Abstract
CircRNAs, a class of endogenous non-coding RNAs with closed-loop structures, have attracted increasing attention because of their good stability, high specificity of tissue expression, long half-life, and highly conserved sequence. CircRNAs have multiple biological functions, including miRNA sponge, transcription regulator, protein translation, interaction with protein, RNA maturation, and so on. These functions indicate the important role of circRNAs in tumorigenesis and malignant progression and their potential as potent diagnostic biomarkers and therapeutic molecules. In recent years, an increasing body of evidence suggests that circRNAs play a crucial role in proliferation, migration, invasion, and apoptosis of lung cancer cells. Therefore, circRNAs have gradually become a research focus in the diagnosis and treatment of lung cancer patients. This review summarizes the classification, biogenesis, and function of circRNAs, and discusses the role of circRNAs in the diagnosis, prognosis, and treatment of lung cancer patients.
Keywords: circRNA, lung cancer, diagnosis, prognosis, treatment
Introduction
According to data reported by a cancer journal, approximately 2.2 million new lung cancer (LC) cases worldwide make it the second cancer (11.4% of all cancers), and approximately 1.8 million deaths place lung cancer at the top of the list of cancer-related deaths (18.0% of all cancer deaths, much more than other cancers).1 Lung cancers are generally divided into two pathological types: 85% of the total cases are non-small cell lung cancer (NSCLC), and 15% of the total cases is small cell lung cancer (SCLC).2 NSCLC can be mainly classified into three types: lung adenocarcinoma (LUAD; 40%), lung squamous cell carcinoma (LUSC; 25%), and large cell lung carcinoma (LCLC; 10%).1 Although advances in diagnosis and treatment have improved the survival rates of lung cancer patients, the 5-year survival rate of advanced lung cancer patients is only 17.7%.3 Moreover, the survival rate of patients with early stage LC is significantly higher than that of patients with advanced LC. Therefore, more effective biomarkers are crucial for early diagnosis and prediction of prognosis.
Circular RNAs (circRNAs), a class of endogenous non-coding RNAs, were first discovered in RNA viruses via electron microscopy in 1976.4 In contrast to linear RNAs, circRNAs have no 5ʹ to 3ʹ polarity and a polyadenylated tail but form a highly stable covalent closed-loop structure, which makes them more stable in tissues and plasma.5 Many studies have confirmed the crucial role of circRNAs in the development and progression of various cancers.6 Furthermore, an increasing body of research shows that abnormal expression of circRNAs plays an important role in the development and progression of LC.7 This suggests that circRNAs can function as potential targets for the diagnosis and prognosis of LC. The current review introduces the classification and biogenesis of circRNAs, and summarizes the roles and corresponding mechanisms of many discovered circRNAs in lung cancer, and discusses the potential value of circRNAs as diagnostic and prognostic biomarkers for lung cancer.
Classification and Biogenesis of circRNAs
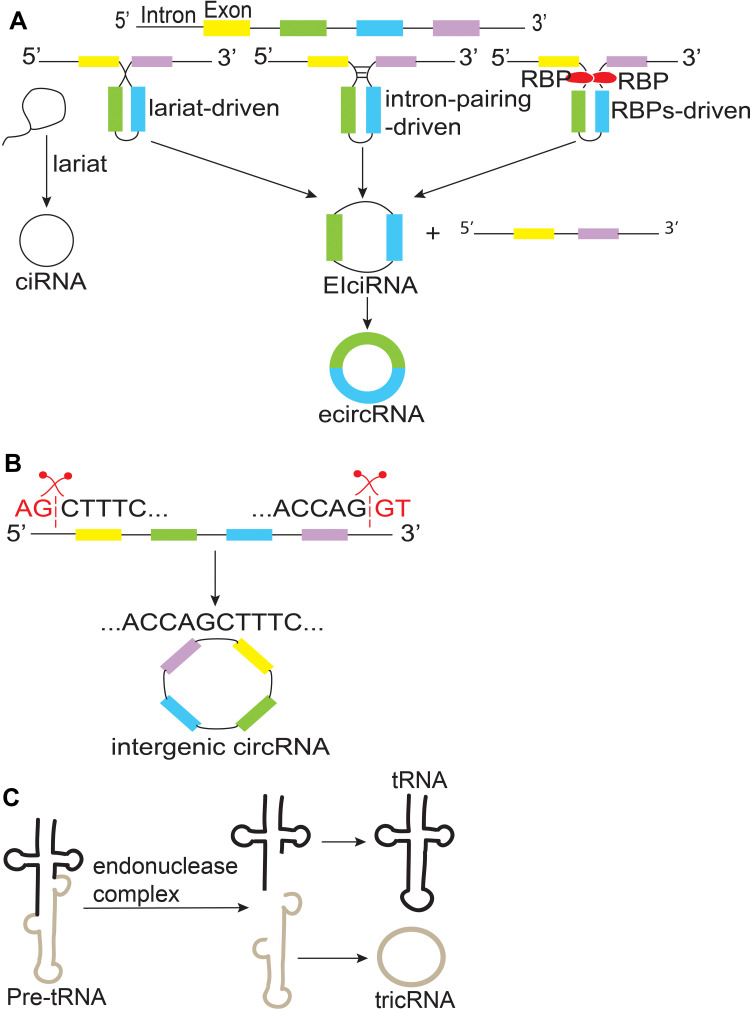
Based on their origins and compositions, circRNA can be mainly classified into six categories (Table 1).
Table 1.
Classification of circRNAs
Exonic circRNAs (ecircRNAs)
EcircRNA consists of one or multiple exons, which is the most common type (more than three-fourths of the whole).8 There are two types of ecircRNA formation, namely, direct back-splicing, and exon skipping.9 The direct back-splicing model includes two mechanisms, intron-pairing-driven, and RNA-binding proteins (RBPs)-driven circularization, while lariat-driven circularization is the main mechanism in the exon-skipping model.10
Intronic circRNA (ciRNAs)
CiRNAs are only composed of introns that have failed to be debranched and abundantly distributed in the nucleus. Furthermore, the processing of ciRNAs mainly depends on consensus sequence motif that might be essential for escaping from de-branching.11
Exonic-Intronic circRNAs (EIciRNAs)
EIciRNAs consist of exons and introns that are retained between the exons.12 That is to say, in circular RNA containing several exons and introns, introns were not removed.
Intergenic circRNAs
The two intronic RNA fragments with GT-AG splicing signal act as splicing donor and acceptor to form the entire circRNA.13
tRNA Intronic circRNAs (tricRNAs)
Pre-tRNAs, which containing introns, are cut by endonuclease complex and forming a special type of intronic circRNAs and mature tRNAs.14
Antisense circRNAs
Mechanically speaking, they are also a type of ecircRNAs, but are formed from antisense non-coding RNAs.15
Schematic of circRNAs biogenesis is shown in Figure 1.
Figure 1.
Biogenesis of circRNAs. (A) CircRNAs are mainly produced by three mechanisms: lariat-driven, intron-pairing-driven, RNA-binding proteins (RBPs)-driven circularization. (B) Inter-genic circRNAs are spliced by two intronic circRNA fragments containing GT-AG splicing signals. (C) TricRNAs are produced from pre-tRNAs that containing introns.
Functions of circRNAs
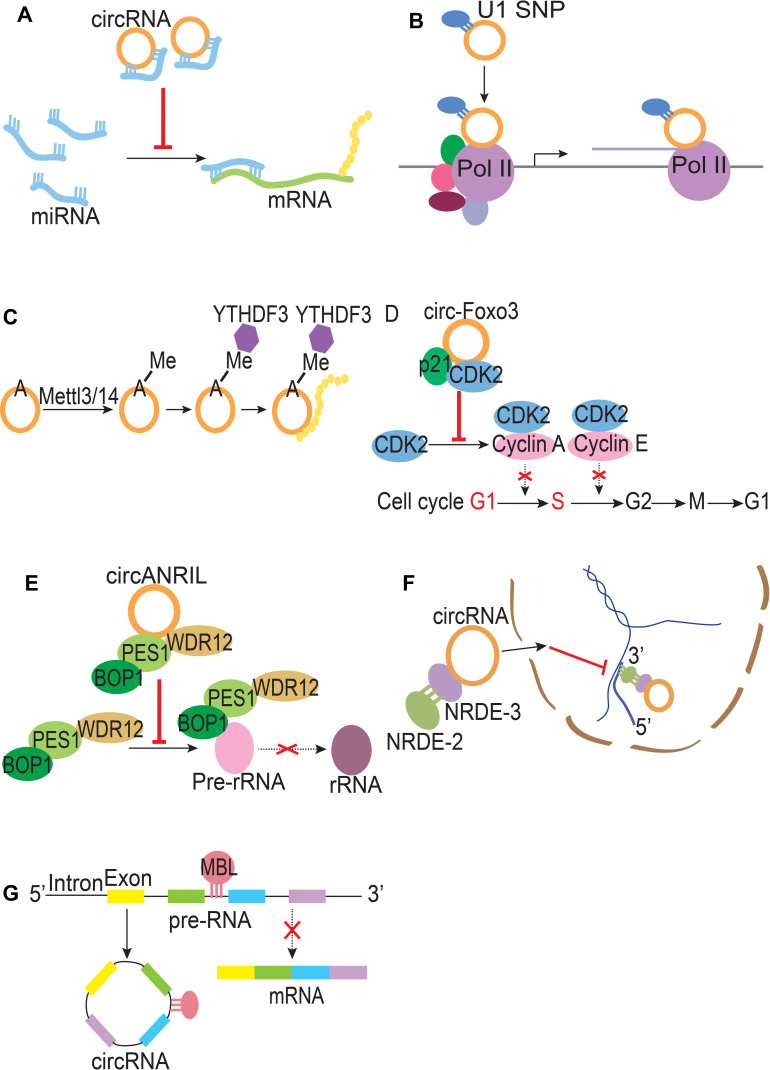
miRNA Sponge
MiRNA is an important negative regulator of protein expression, which inhibits the translation of target gene mRNA or promotes the degradation of mRNA by binding to mRNA.18 Many studies have demonstrated that circRNAs can act as competitive endogenous RNAs (ceRNAs), namely miRNA sponges, which bind to miRNAs by means of microRNA response elements (MREs) to inhibit the function of mRNAs.19,20 For example, CDR1As were observed to act as miRNA sponges, with more than 70 conserved binding sites of miRNA-7, and overexpression of ciRS-7 competitive binding to miR-7 and facilitated malignant progression in esophageal squamous cell carcinoma (ESCC).21 Moreover, circRNA-FOXO3 inhibits proliferation, migration, and invasion of NSCLC cells through specifically sponging miR-155 and abrogating the inhibition of FOXO3 gene.22 In addition, circ-HIPK3 can both facilitate tumor progression by regulating miR-421/ZIC5 axis in glioma and promote epithelial–mesenchymal transition (EMT) of cervical cancer through sponging miR-338-3p to release HIF-1α gene.23,24 It can also inhibit tumorigenesis of hepatocellular carcinoma via the miR-582-3p/DLX2 axis.25 Therefore, one circRNA can sponge multiple miRNAs to play a role in promoting or inhibiting tumorigenesis in different cancers.
Translation Regulator
Further evidence has suggested that circRNAs can be involved in regulating gene transcription. Compared with ecircRNAs, which are mainly located in the cytoplasm, EIciRNAs and ciRNAs are predominantly located in the nucleus and do not sponge miRNA, so they are normally involved in regulating gene expression levels. ElciRNAs may form ElciRNA-U1 snRNA complexes through specific RNA–RNA interactions with U1 snRNA, and then interact with the promoters of the Pol II transcription complex to promote gene expression.12 Similar to EIciRNAs, cirRNAs also can interact with polymerase II complex, such as c-siRT7, which down-regulates the expression of the corresponding genes ANKRD52 and SIRT7 by interacting with the polymerase II complex.11 Thus, circRNA plays a crucial role in the regulation of gene expression.
Protein Translation
CircRNAs were initially considered to be non-coding RNAs, but more and more evidence has emerged that some circRNAs can also encode proteins. As long as circRNA has a complete and sufficiently long open reading framework, and with the help of some necessary regulatory elements, such as N6-methyladenosine (m6A) modifications and the internal ribosome entry site (IRES) element, it can be translated into proteins, but the translation efficiency may be slightly lower than that of linear RNA.26–28 As early as 1986, it was found that the circRNA of the hepatitis δ virus can generate a kind of protein with 122 amino acids.29 Then, circ-ZnF609 was found to translate proteins into mouse myoblasts when driven by IRES.30 In addition, other studies have indicated that circular RNA molecules can be translated into protein even without any IRES sequence, poly A, or cap structure.31 All the above findings prove that circRNAs do have the ability to encode proteins.
Interaction with Protein
CircRNAs can bind to and interact with RNA-binding proteins (RBPs) through conserved protein-binding sites. For instance, CDR1as and circSry can bind with the miRNA effector AGO to cleave it and eventually promote its degradation.15 CDK2 is essential for G1-S phase transition, the formation of circ-Foxo3–p21–CDK2 complex would inhibit the function of CDK2; then, cell cycle progression would be arrested in G1 phase.32 In addition, circ-Foxo3 could bind to senescence-related proteins ID1 and E2F1 and stress-related proteins HIF1α and FAK to decrease levels of these proteins in the nucleus, block the anti-senescent function of these proteins and promote cellular senescence.33 In summary, the interaction between circRNAs and proteins can alter the biological activity of proteins to affect their biological function or change the subcellular localization of proteins.
RNA Maturation
A variety of circRNA–protein interactions can be used to control ribosomal RNA maturation. For example, circANRIL binding to the C-terminal lysine-rich domain of PES1 hinders pre-rRNA binding and exonuclease-mediated rRNA maturation, thereby circANRIL impairs ribosome biogenesis, leading to activation of p53, promoting cell apoptosis and decreasing cell proliferation.17 Consequently, circRNAs can be involved in regulating rRNA maturation.
Inhibit RNA Polymerase II Elongation
Acting as endogenous small regulatory RNAs, circRNAs can interfere with gene expression in the nucleus. The complex formed by circRNAs with NRDE-3 associates with NRDE-2 and recruits it into the nucleus, where it inhibits RNAP II during the elongation phase of transcription.34 These nuclear-localized circRNAs direct an NRDE-2-dependent silencing of pre-mRNAs 3ʹ to sites of RNAi, thus inhibiting gene expression during the elongation phase of transcription. This is also a part of the regulation of gene expression.
Modulate Linear Splicing
CircRNAs are generally produced by pre-mRNA spliceosome.35 CircRNAs production competes with linear splicing of flanking exons, and they can mutually regulate each other by competing for splice sites. Splicing factor-mediated exon circularization replaces linear splicing, thereby reducing the production of its mRNA. In brief, these competition effects might regulate the levels of both circRNAs and mRNAs.
The functions of circRNA are shown in Figure 2
Figure 2.
Function of circRNAs. (A) miRNA sponge (B) Translation regulator (C) Protein translation (D) Protein sponge (E) RNA maturation (F) Inhibit RNA polymerase II elongation (G) Modulate linear splicing.
CircRNAs in Lung Cancer
Advanced studies suggest that circRNAs may play an important role in the progression and development of lung cancer. CircRNAs can be involved in regulating the proliferation, migration, invasion, and apoptosis of tumor cells, and play a crucial role in the diagnosis, prognosis, and treatment of lung cancer.
CircRNAs as Potential Diagnostic Biomarkers in Lung Cancer
Compared with other non-coding RNAs, circRNAs have higher tolerance to RNA exonuclease due to their covalently closed structure.36 Therefore, circRNAs can function as potential diagnostic biomarkers for lung cancer by virtue of their stable structure, high abundance, and tissue-specific expression (Table 2).
Table 2.
CircRNAs as Potential Diagnostic Biomarkers in Lung Cancer
| circRNA | Function | Mechanism | Expression Pattern | Cell Characteristics | Clinical Characteristics | References |
|---|---|---|---|---|---|---|
| circ-ITCH | miRNA sponge | circ-ITCH↓- miR-7↑/ miR-214↑-Wnt/β-catenin↑ | Down | Proliferation↓ | Age (≥60), TNM | [37] |
| circPVT1 | miRNA sponge | circPVT1↑-miR-125b↓-E2F2 pathway↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | Distant metastasis | [38] |
| circMET | miRNA sponge | circMET↑-miR-145-5p↓-CXCL3↑ | Up | Proliferation↑, migration↑, invasion↑ | TNM, LNM, tumor differentiation, OS | [39] |
| circGFRA1 | miRNA sponge | circGFRA1↑-miR-188-3p↓-PI3K/AKT↑ | Up | Proliferation↑ | Unknown | [40] |
| hsa_circ_0013958 | miRNA sponge | hsa_circ_0013958↑-miR-134↓-cyclin D1↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | TNM, LNM | [41] |
| circ_0005280 | Unknown | Unknown - | Down | Unknown | Age (≥60), tumor size | [62] |
| circRNA100146 | miRNA sponge | circRNA100146↑- miR-361-3p ↓/miR-615-5p↓-SF3B3↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | Unknown | [66] |
| hsa_circ_0001946 | miRNA sponge | hsa_circ_0001946↓-NER signaling pathway↑ | Down | Proliferation↓, migration↓, invasion↓, apoptosis↑, cisplatin sensitivity↑ | Unknown | [70] |
| hsa_circ_0030998 | miRNA sponge | hsa_circ_0030998↓-miR-558↑-MMP1/MMP17↓ | Down | Proliferation↓, migration↓, invasion↓, Taxol resistance↓ | TNM, distal metastasis | [72] |
| circFARSA | miRNA sponge | circFARSA↑-miR-330-5p↓/miR-326↓-FASN↑ | Up | Migration↑, invasion↑ | Gender | [73] |
| circ-CCS | miRNA sponge | circ-CCS↑-miR-383↓-E2F7↑ | Up | Proliferation↑, migration↑, apoptosis↓ | TNM, LNM, tumor size | [77] |
| circ-IGF1R | miRNA sponge | circ-IGF1R↓–miR-1270↑–VANGL2↓ | Down | Migration↓, invasion↓ | LNM, tumor size | [82] |
| circRNA_102179 | miRNA sponge | circRNA_102179↑-miR-330-5p↓-HMGB3↑ | Up | Proliferation↑, migration↑, invasion↑ | Unknown | [95] |
| circSATB2 | miRNA sponge | circSATB2↑-miR-326↓-FSCN1↑ | Up | Proliferation↑, migration↑, invasion↑ | LNM | [96] |
| circ-ZKSCAN1 | miRNA sponge | circ-ZKSCAN1↑-miR-330-5p↓-FAM83A↑ | Up | Proliferation↑, migration↑ | OS, tumor size, clinical stage | [97] |
| hsa_circ_0007059 | miRNA sponge | hsa_circ_0007059↓-miR-378↑-Wnt/β-catenin↑/ERK1/2↑ | Down | Proliferation↓, EMT↓, apoptosis↑ | TNM, LNM | [99] |
| circ-PITX1 | miRNA sponge | circ-PITX1↑-miR-1248↓-CCND2↑ | Up | Proliferation↑, migration↑, invasion↑, glycolysis↑, glutamine metabolism↑, apoptosis↓ | Unknown | [126] |
| circ_0000429 | miRNA sponge | circ_0000429↑-miR-1197↓-MADD↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | Unknown | [132] |
| circ_0001287 | miRNA sponge | circ_0001287↓-miR-21↑-PTEN↓ | Down | Proliferation↓, migration↓, invasion↓, radio-resistance↓ | N status, histological grade | [136] |
| hsa_circ_0000064 | Unknown | hsa_circ_0000064↑-caspase-3/9↑/ bax↑/p21↑/CDK6↑/cyclin D1↑/bcl-2↓/MMP-2/9↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | TNM, LNM, T stage | [147] |
Notes: ↑, up-regulated; ↓, down-regulated.
Abbreviations:TNM, the high stage of tumor node metastasis; LNM, lymph node metastasis; OS, overall survival.
circ-ITCH
Circ-ITCH can sponge of miR-7 and miR-214 and inhibit the activation of the Wnt/β-catenin pathway in lung cancer, and thus regulate lung cancer cell proliferation.37 Circ-ITCH serves as epigenetic miRNA sponges to competitively inhibit the binding between miRNA and ITCH, thereby increasing ITCH expression. ITCH can promote proteasome degradation and inhibit Wnt/β-catenin pathway. Thus, it can be concluded that circ-ITCH can act as a tumor suppressor gene in lung cancer by controlling miRNA activity, which increases the concentration of ITCH and results in suppression of the canonical Wnt/β-catenin pathway. Consequently, the related research of circ-ITCH is important for the diagnosis and treatment of cancer.
circPVT1
CircPVT1 can sponge miR-125b and promote E2F2 expression in NSCLC.38 C-Fos upregulated circPVT1 in NSCLC and decreased the Ago2-based activity of miR-125b, which increased the E2F2 and the downstream effector expression. Overexpressed E2F2 is crucial for cell cycle regulation in NSCLC and is significantly associated with poor prognosis. In summary, c-Fos-activated circPVT1 acts as a ceRNA and may be considered as a diagnostic biomarker and therapeutic target for NSCLC patients.
circMET
CircMET primarily sponge miR-145-5p to regulate CXCL3 expression in NSCLC cells39 and is significantly upregulated in NSCLC tissues. Thus, circMET promotes the expression level of CXCL3. CXCL3 is a well-known oncogene, which has been confirmed to promote cancer progression. Taken together, circMET might be considered a novel diagnostic biomarker and potential therapeutic target for NSCLC treatment.
circGFRA1
The expression of circGFRA1 in NSCLC tissues increased, consistent with the previous data in breast cancer and ovarian cancer, and was negatively correlated with the expression of miR-188-3p.40 CircGFRA1 acts as a ceRNA to sponge miR-188-3p, and the circGFRA1/miR-188-3p axis may regulate the proliferation of NSCLC cells through the PI3K/AKT signaling pathway, which is a classical oncogenic signaling pathway. Consequently, circGFRA1 plays a crucial role in NSCLC, and might be a potential diagnostic biomarker and therapeutic target for NSCLC.
hsa_circ_0013958
hsa_circ_0013958 could act as a sponge of miR-134 and inhibit miR-134 activity.41 Studies have shown that miR-134 could inhibit the expression of CCND1. It promotes G1-S progression by sequentially phosphorylating retinoblastoma proteins, thereby promoting the initiation and progression of tumor cells. In conclusion, the up-regulated expression of hsa_circ_0013958 is closely related to the tumorigenesis of NSCLC and could be used as a potential diagnostic biomarker for NSCLC, especially for early lung adenocarcinoma.
CircRNAs as Potential Prognostic Biomarkers in Lung Cancer
Prognostic assessment plays a momentous role in the treatment of lung cancer, which can help prolong the survival of lung cancer patients. Hence, circRNAs have gradually gained some value as potential prognostic biomarkers for lung cancer42 (Table 3).
Table 3.
CircRNAs as Potential Prognostic Biomarkers in Lung Cancer
| circRNA | Function | Mechanism | Expression Pattern | Cell Characteristics | Clinical Characteristics | References |
|---|---|---|---|---|---|---|
| circFGFR3 | miRNA sponge | circFGFR3↑-miR‐22‐3p↓-Gal‐1↑/p‐AKT↑/p‐ERK1/2↑ | Up | Proliferation↑, invasion↑ | TNM, LNM, tumor size, tumor differentiation, OS | [43] |
| circ_0003645 | miRNA sponge | circ_0003645↑-miR-1179↓-TMEM14A↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | TNM, LNM, OS | [46] |
| CDR1as | miRNA sponge | CDR1as↑-miR-7↓-EGFR↑/CCNE1↑/PIK3CD↑ | Up | Proliferation↑, apoptosis↓ | TNM, LNM, OS, tumor size | [47,59] |
| circ_POLA2 | miRNA sponge | circ_POLA2↑-miR-326↓-GNB1↑ | Up | Unknown | Distant metastasis, TNM, OS | [48] |
| circ-FOXM1 | miRNA sponge | circ-FOXM1↑-miR-1304-5p↓-PPDPF↑/MACC1↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | TNM, LNM | [49] |
| circPIP5K1A | miRNA sponge | circPIP5K1A↑-miR‐600↓-HIF‐1α↑ circPIP5K1A↑-miR‑101↓-ABCC1↑ |
Up | Proliferation↑, migration↑, invasion↑, apoptosis↓, EMT↑, cisplatin sensitivity↓ | Unknown | [86,87] |
| circRNA_010763 | miRNA sponge | circRNA_010763↑-miR-715↓-c-Myc↑ | Up | Proliferation↑, migration↑, invasion↑ | Unknown | [93] |
| circRNA_100876 | Unknown | Unknown | Up | Unknown | LNM, tumor staging, OS | [101] |
| circ-ANXA7 | miRNA sponge | circ-ANXA7↑- miR-331↓-LAD1↑ | Up | Proliferation↑, migration↑, invasion↑ | TNM, LNM, tumor size, recurrence status, OS | [119] |
| circ‑PTEN | miRNA sponge | circ‑PTEN↓-miR-155↑/miR-330-3p↑-PTEN↓ | Down | Proliferation↓ | TNM, tumor size, OS | [140] |
| hsa_circ_0008003 | miRNA sponge | hsa_circ_0008003↑-miR-488↓-ZNF281↑ | Up | Proliferation↑, migration↑, invasion↑ | TNM, LNM | [142] |
| circ-MTHFD2 | Unknown | Unknown | Up | Unknown | TNM, LNM, tumor size, recurrence, smoking history, OS | [157] |
Notes: ↑, up-regulated; ↓, down-regulated.
Abbreviations: TNM, the high stage of tumor node metastasis; LNM, lymph node metastasis; OS, overall survival.
circFGFR3
The high expression of circFGFR3 promotes the invasion and proliferation by sponging miR‐22‐3p to upregulate the Gal‐1, p‐AKT, and p‐ERK1/2 expressions in NSCLC cells.43 miR‐22‐3p acts as a tumor suppressor gene in multiple cancers, including NSCLC. Gal‐1, p‐AKT, and p‐ERK1/2 are downstream regulators of miR‐22‐3p in NSCLC cells. The AKT and ERK1/2 pathways activated abnormally by forced expression of Gal‐1 are closely related to tumor proliferation and invasion.44,45 Therefore, circFGFR3 may upregulate the Gal‐1 expression and activate the AKT and ERK1/2 pathways to promote NSCLC cell invasion and proliferation, and it may be a novel biomarker for the prognosis of lung cancer.
circ_0003645
Circ_0003645 was both increased in the NSCLC cells and tissues, and functions as an oncogene in NSCLC.46 Circ_0003645 could upregulate TMEM14A expression by acting as a ceRNA to sponge miR-1179. Circ_0003645 affects cell growth, apoptosis, and metastatic properties, and thus participates in the initiation and progression of NSCLC. Elevated circ_0003645 expression in NSCLC tissues was related to advanced TNM stages and positive lymph node invasion. Additionally, a high expression of circ_0003645 results in a worse overall survival. Therefore, circ_0003645 may possess significant potential as a prognostic predictor and therapeutic target for patients with NSCLC.
CDR1as
Plenty of research evidence has suggested that circRNA CDR1as could function as an oncogene through sponge of tumor suppressor miR-7.21 Tumor suppressor miR-7 can induce apoptosis and G1/S arrest, while CDR1AS can negatively regulate the antitumor effects of miR-7.47 The cell growth-related target gene expression of miR-7, EGFR, CCNE1, and PIK3CD can be remarkably elevated by the overexpression of CDR1as. That is to say, CDR1as could promote cell growth via the miR-7/EGFR/CCNE1/PIK3CD signaling pathway in NSCLC. Consequently, CDR1as may serve as a novel prognostic marker for NSCLC.
circ_POLA2
Circ_POLA2 is significantly upregulated in lung cancer cells and tissues, and promotes lung cancer progression via miR-326/GNB1 axis.48 GNB1 is a direct target of miR-326 and necessary for the miR-326-mediated suppression on the stemness of lung cancer cells. In conclusion, circ_POLA2 plays a potential regulatory role via regulating the miR-326/GNB1 axis in lung cancer cell stemness. Meanwhile, lung cancer patients with a high expression of circ_POLA2 exhibited a worse overall survival. Thus, circ_POLA2 may act as a potential prognostic biomarker and therapeutic target for lung cancer treatment.
circ-FOXM1 (hsa_circ_0025033)
The expression of circ-FOXM1 is remarkably increased in NSCLC tissues and cell lines and promotes NSCLC cell progression.49 Circ-FOXM1 can upregulate PPDPF and MACC1 expression via acting as a sponge of miR-1304-5p to facilitate cell growth and invasion. Previously, studies indicated that PPDPF and MACC1 were elevated and involved in tumor invasion and metastasis in different types of malignancies.50,51 In addition, the high expression of circ-FOXM1 was tightly connected to advanced TNM stages, lymph node invasion, and poor prognosis. To sum up, circ-FOXM1 may play a crucial role in the progression of NSCLC, which could function as a novel potential prognostic marker and therapeutic target for NSCLC.
CircRNAs as Potential Therapeutic Target in Lung Cancer
With further research on circRNAs, many circRNAs have been proven to be involved in the initiation and progression of lung cancer, and their potential targets and mechanisms in the treatment of lung cancer have also been gradually known to the public. The correlational research of circRNAs may contribute to further breakthroughs in the treatment of lung cancer (Table 4).
Table 4.
CircRNAs as Potential Therapeutic Target in Lung Cancer
| circRNA | Function | Mechanism | Expression Pattern | Cell Characteristics | Clinical Characteristics | References |
|---|---|---|---|---|---|---|
| circ_0003998 | miRNA sponge | circ_0003998↑- miR-136-5p↓-CORO1C↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓, dtx sensitivity↓ | TNM, LNM, tumor metastasis | [52] |
| circRNA_103762 | Unknown | circRNA_103762↑-CHOP↓ | Up | Proliferation↑, migration↑, invasion↑, mdr↑ | OS | [53] |
| hsa_circ_0020123 | miRNA sponge | hsa_circ_0020123↑-miR-144↓-ZEB1↑/EZH2↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | TNM, LNM, tumor differentiation, OS | [56] |
| circSEC31A | miRNA sponge | circSEC31A↑-miR-376a↓-SEC31A↑ | Up | Migration↑, invasion↑, glycolysis↑, apoptosis↓ | TNM, LNM, tumor size | [57] |
| circCDYL | miRNA sponge | circCDYL↓-miR-185-5p↑/TNRC6A↓-ERK1/2↑ | Down | Proliferation↓, apoptosis↑ | Unknown | [58] |
| circRNA-FOXO3 | miRNA sponge | circRNA-FOXO3↓-miR-155↑-FOXO3↓ | Down | Proliferation↓, migration↓, invasion↓, apoptosis↑ | Unknown | [22] |
| circFADS2 | miRNA sponge | circFADS2↑-miR-498↓-HMGA2↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | TNM, LNM, tumor differentiation, OS | [60,61] |
| hsa_circ_100395 | miRNA sponge | hsa_circ_100395↓-miR-1228↑- TCF21↓ | Down | Proliferation↓, migration↓, invasion↓ | TNM, LNM, OS | [63] |
| circRNA_102231 | Unknown | Unknown | Up | Proliferation↑, migration↑, proliferation↑, | TNM, LNM | [64] |
| circUBAP2 | miRNA sponge | circUBAP2↑-miR-3182↓- KLF4↑ | Up | Proliferation↑, migration↑, invasion↑, chemo-resistance↑ | LNM, disease stage | [65] |
| hsa_circ_0008305 (circPTK2) | miRNA sponge | circPTK2↓-miR-429↑/miR-200b-3p↑-TIF1γ↓ | Down | Invasion↓, migration↓, emt↓ | Unknown | [67] |
| circFGFR1 | miRNA sponge | circFGFR1↑- miR-381-3p↓-CXCR4↑ | Up | Proliferation↑, migration↑, invasion↑ | Tumor size | [68] |
| circ-ENO1 | miRNA sponge | circ-ENO1↑-miR-22-3p↓-ENO1↑ | Up | Proliferation↑, migration↑, apoptosis↓, glycolysis↑, emt↑ | Unknown | [69] |
| hsa_circRNA_103809 | miRNA sponge | hsa_circRNA_103809↑-miR-4302↓-ZNF121↑/MYC↑ hsa_circRNA_103809↑-miR-377-3p↓-GOT1↑ |
Up | Proliferation↑, invasion↑, apoptosis↓, cisplatin resistance↑ | Unknown | [71,158] |
| circ_0026134 | miRNA sponge | circ_0026134↑-miR-1256↓/miR-1287↓-TCTN1↑/GAGE1↑ | Up | Proliferation↑, migration↑, invasion↑ | Unknown | [74] |
| circ0001320 | miRNA sponge | circ0001320↓- miR-558↑-TNFAIP1↓/ TPM1↓ | Down | Proliferation↓, invasion↓, apoptosis↑ | Unknown | [75] |
| circ-BANP | miRNA sponge | circ-BANP↑- miR-503↓-LARP1↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | Clinical stages, LNM, OS | [76] |
| circHIPK3 | miRNA sponge | circHIPK3↑-miR124-3p↓-STAT3↓-PRKAA↑(STK11 mutant) circHIPK3↑-miR-107↓-BDNF↑ circHIPK3↑-miR-381-3p↓-AKT/mTOR↑ circHIPK3↑- miR-149↓- FOXM1↑ |
Up | Proliferation↑, migration↑, invasion↑, glycolysis↑, apoptosis↓ | TNM, LNM, tumor size | [78–81] |
| circMAN2B2 | miRNA sponge | circMAN2B2↑-miR-1275↓-FOXK1↑ | Up | Proliferation↑, invasion↑ | Unknown | [83] |
| circNDUFB2 | Protein scaffold | circNDUFB2↓-IGF2BPs↑(m6A-dependent) | Down | Proliferation↓, migration↓, invasion↓, immune responses↑ | LNM, tumor size | [84] |
| circ-PAX2 | miRNA sponge | circ-PAX2↑- miR-186↓ | Up | Proliferation↑, migration↑, apoptosis↓ | TNM | [85] |
| circPTPRA | miRNA sponge | circPTPRA↓-miR-96-5p↑-RASSF8↓/E-cadherin↓ | Down | Migration↓, invasion↓, emt↓ | OS | [88] |
| circBIRC6 | miRNA sponge | circBIRC6↑-miR-145↓- FSCN1↑/S6K1↑ | Up | Proliferation↑, migration↑, invasion↑ | Unknown | [89] |
| circCDR1as | miRNA sponge | circCDR1as↑-miR-219a-5p↓-SOX5↑ circCDR1as↑-miR-641↓-HOXA9↑ |
Up | Proliferation↑, migration↑, invasion↑, apoptosis↓, stemness↑, cisplatin resistance↑ | Unknown | [90,91] |
| circRNA_001010 | miRNA sponge | circRNA_001010↑-miR-5112↓-CDK4↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | Unknown | [92] |
| hsa_circRNA_101237 | miRNA sponge | hsa_circRNA_101237↑-miR-490-3p↓-MAPK1↑ | Up | Proliferation↑, migration↑, invasion↑ | TNM, LNM, OS, tumor size | [94] |
| circ_0002483 | miRNA sponge | circ_0002483↓- miR-182-5p↑-GRB2↓/FOXO1↓/FOXO3↓ | Down | Proliferation↓, invasion↓, taxol sensitivity↑ | OS | [98] |
| hsa_circ_0012673 | miRNA sponge | hsa_circ_0012673↑-miR-320a↓-LIMK18521↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓, emt↑ | Unknown | [100] |
| circTADA2A | miRNA sponge | propofol↑-circTADA2A↓-miR-455-3p↑-FOXM1↓ | Down | Proliferation↑, migration↑, invasion↑, aerobic glycolysis↑ | Unknown | [102] |
| hsa_circ_0002130 | miRNA sponge | hsa_circ_0002130↑-miR-498↓- GLUT1↑/HK2↑/LDHA↑ | Up | Proliferation↑, glycolysis↑, apoptosis↓, osimertinib-resistant↑ | Unknown | [103] |
| circ_0000376 | miRNA sponge | circ_0000376↑-miR-1182↓-NOVA2↑ | Up | Migration↑, invasion↑, glycolysis↑ | TNM, LNM, tumor size | [104] |
| circ_0000735 | miRNA sponge | circ_0000735↑-miR-635↓-FAM83F↑ circ_0000735↑-miR-940↓-BMPER↑ |
Up | Proliferation↑, migration↑, invasion↑, glycolysis↑, apoptosis↓ | TNM, LNM, OS | [105,106] |
| circ_0014130 | miRNA sponge | circ_0014130↑-miR-142-5p↓- IGF-1↑ circ_0014130↑- miR-136-5p↓- BCL2↑ |
Up | Proliferation↑, apoptosis↓ | Tumor size, distant metastasis | [107,108] |
| circ_0016760 | miRNA sponge | circ_0016760↑-miR-4295↓-E2F3↑ circ_0016760↑-miR-1287↓-GAGE1↑ |
Up | Proliferation↑, migration↑, invasion↑, glycolysis↑, apoptosis↓ | TNM, LNM, OS | [109,110] |
| circ_0020123 | miRNA sponge | circ_0020123↑-miR-142-3p↓-ZFX↑ circ_0020123↑- miR-488-3p↓- ADAM9↑ circ_0020123↑-miR-590-5p↓-THBS2↑ |
Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | TNM, LNM | [56,111,112] |
| circ_100565 | miRNA sponge | circ_100565↑- miR-506-3p↓- HMGA2↑ circRNA_100565↑-miR-337↓-3p-ADAM28↑ |
Up | Proliferation↑, migration↑, invasion↑, autophagy↑, apoptosis↓, cisplatin resistance↑ | TNM, LNM, OS | [113,133] |
| circ_ZFR | miRNA sponge | circ_ZFR↑-miR-195-5p↓-KPNA4↑ circ_ZFR↑-miR-101-3p↓-CUL4B↑ |
Up | Proliferation↑, migration↑, invasion↑, apoptosis↓, ptx resistance↑ | Unknown | [114,115] |
| circABCB10 | miRNA sponge | circABCB10↑-miR‐1252↓-FOXR2↑ circABCB10↑-miR-584-5p↓- E2F5↑ |
Up | Proliferation↑, migration↑, invasion↑ | OS | [116,117] |
| circAGFG1 | miRNA sponge | circAGFG1↑-miR-203↓-ZNF281↑ | Up | Proliferation↑, migration↑, invasion↑ | Unknown | [118] |
| circDCUN1D4 | RBPs | circDCUN1D4↓-TXNIP↓/HuR↓ | Down | Migration↓, invasion↓, glycolysis↓ | LNM, OS | [120] |
| circDENND2A | miRNA sponge | circDENND2A↑-miR-34a↓-CCNE1 | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | Unknown | [121] |
| circEPSTI1 | miRNA sponge | circEPSTI1↑-miR-145↓-HMGB3↑ circEPSTI1↑- miR-1248↓- TRIM24↑ |
Up | Proliferation↑, migration↑, invasion↑ | Unknown | [122,123] |
| circFOXP1 | miRNA sponge | circFOXP1↑-miR-185-5p↓- WNT1↑ | Up | Proliferation↑, apoptosis↓ | Unknown | [124] |
| circNFIX | miRNA sponge | circNFIX↑-miR-212-3p↓-ADAM10↑ | Up | Proliferation↑, migration↑, invasion↑, glycolysis↑ | TNM, distant metastasis | [125] |
| circ_EPB41L2 | miRNA sponge | circ_EPB41L2↓-miR-211-5p↑-CDH4↓ | Down | Proliferation↓, migration↓, invasion↓ | Unknown | [127] |
| hsa_circ_0018414 | miRNA sponge | hsa_circ_0018414↓-miR-6807↑-3p-DKK1↓ | Down | Proliferation↓, stemness↓, apoptosis↑ | Unknown | [128] |
| hsa_circ_0087862 | miRNA sponge | hsa_circ_0087862↑-miR-1253↓-RAB3D↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | TNM, LNM, tumor size, OS | [129] |
| circZNF609 | miRNA sponge | circZNF609↑-miR-623↓-FOXM1↑ circZNF609↑-miR‐142‐3p↓-GNB2↑ |
Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | TNM, LNM, OS | [130,131] |
| circ-SOX4 | miRNA sponge | circ-SOX4↑-miR-1270↓-PLAGL2↑/WNT↑ | Up | Proliferation↑, migration↑, invasion↑ | Unknown | [134] |
| circ_0000284 | miRNA sponge | circ_0000284↑-miR-377-3p↓-PD-L1↑ | Up | Proliferation↑, migration↑, invasion↑ | LNM, OS, tumor stage | [135] |
| circ_0074027 | miRNA sponge | circ_0074027↑- miR‐185‐3p↓-BRD4↑/MADD↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | TNM, tumor differentiation, OS | [137] |
| circMAGI3 | miRNA sponge | circMAGI3↑-miR-515-5p↓-HDGF↑ | Up | Proliferation↑, glycolysis↑ | TNM, OS | [138] |
| circ-PRMT5 | miRNA sponge | circ-PRMT5↑-miR-377/382/498↓- EZH2↑ | Up | Proliferation↑ | TNM, LNM, tumor size, OS | [139] |
| hsa_circ_0006571 | miRNA sponge | hsa_circ_0006571↑-miR-138↓-Sirt1↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓, emt↑ | spinal metastasis | [141] |
| circSMARCA5 | miRNA sponge | circSMARCA5↓-miR-19b-3p↑-HOXA9↓ | Down | Proliferation↓, migration↓, invasion↓ | Unknown | [143] |
| circTUBA1C | miRNA sponge | circTUBA1C↑- miR-143-3p↓ | Up | Proliferation↑, apoptosis↓ | Unknown | [144] |
| hsa_circ_0007580 | miRNA sponge | hsa_circ_0007580↑-miR-545-3p↓-PRKCA↑ | Up | Invasion↑, apoptosis↓ | OS | [145] |
| circRNA_103993 | miRNA sponge | circRNA_103993↑-miR-1271↓- ERG↑ | Up | Proliferation↑, apoptosis↓ | Unknown | [146] |
| hsa_circ_0001073 | miRNA sponge | hsa_circ_0001073↓-miR-626↑-LIFR↓ | Down | Proliferation↓, invasion↓, apoptosis↑ | Unknown | [148] |
| hsa_circ_0010235 | miRNA sponge | hsa_circ_0010235↑-miR-433-3p↓-TIPRL↑ | Up | Proliferation↑, migration↑, invasion↑, autophagy↑, apoptosis↓ | TNM, LNM, tumor size, OS, recurrence, smoking history | [149] |
| hsa_circ_0038646 | miRNA sponge | hsa_circ_0038646↑-miR-331-3p↓-GRIK3↑ | Up | Proliferation↑, migration↑ | Unknown | [150] |
| hsa_circ_11780 | miRNA sponge | hsa_circ_11780↓-miR-544a↑-FBXW7↓ | Down | Proliferation↓, migration↓, invasion↓ | Tumor size, distant metastasis, OS | [151] |
| hsa_circ_0002874 | miRNA sponge | hsa_circ_0002874↑-miR-1273f↓-MDM2/P53↑ | Up | Apoptosis↓, paclitaxel resistance↑ | TNM, histology | [152] |
| circCCDC66 | miRNA sponge | circCCDC66↑-miR-33a-5p↓-KPNA4↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | Unknown | [153] |
| circP4HB | miRNA sponge | circP4HB↑-miR-133a-5p↓- vimentin↑ | Up | Migration↑, invasion↑, emt↑ | OS | [154] |
| circARHGAP10 | miRNA sponge | circARHGAP10↑-miR-150-5p↓-GLUT1↑ | Up | Proliferation↑, migration↑, invasion↑, emt↑ | Unknown | [155] |
| circVANGL1 | miRNA sponge | circVANGL1↑-miR-195↓-Bcl-2↑ | Up | Proliferation↑, migration↑, invasion↑, apoptosis↓ | TNM, tumor size, OS | [156] |
Notes: ↑, up-regulated; ↓, down-regulated.
Abbreviations: TNM, the high stage of tumor node metastasis; LNM, lymph node metastasis; OS, overall survival; MDR, multidrug resistance; EMT, epithelial–mesenchymal transition.
circ_0003998
The expression of circ_0003998 has been verified to be associated with NSCLC resistance to docetaxel (DTX).52 The expression level of circ_0003998 was originally significantly increased in DTX-resistant NSCLC tissues and cells, while knockdown of circ_0003998 inhibited cell colony formation and enhanced apoptosis and DTX sensitivity of DTX-resistant NSCLC cells in vitro and in vivo. Meanwhile, circ_0003998 directly sponged miR-136-5p, and CORO1C was a functionally crucial target of miR-136-5p in regulating DTX-resistant NSCLC cell colony formation, apoptosis, and DTX sensitivity. In summary, circ_0003998 modulated CORO1C expression by acting as a ceRNA of miR-136-5p to regulate DTX-resistant NSCLC cell colony formation, apoptosis, and DTX sensitivity at least partially, revealing the potential of circ_0003998 as a therapeutic target for chemoresistant NSCLC.
circRNA_103762
CircRNA_103762 was remarkably highly expressed in NSCLC tissues and cell lines and circRNA_103762 acts as an oncogene in NSCLC.53 A series of cell experiments in vitro determined that the upregulation of circRNA_103762 was associated with multidrug resistance (MDR). The circRNA_103762 was upregulated in NSCLC patients after cisplatin chemotherapy, and downregulation of circRNA_103762 expression can reduce IC50 values of different drugs. Furthermore, circRNA_103762 enhanced MDR by inhibiting CHOP expression in NSCLC cells, which has been pointed out to be related to tumor in early reports, and its expression can be induced by chemotherapeutic drugs and is associated with MDR.54,55 Based on the above results, the correlation between circRNA_103762 and MDR of NSCLC provides new ideas and strategies for the therapy of NSCLC.
hsa_circ_0020123
The expression level of hsa_circ_0020123 in NSCLC tissues was significantly higher than that in normal tissues, and higher hsa_circ_0020123 expression level was associated with a poorer differentiation degree, lymph node metastasis, a higher TNM stage, and a shorter overall survival.56 hsa_circ_0020123 facilitates the growth and metastasis of NSCLC cells through sponging miR-144 to suppress their functions. Thence, the well-known oncogenes EZH2 and ZEB1, which were targeted by miR-144, were upregulated by high-level hsa_circ_0020123. It is worth noting that knockdown hsa_circ_0020123 could dramatically inhibit NSCLC growth and metastasis in nude mice test. Therefore, it can be concluded that hsa_circ_0020123 might serve as a promising prognostic predictor and therapeutic target for NSCLC treatment.
circSEC31A
CircSEC31A and SEC31A were significantly highly expressed in NSCLC cells and tissues, and circSEC31A expression levels were closely associated with tumor size, TNM stage, and lymphatic metastasis.57 CircSEC31A could promote NSCLC cell migration, invasion, glycolysis, and apoptosis by sponging miR-376a, and SEC31A was directly targeted and inhibited by miR-376a in NSCLC Cells. That is to say, circSEC31A promoted NSCLC malignant progression through modulating SEC31A expression by acting as a miR-376a sponge. Conversely, knockdown of circSEC31A weakened tumor growth and thus suppressed NSCLC malignant progression. Based on the above results, we draw the conclusion that circSEC31A may serve as a novel molecular target for NSCLC therapy.
circCDYL
CircCDYL was decreased in NSCLC patients’ tissues and plasma, also downregulated in NSCLC cell lines.58 However, the overexpression of circCDYL can inhibit proliferation and induce apoptosis in NSCLC cells. The downregulated circCDYL binds to miR-185-5p, and miR-185-5p was upregulated in NSCLC. After that, TNRC6A is a downstream target of miR-185-5p and mRNA and protein levels were downregulated in NSCLC. Further, circCDYL repressed the phosphorylation of ERK1/2, which was induced by miR-185-5p or si-TNRC6A. Taken together, circCDYL inhibits ERK1/2 signaling pathway by targeting miR-185-5p/TNRC6A to block NSCLC malignant progression. Therefore, circCDYL possesses the potential to be a promising therapeutic target for NSCLC treatment.
CircRNAs Associated with Chemotherapy and Radiotherapy
At present, chemotherapy plays an indispensable role in the treatment of lung cancer, especially preoperative neoadjuvant chemotherapy to shrink tumor or reduce tumor stage and postoperative adjuvant chemotherapy to prevent recurrence and metastasis, but chemotherapy resistance in cancer cells becomes a fundamental challenge. Our common chemotherapy drugs for lung cancer include cisplatin, Taxol, 5-Fu, MIT, and so on. Cisplatin, a kind of heavy metal drug, is one of the most commonly used chemotherapy drugs at present, hsa_circRNA_103809,158 circ_PIP5K1A,87 circRNA CDR1as,91 circRNA_100565133 can promote cisplatin resistance. While hsa_circ_000194670 can enhance cisplatin sensitivity. Taxol, which is extracted from the bark of the Pacific Yew tree, is also one of the most commonly used chemotherapy drugs. Circ_0003998,52 circ_ZFR115 can reduce Taxol sensitivity. While hsa_circ_0030998,72 circ_000248398 can actually reduce Taxol resistance. Other common resistant chemotherapy drugs, such as osimertinib, an effective EGFR-tyrosine kinase inhibitor for advanced NSCLC patients, have also been found to promote resistance by hsa_circ_0002130.103 In addition, other studies have found that circRNA_103762 can even promote multidrug resistance (MDR).53 Consequently, circRNAs are one of the important breakthroughs in solving chemotherapy resistance.
Radiotherapy is a relatively common and effective treatment method, especially for patients with advanced lung cancer, radiotherapy can effectively control the further growth of tumor, relieve a series of symptoms caused by lung cancer, and control bone metastasis and brain metastasis. For patients with early lung cancer, radiotherapy can be considered in the case of inoperable, and it works very well. Studies have shown that circRNAs can modulate radiotherapy resistance. For example, circ_0001287 can reduce radiotherapy resistance, thus providing new clues for the treatment of lung cancer.136 Therefore, circRNAs are closely related to the solution of radiotherapy resistance.
Conclusion and Perspective
Lung cancer, the second most common cancer and the leading cause of cancer-related deaths, still has so many difficult problems to solve in its diagnosis and treatment. Appearance of circRNAs points out a new direction for the study of lung cancer. Because of its unique structure, circRNAs have advantages in the diagnosis, prognosis, and treatment of lung cancer.
Compared with other protein or non-coding RNA biomarkers, circRNAs have higher sensitivity via their stable structure and high abundance, and higher specificity via their tissue-specific expression. Moreover, some abundant expression has been found in blood, which is conducive to Clinical Blood Testing of circRNA, making circRNA an ideal biomarker.
At the same time, the application of circRNAs in lung cancer still has plenty of limitations. Firstly, although many reports have mentioned that the abnormal expression of circRNAs may be related to lung cancer TNM stage, a relatively accurate diagnostic standard value has not been established to measure the relationship between abnormal expression level of circRNAs and TNM stage. This is still a shortcoming in the early diagnosis of lung cancer. Therefore, a large amount of experimental data are needed to support the staging or early diagnosis of lung cancer. Secondly, a certain amount of circRNAs are still unstable and have a short half-life in the blood, and can only be maintained in a relatively stable content in lung cancer tissues or cells, which has formed a certain obstacle to the non-invasive assisted diagnosis of lung cancer. Finally, as a novel diagnostic marker that has not been widely recognized, the technology, cost, and efficiency of the detection and identification of circRNAs remain to be solved, which increases the difficulty in the clinical application and popularization of circRNAs.
In addition, lung cancer is a kind of tumor with extremely high malignancy, and effective prognostic analysis can help prolong the survival rate of lung cancer patients. It is of concern that the abnormal expression of many circRNAs can affect the prognosis of lung cancer. Based on the comparison of preoperative and postoperative expression, or the change of expression during treatment, the expression level of circRNAs may be able to assess the therapeutic effect and determine tumor growth to some extent.
Target therapy is the frontier of lung cancer treatment. The regulatory mechanism of circRNAs related to lung cancer, especially miRNA sponge, which has been studied extensively, provides effective targets for target therapy of lung cancer. In terms of high-expression circRNAs, exogenous fully complementary siRNA can be introduced to bind to specific back-splice junction of targeted circRNAs and degrade the circRNAs. Additionally, antisense oligonucleotide binding to specific splicing signals of targeted circRNAs can also be introduced into pre-mRNA to interfere with the circRNAs production. In clinical applications of circRNA therapeutics, circRNAs have not been used as a single tumor therapeutic target, more as an adjuvant therapy. However, with the continuous exploration of the molecular mechanism of circRNA, circRNA may play a unique role in lung cancer treatment in the future. In the case of chemotherapy, there are various chemotherapy drugs for lung cancer, and evidences have suggested that diverse circRNAs can enhance or reduce chemotherapy resistance in lung cancer. Even in radiotherapy, a few circRNAs can affect the radiotherapy effect of lung cancer.
In conclusion, although there have been many research achievements on circRNAs, the research on circRNAs is still in its infancy, and their functional role in tumorigenesis is still largely unknown. The evidence that has been found so far makes circRNAs not only as valuable diagnostic and prognostic biomarkers but also as promising therapeutic targets in lung cancer treatment.
Acknowledgments
We thank the authors of references for providing the data for the final manuscript.
Funding Statement
This publication was supported by the National Natural Science Foundation of China (No.31600662) and Haiyan Foundation from Harbin Medical University Cancer Hospital (No. JJMS2020-02) to H.X.
Disclosure
The authors declare no conflict of interest.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594. doi: 10.1016/S0025-6196(11)60735-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ettinger DS, Wood DE, Aisner DL, et al. Non-small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15(4):504–535. doi: 10.6004/jnccn.2017.0050 [DOI] [PubMed] [Google Scholar]
- 4.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 6.Chen RX, Liu HL, Yang LL, et al. Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. Eur Rev Med Pharmacol Sci. 2019;23(20):8771–8778. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Wei S, Wang X, Zhu X, Han S. Progress in research on the role of circular RNAs in lung cancer. World J Surg Oncol. 2018;16(1):215. doi: 10.1186/s12957-018-1515-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Zhu M, Pan J, Chen C, Xia S, Song Y. Circular RNAs: a rising star in respiratory diseases. Respir Res. 2019;20(1):3. doi: 10.1186/s12931-018-0962-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu W, Bi ZY, Chen ZL, et al. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. 2018;427:18–27. doi: 10.1016/j.canlet.2018.04.006 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017 [DOI] [PubMed] [Google Scholar]
- 12.Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959 [DOI] [PubMed] [Google Scholar]
- 13.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt CA, Matera AG. tRNA introns: presence, processing, and purpose. Wiley Interdiscip Rev RNA. 2020;11(3):e1583. doi: 10.1002/wrna.1583 [DOI] [PubMed] [Google Scholar]
- 15.Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–4422. doi: 10.1038/emboj.2011.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi: 10.1016/j.jhep.2018.01.012 [DOI] [PubMed] [Google Scholar]
- 17.Holdt LM, Stahringer A, Sass K, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015 [DOI] [PubMed] [Google Scholar]
- 19.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 20.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 21.Li RC, Ke S, Meng FK, et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9(8):838. doi: 10.1038/s41419-018-0852-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Zhao H, Zhang L. Identification of the tumor suppressive function of circular RNA FOXO3 in nonsmall cell lung cancer through sponging miR155. Mol Med Rep. 2018;17(6):7692–7700. doi: 10.3892/mmr.2018.8830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han C, Wang S, Wang H, Zhang J. Exosomal Circ-HIPK3 facilitates tumor progression and temozolomide resistance by regulating miR-421/ZIC5 axis in glioma. Cancer Biother Radiopharm. 2020. doi: 10.1089/cbr.2019.3492 [DOI] [PubMed] [Google Scholar]
- 24.Qian W, Huang T, Feng W. Circular RNA HIPK3 promotes EMT of cervical cancer through sponging miR-338-3p to up-regulate HIF-1alpha. Cancer Manag Res. 2020;12:177–187. doi: 10.2147/CMAR.S232235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Dai Q, Zheng L, Yuan X, Pan S, Deng J. Knockdown of circ_HIPK3 inhibits tumorigenesis of hepatocellular carcinoma via the miR-582-3p/DLX2 axis. Biochem Biophys Res Commun. 2020;533(3):501–509. doi: 10.1016/j.bbrc.2020.09.050 [DOI] [PubMed] [Google Scholar]
- 26.Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21(2):172–179. doi: 10.1261/rna.048272.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268(5209):415–417. doi: 10.1126/science.7536344 [DOI] [PubMed] [Google Scholar]
- 29.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323(6088):558–560. doi: 10.1038/323558a0 [DOI] [PubMed] [Google Scholar]
- 30.Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37 e9. doi: 10.1016/j.molcel.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abe N, Matsumoto K, Nishihara M, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):1402–1412. [DOI] [PubMed] [Google Scholar]
- 34.Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465(7301):1097–1101. doi: 10.1038/nature09095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, Xie L, Han L, et al. Circular RNAs: regulators of cancer-related signaling pathways and potential diagnostic biomarkers for human cancers. Theranostics. 2017;7(12):3106–3117. doi: 10.7150/thno.19016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/beta-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi: 10.1155/2016/1579490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Zhang Z, Jiang H, et al. Circular RNA circPVT1 promotes proliferation and invasion through sponging miR-125b and activating E2F2 signaling in non-small cell lung cancer. Cell Physiol Biochem. 2018;51(5):2324–2340. doi: 10.1159/000495876 [DOI] [PubMed] [Google Scholar]
- 39.Pei X, Chen SW, Long X, et al. circMET promotes NSCLC cell proliferation, metastasis, and immune evasion by regulating the Mir-145-5p/CXCL3 axis. Aging. 2020;12(13):13038–13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao J, Xu G, Zhu L, Zheng H. circGFRA1 enhances NSCLC progression by sponging miR-188-3p. Onco Targets Ther. 2020;13:549–558. doi: 10.2147/OTT.S230795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu X, Wang X, Wei S, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. doi: 10.1111/febs.14132 [DOI] [PubMed] [Google Scholar]
- 42.de Fraipont F, Gazzeri S, Cho WC, Eymin B. Circular RNAs and RNA splice variants as biomarkers for prognosis and therapeutic response in the liquid biopsies of lung cancer patients. Front Genet. 2019;10:390. doi: 10.3389/fgene.2019.00390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu BQ, Zhang PF, Xiong D, et al. CircRNA fibroblast growth factor receptor 3 promotes tumor progression in non-small cell lung cancer by regulating Galectin-1-AKT/ERK1/2 signaling. J Cell Physiol. 2019;234(7):11256–11264. doi: 10.1002/jcp.27783 [DOI] [PubMed] [Google Scholar]
- 44.Chung LY, Tang SJ, Sun GH, et al. Galectin-1 promotes lung cancer progression and chemoresistance by upregulating p38 MAPK, ERK, and cyclooxygenase-2. Clin Cancer Res. 2012;18(15):4037–4047. doi: 10.1158/1078-0432.CCR-11-3348 [DOI] [PubMed] [Google Scholar]
- 45.Zhang PF, Li KS, Shen YH, et al. Galectin-1 induces hepatocellular carcinoma EMT and sorafenib resistance by activating FAK/PI3K/AKT signaling. Cell Death Dis. 2016;7:e2201. doi: 10.1038/cddis.2015.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.An J, Shi H, Zhang N, Song S. Elevation of circular RNA circ_0003645 forecasts unfavorable prognosis and facilitates cell progression via miR-1179/TMEM14A pathway in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;511(4):921–925. doi: 10.1016/j.bbrc.2019.03.011 [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Yang D, Wei Y. Overexpressed CDR1as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. Onco Targets Ther. 2018;11:3979–3987. doi: 10.2147/OTT.S158316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan Z, Bai Y, Zhang Q, Qian P. CircRNA circ_POLA2 promotes lung cancer cell stemness via regulating the miR-326/GNB1 axis. Environ Toxicol. 2020;35(10):1146–1156. doi: 10.1002/tox.22980 [DOI] [PubMed] [Google Scholar]
- 49.Liu G, Shi H, Deng L, et al. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;513(1):207–212. doi: 10.1016/j.bbrc.2019.03.213 [DOI] [PubMed] [Google Scholar]
- 50.Mao Z, Li X, Ma X, Wang X, Zhang J, Fan X. Pancreatic progenitor cell differentiation and proliferation factor predicts poor prognosis in hepatocellular carcinoma. Medicine. 2019;98(9):e14552. doi: 10.1097/MD.0000000000014552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stein U, Walther W, Arlt F, et al. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat Med. 2009;15(1):59–67. doi: 10.1038/nm.1889 [DOI] [PubMed] [Google Scholar]
- 52.Zhang W, Song C, Ren X. Circ_0003998 regulates the progression and docetaxel sensitivity of DTX-resistant non-small cell lung cancer cells by the miR-136-5p/CORO1C axis. Technol Cancer Res Treat. 2021;20:1533033821990040. doi: 10.1177/1533033821990040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao G, Huang W, Zhan Y, Li J, Tong W. CircRNA_103762 promotes multidrug resistance in NSCLC by targeting DNA damage inducible transcript 3 (CHOP). J Clin Lab Anal. 2020;34(6):e23252. doi: 10.1002/jcla.23252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dilshara MG, Jayasooriya RG, Park SR, Choi YH, Choi IW, Kim GY. Caffeic acid phenethyl ester enhances TRAIL-mediated apoptosis via CHOP-induced death receptor 5 upregulation in hepatocarcinoma Hep3B cells. Mol Cell Biochem. 2016;418(1–2):13–20. doi: 10.1007/s11010-016-2726-x [DOI] [PubMed] [Google Scholar]
- 55.Tan W, Liao Y, Qiu Y, et al. miRNA 146a promotes chemotherapy resistance in lung cancer cells by targeting DNA damage inducible transcript 3 (CHOP). Cancer Lett. 2018;428:55–68. doi: 10.1016/j.canlet.2018.04.028 [DOI] [PubMed] [Google Scholar]
- 56.Qu D, Yan B, Xin R, Ma T. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res. 2018;8(8):1387–1402. [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng F, Yu J, Zhang X, Dai Z, Fang A. CircSEC31A promotes the malignant progression of non-small cell lung cancer through regulating SEC31A expression via sponging miR-376a. Cancer Manag Res. 2020;12:11527–11539. doi: 10.2147/CMAR.S280124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bian WX, Xue F, Wang LY, Xing XF. Circular RNA CircCDYL regulates proliferation and apoptosis in non-small cell lung cancer cells by sponging miR-185-5p and upregulating TNRC6A. Cancer Manag Res. 2021;13:633–642. doi: 10.2147/CMAR.S280315 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Yan B, Zhang W, Mao XW, Jiang LY. Circular RNA ciRS-7 correlates with advance disease and poor prognosis, and its down-regulation inhibits cells proliferation while induces cells apoptosis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22(24):8712–8721. [DOI] [PubMed] [Google Scholar]
- 60.Zhao F, Han Y, Liu Z, Zhao Z, Li Z, Jia K. circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Biosci Rep. 2018;38:4. doi: 10.1042/BSR20180570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao N, Wang FX, Wang G, Zhao QS. Targeting the HMGA2 oncogene by miR-498 inhibits non-small cell lung cancer biological behaviors. Eur Rev Med Pharmacol Sci. 2018;22(6):1693–1699. [DOI] [PubMed] [Google Scholar]
- 62.Li L, Du M, Wang C, He P. Reduced expression of circRNA novel_circ_0005280 and its clinical value in the diagnosis of non-small cell lung cancer. J Thorac Dis. 2020;12(12):7281–7289. doi: 10.21037/jtd-20-2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen D, Ma W, Ke Z, Xie F. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17(16):2080–2090. doi: 10.1080/15384101.2018.1515553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zong L, Sun Q, Zhang H, et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacother. 2018;102:639–644. doi: 10.1016/j.biopha.2018.03.084 [DOI] [PubMed] [Google Scholar]
- 65.Zheng G, Huang J, Chen W, You P, Ding Y, Tu P. circUBAP2 exacerbates malignant capabilities of NSCLC by targeting KLF4 through miR-3182 modulation. Aging. 2021;13(8):11083–11095. doi: 10.18632/aging.202745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen L, Nan A, Zhang N, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019;18(1):13. doi: 10.1186/s12943-019-0943-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L, Tong X, Zhou Z, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol Cancer. 2018;17(1):140. doi: 10.1186/s12943-018-0889-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang PF, Pei X, Li KS, et al. Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol Cancer. 2019;18(1):179. doi: 10.1186/s12943-019-1111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou J, Zhang S, Chen Z, He Z, Xu Y, Li Z. CircRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis. 2019;10(12):885. doi: 10.1038/s41419-019-2127-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang MS, Liu JY, Xia XB, et al. Hsa_circ_0001946 inhibits lung cancer progression and mediates cisplatin sensitivity in non-small cell lung cancer via the nucleotide excision repair signaling pathway. Front Oncol. 2019;9:508. doi: 10.3389/fonc.2019.00508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. doi: 10.1016/j.bbrc.2018.04.172 [DOI] [PubMed] [Google Scholar]
- 72.Li X, Feng Y, Yang B, et al. A novel circular RNA, hsa_circ_0030998 suppresses lung cancer tumorigenesis and Taxol resistance by sponging miR-558. Mol Oncol. 2021;15:2235–2248. doi: 10.1002/1878-0261.12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hang D, Zhou J, Qin N, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7(6):2783–2791. doi: 10.1002/cam4.1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chang H, Qu J, Wang J, Liang X, Sun W. Circular RNA circ_0026134 regulates non-small cell lung cancer cell proliferation and invasion via sponging miR-1256 and miR-1287. Biomed Pharmacother. 2019;112:108743. doi: 10.1016/j.biopha.2019.108743 [DOI] [PubMed] [Google Scholar]
- 75.Mao Y, He JX, Zhu M, Dong YQ, He JX. Circ0001320 inhibits lung cancer cell growth and invasion by regulating TNFAIP1 and TPM1 expression through sponging miR-558. Hum Cell. 2021;34(2):468–477. doi: 10.1007/s13577-020-00453-4 [DOI] [PubMed] [Google Scholar]
- 76.Han J, Zhao G, Ma X, et al. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun. 2018;503(4):2429–2435. doi: 10.1016/j.bbrc.2018.06.172 [DOI] [PubMed] [Google Scholar]
- 77.Yuan Y, Zhou X, Kang Y, et al. Circ-CCS is identified as a cancer-promoting circRNA in lung cancer partly by regulating the miR-383/E2F7 axis. Life Sci. 2021;267:118955. doi: 10.1016/j.lfs.2020.118955 [DOI] [PubMed] [Google Scholar]
- 78.Chen X, Mao R, Su W, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy. 2020;16(4):659–671. doi: 10.1080/15548627.2019.1634945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hong W, Zhang Y, Ding J, Yang Q, Xie H, Gao X. circHIPK3 acts as competing endogenous RNA and promotes non-small-cell lung cancer progression through the miR-107/BDNF signaling pathway. Biomed Res Int. 2020;2020:6075902. doi: 10.1155/2020/6075902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gu F, Zhang J, Yan L, Li D. CircHIPK3/miR-381-3p axis modulates proliferation, migration, and glycolysis of lung cancer cells by regulating the AKT/mTOR signaling pathway. Open Life Sci. 2020;15(1):683–695. doi: 10.1515/biol-2020-0070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu H, Han X, Ren J, Ren K, Li Z, Sun Z. Circular RNA HIPK3 induces cell proliferation and inhibits apoptosis in non-small cell lung cancer through sponging miR-149. Cancer Biol Ther. 2020;21(2):113–121. doi: 10.1080/15384047.2019.1669995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu Z, Xiang W, Chen W, et al. Circ-IGF1R inhibits cell invasion and migration in non-small cell lung cancer. Thorac Cancer. 2020;11(4):875–887. doi: 10.1111/1759-7714.13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ma X, Yang X, Bao W, et al. Circular RNA circMAN2B2 facilitates lung cancer cell proliferation and invasion via miR-1275/FOXK1 axis. Biochem Biophys Res Commun. 2018;498(4):1009–1015. doi: 10.1016/j.bbrc.2018.03.105 [DOI] [PubMed] [Google Scholar]
- 84.Li B, Zhu L, Lu C, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12(1):295. doi: 10.1038/s41467-020-20527-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang L, Xu C, Wang C, et al. Circ-PAX2 promotes proliferation and metastasis by absorbing miR-186 in lung cancer cells. Int J Clin Exp Pathol. 2018;11(7):3793–3801. [PMC free article] [PubMed] [Google Scholar]
- 86.Chi Y, Luo Q, Song Y, et al. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1alpha regulation. J Cell Biochem. 2019;120(11):19019–19030. doi: 10.1002/jcb.29225 [DOI] [PubMed] [Google Scholar]
- 87.Shao N, Song L, Sun X. Exosomal circ_PIP5K1A regulates the progression of non-small cell lung cancer and cisplatin sensitivity by miR-101/ABCC1 axis. Mol Cell Biochem. 2021;476(6):2253–2267. doi: 10.1007/s11010-021-04083-8 [DOI] [PubMed] [Google Scholar]
- 88.Wei S, Zheng Y, Jiang Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193. doi: 10.1016/j.ebiom.2019.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang H, Zhao M, Zhao L, Li P, Duan Y, Li G. CircRNA BIRC6 promotes non-small cell lung cancer cell progression by sponging microRNA-145. Cell Oncol. 2020;43(3):477–488. doi: 10.1007/s13402-020-00503-x [DOI] [PubMed] [Google Scholar]
- 90.Li Y, Zhang J, Pan S, Zhou J, Diao X, Liu S. CircRNA CDR1as knockdown inhibits progression of non-small-cell lung cancer by regulating miR-219a-5p/SOX5 axis. Thorac Cancer. 2020;11(3):537–548. doi: 10.1111/1759-7714.13274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao Y, Zheng R, Chen J, Ning D. CircRNA CDR1as/miR-641/HOXA9 pathway regulated stemness contributes to cisplatin resistance in non-small cell lung cancer (NSCLC). Cancer Cell Int. 2020;20:289. doi: 10.1186/s12935-020-01390-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Q, Kang PM. CircRNA_001010 adsorbs miR-5112 in a sponge form to promote proliferation and metastasis of non-small cell lung cancer (NSCLC). Eur Rev Med Pharmacol Sci. 2020;24(8):4271–4280. [DOI] [PubMed] [Google Scholar]
- 93.Zhang P, Xue XF, Ling XY, et al. CircRNA_010763 promotes growth and invasion of lung cancer through serving as a molecular sponge of miR-715 to induce c-Myc expression. Eur Rev Med Pharmacol Sci. 2020;24(13):7310–7319. [DOI] [PubMed] [Google Scholar]
- 94.Zhang ZY, Gao XH, Ma MY, Zhao CL, Zhang YL, Guo SS. CircRNA_101237 promotes NSCLC progression via the miRNA-490-3p/MAPK1 axis. Sci Rep. 2020;10(1):9024. doi: 10.1038/s41598-020-65920-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou ZF, Wei Z, Yao JC, et al. CircRNA_102179 promotes the proliferation, migration and invasion in non-small cell lung cancer cells by regulating miR-330-5p/HMGB3 axis. Pathol Res Pract. 2020;216(11):153144. doi: 10.1016/j.prp.2020.153144 [DOI] [PubMed] [Google Scholar]
- 96.Zhang N, Nan A, Chen L, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19(1):101. doi: 10.1186/s12943-020-01221-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Y, Xu R, Zhang D, et al. Circ-ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR-330-5p to promote non-small cell lung cancer progression. Transl Lung Cancer Res. 2019;8(6):862–875. doi: 10.21037/tlcr.2019.11.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li X, Yang B, Ren H, et al. Hsa_circ_0002483 inhibited the progression and enhanced the Taxol sensitivity of non-small cell lung cancer by targeting miR-182-5p. Cell Death Dis. 2019;10(12):953. doi: 10.1038/s41419-019-2180-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gao S, Yu Y, Liu L, Meng J, Li G. Circular RNA hsa_circ_0007059 restrains proliferation and epithelial-mesenchymal transition in lung cancer cells via inhibiting microRNA-378. Life Sci. 2019;233:116692. doi: 10.1016/j.lfs.2019.116692 [DOI] [PubMed] [Google Scholar]
- 100.Qin H, Liu J, Du ZH, Hu R, Yu YK, Wang QA. Circular RNA hsa_circ_0012673 facilitates lung cancer cell proliferation and invasion via miR-320a/LIMK18521 axis. Eur Rev Med Pharmacol Sci. 2020;24(4):1841–1852. [DOI] [PubMed] [Google Scholar]
- 101.Yao JT, Zhao SH, Liu QP, et al. Over-expression of CircRNA_100876 in non-small cell lung cancer and its prognostic value. Pathol Res Pract. 2017;213(5):453–456. doi: 10.1016/j.prp.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 102.Zhao H, Wei H, He J, et al. Propofol disrupts cell carcinogenesis and aerobic glycolysis by regulating circTADA2A/miR-455-3p/FOXM1 axis in lung cancer. Cell Cycle. 2020;19(19):2538–2552. doi: 10.1080/15384101.2020.1810393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ma J, Qi G, Li L. A novel serum exosomes-based biomarker hsa_circ_0002130 facilitates osimertinib-resistance in non-small cell lung cancer by sponging miR-498. Onco Targets Ther. 2020;13:5293–5307. doi: 10.2147/OTT.S243214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li C, Liu H, Niu Q, Gao J. Circ_0000376, a novel circRNA, promotes the progression of non-small cell lung cancer through regulating the miR-1182/NOVA2 network. Cancer Manag Res. 2020;12:7635–7647. doi: 10.2147/CMAR.S258340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tai G, Zhang M, Liu F. Circ_0000735 enhances the proliferation, metastasis and glycolysis of non-small cell lung cancer by regulating the miR-635/FAM83F axis. Exp Lung Res. 2021;47(3):136–148. doi: 10.1080/01902148.2021.1881188 [DOI] [PubMed] [Google Scholar]
- 106.Huang W, Xu X, Liu M, Cui W, Peng G. Downregulation of Hsa_circ_0000735 inhibits the proliferation, migration, invasion, and glycolysis in non-small-cell lung cancer by targeting miR-940/BMPER axis. Onco Targets Ther. 2020;13:8427–8439. doi: 10.2147/OTT.S253474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang M, Shi J, Jiang H, Xu K, Huang Z. Circ_0014130 participates in the proliferation and apoptosis of nonsmall cell lung cancer cells via the miR-142-5p/IGF-1 axis. Cancer Biother Radiopharm. 2020;35(3):233–240. doi: 10.1089/cbr.2019.2965 [DOI] [PubMed] [Google Scholar]
- 108.Geng Y, Bao Y, Zhang W, Deng L, Su D, Zheng H. Circular RNA hsa_circ_0014130 inhibits apoptosis in non-small cell lung cancer by sponging miR-136-5p and upregulating BCL2. Mol Cancer Res. 2020;18(5):748–756. doi: 10.1158/1541-7786.MCR-19-0998 [DOI] [PubMed] [Google Scholar]
- 109.Li Y, Hu J, Li L, et al. Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem Biophys Res Commun. 2018;503(3):2089–2094. doi: 10.1016/j.bbrc.2018.07.164 [DOI] [PubMed] [Google Scholar]
- 110.Yan X, Wang T, Wang J. Circ_0016760 acts as a sponge of MicroRNA-4295 to enhance E2F transcription factor 3 expression and facilitates cell proliferation and glycolysis in nonsmall cell lung cancer. Cancer Biother Radiopharm. 2020. doi: 10.1089/cbr.2020.3621 [DOI] [PubMed] [Google Scholar]
- 111.Wan J, Hao L, Zheng X, Li Z. Circular RNA circ_0020123 promotes non-small cell lung cancer progression by acting as a ceRNA for miR-488-3p to regulate ADAM9 expression. Biochem Biophys Res Commun. 2019;515(2):303–309. doi: 10.1016/j.bbrc.2019.05.158 [DOI] [PubMed] [Google Scholar]
- 112.Lu J, Ma X, Lin J, Hou P. Circ_0020123 increases ZFX expression to facilitate non-small cell lung cancer progression by sponging miR-142-3p. Cancer Manag Res. 2021;13:1687–1698. doi: 10.2147/CMAR.S295595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li L, Wei H, Zhang H, Xu F, Che G. Circ_100565 promotes proliferation, migration and invasion in non-small cell lung cancer through upregulating HMGA2 via sponging miR-506-3p. Cancer Cell Int. 2020;20:160. doi: 10.1186/s12935-020-01241-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang H, Wang X, Hu B, Zhang F, Wei H, Li L. Circular RNA ZFR accelerates non-small cell lung cancer progression by acting as a miR-101-3p sponge to enhance CUL4B expression. Artif Cells Nanomed Biotechnol. 2019;47(1):3410–3416. doi: 10.1080/21691401.2019.1652623 [DOI] [PubMed] [Google Scholar]
- 115.Li J, Fan R, Xiao H. Circ_ZFR contributes to the paclitaxel resistance and progression of non-small cell lung cancer by upregulating KPNA4 through sponging miR-195-5p. Cancer Cell Int. 2021;21(1):15. doi: 10.1186/s12935-020-01702-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tian X, Zhang L, Jiao Y, Chen J, Shan Y, Yang W. CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR-1252/FOXR2 axis. J Cell Biochem. 2019;120(3):3765–3772. doi: 10.1002/jcb.27657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ma D, Qin Y, Huang C, et al. Circular RNA ABCB10 promotes non-small cell lung cancer progression by increasing E2F5 expression through sponging miR-584-5p. Cell Cycle. 2020;19(13):1611–1620. doi: 10.1080/15384101.2020.1761617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xue YB, Ding MQ, Xue L, Luo JH. CircAGFG1 sponges miR-203 to promote EMT and metastasis of non-small-cell lung cancer by upregulating ZNF281 expression. Thorac Cancer. 2019;10(8):1692–1701. doi: 10.1111/1759-7714.13131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Y. circ-ANXA7 facilitates lung adenocarcinoma progression via miR-331/LAD1 axis. Cancer Cell Int. 2021;21(1):85. doi: 10.1186/s12935-021-01791-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liang Y, Wang H, Chen B, et al. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol Ther Nucleic Acids. 2021;23:355–368. doi: 10.1016/j.omtn.2020.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang Y, Shan C, Chen Y, et al. CircDENND2A promotes non-small cell lung cancer progression via regulating MiR-34a/CCNE1 signaling. Front Genet. 2020;11:987. doi: 10.3389/fgene.2020.00987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xie Y, Wang L, Yang D. CircEPSTI1 promotes the progression of non-small cell lung cancer through miR-145/HMGB3 axis. Cancer Manag Res. 2020;12:6827–6836. doi: 10.2147/CMAR.S252893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang T, Li M, Li H, Shi P, Liu J, Chen M. Downregulation of circEPSTI1 represses the proliferation and invasion of non-small cell lung cancer by inhibiting TRIM24 via miR-1248 upregulation. Biochem Biophys Res Commun. 2020;530(1):348–354. doi: 10.1016/j.bbrc.2020.06.106 [DOI] [PubMed] [Google Scholar]
- 124.Li O, Kang J, Zhang JJ, et al. Circle RNA FOXP1 promotes cell proliferation in lung cancer by regulating miR-185-5p/Wnt1 signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(12):6767–6778. [DOI] [PubMed] [Google Scholar]
- 125.Lu J, Zhu Y, Qin Y, Chen Y. CircNFIX acts as a miR-212-3p sponge to enhance the malignant progression of non-small cell lung cancer by up-regulating ADAM10. Cancer Manag Res. 2020;12:9577–9587. doi: 10.2147/CMAR.S272309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yue Q, Xu Y, Deng X, et al. Circ-PITX1 promotes the progression of non-small cell lung cancer through regulating the miR-1248/CCND2 axis. Onco Targets Ther. 2021;14:1807–1819. doi: 10.2147/OTT.S286820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang SJ, Ma J, Wu JC, Hao ZZ, Zhang YN, Zhang YJ. CircRNA EPB41L2 inhibits tumorigenicity of lung adenocarcinoma through regulating CDH4 by miR-211-5p. Eur Rev Med Pharmacol Sci. 2020;24(7):3749–3760. [DOI] [PubMed] [Google Scholar]
- 128.Yao Y, Zhou Y, Hua Q. circRNA hsa_circ_0018414 inhibits the progression of LUAD by sponging miR-6807-3p and upregulating DKK1. Mol Ther Nucleic Acids. 2021;23:783–796. doi: 10.1016/j.omtn.2020.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li L, Wan K, Xiong L, Liang S, Tou F, Guo S. CircRNA hsa_circ_0087862 acts as an oncogene in non-small cell lung cancer by targeting miR-1253/RAB3D axis. Onco Targets Ther. 2020;13:2873–2886. doi: 10.2147/OTT.S243533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang F, Li X, Jia X, Geng L. CircRNA ZNF609 knockdown represses the development of non-small cell lung cancer via miR-623/FOXM1 axis. Cancer Manag Res. 2021;13:1029–1039. doi: 10.2147/CMAR.S282162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu S, Yang N, Jiang X, Wang J, Dong J, Gao Y. FUS-induced circular RNA ZNF609 promotes tumorigenesis and progression via sponging miR-142-3p in lung cancer. J Cell Physiol. 2021;236(1):79–92. doi: 10.1002/jcp.29481 [DOI] [PubMed] [Google Scholar]
- 132.Wang JY, Zhang F, Hong L, Wei SJ. CircRNA_0000429 regulates development of NSCLC by acting as a sponge of miR-1197 to control MADD. Cancer Manag Res. 2021;13:861–870. doi: 10.2147/CMAR.S270790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhong Y, Lin H, Li Q, Liu C, Shen J. CircRNA_100565 contributes to cisplatin resistance of NSCLC cells by regulating proliferation, apoptosis and autophagy via miR-337-3p/ADAM28 axis. Cancer Biomark. 2021;30(2):261–273. doi: 10.3233/CBM-201705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gao N, Ye B. Circ-SOX4 drives the tumorigenesis and development of lung adenocarcinoma via sponging miR-1270 and modulating PLAGL2 to activate WNT signaling pathway. Cancer Cell Int. 2020;20:2. doi: 10.1186/s12935-019-1065-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li L, Zhang Q, Lian K. Circular RNA circ_0000284 plays an oncogenic role in the progression of non-small cell lung cancer through the miR-377-3p-mediated PD-L1 promotion. Cancer Cell Int. 2020;20:247. doi: 10.1186/s12935-020-01310-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang CC, Li Y, Feng XZ, Li DB. Circular RNA circ_0001287 inhibits the proliferation, metastasis, and radiosensitivity of non-small cell lung cancer cells by sponging microRNA miR-21 and up-regulating phosphatase and tensin homolog expression. Bioengineered. 2021;12(1):414–425. doi: 10.1080/21655979.2021.1872191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gao P, Wang Z, Hu Z, Jiao X, Yao Y. Circular RNA circ_0074027 indicates a poor prognosis for NSCLC patients and modulates cell proliferation, apoptosis, and invasion via miR-185-3p mediated BRD4/MADD activation. J Cell Biochem. 2020;121(3):2632–2642. doi: 10.1002/jcb.29484 [DOI] [PubMed] [Google Scholar]
- 138.Guo F, Li S, Guo C, et al. Circular RNA circMAGI3 accelerates the glycolysis of non-small cell lung cancer through miR-515-5p/HDGF. Am J Transl Res. 2020;12(7):3953–3963. [PMC free article] [PubMed] [Google Scholar]
- 139.Wang Y, Li Y, He H, Wang F. Circular RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation through upregulating EZH2 via sponging miR-377/382/498. Gene. 2019;720:144099. doi: 10.1016/j.gene.2019.144099 [DOI] [PubMed] [Google Scholar]
- 140.Wang Y, Wang Z, Lu J, Zhang H. Circular RNA circ-PTEN elevates PTEN inhibiting the proliferation of non-small cell lung cancer cells. Hum Cell. 2021:1–11. doi: 10.1007/s13577-021-00526-y [DOI] [PubMed] [Google Scholar]
- 141.Wang HL, Wang HR, Liang Y, et al. Hsa_circ_0006571 promotes spinal metastasis through sponging microRNA-138 to regulate sirtuin 1 expression in lung adenocarcinoma. Transl Lung Cancer Res. 2020;9(6):2411–2427. doi: 10.21037/tlcr-20-1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gu R, Shao K, Xu Q, Zhao X, Qiu H, Hu H. Circular RNA hsa_circ_0008003 facilitates tumorigenesis and development of non-small cell lung carcinoma via modulating miR-488/ZNF281 axis. J Cell Mol Med. 2020;34:1174–1184. doi: 10.1111/jcmm.15987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang Y, Li H, Lu H, Qin Y. Circular RNA SMARCA5 inhibits the proliferation, migration, and invasion of non-small cell lung cancer by miR-19b-3p/HOXA9 axis. Onco Targets Ther. 2019;12:7055–7065. doi: 10.2147/OTT.S216320 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 144.Yang J, Jia Y, Wang B, et al. Circular RNA TUBA1C accelerates the progression of non-small-cell lung cancer by sponging miR-143-3p. Cell Signal. 2020;74:109693. doi: 10.1016/j.cellsig.2020.109693 [DOI] [PubMed] [Google Scholar]
- 145.Chen S, Lu S, Yao Y, et al. Downregulation of hsa_circ_0007580 inhibits non-small cell lung cancer tumorigenesis by reducing miR-545-3p sponging. Aging. 2020;12(14):14329–14340. doi: 10.18632/aging.103472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lv YS, Wang C, Li LX, Han S, Li Y. Effects of circRNA_103993 on the proliferation and apoptosis of NSCLC cells through miR-1271/ERG signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(16):8384–8393. [DOI] [PubMed] [Google Scholar]
- 147.Luo YH, Zhu XZ, Huang KW, et al. Emerging roles of circular RNA hsa_circ_0000064 in the proliferation and metastasis of lung cancer. Biomed Pharmacother. 2017;96:892–898. doi: 10.1016/j.biopha.2017.12.015 [DOI] [PubMed] [Google Scholar]
- 148.Liu Q, Cao G, Wan Y, Xu C, He Y, Li G. Hsa_circ_0001073 targets miR-626/LIFR axis to inhibit lung cancer progression. Environ Toxicol. 2021;36(6):1052–1060. doi: 10.1002/tox.23104 [DOI] [PubMed] [Google Scholar]
- 149.Zhang F, Cheng R, Li P, Lu C, Zhang G. Hsa_circ_0010235 functions as an oncogenic drive in non-small cell lung cancer by modulating miR-433-3p/TIPRL axis. Cancer Cell Int. 2021;21(1):73. doi: 10.1186/s12935-021-01764-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Du H, He Z, Feng F, et al. Hsa_circ_0038646 promotes cell proliferation and migration in colorectal cancer via miR-331-3p/GRIK3. Oncol Lett. 2020;20(1):266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu Y, Yang C, Cao C, Li Q, Jin X, Shi H. Hsa_circ_RNA_0011780 represses the proliferation and metastasis of non-small cell lung cancer by decreasing FBXW7 via targeting miR-544a. Onco Targets Ther. 2020;13:745–755. doi: 10.2147/OTT.S236162 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 152.Xu J, Ni L, Zhao F, et al. Overexpression of hsa_circ_0002874 promotes resistance of non-small cell lung cancer to paclitaxel by modulating miR-1273f/MDM2/p53 pathway. Aging. 2021;13(4):5986–6009. doi: 10.18632/aging.202521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wang Y, Zhao W, Zhang S. STAT3-induced upregulation of circCCDC66 facilitates the progression of non-small cell lung cancer by targeting miR-33a-5p/KPNA4 axis. Biomed Pharmacother. 2020;126:110019. doi: 10.1016/j.biopha.2020.110019 [DOI] [PubMed] [Google Scholar]
- 154.Wang T, Wang X, Du Q, et al. The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun. 2019;513(4):904–911. doi: 10.1016/j.bbrc.2019.04.108 [DOI] [PubMed] [Google Scholar]
- 155.Jin M, Shi C, Yang C, Liu J, Huang G. Upregulated circRNA ARHGAP10 predicts an unfavorable prognosis in NSCLC through regulation of the miR-150-5p/GLUT-1 axis. Mol Ther Nucleic Acids. 2019;18:219–231. doi: 10.1016/j.omtn.2019.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 156.Wang L, Ma H, Kong W, Liu B, Zhang X. Up-regulated circular RNA VANGL1 contributes to progression of non-small cell lung cancer through inhibition of miR-195 and activation of Bcl-2. Biosci Rep. 2019;39(6):BSR20182433. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 157.Geng QQ, Wu QF, Zhang Y, Zhang GJ, Fu JK, Chen NZ. Clinical significance of circ-MTHFD2 in diagnosis, pathological staging and prognosis of NSCLC. Eur Rev Med Pharmacol Sci. 2020;24(18):9473–9479. [DOI] [PubMed] [Google Scholar]
- 158.Zhu X, Han J, Lan H, Lin Q, Wang Y, Sun X. A novel circular RNA hsa_circRNA_103809/miR-377-3p/GOT1 pathway regulates cisplatin-resistance in non-small cell lung cancer (NSCLC). BMC Cancer. 2020;20(1):1190. doi: 10.1186/s12885-020-07680-w [DOI] [PMC free article] [PubMed] [Google Scholar]