Summary:
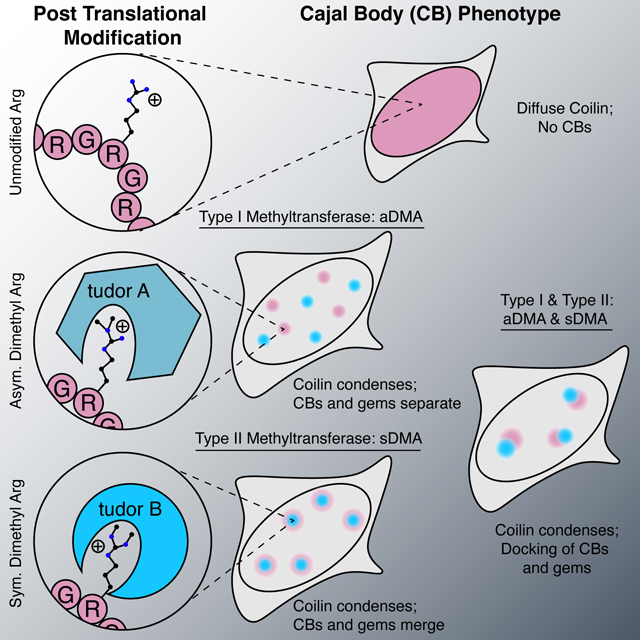
Biomolecular condensation is a widespread mechanism of cellular compartmentalization. Because the ‘survival of motor neuron protein’ (SMN) is implicated in the formation of three different membraneless organelles (MLOs), we hypothesized that SMN promotes condensation. Unexpectedly, we found that SMN’s globular tudor domain was sufficient for dimerization-induced condensation in vivo, while its two intrinsically disordered regions (IDRs) were not. Binding to dimethylarginine (DMA) modified protein ligands was required for condensate formation by the tudor domains in SMN and at least seven other fly and human proteins. Remarkably, asymmetric versus symmetric DMA determined whether two distinct nuclear MLOs – gems and Cajal bodies – were separate or “docked” to one another. This substructure depended on the presence of either asymmetric or symmetric DMA as visualized with sub-diffraction microscopy. Thus, DMA-tudor interaction modules – combinations of tudor domains bound to their DMA ligand(s) – represent versatile yet specific regulators of MLO assembly, composition, and morphology.
Keywords: Biomolecular condensation, membraneless organelle (MLO), tudor domains, dimethylarginine (DMA), post-translational modification, Cajal body, nuclear gem
eTOC/In Brief
The SMN tudor domain is sufficient for condensation by binding to dimethylarginine (DMA) modified protein ligands. Asymmetric versus symmetric DMA determines whether gems and Cajal bodies were separate or “docked” to one another.
Graphical Abstract
Introduction:
In the past ten years, molecular condensation of proteins and RNA has emerged as a prominent mode of subcellular organization (Corbet and Parker, 2020; Courchaine et al., 2016; Shin and Brangwynne, 2017). Our understanding of macromolecular structures formed by condensation has developed from descriptive characterization of organelles like P-granules and nucleoli to mechanistic models of how specific molecules promote phase transitions (Brangwynne et al., 2009; Brangwynne et al., 2011; Pak et al., 2016; Wang et al., 2018). While the mesoscale theoretical model of condensation has proven useful in many cases, an expansion of that conceptual framework has been called for to account for the specificity, selectivity, and function of membraneless organelles (MLOs) (Alberti et al., 2019; Banani et al., 2016; McSwiggen et al., 2019; Peng and Weber, 2019). The necessity for a mechanistic understanding of these phenomena is particularly evident in the nucleus where MLOs – nucleoli, Cajal bodies, gems, histone locus bodies, speckles and others – show considerable overlap in composition but nevertheless do not fuse into one body, instead maintaining their independence (Machyna et al., 2013; Strom and Brangwynne, 2019).
Those calls have been answered by significant steps forward in identifying key molecular features now known to promote the formation of condensates. Early in vitro work established that multivalent binding is a key attribute of condensate formation, and those results have led to a model of Arp2/3 actin regulatory condensates (Case et al., 2019; Li et al., 2012). Around the same time, intrinsically disordered regions (IDRs) in RNA binding proteins were shown to promote condensate formation (Kato et al., 2012; Kwon et al., 2013). These observations opened the floodgates for work seeking to explain how IDRs are encoded to produce condensates with the evident specificity and regulation seen for in vivo MLOs, resulting in a framework where IDRs consist of “sticker” interaction sites interspersed with flexible “spacers” (Choi et al., 2020). The capacity for amino acid residues to act as “stickers” provides a general explanation for why many IDRs readily phase separate in vitro (Lin et al., 2015; Nott et al., 2015; Patel et al., 2015; Sheu-Gruttadauria and MacRae, 2018; Wang et al., 2018). However, these studies have underscored the need to evaluate proteins suspected of promoting condensation on a case-by-case basis. Indeed, the IDRs of G3BP1 were recently reported to each contribute a unique effect on the condensation of stress granules (Guillen-Boixet et al., 2020; Sanders et al., 2020). Equipped with a wealth of in vitro data and an emerging model for biomolecular condensation, the field is turning to address the mechanisms governing the specificity of condensate formation in the context of in vivo endogenous MLOs.
This study focuses on the ‘survival of motor neuron protein’ (SMN), which is an essential component in the biogenesis of small nuclear ribonucleoproteins (snRNPs) that are required for splicing (Pellizzoni et al., 1998; Zhang et al., 2011). SMN deficiency results in the fatal childhood disease, spinal muscular atrophy (SMA); yet SMN’s snRNP biogenesis role does not adequately explain the disease phenotype, because splicing is a required activity of every living cell (Buhler et al., 1999; Pellizzoni et al., 1998). SMN depletion results in the loss of three membraneless compartments – gems, Cajal bodies, and U-bodies (Girard et al., 2006; Lee et al., 2009; Lemm et al., 2006; Raimer et al., 2017; Shpargel et al., 2003; Strzelecka et al., 2010a). Cytoplasmic SMN promotes the assembly of the Sm ring on snRNPs but its molecular function after import to the nucleus is less clear (Buhler et al., 1999; Meister et al., 2001; Renvoise et al., 2006). How loss of cellular compartmentalization may contribute to SMA is currently unknown, but compromised Cajal body integrity has been reported in patient tissue (Tapia et al., 2012). We therefore sought to understand how SMN supports the assembly of diverse MLOs.
Here, we examine the role of SMN in the formation of MLOs by isolating each region of the SMN protein to assess its condensation potential. To do so, we repurposed the ‘optodroplet’ assay in which light is used to induce dimerization of protein domain candidates for condensation (Shin et al., 2017). Unexpectedly, we found that the critical domain for condensation of SMN was its globular tudor domain, not its IDRs. We developed a new analysis of optodroplet data in order to make quantitative comparisons between condensation that accounts for the expression level of each protein. This allowed us to show that dimerization induced condensation of the SMN tudor domain depends on binding to the post-translational modification dimethyl arginine (DMA), and that this property is shared amongst numerous additional tudor domains. Finally, using specific inhibitors of DMA synthesis, we found that it modulates the specific composition of two endogenous MLOs, Cajal bodies and gems. Our structure-function analysis of MLO formation reveals that the DMA-tudor module defines the specific composition of certain in vivo condensate MLOs.
Results:
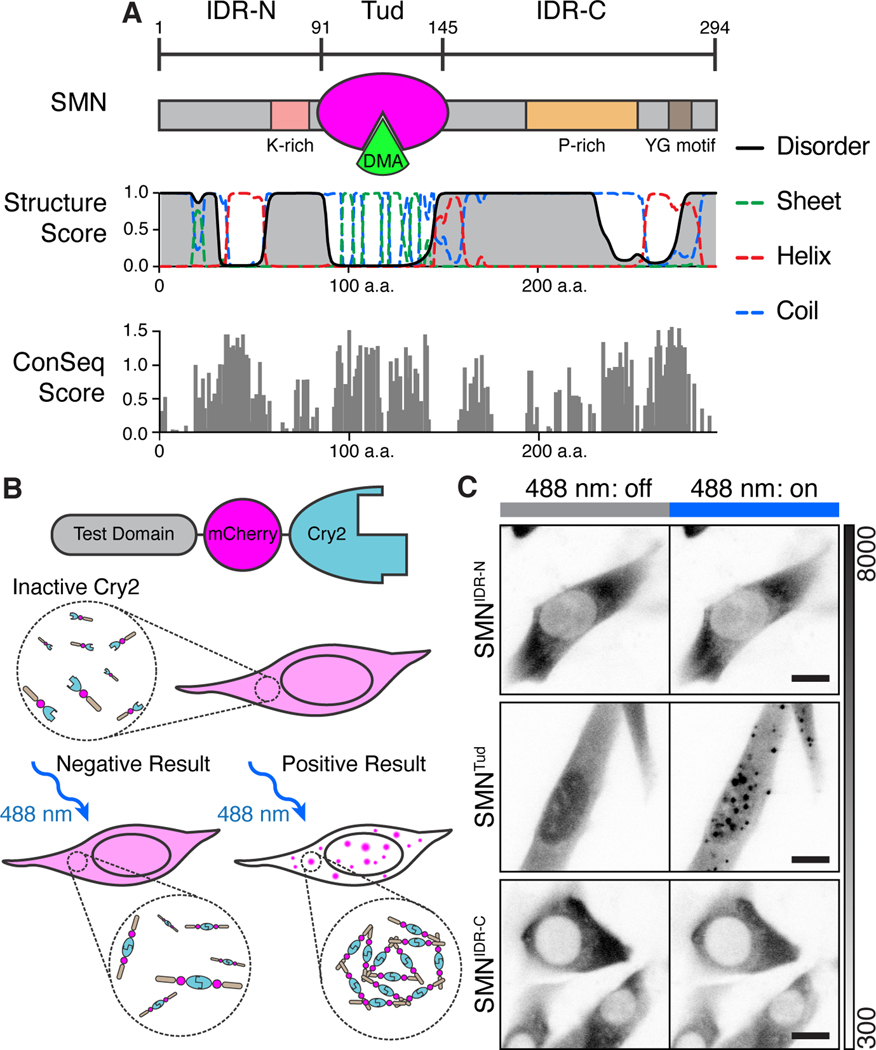
SMN has three regions as distinguished by overall secondary structure (Figure 1A). N-terminal and C-terminal IDRs flank a single tudor domain that binds symmetric DMA on snRNP proteins and other ligands (Friesen et al., 2001; Selenko et al., 2001; Sprangers et al., 2003; Wang and Dreyfuss, 2001; Zhang et al., 2011). The N-terminus contains a lysine-rich region, while the C-terminus has a proline-rich tract and a tyrosine/glycine repeat motif; these IDRs are involved in SMN complex formation in the cytoplasm (Grimm et al., 2013; Zhang et al., 2011). These features show high conservation amongst animal variants of SMN, and the IDRs resemble those that either form condensates or modulate the phase behavior of condensates once formed (Berezin et al., 2004; Kato et al., 2012; Kwon et al., 2013; Sanders et al., 2020; Wang et al., 2018). While some IDRs can form droplets spontaneously in vitro, they often require multimerization to overcome nonspecific interactions and condense in vivo (Protter et al., 2018; Shin et al., 2017). Thus, we fused each of the three SMN regions to the light-activated dimerization domain Cry2. This approach was previously exploited to form light-dependent ‘optodroplets’ with IDRs expressed in NIH-3T3 cells (Figure 1B) (Shin et al., 2017). Providing negative and positive controls, respectively, mCherry-Cry2 did not form clusters upon light-induced dimerization, whereas FUSIDR and hnRNP-A1IDR did (Figure S1A, Movie S1). As expected, full-length SMN-Cry2 formed some clusters without light induction with additional clusters formed upon activation (Figure S1B, Movie S2). We used this system to determine whether any portion of SMN directly promotes condensate formation in vivo.
Figure 1. The multimerized SMN tudor domain forms condensates in vivo.
A) Schematic representation of SMN domain architecture and accompanying secondary structure prediction. The tudor (Tud, magenta) domain binds DMA (green). Structure score is a unitless value for secondary structural properties predicted by the RaptorX algorithm. ConSeq scores are displayed for SMN residues aligned using Clustal Omega and computed using ConSeq (Berezin et al., 2004). The absolute value of conservation scores for the 50% most conserved residues are displayed. B) Diagram of the “optodroplet” condensation assay. Cry2 dimerizes upon blue light activation (488 nm); without added molecular interaction contributed by the test domain, condensation will not occur (Shin et al., 2017). If the test domain provides interactions to increase valency, condensation is observed as mCherry fluorescent foci. C) Micrographs of live cells undergoing blue light activation of Cry2 (180 s, blue bar). Grayscale bar given in analog-digital units. Scale bar = 10 μm.
Although we expected SMN’s IDRs to play a role in condensation, only its tudor domain (SMNTud) formed prominent clusters throughout the cell upon blue light activation (Figure 1C; Figure S1C, Movie S3). These clusters were heterogenous and dynamic; when photobleached, they showed 42 – 100% recovery with time constants of 30 ± 22 s (mean ± SD, n=10) (Figure S1D). When Cry2 was allowed to deactivate after cluster formation, SMNTud clusters dissipated within a few minutes (Figure S1E). Cluster fusion was infrequent but observable (Figure S1F). Together these results show that SMNTud clusters rapidly reorganize their internal molecular composition, consistent with liquid-like condensation rather than stable aggregates. We conclude that SMNTud is sufficient for the formation of dynamic condensates upon light-induced multimerization.
This finding was striking because tudor domains are small (60 amino acids), structured, and lack any obvious multivalency. We therefore asked whether condensate formation by SMNTud depends on the presence of its ligands, which are DMA modified proteins. To do so, we developed an image analysis method to quantify condensate formation as a function of protein expression level, which is measured as each cell’s mean mCherry fluorescence. Our clustering metric is based on variance of intensity for each pixel throughout the movie recorded during Cry2 activation (Figure S2A). Correcting for the noise model of the camera and artefactual effects on variance, we obtained a clustering metric for each cell by averaging all pixels within segmented cell-by-cell masks and plotted it against expression level (STAR Methods, Figure S2B&C). We determined a statistical threshold (Mann-Whitney U test) for the clustering metric of each cell relative to the mCherry-Cry2 negative control and found it to be at a clustering metric of approximately 49.44 ADU (Figure S2C).
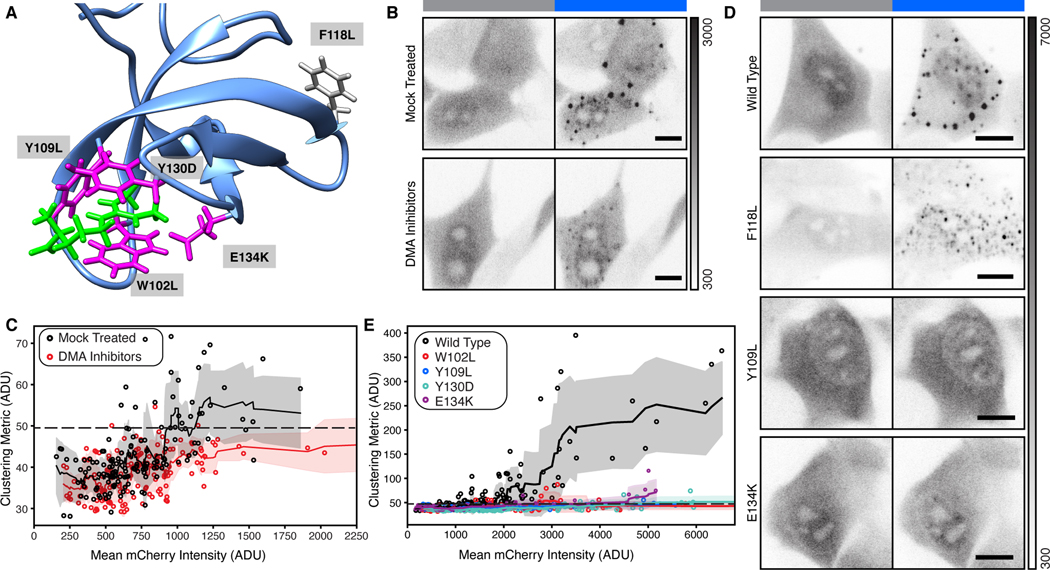
DMA modifications can either be asymmetric or symmetric (aDMA or sDMA), and each is produced by separate sets of methyltransferase enzymes (Figure S2D) (Branscombe et al., 2001; Tang et al., 2000). The SMN tudor domain has a higher affinity for sDMA but recognizes both sDMA and aDMA (Kd = 0.476 mM and Kd = 1.025 mM respectively) (Tripsianes et al., 2011). Accordingly, we chose two small-molecule inhibitors, MS-023 and EPZ015666, to deplete DMA modifications in NIH-3T3 cells (Figure S2E). MS-023 is a specific inhibitor of five type I methyltransferases that synthesize aDMA (Eram et al., 2016). EPZ015666 is a specific inhibitor of protein methyltransferase 5, which synthesizes most sDMA (Chan-Penebre et al., 2015). We found that prolonged DMA inhibition substantially reduced the number of cells that form condensates above our significance threshold, despite the prevalence of residual DMA modified proteins after the 48-hour treatment (Figure 2A-C; Figure S2E&F). From this we conclude that condensation of SMNTud depends on the availability of endogenous DMA ligands.
Figure 2. Formation of condensates by SMNTud depends on binding to DMA.
A) Published solution structure of the SMN tudor domain (blue) bound to sDMA (magenta). Three aromatic amino acids that make up the binding pocket and one associated with SMA are shown in green. An aromatic residue (F118L) not involved in DMA binding is shown in gray. PDB: 4A4E (Tripsianes et al., 2011). B & D) Micrographs of live cells with SMNTud either untreated or treated with DMA inhibitors (B), SMNTud wild-type, a null mutation F118L, a mutation to the aromatic DMA binding cage Y109L, and an SMA-associated mutation E134K (D). Grayscale bar given in analog-digital units. Scale bars = 10 μm. C & E) Quantification of live cells untreated or treated with DMA inhibitors (C), cells expressing wild-type or mutant SMNTud (E). Mean mCherry intensity and the cluster metric are given in analog-digital units (ADU). Solid line with shading is a rolling mean and standard deviation of 10 points. Each point is one cell. Dashed line represents a significance threshold relative to mCherry-Cry2 where a = 0.2 for the Mann-Whitney U test.
To verify that the loss of condensation is due to specific recognition of DMA by SMNTud, we tested a series of mutants in the optodroplet assay. We chose three individually mutated amino acids in the aromatic cage that accepts DMA (W102L, Y109L, & Y130D), a disease-causing mutation associated with SMA (E134K) that also disrupts DMA binding, and an uninvolved phenylalanine as a control (F118L) (Figure 2A) (Buhler et al., 1999; Pellizzoni et al., 1999; Tripsianes et al., 2011). All four mutations that disrupt DMA-binding virtually eliminated condensation, but F118L did not (Figure 2D&E; Figure S3). From these combined data, we conclude that condensation of SMNTud depends on binding to DMA ligands.
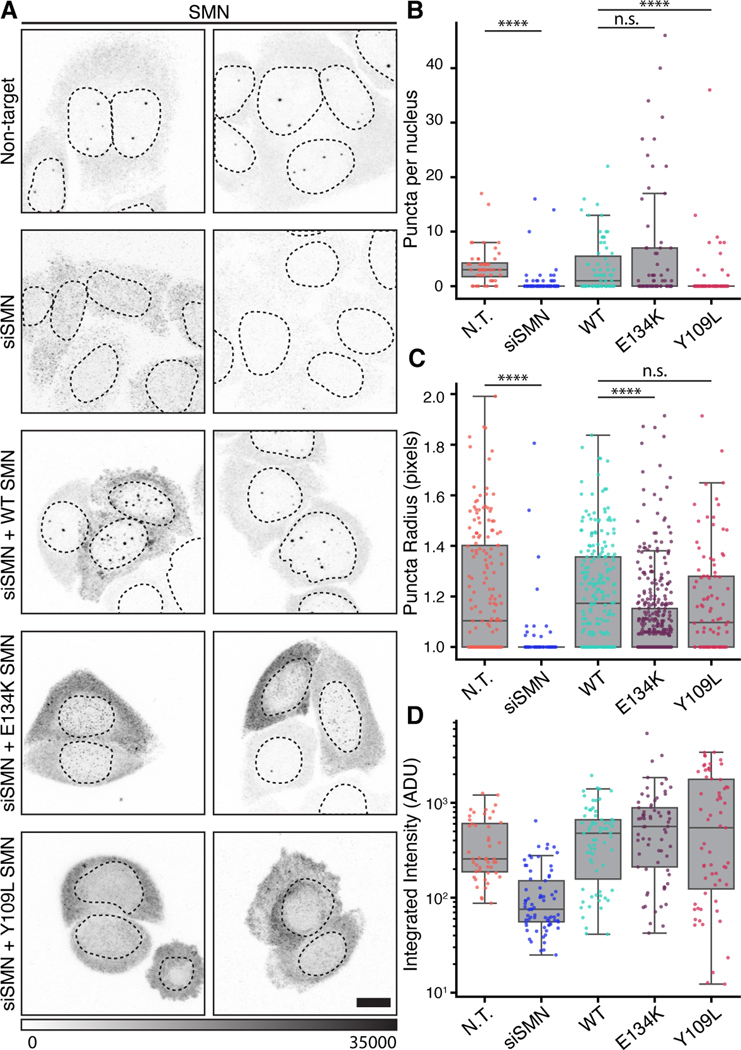
These findings prompted us to reexamine prior reports related to SMN’s role in MLOs and ask whether the same mutations prevent SMN from forming endogenous condensates. SMN depletion has been shown to disrupt nuclear MLOs and ectopic expression of SMN truncations and mutations can induce defects in tissue culture models (Girard et al., 2006; Lemm et al., 2006; Renvoise et al., 2006; Shpargel and Matera, 2005; Strzelecka et al., 2010a). However, the role of the SMN tudor domain has not been studied in the context of a knockdown and rescue despite the association of the E134K mutation with SMA (Pellizzoni et al., 1999). NIH-3T3 cells are a good model for optodroplet studies of SMN because they mostly lack MLOs that contain SMN, avoiding confounding effects of a preexisting assembly (Figure S4A&B). However, we wished to address the requirement for the tudor domain in the assembly activity of SMN. Instead of using NIH-3T3 cells, which lack SMN MLOs, we used HeLa cells, which normally display prominent SMN-rich MLOs known as gems; we then attempted to rescue the formation of gems in vivo by transfecting siRNA-resistant wild-type and mutant SMN constructs (Figure 3A, Figure S4C).
Figure 3. Integrity of the Tudor domain is required for formation of endogenous nuclear MLOs by full-length SMN.
A) Images comparing HeLa cells transfected with non-targeting oligo, siSMN oligo pool, wild type (WT) rescue construct, and mutant rescue constructs. Grayscale bar given in analog-digital units. Scale bar= 10 μm. B) Quantification of SMN-containing MLOs per nucleus in each condition shown in (A). Kruskal-Wallace test for multiple comparisons, p = 2.19 (10−12) C) Quantification of SMN condensate radius given in pixels for each condition shown in (A). Kruskal-Wallace test for multiple comparisons, p = 9.33 (10−15). D) Integrated fluorescence intensity of labeled SMN in cells for each condition, units given as analog-digital units (ADU). Individual samples were compared with Dunn’s post hoc analysis and indicated as not significant (n. s.) where p > 0.05 and **** where p < 0.0001.
Expression of full-length, wild-type SMN in cells depleted of endogenous SMN restored nuclear puncta, which likely correspond to gems (Figure 3A) (Young et al., 2001). In contrast, attempts to rescue the loss of wild-type puncta with SMN constructs bearing mutations to the tudor domain (E134K and Y109L) failed (Figure 3A). Quantification of these data demonstrates that the E134K mutation limited SMN to forming significantly smaller puncta that can be quite numerous, while Y109L nearly eliminated the punctate pattern in most nuclei (Figure 3B&C, Figure S4H). These observations were independent of whether SMN localization was assayed by primary SMN antibodies or via a myc epitope tag included in the rescue construct (Figure S4D). Careful examination of our confocal data also indicates that SMN was still able to reach the nuclear compartment, ruling out localization alone as the cause of these effects (Figure S4E&F). We note that SMN mutants had partial effects on the formation of coilin-rich Cajal bodies, implying that the assembly of these proteins may not have a strict dependency on one another (Figure S4G). Thus, the ability of full-length SMN to form gems is strongly compromised by these two mutations that reduce the affinity of the SMN tudor domain for DMA.
Tudor domains, including the SMN tudor, are known to bind DMA on multiple target proteins (Liu et al., 2012; Tripsianes et al., 2011). Therefore, we began testing known interactors of SMN for co-condensation with SMNTud. DMA modified Sm proteins in snRNPs are canonical interactors of SMN, making them obvious first candidates (Friesen et al., 2001). However, no snRNP marker colocalized with SMNTud condensates, suggesting that Sm proteins do not participate in optodroplet formation by SMNTud (Figure S5A-F). In contrast, endogenous coilin – the DMA-modified scaffolding protein of Cajal bodies – was detected in SMNTud nuclear condensates (Figure S5G). These nuclear condensates recruit coilin from a soluble pool, because the NIH-3T3 cells used do not have canonical (0.5 – 1.0 μm) Cajal bodies, though they do express coilin (Figure S4A&B). To determine if coilin association is essential for condensate formation by SMNTud, Coil knockout mouse embryonic fibroblasts were transduced with the Cry2 construct, and SMNTud condensates still formed in response to blue light (Figure S5H). These observations confirm that SMNTud can recruit coilin, a DMA-modified ligand, to condensates. We conclude that coilin is a non-essential ligand of SMNTud in the context of this assay: it partitions into SMNTud condensates but is not necessary for their formation.
To better understand the role of DMA ligands and to determine whether condensation of tudor domains is common, we turned to the eponymous protein of this domain family: D. melanogaster Tudor. The proteins Tudor, Aubergine, Vasa, and the oskar RNA make up the germ plasm condensate in flies (Trcek and Lehmann, 2019). DMA in the Aubergine N-terminus is required for germ plasm localization, and the binding between this region and the eleventh tudor domain of Tudor has been studied by co-crystallization (Liu et al., 2010). We generated a Cry2 construct with this tudor domain plus flanking sequence known to increase specificity (dmTudorTud) and a construct with three repeats of the Aubergine N-terminus fused to GFP (Aub3-GFP) (Figure 4A, Figure S6A). These constructs were introduced into NIH-3T3 cells. DmTudorTud was able to form condensates in the nucleus of NIH-3T3 cells without the expression of Aubergine showing that endogenous mouse DMA ligands suffice (Figure 4B). Accordingly, treatment with the sDMA inhibitor EPZ015666 prevented condensation (Figure 4C). The requirement for sDMA was further confirmed by the presence of SYM10 and SYM 11 epitopes in dmTudorTud condensates, two antibodies that recognize a constellation of sDMA modified sequences (Figure 4D). Co-expression with Aub3-GFP, predominantly modified by aDMA, blocked condensate formation (Figure 4E&F), suggesting that Aub3-GFP can compete with the endogenous proteins modified by sDMA. When lysine was substituted for arginine in Aub3-GFP, condensation was restored, confirming the necessity of the DMA-tudor interaction. Finally, the endogenous proteins that allow dmTudorTud to condense must be nuclear, because no condensation effect was seen in the cytoplasm. The recognition of a mouse protein by dmTudorTud likely reflects the previously recognized promiscuity of tudor domain recognition of DMA targets (Liu et al., 2012). Furthermore, it appears that the balance between sDMA and aDMA determines the degree of assembly mediated by the DMA- dmTudorTud module.
Figure 4. Methylated ligands partition into and compete for tudor domain binding sites.
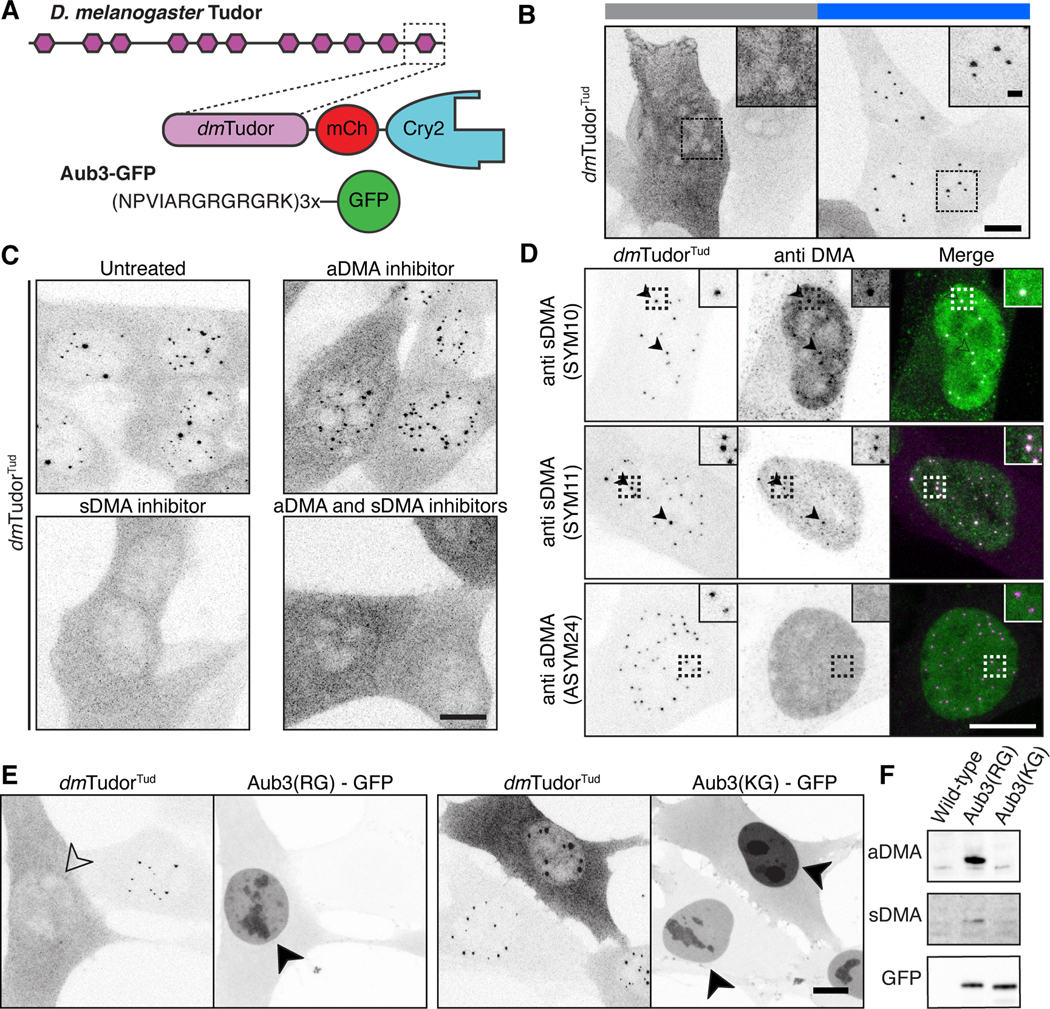
A) Schematic of D. melanogaster Tudor, dmTudorTud and its ligand Aub3-GFP (Liu et al., 2010). B) Fixed NIH-3T3 cells expressing only dmTudorTud in Cry2-inactive and Cry2-active states, revealing condensation property of dmTudorTud. C) Fixed cells expressing dmTudorTud, either untreated, treated with MS-023, EPZ015666, or both inhibitors. D) dmTudorTud counterstained for sDMA (SYM10 & SYM11) or aDMA (ASYM24). Arrowheads indicate colocalized condensate and counterstain. Dotted line insets are enlarged twofold. E) Fixed cells expressing dmTudorTud and either Aub3-GFP or the non-binding, control peptide with R to K mutations (KG). Condensates are inhibited by expression of intact Aub3-GFP (open arrowhead); both Aub3-GFP proteins concentrate in the nuclei (filled arrowheads) of transfected cells that lack condensates. F) Western blot of whole cell lysates of cells expressing Aub3(RG)-GFP, or Aub3(KG)-GFP. Each panel shows the range of ~25–40 kDa. Cry2 is active except where noted in panel B. Scale bars = 10 μm, inset scale bars = 2 μm.
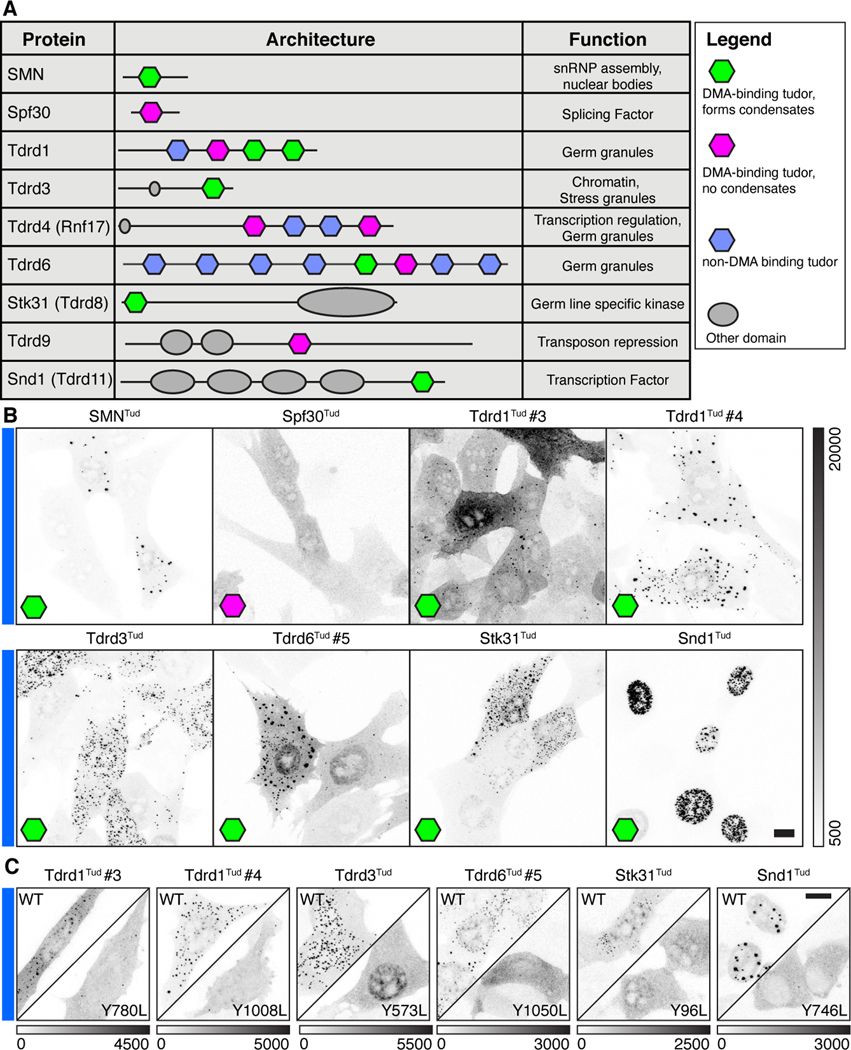
Given two examples of tudor domains that form condensates from two species, we asked if this property could be more broadly generalized. The human proteome has fifty-five annotated tudor domains from twenty-eight different proteins in the Uniprot database. Because each tudor domain binds a discrete set of DMA ligands due to amino acids surrounding the modified arginine, the condensation property observed for SMNTud could be unique (Liu et al., 2012; Tripsianes et al., 2011). Alternatively, the tendency to condense could be shared with other tudor domains. To test for generality, we selected a panel of twelve domains from nine different human proteins from diverse biochemical processes (Figure 5A). The twelve selected have the four aromatic amino acids required to form the binding pocket for DMA (Figure S6D). Of these domains, we provide evidence for six that formed condensates (Figure 5B, Figure S6E-G). As constructed, the assay is best suited to rule-in rather than rule-out domains, so we employed the clustering metric in live cells to assess whether expression level alone dictates whether a tudor domain makes condensates (Figure S6A). We compared SMNTud and Spf30Tud and found that Spf30Tud did not condense across a range of concentrations in spite of sequence, structural and interactome similarity to SMNTud (Figure S6C) (Tripsianes et al., 2011). To validate our findings for tudor domains that do form condensates, we repeated our experiment expressing tudor-Cry2 constructs with mutations in the consensus second tyrosine of the DMA binding motif (Figure S6D). All of these mutations profoundly reduced condensate formation (Figure 5C). Intriguingly, we found that SYM10 DMA epitopes reside in some but not all Tdrd8Tud condensates, aDMA inhibitors eliminate Tdrd3Tud cytoplasmic condensates but not nuclear condensates, and Snd1Tud localization itself changes when it can no longer bind DMA (Figure 5C, Figure S6F-H). Taken together, these data suggest that the ability of any tudor domain to mediate condensation may depend on the localization, availability, expression level, and methylation status of suitable partner molecules.
Figure 5. Condensation is a shared property of multiple human tudor domains.
(A) Table of proteins and schematics of human tudor domain proteins tested for condensation,with tudor domains containing either an intact binding site for DMA that form condensates (green), DMA binding tudor domains that do not form condensates (pink), or tudor domains lacking the DMA binding pocket (blue). Domain architecture and function correspond to Uniprot annotations. B) Fixed NIH-3T3 cells expressing Tudor-Cry2 constructs under Cry2-active conditions, corresponding to eight of the tudor domains above (see Figure S6 for more details). C) Fixed NIH-3T3 cells expressing Tudor-Cry2 constructs under Cry2-active conditions where the designated tudor domain is either wild-type (WT) or mutated as indicated to disable the aromatic DMA-binding cage. Grayscale bars given in analog-digital units. Scale bars = 10 μm.
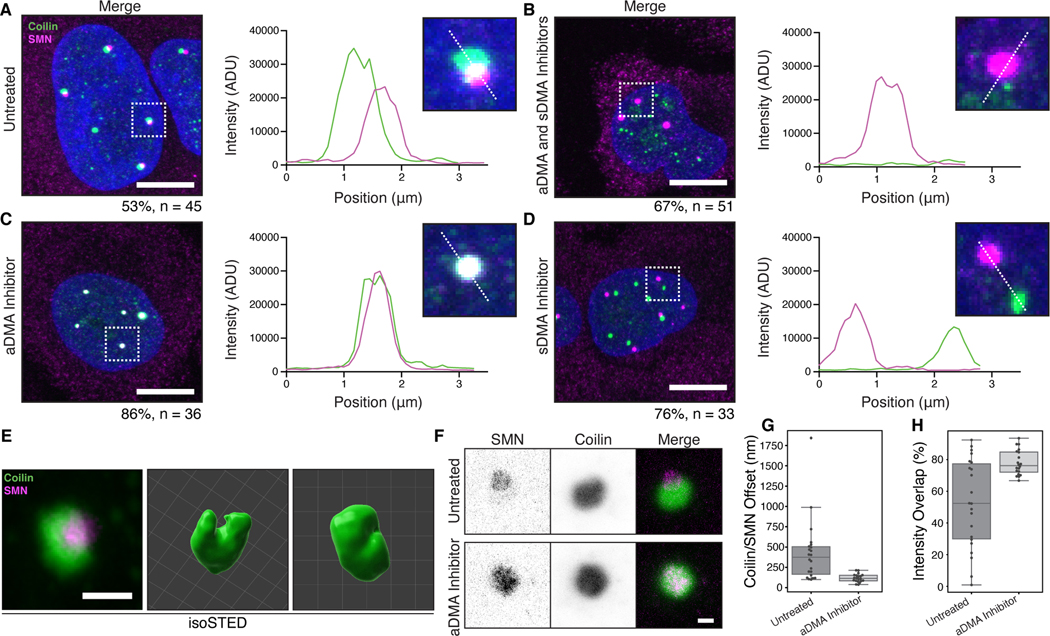
To better understand how DMA-tudor interactions affect endogenous MLOs, we asked what specific effects arginine dimethylation has on the Cajal body. HeLa cells were treated with the specific inhibitors of DMA synthesis, MS-023 and EPZ015666, and monitored for effects on Cajal bodies. Typically, HeLa cell Cajal bodies contain partially overlapping coilin and SMN domains (Figure 6A). The direction of these shifts is random but consistently observed ruling out chromatic aberration as their source (Figure S7A). Inhibition by both inhibitors results in the disassembly of Cajal bodies where residual coilin puncta lack the trimethylguanosine snRNP marker, in agreement with previous findings using a non-specific methylation inhibitor (Figure 6B; Figure S7B&C) (Hebert et al., 2002). We note that SMN-containing nuclear puncta remain, which are likely gems that can form though recognition of residual DMA modified proteins ( see Figure S2E) (Young et al., 2001). Thus, the integrity of the Cajal body nuclear organelle depends on modification of arginine, presumably on coilin.
Figure 6. Specificity of MLO composition depends on DMA modification.
A-D) Wild-type HeLa cells that are A) untreated, B) treated with MS-023 and EPZ015666, C) treated with only MS-023, or D) treated with only EPZ015666, and stained for SMN (magenta) and coilin (green) with accompanying inset and line profile plots. Cajal bodies and gems completely overlap when only sDMA is present; they are completely separate when only aDMA is present. Percentage of cells with the displayed phenotype out of total given below image. Scale bars = 10 μm. E) Micrographs generated with isoSTED nanoscopy of Cajal bodies. Volume renderings generated with Imaris. Scale bar = 500 nm. Grid lines = 200 nm. F) Representative lateral depletion STED micrographs of Cajal bodies in untreated HeLa cells, or cells treated with MS-023. Scale bar = 500 nm. G) Offsets between SMN and Coilin fluorescence intensity-weighted center of mass for Cajal bodies from untreated or MS-023 treated HeLa cells (n = 20 Cajal bodies per condition). H) Percent of intensity weighted overlap of SMN and Coilin fluorescence for Cajal bodies from untreated or MS-023 treated HeLa cells (n = 20 Cajal bodies per condition).
Treatment of cells with individual inhibitors revealed an important requirement for the organization of Cajal bodies, gems, and their specificity. Inhibition of aDMA synthesis merges coilin and SMN into one fully overlapping body (Figure 6C). In contrast, inhibition of sDMA synthesis induces the complete separation of the coilin and SMN domains into two distinct bodies (Figure 6D). Importantly, trimethylguanosine continues to colocalize with coilin in both conditions, indicating snRNPs are still present with coilin in Cajal bodies with both individual inhibitors (Figure S7B-E). Thus, the Cajal body is dependent on arginine methylation overall, and symmetrical dimethylarginine – the highest affinity SMN tudor domain ligand (Tripsianes et al., 2011)– is required for the merging of Cajal bodies and gems into a single nuclear body.
To further understand the relationship between these two proteins and their spatial regulation by DMA, we turned to stimulated emission depletion microscopy (STED) to assess the arrangement of SMN and coilin below the diffraction limit. Using a STED modality with isotropic resolution of about 32 nm, we found that the coilin-containing structure in untreated HeLa cells often has a cleft within which SMN resides (Figure 6E, Figure S7F). Assessing twenty Cajal bodies from twenty different cells per condition using lateral depletion STED, we observe that the offset between SMN and coilin is substantially diminished by inhibiting aDMA synthesis, and that the coilin cleft is no longer apparent (Figure 6F&G, Figure S7G&H). We suggest that normal levels of sDMA and aDMA appear to promote “docking” of the Cajal body to gems, where SMN and coilin adopt a ball-in-socket configuration. These two organelles can either share components or retain distinct identities based on the specific form of arginine methylation. Taken together, these observations demonstrate that the DMA-tudor module controls the compositional specificity of these nuclear MLOs.
Discussion:
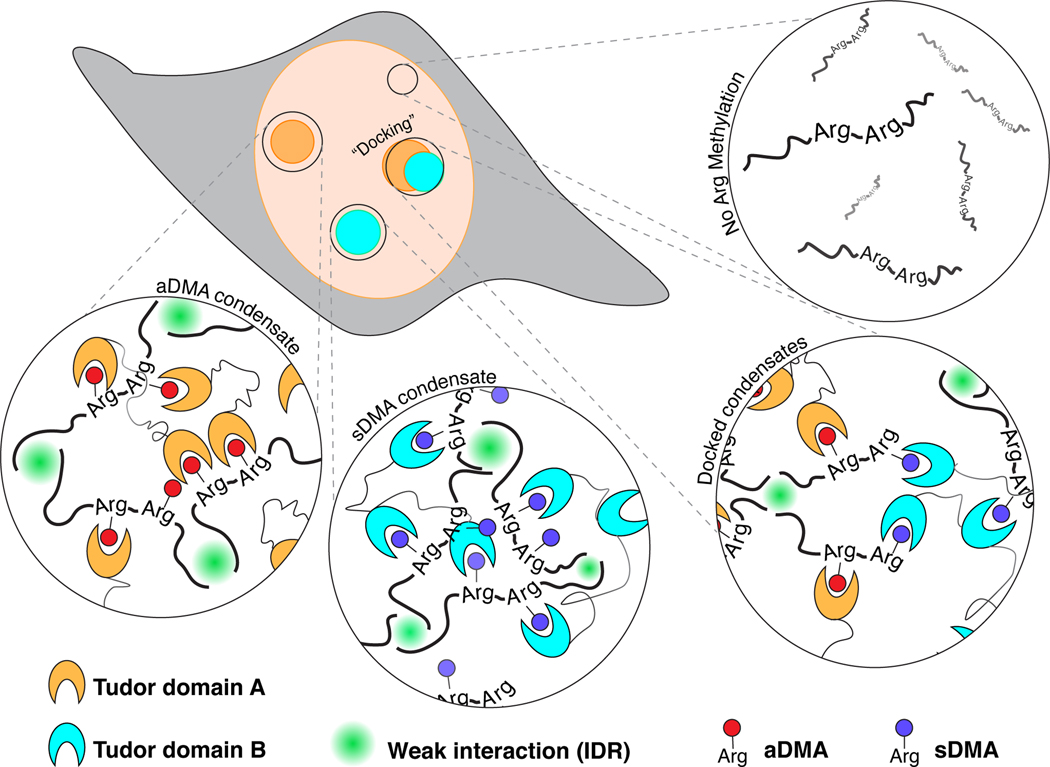
Two key concepts emerge from this study. First, globular tudor domains promote condensation in vivo, by binding specifically to ligands bearing the modified amino acid dimethylarginine or DMA. We have named this active unit the “DMA-tudor module” (Figure 7). Second, the particular constellation of DMA-tudor modules comprising aDMA and sDMA ligands determines the integrity of endogenous membraneless organelles (MLOs) exemplified by Cajal bodies and gems. Specifically, Cajal bodies require aDMA and sDMA, and DMA determines whether Cajal bodies and gems associate with one another. It was previously assumed that these MLOs mix their contents (Hebert and Matera, 2000), and that Cajal bodies form by biomolecular condensation; this study provides evidence for biomolecular condensation and mechanistic insights into the assembly and composition of this MLO (Corbet and Parker, 2020; Hyman et al., 2014; Shin and Brangwynne, 2017). Surprisingly, STED microscopy indicates these MLOs remain distinct and instead “dock” with one another when aDMA and sDMA are both present (Figure 7). Finally, the docking principle reveals rules governing the specificity of MLO composition controlled by post-translational modifications. We provide evidence that additional DMA-tudor modules involving tudor domain-containing proteins with vastly different functions have the capacity to mediate condensation in other contexts, such as chromatin and germ plasm.
Figure 7. Proposed model for MLO formation by DMA-tudor modules.
Working model of DMA modifications bound by tudor domains and how these DMA-tudor modules control the composition of MLOs. Unmodified arginine does not support assembly, while aDMA (red), and sDMA (blue) allow for assembly with tudor domain proteins (cyan and orange) in trans. If a protein bears both modifications, the two distinct bodies may “dock” with one another.
The DMA-tudor module is set apart from other specific interactions in condensates because it is based on a post-translational modification. In this way, it attains switch-like properties akin to the module formed by SH2 domains and phospho-tyrosine in Nck actin signaling condensates (Banjade et al., 2015). However, DMA-tudor condensates are not nucleated on a two-dimensional lipid membrane and do not change the charge of the modified amino acid. Nuclear MLOs like nucleoli and Cajal bodies are dynamic, disassembling during mitosis or cellular stress and reassembling quickly thereafter (Boulon et al., 2010; Strzelecka et al., 2010a). More broadly, remodeling of condensates in response to cell signaling has been proposed as a key mechanism of transcriptional regulation via the phosphorylation of the RNA polymerase II C-terminal domain (Guo et al., 2019; Kwon et al., 2013). The modification of arginine has already been proposed as a way by which cells may modulate condensation by IDRs in cis (Nott et al., 2015; Ryan et al., 2018); this model explains condensate formation by the RGG regions of FUS, which is suppressed by aDMA (Hofweber et al., 2018; Qamar et al., 2018). Our results demonstrate that in vivo the situation can become far more complex beyond changing the chemical properties of the modified residues. DMA recruits the interaction of tudor domain proteins in trans and may promote or antagonize condensation depending on the properties of the tudor domain protein. One demethylase for aDMA has been identified, making the modification reversible and potentially dynamic (Chang et al., 2007). Much is still unknown about how methyltransferases either differentiate between – or compete for – substrates for aDMA or sDMA modification.
Though we focused on tudor domains most likely to bind DMA, the tudor superfamily contains a spectrum of domains that bind other ligands or could bind DMA in a non-canonical manner (Chen et al., 2011). How DMA acts to regulate many condensates in vivo is unexplored. DMA modifications have been found on a vast and growing list of proteins including histones, RNA polymerase II, and G3BP1 (Chitiprolu et al., 2018; Tsai et al., 2016; Zhao et al., 2016). These proteins are associated with chromatin, transcription, and stress granules, respectively. Notably, all are systems reported to act as condensates in vivo, raising the possibility that tudor domain proteins and DMA play regulatory roles in their assembly and/or disassembly (Gibson et al., 2019; Guo et al., 2019; Molliex et al., 2015). The coactivator Tdrd3 uses its tudor domain to read aDMA modifications on core histone tails (H3R17me2a and H4R3me2a) and the CTD of RNA polymerase II at transcription start sites (Yang et al., 2010; Yang et al., 2014). During cellular stress, Tdrd3’s tudor domain targets the protein to stress granules (Goulet et al., 2008). Moreover, other tudor-domain regulators recognize DMA on histone tails (Lu and Wang, 2013). Based on our findings, we speculate that these activities may involve condensation by tudor domains, which could also behave in a switch-like manner depending on the methylation state of the histone tails.
The Cry2 assay, combined here with analysis that relates condensation to protein expression level, reveals that a modest increase in valency through light-activated dimerization of tudor domains is enough to induce condensation (Bugaj et al., 2013; Shin et al., 2017). It is therefore striking that tudor domain proteins can have eight or more tudor domains in addition to other interaction modules (Gan et al., 2019). The inherent multivalency of proteins like Tdrd4 and Tdrd6 implies a mechanism for germ line condensate formation, supported by our study of the original “Tudor” protein from D. melanogaster. Modified proteins often have multiple modified residues, offering avidity as an explanation for how relatively low affinity individual interactions allow for assembly (Tripsianes et al., 2011). Moreover, we show nuclear localization-specific condensation by dmTudorTud and a lack of DMA-containing snRNPs in SMNTud condensates. Although there are many gaps in our knowledge about tudor domains and their ligands, each tudor domain exerts selectivity over the ligands bound, and not every methylated ligand is in every condensate. Since condensation is dependent on the concentration of the active unit required to separate from the bulk phase, the concentration of tudor domains and their available ligands likely determine when and whether condensates form (Elbaum-Garfinkle et al., 2015; Li et al., 2012; Pak et al., 2016). Thus, our data should be viewed as ruling-in certain tudor domains for condensation at the concentrations expressed stably in NIH-3T3 cells. Others may form condensates at higher concentrations or in the presence of other ligands. Thus, we discovered a role for tudor domain ligands and their regulation. Taken together, the DMA-tudor module has the properties of a highly versatile mechanism for cellular condensation.
At a minimum, two developmentally important MLOs – the Cajal body and germ plasm – are controlled by the DMA modification (Hebert et al., 2002; Liu et al., 2010; Nott et al., 2015). Our findings explain how DMA modification is able to control the composition of the Cajal body and its relationship to gems. Prior studies had observed that coilin methylation is required for its interaction with SMN, and inhibition of all cellular methylation disrupts Cajal body assembly (Hebert et al., 2002). Our work connects the current understanding of condensation to the past observation that coilin must bear a methylated arginine-glycine motif to form a Cajal body (Hebert et al., 2001). We additionally build on prior observations that depletion of SMN reduces the number of gems and Cajal bodies (Girard et al., 2006; Lemm et al., 2006; Strzelecka et al., 2010b), showing that the tudor domain mutations placed in the context of the full length SMN protein cannot restore the proper constellation of MLOs in cells. Partial complementation of Cajal body defects by expression of SMN tudor domain mutants may be possible because the SMN IDRs in the N– and C-termini support protein-protein interactions including SMN oligomerization (Grimm et al., 2013; Zhang et al., 2011).
STED microscopy revealed substructure within the Cajal body that relates DNA-tudor module interactions to the specific composition of the MLO. Specifically, a “ball-in-socket arrangement of SMN (ball) and coilin (socket) resolves major domains of the compartment that have been hinted at previously (Novotny et al., 2015). Unperturbed, Cajal bodies and gems are “docked” to one another in this configuration, which is disrupted by aDMA depletion. Conversely, sDMA promotes the fusion of SMN-rich gems with Cajal bodies, showing that the precise form of this post translational mark dictates the occupancy of the body. These observations imply that an equilibrium between aDMA and sDMA exists in HeLa cells and provokes speculation that the interface between SMN and coilin might be controlled by the proportion of each modification. Such docked arrangements may apply to other MLOs, such as Cajal bodies and nucleoli under stress conditions, the transcriptional machinery, cytoplasmic RNPs, and unfolded protein condensates (Boulon et al., 2010; Cho et al., 2018; Mateju et al., 2017; Moon et al., 2019). The creation of interfaces between different protein condensation states may be critical to certain chemical events in cells, such as pre-mRNA splicing (Gordon et al., 2020; Liao and Regev, 2021). Our discovery of a mechanistic explanation for substructure in the Cajal body prompts future investigation of the molecules belonging to gems, Cajal bodies, nucleoli, and histone locus bodies – four distinct MLOs with known overlaps in composition (Machyna et al., 2013).
Considerable effort has gone into developing the most appropriate theoretical framework to understand condensates (Banani et al., 2017; Choi et al., 2020; Hyman et al., 2014; Lyon et al., 2020; Wang et al., 2018). In the case of DMA-tudor, the “polyphasic linkage” framework is most applicable (Wyman and Gill, 1980). Taking either tudor domain proteins to be scaffolds and DMA-modified proteins to be ligands, the resulting condensates would have tunable properties based on the degree of post-translational modification (Ruff et al., 2021). Our study does not address the specific effects of DMA modification on the critical concentrations of either SMN or its ligands. Such an analysis on SMN or another tudor domain protein is an important future step towards understanding how evolution has selected for specific condensation conditions. Several motifs in SMN’s IDRs are conserved in animals, particularly amongst those species whose SMN can rescue SMA model mice (Figure 1A) (Osman et al., 2019). Taking the view that the SMN IDRs may act as spacers or modulators of tudor condensation activity, a detailed biophysical analysis of this protein is now possible.
The rules governing specificity and composition of condensates have been intensely sought after. While IDRs contribute important interactions to the formation of condensates, it is becoming clear that specific interactions are also required to form endogenous organelles via phase separation (Wang et al., 2018). The base pairing and secondary structure of RNA has recently been shown to play an important role (Langdon et al., 2018; Van Treeck et al., 2018). In vitro studies have shown that molecular recognition between folded domains can promote occupancy in a condensate (Ditlev et al., 2018). Clearly, the prolific nature of both tudor domain-containing proteins and DMA-modified proteins leaves open many opportunities for this mechanism to play out throughout the cell. Our study reveals that DMA-tudor modules provide the requisite specificity for the formation of endogenous MLOs and enforce a distinct substructure. We speculate that biomolecular condensation in vivo can be driven by specific interactions of each tudor domain with its DMA ligands. Furthermore, the switch-like binding changes induced by the symmetry of the installed methyl groups introduces versatility and regulation, which can in turn define the identity and specific composition of MLOs formed through the activity of DMA-tudor modules.
Limitations of the Study
This study reveals several important questions with answers lying beyond the scope of the experiments presented here. First, while we were able to leverage differences in tissue culture cells to maximize the impact of our conclusions, how these results generalize to other cell models and tissues remains to be tested. We speculate that the activity of the methyltransferases in specific cell types is a key variable that must be addressed to understand MLO formation. Related to this, our study does not address the identity or stoichiometry of DMA-modified ligands. We were limited to a relatively small selection of small-molecule inhibitors of DMA synthesis, and until more is known about how the methyltransferases work together to regulate assembly via the DMA-tudor module, we can only speculate on the underlying mechanism that allows for residual SMN condensation in our assays.
STAR Methods text:
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Karla M. Neugebauer (karla.neugebauer@yale.edu).
Materials Availability
Plasmids generated in this study are available upon request.
Data and Code Availability
Raw microscopy data for optodroplet experiments made available at data.4dnucleome.org; accession numbers given in the Key Resources Table. Image analysis code available at github.com/bewersdorflab/quant-condensate.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti mCherry | Invitrogen | #PA5-34974; RRID: AB_2552323 |
| Anti coilin (H-300) | Santa Cruz Biotechnology | #sc-32860; RRID: AB_2081431 |
| Anti-coilin | Abcam | #ab210785 |
| Anti sDMA (SYM10) | Millipore Sigma | #07-412; RRID: AB_11212396 |
| Anti sDMA (SYM11) | Millipore Sigma | #07-413; RRID: 310595 |
| Anti aDMA (ASYM24) | Millipore Sigma | #07-414; RRID: AB_310596 |
| Anti SMN (2B1) | Abcam | #ab5831; RRID: 305144 |
| Anti GFP | Invitrogen | #A11122; RRID: AB_221569 |
| Anti GAPDH | Santa Cruz Biotechnology | #sc-25778; RRID: AB_10167668 |
| Anti Sm (Y12) | Laboratory of Joan Steitz; Pettersson et al., 1984 | N/A |
| Anti 2,2,7-Trimethylguanosine (K121) | Calbiochem | #NA02; RRID: AB_213109 |
| Anti SNRPC | Abcam | #ab82862; RRID: AB_2193870 |
| Anti U1-70K (CB7) | Laboratory of Doug Black; Listerman et al., 2006 | N/A |
| Anti-rabbit IgG Horseradish Peroxidase-linked | GE HealthCrae | #NA934; RRID: AB_772206 |
| Alexa Fluor 488-conjugated Donkey Anti-Rabbit | Jackson ImmunoResearch | #711-545-152; RRID: AB_2313584 |
| Alexa Fluor 488-conjugated Donkey Anti-Mouse | Jackson ImmunoResearch | #715-545-150; RRID: AB_2340850 |
| Anti-Mouse IgG - Atto 594 | Sigma | #76085; RRID: AB_1137653 |
| Anti-Rabbit IgG - Atto 594 | Sigma | #77671; RRID: AB_1137663 |
| Anti-Mouse IgG - Atto 647N | Sigma | #50185; RRID: AB_1137661 |
| Anti-Rabbit IgG - Atto 647N | Sigma | #40839; RRID: AB_1137669 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| MS-023 | Sigma | SML1555 |
| EPZ015666 | Sigma | SML1421 |
|
| ||
| Deposited data | ||
|
| ||
| Optodroplet Live Imaging Dataset - mCherry Control |
This Study | 4DNucleome: 4DNESVT8QSLU |
| Optodroplet Live Imaging Dataset- SMNTud | This Study | 4DNucleome: 4DNES9JHBEZC |
| Optodroplet Live Imaging Dataset - hnRNPA1IDR | This Study | 4DNucleome: 4DNES49WH1QD |
| Optodroplet Live Imaging Dataset - SMNTud Mock | This Study | 4DNucleome: 4DNESPZNHYSY |
| Optodroplet Live Imaging Dataset- SMNTud Treated with MS023 and EPZ015666 |
This Study | 4DNucleome: 4DNESM8F8PP5 |
| Optodroplet Live Imaging Dataset - SMNTud W102L |
This Study | 4DNucleome: 4DNESGOUKUHY |
| Optodroplet Live Imaging Dataset- SMNTud Y109L | This Study | 4DNucleome: 4DNESRORWX8M |
| Optodroplet Live Imaging Dataset- SMNTud Y130D | This Study | 4DNucleome: 4DNESGSHWGUK |
| Optodroplet Live Imaging Dataset - SMNTud E134K | This Study | 4DNucleome: 4DNESQJS4P53 |
| Optodroplet Live Imaging Dataset - SMNTud F118L | This Study | 4DNucleome: 4DNESLBSY7UC |
| Optodroplet Live Imaging Dataset - Spf30Tud | This Study | 4DNucleome: 4DNESZZ8T7O9 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HeLa-Kyoto | Neugebauer Lab | RRID: CVCL_1922 |
| NIH 3T3 | ATCC | CRL-1658 |
| HEK293FT | ThermoFisher | R70007 |
| Coil/ mouse embryonic fibroblasts | Laboratory of Greg Matera; Tucker et al., 2001 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| siSMN #1 | Thermo | #4427037 s445227 |
| siSMN #2 | Thermo | #4427037 s531465 |
| siSMN #3 | Thermo | #4427037 s531467 |
|
| ||
| Recombinant DNA | ||
|
| ||
| mCh-Cry2 | Laboratory of Clifford Brangwynne | N/A |
| FUSIDR | Laboratory of Clifford Brangwynne | N/A |
| hnRNPA1IDR | Laboratory of Clifford Brangwynne | N/A |
| SMN-Cry2 | This study | N/A |
| SMNIDR-N | This study | N/A |
| SMNTud | This study | N/A |
| SMNIDR-C | This study | N/A |
| dmTudorTud | This study | N/A |
| Aub3 | This study | N/A |
| Aub3(KG) | This study | N/A |
| Spf30Tud | This study | N/A |
| Tdrd1Tud#2 | This study | N/A |
| Tdrd1Tud#3 | This study | N/A |
| Tdrd1Tud#4 | This study | N/A |
| Tdrd3Tud | This study | N/A |
| Tdrd4Tud#1 | This study | N/A |
| Tdrd4Tud#4 | This study | N/A |
| Tdrd6Tud#5 | This study | N/A |
| Tdrd6Tud#6 | This study | N/A |
| Tdrd8Tud | This study | N/A |
| Tdrd9Tud | This study | N/A |
| Snd1Tud | This study | N/A |
| Recoded SMN-Myc | This study | N/A |
| Recoded SMN-Myc (E134K) | This study | N/A |
| Recoded SMN-Myc (Y109L) | This study | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Python Microscopy (PYME) | David Baddeley | https://Python-microscopy.org |
| FIJI | Schindelin et al., 2012 | https://Fiji.sc |
| Cell Profiler | Carpenter et al., 2006 | https://Cellprofiler.org |
| RaptorX | Kallberg et al., 2012 | https://Raptorx.uchicago.edu |
| Clustal Omega | Sievers et al., 2011 | https://Ebi.ac.uk |
| Imspector | Abberior Instruments | imspector.abberior-instruments.com |
| ConSurf | Berezin et al., 2004 | https://Consurf.tau.ac.il |
| Imaris | Oxford Instruments | https://Imaris.oxinst.com |
Experimental Model and Subject Details
Tissue Culture Cell Lines
NIH-3T3 cells were obtained from ATCC (# CRL-1658), HeLa cells are from the Kyoto lineage (RRID: CVCL_1922), and coil−/−-Mouse Embryonic Fibroblasts (MEF) were a gift from Greg Matera (Tucker et al., 2001). 293FT cells were obtained from Thermo (#R70007) but were not used as a model and only for packaging lentivirus. Cells were cultured in DMEM supplemented with 10% FBS, penicillin, and streptomycin (Gibco). All cells were incubated at 37°C in a 5% CO2 atmosphere. Cells were regularly screened for the presence of mycoplasma infection.
Method Details
Plasmid Construction
The plasmid system for lentiviral generation included pHR_SFFV (generated by Wendell Lim, Addgene #79121), pMD2.G (generated by Didier Trono, Addgene # 12259), and pCMVR 8.74 (generated by Didier Trono, Addgene # 22036). pHR_SFFV containing the IDRs of FUS and hnRNP-A1 were gifts from Cliff Brangwynne (Princeton University). Constructs in Table S1 below were generated with the InFusion HD kit (Takara) by replacing the sequence upstream of mCherry-Cry2 in pHR_SFFV-hnRNPA1-mCherry-Cry2. Aubergine peptide constructs were generated by inserting sequence upstream of GFP in pLV-EGFP (generated by Pantelis Tsoulfas, Addgene #36083, modified by David Phizicky) and expressed from the eIF4a promoter. Point mutations to SMNTud were generated by site-directed mutagenesis.
Cell Culture and Cry2 line generation
Pools of cells stably expressing Cry2 constructs were generated by first transfecting either pHR_SFFV or pLV-EGFP with the desired insert, pMD2.G, and pCMV R8.74 into confluent 293FT. Transfection was performed with the Fugene HD reagent (Promega). After 48–72 h, viral supernatant was harvested, filtered, and applied to NIH-3T3 or coil−/−-MEF (Tucker et al., 2001). For live cell imaging, NIH-3T3 cells were cultured in glass-bottomed 35 mm dishes (MatTek). Just before imaging, the media was replaced with Live Cell Imaging Solution (Invitrogen) supplemented with 20 mM glucose and 1 μg/mL Hoechst 33342 (Invitrogen).
Live Cell Imaging of Cry2 Condensates
All live cell imaging was carried out on a Bruker Opterra II Swept Field instrument. The system allows for simultaneous imaging of mCherry fluorescence with the 561 nm laser and field of view activation of Cry2 with the 488 nm laser. The instrument is equipped with a Photometrics Evolve 512 Delta EMCCD camera. A standard protocol was used to make all Cry2 measurements. A PlanApo 60X 1.2 NA water immersion objective (Nikon) and a slit aperture of 70 μm was used to image at four frames per second. Four-frame averaging resulted in an overall frame rate of one frame per second. The 561 nm laser was set to 80% of full power and the 488 nm was set to 25% of full power. Both were passed through a 20% neutral density filter. We measured power output at the objective to be 80–90 μW for 561 nm and 20–25 μW for 488 nm with a Thorlabs PM100D power meter and a S170C sensor.
For each field of view, 10 s of inactive Cry2 were recorded, followed by turning on the 488 nm laser to capture 180 s of active Cry2. At the end of the series, a reference image of nuclear Hoechst staining was collected using the 405 nm laser. A stage-top incubator was employed to maintain cells at 37°C throughout imaging.
Fluorescence Recovery after Photobleaching (FRAP)
FRAP measurements were taken using identical imaging conditions as above with the following modifications. The objective was changed to a PlanApo 100X 1.4 NA oil immersion objective (Nikon). Frame averaging was replaced with a maximum projection of 7–11 z-steps at each time point, 0.25 s exposure per frame. After 180 s of activation, a single condensate was photobleached using a pulse of 405 nm laser light. The background was subtracted from a bleached region of interest containing a condensate using an unclustered region of the same cell. The intensity was then normalized to the maximum and minimum intensity values. Each recovery curve was fit using a one-phase exponential to estimate the mobile fraction and characteristic recovery time (1/e recovery, tau).
DMA Inhibitor Treatment
DMA inhibition was achieved by either individual or simultaneous treatment with two drugs: MS-023 (Sigma) and EPZ015666 (Sigma) (Chan-Penebre et al., 2015; Eram et al., 2016). Stock solutions were prepared in DMSO at 50 mM and all subsequent dilutions were done in cell media as described above. MS-023 was added to media at a final concentration of 1.0 μM, and EPZ015666 was added at a final concentration of 5.0 μM. After 48 h of drug treatment, imaging was carried out as above, with drug added to the same concentration in the imaging media.
Fixed cell imaging
Cells were grown on No. 1.5 coverslips (Zeiss) in six-well plates and fixed in 4% paraformaldehyde (Sigma), blocked in 3% Bovine Serum Albumin (Sigma) and permeabilized by 0.1% Triton X-100 (American Bioanalytical). If immunofluorescence was performed, staining was conducted in blocking buffer with the antibodies listed in Table S2. For Cry2 activation prior to fixation, we built a custom illumination array designed for six-well plates. Six blue LEDs (470 nm, 3.2 V, Digi-Key 150080BS75000) were arrayed to illuminate each well in a six-well plate for 5 min (Extended Data Fig. 10b). Illumination was maintained during the 10 min fixation step. Inactive Cry2 samples were maintained in the dark for 10 min and fixed in the same manner. Imaging was done on a Leica Sp8 Laser Scanning Confocal.
SMN Knockdown and Rescue
SMN depletion was achieved by transfection of a pool of Silencer Select siRNAs (Thermo, cat# 4427037, siRNA codes s445227, s531465, s531467) using Lipofectamine RNAiMAX (Thermo). Rescue construct expression was achieved by transfecting degradation resistant constructs for SMN after 24 hrs of knockdown depletion using Lipofectamine 3000 (Thermo). Cells were fixed as described above after 48 total hours of knockdown depletion. The expression of the SMN rescue construct was assessed immunofluorescence staining for SMN and for a c-terminal myc epitope tag. No difference in patterning was found between the two stains.
Western Blotting
Western blots were carried out by preparing whole cell lysate in 0.1% SDS, 0.5% sodium deoxycholate, 150 mM NaCl, 10 mM EDTA, and 10 mM Tris (pH 8.0). Lysates were denatured and reduced by adding NuPAGE LDS sample buffer (Thermo) and heating for 10 min at 85°C. Samples were run out on a 4–12% acrylamide Bis-Tris gel (Invitrogen) and transferred to 0.2 μm nitrocellulose membranes (BioRad). After blocking, membranes were stained with the antibodies listed in Table S2.
STED Microscopy
Two modalities of STED microscopy were used in this study: lateral depletion profile STED and three dimensional isoSTED. Two-dimensional STED was carried out using a Leica Sp8 Laser Scanning Confocal enabled with a 775nm depletion laser. Atto 594 and Atto 647N secondary antibodies were used to facilitate two color STED. For isoSTED nanoscopy, The samples were mounted between two 25-mm-diameter, 170-μm-thick coverslips (CSHP-No1.5–25, Bioscience Tools). Mounting medium (Prolong Diamond, Thermo Fisher Scientific) was used to supply anti-fade protection and to reduce the refractive index difference between the sample and the objective’s immersion silicon oil. The samples were imaged with a custom-built isoSTED nanoscope as previously described (Hao et al., 2020). Briefly, depletion laser beams interfere at the common focus of two opposing objectives (100 ×/1.35NA, silicon oil immersion, Olympus) in a 4Pi geometry to create a three-dimensional isotropic zero-intensity minimum at the center of the focus (Bewersdorf et al., 2006; Gugel et al., 2004; Schmidt et al., 2008). The instrument was equipped with a pulsed excitation laser at a wavelength of 650 nm (LDH-P-C-650m, PicoQuant) and an 80-MHz pulsed depletion laser with a pulse length of ~600 ps and wavelength of 775 nm (Kantana HP, OneFive). A 16-kHz resonance mirror (SC-30, EOPC) scans the combined laser beams along the fast scanning axis, while two synchronized galvanometer scanning mirror (dynAXIS XS, SCANLAB) scan the beams along the slow scanning axis. Fluorescence was detected by a single-photon counting avalanche photodiode (APD, SPCM-ACRH-13-FC, Excelitas). All data were acquired with 500 line accumulations, ~10 μW of the 650 nm laser, and ~60 mW of the 775 nm laser (power measured at the back focal plane of each objective lens). Z stacks were acquired from 40 optical sections at 25-nm intervals in a field of view of 256 by 256 pixels and a pixel size of 19.5 nm.
Quantification and Statistical Analysis
Image Analysis: Segmentation
Quantification of condensate formation on a per-cell basis requires accurate segmentation of cells within a field of view. A cell profiler pipeline was used to process movies into cell masks. Binary nuclear masks were generated by segmenting the Hoechst image associated with each sequence. Binary cellular masks were generated by performing an Otsu threshold and watershed transform seeded by the nuclear masks on the mCherry signal from inactive Cry2 cells. Masks defined by automated segmentation were manually checked for accuracy; cells with substantial movement from the mask were excluded from analysis.
Image Analysis: Intensity correction
We use a temporal variance metric to quantify clustering without imposing a model on the time-dependence or shape, in an expression-level independent manner. Fluorophores moving into or out of a pixel will increase the temporal variance, substantially for pixels seeing large fluxes of fluorophores. However, as photon-counting processes are subject to shot-noise, the temporal variance of a pixel will also increase with baseline intensity increases, which would confound comparison between cells of differing expression levels. This effect can be conveniently normalized using the temporal mean, since the variance and expectation value of a Poisson process are equal. Several effects which deviate our raw analog-digital counts from this model were therefore accounted for before calculating the clustering metric.
First, the per-pixel analog-digital offset of the camera was characterized and subtracted from all images. We often observe very bright spots appearing on single frames at random positions, which we interpret to be particles or cosmic rays impacting the EMCCD. Such large spikes in intensity can bias variance-based calculations, and because these spikes sometimes occur within cell masks, we filter them using a temporal sliding-window approach (Extended Data Fig.10a). We iterate through time and calculate the median-absolute deviation (MAD) at each xy-pixel within a 10-frame window, where the MAD of a sequence k is simply median(|k − median(k)|). Values within this window were identified as spikes if they were both larger than 15 times the MAD and were increases on the previous frame larger than 1500 ADU. Spikes were replaced with the median value of the window, then the window was advanced one frame, and the process was repeated until the last frame was included in the window. MAD was employed to determine the variability within a window as it is a robust variability measure and does not increase substantially due to the very spikes we are trying to filter, unlike the standard deviation or variance.
Finally, we performed a mean-normalization to remove effects of read-out laser intensity fluctuation, bleaching, etc., by multiplying j frame by a factor β = 〈I(t = 0)〉x,y / 〈I(tj)〉x,y where 〈k〉x denotes an average k of taken over x.
Image Analysis: Clustering Metric
We calculated the temporal variance and average for each pixel during Cry2-activation in live cells. In order to arrive at a metric which is theoretically independent of baseline signal magnitude, and therefore expression levels, we normalized the temporal variance by the mean. This is a convenient normalization for data influenced by shot noise because the variance of a Poisson process is equal to its mean. The clustering metric is given by
where Ci,j, (t) is the corrected intensity at pixel x,y as a function of time, and m denotes the set of x,y pixels within a mask. An intermediary variance-over-mean image can be generated where individual clusters are visible. Using fluorescence intensity as a proxy for concentration, the clustering metric can be plotted against baseline intensity so that comparisons between constructs may be made (Extended Data Fig. 2d,e).
The clustering metric for a homogeneous Poisson process should be unity, however our camera data is not only subject to shot noise, but also read noise and noise due to the stochastic nature of electron multiplication (Robbins and Hadwen, 2003). A more complete camera noise model may be necessary for comparison of results from different microscopes or camera settings.
Clustering classification
While the clustering metric allows for comparison between the degree of clustering for individual cells, the expression efficiency and/or sampling varies between constructs, complicating statistical analysis between them. To remove the influence of data which did not experience significant clustering, we performed a Mann-Whitney U test between the clustering metric distribution of the negative control (mCherry-Cry2) and each individual clustering metric from other distributions, classifying cells which reject the null hypothesis with α = 0.2 as cells which experience clustering. We then calculated the median clustering metric for a construct using only its cluster-positive cells and used that to compute differences in clustering magnitude.
Image Analysis: STED Quantification
STED and confocal image stacks of Coilin and SMN were segmented using a linear-bin Otsu threshold applied to the entire volume of each channel after smoothing by a Gaussian filter (sigma of 40 nm in x and y, 43.87 nm in z). The STED mask in both colors was taken to be the intersection of the smoothed and thresholded STED channels and their paired confocal channel masks. The original STED images were then analyzed using these masks. The intensity-weighted and masked center of mass was calculated in 3D for both Coilin and SMN, and the Euclidean distance separation calculated as the offset. To calculate the fractional intensity overlap, the intersection of Coilin and SMN masks were taken, and the intensity in this region was summed over both channels and divided by the combined sum of the intensity in each of the individual channels within their own mask.
Image Analysis: SMN Condensate Quantification
Images were segmented to identify nuclei using Hoechst stain and condensates were identified from SMN immunofluorescence via otsu thresholding and object identification in CellProfiler. Nuclei masks were shrunken by three pixels in all directions to avoid confounding cytoplasmic signal and condensates were then matched to their corresponding nuclei. Cells were not filtered out by expression before performing analysis. Condensates were counted and analyzed for size by computing the mean radius of each. The expression level was assessed by expanding nuclear masks out to the cell periphery using a secondary Otsu threshold on SMN signal and then integrating signal over the whole-cell mask. To mitigate multiple comparison problems, SMN condensate number and size distributions were first analyzed for significant variation using the Kruskal-Wallace H test and then followed by pairwise post hoc analysis using Dunn’s test, where appropriate.
Image Analysis: Tdrd3Tud and Snd1Tud DMA Inhibitor Quantification
Tdrd3Tud samples were scored for the percentage of cells with cytoplasmic condensates out of those with nuclear condensates. From Snd1Tud samples, one nucleus with condensates was chosen at random from each image collected per condition and a line trace was drawn across the nuclear diameter. The fifth and ninety-fifth percentile of values in each scan were computed and a relative dynamic range was computed by normalizing the difference between these percentiles by the mean intensity of the scan. The Mann-Whitney U test was used as a non-parametric test of statistical significance between samples.
Software
Images were prepared for figures and FRAP calculations were done using ImageJ (FIJI) version 2.0.0-rc-69/1.52p (Schindelin et al., 2012). Image segmentation SMN condensate measurements were done using Cell Profiler version 4.0.7 (Carpenter et al., 2006). All other image analysis was done using PYME version 20.12.10 which is available at python-microscopy.org. The plugin used to compute clustering metrics can be found at github.com/bewersdorflab/quant-condensate. Structure prediction was performed on the RaptorX Property Prediction server using default parameters (Kallberg et al., 2012). Tudor domains were aligned by extracting domain annotations from Uniprot.org and aligning sequences with Clustal Omega using the default parameters (Sievers et al., 2011) SMN conservation score was computed from a Clustal Omega alignment using the ConSurf online server (Berezin et al., 2004).
Image Display
All images within a figure are contrast matched, and an intensity grayscale or color bar is given for reference where applicable. All live cell images are micrographs from single z-slice movies except where otherwise noted. Images and quantification comparisons are only made between samples taken with identical microscope calibrations. Confocal images of fixed samples are maximum intensity projections of z-stacks. The two-dimensional rendering of isoSTED data was generated using Imspector and the three-dimensional renderings were generated with Imaris.
Supplementary Material
A) Micrographs of live cells expressing mCherry-Cry2 alone or fused to hnRNP-A1IDR or FUSIDR. mCherry alone does not cluster while the IDRs do. Results produced using published constructs and protocols (Shin et al., 2017). B) Micrographs of live cells expressing full length SMN-mCherry-Cry2. C) Micrographs of live cells expressing fragments of SMN fused with mCherry-Cry2. Only the tudor domain forms clusters when Cry2 is active. Grayscale bars are in analog-digital units. Scale bar = 10 μm. D) Fluorescence recovery after photobleaching for ten SMNTud condensates with corresponding tau values from single exponential fits. Example images from blue trace provided below. E) Examples of condensate dissipation after Cry2 inactivation in live cells. Blue light pulse is 30 seconds. Scale bars = 10 μm. F) Example of two SMNTud condensates fusing. Scale bar = 2 μm.
(A) Live cells are recorded for 10 seconds without Cry2 activation and then 180 seconds with blue light which activates Cry2. The mCherry signal before activation allows for segmentation of individual cells and serves as a proxy for expression level. The per-pixel temporal variance and mean are computed for the Cry2-active period. These values are used to construct the per-pixel clustering metric (right). Per-cell values are calculated by averaging over each cell mask. Scale bar = 10 μm. Grayscale bar (left) indicates fluorescence intensity, color bar (right) indicates clustering metric, both given in analog-digital units (ADU). B) An example of a single frame spike in intensity filtered out using a MAD-based approach. Arrowhead indicates spike. Before filtering, a high intensity spot is seen inside the region of measurement for the clustering metric. After filtering, the spot is effectively removed. Scale bar = 10 μm. Each frame is a 1.0 s exposure. C) Clustering metrics for SMNTUD and mCherry-Cry2 as a negative control. Solid lines and shading are a rolling mean and standard deviation of 10 points. Each point represents one cell. Dashed line represents a significance threshold relative to mCherry-Cry2 where a = 0.2 for the Mann-Whitney U test. D) Chemical structures of sDMA and aDMA. E) Detection of aDMA and sDMA modified proteins in Western blots of whole cell lysates from NIH-3T3 cells from the same biological replicate treated for 48 h with MS-023, EPZ015666 or both, using antibodies specific for aDMA (ASYM24) or sDMA (SYM10 & SYM11, see Table S2). The effects of the chemical inhibitors were variable, as illustrated by the bands denoted by numbered asterisks: 1) a band only reduced when treated with both drugs, 2) the major band reduced by MS-023, and 3) a band reduced by EPZ015666. GAPDH blot is from separate lanes to indicate relative protein amounts and shown as a reference for both DMA blots. F) Micrographs of live cells expressing SMNTud untreated and treated with DMA Inhibitors MS-023 and EPZ015666 for 48 h. Grayscale bar is in analog-digital units. Scale bar = 10 μm.
A-E) Micrographs of live cells expressing A) SMNTud or SMNTud with point mutations B) W102L, C) Y109L, D) Y130D, and E) E134K that reduce binding to DMA (Tripsianes et al., 2011). F) Micrographs of live cells expressing F118L mutation to SMNTud. Grayscale bars are in analog-digital units and apply to all images in panel. Scale bars = 10 μm. G) Quantification of the condensation effect of SMNTud F118L compared to cells expressing mCherry-Cry2. Units are analog-digital units (ADU). Solid lines and shading are a rolling mean and standard deviation of 10 points. Each point represents one cell. Dashed line represents a significance threshold relative to mCherry-Cry2 where a = 0.2 for the Mann-Whitney U test.
A) Fixed, untreated, wild-type NIH-3T3 cells showing their typical absence of nuclear bodies as detected by immunofluorescence using antibodies specific for SMN and coilin (Table S2). B) Fixed, untreated, wild-type NIH-3T3 cells showing their lack of snRNPs concentrated in nuclear bodies as marked by antibodies specific for trimethylguanosine (Table S2). C) Western blots from two biological replicates indicating depletion of SMN, recognized by monoclonal antibody 2B1 (Table S2) in HeLa lysate after siSMN transfection. D) SMN rescue visualized by staining for the myc epitope tag included in the degradation resistant rescue construct in HeLa cells. E) Single z-slice images taken from stacks used to produce images in Figure 3A. F) Line scans corresponding to (E) showing intensity in analog-digital units (ADU). G) SMN knockdown and rescue HeLa cells co-immunostained for coilin to indicate Cajal body status in each rescue condition. H) Statistical analysis corresponding to Figure 3B&C. Scale bars = 10 μm.
Fixed cells with Cry2-active SMNTud, which is always visualized by mCherry fluorescent signal. Condensates were co-stained for A) mCherry, B) sDMA using antibody SYM10, C) the Sm epitope with Y12, D) the snRNAs’ 5′ end trimethylguanosine cap, E) U1 snRNP-C, and F) U1-70k, both U1 snRNP specific proteins. In b) arrowheads indicate nuclear condensates that stain positively with SYM 10. Scale bar = 10 μm. G) Light-activated and formaldehyde fixed cells expressing SMNTud (magenta) and stained for endogenous coilin (green), showing coilin is a ligand of SMNTud. Hoechst staining shown in blue. H) Micrographs of Cry2-inactive (488 nm: off, grey) and Cry2-active (488 nm: on, blue) SMNTud in coilin knock-out mouse fibroblasts, showing coilin is not required for SMNTud condensation.
A) Western blot of whole cell lysate from NIH-3T3 cells showing expression levels of Cry2 constructs as measured by mCherry staining. All Constructs include mCherry-Cry2. B) LED array used for activating Cry2 in cells plated in six-well plates with coverslips. C) Quantification of the condensation effect in live cells of SMNTud compared to Spf30Tud. Units are analog-digital units (ADU). Solid lines and shading are a rolling mean and standard deviation of 10 points. Each point represents one cell. Dashed line represents a significance threshold relative to mCherry-Cry2 where a = 0.2 for the Mann-Whitney U test. D) Amino acid sequences of tudor domains aligned using Clustal Omega. Green/Cyan highlights indicate aromatic residues in the DMA binding pocket as determined by structural studies. Cyan indicates the position of mutation for non-functional constructs. E) Fixed NIH-3T3 cells expressing Tudor-Cry2 constructs under Cry2-inactive and Cry2-active conditions. F) Fixed NIH-3T3 cells expressing Tdrd8Tud and stained for sDMA with the SYM10 antibody. G) Fixed NIH-3T3 cells expressing Tdrd3Tud undergoing DMA inhibitor treatment as indicated. Percentage indicates fraction of cells with active Cry2 that have cytoplasmic condensates. Arrowheads indicate cells that possess or lack cytoplasmic condensates. Dotted lines indicate nuclear outlines. H) Fixed NIH-3T3 cells expressing Snd1Tud undergoing DMA inhibitor treatment as indicated. Insets show line profiles of the dashed lines in images. Corresponding bar plot displays line profiles quantified by normalizing the difference between the fifth and ninety-fifth percentile of intensity by the mean value for each profile. P-value computed with Mann-Whitney U test. Color bars given in analog-digital units. Scale bars = 10 μm.
A) Two biological replicates of untreated wild-type HeLa cells immunostained for SMN (magenta) and coilin (green). B) Untreated, C) MS-023 and EPZ015666 treated, D) MS-023 treated, and E) EPZ015666 treated HeLa cells stained for trimethylguanosine (TMG, magenta) and coilin (green). Arrowheads in insets indicate the coilin puncta shown in each line profile plot. Scale bar = 10 μm. F) Line profile plots for single slices of isoSTED data used to generate the 3D renderings in Figure 6E. Scale bar = 500 μm. G) Dye-flip control for SMN and coilin lateral depletion STED. Scale bar = 500 nm. H) Single label control for SMN and coilin two dimensional STED. Scale bar = 500 nm. See Table S2 for antibodies.
NIH-3T3 cells expressing mCherry-Cry2. At time 0.0, blue light is turned on to activate Cry2. Time resolution, 1.0 frame per second. Scale bar = 10 μm.
NIH-3T3 cells expressing SMN-Cry2. At time 0.0, blue light is turned on to activate Cry2. Time resolution, 1.0 frame per second. Scale bar = 10 μm.
NIH-3T3 cells expressing SMNTud. At time 0.0, blue light is turned on to activate Cry2. Time resolution, 1.0 frame per second. Scale bar = 10 μm.
Highlights.
The tudor domain of SMN promotes condensation by binding dimethylarginine (DMA)
DMA-tudor condensation is a shared property of numerous other tudor domains
Each tudor domain has a specific dependence on symmetric or asymmetric DMA
aDMA and sDMA levels define the composition and substructure of Cajal bodies
Acknowledgments:
We thank M. Lessard, A. Mennone, C. Emanuel, D. Baddeley, D. Phizicky, and S. Prophet for technical assistance. We thank members of the Neugebauer Lab, J. Steitz, and J. Howard for discussion and feedback on this manuscript. NIH awards U01DA047734 (to J.B.), NINDS-F31NS105379 (to E.M.C), U01CA200147 TCPA-2017-Neugebauer (to K.M.N.). This work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Declaration of Interests:
J.B. is a consultant for Bruker Corp., has licensed IP to Hamamatsu Photonics, and is a founder of Panluminate, Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Alberti S, Gladfelter A, and Mittag T. (2019). Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 176, 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, and Rosen MK (2016). Compositional Control of Phase-Separated Cellular Bodies. Cell 166, 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banjade S, Wu Q, Mittal A, Peeples WB, Pappu RV, and Rosen MK (2015). Conserved interdomain linker promotes phase separation of the multivalent adaptor protein Nck. Proc Natl Acad Sci U S A 112, E6426–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, and Ben-Tal N. (2004). ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics 20, 1322–1324. [DOI] [PubMed] [Google Scholar]
- Bewersdorf J, Schmidt R, and Hell SW (2006). Comparison of I5M and 4Pi-microscopy. J Microsc 222, 105–117. [DOI] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, and Lamond AI (2010). The nucleolus under stress. Mol Cell 40, 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, and Hyman AA (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, Mitchison TJ, and Hyman AA (2011). Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A 108, 4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, and Clarke S. (2001). PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem 276, 32971–32976. [DOI] [PubMed] [Google Scholar]
- Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, and Schaffer DV (2013). Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods 10, 249–252. [DOI] [PubMed] [Google Scholar]
- Buhler D, Raker V, Luhrmann R, and Fischer U. (1999). Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum Mol Genet 8, 2351–2357. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Zhang X, Ditlev JA, and Rosen MK (2019). Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, Johnston LD, Rioux N, Munchhof MJ, Jin L, Jacques SL, et al. (2015). A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol 11, 432–437. [DOI] [PubMed] [Google Scholar]
- Chang B, Chen Y, Zhao Y, and Bruick RK (2007). JMJD6 is a histone arginine demethylase. Science 318, 444–447. [DOI] [PubMed] [Google Scholar]
- Chen C, Nott TJ, Jin J, and Pawson T. (2011). Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol 12, 629–642. [DOI] [PubMed] [Google Scholar]
- Chitiprolu M, Jagow C, Tremblay V, Bondy-Chorney E, Paris G, Savard A, Palidwor G, Barry FA, Zinman L, Keith J, et al. (2018). A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat Commun 9, 2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, and Cisse II (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JM, Holehouse AS, and Pappu RV (2020). Physical Principles Underlying the Complex Biology of Intracellular Phase Transitions. Annu Rev Biophys 49, 107–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbet GA, and Parker R. (2020). RNP Granule Formation: Lessons from P-Bodies and Stress Granules. Cold Spring Harb Symp Quant Biol. [DOI] [PubMed] [Google Scholar]
- Courchaine EM, Lu A, and Neugebauer KM (2016). Droplet organelles? EMBO J 35, 1603–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditlev JA, Case LB, and Rosen MK (2018). Who’s In and Who’s Out-Compositional Control of Biomolecular Condensates. J Mol Biol 430, 4666–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, and Brangwynne CP (2015). The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A 112, 7189–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eram MS, Shen Y, Szewczyk M, Wu H, Senisterra G, Li F, Butler KV, Kaniskan HU, Speed BA, Dela Sena C, et al. (2016). A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases. ACS Chem Biol 11, 772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Massenet S, Paushkin S, Wyce A, and Dreyfuss G. (2001). SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell 7, 1111–1117. [DOI] [PubMed] [Google Scholar]
- Gan B, Chen S, Liu H, Min J, and Liu K. (2019). Structure and function of eTudor domain containing TDRD proteins. Crit Rev Biochem Mol Biol 54, 119–132. [DOI] [PubMed] [Google Scholar]
- Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, and Rosen MK (2019). Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 179, 470–484 e421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C, Neel H, Bertrand E, and Bordonne R. (2006). Depletion of SMN by RNA interference in HeLa cells induces defects in Cajal body formation. Nucleic Acids Res 34, 2925–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon JM, Phizicky DV, and Neugebauer KM (2020). Nuclear mechanisms of gene expression control: pre-mRNA splicing as a life or death decision. Curr Opin Genet Dev 67, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet I, Boisvenue S, Mokas S, Mazroui R, and Cote J. (2008). TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum Mol Genet 17, 3055–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Chari A, Pelz JP, Kuper J, Kisker C, Diederichs K, Stark H, Schindelin H, and Fischer U. (2013). Structural basis of assembly chaperone- mediated snRNP formation. Mol Cell 49, 692–703. [DOI] [PubMed] [Google Scholar]
- Gugel H, Bewersdorf J, Jakobs S, Engelhardt J, Storz R, and Hell SW (2004). Cooperative 4Pi excitation and detection yields sevenfold sharper optical sections in live-cell microscopy. Biophys J 87, 4146–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlussler R, Kim K, Trussina I, Wang J, Mateju D, et al. (2020). RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 181, 346–361 e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall’Agnese A, Hannett NM, Spille JH, Afeyan LK, Zamudio AV, Shrinivas K, et al. (2019). Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X, Allgeyer ES, Antonello J, Watters K, Gerdes JA, Schroeder LK, Bottanelli F, Zhao J, Kidd P, Lessard MD, et al. (2020). 3D Adaptive Optical Nanoscopy for Thick Specimen Imaging at sub-50 nm Resolution. bioRxiv, 2020.2011.2025.398958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, and Matera AG (2000). Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell 11, 4159–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Shpargel KB, Ospina JK, Tucker KE, and Matera AG (2002). Coilin methylation regulates nuclear body formation. Dev Cell 3, 329–337. [DOI] [PubMed] [Google Scholar]
- Hebert MD, Szymczyk PW, Shpargel KB, and Matera AG (2001). Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev 15, 2720–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, Ruepp MD, Simons M, Niessing D, Madl T, et al. (2018). Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell 173, 706–719 e713. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, and Julicher F. (2014). Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol 30, 39–58. [DOI] [PubMed] [Google Scholar]
- Kallberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, and Xu J. (2012). Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7, 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, and McKnight SL (2013). Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, et al. (2018). mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L, Davies SE, and Liu JL (2009). The spinal muscular atrophy protein SMN affects Drosophila germline nuclear organization through the U body-P body pathway. Dev Biol 332, 142–155. [DOI] [PubMed] [Google Scholar]
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, and Luhrmann R. (2006). Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell 17, 3221–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. (2012). Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao SE, and Regev O. (2021). Splicing at the phase-separated nuclear speckle interface: a model. Nucleic Acids Res 49, 636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, and Parker R. (2015). Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 60, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, and Neugebauer KM (2006). Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol 13, 815–822. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, and Xu RM (2010). Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev 24, 1876–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Guo Y, Liu H, Bian C, Lam R, Liu Y, Mackenzie F, Rojas LA, Reinberg D, Bedford MT, et al. (2012). Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30. PLoS One 7, e30375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, and Wang GG (2013). Tudor: a versatile family of histone methylation ‘readers’. Trends Biochem Sci 38, 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon AS, Peeples WB, and Rosen MK (2020). A framework for understanding the functions of biomolecular condensates across scales. Nat Rev Mol Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machyna M, Heyn P, and Neugebauer KM (2013). Cajal bodies: where form meets function. Wiley interdisciplinary reviews RNA 4, 17–34. [DOI] [PubMed] [Google Scholar]
- Mateju D, Franzmann TM, Patel A, Kopach A, Boczek EE, Maharana S, Lee HO, Carra S, Hyman AA, and Alberti S. (2017). An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J 36, 1669–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggen DT, Mir M, Darzacq X, and Tjian R. (2019). Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Hannus S, Plottner O, Baars T, Hartmann E, Fakan S, Laggerbauer B, and Fischer U. (2001). SMNrp is an essential pre-mRNA splicing factor required for the formation of the mature spliceosome. EMBO J 20, 2304–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SL, Morisaki T, Khong A, Lyon K, Parker R, and Stasevich TJ (2019). Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nature cell biology 21, 162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. (2015). Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57, 936–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny I, Malinova A, Stejskalova E, Mateju D, Klimesova K, Roithova A, Sveda M, Knejzlik Z, and Stanek D. (2015). SART3-Dependent Accumulation of Incomplete Spliceosomal snRNPs in Cajal Bodies. Cell Rep. [DOI] [PubMed] [Google Scholar]
- Osman EY, Bolding MR, Villalon E, Kaifer KA, Lorson ZC, Tisdale S, Hao Y, Conant GC, Pires JC, Pellizzoni L, et al. (2019). Functional characterization of SMN evolution in mouse models of SMA. Sci Rep 9, 9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, Ali R, Yunus AA, Liu DR, Pappu RV, and Rosen MK (2016). Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol Cell 63, 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Charroux B, and Dreyfuss G. (1999). SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci U S A 96, 11167–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Kataoka N, Charroux B, and Dreyfuss G. (1998). A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 95, 615–624. [DOI] [PubMed] [Google Scholar]
- Peng A, and Weber SC (2019). Evidence for and against Liquid-Liquid Phase Separation in the Nucleus. Noncoding RNA 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson I, Hinterberger M, Mimori T, Gottlieb E, and Steitz JA (1984). The structure of mammalian small nuclear ribonucleoproteins. Identification of multiple protein components reactive with anti-(U1)ribonucleoprotein and anti-Sm autoantibodies. J Biol Chem 259, 5907–5914. [PubMed] [Google Scholar]
- Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, and Parker R. (2018). Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Rep 22, 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, Phillips EC, Miyashita A, Williams D, Strohl F, et al. (2018). FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-pi Interactions. Cell 173, 720–734 e715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimer AC, Gray KM, and Matera AG (2017). SMN - A chaperone for nuclear RNP social occasions? RNA Biol 14, 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renvoise B, Khoobarry K, Gendron MC, Cibert C, Viollet L, and Lefebvre S. (2006). Distinct domains of the spinal muscular atrophy protein SMN are required for targeting to Cajal bodies in mammalian cells. J Cell Sci 119, 680–692. [DOI] [PubMed] [Google Scholar]
- Robbins MS, and Hadwen BJ (2003). The noise performance of electron multiplying charge-coupled devices. IEEE Transactions on Electron Devices 50, 1227–1232. [Google Scholar]
- Ruff KM, Dar F, and Pappu RV (2021). Ligand effects on phase separation of multivalent macromolecules. Proc Natl Acad Sci U S A 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan VH, Dignon GL, Zerze GH, Chabata CV, Silva R, Conicella AE, Amaya J, Burke KA, Mittal J, and Fawzi NL (2018). Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol Cell 69, 465–479 e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, et al. (2020). Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 181, 306–324 e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Wurm CA, Jakobs S, Engelhardt J, Egner A, and Hell SW (2008). Spherical nanosized focal spot unravels the interior of cells. Nat Methods 5, 539–544. [DOI] [PubMed] [Google Scholar]
- Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, and Sattler M. (2001). SMN tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol 8, 27–31. [DOI] [PubMed] [Google Scholar]
- Sheu-Gruttadauria J, and MacRae IJ (2018). Phase Transitions in the Assembly and Function of Human miRISC. Cell 173, 946–957 e916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, and Brangwynne CP (2017). Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell 168, 159–171 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357. [DOI] [PubMed] [Google Scholar]
- Shpargel KB, and Matera AG (2005). Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A 102, 17372–17377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, Ospina JK, Tucker KE, Matera AG, and Hebert MD (2003). Control of Cajal body number is mediated by the coilin C-terminus. J Cell Sci 116, 303–312. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprangers R, Selenko P, Sattler M, Sinning I, and Groves MR (2003). Definition of domain boundaries and crystallization of the SMN Tudor domain. Acta Crystallogr D Biol Crystallogr 59, 366–368. [DOI] [PubMed] [Google Scholar]
- Strom AR, and Brangwynne CP (2019). The liquid nucleome - phase transitions in the nucleus at a glance. J Cell Sci 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka M, Oates AC, and Neugebauer KM (2010a). Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus 1, 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, and Neugebauer KM (2010b). Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol 17, 403–409. [DOI] [PubMed] [Google Scholar]
- Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, and Herschman HR (2000). PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem 275, 7723–7730. [DOI] [PubMed] [Google Scholar]
- Tapia O, Bengoechea R, Palanca A, Arteaga R, Val-Bernal JF, Tizzano EF, Berciano MT, and Lafarga M. (2012). Reorganization of Cajal bodies and nucleolar targeting of coilin in motor neurons of type I spinal muscular atrophy. Histochem Cell Biol 137, 657–667. [DOI] [PubMed] [Google Scholar]
- Trcek T, and Lehmann R. (2019). Germ granules in Drosophila. Traffic 20, 650–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, Neugebauer KM, and Sattler M. (2011). Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol 18, 1414–1420. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Gayatri S, Reineke LC, Sbardella G, Bedford MT, and Lloyd RE (2016). Arginine Demethylation of G3BP1 Promotes Stress Granule Assembly. J Biol Chem 291, 22671–22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, and Matera AG (2001). Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol 154, 293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, and Parker R. (2018). RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci U S A 115, 2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. (2018). A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 174, 688–699 e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, and Dreyfuss G. (2001). Characterization of functional domains of the SMN protein in vivo. J Biol Chem 276, 45387–45393. [DOI] [PubMed] [Google Scholar]
- Wyman J, and Gill SJ (1980). Ligand-linked phase changes in a biological system: applications to sickle cell hemoglobin. Proc Natl Acad Sci U S A 77, 5239–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, and Bedford MT (2010). TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell 40, 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, and Bedford MT (2014). Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol Cell 53, 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PJ, Le TT, Dunckley M, Nguyen TM, Burghes AH, and Morris GE (2001). Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp Cell Res 265, 252–261. [DOI] [PubMed] [Google Scholar]
- Zhang R, So BR, Li P, Yong J, Glisovic T, Wan L, and Dreyfuss G. (2011). Structure of a key intermediate of the SMN complex reveals Gemin2’s crucial function in snRNP assembly. Cell 146, 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, Zhong G, Liu K, Li W, Moffat J, et al. (2016). SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 529, 48–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Micrographs of live cells expressing mCherry-Cry2 alone or fused to hnRNP-A1IDR or FUSIDR. mCherry alone does not cluster while the IDRs do. Results produced using published constructs and protocols (Shin et al., 2017). B) Micrographs of live cells expressing full length SMN-mCherry-Cry2. C) Micrographs of live cells expressing fragments of SMN fused with mCherry-Cry2. Only the tudor domain forms clusters when Cry2 is active. Grayscale bars are in analog-digital units. Scale bar = 10 μm. D) Fluorescence recovery after photobleaching for ten SMNTud condensates with corresponding tau values from single exponential fits. Example images from blue trace provided below. E) Examples of condensate dissipation after Cry2 inactivation in live cells. Blue light pulse is 30 seconds. Scale bars = 10 μm. F) Example of two SMNTud condensates fusing. Scale bar = 2 μm.
(A) Live cells are recorded for 10 seconds without Cry2 activation and then 180 seconds with blue light which activates Cry2. The mCherry signal before activation allows for segmentation of individual cells and serves as a proxy for expression level. The per-pixel temporal variance and mean are computed for the Cry2-active period. These values are used to construct the per-pixel clustering metric (right). Per-cell values are calculated by averaging over each cell mask. Scale bar = 10 μm. Grayscale bar (left) indicates fluorescence intensity, color bar (right) indicates clustering metric, both given in analog-digital units (ADU). B) An example of a single frame spike in intensity filtered out using a MAD-based approach. Arrowhead indicates spike. Before filtering, a high intensity spot is seen inside the region of measurement for the clustering metric. After filtering, the spot is effectively removed. Scale bar = 10 μm. Each frame is a 1.0 s exposure. C) Clustering metrics for SMNTUD and mCherry-Cry2 as a negative control. Solid lines and shading are a rolling mean and standard deviation of 10 points. Each point represents one cell. Dashed line represents a significance threshold relative to mCherry-Cry2 where a = 0.2 for the Mann-Whitney U test. D) Chemical structures of sDMA and aDMA. E) Detection of aDMA and sDMA modified proteins in Western blots of whole cell lysates from NIH-3T3 cells from the same biological replicate treated for 48 h with MS-023, EPZ015666 or both, using antibodies specific for aDMA (ASYM24) or sDMA (SYM10 & SYM11, see Table S2). The effects of the chemical inhibitors were variable, as illustrated by the bands denoted by numbered asterisks: 1) a band only reduced when treated with both drugs, 2) the major band reduced by MS-023, and 3) a band reduced by EPZ015666. GAPDH blot is from separate lanes to indicate relative protein amounts and shown as a reference for both DMA blots. F) Micrographs of live cells expressing SMNTud untreated and treated with DMA Inhibitors MS-023 and EPZ015666 for 48 h. Grayscale bar is in analog-digital units. Scale bar = 10 μm.
A-E) Micrographs of live cells expressing A) SMNTud or SMNTud with point mutations B) W102L, C) Y109L, D) Y130D, and E) E134K that reduce binding to DMA (Tripsianes et al., 2011). F) Micrographs of live cells expressing F118L mutation to SMNTud. Grayscale bars are in analog-digital units and apply to all images in panel. Scale bars = 10 μm. G) Quantification of the condensation effect of SMNTud F118L compared to cells expressing mCherry-Cry2. Units are analog-digital units (ADU). Solid lines and shading are a rolling mean and standard deviation of 10 points. Each point represents one cell. Dashed line represents a significance threshold relative to mCherry-Cry2 where a = 0.2 for the Mann-Whitney U test.
A) Fixed, untreated, wild-type NIH-3T3 cells showing their typical absence of nuclear bodies as detected by immunofluorescence using antibodies specific for SMN and coilin (Table S2). B) Fixed, untreated, wild-type NIH-3T3 cells showing their lack of snRNPs concentrated in nuclear bodies as marked by antibodies specific for trimethylguanosine (Table S2). C) Western blots from two biological replicates indicating depletion of SMN, recognized by monoclonal antibody 2B1 (Table S2) in HeLa lysate after siSMN transfection. D) SMN rescue visualized by staining for the myc epitope tag included in the degradation resistant rescue construct in HeLa cells. E) Single z-slice images taken from stacks used to produce images in Figure 3A. F) Line scans corresponding to (E) showing intensity in analog-digital units (ADU). G) SMN knockdown and rescue HeLa cells co-immunostained for coilin to indicate Cajal body status in each rescue condition. H) Statistical analysis corresponding to Figure 3B&C. Scale bars = 10 μm.
Fixed cells with Cry2-active SMNTud, which is always visualized by mCherry fluorescent signal. Condensates were co-stained for A) mCherry, B) sDMA using antibody SYM10, C) the Sm epitope with Y12, D) the snRNAs’ 5′ end trimethylguanosine cap, E) U1 snRNP-C, and F) U1-70k, both U1 snRNP specific proteins. In b) arrowheads indicate nuclear condensates that stain positively with SYM 10. Scale bar = 10 μm. G) Light-activated and formaldehyde fixed cells expressing SMNTud (magenta) and stained for endogenous coilin (green), showing coilin is a ligand of SMNTud. Hoechst staining shown in blue. H) Micrographs of Cry2-inactive (488 nm: off, grey) and Cry2-active (488 nm: on, blue) SMNTud in coilin knock-out mouse fibroblasts, showing coilin is not required for SMNTud condensation.
A) Western blot of whole cell lysate from NIH-3T3 cells showing expression levels of Cry2 constructs as measured by mCherry staining. All Constructs include mCherry-Cry2. B) LED array used for activating Cry2 in cells plated in six-well plates with coverslips. C) Quantification of the condensation effect in live cells of SMNTud compared to Spf30Tud. Units are analog-digital units (ADU). Solid lines and shading are a rolling mean and standard deviation of 10 points. Each point represents one cell. Dashed line represents a significance threshold relative to mCherry-Cry2 where a = 0.2 for the Mann-Whitney U test. D) Amino acid sequences of tudor domains aligned using Clustal Omega. Green/Cyan highlights indicate aromatic residues in the DMA binding pocket as determined by structural studies. Cyan indicates the position of mutation for non-functional constructs. E) Fixed NIH-3T3 cells expressing Tudor-Cry2 constructs under Cry2-inactive and Cry2-active conditions. F) Fixed NIH-3T3 cells expressing Tdrd8Tud and stained for sDMA with the SYM10 antibody. G) Fixed NIH-3T3 cells expressing Tdrd3Tud undergoing DMA inhibitor treatment as indicated. Percentage indicates fraction of cells with active Cry2 that have cytoplasmic condensates. Arrowheads indicate cells that possess or lack cytoplasmic condensates. Dotted lines indicate nuclear outlines. H) Fixed NIH-3T3 cells expressing Snd1Tud undergoing DMA inhibitor treatment as indicated. Insets show line profiles of the dashed lines in images. Corresponding bar plot displays line profiles quantified by normalizing the difference between the fifth and ninety-fifth percentile of intensity by the mean value for each profile. P-value computed with Mann-Whitney U test. Color bars given in analog-digital units. Scale bars = 10 μm.
A) Two biological replicates of untreated wild-type HeLa cells immunostained for SMN (magenta) and coilin (green). B) Untreated, C) MS-023 and EPZ015666 treated, D) MS-023 treated, and E) EPZ015666 treated HeLa cells stained for trimethylguanosine (TMG, magenta) and coilin (green). Arrowheads in insets indicate the coilin puncta shown in each line profile plot. Scale bar = 10 μm. F) Line profile plots for single slices of isoSTED data used to generate the 3D renderings in Figure 6E. Scale bar = 500 μm. G) Dye-flip control for SMN and coilin lateral depletion STED. Scale bar = 500 nm. H) Single label control for SMN and coilin two dimensional STED. Scale bar = 500 nm. See Table S2 for antibodies.
NIH-3T3 cells expressing mCherry-Cry2. At time 0.0, blue light is turned on to activate Cry2. Time resolution, 1.0 frame per second. Scale bar = 10 μm.
NIH-3T3 cells expressing SMN-Cry2. At time 0.0, blue light is turned on to activate Cry2. Time resolution, 1.0 frame per second. Scale bar = 10 μm.
NIH-3T3 cells expressing SMNTud. At time 0.0, blue light is turned on to activate Cry2. Time resolution, 1.0 frame per second. Scale bar = 10 μm.
Data Availability Statement
Raw microscopy data for optodroplet experiments made available at data.4dnucleome.org; accession numbers given in the Key Resources Table. Image analysis code available at github.com/bewersdorflab/quant-condensate.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti mCherry | Invitrogen | #PA5-34974; RRID: AB_2552323 |
| Anti coilin (H-300) | Santa Cruz Biotechnology | #sc-32860; RRID: AB_2081431 |
| Anti-coilin | Abcam | #ab210785 |
| Anti sDMA (SYM10) | Millipore Sigma | #07-412; RRID: AB_11212396 |
| Anti sDMA (SYM11) | Millipore Sigma | #07-413; RRID: 310595 |
| Anti aDMA (ASYM24) | Millipore Sigma | #07-414; RRID: AB_310596 |
| Anti SMN (2B1) | Abcam | #ab5831; RRID: 305144 |
| Anti GFP | Invitrogen | #A11122; RRID: AB_221569 |
| Anti GAPDH | Santa Cruz Biotechnology | #sc-25778; RRID: AB_10167668 |
| Anti Sm (Y12) | Laboratory of Joan Steitz; Pettersson et al., 1984 | N/A |
| Anti 2,2,7-Trimethylguanosine (K121) | Calbiochem | #NA02; RRID: AB_213109 |
| Anti SNRPC | Abcam | #ab82862; RRID: AB_2193870 |
| Anti U1-70K (CB7) | Laboratory of Doug Black; Listerman et al., 2006 | N/A |
| Anti-rabbit IgG Horseradish Peroxidase-linked | GE HealthCrae | #NA934; RRID: AB_772206 |
| Alexa Fluor 488-conjugated Donkey Anti-Rabbit | Jackson ImmunoResearch | #711-545-152; RRID: AB_2313584 |
| Alexa Fluor 488-conjugated Donkey Anti-Mouse | Jackson ImmunoResearch | #715-545-150; RRID: AB_2340850 |
| Anti-Mouse IgG - Atto 594 | Sigma | #76085; RRID: AB_1137653 |
| Anti-Rabbit IgG - Atto 594 | Sigma | #77671; RRID: AB_1137663 |
| Anti-Mouse IgG - Atto 647N | Sigma | #50185; RRID: AB_1137661 |
| Anti-Rabbit IgG - Atto 647N | Sigma | #40839; RRID: AB_1137669 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| MS-023 | Sigma | SML1555 |
| EPZ015666 | Sigma | SML1421 |
|
| ||
| Deposited data | ||
|
| ||
| Optodroplet Live Imaging Dataset - mCherry Control |
This Study | 4DNucleome: 4DNESVT8QSLU |
| Optodroplet Live Imaging Dataset- SMNTud | This Study | 4DNucleome: 4DNES9JHBEZC |
| Optodroplet Live Imaging Dataset - hnRNPA1IDR | This Study | 4DNucleome: 4DNES49WH1QD |
| Optodroplet Live Imaging Dataset - SMNTud Mock | This Study | 4DNucleome: 4DNESPZNHYSY |
| Optodroplet Live Imaging Dataset- SMNTud Treated with MS023 and EPZ015666 |
This Study | 4DNucleome: 4DNESM8F8PP5 |
| Optodroplet Live Imaging Dataset - SMNTud W102L |
This Study | 4DNucleome: 4DNESGOUKUHY |
| Optodroplet Live Imaging Dataset- SMNTud Y109L | This Study | 4DNucleome: 4DNESRORWX8M |
| Optodroplet Live Imaging Dataset- SMNTud Y130D | This Study | 4DNucleome: 4DNESGSHWGUK |
| Optodroplet Live Imaging Dataset - SMNTud E134K | This Study | 4DNucleome: 4DNESQJS4P53 |
| Optodroplet Live Imaging Dataset - SMNTud F118L | This Study | 4DNucleome: 4DNESLBSY7UC |
| Optodroplet Live Imaging Dataset - Spf30Tud | This Study | 4DNucleome: 4DNESZZ8T7O9 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HeLa-Kyoto | Neugebauer Lab | RRID: CVCL_1922 |
| NIH 3T3 | ATCC | CRL-1658 |
| HEK293FT | ThermoFisher | R70007 |
| Coil/ mouse embryonic fibroblasts | Laboratory of Greg Matera; Tucker et al., 2001 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| siSMN #1 | Thermo | #4427037 s445227 |
| siSMN #2 | Thermo | #4427037 s531465 |
| siSMN #3 | Thermo | #4427037 s531467 |
|
| ||
| Recombinant DNA | ||
|
| ||
| mCh-Cry2 | Laboratory of Clifford Brangwynne | N/A |
| FUSIDR | Laboratory of Clifford Brangwynne | N/A |
| hnRNPA1IDR | Laboratory of Clifford Brangwynne | N/A |
| SMN-Cry2 | This study | N/A |
| SMNIDR-N | This study | N/A |
| SMNTud | This study | N/A |
| SMNIDR-C | This study | N/A |
| dmTudorTud | This study | N/A |
| Aub3 | This study | N/A |
| Aub3(KG) | This study | N/A |
| Spf30Tud | This study | N/A |
| Tdrd1Tud#2 | This study | N/A |
| Tdrd1Tud#3 | This study | N/A |
| Tdrd1Tud#4 | This study | N/A |
| Tdrd3Tud | This study | N/A |
| Tdrd4Tud#1 | This study | N/A |
| Tdrd4Tud#4 | This study | N/A |
| Tdrd6Tud#5 | This study | N/A |
| Tdrd6Tud#6 | This study | N/A |
| Tdrd8Tud | This study | N/A |
| Tdrd9Tud | This study | N/A |
| Snd1Tud | This study | N/A |
| Recoded SMN-Myc | This study | N/A |
| Recoded SMN-Myc (E134K) | This study | N/A |
| Recoded SMN-Myc (Y109L) | This study | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Python Microscopy (PYME) | David Baddeley | https://Python-microscopy.org |
| FIJI | Schindelin et al., 2012 | https://Fiji.sc |
| Cell Profiler | Carpenter et al., 2006 | https://Cellprofiler.org |
| RaptorX | Kallberg et al., 2012 | https://Raptorx.uchicago.edu |
| Clustal Omega | Sievers et al., 2011 | https://Ebi.ac.uk |
| Imspector | Abberior Instruments | imspector.abberior-instruments.com |
| ConSurf | Berezin et al., 2004 | https://Consurf.tau.ac.il |
| Imaris | Oxford Instruments | https://Imaris.oxinst.com |