Abstract
Background
A hallmark of heart failure is cardiac fibrosis, which results from the injury‐induced differentiation response of resident fibroblasts to myofibroblasts that deposit extracellular matrix. During myofibroblast differentiation, fibroblasts progress through polarization stages of early proinflammation, intermediate proliferation, and late maturation, but the regulators of this progression are poorly understood. Planar cell polarity receptors, receptor tyrosine kinase–like orphan receptor 1 and 2 (Ror1/2), can function to promote cell differentiation and transformation. In this study, we investigated the role of the Ror1/2 in a model of heart failure with emphasis on myofibroblast differentiation.
Methods and Results
The role of Ror1/2 during cardiac myofibroblast differentiation was studied in cell culture models of primary murine cardiac fibroblast activation and in knockout mouse models that underwent transverse aortic constriction surgery to induce cardiac injury by pressure overload. Expression of Ror1 and Ror2 were robustly and exclusively induced in fibroblasts in hearts after transverse aortic constriction surgery, and both were rapidly upregulated after early activation of primary murine cardiac fibroblasts in culture. Cultured fibroblasts isolated from Ror1/2 knockout mice displayed a proinflammatory phenotype indicative of impaired myofibroblast differentiation. Although the combined ablation of Ror1/2 in mice did not result in a detectable baseline phenotype, transverse aortic constriction surgery led to the death of all mice by day 6 that was associated with myocardial hyperinflammation and vascular leakage.
Conclusions
Together, these results show that Ror1/2 are essential for the progression of myofibroblast differentiation and for the adaptive remodeling of the heart in response to pressure overload.
Keywords: fibroblasts, fibrosis, heart failure, inflammation, myocardial inflammation
Subject Categories: Cell Signalling/Signal Transduction, Fibrosis, Inflammation, Mechanisms, Heart Failure
Nonstandard Abbreviations and Acronyms
- LacZ
beta‐galactosidase
- Ror1/2
receptor tyrosine kinase like orphan receptor 1 and 2
- TAC
transverse aortic constriction
- TGF‐β
transforming growth factor beta
- Ubc‐CreERT2
ubiquitin C‐driven Cre recombinase with tamoxifen‐inducible mutant human estrogen receptor (ERT2)
- Vangl1/2
Vang‐like 1 and 2 ;
Clinical Perspective
What Is New?
This study demonstrated that planar cell polarity receptors, receptor tyrosine kinase–like orphan receptor 1 and 2, are key mediators of early fibroblast induction in myocardial tissue that regulate the proinflammatory environment in a mouse model of pressure overload–induced heart failure.
What Are the Clinical Implications?
A better understanding of the roles of receptor tyrosine kinase–like orphan receptor 1 and 2 in the control of fibroblast activation may provide opportunities to control myocardial inflammation and ultimately lead to better outcomes in patients with heart failure.
Excessive fibrosis during pathological cardiac remodeling is a hallmark of heart failure and the response to cardiac injury. Cardiac fibrosis is the result of resident cardiac fibroblasts that undergo myofibroblast differentiation to promote inflammation and secrete extracellular matrix in response to various forms of cardiac injury.1, 2, 3, 4 Although injury‐induced inflammation and matrix deposition is an adaptation to acute cardiac injury that can prevent the heart from rupture,5, 6 excessive fibrotic deposition leads to ventricle stiffness that impairs cardiac function.7, 8, 9 Recently, myofibroblast differentiation has been characterized as a transition through what is referred to as 3 phenotypic polarization stages: an initial proinflammatory phenotype, an intermediate proliferative phenotype, and a final mature phenotype.10 These polarization stages have specific functional differences. The proinflammatory fibroblast recruits inflammatory cells to the cardiac tissue in response to injury, the proliferative fibroblast undergoes cell division and deposits extracellular matrix in response to transforming growth factor beta (TGF‐β), and the mature myofibroblast maintains and strengthens the fibrotic deposits by expressing matrix remodeling proteins.11 A recent study using single‐cell RNA sequencing to analyze murine interstitial cells after myocardial infarction revealed fibroblast populations consistent with these polarization states.12 Despite its importance, we have limited knowledge of the factors that control the passage of myofibroblast through the different stages of differentiation.
The receptor tyrosine kinase–Like orphan receptors 1 and 2 (Ror1 and Ror2) have been evaluated for their roles in development and cell transformation.13, 14, 15 These membrane proteins are most highly expressed in developing tissues,16 and mice lacking Ror1 display skeletal defects,17 whereas mice lacking Ror2 have impaired heart and limb development.18 Ror1 and Ror2 expression can be reactivated in various cancers,19, 20, 21, 22, 23, 24 and knockdown or Ror1, Ror2, or both Ror1/2 in various transformed cell lines reduces their proliferation and migration.25, 26, 27, 28 Additionally, Ror1 expression in satellite cells promotes proliferation and skeletal muscle regeneration after injury.29 These functions of Ror1/2 may be linked to the ability of this receptor system to control the planar cell polarity signaling pathway. Planar cell polarity is the asymmetrical alignment of cells to coordinate directionality with neighboring cells and extracellular matrix.30 Proteins in the planar cell polarity pathway, including Ror1/2 and others (tyrosine‐protein kinase–like 7; prickle homologue 1; Vang‐like [Vangl] 1 and 2, disheveled1, 2, and 3), regulate cell polarity by organizing the actin cytoskeleton and segregating proteins to opposite sides of the cell.31, 32 In specific cell types, planar cell polarity can regulate proliferation, migration, and cell differentiation.33, 34, 35, 36, 37, 38 Thus, it is tempting to speculate that planar cell polarity‐mediated actin organization may be critical to myofibroblast function, and that it also functions as an integral step in the actin alignment‐mediated signal transduction that promotes locomotion, contraction, and matrix reorganization during fibroblast differentiation.39
Components of the planar cell polarity pathway have been implicated in clinical and experiment heart failure.40, 41, 42, 43, 44, 45, 46 Most recently, Ror1 has been reported to be robustly upregulated in ischemic cardiomyopathy,47 and Ror2 has been shown to be upregulated in right ventricular remodeling and heart failure.48 However, the role of Ror1/2 cell surface receptors in cardiac remodeling has not been addressed. While many of the functions attributed to Ror1 and Ror2 are shared by the process of cardiac fibroblast activation and differentiation, the roles of Ror1 and Ror2 in myofibroblasts have not been investigated previously. In the course of our studies, we found that Ror1 and Ror2 were generally expressed at low levels in the nonchallenged adult mouse tissues. However, the expression of these proteins become markedly increased in activated fibroblast in response to injurious stimuli. Thus, in this study, we investigated the role of Ror1 and Ror2 in cardiac remodeling through in vivo mouse models of heart failure and in vitro cell culture models of cardiac myofibroblast differentiation.
Methods
Data and Method Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Mouse Strains
All animal experiments were approved by the Animal Care and Use Committees at Boston University and the University of Virginia. Ror1/2fl/fl+ubiquitin C‐driven Cre recombinase with tamoxifen‐inducible mutant human estrogen receptor (Ubc‐CreERT2) mice were generated by combining Ror1 flox (Jackson Labs #018353,49), Ror2 flox (Jackson Labs #018354,49), and Ubc‐CreERT2 (Jackson Labs #008085,50) alleles, which are all in the B6 mouse background. Additionally, Ror2 expression β‐galactosidase (LacZ) reporter mice (Ror2‐LacZ,51) were used. Three mice per condition were used for fibroblast isolation experiments and 5 mice per condition were used for transverse aortic constriction experiments.
Transverse Aortic Constriction Surgery
Cardiac pressure overload by transverse aortic constriction (TAC) was performed as previously described.43 Briefly, surgery was performed on anesthetized mice where the aortic arch was accessed and constricted between the brachiocephalic artery and left common carotid artery. A 27‐gauge spacer was placed parallel to the transverse aorta, and 8‐0 vicryl suture (ETHICON Catalog #J401G) was used to ligate the aorta. The spacer was removed, the surgical wounds were sutured, and the mouse was allowed to recover. Successful TAC surgery was confirmed by initial expansion of the brachiocephalic artery and subsequent echocardiography measurements (FUJIFILM VisualSonics, Vevo 2100) at 0, 7, 14, and 28 days after TAC. Fractional shortening was quantified by M‐mode images obtained by short‐axis view visualizing both papillary muscles. Cardiac sectioning was performed by cardiac tissue isolation, fixation with 10% neutral buffered formalin for 24 hours, tissue dehydration, then paraffin embedding and sectioning. Cardiac tissue sections were either stained for 5‐bromo‐4‐chloro‐3‐indolyl‐b‐galactosidase (Xgal; Roche Catalog #XGAL‐RO) or immunostained with rabbit antimouse polyclonal Ror2 (provided by Dr Yasuhiro Minami, Kobe University) with DAB visualization (Vector Laboratories, Catalog #SK‐4100), and counterstained with hematoxylin and eosin (Sigma‐Aldrich, Catalog #HHS128‐4L and #HT110180‐2.5L, respectively). Protein lysates from TAC heart samples were isolated from cardiac tissue by 1% Triton‐X supplemented with protease inhibitors and probed for rabbit antimouse polyclonal Ror1 (provided by Dr Michael Greenberg,49) rabbit antimouse monoclonal Ror2 (Cell Signaling Technology Catalog #88639), or β‐actin (Cell Signaling Technology Catalog #4970) by western blot. Cardiac myocytes and cardiac fibroblasts were isolation in a langendorff apparatus as previously described.52 These primary cells were either used for gene expression analysis after RNA isolation and purification using primers listed in Table S1 from previous publications,53, 54, 55, 56, 57 or analyzed by flow cytometry using the following fluorescently conjugated antibodies: CD45 (BioLegend Catalog #103126), mEF‐SK4 (Miltenyi Biotech Catalog #103‐102‐352), Ror2 (R&D Systems Catalog #AF2064).
Primary Cardiac Cell Isolation and Fibroblast Passaging
Interstitial cells of cardiac tissue were isolated by digestion and cell‐type separation, based on previously published methods.58 Cardiac tissue, either healthy, sham‐surgery, or TAC surgery, was isolated from euthanized mice, minced with scissors, and digested in digestion media (HBSS with 0.1% trypsin and 100 U/mL collagenase type II) for 80 minutes. CD31+ endothelial cells and CD45+ leukocytes were separated and purified by fluorescence‐activated cvell sorting, with BV421 rat antimouse CD31 antibody (BD Biosciences Catalog #562939) and PerCP rat antimouse CD45 antibody (BD Biosciences Catalog #561047). Fibroblasts were purified by plating on plastic tissue culture dishes for 2 hours in fibroblast growth medium 3 (PromoCell Catalog #c‐23130), then media was refed to eliminate cells that had not stuck, leaving the fibroblasts on the tissue culture plate. Genes were assessed by RNA isolation (Qiagen RNeasy Kit Catalog #74104), reverse transcriptase conversion to DNA (Applied Biosystems cDNA Reverse Transcription Kit Catalog #4368814), and SYBR green detection (Applied Biosystems Fast SYBR Green Master Mix Catalog #43‐856‐16) with a quantitative PCR machine (Applied Biosystems QuantStudio 6). Primers for quantitative PCR are listed in Table S1. Protein lysates were isolated by RIPA protein lysate buffer supplemented with protease inhibitors and probed for Ror1, Ror2, or β‐actin with antibodies listed above, or interleukin‐6 (Abcam Catalog #ab179570). Cell proliferation of isolated cardiac fibroblasts was assessed by 5‐ethynyl‐2′‐deoxyuridine EdU incorporation assay (Click‐iT Plus EdU Flow Cytometry Assay Kit, Thermo Fisher Catalog #C10634) per manufacturer's instructions with EdU incubation for 2 hours. Fibroblasts were further passaged and cultured in fibroblast growth medium 3.
Myofibroblast Induction
Isolated cardiac fibroblasts at passage (P)2 were treated with 10 ng/mL TGF‐β1 (R&D Systems Catalog #240‐B) in fibroblast growth medium 3 for 4 days to assess myofibroblast differentiation. Gene expression was assessed as described above using primers listed in Table S1. Immunofluorescent staining of Acta2 (Cell Signaling Technologies Catalog #36110) and DAPI (Thermo Fisher Catalog #62248) was performed by culturing cells in 4‐well chamber slides (Thermo Fisher Catalog #154526PK) with 10 ng/mL TGF‐β1 in fibroblast growth medium 3 for 4 days, fixation with 4% paraformaldehyde for 20 minutes, permeabilization with PBS‐T (PBS+0.1% Tween20), incubation with fluorescent Acta2 antibody, then counterstain with DAPI. Immunostained cells were imaged by confocal microscopy (Leica SP8). Acta2 fiber alignment and Acta2 fluorescence intensity were quantified by ImageJ (version 2.0.0).
Bulk RNA Sequencing
Primary cardiac fibroblasts were isolated as described above from either control mice (Ror1/2fl/fl) or Ror1/2 knockout mice (Ror1/2fl/fl+Ubc‐CreERT2) after injection with tamoxifen (Sigma‐Aldrich Catalog #T5648) when mice were 6 to 8 weeks old. Each isolation pooled 4 hearts from mice of the same litter, with the same male:female ratio variation between control and Ror1/2 knockout samples. Mice were bred between a Ror1/2fl/fl genotype and a Ror1/2fl/fl+Ubc‐CreERT2 genotype to generate ≈50% control mice and ≈50% Ror1/2 knockout mice per litter, allowing for littermate paired samples. Specifically, isolations of control and Ror1/2 knockout mice were performed on 4 different litters with paired samples from the same litters (Ex: control sample 1 and Ror1/2 knockout sample 1 used hearts from mice of the same litter). Primary cardiac fibroblasts were grown for 9 days in culture, then RNA lysate was isolated and purified. Purified RNA was submitted to the University of Virginia Genome Analysis and Technology Core for whole transcriptome sequencing by first library preparation with NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB Catalog #E7760) and then sequencing by Illumina NextSeq 500 Sequencing System for paired‐end 75‐base pair reads. Programs were used with standard inputs for bioinformatic analysis: raw read data was quality checked with FastQC (Babraham Bioinformatics), preprocessed to filter out Illumina primer sequences, and aligned with Kallisto59 to generate estimated counts, normalized by log2 transformation and filtered low‐abundance genes with Sleuth,60 analyzed for gene expression by transcripts per million reads and differential expression with Sleuth by the likelihood ratio test, and analyzed for gene ontology term enrichment with Generally Accepted Gene‐set Enrichment.61 Aligned and normalized sequencing results and gene ontology term enrichment results are provided in Data S1 and S2.
Immunophenotyping of Cardiac Tissue
Cardiac tissue was isolated after sham or TAC surgery. Tissue was digested with collagenase I (450 U/mL), collagenase XI (125 U/mL), DNase I (60 U/mL), and hyaluronidase (60 U/mL), and the isolated cells from digested tissue were immunostained with fluorescently conjugated antibodies against CD45‐Pacific Blue, 30‐F11 (BioLegend Catalog #103126), Ly6G‐PE, 1A8 (BioLegend Catalog #127618), CD11b‐APC‐Cy7, M1/70 (BioLegend Catalog #101226), F4/80‐PE‐Cy7, BM8 (BioLegend Catalog #123114), and Ly6C‐FITC, HK1.4 (BioLegend Catalog #128006). Dead cells were excluded by staining with DAPI. Immunostained fluorescent cells were analyzed by flow cytometry using a BD LSR II flow cytometer (BD Bioscience). Additionally, isolated cardiac tissue was lysed and analyzed for inflammatory cytokines interleukin‐1β, inrerleukin‐6, and C‐C motif chemokine ligand 2 by quantitative reverse transcription PCR. Real‐time PCR primers are listed in Table S1.
Evans Blue Staining
Vascular permeability of the heart after pressure overload was evaluated by the extent of the leakage of Evans blue dye (Sigma‐Aldrich Catalog #E2129). Mice were euthanized 30 minutes after tail‐vein injection of 1% w/v in 0.9% saline. Dyes were allowed to circulate throughout the body during this period.
Statistical Analysis
Analysis of means between groups with a sample size of 4 to 6 in each group was performed using either a Mann‐Whitney test for comparison between 2 groups or a Kruskal‐Wallis test followed by a Dunn's multiple comparison corrected post hoc test for comparison among 3 or more groups by Prism (GraphPad Software, Inc., San Diego, CA). Otherwise, statistical analysis of means between groups was performed using either a standard 2‐tail Student t test or a 2‐way ANOVA test followed by a Tukey's multiple comparison corrected post hoc test by Prism. Statistical analysis of gene set enrichment in gene ontology analysis used the statistical tests in the Generally Accepted Gene‐set Enrichment package. Other statistical tests are described in figure legends. All statistical analysis of RNA sequencing data sets was performed through computational analysis packages, which contain statistical corrections for large data sets.
Results
Transverse Aortic Constriction Induces Early Ror1/2 Expression in Cardiac Tissue
Cardiac pressure overload in the mouse model of TAC induces fibroblast activation and myocardial remodeling, including initial inflammation (1–3 days after TAC), extracellular matrix remodeling (3–14 days after TAC), and eventual heart failure (14–28 days after TAC).62 Thus, cardiac tissue was analyzed before and after TAC surgery to investigate the expression patterns of Ror1 and Ror2 during the remodeling time course. The TAC surgery model was initially performed on Ror2‐LacZ mice that has the LacZ reporter gene knocked in to the Ror2 locus.51 There was no detectable Ror2‐mediated LacZ expression in uninjured, or sham‐operated mice (Figure 1A and not shown), but β‐galactosidase expression was markedly induced at 7 days after TAC (Figure 1B). Immunohistochemical analysis revealed that Ror2 protein was concentrated in cells of the interstitial space of the myocardial tissue at 7 days after TAC (Figure 1C). Next, we determined the time course of Ror1 and Ror2 protein induction after TAC surgery by western blot analysis of protein lysates from cardiac tissue. The expression of both Ror1 and Ror2 protein was increased by 3 days after TAC, peaking at 7 days, and then decreasing at 14 and 28 days after TAC (Figure 1D).
Figure 1. Ror1 and Ror2 expression after transverse aortic constriction in mice.

Cells expressing Ror2 in cardiac tissue were visualized in Ror2‐LacZ mice 28 days after (A) sham surgery or (B) TAC surgery. C, Protein localization was visualized in cardiac tissue 7 days after TAC by immunohistochemistry for Ror2. D, Ror1 and Ror2 protein expression was measured by western blot at various days after TAC surgery (β‐actin as a loading control). Images are representative of >3 replicates per experiment. E, Endothelial cells, leukocytes, and fibroblasts were isolated 7 days after sham or TAC surgery, and RNA expression of relevant genes was quantified (bars represent mean±SD, sample size=4 per group, Kruskal‐Wallis P values: Fsp1=0.0013, Acta2=0.0004, Postn=0.0014, Ror1=0.0016, Ror2=0.0002). A.U., arbitrary units; ICH, immunohistochemistry; LacZ, β‐galactosidase; Ror, receptor tyrosine kinase–like orphan receptor; and TAC, transverse aortic constriction. *p<0.05.
To determine the cell type(s) that express Ror1 and Ror2, RNA was isolated from endothelial cells, leukocytes, and fibroblasts from sham‐operated and TAC‐treated hearts 7 days after surgery, and quantitative PCR was performed to detect the levels of relevant transcripts. Ror1 and Ror2 transcript expression was detected in the activated cardiac fibroblasts at 7 days after TAC, but not in the endothelial or leukocyte cell populations (Figure 1E, Figure S1). Ror2 protein expression was also detected in fibroblasts after TAC surgery by flow cytometry (Figure S2). Overall, the timing, location, and cell‐specific gene expression patterns suggest that both Ror1 and Ror2 are induced during the activation and expansion of cardiac fibroblasts that occurs in response to pressure overload–induced myocardial remodeling.
Ror1 and Ror2 Are Induced During Early Cardiac Fibroblast Activation
The induction of Ror1/2 during myofibroblast differentiation was also investigated in cultured cells using isolated murine cardiac fibroblasts. Fibroblasts from wild‐type murine hearts were attached to cell culture plates, flattened, and expanded over 9 days after isolation (Figure S3). RNA expression of key genes increased at different rates during this activation time course (Figure 2A). As expected, Fsp1 (fibroblast‐specific protein 1) was induced by day 3 and the fibroblast activation genes Slug and Snail increased at day 9. Both Ror1 and Ror2 increased over this time course, but the induction of Ror2 preceded that of Ror1. The planar cell polarity protein transcript tyrosine‐protein kinase–like 7 increased by day 3 and continued to increase at days 6 and 9, but Prickle and Vangl2 displayed no statistically significant change in expression. Protein analysis of Ror1 and Ror2 by western immunoblot in cardiac fibroblasts at day 0 and day 9 after isolation confirm the upregulation of both proteins in cell culture (Figure S4).
Figure 2. Induction of Ror1 and Ror2 during early fibroblast activation and induction of myofibroblast differentiation of murine cardiac fibroblasts.

Murine cardiac fibroblasts were isolated from healthy cardiac tissue of C57Bl6 mice. A, RNA expression of genes associated with fibroblast identity, fibroblast activation, Ror1/2 receptors, and planar cell polarity was quantified over time (bars represent mean±SD, sample size=3 per group, Kruskal‐Wallis P values: Fsp1=0.0003, Slug=0.0014, Snail=0.0003, Ror1=0.0014, Ror2=0.0014, Ptk7=<0.0001, Prickle=0.0020, Vangl2=0.0137). Friedman test P values for overall effect of time: Fsp1=0.002, Slug=0.017, Snail=0.002, Ror1=0.017, Ror2=0.148, Ptk7=0.002, Prickle=0.017, Vangl2=0.054. Cardiac myofibroblasts at passage 2 were stained for ⍺‐smooth muscle actin fibers after (B) control and (C) TGF‐β1 treatment. D, RNA expression of genes associated with myofibroblast differentiation, inflammation, and Ror1/2 receptors was quantified in control and TGF‐β1 treated fibroblasts (bars represent mean±SD, sample size=3 per group, Student t test P values: Acta2=0.0078, Postn=0.0379, IL‐6=0.0487, Tnfa=0.0939, Ror1=0.9921, Ror2=0.2313). A.U., arbitrary units; DAPI, 4′,6‐diamidino‐2‐phenylindole; Ror indicates receptor tyrosine kinase–like orphan receptor; and TGF‐β1, transforming growth factor beta. *p<0.05.
Next, myofibroblast differentiation was stimulated in these cultures by treatment with TGF‐β1. Myofibroblast differentiation could be detected by cell enlargement and SMα‐actin filament elongation (Figure 2B and 2C). However, in contrast to the earlier stage of fibroblast activation, TGF‐β1 treatment did not lead to a further increase in Ror1 or Ror2 expression. On the other hand, hallmarks of myofibroblast differentiation could be observed following TGF‐β1 treatment including the increased transcript expression of myofibroblast differentiation proteins Acta2 and Postn, and decreased expression of the cytokines interleukin‐6 and tumor necrosis factor‐α (Figure 2D). Together, these data suggest that the induction of Ror1/2 is an early event during fibroblast activation, and that their expression is not altered at the later stages of myofibroblast differentiation.
Ror1/2 Double‐Knockout Fibroblasts Exhibited an Immature Cardiac Myofibroblast Phenotype
We next investigated the role of Ror1/2 in fibroblast activation in the fibroblast cell culture system using primary cardiac fibroblast cells isolated from control or Ror1/2 knockout mice. Ror1/2 was eliminated in adult mice through an inducible transgenic knockout strategy by crossing Ror1/2fl/fl and Ubc‐CreERT2 mice and treating the progeny with tamoxifen for 2 weeks. These mice displayed no observable phenotype, and they had a normal cardiac phenotype 3 months after the induction of gene ablation (Figure S5). Primary cardiac fibroblasts were isolated from control or Ror1/2 knockout mice and grown to confluence. RNA was then isolated, and next‐generation RNA sequencing was performed. Many genes associated with a manually curated network of fibrosis and fibroblasts63, 64 were differentially regulated between control and Ror1/2 knockout fibroblasts (Figure 3A). As expected, cells were largely void of Ror1 and Ror2 transcripts, and the planar cell polarity gene Ptk7, Vangl1, Vangl2, Prickle1, Dvl1, Dvl2, Dvl3) were generally reduced in the Ror1/2 knockout fibroblasts. Notably, Ror1/2 knockout fibroblasts showed an increase in inflammatory cytokine gene expression (Il1a, Il6, Tnfa, Ccl2) and a decrease in transcripts associated with cell proliferation (Pcna, Mki67, Cdk1, Cdk2, Cdk4, Cdk6). Additionally, transcripts associated with the promotion of fibrosis in the extracellular signal‐related kinase 1/2 signaling and matrix remodeling pathways were downregulated (Map2k3, Map3k1, Map3k5, Mapk14, Mapk8, Timp1, Timp2), and transcripts associated with the inhibition of fibrosis in the matrix remodeling and natriuretic signaling pathways were upregulated (Mmp2, Mmp9, Mmp14, Nppa, Nppb, Npr1, Npr2, Npr3). Analysis of the top 10 upregulated and downregulated gene ontology terms between control and Ror1/2 knockout fibroblasts revealed the strong induction of inflammation‐related pathways and an inhibition of proliferation‐related and microtubule organization‐related pathways (Figure 3B). The specific genes included in these groupings were highly differentially expressed between control and Ror1/2 knockout fibroblasts, along with planar cell polarity genes, inflammatory genes, and proliferation gene Mki67 also shown (Figure 3C). The reduction in cell proliferation in the Ror1/2 knockout fibroblasts was documented by EdU incorporation assay, showing ≈10% reduction in EdU+ cells during the 2‐hour Edu incubation period (Figure 3D). To the extent that the cell culture system models the early activation of fibroblasts, Ror1/2 dual deficiency led to the upregulation of inflammation pathways and downregulation of pathways associated with matrix production, proliferation, and microtubule organization. Collectively, these data are consistent with a cellular fibroblast phenotype that appears to be stalled in the proinflammatory polarization state, suggesting that Ror1 and Ror2 are required for the progression of fibroblasts from this immature state to the intermediate proliferative state.10
Figure 3. Ror1/2‐mediated early fibroblast activation in primary murine cardiac fibroblasts.

Primary cardiac fibroblasts were isolated from healthy control and transgenic Ror1/2 double‐knockout mice transcriptional phenotype was assessed by bulk RNA sequencing. A, RNA transcript reads of specific genes related to planar cell polarity, inflammatory cytokines, proliferation and cell division, extracellular signal‐related kinase 1/2 signaling, matrix remodeling, natriuretic signaling, and myofibroblast differentiation were quantified as normalized Z score (4 samples for each genotype). B, Gene ontology analysis of differential gene expression between control and Ror1/2 knockout cardiac fibroblasts showed the top 10 upregulated and downregulated terms by q value, with terms grouped by color: cell activation in orange, inflammation in red, proliferation in green, and microtubule regulation in black (bars represent mean±SEM, q values obtained through Generally Applicable Gene‐set Enrichment for Pathway Analysis, or GAGE, statistical algorithms). C, Significance vs fold change of each gene between control and Ror1/2 knockout samples was visualized by volcano plot, with genes in each gene ontology term highlighted in corresponding colors (all other genes in gray), and specific genes related to planar cell polarity, inflammation, and proliferation were highlighted and labeled. D, 5‐ethynyl‐2′‐deoxyuridine (EdU) incorporation assay after 2 hours of EdU incubation determined rate of proliferation in control and Ror1/2 knockout cardiac fibroblasts (bars represent mean±SD, sample size=3 per group, Student t test P=0.0054). ERK, iextracellular signal‐regulated kinase; GO, gene ontology; KO, knock out; pos reg, positive regulation; and Ror indicates receptor tyrosine kinase–like orphan receptor. *p<0.05.
Ror1/2 Double‐Knockout Fibroblasts Are Less Responsive to TGF‐β1–Induced Myofibroblast Maturation
Previous findings showed that activated fibroblasts in the early proinflammatory state are less responsive to TGF‐β stimulation than fibroblasts in the intermediate proliferative state.10 Thus, to investigate whether the Ror1/2 knockout fibroblasts are functionally stalled in an initial proinflammatory phenotype, we compared the responsiveness of control and Ror1/2‐knockout fibroblasts to TGF‐β1–induced myofibroblast differentiation. In control fibroblasts, TGF‐β1 treatment led to gene expression changes that are consistent with myofibroblast differentiation (increased Acta2 and Postn, decreased interleukin‐6). However, Ror1/2 knockout fibroblasts displayed diminished Acta2 and Postn induction and no decrease in interleukin‐6 after TGF‐β1 treatment when compared with control fibroblasts (Figure 4A, Figure S6A and S6B). Western blot analysis of interleukin‐6 protein levels confirmed that interleukin‐6 protein is upregulated in Ror1/2 knockout fibroblasts and that TGF‐β1 treatment reduced interleukin‐6 expression in control but not in Ror1/2 knockout fibroblasts (Figure 4B and 4C).
Figure 4. Ror1/2‐mediated response to myofibroblast differentiation induced by TGF‐β1.

Myofibroblast differentiation was induced by treatment with 10 ng/mL TGF‐β1 for 4 days. A, RNA expression of myofibroblast‐related (Acta2 and Postn) and inflammation‐related (interleukin‐6) genes was quantified in control and Ror1/2 knockout fibroblasts (bars represent mean±SD, sample size=3 per group, Kruskal‐Wallis P values: Acta2=0.0006, Postn<0.0001, Il6=0.0006). B, Protein levels of interleukin‐6 were visualized by Western blot in control and Ror1/2 knockout fibroblasts after TGF‐β1 induction (representative blot shown), quantification of 3 replicates in (C) (bars represent mean±SD, sample size=3 per group, Kruskal‐Wallis P=0.0006). D, ⍺‐Smooth muscle actin filaments were visualized by immunofluorescent staining of subconfluent cells in control and Ror1/2 knockout fibroblasts, and (E) alignment of ⍺‐smooth muscle actin filaments was quantified and normalized per cell (461 filament angles in control, 717 filament angles in Ror1/2 knockout, lines on graph represent contingency table of 10‐degree bins for 19 total bins, chi‐square P value for testing numbers in bins <0.0001). A.U., arbitrary units; KO, knock out; ns, not significant; Ror, receptor tyrosine kinase–like orphan receptor; and TGF‐β1, transforming growth factor beta. *p<0.05, ****p<0.0001.
Next, TGF‐β1–treated control and Ror1/2 knockout fibroblasts were visualized after Acta2 immunostaining. Although treatment with TGF‐β1–induced similar fibroblast cell enlargement between control and Ror1/2 knockout fibroblasts (control=0.073±0.016 mm2, Ror1/2 knockout=0.071±0.010 mm2; P=0.92), the Acta2 fibers appeared misaligned and disorganized in the Ror1/2 knockout fibroblasts compared with the Acta2 fiber alignment in the control fibroblasts (Figure 4D). Quantification of the directional angle of individual Acta2 fibers within each cell showed that the Acta2 fibers in the Ror1/2 knockout fibroblasts were significantly less aligned with each other than the Acta2 fibers in the control fibroblasts (Figure 4E). Quantification of overall Acta2 fluorescent intensity revealed that Ror1/2 deficiency led to diminished TGF‐β1–induced Acta2 expression at the protein level (Figure S6C). Taken together, these results show that Ror1/2 knockout fibroblasts are less responsive to TGF‐β1, a feature that is consistent with the notion that these cells are in the less mature, proinflammatory state. These results further suggest that the planar cell polarity signaling pathway may be critical for actin alignment during fibroblast maturation.
TAC Induces Hyperinflammation, Rapid Heart Failure, and Death in Ror1/2 Double‐Knockout Mice
To investigate the role of Ror1/2 in cardiac tissue remodeling in vivo, the inducible Ror1/2 double‐knockout mice (Ror1/2fl/fl+Ubc‐CreERT2) were treated with tamoxifen for 2 weeks and subjected to TAC. Ror1/2 double‐knockout mice displayed a rapid decline in heart function after TAC, as observed by echocardiography measurements at 3 days after TAC (Figure 5A and 5B). This rapid development of systolic dysfunction was associated with death of the Ror1/2 double‐knockout mice between 4 and 6 days after TAC (Figure 5C). Consistent with the heart failure phenotype, the TAC‐treated, Ror1/2 double‐knockout mice displayed marked increases in heart and lung weights (Figure S7).
Figure 5. Early heart failure in transgenic Ror1/2 double‐knockout mice after TAC surgery.
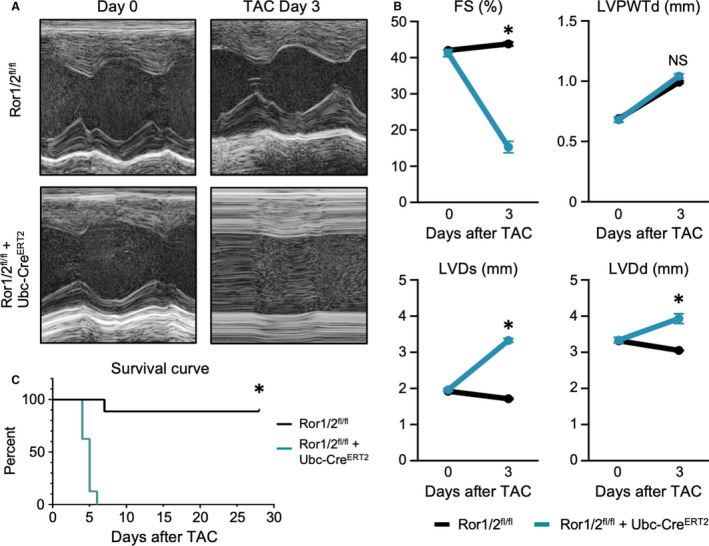
A, Transgenic Ror1/2 double‐knockout and control mice were subjected to TAC surgery, and cardiac output was imaged by echocardiography (n=9 mice were used per group in each experimental condition). B, Cardiac output factors were quantified: fractional shortening (FS), left ventricular posterior wall thickness at end diastole (LVPWTd), left ventricular diameter at end systole (LVDs), and left ventricular diameter at end diastole (LVDd) (graphs represent mean±SEM, sample size=5, Mann‐Whitney P values at day 3: FS, %=0.0079, LVPWTd, mm=0.8413, LVDs, mm=0.0079, LVDd, mm=0.0079). C, Survival of Ror1/2 double‐knockout mice and control mice after TAC surgery was recorded each day (sample size=10 per group, log‐rank [Mantel‐Cox] test P<0.0001). Ror indicates receptor tyrosine kinase–like orphan receptor; TAC, transverse aortic constriction; and Ubc‐CreERT2, ubiquitin C‐driven Cre recombinase with tamoxifen‐inducible mutant human estrogen receptor. *p<0.05.
Hematoxylin and eosin staining of heart sections revealed a massive inflammatory infiltrate in the Ror1/2 double‐knockout mice after TAC (Figure 6A). Inflammatory cell infiltration was further characterized by flow cytometry analysis of cells isolated from digested cardiac tissue at 3 days after TAC. This analysis revealed a large increase in the number of CD11b+Ly6G+ neutrophils, CD11b+Ly6G‐F4/80+ macrophages, and CD11b+Ly6G‐Ly6C+ monocytes in the Ror1/2 double‐knockout mice compared with wild‐type mice treated by TAC (Figure 6B and 6C). In Ror1/2 double‐knockout mice, TAC also caused a large induction of proinflammatory cytokine gene transcripts including Il1b, Il6, and Ccl2 (Figure 6D). Notably, vascular leakage could also be detected after TAC in the Ror1/2 double‐knockout mice as indicated by the permeability of the myocardium to Evans Blue dye (Figure 6E). These data reveal that the hearts of TAC‐treated, Ror1/2 double‐knockout mice are hyperinflammatory. This phenotype is consistent with the notion that Ror1/2 is essential for the progression of fibroblast maturation from the initial proinflammatory stage during their activation sequence.
Figure 6. Inflammation response in transgenic Ror1/2 double‐knockout mice after TAC surgery.

Inflammatory profile of control and transgenic Ror1/2 double‐knockout mice was assessed after TAC or sham surgery (n=5 mice were used per group in each experimental condition). A, Hematoxylin and eosin staining of cardiac tissues 3 days after TAC were imaged. B, Cells were isolated from cardiac tissue and relative quantity of leukocyte populations were determined by flow cytometry, (C) quantified by absolute number (bars represent mean±SEM, sample size=5 per group, Mann‐Whitney P values: neutrophil=0.0079, monocyte=0.0079, macrophage=0.0079). D, Gene expression of proinflammatory cytokines in cardiac lysate was measured (bars represent mean±SEM, sample size=5 per group, Kruskal‐Wallis P values: Il1b=0.0004, Il6=0.0030, Ccl2=0.0011). E, Vascular permeability at 1 day after TAC was assessed by Evans Blue dye injection, with vascular leakage visualized by blue dye in the cardiac tissue. Ror indicates receptor tyrosine kinase–like orphan receptor; TAC, transverse aortic constriction; and Ubc‐CreERT2, ubiquitous cre recombinase estrogen receptor (tamoxifen‐inducible). *p<0.05.
Discussion
Through these experiments, we examined the roles of the cell surface receptors Ror1 and Ror2 in cardiac myofibroblast differentiation. We found that Ror1 and Ror2 are upregulated in fibroblasts early after activation, both in vivo by pressure overload cardiac injury induced by TAC surgery and in cardiac fibroblast cell culture. Multiple lines of data indicate that dual Ror1/2 deficiency impairs myofibroblast differentiation. Specifically, cultured Ror1/2 knockout fibroblasts exhibited an immature, proinflammatory state, and they were impaired in their response to TGF‐β1 stimulation. Additionally, while mice lacking Ror1/2 did not exhibit a baseline phenotype, they were unable to acutely adapt to pressure overload cardiac injury. In response to TAC, these mice underwent profound decompensated remodeling by 3 days, and they typically died within 6 days of surgery. Collectively, these results suggest that the early induction of Ror1/2 in fibroblasts is essential for the appropriate myofibroblast differentiation and required for the myocardium to adapt to the initial stages of pressure overload injury.
The phenotype of the Ror1/2 double‐knockout mice highlighted a critical role for Ror1/2 in controlling the inflammatory phenotype of fibroblasts during the early stages of cardiac remodeling. The rapid upregulation of cytokines in response to TAC injury leads to the infiltration of neutrophils and monocyte‐derived macrophages.43, 65 Several recent studies have highlighted the role of fibroblasts in promoting inflammation during the initial phases of the cardiac remodeling response to myocardial infarction10, 12 and pressure overload.66, 67 Our results expand on these studies by showing that fibroblasts lacking Ror1/2 appear to be phenotypically similar to fibroblasts in the transient, proinflammatory state that occurs early in the fibroblast differentiation continuum following experimental myocardial infarction.10 Transcriptomic analyses of cultured cardiac fibroblasts revealed that Ror1/2 deficiency was associated with a decrease in pathways that promote fibrosis (proliferation genes, extracellular signal‐related kinase 1/2 signaling genes, tissue inhibitors of matrix metalloproteinases, and mothers against decapentaplegic homolog 2/3) and an increase in pathways that inhibit fibrosis (inflammatory cytokines, matrix metalloproteinases, and natriuretic signaling genes). These differences in gene expression suggest that the Ror1/2 knockout fibroblasts are in an immature state that is proinflammatory, leukocyte‐recruiting and less responsive to fibrotic stimuli. We confirmed this by showing that Ror1/2 knockout fibroblasts were less responsive to TGF‐β1 compared with control fibroblasts, displaying reduced induction of Acta2 and Postn gene expression and diminished repression of Il6 gene expression. Thus, we propose that Ror1/2 deficiency causes activated fibroblasts to stall in an early proinflammatory state of differentiation such that they are unable to efficiently transition to the intermediate proliferative state and mature homeostatic state. Consistent with this hypothesis, we found that Ror1/2 double‐knockout mice exhibit a hyperinflammatory phenotype associated with elevated cytokine transcript expression, exuberant inflammatory cell infiltration, and vascular leakage within 3 days of TAC surgery. We speculate that the premature cardiac mortality observed in Ror1/2 knockout mice after TAC is attributable to fibroblast‐mediated vascular dysfunction that leads to excessive leukocyte recruitment and the subsequent impairment of cardiac contraction. This hypothesis can be rigorously tested by models that ablate these genes in specific fibroblast subtypes. While recent studies using single‐cell RNA sequencing have highlighted the large diversity of fibroblast cell types in the heart,12, 68, 69 our study does not identify the fibroblast subtype(s) that upregulate Ror1/2. Future studies should address this important issue. Collectively, the results in this study highlight the critical role of early Ror1/2 induction in the control of the early inflammatory response in the injured myocardium, and they suggest that hyperinflammatory activated fibroblasts can have a detrimental role in myocardial remodeling.
In myofibroblasts, appropriate α‐smooth muscle actin stress fiber organization is required for the transduction of signaling responses to external stimuli,39 and the misalignment of α‐smooth muscle actin stress fibers is associated with dysregulated myofibroblast differentiation.70, 71 Consistent with the notion that Ror1/2 induction is essential for myofibroblast differentiation, we find that Ror1/2 deficiency leads to α‐smooth muscle actin filament misalignment in TGF‐β1‐treated fibroblasts. Ror1/2 signaling can control stress fiber alignment, myofibroblast differentiation, and other cellular phenotypes through the regulation of upstream and downstream planar cell polarity components.13, 40, 43, 46, 56, 72, 73, 74 This system involves the participation of inner plasma membrane proteins associated with Ror1/2 that control cell polarity and asymmetric cell division through the regulation of actin filament organization by the action of the small guanosine triphosphatases RhoA and Rac.31, 32 Our work is consistent with other studies that have implicated planar cell polarity proteins in myofibroblast differentiation, including the reported induction of Frizzled2 in myofibroblasts after experimental myocardial infarction75 and hypoxia‐mediated suppression of myofibroblast differentiation through RhoA inhibition.76 Furthermore, ablation of mothers against decapentaplegic homolog 3, a key signaling protein downstream of TGF‐β cardiac fibroblasts,77 will lead to the suppression of RhoA and a disruption of actin alignment.71 The coregulation of Ror1/2 and components of planar cell polarity signaling in activated cardiac fibroblasts suggests that asymmetrical cell division plays a role in myofibroblast differentiation. Consistent with this notion, the planar cell polarity pathway has been shown to influence actin filament assembly, cell structure, and asymmetrical cell division in various cell types and organisms.41, 78, 79, 80, 81, 82 Together, these results suggest a potential role of planar cell polarity signaling in mediating stress fiber organization and TGF‐β responses in activated cardiac fibroblasts.
We acknowledge that Ror1/2 double‐deficient mice were constructed using a global knockout strategy that employed ubiquitin‐CreERT2. As discussed, these mice lack a baseline phenotype, yet undergo rapid decompensated heart failure and death in response to pressure overload. Although we cannot rule out the contribution of other cell types lacking Ror1/2 to this phenotype, we note that Ror1/2 expression appears specific to activated cardiac fibroblasts under these conditions. Our experimental evidence in support of this include that expression of Ror1/2 was greatly increased after TAC‐induced cardiac injury in the interstitial cells of the myocardium, and the TAC‐induced upregulation of Ror1/2 gene expression was specific to fibroblasts with no Ror1/2 induction in endothelial cells or leukocytes. Consistent with these findings, a number of other studies have reported Ror1 and Ror2 expression in activated fibroblasts or fibroblast‐like cells,12, 83, 84, 85, 86 and a recent proteomic analysis of the human heart reported that ROR1 expression was 200‐fold higher in than cardiac fibroblasts than other cardiac cell types.87 In addition, studies with isolated fibroblasts further corroborate the in vivo observations. These in vitro studies documented the robust expression of Ror1/2 and associated planar cell polarity protein transcripts (tyrosine‐protein kinase–like 7; Vangl1 and 2; prickle homologue 1; and disheveled 1, 2 and 3) in activated cardiac fibroblasts, and showed that the ablation of Ror1/2 leads to a proinflammatory phenotype that is consistent with the cardiac phenotype of the Ror1/2‐deficient mouse following TAC surgery.
Conclusions
Cardiac fibroblasts are sentinel cells in the heart that respond to early injury by adopting a proinflammatory and leukocyte‐recruiting phenotype, followed by their transition to a reparative/proliferative phase that is proangiogenic and profibrotic, and followed by a homeostatic phase. Our study reveals the critical role that the cell surface receptors Ror1 and Ror2 play in allowing myofibroblasts to appropriately transition through these phases. While the deficiency of these proteins has no detectable baseline phenotype, Ror1/2‐deficient fibroblasts activated by pressure overload appears stalled in the early phase of the fibroblast differentiation continuum—a stage of differentiation that is highly proinflammatory. Further analysis of the injury‐induced, hyperinflammatory phenotype of the Ror1/2 double‐knockout mice may provide a greater window of understanding of how excessive inflammation contributes to pathological cardiac remodeling. Elevated inflammation is predictive of worse outcomes in patients with heart failure,88, 89 and recent clinical trials of anti‐inflammatory therapies targeting interleukin‐1β and interleukin‐1R1 have shown promising results in the treatment of this condition in some patient groups.90, 91, 92 Our results suggest that impairments in planar cell polarity‐mediated regulation of myofibroblast differentiation could have a causative role in the development of excessive inflammation that is associated with heart failure. Therefore, a better understanding of the molecular mechanisms that regulate the progression of myofibroblast differentiation could provide opportunities for the development of therapies that more effectively reduce cardiac inflammation.
Sources of Funding
This study was supported by grants to Dr Chavkin (NIH T32 HL007224, NIH T32 HL007284), Dr S. Sano (NIH R01 HL152174), Dr Wang (China Scholarship Council), Dr Ogawa (Japan Heart Foundation), A. Nelson (NIH T32 HL007284), Dr Saucerman (NIH R01 HL137755), Dr Hirschi (R01 HL146056, U2EB017103), Dr Walsh (NIH R01 HL138014, 139819 and HL141256), and Drs Gokce and Walsh (NIH R01 HL142650).
Disclosures
None.
Supporting information
Data S1–S2
Table S1
Figures S1–S7
Acknowledgments
The authors thank the University of Virginia Genome Analysis and Technology Core for advice and technical support on bulk RNA sequencing, and the University of Virginia Health Sciences Library and Research Computing Center for assistance with computational analysis.
(J Am Heart Assoc. 2021;10:e019904. DOI: 10.1161/JAHA.120.019904.)
Preprint posted on BioRxiv March 2, 2021. doi: https://doi.org/10.1101/2021.03.02.433549.
Supplementary Material for this article is available at https://www.ahajournals.org/doi/suppl/10.1161/JAHA.120.019904
For Sources of Funding and Disclosures, see page 13.
References
- 1.Fan D, Takawale A, Lee J, Kassiri Z. Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease. Fibrogenesis Tissue Repair. 2012;5:15. DOI: 10.1186/1755-1536-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van den Borne SW, Diez J, Blankesteijn WM, Verjans J, Hofstra L, Narula J. Myocardial remodeling after infarction: the role of myofibroblasts. Nat Rev Cardiol. 2010;7:30–37. DOI: 10.1038/nrcardio.2009.199. [DOI] [PubMed] [Google Scholar]
- 3.Tallquist MD, Molkentin JD. Redefining the identity of cardiac fibroblasts. Nat Rev Cardiol. 2017;14:484–491. DOI: 10.1038/nrcardio.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, J. Lin S‐C, Aronow BJ, Tallquist MD, et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat Commun. 2016;7:12260. DOI: 10.1038/ncomms12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oka T, Xu J, Kaiser RA, Melendez J, Hambleton M, Sargent MA, Lorts A, Brunskill EW, Dorn GW, Conway SJ, et al. Genetic manipulation of periostin expression reveals a role in cardiac hypertrophy and ventricular remodeling. Circ Res. 2007;101:313–321. DOI: 10.1161/CIRCRESAHA.107.149047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maruyama S, Nakamura K, Papanicolaou KN, Sano S, Shimizu I, Asaumi Y, van den Hoff MJ, Ouchi N, Recchia FA, Walsh K. Follistatin‐like 1 promotes cardiac fibroblast activation and protects the heart from rupture. EMBO Mol Med. 2016;8:949–966. DOI: 10.15252/emmm.201506151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson WJ, Clarke SA, Quinn TA, Holmes JW. Physiological implications of myocardial scar structure. Compr Physiol. 2015;5:1877–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber KT, Brilla CG, Janicki JS. Myocardial fibrosis: functional significance and regulatory factors. Cardiovasc Res. 1993;27:341–348. DOI: 10.1093/cvr/27.3.341. [DOI] [PubMed] [Google Scholar]
- 9.Frangogiannis NG. The extracellular matrix in ischemic and nonischemic heart failure. Circ Res. 2019;125:117–146. DOI: 10.1161/CIRCRESAHA.119.311148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mouton AJ, Ma Y, Rivera Gonzalez OJ, Daseke MJ II, Flynn ER, Freeman TC, Garrett MR, DeLeon‐Pennell KY, Lindsey ML. Fibroblast polarization over the myocardial infarction time continuum shifts roles from inflammation to angiogenesis. Basic Res Cardiol. 2019;114:6. DOI: 10.1007/s00395-019-0715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daseke MJ, Tenkorang MAA, Chalise U, Konfrst SR, Lindsey ML. Cardiac fibroblast activation during myocardial infarction wound healing: fibroblast polarization after MI. Matrix Biol. 2020;91‐92:109–116. DOI: 10.1016/j.matbio.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farbehi N, Patrick R, Dorison A, Xaymardan M, Janbandhu V, Wystub‐Lis K, Ho JWK, Nordon RE, Harvey RP. Single‐cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. ELife. 2019;8:e43882. DOI: 10.7554/eLife.43882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Rubin B, Bodine PV, Billiard J. Wnt5a induces homodimerization and activation of Ror2 receptor tyrosine kinase. J Cell Biochem. 2008;105:497–502. DOI: 10.1002/jcb.21848. [DOI] [PubMed] [Google Scholar]
- 14.Katoh M. WNT/PCP signaling pathway and human cancer (review). Oncol Rep. 2005;14:1583–1588. DOI: 10.3892/or.14.6.1583. [DOI] [PubMed] [Google Scholar]
- 15.Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. DOI: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuda T, Nomi M, Ikeya M, Kani S, Oishi I, Terashima T, Takada S, Minami Y. Expression of the receptor tyrosine kinase genes, Ror1 and Ror2, during mouse development. Mech Dev. 2001;105:153–156. DOI: 10.1016/S0925-4773(01)00383-5. [DOI] [PubMed] [Google Scholar]
- 17.Lyashenko N, Weissenbock M, Sharir A, Erben RG, Minami Y, Hartmann C. Mice lacking the orphan receptor Ror1 have distinct skeletal abnormalities and are growth retarded. Dev Dyn. 2010;239:2266–2277. DOI: 10.1002/dvdy.22362. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi S, Takeda K, Oishi I, Nomi M, Ikeya M, Itoh K, Tamura S, Ueda T, Hatta T, Otani H, et al. Mouse Ror2 receptor tyrosine kinase is required for the heart development and limb formation. Genes Cells. 2000;5:71–78. DOI: 10.1046/j.1365-2443.2000.00300.x. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi M, Shibuya Y, Takeuchi J, Murata M, Suzuki H, Yokoo S, Umeda M, Minami Y, Komori T. Ror2 expression in squamous cell carcinoma and epithelial dysplasia of the oral cavity. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;107:398–406. DOI: 10.1016/j.tripleo.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 20.Daneshmanesh AH, Porwit A, Hojjat‐Farsangi M, Jeddi‐Tehrani M, Tamm KP, Grandér D, Lehmann S, Norin S, Shokri F, Rabbani H, et al. Orphan receptor tyrosine kinases ROR1 and ROR2 in hematological malignancies. Leuk Lymphoma. 2013;54:843–850. DOI: 10.3109/10428194.2012.731599. [DOI] [PubMed] [Google Scholar]
- 21.Henry CE, Emmanuel C, Lambie N, Loo C, Kan B, Kennedy CJ, de Fazio A, Hacker NF, Ford CE. Distinct patterns of stromal and tumor expression of ROR1 and ROR2 in histological subtypes of epithelial ovarian cancer. Transl Oncol. 2017;10:346–356. DOI: 10.1016/j.tranon.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayerlová M, Menck K, Klemm F, Wolff A, Pukrop T, Binder C, Beißbarth T, Bleckmann A. Ror2 signaling and its relevance in breast cancer progression. Front Oncol. 2017;7:135. DOI: 10.3389/fonc.2017.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry CE, Llamosas E, Daniels B, Coopes A, Tang K, Ford CE. ROR1 and ROR2 play distinct and opposing roles in endometrial cancer. Gynecol Oncol. 2018;148:576–584. DOI: 10.1016/j.ygyno.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 24.Liu D, Gunther K, Enriquez LA, Daniels B, O'Mara TA, Tang K, Spurdle AB, Ford CE. ROR1 is upregulated in endometrial cancer and represents a novel therapeutic target. Sci Rep. 2020;10:13906. DOI: 10.1038/s41598-020-70924-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry C, Hacker N, Ford C. Silencing ROR1 and ROR2 inhibits invasion and adhesion in an organotypic model of ovarian cancer metastasis. Oncotarget. 2017;8:112727–112738. DOI: 10.18632/oncotarget.22559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saji T, Nishita M, Ogawa H, Doi T, Sakai Y, Maniwa Y, Minami Y. Critical role of the ror‐family of receptor tyrosine kinases in invasion and proliferation of malignant pleural mesothelioma cells. Genes Cells. 2018;23:606–613. DOI: 10.1111/gtc.12599. [DOI] [PubMed] [Google Scholar]
- 27.Yu J, Chen L, Cui B, Widhopf GF II, Shen Z, Wu R, Zhang L, Zhang S, Briggs SP, Kipps TJ. Wnt5a induces ROR1/ROR2 heterooligomerization to enhance leukemia chemotaxis and proliferation. J Clin Invest. 2016;126:585–598. DOI: 10.1172/JCI83535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S, Chen L, Cui B, Chuang HY, Yu J, Wang‐Rodriguez J, Tang L, Chen G, Basak GW, Kipps TJ. ROR1 is expressed in human breast cancer and associated with enhanced tumor‐cell growth. PLoS One. 2012;7:e31127. DOI: 10.1371/journal.pone.0031127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamizaki K, Doi R, Hayashi M, Saji T, Kanagawa M, Toda T, Fukada SI, Ho HH, Greenberg ME, Endo M, et al. The Ror1 receptor tyrosine kinase plays a critical role in regulating satellite cell proliferation during regeneration of injured muscle. J Biol Chem. 2017;292:15939–15951. DOI: 10.1074/jbc.M117.785709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butler MT, Wallingford JB. Planar cell polarity in development and disease. Nat Rev Mol Cell Biol. 2017;18:375–388. DOI: 10.1038/nrm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seifert JR, Mlodzik M. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet. 2007;8:126–138. DOI: 10.1038/nrg2042. [DOI] [PubMed] [Google Scholar]
- 32.Gao BO, Song H, Bishop K, Elliot G, Garrett L, English MA, Andre P, Robinson J, Sood R, Minami Y, et al. Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev Cell. 2011;20:163–176. DOI: 10.1016/j.devcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradley EW, Drissi MH. Wnt5b regulates mesenchymal cell aggregation and chondrocyte differentiation through the planar cell polarity pathway. J Cell Physiol. 2011;226:1683–1693. DOI: 10.1002/jcp.22499. [DOI] [PubMed] [Google Scholar]
- 34.Brzoska HL, d'Esposito AM, Kolatsi‐Joannou M, Patel V, Igarashi P, Lei Y, Finnell RH, Lythgoe MF, Woolf AS, Papakrivopoulou E, et al. Planar cell polarity genes Celsr1 and Vangl2 are necessary for kidney growth, differentiation, and rostrocaudal patterning. Kidney Int. 2016;90:1274–1284. DOI: 10.1016/j.kint.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cortijo C, Gouzi M, Tissir F, Grapin‐Botton A. Planar cell polarity controls pancreatic beta cell differentiation and glucose homeostasis. Cell Rep. 2012;2:1593–1606. DOI: 10.1016/j.celrep.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vladar EK, Nayak JV, Milla CE, Axelrod JD. Airway epithelial homeostasis and planar cell polarity signaling depend on multiciliated cell differentiation. JCI Insight. 2016;1:e88027. DOI: 10.1172/jci.insight.88027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wada H, Okamoto H. Roles of planar cell polarity pathway genes for neural migration and differentiation. Dev Growth Differ. 2009;51:233–240. DOI: 10.1111/j.1440-169X.2009.01092.x. [DOI] [PubMed] [Google Scholar]
- 38.Yang X, Qian X, Ma R, Wang X, Yang J, Luo W, Chen P, Chi F, Ren D. Establishment of planar cell polarity is coupled to regional cell cycle exit and cell differentiation in the mouse utricle. Sci Rep. 2017;7:43021. DOI: 10.1038/srep43021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandbo N, Dulin N. Actin cytoskeleton in myofibroblast differentiation: ultrastructure defining form and driving function. Transl Res. 2011;158:181–196. DOI: 10.1016/j.trsl.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura K, Sano S, Fuster JJ, Kikuchi R, Shimizu I, Ohshima K, Katanasaka Y, Ouchi N, Walsh K. Secreted frizzled‐related protein 5 diminishes cardiac inflammation and protects the heart from ischemia/reperfusion injury. J Biol Chem. 2016;291:2566–2575. DOI: 10.1074/jbc.M115.693937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abraityte A, Vinge LE, Askevold ET, Lekva T, Michelsen AE, Ranheim T, Alfsnes K, Fiane A, Aakhus S, Lunde IG, et al. Wnt5a is elevated in heart failure and affects cardiac fibroblast function. J Mol Med (Berl). 2017;95:767–777. DOI: 10.1007/s00109-017-1529-1. [DOI] [PubMed] [Google Scholar]
- 42.Mizutani M, Wu JC, Nusse R. Fibrosis of the neonatal mouse heart after cryoinjury is accompanied by Wnt signaling activation and epicardial‐to‐mesenchymal transition. J Am Heart Assoc. 2016;5:e002457. DOI: 10.1161/JAHA.115.002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Sano S, Oshima K, Sano M, Watanabe Y, Katanasaka Y, Yura Y, Jung C, Anzai A, Swirski FK, et al. Wnt5a‐mediated neutrophil recruitment has an obligatory role in pressure overload–induced cardiac dysfunction. Circulation. 2019;140:487–499. DOI: 10.1161/CIRCULATIONAHA.118.038820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong S, Du Y, Ji Q, Dong R, Cao J, Wang Z, Li W, Zeng M, Chen H, Zhu X, et al. Expression of Sfrp5/Wnt5a in human epicardial adipose tissue and their relationship with coronary artery disease. Life Sci. 2020;245:117338. DOI: 10.1016/j.lfs.2020.117338. [DOI] [PubMed] [Google Scholar]
- 45.Abraityte A, Lunde IG, Askevold ET, Michelsen AE, Christensen G, Aukrust P, Yndestad A, Fiane A, Andreassen A, Aakhus S, et al. Wnt5a is associated with right ventricular dysfunction and adverse outcome in dilated cardiomyopathy. Sci Rep. 2017;7:3490. DOI: 10.1038/s41598-017-03625-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hagenmueller M, Riffel JH, Bernhold E, Fan J, Katus HA, Hardt SE. Dapper‐1 is essential for Wnt5a induced cardiomyocyte hypertrophy by regulating the Wnt/PCP pathway. FEBS Lett. 2014;588:2230–2237. DOI: 10.1016/j.febslet.2014.05.039. [DOI] [PubMed] [Google Scholar]
- 47.Heliste J, Jokilammi A, Paatero I, Chakroborty D, Stark C, Savunen T, Laaksonen M, Elenius K. Receptor tyrosine kinase profiling of ischemic heart identifies ROR1 as a potential therapeutic target. BMC Cardiovasc Disord. 2018;18:196. DOI: 10.1186/s12872-018-0933-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edwards JJ, Brandimarto J, Hu D‐Q, Jeong S, Yucel N, Li LI, Bedi KC, Wada S, Murashige D, Hwang HTV, et al. Noncanonical WNT activation in human right ventricular heart failure. Front Cardiovasc Med. 2020;7:582407. DOI: 10.3389/fcvm.2020.582407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho HY, Susman MW, Bikoff JB, Ryu YK, Jonas AM, Hu L, Kuruvilla R, Greenberg ME. Wnt5a‐Ror‐dishevelled signaling constitutes a core developmental pathway that controls tissue morphogenesis. Proc Natl Acad Sci USA. 2012;109:4044–4051. DOI: 10.1073/pnas.1200421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruzankina Y, Pinzon‐Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age‐related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. DOI: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeChiara TM, Kimble RB, Poueymirou WT, Rojas J, Masiakowski P, Valenzuela DM, Yancopoulos GD. Ror2, encoding a receptor‐like tyrosine kinase, is required for cartilage and growth plate development. Nat Genet. 2000;24:271–274. DOI: 10.1038/73488. [DOI] [PubMed] [Google Scholar]
- 52.Papanicolaou KN, Streicher JM, Ishikawa TO, Herschman H, Wang Y, Walsh K. Preserved heart function and maintained response to cardiac stresses in a genetic model of cardiomyocyte‐targeted deficiency of cyclooxygenase‐2. J Mol Cell Cardiol. 2010;49:196–209. DOI: 10.1016/j.yjmcc.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Acharya A, Baek ST, Huang G, Eskiocak B, Goetsch S, Sung CY, Banfi S, Sauer MF, Olsen GS, Duffield JS, et al. The bHLH transcription factor Tcf21 is required for lineage‐specific EMT of cardiac fibroblast progenitors. Development. 2012;139:2139–2149. DOI: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okuda H, Miyata S, Mori Y, Tohyama M. Mouse Prickle1 and Prickle2 are expressed in postmitotic neurons and promote neurite outgrowth. FEBS Lett. 2007;581:4754–4760. [DOI] [PubMed] [Google Scholar]
- 55.Rocque BL, Babayeva S, Li J, Leung V, Nezvitsky L, Cybulsky AV, Gros P, Torban E. Deficiency of the planar cell polarity protein Vangl2 in podocytes affects glomerular morphogenesis and increases susceptibility to injury. J Am Soc Nephrol. 2015;26:576–586. DOI: 10.1681/ASN.2014040340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fuster JJ, Zuriaga MA, Ngo DT, Farb MG, Aprahamian T, Yamaguchi TP, Gokce N, Walsh K. Noncanonical wnt signaling promotes obesity‐induced adipose tissue inflammation and metabolic dysfunction independent of adipose tissue expansion. Diabetes. 2015;64:1235–1248. DOI: 10.2337/db14-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chavkin NW, Hirschi KK. Single cell analysis in vascular biology. Front Cardiovasc Med. 2020;7:42. DOI: 10.3389/fcvm.2020.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zafiriou MP, Noack C, Unsöld B, Didie M, Pavlova E, Fischer HJ, Reichardt HM, Bergmann MW, El‐Armouche A, Zimmermann WH, et al. Erythropoietin responsive cardiomyogenic cells contribute to heart repair post myocardial infarction. Stem Cells. 2014;32:2480–2491. DOI: 10.1002/stem.1741. [DOI] [PubMed] [Google Scholar]
- 59.Bray NL, Pimentel H, Melsted P, Pachter L. Near‐optimal probabilistic RNA‐seq quantification. Nat Biotechnol. 2016;34:525–527. DOI: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 60.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA‐seq incorporating quantification uncertainty. Nat Methods. 2017;14:687–690. DOI: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 61.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. DOI: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.deAlmeida AC, van Oort RJ, Wehrens XH. Transverse aortic constriction in mice. J Vis Exp. 2010:38: 1729. DOI: 10.3791/1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeigler AC, Richardson WJ, Holmes JW, Saucerman JJ. A computational model of cardiac fibroblast signaling predicts context‐dependent drivers of myofibroblast differentiation. J Mol Cell Cardiol. 2016;94:72–81. DOI: 10.1016/j.yjmcc.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeigler AC, Nelson AR, Chandrabhatla AS, Brazhkina O, Holmes JW, Saucerman JJ. Computational model predicts paracrine and intracellular drivers of fibroblast phenotype after myocardial infarction. Matrix Biol. 2020;91‐92:136–151. DOI: 10.1016/j.matbio.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patel B, Bansal SS, Ismahil MA, Hamid T, Rokosh G, Mack M, Prabhu SD. CCR2 + monocyte‐derived infiltrating macrophages are required for adverse cardiac remodeling during pressure overload. JACC Basic Transl Sci. 2018;3:230–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Unudurthi SD, Nassal DM, Patel NJ, Thomas E, Yu J, Pierson CG, Bansal SS, Mohler PJ, Hund TJ. Fibroblast growth factor‐inducible 14 mediates macrophage infiltration in heart to promote pressure overload‐induced cardiac dysfunction. Life Sci. 2020;247:117440. DOI: 10.1016/j.lfs.2020.117440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzuki K, Satoh K, Ikeda S, Sunamura S, Otsuki T, Satoh T, Kikuchi N, Omura J, Kurosawa R, Nogi M, et al. Basigin promotes cardiac fibrosis and failure in response to chronic pressure overload in mice. Arterioscler Thromb Vasc Biol. 2016;36:636–646. DOI: 10.1161/ATVBAHA.115.306686. [DOI] [PubMed] [Google Scholar]
- 68.Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA, Pinto AR. Single‐cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 2018;22:600–610. DOI: 10.1016/j.celrep.2017.12.072. [DOI] [PubMed] [Google Scholar]
- 69.Gladka MM, Molenaar B, de Ruiter H, van der Elst S, Tsui H, Versteeg D, Lacraz GPA, Huibers MMH, van Oudenaarden A, van Rooij E. Single‐cell sequencing of the healthy and diseased heart reveals cytoskeleton‐associated protein 4 as a new modulator of fibroblasts activation. Circulation. 2018;138:166–180. DOI: 10.1161/CIRCULATIONAHA.117.030742. [DOI] [PubMed] [Google Scholar]
- 70.Wang YS, Li SH, Guo J, Mihic A, Wu J, Sun L, Davis K, Weisel RD, Li RK. Role of miR‐145 in cardiac myofibroblast differentiation. J Mol Cell Cardiol. 2014;66:94–105. DOI: 10.1016/j.yjmcc.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 71.Huang S, Chen B, Su Y, Alex L, Humeres C, Shinde AV, Conway SJ, Frangogiannis NG. Distinct roles of myofibroblast‐specific Smad2 and Smad3 signaling in repair and remodeling of the infarcted heart. J Mol Cell Cardiol. 2019;132:84–97. DOI: 10.1016/j.yjmcc.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiao Q, Chen Z, Jin X, Mao R, Chen Z. The many postures of noncanonical Wnt signaling in development and diseases. Biomed Pharmacother. 2017;93:359–369. DOI: 10.1016/j.biopha.2017.06.061. [DOI] [PubMed] [Google Scholar]
- 73.Abdolmaleki F, Ahmadpour‐Yazdi H, Hayat SMG, Gheibi N, Johnston TP, Sahebkar A. Wnt network: a brief review of pathways and multifunctional components. Crit Rev Eukaryot Gene Expr. 2020;30:1–18. DOI: 10.1615/CritRevEukaryotGeneExpr.2019025774. [DOI] [PubMed] [Google Scholar]
- 74.Kikuchi R, Nakamura K, MacLauchlan S, Ngo D‐M, Shimizu I, Fuster JJ, Katanasaka Y, Yoshida S, Qiu Y, Yamaguchi TP, et al. An antiangiogenic isoform of VEGF‐A contributes to impaired vascularization in peripheral artery disease. Nat Med. 2014;20:1464–1471. DOI: 10.1038/nm.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blankesteijn WM, Essers‐Janssen YP, Verluyten MJ, Daemen MJ, Smits JF. A homologue of drosophila tissue polarity gene frizzled is expressed in migrating myofibroblasts in the infarcted rat heart. Nat Med. 1997;3:541–544. DOI: 10.1038/nm0597-541. [DOI] [PubMed] [Google Scholar]
- 76.Leinhos L, Peters J, Krull S, Helbig L, Vogler M, Levay M, van Belle GJ, Ridley AJ, Lutz S, Katschinski DM, et al. Hypoxia suppresses myofibroblast differentiation by changing RhoA activity. J Cell Sci. 2019;132:jcs223230. DOI: 10.1242/jcs.223230. [DOI] [PubMed] [Google Scholar]
- 77.Khalil H, Kanisicak O, Prasad V, Correll RN, Fu X, Schips T, Vagnozzi RJ, Liu R, Huynh T, Lee SJ, et al. Fibroblast‐specific TGF‐beta‐Smad2/3 signaling underlies cardiac fibrosis. J Clin Invest. 2017;127:3770–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Winter CG, Wang B, Ballew A, Royou A, Karess R, Axelrod JD, Luo L. Drosophila Rho‐associated kinase (Drok) links frizzled‐mediated planar cell polarity signaling to the actin cytoskeleton. Cell. 2001;105:81–91. DOI: 10.1016/S0092-8674(01)00298-7. [DOI] [PubMed] [Google Scholar]
- 79.Gong Y, Mo C, Fraser SE. Planar cell polarity signalling controls cell division orientation during zebrafish gastrulation. Nature. 2004;430:689–693. DOI: 10.1038/nature02796. [DOI] [PubMed] [Google Scholar]
- 80.Devenport D, Fuchs E. Planar polarization in embryonic epidermis orchestrates global asymmetric morphogenesis of hair follicles. Nat Cell Biol. 2008;10:1257–1268. DOI: 10.1038/ncb1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Witze ES, Litman ES, Argast GM, Moon RT, Ahn NG. Wnt5a control of cell polarity and directional movement by polarized redistribution of adhesion receptors. Science. 2008;320:365–369. DOI: 10.1126/science.1151250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee J, Andreeva A, Sipe CW, Liu L, Cheng A, Lu X. PTK7 regulates myosin II activity to orient planar polarity in the mammalian auditory epithelium. Curr Biol. 2012;22:956–966. DOI: 10.1016/j.cub.2012.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X, Yamagata K, Nishita M, Endo M, Arfian N, Rikitake Y, Emoto N, Hirata K, Tanaka Y, Minami Y. Activation of Wnt5a‐Ror2 signaling associated with epithelial‐to‐mesenchymal transition of tubular epithelial cells during renal fibrosis. Genes Cells. 2013;18:608–619. DOI: 10.1111/gtc.12064. [DOI] [PubMed] [Google Scholar]
- 84.Takahashi D, Suzuki H, Kakei Y, Yamakoshi K, Minami Y, Komori T, Nishita M. Expression of Ror2 associated with fibrosis of the submandibular gland. Cell Struct Funct. 2017;42:159–167. DOI: 10.1247/csf.17019. [DOI] [PubMed] [Google Scholar]
- 85.Li C, Smith SM, Peinado N, Gao F, Li W, Lee MK, Zhou B, Bellusci S, Pryhuber GS, Ho H‐Y, et al. WNT5A‐ROR signaling is essential for alveologenesis. Cells. 2020;9:384. DOI: 10.3390/cells9020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wilson DH, Jarman EJ, Mellin RP, Wilson ML, Waddell SH, Tsokkou P, Younger NT, Raven A, Bhalla SR, Noll ATR, et al. Non‐canonical wnt signalling regulates scarring in biliary disease via the planar cell polarity receptors. Nat Commun. 2020;11:445. DOI: 10.1038/s41467-020-14283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Doll S, Dreßen M, Geyer PE, Itzhak DN, Braun C, Doppler SA, Meier F, Deutsch M‐A, Lahm H, Lange R, et al. Region and cell‐type resolved quantitative proteomic map of the human heart. Nat Commun. 2017;8:1469. DOI: 10.1038/s41467-017-01747-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kardys I, Knetsch AM, Bleumink GS, Deckers JW, Hofman A, Stricker BH, Witteman JC. C‐reactive protein and risk of heart failure. The Rotterdam Study. Am Heart J. 2006;152:514–520. DOI: 10.1016/j.ahj.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 89.Kalogeropoulos A, Georgiopoulou V, Psaty BM, Rodondi N, Smith AL, Harrison DG, Liu Y, Hoffmann U, Bauer DC, Newman AB, et al. Inflammatory markers and incident heart failure risk in older adults: the Health ABC (Health, Aging, and Body Composition) study. J Am Coll Cardiol. 2010;55:2129–2137. DOI: 10.1016/j.jacc.2009.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Everett BM, Cornel JH, Lainscak M, Anker SD, Abbate A, Thuren T, Libby P, Glynn RJ, Ridker PM. Anti‐inflammatory therapy with canakinumab for the prevention of hospitalization for heart failure. Circulation. 2019;139:1289–1299. DOI: 10.1161/CIRCULATIONAHA.118.038010. [DOI] [PubMed] [Google Scholar]
- 91.Abbate A, Toldo S, Marchetti C, Kron J, Van Tassell BW, Dinarello CA. Interleukin‐1 and the inflammasome as therapeutic targets in cardiovascular disease. Circ Res. 2020;126:1260–1280. DOI: 10.1161/CIRCRESAHA.120.315937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Van Tassell BW, Canada J, Carbone S, Trankle C, Buckley L, Oddi Erdle C, Abouzaki NA, Dixon D, Kadariya D, Christopher S, et al. Interleukin‐1 blockade in recently decompensated systolic heart failure: results from REDHART (Recently Decompensated Heart Failure Anakinra Response Trial). Circ Heart Fail. 2017;10:e004373. DOI: 10.1161/CIRCHEARTFAILURE.117.004373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1–S2
Table S1
Figures S1–S7


