter participates in the recruitment of the ESCRT cellular proteins necessary for PTAP- and YPX(n)L-mediated budding [63]. Indeed, the deletion of the NC domain decreased in cells the amount of the complexes formed by Pr55Gag and TSG101 [64]. Similarly, mutation of NC resulted in the release of DNA-containing viruses, which was the consequence of budding defects mainly associated with deficiency in the recruitment of TSG101 [65]. Moreover, the interactions between NC and the ESCRT-III-associated factor ALIX were found to be promoted by ZFs, as mutations of these motifs resulted in defective production of viral particles [66]. Interestingly, TSG101 was found to substitute for the distal ZnF2, since chimeric Gag-ΔZF2-TSG101 were found to rescue budding [67], thus confirming the role of those motifs in the recruitment of ESCRT machinery.
3. Identification of the PPxY L Domain within RSV p2b Domain of Pr76Gag
Mutagenesis analysis on the Gag polyprotein (Pr76Gag) of the avian alpha-retrovirus RSV revealed that the p2b spacer peptide located between the MA and CA domains is necessary for the release of viral particles [14] (Figure 1). In line with this idea, truncation of the p2b peptide caused severe release defects, showing a phenotype analogous to what was observed for HIV-1 p6 mutants in which viral budding was arrested at the last stage [13]. The function of p2b in virus release was then attributed to a Pro-rich L domain that is located between the MA and the CA domains. This L domain was later finely mapped to the highly conserved amino acid sequence PPPPYV, and defects caused by its deletion can be rescued by its replacement at several other positions in RSV Gag [68]. Moreover, this RSV L domain was observed to specifically interact with the E3 ubiquitin protein ligases NEDD4-like family ([69], and see Section 6.3). In a further step, these PPxY L domains (where x could be any residue, even if it is often a Pro) were also found in several other viruses [8], except for Gag proteins of lentiviruses including HIV-1 [14]. Interestingly, by using RSV Gag chimeras, researchers showed that the L domain activity carried by p2b could be functionally replaced with the C-terminal HIV-1 p6 sequence, and these domains could function independently of their position in the precursor since they can be located either internally or at the C-terminus [15]. Thus, these findings suggested for the first time that, despite their differences, L domains share the same function as they both ensure the release of viral particles.
4. Identification of the YPXnL L Domain within the EIAV p9 Domain of Pr55Gag
Similarly to the HIV-1 p6 domain, the p9 domain located at the C-terminus of the EIAV Gag protein (named EIAV Pr55Gag) (Figure 1) was found to rescue the RSV viral particle release defect induced by p2b deletion, thus suggesting the presence of a L domain in p9 [15]. Interestingly, unlike other lentiviruses, the EIAV p9 domain does not contain a P(T/S)AP L motif, and moreover, no sequence homology between EIAV p9 and its functional homologues RSV p2b and HIV-1 p6 was identified. Thereafter, mutagenesis analysis revealed that substitutions of each residue Y23, P24, or L26 with A within p9 abolished viral release, while other mutations within the same domain did not display any effect on this step. This analysis demonstrated that the p9 L domain activity is supported by a YPXL motif [16], which was then identified in several other viruses [8] in the more generic YPXnL form (where Xn can vary in amino acid sequence and length). Remarkably, budding assay of EIAV/HIV-1 and EIAV/RSV chimeric Gag polyprotein demonstrated that p6 and p2b, respectively, can replace the EIAV p9 domain. These data, combined with similar observations on the substitution of the p2b domain of RSV precursor by p6 and p9 domains [15], confirmed the notion these L domains are mostly interchangeable.
5. Functional Exchangeability and Multiplicity of L Domains
The functional exchangeability of L domains has indeed been widely demonstrated in several viral contexts [15,16,70,71,72,73,74]. In addition to HIV-1, EIAV, and RSV, similar experiments have also been carried out with gamma-retrovirus. The MLV Gag polyprotein (Pr65Gag) (Figure 1) indeed contains a PPPY L motif in its p12 domain, located between the MA and CA domains, similarly to RSV. Interestingly, in a PPPY-defective MLV Pr65Gag protein, the insertion of the p12 domain containing the PPPY motif and its flanking sequences at different positions within the precursor can partially or completely restore the release of viral particles. Moreover, in these same PPYP-deficient Pr65Gag, insertion of the RSV PPYP L domain or the HIV-1 PTAP domain within the MA domain of MLV Pr65Gag was found to rescue the assembly defects [70]. In addition, L domains were found to be functionally exchangeable also with viruses outside the Retroviridae family. The first description of L domains and their function in other viral families was reported in Rhabdovirus [71], where a PPxY conserved motif with analogous properties to those observed for the L domain in RSV p2b, was identified in matrix (M) of Rabbies virus and of vesicular stomatitis virus (VSV) [75] (Figure 2). Notably, the N-terminal part of the VSV M protein, which includes the PPxY motif, was found to replace the MA-p2b region of RSV Pr76Gag, and since mutations in this motif severely impaired the budding of the M-Gag chimera, this PPxY domain was assigned as the VSV L domain [71]. Similarly, the L domain of Ebola virus (EBOV), which consists in an overlapping of P(T/S)AP and PPxY motifs within the viral VP40 protein (Figure 2), can restore the budding of a minimal assembly system in which HIV-1 Gag lacks the natural NC-p1 and p6 domains [76].
Overall, the regulation of viral release mediated by L domains is highly conserved among different groups of viruses, and new motifs acting as L domains were recently identified using the ability of the L domains to be mostly functionally interchangeable. That was, for example, the case of the FPIV motif identified within the M protein of several paramyxoviruses (such as the parainfluenza Virus 5 (PIV5), the mumps virus (MV), and the Newcastle disease virus (NDV)), which was observed to be necessary for viral budding [72,77,78]. Interestingly, when fused to the C-terminal end of HIV-1 Gag proteins lacking the PTAP L domain, the PIV5 FPIV motif was able to partially rescue the budding defect induced by the lack of PTAP [72]. However, mutagenesis experiments suggested a more general sequence O-P-x-V, where the Pro residue was found to be particularly critical for function as its substitution led to poor budding [72]. More recently, a PLPPV motif within the p8 domain of the mouse mammary tumor virus (MMTV) Gag protein (Pr77Gag) has been described as a fourth type of retroviral L domain, which can restore the budding defect of EIAV induced by the deletion of the YPXnL domain within the precursor [73]. However, whether this motif is effectively a new type of retroviral L domain is yet to be confirmed, and its precise role in viral budding remains to be fully characterized. Finally, the LXXL motif, which is highly conserved in p2 domain of FIV Pr50Gag precursor (Figure 1), is known to participate to ALIX-mediated release and to interact with clathrin adaptor proteins for budding [18,79,80].
While some viruses such as EIAV rely on one single L domain to ensure their budding, retroviruses such as FIV display two L domains (PSAP and LXXL) within their retroviral precursor. This is indeed a common feature amongst viruses that frequently use a combination of multiple L domains to promote virions egress. For instance, the MLV Gag polyprotein contains three putative late domains (PSAP, LYPAL, and PPPY) located in the MA domain and in p12 (Figure 1) [20]. Similarly, the HIV-1 p6 domain of Pr55Gag contains also a YPXnL motif, which is located downstream to the PTAP domain [62,81]. However, if the PTAP motif is conserved in all subtypes, the HIV-1 subtype C (HIV-1C) naturally lacks the YPXnL L domain [82]. Interestingly, in HIV-1C-infected patients in therapy failure, a tetra-peptide PYXE (where X is either Arg, Lys, or Gln) was found to replace YPXnL [83]. In this context, this sequence was found to fulfill the role of YPXnL in viral budding in order to increase the replication fitness, as well as to decrease the sensitivity to certain antiretroviral drugs such as the protease inhibitor lopinavir [84,85]. Moreover, viruses such as M-PMV, HTLV-I, EBOV, marburg virus (MARV), and Lassa fever virus (LASV) contain both PPxY and P(T/S)AP L domains (Figure 2) [86,87,88,89,90,91], while others such as RSV contain both YPXnL and PPxY L domains [92]. In the VP40 (or M) protein of EBOV, both domains are required for efficient budding. Interestingly, when these same domains were transplanted in MLV, the two motifs overall contributed to viral budding, while in the HIV-1 context, the PPxY-type L domains were mostly inactive, and L-domain activity was entirely due to PTAP. These context-dependent effects of L-domain function may reflect requirements for distinct host-driven functions during the viral assembly [76,88,93]. Moreover, multiple late domains typically exhibit a hierarchy of importance, considering that mutations in one of the L domains display a more deleterious effect on budding than the other. For instance, in HTLV-1, the viral precursor Pr53Gag carries both PPPY and PTAP L domains (Figure 1), and if PPPY motif cannot be substituted with PTAP or YPXnL domains without affecting viral budding, the replacement of the PTAP domain with either the PPPY or YPXnL motifs has no influence on the release of viral particles [88]. Nevertheless, mutations of either PPPY or PTAP L domains with APPY or PTRP motifs, respectively, impaired viral budding, demonstrating that these L domains do not display redundancy and both play an active role in virus release [88]. Indeed, electron microscopy analysis revealed that mutations in the PPPY L domain of HTLV-1 or of M-PMV cause early budding defects with consequent viral particle accumulation underneath the PM without membrane curvature induction [86,94], while the mutation of P(T/S)AP blocks the release of viruses at a later step [86,95]. Similarly, three different motifs, namely, P(T/S)AP, PPxY, and YPX(n)L, were proposed to play a role in the release of human exogenous retrovirus-K (HML-2). However, further analysis clarified that viral release was predominantly found to be mediated by PTAP motif, as well as by two auxiliary YPX(n)L motifs in the p15 domain of the Gag precursor (Figure 1) [22]. The notion of a hierarchy in L domains function was also supported by the case of the porcine endogenous retrovirus (PERV). Indeed even though its Gag protein contains the two L domains PPPY and P(F/S)AP (Figure 1), the dominant L domain involved in PERV release was found to be PPPY [21]. Taken together, these finding indicate that L domains exhibit distinct functions for viral egress. Of note is the case of the PFV, for which three potential L-domain motifs (PSAP, PPPI, and YEIL) were proposed, considering the homology of these sequences with other viral L domains (Figure 1) [25]. Mutation of the PSAP domain suggested that this domain corresponds to a conventional L domain, and two hybrid analyses revealed that the interaction between PFV Gag and TSG101 is indeed mediated by this sequence. In contrast, the inactivation of PPPI suggested an unconventional mechanism to facilitate PFV egress [25], and mutant viral particles displayed reduced infectivity. Similarly, mutation of the conserved YEIL motif revealed no classical L domain function but resulted in a reduced rate of Gag processing by the viral protease, as well as in the release of noninfectious VLPs [96]. Finally, viruses possessing multiple L domains can also change the L-domain usage to replicate in various cells as M-PMV. Indeed, it was found that the PPxY constitutes a major L-domain in several cell lines, while the PSAP sequence was shown to function as a L-domain in HeLa cells, and it is thought to be dispensable for viral production in 293T and COS-7 cells [23].
6. Interplay between L Domains and ESCRT Machinery
Historically, the first in vitro and in vivo evidence that L domains within retroviral Gag interact with host factors involved in the ubiquination machinery were provided by Leis and Carter and Leis labs [61,69,97], thus indicating an important link between this cellular pathway and retroviral budding. Indeed, the NEDD4-like family of E3 ubiquitin protein ligases was found to specifically interact with the RSV PPPPYV L domains [69,97], while the HIV-1 PTAPP L domain binds the homologue of ubiquitin-conjugating (E2) enzyme TSG101 [61]. Viral particle release was then found to be ensured by the specific interaction between L domains and the ESCRT machinery, which is known to remodel membrane. ESCRT proteins have been originally characterized in yeast for their role in multivesicular bodies (MVBs) biogenesis (for a review, see [98]) and are functionally conserved throughout several archaeal species and eukaryotes [99]. In metazoans, it is now well established that ESCRT proteins, beyond their role in the biogenesis of MVBs and in cytokinetic abscission [100,101], are also involved in many remodeling processes, including vesicle and virus budding from PM, endolysosomal membrane and PM repair, neuronal pruning, nuclear envelope maintenance, and autophagy (for a review, see [9]). In humans, the core of the ESCRT machinery is composed of the stalk-shaped hetero-tetramer ESCRT-I (TSG101, VPS28, VPS37A-D, and MVB12/UBAP1), the Y-shaped hetero-tetramer ESCRT-II (two copies of EAP20, one copy of EAP30 and EAP45), the ESCRT-III complex formed by IST1 and the charged multivesicular body proteins (CHMP) 1 to 7 (CHMP1A/B, CHMP2A/B, CHMP3, CHMP4A-C, CHMP5, CHMP6, and CHMP7), the ESCRT-associated protein ALIX, and the AAA ATPase VPS4 (for a detailed list of the homolog components of the ESCRT machinery in yeast, see [10]). The recruitment of the ESCRT factors to their proper sites of action at membranes is enabled by targeting adapter proteins including the HRS/STAM complex (also named ESCRT-0), the centrosomal protein 55 (CEP55), LEM domain 2 protein (LEMD2), and arrestin domain-containing protein 1 (ARRDC1) (for a review, see [9]). Remarkably, P(T/S)AP motifs are used by several of these adapter proteins, including the HRS subunit of ESCRT-0 [102] and ARRDC1 [103], to recruit the ESCRT machinery via the ubiquitin E2 variant (UEV) domain of TSG101. Similarly, the P(T/S)AP L domain in HIV-1 and EBOV recruits the ESCRT-I factor TSG101 at viral assembly sites by direct binding to its UEV domain [61,104,105,106,107]. Moreover, the expression of an N-terminal fragment of TSG101 containing UEV was found to inhibit HIV-1 and FIV release, and in this sense, the FIV PSAP motif behaves similarly to the PTAP motif of HIV-1 [108]. Interestingly, a PTAP motif duplication reported in some HIV-1-infected patients [109,110,111] was associated with an enhanced interaction with TSG101 [112], as well as with an increased retroviral replication fitness and a decreased susceptibility for protease inhibitors [84,112,113]. Other L domains such as the HIV-1 YPXnL recruits the ESCRT-III-associated factor ALIX by direct binding to its V domain [62,79,114,115], while the HTLV-1 PPxY motif binds to the WW domain of members of the HECT E3 ubiquitin ligases NEDD4 family (see Section 6.3) [69,116] (Figure 4). Moreover, if the ESCRT-I and ESCRT-II complexes constitute early acting factors that coordinate ESCRT-III recruitment and membrane curvature, the ESCRT-III polymerization and depolymerization are thought to be the main driving force for membrane remodeling and fission [117,118,119,120,121,122], although the exact mechanism by which these components mediate this process is still unclear. In cytoplasm, ESCRT-III proteins exist in a monomeric closed autoinhibited state [123,124,125] that is overcome by binding with the upstream ESCRT components [126,127,128]. This leads to the membrane binding and to interaction with the VPS4 ATPase that plays a key role in ESCRT-III-mediated membrane scission by remodeling and disassembling ESCRT-III filaments [117,119,129,130,131]. Therefore, VPS4, together with the ESCRT-III complex, corresponds to late acting factors [9] that are usurped by viruses for viral egress.
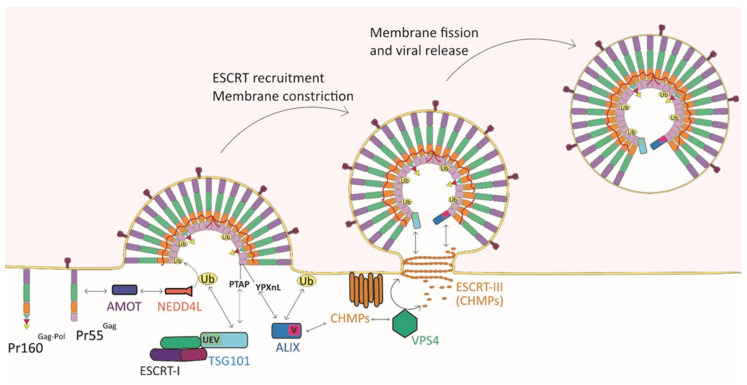
Figure 4.
The recruitment of the ESCRT cellular factors by L domains in HIV Pr55Gag ensures viral particles release. The cellular factor angiomotin (AMOT) promotes the recruitment of the ubiquitin ligase NEDD4L at HIV-1 budding sites. The ESCRT machinery is then recruited by NEDD4 family, and this interaction seems to be related to the ubiquitination of viral structural proteins. The P(T/S)AP L domain of Pr55Gag recruits the ESCRT-I factor TSG101 at viral assembly sites by direct binding to its ubiquitin E2 variant (UEV) domain, and the YPXnL motif recruits the ESCRT-III-associated factor ALIX by binding to its V domain. The ESCRT-III proteins drive the interaction with the VPS4 ATPase. This late-acting factor leads to membrane remodeling and its fission, and it drives the disassembling of the ESCRT-III filaments. The process ends with the release of the newly formed viral particle.
6.1. L Domain Interaction with the ESCRT-I
The role of ESCRT-I in viral egress was exhaustively characterized in the case of HIV-1, and the interaction between two ESCRT-I factors such as the C-terminal domain of TSG101 and the N-terminal domain of VPS28 [132] is thought to be essential to ensure HIV-1 budding. Indeed, fusion of TSG101 mutated in its VPS28 binding domain to an HIV-1 Gag protein deleted of the p6 domain (also named Gag∆p6) [133], as well as the same TSG101 mutants expressed in cells depleted of endogenous TSG101 [134], failed to rescue viral budding. Conversely, fusion of TSG101, VPS37B, or VPS37C to the C-terminus of Gag∆p6 efficiently rescued VLP budding, despite L domain truncation [106,134,135]. In addition, knockdown experiments of TSG101 using siRNAs led to a defective HIV-1 budding, which can be rescued by TSG101 trans-complementation [104], while the overexpression of the UEV domain of TSG101 inhibited HIV-1 release [105]. The ESCRT-I UBAP1 factor was also found to promote HIV-1 budding when fused to the C-terminus of a Gag protein lacking L domains, although its depletion by siRNAs did not impact on virus release [136]. Similarly, depletion or overexpression of the ESCRT-I MVB12 had no direct effect on virus egress, even if the infectivity and virion morphogenesis resulted to be defective [137]. Furthermore, depletion of ESCRT-II had little effect on HIV-1 release but blocked the ASV (avian sarcoma virus) budding, and conversely, depletion of ESCRT-I had little effect on ASV release but blocked HIV-1 budding, suggesting that ASV and HIV-1 Gag proteins use different combinations of ESCRT proteins to promote budding [74]. Similarly, RSV and HIV-1 Gag particle release is achieved through independent ESCRT-mediated pathways [138], even though they are linked through TSG101–NEDD4 interaction. However, while the importance of ESCRT-I factors in HIV-1 budding is well established, the role of ESCRT-II factors is yet to be clarified [74,139,140,141,142], and further investigations will then be necessary to fully understand their function in HIV-1 cycle. In the context of HIV-2, TSG101 and Gag precursor (Pr57Gag) were also found to interact in vitro and ex vivo via the PTAPP motif in the p6 domain of the precursor (Figure 1) and via the N-terminal Ubc-conjugation homology domain of TSG101. Moreover, the overexpression of TSG101 resulted in an increased level of ubiquitination of Pr57Gag, thus indicating the involvement of the cell ubiquitination machinery during the late stages of the viral life cycle [17]. A PSAP L-domain binding TSG101 was also identified in the p2 domain of the FIV Pr50Gag precursor (Figure 1). Interestingly, the Bro1 domain of human ALIX was found to rescue FIV mutants lacking TSG101-interacting motif. However, in contrast to HIV-1, mutations of ZFs in NC domain did not impair FIV rescue, suggesting conserved and divergent mechanisms employed by lentiviruses to achieve viral budding [19].
6.2. L Domain Interaction with ALIX
The role of ESCRT-III-associated ALIX protein in YPXnL-mediated viral budding was first determined in the EIAV context, where yeast two hybrid and GST pull down assays showed that the p9 domain of EIAV directly interacts with ALIX, and that this interaction would depend on the YPDL sequence, as substitutions in this motif inhibited the interaction between p9 and ALIX [62,143]. Moreover, the overexpression of truncated ALIX in its Bro1 domain, or the depletion of endogenous ALIX using siRNA knockdown, inhibited p9-mediated EIAV budding. Similar effects were observed for HIV-1 constructs, where p6 domain was replaced by the EIAV p9 domain [143]. However, even though it has been well established that ALIX plays a crucial role in EIAV budding, its function in HIV-1 egress has been formerly considered as secondary. Indeed, the second L domain of HIV-1 (i.e., YPXnL) in p6 was found to provide an alternative pathway for viral egress by interacting with the V-domain of ALIX [62], which in turn leads to the recruitment of ESCRT-III at budding sites via a direct interaction between its Bro1 domain and the C-terminal domain of CHMP4 [114,144]. However, early findings suggested that this interaction plays a minor role in HIV-1 release in comparison to PTAP–TSG101 interaction. Moreover, in contrast to EIAV budding mediated by the p9 domain, depletion of ALIX with siRNA only slightly affected p6-mediated release of HIV-1 from P4/R5 cells, and the overexpression of ALIX truncated in its Bro1 domain did not drastically inhibit HIV-1 p6-mediated budding from P4/R5 or 293T cells [143]. In addition, deletion or mutations of the YPXnL L domain within p6 did not completely impaired HIV-1 budding from COS or HeLa cells [13,58,59,115]. On the other hand, overexpression of the V domain of ALIX considerably inhibited the release of HIV-1 and EIAV virions form HeLa or COS-1 cells [79,115,145], suggesting that HIV-1 release displays cell type dependence [59,146]. This notion was also supported by the fact that although the TSG101-P(T/S)AP interaction has been shown to play a more important role than the ALIX-YPXnL interaction in HIV-1 budding from various epithelial cell lines, YPXnL mutations were found to play a major role in Jurkat T cells [146], and similarly, in HAP1 cells, the YPXnL–ALIX interaction was found to mainly regulate HIV-1 particle production [141].
Moreover, the overexpression of ALIX was shown to rescue the budding of HIV-1 PTAP mutants, and this would depend on the interactions occurring between its Bro1 domain and CHMP4B [114,147]. Moreover, substitutions in the YPXnL motif that abrogates ALIX interaction with Pr55Gag are thought to impair the recruitment of the ESCRT-III machinery, presumably due to catastrophic disassembly of the ESCRT components [148]. Altogether, these findings show the implication of ALIX in the final stages of ESCRT-mediated budding in the HIV-1 context, supporting the idea that ALIX is recruited to the membrane neck at the end of viral particle assembly [149] and mainly acts as a scaffold for ESCRT-III polymerization during viral budding (for a review, see [150]).
As mentioned above, the HIV-1 subtype C (HIV-1C) naturally lacks the YPXnL L domain, and in this case, the overexpression of the V domain of ALIX did not affect viral particle release [82]. In this context, the PYXE domain that replaces the YPXnL motif in patients in therapy failure [83] was found to interact with ALIX, thus behaving as a L domain to enhance viral budding [84,85]. Similarly, precursors of some simian immunodeficiency virus (SIV) (such as the SIV rhesus macaque and the SIV African green monkeys) lack YPXnL motif, although they can bind ALIX. Indeed, crystal structures revealed that in this case anchoring Tyr and nearby hydrophobic residues lead to the interaction between the precursors and the V domain of ALIX, thus revealing how lentiviruses employ diverse sequences to bind ALIX to efficiently promote virus budding [151].
Finally, the suppression of ALIX and TSG101 mediated by RNA interference (RNAi) was also enabled to clarify the contribution of these factors in the production of a DNA virus, the herpes simplex virus type 1 (HSV-1). Indeed, these findings suggested the presence of alternative mechanisms to recruit the ESCRT-III proteins that provide the scission of the membrane and thus the release viral particles [152].
6.3. L Domain Interaction with E3 Ubiquitin Ligase NEDD4 Family and the Ubiquitination Machinery
The overexpression of the HECT ubiquitin E3 ligase NEDD4L was observed to stimulate the release of HIV-1 constructs lacking TSG101- and ALIX-binding L domains, increasing infectious titers over 20-fold [153]. Moreover, efficient M-PMV budding was found to be related to the recruitment of NEDD4-like proteins by a PPXY L domain [153], thus indicating an alternative pathway to recruit the ESCRT machinery at viral budding sites [69,116]. The recruitment of the ESCRT machinery by E3 ubiquitin ligases NEDD4 family is linked to the ubiquitination [61,69,154,155,156,157]. Indeed all retroviral Gag polyproteins, except Gag from spumaviruses [158], are ubiquitinated (for a review, see [159]), and ESCRT-0, ESCRT-I, and ESCRT-II recognize and interact with ubiquitylated cargos (for a review, see [160]). Moreover, NEDD4 interacts with the RSV PPxY L domain via its WW domain [69,161], and overexpression of WW domain impaired RSV budding [69], similarly to the mutations occurring in the HECT catalytic site [97]. Likewise, the PPxY motif of HTLV-1 Pr53Gag precursor interacts with the WW domains of several members of NEDD4 family (including NEDD4, WWP1, and ITCH) (for a review, see [161]). The recruitment of NEDD4 by the PPxY L domain was found to promote Pr53Gag ubiquitination, and at this level, TSG101 is then recruited by the second PTAP L domain, thereby ensuring viral budding [95]. Similarly, MLV release is promoted by the interaction occurring between WWP1 ubiquitin ligase and the viral PPxY L domain [116]. Furthermore, the depletion of free cellular ubiquitin by proteasome inhibitors resulted in an inhibited RSV egress, and importantly this phenotype was rescued by ubiquitin overexpression or by ubiquitin fusion to RSV [26]. Interestingly, in the EIAV context, the fusion of ubiquitin to the C-terminus of EIAV Pr55Gag precursor lacking the YPxnL L domain led to overcoming of the release defects [154], thus demonstrating that ubiquitin can functionally counterbalance the absence of retroviral L domains. All these findings confirm that ubiquitination is a part of retroviral budding machinery.
The interaction between PPxY and the WW domain in HECT E3 ubiquitin ligases also regulates the budding of several viruses outside the Retroviridae, as for instance rhabdoviruses and filoviruses [162]. These viruses possess a PPxY L domain in their matrix protein (M or VP40, respectively) that functionally interacts with the WW domains of E3 ubiquitin ligases [161,163,164,165,166]. It was shown that VP40 protein of EBOV interacts with WWP1, and its ubiquitination enhances viral budding. Moreover, WWP1 depletion by siRNA was observed to induce the inhibition of VLP budding [167]. Interestingly, although HIV-1 Pr55Gag does not contain a PPxY L domain, the overexpression of catalytically active NEDD4-2 (NEDD4L) E3 ubiquitin ligase can rescue the budding of an HIV-1 construct where both PTAP and YPXnL L motifs were deleted [153,168]. Furthermore, the recruitment of NEDD4L at HIV-1 budding sites was found to be promoted by the cellular factor angiomotin (AMOT) [169] (Figure 4). Finally, other cellular proteins were found to provide an alternative access to ESCRT-III recruitment and act as adaptors, such as, for example, the arrestin-related trafficking (ART) proteins [170].
7. Quasi-Enveloped RNA Viruses and the ESCRT Pathway
It was recently shown that several non-enveloped viruses such as hepatitis A virus (HAV), poliovirus, rotavirus, and norovirus [5,6,7] can be referred to as quasi-enveloped viruses since they enclose themselves in vesicles derived from host membranes and exit cells non-lytically (for a review, see [171]). Interestingly, several studies have indicated that the formation of quasi-enveloped HAV virions (eHAV) also depends on the ESCRT components ALIX, VPS4, CHMP2, and IST1 [5,172,173,174]. Notably, one of the HAV capsid major structural proteins VP2 encodes two YPXnL motifs that interact with ALIX [5,172]. Mutational analysis revealed that single substitution of Leu for Ala within either YPXnL motifs resulted in reduction of eHAV release, and that dual substitutions of Leu for Ala in both motifs impaired eHAV budding, but not the assembly of infectious intracellular particles [172]. In those double mutants, the failure in eHAV releasing correlates with the loss of interaction between the capsid and ALIX [172], and similar budding defects were observed when ALIX was depleted by RNAi [5]. Interestingly, eHAV capsid contains a pX domain on its surface carrying several ubiquitination sites that could promote the ESCRT recruitment [171], and pX was found to interact with the V domain of ALIX, thus participating to eHAV biogenesis, although no defined ALIX-binding motif has been identified within pX thus far [174]. Altogether, these findings suggest that HAV recruits ALIX to bud into membranous vesicles, thus facilitating viral protection from neutralizing antibodies [5] and viral spreading as it was demonstrated for rotaviruses, noroviruses, and enteroviruses [6,7].
8. Conclusions
Enveloped RNA viruses derive their envelope from the host as they bud through cellular membranes, and viral budding requires the deformation of the cellular membrane followed by a fission event allowing the generation of distinct cellular and viral membranes. Many viruses therefore hijack the cellular ESCRT machinery to remodel the membranes at viral budding sites, thereby enabling the release of infectious enveloped viral particles. To recruit the ESCRT factors, enveloped viruses use conserved viral assembly L domains encoded by their multifunctional structural proteins. Three types of L domains have been characterized thus far: P(T/S)AP, YPXnL, and PPxY, and two other types, FPIV and PLPPV, have been reported in the M protein of paramyxoviruses [72,77,78] and in the MMTV Pr77Gag precursor [73], respectively, even though the cellular partners of these two last motifs remain to be identified. In addition, it was shown that quasi-enveloped viruses, such as HAV, that can exit cells non-lytically within vesicles, encode two YPXnL motifs in their structural protein VP2 that interact with ALIX, and use the ESCRT pathway to promote their budding [5,172,173,174]. As the mechanisms for ESCRT recruitment through the action of L domains and consequent budding are well conserved processes between different RNA virus families, a better comprehension will be determinant for the development of new broad-spectrum antiviral drugs directed against viral release. For example, it was observed that compounds that block the PPxY–NEDD4 interaction also efficiently inhibited the egress of MARV [175,176]. Similarly, recent studies have shown that prazole compounds can inhibit the release of some enveloped viruses, including HIV-1, EBOV, Mayaro virus (MAYV), and HSV by covalently binding the UEV domain of TSG101, which results in the disruption of TSG101–ubiquitin interaction [177,178,179]. Such a class of inhibitors interfering with the budding machinery could thus resultingly be extremely useful for impairing the production of newly emerging RNA viruses for which no therapeutics are available.
Author Contributions
S.B. supervised the project; L.W. wrote the manuscript with contributions from S.B. and J.-C.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Agence Nationale de Recherche sur le SIDA et les hépatites virales (ANRS), SIDACTION, and IdEx (Initiative d’Excellence, Université de Strasbourg, France). L.W. is supported by a fellowship from the French Ministry of Research and Higher Education.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Koonin E.V., Dolja V.V., Krupovic M. Origins and Evolution of Viruses of Eukaryotes: The Ultimate Modularity. Virology. 2015;479:2–25. doi: 10.1016/j.virol.2015.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolf Y.I., Kazlauskas D., Iranzo J., Lucía-Sanz A., Kuhn J.H., Krupovic M., Dolja V.V., Koonin E.V. Origins and Evolution of the Global RNA Virome. mBio. 2018;9:e02329-18. doi: 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrasco-Hernandez R., Jácome R., López Vidal Y., Ponce de León S. Are RNA Viruses Candidate Agents for the Next Global Pandemic? A Review. ILAR J. 2017;58:343–358. doi: 10.1093/ilar/ilx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Global HIV & AIDS Statistics—Fact Sheet. [(accessed on 27 June 2021)]; Available online: https://www.unaids.org/en/resources/fact-sheet.
- 5.Feng Z., Hensley L., McKnight K.L., Hu F., Madden V., Ping L., Jeong S.-H., Walker C., Lanford R.E., Lemon S.M. A Pathogenic Picornavirus Acquires an Envelope by Hijacking Cellular Membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y.-H., Du W., Hagemeijer M.C., Takvorian P.M., Pau C., Cali A., Brantner C.A., Stempinski E.S., Connelly P.S., Ma H.-C., et al. Phosphatidylserine Vesicles Enable Efficient En Bloc Transmission of Enteroviruses. Cell. 2015;160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santiana M., Ghosh S., Ho B.A., Rajasekaran V., Du W.-L., Mutsafi Y., De Jésus-Diaz D.A., Sosnovtsev S.V., Levenson E.A., Parra G.I., et al. Vesicle-Cloaked Virus Clusters Are Optimal Units for Inter-Organismal Viral Transmission. Cell Host Microbe. 2018;24:208–220.e8. doi: 10.1016/j.chom.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Votteler J., Sundquist W.I. Virus Budding and the ESCRT Pathway. Cell Host Microbe. 2013;14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vietri M., Radulovic M., Stenmark H. The Many Functions of ESCRTs. Nat. Rev. Mol. Cell. Bio. 2020;21:25–42. doi: 10.1038/s41580-019-0177-4. [DOI] [PubMed] [Google Scholar]
- 10.Christ L., Raiborg C., Wenzel E.M., Campsteijn C., Stenmark H. Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem. Sci. 2017;42:42–56. doi: 10.1016/j.tibs.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 11.Schöneberg J., Lee I.-H., Iwasa J.H., Hurley J.H. Reverse-Topology Membrane Scission by the ESCRT Proteins. Nat. Rev. Mol. Cell. Bio. 2017;18:5–17. doi: 10.1038/nrm.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed E.O. Viral Late Domains. J. Virol. 2002;76:4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Göttlinger H.G., Dorfman T., Sodroski J.G., Haseltine W.A. Effect of Mutations Affecting the P6 Gag Protein on Human Immunodeficiency Virus Particle Release. Proc. Natl. Acad. Sci. USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wills J.W., Cameron C.E., Wilson C.B., Xiang Y., Bennett R.P., Leis J. An Assembly Domain of the Rous Sarcoma Virus Gag Protein Required Late in Budding. J. Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parent L.J., Bennett R.P., Craven R.C., Nelle T.D., Krishna N.K., Bowzard J.B., Wilson C.B., Puffer B.A., Montelaro R.C., Wills J.W. Positionally Independent and Exchangeable Late Budding Functions of the Rous Sarcoma Virus and Human Immunodeficiency Virus Gag Proteins. J. Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puffer B.A., Parent L.J., Wills J.W., Montelaro R.C. Equine Infectious Anemia Virus Utilizes a YXXL Motif within the Late Assembly Domain of the Gag P9 Protein. J. Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers E.L., Allen J.F. Tsg101, an Inactive Homologue of Ubiquitin Ligase E2, Interacts Specifically with Human Immunodeficiency Virus Type 2 Gag Polyprotein and Results in Increased Levels of Ubiquitinated Gag. J. Virol. 2002;76:11226–11235. doi: 10.1128/JVI.76.22.11226-11235.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burkala E., Poss M. Evolution of Feline Immunodeficiency Virus Gag Proteins. Virus Genes. 2007;35:251–264. doi: 10.1007/s11262-006-0058-8. [DOI] [PubMed] [Google Scholar]
- 19.Del Vecchio C., Celestino M., Celegato M., Palù G., Parolin C., Bouamr F., Calistri A. Alix-Mediated Rescue of Feline Immunodeficiency Virus Budding Differs from That Observed with Human Immunodeficiency Virus. J. Virol. 2020;94:e02019-19. doi: 10.1128/JVI.02019-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartusch C., Prange R. ESCRT Requirements for Murine Leukemia Virus Release. Viruses. 2016;8:103. doi: 10.3390/v8040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcucci K.T., Martina Y., Harrison F., Wilson C.A., Salomon D.R. Functional Hierarchy of Two L Domains in Porcine Endogenous Retrovirus (PERV) That Influence Release and Infectivity. Virology. 2008;375:637–645. doi: 10.1016/j.virol.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chudak C., Beimforde N., George M., Zimmermann A., Lausch V., Hanke K., Bannert N. Identification of Late Assembly Domains of the Human Endogenous Retrovirus-K(HML-2) Retrovirology. 2013;10:140. doi: 10.1186/1742-4690-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narahara C., Yasuda J. Roles of the Three L-Domains in β-Retrovirus Budding. Microbiol. Immunol. 2015;59:545–554. doi: 10.1111/1348-0421.12285. [DOI] [PubMed] [Google Scholar]
- 24.Hütter S., Zurnic I., Lindemann D. Foamy Virus Budding and Release. Viruses. 2013;5:1075. doi: 10.3390/v5041075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patton G.S., Morris S.A., Chung W., Bieniasz P.D., McClure M.O. Identification of Domains in Gag Important for Prototypic Foamy Virus Egress. J. Virol. 2005;79:6392–6399. doi: 10.1128/JVI.79.10.6392-6399.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patnaik A., Chau V., Wills J.W. Ubiquitin Is Part of the Retrovirus Budding Machinery. Proc. Natl. Acad. Sci. USA. 2000;97:13069–13074. doi: 10.1073/pnas.97.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schubert U., Ott D.E., Chertova E.N., Welker R., Tessmer U., Princiotta M.F., Bennink J.R., Kräusslich H.-G., Yewdell J.W. Proteasome Inhibition Interferes with Gag Polyprotein Processing, Release, and Maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA. 2000;97:13057–13062. doi: 10.1073/pnas.97.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strack B., Calistri A., Accola M.A., Palù G., Göttlinger H.G. A Role for Ubiquitin Ligase Recruitment in Retrovirus Release. Proc. Natl. Acad. Sci. USA. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogt V.M. Ubiquitin in Retrovirus Assembly: Actor or Bystander? Proc. Natl. Acad. Sci. USA. 2000;97:12945–12947. doi: 10.1073/pnas.97.24.12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruce E.A., Medcalf L., Crump C.M., Noton S.L., Stuart A.D., Wise H.M., Elton D., Bowers K., Digard P. Budding of Filamentous and Non-Filamentous Influenza A Virus Occurs via a VPS4 and VPS28-Independent Pathway. Virology. 2009;390:268–278. doi: 10.1016/j.virol.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 31.Rossman J.S., Jing X., Leser G.P., Lamb R.A. Influenza Virus M2 Protein Mediates ESCRT-Independent Membrane Scission. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weissenhorn W., Poudevigne E., Effantin G., Bassereau P. How to Get out: SsRNA Enveloped Viruses and Membrane Fission. Curr. Opin. Virol. 2013;3:159–167. doi: 10.1016/j.coviro.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waheed A.A., Freed E.O. The Role of Lipids in Retrovirus Replication. Viruses. 2010;2:1146. doi: 10.3390/v2051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundquist W.I., Kräusslich H.-G. HIV-1 Assembly, Budding, and Maturation. Cold Spring Harb. Perspect. Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freed E.O. HIV-1 Assembly, Release and Maturation. Nat. Rev. Microbiol. 2015;13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lingappa J.R., Reed J.C., Tanaka M., Chutiraka K., Robinson B.A. How HIV-1 Gag Assembles in Cells: Putting Together Pieces of the Puzzle. Virus Res. 2014;193:89–107. doi: 10.1016/j.virusres.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rheinemann L., Sundquist W.I. Virus Budding. Encycl. Virol. 2021:519–528. doi: 10.1016/B978-0-12-814515-9.00023-0. [DOI] [Google Scholar]
- 38.Olson E.D., Musier-Forsyth K. Retroviral Gag Protein-RNA Interactions: Implications for Specific Genomic RNA Packaging and Virion Assembly. Semin. Cell Dev. Biol. 2019;86:129–139. doi: 10.1016/j.semcdb.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mailler E., Bernacchi S., Marquet R., Paillart J.-C., Vivet-Boudou V., Smyth R.P. The Life-Cycle of the HIV-1 Gag–RNA Complex. Viruses. 2016;8:248. doi: 10.3390/v8090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ono A., Freed E.O. Plasma Membrane Rafts Play a Critical Role in HIV-1 Assembly and Release. Proc. Natl. Acad. Sci. USA. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ono A., Ablan S.D., Lockett S.J., Nagashima K., Freed E.O. Phosphatidylinositol (4,5) Bisphosphate Regulates HIV-1 Gag Targeting to the Plasma Membrane. Proc. Natl. Acad. Sci. USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paillart J.C., Göttlinger H.G. Opposing Effects of Human Immunodeficiency Virus Type 1 Matrix Mutations Support a Myristyl Switch Model of Gag Membrane Targeting. J. Virol. 1999;73:2604–2612. doi: 10.1128/JVI.73.4.2604-2612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang C., Loeliger E., Luncsford P., Kinde I., Beckett D., Summers M.F. Entropic Switch Regulates Myristate Exposure in the HIV-1 Matrix Protein. Proc. Natl. Acad. Sci. USA. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saad J.S., Miller J., Tai J., Kim A., Ghanam R.H., Summers M.F. Structural Basis for Targeting HIV-1 Gag Proteins to the Plasma Membrane for Virus Assembly. Proc. Natl. Acad. Sci. USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datta S.A.K., Temeselew L.G., Crist R.M., Soheilian F., Kamata A., Mirro J., Harvin D., Nagashima K., Cachau R.E., Rein A. On the Role of the SP1 Domain in HIV-1 Particle Assembly: A Molecular Switch? J. Virol. 2011;85:4111–4121. doi: 10.1128/JVI.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo X., Roldan A., Hu J., Wainberg M.A., Liang C. Mutation of the SP1 Sequence Impairs Both Multimerization and Membrane-Binding Activities of Human Immunodeficiency Virus Type 1 Gag. J. Virol. 2005;79:1803–1812. doi: 10.1128/JVI.79.3.1803-1812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schur F.K.M., Obr M., Hagen W.J.H., Wan W., Jakobi A.J., Kirkpatrick J.M., Sachse C., Kräusslich H.-G., Briggs J.A.G. An Atomic Model of HIV-1 Capsid-SP1 Reveals Structures Regulating Assembly and Maturation. Science. 2016;353:506–508. doi: 10.1126/science.aaf9620. [DOI] [PubMed] [Google Scholar]
- 48.Wagner J.M., Zadrozny K.K., Chrustowicz J., Purdy M.D., Yeager M., Ganser-Pornillos B.K., Pornillos O. Crystal Structure of an HIV Assembly and Maturation Switch. eLife. 2016;5:e17063. doi: 10.7554/eLife.17063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernacchi S., Abd El-Wahab E.W., Dubois N., Hijnen M., Smyth R.P., Mak J., Marquet R., Paillart J.-C. HIV-1 Pr55Gag Binds Genomic and Spliced RNAs with Different Affinity and Stoichiometry. RNA Biol. 2016;14:90–103. doi: 10.1080/15476286.2016.1256533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferrer M., Clerté C., Chamontin C., Basyuk E., Lainé S., Hottin J., Bertrand E., Margeat E., Mougel M. Imaging HIV-1 RNA Dimerization in Cells by Multicolor Super-Resolution and Fluctuation Microscopies. Nucleic Acids Res. 2016;44:7922–7934. doi: 10.1093/nar/gkw511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nikolaitchik O.A., Dilley K.A., Fu W., Gorelick R.J., Tai S.-H.S., Soheilian F., Ptak R.G., Nagashima K., Pathak V.K., Hu W.-S. Dimeric RNA Recognition Regulates HIV-1 Genome Packaging. PLoS Pathog. 2013;9:e1003249. doi: 10.1371/journal.ppat.1003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown J.D., Kharytonchyk S., Chaudry I., Iyer A.S., Carter H., Becker G., Desai Y., Glang L., Choi S.H., Singh K., et al. Structural Basis for Transcriptional Start Site Control of HIV-1 RNA Fate. Science. 2020;368:413–417. doi: 10.1126/science.aaz7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dannull J., Surovoy A., Jung G., Moelling K. Specific Binding of HIV-1 Nucleocapsid Protein to PSI RNA in Vitro Requires N-Terminal Zinc Finger and Flanking Basic Amino Acid Residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abd El-Wahab E.W., Smyth R.P., Mailler E., Bernacchi S., Vivet-Boudou V., Hijnen M., Jossinet F., Mak J., Paillart J.-C., Marquet R. Specific Recognition of the HIV-1 Genomic RNA by the Gag Precursor. Nat. Commun. 2014;5:4304. doi: 10.1038/ncomms5304. [DOI] [PubMed] [Google Scholar]
- 55.Boutant E., Bonzi J., Anton H., Nasim M.B., Cathagne R., Réal E., Dujardin D., Carl P., Didier P., Paillart J.-C., et al. Zinc Fingers in HIV-1 Gag Precursor Are Not Equivalent for GRNA Recruitment at the Plasma Membrane. Biophys. J. 2020;119:419–433. doi: 10.1016/j.bpj.2020.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Marco A., Heuser A.-M., Glass B., Kräusslich H.-G., Müller B., Briggs J.A.G. Role of the SP2 Domain and Its Proteolytic Cleavage in HIV-1 Structural Maturation and Infectivity. J. Virol. 2012;86:13708–13716. doi: 10.1128/JVI.01704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dubois N., Khoo K.K., Ghossein S., Seissler T., Wolff P., McKinstry W.J., Mak J., Paillart J.-C., Marquet R., Bernacchi S. The C-Terminal P6 Domain of the HIV-1 Pr55Gag Precursor Is Required for Specific Binding to the Genomic RNA. RNA Biol. 2018;15:923–936. doi: 10.1080/15476286.2018.1481696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang M., Orenstein J.M., Martin M.A., Freed E.O. P6Gag Is Required for Particle Production from Full-Length Human Immunodeficiency Virus Type 1 Molecular Clones Expressing Protease. J. Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Demirov D.G., Orenstein J.M., Freed E.O. The Late Domain of Human Immunodeficiency Virus Type 1 P6 Promotes Virus Release in a Cell Type-Dependent Manner. J. Virol. 2002;76:105–117. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Demirov D.G., Freed E.O. Retrovirus Budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 61.VerPlank L., Bouamr F., LaGrassa T.J., Agresta B., Kikonyogo A., Leis J., Carter C.A. Tsg101, a Homologue of Ubiquitin-Conjugating (E2) Enzymes, Binds the L Domain in HIV Type 1 Pr55(Gag) Proc. Natl. Acad. Sci. USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strack B., Calistri A., Craig S., Popova E., Göttlinger H.G. AIP1/ALIX Is a Binding Partner for HIV-1 P6 and EIAV P9 Functioning in Virus Budding. Cell. 2003;114:689–699. doi: 10.1016/S0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 63.Dussupt V., Javid M.P., Abou-Jaoudé G., Jadwin J.A., de La Cruz J., Nagashima K., Bouamr F. The Nucleocapsid Region of HIV-1 Gag Cooperates with the PTAP and LYPXnL Late Domains to Recruit the Cellular Machinery Necessary for Viral Budding. PLoS Pathog. 2009;5:e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.El Meshri S.E., Boutant E., Mouhand A., Thomas A., Larue V., Richert L., Vivet-Boudou V., Mély Y., Tisné C., Muriaux D., et al. The NC Domain of HIV-1 Gag Contributes to the Interaction of Gag with TSG101. BBA Gen. Subj. 2018;1862:1421–1431. doi: 10.1016/j.bbagen.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 65.Chamontin C., Rassam P., Ferrer M., Racine P.-J., Neyret A., Lainé S., Milhiet P.-E., Mougel M. HIV-1 Nucleocapsid and ESCRT-Component Tsg101 Interplay Prevents HIV from Turning into a DNA-Containing Virus. Nucleic Acids Res. 2015;43:336–347. doi: 10.1093/nar/gku1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Popov S., Popova E., Inoue M., Göttlinger H.G. Human Immunodeficiency Virus Type 1 Gag Engages the Bro1 Domain of ALIX/AIP1 through the Nucleocapsid. J. Virol. 2008;82:1389–1398. doi: 10.1128/JVI.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watanabe S.M., Strickland M., Tjandra N., Carter C.A. RNA Binding Suppresses Tsg101 Recognition of Ub-Modified Gag and Facilitates Recruitment to the Plasma Membrane. Viruses. 2020;12:447. doi: 10.3390/v12040447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiang Y., Cameron C.E., Wills J.W., Leis J. Fine Mapping and Characterization of the Rous Sarcoma Virus Pr76gag Late Assembly Domain. J. Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kikonyogo A., Bouamr F., Vana M.L., Xiang Y., Aiyar A., Carter C., Leis J. Proteins Related to the Nedd4 Family of Ubiquitin Protein Ligases Interact with the L Domain of Rous Sarcoma Virus and Are Required for Gag Budding from Cells. Proc. Natl. Acad. Sci. USA. 2001;98:11199–11204. doi: 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan B., Campbell S., Bacharach E., Rein A., Goff S.P. Infectivity of Moloney Murine Leukemia Virus Defective in Late Assembly Events Is Restored by Late Assembly Domains of Other Retroviruses. J. Virol. 2000;74:7250–7260. doi: 10.1128/JVI.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Craven R.C., Harty R.N., Paragas J., Palese P., Wills J.W. Late Domain Function Identified in the Vesicular Stomatitis Virus M Protein by Use of Rhabdovirus-Retrovirus Chimeras. J. Virol. 1999;73:3359–3365. doi: 10.1128/JVI.73.4.3359-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmitt A.P., Leser G.P., Morita E., Sundquist W.I., Lamb R.A. Evidence for a New Viral Late-Domain Core Sequence, FPIV, Necessary for Budding of a Paramyxovirus. J. Virol. 2005;79:2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coren L.V., Nagashima K., Ott D.E. A PLPPV Sequence in the P8 Region of Gag Provides Late Domain Function for Mouse Mammary Tumor Virus. Virology. 2019;535:272–278. doi: 10.1016/j.virol.2019.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pincetic A., Medina G., Carter C., Leis J. Avian Sarcoma Virus and Human Immunodeficiency Virus, Type 1 Use Different Subsets of ESCRT Proteins to Facilitate the Budding Process. J. Biol. Chem. 2008;283:29822–29830. doi: 10.1074/jbc.M804157200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harty R.N., Paragas J., Sudol M., Palese P. A Proline-Rich Motif within the Matrix Protein of Vesicular Stomatitis Virus and Rabies Virus Interacts with WW Domains of Cellular Proteins: Implications for Viral Budding. J. Virol. 1999;73:2921–2929. doi: 10.1128/JVI.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Strack B., Calistri A., Göttlinger H.G. Late Assembly Domain Function Can Exhibit Context Dependence and Involves Ubiquitin Residues Implicated in Endocytosis. J. Virol. 2002;76:5472–5479. doi: 10.1128/JVI.76.11.5472-5479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li M., Schmitt P.T., Li Z., McCrory T.S., He B., Schmitt A.P. Mumps Virus Matrix, Fusion, and Nucleocapsid Proteins Cooperate for Efficient Production of Virus-Like Particles. J. Virol. 2009;83:7261–7272. doi: 10.1128/JVI.00421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duan Z., Hu Z., Zhu J., Xu H., Chen J., Liu H., Hu S., Liu X. Mutations in the FPIV Motif of Newcastle Disease Virus Matrix Protein Attenuate Virus Replication and Reduce Virus Budding. Arch. Virol. 2014;159:1813–1819. doi: 10.1007/s00705-014-1998-2. [DOI] [PubMed] [Google Scholar]
- 79.Lee S., Joshi A., Nagashima K., Freed E.O., Hurley J.H. Structural Basis for Viral Late-Domain Binding to Alix. Nat. Struct. Mol. Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhai Q., Fisher R.D., Chung H.-Y., Myszka D.G., Sundquist W.I., Hill C.P. Structural and Functional Studies of ALIX Interactions with YPX n L Late Domains of HIV-1 and EIAV. Nat. Struct. Mol. Biol. 2008;15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- 81.Martin-Serrano J., Bieniasz P.D. A Bipartite Late-Budding Domain in Human Immunodeficiency Virus Type 1. J. Virol. 2003;77:12373–12377. doi: 10.1128/JVI.77.22.12373-12377.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patil A., Bhattacharya J. Natural Deletion of L35Y36 in P6 Gag Eliminate LYPXnL/ALIX Auxiliary Virus Release Pathway in HIV-1 Subtype C. Virus Res. 2012;170:154–158. doi: 10.1016/j.virusres.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 83.Neogi U., Rao S.D., Bontell I., Verheyen J., Rao V.R., Gore S.C., Soni N., Shet A., Schülter E., Ekstrand M.L., et al. Novel Tetra-Peptide Insertion in Gag-P6 ALIX-Binding Motif in HIV-1 Subtype C Associated with Protease Inhibitor Failure in Indian Patients. AIDS. 2014;28:2319–2322. doi: 10.1097/QAD.0000000000000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Domselaar R., Njenda D.T., Rao R., Sönnerborg A., Singh K., Neogi U. HIV-1 Subtype C with PYxE Insertion Has Enhanced Binding of Gag-P6 to Host Cell Protein ALIX and Increased Replication Fitness. J. Virol. 2019;93:e00077-19. doi: 10.1128/JVI.00077-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chaturbhuj D., Patil A., Gangakhedkar R. PYRE Insertion within HIV-1 Subtype C P6-Gag Functions as an ALIX-Dependent Late Domain. Sci. Rep. 2018;8:8917. doi: 10.1038/s41598-018-27162-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gottwein E., Bodem J., Müller B., Schmechel A., Zentgraf H., Kräusslich H.-G. The Mason-Pfizer Monkey Virus PPPY and PSAP Motifs Both Contribute to Virus Release. J. Virol. 2003;77:9474–9485. doi: 10.1128/JVI.77.17.9474-9485.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bouamr F., Melillo J.A., Wang M.Q., Nagashima K., de Los Santos M., Rein A., Goff S.P. PPPYEPTAP Motif Is the Late Domain of Human T-Cell Leukemia Virus Type 1 Gag and Mediates Its Functional Interaction with Cellular Proteins Nedd4 and Tsg101. J. Virol. 2003;77:11882–11895. doi: 10.1128/JVI.77.22.11882-11895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang H., Machesky N.J., Mansky L.M. Both the PPPY and PTAP Motifs Are Involved in Human T-Cell Leukemia Virus Type 1 Particle Release. J. Virol. 2004;78:1503–1512. doi: 10.1128/JVI.78.3.1503-1512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Licata J.M., Simpson-Holley M., Wright N.T., Han Z., Paragas J., Harty R.N. Overlapping Motifs (PTAP and PPEY) within the Ebola Virus VP40 Protein Function Independently as Late Budding Domains: Involvement of Host Proteins TSG101 and VPS-4. J. Virol. 2003;77:1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Perez M., Craven R.C., de la Torre J.C. The Small RING Finger Protein Z Drives Arenavirus Budding: Implications for Antiviral Strategies. Proc. Natl. Acad. Sci. USA. 2003;100:12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dolnik O., Kolesnikova L., Stevermann L., Becker S. Tsg101 Is Recruited by a Late Domain of the Nucleocapsid Protein to Support Budding of Marburg Virus-like Particles. J. Virol. 2010;84:7847–7856. doi: 10.1128/JVI.00476-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dilley K.A., Gregory D., Johnson M.C., Vogt V.M. An LYPSL Late Domain in the Gag Protein Contributes to the Efficient Release and Replication of Rous Sarcoma Virus. J. Virol. 2010;84:6276–6287. doi: 10.1128/JVI.00238-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ott D.E., Coren L.V., Gagliardi T.D., Nagashima K. Heterologous Late-Domain Sequences Have Various Abilities To Promote Budding of Human Immunodeficiency Virus Type 1. J. Virol. 2005;79:9038–9045. doi: 10.1128/JVI.79.14.9038-9045.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Le Blanc I., Prévost M.-C., Dokhélar M.-C., Rosenberg A.R. The PPPY Motif of Human T-Cell Leukemia Virus Type 1 Gag Protein Is Required Early in the Budding Process. J. Virol. 2002;76:10024–10029. doi: 10.1128/JVI.76.19.10024-10029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blot V., Perugi F., Gay B., Prévost M.-C., Briant L., Tangy F., Abriel H., Staub O., Dokhélar M.-C., Pique C. Nedd4.1-Mediated Ubiquitination and Subsequent Recruitment of Tsg101 Ensure HTLV-1 Gag Trafficking towards the Multivesicular Body Pathway Prior to Virus Budding. J. Cell Sci. 2004;117:2357–2367. doi: 10.1242/jcs.01095. [DOI] [PubMed] [Google Scholar]
- 96.Stange A., Mannigel I., Peters K., Heinkelein M., Stanke N., Cartellieri M., Göttlinger H., Rethwilm A., Zentgraf H., Lindemann D. Characterization of Prototype Foamy Virus Gag Late Assembly Domain Motifs and Their Role in Particle Egress and Infectivity. J. Virol. 2005;79:5466–5476. doi: 10.1128/JVI.79.9.5466-5476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vana M.L., Tang Y., Chen A., Medina G., Carter C., Leis J. Role of Nedd4 and Ubiquitination of Rous Sarcoma Virus Gag in Budding of Virus-Like Particles from Cells. J. Virol. 2004;78:13943–13953. doi: 10.1128/JVI.78.24.13943-13953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Henne W.M., Buchkovich N.J., Emr S.D. The ESCRT Pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 99.Leung K.F., Dacks J.B., Field M.C. Evolution of the Multivesicular Body ESCRT Machinery; Retention Across the Eukaryotic Lineage. Traffic. 2008;9:1698–1716. doi: 10.1111/j.1600-0854.2008.00797.x. [DOI] [PubMed] [Google Scholar]
- 100.Carlton J.G., Martin-Serrano J. Parallels between Cytokinesis and Retroviral Budding: A Role for the ESCRT Machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 101.Morita E., Sandrin V., Chung H.-Y., Morham S.G., Gygi S.P., Rodesch C.K., Sundquist W.I. Human ESCRT and ALIX Proteins Interact with Proteins of the Midbody and Function in Cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bache K.G., Brech A., Mehlum A., Stenmark H. Hrs Regulates Multivesicular Body Formation via ESCRT Recruitment to Endosomes. J. Cell Biol. 2003;162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nabhan J.F., Hu R., Oh R.S., Cohen S.N., Lu Q. Formation and Release of Arrestin Domain-Containing Protein 1-Mediated Microvesicles (ARMMs) at Plasma Membrane by Recruitment of TSG101 Protein. Proc. Natl. Acad. Sci. USA. 2012;109:4146–4151. doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Garrus J.E., von Schwedler U.K., Pornillos O.W., Morham S.G., Zavitz K.H., Wang H.E., Wettstein D.A., Stray K.M., Côté M., Rich R.L., et al. Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding. Cell. 2001;107:55–65. doi: 10.1016/S0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 105.Demirov D.G., Ono A., Orenstein J.M., Freed E.O. Overexpression of the N-Terminal Domain of TSG101 Inhibits HIV-1 Budding by Blocking Late Domain Function. Proc. Natl. Acad. Sci. USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Martin-Serrano J., Zang T., Bieniasz P.D. HIV-1 and Ebola Virus Encode Small Peptide Motifs That Recruit Tsg101 to Sites of Particle Assembly to Facilitate Egress. Nat. Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 107.Pornillos O., Alam S.L., Rich R.L., Myszka D.G., Davis D.R., Sundquist W.I. Structure and Functional Interactions of the Tsg101 UEV Domain. EMBO J. 2002;21:2397–2406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Luttge B.G., Panchal P., Puri V., Checkley M.A., Freed E.O. Mutations in the Feline Immunodeficiency Virus Envelope Glycoprotein Confer Resistance to a Dominant–Negative Fragment of Tsg101 by Enhancing Infectivity and Cell-to-Cell Virus Transmission. BBA-Biomembranes. 2014;1838:1143–1152. doi: 10.1016/j.bbamem.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sharma S., Aralaguppe S.G., Abrahams M.-R., Williamson C., Gray C., Balakrishnan P., Saravanan S., Murugavel K.G., Solomon S., Ranga U. The PTAP Sequence Duplication in HIV-1 Subtype C Gag P6 in Drug-Naive Subjects of India and South Africa. BMC Infect. Dis. 2017;17:95. doi: 10.1186/s12879-017-2184-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Martins A.N., Arruda M.B., Pires A.F., Tanuri A., Brindeiro R.M. Accumulation of P(T/S)AP Late Domain Duplications in HIV Type 1 Subtypes B, C, and F Derived from Individuals Failing ARV Therapy and ARV Drug-Naive Patients. AIDS Res. Hum. Retrovir. 2011;27:687–692. doi: 10.1089/aid.2010.0282. [DOI] [PubMed] [Google Scholar]
- 111.Flys T., Marlowe N., Hackett J., Parkin N., Schumaker M., Holzmayer V., Hay P., Eshleman S.H. Analysis of PTAP Duplications in the Gag P6 Region of Subtype C HIV Type 1. AIDS Res. Hum. Retroviruses. 2005;21:739–741. doi: 10.1089/aid.2005.21.739. [DOI] [PubMed] [Google Scholar]
- 112.Sharma S., Arunachalam P.S., Menon M., Ragupathy V., Satya R.V., Jebaraj J., Aralaguppe S.G., Rao C., Pal S., Saravanan S., et al. PTAP Motif Duplication in the P6 Gag Protein Confers a Replication Advantage on HIV-1 Subtype C. J. Biol. Chem. 2018;293:11687–11708. doi: 10.1074/jbc.M117.815829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Martins A.N., Waheed A.A., Ablan S.D., Huang W., Newton A., Petropoulos C.J., Brindeiro R.D.M., Freed E.O. Elucidation of the Molecular Mechanism Driving Duplication of the HIV-1 PTAP Late Domain. J. Virol. 2016;90:768–779. doi: 10.1128/JVI.01640-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fisher R.D., Chung H.-Y., Zhai Q., Robinson H., Sundquist W.I., Hill C.P. Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 115.Munshi U.M., Kim J., Nagashima K., Hurley J.H., Freed E.O. An Alix Fragment Potently Inhibits HIV-1 Budding: Characterization of Binding to Retroviral YPXL Late Domains. J. Biol. Chem. 2007;282:3847–3855. doi: 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- 116.Martin-Serrano J., Eastman S.W., Chung W., Bieniasz P.D. HECT Ubiquitin Ligases Link Viral and Cellular PPXY Motifs to the Vacuolar Protein-Sorting Pathway. J. Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Adell M.A.Y., Vogel G.F., Pakdel M., Müller M., Lindner H., Hess M.W., Teis D. Coordinated Binding of Vps4 to ESCRT-III Drives Membrane Neck Constriction during MVB Vesicle Formation. J. Cell Biol. 2014;205:33–49. doi: 10.1083/jcb.201310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fabrikant G., Lata S., Riches J.D., Briggs J.A.G., Weissenhorn W., Kozlov M.M. Computational Model of Membrane Fission Catalyzed by ESCRT-III. PLoS Comput. Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McCullough J., Clippinger A.K., Talledge N., Skowyra M.L., Saunders M.G., Naismith T.V., Colf L.A., Afonine P., Arthur C., Sundquist W.I., et al. Structure and Membrane Remodeling Activity of ESCRT-III Helical Polymers. Science. 2015;350:1548–1551. doi: 10.1126/science.aad8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chiaruttini N., Redondo-Morata L., Colom A., Humbert F., Lenz M., Scheuring S., Roux A. Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell. 2015;163:866–879. doi: 10.1016/j.cell.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shen Q.-T., Schuh A.L., Zheng Y., Quinney K., Wang L., Hanna M., Mitchell J.C., Otegui M.S., Ahlquist P., Cui Q., et al. Structural Analysis and Modeling Reveals New Mechanisms Governing ESCRT-III Spiral Filament Assembly. J. Cell Biol. 2014;206:763–777. doi: 10.1083/jcb.201403108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hanson P.I., Roth R., Lin Y., Heuser J.E. Plasma Membrane Deformation by Circular Arrays of ESCRT-III Protein Filaments. J. Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zamborlini A., Usami Y., Radoshitzky S.R., Popova E., Palu G., Göttlinger H. Release of Autoinhibition Converts ESCRT-III Components into Potent Inhibitors of HIV-1 Budding. Proc. Natl. Acad. Sci. USA. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shim S., Kimpler L.A., Hanson P.I. Structure/Function Analysis of Four Core ESCRT-III Proteins Reveals Common Regulatory Role for Extreme C-Terminal Domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- 125.Bajorek M., Schubert H.L., McCullough J., Langelier C., Eckert D.M., Stubblefield W.-M.B., Uter N.T., Myszka D.G., Hill C.P., Sundquist W.I. Structural Basis for ESCRT-III Protein Autoinhibition. Nat. Struct. Mol. Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tang S., Buchkovich N.J., Henne W.M., Banjade S., Kim Y.J., Emr S.D. ESCRT-III Activation by Parallel Action of ESCRT-I/II and ESCRT-0/Bro1 during MVB Biogenesis. eLife. 2016;5:e15507. doi: 10.7554/eLife.15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Teis D., Saksena S., Judson B.L., Emr S.D. ESCRT-II Coordinates the Assembly of ESCRT-III Filaments for Cargo Sorting and Multivesicular Body Vesicle Formation. EMBO J. 2010;29:871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Christ L., Wenzel E.M., Liestøl K., Raiborg C., Campsteijn C., Stenmark H. ALIX and ESCRT-I/II Function as Parallel ESCRT-III Recruiters in Cytokinetic Abscission. J. Cell Biol. 2016;212:499–513. doi: 10.1083/jcb.201507009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McCullough J., Frost A., Sundquist W.I. Structures, Functions, and Dynamics of ESCRT-III/Vps4 Membrane Remodeling and Fission Complexes. Annu. Rev. Cell Dev. Biol. 2018;34:85–109. doi: 10.1146/annurev-cellbio-100616-060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pfitzner A.-K., Mercier V., Jiang X., Filseck J.M., von Baum B., Šarić A., Roux A. An ESCRT-III Polymerization Sequence Drives Membrane Deformation and Fission. Cell. 2020;182:1140–1155.e18. doi: 10.1016/j.cell.2020.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mierzwa B.E., Chiaruttini N., Redondo-Morata L., Moser von Filseck J., König J., Larios J., Poser I., Müller-Reichert T., Scheuring S., Roux A., et al. Dynamic Subunit Turnover in ESCRT-III Assemblies Is Regulated by Vps4 to Mediate Membrane Remodelling during Cytokinesis. Nat. Cell Biol. 2017;19:787–798. doi: 10.1038/ncb3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Flower T.G., Takahashi Y., Hudait A., Rose K., Tjahjono N., Pak A.J., Yokom A.L., Liang X., Wang H.-G., Bouamr F., et al. A Helical Assembly of Human ESCRT-I Scaffolds Reverse-Topology Membrane Scission. Nat. Struct. Mol. Biol. 2020;27:570–580. doi: 10.1038/s41594-020-0426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Martin-Serrano J., Zang T., Bieniasz P.D. Role of ESCRT-I in Retroviral Budding. J. Virol. 2003;77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stuchell M.D., Garrus J.E., Müller B., Stray K.M., Ghaffarian S., McKinnon R., Kräusslich H.-G., Morham S.G., Sundquist W.I. The Human Endosomal Sorting Complex Required for Transport (ESCRT-I) and Its Role in HIV-1 Budding. J. Biol. Chem. 2004;279:36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- 135.Eastman S.W., Martin-Serrano J., Chung W., Zang T., Bieniasz P.D. Identification of Human VPS37C, a Component of Endosomal Sorting Complex Required for Transport-I Important for Viral Budding. J. Biol. Chem. 2005;280:628–636. doi: 10.1074/jbc.M410384200. [DOI] [PubMed] [Google Scholar]
- 136.Agromayor M., Soler N., Caballe A., Kueck T., Freund S.M., Allen M.D., Bycroft M., Perisic O., Ye Y., McDonald B., et al. The UBAP1 Subunit of ESCRT-I Interacts with Ubiquitin via a SOUBA Domain. Structure. 2012;20:414–428. doi: 10.1016/j.str.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Morita E., Sandrin V., Alam S.L., Eckert D.M., Gygi S.P., Sundquist W.I. Identification of Human MVB12 Proteins as ESCRT-I Subunits That Function in HIV Budding. Cell Host Microbe. 2007;2:41–53. doi: 10.1016/j.chom.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Medina G., Zhang Y., Tang Y., Gottwein E., Vana M.L., Bouamr F., Leis J., Carter C.A. The Functionally Exchangeable L Domains in RSV and HIV-1 Gag Direct Particle Release Through Pathways Linked by Tsg101. Traffic. 2005;6:880–894. doi: 10.1111/j.1600-0854.2005.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Langelier C., Schwedler U.K., von Fisher R.D., Domenico I.D., White P.L., Hill C.P., Kaplan J., Ward D., Sundquist W.I. Human ESCRT-II Complex and Its Role in Human Immunodeficiency Virus Type 1 Release. J. Virol. 2006;80:9465–9480. doi: 10.1128/JVI.01049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Meng B., Ip N.C.Y., Prestwood L.J., Abbink T.E.M., Lever A.M.L. Evidence That the Endosomal Sorting Complex Required for Transport-II (ESCRT-II) Is Required for Efficient Human Immunodeficiency Virus-1 (HIV-1) Production. Retrovirology. 2015;12:72. doi: 10.1186/s12977-015-0197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meng B., Ip N.C.Y., Abbink T.E.M., Kenyon J.C., Lever A.M.L. ESCRT-II Functions by Linking to ESCRT-I in Human Immunodeficiency Virus-1 Budding. Cell Microbiol. 2020;22:e13161. doi: 10.1111/cmi.13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Meng B., Lever A.M.L. The Interplay between ESCRT and Viral Factors in the Enveloped Virus Life Cycle. Viruses. 2021;13:324. doi: 10.3390/v13020324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Martin-Serrano J., Yaravoy A., Perez-Caballero D., Bieniasz P.D. Divergent Retroviral Late-Budding Domains Recruit Vacuolar Protein Sorting Factors by Using Alternative Adaptor Proteins. Proc. Natl. Acad. Sci. USA. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.McCullough J., Fisher R.D., Whitby F.G., Sundquist W.I., Hill C.P. ALIX-CHMP4 Interactions in the Human ESCRT Pathway. Proc. Natl. Acad. Sci. USA. 2008;105:7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen C., Vincent O., Jin J., Weisz O.A., Montelaro R.C. Functions of Early (AP-2) and Late (AIP1/ALIX) Endocytic Proteins in Equine Infectious Anemia Virus Budding. J. Biol. Chem. 2005;280:40474–40480. doi: 10.1074/jbc.M509317200. [DOI] [PubMed] [Google Scholar]
- 146.Fujii K., Munshi U.M., Ablan S.D., Demirov D.G., Soheilian F., Nagashima K., Stephen A.G., Fisher R.J., Freed E.O. Functional Role of Alix in Hiv-1 Replication. Virology. 2009;391:284–292. doi: 10.1016/j.virol.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Usami Y., Popov S., Göttlinger H.G. Potent Rescue of Human Immunodeficiency Virus Type 1 Late Domain Mutants by ALIX/AIP1 Depends on Its CHMP4 Binding Site. J. Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gupta S., Bendjennat M., Saffarian S. Abrogating ALIX Interactions Results in Stuttering of the ESCRT Machinery. Viruses. 2020;12:1032. doi: 10.3390/v12091032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ku P.-I., Bendjennat M., Ballew J., Landesman M.B., Saffarian S. ALIX Is Recruited Temporarily into HIV-1 Budding Sites at the End of Gag Assembly. PLoS ONE. 2014;9:e96950. doi: 10.1371/journal.pone.0096950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rose K.M., Hirsch V.M., Bouamr F. Budding of a Retrovirus: Some Assemblies Required. Viruses. 2020;12:1188. doi: 10.3390/v12101188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhai Q., Landesman M.B., Robinson H., Sundquist W.I., Hill C.P. Identification and Structural Characterization of the ALIX-Binding Late Domains of Simian Immunodeficiency Virus SIVmac239 and SIVagmTan-1. J. Virol. 2011;85:632–637. doi: 10.1128/JVI.01683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pawliczek T., Crump C.M. Herpes Simplex Virus Type 1 Production Requires a Functional ESCRT-III Complex but Is Independent of TSG101 and ALIX Expression. J. Virol. 2009;83:11254–11264. doi: 10.1128/JVI.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chung H.-Y., Morita E., Schwedler U., von Müller B., Kräusslich H.-G., Sundquist W.I. NEDD4L Overexpression Rescues the Release and Infectivity of Human Immunodeficiency Virus Type 1 Constructs Lacking PTAP and YPXL Late Domains. J. Virol. 2008;82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Joshi A., Munshi U., Ablan S.D., Nagashima K., Freed E.O. Functional Replacement of a Retroviral Late Domain by Ubiquitin Fusion. Traffic. 2008;9:1972–1983. doi: 10.1111/j.1600-0854.2008.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dowlatshahi D.P., Sandrin V., Vivona S., Shaler T.A., Kaiser S.E., Melandri F., Sundquist W.I., Kopito R.R. ALIX Is a Lys63-Specific Polyubiquitin Binding Protein That Functions in Retrovirus Budding. Dev. Cell. 2012;23:1247–1254. doi: 10.1016/j.devcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Keren-Kaplan T., Attali I., Estrin M., Kuo L.S., Farkash E., Jerabek-Willemsen M., Blutraich N., Artzi S., Peri A., Freed E.O., et al. Structure-Based in Silico Identification of Ubiquitin-Binding Domains Provides Insights into the ALIX-V:Ubiquitin Complex and Retrovirus Budding. EMBO J. 2013;32:538–551. doi: 10.1038/emboj.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Okumura A., Lu G., Pitha-Rowe I., Pitha P.M. Innate Antiviral Response Targets HIV-1 Release by the Induction of Ubiquitin-like Protein ISG15. Proc. Natl. Acad. Sci. USA. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhadina M., McClure M.O., Johnson M.C., Bieniasz P.D. Ubiquitin-Dependent Virus Particle Budding without Viral Protein Ubiquitination. Proc. Natl. Acad. Sci. USA. 2007;104:20031–20036. doi: 10.1073/pnas.0708002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bussienne C., Marquet R., Paillart J.-C., Bernacchi S. Post-Translational Modifications of Retroviral HIV-1 Gag Precursors: An Overview of Their Biological Role. Int. J. Mol. Sci. 2021;22:2871. doi: 10.3390/ijms22062871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Raiborg C., Stenmark H. The ESCRT Machinery in Endosomal Sorting of Ubiquitylated Membrane Proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 161.Shepley-McTaggart A., Fan H., Sudol M., Harty R.N. Viruses Go Modular. J. Biol. Chem. 2020;295:4604–4616. doi: 10.1074/jbc.REV119.012414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Han Z., Sagum C.A., Bedford M.T., Sidhu S.S., Sudol M., Harty R.N. ITCH E3 Ubiquitin Ligase Interacts with Ebola Virus VP40 To Regulate Budding. J. Virol. 2016;90:9163–9171. doi: 10.1128/JVI.01078-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Harty R.N., Brown M.E., Wang G., Huibregtse J., Hayes F.P. A PPxY Motif within the VP40 Protein of Ebola Virus Interacts Physically and Functionally with a Ubiquitin Ligase: Implications for Filovirus Budding. Proc. Natl. Acad. Sci. USA. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Harty R.N., Brown M.E., McGettigan J.P., Wang G., Jayakar H.R., Huibregtse J.M., Whitt M.A., Schnell M.J. Rhabdoviruses and the Cellular Ubiquitin-Proteasome System: A Budding Interaction. J. Virol. 2001;75:10623–10629. doi: 10.1128/JVI.75.22.10623-10629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Timmins J., Schoehn G., Ricard-Blum S., Scianimanico S., Vernet T., Ruigrok R.W.H., Weissenhorn W. Ebola Virus Matrix Protein VP40 Interaction with Human Cellular Factors Tsg101 and Nedd4. J. Mol. Biol. 2003;326:493–502. doi: 10.1016/S0022-2836(02)01406-7. [DOI] [PubMed] [Google Scholar]
- 166.Urata S., Noda T., Kawaoka Y., Morikawa S., Yokosawa H., Yasuda J. Interaction of Tsg101 with Marburg Virus VP40 Depends on the PPPY Motif, but Not the PT/SAP Motif as in the Case of Ebola Virus, and Tsg101 Plays a Critical Role in the Budding of Marburg Virus-like Particles Induced by VP40, NP, and GP. J. Virol. 2007;81:4895–4899. doi: 10.1128/JVI.02829-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Han Z., Sagum C.A., Takizawa F., Ruthel G., Berry C.T., Kong J., Sunyer J.O., Freedman B.D., Bedford M.T., Sidhu S.S., et al. Ubiquitin Ligase WWP1 Interacts with Ebola Virus VP40 To Regulate Egress. J. Virol. 2017;91:e00812-17. doi: 10.1128/JVI.00812-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Usami Y., Popov S., Popova E., Göttlinger H.G. Efficient and Specific Rescue of Human Immunodeficiency Virus Type 1 Budding Defects by a Nedd4-Like Ubiquitin Ligase. J. Virol. 2008;82:4898–4907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mercenne G., Alam S.L., Arii J., Lalonde M.S., Sundquist W.I. Angiomotin Functions in HIV-1 Assembly and Budding. eLife. 2015;4:e03778. doi: 10.7554/eLife.03778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rauch S., Martin-Serrano J. Multiple Interactions between the ESCRT Machinery and Arrestin-Related Proteins: Implications for PPXY-Dependent Budding. J. Virol. 2011;85:3546–3556. doi: 10.1128/JVI.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Feng Z., Hirai-Yuki A., McKnight K.L., Lemon S.M. Naked Viruses That Aren’t Always Naked: Quasi-Enveloped Agents of Acute Hepatitis. Annu. Rev. Virol. 2014;1:539–560. doi: 10.1146/annurev-virology-031413-085359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.González-López O., Rivera-Serrano E.E., Hu F., Hensley L., McKnight K.L., Ren J., Stuart D.I., Fry E.E., Lemon S.M. Redundant Late Domain Functions of Tandem VP2 YPX3L Motifs in Nonlytic Cellular Egress of Quasi-Enveloped Hepatitis A Virus. J. Virol. 2018;92:e01308-18. doi: 10.1128/JVI.01308-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.McKnight K.L., Xie L., González-López O., Rivera-Serrano E.E., Chen X., Lemon S.M. Protein Composition of the Hepatitis A Virus Quasi-Envelope. Proc. Natl. Acad. Sci. USA. 2017;114:6587–6592. doi: 10.1073/pnas.1619519114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jiang W., Ma P., Deng L., Liu Z., Wang X., Liu X., Long G. Hepatitis A Virus Structural Protein PX Interacts with ALIX and Promotes the Secretion of Virions and Foreign Proteins through Exosome-like Vesicles. J. Extracell. Vesicles. 2020;9:1716513. doi: 10.1080/20013078.2020.1716513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Han Z., Lu J., Liu Y., Davis B., Lee M.S., Olson M.A., Ruthel G., Freedman B.D., Schnell M.J., Wrobel J.E., et al. Small-Molecule Probes Targeting the Viral PPxY-Host Nedd4 Interface Block Egress of a Broad Range of RNA Viruses. J. Virol. 2014;88:7294–7306. doi: 10.1128/JVI.00591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Han Z., Ye H., Liang J., Shepley-McTaggart A., Wrobel J.E., Reitz A.B., Whigham A., Kavelish K.N., Saporito M.S., Freedman B.D., et al. Compound FC-10696 Inhibits Egress and Spread of Marburg Virus. bioRxiv. 2021 doi: 10.1101/2021.01.15.426918. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Strickland M., Ehrlich L.S., Watanabe S., Khan M., Strub M.-P., Luan C.-H., Powell M.D., Leis J., Tjandra N., Carter C.A. Tsg101 Chaperone Function Revealed by HIV-1 Assembly Inhibitors. Nat. Commun. 2017;8:1391. doi: 10.1038/s41467-017-01426-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Watanabe S.M., Ehrlich L.S., Strickland M., Li X., Soloveva V., Goff A.J., Stauft C.B., Bhaduri-McIntosh S., Tjandra N., Carter C. Selective Targeting of Virus Replication by Proton Pump Inhibitors. Sci. Rep. 2020;10:4003. doi: 10.1038/s41598-020-60544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Leis J., Luan C.-H., Audia J.E., Dunne S.F., Heath C.M. Ilaprazole and Other Novel Prazole-Based Compounds That Bind Tsg101 Inhibit Viral Budding of HSV-1/2 and HIV from Cells. J. Virol. 2021;95:e00190-21. doi: 10.1128/JVI.00190-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.