Abstract
Genetic predispositions and environmental influences both play an important role in adolescent externalizing behavior; however, they are not always independent. To elucidate gene-environment interplay, we examined the interrelationships between externalizing polygenic risk scores, parental knowledge, and peer substance use in impacting adolescent externalizing behavior across two time-points in a high-risk longitudinal sample of 1,200 adolescents (764 European and 436 African ancestry; Mage = 12.99) from the Collaborative Study on the Genetics of Alcoholism. Results from multivariate path analysis indicated that externalizing polygenic scores were directly associated with adolescent externalizing behavior but also indirectly via peer substance use, in the European ancestry sample. No significant polygenic association nor indirect effects of genetic risk were observed in the African ancestry group, likely due to more limited power. Our findings underscore the importance of gene-environment interplay and suggest peer substance use may be a mechanism through which genetic risk influences adolescent externalizing behavior.
Keywords: Adolescent externalizing, polygenic score, gene-environment interplay, parenting, peers
Externalizing behavior, which broadly refers to a constellation of behaviors characterized by behavioral disinhibition such as substance use and antisocial behaviors, is one of the most common and persistent forms of behavioral health problems in youth. It is associated with a wide range of psychosocial outcomes with far-reaching consequences, including poor academic achievement (Masten et al. 2005), increased risk for developing substance use problems and psychiatric disorders (Fergusson et al. 2007; Reef et al. 2011), and impaired psychosocial functioning (Bongers et al. 2007; Narusyte et al. 2017). Twin and family studies indicate that genetic influences account for approximately 50% of the variation (range ~20–80%) for each individual phenotype in the externalizing spectrum in the population (Barr and Dick 2020). Importantly, individual externalizing-related phenotypes load on a shared genetic factor, which is highly heritable (~80%; Kendler et al. 2003; Krueger et al. 2002; Young et al. 2000). Gene identification efforts for externalizing behaviors and related disorders have rapidly progressed in recent years, with various consortia conducting large-scale genome-wide association studies (GWAS) in order to detect variants associated with externalizing related phenotypes (e.g., Karlsson Linnér et al. 2019; Karlsson Linnér et al. in press; Liu et al. 2019). Using results from these GWAS, we can aggregate effects of common genetic variants across the genome and calculate polygenic scores in independent samples (Bogdan et al. 2018; Purcell et al. 2009). Studying these genetic risk scores in deeply phenotyped, longitudinal samples allows us to unpack the developmental processes by which genetic risk unfolds. Understanding the pathways by which genetic factors confer risk on adolescent externalizing behavior can aid in identifying targeted areas for prevention and intervention.
There are many routes by which genetic risk may influence behavioral development. Genetic risk could directly impact the behavior, likely by influencing brain development (Bos et al. 2018), gene expression, and associated regulatory pathways. In addition, the effects of genetic risk could operate indirectly via environmental pathways. In other words, the effects of genetic variation might depend on transaction with environmental processes (Tucker-Drob et al. 2013). Many “environmental” factors show evidence of genetic influence, including parenting practices, family environment, and peer experiences, with heritabilities in the range of 15–35% (Kendler and Baker 2007). The heritability of environmental factors reflects, in part, the fact that genetic predispositions are associated with the environments that individuals are born into or select into, the way they interpret environmental events, and the reactions they evoke from the world (Plomin et al. 1977; Scarr and McCartney 1983). Thus, genetic predispositions shape behaviors partially through genetic differences in exposure to particular environments (Avinun and Knafo 2014; Beaver and Wright 2007). This process, whereby individuals create or select into particular environments that are associated with their genetic predispositions, is known as gene-environment correlation (rGE; Jaffee and Price 2007; Rutter and Silberg 2002).
There are three main types of rGE processes: passive, evocative (reactive), and active. In passive rGE, genetically related parents pass on genes and also provide the child’s environment, both of which influence the child’s behaviors. For example, parents may pass on genes that are involved in the development of externalizing behaviors and also practice parenting strategies that are influenced by the same genetic factors. In evocative rGE, individuals’ genetic predispositions evoke a response from their environment, such as when parenting is a response to a child’s genetically influenced behavior. In active rGE, individuals select and shape their environments based on their genetic predispositions. For example, a child who has inherited a genetic propensity for externalizing may choose to spend time with others who encourage such behaviors. Although much attention has focused in recent years on characterizing gene-environment interaction (Dick et al. 2015; Dick and Kendler 2012), it is possible that these genetically influenced environmental pathways also play a considerable role in influencing behavioral outcomes (Tucker-Drob et al. 2013). There has been renewed interest in this area by recent demonstrations of genetic nurturance (Kong et al. 2018; Wertz et al. 2019) and indirect genetic effects (Xia et al. 2020) as indicated via genotypic correlations.
Parenting and family processes provide an important context for adolescent development. Parenting behaviors such as parental knowledge, monitoring, and support are associated with externalizing behaviors in adolescence (Racz and McMahon 2011). The association between parenting and adolescent externalizing behaviors is in part genetically influenced (Narusyte et al. 2011; Neiderhiser et al. 1999). Research and meta-analytic evidence show that genetically based differences among youth may evoke different responses from their parents (evocative rGE; Avinun and Knafo 2014; Klahr and Burt 2014). Twin and family studies demonstrate that children’s genetic predispositions contribute to parenting behaviors, such as discipline (Button et al. 2008), monitoring (Neiderhiser et al. 2007), and criticism (Narusyte et al. 2011). Moreover, evocative rGE has been shown to be involved in the relationship between parenting and adolescent externalizing, such that a genetic predisposition towards externalizing is associated with greater environmental risk by evoking a negative response from parents such as less parental knowledge of the child’s whereabouts and more parent-child relationship problems (Burt et al. 2005; Samek et al. 2014; Wertz et al. 2016). Studies with measured genotype data similarly show, for example, that children’s genetic risk for aggression can evoke negative parenting behaviors, as indexed by less parental monitoring (Elam et al. 2017). There is also evidence that problematic childhood behaviors are predictive of subsequent lower parental knowledge (Kerr and Stattin 2003; Kerr et al. 2008) because these youth may choose to disclose less about their whereabouts and parents may feel unable to manage the child’s misbehavior or withdraw (Racz and McMahon 2011), resulting in less parental knowledge. In turn, research consistently shows that a low level of parental knowledge and/or monitoring is a robust risk factor for poor psychosocial adjustment during adolescence, including externalizing behavior (Lac and Crano 2009; Lopez-Tamayo et al. 2016; Mason et al. 1994; Racz and McMahon 2011; Yap et al. 2017). Taken together, this suggests that parenting may play an important mediating role in linking children’s genetic predispositions and adolescent externalizing behaviors.
Peers also serve as an important context for adolescent development. Adolescents do not randomly choose friends; twin data show that genetic factors contribute to affiliation with deviant peers in adolescence (Tarantino et al. 2014). Research also indicates that genetic factors influence adolescent exposure to peers exhibiting externalizing behavior, such as alcohol and cigarette use (Fowler et al. 2007; Harden et al. 2008). Adolescents with genetic predispositions toward higher sensation seeking or delinquent behaviors are more likely to be affiliated with deviant peers (Mann et al. 2016; TenEyck and Barnes 2015). Collectively, this suggests that children may seek out affiliation with peers who have similar externalizing tendencies, thereby actively influencing the environments they experience. Accordingly, gene-environment correlation processes in relation to peers may also serve as a mechanism by which genetic factors influence adolescent externalizing behavior, as substance-using peers may provide opportunities and reinforcement for adolescent externalizing behavior (Dodge et al. 2009; Hawkins et al. 1992; Pratt et al. 2010).
Here, we report longitudinal analyses of some of the processes by which genetic risk influences externalizing behavior across adolescence. In this study, we used the results from the Externalizing Consortium’s multivariate GWAS (Karlsson Linnér et al. in press), the largest analysis to date on externalizing spectrum behavior (with data from ~1.5 million individuals) to calculate genome-wide polygenic scores in our independent, high-risk sample of adolescents of European and African ancestry. Given that rates of externalizing behaviors in adolescents vary across racial/ethnic groups (McLaughlin et al. 2007) and there are important differences in genetic diversity, allele frequencies, and linkage disequilibrium patterns between individuals of European and African ancestry (Campbell and Tishkoff 2008; Cardon and Palmer 2003), we stratified the sample by analyzing separately within European ancestry and African ancestry groups. We examined whether adolescents’ genetic predispositions, as characterized by genome-wide polygenic scores, have direct effects on externalizing behavior but also influence externalizing behavior via gene-environment correlation processes. We operationalized adolescent externalizing with both non-clinical indictors such as substance use behaviors and clinical indicators of externalizing problems including conduct disorder and oppositional defiant disorder criteria. We focused on the role of parent/family and peers as potential mechanisms through which genetic risk may influence adolescent externalizing behavior, as these are two salient developmental contexts for adolescent externalizing behavior (Dishion 2000; Hawkins et al. 1992; Pinquart 2017). We used repeated measures of externalizing behavior, parental knowledge, and perceived peer substance use across two time points (approximately two years apart) in adolescence to examine the following pre-registered (https://osf.io/d65au/) hypotheses:
1. Adolescents’ genetic predispositions toward externalizing behavior, as indexed by a polygenic score, would be associated with reported adolescent externalizing behavior, parental knowledge, and peer substance use at the baseline assessment (Time 1) such that higher externalizing polygenic scores would be associated with greater externalizing behavior and peer substance use, and lower parental knowledge.
2. Each of the domains (i.e., adolescent externalizing behavior; parental knowledge; and peer substance use) at Time 1 would predict variation in that same outcome at follow-up (Time 2).
3. Each of the domains at Time 1 would partly mediate the associations between externalizing polygenic score and adolescent externalizing behavior at Time 2.
In exploratory analyses, we examined whether there were sex differences in pathways of risk, and no specific hypotheses were made.
Methods
Sample
Data were drawn from the Collaborative Study on the Genetics of Alcoholism (COGA) Prospective Study (Bucholz et al. 2017). COGA is a diverse, multi-site, multi-generational family-based study of genetic and environmental factors for alcohol use disorders (Begleiter et al. 1995; Reich et al. 1998). Alcohol-dependent probands were ascertained through alcohol treatment programs at seven U.S. sites. In addition, a group of community comparison families were recruited into the study. Beginning in 2004, a sample of adolescent and young adult offspring (ages 12–22) of prior adult COGA participants were recruited into the COGA Prospective Study to study the development of alcohol use disorders and related problems (Bucholz et al. 2017). Prospective Study participants were interviewed at enrollment and followed up at approximately biennial intervals. The Institutional Review Board at all sites approved this study, and written consent (and assent for adolescents) was obtained from all participants.
The present study included adolescents (aged 12–17) for whom the following apply: (1) had GWAS data available, (2) completed the adolescent version of the Semi-Structured Assessment for the Genetics of Alcoholism for Children Interview (Bucholz et al. 1994; Kuperman et al. 2013) during their baseline and first follow-up (approximately two years after the baseline) assessments, (3) were under age 18 at their first follow-up assessment, and (4) were of European or African ancestry as determined by genetic ancestry principal component analysis. This strategy resulted in an analytic sample of 1,200 adolescents (49.9% male) from 458 COGA extended families. Youth from both case and comparison families were included. Specifically, this analytic sample included 764 European (EA; Mage = 12.99, SD = 1.13 at baseline assessment; Mage = 15.05, SD = 1.23 at Time 2; 50.0% male) and 436 African (AA; Mage = 12.99, SD = 1.11 at baseline assessment; Mage = 15.11, SD = 1.23 at Time 2; 49.8% male) ancestry participants. No differences were observed between this analytic sample and the whole adolescent sample at baseline in externalizing behaviors. However, this analytic sample was younger at baseline (t = −7.23, p < .01) and had lower peer substance use (t = −3.35, p < .01) at baseline.
This is a relatively early adolescent sample, as 46.9% of the sample was aged 12 at their baseline assessment. The interval between participants’ baseline assessment and their first follow-up assessment was, on average, 2.08 years (SD = .55). For simplicity, we refer to the baseline assessment and first follow-up assessment as Time 1 (T1) and Time 2 (T2), respectively, from this point forward.
Measures
Externalizing Behavior Score.
We created a composite measure of externalizing behaviors/disorders using the first principal component extracted from a principal component analysis. We used both non-clinical and clinical indicators, as age-appropriate, measured in the C-SSAGA interview: (1) alcohol use, (2) marijuana use, (3) cigarette use, (4) DSM-IV clinical criterion counts (American Psychiatric Association 1987) of conduct disorder (CD) criteria, and (5) DSM-IV oppositional defiant disorder (ODD) criterion counts. DSM-IV criterion counts were obtained from the C-SSAGA (Bucholz et al. 1994; Hesselbrock et al. 1999). Alcohol use was measured in the C-SSAGA by asking individuals to report the frequency of past-year drinking on a 12-point scale from 1 (about 1 to 2 days a year) to 12 (every day). Non-drinkers were coded as zero. Marijuana use was assessed by asking participants to report the number of times they used marijuana in the last 12 months. Cigarette use was coded as 1 = past-year use, and 0 = no cigarette use in the past year. We used developmentally-appropriate substance use variables, including alcohol, marijuana, and cigarette use, to capture more variability in risky behavior in the young sample, rather than using the more severe clinical-level substance use problems as indices of externalizing behaviors, which would yield floor effects in our early adolescent sample. We included a count of DSM-IV CD and ODD symptoms because they index externalizing behavior and problems in youth. Descriptive statistics for externalizing indicators are summarized in Table 1. At T1, the first principal component accounted for approximately 44% of the common variance among the externalizing variables with the following loadings: alcohol use, 0.72; marijuana use, 0.69; cigarette use, 0.75; CD, 0.62; and ODD, 0.49. At T2, the first principal component accounted for approximately 43% of the common variance among the externalizing variables with the following loadings: alcohol use, 0.71; marijuana use, 0.68; cigarette use, 0.70; CD, 0.67; and ODD, 0.51.
Table 1.
Descriptive statistics for externalizing indicators
| Time 1 | Time 2 | |||
|---|---|---|---|---|
| Indicators | Mean | SD | Mean | SD |
| Alcohol use | .23 | .95 | .92 | 1.84 |
| Marijuana use | 2.66 | 24.82 | 12.08 | 53.26 |
| Cigarette usea | 3.2% | - | 9.5% | - |
| Conduct disorder criterion count | .71 | 1.18 | .66 | 1.20 |
| Oppositional defiant disorder criterion count | .98 | 1.57 | .89 | 1.52 |
Note. Abbreviations: SD = standard deviation.
For binary variable, proportion of response category = 1 were reported. Alcohol use was measured by asking individuals to report the frequency of past-year use on a 12-point scale. Non-drinkers were coded as zero. Marijuana use was assessed by asking participants to report the number of times they used marijuana in the last 12 months. Cigarette use was coded as 1 = past-year use, and 0 = no cigarette use in the past year.
Parental Knowledge.
Parental knowledge was assessed via participants’ responses to three questions (how much their parent figures know about their plans, their interests, and where and with whom they spend time when not at home) adapted from Chassin et al. (1993), and included as part of the C-SSAGA. Responses were rated on a 4-point scale from 1 (always) to 4 (rarely). Cronbach’s alphas were .70 and .76 for T1 and T2, respectively. Items were reversed coded and averaged, and higher scores indicated higher parental knowledge.
Peer Substance Use.
Perception of peer substance use was measured using 4 items adapted from the FinnTwin12 study (Kaprio et al. 2002), included as part of the C-SSAGA. These questions asked participants about how many of their best friends smoke, use alcohol, use marijuana, and use other drugs. Responses were rated on a 4-point scale from 1 (none of them) to 4 (all of them). Cronbach’s alphas were .77 and .79 for T1 and T2, respectively. Items were averaged, and higher scores indicated higher levels of perceived peer substance use.
Genotyping and Externalizing Polygenic Scores.
Participants’ DNA samples were genotyped using the Illumina Human1M array (Illumina, San Diego, CA), the Illumina Human OmniExpress 12V1 array (Illumina), the Illumina 2.5M array (Illumina) or the Smokescreen genotyping array (Biorealm LLC, Walnut, CA; Baurley et al. 2016). A detailed description of data processing, quality control, and imputation is available elsewhere (Lai et al. 2019). Data were imputed to 1000 Genome Phase 3, and single nucleotide polymorphisms (SNPs) with a genotyping rate < 0.95, that violated Hardy-Weinberg equilibrium (p < 10−6), or had minor allele frequency (MAF) < 0.01 were excluded from analysis.
Genetic risk for externalizing problems was assessed by constructing polygenic scores (PGS), which are aggregate measures of the number of risk alleles individuals carry, each weighted by effect sizes from GWAS summary statistics. We used PRS-CS auto version (Ge et al. 2019) to calculate the externalizing polygenic scores. This approach employs a Bayesian regression and continuous shrinkage method to correct for the non-independence among nearby SNPs in the genome (i.e. linkage disequilibrium, LD). In keeping with the recommendations of the PRS-CS developers, we limited the SNPs included in the PGS to HapMap3 SNPs that overlapped between the original GWAS summary statistics and the LD reference panel (1000 Genomes Phase III reference panel).
We calculated externalizing polygenic scores using estimates from a multivariate genomic analysis of externalizing behaviors/problems (which included alcohol problems, attention-deficit/hyperactivity disorder (ADHD), lifetime cannabis use, age of first sexual intercourse, number of sexual partners, general risk tolerance, and lifetime smoking initiation) with an effective sample size of ~1.5 million individuals of European ancestry, from the international Externalizing Consortium(Karlsson Linnér et al. in press). For a detailed description of the samples and phenotypes that went into the multivariate genomic modeling, please see Karlsson Linnér (in press) and its pre-registered analysis plan (https://osf.io/pmyq8/).
Our sample included participants of European and African ancestry. For the European ancestry sample, we used estimates from the above referenced discovery GWAS to derive the externalizing polygenic scores. For the African ancestry sample, we calculated externalizing polygenic scores using two methods, given prior evidence that PGS are most predictive when individuals in the discovery GWAS and the target sample are matched on ancestral background (Martin et al. 2017; Peterson et al. 2019). First, we created the PGS based on the weights from the discovery GWAS of European ancestry individuals (Karlsson Linnér et al. in press). Second, we used a multi-ethnic polygenic scoring approach (Márquez-Luna et al. 2017) to calculate PGS for the African ancestry sample. This multi-ethnic scoring approach combines GWAS results from the European ancestry discovery sample with training GWAS data from the target (i.e., African ancestry) population. Specifically, we combined GWAS results weights from Karlson Linnér et al. (in press) with results from a 10-fold GWAS method of a latent externalizing factor constructed by a phenotypic externalizing factor that corresponded with the seven indicator phenotypes in the discovery multivariate analyses (Karlsson Linnér et al. in press) in the COGA sample of African ancestry individuals (including all COGA African ancestry individuals, not limiting to the subset of African adolescents in the current study; n = 3379). Thus, for our African ancestry sample, we calculated two versions of the externalizing polygenic scores. The externalizing polygenic score that accounted for the largest amount of variance in adolescent externalizing behavior scores in the African ancestry sample was advanced to subsequent analyses of that sample, per our pre-registered analytic plan. We recognize the limitations of this approach and understand that it is less than ideal; however, we believe it is an ethical imperative to include African Americans in genetic studies like this one.
In order to account for population stratification, we regressed all polygenic scores on the first 10 genetic ancestry principal components (PC1-PC10) and saved the residualized PGS. Analyses were conducted separately by ancestral background. We used standardized, residualized polygenic scores in all subsequent analyses.
Covariates.
We included participants’ age and sex as covariates given their demonstrated associations with adolescent externalizing behaviors.
Analytic Strategy
We first examined descriptive statistics and correlations among study variables. We then conducted regression analyses to examine how well the externalizing polygenic scores could explain variability in the externalizing scores in our sample using the change in R2 above a baseline model with sex, age, and the first 10 genetic ancestry principal components.
Next, we conducted path analysis (see Figure 1 for the conceptual model). Externalizing PGS was specified to have a direct genetic effect on T1 externalizing (as indicated by path a in Figure 1), a direct genetic effect on T1 parental knowledge (path b), and a direct genetic effect on T1 peer substance use (path c). Paths b and c test for evidence of gene-environment correlations. In addition, adolescent externalizing behavior, parental knowledge, and peer substance use were specified to be correlated within T1. Externalizing PGS was specified to predict T2 externalizing behavior directly (not shown), and indirectly through T1 parental knowledge (path d) and T1 peer substance use (path e), while controlling for previous levels of externalizing behavior at T1. Additional cross-domain paths between T1 and T2 were also included: associations between T1 externalizing behavior and T2 parental knowledge (path f) and T2 peer substance use (path g); association between T1 parental knowledge and T2 peer substance use (path h); and association between T1 peer substance use and T2 parental knowledge (path i).
Figure 1.
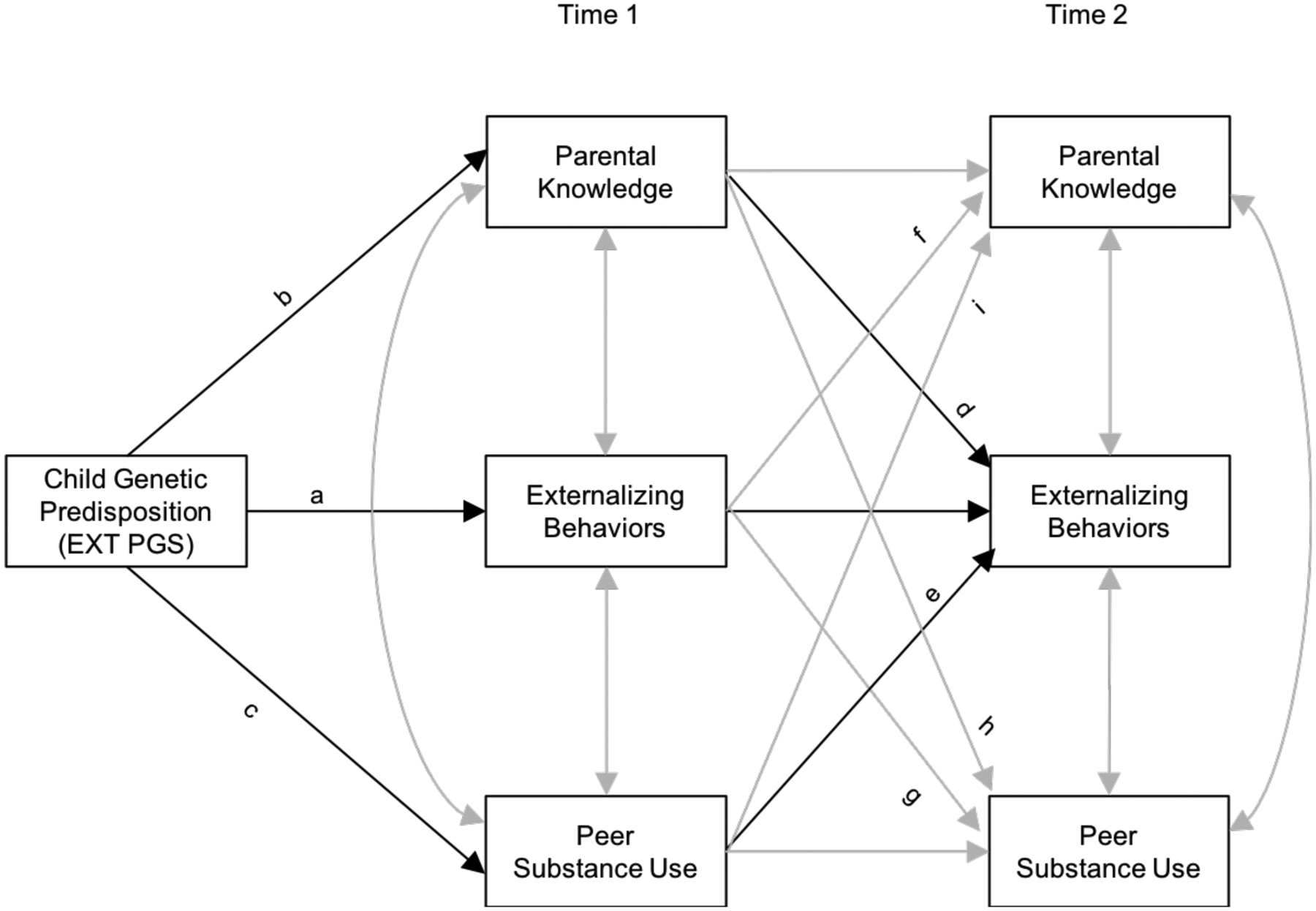
Conceptual model of the associations among externalizing polygenic score, parental knowledge, peer substance use, and adolescent externalizing behaviors and their interplay
Note. a = direct genetic effect on Time 1 (T1) externalizing behavior. b = direct genetic effect on T1 parental knowledge. c = direct genetic effect on T1 peer substance use. Paths b and c indicate evidence of gene-environment correlations. d = direct effect of T1 parental knowledge on T2 externalizing behavior. e = direct effect of T1 peer substance use on T2 externalizing behavior. f = effect of T1 externalizing behavior on T2 parental knowledge. g = effect of T1 externalizing behavior on T2 peer substance use. h = effect of T1 parental knowledge on T2 peer substance use. i = effect of T1 peer substance use on T2 parental knowledge.
Adolescents’ age and sex were included as covariates for adolescent externalizing behaviors, parental knowledge, and peer substance use at both time points. We used the MODEL INDIRECT command in Mplus to examine indirect effects of externalizing PGS on T2 externalizing behaviors via T1 parental knowledge and T1 peer substance use. This provides a test of specific indirect effects in addition to the total indirect and direct effects of externalizing PGS on adolescent externalizing behaviors. Since multiple indirect pathways were examined simultaneously, specific indirect effects reflect each of the specific pathways (e.g., EXT PGS→T1 parental knowledge →T2 externalizing behaviors) while also accounting for the shared associations between them. We evaluated indirect effects using bias-corrected bootstrapped (1000 times) 95% confidence intervals. Confidence intervals not including zero would provide evidence of statistically significant indirect effects.
Following this primary path modeling analysis, we also conducted exploratory analyses to examine potential differences in path coefficients across adolescent sex. We used a multigroup model by removing sex from the path model and compared a model with paths constrained to equality with another model that had paths freely estimated between males and females. Wald test of parameter equalities was used to test whether there were significant sex differences in observed effects.
We conducted analyses using Mplus version 8.3 (Muthén and Muthén 1998–2012). Full information maximum likelihood estimation method was used to account for missing data. Clustering within families was accounted for using the CLUSTER command in Mplus. We conducted analyses separately by ancestry in order to minimize the issue of population stratification.
Results
Descriptive statistics and zero-order correlations for the study variables are presented in Table 2. Among EAs, positive correlations were observed between EXT PGS and externalizing behavior and peer substance use at both T1 and T2, and negative correlations were observed between EXT PGS and parental knowledge at both time points. T1 parental knowledge and T1 peer substance use were also correlated with T2 externalizing behavior. Among AAs, EXT PGS was not significantly correlated with any of the study variables. However, the pattern of correlations among the phenotypic variables were consistent with the correlations observed among EAs.
Table 2.
Descriptive statistics and zero-order correlations among study variables for the European ancestry sample (below diagonal) and African ancestry sample (above diagonal)
| Measure | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| 1. EXT PGS | – | .08 | .02 | −.07 | .00 | .03 | .00 |
| 2. T1 Externalizing score | .17 | – | −.29 | .57 | .57 | −.31 | .36 |
| 3. T1 Parental knowledge | −.12 | −.48 | – | −.26 | −.25 | .47 | −.21 |
| 4. T1 Peer substance use | .14 | .68 | −.41 | – | .36 | −.24 | .46 |
| 5. T2 Externalizing score | .23 | .55 | −.36 | .48 | – | −.41 | .52 |
| 6. T2 Parental knowledge | −.12 | −.27 | .47 | −.21 | −.41 | – | −.33 |
| 7. T2 Peer substance use | .19 | .45 | −.34 | .57 | .71 | −.38 | – |
| EA Mean | 0.00 | −0.22 | 3.49 | 1.13 | −0.02 | 3.46 | 1.34 |
| EA SD | 0.99 | 0.79 | 0.57 | 0.32 | 1.00 | 0.57 | 0.51 |
| AA Mean | 0.00 | −0.07 | 3.28 | 1.14 | 0.07 | 3.20 | 1.35 |
| AA SD | 0.99 | 0.67 | 0.73 | 0.30 | 1.04 | 0.76 | 0.48 |
Note. Abbreviations: SD = standard deviation. EXT PGS = Externalizing polygenic score. T1 = Time 1. T2 = Time 2. EA = participants of European ancestry. AA = participants of African ancestry. Correlations for the European ancestry sample are presented in the lower diagonal, and correlations for the African ancestry sample are presented in the upper diagonal. In the African ancestry sample, EXT PGS was constructed based on the weights from the discovery GWAS of European ancestry individuals. Bolded correlations indicate p < .05
Next, we conducted linear regressions to assess the predictive power of EXT PGS in explaining variation in adolescent externalizing behavior scores in our sample. Among EAs, the externalizing polygenic score was associated with both T1 externalizing behavior score (ΔR2 = .03, p < .01) and T2 externalizing behavior score (ΔR2 = .05, p < .01). Among AAs, neither the polygenic score constructed using the EA weights nor the multi-ethnic polygenic score was associated with externalizing behavior score (ΔR2 = .001–.007, all ns). Because the multi-ethnic polygenic score did not improve the predictive ability in the AA sample, we carried forward the polygenic score constructed using the weights from results of the GWAS based on samples of European ancestry.
Path modeling results
European ancestry participants.
Figure 2 summarizes the results of the path analysis. Path modeling results are shown in Table 3, which includes the coefficients and p-values for the paths shown in Figure 1, as well as indirect effects. As hypothesized, EXT PGS was positively associated with externalizing behavior and peer substance use at T1, and negatively associated with parental knowledge at T1. All stability paths between T1 and T2 (e.g., T1 parental knowledge and T2 parental knowledge) were significant (ps < .01). Further, within-time covariation paths (e.g., T1 parental knowledge with T1 externalizing behavior) were all significant (ps < .01). Controlling for covariation links among domains within time or stability within domains between T1 and T2, peer substance use at T1 predicted T2 externalizing behavior (p < .01). T1 parental knowledge was, however, not significantly associated with T2 externalizing behavior (p = .07). T1 externalizing behavior was not associated with subsequent parental knowledge nor peer substance use at T2. However, T1 parental knowledge predicted lower levels of perceived peer substance use at T2.
Figure 2.
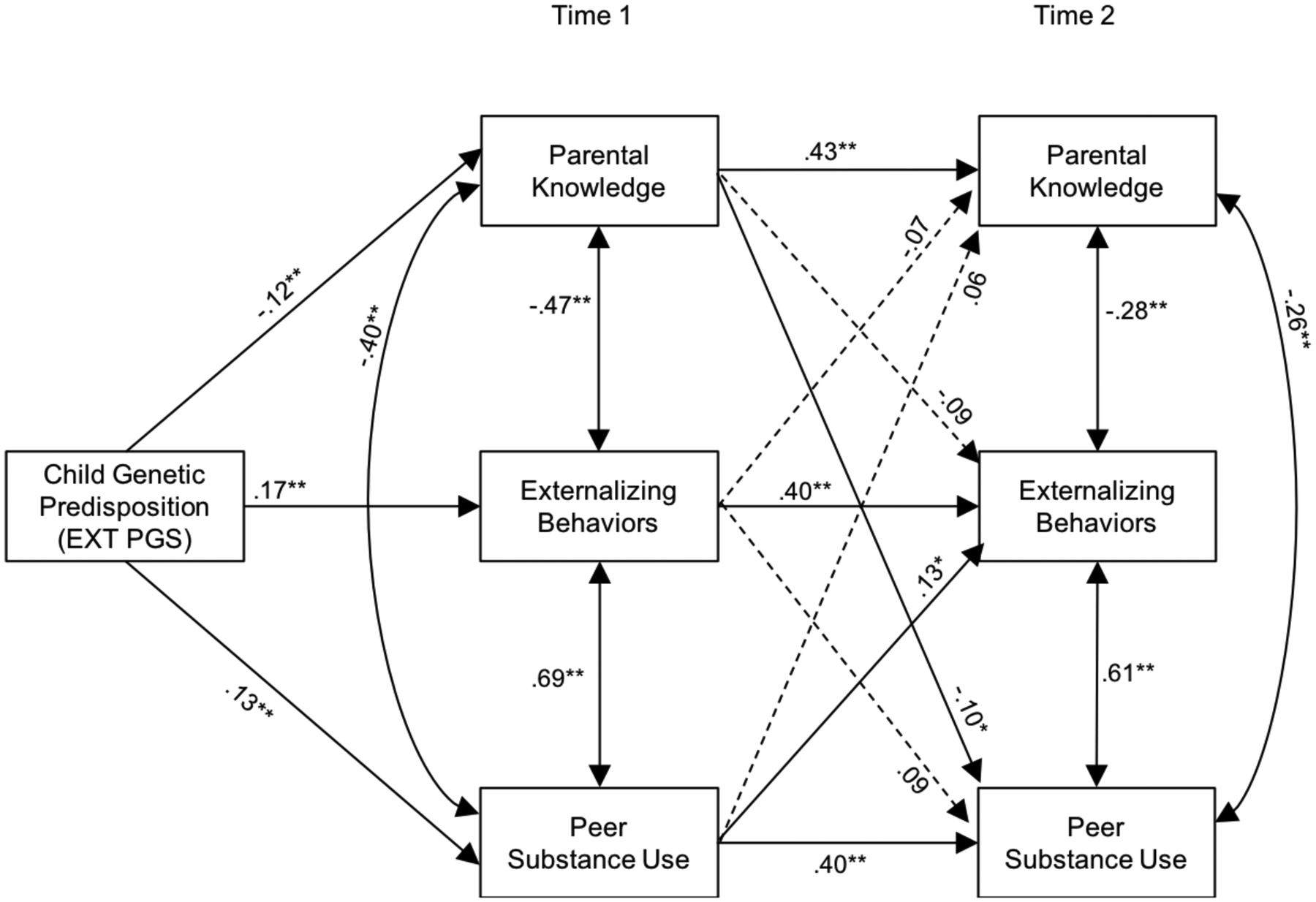
Path model linking the associations among externalizing polygenic scores, parental knowledge, peer substance use, and adolescent externalizing behaviors at Time 1 and Time 2 among participants of European ancestry
Note. Standardized path coefficients are presented. The EXT PGS included in the analyses was regressed on the ten genetic ancestry principal components. To simplify the figure, covariates (age, sex) are not illustrated. Dashed lines represent nonsignificant paths.
*p < .05, **p < .01.
Table 3.
Path coefficients, direct and indirect effects from the path model examining parental knowledge and peer substance use as indirect pathways linking externalizing polygenic scores and adolescent externalizing behaviors among participants of European ancestry
| Paths | B | SE | β | p |
|---|---|---|---|---|
| EXT PGS → T1 EXT | .14 | .03 | .17 | <.001 |
| EXT PGS → T1 par knowledge | −.07 | .02 | −.12 | .004 |
| EXT PGS → T1 peer sub use | .04 | .01 | .13 | <.001 |
| EXT PGS → T2 EXT | .14 | .03 | .14 | <.001 |
| EXT PGS → T2 par knowledge | −.04 | .02 | −.06 | .047 |
| EXT PGS → T2 peer sub use | .06 | .02 | .11 | <.001 |
| T1 EXT → T2 EXT | .51 | .10 | .40 | <.001 |
| T1 EXT → T2 par knowledge | −.05 | .06 | −.07 | .421 |
| T1 EXT → T2 peer sub use | .06 | .05 | .09 | .226 |
| T1 par monitor → T2 EXT | −.16 | .09 | −.09 | .070 |
| T1 par monitor → T2 par knowledge | .43 | .05 | .43 | <.001 |
| T1 par monitor → T2 peer sub use | −.09 | .04 | −.10 | .020 |
| T1 peer sub use → T2 EXT | .42 | .16 | .13 | <.001 |
| T1 peer sub use → T2 par knowledge | .11 | .09 | .06 | .263 |
| T1 peer sub use → T2 peer sub use | .63 | .09 | .40 | <.001 |
| Indirect Effects | B | 95% CI | β | p |
| EXT PGS → T1 EXT → T2 EXT | .070 | [.030, .123] | .068 | .003 |
| EXT PGS → T1 par knowledge → T2 EXT | .011 | [.000, .032] | .011 | .145 |
| EXT PGS → T1 peer sub use → T2 EXT | .018 | [.004, .039] | .017 | .043 |
Note. Abbreviations: EXT PGS = externalizing polygenic score. T1 = Time 1. T2 = Time 2. EXT = externalizing behaviors. Par knowledge = parental knowledge. Peer sub use = peer substance use. CI = confidence intervals. The EXT PGS included in the analyses was regressed on the ten genetic ancestry principal components. Age and sex were included as covariates. Statistically significant coefficients/effects are bolded.
Consistent with our hypothesis, results indicated that there were significant indirect effects of EXT PGS on T2 externalizing behavior via T1 variables. Specifically, EXT PGS was associated with higher T2 externalizing behavior via higher levels of peer substance use at T1 (B = .02, 95% CI [.004, .039], p < .05). Further, as expected, EXT PGS was also associated with higher T2 externalizing behavior via more externalizing behaviors at T1 (B = .07, 95% CI [.030, .123], p < .01). There was no significant indirect effect of EXT PGS on T2 externalizing through T1 parental knowledge (B = .01, 95% CI [.000, .032], p = .145).
In an exploratory analysis, we examined potential sex differences by conducting a multigroup analysis. Results from the Wald test of constraining all paths linking EXT PGS, externalizing behavior, parental knowledge, and peer substance use to be equal across males and females was not significant (χ2 = 9.38, df = 12, p = .67), suggesting no evidence for sex differences in the effects.
African ancestry participants.
Figure 3 summarizes the results of the path analysis, and results are shown in Table 4. Contrary to our hypothesis, EXT PGS was not associated with externalizing behavior, parental knowledge, and peer substance use at T1. However, consistent with our expectation, all stability paths between T1 and T2 (e.g., T1 parental knowledge and T2 parental knowledge) were significant (ps < .01). Further, within time covariation paths (e.g., T1 parental knowledge with T1 externalizing behavior) were all significant (ps < .01) in the expected direction. Controlling for covariation links among domains within time or stability within domains between T1 and T2, T1 parental knowledge predicted lower T2 externalizing behavior (p < .05). T1 peer substance use was, however, not significantly associated with T2 externalizing behavior. T1 externalizing behavior predicted lower levels of parental knowledge at T2 and higher levels of peer substance use at T2. There was no evidence of indirect effects of EXT PGS on T2 externalizing behavior via any of the T1 variables.
Figure 3.
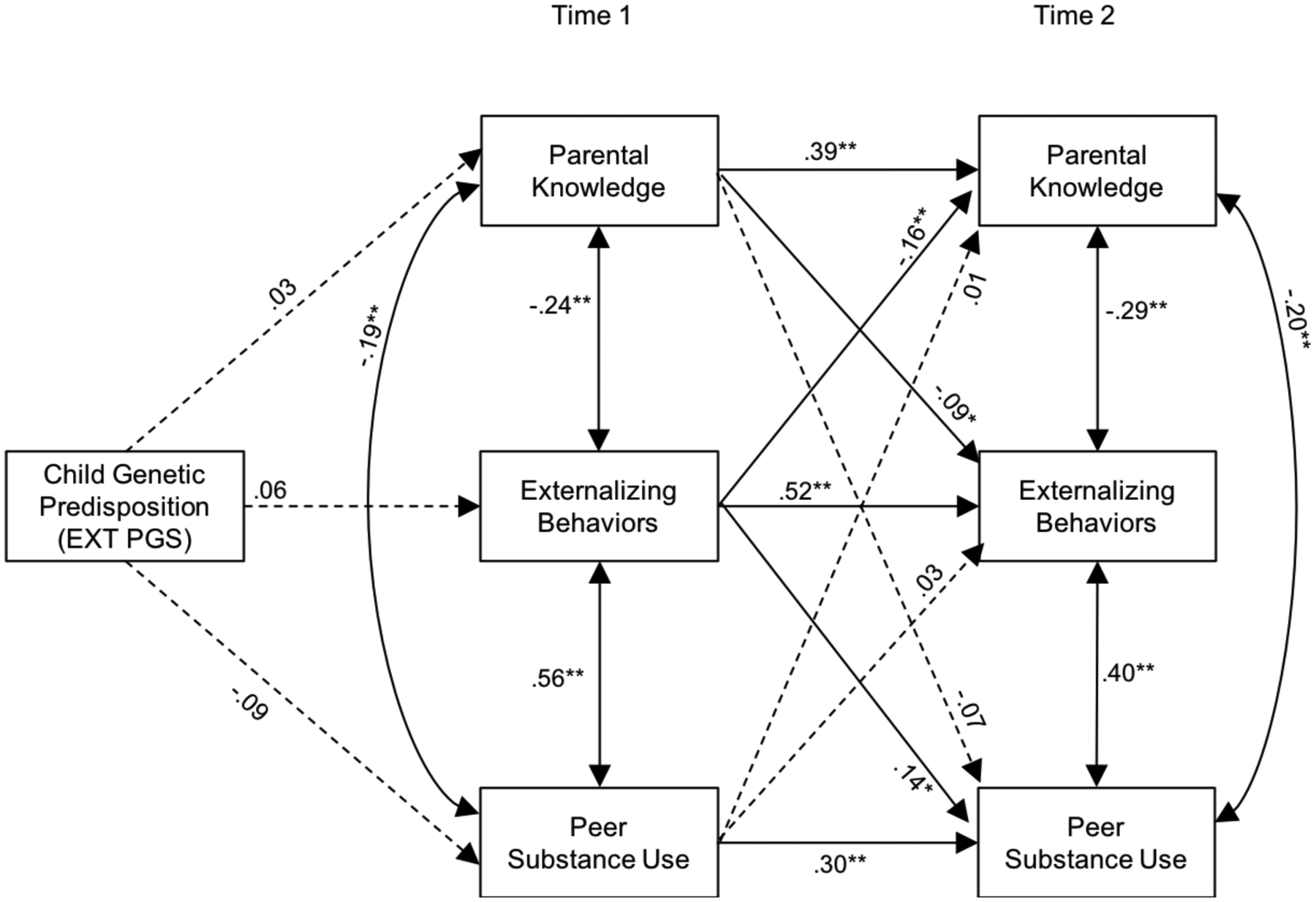
Path model linking the associations among externalizing polygenic scores, parental knowledge, peer substance use, and adolescent externalizing behaviors at Time 1 and Time 2 among participants of African ancestry
Note. Standardized path coefficients are presented. The EXT PGS included in the analyses was regressed on the ten genetic ancestry principal components. To simplify the figure, covariates (age, sex) are not illustrated. Dashed lines represent nonsignificant paths.
*p < .05, **p < .01.
Table 4.
Path coefficients, direct and indirect effects from the path model examining parental knowledge and peer substance use as indirect pathways linking externalizing polygenic scores and adolescent externalizing behaviors among participants of African ancestry
| Paths | B | SE | β | p |
|---|---|---|---|---|
| EXT PGS → T1 EXT | .04 | .04 | .06 | .266 |
| EXT PGS → T1 par knowledge | .02 | .04 | .03 | .512 |
| EXT PGS → T1 peer sub use | −.03 | .02 | −.09 | .094 |
| EXT PGS → T2 EXT | −.04 | .04 | −.03 | .369 |
| EXT PGS → T2 par knowledge | .03 | .03 | .04 | .300 |
| EXT PGS → T2 peer sub use | .01 | .02 | .05 | .218 |
| T1 EXT → T2 EXT | .79 | .12 | .52 | <.001 |
| T1 EXT → T2 par knowledge | −.18 | .07 | −.16 | .008 |
| T1 EXT → T2 peer sub use | .10 | .05 | .14 | .030 |
| T1 par monitor → T2 EXT | −.13 | .06 | −.09 | .040 |
| T1 par monitor → T2 par knowledge | .41 | .05 | .39 | <.001 |
| T1 par monitor → T2 peer sub use | −.04 | .04 | −.07 | .213 |
| T1 peer sub use → T2 EXT | .10 | .27 | .03 | .712 |
| T1 peer sub use → T2 par knowledge | .02 | .14 | .01 | .896 |
| T1 peer sub use → T2 peer sub use | .48 | .09 | .30 | <.001 |
| Indirect Effects | B | 95% CI | β | p |
| EXT PGS → T1 EXT → T2 EXT | .032 | [−.026, .084] | .030 | .248 |
| EXT PGS → T1 par knowledge → T2 EXT | −.003 | [−.016, .005] | −.003 | .562 |
| EXT PGS → T1 peer sub use → T2 EXT | −.003 | [−.020, .005] | −.003 | .733 |
Note. Abbreviations: EXT PGS = externalizing polygenic score. T1 = Time 1. T2 = Time 2. EXT = externalizing behaviors. Par knowledge = parental knowledge. Peer sub use = peer substance use. CI = confidence intervals. The EXT PGS included in the analyses was regressed on the ten genetic ancestry principal components. Age and sex were included as covariates. Statistically significant coefficients/effects are bolded.
Discussion
The main goal of the current study was to use longitudinal data to examine the processes by which genetic risk for externalizing behavior unfolds. We focused on direct genetic effects, as well as the possible role of indirect genetically influenced environmental pathways involving family and peer contexts on influencing adolescent externalizing behavior. We tested whether parental knowledge and peer substance use accounted for, in part, the association of genetic predispositions with adolescent externalizing behavior. We considered parental knowledge and peer substance use mediating pathways simultaneously in order to examine the unique effects of each specific pathway. The present study adds to the literature in understanding the mechanisms through which genetic risk, as operationalized using polygenic scores, unfolds to influence adolescent development and behavior.
We found that polygenic scores derived from results of a large multivariate genomic analysis of externalizing spectrum behavior/disorders (Karlsson Linnér et al. in press) were associated with adolescent externalizing behavior in an independent sample of European ancestry. Our polygenic scores were derived using results from a multivariate genomic approach of externalizing behaviors/problems that included a range of clinical and non-clinical traits related to the externalizing spectrum. Notably, the phenotypic adolescent externalizing behavior in our sample was derived using age-appropriate, similar externalizing phenotypes, but not directly overlapping with the externalizing phenotypes in the discovery GWAS (Karlsson Linnér et al. in press). The observed polygenic association is consistent with evidence from twin and family studies that much of the genetic influences on any externalizing psychopathology/behavior is broadly shared with other externalizing spectrum behavior/problems (Kendler et al. 2003; Krueger et al. 2002; Young et al. 2000). In addition, as hypothesized, externalizing polygenic score predicted greater peer substance use, which in turn was associated with adolescents’ subsequent externalizing behavior, while controlling for prior level of externalizing behavior. Our results indicated that peer substance use serves as a mediating pathway of genetic influences on adolescent externalizing behavior, presumably reflecting active and evocative gene-environment correlation processes as adolescents select and shape their environments (Scarr and McCartney 1983). These findings are consistent with previous research, with evidence from mostly twin and family studies, showing that children’s genetic predispositions are associated with their peer group selections/ affiliation (Elam et al. 2017; Fowler et al. 2007; Harden et al. 2008; Mann et al. 2016; TenEyck and Barnes 2015), as well as with the robust relationship between peer substance use and adolescents externalizing behavior (Hawkins et al. 1992; Pratt et al. 2010). It is also possible that adolescents with a genetic predisposition toward externalizing may be considered “more fun” for other like-mind peers, making other children with similar externalizing tendencies more likely to want to spend time with them. Taken together, our results demonstrate that genetic risk conferred not only direct effects on adolescent externalizing behavior but also impacted subsequent externalizing behavior indirectly through perceived peer substance use. This underscores the importance of gene-environment interplay, and highlights gene-environment correlations, particularly in relation to peers, as important mechanisms by which genetic risk influence behavior and psychosocial outcomes in adolescence. Additional research is needed to consider other psychosocial mechanisms underlying genetic risk for adolescent outcomes.
We also found that adolescent’ genetic predispositions for externalizing behavior were associated with parental knowledge, providing some evidence of evocative gene-environment correlation (Klahr and Burt 2014). This is consistent with prior literature indicating that children’s genetic predispositions contribute to parenting behaviors and could evoke, for example, less parental monitoring and less knowledge (Elam et al. 2017; Samek et al. 2014; Wertz et al. 2016). Higher genetic risk for externalizing was associated with lower parental knowledge, and this inverse relationship likely reflects lower levels of adolescent self-disclosure of their activities and whereabouts and perhaps poorer quality of parent-adolescent relationship (e.g., less enjoyable parent-child interactions, less parental involvement).
Contrary to our expectations, there was no indirect effect of genetic risk on adolescent externalizing at T2 via parental knowledge from T1. This seems to be consistent with some evidence from the twin literature that suggests parental knowledge is associated with adolescent externalizing via a direct environmental influence independent of genetic influences (Marceau et al. 2015). It is also possible that the lack of significant indirect effects via parenting from T1 may mean that parental knowledge may have more of a concurrent effect than a lagged effect. In fact, in looking at the within-time correlations, the magnitude of correlation between T1 parental knowledge and T1 externalizing was smaller as compared to the correlation between T1 peer substance use and T1 externalizing, and, thus, there is even less predictive variance to be carried over. It is interesting to also note that higher parental knowledge at T1 was associated with decreased perceived peer substance use at T2, which is correlated with T2 externalizing behavior. Thus, it is possible that the lagged effect of parental knowledge might not be operating on externalizing behavior per se, but through the discouragement of affiliations with deviant peers. More generally, the lack of indirect effect via parental knowledge may in part also reflect that youth spend less time with parents during adolescence (Hill et al. 2007; Larson et al. 1996) as adolescents increasingly gain autonomy to shape their social worlds, with a consequent increase in the influence of peers.
In our analysis of the smaller African ancestry subsample, we found no significant association between externalizing polygenic scores and adolescent externalizing behavior, nor associations between externalizing polygenic score and T1 parental knowledge and T1 peer substance use. The lack of predictive polygenic association is likely due in part to the smaller sample size and thereby reduced power, and in part to the fact that the discovery sample of our externalizing polygenic score included only individuals of European ancestry (Karlsson Linnér et al. in press). Polygenic scores do not translate well across ancestries (Martin et al. 2017), and populations of non-European ancestry, particularly those of African ancestry, have been historically underrepresented in gene identification efforts (Popejoy and Fullerton 2016). This is a limitation of the field as a whole, and represents a serious issue for studies with diverse populations. Although we attempted to address this issue by also implementing a multi-ethnic polygenic scoring approach (Márquez-Luna et al. 2017), the predictive power of the polygenic score constructed using this approach in the African ancestry subsample was still severely attenuated when compared to the results observed in the European ancestry subsample. Newer statistical methods and approaches to better account for differences in allele frequencies and LD patterns in order to calculate polygenic scores across populations are under development (e.g., Liang et al. 2020). However, there is a critical need to prioritize diversity and increase representation of non-European population in genetic research in order to ensure that all racial/ethnic groups benefit equally from advances related to genetic findings (Martin et al. 2019).
Although there were no significant genotypic effects in our analysis of African ancestry subsample, interesting cross-phenotypic correlations between T1 and T2 emerged. In contrast to the link observed in the European ancestry subsample, T1 peer substance use was not correlated with T2 externalizing behavior in the African ancestry sample. T1 parental knowledge was, however, predictive of subsequent adolescent externalizing behavior at T2. This is consistent with the prior literature that shows that a high level of parental knowledge is a protective factor for adolescent externalizing behavior (Lac and Crano 2009; Lopez-Tamayo et al. 2016; Mason et al. 1994; Racz and McMahon 2011; Yap et al. 2017). Additionally, there was an inverse relationship between T1 adolescent externalizing behavior and T2 parental knowledge. This provides some evidence that youth behavioral problems could predict a change in parenting behavior (i.e., less parental knowledge). One explanation of this relationship could be because youth with behavioral problems are less likely to disclose details of their lives to parents. Additionally, parents may feel that they are unable to manage their high externalizing child and start to give up and withdraw, resulting in less parental knowledge (Kerr et al. 2008; Racz and McMahon 2011). In addition, T1 externalizing behavior also predicted subsequent levels of parental knowledge at T2. This suggests the bidirectional associations between adolescent externalizing behavior and parenting (Pardini 2008), highlighting the effects of the child on parents as well. Collectively, our findings suggest potential ethnic/racial differences in vulnerability such that European American youth may be more susceptible to peer influences (e.g., peer substance use), whereby parents continue to exert a more important role in youth outcomes further into adolescence in African American youth (Wallace and Muroff 2002). Future research is needed to take a cultural genomics approach by examining the interplay between genes and environments across and within different cultural groups (Causadias and Korous 2018), as the salience of different pathways/mechanisms of genetic risk may vary across different racial/ethnic background.
Our results should be interpreted within the context of the following limitations. First, COGA is a high-risk sample with most participants from extended families enriched for alcohol use disorders. Findings from this study may not generalize to samples with different recruitment/ascertainment strategies (Savage et al. 2018). Replicating our findings in community and population-based samples is an important next step. Second, all of our measures were based on self-report data. It is possible that adolescents’ externalizing traits or their genetic predispositions led to biased reports of perceived peer substance use (Bauman and Ennett 1996) and parental knowledge. Third, our measure of parental knowledge tapped adolescent perceptions rather than actual parental knowledge or specifically implemented parental monitoring behavior/strategies and control (Kerr et al. 2010; Stattin and Kerr 2000). Future work is needed to replicate our findings and to use multi-informant data (e.g., adolescent report, parent report) and consider other dimensions of parenting. In addition, our measure of parental knowledge was not specific to maternal or paternal knowledge. Overall, findings of the current study appear to generalize to males and females. However, it is possible that sex of the parent and sex of the child may interact to influence the relationships between parenting and adolescent externalizing (Finan et al. 2015; Trudeau et al. 2012).
Fourth, we found an association between child’s genetic predispositions and parental knowledge, suggestive of evocative gene-environment correlation. However, there could be additional alternative explanations, such as passive rGE. For example, this relationship could also in part indicate that parents themselves were high on externalizing symptomatology (genetically influenced characteristics), which contributed to lower levels of parental knowledge, as research suggests that parental psychopathology negatively influences parenting (Berg-Nielsen et al. 2002; Cummings et al. 2005). Future work with assessments of parental psychopathology will be needed to tease apart the pathways by which risk unfolds. Fifth, we focused only on peer substance use because it is a commonly studied aspect of peer influences on externalizing behaviors. Our sample was, however, limited in the peer measures that were collected. Future research can consider the role of broader peer influences, including delinquency and deviance.
Sixth, we examined peer substance use and parental knowledge as two plausible pathways underlying the mechanisms through which genetic risks influence externalizing in adolescence. This study offers an illustration of broad strokes of patterns representing the interplay between polygenic risk, parenting, peers, and adolescent externalizing behavior, but these broad patterns require closer examination in future studies, both to replicate these findings and consider other explanatory pathways and shared processes from multiple levels and contexts that may underpin the genetic influences on adolescent behavior and outcomes. Finally, our study focused on adolescence, and by T1 in this study, the constructs examined were already showing strong interconnections. This suggests that many of the gene-environment correlation processes began earlier in development. Future research should consider earlier antecedents and rGE processes at younger ages to further delineate how genetic risk unfolds across development and interfaces with the environment.
In conclusion, the present study adds to the literature by unpacking the cascading developmental processes by which genetic factors confer risk for adolescent behavior. Our results show that polygenic scores from a large-scale multivariate genomic analyses of externalizing spectrum behavior/disorders predicted externalizing behavior among a sample of independent adolescents. Among individuals of European ancestry, in addition to direct genetic effects on adolescent externalizing behavior, our findings show that peer substance use serves as an important mediating pathway by which genetic influences further convey risk for future adolescent externalizing behavior. Externalizing polygenic score predicted higher levels of adolescent externalizing behavior indirectly via peer substance use. More generally, our results indicate that genetically at-risk youth carry risk that is exacerbated through a combination of gene-environment processes. This research underscores the importance of considering the complex interplay between genes and environments when considering etiology and prevention/intervention for externalizing behavior because gene-environment correlation processes serve as mechanisms by which genetic influences, in part, impact adolescent behavioral problems. Understanding the mechanisms of genetic risk for problematic behaviors in youth will be critical for identifying targets for effective prevention and intervention efforts. Additionally, our findings underscore the need for expanded genetic study in diverse populations.
Acknowledgements
The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, T. Foroud; Scientific Director, A. Agrawal; Translational Director, D. Dick, includes eleven different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, T. Foroud, J. Nurnberger Jr., Y. Liu); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz, J. Meyers, C. Kamarajan, A. Pandey); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz, A. Agrawal); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield, A. Brooks, R. Hart); The Children’s Hospital of Philadelphia, University of Pennsylvania (L. Almasy); Virginia Commonwealth University (D. Dick, J. Salvatore); Icahn School of Medicine at Mount Sinai (A. Goate, M. Kapoor, P. Slesinger); and Howard University (D. Scott). Other COGA collaborators include: L. Bauer (University of Connecticut); L. Wetherill, X. Xuei, D. Lai, S. O’Connor, M. Plawecki, S. Lourens (Indiana University); L. Acion (University of Iowa); G. Chan (University of Iowa; University of Connecticut); D.B. Chorlian, J. Zhang, S. Kinreich, G. Pandey (SUNY Downstate); M. Chao (Icahn School of Medicine at Mount Sinai); A. Anokhin, V. McCutcheon, S. Saccone (Washington University); F. Aliev, P. Barr (Virginia Commonwealth University); H. Chin and A. Parsian are the NIAAA Staff Collaborators.
We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
The Externalizing Consortium: Principal Investigators: Danielle M. Dick, Philipp Koellinger, K. Paige Harden, Abraham A. Palmer. Lead Analysts: Richard Karlsson Linnér, Travis T. Mallard, Peter B. Barr, Sandra Sanchez-Roige. Significant Contributor: Irwin Waldman.
The Externalizing Consortium has been supported by the National Institute of Alcohol Abuse and Alcoholism (R01AA015146 - administrative supplement), and the National Institute on Drug Abuse (R01DA050721). Additional funding for investigator effort has been provided by K02AA018755, U10AA008401, P50AA022537, as well as a European Research Council Consolidator Grant (647648 EdGe). The content is solely the responsibility of the authors and does not necessarily represent the official views of the above funding bodies. The Externalizing Consortium would like to thank the following for making its research possible: 23andMe, Add Health, Vanderbilt University Medical Center’s BioVU, Collaborative Study on the Genetics of Alcoholism (COGA), the Psychiatric Genomics Consortium’s Substance Use Disorders working group, UK10K Consortium, UK Biobank, and Philadelphia Neurodevelopmental Cohort.
Funding
This study was funded by the National Institutes of Health through the National Institute on Alcohol Abuse and Alcoholism (U10AA008401, K02AA018755, K01AA024152) and the National Institute on Drug Abuse (R01DA050721).
Footnotes
Conflict of interest
The authors have no conflicts of interest to report.
Compliance with ethical standards
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. No animal subjects were used in the studies and no human experimental protocols were carried out. Informed consent was obtained from all individual participants included in the study.
References
- American Psychiatric Association (1987) Diagnostic and Statistical Manual of Mental Disorders. American Psychiatric Association, Washington, DC [Google Scholar]
- Avinun R, Knafo A (2014) Parenting as a Reaction Evoked by Children’s Genotype: A Meta-Analysis of Children-as-Twins Studies. Personality and Social Psychology Review 18(1):87–102 [DOI] [PubMed] [Google Scholar]
- Barr PB, Dick DM (2020) The Genetics of Externalizing Problems. In: de Wit H, Jentsch JD (eds) Recent Advances in Research on Impulsivity and Impulsive Behaviors. Springer International Publishing, Cham, pp 93–112 [Google Scholar]
- Bauman KE, Ennett ST (1996) On the importance of peer influence for adolescent drug use: commonly neglected considerations. Addiction 91(2):185–198 [PubMed] [Google Scholar]
- Baurley JW, Edlund CK, Pardamean CI, Conti DV, Bergen AW (2016) Smokescreen: a targeted genotyping array for addiction research. BMC Genomics 17(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaver KM, Wright JP (2007) A Child Effects Explanation for the Association Between Family Risk and Involvement in an Antisocial Lifestyle. Journal of adolescent research 22(6):640–664 [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li T-K, Schuckit MA, Edenberg HJ, Rice JP (1995) The collaborative study on the genetics of alcoholism. Alcohol Health and Research World 19228–228 [Google Scholar]
- Berg-Nielsen TS, Vikan A, Dahl AA (2002) Parenting Related to Child and Parental Psychopathology: A Descriptive Review of the Literature. Clinical Child Psychology and Psychiatry 7(4):529–552 [Google Scholar]
- Bogdan R, Baranger DAA, Agrawal A (2018) Polygenic Risk Scores in Clinical Psychology: Bridging Genomic Risk to Individual Differences. Annual Review of Clinical Psychology 14(1):119–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers IL, Koot HM, van der Ende J, Verhulst FC (2007) Predicting young adult social functioning from developmental trajectories of externalizing behaviour. Psychological Medicine 38(7):989–999 [DOI] [PubMed] [Google Scholar]
- Bos MGN, Wierenga LM, Blankenstein NE, Schreuders E, Tamnes CK, Crone EA (2018) Longitudinal structural brain development and externalizing behavior in adolescence. Journal of Child Psychology & Psychiatry 59(10):1061–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Reich T, Schmidt I, Schuckit MA (1994) A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. Journal of Studies on Alcohol 55(2):149–158 [DOI] [PubMed] [Google Scholar]
- Bucholz KK, McCutcheon VV, Agrawal A, Dick DM, Hesselbrock VM, Kramer JR, Kuperman S, Nurnberger JI Jr, Salvatore JE, Schuckit MA, Bierut LJ, Foroud TM, Chan G, Hesselbrock M, Meyers JL, Edenberg HJ, Porjesz B (2017) Comparison of Parent, Peer, Psychiatric, and Cannabis Use Influences Across Stages of Offspring Alcohol Involvement: Evidence from the COGA Prospective Study. Alcoholism: Clinical and Experimental Research 41(2):359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt SA, McGue M, Krueger RF, Iacono WG (2005) How are parent–child conflict and childhood externalizing symptoms related over time? Results from a genetically informative cross-lagged study. Development and Psychopathology 17(1):145–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Button TMM, Lau JYF, Maughan B, Eley TC (2008) Parental punitive discipline, negative life events and gene–environment interplay in the development of externalizing behavior. Psychological Medicine 38(1):29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MC, Tishkoff SA (2008) African Genetic Diversity: Implications for Human Demographic History, Modern Human Origins, and Complex Disease Mapping. Annual Review of Genomics and Human Genetics 9(1):403–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ (2003) Population stratification and spurious allelic association. The Lancet 361(9357):598–604 [DOI] [PubMed] [Google Scholar]
- Causadias JM, Korous KM (2018) How are genes related to culture? Introduction to the field of cultural genomics. In: Causadias JM, Telzer EH, Gonzales NA (eds) The Handbook of Culture and Biology. John Wiley & Sons, New York, pp 153–177 [Google Scholar]
- Chassin L, Pillow DR, Curran PJ, Molina BSG, Barrera M Jr. (1993) Relation of parental alcoholism to early adolescent substance use: A test of three mediating mechanisms. Journal of Abnormal Psychology 102(1):3–19 [DOI] [PubMed] [Google Scholar]
- Cummings ME, Keller PS, Davies PT (2005) Towards a family process model of maternal and paternal depressive symptoms: exploring multiple relations with child and family functioning. Journal of Child Psychology and Psychiatry 46(5):479–489 [DOI] [PubMed] [Google Scholar]
- Dick DM, Agrawal A, Keller MC, Adkins A, Aliev F, Monroe S, Hewitt JK, Kendler KS, Sher KJ (2015) Candidate Gene–Environment Interaction Research: Reflections and Recommendations. Perspectives on Psychological Science 10(1):37–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Kendler KS (2012) The Impact of Gene-Environment Interaction on Alcohol Use Disorders. Alcohol Research: Current Reviews 34(3):318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishion TJ (2000) Cross-setting consistency in early adolescent psychopathology: deviant friendships and problem behavior sequelae. Journal of Personality 68(6):1109–1126 [DOI] [PubMed] [Google Scholar]
- Dodge KA, Malone PS, Lansford JE, Miller S, Pettit GS, Bates JE (2009) A dynamic cascade model of the development of substance-use onset. Monographs of the Society for Research in Child Development 74(3):vii–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elam KK, Chassin L, Lemery-Chalfant K, Pandika D, Wang FL, Bountress K, Dick D, Agrawal A (2017) Affiliation with substance-using peers: Examining gene-environment correlations among parent monitoring, polygenic risk, and children’s impulsivity. Developmental Psychobiology 59(5):561–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergusson DM, Horwood LJ, Ridder EM (2007) Conduct and attentional problems in childhood and adolescence and later substance use, abuse and dependence: Results of a 25-year longitudinal study. Drug and Alcohol Dependence 88S14–S26 [DOI] [PubMed] [Google Scholar]
- Finan LJ, Schulz J, Gordon MS, Ohannessian CM (2015) Parental problem drinking and adolescent externalizing behaviors: The mediating role of family functioning. Journal of Adolescence 43100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler T, Shelton K, Lifford K, Rice F, McBride A, Nikolov I, Neale MC, Harold G, Thapar A, Van Den Bree MBM (2007) Genetic and environmental influences on the relationship between peer alcohol use and own alcohol use in adolescents. Addiction 102(6):894–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW (2019) Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nature Communications 10(1):1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden KP, Hill JE, Turkheimer E, Emery RE (2008) Gene-Environment Correlation and Interaction in Peer Effects on Adolescent Alcohol and Tobacco Use. Behavior Genetics 38(4):339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JD, Catalano RF, Miller JY (1992) Risk and protective factors for alcohol and other drug problems in adolescence and early adulthood: Implications for substance abuse prevention. Psychological Bulletin 112(1):64–105 [DOI] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V (1999) A validity study of the SSAGA-a comparison with the SCAN. Addiction 94(9):1361–1370 [DOI] [PubMed] [Google Scholar]
- Hill NE, Bromell L, Tyson DF, Flint R (2007) Developmental Commentary: Ecological Perspectives on Parental Influences During Adolescence. Journal of clinical child and adolescent psychology 36(3):367–377 [DOI] [PubMed] [Google Scholar]
- Jaffee SR, Price TS (2007) Gene–environment correlations: a review of the evidence and implications for prevention of mental illness. Molecular Psychiatry 12(5):432–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaprio J, Pulkkinen L, Rose RJ (2002) Genetic and Environmental Factors in Health-related Behaviors: Studies on Finnish Twins and Twin Families. Twin Research 5(5):366–371 [DOI] [PubMed] [Google Scholar]
- Karlsson Linnér R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, Lebreton M, Tino SP, Abdellaoui A, Hammerschlag AR, Nivard MG, Okbay A, Rietveld CA, Timshel PN, Trzaskowski M, Vlaming Rd, Zünd CL, Bao Y, Buzdugan L, Caplin AH, Chen C-Y, Eibich P, Fontanillas P, Gonzalez JR, Joshi PK, Karhunen V, Kleinman A, Levin RZ, Lill CM, Meddens GA, Muntané G, Sanchez-Roige S, Rooij FJv, Taskesen E, Wu Y, Zhang F, Agee M, Alipanahi B, Bell RK, Bryc K, Elson SL, Furlotte NA, Huber KE, Litterman NK, McCreight JC, McIntyre MH, Mountain JL, Northover CAM, Pitts SJ, Sathirapongsasuti JF, Sazonova OV, Shelton JF, Shringarpure S, Tian C, Tung JY, Vacic V, Wilson CH, Agbessi M, Ahsan H, Alves I, Andiappan A, Awadalla P, Battle A, Beutner F, Jan Bonder M, Boomsma DI, Christiansen M, Claringbould A, Deelen P, Esko T, Favé M-J, Franke L, Frayling T, Gharib SA, Gibson G, Heijmans B, Hemani G, Jansen R, Kähönen M, Kalnapenkis A, Kasela S, Kettunen J, Kim Y, Kirsten H, Kovacs P, Krohn K, Kronberg-Guzman J, Kukushkina V, Kutalik Z, Lee B, Lehtimäki T, Loeffler M, Marigorta UM, Metspalu A, Milani L, Montgomery GW, Müller-Nurasyid M, Nauck M, Nivard MG, Penninx B, Perola M, Pervjakova N, Pierce B, Powell J, Prokisch H, Psaty BM, Raitakari O, Ring S, Ripatti S, Rotzchke O, Rüeger S, Saha A, Scholz M, Schramm K, Seppälä I, Stumvoll M, Sullivan P,Hoen P-Bt, Teumer A, Thiery J, Tong L, Tönjes A, Dongen Jv, Meurs Jv, Verlouw J, Visscher PM, Völker U, Võsa U, Westra H-J, Yaghootkar H, Yang J, Zeng B, Zhang F, Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, Turley P, Chen G-B, Emilsson V, Meddens SFW, Oskarsson S, Pickrell JK, Thom K, Timshel P, de Vlaming R, Abdellaoui A, Ahluwalia TS, Bacelis J, Baumbach C, Bjornsdottir G, Brandsma JH, Concas MP, Derringer J, Furlotte NA, Galesloot TE, Girotto G, Gupta R, Hall LM, Harris SE, Hofer E, Horikoshi M, Huffman JE, Kaasik K, Kalafati IP, Karlsson R, Kong A, Lahti J, Lee SJvd, de Leeuw C, Lind PA, Lindgren K-O, Liu T, Mangino M, Marten J, Mihailov E, Miller MB, Most PJvd, Oldmeadow C, Payton A, Pervjakova, Peyrot WJ, Qian Y, Raitakari O, Rueedi R, Salvi E, Schmidt B, Schraut KE, Shi J, Smith AV, Poot RA, Pourcain BS, Teumer A, Thorleifsson G, Verweij N, Vuckovic D, Wellmann J, Westra H-J, Yang J, Zhao W, Zhu Z, Alizadeh BZ, Amin N, Bakshi A, Baumeister SE, Biino G, Bønnelykke K, Boyle PA, Campbell H, Cappuccio FP, Davies G, De Neve J-E, Deloukas P, Demuth I, Ding J, Eibich P, Eisele L, Eklund N, Evans DM, Faul JD, Feitosa MF, Forstner AJ, Gandin I, Gunnarsson B, Halldórsson BV, Harris TB, Heath AC, Hocking LJ, Holliday EG, Homuth G, Horan MA, Hottenga J-J, de Jager PL, Joshi PK, Jugessur A, Kaakinen MA, Kähönen M, Kanoni S, Keltigangas-Järvinen L, Kiemeney LALM, Kolcic I, Koskinen S, Kraja AT, Kroh M, Kutalik Z, Latvala A, Launer LJ, Lebreton MP, Levinson DF, Lichtenstein P, Lichtner P, Liewald DCM, Loukola A, Madden PA, Mägi R, Mäki-Opas T, Marioni RE, Marques-Vidal P, Meddens GA, McMahon G, Meisinger C, Meitinger T, Milaneschi Y, Milani L, Montgomery GW, Myhre R, Nelson CP, Nyholt DR, Ollier WER, Palotie A, Paternoster L, Pedersen NL, Petrovic KE, Porteous DJ, Räikkönen K, Ring SM, Robino A, Rostapshova O, Rudan I, Rustichini A, Salomaa V, Sanders AR, Sarin A-P, Schmidt H, Scott RJ, Smith BH, Smith JA, Staessen JA, Steinhagen-Thiessen E, Strauch K, Terracciano A, Tobin MD, Ulivi S, Vaccargiu S, and Me Research T, e QC, International Cannabis C, Social Science Genetic Association C (2019) Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nature Genetics 51(2):245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson Linnér R, Mallard TT, Barr PB, Sanchez-Roige S, Madole JW, Driver MN, Poore HE, de Vlaming R, Grotzinger AD, Tielbeek JJ, Johnson EC, Liu M, Rosenthal SB, Ideker T, Zhou H, Kember RL, Pasman JA, Verweij KJH, Liu DJ, Vrieze S, COGA Collaborators, Kranzler HR, Gelernter J, Harris KM, Tucker-Drob EM, Waldman I, Palmer AA, Harden KP, Koellinger PD, Dick DM (in press)Multivariate genomic analysis of 1.5 million people identifies genes related to addiction, antisocial behavior, and health. Nature Neuroscience [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Baker JH (2007) Genetic influences on measures of the environment: a systematic review. Psychological Medicine 37(5):615–626 [DOI] [PubMed] [Google Scholar]
- Kerr M, Stattin H (2003) Parenting of adolescents: Action or reaction? In: Crouter AC, Booth A (eds) Children’s influence on family dynamics. Erlbaum, Mahwah, NJ, pp 121–151 [Google Scholar]
- Kerr M, Stattin H, Burk WJ (2010) A reinterpretation of parental monitoring in longitudinal perspective. Journal of Research on Adolescence 20(1):39–64 [Google Scholar]
- Kerr M, Stattin H, Pakalniskiene V (2008) Parents React to Adolescent Problem Behaviors by Worrying More and Monitoring Less. InWhat Can Parents Do?, pp 89–112 [Google Scholar]
- Klahr AM, Burt SA (2014) Elucidating the etiology of individual differences in parenting: A meta-analysis of behavioral genetic research. Psychological Bulletin 140(2):544–586 [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young AI, Thorgeirsson TE, Benonisdottir S, Oddsson A, Halldorsson BV, Masson G, Gudbjartsson DF, Helgason A, Bjornsdottir G, Thorsteinsdottir U, Stefansson K (2018) The nature of nurture: Effects of parental genotypes. Science 359(6374):424. [DOI] [PubMed] [Google Scholar]
- Kuperman S, Chan G, Kramer JR, Wetherill L, Bucholz KK, Dick D, Hesselbrock V, Porjesz B, Rangaswamy M, Schuckit M (2013) A Model to Determine the Likely Age of an Adolescent’s First Drink of Alcohol. Pediatrics 131(2):242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lac A, Crano WD (2009) Monitoring Matters: Meta-Analytic Review Reveals the Reliable Linkage of Parental Monitoring With Adolescent Marijuana Use. Perspectives on Psychological Science 4(6):578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D, Wetherill L, Bertelsen S, Carey CE, Kamarajan C, Kapoor M, Meyers JL, Anokhin AP, Bennett DA, Bucholz KK, Chang KK, De Jager PL, Dick DM, Hesselbrock V, Kramer J, Kuperman S, Nurnberger JI, Raj T, Schuckit M, Scott DM (2019) Genome-wide association studies of alcohol dependence, DSM-IV criterion count and individual criteria. Genes, Brain & Behavior 18(6):N.PAG-N.PAG [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson RW, Richards MH, Moneta G, Holmbeck G, Duckett E (1996) Changes in adolescents’ daily interactions with their families from ages 10 to 18: Disengagement and transformation. Developmental psychology 32(4):744–754 [Google Scholar]
- Liang Y, Pividori M, Manichaikul A, Palmer AA, Cox NJ, Wheeler H, Im HK (2020) Polygenic transcriptome risk scores improve portability of polygenic risk scores across ancestries. bioRxiv 2020.2011.2012.373647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, Zhan X, Agee M, Alipanahi B, Auton A, Bell RK, Bryc K, Elson SL, Fontanillas P, Furlotte NA, Hinds DA, Hromatka BS, Huber KE, Kleinman A, Litterman NK, McIntyre MH, Mountain JL, Northover CAM, Sathirapongsasuti JF, Sazonova OV, Shelton JF, Shringarpure S, Tian C, Tung JY, Vacic V, Wilson CH, Pitts SJ, Mitchell A, Skogholt AH, Winsvold BS, Sivertsen B, Stordal E, Morken G, Kallestad H, Heuch I, Zwart J-A, Fjukstad KK, Pedersen LM, Gabrielsen ME, Johnsen MB, Skrove M, Indredavik MS, Drange OK, Bjerkeset O, Børte S, Stensland SØ, Choquet H, Docherty AR, Faul JD, Foerster JR, Fritsche LG, Gabrielsen ME, Gordon SD, Haessler J, Hottenga J-J, Huang H, Jang S-K, Jansen PR, Ling Y, Mägi R, Matoba N, McMahon G, Mulas A, Orrù V, Palviainen T, Pandit A, Reginsson GW, Skogholt AH, Smith JA, Taylor AE, Turman C, Willemsen G, Young H, Young KA, Zajac GJM, Zhao W, Zhou W, Bjornsdottir G, Boardman JD, Boehnke M, Boomsma DI, Chen C, Cucca F, Davies GE, Eaton CB, Ehringer MA, Esko T, Fiorillo E, Gillespie NA, Gudbjartsson DF, Haller T, Harris KM, Heath AC, Hewitt JK, Hickie IB, Hokanson JE, Hopfer CJ, Hunter DJ, Iacono WG, Johnson EO, Kamatani Y, Kardia SLR, Keller MC, Kellis M, Kooperberg C, Kraft P, Krauter KS, Laakso M, Lind PA, Loukola A, Lutz SM, Madden PAF, Martin NG, McGue M, McQueen MB, Medland SE, Metspalu A, Mohlke KL, Nielsen JB, Okada Y, Peters U, Polderman TJC, Posthuma D, Reiner AP, Rice JP, Rimm E, Rose RJ, Runarsdottir V, Stallings MC, Stančáková A, Stefansson H, Thai KK, Tindle HA, Tyrfingsson T, Wall TL, Weir DR, Weisner C, Whitfield JB, Winsvold BS, Yin J, Zuccolo L, Bierut LJ, Hveem K, Lee JJ, Munafò MR, Saccone NL, Willer CJ, Cornelis MC, David SP, Hinds DA, Jorgenson E, Kaprio J, Stitzel JA, Stefansson K, Thorgeirsson TE, Abecasis G, Liu DJ, Vrieze S, andMe Research T, Psychiatry HA-I (2019) Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nature Genetics 51(2):237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Tamayo R, LaVome Robinson W, Lambert SF, Jason LA, Ialongo NS (2016) Parental Monitoring, Association with Externalized Behavior, and Academic Outcomes in Urban African-American Youth: A Moderated Mediation Analysis. American Journal of Community Psychology 57(3/4):366–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann FD, Patterson MW, Grotzinger AD, Kretsch N, Tackett JL, Tucker-Drob EM, Harden KP (2016) Sensation seeking, peer deviance, and genetic influences on adolescent delinquency: Evidence for person-environment correlation and interaction. Journal of Abnormal Psychology 125(5):679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau K, Narusyte J, Lichtenstein P, Ganiban JM, Spotts EL, Reiss D, Neiderhiser JM (2015) Parental knowledge is an environmental influence on adolescent externalizing. Journal of Child Psychology & Psychiatry 56(2):130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez-Luna C, Loh P-R, South Asian Type 2 Diabetes C, The STDC, Price AL (2017) Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genetic Epidemiology 41(8):811–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, Kenny EE (2017) Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. The American Journal of Human Genetics 100(4):635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason CA, Cauce AM, Gonzales N, Hiraga Y, Grove K (1994) An Ecological Model of Externalizing Behaviors in African-American Adolescents: No Family Is an Island. Journal of Research on Adolescence (Lawrence Erlbaum) 4(4):639–655 [Google Scholar]
- Masten AS, Roisman GI, Long JD, Burt KB, Obradović J, Riley JR, Boelcke-Stennes K, Tellegen A (2005) Developmental Cascades: Linking Academic Achievement and Externalizing and Internalizing Symptoms Over 20 Years. Developmental Psychology 41(5):733–746 [DOI] [PubMed] [Google Scholar]
- McLaughlin KA, Hilt LM, Nolen-Hoeksema S (2007) Racial/Ethnic Differences in Internalizing and Externalizing Symptoms in Adolescents. Journal of Abnormal Child Psychology 35(5):801–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthén K, Muthén BO (1998–2012) Mplus user’s guide (7th ed.). Authors, Los Angeles, CA [Google Scholar]
- Narusyte J, Neiderhiser JM, Andershed A-K, D’Onofrio BM, Reiss D, Spotts E, Ganiban J, Lichtenstein P (2011) Parental criticism and externalizing behavior problems in adolescents: The role of environment and genotype–environment correlation. Journal of Abnormal Psychology 120(2):365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusyte J, Ropponen A, Alexanderson K, Svedberg P (2017) Internalizing and externalizing problems in childhood and adolescence as predictors of work incapacity in young adulthood. Social Psychiatry and Psychiatric Epidemiology 52(9):1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiderhiser JM, Reiss D, Hetherington EM, Plomin R (1999) Relationships between parenting and adolescent adjustment over time: Genetic and environmental contributions. Developmental Psychology 35(3):680–692 [DOI] [PubMed] [Google Scholar]
- Neiderhiser JM, Reiss D, Lichtenstein P, Spotts EL, Ganiban J (2007) Father-adolescent relationships and the role of genotype-environment correlation. Journal of Family Psychology 21(4):560–571 [DOI] [PubMed] [Google Scholar]
- Pardini DA (2008) Novel Insights into Longstanding Theories of Bidirectional Parent–Child Influences: Introduction to the Special Section. Journal of Abnormal Child Psychology 36(5):627–631 [DOI] [PubMed] [Google Scholar]
- Peterson RE, Kuchenbaecker K, Walters RK, Chen C-Y, Popejoy AB, Periyasamy S, Lam M, Iyegbe C, Strawbridge RJ, Brick L, Carey CE, Martin AR, Meyers JL, Su J, Chen J, Edwards AC, Kalungi A, Koen N, Majara L, Schwarz E, Smoller JW, Stahl EA, Sullivan PF, Vassos E, Mowry B, Prieto ML, Cuellar-Barboza A, Bigdeli TB, Edenberg HJ, Huang H, Duncan LE (2019) Genome-wide Association Studies in Ancestrally Diverse Populations: Opportunities, Methods, Pitfalls, and Recommendations. Cell 179(3):589–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinquart M (2017) Associations of parenting dimensions and styles with externalizing problems of children and adolescents: An updated meta-analysis. Developmental Psychology 53(5):873–932 [DOI] [PubMed] [Google Scholar]
- Plomin R, DeFries JC, Loehlin JC (1977) Genotype-environment interaction and correlation in the analysis of human behavior. Psychological Bulletin 84(2):309–322 [PubMed] [Google Scholar]
- Popejoy AB, Fullerton SM (2016) Genomics is failing on diversity. Nature 538(7624):161–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt TC, Cullen FT, Sellers CS, Thomas Winfree L, Madensen TD, Daigle LE, Fearn NE, Gau JM (2010) The Empirical Status of Social Learning Theory: A Meta-Analysis. Justice quarterly JQ / 27(6):765–802 [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P (2009) Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460(7256):748–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racz SJ, McMahon RJ (2011) The Relationship Between Parental Knowledge and Monitoring and Child and Adolescent Conduct Problems: A 10-Year Update. Clinical Child and Family Psychology Review 14(4):377–398 [DOI] [PubMed] [Google Scholar]
- Reef J, Diamantopoulou S, van Meurs I, Verhulst FC, van der Ende J (2011) Developmental trajectories of child to adolescent externalizing behavior and adult DSM-IV disorder: results of a 24-year longitudinal study. Social Psychiatry and Psychiatric Epidemiology 46(12):1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li T-K, Conneally PM, Nurnberger JI Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H (1998) Genome-wide search for genes affecting the risk for alcohol dependence. American Journal of Medical Genetics 81(3):207–215 [PubMed] [Google Scholar]
- Rutter M, Silberg J (2002) GENE-ENVIRONMENT INTERPLAY IN RELATION TO EMOTIONAL AND BEHAVIORAL DISTURBANCE. Annual Review of Psychology 53(1):463. [DOI] [PubMed] [Google Scholar]
- Samek DR, Hicks BM, Keyes MA, Bailey J, McGue M, Iacono WG (2014) Gene–environment interplay between parent–child relationship problems and externalizing disorders in adolescence and young adulthood. Psychological Medicine 45(2):333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage JE, Salvatore JE, Aliev F, Edwards AC, Hickman M, Kendler KS, Macleod J, Latvala A, Loukola A, Kaprio J, Rose RJ, Chan G, Hesselbrock V, Webb BT, Adkins A, Bigdeli TB, Riley BP, Dick DM (2018) Polygenic Risk Score Prediction of Alcohol Dependence Symptoms Across Population-Based and Clinically Ascertained Samples. Alcoholism, clinical and experimental research 42(3):520–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarr S, McCartney K (1983) How People Make Their Own Environments: A Theory of Genotype → Environment Effects. Child Development 54(2):424–435 [DOI] [PubMed] [Google Scholar]
- Stattin H, Kerr M (2000) Parental Monitoring: A Reinterpretation. Child development 71(4):1072–1085 [DOI] [PubMed] [Google Scholar]
- Tarantino N, Tully EC, Garcia SE, South S, Iacono WG, McGue M (2014) Genetic and environmental influences on affiliation with deviant peers during adolescence and early adulthood. Developmental Psychology 50(3):663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TenEyck M, Barnes JC (2015) Examining the Impact of Peer Group Selection on Self-Reported Delinquency: A Consideration of Active Gene–Environment Correlation. Criminal Justice and Behavior 42(7):741–762 [Google Scholar]
- Trudeau L, Mason WA, Randall GK, Spoth R, Ralston E (2012) Effects of Parenting and Deviant Peers on Early to Mid-Adolescent Conduct Problems. Journal of Abnormal Child Psychology 40(8):1249–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker-Drob EM, Briley DA, Harden KP (2013) Genetic and Environmental Influences on Cognition Across Development and Context. Current Directions in Psychological Science 22(5):349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JJM, Muroff JR (2002) Preventing substance abuse among African American children and youth: Race differences in risk factor exposure and vulnerability. The Journal of primary prevention 22(3):235–261 [Google Scholar]
- Wertz J, Belsky J, Moffitt TE, Belsky DW, Harrington H, Avinun R, Poulton R, Ramrakha S, Caspi A (2019) Genetics of nurture: A test of the hypothesis that parents’ genetics predict their observed caregiving. Developmental Psychology 55(7):1461–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz J, Nottingham K, Agnew-Blais J, Matthews T, Pariante CM, Moffitt TE, Arseneault L (2016) Parental monitoring and knowledge: Testing bidirectional associations with youths’ antisocial behavior. Development and Psychopathology 28(3):623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Canela-Xandri O, Rawlik K, Tenesa A (2020) Evidence of horizontal indirect genetic effects in humans. Nature Human Behaviour [DOI] [PubMed] [Google Scholar]
- Yap MBH, Cheong TWK, Zaravinos-Tsakos F, Lubman DI, Jorm AF (2017) Modifiable parenting factors associated with adolescent alcohol misuse: a systematic review and meta-analysis of longitudinal studies. Addiction 112(7):1142–1162 [DOI] [PubMed] [Google Scholar]


