Abstract
Virus-like particles (VLPs) have the potential to be used as display platforms to develop vaccines against infectious and non-infectious agents. However, most VLPs used as vaccine display platforms are derived from viruses that infect humans; unfortunately, most humans already have pre-existing antibodies against these platforms and thus, the immunogenicity of these vaccines may be compromised. VLP platforms derived from viruses that infect bacteria (bacteriophages), especially bacteriophages that infect bacteria, which do not colonize humans are less likely to have pre-existing antibodies against the platforms in the human population. In this study, we assessed whether two putative coat proteins (ORF13 and OFR14) derived from a thermophilic bacteriophage (ΦIN93) can be expressed and purified from a mesophilic bacterium such as E. coli. We also assessed whether expressed coat proteins can assemble to form VLPs. Truncated versions of ORF13 and OFR14 were successfully co-expressed in bacteria; the co-expressed truncated proteins formed oval structures that look like VLPs, but their sizes were less than those of an authentic ΦIN93 virus.
Keywords: thermophilic bacteriophage, coat proteins, expression, assembly, virus-like particles
Graphical Abstract
1. Introduction
The genomes of viruses code for structural and non-structural proteins. Non-structural proteins are enzymes required for viral replication while structural proteins are protein components (e.g. capsid or coat protein, envelope protein, membrane protein), which assemble around the viral genome to form a virus particle (virion) [1]. Some of these structural proteins, by themselves or together, can also assemble naturally during the life cycle of a virion, without the viral genome, to form structures known as virus-like particles (VLPs) (reviewed in [2]). VLPs are therefore empty viral shells without the viral genome; thus, they cannot replicate. VLPs can also be generated in the lab by cloning and expression of the coat protein(s) or envelope proteins in a suitable host cell. Overexpression of coat proteins or envelope proteins in a host cell allows the proteins to spontaneously self-assemble into VLPs [3, 4].
VLPs can be derived from animal viruses, plant viruses, human viruses, and even from bacterial viruses (bacteriophages). VLPs have so many applications in science and in medicine; they have been used in pre-clinical studies to deliver drugs/cargo to specific cancer cells, to develop armored RNA (positive controls for diagnostic tests for infectious diseases), and for in vivo imaging (VLPs loaded with fluorophores) [5–7]. VLPs also have so many features that make them suitable for vaccine design. For example, they resemble viruses in terms of size, coat proteins on VLPs are arrangement in a multivalent manner, and VLPs are very immunogenic [8]; thus, VLPs have gained a lot of attention within the last two decades as an excellent approach to develop vaccines [9, 10]. Five VLP-based vaccines, derived from human papillomaviruses (HPVs) and hepatitis B virus (HBV), have been approved by the Food and Drug Administration (FDA) to protect against HPVs [11, 12] and HBV [2] infections, respectively; other VLP-based vaccines against viruses are under development or in clinical trials [13, 14].
VLPs cannot only be used to develop vaccines against the viruses from which the structural proteins were derived from but they can also be used as display platforms to develop chimeric VLP vaccines displaying heterologous antigens from other viruses [15–19], bacteria [20], parasites [21, 22] and even tumor-associated antigens [23]. The goal of chimeric VLPs is to elicit immune responses against the heterologous antigens displayed on the platform and not necessarily the platform. Thus, VLPs have been used to develop candidate vaccines against both microbial and non-microbial diseases [24] ranging from allergy, hypotension, cholesterol [25], to cancer [23].
Despite the success of using VLPs as platforms to develop candidate vaccines, there have been some challenges associated with current VLP platforms. For example, most VLP platforms used for heterologous peptide display (chimeric VLPs) including viral vector vaccine platforms, are derived from viruses that infect humans and/or humans had previously been immunized with VLPs or antigens derived from those viruses. Thus, pre-existing neutralizing antibodies against the VLP-display platforms already exist in the general population and hence, they may compromise the immunogenicity of the display platforms and ultimately the immunogenicity of heterologous antigens displayed on the platforms [26–28]. To address some of these challenges, we assessed whether coat proteins derived from a thermophilic bacteriophage, ΦIN93, can be used to develop new vaccine platforms; i.e., we assessed whether the coat proteins from ΦIN93 can assemble into VLPs.
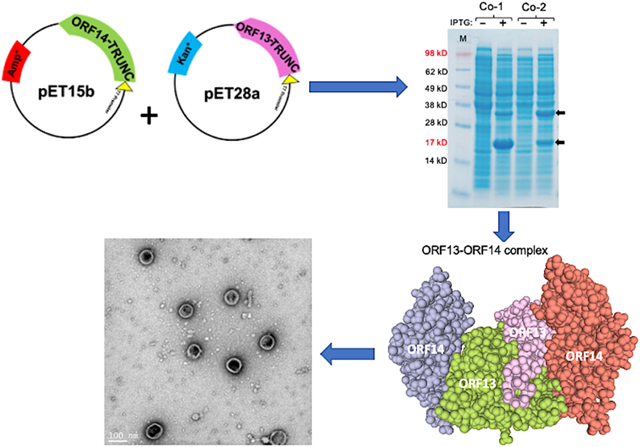
Bacteriophage ΦIN93 was isolated from a thermophilic bacterium, Thermus aquaticus, with an optimum growth temperature of 75°C [29]. The bacteriophage is a double-stranded circular DNA virus with a genome size of ~19.4kbp. Two proteins, open reading frame (ORF) 13 and ORF14, have been identified in the bacteriophage genome as putative coat proteins based on sequence homology with two major coat proteins (VP16 and VP17) from another thermophilic bacteriophage, P23–77 [30]. ORF13 and ORF14 is 80% and 73% identical (in amino acid) to VP16 and VP17 of P23–77, respectively (Figure 1). A recent study has shown that a mixture of VP16, VP17, and VP11 (a minor capsid protein) --which were expressed and purified separately -- can lead to the formation of complexes in vitro [31]; coat protein complexes are a prelude to viral assembly/formation of VLPs. However, no studies have ever assessed the expression of ORF13 and ORF14 from bacteriophage ΦIN93 or assessed the ability of these coat proteins to form complexes/assemble into VLPs. Here, we assessed for the first time, the expression of ORF13 and ORF14 in a mesophilic bacterial expression system, E. coli. We further assessed whether the coat proteins can assemble into VLPs.
Figure 1:

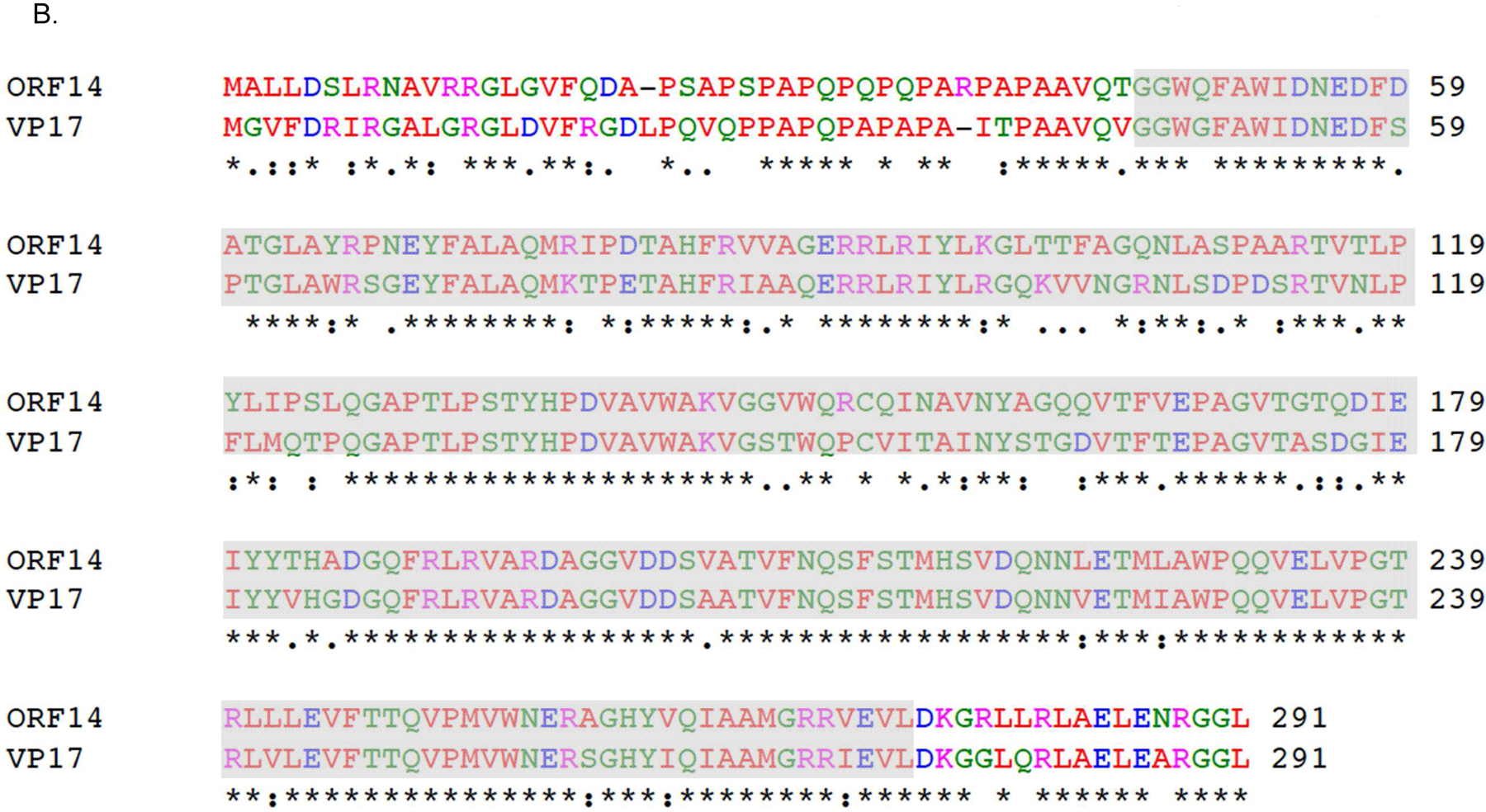
Sequence alignments of coat proteins of bacteriophages ΦIN93 and P23–77. Sequence alignment of: A) ORF13 of bacteriophage ΦIN93 & VP16 of bacteriophage P23–77 and B) ORF14 of bacteriophage ΦIN93 & VP17 of bacteriophage P23–77. Alignments were done using Clustal Omega. Asterisks (*) denote identical amino acids, colons (:) denote amino acids with similar properties and periods (.) denote amino acids with weak similar properties. Amino acids highlighted in gray-background were used in model structures in Figure 4.
2. Materials and Methods
2.1. Generation of expression vectors
The genes encoding putative coat proteins (ORF13 and ORF14) of bacteriophage ΦIN93 were codon-optimized for high-level protein expression in C41 E. coli bacteria and they were synthesized by Epoch Life Science. The complete sequence of each synthesized gene was separately amplified by polymerase chain reaction (PCR) and cloned into pET28a plasmid (Kanamycin resistance) and pET15b plasmid (Ampicillin resistance) using NcoI and BamHI restriction sites; this gave rise to four plasmids, namely: pET15b-ORF13, pET15b-ORF14, pET28a-ORF13, and pET28a-ORF14.
In addition to cloning the complete sequences of ORF13 and OFR14, we also generated truncated versions of each of the two genes. To generate truncated ORF13 and ORF14 genes, DNA sequences representing amino acids (aa) 21–165 and aa 46–271, respectively, were separately amplified by PCR and each truncated gene was cloned into pET28a and pET15b using the same restriction sites above; this gave rise to four additional plasmids, namely: pET15b-ORF13-trunc with aa 21–165, and pET15b-ORF14-trunc with aa 46–271, pET28a-ORF13-trunc with aa 21–165, and pET28a-ORF14-trunc with aa 46–271. In all cases, the genes in each of the constructs were sequenced across restriction sites to confirm the cloning.
2.2. Coat protein expression and purification
For coat protein expression, C41 E. coli cells were transformed separately with each plasmid and single colonies were screened for protein expression as follows. Bacterial cultures from colonies were grown in Luria-Bertani (LB) medium with appropriate antibiotics at 37°C until the cultures reached an optical density (OD)600 of 0.6. Coat protein expression was induced with 0.5mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) overnight at the same temperature. For co-expression of ORF13 and ORF14 in the same bacterial cells, equal concentrations of pET15b-ORF13 and pET28a-ORF14, pET15b-ORF14 and pET28a-ORF13, pET15b-ORF13-trunc and pET28a-ORF14-trunc or pET15b-ORF14-trunc and pET28a-ORF13-trunc were mixed and used to transform C41 cells. Transformed cells were grown in LB medium with 30 mg/ml of kanamycin plus 50mg/ml of ampicillin at 37°C until the culture reached an OD600 of 0.6. Coat protein expression was induced with 1mM IPTG under the same conditions and time as above. To screen for coat protein expression, bacterial cells were pelleted, resuspended in 8M urea, sonicated and resolved on SDS PAGE gels.
Cultures, which showed protein expression, were used to grow large volume cultures (100 ml or 250 ml) for large-scale protein expression and purification. The cultures were grown and induced as above. Cell pellets were then lysed using 0.2% lysozyme solution (with 100 mM NaCl, 10 mM EDTA, 50 mM Tris-HCl pH 8.5, plus 0.05% deoxycholate) and sonicated to help lyse the bacteria. Two ng/ul of DNase and 2 mM of MgCl2 were added to the lysates and the lysates were incubated for 1 hour at 37°C. Lysates were spun at 3,700 rpm (4°C) for 30 minutes and the supernatants collected and resuspended in 50% ammonium sulfate to precipitate coat proteins. The supernatants with precipitated coat proteins were spun at 10,000 rpm (4°C) for 10 minutes. The pellets were resuspended in sepharose column buffer (SCB: 10 mM Tris-HCl pH 7.4, 100 mM NaCl, 0.1 mM MgSO4), spun at the same time and conditions. Supernatants were then loaded on sepharose CL-4B column connected to SCB buffer and fractions were collected and analyzed for coat proteins. Fractionation tubes containing coat proteins were combined and the SCB was exchanged with PBS (1x phosphate buffered saline) using Millipore Amicon™ Ultra-4 Centrifugal Filter Units (100K MWCO). Coat proteins were evaluable for assembly into VLPs under a transmission electron microscope as described below.
2.3. Transmission electron microscopy (TEM)
To assess the potential of coat proteins to assemble into VLPs, TEM was conducted as follows. Purified coat proteins of bacteriophage ΦIN93 or the virus (a gift from Dr. Isao Matsushita) were loaded onto glow-discharged carbon grids for 2 minutes and the grids were stained with 2% uranyl acetate for 2 minutes. Samples were visualized using a FEI 200kV Titan Themis Scanning Transmission Electron Microscope (STEM) or a JEOL JEM-2010 TEM.
2.4. Generation of sera for immunoassays
To generate sera that can react with both ORF13 and ORF14, we expressed ORF13 and ORF14 in pET28a as a recombinant protein with three amino acids linker sequence, glycineserine-serine, in-between the two proteins. Expression and purification were done as described above. To generate sera, 3–4-week-old female Balb/c mice were immunized three times, intramuscularly, each with 10 μg of purified recombinant protein (ORF13+ORF14); all immunizations were done at two-week intervals in the presence of alum adjuvant. Two weeks after the last immunization, whole blood were collected and sera from the blood were used for Western blots (described below) against ORF13 and ORF14 coat proteins. Animal work was conducted in accordance with Michigan Tech Institutional Animal Care and Use Committee (IACUC) guidelines.
2.5. Western blots
One hundred ng of purified ΦIN93 coat proteins, bacteriophage ΦIN93, or control MS2 coat proteins were resolved on an SDS PAGE gel and transferred to polyvinylidene fluoride membranes. The membranes were then blocked overnight with 5% non-fat milk followed by the addition of 1:1,000 dilution of ORF13+ORF14 serum antibodies. The membranes were incubated for 2 hours at room temperature, washed, and 1:5,000 dilution of horseradish peroxide-conjugated goat anti-mouse IgG antibodies were added for 1 hour at the same temperature. The membranes were then washed, developed with enhanced chemiluminescent substrate, and scanned using Luminescent Image Analyzer (LAS-4000).
2.6. Immunolabeling with gold nanoparticles
Immuno-gold labelling was conducted following reported procedures [32]. In brief, purified coat protein samples were loaded onto discharged copper grids and incubated for 5 minutes. Samples were then fixed with 2.5% glutaraldehyde and washed twice with 1X PBS. The fixed samples were blocked with 2% BSA (bovine serum albumin) in PBS for 1 hour and ORF13+ORF14 recombinant sera were added (1:500 dilution) as primary antibody at room temperature for 1 hour. The grids were washed with PBS and gold-conjugated anti-mouse IgG secondary antibodies were added (1:30 dilution) for 1 hour. The grids were washed with deionized water. Finally, negative staining with uranyl acetate was conducted and the samples were observed under the TEM as above.
3. Results
3.1. ORF13 and ORF14 can be successfully expressed in E. coli
To assess whether ORF13 and ORF14 coat proteins from a thermophilic bacteriophage can be expressed in a mesophilic bacterium, E. coli, we first generated expression vectors, pET15b-ORF14 and pET28a-ORF13, each with complete sequences of ORF14 and ORF13, respectively (Figure 2A, top). As shown in Figure 2A (bottom), the coat proteins were successfully expressed in C41 E. coli cells. However, the expression level of ORF13 in pET28a seemed to be higher compared to that of ORF14 in pET15b. To assess if the difference in expression level was due to the expression vectors used or the genes cloned to the expression vectors, we swapped the genes in the expression vectors by cloning ORF13 gene into pET15b and ORF14 gene into pET28a (Figure 2B). The expression level of ORF14 from pET28a vector was still low compared to that of ORF13 from pET15b vector (Figure 2B, bottom), suggesting that the genes and not the expression vectors may be associated with the differences in expression levels.
Figure 2:
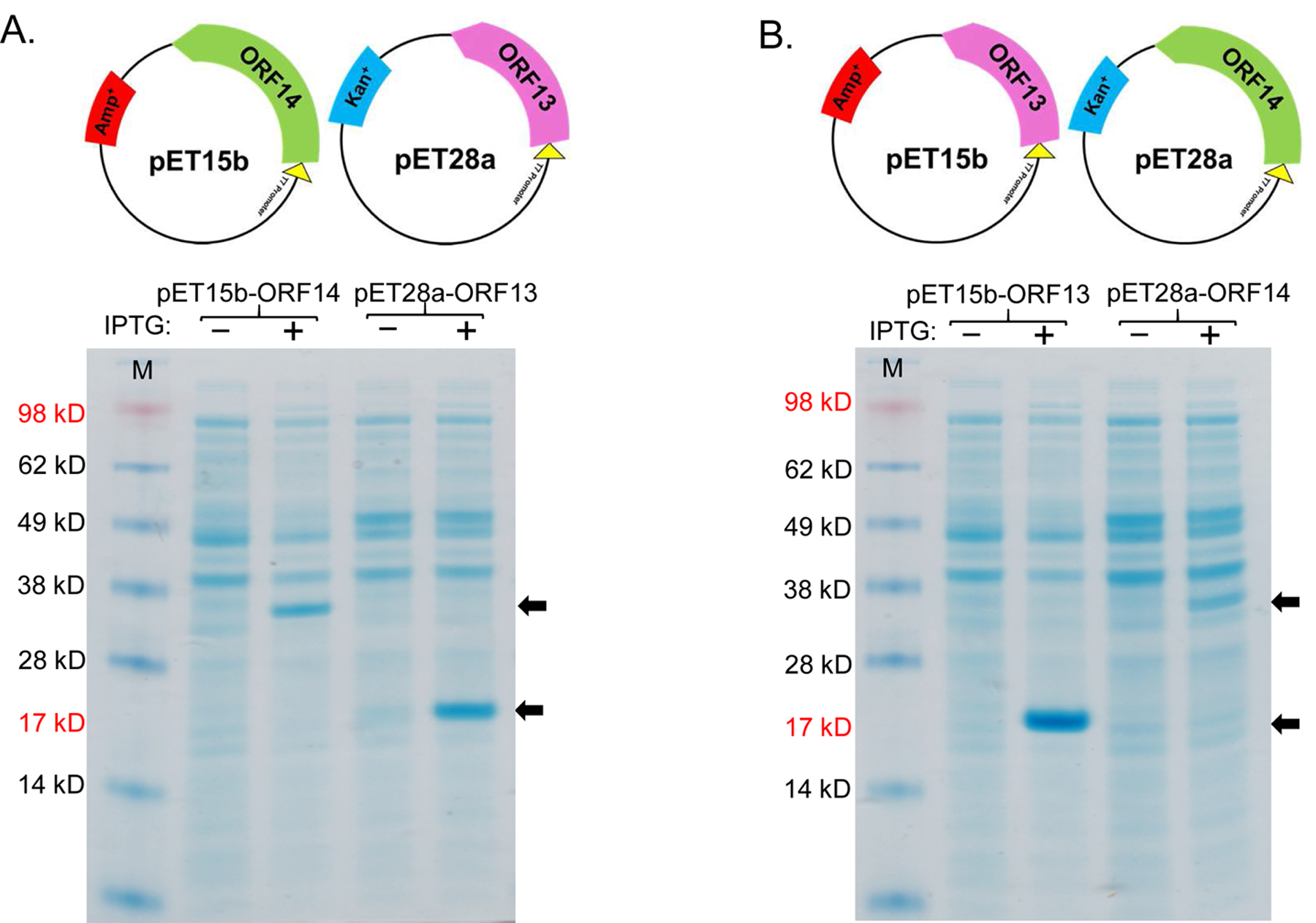
Expression of coat proteins, ORF13 and ORF14, in pET15b and pET28a vectors. A) Plasmids pET15b-ORF14 or pET28a-ORF13 and B) plasmids pET15b-ORF13 or pET28a-ORF14 were used to transformed C41 E. coli cells. The bacteria were grown at 37 °C and protein express was induced with 0.5 mM IPTG. Cell pellets were lysed with 8 M Urea and loaded on SDS-PAGE gel for analysis. Gels were stained with Coomassie blue. Arrows indicate coat protein bands of ORF13 (18.81 KD) and ORF14 (32.01 KD). (−) is uninduced culture and (+) is induced IPTG culture. M=Molecular weight marker.
To assess whether ORF13 and ORF14 coat proteins can be expressed in the same bacterium, on the assumption that the two are required for assembly into VLPs, we co-transformed C41 cells with two different combinations of expression vectors, pET15b-ORF14 and pET28a-ORF13 or pET15b-ORF13 and pET28a-ORF14. Both proteins were successfully co-expressed in bacteria regardless of the combinations used (Figure 3A). In pET15b-ORF14 and pET28a-ORF13 combination, ORF14 was expressed at low levels compared to ORF13; the results were similar to those obtained following expression of the proteins, individually (Figure 2A). In pET15b-ORF13 and pET28a-ORF14 combination, both proteins seemed to be expressed at similar levels (Figure 3A).
Figure 3:
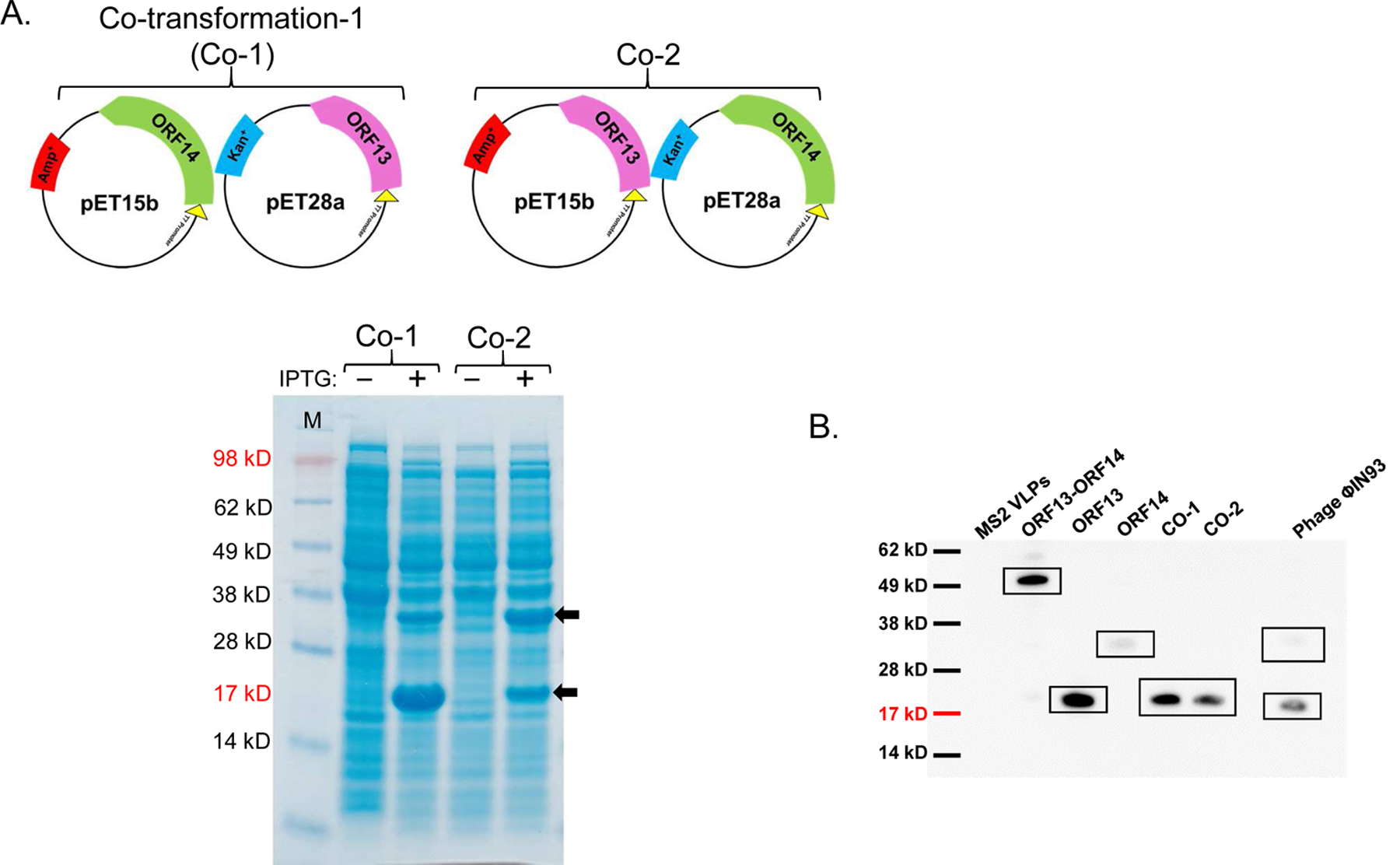
Co-expression of ORF13 and ORF14 proteins in C41 cells. A) Equal concentrations of pET15b-ORF14 and pET28a-ORF13 (Co-1) or pET15b-ORF13 and pET28a-ORF14 (Co-2) were mixed together and the mixtures were then used to separately transform C41 E. coli. The bacteria were grown at 37 °C and co-protein expression was induced with 1 mM IPTG. Cell pellets were lysed with 8 M Urea and loaded on SDS-PAGE gel for analysis. Gels were stained with Coomassie blue. Arrows indicate coat protein bands of ORF13 (18.81 KD) and ORF14 (32.01 KD). (−) is uninduced culture and (+) is induced IPTG culture. M=Molecular weight marker. B) Western Blots: purified ORF13, ORF14 and co-expressed proteins (ORF13 and ORF14) were prepared for SDS-PAGE gel and detected by serum raised from mice immunized with ORF13+ORF14 recombinant protein. Bacteriophage MS2 VLPs were used as a negative control while purified ORF13+ORF14 recombinant protein and ΦIN93 virus were set as positive controls. Expected protein bands are highlighted in black squares.
ORF13 and ORF14 (single- or co-expressed) coat proteins were purified by gel filtration and the coat proteins were confirmed by western blot using recombinant ORF13+ORF14 antibodies. As shown in Figure 3B, single-expressed ORF13 reacted strongly with the antibodies while single-expressed ORF14 reacted weakly. Similarly, when the two coat proteins were co-expressed, only ORF13 coat protein and not ORF14 was visible, suggesting that the ORF14 may be expressed at low levels (similar to our Coomassie blue data). The result was similar when authentic ΦIN93 virus was used as a positive control to probe for ORF14; ORF14 reacted weakly while ORF13 reacted very strongly. To assess whether the expressed coat proteins can assemble into virus-like particles, TEM analyses were conducted using purified coat proteins. Unfortunately, none of the coat proteins, single- or co-expressed (pET28a-ORF13 and pET15b-ORF14 or pET28a-ORF14 and pET15b-ORF13), assembled into VLPs (data not shown).
3.2. Truncated ORF13 and ORF14 coat proteins form structures that resemble VLPs
As mentioned above, ORF13 and ORF14 are 80% and 73% identical (in terms of amino acids) to VP16 and VP17 of bacteriophage P23–77, respectively. The 3D structure of VP16–VP17 complex has previously been predicted [33]; VP16 forms a homodimer in the complex and VP17 coat protein interacts with at least one monomer of VP16 in the complex. Given this information, we decided to predict the 3D structure of ORF13–ORF14 complex based on the structure of VP16–VP17 complex, using SWISS-MODEL and Cn3D [34, 35]. As shown in Figure 4, the 3D structure of ORF13–ORF14 complex is similar to that of VP16–VP17; nevertheless, the software predicted the structure using only amino acids number 21–165 from ORF13 and amino acids 46–271 from ORF14 (the same amino acids range in VP16–VP17 complex; Figure 1). Because the structure was predicted using only amino acids 21–165 and 46–271 but not the complete sequences, we generated truncated versions of ORF13 (aa 21–165) and ORF14 (aa 46–271) and assessed the potential of the coat proteins to assemble into VLPs. ORF13-trunc and ORF14-trunc were successfully expressed, as single proteins or co-expressed proteins, regardless of whether pET15b or pET28a was used as the expression vectors (Figures 5A and B). Co-expressed ORF13-trunc and ORF13+ORF14 were successfully purified from bacterial lysates (Figure 5C). Only purified ORF13-trunc coat protein (expressed individually or co-expressed) but not ORF14-trunc (expressed individually or co-expressed) reacted with recombinant ORF13+ORF14 antibodies (Figure 5D); there was only a very faint band of ORF14-trunc in co-expression of pET15b-ORF13-trunc and pET28a-ORF14-trunc. these results are consistent with those of coat proteins expressed with full-length proteins above (Figure 3B).
Figure 4:
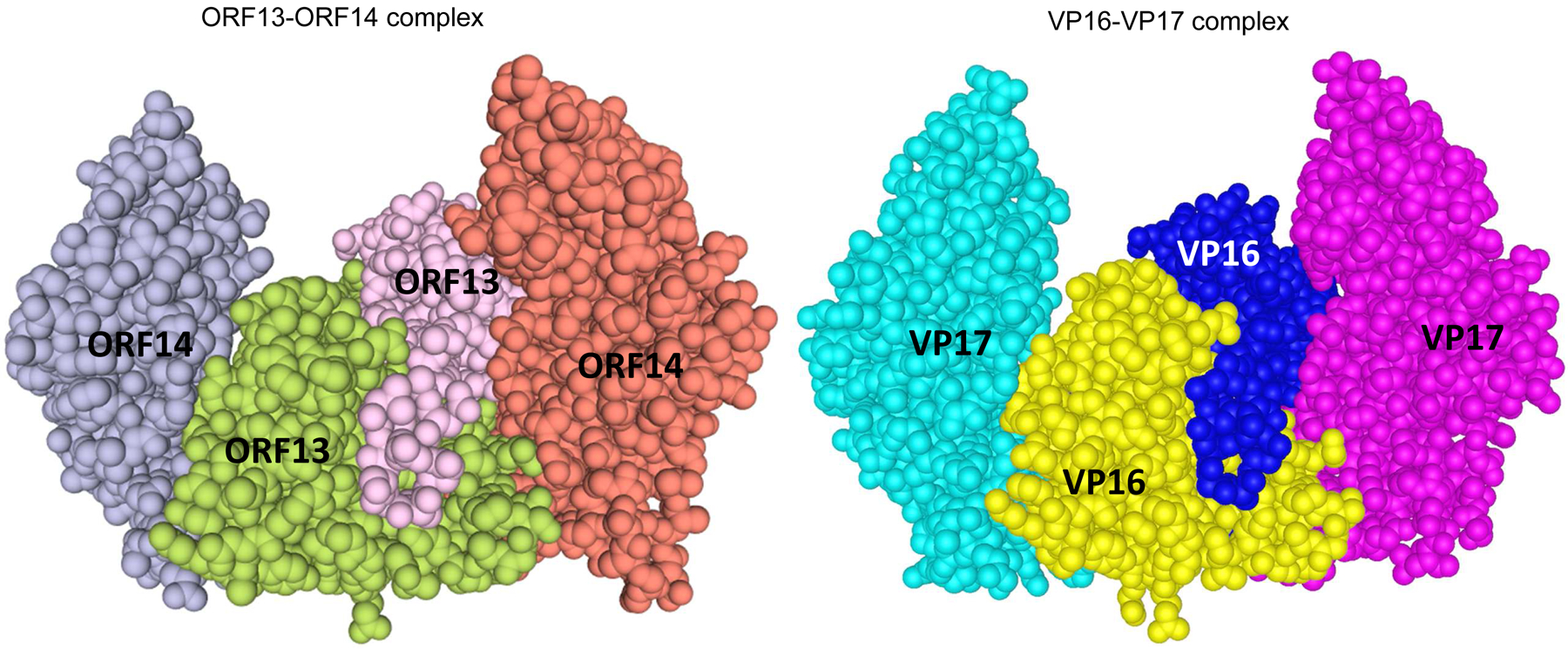
Three dimensional (3D) structures of coat proteins of bacteriophages ΦIN93 and P23–77. The 3D structure of ORF13-ORF14 complex (left) from bacteriophage ΦIN93 was modeled, based on sequences and the 3D structure of VP16–VP17 complex (right; Cn3D view) from bacteriophage P23–77 as template, using Swiss-Model software. Homodimers of ORF13 (middle) and VP16 (middle) interact with each of the two copies of ORF14 and VP17, respectively. Both structures were modeled using amino acids 21–165 from ORF13 or VP16 and amino acids 46–271 from ORF14 or VP17 (highlighted in gray-background in Figure 1).
Figure 5:
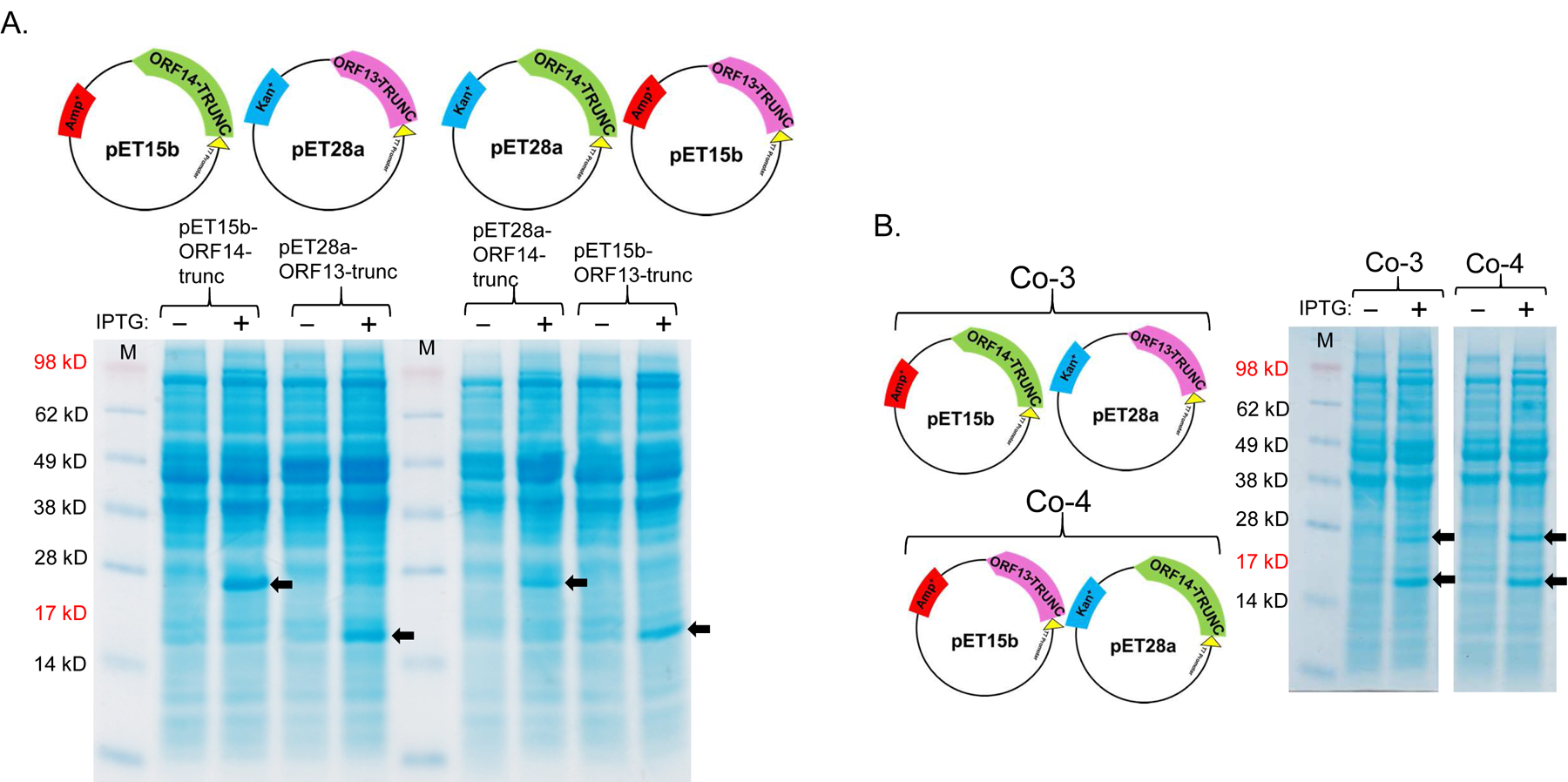
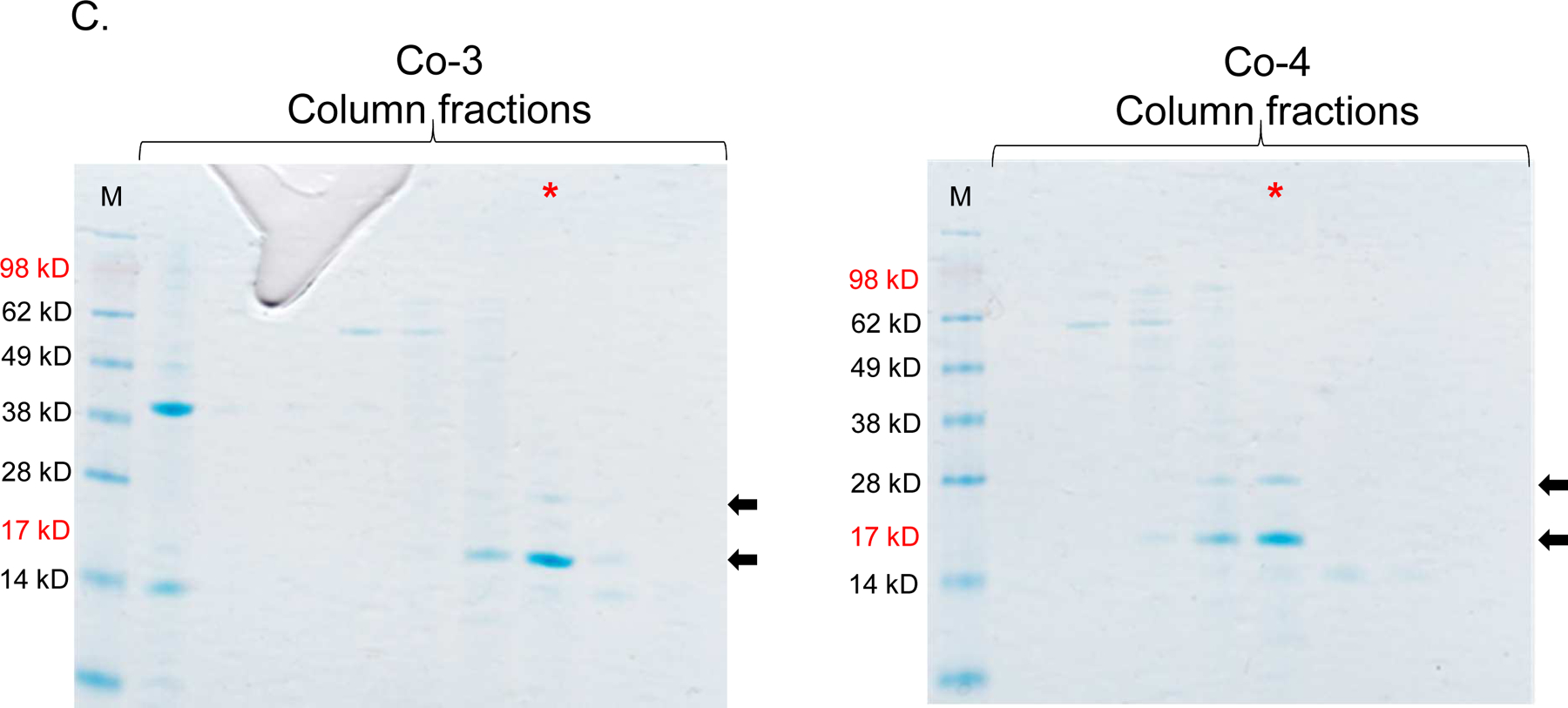
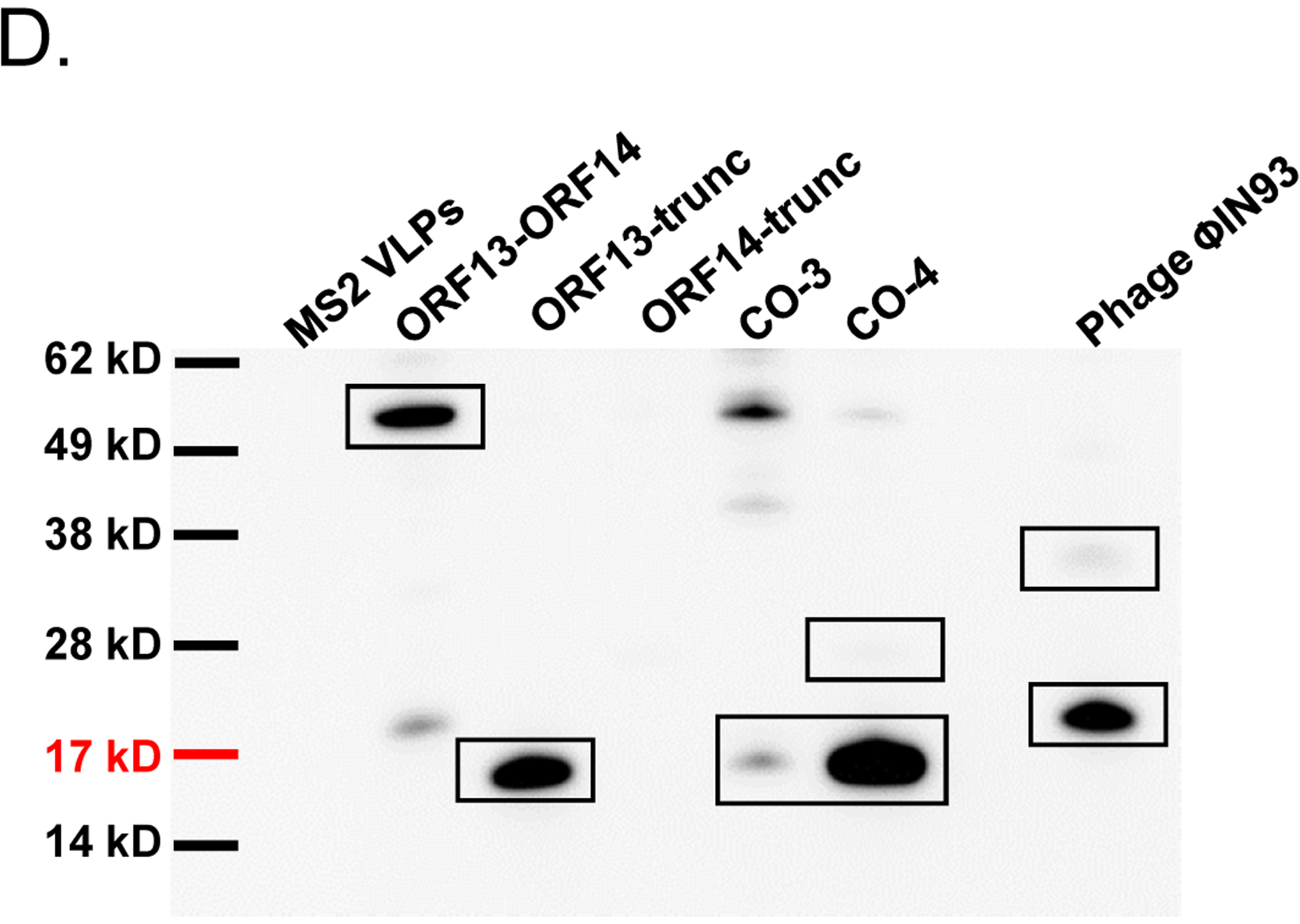
Single-expression and co-expression of truncated ORF13 (ORF13-trunc) and truncated ORF14 (ORF14-trunc) proteins in C41 cells. A) Plasmids pET15b-ORF14-trunc or pET28a-ORF13-trunc were used to separately transform C41 E. coli cells and the bacteria were grown at 37 °C. Protein expression was induced with 0.5 mM IPTG. B) Equal concentrations of plasmids pET15b-ORF14-trunc and pET28a-ORF13-trunc (Co-3) or pET15b-ORF13-trunc and pET28a-ORF14-trunc (Co-4) were mixed and used to transform C41 E. coli cells. The bacteria were grown at 37 °C and co-protein expression was induced with 1 mM IPTG. Cell pellets were lysed with 8 M Urea and loaded on SDS-PAGE gel for analysis. Gels were stained with Coomassie blue. Arrows indicate protein bands, ORF13-trunc (16.06 kD) and ORF14-trunc (24.97 kD). (−) is uninduced culture and (+) is induced IPTG culture. M=Molecular weight marker. C) Portions of column fractions of Co-3 and Co-4 were loaded to SDS-PAGE gels followed by Coomassie blue staining. Arrows indicate protein bands (ORF13-trunc and ORF14-trunc) of interest. Fractions with high purity (for example, those highlighted in red asterisk) were buffer exchanged and used for western blotting & TEM analysis. D) Western blots: ORF13-trunc, ORF14-trunc and co-expressed proteins (ORF13-trunc and ORF14-trunc) were prepared for SDS-PAGE gel and detected by serum raised from mice mentioned above. The phage MS2 VLPs were used as a negative control while the purified ORF13–ORF14 recombinant proteins and the phage ΦIN93 were used as the positive controls. Expected protein bands are highlighted in black squares.
To assess whether the expressed coat proteins can assemble into VLPs, TEM analyses were conducted using purified coat proteins. As shown in Figure 6A, single-expressed ORF13-trunc or ORF14-trunc gave rise to very few structures that resemble VLPs, but their sizes were very small compared to control ΦIN93 virus, which is ~130 nm (Figure 6B). The co-expressed version of the truncated coat proteins also seemed to form structures that resemble VLPs (Figure 6C); their sizes (~75 nm to ~100 nm) were closer to those of authentic ΦIN93 virus.
Figure 6:
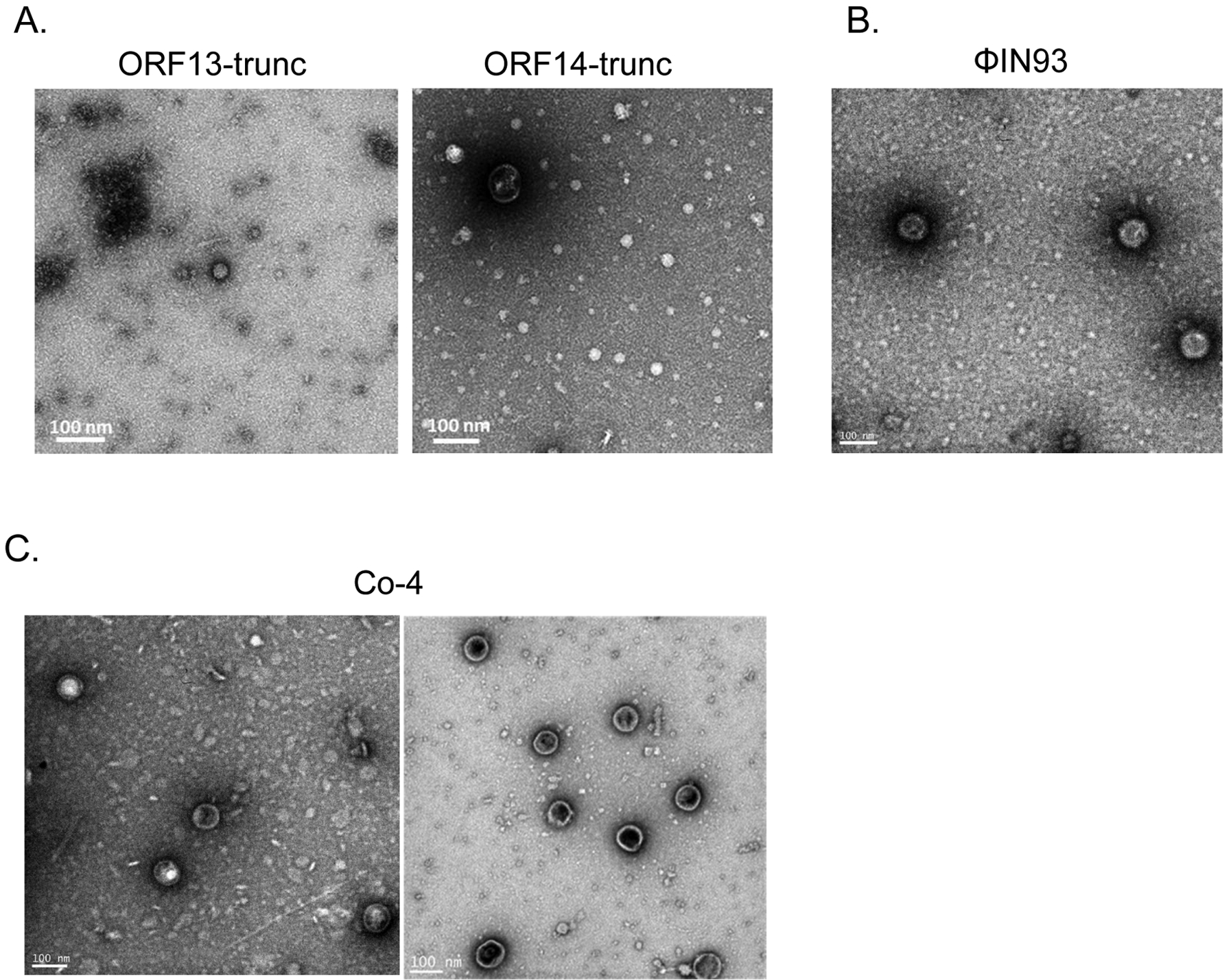
Assessing the assembly of coat proteins of single-expressed and co-expressed ORF13-trunc and ORF14-trunc. A) TEM of purified ORF13-trunc and purified ORF14-trunc. B) TEM of bacteriophage ΦIN93. C) TEM of purified co-expressed truncated coat proteins (Co-4; ORF13-trunc and ORF14-trunc expressed from pET15b and pET28a, respectively). TEM was conducted at 25,000 X magnification.
To determine if the structures in Figure 6 contain ORF13-truc and ORF14-truc coat proteins (in other words, if they were virus-like particles), we did immunolabeling with gold nanoparticles followed by TEM. Most of the gold nanoparticles bound to ΦIN93 virus while only a few of the nanoparticles bound to samples from the co-expressed ORF13-truc and ORF14-truc coat proteins (Figure 7).
Figure 7:
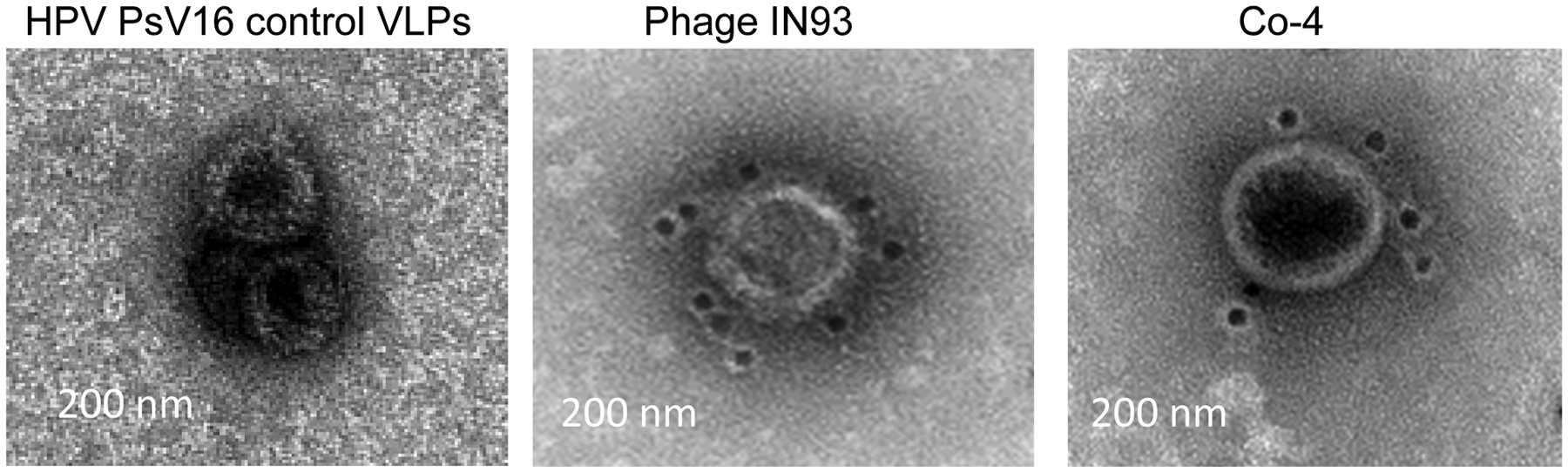
Transmission electron microscopy images of gold-immunolabeled ΦIN93 phage and co-expressed ORF13-trunc and ORF14-trunc. Purified co-expressed ORF13-trunc and ORF14-trunc samples were stained with sera against ORF13-ORF14, and gold-conjugated secondary antibodies were used followed by negative staining for TEM. Phage ΦIN93 was used as a positive control while HPV pseudovirus VLPs were used as a negative control. TEM was conducted at 25,000 X magnification.
4. Discussion
Two putative coat proteins (ORF13 and ORF14) from thermophilic bacteriophage ΦIN93 are 80% and 73% identical to two major coat proteins (VP16 and VP17) from thermophilic bacteriophage P23–77, respectively [30]. While VP16 and VP17 coat proteins have been expressed in previous studies [31, 33], the expressions of ORF13 and ORF14 have never been explored. Here, we cloned and assessed the expression of ORF13 and ORF14 individually and in combination (co-expression) in mesophilic bacteria (E. coli) with the long term-goal of assessing whether the coat proteins can assemble into VLPs. ORF13 and ORF14 coat proteins were successfully expressed, as separate proteins, in C41 E. coli cells. Moreover, the coat proteins could also be co-expressed in the same bacteria by using two plasmids that are incompatible (pET28a and pET15b; have the same origin of replication, pBR322) by simply applying two different selective pressures (two antibiotics resistance; kanamycin and ampicillin) on the bacterial cells for 24 hours. The co-expression of ORF13 and ORF14 using two incompatible plasmids are consistent with a previous study, which showed that two proteins can be co-expressed from incompatible plasmids by using vectors with different antibiotic resistant genes [36]. While ORF13 and ORF14 were successfully expressed (individually or together), the expression of ORF14 in all cases was less compared to that of ORF13, regardless of whether the protein was expressed using pET28a plasmid or pET15b plasmid. In the co-expression study, especially Co-1 with pET28a expressing ORF13 and pET15b expressing ORF14 (Figure 3A, left), we initially thought that the bacterial cells expressing the proteins might have only pET28a-ORF13 plasmid (i.e., pET15b-ORF14 plasmid might have been lost from the bacteria due to plasmid incompatibility). To assess if both expression plasmids were maintained in the bacteria during expression, bacterial cultures expressing the coat proteins were induced and aliquots (equal volumes) of the cultures were used as templates for PCR. As shown in Figure 8A, the bands suggested that both plasmid combinations (pET15b-ORF14 and pET28a-ORF13 or pET15b-ORF13 and pET28a-ORF14) were maintained in the bacterial cells and consequently the proteins were being expressed. Given the low levels of ORF14 expression, we speculate that the protein might be expressed, normally, at low levels in the virus. This view is supported by our Western blot data using ΦIN93 virus. In our Western blot data (Figures 3B and 5D), ORF14 band in the lane with positive control ΦIN93 virus was very faint compared to ORF13 band. The view (low expression) is further supported by data from a related coat protein, VP17 from P23–77, which is 73% identical to ORF14; in a Master’s Thesis, University of Jyväskylä (Finland), it was observed that VP17 was expressed at low levels compared to VP16 following co-expression of VP17 and VP16 using a bicistronic VP16–VP17 transcription unit [37]. Taken together, our data suggest that ORF14 is expressed at low levels compared to ORF13. Alternatively, the protein could be expressed at normal levels; if that was the case, then the faint bands of ORF14 observed in Western blots could be due to the fact that anti-ORF-14 antibodies in sera from immunized mice had low reactivity with the antigen. If the former (low expression) is correct, that would suggest that less proportion of ORF14 may be required for viral assembly compared to ORF13; this has been suggested for the related protein (VP17) from bacteriophage P23–77. It has been suggested that 540 copies of VP17 versus 1080 copies of VP16 are required for bacteriophage P23–77 to assemble into a virus particle [31, 33].
Figure 8:
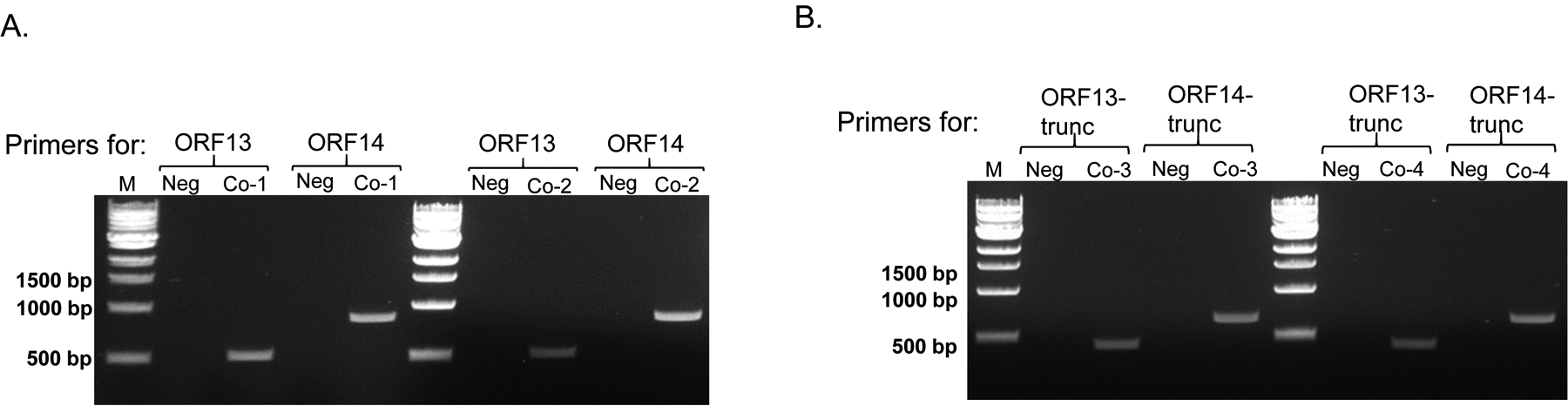
PCR amplifications of co-expressed ORF13 & ORF14 (complete gene sequences and truncated versions). A) Induced cultures from Figure 3A (Co-1 and Co-2 with complete gene sequences) were use as templates for PCR. The presence of ORF13 or ORF14 was tested using primers for each gene. Sizes of ORF13 and ORF14 are ~579 base pairs and ~876 base pairs, respectively. B) Induced cultures from Figure 5B (Co-3 and Co-4 with truncated versions of genes) were used as templates for PCR. The presence of ORF13 or ORF14 was tested using primers for each gene. Neg: negative control untransformed E. coli culture. Sizes of ORF13-trunc and ORF14-trunc are ~441 base pairs and ~684 base pairs, respectively. M=Molecular weight marker.
Despite the low levels of ORF14 expression, we assessed whether ORF13 and ORF14 could assemble to VLPs; unfortunately, we did not observe any structures that look like VLPs. At the moment, it is unclear whether additional protein(s) is required to form complexes/VLPs as have been identified with bacteriophage P23–77. A mixture of VP16 and VP17 with the minor capsid protein (VP11) of P23–77 has been shown to form complexes in vitro (which is a prelude to the formation of VLPs). However, a sequence in bacteriophage ΦIN93 that is homologous to VP11 (minor capsid protein of P23–77) has not be identified, suggesting that the capsid protein of ΦIN93 may be composed of only two coat proteins, ORF13 and ORF14. This may not be surprising given the fact that in some viruses (e.g., adeno-associated virus type 2), only two coat proteins out of 3 or more coat proteins are sufficient to form VLPs [4, 38].
As an alternative to co-expressing full length ORF13 and ORF14 coat proteins to evaluate assemble into VLPs, we generated truncated forms of the proteins. The truncated proteins were based on amino acid sequences that were used by Swiss-Model software to predict ORF13-ORF14 complex structure (Figures 1 and 4). The truncated versions of the proteins (individually or together) were expressed; PCR also confirmed that the expression plasmids for truncated ORF13 and ORF14 were both maintained in the same bacterial cells (Figure 8B). At the moment, it is still not clear whether the oval structures observed with co-expressed ORF13-trunc and ORF14-trunc are VLPs or not; their sizes (~75 nm to ~100 nm) were less than those previously published for authentic ΦIN93 virus (130nm). Thus, additional studies are needed to confirm this. VLPs derived from thermophilic bacteriophage ΦIN93 have great potentials for vaccine development.
Research Highlights.
Expression of coat proteins (ORF13 & OFR14) from a thermophilic bacteriophage, ΦIN93
ORF13 and OFR14 were successfully expressed separately in a mesophilic bacterium
ORF13 and OFR14 were co-expressed using two incompatible plasmids
Co-expressing ORF13-trunc & ORF14-trunc gave rise to structures that resemble VLPs
Acknowledgement
This work was supported by grant number 1R15AI146982-01A1 from the US National Institutes of Health (National Institute of Allergy and Infectious Diseases). The content is solely the responsibility of the authors and does not necessarily represent the views of the National Institutes of Health. We would like to thank Dr. Isao Matsushita (Osaka Gas Co., Ltd., Energy Technology Laboratories, Osaka, Japan) for providing us with bacteriophage ΦIN93 virus. We would also like to thank Owen Mills and Dr. Pinaki Mukherjee at Applied Chemical and Morphological Analysis Laboratory at Michigan Tech for help with TEM imaging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors do not have any conflict to declare
References
- 1.Howley M and Knipe M, Fields virology. 6 ed. Vol. 1. 2013: Philadelphia: : Wolters Kluwer Health/Lippincott Williams & Wilkins. 2664. [Google Scholar]
- 2.Gerlich WH, Medical virology of hepatitis B: how it began and where we are now. Virol J, 2013. 10: p. 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeltins A, Construction and characterization of virus-like particles: a review. Mol Biotechnol, 2013. 53(1): p. 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palucha A, et al. , Virus-like particles: models for assembly studies and foreign epitope carriers. Prog Nucleic Acid Res Mol Biol, 2005. 80: p. 135–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pumpens P, et al. , The True Story and Advantages of RNA Phage Capsids as Nanotools. Intervirology, 2016. 59(2): p. 74–110. [DOI] [PubMed] [Google Scholar]
- 6.Shirbaghaee Z and Bolhassani A, Different applications of virus-like particles in biology and medicine: Vaccination and delivery systems. Biopolymers, 2016. 105(3): p. 113–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan D, et al. , The application of virus-like particles as vaccines and biological vehicles. Appl Microbiol Biotechnol, 2015. 99(24): p. 10415–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yadav R, Zhai L, and Tumban E, Virus-like Particle-Based L2 Vaccines against HPVs: Where Are We Today? Viruses, 2019. 12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chroboczek J, Szurgot I, and Szolajska E, Virus-like particles as vaccine. Acta Biochim Pol, 2014. 61(3): p. 531–9. [PubMed] [Google Scholar]
- 10.Tao P, et al. , Bacteriophage T4 nanoparticles for vaccine delivery against infectious diseases. Adv Drug Deliv Rev, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyler M, Tumban E, and Chackerian B, Second-generation prophylactic HPV vaccines: successes and challenges. Expert Rev Vaccines, 2014. 13(2): p. 247–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhai L and Tumban E, Gardasil-9: A global survey of projected efficacy. Antiviral Res, 2016. 130: p. 101–9. [DOI] [PubMed] [Google Scholar]
- 13.ClinicalTrials.gov. ClinicalTrials.gov- virus-like particles. [cited2018 June 11]; Available from: https://www.clinicaltrials.gov/ct2/results?cond=virus-like+particles&term=&cntry=&state=&city=&dist=.
- 14.Zhao C, Ao Z, and Yao X, Current Advances in Virus-Like Particles as a Vaccination Approach against HIV Infection. Vaccines (Basel), 2016. 4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei S, et al. , Neutralization effects of antibody elicited by chimeric HBV S antigen viral-like particles presenting HCV neutralization epitopes. Vaccine, 2018. 36(17): p. 2273–2281. [DOI] [PubMed] [Google Scholar]
- 16.Basu R, et al. , Immunization with phage virus-like particles displaying Zika virus potential B-cell epitopes neutralizes Zika virus infection of monkey kidney cells. Vaccine, 2018. 36(10): p. 1256–1264. [DOI] [PubMed] [Google Scholar]
- 17.Zhai L, et al. , A novel candidate HPV vaccine: MS2 phage VLP displaying a tandem HPV L2 peptide offers similar protection in mice to Gardasil-9. Antiviral Res, 2017. 147: p. 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berkower I, et al. , Assembly, structure, and antigenic properties of virus-like particles rich in HIV-1 envelope gp120. Virology, 2004. 321(1): p. 75–86. [DOI] [PubMed] [Google Scholar]
- 19.Ramasamy V, et al. , A tetravalent virus-like particle vaccine designed to display domain III of dengue envelope proteins induces multi-serotype neutralizing antibodies in mice and macaques which confer protection against antibody dependent enhancement in AG129 mice. PLoS Negl Trop Dis, 2018. 12(1): p. e0006191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daly SM, et al. , VLP-based vaccine induces immune control of Staphylococcus aureus virulence regulation. Sci Rep, 2017. 7(1): p. 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins KA, et al. , Enhancing protective immunity to malaria with a highly immunogenic virus-like particle vaccine. Sci Rep, 2017. 7: p. 46621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crossey E, et al. , Identification of an Immunogenic Mimic of a Conserved Epitope on the Plasmodium falciparum Blood Stage Antigen AMA1 Using Virus-Like Particle (VLP) Peptide Display. PLoS One, 2015. 10(7): p. e0132560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong HK, Tan WS, and Ho KL, Virus like particles as a platform for cancer vaccine development. PeerJ, 2017. 5: p. e4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chackerian B and Frietze KM, Moving towards a new class of vaccines for non-infectious chronic diseases. Expert Rev Vaccines, 2016. 15(5): p. 561–3. [DOI] [PubMed] [Google Scholar]
- 25.Crossey E, et al. , A cholesterol-lowering VLP vaccine that targets PCSK9. Vaccine, 2015. 33(43): p. 5747–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knuchel MC, et al. , Relevance of a pre-existing measles immunity prior immunization with a recombinant measles virus vector. Hum Vaccin Immunother, 2013. 9(3): p. 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zak DE, et al. , Merck Ad5/HIV induces broad innate immune activation that predicts CD8(+) T-cell responses but is attenuated by preexisting Ad5 immunity. Proc Natl Acad Sci U S A, 2012. 109(50): p. E3503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tumban E, Lead SARS-CoV-2 Candidate Vaccines: Expectations from Phase III Trials and Recommendations Post-Vaccine Approval. Viruses, 2020. 13(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsushita I and Yamashita N, Isolation and Characterization of Bacteriophage Induced from a New Isolate of Thermus aquaticus. Microbiol. Cult. Coll, 1995. 11(2): p. 133–138. [Google Scholar]
- 30.Pawlowski A, et al. , Gammasphaerolipovirus, a newly proposed bacteriophage genus, unifies viruses of halophilic archaea and thermophilic bacteria within the novel family Sphaerolipoviridae. Arch Virol, 2014. 159(6): p. 1541–54. [DOI] [PubMed] [Google Scholar]
- 31.Pawlowski A, et al. , The Minor Capsid Protein VP11 of Thermophilic Bacteriophage P23–77 Facilitates Virus Assembly by Using Lipid-Protein Interactions. J Virol, 2015. 89(15): p. 7593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nika L, et al. , Expression of full-length HER2 protein in Sf9 insect cells and its presentation on the surface of budded virus-like particles. Protein Expr Purif, 2017. 136: p. 27–38. [DOI] [PubMed] [Google Scholar]
- 33.Rissanen I, et al. , Bacteriophage P23–77 capsid protein structures reveal the archetype of an ancient branch from a major virus lineage. Structure, 2013. 21(5): p. 718–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, et al. , Cn3D: sequence and structure views for Entrez. Trends Biochem Sci, 2000. 25(6): p. 300–2. [DOI] [PubMed] [Google Scholar]
- 35.O’Shea JJ, Ma A, and Lipsky P, Cytokines and autoimmunity. Nat Rev Immunol, 2002. 2(1): p. 37–45. [DOI] [PubMed] [Google Scholar]
- 36.Yang W, et al. , A new method for protein coexpression in Escherichia coli using two incompatible plasmids. Protein Expr Purif, 2001. 22(3): p. 472–8. [DOI] [PubMed] [Google Scholar]
- 37.Rissanen I Purification and crystallization of the two major coat proteins of bacteriophage P23–77 for X-ray crystallography. 2009. [cited 2019 March 12]; Available from: https://jyx.jyu.fi/bitstream/handle/123456789/23246/1/URN%3ANBN%3Afi%3Ajyu-201004191538.pdf.
- 38.Backovic A, et al. , Capsid protein expression and adeno-associated virus like particles assembly in Saccharomyces cerevisiae. Microb Cell Fact, 2012. 11: p. 124. [DOI] [PMC free article] [PubMed] [Google Scholar]


