Abstract
Mast cells (MCs) are pro-inflammatory tissue-resident immune cells that play a key role in inflammation. MCs circulate in peripheral blood as progenitors and undergo terminal differentiation in the tissue microenvironment where they can remain for many years. This in situ maturation results in tissue- and species-specific MC phenotypes, culminating in significant variability in response to environmental stimuli. There are many challenges associated with studying mature tissue-derived MCs, particularly in humans. In cases where cultured MCs are able to differentiate in two-dimensional in vitro cultures, there remains an inability for full maturation. Extracellular matrix (ECM) scaffolds provide for a more physiologically relevant environment for cells in vitro and have been shown to modulate the response of other immune cells such as T cells, monocytes, and macrophages. To improve current in vitro testing platforms of MCs and to assess future use of ECM scaffolds for MC regulation, we studied the in vitro response of human MCs cultured on decellularized porcine dermis hydrogels (dermis extracellular matrix hydrogel [dECM-H]). This study investigated the effect of dECM-H on cellular metabolic activity, cell viability, and receptor expression compared to collagen type I hydrogel (Collagen-H). Human MCs showed different metabolic activity when cultured in the dECM-H and also upregulated immunoglobulin E (IgE) receptors associated with MC maturation/activation compared to collagen type I. These results suggest an overall benefit in the long-term culture of human MCs in the dECM-H compared to Collagen-H providing important steps toward a model that is more representative of in vivo conditions.
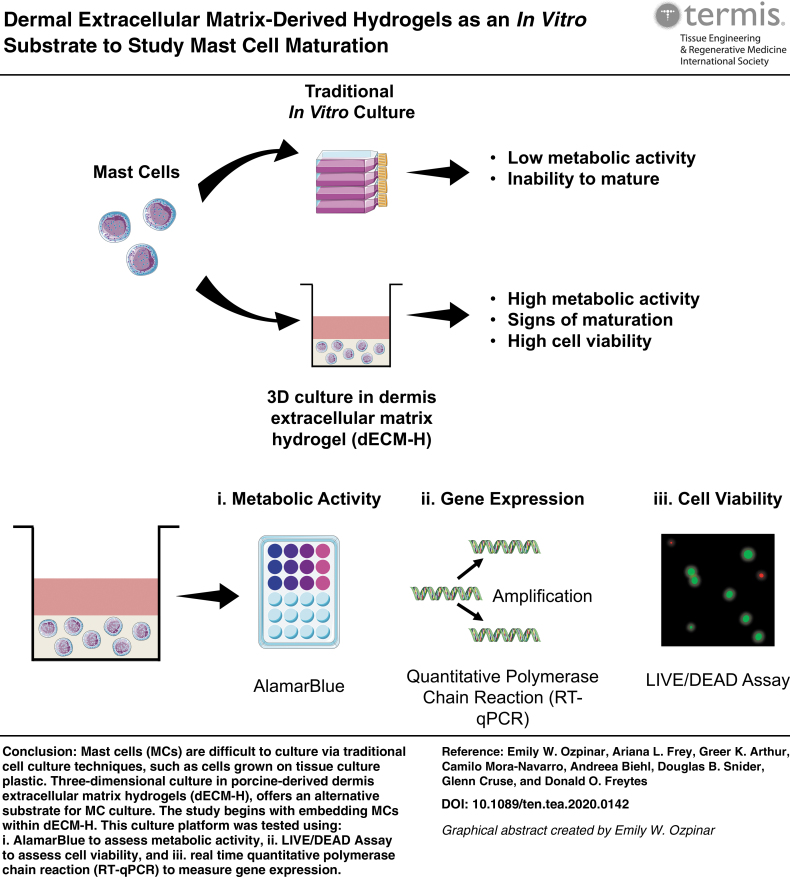
Graphical abstract
Impact statement
Mast cells (MCs) are difficult to culture in vitro as current culture conditions and substrates fail to promote similar phenotypic features observed in vivo. Extracellular matrix (ECM)-based biomaterials offer three-dimensional, tissue-specific environments that more closely resemble in vivo conditions. Our study explores the use of dermal ECM hydrogels for MC culture and shows significant upregulation of metabolic activity, cell viability, and gene expression of markers associated with MC maturation or activation compared to collagen type I-hydrogel and tissue culture plastic controls at 7 days. These results are among the first to describe MC behavior in response to ECM hydrogels.
Keywords: extracellular matrix, mast cell, allergy, metabolism
Introduction
Mast cells (MCs) are specialized, pro-inflammatory immune cells that form heterogeneous populations in all vascularized tissues. MCs are responsible for triggering inflammation to combat infections, envenomation, and cancer progression.1–4 MCs also facilitate tissue repair by regulating angiogenesis, reepithelialization, and scar formation.5 The ability of MCs to contribute to tissue homeostasis is due, in part, to the diversity of surface receptor expression, such as the high affinity immunoglobulin E (IgE) receptor (Fc fragment of IgE receptor 1 [FcɛRI]) and various G protein-coupled and Toll-like receptors. This diversity of receptors confers reactivity to a broad range of environmental and immunological stimuli (reviewed in Bradding and Cruse6 and Bradding and Arthur7). Other receptors mediate the adhesion of MCs to the extracellular matrix (ECM) and their interaction with neighboring cells, either directly or indirectly.8–12
Importantly, the receptor repertoire expressed on MCs differs significantly between tissues, conferring a tissue-specific phenotype that results in considerable variability in response to environmental stimuli (reviewed in Bradding and Cruse6 and Bradding and Arthur7).13–17 This heterogeneity exists as both alterations in the level of receptor expression, as well as examples of specific receptor expression, in some tissues that lack expression in MCs at other tissue sites.
Maladaptation and inappropriate activation of protective MC responses contribute to the pathophysiology of inflammatory diseases such as asthma and allergy and fibrotic diseases such as idiopathic pulmonary fibrosis.18 However, while MCs are key to allergic inflammation in a wide range of tissues, their precise role during inflammation is not clear. This is partly due to the transcriptional and phenotypic heterogeneity of mature tissue-resident MCs,19–22 which limits our ability to conduct physiologically relevant experiments in MC-related allergy research and neoplastic MC disorders such as mastocytosis and mast cell tumors (MCTs). Although mouse MCs are frequently used in MC and allergy research, human and mouse MCs differ widely in phenotype and there is often poor translation of mouse studies into human cells.6,23–25
Studying primary human MCs is preferable, but since MCs are purely tissue-resident in vivo, the phenotypic plasticity and transcriptional alterations induced by isolating the cells from tissue and altering the microenvironment during culture complicate the ex vivo study of primary human cells.26–29 The magnitude of these alterations could be limited by studying ex vivo MCs immediately after isolation, but the isolation process requires enzymatic digestion and mechanical force, which activates MCs and damages surface proteins, possibly resulting in hyporesponsive cells. To avoid this, MCs can be cultured from CD34+ precursors in vitro, but MCs do not fully differentiate in vitro or recapitulate ex vivo tissue-derived MCs. Thus, a conundrum of MC research is that in vivo (i.e., mouse), ex vivo, and in vitro systems can give different MC responses, and none may be true representations of human MC in vivo response.
An alternative method that we have explored in this study, and which may represent a low-cost and effective platform for translational MC culture, is the use of three-dimensional (3D) culture using tissue-specific ECM hydrogels. ECM is the supportive network comprised predominantly of tissue-specific proteins that provide structural and biochemical support to local cells, which can be harvested and isolated from tissues through a process known as decellularization.30,31 Of relevance to the challenges associated with MC research, previous studies have suggested that ECM-based biomaterials may affect cellular responses and maturation. For example, ECM scaffolds have been shown to influence adult and pluripotent stem cell differentiation in a tissue-specific manner.32–37 ECM scaffolds have also been shown to affect the responses of T cells and macrophages in vitro and in vivo with profound implication on the overall host tissue response.38–41 A multitude of studies have characterized the in vitro and in vivo response of macrophages toward ECM scaffolds, showing a shift in macrophage polarization toward pro-healing phenotypes.42,43 Given the potential role of MCs in disease and inflammation, understanding the interactions between MCs and ECM scaffolds could open new therapeutic avenues, as well as superior in vitro testing platforms.
Our goal was to derive and characterize an ECM-based hydrogel from porcine dermis (dermis extracellular matrix hydrogel [dECM-H]) and determine its effect on the metabolic activity and gene expression of a human line that is frequently used in MC research. We measured metabolic activity, cell viability, and IgE receptor expression to determine how dECM-H affected MC behavior and maturation.
Although some studies have explored MC behavior in hydrogels,44–46 to the best of our knowledge, the effect of tissue-derived ECM scaffolds on MC maturation and in vitro behavior has not yet been examined. Consequently, there is a need to investigate the ECM hydrogel (dECM-H) as a 3D culture platform for MCs, as well as its effects on MC behavior and receptor expression. This study marks the first step towards developing a low-cost and effective platform for MC culture that promotes maturation and tissue-specific MC behavior.
Materials and Methods
Deriving dECM-H
dECM-H was obtained by adapting a novel decellularization technique recently published by our research group.47 Full-thickness porcine skin was acquired from the dorsolateral flank of an ∼6-month-old Yorkshire pig provided by the NCSU Swine Education Unit. The skin was washed thoroughly with tap water, patted dry, cut into 3 × 7 cm rectangles, and placed at −80°C for at least 48 h. The skin sheets were thawed, and subcutaneous fat and hair were mechanically removed using a scalpel.
The skin was lyophilized overnight and ground into a powder. One gram of powder was agitated in a series of washes in the following solutions: dH2O rinse, 2 × phosphate buffered saline (PBS) for 5 min, 0.02% trypsin for 30 min, dH2O rinse, 2 × PBS for 5 min, 3% Tween-20 for 30 min, dH2O rinse, 2 × PBS for 5 min, 4% sodium deoxycholate for 30 min, dH2O rinse, 2 × PBS for 5 min, 0.1% peracetic acid for 30 min, dH2O rinse, and 1 × PBS for 5 min.
The samples were then lyophilized overnight. The dECM powder was digested at 10 mg/mL with 0.67 mg/mL of pepsin and 0.1 M HCl for 24 h on a stirring plate.48 dECM-H was formed by adjusting the salt concentration with 1 × PBS and 10 × PBS and then neutralizing the dECM digest with 0.1 N NaOH to a final dECM concentration of 6 mg/mL. The dECM digest was then cured by aliquoting into tissue culture treated plates and placing at 37°C for 30–45 min. FibriCol (Cat. No. 5133; Advanced Biomatrix, Carlsbad, CA), a collagen type I > 97% solution obtained from bovine hides, was used as a control (collagen type I hydrogel [Collagen-H]) following the manufacturers' instructions using dH2O, 10 × PBS, and 0.1 N NaOH.
Histological staining
To characterize the compositional changes before and after decellularization, lyophilized native dermis (dNative) and dECM powders were fixed overnight using 4% paraformaldehyde and then transferred to 70% ethanol. Samples were stained using hematoxylin and eosin (H&E) and Gomori's trichrome staining. The histology was performed at the Histology Laboratory in the College of Veterinary Medicine at North Carolina State University. Images were taken on an EVOS FL Auto imaging system (Thermo Fisher Scientific, Waltham, MA).
DNA quantification
Double-stranded DNA (dsDNA) removal quantification was performed using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies, Carlsbad, CA). Lyophilized samples before and after decellularization (∼3 mg) were stored in 200 μL TE buffer provided by the Quant-iT PicoGreen dsDNA Assay Kit and frozen at −20°C for at least 24 h. Samples were thawed, digested in a 1:10 proteinase K solution (Qiagen, Hilden, Germany) in Buffer ATL (Qiagen) overnight at 60°C, and then brought up to 1 mL with TE buffer. Then, a second dilution (1:100) was done to bring the sample into working conditions.
The Quant-iT PicoGreen dsDNA Assay Kit was used according to the manufacturer's protocol. Samples of 100 μL were read using an Infinite M200 Pro plate reader (Tecan, Männedorf, Switzerland); fluorescence was measured at 520 nm with excitation at 480 nm. The results for the standard curve, as well as the sample, were read in duplicates. Prism 8 was used to graph the data and perform statistics. Significance for decellularization efficiency was measured using an unpaired two-tailed t-test, where α = 0.05.
Proteomic analysis
Samples for proteomics were prepared as previously described47 and submitted to the NCSU Molecular Education, Technology, and Research Innovation Center (METRIC). Proteome Discoverer (Thermo Fisher Scientific) was used to analyze mass spectrometry results, and the proteins were categorized using the protein analysis through evolutionary relationships (PANTHER) Classification System (University of Southern California, Los Angeles, CA). Proteins that were classified as ECM proteins (PC00102) were extracted and organized into PRISM 8 for figures.
Gelation kinetics
Turbidimetric analysis was used to measure gelation kinetics of the hydrogels. After hydrogel preparation, 100 μL of hydrogel/well was aliquoted in a 96-well plate and kept on ice. The plate was placed in a Synergy Neo2 Multi-Mode Microplate Reader (Bio-Tek, Winooski, VT) warmed at 37°C. Absorbance at 405 nm was then measured every 2 min for 90 min, as the hydrogels “solidify” as collagen fibers self-assemble, thus increasing its turbidity and absorbance values.
Data were then normalized as previously described using Equation (1), spreading the values between 0 and 1, to allow for more accurate comparison between hydrogels. In the equation, At is the experimental measurement for absorbance at timepoint t, Amin is the minimal absorbance recorded, and Amax is the maximum absorbance recorded.49 The time required for the hydrogels to reach 50% of the maximum optical density turbidity was calculated (t50). Data were graphed in Prism 8.
| (1) |
Atomic force microscopy
Stiffness of the hydrogels was measured by atomic force microscopy (AFM) to describe the microenvironment the cells would sense. dECM-H and Collagen-H were formed as previously described on microscope slides and immersed in 1 × PBS.49 Measurements of Young's moduli were taken on an Asylum Research MFP-3D-BIO (Oxford Instruments, Santa Barbara, CA) using sQube polystyrene cantilevers partially coated with gold with a diameter of 6.1 μm (NanoAndMore USA Corp, Watsonville, CA). Three dECM-H and three Collagen-H were measured as three separate 10 × 10 μm force maps containing 256 points. Data were graphed in Prism 8, where points ≥50 Pa were plotted by relative frequency. Spatial averages were calculated by averaging all the points ≥50 Pa in the three hydrogels (for each dECM-H and Collagen-H) together.
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
The protein content between dECM-H and Collagen-H was compared using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Samples of diluted dECM-H, Collagen-H, and Pepsin HCl of same protein concentrations (1 mg/mL) were loaded into a 4–20% Mini-PROTEAN® TGX stain-Free™ Precast Protein Gel (Bio-Rad, Hercules, CA). Pepsin protein was used as a control, as it is used to digest dECM into dECM-H. The standard used was Precision Plus Protein™ Unstained Protein Standard, Strep-tagged recombinant (Bio-Rad).
Images of the gel were taken on a ChemiDoc Imaging System using self-staining settings (Bio-Rad). The gel was then further stained with Coomassie Brilliant Blue R-250 Staining Solution (Bio-Rad) overnight and washed several times before being imaged with a Canon EOS Rebel T7 DSLR Camera with a 18–55 mm lens (Canon, Tokyo, Japan).
Cell culture
The LAD2 human MC line was obtained from Drs. Dean Metcalfe and Arnold Kirshenbaum from the Laboratory of Allergic Diseases, National Institute of Allergy and Infectious Diseases (NIAID; National Institutes of Health [NIH], Bethesda, MD). LAD2 cells were cultured as previously described50–52 in StemPro-34 medium containing 13 mL supplement, penicillin (100 units/mL) and streptomycin (100 μg/mL; Life Technologies), and 100 ng/mL recombinant human stem cell factor (SCF; R&D, Abingdon, United Kingdom). Half of the medium was replaced with fresh medium and SCF every 7 days. The culture of HMC-1 and HRMC cell lines is described in Supplementary Data.
Transforming growth factor beta 1 stimulation and enzyme-linked immunosorbent assay
LAD2 cells were seeded in 12-well plates at a density of 5 × 105 cells per well in 2 mL of media. Human transforming growth factor beta 1 (TGF-β1; Peprotech, Rocky Hill, NJ) was added at 0.1 μg/mL final concentration to half of the samples (n = 3). Samples were collected at 24 h (n = 5) and 48 h (n = 6) for real-time quantitative polymerase chain reaction (RT-qPCR) and analyzed by the comparative CT method (method described in the following section). The ΔCT of samples stimulated with TGF-β1 was divided by the ΔCT of the nonstimulated cells (“No TGF-β1”) to give ΔΔCT. Results were graphed in Prism 8, and one-sample t-tests were conducted with μo = 1.00 and where α = 0.05.
TGF-β1 in dECM-H was measured using a Human/Mouse TGF-β1 Uncoated Enzyme-linked Immunosorbent Assay (ELISA) Kit (Invitrogen, Carlsbad, CA) according to the manufacturers' instructions. Lyophilized dECM powder (n = 3) was digested at 10 mg/mL using methods previously mentioned in Deriving dECM-H section. Collagen-H (n = 3) was also prepared according to manufacturer's instructions. Hydrogels were then diluted to concentrations of 1 mg/mL and measured with the ELISA. Absorbance was measured using an Infinite M200 Pro plate reader at 450 nm, with a reference wavelength of 570 nm. The values for Collagen-H were subtracted from the dECM-H samples as background.
Real-time quantitative polymerase chain reaction
dECM-H and Collagen-H were each seeded onto a 24-well plate at 100 μL per well. The hydrogel was coated evenly across the bottom of the well and allowed to self-assemble for ∼45 min at 37°C before adding LAD2 cells on top of each gel, along with the tissue culture plastic (TCP) control, at a density of 1.5 × 105 cells per well. Hydrogels and cells on TCP (n = 3 for each) were collected and lysed in TRK lysis buffer, provided by the E.Z.N.A. Total RNA Kit (Omega Bio-Tek, Norcross, GA), with 2-mercaptoethanol. RNA was cleaned and further concentrated using the RNeasy MinElute Cleanup Kit (Qiagen), and concentrations were measured using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific).
Complementary DNA (cDNA) was made with the GoScript Reverse Transcriptase Kit (Promega, Madison, WI) according to the manufacturers' instructions using the SimpliAmp Thermocycler (Applied Biosystems, Foster City, CA). RT-qPCRs were run on a QuantStudio 3 Real-Time PCR System (Applied Biosystems) using SYBR Select Master Mix (Applied Biosystems). Primers were obtained from Integrated DNA Technologies (Coralville, IA) and are listed in Table 1. Data were analyzed using QuantStudio 3 and 5 qPCR Data Analysis Software, and ΔCT values were calculated in Microsoft Excel v 16.30 using the comparative CT method.53 Data were then graphed and analyzed in Prism 8 through two-way ordinary analysis of variance (ANOVA) using Tukey's statistical hypothesis testing.
Table 1.
Real Time Quantitative Polymerase Chain Reaction Primers
| Gene | Species | Forward sequence | Reverse sequence |
|---|---|---|---|
| GAPDH | Human | AAGGTGAAGGTCGGAGTCAAC | GGGGTCATTGATGGCAACAATA |
| c-KIT | Human | TCATGGTCGGATCACAAAGA | AGGGGCTGCTTCCTAAAGAG |
| FcɛR1α | Human | TGTGGCAGCTGGACTATGAG | GAAATGTGACCTGCTGCTGA |
| FcɛR1β | Human | AATCTTGCTCTCCCACAGGA | TGTGTTACCCCCAGGAACTC |
| FcɛR1γ | Human | GGAGAGCCTCAGCTCTGCTA | TGGTGGTTTCTCATGCTTCA |
| TGF-β1 | Human | GGCCAGATCCTGTCCAAGC | GTGGGTTTCCACCATTAGCAC |
Cell viability assay
LAD2 cells were seeded on top of self-assembled dECM-H and Collagen-H at a density of 1 × 106 cells/mL in 100 μL of cultured medium per well. The hydrogels with LAD2s were stained at 24, 48, and 168 h (7 days) timepoints using the LIVE/DEAD Viability/Cytotoxicity Kit for mammalian cells (Life Technologies) according to the manufacturer's protocol. Live cells were stained green by calcein-AM, while dead cells were stained red by ethidium homodimer-1. Samples were imaged using fluorescence microscopy with a Revolve microscope (Echo, San Diego, CA).
alamarBlue® assay
LAD2s were seeded onto hydrogels as described previously in Cell Viability Assay section. At 24, 48, and 168 h timepoints, 10 μL alamarBlue Cell Viability Reagent (Invitrogen) was added directly to each of the hydrogels (n = 3 for each). The plate was incubated for 4 h at 37°C, and samples were read using an Infinite M200 Pro plate reader. Absorbance was quantified at 570 nm with a reference wavelength of 600 nm.
Overlap between 570 and 600 nm was accounted for using a RO variable, based off concepts published in Goegan et al.,54 for each hydrogel type. This was calculated using Equation (2), where AO(570) is the absorbance reading at 570 nm for collagen-H or dECM-H with no cells, and AO(600) is the absorbance reading at 600 nm for collagen-H or dECM-H with no cells. Percentage of alamarBlue reagent reduced (%R570) was calculated using Equation (3), where A570 is the absorbance reading at 570 nm and A600 is the absorbance reading at 600 nm. Results were graphed in Prism 8 using two-way ordinary ANOVA with Tukey's multiple comparison testing, where α = 0.05.
| (2) |
| (3) |
Results
dECM-H derivation and characterization
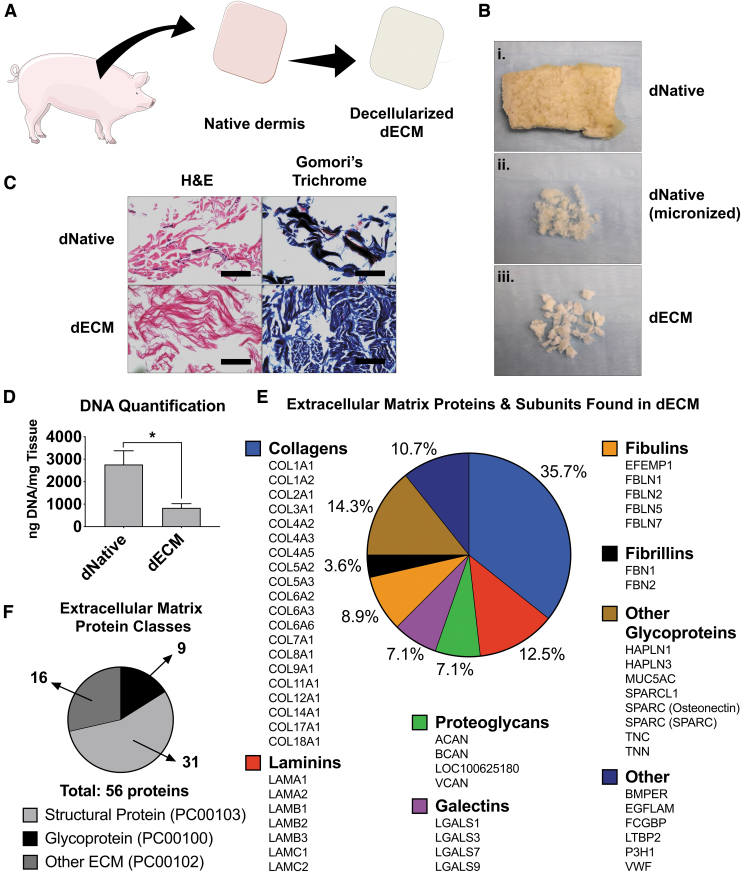
The overall decellularization and hydrogel formation are summarized in Figure 1A and B (decellularization) and Figure 2A (hydrogel formation). H&E staining showed a reduction in the number of nuclei (stained in purple) present between dNative and the dECM (Fig. 1C). Gomori's blue staining showed a reduction in, what is most likely, keratin and cellular cytoplasm (red-colored areas) and nuclei (black), but retention of collagen fiber (blue) areas within the tissue after decellularization. These results indicate a retention of important ECM components. DNA quantification showed a significant (p = 0.04) decrease in DNA content between dNative (2765.42 ng/mL ±610.57) and dECM (833.70 ng/mL ±188.69) (Fig. 1D).
FIG. 1.
ECM derivation and characterization. (A) General schematic of the derivation of dECM. (B) Progression of decellularization. The top image is a piece of lyophilized native porcine dermis (dNative) (B.i), the center image is native porcine dermis after micronization (B.ii), and the bottom image is after decellularization and lyophilization (B.iii). (C) Histological staining (hematoxylin and eosin and Gomori's trichrome) before (dNative) and after (dECM) decellularization, showing maintenance of crucial ECM components. Scale bar = 100 μm. (D) DNA quantification using PicoGreen assay showing significant decrease of DNA content in dECM compared to dNative. *p < 0.05. (E) Breakdown of the 56 proteins found in dECM through mass spectrometry that are organized within ECM protein classes (found in the PANTHER classification system). (F) Class breakdown of the 56 proteins as sorted by their PANTHER classification. dECM, dermis extracellular matrix; dNative, native dermis; ECM, extracellular matrix. Color images are available online.
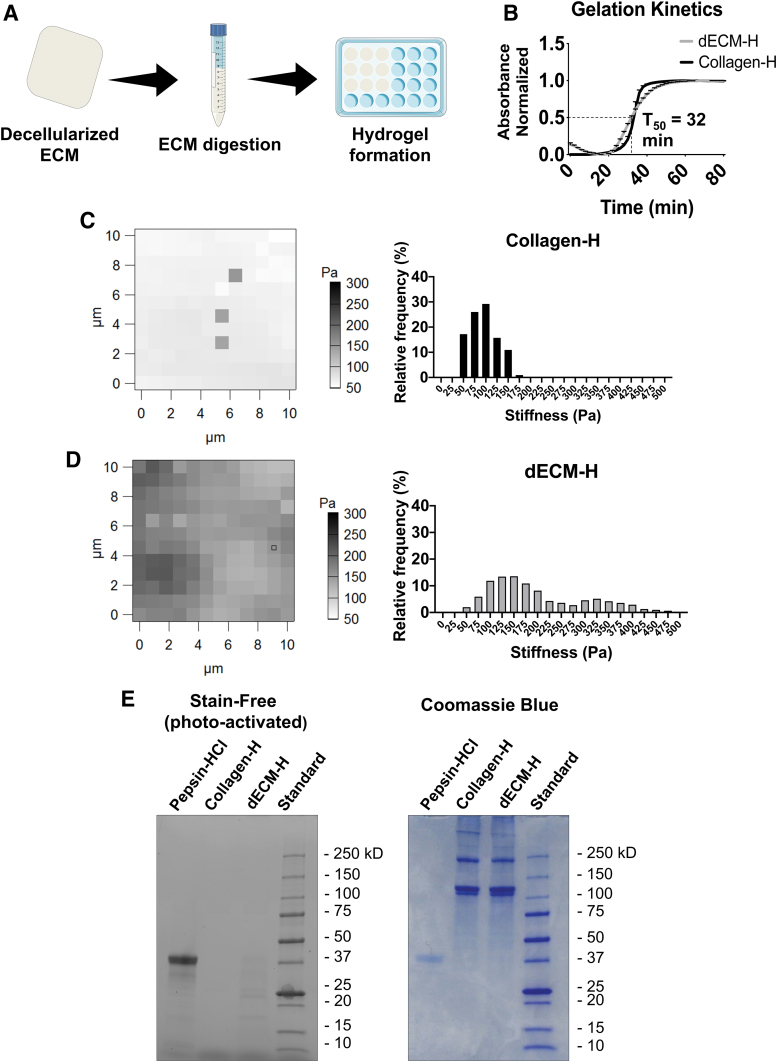
FIG. 2.
dECM-H formation and characterization. (A) General schematic of the derivation of dECM. (B) Gelation kinetics of dECM-H and Collagen-H, where t50 is the time it takes for dECM-H to reach 50% of maximum absorbance. (C) Force maps generated from AFM and the relative frequency of stiffness seen in Collagen-H. (D) Force maps generated from AFM and the relative frequency of stiffnesses seen in dECM-H. (E) SDS-PAGE results on a stain-free gel, which was then stained further with Coomassie Blue. AFM, atomic force microscopy; Collagen-H, collagen type I hydrogel; dECM-H, dermis extracellular matrix hydrogel; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Color images are available online.
A proteomic discover workflow was used to identify the protein composition of the dECM as previously reported.47 Gene ontology analysis was performed with a total of 3068 proteins submitted. The analysis reported 1749 hits, with 56 protein identified hits in PC00102 (Fig. 1F). Nine proteins from ECM glycoproteins (PC00100) and 31 from ECM structural protein (PC00103) classes were identified, as defined by the PANTHER classification system. Analyzing the composition based on the ECM proteins presented in the material resulted in a combination of collagens, laminins, proteoglycans, galectins, fibulins, fibrillins, and other glycoproteins (Fig. 1E). Altogether, these data are evidence of the complex protein composition of dECM.
Derivation of dECM-H resulted in successful hydrogel formation (Supplementary Fig. S1A). Gelation kinetics was measured by turbidimetric analysis, which is a commonly used method used to quantify the self-assembly of ECM hydrogels.55–57 Using a spectrometer to measure absorbance, formation of dECM-H and Collagen-H was confirmed (Fig. 2B). t50, where the dECM-H reaches 50% of the total absorbance, was 32 min, while t95, where dECM-H reaches 95%, was 50 min. For Collagen-H, t50 was 34 min, while t95 was 42 min.
AFM data at 750 pts on the Collagen-H showed a median stiffness of 103.07 Pa and a mean stiffness of 96.02 ± 1.15 Pa (Fig. 2C). AFM data at 1,708 pts on the dECM-H showed a median stiffness of 169.26 Pa and a mean stiffness of 199.73 ± 2.44 Pa (Fig. 2D). These values are consistent with previously reported values from ECM hydrogels derived from other porcine tissues.49 Spatial heterogeneity was also observed in dECM-H and Collagen-H, with a larger range in dECM-H. This could be due to the cantilever hitting collagen fibrils (in both Collagen-H and dECM-H) or to the undigested particle size of dECM (which can be observed in Fig. 1B.iii). Both dECM-H and Collagen-H were able to maintain its hydrogel structure when seeded with LAD2 over 168 h. However, the ability of dECM-H to hold its shape was superior to Collagen-H, which fell apart when removed from a 96-well plate after culture (Supplementary Fig. S1B).
As evident from the proteomics data, collagens make up a large portion of dECM. SDS-PAGE was then used to compare the differences in composition between dECM-H and Collagen-H (Fig. 2E). In the self-staining gel, there were faint bands on the smaller protein size range in the dECM-H, which could indicate the presence of smaller proteins. The gel was further stained with Coomassie Brilliant Blue, which showed similar size bands between Collagen-H and dECM-H. These results suggest that collagen type I is maintained in dECM-H throughout the digestion process.
TGF-β1 stimulation
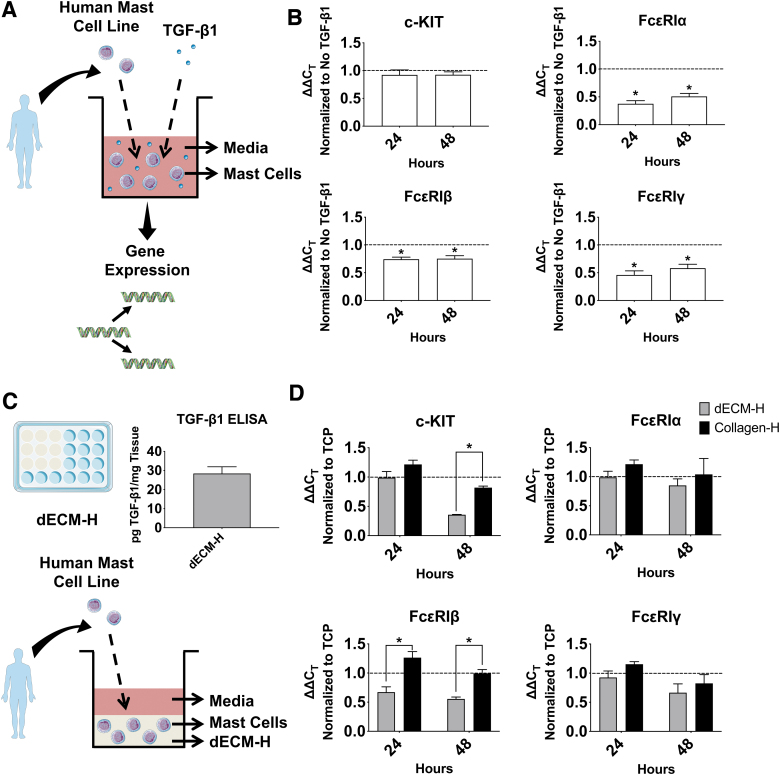
It has been previously shown that endogenous TGF-β1 can downregulate IgE receptor expression in MCs.58–60 To confirm this, LAD2s were stimulated with TGF-β1 for 24 and 48 h (Fig. 3A) and normalized to LAD2s that were not stimulated. A figure of gene expression normalized to the housekeeping gene, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), is included in Supplementary Figure S2B. LAD2s cultured on TCP that were stimulated TGF-β1 expressed similar levels of c-KIT to LAD2s not stimulated with TGF-β1 (Fig. 3B). However, for the three chains of FcɛRI (FcɛRIα, FcɛRIβ, and FcɛRIγ), gene expression of LAD2s stimulated with TGF-β1 was downregulated compared to the No TGF-β1 control. For all four genes, there was no significant difference between expression at 24 and 48 h.
FIG. 3.
TGF-β1 stimulation on human mast cells and c-KIT and IgE receptor expression in human mast cells in dECM-H. (A) Schematic of the experiment targeting TGF-β1. (B) RT-qPCR results of LAD2s cultured with and without TGF-β1, normalized first to GAPDH as a housekeeping gene and then taken as a ratio to LAD2s cultured without TGF-β1. The dotted line identifies 1.0 as an equal amount of expression between LAD2s cultured with and without TGF-β1. *p < 0.05 when the CT value of 24 or 48 h is compared to that of the No TGF-β1 control at the same timepoint (dotted line). (C) Experimental design of how cKIT and IgE receptor expression was measured. The bar graph on the right is the TGF-β1 ELISA results of three separate hydrogels from decellularized matrix. Collagen-H was used as background subtraction. (D) RT-qPCR results of LAD2s cultured in dECM-H and collagen-H. Results were normalized to GAPDH as a housekeeping gene at 24 and 48 h and then to TCP. The dotted line identifies 1.0 as an equal amount of expression between LAD2s cultured on hydrogel and TCP. *p < 0.05 when the CT value of dECM-H compared to Collagen-H at the same timepoint. ELISA, enzyme-linked immunosorbent assay; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IgE, immunoglobulin E; RT-qPCR, real time quantitative polymerase chain reaction; TCP, tissue culture plastic; TGF-β1, transforming growth factor beta 1. Color images are available online.
To find out whether or not dECM-H (with no cells) contains TGF-β1, we measured an average of 28.40 ± 3.54 pg of TGF-β1/mg of dECM-H using ELISA (Fig. 3C). This amount is low compared to previous studies that measured TGF-β1 in human skin biopsies.61,62 It is possible that the decellularization or digestion process removes considerable amounts of TGF-β1.
Gene expression of c-KIT and the three FcɛRI chains at 24 and 48 h after addition to the dECM-H or Collagen-H was measured to determine how regulation of these genes compared to TGF-β1 stimulation (Fig. 3D). These values were normalized to TCP to compare the effect of the hydrogels. c-KIT expression at 48 h was significantly reduced (p < 0.01) in dECM-H. FcɛRIβ also demonstrated significant downregulation (p < 0.01) in dECM-H compared to Collagen-H for both 24 and 48 h. While there was no significance for FcɛRIα and FcɛRIγ at either 24 or 48 h, there was still a trend toward a slight downregulation of expression in dECM-H compared to Collagen-H.
LAD2 cell viability and metabolic activity
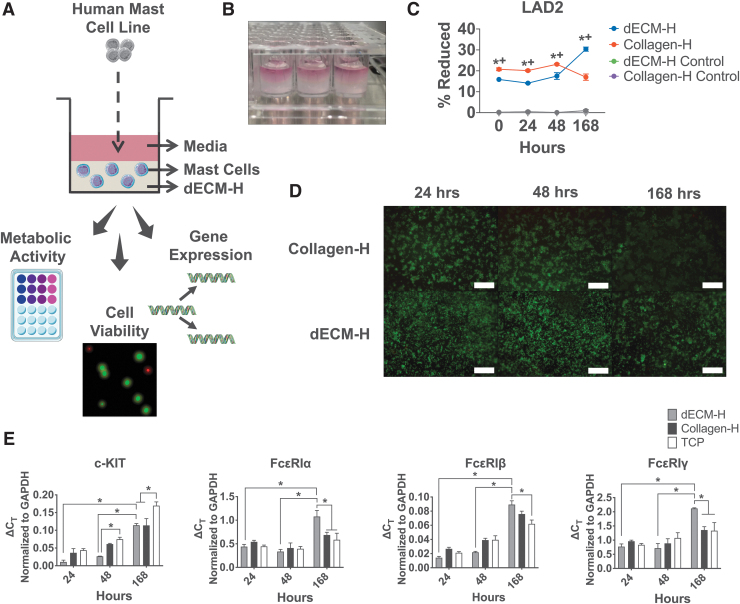
Following characterization of the hydrogels, LAD2 cells were assessed in situ for metabolic activity using alamarBlue (Fig. 4C) and cell viability using LIVE/DEAD assay (Fig. 4D). The alamarBlue assay showed that the percent of reduced alamarBlue reagent of LAD2 cells in Collagen-H was significantly higher than cells in dECM-H at time 0, 24, and 48 h. However, at 168 h, LAD2 cells seeded into dECM-H (30.36% ± 0.98) exhibited significantly (p < 0.01) more metabolic activity compared to Collagen-H (16.99% ± 1.49). This trend was also observed with the human neoplastic HMC-1 cell line (Supplementary Fig. S3B) and the canine tumor-derived neoplastic HRMC cell line (Supplementary Fig. S4E).
FIG. 4.
Human mast cell behavior in dECM-H. (A) Schematic of the seeding of human LAD2 cells into dECM-H and their outputs. (B) Image of dECM-H formed in a 96-well plate with media on top. (C) Metabolic activity of LAD2s seeded in dECM-H and Collagen-H, along with their respective controls (hydrogels with no cells). *p < 0.05 when comparing dECM-H to Collagen-H. +p < 0.05 when comparing dECM-H to controls. (D) Cell viability using LIVE/DEAD assay, where live cells are in green and dead cells are in red. Scale bars = 320 μm. (E) RT-qPCR results of LAD2s cultured in dECM-H, Collagen-H, and TCP normalized to GAPDH as a housekeeping gene at 168 h after seeding. *p < 0.05. Color images are available online.
The cell viability assay was conducted at 24, 48, and 168 h after seeding. At all timepoints, LAD2s showed a majority of cells alive (stained green) and with relatively steady levels of confluence. However, as time increased, the green signal diminished in Collagen-H as opposed to the signal in dECM-H. There is also significant clumping of the cells in Collagen-H at all timepoints, which is not observed in the cells in dECM-H. The results from this assay demonstrate that LAD2 survives well in dECM-H.
Human MC receptor expression
MCs are tissue-resident cells that are recruited into tissues as progenitors and only mature once they enter their target tissue. Therefore, receptor expression in mature MCs varies according to the tissue of residence and local factors (reviewed in Bradding and Cruse6 and Bradding and Arthur7). The cardinal receptors on MCs that regulate many of their functions are the high affinity IgE receptor, FcɛRI, and the receptor tyrosine kinase, KIT. Therefore, the level of expression of these receptors can be an indicator of MC phenotype. Having established that dECM-H promoted LAD2 metabolism for 7 days, we proposed that this effect was a consequence of MC maturation or altered phenotype. MCs circulate in the blood as progenitor cells that migrate into tissue where they mature and develop the expression of FcɛRI.6 Higher expression of FcɛRI develops as MCs mature from precursors63 and may thus be an indicator of greater MC maturity or represent a more responsive MC population. The human FcɛRI complex is generally a tetrameric complex comprising of an α subunit that binds IgE, a dimer of γ subunits that provide the majority of the signaling potential, and 4 transmembrane protein β subunits that traffic the complex to the plasma membrane.64 Using RT-qPCR (Fig. 4E) to establish the expression of each of the genes encoding the subunits of FcɛRI and c-KIT (that encodes CD117) as we had performed at earlier timepoints (Fig. 3D), we measured receptor expression at 168 h after addition to dECM-H to establish if differences in receptor gene expression were associated with the increased metabolic activity at 168 h (Fig. 4C). Gene expression of c-KIT in LAD2 cells cultured in dECM-H at 168 h (0.11 ± 0.01), along with Collagen-H at 168 h (0.11 ± 0.02), was downregulated (p < 0.01) compared to the TCP control (0.17 ± <0.01). However, dECM-H at 168 h was upregulated compared to dECM-H at 24 h (0.01 ± 0.01) and dECM-H at 48 h (0.03 ± <0.01).
In contrast to the downregulation of c-KIT expression of dECM-H compared to controls, the RT-qPCR results for the three chains of FcɛRI (FcɛRIα, FcɛRIβ, and FcɛRIγ) demonstrate significant upregulation at the later timepoint to the controls (Fig. 4E). FcɛRIα gene expression of LAD2 cells cultured in dECM-H at 168 h (1.08 ± 0.13) was significantly increased (p < 0.01) compared to Collagen-H (0.69 ± 0.06) and TCP (0.59 ± 0.14; p < 0.01), as well as to dECM-H at 24 h (0.44 ± 0.05) and dECM-H at 48 h (0.34 ± 0.05). FcɛRIα expression in LAD2 cells cultured in dECM-H at 168 h (0.09 ± 0.01) was also significantly higher than dECM-H at 24 h (0.01 ± <0.01) and dECM-H at 48 h (0.02 ± <0.01), as well as TCP (0.06 ± 0.01, p < 0.01), but not to Collagen-H at 168 h (0.08 ± <0.01, p = 0.28). Finally, FcɛRIα gene expression of LAD2 cells cultured in dECM-H at 168 h (2.12 ± 0.02) was significantly increased (p < 0.01) compared to Collagen-H at 168 h (1.36 ± 0.12) and TCP (1.33 ± 0.29) and additionally higher than dECM-H at 24 h (0.77 ± 0.09) and dECM-H at 48 h (0.72 ± 0.16). These results suggest that dECM-H promotes expression of FcɛRI IgE receptor chain subunits compared to Collagen-H and TCP controls while downregulating c-KIT expression.
Discussion
The present study focuses on the relatively unknown interactions between ECM hydrogels and MCs. ECM scaffolds have been shown to modulate immune cells through the promotion of a Th2 phenotype on T cells41 and M2-like response on macrophages.38–40 However, to the best of our knowledge, there is not much known about the in vitro interactions between MCs and ECM scaffolds. ECM scaffolds could represent an ideal culture substrate to modulate the MC response in vivo or to create novel in vitro testing platforms to study MC biology. MCs circulate in blood as progenitors and only mature once they are in tissues, where they reside for several years.6 This results in marked heterogeneity of MCs in vivo, a feature that makes the study of MCs, particularly human MCs, difficult to achieve in vivo in animal models, ex vivo and in vitro.
Derivation of MCs from primary tissues, such as the dermis, lung, and gastrointestinal tract, yields low numbers. Furthermore, the harsh mechanical and biochemical conditions required to isolate the cells from tissue can damage surface proteins and activate the cells. An alternative approach is to culture MCs from MC precursors found in peripheral blood or bone marrow. However, this approach results in an immature MC population with an artificial “cultured” phenotype that is driven by the addition of limited cocktails of cytokines during culture.
Cell lines are available, but often do not express FcɛRI, a key receptor for MC biology. The human LAD2 MC line derived from an adult patient with MC sarcoma50 does express high levels of FcɛRI and is therefore commonly used in research for MC-associated diseases.65 However, in vitro data from LAD2 MCs must be interpreted with care because, as with any cell line used, it is not clear how the transformed nature of these cells affects the cellular phenotype.
In this study, dECM-H was used as an in vitro culture substrate, offering a tissue-specific 3D environment to enhance MC culture and provide a more physiologically relevant environment. Porcine-derived ECM was chosen given its relative abundance and ease of procurement as by-products of the meatpacking industry. Decellularized porcine and human tissues have also been found to be similar in terms of mechanical properties, protein composition, and potential cellular interactions.66,67 However, while healthy porcine tissue is inexpensive and easily procured, human tissue is expensive to handle and difficult to procure, as healthy tissue is preferably sourced for transplantation and tissues rejected for transplantation may not be the best for decellularization. Dermal tissue was chosen over other ECM types because a number of human and animal MC-associated diseases, such as dermatitis, mastocytosis, and canine MCTs, occur frequently in the skin.6 However, ECM isolated from other tissues could be utilized for MC-driven diseases, such as lung for asthma.
We report significant differences in human cells (LAD2) when embedded in dECM-H in comparison to our control substrates, Collagen-H and TCP. While dECM-H and Collagen-H exhibit similar gelation kinetics profiles, dECM-H has some unique properties that may explain the MC behavior observed in this study. The first of these differences is the average surface stiffness between dECM-H and Collagen-H (Fig. 2C, D). The results show that dECM-H had, on average, stiffer regions than Collagen-H. The reasons for these differences remain unknown, but differences in composition and particulate size could help explain varying levels of stiffness measured. It is well known that cell-level matrix stiffness can affect the shape, function, and differentiation of many different cell types68–70 and could have an effect on MCs. Reports show how MCs themselves respond differently to substrates of varying levels of stiffness affecting processes such as migration activity71 and cell shape.45 ECM-derived hydrogels are soft substrates and possess different mechanical properties than the source tissue. Therefore, these hydrogels cannot recapitulate the native stiffness of the tissue, but they harness the compositional cues inherent within the matrix. Further studies will determine the overall effects of hydrogel stiffness on MC maturation and function in vitro. Nevertheless, the changes in stiffness might not fully explain the short- and long-term response of MCs when cultured on dECM-H observed in the present study. This will also be explored in future studies.
Another possible factor modulating MC behavior could be the presence of TGF-β1 within the dECM-H (Fig. 3C). TGF-β1 has been detected in other porcine ECM tissues72 and plays an important role in the mammalian immune system.73 Previous studies have shown that LAD2 cells expressed lower levels of FcɛRIβ in vitro compared to human skin-derived MCs.74 It has also been shown that MCs secrete TGF-β1 when stimulated with IgE.75 In turn, MCs respond to exogenous TGF-β1 by decreasing FcɛRI expression,58 inhibiting proliferation,76 and downregulating histamine and tumor necrosis factor alpha (TNF-α).77 TGF-β1 has been shown to regulate c-KIT expression78 and SCF, the ligand for c-KIT.79
In the present study, LAD2 cells showed initial downregulation of FcɛRI receptor chains as a result of TGF-β1 stimulation in vitro confirming previous reports. However, c-KIT was not significantly affected. When LAD2 cells were cultured within dECM-H, only c-KIT and FcɛRIβ were significantly downregulated compared to cells cultured on Collagen-H. The data suggest a possible influence of TGF-β1 within the dECM-H during the initial 48 h of culture although further testing is needed to elucidate the exact role of dECM-derived TGF-β1 on MCs.
Since MCs cultured in vitro do not show similar gene expression profiles to MCs found in vivo, our study focused on examining whether dECM-H could provide a superior in vitro environment to promote MC maturation, viability, and more appropriate, physiologically relevant responses. Metabolic activity (Fig. 4C) was significantly upregulated at 7 days (a finding we replicated with the human neoplastic cell line, HMC-1 [Supplementary Fig. S3], and canine neoplastic HRMCs [Supplementary Fig. S4E]). In a cell viability assay, LAD2s became clumped and showed a diminished live signal at 168 h when in culture with Collagen-H, but maintained strong viability and resisted clumping in dECM-H. We also found evidence of tumor progression in neoplastic HRMCs where histological staining revealed multinucleated cells and cells with bizarre nuclei and mitotic figures, corresponding with the Kiupel grading system for canine MCTs (Supplementary Fig. S4B–D). Histological staining and analysis is described in Supplementary Data.
MC development and regulation are heavily dependent on c-KIT, but as a proto-oncogene, constitutive activation due to mutations in c-KIT can lead to human mastocytosis or canine MCTs.80–82 Since HRMCs and HMC-1s have clinically relevant c-KIT mutations resulting in constitutive activation of the receptor tyrosine kinase KIT, which leads to neoplastic MC growth, we were interested in the effect dECM-H would have on c-KIT expression. The downregulation in both hydrogels at 168 h (Fig. 4E) shows that perhaps the 3D environment could play a larger role on the long-term response of LAD2s than the presence of TGF-β1.
Given the metabolic changes observed at this timepoint, gene expression at 168 h of all three FcɛRI chains was measured. In MCs, IgE antibodies bind to FcɛRI, causing downstream signaling cascades and subsequent MC activation. As the FcɛRI receptor consists of α, β, and γ subunits,83 gene expression of all three chains was measured, and the results indicate upregulated expression of all three when LAD2s were cultured in dECM-H (Fig. 4E). This suggests that, in comparison to c-KIT expression, this upregulation is due to the culture of dECM-H specifically and not just the 3D environment that the hydrogel provides.
While c-KIT and IgE receptor expression (FcɛRI) alone may not be sufficient to characterize MCs as fully mature, there are other markers of MC maturation that will be the focus of future studies, such as measuring granule contents for tryptase, chymase, heparin, intracellular calcium, or cytokine production and degranulation. In addition, due to the heterogeneity of receptors that MCs can express, it would be worthwhile to measure expression of receptors that mediate adhesion to the ECM.
One limitation of this study is the inherent variability found in ECM scaffolds and ECM hydrogels. Although we used quality control parameters, such as dsDNA quantification, histology, and standard operating procedures during this study, ECM hydrogels can vary in composition depending on the animal source (animal age, etc.) and decellularization protocol used. Particle size of dECM after decellularization and lyophilization can also vary and be difficult to control. Synthetic hydrogels can offer a more standardized product, but do not offer the unique biological activity that is found in ECM biomaterials.
To account for this, we used an automated decellularization method developed in our laboratory47 that is considerably shorter than previously reported methods,84,85 yet retains crucial ECM components (Fig. 1C, E) with sufficient DNA content removal (Fig. 1D). Shortening this process allows for cost savings on reagents and time and helps mitigate potential variations in the decellularization process by consolidating the steps. Improperly decellularized tissues could influence downstream processes and cell survival, so the automated process allows for proper and thorough rinsing of the tissue from residual reagents that could elicit immunogenic responses or other negative effects.
An additional limitation of this study is the use of immortalized cell lines. Consequently, future studies will focus on primary cell sources. Regardless of these limitations, this study reports on the potential use of dECM-H as a 3D culture platform for MCs and demonstrates significant potential to study MC maturation and phenotype with improved in vitro cell culture of primary human MCs, thus advancing current MC culture techniques for research into MC-associated diseases.
Conclusions
dECM-H can be used as a 3D in vitro platform to study human and canine MC culture. dECM-H promotes increased metabolic activity, cell viability, and expression of IgE receptor chains under prolonged culture times. The present study shows the benefits of dECM-H as a substrate for advancing the translation of findings from in vitro MC studies and the development of new testing platforms for therapeutics targeting MC-associated diseases.
Supplementary Material
Acknowledgments
All sample preparation and mass spectrometry measurements were done by the NCSU METRIC. A special thanks goes to Lauren Ehrhardt-Humbert and Dr. Barry A. Hedgespeth for their assistance with cell culture and Dr. Daniel Chester and Dr. Seema Nandi for their assistance with the AFM. The figures in this publication were made in part with Servier Medical ART (SMART).
Disclosure Statement
Dr. Cruse has research support from Hoth Therapeutics for a project not directly related to the research reported in this publication and also serves on their Scientific Advisory Board. The research findings included in this publication were not funded by Hoth Therapeutics and may not necessarily be related to the interests of Hoth Therapeutics. The terms of this arrangement have been reviewed and approved by NC State University in accordance with its policy on objectivity in research. The remaining authors declare no competing financial interests.
Funding Information
We thank our funding sources: UNC-CH/NCSU Joint Department of Biomedical Engineering and its Abrams Scholar Program, the NCSU Office of Undergraduate Research (OUR), NCSU Center for Human Health and the Environment (NIH, NIEHS Award No. P30ES025128), the Comparative Medicine Institute (CMI) at NCSU, especially the CMI's Summer Interdisciplinary Research Initiative (SIRI), Young Scholar Award (YSA) and Comparative Medicine and Translational Research Training Program (CMTRTP; NIH 2T32OD011130-11) programs, and the NIH, NIAID (Award No. R01AI143985).
Supplementary Material
References
- 1.Galli, S.J.The mast cell-IgE paradox: from homeostasis to anaphylaxis. Am J Pathol 186,212, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trivedi, N.H., Guentzel, M.N., Rodriguez, A.R., Yu, J.J., Forsthuber, T.G., and Arulanandam, B.P.. Mast cells: multitalented facilitators of protection against bacterial pathogens. Expert Rev Clin Immunol 9,129, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welsh, T.J., Green, R.H., Richardson, D., Waller, D.A., O'Byrne, K.J., and Bradding, P.. Macrophage and mast-cell invasion of tumor cell islets confers a marked survival advantage in non-small-cell lung cancer. J Clin Oncol 23,8959, 2005 [DOI] [PubMed] [Google Scholar]
- 4.Shelburne, C.P., and Abraham, S.N.. The mast cell in innate and adaptive immunity. Adv Exp Med Biol 716,162, 2011 [DOI] [PubMed] [Google Scholar]
- 5.Wulff, B.C., and Wilgus, T.A.. Mast cell activity in the healing wound: more than meets the eye?. Exp Dermatol 22,507, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradding, P., and Cruse, G.. Mast cells: biological properties and role in health and allergic diseases. In: Kay, B., Kaplan, A., Bousquet, J., and Holt, P., eds. Allergy and Allergic Diseases, 2nd Edition. Hoboken, NJ: Wiley-Blackwell, 2008, pp. 217–257 [Google Scholar]
- 7.Bradding, P., and Arthur, G.. Mast cells in asthma—state of the art. Clin Exp Allergy 46,194, 2016 [DOI] [PubMed] [Google Scholar]
- 8.Hollins, F., Kaur, D., Yang, W., et al. . Human airway smooth muscle promotes human lung mast cell survival, proliferation, and constitutive activation: cooperative roles for CADM1, stem cell factor, and IL-6. J Immunol 181,2772, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moiseeva, E.P., Roach, K.M., Leyland, M.L., and Bradding P.. CADM1 is a key receptor mediating human mast cell adhesion to human lung fibroblasts and airway smooth muscle cells. PLoS One 8,e61579, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moiseeva, E.P., Leyland, M.L., and Bradding P.. CADM1 isoforms differentially regulate human mast cell survival and homotypic adhesion. Cell Mol Life Sci 69,2751, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moiseeva, E.P., Straatman, K.R., Leyland, M.L., and Bradding, P.. CADM1 controls actin cytoskeleton assembly and regulates extracellular matrix adhesion in human mast cells. PLoS One 9,e85980, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang, W., Kaur, D., Okayama, Y., et al. . Human lung mast cells adhere to human airway smooth muscle, in part, via tumor suppressor in lung cancer-1. J Immunol 176,1238, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Varricchi, G., Pecoraro, A., and Loffredo, S.. Heterogeneity of human mast cells with respect to MRGPRX2 receptor expression and function. Front Cell Neurosci 13,299, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersson, C.K., Mori, M., Bjermer, L., Lofdahl, C.G., and Erjefalt, J.S.. Novel site-specific mast cell subpopulations in the human lung. Thorax 64,297, 2009 [DOI] [PubMed] [Google Scholar]
- 15.Balzar, S., Strand, M., Rhodes, D., and Wenzel S.E.. IgE expression pattern in lung: relation to systemic IgE and asthma phenotypes. J Allergy Clin Immunol 119,855, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujisawa, D., Kashiwakura, J., Kita, H., et al. . Expression of Mas-related gene X2 on mast cells is upregulated in the skin of patients with severe chronic urticaria. J Allergy Clin Immunol 134,622, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Cildir, G., Pant, H., Lopez, A.F., and Tergaonkar, V.. The transcriptional program, functional heterogeneity, and clinical targeting of mast cells. J Exp Med 214,2491, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruse, G., and Bradding, P.. Mast cells in airway diseases and interstitial lung disease. Eur J Pharmacol 778,125, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurashima, Y., Amiya, T., Fujisawa, K., et al. . The enzyme Cyp26b1 mediates inhibition of mast cell activation by fibroblasts to maintain skin-barrier homeostasis. Immunity 40,530, 2014 [DOI] [PubMed] [Google Scholar]
- 20.Dwyer, D.F., Barrett, N.A., and Austen, K.F.; Immunological Genome Project Consortium.. Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat Immunol 17,878, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weidner, N., and Austen K.F.. Heterogeneity of mast cells at multiple body sites. Fluorescent determination of avidin binding and immunofluorescent determination of chymase, tryptase, and carboxypeptidase content. Pathol Res Pract 189,156, 1993 [DOI] [PubMed] [Google Scholar]
- 22.Xing, W., Austen, K.F., Gurish, M.F., and Jones, T.G.. Protease phenotype of constitutive connective tissue and of induced mucosal mast cells in mice is regulated by the tissue. Proc Natl Acad Sci U S A 108,14210, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tkaczyk, C., Okayama, Y., Metcalfe, D.D., and Gilfillan, A.M.. Fcγ receptors on mast cells: activatory and inhibitory regulation of mediator release. Int Arch Allergy Immunol 133,305, 2004 [DOI] [PubMed] [Google Scholar]
- 24.Bischoff, S.C.Role of mast cells in allergic and non-allergic immune responses: comparison of human and murine data. Nat Rev Immunol 7,93, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Dillahunt, S.E., Sargent, J.L., Suzuki, R., et al. . Usage of sphingosine kinase isoforms in mast cells is species and/or cell type determined. J Immunol 190,2058, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monticelli, S., and Leoni, C.. Epigenetic and transcriptional control of mast cell responses. F1000Res 6,2064, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guhl, S., Artuc, M., Neou, A., Babina, M., and Zuberbier, T.. Long-term cultured human skin mast cells are suitable for pharmacological studies of anti-allergic drugs due to high responsiveness to FcɛRI cross-linking. Biosci Biotechnol Biochem 75,382, 2011 [DOI] [PubMed] [Google Scholar]
- 28.Guhl, S., Neou, A., Artuc, M., Zuberbier, T., and Babina, M.. Skin mast cells develop non-synchronized changes in typical lineage characteristics upon culture. Exp Dermatol 23,933, 2014 [DOI] [PubMed] [Google Scholar]
- 29.Motakis, E., Guhl, S., Ishizu, Y., et al. . Redefinition of the human mast cell transcriptome by deep-CAGE sequencing. Blood 123,e58, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert, T.W., Sellaro, T.L., and Badylak, S.F.. Decellularization of tissues and organs. Biomaterials 27,3675, 2006 [DOI] [PubMed] [Google Scholar]
- 31.Crapo, P.M., Gilbert, T.W., and Badylak, S.F.. An overview of tissue and whole organ decellularization processes. Biomaterials 32,3233, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Neill, J.D., Freytes, D.O., Anandappa, A.J., Oliver, J.A., and Vunjak-Novakovic, G.V.. The regulation of growth and metabolism of kidney stem cells with regional specificity using extracellular matrix derived from kidney. Biomaterials 34,9830, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duan, Y., Liu, Z., O'Neill, J., Wan, L.Q., Freytes, D.O., and Vunjak-Novakovic, G.. Hybrid gel composed of native heart matrix and collagen induces cardiac differentiation of human embryonic stem cells without supplemental growth factors. J Cardiovasc Transl Res 4,605, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, D., Zhang, Y., Zhang, Y., et al. . Tissue-specific extracellular matrix enhances skeletal muscle precursor cell expansion and differentiation for potential application in cell therapy. Tissue Eng Part A 23,784, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agmon, G., and Christman, K.L.. Controlling stem cell behavior with decellularized extracellular matrix scaffolds. Curr Opin Solid State Mater Sci 20,193, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaffney, L., Wrona, E.A., and Freytes, D.O.. Potential synergistic effects of stem cells and extracellular matrix scaffolds. ACS Biomater Sci Eng 4,1208, 2018 [Google Scholar]
- 37.Hoshiba, T., Chen, G.P., Endo, C., et al. . Decellularized extracellular matrix as an in vitro model to study the comprehensive roles of the ECM in stem cell differentiation. Stem Cells Int 2016,6397820, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huleihel, L., Dziki, J.L., Bartolacci, J.G., et al. . Macrophage phenotype in response to ECM bioscaffolds. Semin Immunol 29,2, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, R.M., Johnson, T.D., He, J., et al. . Humanized mouse model for assessing the human immune response to xenogeneic and allogeneic decellularized biomaterials. Biomaterials 129,98, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dziki, J.L., Wang, D.S., Pineda, C., Sicari, B.M., Rausch, T., and Badylak, S.F.. Solubilized extracellular matrix bioscaffolds derived from diverse source tissues differentially influence macrophage phenotype. J Biomed Mater Res A 105,138, 2017 [DOI] [PubMed] [Google Scholar]
- 41.Sadtler, K., Estrellas, K., Allen, B.W., et al. . Developing a pro-regenerative biomaterial scaffold microenvironment requires T helper 2 cells. Science 352,366, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf, M.T., Dearth, C.L., Ranallo, C.A., et al. . Macrophage polarization in response to ECM coated polypropylene mesh. Biomaterials 35,6838, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sicari, B.M., Dziki J.L., Siu, B.F., Medberry, C.J., Dearth, C.L., and Badylak, S.F.. The promotion of a constructive macrophage phenotype by solubilized extracellular matrix. Biomaterials 35,8605, 2014 [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto, T., Hartmann, K., Eckes, B., and Krieg T.. Mast cells enhance contraction of three-dimensional collagen lattices by fibroblasts by cell-cell interaction: role of stem cell factor/c-kit. Immunology 99,435, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiki, A., Inoh, Y., Yokawa, S., and Furuno, T.. Inhibition of degranulation in mast cells attached to a hydrogel through defective microtubule tracts. Exp Cell Res 381,248, 2019 [DOI] [PubMed] [Google Scholar]
- 46.Derakhshan, T., Bhowmick, R., Meinkoth, J.H., Ritchey, J.W., and Gappa-Fahlenkamp, H.. Human mast cell development from hematopoietic stem cells in a connective tissue-equivalent model. Tissue Eng Part A 25,1564, 2019 [DOI] [PubMed] [Google Scholar]
- 47.Badileanu, A., Mora Navarro, C., Gracioso Martins, A.M., et al. . Fast automated approach for the derivation of acellular extracellular matrix scaffolds from porcine soft tissues. ACS Biomater Sci Eng 6,4200, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freytes, D.O., Martin, J., Velankar, S.S., Lee, A.S., and Badylak, S.F.. Preparation and rheological characterization of a gel form of the porcine urinary bladder matrix. Biomaterials 29,1630, 2008 [DOI] [PubMed] [Google Scholar]
- 49.Mora-Navarro, C., Badileanu, A., Martins, A.M.G., et al.Porcine vocal fold lamina propria-derived biomaterials modulate TGF-beta 1-mediated fibroblast activation in vitro. ACS Biomater Sci Eng 6,1690, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kirshenbaum, A.S., Akin, C., Wu, Y., et al. . Characterization of novel stem cell factor responsive human mast cell lines LAD 1 and 2 established from a patient with mast cell sarcoma/leukemia; activation following aggregation of FcepsilonRI or FcgammaRI. Leuk Res 27,677, 2003 [DOI] [PubMed] [Google Scholar]
- 51.Cruse, G., Beaven, M.A., Music, S.C., Bradding, P., Gilfillan, A.M., and Metcalfe, D.D.. The CD20 homologue MS4A4 directs trafficking of KIT toward clathrin-independent endocytosis pathways and thus regulates receptor signaling and recycling. Mol Biol Cell 26,1711, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cruse, G., Beaven, M.A., Ashmole, I., Bradding, P., Gilfillan, A.M., and Metcalfe, D.D.. A truncated splice-variant of the FcepsilonRIbeta receptor subunit is critical for microtubule formation and degranulation in mast cells. Immunity 38,906, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmittgen, T.D., and Livak, K.J.. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3,1101, 2008 [DOI] [PubMed] [Google Scholar]
- 54.Goegan, P., Johnson, G., and Vincent, R.. Effects of serum protein and colloid on the alamarBlue assay in cell cultures. Toxicol in Vitro 9,257, 1995 [DOI] [PubMed] [Google Scholar]
- 55.Gaetani, R., Aude, S., DeMaddalena, L.L., et al. . Evaluation of different decellularization protocols on the generation of pancreas-derived hydrogels. Tissue Eng Part C Methods 24,697, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sackett, S.D., Tremmel, D.M., Ma, F., et al. . Extracellular matrix scaffold and hydrogel derived from decellularized and delipidized human pancreas. Sci Rep 8,10452, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernandez-Perez, J., and Ahearne, M.. The impact of decellularization methods on extracellular matrix derived hydrogels. Sci Rep 9,14933, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gomez, G., Ramirez, C.D., Rivera, J., et al. TGF-beta 1 inhibits mast cell Fc epsilon RI expression, J Immunol 174, 5987, 2005 [DOI] [PMC free article] [PubMed]
- 59.Kashyap, M., Bailey, D.P., Gomez, G., Rivera, J., Huff, T.F., and Ryan, J.J.. TGF-beta1 inhibits late-stage mast cell maturation. Exp Hematol 33,1281, 2005 [DOI] [PubMed] [Google Scholar]
- 60.Caslin, H.L., Kiwanuka, K.N., Haque, T.T., et al. . Controlling mast cell activation and homeostasis: work influenced by Bill Paul that continues today. Front Immunol 9,868, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zaher, H., Shaker, O.G., El-Komy, M.H.M., El-Tawdi, A., Fawzi, M., and Kadry, D.. Serum and tissue expression of transforming growth factor beta 1 in psoriasis. J Eur Acad Dermatol Venereol 23,406, 2009 [DOI] [PubMed] [Google Scholar]
- 62.Hawinkels, L.J., Verspaget, H.W., van Duijn, W., et al. . Tissue level, activation and cellular localisation of TGF-beta1 and association with survival in gastric cancer patients. Br J Cancer 97,398, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rottem, M., Barbieri, S., Kinet, J.P., and Metcalfe, D.D.. Kinetics of the appearance of Fc epsilon RI-bearing cells in interleukin-3-dependent mouse bone marrow cultures: correlation with histamine content and mast cell maturation. Blood 79,972, 1992 [PubMed] [Google Scholar]
- 64.Kinet, J.P.The high-affinity IgE receptor (Fc epsilon RI): from physiology to pathology. Annu Rev Immunol 17,931, 1999 [DOI] [PubMed] [Google Scholar]
- 65.Kirshenbaum, A.S., Petrik, A., Walsh, R., et al. . A ten-year retrospective analysis of the distribution, use and phenotypic characteristics of the LAD2 human mast cell line. Int Arch Allergy Immunol 164,265, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson, T.D., Dequach, J.A., Gaetani, R., et al. . Human versus porcine tissue sourcing for an injectable myocardial matrix hydrogel. Biomater Sci 2014,60283D, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Neill, J.D., Anfang, R., Anandappa, A., et al. . Decellularization of human and porcine lung tissues for pulmonary tissue engineering. Ann Thorac Surg 96, 1046; discussion 1055, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Discher, D.E., Janmey, P., and Wang, Y.L.. Tissue cells feel and respond to the stiffness of their substrate. Science 310,1139, 2005 [DOI] [PubMed] [Google Scholar]
- 69.Xia, T.T., Liu, W.Q., and Yang, L.. A review of gradient stiffness hydrogels used in tissue engineering and regenerative medicine. J Biomed Mater Res Part A 105,1799, 2017 [DOI] [PubMed] [Google Scholar]
- 70.Handorf, A.M., Zhou, Y., Halanski, M.A., and Li, W.J.. Tissue stiffness dictates development, homeostasis, and disease progression. Organogenesis 11,1, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang, H.W., Liu, X.Y., Shen, Z.F., et al., An investigation of the distribution and location of mast cells affected by the stiffness of substrates as a mechanical niche. Int J Biol Sci 14, 1142, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McDevitt, C.A., Wildey, G.M., and Cutrone, R.M.. Transforming growth factor-beta1 in a sterilized tissue derived from the pig small intestine submucosa. J Biomed Mater Res A 67,637, 2003 [DOI] [PubMed] [Google Scholar]
- 73.Sanjabi, S., Oh, S.A., and Li, M.O.. Regulation of the immune response by TGF-beta: from conception to autoimmunity and infection. Cold Spring Harb Perspect Biol 9,a022236, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guhl, S., Babina, M., Neou, A., Zuberbier, T., and Artuc, M.. Mast cell lines HMC-1 and LAD2 in comparison with mature human skin mast cells—drastically reduced levels of tryptase and chymase in mast cell lines. Exp Dermatol 19,845, 2010 [DOI] [PubMed] [Google Scholar]
- 75.Cho, S.H., Lee, S.H., Kato, A., et al. . Cross-talk between human mast cells and bronchial epithelial cells in plasminogen activator inhibitor-1 production via transforming growth factor-beta1. Am J Respir Cell Mol Biol 52,88, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olsson, N., Piek, E., ten Dijke, P., and Nilsson, G.. Human mast cell migration in response to members of the transforming growth factor-beta family. J Leukoc Biol 67,350, 2000 [DOI] [PubMed] [Google Scholar]
- 77.Bissonnette, E.Y., Enciso, J.A., and Befus, A.D.. TGF-beta1 inhibits the release of histamine and tumor necrosis factor-alpha from mast cells through an autocrine pathway. Am J Respir Cell Mol Biol 16,275, 1997 [DOI] [PubMed] [Google Scholar]
- 78.Zhao, W., Gomez, G., Yu, S.H., Ryan, J.J., and Schwartz, L.B.. TGF-beta1 attenuates mediator release and de novo kit expression by human skin mast cells through a Smad-dependent pathway. J Immunol 181,7263, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rojas, A., Zhang, P., Wang, Y., et al. . A positive TGF-beta/c-KIT feedback loop drives tumor progression in advanced primary liver cancer. Neoplasia 18,371, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chatterjee, A., Ghosh, J., and Kapur, R.. Mastocytosis: a mutated KIT receptor induced myeloproliferative disorder. Oncotarget 6,18250, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cruse, G., Metcalfe, D.D., and Olivera, A.. Functional deregulation of KIT: link to mast cell proliferative diseases and other neoplasms. Immunol Allergy Clin North Am 34,219, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takeuchi, Y., Fujino, Y., Watanabe, M., et al. . Aberrant autophosphorylation of c-Kit receptor in canine mast cell tumor cell lines. Vet Immunol Immunopathol 137,208, 2010 [DOI] [PubMed] [Google Scholar]
- 83.Garman, S.C., Kinet, J.P., and Jardetzky T.S.. Crystal structure of the human high-affinity IgE receptor. Cell 95,951, 1998 [DOI] [PubMed] [Google Scholar]
- 84.Reing, J.E., Brown, B.N., Daly, K.A., et al. . The effects of processing methods upon mechanical and biologic properties of porcine dermal extracellular matrix scaffolds. Biomaterials 31,8626, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wolf, M.T., Daly, K.A., Brennan-Pierce, E.P., et al. . A hydrogel derived from decellularized dermal extracellular matrix. Biomaterials 33,7028, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.