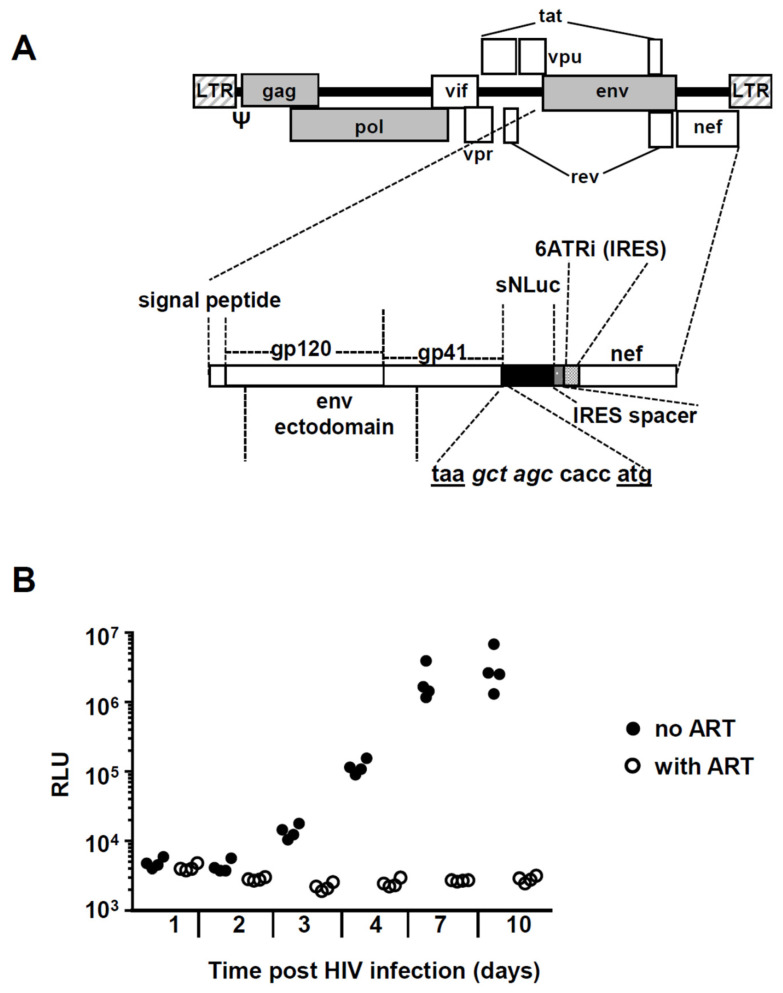
Figure 2.
Experimental strategy to quantify in vitro HIV-1 replication in PBMC. (A) Simplified schema of the replication-competent R5-tropic HIV-1 reporter virus vNL-sNLuc.6ATRi-B-Bal.ecto. The vector was based on the NL-Bal.Env.ecto-derived molecular clone of HIV-1 expressing Renilla luciferase previously generated by Ochsenbauer and Astronomo et al. [50,51]. The reporter gene (indicated as the black area) is a secreted form of nanoluciferase (encoded by the reporter gene sNLuc); nef expression is regulated by an internal ribosome entry site (IRES). The sNLuc.IRES cassette was inserted between the NL.Bal.ecto env and nef genes; 6ATRi is a truncation (“TR”) fragment derived from the encephalomyocarditis virus (EMCV) IRES and contains the “wild type” (A)6 (i.e., “6A”) bifurcation loop. The nucleotide sequence shows the junction between the stop codon of env (taa) and the start codon (atg) of sNLuc, and contains a Nhe I restriction site (gct agc) and the translation initiation Kozak sequence (ccacc). The construct contains a 26 nt “IRES spacer” between the sNLuc gene and the IRES element, as previously described [51]. (B) Example of an in vitro HIV-1 infection experiment. PBMC were infected at MOI 0.2 in the presence (empty circles) or absence (full circles) of 1 µM raltegravir and 1 µM efavirenz. Aliquots from supernatants were taken at the indicated time points (x-axis) and nanoluciferase activity was determined using the Promega Nano-Glo© kit (y-axis, RLU: relative light units).