Abstract
All aerobic organisms are susceptible to damage by reactive oxygen species (ROS). ROS-induced damage has been associated with aging and diseases such as metabolic syndrome and cancer. However, not all organisms develop these diseases, nor do they age at the same rate; this is partially due to resistance to oxidative stress, a quantitative trait attributable to the interaction of factors including genetics and environmental. Drosophila melanogaster represents an ideal system to study how genetic variation can affect resistance to oxidative stress. In this work, oxidative stress (total and mitochondrial ROS), antioxidant response, and Cap 'n' collar isoform C and Spineless gene expression, one pesticide resistant (Oregon R(R)-flare) and wild-type (Canton-S) strains of D. melanogaster, were analyzed to test resistance to basal oxidative stress. ROS, catalase, and superoxide dismutase were determined by flow cytometry, and Cap 'n' collar isoform C and Spineless expression by qRT-PCR. The intensity of oxidative stress due to the pro-oxidant zearalenone in both was evaluated by flow cytometry. Data confirm expected differences in oxidative stress between strains that differ in Cyp450s levels. The Oregon (R)R-flare showed greater ROS, total and mitochondrial, compared to Canton-S. Regarding oxidative stress genes expression Cap 'n' collar isoform C and Spineless (Ss), Oregon R(R)-flare strain showed higher expression. In terms of response to zearalenone mycotoxin, Canton-S showed higher ROS concentration. Our data show variation in the resistance to oxidative stress among these strains of D. melanogaster.
Keywords: Canton-S, Oregon (R)R-flare, antioxidant response, Drosophila melanogaster, oxidative stress
Introduction
Oxygen-free radicals and other reactive oxygen species (ROS) are formed by all aerobic organisms, and although they play an important role as mediators in multiple intracellular signals that regulate physiological responses [1], their overproduction results in phenomena such as aging [2]. Besides, overproduction is also involved in different chronic pathological states such as diabetes, neurodegenerative and cardiac diseases, chronic renal disease, and even cancer [3–6].
The association of oxidative damage in human disease is known as oxidative stress, and currently it is considered as an imbalance between oxidants and antioxidants in favor of oxidants, leading to interruption of redox signals and control of molecular damage [7].
The origin of this imbalance is complex and may involve the overproduction of mitochondrial ROS (mROS) due to alterations in these organelles [8], or exposure to pro-oxidant agents [9]; in addition, the genetic background of individuals makes them susceptible to oxidative stress imbalance, for example, it is known that genetic variants can be associated with the severity of type 2 diabetes [10] and Alzheimer’s disease [11]. Also, variants in genes related to oxidative stress can be related to the origin of cancer, as well as to the response to treatment [12].
Model organisms show how genetic variants affect the oxidative stress or exposure to pro-oxidant agents, i.e. multiple genetic variants have been associated with longevity and resistance to oxidative stress, which may improve human health. Some variants are related to antioxidant enzymes such as catalase (CAT) and superoxide dismutase (SOD), or regulators of xenobiotic metabolism (Cytochromes P450), which have pleiotropic effects that increase resistance to oxidative stress [13, 14]. Drosophila melanogaster genetic variants have shown an increase in life span and resistance to toxic agents [15–17]. One of these pesticide-resistant strains is Oregon R(R)-flare, used in the Drosophila wing spot genotoxic test and contain the gene Cyp6g1 (48E7-48E7) with the dominant mutation Rst(2) DDT that codes for the inducer protein of genes located on 2R: Cyp6a2 (42D1-42D1), Cyp6a8, and Cyp6a9 (51D1-51D1), leading to higher expression of Cyp450s and has been previously related to the resistance to several insecticides [18, 19]. However, variants in Oregon R(R)-flare involve antioxidant enzymes expression and genes that regulate other mechanisms, which could result in disparity in the basal ROS concentration and its resistance to pro-oxidant challenges [15, 16].
In this work, the concentration of cytosolic antioxidant enzymes (SOD and CAT), and the expression of genes regulating the antioxidant response (Spineless and Cap “n” collar isoform C) in both strains were evaluated, also the total ROS and mROS concentrations, and the differences between strains in a pro-oxidant environment (zearalenone) were compared to reveal such differences between all those parameters in both strains.
Materials and Methods
Strains
Drosophila melanogaster, Oregon R(R)-flare strain (OR1; OR2; flr3/TM3, BdS) was originally and kindly donated by Prof. Ulrich Graf (ETH, University of Zurich, Switzerland), and D. melanogaster wild-type Canton-S strain was donated by Prof. Norma Velázquez Ulloa (Lewis and Clark University, Portland, Oregon).
Egg collection and preparation of the larvae
Eggs of both strains were grown at 25°C in dark conditions with a relative humidity of 60 to 80% inside culture bottles, containing a layer of fermented baker’s yeast supplemented with 3 g of sucrose. After 4 days (96 ± 4 h), larvae were washed from the bottles with purified tap water (25°C) through a fine mesh stainless steel filter.
Pro-oxidant treatment
Oregon R(R)-flare and Canton-S larvae were transferred into bottles containing 3 g of D. melanogaster instant medium (Carolina Biological Supply Co, NC, USA), supplemented with 12 ml of ZEN (20 μM concentration that has been documented induces increase in cells’ concentration of ROS) [20] or deionized water, as control. Bottles were kept at 25°C, at a relative humidity of 65%, and light–dark cycles 12:12 h.
Cell suspension preparation
Cells of larvae’ midgut tissue were prepared as described [21]. The midgut of 20 larvae of each strain was incubated with collagenase (.5 mg/mL) (Sigma-Aldrich 9001-1-12-1) for 15 min at 24°C. The cell suspension was passed through a nylon membrane (85 μm) and then suspended in phosphate buffer (PBS, pH 7.4, 4°C). Then, the cell suspension was processed for detection of ROS.
Quantification of ROS
Intracellular ROS from midgut cells of each strain was estimated by flow cytometry using 2′,7′-dichlorodihydrofluorescein diacetate (DCF-DA; Sigma Chemicals, D6883) [22]. Samples were measured on CytoFlex equipment (Beckman coulter), and data were analyzed using FlowJo software (Veritas software).
Mitochondrial extraction
A total of 5 × 107 cells were centrifuged at 800 rpm for 10 min at 4°C, the pellet was suspended in cold SEM buffer (10 mM MOPS, pH 7.5, 4°C, 320 mM sucrose, and 1 mM EDTA), the suspension was homogenized with a Dounce device. The lysate was centrifuged at 3000 rpm at 4°C to eliminate debris, the supernatant was collected and centrifuged at 12 000 rpm for 15 min at 4°C. The pellet was washed with 500 μL SEM buffer and centrifuged at 12 000 rpm for 15 min at 4°C and suspended in 50 μL SEM buffer to obtain a mitochondrial suspension.
Evaluation of mitochondrial integrity
Mitochondrial membrane integrity was measured by staining the suspension with Rhodamine 123 (Sigma chemicals, R8004). The samples were acquired in CytoFlex equipment, and data analyzed using FlowJo software.
Determination of mROS
mROS production in the midgut cells was estimated by flow cytometry using DCF-DA in the mitochondrial suspension. The samples were acquired on CytoFlex equipment, and data analyzed using FlowJo software.
Antioxidant enzyme detection
Quantification of cytosolic antioxidant enzymes was obtained by staining the cell suspension with specific antibodies against catalase (CAT) (GenTex, GTX110704) and superoxide dismutase (SOD) (abcam, ab13534). A total of 1 × 106 cells in saline buffer were incubated with 1 μL of the corresponding antibody, and then incubated for 30 min in the dark. The samples were collected in a CytoFlex Kit, and data analyzed using the FlowJo software.
Expression of antioxidant response regulatory genes
Expression of antioxidant response regulators was done by qRT-PCR quantifying mRNA of Cap “n” collar isoform C (CnCC) and Spineless (Ss) genes. Total RNA was extracted using the Trizol method (Invitrogen, CA, USA). About, 2 μg of RNA was reverse transcribed into cDNA using the enzyme M-MLV RT (Promega, WI, USA) at 42°C, and using Oligo-dT15 (Promega). The Maxima SYBR Green qRT-PCR Kit (Thermo Scientific, OR, USA) was used for qRT-PCR analysis. For every 20 μL reaction, 2 μL of cDNA were used, which were mixed with 10 μL of 2 SYBR Green reaction mix, .5 μL of specific oligonucleotides (10 pmol/ μL), and 7 μL of RNA-free water. The reaction conditions were 95°C for 15 min followed by 40 cycles of 95°C for 45 s, and 60°C for 1 min, these reactions were performed in triplicate. The sequence of oligonucleotides used was CnCC: 5`-GGTGGACTACGGACTACAA-3` and 5`-AGCGATGGAGCAGGTAATC-3`; Spineless (Ss): 5`-ACTGGATCGACTGAGCATTC-3` and 5`-CGTGCCTGTAGCCGTCAT-3`. The quantification was done following Pfaffl [23].
Chemicals and reagents
Collagenase (CAS-No. 9001-1-12-1), dichlorofluorescein diacetate (DCF-DA; CAS-No. D6883), rhodamine 123 (CAS-No. R8004), and zearalenone (CAS-No. 17924–92-42) were purchased from Sigma Aldrich (St. Louis, MO, USA). Formula 4–24® Instant Drosophila medium was purchased from Carolina Biological Supply Co. (NC, USA). Rabbit polyclonal antibodies to catalase (CAT; CAS-No. GTX110704) was purchased from GeneTex, Inc., North America; rabbit polyclonal antibodies to superoxide dismutase (SOD, Cat. No. ab13534) was purchased from abcam plc (Cambridge, UK). TRIzol™ LS (Cat. No. 10296028) was purchased from Invitrogen (CA, USA). Maxima SYBR Green/ROX qRT-PCR Kit (Cat. No. K0221) was purchased from ThermoFisher Sci. (OR, USA). M-MLV reverse transcriptase was purchased from Promega. (WI, USA).
Statistical analysis
Data were analyzed using GraphPad Prism v6.0 software. Results represent the mean of at least three independent experiments; bar ± S.D. Two-way analysis of variance (treatment by time) (ANOVA test) was used for statistical analysis of differences in values among multiple groups. A Student’s t-test was used to determine the statistical significance of differences in values between two groups (analysis between each two different treatments at each time). Statistical significance was defined as a P value <.05.
Results
Determination of ROS and mROS
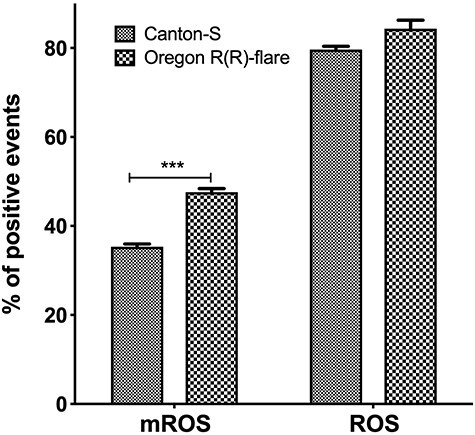
The first parameter evaluated was the basal mROS by flow cytometry in both strains, before this quantification, we evaluated the integrity of the isolated mitochondria, by staining with Rhodamine 123, which showed that 85% of the isolated mitochondria remained viable to perform the quantification of mROS. When comparing the number of mROS-positive mitochondria between strains, we found 35.3% in Canton S, while it was 47.6% in Oregon R(R)-flare, a 34% increase of positive mROS mitochondria (Fig. 1 left side) of this strain. As for the total number of ROS positive cells by DCF-DA staining, no difference was observed between strains, showing 80–82% positive cells (Fig. 1 right side).
Figure 1.
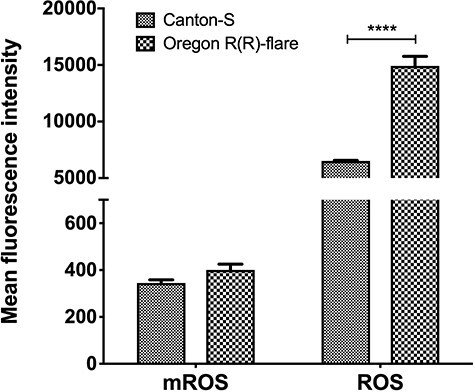
Later, to compare the relative concentration of basal ROS and mROS between strains the average intensity of fluorescence (MFI) obtained in the cytometry was used. The analysis showed more than twice the concentration of ROS in Oregon R(R)-flare than in Canton-S (Fig. 2 right side). For mROS concentration, the comparison of MFIs showed no statistical difference between strains (Fig. 2 left side).
Figure 2.
Quantification of CAT and SOD
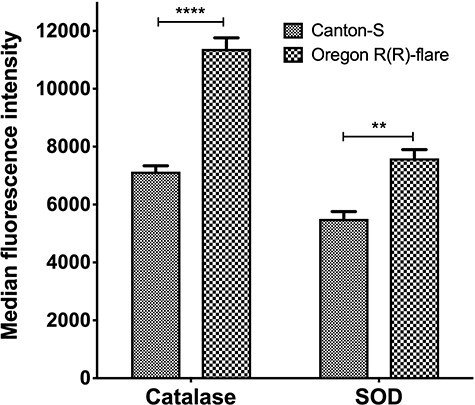
To evaluate and compare the cellular mechanisms that regulate ROS concentration in both strains, we quantified CAT and SOD through specific antibodies and flow cytometry. The data show that CAT expressed 59% more concentration in Oregon R(R)-flare compared to Canton-S strain, for SOD the increase was 38% more in Oregon R(R)-flare than in Canton-S strain (Fig. 3).
Figure 3.
Quantification of ROS regulatory genes
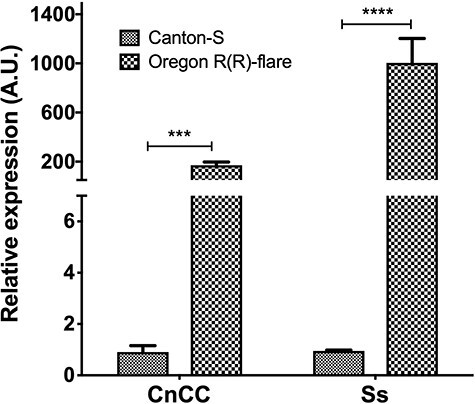
In addition to cytosolic antioxidant enzymes, other important mechanisms of cellular response to ROS are the activation of alternative antioxidant mechanisms such as expression of CnCC and Ss genes. Then, the relative concentration of mRNA of these regulators in midgut cells of both strains were evaluated. The expression of CnCC gene was more than 150 times higher in Oregon R(R)-flare than in Canton-S (Fig. 4 left side). While for the regulatory gene Ss, the difference is even greater, 9000 times more mRNA concentration in Oregon R(R)-flare than in Canton-S (Fig. 4 right side).
Figure 4.
Response to a pro-oxidant environment
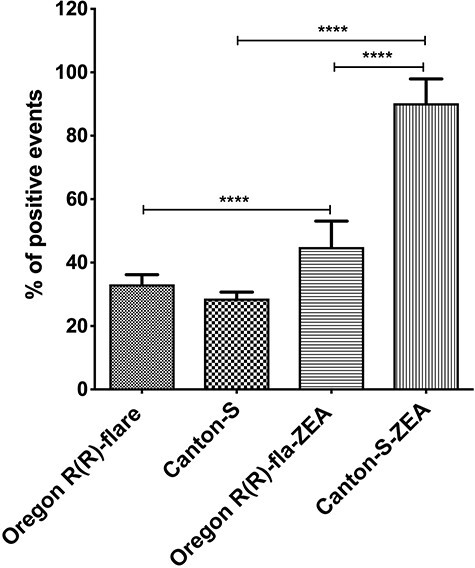
Drosophila melanogaster strains larvae were exposed to pro-oxidant environment generated by ZEN, which is an inducer of oxidative stress in different cells [20, 24], then midgut cells were isolated and total ROS generation was measured by strain. Both strains showed increased ROS positive cells compared to the control samples; however, in Canton-S strain, ROS generation was higher since the treatment with ZEN increased ROS ~ 2-fold related to unexposed larvae, while Oregon R(R)-flare increased .35 times more than its control (Fig. 5).
Figure 5.
Discussion
Our work compared the mechanisms of response to a pro-oxidant environment (oxidative stress) of two strains of Drosophila melanogaster, one with a wild-type genetic background (Canton-S) and another with a documented resistance to insecticides (Oregon R(R)-flare) and high levels of xenobiotic metabolism Phase I Cyp450s proteins [18, 19]; all this describe whether there are differences in oxidative stress resistance among strains, which makes one or other strain more susceptible or resistant to alterations generated by such stress, and putatively associated with genetic background differences. Accordingly, the basal total and mitochondrial ROS clearly suggest the susceptibility to aging and damages generated by oxidative stress [25], but also differences between strains and genetic variants of different organisms [26–28].
ROS comparison between Canton-S and Oregon R(R)-flare strains showed no significant difference in the number of midgut cells positive for DCFA staining; however, it showed higher total ROS in the Oregon R(R)-flare strain, which suggest that the pesticide resistant strain carries a higher baseline state of oxidative stress that is correlated with expressing more CAT and SOD, compared with Canton-S strain. In addition, the basal number of mitochondria positive for ROS is also higher in the Oregon R(R)-flare strain, which could be involved in a mitochondrial origin of oxidative stress, as have been described for neurodegenerative and metabolic disorders [29, 30].
Furthermore, mitochondrial mutations increase mROS to total oxidative stress, even though this is controversial related to aging [31]. This scenario suggests that the differences between strains could be found in mitochondrial DNA, in addition to the nuclear genetic markers already reported [18].
The difference in ROS concentration and mROS-positive mitochondria could be related to pesticide resistance as reported in different insects [32, 33]; however, this higher stress in the resistant strain should be compensated by increased regulatory mechanisms of oxidative stress, such as cytosolic antioxidant enzymes [33] and oxidative stress regulators [34]. This difference was observed in the Oregon R(R)-flare strain showing a higher concentration of CAT and SOD, and a higher quantity of mRNA from CnCC and Ss genes. The CnCC and Ss genes are transcription factors that regulate the expression of several detoxification genes responsible for counteracting oxidative stress, of metabolism, signal-sensing domain related to pathways during hypoxia, circadian rhythms signaling, or toxins, and even the function of stem cells [35–37]. Under non-stress conditions, CnCC protein is bound to a cytosolic Keap1 inhibitor. Keap1 suppresses CnCC activity by sequestering it in the cytoplasm, so Keap1 can function as another oxidant sensor that could be evaluated in future studies as another element in the regulation of the redox state in these strains [38].
Our results confirm the idea that insecticide resistance is not restricted only to the activity of Cyp450s [19], and as recently described by Lu and collaborators [39], this phenomenon of tolerance depends on a mechanism that adds the high levels of ROS, the overexpression of regulators such as CnCC and the already known response of Cyp450s; therefore, a higher baseline state of oxidative stress in Oregon R(R)-flare strain as well as the constant overexpression not only of the antioxidant CnCC and Ss regulators but also of its response elements downstream such as CAT and SOD [40], and mitochondria’s ROS high production could correspond to the mechanisms that confer the pesticide resistance reported to the Oregon R(R)-flare strain.
Added to the above, the better antioxidant response of the Oregon R(R)-flare strain to the pro-oxidant environment reinforces the idea that all these cells’ strategies: the elevated levels of antioxidant agents, and a higher baseline state of oxidative stresses are an advantage over toxic agents such as insecticides or any other pro-oxidant agent as the mycotoxin ZEN.
Conclusion
Our data show a clear difference in the oxidative state and the oxidative response of both strains, besides that these differences may correspond to variations in the genetic background of both strains, which may include the mDNA, in addition, these differences reveal a possible mechanism for pesticide resistance that not only depends on the activity of Cyp450s but also on a higher basal state of total ROS and mROS, of cytosolic antioxidant enzymes, and overexpression of two regulators of the antioxidant response, proposal that should be corroborated by further research.
Contributor Information
Santiago Cristobal Sigrist-Flores, Laboratorio de Inmunología (UMF), Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Barrios N° 1, Los Reyes Iztacala, Tlalnepantla, C.P. 54090, Estado de México, México.
Laura Castañeda-Partida, Toxicología Genética, Biología, Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Barrios N° 1, Los Reyes Iztacala, Tlalnepantla C.P. 54090, Estado de México, México.
Myriam Campos-Aguilar, Laboratorio de Inmunología (UMF), Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Barrios N° 1, Los Reyes Iztacala, Tlalnepantla, C.P. 54090, Estado de México, México.
Luis Felipe Santos-Cruz, Toxicología Genética, Biología, Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Barrios N° 1, Los Reyes Iztacala, Tlalnepantla C.P. 54090, Estado de México, México.
Aranza Miranda-Gutierrez, Laboratorio de Inmunología (UMF), Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Barrios N° 1, Los Reyes Iztacala, Tlalnepantla, C.P. 54090, Estado de México, México.
I A Gallardo-Ortíz, Unidad de Biomedicina, Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Barrios N° 1, Los Reyes Iztacala, Tlalnepantla C.P. 54090, Estado de México, México.
R Villalobos-Molina, Unidad de Biomedicina, Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Barrios N° 1, Los Reyes Iztacala, Tlalnepantla C.P. 54090, Estado de México, México.
Irma Elena Dueñas-García, Toxicología Genética, Biología, Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Barrios N° 1, Los Reyes Iztacala, Tlalnepantla C.P. 54090, Estado de México, México.
María Eugenia Heres-Pulido, Toxicología Genética, Biología, Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Barrios N° 1, Los Reyes Iztacala, Tlalnepantla C.P. 54090, Estado de México, México.
Elías Piedra-Ibarra, Fisiología Vegetal (UBIPRO), Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Barrios N° 1, Los Reyes Iztacala, Tlalnepantla C.P. 54090, Estado de México, México.
Víctor Hugo Rosales-García, Laboratorios Nacionales de Servicios Experimentales, Centro de Investigacion y de Estudios Avanzados del Instituto Politécnico Nacional. Ciudad de México, La Laguna Ticoman, Gustavo A. Madero, 07340 Mexico City, México.
Rafael Jimenez-Flores, Laboratorio de Inmunología (UMF), Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Barrios N° 1, Los Reyes Iztacala, Tlalnepantla, C.P. 54090, Estado de México, México.
Alberto Ponciano-Gómez, Laboratorio de Inmunología (UMF), Facultad de Estudios Superiores Iztacala, Universidad Nacional Autónoma de México, Los Barrios N° 1, Los Reyes Iztacala, Tlalnepantla, C.P. 54090, Estado de México, México.
Conflict of interest statement
All the authors declare that they have no conflict of interest.
Funding
This project was supported by DGAPA-PAPIIT-IN227619, UNAM.
References
- 1.Reczek CR, Chandel NS. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2015;33:8–13. 10.1016/j.ceb.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liguori I, Russo G, Curcio F et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging 2018;13:757–72. 10.2147/CIA.S158513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asmat U, Abad K, Ismail K (2016) Diabetes mellitus and oxidative stress—A concise review. Saudi Pharm. J. 24(5):547–553. doi: 10.1016/j.jsps.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elfawy HA, Das B. Crosstalk between mitochondrial dysfunction, oxidative stress, and age related neurodegenerative disease: Etiologies and therapeutic strategies. Life Sci. 2019;218:165–84. 10.1016/j.lfs.2018.12.029. [DOI] [PubMed] [Google Scholar]
- 5.Incalza MA, D’Oria R, Natalicchio A et al. Oxidative stress and reactive oxygen species in endothelial dysfunction associated with cardiovascular and metabolic diseases. Vascul. Pharmacol. 2018;100:1–19. 10.1016/j.vph.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Srinivas US, Tan BWQ, Vellayappan BA, Jeyasekharan AD. ROS and the DNA damage response in cancer. Redox Biol. 2019;25:101084. 10.1016/j.redox.2018.101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sies H (2018) On the history of oxidative stress: Concept and some aspects of current development. Curr. Opin. Toxicol. 7: 122–126. 10.1016/j.cotox.2018.01.002 [DOI] [Google Scholar]
- 8.Lopez-Fabuel I, Martin-Martin L, Resch-Beusher M et al. Mitochondrial respiratory chain disorganization in Parkinson’s disease-relevant PINK1 and DJ1 mutants. Neurochem Int. 2017;109:101–5. 10.1016/j.neuint.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 9.Zheng W, Wang B, Si M et al. Zearalenone altered the cytoskeletal structure via ER stress-autophagy-oxidative stress pathway in mouse TM4 Sertoli cells. Sci Rep. 2018;8:3320. 10.1038/s41598-018-21567-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim M, Kim M, Huang L et al. Genetic risk score of common genetic variants for impaired fasting glucose and newly diagnosed type 2 diabetes influences oxidative stress. Sci Rep 2018;8:7828. 10.1038/s41598-018-26106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowalska M, Wize K, Prendecki M et al. Genetic variants and oxidative stress in Alzheimer’s Disease. Curr Alzheimer Res 2020;17:208–23. 10.2174/1567205017666200224121447. [DOI] [PubMed] [Google Scholar]
- 12.Klaunig JE, Wang Z. Oxidative stress in carcinogenesis. Curr. Opin. Toxicol. 2018;7:116–21. 10.1016/j.cotox.2017.11.014. [DOI] [Google Scholar]
- 13.Balmus IM, Ciobica A, Antioch I et al. Oxidative stress implications in the affective disorders: main biomarkers, animal models relevance, genetic perspectives, and antioxidant approaches. Oxid. Med. Cell. Longev. 2016;2016:3975101. 10.1155/2016/3975101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YJ, Seroude L, Benzer S (1998) Extended life-span and stress resistance in the Drosophila mutant methuselah. Science 282(5390):943–946. 10.1126/science.282.5390.943 [DOI] [PubMed] [Google Scholar]
- 15.Orr WC, Sohal RS (1994) Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science 263(5150):1128–30. 10.1126/science.8108730 [DOI] [PubMed] [Google Scholar]
- 16.Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, Boulianne GL (1998) Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 19(2):171–174. 10.1038/534 [DOI] [PubMed] [Google Scholar]
- 17.Hällström I (1985) Genetic regulation of the cytochrome P-450 system in Drosophila melanogaster. II. Localization of some genes regulating cytochrome P-450 activity. Chem Biol Interact. 56(2–3):173–184. 10.1016/0009-2797(85)90004-3 [DOI] [PubMed] [Google Scholar]
- 18.Vázquez-Gómez G, Sánchez-Santos A, Vázquez-Medrano J, Quintanar-Zúñiga R, Monsalvo-Reyes AC, Piedra-Ibarra E, Dueñas-García IE, Castañeda-Partida L, Graf U, Heres-Pulido ME (2010) Sulforaphane modulates the expression of Cyp6a2 and Cyp6g1 in larvae of the ST and HB crosses of the Drosophila wing spot test and is genotoxic in the ST cross. Food Chem Toxicol. 48(12): 3333–3339. 10.1016/j.fct.2010.08.038 [DOI] [PubMed] [Google Scholar]
- 19.Daborn PJ, Lumb C, Boey A, Wong W, ffrench-Constant RH, Batterham P (2007) Evaluating the insecticide resistance potential of eight Drosophila melanogaster cytochrome P450 genes by transgenic over-expression. Insect Biochem Mol Biol. 37(5):512–519. 10.1016/j.ibmb.2007.02.008 [DOI] [PubMed] [Google Scholar]
- 20.Zheng W-L, Wang B-J, Wang L et al. ROS-mediated cell cycle arrest and apoptosis induced by zearalenone in mouse sertoli cells via ER Stress and the ATP/AMPK pathway. Toxins (Basel) 2018;10:24. 10.3390/toxins10010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey V, Turm H, Bekenstein U et al. A new in vivo model of pantothenate kinase-associated neurodegeneration reveals a surprising role for transcriptional regulation in pathogenesis. Front Cell Neurosci. 2013;7:146. 10.3389/fncel.2013.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eruslanov E, Kusmartsev S. Identification of ROS using oxidized DCFDA and flow-cytometry. Methods Mol Biol. 2010;594:57–72. 10.1007/978-1-60761-411-1_4. [DOI] [PubMed] [Google Scholar]
- 23.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29:e45. 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Li M, Zhang W et al. Protective Effect of N-Acetylcysteine against Oxidative Stress Induced by Zearalenone via Mitochondrial Apoptosis Pathway in SIEC02 Cells. Toxins 2018;10:407. 10.3390/toxins10100407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luceri C, Bigagli E, Pietro FA et al. Aging related changes in circulating reactive oxygen species (ROS) and protein carbonyls are indicative of liver oxidative injury. Toxicol Reports. 2018;5:141–5. 10.1016/j.toxrep.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mondal S, Kumar V, Singh SP. Oxidative stress measurement in different morphological forms of wild-type and mutant cyanobacterial strains: Overcoming the limitation of fluorescence microscope-based method. Ecotoxicol Environ Saf. 2020;200:110730. 10.1016/j.ecoenv.2020.110730. [DOI] [PubMed] [Google Scholar]
- 27.Wang IJ, Karmaus WJJ (2017) Oxidative stress-related genetic variants may modify associations of phthalate exposures with asthma. Int J Environ Res Public Health. 14(2):162. 10.3390/ijerph14020162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D, Wencheng L, Matsuyama Y, Cho K, Yamasaki Y, Takeshita S, Yamaguchi K, Oda T (2019) Extremely high level of reactive oxygen species (ROS) production in a newly isolated strain of the dinoflagellate Karenia mikimotoi. Eur J Phycol. 54(4):632–640. 10.1080/09670262.2019.1632936 [DOI] [Google Scholar]
- 29.Islam MT (2017) Oxidative stress and mitochondrial dysfunction-linked neurodegenerative disorders. Neurol. Res. 39(1):73–82. doi: 10.1080/01616412.2016.1251711 [DOI] [PubMed] [Google Scholar]
- 30.Stepien KM, Heaton R, Rankin S, Murphy A, Bentley J, Sexton D, Hargreaves IP (2017) Evidence of oxidative stress and secondary mitochondrial dysfunction in metabolic and non-metabolic disorders. J Clin Med. 6(7):71. 10.3390/jcm6070071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zsurka G, Peeva V, Kotlyar A, Kunz WS (2018) Is there still any role for oxidative stress in mitochondrial DNA-dependent aging? Genes (Basel) 9(4): 175. doi: 10.3390/genes9040175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Champion CJ, Xu J. Redox state affects fecundity and insecticide susceptibility in Anopheles gambiae. Sci Rep. 2018;8:13054. 10.1038/s41598-018-31360-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliver S V., Brooke BD (2016) The role of oxidative stress in the longevity and insecticide resistance phenotype of the major malaria vectors Anopheles arabiensis and Anopheles funestus. PLoS One. 11(3):e0151049. 10.1371/journal.pone.0151049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bottino-Rojas V, Talyuli OAC, Carrara L, Martins AJ, James AA, Oliveira PL, Paiva-Silva GO (2018) The redox-sensing gene Nrf2 affects intestinal homeostasis, insecticide resistance, and Zika virus susceptibility in the mosquito Aedes aegypti. J Biol Chem. 293(23):9053–9063. 10.1074/jbc.RA117.001589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi L, Ya S, Liu MF et al. Transcription factor CncC potentially regulates the expression of multiple detoxification genes that mediate indoxacarb resistance in Spodoptera litura. Insect Science 2020;1744–7917. 10.1111/1744-7917.12860. [DOI] [PubMed] [Google Scholar]
- 36.van der Burg KRL, Lewis JJ, Martin A, Nijhout HF, Danko CG, Reed RD. 2019. Contrasting roles of transcription factors spineless and EcR in the highly dynamic chromatin landscape of butterfly wing metamorphosis. Cell Rep 27(4):1027–1038.e3. doi: 10.1016/j.celrep.2019.03.092. [DOI] [PubMed] [Google Scholar]
- 37.Dai X, Yan X, Wintergerst KA, Cai L, Keller BB, Tan Y (2020) Nrf2: redox and metabolic regulator of stem cell state and function. Trends Mol. Med. 26(2): 185–200. 10.1016/j.molmed.2019.09.007 [DOI] [PubMed] [Google Scholar]
- 38.Misra JR, Lam G, Thummel CS (2013) Constitutive activation of the Nrf2/Keap1 pathway in insecticide-resistant strains of Drosophila. Insect Biochem Mol Biol. 43(12):1116–1124. 10.1016/j.ibmb.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu K, Cheng Y, Li W et al. Activation of CncC pathway by ROS burst regulates cytochrome P450 CYP6AB12 responsible for λ-cyhalothrin tolerance in Spodoptera litura. J. Hazardous Materials 2020;387:121698. 10.1016/j.jhazmat.2019.121698. [DOI] [PubMed] [Google Scholar]
- 40.Yi Y, Xu W, Fan Y, Wang H-X. Drosophila as an emerging model organism for studies of food-derived antioxidants. Food Res. Int. 2021;143:110307. 10.1016/j.foodres.2021.110307. [DOI] [PubMed] [Google Scholar]


