Abstract
Yeast cells overexpressing the Ser/Thr protein phosphatase Ppz1 display a slow-growth phenotype. These cells recover slowly from α-factor or nutrient depletion-induced G1 arrest, showing a considerable delay in bud emergence as well as in the expression of the G1 cyclins Cln2 and Clb5. Therefore, an excess of the Ppz1 phosphatase interferes with the normal transition from G1 to S phase. The growth defect is rescued by overexpression of the HAL3/SIS2 gene, encoding a negative regulator of Ppz1. High-copy-number expression of HAL3/SIS2 has been reported to improve cell growth and to increase expression of G1 cyclins in sit4 phosphatase mutants. We show here that the described effects of HAL3/SIS2 on sit4 mutants are fully mediated by the Ppz1 phosphatase. The growth defect caused by overexpression of PPZ1 is intensified in strains with low G1 cyclin levels (such as bck2Δ or cln3Δ mutants), whereas mutation of PPZ1 rescues the synthetic lethal phenotype of sit4 cln3 mutants. These results reveal a role for Ppz1 as a regulatory component of the yeast cell cycle, reinforce the notion that Hal3/Sis2 serves as a negative modulator of the biological functions of Ppz1, and indicate that the Sit4 and Ppz1 Ser/Thr phosphatases play opposite roles in control of the G1/S transition.
Regulation of the eukaryotic cell cycle is a complex process that involves two major control points: G1/S, which determines DNA replication; and G2/M, which regulates entry into mitosis. In the budding yeast Saccharomyces cerevisiae, commitment to a new round of cell duplication occurs at a control point called Start. Execution of Start demands a sufficient level of G1 cyclin/Cdc28 protein kinase activity and is a requirement for DNA synthesis, bud formation, and replication of the spindle pole body (9, 23).
The S. cerevisiae gene SIT4 encodes a Ser/Thr protein phosphatase related to type 2A enzymes (1). The phenotype of sit4 cells depends on the polymorphic SSD1 locus. Deletion of SIT4 in the absence of SSD1 (or in the presence of certain alleles of this gene, termed ssd1-d) yields inviable cells. On the contrary, sit4Δ cells are viable in an ssd1-v background (“v” for “viable”) although they grow slowly and are enriched in unbudded cells (36). Several lines of evidence indicate that Sit4p is required in late G1 for progression into S phase (36, 37). SIT4 is required for late G1 expression of SWI4, CLN1, and CLN2 (and, therefore, necessary for efficient DNA synthesis) in a pathway that is additive to that of CLN3 (14). Although the link between Sit4 and cyclin activation is not well understood, the Bck2 gene product has been proposed to function in a branch of the SIT4 pathway (11). As expected, bck2 mutants require Cln3 in order to complete the cell cycle (11, 13). It is not clearly established whether Bck2 acts upstream or downstream Sit4 (11).
SIT4 seems to be required also for bud emergence, because Sit4-deficient cells provided with CLN2 from a SIT4-independent promoter can replicate DNA but still are blocked for bud initiation (14). In this regard, it has been described that the mutation of sit4 in cells lacking BEM2, a gene that encodes a GTPase-activating protein for the Rho1 small GTPase and that has a role in bud emergence and cell cycle-related cytoskeletal reorganization (6, 20, 39), results in lethality (10, 20). The function(s) carried out by the Sit4 protein may have been conserved through evolution, as Drosophila PPV and human PP6 phosphatases have been proposed as functional homologs of the Sit4 phosphatase (3, 25).
SIS2 encodes a protein with a very acidic COOH-terminal region that was identified by its ability, when expressed at high copy number, to dramatically increase the growth rate of sit4 mutants (10). High levels of SIS2 are able to increase expression of SWI4, CLN1, and CLN2 in sit4 mutants, although they are unable to rescue the lethal phenotype of the sit4Δ mutation in an ssd1-d background. Deletion of SIS2 caused no evident growth defect (see below), although the SIS2 gene is essential in the absence of SIT4 function. It has been postulated that SIS2 plays a role in a pathway parallel to that of SIT4 (10).
SIS2 is allelic to HAL3, a gene identified by its ability to confer tolerance to high levels of sodium and lithium to yeast cells (15). Cells lacking SIS2/HAL3 are hypersensitive to sodium and lithium because in these mutants the expression of the ENA1 gene, encoding the ATPase responsible for the efficient output of sodium, is not induced at the appropriate levels in response to salt stress. The expression of ENA1 is also under the negative control of the Ser/Thr protein phosphatase Ppz1, a type 1-related phosphatase with a large NH2-terminal extension (29, 31). In addition of its role in salt tolerance, the Ppz1 phosphatase probably functions in conjunction with the protein kinase C (PKC)-activated mitogen-activated protein (MAP) kinase pathway (21, 30), which is involved in the proper construction of the cell wall (see references 5 and 22 for reviews).
We observed some time ago that strong overexpression of PPZ1 (driven by the GAL1 promoter from a multicopy plasmid) dramatically blocked cell growth in a wild-type background (7) and that high-copy-number expression of PPZ1 from its own promoter yields cells that display a slow-growth phenotype with increased number of unbudded cells (8). Recently we obtained genetic and biochemical evidence that Hal3/Sis2 can act as a negative regulatory subunit of Ppz1 (8). The observation that an excess of Ppz1 could alter cell growth, together with the existence of a functional link between HAL3/SIS2 and SIT4, prompted us to consider the possibility that the Ppz1 phosphatase is involved in cell cycle regulation. In this report, we present evidence suggesting that Ppz1 plays a role in the G1/S transition that is opposed to some of the functions of the Sit4 Ser/Thr phosphatase.
MATERIALS AND METHODS
Growth of Escherichia coli and yeast strains.
E. coli NM522 or DH5α was used as a host for DNA cloning. Bacterial cells were grown at 37°C in LB medium containing ampicillin (50 μg/ml), when needed, for plasmid selection. Yeast cells were grown at 28°C in YPD medium or, when indicated, in complete minimal (CM) synthetic medium (35). Unless otherwise stated, yeast strains generated in this work (Table 1) derive from S. cerevisiae JA-100 (MATa PPZ1 PPZ2 HAL3 ura 3-52 leu2-3,112 trp1-1 his4 can-1r).
TABLE 1.
Yeast strains used in this work
| Straina | Relevant genotype | Reference |
|---|---|---|
| JA-100 | MATa PPZ1 SIT4 HAL3 BEM2 | 8 |
| JA-300 | MATα PPZ1 SIT4 HAL3 BEM2 | This work |
| JA-101 | MATa ppz1::URA3 | 8 |
| JA-103 | MATa ppz2::TRP1 | 8 |
| JA-104 | MATa hal3::LEU2 | 8 |
| JA-110 | MATa sit4::TRP1 | This work |
| JA-111 | MATα sit4::TRP1 | This work |
| JA-112 | MATa ppz1::URA3 sit4::TRP1 | This work |
| JA-113 | MATa ppz2::TRP1 sit4::kanMX4 | This work |
| JA-114 | MATα ppz1::URA3 sit4::TRP1 | This work |
| JA-115 | MATa ppz1::URA3 hal3::LEU2 | This work |
| JA-120 | MATa/MATα PPZ1/ppz1::URA3 SIT4/sit4::TRP1 HAL3/hal3::LEU2 | This work |
| JA-121 | MATa/MATα PPZ1/ppz1::URA3 SIT4/sit4::TRP1 HAL3/hal3::LEU2 BEM2/bem2::kanMX4 | This work |
| JA-130 | MATα bem2::kanMX4 | This work |
| JA-131 | MATα ppz1::URA3 bem2::kanMX4 | This work |
| JA-140 | MATa sit4::TRP1 bem2::kanMX4 | This work |
| JA-150 | MATα hal3::LEU2 bem2::kanMX4 | This work |
| JA-151 | MATα ppz1::URA3 sit4::TRP1 bem2::kanMX4 | This work |
| JA-152 | MATa ppz1::URA3 hal3::LEU2 bem2::kanMX4 | This work |
| JA-153 | MATa ppz1::URA3 sit4::TRP1 hal3::LEU2 bem2::kanMX4 | This work |
| JA-170 | MATa bck2::TRP1 | This work |
| JA-171 | MATa bck2::TRP1 hal3::LEU2 | This work |
| CML211 | MATa cln3:LEU2 | This work |
| JA-190 | MATa/MATα PPZ1/ppz1::URA3 SIT4/sit4::TRP1 CLN3/cln3::LEU2 | This work |
| JA-195 | MATa ppz1::URA3 sit4::TRP1 cln3::LEU2 | This work |
| JA-500 | MATa/MATα PPZ1/ppz1::URA3 SIT4/sit4::TRP1 HAL3/hal3::LEU2 | This work |
All strains except of JA-500 derive from strain 1788 (diploid, homozygous for ura3-52 leu2-3,112 trp1-1 his4 can-1′). Strain JA-170 was obtained by tetrad analysis of the diploid strain DL763 (21), which has also a 1788 background. Strain JA-500 is a W303 derivative (diploid, homozygous for ura3-1 leu2-3,112 trp1Δ2 his3-11, 15 ade2-1 can1-100 ssd1-d2).
Recombinant DNA techniques, gene disruptions, and plasmids.
E. coli and S. cerevisiae cells were transformed by using standard techniques as previously described (8). Restriction reactions, DNA ligations, and other standard recombinant DNA techniques were carried out as described elsewhere (33).
Gene disruptions were performed by the one-step technique (32). The PPZ1 disruption was as described elsewhere (29), and HAL3 was interrupted with the LEU2 marker as described in reference 15. The PPZ2 gene was disrupted by replacing a 0.95-kbp HpaI-XhoI fragment containing about 100 bp of 5′ untranslated region plus most of the NH2-terminal coding region by the 0.9-kbp SalI-SmaI fragment containing the TRP1 gene, obtained from vector YDp-W (4). The SIT4 gene was interrupted with the TRP1 marker as described in reference 28. Alternatively, SIT4 was disrupted with the kanamycin gene as follows. The kanMX4 module containing the kanamycin resistance gene was recovered from plasmid pFA6a-kanMX4 (38) by digestion with SmaI and SpeI and was cloned into plasmid pUC19, previously digested with SmaI and XbaI. The insert was recovered by digestion with BglII and SphI and cloned into these same sites present in the coding region of the SIT4 gene in plasmid pJA-1 (28). The construct was linearized by digestion with BamHI, and the 2.75-kbp fragment was used for integration at the SIT4 locus. Positive clones were selected by resistance to geneticin. The BEM2 gene was also disrupted with the kanMX module as follows. Plasmid pCT3-H2, which contains a 9.3-kbp insert including the entire BEM2 gene (6), was digested with EcoRI and PstI, and the 4.3-kbp fragment was recovered and ligated into the same sites of plasmid pUC19, to give pUC19-BEM2(4.3). Plasmid pFA6a-kanMX4 was linearized by EcoRI digestion, blunt ended with the Klenow enzyme, and then digested with BglII. The released fragment, containing the kanMX4 module, was ligated into pUC19-BEM2(4.3). previously digested with BamHI and HincII. The resulting construct was then digested with BglII and EcoRI (present in the polylinker of pUC19), and the 2.95-kbp fragment was used for yeast transformation. For disruption of CLN3, a 2.1-kbp fragment (XhoI-HpaI) containing the whole open reading frame was replaced by the LEU2 gene. All disruptions were verified by PCR.
The construction of plasmids YEplac181-PPZ1, YCplac111-PPZ1, and pYES2-PPZ1 was previously described (7). Plasmid YEplac195-PPZ1 was made by recovering the insert from YEplac181-PPZ1 by digestion with BamHI and HindIII and ligating it into the same sites of plasmid YEplac195, which carries a URA3 marker (17). Construction of plasmid YEp351-HAL3 was described previously (15). Plasmid YEp-PPZ2 corresponds to the entire genomic clone described in reference 21.
Other methods.
Yeast cells were arrested at G1 phase by treatment with α-factor or by nitrogen deprivation. For determination of the budding index after release from α-factor arrest, cultures (5 ml) were grown to an optical density at 660 nm (OD660) of 0.3 to 0.4; then α-factor (Sigma) was added to a final concentration of 20 μg/ml, and growth resumed for 2.5 to 3 h. Cells were recovered by centrifugation, washed in fresh medium, and resuspended in 1/10 of the original volume. Samples were taken at different times, and at least 200 to 300 cells were counted for each time point. Alternatively, for flow cytometry analysis or RNA preparation, initial cultures (optical density of 0.4 to 0.6) were filtered and resuspended in prewarmed medium containing the pheromone (10 μg/ml). After 2.5 to 3 h, the cultures were filtered and resuspended in prewarmed fresh medium to give an optical density of 0.5 to 1. Samples were taken at different times and processed for flow cytometry and RNA preparation as described in reference 16. Nitrogen deprivation experiments were carried out essentially as described in reference 16. Yeast cells were arrested at the G2/M transition as follows. Cultures (5 ml) were grown on CM medium lacking uracil at an optical density of 0.1 to 0.2, nocodazole (10 μg/ml) was added, and growth resumed for 3 h. Cell were washed and resuspended in fresh medium, and samples were taken every 15 min and processed for microscopic observation and flow cytometry.
RESULTS
Overexpression of PPZ1 causes a delay in the G1/S transition.
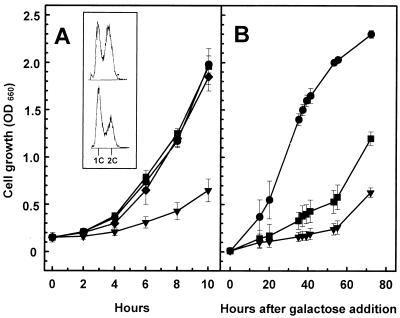
We have reported that yeast strains overexpressing PPZ1 from the GAL1 promoter show a severe growth defect (7). Although less pronounced, this defect is evident even when PPZ1 is expressed from its own promoter from a multicopy plasmid (Fig. 1A). Early-exponential-phase cultures (OD660 of 0.4 to 0.5) of cells overexpressing PPZ1 showed a marked increase in small budded cells compared with cells with normal levels of the phosphatase (61% versus 39%). Furthermore, flow cytometry analysis of these samples (Fig. 1A, inset) indicated that the culture of cells overexpressing PPZ1 was enriched in 1C cells, indicative of an specific delay in G1 phase. In contrast, high-copy-number expression of PPZ2 did not affect cell growth (Fig. 1A). A noteworthy observation was that the Ppz1-induced growth defect was completely rescued by high-copy-number expression of the gene HAL3/SIS2, which encodes a protein recently identified in our laboratory as a negative regulator of Ppz1 in salt tolerance. High-copy-number expression of PPZ1 results in a temperature-sensitive phenotype, since growth of the cells at 37°C is rather slow, with increased number of small budded cells and a higher fraction of 1C cells (not shown). This phenotype is also rescued by increased dosage of HAL3. Overexpression of HAL3 from a high-copy-number plasmid was also able to ameliorate the very poor growth of cells strongly overexpressing PPZ1 from the GAL1 promoter (Fig. 1B).
FIG. 1.
Overexpression of the Ser/Thr phosphatase Ppz1 produces a phenotype of slow growth that is rescued by overexpression of Hal3. (A) Wild-type strain JA-100 was transformed with plasmid YEplac195 (●), plasmid YEplac195-PPZ1 (▾), plasmid YEp-PPZ2 (■), or plasmids YEplac195-PPZ1 and YEp351-HAL3 (⧫). Positive clones were grown overnight in CM medium lacking uracil, diluted to an OD660 of 0.15, and then grown for the indicated times in the same medium. Data are means ± SEM of three experiments. The inset shows the flow cytometry analysis of cells carrying the empty plasmid YEplac195 (upper profile) or plasmid YEplac195-PPZ1 (lower profile). Samples were taken at equivalent optical densities to avoid interferences due to the availability of nutrients. (B) Wild-type JA-100 cells were transformed with plasmid pYES2 (●), plasmid pYES2-PPZ1 (▾), or plasmid pYES2-PPZ1 plus plasmid YEp351-HAL3 (■). Cells were grown overnight on CM medium lacking uracil in the presence of 2% glucose; then an aliquot was inoculated (to achieve a starting optical density of 0.01) in minimal medium lacking uracil and containing 2% galactose. Growth was resumed, and samples were taken at the indicated times. Data are means ± SEM of four experiments.
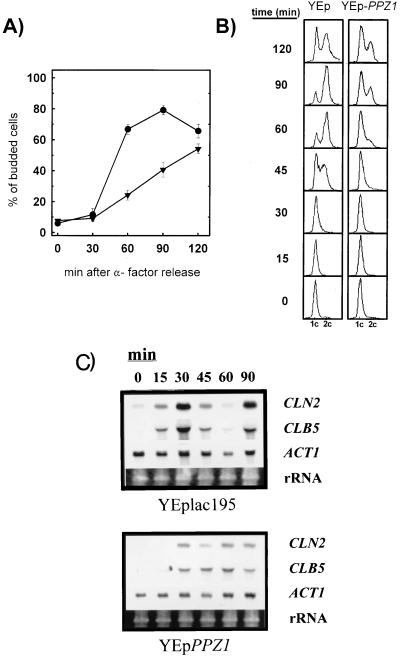
To characterize the nature of the observed growth defect, we synchronized cells in G1 by incubation with α-factor and then released the cells from the pheromone-induced arrest. As shown in Fig. 2A, α-factor arrest of wild-type cells results in a very low percentage of budded cells that increases rapidly when the pheromone is removed. However, cells overexpressing PPZ1 show a very slow recovery from G1 arrest, as deduced by the very low budding index of the cultures. A similar behavior was observed when these cells were arrested in G1 by nitrogen starvation instead of pheromone treatment (not shown). The delay of PPZ1-overexpressing cells in recovering from α-factor-induced G1 arrest is also evident when DNA contents are monitored by flow cytometry (Fig. 2B). Therefore, high levels of the Ppz1 phosphatase seem to interfere with two essential processes in the G1-to-S transition: budding initiation and DNA synthesis. Evaluation of the levels of two different G1 cyclins by Northern blot analysis confirmed this observation (Fig. 2C): the expression of both CLN2 and CLB5 is delayed in PPZ1-overexpressing cells, and the levels attained are also lower than those of cells with normal levels of the phosphatase. On the contrary, an excess of Ppz1 seems without effect at the G2/M cell cycle transition point because after nocodazole treatment, control cells and cells overexpressing PPZ1 resumed cell cycle in similar fashions (not shown). Taken together, these results indicate that high levels of Ppz1p result in a defect in the G1/S transition.
FIG. 2.
Overexpression of Ppz1 causes a delay in G1/S transition and G1 cyclin expression. (A) JA-100 cells carrying plasmid YEplac181 (●) or YEplac181-PPZ1 (▾) were arrested in G1 phase by incubation with α-factor (20 μg/ml) for 3 h. Cells were collected, washed, and resuspended in fresh CM medium lacking leucine. Samples were taken at the indicated times, and the budding index of the culture was determined by microscopic observation. Data are means ± SEM of four experiments. (B) Control cells (with plasmid YEplac181; left) and cells overexpressing Ppz1 (with plasmid YEplac181-PPZ1; right) were grown as indicated, and samples were taken for DNA content determination by flow cytometry. (C) Cells were arrested as indicated. After release, samples of cells were taken at the indicated times and total RNA was isolated. Equivalent amounts (5 μg) of total RNA were run in formaldehyde-agarose gels, transferred to membranes, and hybridized with the indicated DNA probes. Hybridization with the ACT1 probe and the ethidium bromide staining of rRNA are shown for comparison.
Lack of PPZ1 improves growth of a sit4 mutant.
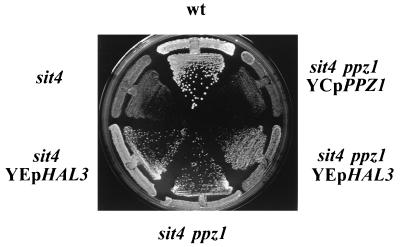
We have shown that high-copy-number expression of HAL3/SIS2 rescues the growth defect induced by overexpression of PPZ1. Hal3/Sis2 is a protein that interacts with Ppz1 and negatively regulates a number of functions of this phosphatase. On the other hand, HAL3/SIS2 was identified as a gene that in high copy number could improve the growth of sit4 mutants. Therefore, we considered the possibility that the effects described for HAL3/SIS2 in Sit4-deficient cells result from attenuation of the function of the Ppz1 phosphatase. To test this hypothesis, we constructed cells (with an ssd1-v background) lacking PPZ1, SIT4, or both genes. As shown in Fig. 3, sit4 cells are viable but display a slow-growth phenotype that is clearly ameliorated by disruption of PPZ1. Furthermore, the potency of this effect is roughly the same as that obtained by overexpressing HAL3/SIS2, and the two effects are not additive. Interestingly, deletion of PPZ2 resulted only in a slight improvement of growth of sit4 mutants, indicating that the lack of Ppz2 was clearly less effective than the absence of Ppz1 (not shown). The positive effect of the deletion of PPZ1 on the growth rate of sit4 mutants has also been verified in two additional strains (DBY746 and UTL-7A), in which the deletion of SIT4 is not lethal (data not shown). These results are compatible with the notion that the described effect of the overexpression of HAL3/SIS2 on the growth of sit4 mutants is mediated by Ppz1.
FIG. 3.
Deletion of PPZ1 mimics the effect of overexpression of HAL3 in sit4 cells. The following strains were streaked on CM plates lacking leucine: wild-type JA-100, JA-110 (PPZ1 sit4), JA-110 containing plasmid YEp351-HAL3, JA-112 (ppz1 sit4), JA-112 with plasmid YEp351-HAL3, and JA-112 with plasmid YCp111-PPZ1. Strains without an specific plasmid were transformed with YEplac181 to allow growth in the selective medium. Growth was scored after 3 days.
Deletion of HAL3/SIS2 has virtually no effect on cell growth under normal conditions. However, it has been documented that the HAL3/SIS2 gene is essential for viability in the absence of SIT4, since hal3/sis2 and sit4 deletions have been reported to be synthetically lethal (10). We considered that if the effect of Hal3/Sis2 is mediated by Ppz1, the deletion of PPZ1 in a hal3/sis2 sit4 background should yield viable cells, with a growth phenotype resembling to that of sit4 cells. To test this possibility, we crossed strain JA-115 (MATa ppz1::URA3 hal3::LEU2) with strain JA-111 (MATα sit4::TRP1), to create the diploid strain JA-120, which was induced to sporulate. In 10 of 26 tetrads analyzed, only three spores could germinate or grow, and the genotype deduced for the nonviable spores corresponded to the combination of the sit4 and hal3 mutations, thus confirming the synthetic lethality of these mutations. However, in 11 tetrads we recovered the combination of markers corresponding to the triple mutation sit4 ppz1 hal3, and these cells had a growth rate similar to that of sit4 ppz1 double mutants (not shown). Therefore, deletion of ppz1 rescues the lethal phenotype of the sit4 hal3 mutation. This result is in agreement with the notion that the function(s) of Hal3 necessary for viability in the absence of SIT4 is mediated by Ppz1.
It is known that the phenotype of sit4 deletants depends on the unlinked polymorphic SSD1 locus (36). Thus, in an ssd1-d2 strain (as in W303 derivatives) or in an ssd1Δ strain, the deletion of SIT4 is lethal, and it has been reported that the overexpression of HAL3/SIS2 does not rescue this lethal phenotype. Consequently, our hypothesis was that if the effects of Hal3/Sis2 are mediated by Ppz1, the deletion of this phosphatase in an ssd1-d background would not rescue the lethality caused by disruption of SIT4. To test this, diploid W303 cells heterozygous for the sit4 and ppz1 deletions were constructed (strain JA-500), and their sporulation was induced. Consistent with our hypothesis, after analysis of 36 tetrads, we failed to obtain cells carrying the combination of the sit4 and ppz1 deletions.
Deletion of PPZ1 accelerates the slow recovery from α-factor G1 arrest of sit4 cells.
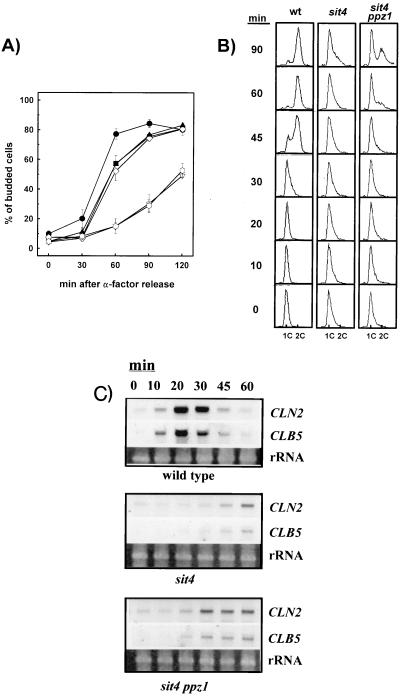
Cells lacking functional Sit4 present a expanded G1 phase and show a delayed entry in cell cycle after α-factor G1 arrest. This situation can be monitored by determining the budding index of the cultures after removal of the pheromone. As shown in Fig. 4A, deletion of PPZ1 accelerates the slow recovery from the α-factor G1 arrest that is characteristic of sit4 mutants, and this effect is completely suppressed by expression of PPZ1 from a low-copy-number, centromeric plasmid. Interestingly, the enhanced recovery induced by deletion of PPZ1 is virtually identical in potency to that the obtained by overexpression of HAL3/SIS2, and the two effects are not additive. It is worth noting that neither the deletion of PPZ1 nor the overexpression of HAL3/SIS2 is able to fully restore the wild-type phenotype.
FIG. 4.
Deletion of PPZ1 accelerates the entry into G1/S transition of a sit4 mutant. (A) Strains JA-100 (wild type; ●), JA-110 (sit4; ▿), JA-110 containing plasmid YEp351-HAL3 (■), JA-112 (ppz1 sit4; ◊), JA-112 containing plasmid YEp351-HAL3 (▴), and JA-112 containing plasmid YCplac111-PPZ1 ( ) were arrested in G1 phase by incubation with α-factor and released from arrest as indicated. Cells were taken at the indicated times, and the budding index was determined by microscopic counting. Data are means ± SEM of six experiments. (B) Analysis of DNA content of wild-type (wt), sit4 (strain JA-110), and sit4 ppz1 (strain A-112) cells by flow cytometry. Cells were arrested as described above, and samples were taken at different times after release from α-factor arrest. (C) The indicated strains were released form α-factor arrest, and samples were taken at various times. Total RNA was prepared, electrophoresed, transferred to membranes, and hybridized with the indicated probes. Ethidium bromide staining of rRNAs is shown for comparison.
) were arrested in G1 phase by incubation with α-factor and released from arrest as indicated. Cells were taken at the indicated times, and the budding index was determined by microscopic counting. Data are means ± SEM of six experiments. (B) Analysis of DNA content of wild-type (wt), sit4 (strain JA-110), and sit4 ppz1 (strain A-112) cells by flow cytometry. Cells were arrested as described above, and samples were taken at different times after release from α-factor arrest. (C) The indicated strains were released form α-factor arrest, and samples were taken at various times. Total RNA was prepared, electrophoresed, transferred to membranes, and hybridized with the indicated probes. Ethidium bromide staining of rRNAs is shown for comparison.
We monitored the delay of sit4 mutants in reaching S phase by flow cytometry (Fig. 4B) and observed that deletion of PPZ1 accelerates the entry of sit4 cells in S phase. When the levels of G1 cyclins were monitored (Fig. 4C), we observed that lack of Ppz1 in a sit4 background resulted in an earlier expression of CLN2 and CLB5. In both cases, however, the mutation of PPZ1 alleviates the sit4 defect but does not fully restore the wild-type behavior. These results suggest that the Ppz1 phosphatase may counteract some but not all of the effects of the Sit4 phosphatase on the cell cycle.
Increased expression of PPZ1 results in a strong growth defect in strains with low G1 cyclin levels.
Bck2 has been postulated to be involved in the SIT4 pathway for the activation of G1 cyclin expression. To better understand the relationships between BCK2, SIT4, and PPZ1, we tested the effect of increased PPZ1 function in the absence of BCK2. We observed that whereas lack of Hal3 does not significantly alter cell growth in an otherwise wild-type background, deletion of HAL3 results in poor growth in the absence of BCK2 (not shown). This effect on cell growth is consistent with an increased Ppz1 function. To directly test this hypothesis, we transformed bck2 cells with a multicopy plasmid containing the PPZ1 gene. As observed in Fig. 5A, overexpression of PPZ1 drastically reduces cell growth of bck2 cells.
FIG. 5.
Ppz1 function is not mediated by BCK2 or CLN3. (A) Strains JA-100 (wild type [wt]), CML211 (cln3Δ), and JA-170 (bck2Δ) were transformed with plasmid YEplac195 (upper half) or YEplac195-PPZ1 (lower half). Positive clones were streaked on CM plates lacking uracil and grown at 28°C for 2 days. (B) Strain JA-114 (MATα sit4Δ pz1Δ) was crossed with strain CML211 (MATa cln3Δ). The resulting diploid strain, JA-190, was sporulated, and ppz1 sit4 cln3 triple (but not sit4 cln3 double) mutants were recovered. Growth of the triple mutant was compared with that of cln3 cells (which grow nearly like a wild-type strain), sit4 cells, and sit ppz4 cells by streaking the cultures in YPD plates and incubating them at 28°C for 2 days.
Both SIT4 and BCK2 have been integrated in a pathway that is additive to CLN3 function for G1 cyclin expression. We then considered the possibility that Ppz1 exerts its effect on the G1/S transition by affecting CLN3 function. To test this, we transformed a cln3Δ strain with plasmid YEplac195-PPZ1. As shown in Fig. 5A, high-copy-number expression of PPZ1 in the absence of CLN3 results in an extremely poor growth (resembling to that obtained by overexpression of PPZ1 in a bck2 mutant). This finding suggests that the effect of the phosphatase is not mediated by Cln3 and that, in fact, the absence of CLN3 strongly aggravates the growth defect resulting from high levels of Ppz1.
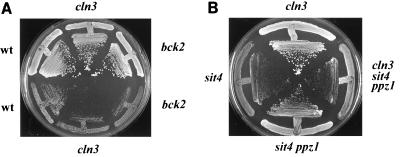
Mutation of PPZ1 rescues the lethal phenotype of the sit4 cln3 mutation.
It has been postulated (14) that Sit4 and Cln3 provide parallel pathways for the accumulation of CLN1, CLN2, and HCS26 mRNAs. As a consequence, sit4Δ cln3Δ cells are virtually inviable. We postulated that if the role of Ppz1 is opposed to some functions regulated by Sit4 and Cln3 (and is not mediated by these components), lack of PPZ1 might allow growth of cells lacking both Sit4 and Cln3. This was tested by crossing strain JA-114 (MATα sit4Δ ppz1Δ) with strain CML211 (MATa cln3Δ). The resulting diploid strain (JA-190) was then sporulated, and 22 tetrads were dissected. The results confirmed the inviability of a sit4Δ cln3Δ mutant. However, nine spores gave rise to colonies that showed the combination of markers corresponding to the triple sit4Δ ppz1Δ cln3Δ mutation. Growth of this strain in comparison with that of sit4 and sit4 ppz1 strains (Fig. 5B) supports the notion that Cln3 does not mediate the function of Ppz1.
Analysis of the genetic interactions between PPZ1, BEM2, and SIT4.
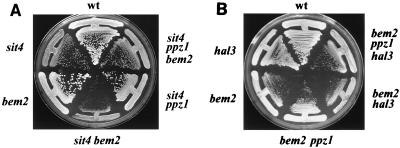
In addition to a defect in G1 cyclin expression, lack of sit4 results in a defect in bud formation (14). It has been described that null mutations in the SIT4 and BEM2 genes are synthetically lethal. To explore the possibility that deletion of PPZ1 can rescue this lethal phenotype, we used the diploid strain JA-121, which is heterozygotic for the ppz1, bem2, and sit4 mutations, and induced its sporulation. To our surprise, we were able to recover viable double sit4 bem2 mutants, although these cells grew very slowly, even in comparison with sit4 mutants (Fig. 6A). This could be explained by the fact that the defect of the bem2 mutation in our genetic background is relatively mild. It has been previously reported that the genetic background (in particular the presence of specific alleles of the gene SSD1) is a major determinant for the severity of the bem2 phenotype (20). As can be observed, deletion of PPZ1 improves the growth of sit4 bem2 cells. However, these cells are still defective in growth compared to sit4 ppz1 mutants.
FIG. 6.
Genetic interactions between the sit4, bem2, and ppz1 mutations. (A) Strains JA-100 (wild type [wt]), JA-110 (sit4), JA-130 (bem2), JA-140 (sit4 bem2), JA-112 (sit4 ppz1), and JA-151 (sit4 ppz1 bem2) were grown at 28°C on YPD plates. Growth was scored after 2 days. (B) Strains JA-100 (wild type [wt]), JA-130 (bem2), JA-131 (ppz1 bem2), JA-150 (bem2 hal3), and JA-152 (ppz1 bem2 hal3) were streaked on YPD plates and grown at 37°C for 2 days.
To test a possible genetic interaction between BEM2 and PPZ1, we grew cells at 37°C (to aggravate the relatively mild bem2 phenotype observed in our genetic background). As can be observed in Fig. 6B, deletion of PPZ1 does not rescue the growth defect of bem2 mutants (or the characteristic morphology of these cells [not shown]). However, a double bem2 hal3 mutant shows a growth defect stronger than that of bem2 cells, suggesting an additive effect of the two mutations. The notion that the effect of lack of Hal3 on a bem2 background is the result of an increased Ppz1 functional activity is confirmed by the observation that the additional growth defect is abolished by mutation of the PPZ1 gene.
DISCUSSION
In this report, we present evidence that Ppz1, a Ser/Thr phosphatase initially identified as involved in the maintenance of cell integrity and the tolerance to sodium cations (21, 30, 31), is also a regulatory component of the G1/S transition in the yeast cell cycle. Preliminary observations suggested that Ppz1 may be related to certain cell growth processes, since overexpression of the gene caused a dramatic growth defect (7). We show here that an excess of Ppz1 results in delayed bud emergence, CLN2 and CLB5 cyclin expression, and DNA synthesis, all characteristics of a delay in the G1/S transition. sit4 mutants also present a delayed G1/S transition, as a result of a defect in G1 cyclin expression and bud emergence (14, 36, 37) that can be relieved by overexpression of SIS2/HAL3 (10). A key point that suggested a possible link between the functions of Ppz1 and Sit4 was our recent finding that Hal3/Sis2 can act as a negative regulatory subunit of Ppz1 (8). This led us to suspect that the observed growth defect attributed to an excess of Ppz1 may be due to a negative effect of the phosphatase on the cell cycle and to postulate that the rescue of the sit4 phenotype by high levels of Hal3 may be a consequence of a Hal3-mediated attenuation of Ppz1 function(s). The fact that an increased HAL3 dosage is able to cure the growth defect due to overexpression of PPZ1 confirmed the negative role of Hal3 on the cell cycle-related function(s) of Ppz1. Therefore, the emerging scenario is that Hal3 is a negative regulatory component of all known Ppz1 functions, since it appears to regulate the biological activity of Ppz1 with respect to sodium tolerance, cell integrity (in connection with the PKC-activated MAP kinase pathway), and cell cycle (references 8 and 31 and this work).
Furthermore, we demonstrate in different ways that Ppz1 plays a role in the G1/S transition that is opposed to some of the functions attributed to Sit4 and that the previously described effect of high-copy-number expression of HAL3 (10) on sit4 mutants can be explained through the inactivation of Ppz1. This role in cell cycle might be very specific for Ppz1, because we have observed that high-copy-number expression of PPZ2, a phosphatase highly related to Ppz1 (19, 21), does not affect cell growth and that the deletion of PPZ2 is fairly ineffective in curing the sit4 cell growth defect. This finding suggests that Ppz1 and Ppz2 are not functionally identical and that the role of Ppz1 in the cell cycle cannot be fulfilled by Ppz2. This situation is reminiscent of the virtual absence of a salt tolerance phenotype in a ppz2 deletant, compared with the hypertolerance of the ppz1 mutant (8, 31).
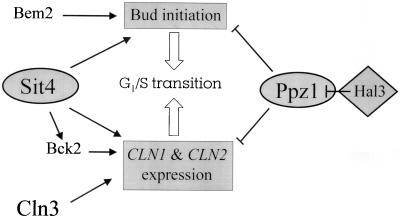
Two independent pathways have been proposed for the activation of G1 cyclin transcription (Fig. 7), one involving Sit4 and Bck2 and the other involving Cln3 (11, 13). The gene BCK2 is required for CLN1 and CLN2 gene expression, although its absence has no evident phenotypic effect in an otherwise wild-type background (11, 13, 21) and does not aggravate the slow-growth sit4 phenotype (11). We have observed that deletion of HAL3 (as well as high-copy-number expression of PPZ1) in a bck2 background yields cells that, while displaying a slow-growth phenotype, are viable. This is in contrast with the lethal phenotype (confirmed in this report) ascribed to the hal3 sit4 mutation. Therefore, the weaker phenotype of the hal3 bck2 mutant suggest that Bck2 functions downstream Sit4, in a branch on the Sit4 pathway for CLN activation (11). sit4 cln3 double mutants have been reported to be inviable, most probably as a result of a defect in the activation in cyclin transcription (14). Our observations that lack of PPZ1 allows growth of a sit4Δ cln3Δ strain, in combination with the results described above, strongly suggest that Ppz1 is a novel component of a pathway that negatively regulates G1 cyclin transcription. Similarly to the lack of CLN3 or BCK2, the effect of the absence of this component is barely detectable in otherwise wild-type cells, but it is readily observed in cells lacking additional positive components of the cyclin-activating pathway. In contrast, the excess of Ppz1 function can be easily detected in wild-type cells, although the effect is stronger in cells deficient in some components of these activating pathways (such as Bck2 or Cln3).
FIG. 7.
Schematic diagram of the role of the Ser/Thr phosphatase Ppz1 in cell cycle regulation. See Discussion for details.
The growth defect of the sit4 mutants cannot be fully attributed to their defective G1 cyclin expression. In these mutants, the expression of CLN2 from a SIT4-independent promoter cannot restore growth because although it allows DNA replication to progress, cells are still blocked for bud initiation (10, 14). Therefore, the lack of Sit4 negatively affects other processes, such as bud initiation, at late G1 phase. These processes would be also stimulated by high-copy-number expression of HAL3/SIS2 (10) and, therefore, negatively regulated by Ppz1. Our data suggest that the effect of lack of Ppz1 in alleviating the budding defect of sit4 mutants is even more dramatic that the observed restoration of cyclin levels, indicating that Ppz1 must have a role in the budding process. Bem2 has been recognized as an important constituent of the regulatory mechanisms for polarized cell growth and bud emergence (6, 20, 39), and it has been shown to display genetic interactions with SIT4, since sit4 and bem2 mutations have been described as synthetically lethal (10, 20). We observe that the deletion of PPZ1 is unable to rescue the morphological defects and the slow growth at 37°C of bem2 mutants, whereas the deletion of HAL3 (which would result in increased Ppz1 function) aggravates the growth defect of bem2 cells. All of these results suggest that the possible role of Ppz1 in bud emergence is not mediated by Bem2. Our working model for Ppz1 function in cell cycle is delineated in Fig. 7. Ppz1 should negatively regulate G1 cyclin expression through a mechanism that would not be mediated by Sit4, Bck2, or Cln3. In addition, Ppz1 should negatively regulate certain processes that affect proper budding. Although the precise nature of these processes remains to be elucidated, our data suggest that they are probably unrelated to those mediated by Bem2. In any case, the regulatory effect of Ppz1 on the cell cycle seems to be restricted to the G1/S transition, and it is opposed to the function(s) of Sit4. Therefore, it would be interesting to identify biological targets for both Sit4 and Ppz1 phosphatases. We have observed that the bidimensional electrophoretic pattern of proteins in sit4 mutants markedly differs from that of wild-type cells. However, lack of Ppz1 barely alters the protein pattern, and the ppz1 sit4 double mutant display a pattern very similar to that of sit4 cells (27). This finding suggest that the lack of Ppz1 might allow restoration of growth in sit4 cells by affecting very specific biological targets.
An aspect most suggestive of the involvement of Ppz1 in the cell cycle is that this phosphatase has been previously shown to participate in other important cell functions, such as the maintenance of cell integrity (21, 30), in connection with the PKC-activated SLT2/MPK1 pathway involved in bud emergence and cell surface growth, and the regulation of sodium tolerance (31). It has been shown that the pathway leading to the activation of the Slt2/Mpk1 MAP kinase is activated in turn at the G1/S transition (18, 26). On the other hand, it seems reasonable to postulate that the maintenance of proper intracellular ion concentrations may be required for the cell to take the decision to progress in the cell cycle, as it has been shown that many environmental stresses cause a delay in cell cycle progression (24). It appears that Ppz1p may be a component of a pathway that would integrate different stimuli relevant for cell cycle progression. It is worth stressing that functional homologs of Sit4 have been found in Drosophila and humans (25, 3), suggesting a strongly conserved function. Although Ppz phosphatases have been found so far only in S. cerevisiae (19, 21, 29), Schizosaccharomyces pombe (2), and Neurospora crassa (12), the recent discovery of Hal3 homologs in plants (34) and the evidence from the data banks that proteins similar to Hal3 can be found in animals suggest that the existence of Ppz phosphatase-mediated regulatory pathways may not be restricted to fungi.
ACKNOWLEDGMENTS
We thank M. Zaguirre and A. Vilalta for skillful technical help, V. J. Cid and M. Sánchez for the BEM2 gene, R. Serrano for the HAL3 gene, and D. L. Levin for the PPZ2 gene. We gratefully acknowledge the advice of I. López Calderón regarding the genetic analysis of some mutants, as well as the support of E. Simón and J. Torres at some phases of the work.
This work was supported by grants PB95-0663 and PB94-0511 from the Dirección General de Investigación Científica y Técnica (Ministry of Education, Spain), to J.A. and M.A., respectively, and SGR97-127 from the Comissió Interdepartamental de Ciència i Tecnologia (Generalitat de Catalunya) to J.A. E.G. is recipient of a postdoctoral research contract from the Ministry of Education, Spain.
REFERENCES
- 1.Arndt K T, Styles C A, Fink G R. A suppressor of a HIS4 transcriptional defect encodes a protein with homology to the catalytic subunit of protein phosphatases. Cell. 1989;56:527–537. doi: 10.1016/0092-8674(89)90576-x. [DOI] [PubMed] [Google Scholar]
- 2.Balcells L, Gómez N, Casamayor A, Clotet J, Ariño J. Regulation of salt tolerance in fission yeast by a protein-phosphatase-Z-like Ser/Thr protein phosphatase. Eur J Biochem. 1997;250:2476–2483. doi: 10.1111/j.1432-1033.1997.0476a.x. [DOI] [PubMed] [Google Scholar]
- 3.Bastians H, Ponstingl H. The novel human protein serine/threonine phosphatase 6 is a functional homologue of budding yeast Sit4p and fission yeast ppe1, which are involved in cell cycle regulation. J Cell Sci. 1996;109:2865–2874. doi: 10.1242/jcs.109.12.2865. [DOI] [PubMed] [Google Scholar]
- 4.Berben G, Dumont J, Gilliquet V, Bolle P, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 5.Cid V J, Durán A, Del Rey F, Snyder M, Nombela C, Sánchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cid V J, Cenamor R, Sánchez M, Nombela C. A mutation in the Rho1-GAP-encoding gene BEM2 of Saccharomyces cerevisiae affects morphogenesis and cell wall functionality. Microbiology. 1998;144:25–36. doi: 10.1099/00221287-144-1-25. [DOI] [PubMed] [Google Scholar]
- 7.Clotet J, Posas F, De Nadal E, Ariño J. The NH2-terminal extension of protein phosphatase PPZ1 has an essential functional role. J Biol Chem. 1996;271:26349–26355. doi: 10.1074/jbc.271.42.26349. [DOI] [PubMed] [Google Scholar]
- 8.De Nadal E, Clotet J, Posas F, Serrano R, Gómez N, Ariño J. The yeast halotolerance determinant Hal3p is an inhibitory subunit of the Ppz1p Ser/Thr protein phosphatase. Proc Natl Acad Sci USA. 1998;95:7357–7362. doi: 10.1073/pnas.95.13.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deshaies R J. Phosphorylation and proteolisis: partners in the regulation of cell division in budding yeast. Curr Opin Genet Dev. 1997;7:7–16. doi: 10.1016/s0959-437x(97)80103-7. [DOI] [PubMed] [Google Scholar]
- 10.Di Como C J, Bose R, Arndt K T. Overexpression of SIS2, which contains an extremely acidic region, increases the expression of SWI4, CLN1 and CLN2 in sit4 mutants. Genetics. 1995;139:95–107. doi: 10.1093/genetics/139.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Como C J, Chang H, Arndt K T. Activation of CLN1 and CLN2 G1 cyclin gene expression by BCK2. Mol Cell Biol. 1995;15:1835–1846. doi: 10.1128/mcb.15.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dombràdi, V. Personal communication.
- 13.Epstein C B, Cross F R. Genes that can bypass the CLN requirements for Saccharomyces cerevisiae cell cycle start. Mol Cell Biol. 1994;14:2041–2047. doi: 10.1128/mcb.14.3.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernández-Sarabia M J, Sutton A, Zhong T, Arndt K T. SIT4 protein phosphatase is required for the normal accumulation of SWI4, CLN1, CLN2, and HCS26 RNAs during late G1. Genes Dev. 1992;6:2417–2428. doi: 10.1101/gad.6.12a.2417. [DOI] [PubMed] [Google Scholar]
- 15.Ferrando A, Kron S J, Rios G, Fink G R, Serrano R. Regulation of cation transport in Saccharomyces cerevisiae by the salt tolerance gene HAL3. Mol Cell Biol. 1995;15:5470–5481. doi: 10.1128/mcb.15.10.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallego C, Gari E, Colomina N, Herrero E, Aldea M. The Cln3 cyclin is down-regulated by translational repression and degradation during the G1 arrest caused by nitrogen deprivation in budding yeast. EMBO J. 1997;16:7196–7206. doi: 10.1093/emboj/16.23.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 18.Gray J V, Ogas J P, Kamada Y, Stone M, Levin D E, Herskowitz I. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 1997;17:4924–4937. doi: 10.1093/emboj/16.16.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes V, Muller A, Stark M J, Cohen P T. Both isoforms of protein phosphatase Z are essential for the maintenance of cell size and integrity in Saccharomyces cerevisiae in response to osmotic stress. Eur J Biochem. 1993;218:269–279. doi: 10.1111/j.1432-1033.1993.tb18142.x. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y J, Francisco L, Chen G C, Marcotte E, Chan C S. Control of cellular morphogenesis by the Ipl2/Bem2 GTPase-activating protein: possible role of protein phosphorylation. J Cell Biol. 1994;127:1381–1394. doi: 10.1083/jcb.127.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee K S, Hines L K, Levin D E. A pair of functionally redundant yeast genes (PPZ1 and PPZ2) encoding type 1-related protein phosphatases function within the PKC1-mediated pathway. Mol Cell Biol. 1993;13:5843–5853. doi: 10.1128/mcb.13.9.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin D E, Bowers B, Chen C Y, Kamada Y, Watanabe M. Dissecting the protein kinase C/MAP kinase signalling pathway of Saccharomyces cerevisiae. Cell Mol Biol Res. 1994;40:229–239. [PubMed] [Google Scholar]
- 23.Lew D J, Reed S I. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 24.Lew D J, Weinert T, Pringle J R. Cell cycle control in Saccharomyces cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. 3. Cell cycle and cell biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 607–696. [Google Scholar]
- 25.Mann D J, Dombràdi V, Cohen P T. Drosophila protein phosphatase V functionally complements a SIT4 mutant in Saccharomyces cerevisiae and its amino-terminal region can confer this complementation to a heterologous phosphatase catalytic domain. EMBO J. 1993;12:4833–4842. doi: 10.1002/j.1460-2075.1993.tb06173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marini N J, Meldrum E, Buehrer B, Hubberstey A V, Stone D E, Traynor-Kaplan A, Reed S I. A pathway in the yeast cell division cycle linking protein kinase C (Pkc1) to activation of Cdc28 at START. EMBO J. 1996;15:3040–3052. [PMC free article] [PubMed] [Google Scholar]
- 27.Martín-Pérez, J., and J. Ariño. Unpublished results.
- 28.Posas F, Clotet J, Ariño J. Saccharomyces cerevisiae gene SIT4 is involved in the control of glycogen metabolism. FEBS Lett. 1991;279:341–345. doi: 10.1016/0014-5793(91)80183-4. [DOI] [PubMed] [Google Scholar]
- 29.Posas F, Casamayor A, Morral N, Ariño J. Molecular cloning and analysis of a yeast protein phosphatase with an unusual amino-terminal region. J Biol Chem. 1992;267:11734–11740. [PubMed] [Google Scholar]
- 30.Posas F, Casamayor A, Ariño J. The PPZ protein phosphatases are involved in the maintenance of osmotic stability of yeast cells. FEBS Lett. 1993;318:282–286. doi: 10.1016/0014-5793(93)80529-4. [DOI] [PubMed] [Google Scholar]
- 31.Posas F, Camps M, Ariño J. The PPZ protein phosphatases are important determinants of salt tolerance in yeast cells. J Biol Chem. 1995;270:13036–13041. doi: 10.1074/jbc.270.22.13036. [DOI] [PubMed] [Google Scholar]
- 32.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Serrano, R., and F. Culiáñez-Maciá. Personal communication.
- 35.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 36.Sutton A, Immanuel D, Arndt K T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutton A, Lin F, Arndt K T. The SIT4 protein phosphatase is required in late G1 for progression into S phase. Cold Spring Harbor Symp Quant Biol. 1991;56:75–81. doi: 10.1101/sqb.1991.056.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 39.Wang T, Bretscher A. The Rho-GAP encoded by BEM2 regulates cytoskeletal structure in budding yeast. Mol Biol Cell. 1995;6:1011–1024. doi: 10.1091/mbc.6.8.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]