Abstract
Life span determination in normal human cells may be regulated by nucleoprotein structures called telomeres, the physical ends of eukaryotic chromosomes. Telomeres have been shown to be essential for chromosome stability and function and to shorten with each cell division in normal human cells in culture and with age in vivo. Reversal of telomere shortening by the forced expression of telomerase in normal cells has been shown to elongate telomeres and extend the replicative life span (H. Vaziri and S. Benchimol, Curr. Biol. 8:279–282, 1998; A. G. Bodnar et al., Science 279:349–352, 1998). Extension of the life span as a consequence of the functional inactivation of p53 is frequently associated with loss of genomic stability. Analysis of telomerase-induced extended-life-span fibroblast (TIELF) cells by G banding and spectral karyotyping indicated that forced extension of the life span by telomerase led to the transient formation of aberrant structures, which were subsequently resolved in higher passages. However, the p53-dependent G1 checkpoint was intact as assessed by functional activation of p53 protein in response to ionizing radiation and subsequent p53-mediated induction of p21Waf1/Cip1/Sdi1. TIELF cells were not tumorigenic and had a normal DNA strand break rejoining activity and normal radiosensitivity in response to ionizing radiation.
The finite life span of normal human cells (16) is thought to be caused by the shortening of telomeres. Telomeres are nucleoprotein structures that protect the ends of eukaryotic chromosomes (4) and are composed of tandem G-rich repeats maintained by telomerase, an RNA-protein complex which synthesizes telomeric repeats de novo (9). Vertebrate telomeres contain tandem (TTAGGG)n repeats (24) which are bound to a unique family of telomere binding proteins (6). Due to incomplete replication of the DNA termini (27), human somatic cells lose telomeric DNA each time they divide (1, 11, 15, 33). The telomere hypothesis proposes that the shortening of telomeres of one or more chromosomes will eventually lead to senescence (10, 12, 27) and that the expression of telomerase activity in cancer cells may be required for cell immortality (7, 19, 22).
In yeast and euplotes the catalytic domain subunit of telomerase is required for in vivo telomere maintenance (20). The human telomerase complex contains at least two components, the RNA template component hTR (8) and a conserved catalytic subunit, hTERT (13, 18, 21, 25).
Recently it has been shown that hTR and hTERT are sufficient for reconstitution of telomerase activity (2, 35) and that forced expression of hTERT in normal human cells leads to reconstitution of telomerase activity, telomere elongation, and extended replicative potential (5, 32).
Normal human diploid fibroblasts are chromosomally stable during most of their life span; however, near senescence they accumulate a significant number of chromosomal aberrations, including a large number of dicentric and ring chromosomes (3, 30). The extension of the replicative life span and the bypass of senescence by DNA tumor viruses or p53 alterations (28) are frequently associated with chromosomal instability (37) and the formation of dicentric chromosomes near crisis (7). Resolution of these aberrant structures is associated with reactivation of telomerase and maintenance of telomeres (7). These findings suggest that critical telomere shortening is associated with chromosomal instability and that telomere elongation may be associated with chromosome stabilization. Furthermore, telomerase has also been directly implicated in the healing of broken chromosome ends (14, 23, 36). The unique phenomenon of life span extension by telomerase enables us to use an isogenic system to directly test the role of telomerase and telomeres in the maintenance of genomic integrity during life span extension or the induction of DNA damage. Furthermore, genomic stability in telomerase-induced extended-life-span fibroblast (TIELF) cells has significant implications for their use in cell- and gene-based therapeutic strategies to overcome the senescence barrier in vivo.
In this work, we investigated the possibility that forced expression of telomerase, leading to the generation of long telomeres and the extension of the cellular life span, may result in genomic instability and checkpoint-related defects in TIELF cells. Our results indicate that TIELF cells are genetically stable, are nontumorigenic, have an intact p53-dependent G1 checkpoint in response to DNA damage, and can rejoin double-strand DNA breaks normally.
MATERIALS AND METHODS
Cell cultures and retroviral infections.
The BJ neonatal human fibroblast cell strain was grown in α-minimal essential medium supplemented with 10% fetal bovine serum, and viral infections with pBabe and pBabest2 were performed as described previously (32). The pBNhEST2HA virus was constructed by excision of the EcoRI/SalI fragment of hTERT in plasmid pCINeo-hEST2HA and its ligation into the pBabe plasmid backbone. Plasmid pBA143 carried a dominant-negative mutant of p53 protein (p53Ala143) which was subcloned into a pBabe-Ires-EGFP construct and subsequently used to infect TIELF cells as described previously (32). Fluorescence-activated cell sorter-sorted EGFP+ cells were subsequently subjected to 6 Gy of radiation and cell cycle analysis.
Comet assay.
Briefly, 2 × 104 exponentially growing cells were trypsinized, suspended in phosphate-buffered saline (PBS), and irradiated on ice. After irradiation an aliquot of 1.5 ml of 1% low-melting-point agarose held at 50°C was added to the tube, and the suspension was rapidly pipetted onto a glass microscope slide. For a neutral comet assay, cells were lysed at 55°C for 2 h in buffer containing 30 mM EDTA and 0.5% sodium dodecyl sulfate (pH 8.5) and were washed by submersion in TBE buffer (90 mM Tris, 2 mM EDTA, 90 mM boric acid [pH 8.5]) for 3 h with three changes of buffer. This was followed by electrophoresis in fresh TBE buffer at 0.6 V/cm for 25 min. Then the slides were rinsed with distilled water and stained for 30 min in 2.5 μg of propidium iodide (PI)/ml.
The individual nuclei with broken DNA drawn out of the nucleus by electrophoresis form “comet-like” structures. The comets were digitized, and the tail moment was calculated as described elsewhere (26) by Northern Eclipse software (Empix). One hundred comets from each sample were analyzed for each dose or time point. No attempt was made to select comets, other than to avoid obvious debris or comets that were spaced too closely or overlapped. The normalized tail moment was used as a parameter to indicate DNA damage (26). Three independent experiments were done, and the mean ± standard deviation was expressed in the final plot.
Radiation survival assay.
Cells were irradiated with 60Co gamma rays at a fractionated low dose rate of 0.025 Gy/min at 37°C. This dose was chosen to be low enough that repair of radiation damage would be possible. Two million cells were grown in T75 flasks until they reached confluency. Following irradiation the cells were held at 37°C for 24 h before being trypsinized, counted, diluted, and plated for colony formation. Survival was calculated by using the cell count obtained just prior to plating. The cells were plated for a colony formation assay at three consecutive 10-fold dilutions and then were incubated for 10 to 14 days before being stained with methylene blue in 50% ethanol. Colonies containing more than 50 cells were counted as survivors, and percent survival was calculated as the ratio of plating efficiency for the treated group to that for an untreated control.
Cell cycle analysis.
Cells were fixed with 70% ethanol, washed in PBS, and resuspended in PBS with 0.12% Triton X-100–0.12 mM EDTA containing 50 μg of RNase. After the addition of PI (at 50 μg/ml), the DNA content was measured without gating. The cell cycle was quantitated by using the fully automated MODFIT program, and the G1/S ratio was calculated. Results of three independent experiments were averaged and plotted.
RESULTS
Analysis of telomeric DNA by terminal restriction fragment (TRF) analysis and telomere fluorescent in situ hybridization (FISH).
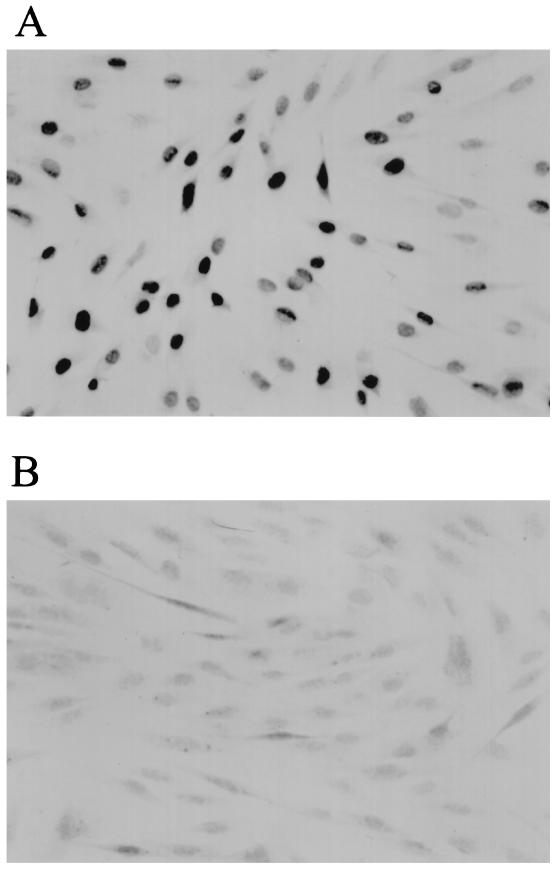
TIELF cells were generated by infection of BJ cells (expressing endogenous hTR) at population doubling (PD)50 with pBabest2 retrovirus expressing hTERT as described previously (32). In this experiment, BJ cells infected with the control virus (pBabe) senesced at approximately PD 80. Hence, hTERT-expressing cells with PDs beyond this value are considered TIELF cells. TIELF cells expressed the hTERT protein when assayed by immunohistochemistry with a rabbit polyclonal antibody (Fig. 1A). No staining was observed in the parental strain expressing the control construct (Fig. 1B). A control pBNhEST2HA virus carrying a C-terminally tagged cDNA for hTERT was also used in these experiments. This construct was unable to elongate telomeres or extend the life span despite full reconstitution of telomerase activity in BJ cells (data not shown). This indicates that the hemagglutinin tag interferes with telomere elongation in vivo and that telomerase activity per se is not sufficient for life span extension.
FIG. 1.
hTERT staining of TIELF90 cells (A) and control BJ cells (B). Confluent BJ and TIELF cells were fixed and stained with a polyclonal rabbit antibody against hTERT by using the Vectastain ABC system. A strong and punctate nuclear signal was evident in TIELF cells.
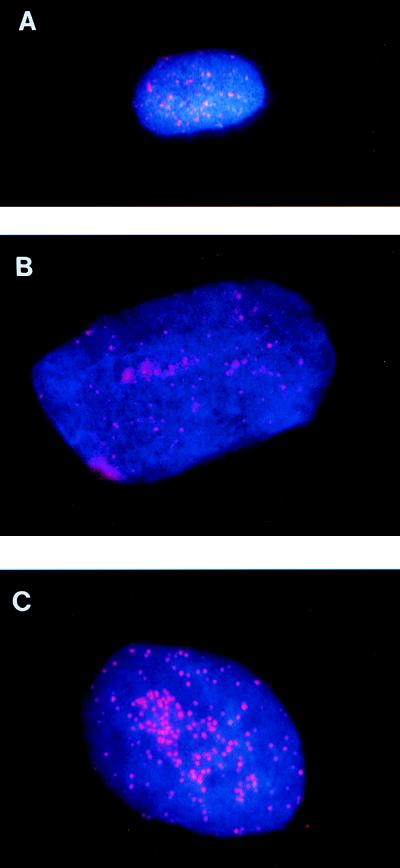
Analysis of telomeric DNA in nontagged hTERT-expressing cells indicated that after hTERT expression, the mean TRF length increased at a rate of approximately +116 bp/PD (average of two independent experiments) between PD 59 and PD 112. TIELF cells at PD 112 (TIELF112 cells) had a mean TRF length of 16.8 ± 1 kb, which was comparable to, and slightly longer than, germ line (data not shown). Analysis of mean TRF length was performed by TeloRun Software (1, 7, 12, 33). Analysis of telomeric DNA on individual telomeres by FISH (Fig. 2) revealed that (i) young cells (Fig. 2A) have a substantially more intense signal than old cells (Fig. 2B) and (ii) the telomeric signals on most chromosomes in TIELF cells are more homogeneous and intense than those on either young or old cells (Fig. 2C).
FIG. 2.
Analysis of telomeric DNA by telomere FISH during life span extension of TIELF cells. Interphase telomeres were analyzed by telomere FISH with superimposed 4′,6-diamidino-2-phenylindole (DAPI) staining. The telomere signal intensity diminished with increasing passages and was significantly restored in TIELF cells. Young BJ31 (A), old BJ78 (B), and TIELF (C) cells were grown on the same chamber slide. A telomere-specific probe (Oncor) was used to detect (TTAGGG)n repeats as previously described (17) and according to Oncor’s protocols.
Chromosomal stability in TIELF cells.
The question of chromosomal stability in TIELF cells was addressed by cytogenetic G-banding analysis of metaphase chromosomes in mass cultures of young control BJ31 cells (BJ cells at PD31, without telomerase activity), BJ66 cells (telomerase-positive cells at PD 66; TRF length, 11.9 ± 0.8 kbp), TIELF92 cells (TRF length, 14.1 ± 0.6 kbp), and TIELF140 cells. Analysis of metaphase spreads by G banding revealed no significant differences between BJ31, BJ66 (n = 1 of 105), and TIELF cells (n = 4 of 94; 50 of these metaphases were obtained from TIELF140 cells) (P = 0.2 by a two-tailed Fisher’s exact test). All aberrations were found in TIELF92 cells (4 of 44), and no aberrations were found in TIELF140 cells (0 of 50). Comparison of TIELF92 aberrations (4 of 44) to aberrations in control BJ cells (0 of 55) by Fisher’s exact test showed a significant difference (P = 0.036). However, no further aberrations or significant differences were observed when TIELF cells were passaged further (aberrations were found in 0 of 50 TIELF140 cells versus 1 of 50 BJ31 cells). (The nonclonal chromosomal aberrations which were detected in TIELF92 cells included 5p+, 2q+, and balanced translocation t(12;14) (Fig. 3). A more detailed analysis of possible chromosomal aberrations was performed by spectral karyotyping (SKY) (29). This analysis uses colored fluorescent chromosome-specific paints that provide a complete analysis of the human chromosomal complement. Thus, chromosomal rearrangements can be identified by the juxtaposition of different colors along a single chromosome. The SKY analysis confirmed the results of the G-banding analysis (Fig. 3) and revealed one additional structural change, 12p−, in TIELF cells (SKY analysis is more sensitive than G banding). Even with the addition of this new aberration to our previous data set, no significant statistical difference between BJ (n = 1 of 105) and TIELF (n = 5 of 94) cells was present (P = 0.1 by a two-tailed Fisher’s exact test). None of the aberrations detected were identical, and hence they do not represent clonal changes. The frequency of aberrations found in TIELF cells (5%) is comparable to that in other normal fibroblast strains (3 to 8%) (37). We also analyzed numerical changes as a measure of chromosomal stability, using a chr8-specific centromeric probe (D8Z2; Vysis Inc) by interphase FISH analysis. Analysis of 400 cells from each strain (1,200 total) did not reveal any evidence of aneuploidy. Percentages of aneuploid cells were as follows: for BJ31 cells, 7%; for BJ66 cells, 8%; and for TIELF cells, 5%). Further analysis of two independent clones from our previous studies (32) (TIELF clone 1 and TIELF clone 2) at PD 154 by G banding (n = 50) revealed 3 and 4% aberrant chromosomes, frequencies comparable to that in young BJ31 cells (3%).
FIG. 3.
G-banding and SKY analyses of metaphases from TIELF cells containing a t(12;14) translocation (arrow) and other aberrations. Structural chromosome aberrations were detected in metaphases from TIELF cultures. Cells were grown as described elsewhere (32) and were split 1:16 in growth medium without G418 for 24 h before any treatment. Well-spread G-banded metaphases (550 band level) from BJ31, BJ66, and TIELF strains were analyzed. These were coded prior to analysis and scored double-blind. (A) G banding of the t(12;14). (B and C) SKY analysis of the t(12;14). (D) SKY and G banding of cells shown in panel A. (E) 2q+, structural aberration shown by the solid bar. (F) 5p+ aberration. (G) 12p− deletion. A SKY hybridization and detection kit (SD-200 Bio system; Applied Spectral Imaging Inc.) was used to visualize all human chromosomes in 23 to 24 colors. Chromosomes were analyzed by a combination of Fourier spectroscopy, charge-coupled device imaging, and computerization to excite and measure the emission spectra simultaneously for all dyes in the spectral range and from all points in the metaphase spread (29). Images were analyzed by using SKYVIEW software.
DNA strand break rejoining activity and radiation sensitivity are intact in TIELF cells.
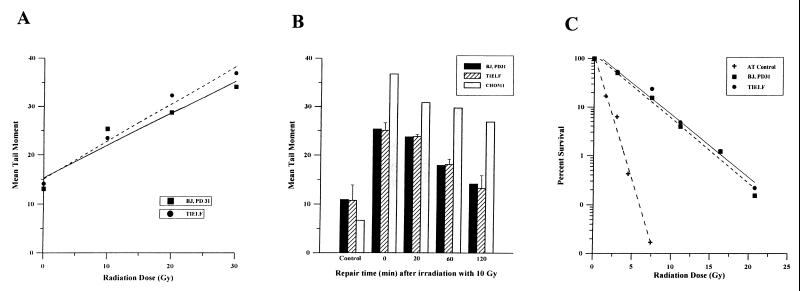
To determine if TIELF cells have a deficiency in DNA repair, the following experiments were performed. TIELF and BJ31 control cells were exposed to increasing doses of ionizing radiation (0 to 30 Gy), and DNA strand breaks and rejoining were analyzed by the comet assay (26). There was a linear relationship between the tail moment (a measure of the DNA double-strand breaks) and the radiation dose, and this relationship was identical in BJ31 and TIELF cells (Fig. 4A). Cells were tested for their ability to rejoin DNA double-strand breaks following 10 Gy of ionizing radiation. The normalized tail moments, measured at different times after irradiation, were identical in BJ31 and TIELF cells (Fig. 4B). Similar results were also obtained for single-strand break rejoining (data not shown).
FIG. 4.
Responses of BJ31 and TIELF cells to ionizing radiation: generation of DNA double-strand breaks (A), rejoining kinetics of double-strand breaks (B), and clonogenic survival after low-dose rate irradiation (C). In all assays BJ31 and TIELF cells behave identically. (A) Ionizing radiation dose versus tail moment of BJ31 and TIELF cells as measured immediately after irradiation by the neutral comet assay. (B) Mean tail moment as a function of time after a dose of 10 Gy in BJ31, TIELF, and positive-control CHO511 cells (Ku-70 deficient), which have impaired DNA repair and show higher tail moments. (C) Clonogenic survival as a function of ionizing radiation dose. Cells were irradiated with 60Co gamma rays at a low dose rate of 0.025 Gy/min at 37°C. This dose rate was chosen to be low enough that cell survival could be influenced by repair of radiation damage occurring during the treatment. Two million cells were grown to confluency. Following irradiation the cells were held at 37°C for 24 h before being trypsinized, counted, diluted, and plated for colony formation. Survival was calculated by using the cell count obtained just prior to plating. The cells were plated for a colony formation assay and then incubated for 10 to 14 days before being stained. Colonies containing more than 50 cells were counted. Survival was expressed as the ratio of the plating efficiency of the irradiated cells to that of control cells.
Finally, we used continuous low-dose ionizing radiation to measure long-term clonogenic survival and relative radiosensitivities. There was no difference between the survival of BJ31 cells and that of TIELF cells at these biologically relevant doses of ionizing radiation (Fig. 4C).
Preserved radiation-induced p53-dependent G1 checkpoint and lack of tumorigenicity in TIELF cells.
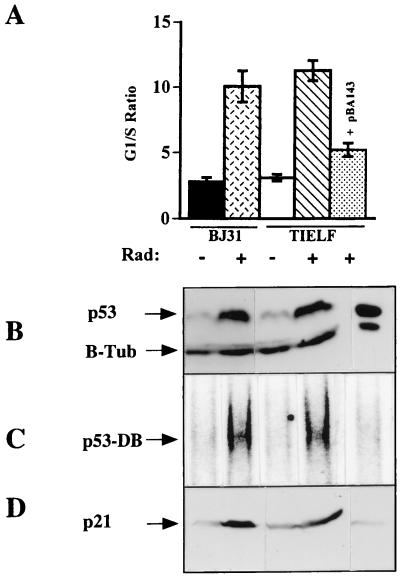
To determine if the DNA damage G1 checkpoint was retained in TIELF cells, cell cycle analysis was performed 24 h after exposure to 6 Gy of ionizing radiation, and the results of three experiments were obtained (Fig. 5). Both the BJ31 and the TIELF cells undergo a G1 arrest as measured by the increased G1/S ratio. There was no significant difference in the G1/S ratio between the two cell strains (G1/S ratios, 10 for BJ cells and 11 for TIELF cells). Therefore the radiation-induced G1 checkpoint appears to be intact in both strains. Consistent with the wild-type function of p53 protein in their parental BJ cells (34), TIELF cells were able to induce p53 protein (Fig. 5B) in response to 6 Gy of ionizing radiation and to upregulate its major downstream transcriptional target, p21Waf1/Cip1/Sdi1 (Fig. 5D). Since simple upregulation of the p53 protein level in response to DNA damage is not a measure of its functionality, we measured the specific DNA binding activity of p53 protein by an electromobility shift assay (EMSA) (34). After normalization of the increased level of p53 protein postradiation to that of unirradiated cells, EMSA was performed and detected an increase in the specific activity of p53 protein independent of its level in both BJ31 and TIELF cells (Fig. 5C). The increase in the specific DNA binding activity of p53 protein was sufficient to upregulate p21Waf1/Cip1/Sdi1 protein (Fig. 5D) in both strains. This induction was p53 dependent, since expression of a dominant-negative form of p53 protein in TIELF cells (p53Ala143) was able to reduce its DNA binding activity and led to reduced induction of p21Waf1/Cip1/Sdi1 (Fig. 5D) and a decreased G1/S ratio (Fig. 5A).
FIG. 5.
(A) Cell cycle analysis of BJ31 and TIELF cells in response to ionizing radiation. Cells were fixed with 70% ethanol, washed in PBS, and resuspended in PBS with 0.1% Triton X-100–0.12 mM EDTA containing 50 μg of RNase. After the addition of PI (50 μg/ml), the DNA content was measured without gating. The cell cycle was quantitated by using the fully automated MODFIT program, and the G1/S ratio was calculated. Solid bar, untreated BJ31 cells;crosshatched bar, BJ31 cells exposed to 6 Gy of ionizing radiation; open bar, untreated TIELF cells; hatched bar, TIELF cells exposed to 6 Gy of ionizing radiation; stippled bar, TIELF cells carrying pBA143, a dominant-negative form of p53, exposed to 6 Gy of ionizing radiation. Results of three experiments were pooled, and the means and standard deviations were plotted. (B) Western blot of p53 and beta-tubulin (B-Tub) proteins. TIELF and BJ cells were exposed to 6 Gy of ionizing radiation. Three hours postradiation, equal amounts of protein lysate were resolved and p53 protein was detected with a DO-1 antibody as previously described (34). A beta-tubulin antibody was used as a loading control. (C) DNA binding activity of p53 protein as measured by an EMSA as described previously (34). (D) Western blot analysis of p21Waf1/Cip1/Sdi1 as measured by an SC-817 antibody.
We investigated as well the tumorigenic potential of TIELF cells by injection of 2 × 106 BJ31 and TIELF cells into the leg muscles of six CB17 scid mice. No tumors were formed after 160 days.
DISCUSSION
It has been proposed that telomere shortening during replicative aging of normal cells eventually generates antiproliferative signals which activate p53 protein and subsequent G1 arrest, observed at senescence (34). In the presence of factors, such as simian virus 40 large T antigen (SV40LT) or E6, which are known to bind and inactivate p53 protein, the telomere-shortening signal is bypassed, leading to more telomere shortening and extension of the life span and subsequent genomic instability (7). Extension of the life span by forced telomerase expression, however, may be fundamentally different from that of SV40LT or other DNA tumor viruses. Telomerase expression prevents the antiproliferative signal generated by telomere shortening at senescence, possibly by telomere elongation. We suggest that prevention of the p53-mediated antiproliferative signal in response to telomere shortening allows cells to divide further. Extension of the life span by SV40LT, however, relies on inactivation of p53 and pRb proteins, and telomere shortening continues to persist until crisis. This suggests that life span extension by forced telomerase expression may not interfere with the p53-dependent signaling pathway, and therefore TIELF cells would retain their genomic integrity (31) at least by 60 PD postsenescence.
Cytogenetic analysis of TIELF cells by G-banding and SKY analyses revealed that at PD92 TIELF cells have a significantly higher number of aberrations than their young parental BJ cells. However, subsequent passaging of TIELF cells to PD 140 led to resolution of these nonclonal aberrations. The molecular mechanism behind the formation and resolution of these aberrations is not clear. The number of aberrations in TIELF cells was also measured by using a chr8-specific centromeric probe by interphase FISH of 400 TIELF cells and 800 control cells. We were unable to detect a significant number of aberrations compared to that in controls. In agreement with these results, TIELF cells had an intact p53-dependent G1 checkpoint in response to ionizing radiation as measured by four criteria: (i) an increased G1/S ratio postradiation, (ii) increased levels of both p53 protein and its specific DNA binding activity, (iii) increased levels of p21Waf1/Cip1/Sdi1, the downstream target of p53 protein, and (iv) the abrogation of the G1 checkpoint and the reduction of p53 protein activity and of the level of p21Waf1/Cip1/Sdi1 by dominant-negative expression of p53Ala143 in TIELF cells.
Telomerase is able to elongate DNA termini that are not complementary to its RNA template sequence (14) and is implicated in the healing of chromosome breaks in alpha thalassemia (14, 23, 36) by direct addition of (TTAGGG)n repeats to stabilize the ends. To investigate the probability that telomerase is involved in the repair of double-strand DNA breaks in response to DNA damage, we subjected telomerase-negative BJ cells and TIELF cells to varying doses of ionizing radiation; we then measured the DNA strand break rejoining ability of these cells by the comet assay and their radiosensitivity by a long-term colony survival assay. We were unable to detect a measurable difference in survival between BJ and TIELF strains. Although one cannot rule out the possibility that telomerase is involved in the addition of (TTAGGG)n repeats to sites of double-strand breaks, we cannot detect a measurable physiological difference between the two cell strains in the presence or absence of telomerase in response to ionizing radiation. Furthermore, these results indicate that extension of the life span does not affect DNA strand rejoining activity or long-term survival of cells in response to DNA damage.
We conclude on the basis of five criteria, cytogenetic analysis, radiation sensitivity, DNA break rejoining activity, radiation-induced p53-dependent G1 checkpoint, and tumorigenicity, that TIELF cells appear similar to their normal young counterparts. The preservation of a normal phenotype in TIELF cells makes it possible to generate difficult-to-establish cell strains for further genetic alterations and manipulation by homologous targeted recombination. These modified cells can further be used for different cell and gene therapy applications to overcome potential senescence in vivo.
ACKNOWLEDGMENTS
This work was supported by the Medical Research Council of Canada and the National Cancer Institute of Canada.
We thank Robert Weinberg for pCI-Neo-hEST2HA and Solomon Minkin for help with statistical analysis.
REFERENCES
- 1.Allsopp R C, Vaziri H, Patterson C, Goldstein S, Younglai E V, Futcher A B, Greider C W, Harley C B. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beattie T L, Zhou W, Robinson M O, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 3.Benn P. Specific chromosome alterations in senescent fibroblast cell lines derived from human embryos. Am J Hum Genet. 1976;28:465–473. [PMC free article] [PubMed] [Google Scholar]
- 4.Blackburn E H, Greider C W. Telomeres. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 5.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 6.Chong L, van Steensel B, Broccoli D, Erdjument H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 7.Counter C M, Avilion A A, LeFeuvre C E, Stewart N G, Greider C W, Harley C B, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng J, Funk W D, Wang S, Weinrich S L, Avilion A A, Chiu C, Adams R R, Chang E, Allsopp R C, Yu J, Siyuan L, West M D, Harley C B, Andrews W H, Greider C W, Villeponteau B. RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 9.Greider C W, Blackburn E H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 10.Harley C B. Telomere loss: mitotic clock or genetic time bomb? Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 11.Harley C B, Futcher A B, Greider C W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 12.Harley C B, Vaziri H, Counter C M, Allsopp R C. The telomere hypothesis of cellular aging. Exp Gerontol. 1992;27:375–382. doi: 10.1016/0531-5565(92)90068-b. [DOI] [PubMed] [Google Scholar]
- 13.Harrington L, Zhou W, McPhail T, Oulton R, Yeung D S, Mar V, Bass M B, Robinson M O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington L A, Greider C W. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- 15.Hastie N D, Dempster M, Dunlop M G, Thompson A M, Green D K, Allshire R C. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 16.Hayflick L, Moorhead P. The serial cultivation of human diploid strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 17.Henderson S, Allsopp R C, Spector D, Wang S, Harley C B. In situ analysis of changes in telomere size during replicative aging and cell transformation. J Cell Biol. 1996;134:1–12. doi: 10.1083/jcb.134.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilian A, Bowtell D L, Abud H E, Hime G R, Venter D J, Keese P K, Duncan E L, Reddel R R, Jefferson R A. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 19.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L, Coviello G M, Wright W E, Weinrich S L, Shay J W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 20.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 21.Meyerson M, Counter C M, Ng Eaton E, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, Bacchetti S, Haber D A, Weinberg R A. hEST2, the putative human telomerase catalytic subunit gene, is upregulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 22.Morin G B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 23.Morin G B. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 24.Moyzis R K, Buckingham J M, Cram L S, Dani M, Deaven L L, Jones M D, Meyne J, Ratliff R L, Wu J R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 26.Olive P L, Banath J P, Durand R E. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the “comet assay.”. Radiat Res. 1990;122:86–94. [PubMed] [Google Scholar]
- 27.Olovnikov A M. A theory of marginotomy. Dokl Biochem. 1971;201:394–397. [Google Scholar]
- 28.Rogan E M, Bryan T M, Hukku B, Maclean K, Chang A C, Moy E L, Englezou A, Warneford S G, Dalla-Pozza L, Reddel R R. Alterations in p53 and p16INK4 expression and telomere length during spontaneous immortalization of Li-Fraumeni syndrome fibroblasts. Mol Cell Biol. 1995;15:4745–4753. doi: 10.1128/mcb.15.9.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrock E, du Manoir S, Veldman T, Schoell B, Weinberg J, Ferguson M A, Ning Y, Ledbetter D H, Bar-Am I, Soeksen D, Garini Y, Ried T. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 30.Sherwood S W, Rush D, Ellsworth J L, Schimke R T. Defining cellular senescence in IMR-90 cells: a flow cytometric analysis. Proc Natl Acad Sci USA. 1988;85:9086–9090. doi: 10.1073/pnas.85.23.9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaziri H. Extension of life span by telomerase activation: a revolution in cultural senescence. J Anti-Aging Med. 1998;1:125–130. [Google Scholar]
- 32.Vaziri H, Benchimol S. Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol. 1998;8:279–282. doi: 10.1016/s0960-9822(98)70109-5. [DOI] [PubMed] [Google Scholar]
- 33.Vaziri H, Schachter F, Uchida I, Wei L, Zhu X, Effros R, Cohen D, Harley C B. Loss of telomeric DNA during aging of normal and trisomy 21 human lymphocytes. Am J Hum Genet. 1993;52:661–667. [PMC free article] [PubMed] [Google Scholar]
- 34.Vaziri H, West M D, Allsopp R C, Davison T S, Wu Y S, Arrowsmith C H, Poirier G G, Benchimol S. ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the posttranslational activation of p53 protein involving poly(ADP-ribose) polymerase. EMBO J. 1997;16:6018–6033. doi: 10.1093/emboj/16.19.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinrich S, Pruzan L, R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichtsteiner S, Kim N W, Trager J B, Taylor R D, Carlos R, Andrews W H, Wright W E, Shay J W, Harley C B, Morin G B. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 36.Wilkie A O, Lamb J, Harris P C, Finney R D, Higgs D R. A truncated human chromosome 16 associated with alpha thalassaemia is stabilized by addition of telomeric repeats (TTAGGG)n. Nature. 1990;346:868–871. doi: 10.1038/346868a0. [DOI] [PubMed] [Google Scholar]
- 37.Wolman S R, Hirschhorn K, Todaro G J. Early chromosomal changes in SV40-infected human fibroblast cultures. Cytogenetics. 1964;3:45–61. doi: 10.1159/000129797. [DOI] [PubMed] [Google Scholar]